Abstract
MARCH5 is a RING finger E3 ligase involved in mitochondrial integrity, cellular protein homeostasis, and the regulation of mitochondrial fusion and fission. To determine the function of MARCH5 during development, we assessed transcript expression in zebrafish embryos. We found that march5 transcripts were of maternal origin and evenly distributed at the 1-cell stage, except for the mid-blastula transition, with expression predominantly in the developing central nervous system at later stages of embryogenesis. Overexpression of march5 impaired convergent extension movement during gastrulation, resulting in reduced patterning along the dorsoventral axis and alterations in the ventral cell types. Overexpression and knockdown of march5 disrupted the organization of the developing telencephalon and diencephalon. Lastly, we found that the transcription of march5 was tightly regulated by the transcriptional regulators CHOP, C/EBPα, Staf, Znf143a, and Znf76. These results demonstrate the essential role of March5 in the development of zebrafish embryos.
Keywords: convergence and extension movement, diencephalon, March5/MITOL, telencephalon, ubiquitin proteasome system
INTRODUCTION
Membrane-associated RING-CH protein 5 (MARCH5) is an E3 ubiquitin ligase located in the mitochondrial outer membrane (with the E3 ligase domain facing the cytoplasm) and is involved in a variety of mitochondrial and cellular processes (for review; Nagashima et al., 2014). For example, MARCH5 ubiquitylates the mitochondrial outer membrane proteins involved in mitochondrial fusion such as mitofusin 1 (Mfn1), which regulates mitochondrial docking and fusion, and Mfn2, which stabilizes the interactions between mitochondria. Increased expression of Mfn1 in MARCH5-depleted cells enhances mitochondrial fusion but interferes with fission (Chen et al., 2003; Park and Cho, 2012; Park et al., 2010). Ubiquitylation of Mfn2 generates a mitochondrion-associated endoplasmic reticulum membrane domain for Mfn2 oligomerization, enabling Mfn2 proteins to interact and transfer Ca2+ from the endoplasmic reticulum to the mitochondrion (De Brito and Scorrano, 2008; Sugiura et al., 2013; Szabadkai et al., 2006).
Another target of MARCH5, cytoplasmic dynamin-related protein 1 (Drp1), regulates mitochondrial fission. Drp1 transiently interacts with tetratricopeptide repeats in the outer-membrane-associated protein Fis1 via cytosolic adaptor proteins Mdv1 and Caf4 (Lackner et al., 2009; Mears et al., 2011) while MARCH5 counteracts mitochondrial fission by marking Drp1 and Fis1 for degradation (Nakamura et al., 2006; Yonashiro et al., 2006). MARCH5 also negatively regulates mitochondrial fission via stress-induced degradation of mitochondrial dynamics protein 49 kDa (MiD49) (Cherok et al., 2017; Xu et al., 2016). MiD49 is located in the mitochondrial outer membrane and recruits Drp1 to the mitochondrial surface rather than the peroxisomal surface; thus, MiD49 is thought to facilitate Drp1-directed mitochondrial fission (Palmer et al., 2011; Zhao et al., 2011).
MARCH5 marks mitophagy receptor FUN14 domain-containing protein 1 for degradation in response to hypoxia (Wu et al., 2017). MARCH5 also regulates mitochondrial transport via the degradation of abnormal proteins, such as microtubule-associated protein 1B, which interfere with dynein motor function and block the transport of mitochondria along axonal microtubules (Yonashiro et al., 2012).
Despite the many reports describing the various molecular functions of MARCH5 in mammalian cells, its embryological functions have yet to be described. Therefore, we conducted whole-mount in situ hybridization (WISH) and induced ectopic expression and knockdown of march5 in zebrafish embryos to investigate the role of MARCH5 in vertebrate embryogenesis.
MATERIALS AND METHODS
Zebrafish care and embryos
Wild-type (WT) zebrafish were obtained from Korea Zebrafish Organogenesis Mutant Bank (ZOMB) and grown at 28.5°C. Embryos were obtained through natural spawning and raised, and staged as described previously (Kimmel et al., 1995; Westerfield, 2000). Embryonic pigmentation was blocked by treating the embryos with 0.002% phenylthiourea after onset of somitogenesis.
Sequence analysis
March5 sequence similarity searches to identify homologous sequences were performed as described previously (Kim et al., 2008) and phylogenic analysis of March5 was conducted at http://www.treefam.org.
Molecular cloning
Total RNA was isolated from the embryos at various stages using the easy BLUE total RNA extraction Kit (iNtRON Bio, Korea) according to the manufacturer’s guidelines. cDNA was synthesized using Superscript III reverse transcriptase (Invitrogen, USA) as described in (Kumar et al., 2017). For overexpression studies, the open reading frame (ORF) of march5 was subcloned into the pcGlobin2 vector between the restriction sites for Xho1 and EcoR1 (Ro et al., 2004).
Morpholino, in vitro transcription, and microinjections
Splicing-blocking morpholinos (E1/I1: 5′TTTGTTTCTTTCACTTACCTGTCCACG3′) were purchased from Gene-Tools (USA), and dissolved in water. march5-specific morpholinos (0.8 to 1 ng) or control morpholinos were injected into zebrafish embryos at the 1-cell stage. march5 mRNA was synthesized using the Ambion mMESSAGE mMACHINE kit according to the manufacturer’s instructions. mRNAs were dissolved in nuclease-free water and diluted in 0.5% phenol red solution for microinjection via a Picopump microinjection device (World Precision Instruments, USA).
Whole-mount in situ hybridization (WISH)
Embryos were fixed in 4% paraformaldehyde (PFA) overnight, and dehydrated in 100% methanol. Embryos after 24 h post-fertilization (hpf) were digested with 10 μg/ml protease K (Thermo Scientific, USA). WISH was performed with minor modifications as described in (Kumar et al., 2019; Thisse et al., 1993). Antisense probes of march5 were synthesized from a specific region within the ORF. Antisense probes of march5 were synthesized using the DIG RNA Labeling Kit (SP6/T7) (Roche, USA).
Cell culture
HEK293T (human embryonic kidney 293T) cells were obtained from KCLB (Korean Cell Line Bank, Korea) for the Dual-Luciferases assay. HEK 293T cells were cultured in Dulbecco’s modified Eagle medium (DMEM; Welgene, Korea) containing 10% fetal bovine serum (FBS; Welgene) and 1% Antibiotic-Antimycotic solution (Gibco, USA).
Luciferase activity assay
Cells were harvested and lysates were collected 24 h post transfection. Firefly and Renilla luciferase activities were measured using a Dual-Luciferase Reporter Assay System (Promega, USA) according to the manufacturer’s instructions. Relative luciferase activities are the ratios of the activity of firefly luciferase to that of the Renilla luciferase control.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from WT and march5 morpholino injected zebrafish embryos at 4.7 hpf was extracted by RNAiso Plus reagent (TaKaRa Bio, Japan), according to the manufacturer’s protocol. The PrimeScript First strand cDNA synthesis kit (TaKaRa Bio) was applied for cDNA generation. qPCR was performed using SYBR-Green qPCR mix (TaKaRa Bio) in a Thermal Cycler Dice Real-Time system TP950 1 Set (TaKaRa Bio). Each assay was performed in triplicate. The specificity of the amplified products was determined by corresponding melting curve analysis. Primers used for RT-qPCR are listed in Supplementary Table S1.
Statistical analysis
All data are presented as mean ± SD. Statistically significant differences between the two groups were determined using the two-tailed Student’s t-test. One-way ANOVA was used for comparisons among multiple groups. P < 0.05 was considered to indicate a statistically significant difference. Data analysis were carried out using SPSS 17.0 (IBM, USA).
RESULTS
march5 is maternally expressed and restricted to the optic vesicles, telencephalon, midbrain, and hindbrain in embryos at 18 hpf
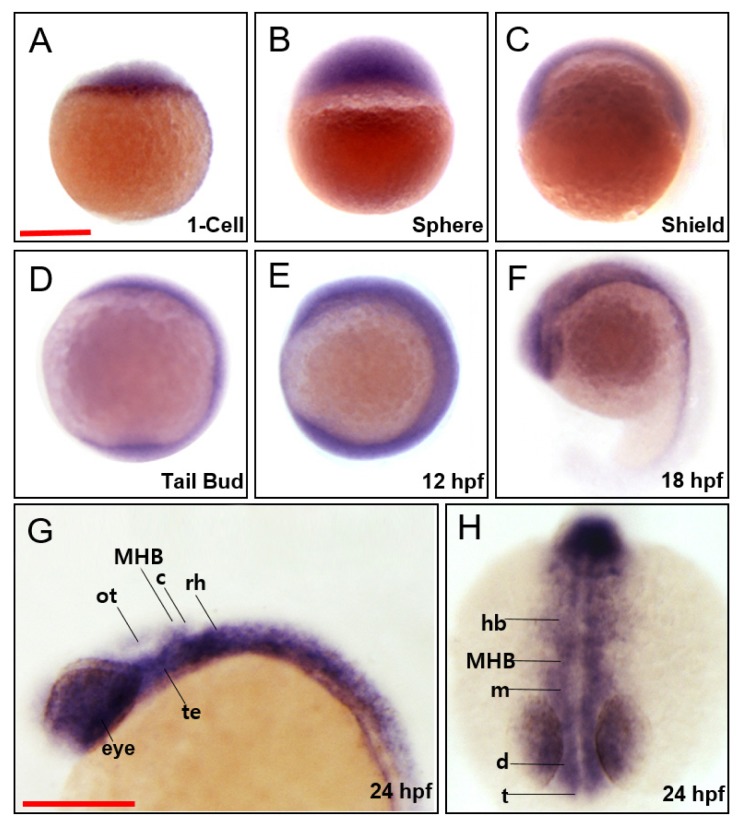
To investigate the contribution of march5 to vertebrate embryogenesis, we examined spatiotemporal expression in zebrafish embryos at various developmental stages. march5 was expressed both maternally and zygotically because march5 transcripts were detected by reverse transcriptase polymerase chain reaction (RT-PCR) at the 1-cell stage, and the expression persisted until 24 hpf. WISH confirmed that march5 is expressed at the 1-cell stage (Fig. 1A) and revealed evenly distributed transcripts in zygotes at the sphere and shield stages (Figs. 1B and 1C). march5 transcripts were localized to the central nervous system at 10 and 12 hpf (Figs. 1D and 1E) and restricted to the optic vesicles, telencephalon, midbrain, and hindbrain at 18 hpf (Fig. 1F). This pattern remained at 24 hpf, with march5 transcripts restricted to the eyes, telencephalon, diencephalon, tegmentum, optic tectum, cerebellum, and rhombomeres (Figs. 1G and 1H). Thus, march5 likely contributes to the development of these regions.
Fig. 1. Spatiotemporal expression patterns of zebrafish march5.
WISH analysis of march5 at 1 cell through 24 hpf. (A) march5 was expressed in the blastodisc at 1-cell stage, indicating it is maternally expressed. (B) After MBT (512-cell), march5 was expressed in deep cell layer (DEL) and enveloping layer (EVL) except I-YSL (yolk syncytial layer). (C) march5 was expressed in both ventral & dorsal region at shield stage. (D–F) The transcripts were abundant in the central nervous system at 10 hpf through 18 hpf. (F) At 18 somite, march5 transcripts were distributed in the precursor region of brain along the AP axis. (G and H) march5 expression patterns at 24 hpf zebrafish embryo stage. Lateral (G) and anterior view (H) of the embryos labeled with march5 antisense probe at 24 hpf. march5 was expressed in the forebrain through the notochord including the telencephalon, diencephalon, tegmentum, optic tectum, cerebellum and rhombomere. All embryos were collected synchronously from WT zebrafish for WISH analysis at the corresponding stages. MHB, midbrain hindbrain boundary; ot, optic tectum; c, cerebellum; rh, rhombomere; te, tegmentum; hb, hindbrain; m, midbrain; d, diencephalon; t, telencephalon. (A–H) Scale bars = 250 μm.
Ectopic expression and knockdown of march5 cause developmental defects in the brain and eyes and dorsoventralization of zebrafish embryos
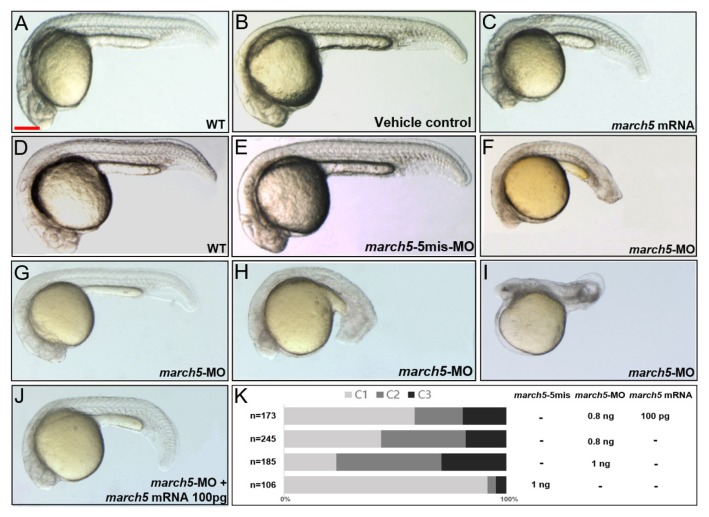
Microinjection of march5 mRNA into zebrafish embryos at the 1-cell stage to induce ectopic expression resulted in notochords that were shorter along the anterior-posterior (A–P) axis but that mediolaterally expanded in comparison to those of control embryos at 24 hpf (Figs. 2A–2C). Interestingly, repression of march5 expression in embryos at the 1-cell stage via microinjection of march5-specific antisense oligonucleotide morpholinos (march5 MOs; 0.8 ng per embryo) also resulted in a shortened A–P axis as well as a reduced eye diameter (Fig. 2F) in comparison to those of 5-mismatch controls (Fig. 2E) and WT embryos at 24 hpf (Fig. 2D). We categorized the knockdown defects into three classes on the basis of their severity: class I (Fig. 2G), embryos with developmental delay but no distinguishable axial defect; class II (Fig. 2H), embryos with reduced brain volume, poor eye development, and shortened body axis; class III (Fig. 2I), embryos with truncated head and no discernible axial structure. As shown in Figure 2K, march5 MOs increased defects in class II and class III embryos in a dose-dependent manner; when the amount of march5 MOs per embryo was increased from 0.8 to 1 ng, class I and II defects increased from 37% and 18% to 57% and 35%, respectively.
Fig. 2. Overexpression and knockdown of march5 in zebrafish embryos.
(A) WT zebrafish embryo, (B) vehicle control injected with the same volume of phenol red dye in distilled water as the volume of the mRNAs injected, and (C) march5 mRNA injected embryo. Microinjection of march5 mRNA (50 pg) into embryos at 1-cell stage for overexpression of march5. (D) WT, (E) 5-mismatch MO, and (F) march5 MO that 0.8 ng of march5 morpholino was injected into embryos at 1-cell stage for knockdown of march5. (G–J) At 24 hpf, embryos injected with march5 MO and coinjected with march5-MO and march5 mRNA. (G–I) march5 morphants are categorized as Class 1 through 3. (J) Rescue with march5 mRNA after injection of march5-MO. (K) The proportion of normal appearing embryos (C1) is increased after recue with march5 mRNA. (A–J) Scale bars = 250 μm.
The defects in the A–P axis of the notochord induced by march5 MOs (0.8 ng per embryo) were rescued by overexpression of march5 (100 pg mRNA per embryo); the reduction in brain volume was partially rescued (Fig. 2J). Together, these data strongly suggest that proper expression of march5 is critical to the development of the A–P axis, notochord, and brain.
Molecular events associated with the developmental defects induced by march5 MOs
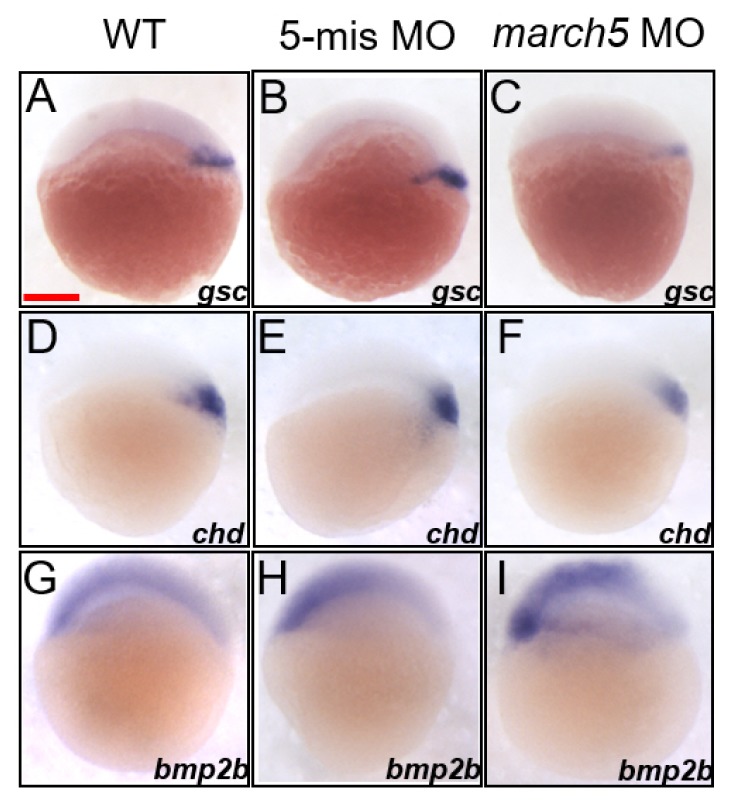
Cellular rearrangements that reshape the blastoderm into a characteristic vertebrate body plan begin at approximately 4 hpf. At 5 hpf, cells at the margin internalize and form the so-called hypoblast, the precursor of the mesoderm and endoderm (Solnica-Krezel, 2005). We thus examined whether an impairment of the normal gastrulation process results in morphological defects, such as a shortened A–P axis in the march5 morphants at 24 hpf (Fig. 2H). We conducted WISH analysis of marker genes known to be critical for cell fate specification, including gsc and chd for the dorsal region, bmp2b for the ventral region, and ntl for the mesoderm (Barth et al., 1999; Fisher and Halpern, 1999; Kishimoto et al., 1997; Schulte-Merker et al., 1994). Transcript levels of the two dorsal markers, gsc and chd were remarkably reduced in the dorsal region, accompanying shrinkage of the two gene-expressing domains (Figs. 3C and 3F) in march5 morphants at 4.7 hpf when compared with those of WT and the 5′-mismatch control (Figs. 3A, 3B, 3D, and 3E). In contrast transcripts level of the ventral marker, bmp2b was robustly elevated in the ventral region to cause significant expansion of bmp2b expressing domain (Fig. 3I) in march5 morphants at 4.7 hpf in comparison to those of WT (Fig. 3G) and the 5′-mismatch control (Fig. 3H). Furthermore, quantitative studies of march5 morphants at 4.7 hpf using RT-qPCR showed similar patterns to the spatiotemporal expression patterns of the two dorsal markers, gsc and chd as well as the ventral marker, bmp2b (Supplementary Fig. S1). These results suggest that March5 contributes to dorsalizing process in zebrafish embryos. It is thus conceivable that proper spatiotemporal expression of march5 is essential for formation of the normal dorsoventral axis in zebrafish embryos.
Fig. 3. Knock-down of march5 expression alters dorso-ventral patterning at 4.7 hpf.
(A–F) WISH analysis of march5 in dorso-ventral axis development using dorsal markers, gsc and chd at 4.7 hpf. WT embryos (A and D), embryos injected with 5-mismatch as control (B and E), and march5 MO (C and F). march5 morphants showed narrower and reduced expression pattern for gsc (C) and the level of chd transcripts was significantly reduced in the dorsal region (D–F). (G–I) Embryos were examined for the expression of ventral marker, bmp2b at 4.7 hpf. Transcripts of bmp2b in WT (G) and 5-mis MO control (H). Injection of march5-MO caused notable expanded bmp2b expression domains, circumference around the germ ring region at 4.7 hpf (I). (A–I) Scale bars = 200 μm.
march5 contributes to convergent extension (CE)
During gastrulation, CE of both ectodermal and mesendodermal cells leads to mediolateral narrowing and A–P extension of the emerging embryonic body axis (Solnica-Krezel, 2005). At the shield stage, cells deep underneath the superficial noninvoluting endocytic marginal (NEM) cell domain involute to form the nascent hypoblast of the embryonic shield, whereas cells within the NEM cell cluster form a loose mass of forerunner cells in front of the blastoderm margin (D’Amico and Cooper, 1997). To examine if MARCH5 is essential for CE, we first analyzed the expression pattern of ntl, a marker of mesendodermal cells in zebrafish embryos with ectopic expression or knockdown of march5. For ectopic expression, 50 pg march5 mRNA was injected into embryos at the 1- to 2-cell stage. WISH for ntl revealed that overexpression of march5 hindered extension of posterior paraxial mesendodermal cells, resulting in a shorter A–P axis and mediolateral expansion at 8 hpf (Fig. 4B) in comparison to that in the control (Fig. 4A). By contrast, knockdown of march5 generated a partial axial mesendoderm, which left the posterior region separated at 8 hpf (Fig. 4D), rather than a complete axial mesoderm, as seen in the controls (Figs. 4A and 4C).
Fig. 4. Knock-down of march5 expression causes changes in expression patterns of the molecular markers, ntl and sox17 at 8 hpf.
(A–D) WISH analysis using ntl as a molecular marker for the convergence and extension (C&E) indicates that loss of march5 function disrupted the processes in C&E governing anterior-posterior patterning. (A) WT embryo. Microinjection of march5 mRNA (50 pg per embryo) into wild-type embryos thickened the C&E (B). march5 5-mismatch control embryos had the similar expression patterns in C&E (C) to that of WT (A). In contrast, knock-down of march5 (0.8 ng morpholino per embryo) splited the body axis (arrowheads) in march5 morphants at 8 hpf (D). (E–H) WISH analysis with sox17 as a molecular marker for the DFCs. sox17 transcripts were present in the DFC of WT (E) and control embryos injected with 5-mismatch (G). Overexpression of march5 (march5 mRNA 50 pg per embryo) extended the area expressing sox17 (F) while knock-down of march5 (0.8 ng of march5 morpholino per embryo) remarkably reduced the level of sox17 transcripts in the DFC (asterisk) of the morphants (H). (A–H) Scale bars = 200 μm.
The nascent hypoblast formed during involution then moves toward the animal pole (Trinkaus, 1993), and at 60% epiboly, the dorsal forerunner cells (DFCs) develop at the leading edge of the blastoderm (D’Amico and Cooper, 1997). To examine if the defective involution of the axial hypoblast disrupts the formation of DFCs in zebrafish embryos, we studied the expression pattern of the marker gene sox17. At 8 hpf, sox17 expression in control embryos revealed the migration of DFCs toward the posterior region in front of the advancing blastoderm margin (Fig. 4E). However, these DFCs were dispersed in embryos overexpressing march5 (Fig. 4F). By contrast, knockdown of march5 substantially reduced the area of sox17 expression (Fig. 4H). In addition, the development of Kupffer’s vesicle was severely retarded in march5 morphants after gastrulation (data not shown). These data suggest that march5 regulates the movement of the hypoblast to initiate DFC formation at the blastoderm margin during gastrulation in zebrafish embryos (Supplementary Fig. S2).
march5 controls patterning of the forebrain and hindbrain fields during gastrulation
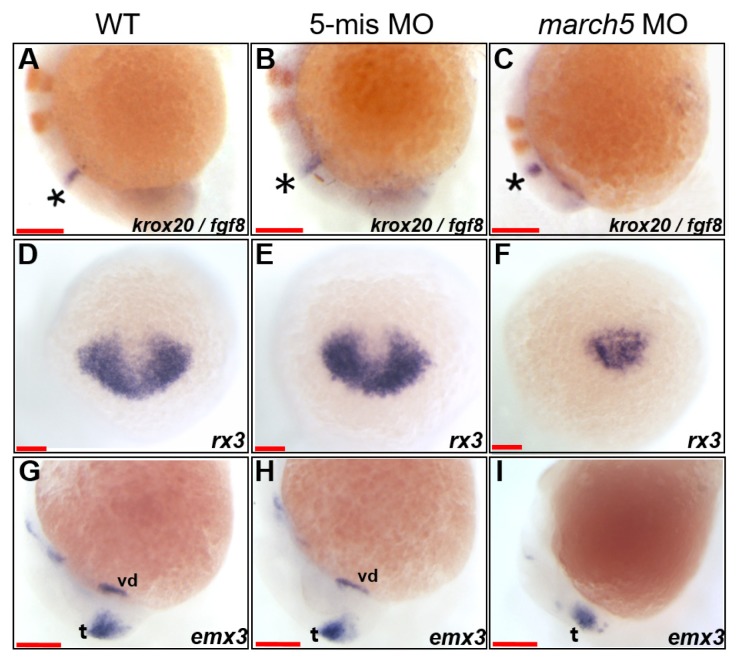
We next examined the embryological consequences of march5 overexpression and knockdown on brain development. Specifically, two-color WISH was performed at 10 and 18 hpf for fgf8 and krox20, markers of the midbrain–hindbrain boundary, hindbrain, and rhombomere (Oxtoby and Jowett, 1993; Reifers et al., 1998). The analysis demonstrated that the distance between the midbrain–hindbrain boundary (marked by an asterisk in Fig. 5C) and rhombomere 3 was shortened along the A–P axis by approximately 3-fold in march5 morphants in comparison to those of WT and 5-mismatch MO controls (Figs. 5A and 5B) at 18 hpf. These results imply that the retarded CE in march5 morphants results in the abnormal patterning of the corresponding developing brain subdomains.
Fig. 5. march5 knock-down reduced expression of rx3 and emx3 in zebrafish embryos.
(A–C) Two color WISH analysis with krox20- and fgf8-specific probes detected changes in their transcripts in the MHB (asterisks) and the hindbrain, the rhombomeres (r) 3 and 5 at 18 hpf. (A) WT embryo, (B) 5-mismatch MO control, and (C) march5 MO (0.8 ng of march5 morpholino per embryo). (D–I) WISH analysis of march5 MO (0.8 ng of march5 morpholino per embryo) using rx3 and emx1 as probes. (D) rx3 transcripts in the presumptive eye field and hypothalamus of WT embryos at 10 hpf. (E) Embryos injected with 5-mismatch showed similar patterns to those of WT embryos. (F) march5 MO shows remarkable reduction of rx3 transcripts in the presumptive eye field and hypothalamus, which abut the telencephalic primordium at 10 hpf. Transcripts of emx3 in WT (G), 5-mismatch control (H), and march5 MO (I) at 24 hpf. emx3 transcripts were present in the telencephalon and ventral diencephalon in WT and 5-mismatch control whereas the ventral diencephalon expression was lost in the corresponding areas of march5 MO. vd, ventral diencephalon; t, telencephalon. (A–I) Scale bars = 100 μm.
We then characterized anterior neural plate development between 10 and 24 hpf by performing WISH for rx3 and emx3, markers of the telencephalon (Chuang et al., 1999; Mathers et al., 1997; Morita et al., 1995; Viktorin et al., 2009). rx3 transcripts were substantially reduced in the presumptive anterior forebrains of march5 morphants at 10 hpf (Fig. 5F) in comparison to those of WT and 5-mismatch MO controls (Figs. 5D and 5E). The near elimination of emx3 at 24 hpf in the ventral forebrain of embryos with march5 knockdown (Fig. 5I) suggests that march5 contributes to the specification of anterior forebrain precursors during gastrulation (Supplementary Fig. S3). However, emx3 expression was only slightly decreased in the telencephalic regions of march5 morphants in comparison to those of WT and 5-mismatch MO controls (Figs. 5G and 5H). Altogether, these data suggest that proper march5 expression is essential for appropriate patterning of the telencephalon.
Transcriptional factors Znf143a and Znf76 regulate march5 expression in 293T cells
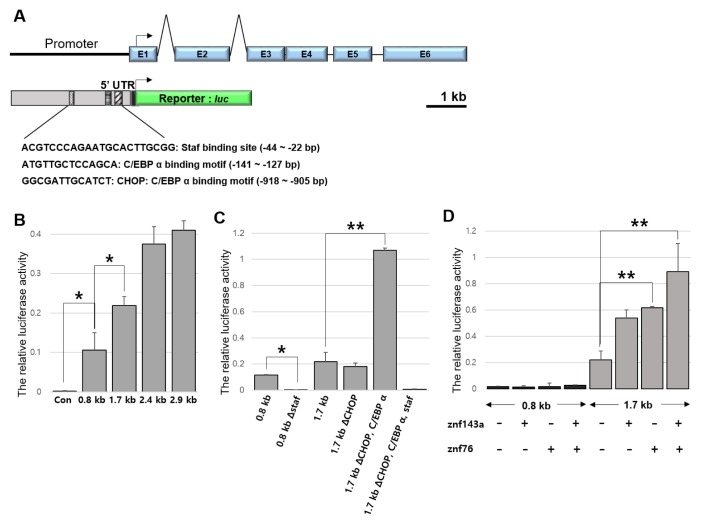
We next examined the regulatory region of march5 for the presence of cis-acting elements conferring transcriptional activity. Specifically, nucleotide sequences in the zebrafish march5 promoter located between −2917 and +296 bp of the transcriptional start site were targeted because this region contains three putative binding sites for Staf (selenocysteine tRNA gene transcription-activating factor), C/EBPα (CCAAT enhancer binding protein alpha), and CHOP (C/EBP homologous protein):C/EBPα (shown in Fig. 6A). To measure the transcriptional activities of these three cis-acting elements, we generated four march5-luc reporter constructs lacking the binding sites for Staf, C/EBPα, and/or CHOP:C/EBPα (Fig. 6A), and expressed them in human embryonic kidney cells (HEK 293T cells). Transcriptional activities of the three reporter constructs containing 1.7 kb, 2.4 kb, or 2.9 kb regions were 2-, 3.5-, 4-fold higher that of the reporter containing the 0.8 kb region (Fig. 6B). The reporter construct containing the 1.7 kb region was chosen because it contains most of the cis- and trans-acting elements, such as Staf, CHOP, and C/EBPα.
Fig. 6. Znf143a and Znf76 modulate march5 expression.
(A) Schematic representation of zebrafish march5 genomic region and reporter construct. Six exons (EX1 to EX6) and five introns are depicted; the translation initiation site is indicated with an arrow. The firefly luciferase (luc) coding region was fused with the zebrafish march5 promoter (−2904 to +296 bp). (B–D) Luciferase activity under the control of the march5 regulatory regions. (B) Luciferase activity of a transiently transfected march5-luc reporter in 293T cells (*P < 0.05 vs Con, as determined by Student’s t-test). (C) Deletion analysis of the zebrafish march5 promoter. Cells were transfected with the Staf, CHOP and CHOP:C/EBPα-deletion promoter-luciferase reporter plasmids. A significant difference between 1.7 kb and 1.7 kbΔCHOP, C/EBPα (**P < 0.01, as determined by Student’s t-test). (D) Effects of Znf143a and Znf76 proteins on the zebrafish march5 promoter activation. Cells were co-transfected with Znf143a and Znf76 expression plasmid (empty pc2+ expression plasmid as control), and Pgl3 promoter-luciferase reporter plasmids fused with the zebrafish march5 promoter (0.8 kb and 1.7 kb). Luciferase activity of march5 promoter in 293T cells is expressed as normalized luciferase activity (to Renilla luciferase activity) relative to empty Pgl3-Basic plasmid (*P < 0.05, **P < 0.01 vs 1.7kb, as determined by Student’s t-test).
To determine if CHOP and C/EBPα transcriptionally regulate march5, we measured the luciferase activities of the constructs with or without CHOP and/or C/EBPα in the 1.7 kb-long region. As shown in Figure 6C, deletion of the binding site for CHOP did not alter the transcriptional activity of the 1.7 kb-long region, whereas deletion of the binding sites for both CHOP and C/EBPα dramatically increased the luciferase activity by more than 5-fold relative to that of the control reporter construct containing both elements. However, deletion of the Staf binding site in addition to the CHOP:C/EBPα sites abolished transcriptional activity, observed as a complete loss of luciferase activity. Taken together, these results demonstrate that Staf functions as a potent transcriptional activator and that CHOP and C/EBPα act as transcriptional repressors of march5. Expression of these three transcriptional regulators may thus coordinate transcription of march5 in zebrafish embryos.
There is notable homology between Staf and the human proteins ZNF76 and ZNF143 (Jung et al., 2019; Ragoussis et al., 1992; Tommerup and Vissing, 1995), which both contain domains similar to the Staf transactivation domains that stimulate transcription from RNA polymerase II (PolII) and PolIII small nuclear RNA (snRNA)-type promoters as well as PolII TATA box-containing mRNA promoters (Myslinski et al., 1998; Schaub et al., 1997). The high degree of sequence conservation between the zinc finger regions in znf76 and znf143 in the zebrafish indicate that both proteins may recognize the same Staf motif within the march5 promoter. Luciferase activity from HEK 293T cells co-transfected with plasmids harboring the march5 promoter and those encoding Znf143a or Znf76 was increased 3-fold, and luciferase activity was increased 4-fold when cells expressed the march5 promoter and both Znf143a and Znf76 (Fig. 6D). Thus, we postulate that march5 is transcriptionally regulated in a finely tuned manner by CHOP, C/EBPα, Staf, Znf143a, and Znf76.
DISCUSSION
MARCH5 is a mitochondrial ubiquitin ligase that regulates mitochondrial integrity and cellular protein homeostasis via the ubiquitin proteasome system (Nagashima et al., 2014). We demonstrate here that March5 also regulates vertebrate embryogenesis because either knockdown or ectopic expression of march5 in zebrafish embryos retarded CE at 8 hpf, resulting in defective development of the ventral diencephalon, midbrain, and midbrain–hindbrain boundary at 24 hpf.
Beginning at 4.3 hpf, cells intercalate radially, contributing to epiboly, while cells at the margins migrate toward the animal pole to form the hypoblast. The various progenitor territories at 6 hpf are not sharply demarcated in the fate maps of zebrafish embryos (Kimmel et al., 1990; Sepich et al., 2005). We thus examined the expression of morphogenetic markers at pre-gastrulation and gastrulation stages. We found that at 4.7 hpf, knockdown of march5 reduced the expression of the dorsal markers but increased the expression of the ventral markers. In zebrafish, the fates of dorsal cells at the shield were once thought to be regulated by inducers expressed by the organizer; however, it is now known that the organizer represses factors secreted from ventral regions of the embryos (Niehrs, 2004), such as bone morphogenetic protein (BMP), Wnt, and Nodal family proteins; For example, ventrally expressed Wnt8 restricts the size of the zebrafish organizer in the late blastula/early gastrula by regulating the expression of the transcriptional repressors Vox and Vent, whereas during late gastrulation, the expression of these genes is maintained by BMP signaling (Ramel and Lekven, 2004; Ramel et al., 2005). In the absence of secreted BMP antagonists, Gsc has chd-independent functions in dorsoventral patterning and elicits complete secondary axes (Fox and Bruce, 2009). We also found that march5 knockdown not only changed level of wnt8 transcripts (Supplementary Fig. S4) but also reduced the transcript levels of gsc and chd (Figs. 3C and 3F).
We demonstrated that march5 was transcriptionally regulated at CHOP and C/EBP binding sites. CHOP is a developmentally regulated nuclear protein that inhibits wnt8-mediated dorsal development in Xenopus embryos (Horndasch et al., 2006). However, CHOP also dimerizes with C/EBP via its leucine zipper domain to function as a dominant negative inhibitor of gene transcription (Ron and Habener, 1992), arresting growth and inducing apoptosis under conditions of endoplasmic reticulum stress or DNA damage (Maytin et al., 2001; Talukder et al., 2002; Ubeda et al., 1996; Wang et al., 1996; Zinszner et al., 1998). Thus, CHOP and C/EBPα may work similarly to represses march5 transcription. By contrast, we found that the binding of Znf76 and Znf143 to the Staf site in the proximal promoter of march5 may induce transcription. Staf, a transcriptional activator originally identified in Xenopus laevis, enhances the transcription of snRNA and snRNA-type genes via Pol II and Pol III (Myslinski et al., 1992; Schuster et al., 1995). The Staf binding site may be a regulator of organ-specific transcription because transgenic mice expressing a Trsp (selenocysteine tRNA gene) transgene lacking the Staf binding site had an ~80% reduction in the levels of Sec tRNA in the brain and muscle but unaltered or elevated levels in other tissues (Carlson et al., 2009). We found that march5 transcription in zebrafish embryos was regulated by Znf76 and Znf143, which bind with similar affinities to Staf responsive elements (Myslinski et al., 1998). It is thus most probable that Znf76 and Znf143, together with C/EBPα and CHOP, modulate march5 transcription to regulate biological processes such as vertebrate embryogenesis.
The data presented here provide insight into the functions of a mitochondrial ubiquitin ligase during vertebrate embryogenesis. We found that expression of march5 must be tightly regulated for proper gastrulation and CE in zebrafish embryos. Our data indicate that Znf76 and Znf143 bind to the Staf region in the march5 promoter and modulate C/EBPα:CHOP regulation of march5 transcription. This is the first report to show that March5 affects the functional connection between convergence and extension processes during gastrulation. The newly identified cis- and trans-regulatory elements provide new insights into the molecular mechanisms of march5 specific transcriptional networks in zebrafish.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by research fund of Chungnam National University.
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
Disclosure
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Barth K.A., Kishimoto Y., Rohr K.B., Seydler C., Schulte-Merker S., Wilson S.W. Bmp activity establishes a gradient of positional information throughout the entire neural plate. Development. 1999;126:4977–4987. doi: 10.1242/dev.126.22.4977. [DOI] [PubMed] [Google Scholar]
- Carlson B.A., Schweizer U., Perella C., Shrimali R.K., Feigenbaum L., Shen L., Speransky S., Floss T., Jeong S.J., Watts J. The selenocysteine tRNA STAF-binding region is essential for adequate selenocysteine tRNA status, selenoprotein expression and early age survival of mice. Biochem J. 2009;418:61–71. doi: 10.1042/BJ20081304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Detmer S.A., Ewald A.J., Griffin E.E., Fraser S.E., Chan D.C. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherok E., Xu S., Li S., Das S., Meltzer W.A., Zalzman M., Wang C., Karbowski M. Novel regulatory roles of Mff and Drp1 in E3 ubiquitin ligase MARCH5–dependent degradation of MiD49 and Mcl1 and control of mitochondrial dynamics. Mol Biol Cell. 2017;28:396–410. doi: 10.1091/mbc.e16-04-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang J.C., Mathers P.H., Raymond P.A. Expression of three Rx homeobox genes in embryonic and adult zebrafish. Mech Dev. 1999;84:195–198. doi: 10.1016/S0925-4773(99)00077-5. [DOI] [PubMed] [Google Scholar]
- D’Amico L.A., Cooper M.S. Spatially distinct domains of cell behavior in the zebrafish organizer region. Biochem Cell Biol. 1997;75:563–577. doi: 10.1139/o97-074. [DOI] [PubMed] [Google Scholar]
- De Brito O.M., Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- Fisher S., Halpern M.E. Patterning the zebrafish axial skeleton requires early chordin function. Nat Genet. 1999;23:442–446. doi: 10.1038/70557. [DOI] [PubMed] [Google Scholar]
- Fox M.D., Bruce A.E. Short-and long-range functions of Goosecoid in zebrafish axis formation are independent of Chordin, Noggin 1 and Follistatin-like 1b. Development. 2009;136:1675–1685. doi: 10.1242/dev.031161. [DOI] [PubMed] [Google Scholar]
- Horndasch M., Lienkamp S., Springer E., Schmitt A., Pavenstädt H., Walz G., Gloy J. The C/EBP homologous protein CHOP (GADD153) is an inhibitor of Wnt/TCF signals. Oncogene. 2006;25:3397–3407. doi: 10.1038/sj.onc.1209380. [DOI] [PubMed] [Google Scholar]
- Jung J., Udhaya Kumar S., Choi I., Huh T.L., Rhee M. Znf76 is associated with development of the eyes, midbrain, MHB, and hindbrain in zebrafish embryos. Anim Cells Syst. 2019;23:26–31. doi: 10.1080/19768354.2018.1557744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.J., Ro H., Huh T.L., Lee C.J., Choi J., Rhee M. A novel Kinesin-like protein, Surhe is associated with dorsalization in the zebrafish embryos. Anim Cells Syst. 2008;12:219–230. doi: 10.1080/19768354.2008.9647176. [DOI] [Google Scholar]
- Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kimmel C.B., Warga R.M., Schilling T.F. Origin and organization of the zebrafish fate map. Development. 1990;108:581–594. doi: 10.1242/dev.108.4.581. [DOI] [PubMed] [Google Scholar]
- Kishimoto Y., Lee K.H., Zon L., Hammerschmidt M., Schulte-Merker S. The molecular nature of zebrafish swirl: BMP2 function is essential during early dorsoventral patterning. Development. 1997;124:4457–4466. doi: 10.1242/dev.124.22.4457. [DOI] [PubMed] [Google Scholar]
- Kumar A., Anuppalle M., Maddirevula S., Huh T.L., Choe J., Rhee M. Peli1b governs the brain patterning via ERK signaling pathways in zebrafish embryos. Gene. 2019;694:1–6. doi: 10.1016/j.gene.2018.12.078. [DOI] [PubMed] [Google Scholar]
- Kumar A., Huh T.L., Choe J., Rhee M. Rnf152 is essential for NeuroD expression and Delta-notch signaling in the zebrafish embryos. Mol Cells. 2017;40:945. doi: 10.14348/molcells.2017.0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner L.L., Horner J.S., Nunnari J. Mechanistic analysis of a dynamin effector. Science. 2009;325:874–877. doi: 10.1126/science.1176921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathers P., Grinberg A., Mahon K., Jamrich M. The Rx homeobox gene is essential for vertebrate eye development. Nature. 1997;387:603–607. doi: 10.1038/42475. [DOI] [PubMed] [Google Scholar]
- Maytin E.V., Ubeda M., Lin J.C., Habener J.F. Stress-inducible transcription factor CHOP/gadd153 induces apoptosis in mammalian cells via p38 kinase-dependent and-independent mechanisms. Exp Cell Res. 2001;267:193–204. doi: 10.1006/excr.2001.5248. [DOI] [PubMed] [Google Scholar]
- Mears J.A., Lackner L.L., Fang S., Ingerman E., Nunnari J., Hinshaw J.E. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat Struct Mol Biol. 2011;18:20–26. doi: 10.1038/nsmb.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T., Nitta H., Kiyama Y., Mori H., Mishina M. Differential expression of two zebrafish emx homeoprotein mRNAs in the developing brain. Neurosci Lett. 1995;198:131–134. doi: 10.1016/0304-3940(95)11988-9. [DOI] [PubMed] [Google Scholar]
- Myslinski E., Krol A., Carbon P. Optimal tRNA (Ser) Sec gene activity requires an upstream SPH motif. Nucleic Acids Res. 1992;20:203–209. doi: 10.1093/nar/20.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myslinski E., Krol A., Carbon P. ZNF76 and ZNF143 are two human homologs of the transcriptional activator Staf. J Biol Chem. 1998;273:21998–22006. doi: 10.1074/jbc.273.34.21998. [DOI] [PubMed] [Google Scholar]
- Nagashima S., Tokuyama T., Yonashiro R., Inatome R., Yanagi S. Roles of mitochondrial ubiquitin ligase MITOL/MARCH5 in mitochondrial dynamics and diseases. J Biochem. 2014;155:273–279. doi: 10.1093/jb/mvu016. [DOI] [PubMed] [Google Scholar]
- Nakamura N., Kimura Y., Tokuda M., Honda S., Hirose S. MARCH-V is a novel mitofusin 2-and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 2006;7:1019–1022. doi: 10.1038/sj.embor.7400790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehrs C. Regionally specific induction by the Spemann–Mangold organizer. Nat Rev Genet. 2004;5:425–434. doi: 10.1038/nrg1347. [DOI] [PubMed] [Google Scholar]
- Oxtoby E., Jowett T. Cloning of the zebrafish krox-20 gene (krx-20) and its expression during hindbrain development. Nucleic Acids Res. 1993;21:1087–1095. doi: 10.1093/nar/21.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C.S., Osellame L.D., Laine D., Koutsopoulos O.S., Frazier A.E., Ryan M.T. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 2011;12:565–573. doi: 10.1038/embor.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.Y., Cho H. Mitofusin 1 is degraded at G2/M phase through ubiquitylation by MARCH5. Cell Div. 2012;7:25. doi: 10.1186/1747-1028-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.Y., Lee S., Karbowski M., Neutzner A., Youle R.J., Cho H. Loss of MARCH5 mitochondrial E3 ubiquitin ligase induces cellular senescence through dynamin-related protein 1 and mitofusin 1. J Cell Sci. 2010;123:619–626. doi: 10.1242/jcs.061481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragoussis J., Senger G., Mockridge I., Sanseau P., Ruddy S., Dudley K., Sheer D., Trowsdale J. A testis-expressed Zn finger gene (ZNF76) in human 6p21.3 centromeric to the MHC is closely linked to the human homolog of the t-complex gene tcp-11. Genomics. 1992;14:673–679. doi: 10.1016/S0888-7543(05)80167-3. [DOI] [PubMed] [Google Scholar]
- Ramel M.C., Buckles G.R., Baker K.D., Lekven A.C. WNT8 and BMP2B co-regulate non-axial mesoderm patterning during zebrafish gastrulation. Dev Biol. 2005;287:237–248. doi: 10.1016/j.ydbio.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Ramel M.C., Lekven A.C. Repression of the vertebrate organizer by Wnt8 is mediated by Vent and Vox. Development. 2004;131:3991–4000. doi: 10.1242/dev.01277. [DOI] [PubMed] [Google Scholar]
- Reifers F., Bohli H., Walsh E.C., Crossley P.H., Stainier D., Brand M. Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesis. Development. 1998;125:2381–2395. doi: 10.1242/dev.125.13.2381. [DOI] [PubMed] [Google Scholar]
- Ro H., Soun K., Kim E.J., Rhee M. Novel vector systems optimized for injecting in vitro-synthesized mRNA into zebrafish embryos. Mol Cells. 2004;17:373–376. [PubMed] [Google Scholar]
- Ron D., Habener J.F. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 1992;6:439–453. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- Schaub M., Myslinski E., Schuster C., Krol A., Carbon P. Staf, a promiscuous activator for enhanced transcription by RNA polymerases II and III. EMBO J. 1997;16:173–181. doi: 10.1093/emboj/16.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte-Merker S., Hammerschmidt M., Beuchle D., Cho K., De Robertis E., Nusslein-Volhard C. Expression of zebrafish goosecoid and no tail gene products in wild-type and mutant no tail embryos. Development. 1994;120:843–852. doi: 10.1242/dev.120.4.843. [DOI] [PubMed] [Google Scholar]
- Schuster C., Myslinski E., Krol A., Carbon P. Staf, a novel zinc finger protein that activates the RNA polymerase III promoter of the selenocysteine tRNA gene. EMBO J. 1995;14:3777–3787. doi: 10.1002/j.1460-2075.1995.tb00047.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepich D.S., Calmelet C., Kiskowski M., Solnica-Krezel L. Initiation of convergence and extension movements of lateral mesoderm during zebrafish gastrulation. Dev Dyn. 2005;234:279–292. doi: 10.1002/dvdy.20507. [DOI] [PubMed] [Google Scholar]
- Solnica-Krezel L. Conserved patterns of cell movements during vertebrate gastrulation. Curr Biol. 2005;15:R213–R228. doi: 10.1016/j.cub.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Sugiura A., Nagashima S., Tokuyama T., Amo T., Matsuki Y., Ishido S., Kudo Y., McBride H.M., Fukuda T., Matsushita N. MITOL regulates endoplasmic reticulum-mitochondria contacts via Mitofusin2. Mol Cell. 2013;51:20–34. doi: 10.1016/j.molcel.2013.04.023. [DOI] [PubMed] [Google Scholar]
- Szabadkai G., Bianchi K., Várnai P., De Stefani D., Wieckowski M.R., Cavagna D., Nagy A.I., Balla T., Rizzuto R. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J Cell Biol. 2006;175:901–911. doi: 10.1083/jcb.200608073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukder A.H., Wang R.A., Kumar R. Expression and transactivating functions of the bZIP transcription factor GADD153 in mammary epithelial cells. Oncogene. 2002;21:4289–4300. doi: 10.1038/sj.onc.1205529. [DOI] [PubMed] [Google Scholar]
- Thisse C., Thisse B., Schilling T., Postlethwait J. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119:1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- Tommerup N., Vissing H. Isolation and fine mapping of 16 novel human zinc finger-encoding cDNAs identify putative candidate genes for developmental and malignant disorders. Genomics. 1995;27:259–264. doi: 10.1006/geno.1995.1040. [DOI] [PubMed] [Google Scholar]
- Trinkaus J. The yolk syncytial layer of Fundulus: its origin and history and its significance for early embryogenesis. J Exp Zool. 1993;265:258–284. doi: 10.1002/jez.1402650308. [DOI] [PubMed] [Google Scholar]
- Ubeda M., Wang X.Z., Zinszner H., Wu I., Habener J.F., Ron D. Stress-induced binding of the transcriptional factor CHOP to a novel DNA control element. Mol Cell Biol. 1996;16:1479–1489. doi: 10.1128/MCB.16.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viktorin G., Chiuchitu C., Rissler M., Varga Z.M., Westerfield M. Emx3 is required for the differentiation of dorsal telencephalic neurons. Dev Dyn. 2009;238:1984–1998. doi: 10.1002/dvdy.22031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.Z., Lawson B., Brewer J.W., Zinszner H., Sanjay A., Mi L.J., Boorstein R., Kreibich G., Hendershot L.M., Ron D. Signals from the stressed endoplasmic reticulum induce C/EBP-homologous protein (CHOP/GADD153) Mol Cell Biol. 1996;16:4273–4280. doi: 10.1128/MCB.16.8.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book: a guide for the laboratory use of zebrafish. 2000 http://zfin.org/zf_info/zfbook/zfbk.html.
- Wu X., Wu F.H., Wu Q., Zhang S., Chen S., Sima M. Phylogenetic and molecular evolutionary analysis of Mitophagy receptors under hypoxic conditions. Front Physiol. 2017;8:539. doi: 10.3389/fphys.2017.00539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Cherok E., Das S., Li S., Roelofs B.A., Ge S.X., Polster B.M., Boyman L., Lederer W.J., Wang C. Mitochondrial E3 ubiquitin ligase MARCH5 controls mitochondrial fission and cell sensitivity to stress-induced apoptosis through regulation of MiD49 protein. Mol Biol Cell. 2016;27:349–359. doi: 10.1091/mbc.e15-09-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonashiro R., Ishido S., Kyo S., Fukuda T., Goto E., Matsuki Y., Ohmura-Hoshino M., Sada K., Hotta H., Yamamura H. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 2006;25:3618–3626. doi: 10.1038/sj.emboj.7601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonashiro R., Kimijima Y., Shimura T., Kawaguchi K., Fukuda T., Inatome R., Yanagi S. Mitochondrial ubiquitin ligase MITOL blocks S-nitrosylated MAP1B-light chain 1-mediated mitochondrial dysfunction and neuronal cell death. Proc Natl Acad Sci U S A. 2012;109:2382–2387. doi: 10.1073/pnas.1114985109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Liu T., Jin S., Wang X., Qu M., Uhlén P., Tomilin N., Shupliakov O., Lendahl U., Nistér M. Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. EMBO J. 2011;30:2762–2778. doi: 10.1038/emboj.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinszner H., Kuroda M., Wang X., Batchvarova N., Lightfoot R.T., Remotti H., Stevens J.L., Ron D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.