Abstract
Based on the endosymbiotic theory, one of the key events that occurred during mitochondrial evolution was an extensive loss of non-essential genes from the protomitochondrial endosymbiont genome and transfer of some of the essential endosymbiont genes to the host nucleus. We have developed an approach to recapitulate various aspects of endosymbiont genome minimization using a synthetic system consisting of E. coli endosymbionts within host yeast cells. As a first step, we identified a number of E. coli auxotrophs of central metabolites that can form viable endosymbionts within yeast cells. These studies provide a platform to identify non-essential biosynthetic pathways that can be deleted in the E. coli endosymbionts to investigate the evolutionary adaptations in the host and endosymbiont during the evolution of mitochondria.
Graphical Abstract

The endosymbiotic theory1 of mitochondria evolution suggests that mitochondria evolved from prokaryotic lineages that entered an archaeal host and were established as endosymbionts.2–3 Several phylogenetic and proteomic studies suggest that all mitochondrial genomes are descended from a common protomitochondrial ancestor.4–8 One of the key events that occurred during mitochondrial evolution is an extensive loss of non-essential genes from the protomitochondrial endosymbiont genome and transfer of some of the essential endosymbiont genes to the host nucleus.9 This resulted in the relatively low gene content of the present-day mitochondrial genome. Nature has used similar strategies to shrink genomes in various intracellular pathogenic organisms, and ground-up approaches to synthetically obtain minimal genomes have also been performed on intracellular pathogens like Mycoplasma.10–13
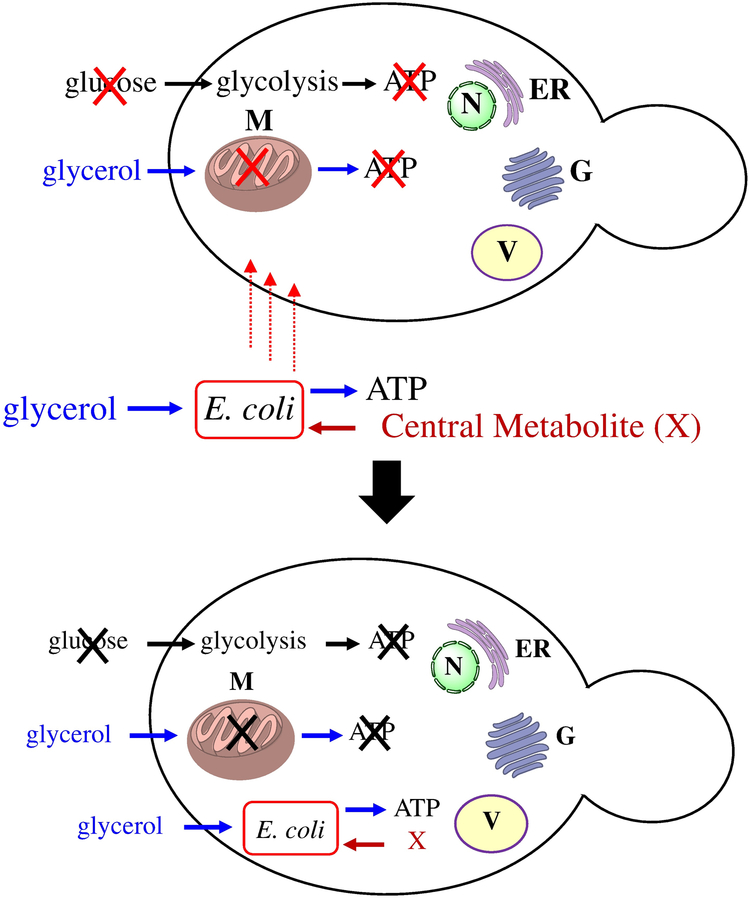
Recently, we developed a synthetic strategy to establish E. coli endosymbionts within host yeast cells to mimic mitochondrial evolution starting from bioenergetically competent and genetically tractable endosymbionts.14 Briefly, our strategy (Figure 1) involved engineering E. coli strains such that they (i) are auxotrophic for a key central metabolite (present in the host cytosol) for which there are endogenous transporters, (ii) express a functional ADP/ATP translocase to provide ATP to the respiration deficient S. cerevisiae mutant and (iii) express SNARE-like proteins to avoid lysosomal degradation as previously described.14–18 A respiration deficient mutant of S. cerevisiae, S. cerevisiae cox2–60 (NB97), was used which is unable to synthesize ATP under non-fermentable growth conditions.19–21 Under selection conditions containing non-fermentable carbon sources, the S. cerevisiae cox2–60 obtains ATP from the engineered E. coli endosymbiont and the S. cerevisiae cox2–60 cytosol in return provides thiamin to the E. coli endosymbiont that is a thiamin auxotroph. The logical next step would be to expand this system towards a minimal endosymbiont genome, similar to mitochondria evolution. We envisioned that the yeast cytosol would provide an ideal “rich medium” for the growth of E. coli endosymbionts thereby allowing us to delete numerous non-essential genes and pathways under these conditions.22–24
Figure 1: Strategy to test permissive E. coli auxotrophs that could establish endosymbiosis with S. cerevisiaecox2–60.
X- key central metabolite provided by the yeast cytosol, M-mitochondria, N-nucleus, ER-endoplasmic reticulum, G-Golgi apparatus, V-vacuole.
Since amino acid biosynthesis is a major component of bacterial metabolism, we began by testing E. coli auxotrophs for the twenty amino acids.25 Similar to the selection platform described above, the S. cerevisiae cox2–60 should obtain ATP from the engineered E. coli endosymbiont that is auxotrophic for a defined amino acid(s), and in return, the S. cerevisiae cox2–60 cytosol was expected to provide the corresponding amino acid(s) to the E. coli endosymbiont. Since E. coli encodes amino acid transporters, the E. coli endosymbiont should be able to acquire amino acids from the yeast cytosol. To this end, E. coli strains that were individual amino acid auxotroph were transformed with a plasmid - pAM136,14 containing ADP/ATP translocase from the intracellular bacterium Protochlamydia amoebophila strain UWE25, and SNAREs - Chlamydia trachomatis incA, Chlamydia caviae incA, Chlamydia trachomatis CT_813 under a PBAD promoter. These strains were then fused to respiration deficient S. cerevisiae cox2–60 (NB97). The S. cerevisiae cox2–60-E. coli chimeras were selected on partial selection medium I (3% glycerol and 0.1% glucose as carbon source) containing predominantly non-fermentable carbon sources. The permissive E. coli amino acid auxotrophs that were able to establish endosymbiotic relationship are listed in Table 1.
Table 1:
List of E. coli auxotrophs that were tested as for their ability to establish endosymbiotic relationship with S. cerevisiae cox2–60
| Strain # | Gene deleted/Strain | Auxotroph | Growth on selection medium I & II |
|---|---|---|---|
| 1 | ilvD | isoleucine, leucine, valine | ++ |
| 2 | trpC | tryptophan | + |
| 3 | serA | Serine | ++ |
| 4 | metA | methionine | ++ |
| 5 | PA340 strain | glutamic acid | ++ |
| 6 | cysE | cysteine | ++ |
| 7 | glnA | glutamine | ++ |
| 8 | argA | arginine | ++ |
| 9 | proA | proline | ++ |
| 10 | lysA | lysine | ++ |
| 11 | thrC | threonine | ++ |
| 12 | hisB | histidine | + |
| 13 | tyrA | tyrosine | − |
| 14 | pheA | phenylalanine | − |
| 15 | DL39 strain | phenylalanine, tyrosine, aspartic acid, leucine, isoleucine, valine | − |
| 16 | KA12 strain | Aromatic amino acids | − |
| 17 | thiC | thiamin | ++ |
| 18 | nadA | NAD | ++ |
| 19 | thiC, nadA | thiamin and NAD | ++ |
| 20 | thiC, nadA, metA | thiamin, NAD, methionine | ++ |
| 21 | thiC, nadA, metA, ilvD | thiamin, NAD, methionine, leucine, isoleucine, valine | ++ |
| 22 | thiC, nadA, metA, ilvD, argA | thiamin, NAD, methionine, leucine, isoleucine, valine, arginine | ++ |
| 23 | thiC, nadA, metA, ilvD, argA, proA | thiamin, NAD, methionine, leucine, isoleucine, valine, arginine, proline | ++ |
| 24 | thiC, nadA, metA, ilvD, argA, proA, glnA | thiamin, NAD, methionine, leucine, isoleucine, valine, arginine, proline, glutamine | + |
| 25 | thiC, ΔilvLXGMEDAYC | thiamin, leucine, isoleucine, valine | ++ |
As can be seen from these experiments, most of the E. coli single amino acid auxotrophs tested are permissive endosymbionts. All of the chimeric strains generated from the permissive E. coli auxotrophs and S. cerevisiae cox2–60 were able to grow on selection medium II (containing only non-fermentable carbon sources, 3% glycerol and no glucose) for multiple rounds of re-plating and re-growth (> 40 generations) unlike the host S. cerevisiae cox2–60. Growth rates for all permissive auxotrophs listed in Table 1 and Figure S19 (except for histidine and tryptophan auxotrophs) were similar to those of S. cerevisiae cox2–60 – E. coli DH10B ΔnadA::gfp-kanR- pAM136 (Figure S19).14 Significantly slower growth rate (>12 h per doubling on selection medium I) was observed for S. cerevisiae cox2–60 – E. coli DH10B ΔtrpC::kanR- pAM136 and S. cerevisiae cox2–60 – E. coli DH10B ΔhisB::kanR- pAM136. Further, we isolated total genomic DNA from the chimeric strains and PCR amplified characteristic marker gene MATa from S. cerevisiae cox2–60 and kanR gene corresponding to the E. coli auxotrophs to confirm the presence of E. coli endosymbionts within S. cerevisiae cox2–60 (see SI for data).
The inability to form endosymbionts with the Phe and Tyr auxotrophs could be due to lower intracellular concentration of phenylalanine and tyrosine in the yeast cytosol26–27. Unfortunately, our efforts either to add tyrosine to the growth medium or over-express aromatic amino acid uptake systems in E. coli (e.g., aroP) were not able to rescue this phenotype. Likewise, at present it is unclear why slower growth rates were observed for E. coli tryptophan and histidine auxotrophs (reported intracellular concentrations in the yeast cytosol: tryptophan ~ 450 μM and histidine ~ 9 mM26–27).
Because we ultimately want to minimize the E. coli genome, we asked whether we could delete the entire operon for one of the permissive auxotrophs. To test this notion, we used a deletion mutant of the isoleucine-valine biosynthetic pathway. We transformed this E. coli strain (strain 25, Table 1) with pAM136, fused it to S. cerevisiae cox2–60, and selected for S. cerevisiae cox2–60 – E. coli ΔthiC::kanR-gfp ΔilvLXGMEDAYC::tetR chimeras on selection medium I (Figure S18). We were able to propagate the chimeras on selection medium I and II for multiple rounds of re-growth, and the chimera grew at a comparable rate to that of S. cerevisiae cox2–60 – E. coli DH10B ΔnadA::gfp-kanR- pAM136 (~ 6 h per doubling on selection medium I).14
Next, we wanted to investigate whether we could generate E. coli strain auxotrophic to multiple metabolites and test the ability of such auxotrophic strains to generate yeast-endosymbionts. We had previously tested thiamin and NAD auxotrophs of E. coli and both of these auxotrophs along with their combination were permissive E. coli endosymbionts.14 We therefore started with a thiamin/NAD double auxotroph of E. coli generated by deleting the thiC and nadA genes. We further deleted the metA gene from this strain to generate E. coli strain 20 (Table 1) that is auxotrophic for thiamin, NAD and methionine.28 As before, this strain was transformed with pAM136 and fused to S. cerevisiae cox2–60. We were able to generate chimeras that propagated on selection medium II (3% glycerol and no glucose) for more than 40 generations of growth, and the growth rate was similar to that of S. cerevisiae cox2–60 – E. coli DH10B ΔnadA::gfp-kanR- pAM136 (Figure S19).14
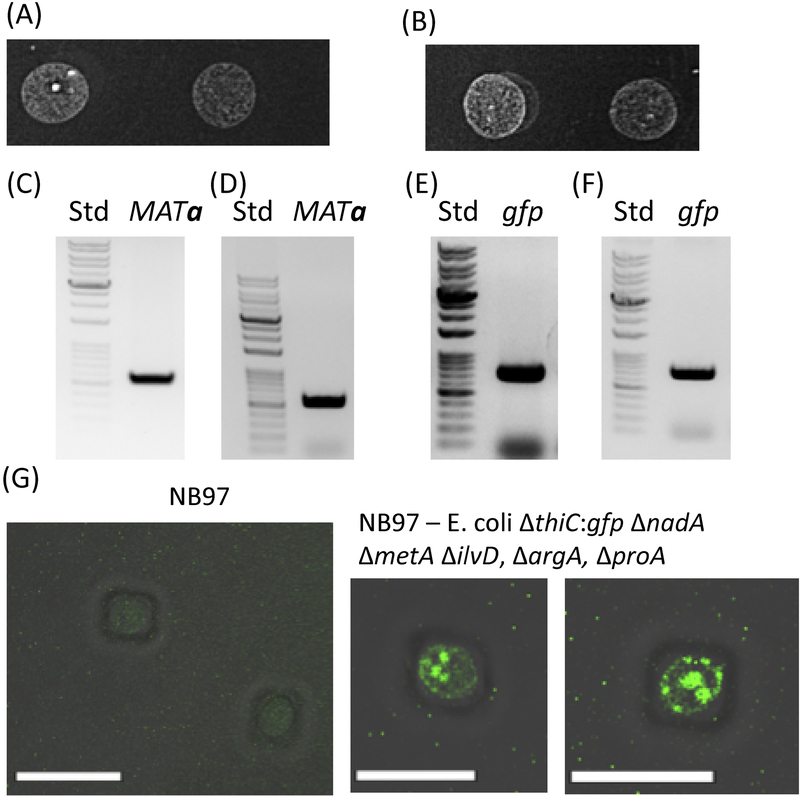
Next, we sequentially generated a series of E. coli strains that were auxotrophic in an increasing number of metabolites up to an E. coli strain auxotrophic to 8 different metabolites (thiamin, NAD, methionine, isoleucine, leucine, valine, arginine and proline, strain 23. All of these auxotrophic strains (strains 20–23 transformed with pAM136) were permissive yeast endosymbionts. The chimeras were able to sustain growth on both Selection Medium I and Selection Medium II, thereby demonstrating a respiration competent phenotype unlike the host strain S. cerevisiae cox2–60 (e.g., Figure 2A and 2B respectively, for chimeras generated from strain 23 of E. coli). Moreover, these chimeric strains showed robust growth on selection medium I (Figure S19).14 We also isolated the total genomic DNA from the chimeric strains (generated from strains 20–23) and PCR amplified the characteristic marker gene MATa from S. cerevisiae cox2–60 and gfp/tetR gene from the E. coli endosymbiont to confirm the presence of the endosymbionts within S. cerevisiae cox2–60 (Figure 2C–F, Figure S13–16). Furthermore, as a representative endosymbiont, the S. cerevisiae cox2–60 - E. coli ΔthiC:gfp ΔnadA ΔmetA ΔilvD, ΔargA, ΔproA – pAM136 chimera was characterized by fluorescence imaging to confirm the presence of the E. coli endosymbionts within the yeast. As can be seen from Figure 2G, we detect the presence of GFP expressing endosymbionts within the host S. cerevisiae cox2–60 cells.
Figure 2: Characterization of Saccharomyces cerevisiae cox2–60 - E. coli DH10B ΔthiC:gfp ΔnadA ΔmetA ΔilvD, ΔargA, ΔproA – pAM136 chimeras.
(A) and (B), Growth observed for chimeras on Selection Medium I and Selection Medium II respectively, thereby demonstrating a respiration competent phenotype. (C) and (D), Detection of NB97-specific MATa mating type by PCR analysis of total DNA samples isolated from chimeras shown in Figure 2A and 2B respectively. Std indicates DNA molecular weight standards (E) and (F) Detection of gfp gene by PCR analysis of DNAs isolated from chimeras shown in Figure 2A and 2B respectively. (G) Detection of GFP expressing E. coli endosymbionts within Saccharomyces cerevisiae cox2–60. No GFP signals are detected in control Saccharomyces cerevisiae cox2–60 cells. Scale bars represent10 μm.
When we started deleting additional genes (e.g., glnA, strain 24) from strain 23 - E. coli ΔthiC:gfp ΔnadA ΔmetA ΔilvD, ΔargA, ΔproA, we observed significantly slower growing chimeras. As a first step, we genome sequenced strain 24 to investigate if there were any genomic rearrangements that might have occurred in strain 24 during our recombineering efforts28 that might have resulted in slower growth of chimeras. We did not detect any such rearrangements when compared to E. coli ΔnadA:gfp that we had previously generated. At this point, we are pursuing E. coli genomic library complementation29 and mutation/selection based evolutionary approaches to attempt to correct the growth defects in the chimeras. Similar approaches are being tested to correct the growth defects in case of chimeras generated from E. coli strains auxotrophic for tyrosine, phenylalanine, tryptophan and histidine.
In conclusion, we were able to identify a number of E. coli auxotrophs corresponding to central metabolites (i.e., amino acids and cofactors) as potential endosymbionts within yeast cells. Further, we generated an E. coli strain auxotrophic to 8 different metabolites, (for which the biosynthetic pathways involve about 40 gene products ~ 46 kb30), and demonstrated the generation of S. cerevisiae cox2–60-E. coli chimeras using this strain. Considering that more than 30 percent of genes in E. coli are functionally assigned to metabolism30, these studies are expected to set up a platform towards the deletion of a significant fraction of non-essential genes in the E. coli endosymbionts in the context of the yeast cytosol. Our eventual goal would be to obtain a minimal E. coli genome within yeast cells. Such studies are expected to provide insights into the evolutionary adaptations that may have been necessary for mitochondrial evolution starting from prokaryotic endosymbionts.
Supplementary Material
ACKNOWLEDGMENT
We would like to acknowledge Kristen Williams for her assistance in manuscript preparation.
Funding Sources
This work was supported by the California Institute of Biomedical Research and NIH (R01GM132071).
Footnotes
Supporting Information. Additional data and experimental details are provided in the supporting information. The Supporting Information is available free of charge on the ACS Publications website.
REFERENCES
- 1.Margulis L, Origin of eukaryotic cells: Evidence and research implications for a theory of the origin and evolution of microbial, plant and animal cells on the precambrian Earth. Yale University Press: 1970. [Google Scholar]
- 2.Kutschera U; Niklas KJ, The modern theory of biological evolution: an expanded synthesis. Naturwissenschaften 2004, 91 (6), 255–276. [DOI] [PubMed] [Google Scholar]
- 3.Gray MW, Mitochondrial Evolution. Cold Spring Harbor Perspectives in Biology 2012, 4 (9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonen L; Cunningham R; Gray M; Doolittle W, Wheat embryo mitochondrial 18S ribosomal RNA: evidence for its prokaryotic nature. Nucleic acids research 1977, 4 (3), 663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams KP; Sobral BW; Dickerman AW, A robust species tree for the alphaproteobacteria. Journal of bacteriology 2007, 189 (13), 4578–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaremba-Niedzwiedzka K; Caceres EF; Saw JH; Bäckström D; Juzokaite L; Vancaester E; Seitz KW; Anantharaman K; Starnawski P; Kjeldsen KU; Stott MB; Nunoura T; Banfield JF; Schramm A; Baker BJ; Spang A; Ettema TJG, Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 2017, 541 (7637), 353–358. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z; Wu M, Phylogenomic reconstruction indicates mitochondrial ancestor was an energy parasite. PLoS One 2014, 9 (10), e110685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martijn J; Vosseberg J; Guy L; Offre P; Ettema TJ, Deep mitochondrial origin outside the sampled alphaproteobacteria. Nature 2018, 557 (7703), 101. [DOI] [PubMed] [Google Scholar]
- 9.Andersson SG; Zomorodipour A; Andersson JO; Sicheritz-Pontén T; Alsmark UCM; Podowski RM; Näslund AK; Eriksson A-S; Winkler HH; Kurland CG, The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 1998, 396 (6707), 133. [DOI] [PubMed] [Google Scholar]
- 10.Fraser CM; Gocayne JD; White O; Adams MD; Clayton RA; Fleischmann RD; Bult CJ; Kerlavage AR; Sutton G; Kelley JM, The minimal gene complement of Mycoplasma genitalium. Science 1995, 270 (5235), 397–404. [DOI] [PubMed] [Google Scholar]
- 11.Corradi N; Pombert J-F; Farinelli L; Didier ES; Keeling PJ, The complete sequence of the smallest known nuclear genome from the microsporidian Encephalitozoon intestinalis. Nature communications 2010, 1, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibson DG; Glass JI; Lartigue C; Noskov VN; Chuang R-Y; Algire MA; Benders GA; Montague MG; Ma L; Moodie MM, Creation of a bacterial cell controlled by a chemically synthesized genome. science 2010, 1190719. [DOI] [PubMed] [Google Scholar]
- 13.Hutchison CA; Chuang R-Y; Noskov VN; Assad-Garcia N; Deerinck TJ; Ellisman MH; Gill J; Kannan K; Karas BJ; Ma L, Design and synthesis of a minimal bacterial genome. Science 2016, 351 (6280), aad6253. [DOI] [PubMed] [Google Scholar]
- 14.Mehta AP; Supekova L; Chen J-H; Pestonjamasp K; Webster P; Ko Y; Henderson SC; McDermott G; Supek F; Schultz PG, Engineering yeast endosymbionts as a step toward the evolution of mitochondria. Proceedings of the National Academy of Sciences 2018, 115 (46), 11796–11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delevoye C; Nilges M; Dehoux P; Paumet F; Perrinet S; Dautry-Varsat A; Subtil A, SNARE protein mimicry by an intracellular bacterium. PLoS pathogens 2008, 4 (3), e1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paumet F; Wesolowski J; Garcia-Diaz A; Delevoye C; Aulner N; Shuman HA; Subtil A; Rothman JE, Intracellular bacteria encode inhibitory SNARE-like proteins. PloS one 2009, 4 (10), e7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wesolowski J; Paumet F, SNARE motif: a common motif used by pathogens to manipulate membrane fusion. Virulence 2010, 1 (4), 319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wesolowski J; Weber MM; Nawrotek A; Dooley CA; Calderon M; Croix CMS; Hackstadt T; Cherfils J; Paumet F, Chlamydia hijacks ARF GTPases to coordinate microtubule posttranslational modifications and Golgi complex positioning. MBio 2017, 8 (3), e02280–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonnefoy N; Bsat N; Fox TD, Mitochondrial translation of Saccharomyces cerevisiae COX2 mRNA is controlled by the nucleotide sequence specifying the pre-Cox2p leader peptide. Molecular and cellular biology 2001, 21 (7), 2359–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Supekova L; Supek F; Greer JE; Schultz PG, A single mutation in the first transmembrane domain of yeast COX2 enables its allotopic expression. Proceedings of the National Academy of Sciences 2010, 107 (11), 5047–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torello AT; Overholtzer MH; Cameron VL; Bonnefoy N; Fox TD, Deletion of the leader peptide of the mitochondrially encoded precursor of Saccharomyces cerevisiae cytochrome c oxidase subunit II. Genetics 1997, 145 (4), 903–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smalley DJ; Whiteley M; Conway T, In search of the minimal Escherichia coli genome. Trends in microbiology 2003, 11 (1), 6–8. [DOI] [PubMed] [Google Scholar]
- 23.Gerdes S; Scholle M; Campbell J; Balazsi G; Ravasz E; Daugherty M; Somera A; Kyrpides N; Anderson I; Gelfand M, Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. Journal of bacteriology 2003, 185 (19), 5673–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juhas M; Reuß DR; Zhu B; Commichau FM, Bacillus subtilis and Escherichia coli essential genes and minimal cell factories after one decade of genome engineering. Microbiology 2014, 160 (11), 2341–2351. [DOI] [PubMed] [Google Scholar]
- 25.Lin MT; Fukazawa R; Miyajima-Nakano Y; Matsushita S; Choi SK; Iwasaki T; Gennis RB, Escherichia coli auxotroph host strains for amino acid-selective isotope labeling of recombinant proteins In Methods in enzymology, Elsevier: 2015; Vol. 565, pp 45–66. [DOI] [PubMed] [Google Scholar]
- 26.Jewison T; Knox C; Neveu V; Djoumbou Y; Guo AC; Lee J; Liu P; Mandal R; Krishnamurthy R; Sinelnikov I, YMDB: the yeast metabolome database. Nucleic acids research 2011, 40 (D1), D815–D820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramirez-Gaona M; Marcu A; Pon A; Guo AC; Sajed T; Wishart NA; Karu N; Djoumbou Feunang Y; Arndt D; Wishart DS, YMDB 2.0: a significantly expanded version of the yeast metabolome database. Nucleic acids research 2016, 45 (D1), D440–D445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Datsenko KA; Wanner BL, One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences 2000, 97 (12), 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reyes LH; Almario MP; Kao KC, Genomic library screens for genes involved in n-butanol tolerance in Escherichia coli. PloS one 2011, 6 (3), e17678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keseler IM; Mackie A; Santos-Zavaleta A; Billington R; Bonavides-Martínez C; Caspi R; Fulcher C; Gama-Castro S; Kothari A; Krummenacker M, The EcoCyc database: reflecting new knowledge about Escherichia coli K-12. Nucleic acids research 2016, 45 (D1), D543–D550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.