Abstract
The first step in treating lung cancer is to establish the stage of the disease, which in turn determines the treatment options and prognosis of the patient. Many factors are involved in lung cancer staging, but all involve anatomical information. However, new approaches, mainly those based on the molecular biology of cancer, have recently changed the paradigm for lung cancer treatment and have not yet been incorporated into staging. In a group of patients of the same stage who receive the same treatment, some may experience unexpected recurrence or metastasis, largely because current staging methods do not reflect the findings of molecular biological studies. In this review, we provide a brief summary of the latest research on lung cancer staging and the molecular events associated with carcinogenesis. We hope that this paper will serve as a bridge between clinicians and basic researchers and aid in our understanding of lung cancer.
Keywords: adenocarcinoma, epigenetic alteration, lung cancer, mutation, staging
LUNG CANCER STAGING
The first American Joint Committee on Cancer (AJCC) lung cancer staging manual was published in 1977 (American Joint Committee, 1977). The TNM was composed of T (tumor), N (lymph node), and M (metastasis) descriptors. Each of them can be in different combinations, each combination grouped on the basis of survival and referred to the same stage. In the three decades since, lung cancer staging has changed dramatically. The most recent manual, the Eight edition, developed by the International Association for the Study of Lung Cancer (IASLC) in 2015 and published by the Union for International Cancer Control (UICC) and the AJCC in 2016, is based on data collected between 1999 and 2010 from 94,708 patients including small cell and non-small cell lung cancer (NSCLC) treated in 35 institutions in 16 countries on five continents (Goldstraw et al., 2016). Table 1 outlines the detailed contents of the current one. Although the dataset is somewhat geographically biased (data from Europe, 49%; Asia, 44%; and North America, 5%), it remains the unique and official staging manual for lung cancer from the AJCC, UICC, and IASLC.
Table 1.
Essentials of T, N, and M descriptors for the lung cancer and individual staging
| T descriptors (primary tumor) | |
| Tis | Carcinoma in situ |
| T1 | T1a(mi) – Minimally invasive adenocarcinoma T1a – tumor ≤ 1 cm in greatest dimension T1b – tumor > 1 cm but ≤ 2 cm in greatest dimension T1c – tumor > 2 cm but ≤ 3 cm in greatest dimension |
| T2 | Invasion of visceral pleura T2a – tumor > 3 cm but ≤ 4 cm in greatest dimension T2b – tumor > 4 cm but ≤ 5 cm in greatest dimension |
| T3 | Separate nodule in the same lobe (primary tumor) Invasion of chest wall (including parietal pleura), phrenic nerve, pericardium Tumor > 5 cm but ≤ 7 cm in greatest dimension |
| T4 | Separate nodule in a same side (primary tumor) but a different lobe Invasion of diaphragm, mediastinum, heart, trachea, and esophagus, etc. Tumor > 7 cm in greatest dimension |
| N descriptors (regional lymph node involvement) | |
| N0 | No lymph node metastasis |
| N1 | Metastasis in ipsilateral peribronchial, hilar, or intrapulmonary lymph node(s) |
| N2 | Metastasis in ipsilateral mediastinal or subcarinal lymph node(s) |
| N3 | Metastasis in scalene, supraclavicular, or contralateral mediastinal, hilar lymph node(s) |
| M descriptors (distant metastasis) | |
| M0 | No distant metastasis |
| M1a | Separate tumor nodule(s) contralateral lobe or pleural/pericardial nodule or malignant pleural/pericardial effusion |
| M1b | Single extrathoracic metastasis |
| M1c | Multiple extrathoracic metastasis in one or more organs |
| Individual staging | |
| Stage 0 | TisN0M0 |
| Stage I | T1, T2a with N0M0 |
| Stage II | T2b, T3 with N0M0 T1, T2 with N1M0 |
| Stage III | T4N0M0 T3N1, T4N1 Any T, N2-3M0 |
| Stage IV | Any T, Any N, M1 |
T (tumor) descriptors
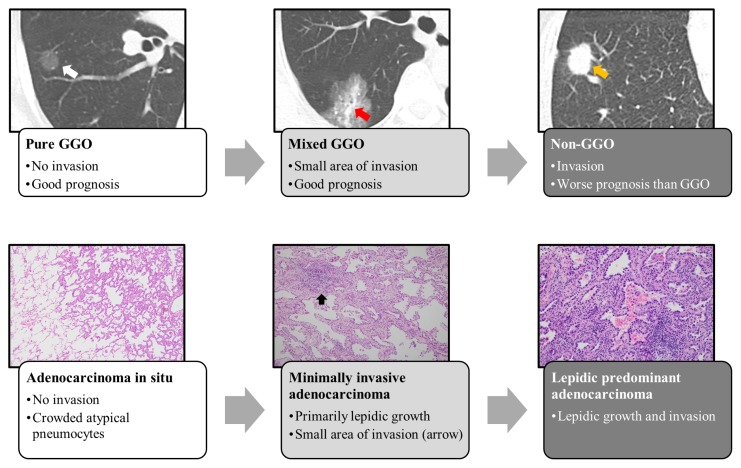
T descriptors consist of Tis, T1a(mi), T1, T2, T3, and T4. What was formerly called bronchioloalveolar carcinoma (BAC) was classified into Tis and T1a(mi); Tis refers to adenocarcinoma in situ (AIS), and T1a(mi) refers to minimally invasive adenocarcinoma (MIA) (Goldstraw et al., 2016). AIS is defined as a lung adenocarcinoma (LUAD) 3 cm or less in size, with a pattern of pure lepidic growth in which tumor cells proliferate along the surface of intact alveolar walls without stromal or vascular invasion. MIA is similar to AIS, but has a pattern of predominantly lepidic growth and an invasive component with a long axis shorter than 0.5 cm. T1 is subdivided into T1a (≤ 1 cm), T1b (> 1 cm but ≤ 2 cm), and T1c (> 2 cm but ≤ 3 cm). T2 is defined as a tumor of 3 to 5 cm in size or a tumor that has invaded the visceral pleura (the outer surface of the lung). T3 refers to tumors of 5 to 7 cm, and T4 is defined as a mass larger than 7 cm or a tumor that has invaded the diaphragm. Detailed information is provided in Table 1.
N descriptors
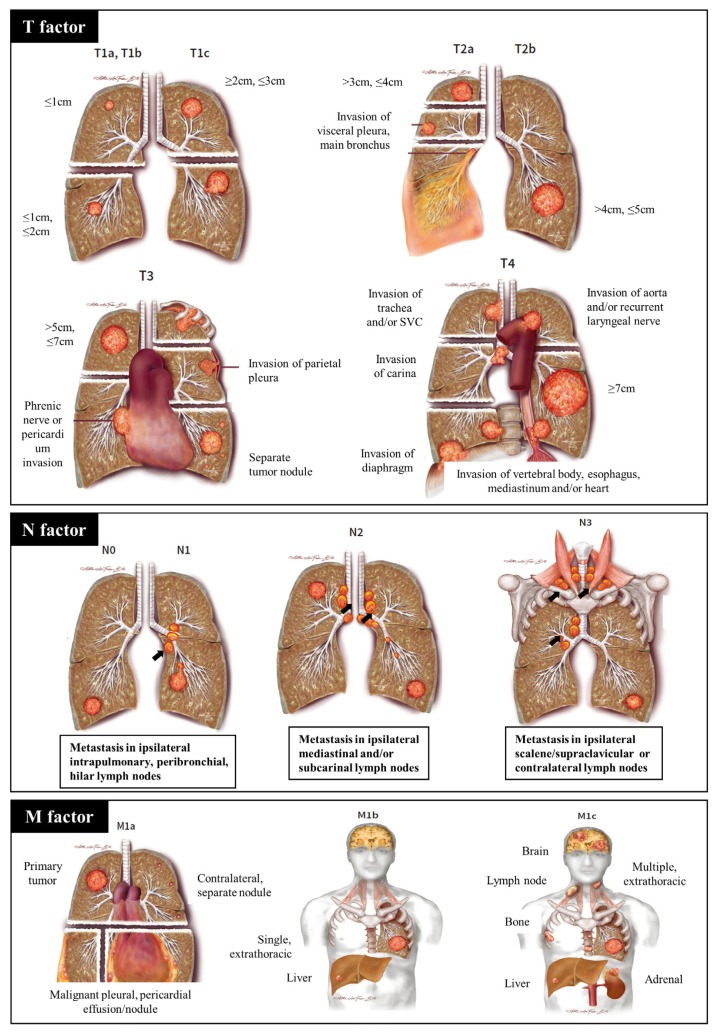
The N descriptors consist of N0, N1, N2, and N3 (Fig. 1). N0 means no metastasis of the intrathoracic lymph node. N1 is defined as metastasis in the ipsilateral (i.e., on the same side as the main tumor), intrapulmonary, and peribronchial lymph nodes. N2 is defined as the presence of metastasis in ipsilateral mediastinal and/or subcarinal lymph nodes. N3 is any kind of lymph node metastasis beyond N2. Usually, the N descriptors are decided not by number, but by the location of the metastatic lymph node(s).
Fig. 1. Atlas of lung cancer staging.
T factor is determined by size, location, invasion, and number of primary tumors. Location of metastatic lymph nodes defines the N factor. The M factor refers to metastasis: malignant pleural or pericardial metastasis is M1a, single extrathoracic metastasis is M1b, and multiple extrathoracic metastasis is M1c. These images and content are from the chapter 6 in the book of Rami-Porta (2016). Permission for use of this material was granted by IASLC.
Although previously, the prognosis of N descriptors was considered to be independent of the number of metastatic lymph nodes, a recent proposal recommends that the number should be considered when determining stage; however, it was not adopted in the current manual (Asamura et al., 2015).
M descriptors
The M descriptors consist of M0, M1a, M1b, and M1c (Fig. 1). M0 indicates no metastasis except those in lymph nodes, whereas M1 refers to distant organ metastasis, and is subdivided into M1a, M1b, and M1c. M1a is defined as intra-thoracic metastasis, including pleural or pericardial effusions (Rami-Porta, 2016). Moreover, separate and contralateral nodules with similar histology to the primary tumor can be classified as M1a. M1b refers to single extra-thoracic metastasis, and M1c to multiple extra-thoracic metastasis.
PATHOLOGIC, GENETIC, AND EPIGENETIC DESCRIPTORS TO CONSIDER FOR CANCER STAGING
As can be seen in the latest version, the criteria for defining individual stage are overall survival rate and anatomic information. T stage is determined by size, location, invasion, and number: how large the malignant tumor is, where the mass is located, whether it invades, and how many tumors are present. N stage is determined by location, i.e., where the metastatic lymph nodes are located. M stage is determined by the location and number: of metastases inside or outside of the chest. The various combinations of T, N, and M, regardless of whether they are determined by computed tomography (CT) imaging or magnetic resonance imaging (MRI), are divided into subgroups based on survival rate. Each subgroup is defined as an independent stage.
However, the current staging system has some limitations. First, until now only anatomical data has been considered. It is natural that the principles of the TNM system classifies cancer cases into groups according to anatomical extent (Rami-Porta, 2016). However, modern medicine has produced a great deal of information that leads to significant differences in the survival rates of lung cancer patients. Next, overall survival, which is used in the current system, can be meaningfully assessed only if the patient has received the best available treatment. In other words, the best survival rate of stage I NSCLC can be guaranteed if the patients have had a curative resection, whereas the best survival rates of stage II or III disease can be guaranteed if the patient had the standard treatment including chemotherapy and/or surgery/radiotherapy. However, if a patient with stage IV lung cancer who receives the latest medicine (tyrosine kinase inhibitor [TKI] or immune check point inhibitor) has the same chance of survival as a patient with stage I lung cancer, it is debatable whether this patient should be considered stage I or stage IV. Finally, the staging system focuses on the disease itself, rather than on the patient. The progression of disease, or stage of lung cancer, is the result of the interaction between the disease and the patient. It is widely accepted that the current TNM classification is a potent prognostic factor; however, the current approach to staging, which does not take the patient into account, is bound to have some limitations (Rami-Porta, 2016). Therefore, we will describe the deficiencies in the current stage in terms of pathology, mutation, and epigenetic alteration (Table 2).
Table 2.
Important factors not addressed in the current staging method
| Title | Factors | Clinical implications |
|---|---|---|
| Histopathologic information | ||
| Grade | Poorly differentiated | Worse prognostic factor for NSCLC (< 20 mm) |
| Mitosis | Mitosis count (> 10/10 HPF) | Worse prognostic factor for stage I NSCLC |
| Mutations of prognosis | ||
| BRAF | V600E | High incidence of axillary lymph node metastasis |
| EGFR | Mutations in exon 18, 19, or 21 | Better prognostic factor for survival |
| KRAS | Mutations in exon 2 | Worse prognostic factor for survival |
| RET | RET rearrangement | Better prognostic factor for survival |
| MET | Gene copy number variations (> 5 copies/cell) | Worse prognostic factors, especially in squamous cell or stage III/IV NSCLC |
| Epigenetic alterations | ||
| RUNX3 | Inactivation | Worse prognostic factor |
| MGMT | Methylation | More developed in stage II–IV |
| RASSF1A | Methylation | Shorter duration of survival |
Histopathologic information
Histologic grade, which is not addressed in the current staging system, is an important prognostic factor for lung cancers sized 20 mm or less (Kobayashi et al., 2007). Histologic grade is categorized into well-, moderately, and poorly differentiated carcinoma according to the degree of structural and cytologic atypia. In LUAD, ‘poorly differentiated’ (PD) is defined as a solid-pattern tumor without any clear gland formation, whereas in a squamous cell carcinoma, PD is defined as a solid-pattern tumor with a low degree of keratinization, intracellular bridges, and squamous pearl formation. PD status is the only independent factor identified as influencing overall survival, disease-specific survival, and disease-free survival in the study cited at the beginning of this section (Kobayashi et al., 2007). Therefore, adjuvant therapy should be considered in patients with PD lung cancer even if they are stage I.
Mitotic index is a strong prognostic factor for stage I LUAD (Duhig et al., 2015). A total of 145 cases of stage 1 LUAD were retrospectively reviewed by pathologists, who analyzed the specimens from these cases for predominant architectural pattern, nuclear grade, mitotic index, and necrosis. Mitotic activity was assessed by counting the number of mitoses within 10 high-power fields (HPFs), with the aim of counting 50 HPFs or five sets of 10 HPFs. For multivariate analysis, mitotic count was categorized as 0 to 10 or > 10 mitoses per 10 HPF. Interestingly, stage (1A vs 1B) was not a prognostic factor in stage 1 LUAD, but mitosis count (over 10 per 10 HPF) was a significantly worse prognostic factor, with a hazard ratio (HR) of 4.58 (95% confidence interval = 1.893-11.11) in multivariate analysis (Vlahos, 2018).
Mutations that influence prognosis
Mutations of genes including B-RAF (McEvoy et al., 2017), EGFR (epidermal growth factor receptor), K-RAS (Cadranel et al., 2012), RET (Tsai et al., 2015), and MET (Cappuzzo et al., 2009; Go et al., 2010) are used in determining the prognosis of lung cancer. In NSCLC patients with B-RAF mutations, metastasis of axillary lymph nodes frequently develops (McEvoy et al., 2017). Axillary lymph nodes are not usually involved in lung cancer; consequently, the incidence of metastasis to these nodes in lung cancer patients is only 0.6% to 0.7% overall (Riquet et al., 1998; Satoh et al., 2009). However, in patients with B-RAF mutations, the incidence is much higher, about 15%. Therefore B-RAF–mutated lung cancer may represent a genetically distinct subgroup, requiring detailed physical examination.
A prospective study of a French cohort reported that EGFR and K-RAS mutations were prognostic factors for advanced lung cancer patients (Cadranel et al., 2012). However, the prognosis differed in two ways depending on whether the EGFR or K-RAS mutation was employed. First, EGFR mutation was a prognostic factor for both overall and progression-free survival, whereas K-RAS mutation was significant only for overall survival. Second, EGFR mutation was a positive prognostic factor with an HR for overall survival of 0.7, whereas K-RAS mutation was a negative prognostic factor with an HR of 1.7. Moreover, because all patients in this study were treated with EGFR-TKI, the physiological significance of K-RAS mutation in TKI-naïve patients remains to be elucidated. However, other studies of EGFR copy number revealed that EGFR amplification had no significant effect on survival (Cappuzzo et al., 2009).
RET rearrangement is a rare genetic change in lung cancer, occurring in 1% to 2% of cases. However, according to a recent report, it is detected in 2.4% of advanced lung cancers with malignant pleural effusion (Tsai et al., 2015). In that study, advanced lung cancer patients with malignant pleural effusion were divided into four groups: RET rearrangement, EGFR mutation, K-RAS mutation, and ALK rearrangement. The median overall survival of patients with RET rearrangement was 22.4 months, versus 21.3 months for patients with EGFR-mutant tumors. Notably, all patients with EGFR mutations were treated with EGFR-TKI, whereas kinase inhibitors were not administered to patients with RET rearrangement. Although the study population was limited to patients with malignant pleural effusion, the results suggest that the oncogenic activity of RET may be modest.
MET, which encodes the receptor for hepatocyte growth factor, is frequently overexpressed in NSCLC. Activation of MET stimulates cell–cell detachment, migration, and invasiveness (Birchmeier et al., 2003). In a retrospective study of 447 NSCLC patients who underwent radical tumor resection, high MET gene copy number (mean ≥ 5 copies per cell) was associated with worse prognosis than low copy number (Cappuzzo et al., 2009). Another study of 180 NSCLC tissue microarrays revealed that MET fluorescence in situ hybridization (FISH) positivity was significantly associated with shorter survival, especially in squamous cell lung cancer (median overall survival, 49 months; P = 0.028) and stage III and IV disease (median overall survival, 23 months; P = 0.034) (Go et al., 2010). Multivariate analysis revealed similar results, but the effect was significant only in squamous cell carcinoma (HR = 2.330, 95% confidence interval = 1.151–4.713) (Go et al., 2010).
Epigenetic alterations
Epigenetic abnormalities, including promoter hypermethylation, are involved in the prognosis of NSCLC. In this context, RUNX3 (RUNX family transcription factor 3), MGMT (O-6-methylguanine DNA methyltransferase), and RASSF1 (Ras association domain family member 1) have been investigated. Inactivation of Runx3 is a crucial event in the development of LUAD. Lee et al. (2013) demonstrated that targeted inactivation of Runx3 induces lung adenomas and markedly shortens the latency of adenocarcinoma formation induced by oncogenic K-RAS. Moreover, aberrant inactivation of RUNX3 is frequently detected in lung cancer tissue and is also associated with poor prognosis in lung cancer patients (Chen et al., 2018; Omar et al., 2012). The DNA repair gene MGMT is inactivated after methylation of p16 gene, and this methylation prevents DNA repair of alkyl adducts, potentially leading to acquisition of somatic mutations in genes such as TP53 (Wistuba et al., 1999). Methylation of MGMT is detected more frequently in stage II to IV adenocarcinoma than in stage I (Pulling et al., 2003). Recent work showed that RASSF1A promoter hypermethylation is a prognostic marker in surgically resected NSCLC (Yanagawa et al., 2007). The authors of that study analyzed 101 (n = 68 in stage I, n = 33 in stage II, III) surgically resected NSCLCs and studied methylation in 10 genes including RASSF1A, RUNX3, DAPK, and CDKN2A (p16). The frequencies of gene methylation were 42% for RASSF1A, 27% for p16, 26% for DAPK, and 25% for RUNX3. In patients with stage I disease, positive RASSF1A or RUNX3 methylation status was associated with poor prognosis.
MOLECULAR EVENTS OF LUNG ADENOCARCINOMA TUMORIGENESIS
Stepwise progression of LUAD and molecular events
LUAD develops into invasive carcinoma through atypical adenomatous hyperplasia (AAH), AIS, and MIA (Noguchi, 2010). Usually, early LUAD exhibits ground-glass opacity (GGO) in chest CT; a case report of a 10-year follow-up of GGO clearly depicted the stepwise progression of LUAD (Fig. 2) (Min et al., 2010). In that report, the authors made three important points. First, the tumor size of GGO does not reflect tumor invasion; instead, the size of the solid portion is a stronger determinant of tumor aggressiveness. GGO can be classified as pure or mixed: the former consists exclusively of GGO, whereas the latter consists of peripheral GGO and a central solid portion (Fig. 2). Second, routine 2-year follow-up may be insufficient for GGO nodules because they grow slowly. Third, positron emission tomography (PET) imaging is not an effective means of detecting pure GGO nodules because in contrast to typical LUAD, these nodules take up very little FDG (fluorine-18 labeled glucose), a marker of elevated metabolism and risk of malignancy. As shown above, because LUAD develops in a stepwise manner, it is associated not only with radiological changes, but also changes in the pathophysiology of the tumor. These changes can be attributed to various molecular events such as driver mutations and epigenetic alteration at each stage.
Fig. 2. Step-wise progression of LUAD.
Radiologically, LUAD develops from GGO (ground-glass opacity) to mixed and non-GGO. Pathologically AIS lesions progresses to MIA when invasion occurs. If the invasion exceeds 5 mm, then the MIA will progress to the next step. White arrow indicates pure GGO, and red arrow indicates the solid portion of a GGO nodule, yellow arrow indicates a non-GGO lung nodule.
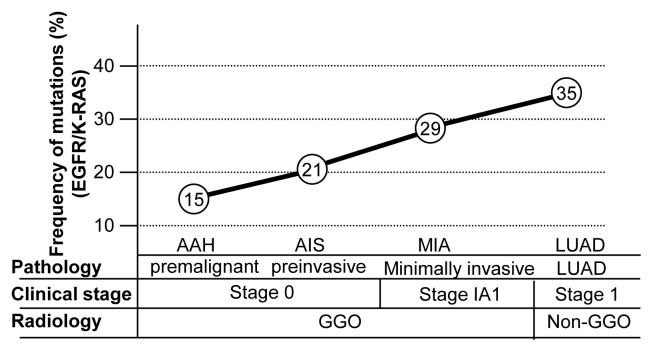
Alterations of EGFR, KRAS, and TP53 have been confirmed as significant driver mutations of LUAD in many studies. Although these mutations are called drivers, this does not necessarily mean they are sufficient for tumor initiation. If they were, then their degree of their expression would have to be constant over the course of the various stages of LUAD. Instead, however, their expression tends to increase at later stages. A study of stage I LUAD in a Korean cohort (Yoo et al., 2010) found that the frequency of EGFR mutation increased with stage up (35% in AAH, 34.9% in AIS, and 49% in MIA). A gene panel study revealed that EGFR and TP53 mutations were present, but were not predominant (Izumchenko et al., 2015): EGFR mutations were detected in 8% of AAH, 20% of AIS, and 75% of MIA. By contrast, KRAS mutations were detected in 12% of AAH, 20% of AIS, and 0% of MIA; and TP53 mutations were detected in 8% of AAH, 7% of AIS, and 35% of MIA. A similar increasing trend in the frequency of oncogenic mutations and TP53 mutation with tumor progression was also observed in a study based on whole-genome allelic imbalance scanning (Matsumoto et al., 2006; Nakanishi et al., 2009). Detailed data are shown in Table 3 and described in Figure 3.
Table 3.
Stepwise progression of LUAD and associated molecular events
| Authors | Population | AAH (%) | AIS (%) | MIA (%) | LUAD (%) | |
|---|---|---|---|---|---|---|
| Pre-malignant to minimally invasive | ||||||
| Yoo et al. (2010) | AAH (n = 20) | EGFR | 35 | 35 | 49 | |
| AIS (n = 43) | ||||||
| MIA (n = 47) | ||||||
| Izumchenko et al. (2015) | AAH (n = 25) | EGFR | 8 | 20 | 75 | |
| AIS (n = 20 zones) | KRAS | 12 | 20 | 0 | ||
| MIA (n = 15 zones) | TP53 | 8 | 7 | 35 | ||
| Nakanishi et al. (2009) | AIS (n = 15) | EGFR | 67 | 68 | ||
| MIA (n = 40) | KRAS | 13 | 8 | |||
| LUAD (n = 17) | TP53 | 13 | 53 | |||
| Matsumoto et al. (2006) | AIS (n = 11) | EGFR | 17 | 17 | ||
| MIA (n = 25) | KRAS | 2 | 10 | |||
| LUAD (n = 6) | TP53 | 0 | 21 | |||
| LUAD | ||||||
| Ahrendt et al. (2003) | Stage I (n = 106) | TP53 | 55 | |||
| Kosaka et al. (2009) | Stage I (n = 127) | EGFR | 51 | |||
| KRAS | 11 | |||||
| Shepherd et al. (2017) | Stage I (n = 569) | KRAS | 63 | |||
| TP53 | 42 | |||||
Fig. 3. Molecular events associated with the stepwise progression of LUAD.
In a series of processes from precancerous lesions to AIS, MIA, and invasive adenocarcinoma, epigenetic alterations such as RUNX3 inactivation occur first (oncogenic initiation), and then driver mutations in genes such as EGFR and K-RAS are activated (oncogenic activation). Finally, the development of TP53 mutation causes invasion and metastasis (oncogenic progression).
As shown in Table 3, not all cases of LUAD have a driver mutation in genes such as EGFR, KRAS, or TP53, although the frequency of driver mutations increases with stage. This phenomenon was also observed in a whole-exome sequencing study of 100 early lung cancers (Jamal-Hanjani et al., 2017). The results showed that mutations of KRAS (n = 26), EGFR (n = 13), and TP53 (n = 29) mainly involved pre-genome doubling and/or clonal somatic events, although a minority were due to post-genome doubling or sub-clonal somatic events. In other words, it is possible that other molecular events occur prior to oncogenic driver mutation (Izumchenko et al., 2015). Therefore, it is necessary to pay attention to epigenetic alterations.
Stepwise progression of LUAD and epigenetic alteration
Epigenetic alteration, including promoter hypermethylation, is a crucial component of cancer initiation and progression (Belinsky, 2004). Hypermethylation of many genes is a general characteristic of the cellular transformations leading to LUAD. In this context, RUNX3, CDKN2A (cyclin-dependent kinase inhibitor 2A), DAPK, MGMT, and RASSF1 have been investigated. RUNX3, which plays indispensable roles in differentiation and also functions as a tumor suppressor, is frequently inactivated in multiple types of cancers (Ito et al., 2015). Furthermore, recent data showed RUNX3 is frequently inactivated by down-regulation or mis-localization in human LUAD (Lee et al., 2010; 2013). In adult mouse lung, inactivation of Runx3 induces lung adenomas of either the mucinous or non-mucinous type and accelerates progression of oncogenic KRAS–induced LUAD (Lee et al., 2013). Development of LUAD is often associated with dysregulation of lung epithelial lineage–determining transcriptional regulators that govern differentiation status, and RUNX3 is required for both bronchiolar and alveolar lineage differentiation (Ito et al., 2015; Lee and Bae, 2016). As a component of the p53-ARF pathways, RUNX3 also plays a critical role in the defense against oncogene activation. When oncogenic K-RAS is constitutively activated, the RUNX3–BRD2 complex is maintained, and expression of ARF and TP53 is prolonged until the G1/S checkpoint (Lee et al., 2013). Thus, the RUNX3–BRD2 complex functions as a sensor for abnormal persistence of RAS activity (Lee et al., 2013). However, inactivation of RUNX3 disturbs these pathways, making cells vulnerable to activation of oncogenes such as K-RAS.
CDKN2A encodes two proteins of the INK4 family, p16 and p14-ARF. Both act as tumor suppressors by regulating the cell cycle. Hypermethylation of CDKN2A can occur early in the genesis of some premalignant lesions (Belinsky et al., 2005), and methylation levels increase during disease progression from basal cell hyperplasia (17%) to squamous cell metaplasia (24%) to carcinoma in situ (50%). Interestingly, inactivation of p16 has been detected in bronchial epithelial cells of cancer-free smokers.
DAPK encodes death-associated protein kinase, a serine/threonine kinase. Methylation of DAPK is detected in approximately 50% of alveolar hyperplasia induced in mouse lungs exposed chronically to NKK (tobacco-specific carcinogen 4-methylnitrosamino-1-(3-pyridyl)-1-butanone) (Pulling et al., 2004). Methylation of MGMT blocks DNA repair of alkyl adducts and can lead to acquisition of somatic mutations in genes such as TP53 (Wistuba et al., 1999). Methylation of MGMT was more developed in stage II to IV adenocarcinoma than stage I (Pulling et al., 2003). CpG island methylation of the RASSF1A promoter region was detected in 72% of small cell lung cancer and 34% of NSCLC (Agathanggelou et al., 2001). Molecular studies revealed that RASSF1A suppresses the invasion and metastatic potential of human NSCLC by inhibiting activation of YAP (Yes-associated protein) through the GEF-H1/RhoB pathway (Dubois et al., 2016).
Epigenetic alteration and mutations according to LUAD progression
Epigenetic alteration of multiple important genes could be a potent cause of early LUAD tumorigenesis (Jones and Baylin, 2007). Lee et al. (2010; 2013) showed that inactivation of Runx3 is frequently detected in mouse and human lung cancer tissue. Disruption of p16 appears to be one of the earliest events involved in enabling clonal expansion (Belinsky, 2005), and DAPK further aids in the development of autonomous premalignant clones. Subsequent loss of MGMT could lead to acquisition of somatic mutations in genes such as TP53, whose inactivation occurs at more advanced stages of pre malignancy (Wistuba et al., 1999). These epigenetic alterations can result in driver mutations in genes such as EGFR and K-RAS, or create an environment that promotes the occurrence of these mutations (oncogenic activation). The activity of driver mutations accelerates cell division. This process is illustrated in Figure 3.
SUMMARY
The current lung cancer staging system is the result of systematic studies using large-scale, long-term follow-up data (Eberhardt et al., 2015; Goldstraw et al., 2016; Rami-Porta et al., 2014; Travis et al., 2016). However, 40 years have passed since the first version of lung cancer staging was published, and it is to be expected that staging using only anatomical data will be subject to certain limitations. For example, among stage IV lung cancer patients, survival rates differ significantly depending on the presence or absence of EGFR mutation (Feldser et al., 2010; Janne et al., 2015; Kobayashi et al., 2005; Rami-Porta et al., 2014; Sequist et al., 2013; Yatabe et al., 2014). Therefore, in order to achieve accurate lung cancer staging, it is necessary to consider a wide range of medical information in addition to anatomy.
LUAD progresses in a stepwise manner. Driver mutations in genes as EGFR, K-RAS, and TP53 are involved in this progression, and epigenetic alterations also play important roles in tumorigenesis. However, not all cases of LUAD have apparent driver mutations. Yatabe et al. (2014) proposed four explanations for why not all LUAD follows a stepwise progression: i) the linear progression hypothesis could be applied to a subset of LUAD, according to a molecular classification; ii) the mutation rates of EGFR and K-RAS are not evenly distributed during progression; iii) some cases of LUAD revealed gene alterations discontinuous to invasive carcinoma; and iv) some lesions do not progress. In regard to driver mutations and epigenetic alterations, it is true that EGFR, KRAS, and TP53 play important roles in lung tumorigenesis, but not all tumors develop by activating these mutations alone, and conversely, not all tumors can be eliminated solely by repressing these changes (Feldser et al., 2010; Janne et al., 2015; Kobayashi et al., 2005; Sequist et al., 2013). Although the mutations described above have been called drivers, the mutations themselves might not be sufficient to drive lung cancer; instead, epigenetic alterations might cause a mutation to act as a true driver. Therefore, future lung cancer treatment should focus on both mutations and epigenetic alterations (Lee et al., 2010; 2013; 2019; Yanagawa et al., 2007).
ACKNOWLEDGMENTS
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (2017R1C1B5015969). S-C Bae is supported by a Creative Research Grant (2014R1A3A2030690) through the National Research Foundation (NRF) of Korea. Y-S Lee is supported by Basic Science Research Program grant 2017R1D1A3B03034076.
Footnotes
Disclosure
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Agathanggelou A., Honorio S., Macartney D.P., Martinez A., Dallol A., Rader J., Fullwood P., Chauhan A., Walker R., Shaw J.A., et al. Methylation associated inactivation of RASSF1A from region 3p21.3 in lung, breast and ovarian tumours. Oncogene. 2001;20:1509–1518. doi: 10.1038/sj.onc.1204175. [DOI] [PubMed] [Google Scholar]
- Ahrendt S.A., Hu Y., Buta M., McDermott M.P., Benoit N., Yang S.C., Wu L., Sidransky D. p53 mutations and survival in stage I non-small-cell lung cancer: results of a prospective study. J Natl Cancer Inst. 2003;95:961–970. doi: 10.1093/jnci/95.13.961. [DOI] [PubMed] [Google Scholar]
- American Joint Committee. Manual for Staging of Cancer 1977. Chicago: American Joint Committee; 1977. [Google Scholar]
- Asamura H., Chansky K., Crowley J., Goldstraw P., Rusch V.W., Vansteenkiste J.F., Watanabe H., Wu Y.L., Zielinski M., Ball D., et al. The International Association for the Study of Lung Cancer Lung Cancer Staging Project: proposals for the revision of the N descriptors in the forthcoming 8th edition of the TNM Classification for lung cancer. J Thorac Oncol. 2015;10:1675–1684. doi: 10.1097/JTO.0000000000000678. [DOI] [PubMed] [Google Scholar]
- Belinsky S.A. Gene-promoter hypermethylation as a biomarker in lung cancer. Nat Rev Cancer. 2004;4:707–717. doi: 10.1038/nrc1432. [DOI] [PubMed] [Google Scholar]
- Belinsky S.A. Silencing of genes by promoter hypermethylation: key event in rodent and human lung cancer. Carcinogenesis. 2005;26:1481–1487. doi: 10.1093/carcin/bgi020. [DOI] [PubMed] [Google Scholar]
- Birchmeier C., Birchmeier W., Gherardi E., Vande Woude G.F. Met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003;4:915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- Cadranel J., Mauguen A., Faller M., Zalcman G., Buisine M.P., Westeel V., Longchampt E., Wislez M., Coudert B., Daniel C., et al. Impact of systematic EGFR and KRAS mutation evaluation on progression-free survival and overall survival in patients with advanced non-small-cell lung cancer treated by erlotinib in a French prospective cohort (ERMETIC project--part 2) J Thorac Oncol. 2012;7:1490–1502. doi: 10.1097/JTO.0b013e318265b2b5. [DOI] [PubMed] [Google Scholar]
- Cappuzzo F., Marchetti A., Skokan M., Rossi E., Gajapathy S., Felicioni L., Del Grammastro M., Sciarrotta M.G., Buttitta F., Incarbone M., et al. Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients. J Clin Oncol. 2009;27:1667–1674. doi: 10.1200/JCO.2008.19.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Deng Y., Shi Y., Zhu W., Cai Y., Xu C., Zhu K., Zheng X., Chen G., Xie Q., et al. Loss of expression rather than cytoplasmic mislocalization of RUNX3 predicts worse outcome in non-small cell lung cancer. Oncol Lett. 2018;15:5043–5055. doi: 10.3892/ol.2018.7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois F., Keller M., Calvayrac O., Soncin F., Hoa L., Hergovich A., Parrini M.C., Mazières J., Vaisse-Lesteven M., Camonis J., et al. RASSF1A suppresses the invasion and metastatic potential of human non-small cell lung cancer cells by inhibiting YAP activation through the GEF-H1/RhoB pathway. Cancer Res. 2016;76:1627–1640. doi: 10.1158/0008-5472.CAN-15-1008. [DOI] [PubMed] [Google Scholar]
- Duhig E.E., Dettrick A., Godbolt D.B., Pauli J., van Zwieten A., Hansen A.R., Yang I.A., Fong K.M., Clarke B.E., Bowman R.V. Mitosis trumps T stage and proposed international association for the study of lung cancer/american thoracic society/european respiratory society classification for prognostic value in resected stage 1 lung adenocarcinoma. J Thorac Oncol. 2015;10:673–681. doi: 10.1097/JTO.0000000000000446. [DOI] [PubMed] [Google Scholar]
- Eberhardt W.E., Mitchell A., Crowley J., Kondo H., Kim Y.T., Turrisi A.3rdGoldstraw P., Rami-Porta R. The IASLC lung cancer staging project: proposals for the revision of the M descriptors in the forthcoming eighth edition of the TNM Classification of lung cancer. J Thorac Oncol. 2015;10:1515–1522. doi: 10.1097/JTO.0000000000000673. [DOI] [PubMed] [Google Scholar]
- Feldser D.M., Kostova K.K., Winslow M.M., Taylor S.E., Cashman C., Whittaker C.A., Sanchez-Rivera F.J., Resnick R., Bronson R., Hemann M.T., et al. Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature. 2010;468:572–575. doi: 10.1038/nature09535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go H., Jeon Y.K., Park H.J., Sung S.W., Seo J.W., Chung D.H. High MET gene copy number leads to shorter survival in patients with non-small cell lung cancer. J Thorac Oncol. 2010;5:305–313. doi: 10.1097/JTO.0b013e3181ce3d1d. [DOI] [PubMed] [Google Scholar]
- Goldstraw P., Chansky K., Crowley J., Rami-Porta R., Asamura H., Eberhardt W.E., Nicholson A.G., Groome P., Mitchell A., Bolejack V. The IASLC lung cancer staging project: proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM Classification for lung cancer. J Thorac Oncol. 2016;11:39–51. doi: 10.1016/j.jtho.2015.09.009. [DOI] [PubMed] [Google Scholar]
- Ito Y., Bae S.C., Chuang L.S. The RUNX family: developmental regulators in cancer. Nat Rev Cancer. 2015;15:81–95. doi: 10.1038/nrc3877. [DOI] [PubMed] [Google Scholar]
- Izumchenko E., Chang X., Brait M., Fertig E., Kagohara L.T., Bedi A., Marchionni L., Agrawal N., Ravi R., Jones S., et al. Targeted sequencing reveals clonal genetic changes in the progression of early lung neoplasms and paired circulating DNA. Nat Commun. 2015;16:8258. doi: 10.1038/ncomms9258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal-Hanjani M., Wilson G.A., McGranahan N., Birkbak N.J., Watkins T.B.K., Veeriah S., Shafi S., Johnson D.H., Mitter R., Rosenthal R., et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017;376:2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- Janne P.A., Yang J.C., Kim D.W., Planchard D., Ohe Y., Ramalingam S.S., Ahn M.J., Kim S.W., Su W.C., Horn L., et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med. 2015;372:1689–1699. doi: 10.1056/NEJMoa1411817. [DOI] [PubMed] [Google Scholar]
- Jones P.A., Baylin S.B. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N., Toyooka S., Soh J., Ichimura K., Yanai H., Suehisa H., Ichihara S., Yamane M., Aoe M., Sano Y., et al. Risk factors for recurrence and unfavorable prognosis in patients with stage I non-small cell lung cancer and a tumor diameter of 20 mm or less. J Thorac Oncol. 2007;2:808–812. doi: 10.1097/JTO.0b013e31814617c7. [DOI] [PubMed] [Google Scholar]
- Kobayashi S., Boggon T.J., Dayaram T., Jänne P.A., Kocher O., Meyerson M., Johnson B.E., Eck M.J., Tenen D.G., Halmos B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- Kosaka T., Yatabe Y., Onozato R., Kuwano H., Mitsudomi T. Prognostic implication of EGFR, KRAS, and TP53 gene mutations in a large cohort of Japanese patients with surgically treated lung adenocarcinoma. J Thorac Oncol. 2009;4:22–29. doi: 10.1097/JTO.0b013e3181914111. [DOI] [PubMed] [Google Scholar]
- Lee J.W., Kim D.M., Jang J.W., Park T.G., Song S.H., Lee Y.S., Chi X.Z., Park I.Y., Hyun J.W., Ito Y., et al. RUNX3 regulates cell cycle-dependent chromatin dynamics by functioning as a pioneer factor of the restriction-point. Nat Commun. 2019;10:1897. doi: 10.1038/s41467-019-09810-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.S., Lee Y.S., Lee J.M., Ito K., Cinghu S., Kim J.H., Jang J.W., Li Y.H., Goh Y.M., Chi X.Z., et al. Runx3 is required for the differentiation of lung epithelial cells and suppression of lung cancer. Oncogene. 2010;29:3349–3361. doi: 10.1038/onc.2010.79. [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Bae S.C. How do K-RAS-activated cells evade cellular defense mechanisms? Oncogene. 2016;35:827–832. doi: 10.1038/onc.2015.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.S., Lee J.W., Jang J.W., Chi X.Z., Kim J.H., Li Y.H., Kim M.K., Kim D.M., Choi B.S., Kim E.G., et al. Runx3 inactivation is a crucial early event in the development of lung adenocarcinoma. Cancer Cell. 2013;24:603–616. doi: 10.1016/j.ccr.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Matsumoto S., Iwakawa R., Kohno T., Suzuki K., Matsuno Y., Yamamoto S., Noguchi M., Shimizu E., Yokota J. Frequent EGFR mutations in noninvasive bronchioloalveolar carcinoma. Int J Cancer. 2006;118:2498–2504. doi: 10.1002/ijc.21670. [DOI] [PubMed] [Google Scholar]
- McEvoy S.H., Halpenny D.F., Viteri-Jusué A., Hayes S.A., Plodkowski A.J., Riely G.J., Ginsberg M.S. Investigation of patterns of nodal metastases in BRAF mutant lung cancer. Lung Cancer. 2017;108:62–65. doi: 10.1016/j.lungcan.2017.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J.H., Lee H.Y., Lee K.S., Han J., Park K., Ahn M.J., Lee S.J. Stepwise evolution from a focal pure pulmonary ground-glass opacity nodule into an invasive lung adenocarcinoma: an observation for more than 10 years. Lung Cancer. 2010;69:123–126. doi: 10.1016/j.lungcan.2010.04.022. [DOI] [PubMed] [Google Scholar]
- Nakanishi H., Matsumoto S., Iwakawa R., Kohno T., Suzuki K., Tsuta K., Matsuno Y., Noguchi M., Shimizu E., Yokota J. Whole genome comparison of allelic imbalance between noninvasive and invasive small-sized lung adenocarcinomas. Cancer Res. 2009;69:1615–1623. doi: 10.1158/0008-5472.CAN-08-3218. [DOI] [PubMed] [Google Scholar]
- Noguchi M. Stepwise progression of pulmonary adenocarcinoma: clinical and molecular implications. Cancer Metastasis Rev. 2010;29:15–21. doi: 10.1007/s10555-010-9210-y. [DOI] [PubMed] [Google Scholar]
- Omar M.F., Ito K., Nga M.E., Soo R., Peh B.K., Ismail T.M., Thakkar B., Soong R., Ito Y., Salto-Tellez M. RUNX3 downregulation in human lung adenocarcinoma is independent of p53, EGFR or KRAS status. Pathol Oncol Res. 2012;18:783–792. doi: 10.1007/s12253-011-9485-5. [DOI] [PubMed] [Google Scholar]
- Pulling L.C., Divine K.K., Klinge D.M., Gilliland F.D., Kang T., Schwartz A.G., Bocklage T.J., Belinsky S.A. Promoter hypermethylation of the O6-methylguanine-DNA methyltransferase gene: more common in lung adenocarcinomas from never-smokers than smokers and associated with tumor progression. Cancer Res. 2003;63:4842–4848. [PubMed] [Google Scholar]
- Pulling L.C., Vuillemenot B.R., Hutt J.A., Devereux T.R., Belinsky S.A. Aberrant promoter hypermethylation of the death-associated protein kinase gene is early and frequent in murine lung tumors induced by cigarette smoke and tobacco carcinogens. Cancer Res. 2004;64:3844–3848. doi: 10.1158/0008-5472.CAN-03-2119. [DOI] [PubMed] [Google Scholar]
- Rami-Porta R. IASLC Staging Manual in Thoracic Oncology. 2nd Edition. North Fort Myers: Editorial Rx Press; 2016. [Google Scholar]
- Rami-Porta R., Bolejack V., Giroux D.J., Chansky K., Crowley J., Asamura H., Goldstraw P. The IASLC lung cancer staging project: the new database to inform the eighth edition of the TNM Classification of lung cancer. J Thorac Oncol. 2014;9:1618–1624. doi: 10.1097/JTO.0000000000000334. [DOI] [PubMed] [Google Scholar]
- Riquet M., Le Pimpec-Barthes F., Danel C. Axillary lymph node metastases from bronchogenic carcinoma. Ann Thorac Surg. 1998;66:920–922. doi: 10.1016/S0003-4975(98)00556-6. [DOI] [PubMed] [Google Scholar]
- Satoh H., Ishikawa H., Kagohashi K., Kurishima K., Sekizawa K. Axillary lymph node metastasis in lung cancer. Med Oncol. 2009;26:147–150. doi: 10.1007/s12032-008-9097-4. [DOI] [PubMed] [Google Scholar]
- Sequist L.V., Yang J.C., Yamamoto N., O’Byrne K., Hirsh V., Mok T., Geater S.L., Orlov S., Tsai C.M., Boyer M., et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol. 2013;31:3327–3334. doi: 10.1200/JCO.2012.44.2806. [DOI] [PubMed] [Google Scholar]
- Shepherd F.A., Lacas B., Le Teuff G., Hainaut P., Jänne P.A., Pignon J.P., Le Chevalier T., Seymour L., Douillard J.Y., Graziano S., et al. Pooled analysis of the prognostic and predictive effects of TP53 comutation status combined with KRAS or EGFR mutation in early-stage resected non-small-cell lung cancer in four trials of adjuvant chemotherapy. J Clin Oncol. 2017;35:2018–2027. doi: 10.1200/JCO.2016.71.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis W.D., Asamura H., Bankier A.A., Beasley M.B., Detterbeck F., Flieder D.B., Goo J.M., MacMahon H., Naidich D., Nicholson A.G., et al. The IASLC lung cancer staging project: proposals for coding T categories for subsolid nodules and assessment of tumor size in part-solid tumors in the forthcoming eighth edition of the TNM Classification of lung cancer. J Thorac Oncol. 2016;11:1204–1223. doi: 10.1016/j.jtho.2016.03.025. [DOI] [PubMed] [Google Scholar]
- Tsai T.H., Wu S.G., Hsieh M.S., Yu C.J., Yang J.C., Shih J.Y. Clinical and prognostic implications of RET rearrangements in metastatic lung adenocarcinoma patients with malignant pleural effusion. Lung Cancer. 2015;88:208–214. doi: 10.1016/j.lungcan.2015.02.018. [DOI] [PubMed] [Google Scholar]
- Vlahos I. Dilemmas in lung cancer staging. Radiol Clin North Am. 2018;56:419–435. doi: 10.1016/j.rcl.2018.01.010. [DOI] [PubMed] [Google Scholar]
- Wistuba I.I., Behrens C., Milchgrub S., Bryant D., Hung J., Minna J.D., Gazdar A.F. Sequential molecular abnormalities are involved in the multistage development of squamous cell lung carcinoma. Oncogene. 1999;21:643–650. doi: 10.1038/sj.onc.1202349. [DOI] [PubMed] [Google Scholar]
- Yanagawa N., Tamura G., Oizumi H., Kanauchi N., Endoh M., Sadahiro M., Motoyama T. Promoter hypermethylation of RASSF1A and RUNX3 genes as an independent prognostic prediction marker in surgically resected non-small cell lung cancers. Lung Cancer. 2007;58:131–138. doi: 10.1016/j.lungcan.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Yatabe Y., Borczuk A.C., Powell C.A. Do all lung adenocarcinomas follow a stepwise progression? Lung Cancer. 2014;74:7–11. doi: 10.1016/j.lungcan.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.B., Chung J.H., Lee H.J., Lee C.T., Jheon S., Sung S.W. Epidermal growth factor receptor mutation and p53 overexpression during the multistage progression of small adenocarcinoma of the lung. J Thorac Oncol. 2010;5:964–969. doi: 10.1097/JTO.0b013e3181dd15c0. [DOI] [PubMed] [Google Scholar]