Abstract
Osteolytic lesions in multiple myeloma are caused by osteoclast-mediated bone resorption and reduced bone formation. A unique feature of myeloma is a failure of bone healing after successful treatment. We observed adipocytes on trabecular bone near the resorbed area in successfully treated patients. Normal marrow adipocytes, when cocultured with myeloma cells, were reprogrammed and produced adipokines that activate osteoclastogenesis and suppress osteoblastogenesis. These adipocytes have reduced expression of peroxisome proliferator- activated receptor γ (PPARγ) mediated by recruitment of polycomb repressive complex 2 (PRC2), which modifies PPARγ promoter methylation at trimethyl lysine-27 histone H3. We confirmed the importance of methylation in the PPARγ promoter by demonstrating that adipocyte-specific knockout of EZH2, a member of the PRC2, prevents adipocyte reprogramming and reverses bone changes in a mouse model. We validated the strong correlation between the frequency of bone lesions and the expression of EZH2 in marrow adipocytes from patients in remission. These results define a role for adipocytes in genesis of myeloma-associated bone disease and that reversal of adipocyte reprogramming has therapeutic implications.
INTRODUCTION
Multiple myeloma is a B cell malignancy characterized by the accumulation of malignant plasma cells in bone marrow (BM) that occurs predominantly in older individuals. More than 80% of patients with myeloma develop osteolytic lesions, which cause pathologic fractures, severe bone pain, spinal cord compression, and hypercalcemia, severely affecting quality of life (1). Myeloma-associated bone disease remains a problem even in treated patients with few or no plasma cells in their marrow, as lytic lesions that develop during the period of active myeloma do not heal (2).
The mechanism of lytic bone disease development in patients with active myeloma is well established, but little is known about why bone lesions do not heal in patients in remission. In those with active myeloma, factors secreted by malignant plasma cells can disrupt the delicate balance of osteoclast-mediated bone resorption and osteoblast-mediated bone formation. Osteoclasts home and adhere to the bone matrix and release osteolytic enzymes that can resorb bone. Differentiation of osteoclasts from hematopoietic monocytic precursors is driven by the cytokines receptor activator of nuclear factor-κB ligand (RANKL) and macrophage colony-stimulating factor (M-CSF). Myeloma cells can stimulate production of these osteolytic cytokines and thus enhance osteoclast differentiation (3, 4). Osteoblasts, on the other hand, are responsible for matrix synthesis by secreting and organizing type I collagen and other proteins; the matrix is then mineralized to form bone. Myeloma cells suppress the maturation of mesenchymal stem cells (MSCs) into osteoblasts via enhanced production of Wnt inhibitors (5). When patients are in remission and thus free of myeloma, normal remodeling of bone is expected to return to heal the lytic lesions. Unfortunately, this does not occur in most patients in remission, indicating that other factors within the marrow microenvironment contribute to the failure of bone healing.
Adipocytes are cells commonly found in the marrow, particularly in older individuals. Marrow adipocytes share some characteristics with other types of adipocytes: They contain membrane-enclosed lipid droplets, serve as major sources of energy reserve, and secrete a variety of adipokines. Although their functional importance in the marrow is unclear, these adipocytes are metabolically and biologically distinct from white, brown, and beige adipocytes. The adipokines produced by marrow adipocytes, including adiponectin and tumor necrosis factor–α (TNFα), can regulate both osteoclast and osteoblast differentiation (6). As people age, their marrow adipose tissue increases and may approach 70% of marrow volume (7). The number of marrow adipocytes is positively correlated with age-induced osteoporosis (8). Adipocytes can secrete low concentrations of RANKL and M-CSF, both of which are responsible for osteoclastogenesis. Deficiency of M-CSF in mice causes the occurrence of osteopetrosis (9). In obese individuals, adipocytes regulate bone integrity and mass via secretion of adipokines (10) and may induce osteoporosis (11). Recent studies have shown that marrow adipocytes participate in the development of malignancies (12, 13). Adipocytes support tumor growth by providing energy to tumor cells (13). Adipocyte-derived adipokines can protect tumor cells against chemotherapy-induced apoptosis in leukemia and myeloma (14, 15); adipokines like adiponectin have a function in suppression of tumor growth, with decreased circulating adiponectin concentrations reported in myeloma, breast, and prostate cancers (16, 17). However, the role of marrow adipocytes in the genesis of tumor-associated bone disease remains unclear.
In this study, we demonstrated that marrow adipocytes contribute to the persistence of myeloma-induced osteolytic lesions in patients in remission via secretion of adipokines that stimulate bone resorption and inhibit bone formation. These results demonstrated that transformed plasma cells can reprogram adipocytes via increased methylation of peroxisome proliferator–activated receptor γ (PPARγ) and altered adipokine production.
RESULTS
Adipocytes isolated from patients with myeloma can stimulate bone resorption
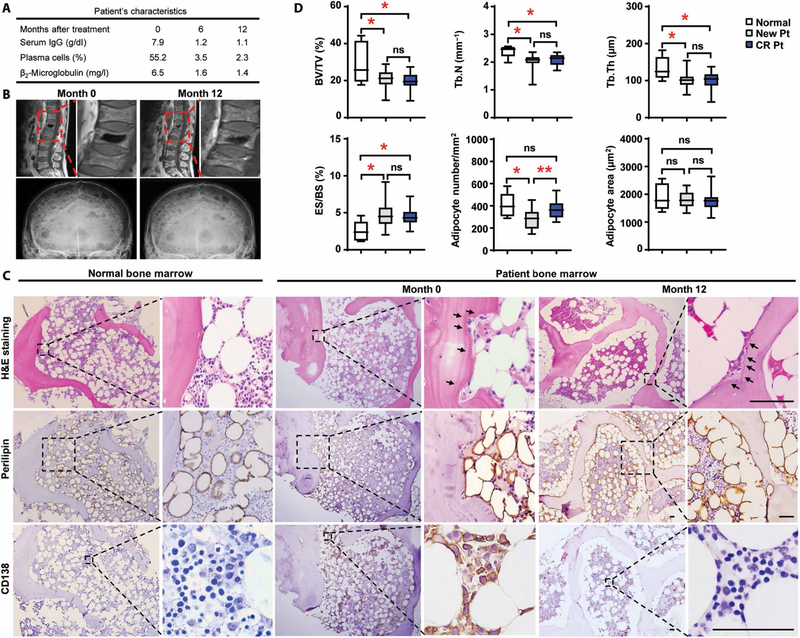
Figure 1 (A and B) describes a common problem in the management of myeloma: failure of bone lesions that develop during the active phase of the disease to heal when the patient is in remission, characterized by normalization of most myeloma-associated abnormalities (1). The causative mechanism for this phenomenon is unclear. Histologic examination of bone biopsy samples obtained from patients with myeloma in remission revealed an increase of fat cells compared to patients with active myeloma, whereas lytic lesions still persisted (Fig. 1, C and D). Although adipocyte numbers and size were comparable with those in normal BM, we observed a higher tendency of juxtaposition of adipocytes on trabecular bone within the marrow cavity, especially the proximity of adipocytes to the resorbed area (Fig. 1, C and D), leading us to believe that the function of adipocytes has already been modified.
Fig. 1. Association of marrow adipocytes with bone lesion in patients with myeloma in remission.
(A to C) Samples were obtained from a patient in complete remission on the date of the diagnosis and at follow-up visits after chemotherapy. (A) Characterization of disease signs, serum immunoglobulin G (IgG) and β2-microglobulin values, and the frequency of marrow-infiltrated plasma cells in the patient with myeloma before (month 0) and after (months 6 and 12) treatment. (B) Representative images of magnetic resonance imaging scanning for lytic lesions in the spine (upper panels) and skull (lower panels). (C) Representative images of hematoxylin and eosin (H&E)–stained sections of normal BM or the BM from the patient with myeloma showing histological changes and images of immunohistochemical staining for perilipin (a lipid droplet marker) or CD138 (a myeloma marker) expression. Arrows, lytic areas. Scale bars, 50 μm. Each experiment was repeated three times. (D) Percentages of BV/TV, Tb.N, Tb.Th, and ES/BS; numbers of adipocytes per mm2 of tissue; and size of adipocytes (μm2) in the sections of normal BM (n = 6) and the BM of patients with newly diagnosed myeloma (New Pt) (n = 17) or patients in remission (CR Pt) (n = 17). ns, not significant; *P < 0.05; **P < 0.01. P values were determined using one-way analysis of variance (ANOVA).
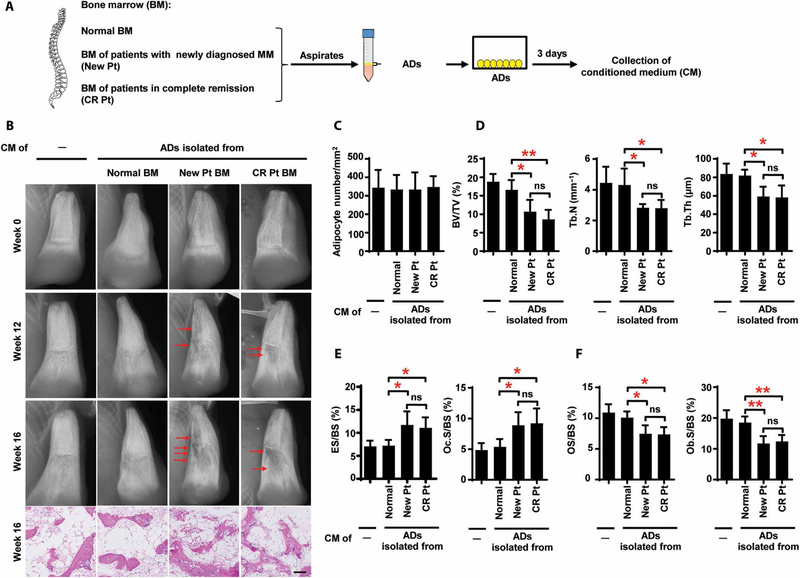
To determine the functional role of marrow adipocytes in bone, we isolated adipocytes from BM aspirates from normal subjects and patients with newly diagnosed myeloma and in complete remission (Fig. 2A and fig. S1). Oil Red O and calcein AM staining demonstrated no difference in cellular morphology or viability between the normal adipocytes and either group of myeloma patient- derived adipocytes (fig. S2). We used a humanized mouse model in which human fetal bone chips were subcutaneously implanted into NOD-scid IL2Rgnull mice. After 6 weeks, we injected aliquots of conditioned medium (CM) from cultured adipocytes directly into human bone chips, three times a week over 16 weeks. The mice that received unconditioned medium served as controls for baseline measurements of bone density. Injection of CM from marrow adipocytes obtained from patients with newly diagnosed myeloma or in complete remission caused multiple large lytic lesions, whereas we observed little resorption in the two control groups of mice treated with unconditioned medium or CM from normal adipocytes (Fig. 2B). The numbers of adipocytes were similar among all groups (Fig. 2C), and the number of lytic lesions did not differ between mice given CM from patients with active myeloma and those given CM from patients in remission (Fig. 2D). Bone histomorphometric analysis demonstrated a lower bone volume/total volume (BV/TV), trabecular number (Tb.N), and trabecular thickness (Tb.Th) in the chips of mice injected with CM from the two patient groups than in those given CM from normal subjects (Fig. 2D). In mice injected with CM of patient-derived adipocytes, we also found higher percentages of bone surface eroded by osteoclasts (ES/BS) and bone surface covered with osteoclasts (Oc.S/BS), lower percentages of osteoid surface (OS/BS), and bone surface lined with osteoblasts (Ob.S/BS; Fig. 2, E and F), indicating that adipocytes from patient BM have an activity to induce osteolytic bone lesions.
Fig. 2. Resorption of bone by marrow adipocytes isolated from patients with myeloma in vivo.
(A) Schematic for collection of the CM from cultures of adipocytes (ADs) isolated from BM. (B) Representative x-rays and images of H&E staining of bone chips from SCID-hu mice injected with the CM. Mice that received unconditioned medium served as controls. Red arrows, bone lesion. (C) Summarized quantification of adipocytes. Analysis of the bone chips using bone histomorphometry shows the percentages of BV/TV, Tb.N, and Tb.Th (D); the percentages of ES/BS and Oc.S/BS (E); and the percentages of OS/BS and Ob.S/BS (F). The data are averages ± SD (five mice per group, three replicate studies). *P < 0.05; **P < 0.01. P values were determined using one-way ANOVA.
Myeloma cells can reprogram normal adipocytes
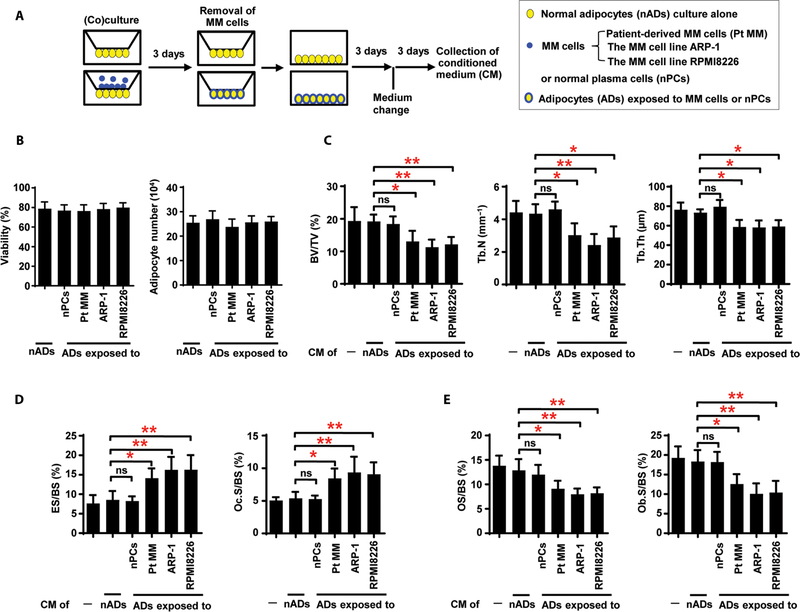
Because the marrow adipocytes isolated from the patients with myeloma but not those from normal subjects resorbed bone, we hypothesized that normal adipocytes acquire this function after exposure to myeloma cells. As schematically shown in Fig. 3A, normal adipocytes were cocultured with patient-derived CD138+ primary myeloma cells or with the human myeloma cell lines before adipocyte purification and additional culture. We found no obvious differences in viability and cell numbers between myeloma-associated adipocytes and normal cells (Fig. 3B). Injection of CM from normal adipocytes exposed to myeloma cells into implanted human bone chips of mice caused more lytic lesions, higher ES/BS and Oc.S/BS, and lower BV/TV, Tb.N, Tb.Th, OS/BS, and Ob.S/BS (Fig. 3, C to E). These findings demonstrate that normal adipocytes, when exposed to myeloma cells, acquire the ability to produce soluble factors that stimulate bone resorption even in the absence of myeloma cells.
Fig. 3. Induction of bone resorption by adipocytes exposed to myeloma cells.
(A) Schematic for collection of adipocyte CM. Normal adipocytes (nADs) derived from healthy human MSCs were cocultured without or with normal plasma cells (nPCs) or myeloma cells. MM, multiple myeloma. (B) Viability and number of adipocytes in culture alone or cocultured with normal plasma cells or myeloma cells. (C to E) Bone histomorphometric analysis of the bone chips from SCID-hu mice injected with the CM shows the percentages of BV/TV, Tb.N, and Tb.Th (C); the percentages of ES/BS and Oc.S/BS (D); and the percentages of OS/BS and Ob.S/BS (E) in the implanted bone chips. The data are averages ± SD (five mice per group, three replicate studies). *P < 0.05; **P < 0.01. P values were determined using one way ANOVA.
Myeloma-associated adipocytes disrupt the balance between osteoclastogenesis and osteoblastogenesis
Normal bone remodeling is maintained by a balance between osteoclast-mediated resorption and osteoblast-mediated matrix synthesis. To examine whether myeloma-associated adipocytes can regulate this balance, we first assessed their effects on osteoclast differentiation. In the absence of RANKL, addition of CM from normal or patient-derived adipocytes to cultures of the precursors of osteoclasts (preOCs) did not induce multinucleated tartrate-resistant acid phosphatase- positive (TRAP+) cell formation, which would indicate osteoclastogenesis (fig. S3, A and B). In the presence of RANKL, culture of preOCs with CM from patient-derived adipocytes increased the numbers of multinuclear TRAP+ cells. Culture of preOCs with CM from normal adipocytes preexposed to myeloma cells stimulated RANKL-induced TRAP+ cell formation (fig. S3C), TRAP 5b secretion (fig. S3D), and osteoclast gene expression (fig. S3E). Culture of preOCs with CM from normal adipocytes or CM from adipocytes preexposed to normal plasma cells could not enhance RANKL-induced osteoclastogenesis.
To assess the effect of CM from myeloma-associated adipocytes on osteoblast formation, we cultured osteoblast progenitors, MSCs, in osteoblast medium with CM. MSCs cultured with CM from marrow adipocytes obtained from newly diagnosed patients or patients in remission had less Alizarin red S staining than cultured with CM from normal adipocytes (fig. S4A). We observed a similar pattern of Alizarin red S staining, alkaline phosphatase activity, and osteoblast gene expression in MSCs cultured with CM from adipocytes preexposed to myeloma cells (fig. S4, B to D), indicating that myeloma-associated adipocytes inhibit osteoblastogenesis.
We used an extramedullary model of bone formation (18) to examine whether myeloma-associated adipocytes regulate the ability of osteoblasts to form new bone ex vivo. MSCs and adipocytes with or without preexposure to myeloma cells were subcutaneously implanted with Matrigel on either flanks of the mice. New bone was observed in mice 8 weeks after implantation with MSCs and normal adipocytes, but the amount of fluorescent labeling was lower in mice implanted with MSCs and myeloma-associated adipocytes (fig. S5A). Similarly, histologic and immunohistochemical staining of subcutaneous tissue samples from the same experiment showed reduced amounts of new bone and osteocalcin+ cells (fig. S5, B to D). In the calvarial mouse model, we generated critical-sized calvarial bone defects in mice (2 mm). We validated that myeloma-associated adipocytes have an ability to reduce new bone formation (fig. S5, E and F). Our aggregated results demonstrated that factors secreted by myeloma-associated adipocytes enhance osteoclastogenesis and suppress osteoblastogenesis.
Adipokine production changes in patients with active myeloma persist during myeloma remission
We next investigated the mechanism by which adipocytes modify bone cell differentiation. Adipokines produced by adipocytes, particularly adiponectin and TNFα, are known to regulate bone resorption and formation (6). Microarray analysis examined the adipokine expression profiles in adipocytes isolated from normal BM and BM of patients in remission. Using the criteria of signal level greater than 10 and the change greater than twofold between the two groups, we identified 6 up-regulated and 12 down-regulated genes (fig. S6A). Quantitative polymerase chain reaction (PCR) analysis confirmed down-regulation of ADIPONECTIN (GBP-28 or Acrp30), ADIPSIN (complement factor D), and VISFATIN (pre-B cell colony-enhancing factor) and up-regulation of TNFα expression (fig. S6B). The observed changes were present not only in adipocytes exposed to patient-derived myeloma cells or myeloma cell lines but also in adipocytes derived from the marrow of patients with newly diagnosed myeloma or in complete remission (fig. S6, C and D).
Addition of antibodies against adiponectin, adipsin, or visfatin or addition of TNFα to preOCs coincubated with CM from normal adipocytes increased TRAP+ cell numbers (fig. S7, A and B), indicating a role for these adipokines in osteoclast differentiation. Addition of CM from myeloma-exposed adipocytes increased TRAP+ cell formation. This effect was partially reversed by the addition of adiponectin, adipsin, or visfatin, and additive effects were observed when all three were combined (fig. S7C). With addition of an anti-TNFα antibody, cultures of preOCs with CM from myeloma-exposed adipocytes inhibited TRAP+ cell formation (fig. S7D). In a parallel series of experiments, we demonstrated similar effects of these adipokines on osteoblastogenesis as assessed using Alizarin red S staining (fig. S8). These experiments demonstrate that these adipokines have biological effects on osteoblast and osteoclast function.
Reduced PPARγ modifies adipokines in myeloma-associated adipocytes
PPARγ is highly expressed in adipose tissue, and its activity is essential for the expression of many adipokines (19). We found that the expression and activity of PPARγ were markedly down-regulated in adipocytes derived from patients in remission and normal adipocytes coincubated with myeloma cells (fig. S9, A to C). When we added the PPARγ agonist troglitazone to the cultured myeloma- exposed adipocytes, we observed an increase of mRNAs for ADIPONECTIN, VISFATIN, and ADIPSIN and decreased TNFα mRNA (fig. S9, D and E). In contrast, addition of the PPARγ antagonist G3335 to normal adipocytes has the reversed effects (fig. S9F). Collectively, these findings confirmed previously reported results that PPARγ regulates adipokine expression (19), and further demonstrate an effect of myeloma on adipocyte PPARγ expression and activity.
Polycomb repressive complex 2 interacts with the PPARγ promoter and down-regulates PPARγ expression via histone trimethylation
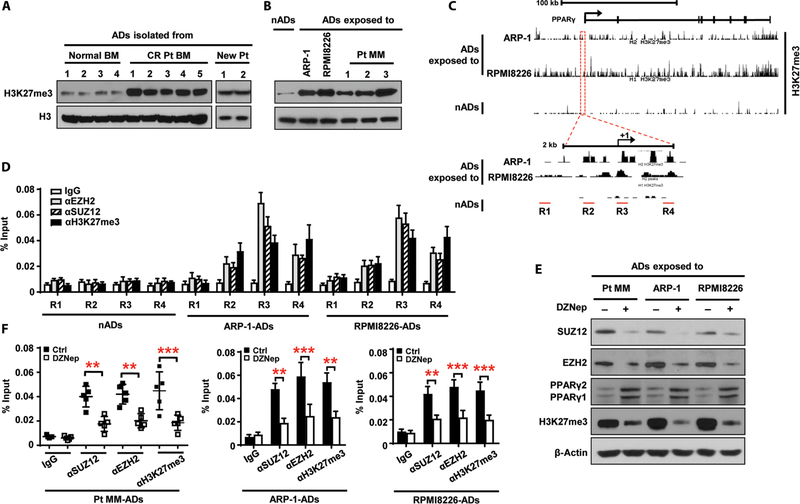
Methylation is a key epigenetic modification that influences tissue-and context-specific gene expression and is associated with gene silencing (20, 21), so we used a microarray to analyze the profile of genes that regulate DNA or histone methylation in adipocytes. We found elevated expression of the enhancer of zeste homolog 2 (EZH2) and suppressor of zeste 12 homolog (SUZ12) genes in patient-derived adipocytes (fig. S10A). We further demonstrated that EZH2 and SUZ12 expression was increased in adipocytes obtained from patients in remission, whereas embryonic ectoderm development, RB binding protein 4, and RB binding protein 7 expression was unchanged (fig. S10B). Western blot analysis of EZH2 and SUZ12 expression in adipocytes from patients in remission (fig. S10C) and normal adipocytes that coincubated with myeloma cells (fig. S10D) showed similar patterns of up-regulation.
EZH2 and SUZ12 are the core subunits of polycomb repressive complex 2 (PRC2), a transcriptional repressor. The only known histone methyltransferase activity possessed by PRC2 targets the regulation of histone H3 lysine-27 trimethylation (H3K27me3) (22). Consistent with this, we observed more H3K27me3 protein in myeloma-associated adipocytes than in normal adipocytes (Fig. 4, A and B). We queried H3K27me3 methylation status with genome-wide chromatin immunoprecipitation (ChIP) sequencing (ChIP-seq). We found an enrichment of H3K27me3 in the PPARγ promoter and gene body in myeloma-exposed adipocytes but not in normal adipocytes (Fig. 4C). To corroborate the effect of H3K27me3 and PRC2 complex on transcriptional initiation, we performed ChIP assays using anti-EZH2, anti-SUZ12, and anti-H3K27me3 antibodies and specific primers for four subregions of the PPARγ promoter (Fig. 4C). Consistent with the ChIP-seq results, H3K27me3 methylation was increased in the R3 and R4 subregions, and EZH2 and SUZ12 were predominantly enriched in R3 (Fig. 4D), indicating interaction of EZH2 and SUZ12 within R3. To show the specificity of this methylation, we treated myeloma-exposed adipocytes with 3-deazaneplanocin A, a PRC2 inhibitor, and found inhibition of expression of SUZ12 and EZH2, resulting in up-regulation of PPARγ expression (Fig. 4E). We observed reduced expression of H3K27me3 (Fig. 4E) and - enrichment of EZH2, SUZ12, and H3K27me3 in R3 (Fig. 4F) in myeloma associated adipocytes than in normal adipocytes (Fig. 4, A and B). We queried H3K27me3 methylation status with genome-wide chromatin immunoprecipitation (ChIP) sequencing (ChIP-seq). We found an enrichment of H3K27me3 in the PPARγ promoter and gene body in myeloma-exposed adipocytes but not in normal adipocytes (Fig. 4C). To corroborate the effect of H3K27me3 and PRC2 complex on transcriptional initiation, we performed ChIP assays using anti-EZH2, anti-SUZ12, and anti-H3K27me3 antibodies and specific primers for four subregions of the PPARγ promoter (Fig. 4C). Consistent with the ChIP-seq results, H3K27me3 methylation was increased in the R3 and R4 subregions, and EZH2 and SUZ12 were predominantly enriched in R3 (Fig. 4D), indicating interaction of EZH2 and SUZ12 within R3. To show the specificity of this methylation, we treated myeloma-exposed adipocytes with 3-deazaneplanocin A, a PRC2 inhibitor, and found inhibition of expression of SUZ12 and EZH2, resulting in up-regulation of PPARγ expression (Fig. 4E). We observed reduced expression of H3K27me3 (Fig. 4E) and enrichment of EZH2, SUZ12, and H3K27me3 in R3 (Fig. 4F) in myeloma-associated adipocytes These results indicate that elevation of the PRC2 complex up-regulates histone methylation of PPARγ at H3K27me3 and thereby down-regulates PPAR expression.
Fig. 4. Up-regulation of histone methylation in the PPARγ gene in myeloma-associated adipocytes.
(A and B) Western blots showing the expression of H3K27me3 in adipocytes isolated from normal BM (n = 4), the BM of patients in complete remission (CR Pt; n = 5), and the BM of patients with newly diagnosed myeloma (New Pt; n = 2) (A) and in normal adipocytes and adipocytes exposed to ARP-1, RPMI8226, or patient-derived myeloma cells (Pt MM; n = 3) (B). The expression of H3 protein served as loading controls. (C) Binding profile of ChIP-seq results for H3K27me3 in normal adipocytes and adipocytes exposed to myeloma cells in the PPARγ gene located in chromosome 3. Lower panel, enlarged view. R1 to R4, subregions covering promoter region and transcriptional starting site of the PPARγ gene. (D) ChIP assay showing the enrichment of EZH2, SUZ12, and H3K27me3 in normal adipocytes and adipocytes exposed to myeloma cells. (E) Western blots showing the expression of EZH2, SUZ12, PPARγ, and H3K27me3 in adipocytes exposed to myeloma cells with or without 1 μM 3-deazaneplanocin A (DZNep) treatment. β-Actin served as a loading control. (F) ChIP assay showing EZH2, SUZ12, and H3K27me3 enrichment in adipocytes exposed to Pt MM (n = 5), ARP-1, or RPMI8226 cells with or without DZNep. Data are averages ± SD. Each experiment was repeated three times. **P < 0.01; ***P < 0.001. All P values were determined using one-way ANOVA.
Specificity protein 1 recruits the PRC2 complex to the PPARγ promoter
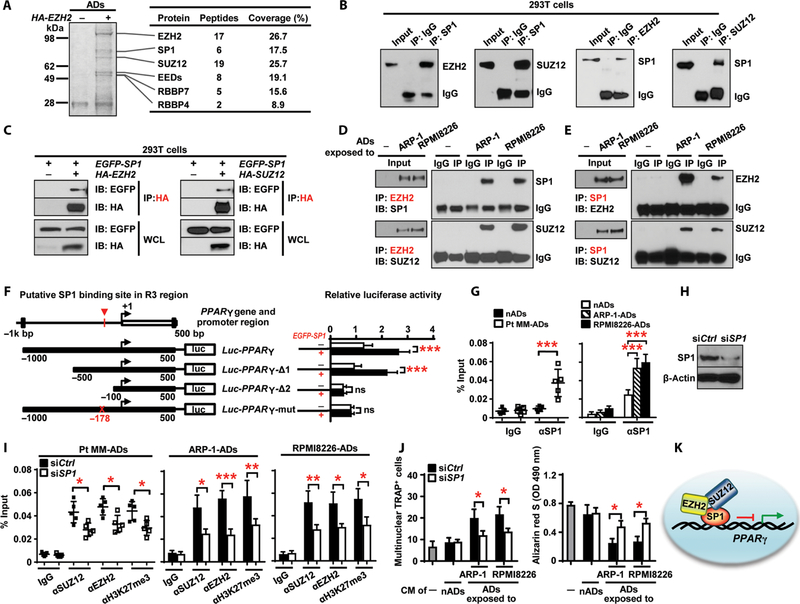
In mammalian cells, PRC2 proteins cannot bind directly to DNA, so a linker is needed to bridge the PRC2 complex to specific DNA regions to regulate methylation (23). To identify this linker, we performed immunoprecipitation of an anti-EZH2 antibody and analysis with mass spectrometry. In addition to the known components of the PRC2 complex, the analysis identified transcriptional factor specificity protein 1 (SP1) as a specific binding partner of EZH2 (Fig. 5A and table S1).
Fig. 5. SP1 bridges the interaction between the PRC2 complex and PPARγ promoter.
(A) Analysis of immunoprecipitates of HA-EZH2 in adipocytes transfected with an HA-EZH2 plasmid using Coomassie blue staining and mass spectrometry. The proteins identified are indicated on the right. (B) Coimmunoprecipitation of EZH2 or SUZ12 with SP1 in HEK293T cells cotransfected with SP1 and either EZH2 or SUZ12 plasmid. (C) Pull-down of HA-EZH2 or HA-SUZ12 with EGFP-SP1 in HEK293T cells. WCL, whole-cell lysate. (D and E) Coimmunoprecipitation of EZH2 (D) or SP1 (E) in normal adipocytes or adipocytes exposed to myeloma cells. Data are representative of trip-licate blots. (F) Schematic of the PPARγ promoter luciferase reporter. Solid boxes, promoter region of PPARγ; red crosses, mutations of four nucleotides. The luciferase activity of Luc-PPARγ constructs was set to 1. (G) ChIP assay showing SP1 enrichment around −178 bp of the PPARγ promoter in normal adipocytes and adipocytes exposed to myeloma cells. (H) Western blotting showing the expression of SP1 in adipocytes transfected with SP1 siRNAs (siSP1). Nontargeted siRNA (siCtrl)–expressing adipocytes served as controls. (I) ChIP assay showing enrichment of SUZ12, EZH2, and H3K27me3 on PPARγ promoter in siCtrl and siSP1 adipocytes cocultured with Pt MM (n = 5), ARP-1, or RPMI8226 cells. (J) Formation of TRAP+ cells from preOCs and Alizarin red S staining for osteoblast differentiation from MSCs, cultured with the CM of normal adipocytes or myeloma-associated adipocytes. The cultures without the CM served as controls. OD, optical density. (K) Schematic of the recruitment of PRC2 complex by SP1 to the PPARγ promoter. Data are averages ± SD. Each experiment was repeated three times. *P < 0.05; **P < 0.01; ***P < 0.001. All P values were determined using one-way ANOVA.
We used coimmunoprecipitation assays to interrogate the interaction between SP1 and PRC2 proteins. First, we cotransfected human embryonic kidney (HEK) 293T cells with plasmids expressing SP1 and either EZH2 or SUZ12 and found SP1 proteins in the immunoprecipitates (Fig. 5B), indicating that SP1 interacts with both EZH2 and SUZ12. Next, we mixed the lysate of hemagglutinin (HA)- tagged EZH2 (HA-EZH2)- or HA-SUZ12–expressing HEK293T cells with the lysate of EGFP-SP1–expressing HEK293T cells and pulled down with an anti-HA antibody. We detected either enhanced green fluorescent protein (EGFP) or HA proteins in the lysates, indicating that SP1 can bind directly to EZH2 and SUZ12 (Fig. 5C). Last, we asked whether the endogenous SP1/PRC2 complex existed in myeloma-exposed adipocytes. We immunoprecipitated adipocyte lysates with an anti-EZH2 antibody and observed the presence of SUZ12 or SP1 proteins in the lysates of myeloma-associated adipocytes but not in lysates of normal adipocytes (Fig. 5D). We also detected EZH2 and SUZ12 proteins in immunoprecipitates using an anti-SP1 antibody (Fig. 5E), indicating that EZH2, SUZ12, and SP1 form a co-repressor complex.
To determine whether SP1 binds to PPARγ promoter, we examined a 1.5-kb region around the PPARγ transcriptional start site. Because we observed the greatest enrichment of H3K27me3, EZH2, and SUZ12 in R3, we generated two truncated forms of the promoter before and after R3. Deletion mapping identified the SP1 binding site between −500 and −100 base pairs (bp) of the PPARγ promoter (Fig. 5F). Mutating a putative SP1 binding motif at −178 bp from GGGCGG to GATAAG confirmed this locus as the SP1 binding site (Fig. 5F). A ChIP assay using an anti-SP1 antibody confirmed enrichment of SP1 in the promoter region of PPARγ around −178 bp in myeloma-exposed adipocytes (Fig. 5G). To determine whether PRC2 proteins bind to the PPARγ promoter via SP1, we knocked down SP1 expression in myeloma-associated adipocytes using specific small interfering RNAs (siRNAs) (Fig. 5H) and observed reduced enrichment of EZH2, SUZ12, and H3K27me3 (Fig. 5I). Knockdown of SP1 in myeloma-associated adipocytes reduced osteoclastogenesis and enhanced osteoblastogenesis (Fig. 5J). These results indicate that SP1 plays a critical role in recruitment of PRC2 proteins to the PPARγ promoter (Fig. 5K) and PPARγ gene methylation.
Adipocytes are reprogrammed by myeloma cells via integrin α6–mediated signaling pathways
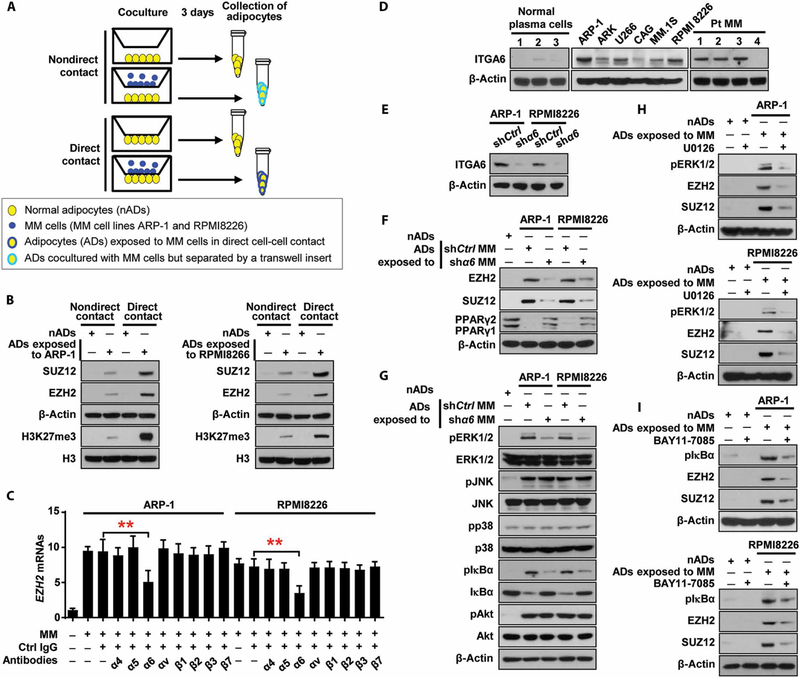
To find out whether the increased expression of EZH2 and SUZ12 was a result of the direct interaction between adipocytes and myeloma cells or through soluble factors secreted from myeloma cells, we used a transwell system (Fig. 6A). We found that the adipocytes from cell-cell cocultures with myeloma cells had much higher expression of EZH2 and SUZ12 than those from cell-separated cocultures (Fig. 6B), indicating that adipocytes are reprogrammed by myeloma cells mostly through a direct cell-cell interaction.
Fig. 6. Myeloma cell α6 enhances PRC2 expression through the ERK1/2 and NF-κB signaling pathways in adipocytes.
(A) Schematic of coculture systems. (B) Western blots showing the expression of EZH2 and SUZ12 in normal adipocytes or adipocytes exposed to myeloma cells through direct or nondirect contact manner. (C) Expression of EZH2 mRNAs in the adipocytes directly contacted with myeloma cells in the presence of blocking antibodies against integrins. (D) Western blots showing the expression of integrin α6 (ITGA6) in normal plasma cells isolated from normal BM (n = 3), primary myeloma cells isolated from patients (n = 4), and myeloma cell lines (n = 6). (E) Expression of ITGA6 in nonspecific or α 6 shRNA–expressing myeloma cells. (F and G) Western blots showing the expression of EZH2, SUZ12, and PPARγ (F) and the expression of nonphosphorylated or phosphorylated (p) kinases in the adipocytes cocultured with nonspecific or α6 shRNA–expressing myeloma cells for 24 hours (G). (H and I) Expression of pERK1/2, pIκBα, EZH2, and SUZ12 in the adipocytes treated without or with U0126 (H) or BAY11–7085 (I). The expression of β-actin served as loading controls. Data are averages ± SD. Each experiment was repeated three times. **P < 0.01. All P values were determined using one-way ANOVA.
Using blocking antibodies, we screened adhesion molecules that are known to be expressed in myeloma cells. As shown in Fig. 6C, addition of the antibody against α6, a subunit of integrin α6β1, but not the antibodies against other adhesion molecules, reduced the expression of EZH2 and SUZ12 in the adipocytes directly contacted with myeloma cells. Most myeloma cells, but not normal plasma cells, highly expressed α6 (Fig. 6D). We found that the adipocytes exposed to α6 short hairpin RNA (shRNA)–myeloma cells in a cell-cell contact manner expressed lower EZH2 and SUZ12 proteins and higher PPARγ proteins than those in the adipocytes exposed to nonspecific shRNA-myeloma cells (Fig. 6, E and F), suggesting that α6 mediates myeloma-induced adipocyte reprogramming.
We examined the profile of α6-mediated signaling pathways and observed up-regulation of phosphorylated extracellular signal–regulated kinase 1/2 (ERK1/2), c-Jun N-terminal kinase (JNK), IκBα, and Akt, but not p38 mitogen-activated protein kinase (MAPK), in adipocytes cocultured with myeloma cells than that in adipocytes cultured alone (Fig. 6G); the knockdown of α6 in myeloma cells deterred the elevated phosphorylation of ERK1/2 and IκBα in adipocytes, but not JNK and Akt (Fig. 6G). To determine the impact of such signaling pathways on PRC2 expression, we added inhibitors specific for ERK1/2 or NF-κB (nuclear factor κB) to cocultures of adipocytes and myeloma cells. As expected, the inhibitors down-regulated the phosphorylated ERK1/2 or IκBα, respectively, and reduced the expression of EZH2 and SUZ12 in the adipocytes (Fig. 6, H and I). Our findings indicate that myeloma cells up-regulate PRC2 expression in adipocytes via α6-mediated ERK1/2 and NF-κB signaling pathways.
Knockout of EZH2 in adipocytes facilitates healingof resorbed bone in a mouse model of myeloma in remission
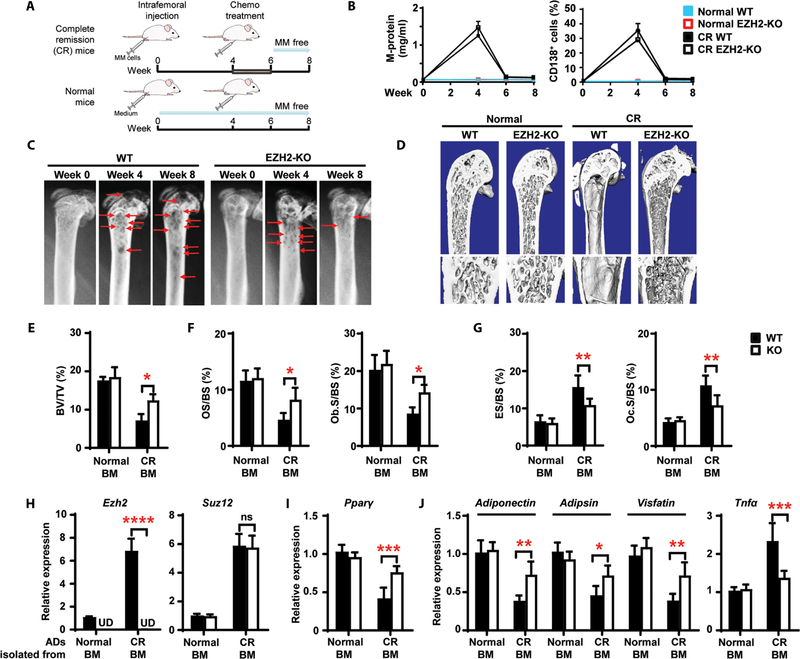
EZH2 is the only protein in the PRC2 complex with methylation enzymatic activity, so we asked whether deletion of EZH2 expression in adipocytes facilitates healing of resorbed bone in myeloma in remission. We first generated conditional adipose-specific Ezh2-knockout mice using a Flp-Cre strategy (fig. S11). To model myeloma in remission, we intrafemorally injected murine myeloma Vk*MYC cells (24), treated the mice with chemotherapy, and evaluated them for resolution of myeloma; mice not injected with myeloma cells served as normal controls (Fig. 7A). Two weeks after treatment, M-protein, an indicator of myeloma burden, fell to an undetectable amount (Fig. 7B), and we observed few CD138+ myeloma cells in marrow or infiltration to other organs (Fig. 7B and fig. S12). This suggests that the drug-treated mice were almost or totally myeloma free. As shown in Fig. 7C, femoral bone lesions developed in both wild-type and knockout mice 4 weeks after injection of Vk*MYC cells. In wild-type mice, severe lytic lesions persisted even after treatment, similar to that seen in human patients in remission, whereas Ezh2-knockout mice in remission had few lytic lesions (Fig. 7C). At 8 weeks, we found almost complete reversal of the bone phenotype in Ezh2-knockout mice (Fig. 7D), with higher percentages of BV/TV (Fig. 7E), OS/BS, and Ob.S/BS (Fig. 7F) and lower percentages of ES/BS and Oc.S/BS (Fig. 7G), confirming the radiographic observations. We found no obvious changes in wild-type or knockout mice not injected with myeloma cells (Fig. 7, E to G).
Fig. 7. Knockout of EZH2 in adipocytes heals resorbed bone in a mouse model of myeloma in remission.
Wild-type (WT) and adipocyte Ezh2-knockout (KO) mice were intrafemorally injected with the murine myeloma cell line Vk*MYC (1 × 106 cells per mouse). After 4 weeks, bortezomib (1 mg/kg) and melphalan (2 mg/kg) were injected intraperitoneally into the mice thrice weekly for 2 weeks. Shown are the experimental schematic (A), the concentrations of M-protein in mouse sera, the percentages of marrow-infiltrated CD138+ myeloma cells (B), representative x-rays (C) of femurs from complete remission (CR) mice, and representative microcomputed tomography images of mouse femurs at week 8 (D). Red arrows, lytic lesions. (E to G) Percentages of BV/TV (E), OS/BS and Ob.S/BS (F), and ES/BS and Oc.S/BS (G) at week 8. (H to J) Relative mRNA expression for Ezh2 and Suz12 (H), Pparγ (I), and the adipokines Adiponectin, Adipsin, Visfatin, and Tnfα (J) in marrow adipocytes at week 8. Data are means ± SD (n = 5 mice per group, three replicate studies). UD, undetectable. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. P values were determined using one-way ANOVA.
We validated the mechanisms of osteolysis induced by myeloma-exposed adipocytes in vivo. In wild-type mice injected with Vk*MYC cells, we found higher Ezh2, Suz12, and Tnfα mRNAs and lower Pparγ, Adiponectin, Adipsin, and Visfatin mRNAs in adipocytes isolated from the BM of mice in remission than in those from normal mice (Fig. 7, H and J, and fig. S11C). In adipocyte-specific Ezh2-knockout mice, there was no detectable expression of Ezh2 in adipocytes, and Suz12 mRNAs did not change (Fig. 7H). During myeloma remission, we observed increased Pparγ and altered adipokine mRNAs in marrow adipocytes in knockout mice as compared with wild-type mice (Fig. 7, I and J). These results indicate that the bone in Ezh2-knockout mice can be remodeled with recovery of PPARγ/adipokine-induced balance of bone resorption and formation in myeloma in remission.
The role of adipocyte reprogramming by myeloma cells is validated in patient samples
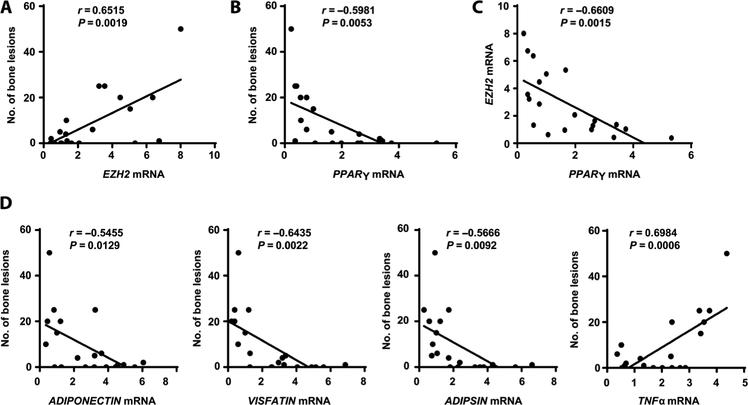
To assess the clinical relevance of marrow adipocytes in patients with myeloma, we collected adipocytes from BM aspirates of 20 patients in remission. We found a robust positive correlation between the frequency of bone lesions and EZH2 mRNAs in marrow adipocytes (Fig. 8A). Correspondingly, we observed a strong negative correlation between the frequency of bone lesions and PPARγ mRNAs (Fig. 8B), suggesting that increased EZH2 and reduced PPARγ expression have a substantial impact on bone healing in patients with myeloma in remission. A head-to-head comparison of the expression of EZH2 and PPARγ in adipocytes also demonstrated a negative correlation (Fig. 8C). Moreover, bone lesions were highly correlated with expression of the PPARγ-downstream adipokine genes ADIPONECTIN, VISFATIN, ADIPSIN, and TNFα (Fig. 8D). These results in humans are consistent with the aforementioned observations in mice regarding the roles of EZH2 and PPARγ in adipocytes.
Fig. 8. Association of PRC2 expression in marrow adipocytes with bone lesions in patients in remission.
Adipocytes were isolated from BM aspirates from 20 patients in remission, randomly selected, some with and some without bone lesions, to provide a representative sample of all patients in remission. Analysis of the number of bone lesions was performed by radiologists who were blinded to the molecular analysis results. Similarly, the laboratory person who performed the molecular analysis was blinded to the bone lesion results. Shown are the correlation coefficients for the numbers of bone lesions in the patients in remission and the mRNAs of EZH2 (A) and PPAR (B) and the correlation coefficient for the expression of EZH2 and PPARγ mRNA (C) in adipocytes isolated from patients’ BM. (D) Correlation coefficients for the numbers of bone lesions in the patients in remission and the expression of ADIPONECTIN, ADIPSIN, VISFATIN, and TNFα mRNA in patients’ BM adipocytes. The correlations were evaluated using Pearson coefficient. r, correlation coefficient. P values were determined using the Pearson correlation coefficient. Each point represents the analysis of a representative sample from the aggregate of adipocytes from one patient sample.
DISCUSSION
A perplexing problem that has plagued the management of patients with myeloma is why myelomatous bone lesions do not heal after successful treatment. The data presented in this article described a role for marrow adipocytes in this process. These data demonstrate that myeloma cells can reprogram adipocytes via altered methylation of the key adipocyte gene PPARγ, damaging the bone that persists even after successful elimination of myeloma. Once reprogrammed by myeloma cells, adipocytes have reduced expression of PPARγ and a modified adipokine secretion profile, causing enhanced osteoclastogenesis and suppression of osteoblastogenesis. This effect is mediated via SP1 recruitment of PRC2 proteins to the promoter region of PPARγ, resulting in induction of histone methylation of PPARγ at H3K27me3 by the co-repressor complex. Further evidence supporting this epigenetic mechanism is the demonstration that targeted deletion of EZH2, a key PRC2 complex component, restores normal bone remodeling in mice. These observations are important because they not only provide insight into a common problem but also suggest a therapeutic strategy for myeloma bone disease.
PPARγ is preferentially expressed in adipose tissue, which is essential for adipogenesis, energy balance, lipid biosynthesis, and adipokine production (19, 25, 26). Epigenetic modulation of PPARγ is known to be important during adipogenesis (27, 28). The H3K4 mono- or di-methyltransferases, mixed-lineage leukemia (MLL) 3/4, directly promote PPARγ expression in adipocyte progenitors (27), whereas the H3K9me3 methyltransferase, euchromatic histone-lysine N-methyltransferase 2 (EHMT2), represses PPARγ expression (28). In the present study, we analyzed the gene profiles of the enzymes responsible for DNA and histone methylation and found that only expression of the PRC2 proteins EZH2 and SUZ12 was up-regulated in myeloma-associated adipocytes, whereas that of MLL3/4 and EHMT2 was unchanged. These findings suggest that myeloma cells enhance the histone methylation of PPARγ at H3K27me3 via the PRC2 complex and that the methylation thus represses the expression and function of PPAR in mature adipocytes.
The mechanism of PRC2 complex binding to DNAs in mammalian cells remains unclear, because it cannot directly bind to the chromatin. Additional linkers, such as transcriptional factors, are required. One of these, SP1, can bind to the consensus sequence 5′-GGGCGG-3′ (GC box element) in the promoter region. In bone homeostasis, SP1 is involved in the progression of ossification by enhancing bone formation (29). This study demonstrates a function of SP1 as a bridge between PRC2 complex and PPARγ promoter. Whereas SP1 can interact with both EZH2 and SUZ12 in the PRC2 complex, it also binds to a unique SP1 site in the promoter of the PPARγ gene. SP1 is expressed in many types of mammalian cells. The interaction of PRC2 with SP1 and recruitment of the PRC2/SP1 complex to SP1-targeted genes may be a general mechanism of gene silencing in eukaryotic systems. In addition, we address the question of how myeloma cells reprogram marrow adipocytes. Our results showed that the expression of EZH2 and SUZ12 in adipocytes was increased when adipocytes interacted directly with myeloma cells through the integrin α6. Myeloma-expressed α6 enhances PRC2 proteins through the ERK1/2 and NF-κB signaling pathways, resulting in reduced PPARγ expression and activity. These findings point to the importance of the physical interaction between myeloma cells and adipocytes for the reprogramming of adipocytes. Besides integrin interaction, the potential role of soluble factors such as exosomes or microvesicles in adipocyte reprogramming needs further investigation.
Collectively, our results demonstrate that marrow adipocytes exposed to myeloma cells enhance osteoclastogenesis and suppress osteoblastogenesis and that methylation of PPARγ by the myeloma cell α6-mediated PRC2 complex contributes to adipocyte reprogramming. There are still limitations in our current study. For example, we purified and identified mature adipocytes using the standard protocols, which depend on the presence of cytoplasmic lipid droplets. Recent studies have shown that delipidation of adipocytes may occur in the advanced stages of patients with malignancies (30), suggesting the potential existence of a subset of adipocytes with non-classical morphology that may function differently. To focus on the function of myeloma-associated adipocytes, we have also taken extra caution to minimize the potential contamination from other stromal cells, which may include atypical adipocytes. In addition, the BM microenvironment is a complex system with many types of cells such as fibroblasts and endothelial cells. We did not address whether these cells could be reprogrammed by myeloma cells, or whether the reprogramed cells could gain a function to resorb bone. Many experiments involved administering CM from adipocytes into implanted bone chips or to in vitro cell culture, which is a reduction-ist approach that does not fully recapitulate native cell interactions.
Currently, there are few choices for the targeted therapy for patients with myeloma bone disease, and bisphosphonates, which are effective at inhibiting bone resorption in patients with myeloma, do not promote restoration of bone healing in patients in remission (2). We believe that targeting the α6/PRC2/SP1/PPARγ signaling pathway is a potential strategy for treatment of myeloma-induced bone disease. Of equal importance, other malignancies, such as breast and lung cancers, often metastasize to bone (31, 32), and our findings thus may also have broader implications for the genesis of osteolytic lesions in these cancers.
MATERIALS AND METHODS
Study design
The purpose of this study was to determine the contribution of marrow adipocytes to the failure of bone healing in multiple myeloma on the basis that surfeit adipocytes were observed in the biopsy samples from patients in remission. We began our studies by collecting the CM of myeloma-associated adipocytes from different sources: adipocytes isolated from BM aspirates of newly diagnosed patients and patients in remission, and normal adipocytes exposed to myeloma cells including those isolated from patient BMs or human myeloma cell lines. Next, we characterized myeloma-associated adipocytes on their effects in bone remodeling, adipokine production, and PPARγ expression. We then investigated the mechanisms underlying myeloma-induced adipocyte reprogramming, focusing on the integrin α6–activated PRC2-mediated histone methylation in the PPARγ gene. Last, we validated our findings in a myeloma remission mouse model and in patient samples.
All patient samples were obtained from the Myeloma Tissue Bank of the University of Texas MD Anderson Cancer Center, and this study was approved by the MD Anderson Institutional Review Board (#PA12–0034). Myeloma disease was characterized by physicians and hematologists; bone lesions were evaluated by radiologists. The investigators who performed the bench studies were blinded to the patient information. The number of patient samples required to achieve a correlation coefficient (r) ≥ 0.7, a power of 80%, and the level of significance at 5% was determined to be at least seven samples. For mouse studies, similar aged mice were randomly assigned into different groups. The sample size, composition of replicates, and the intermediate end point were based on previous knowledge (4). We initially estimated the sample size using power analysis with our previous knowledge on bone histomorphometric analysis. We included five mice per group to ensure a power of 80% to detect the changes in bone between the different groups with two-sided type I error rate controlled at the 0.05 level. The final end point before mice were sacrificed was in accordance with the Institutional Animal Care and Use Committee policies and was predefined. All data were included in the analysis, and the criteria for interpretation were established prospectively. Experiments were performed three times, and the results were verified by repetition over a 3-year period. Primary data are reported in data file S1.
Statistical analysis
Statistical significance was analyzed using the SPSS software program (version 10.0; IBM Corporation) with two-tailed unpaired Student’s t tests for comparison of two groups and one-way ANOVA with Tukey’s multiple comparisons test for comparison of more than two groups. P values less than 0.05 were considered statistically significant. All results were reproduced in at least three independent experiments.
Supplementary Material
Fig. S1. Gating/sorting strategy for adipocytes and characterization of primary adipocytes by fluorescent staining.
Fig. S2. Characterization of marrow adipocytes.
Fig. S3. Myeloma-associated adipocytes enhance osteoclastogenesis in vitro.
Fig. S4. Myeloma-associated adipocytes inhibit osteoblastogenesis in vitro.
Fig. S5. Myeloma-associated adipocytes inhibit in vivo extramedullary bone formation.
Fig. S6. Expression of adipokines in myeloma-associated adipocytes.
Fig. S7. Adipokines secreted from myeloma-associated adipocytes enhance osteoclast differentiation.
Fig. S8. Adipokines secreted from myeloma-associated adipocytes suppress osteoblast differentiation.
Fig. S9. Reduced expression of PPARγ in myeloma-associated adipocytes.
Fig. S10. Elevated expression of the PRC2 proteins EZH2 and SUZ12 in myeloma-associated adipocytes.
Fig. S11. Characterization of adipocyte-targeted Ezh2-knockout mice.
Fig. S12. Distribution of myeloma cells in the Vk*MYC myeloma cell–bearing mice.
Table S1. Identification of SP1 that binds to the PRC2 complex in adipocytes using mass spectrometry.
Table S2. Characteristics of patients with myeloma and donors with normal BM.
Table S3. Primers used in real-time reverse transcription PCR analysis.
Table S4. Primers used in ChIP-PCR.
Data file S1. Primary data (Excel file).
Acknowledgments:
We thank the UT MD Anderson Myeloma Tissue Bank.
Funding: This work was supported by the National Cancer Institute (R01 awards CA190863 and CA193362 to J.Y.) and the American Cancer Society (Research Scholar grant 127337-RSG-15-069-01-TBG to J.Y.). This work was also supported by the NIH/NCI (Core Labs, P30CA016672) for the Small Animal Imaging, Advanced Microscopy Core, and Research Histopathology Facilities.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data associated with this study can be found in the paper or the Supplementary Materials. ChIP-seq data are available in the Gene Expression Omnibus (GEO) database (GSE113642); DNA microarray data are available in the GEO database (GSE113295). Vk*MYC myeloma cell lines are available from P.L.B. under a material transfer agreement with Cornell University.
REFERENCES AND NOTES
- 1.Palumbo A, Anderson K, Multiple myeloma. N. Engl. J. Med 364, 1046–1060 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Zangari M, Suva LJ, The effects of proteasome inhibitors on bone remodeling in multiple myeloma. Bone 86, 131–138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruzzaniti A, Baron R, Molecular regulation of osteoclast activity. Rev. Endocr. Metab. Disord 7, 123–139 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Liu H, Liu Z, Du J, He J, Lin P, Amini B, Starbuck MW, Novane N, Shah JJ, Davis RE, Hou J, Gagel RF, Yang J, Thymidine phosphorylase exerts complex effects on bone resorption and formation in myeloma. Sci. Transl. Med 8, 353ra113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian E, Zhan F, Walker R, Rasmussen E, Ma Y, Barlogie B, Shaughnessy JD Jr., The role of the Wnt-signaling antagonist DKK1 in the development of osteolytic lesions in multiple myeloma. N. Engl. J. Med 349, 2483–2494 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Muruganandan S, Sinal CJ, The impact of bone marrow adipocytes on osteoblast and osteoclast differentiation. IUBMB Life 66, 147–155 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Gimble JM, Robinson CE, Wu X, Kelly KA, The function of adipocytes in the bone marrow stroma: An update. Bone 19, 421–428 (1996). [DOI] [PubMed] [Google Scholar]
- 8.Justesen J, Stenderup K, Ebbesen EN, Mosekilde L, Steiniche T, Kassem M, Adipocyte tissue volume in bone marrow is increased with aging and in patients with osteoporosis. Biogerontology 2, 165–171 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Yoshida H, Hayashi S-I, Kunisada T, Ogawa M, Nishikawa S, Okamura H, Sudo T, Shultz LD, Nishikawa S-I, The murine mutation osteopetrosis is in the coding region of the macrophage colony stimulating factor gene. Nature 345, 442–444 (1990). [DOI] [PubMed] [Google Scholar]
- 10.van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, Schrauwen P, Teule GJ, Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med 360, 1500–1508 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Rosen CJ, Bouxsein ML, Mechanisms of disease: Is osteoporosis the obesity of bone? Nat. Clin. Pract. Rheumatol 2, 35–43 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Morris EV, Edwards CM, Bone marrow adipose tissue: A new player in cancer metastasis to bone. Front. Endocrinol 7, 90 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Templeton ZS, Lie W-R, Wang W, Rosenberg-Hasson Y, Alluri RV, Tamaresis JS, Bachmann MH, Lee K, Maloney WJ, Contag CH, King BL, Breast cancer cell colonization of the human bone marrow adipose tissue niche. Neoplasia 17, 849–861 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z, Xu J, He J, Liu H, Lin P, Wan X, Navone NM, Tong Q, Kwak LW, Orlowski RZ, Yang J, Mature adipocytes in bone marrow protect myeloma cells against chemotherapy through autophagy activation. Oncotarget 6, 34329–34341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caers J, Deleu S, Belaid Z, De Raeve H, Van Valckenborgh E, De Bruyne E, Defresne M-P, Van Riet I, Van Camp B, Vanderkerken K, Neighboring adipocytes participate in the bone marrow microenvironment of multiple myeloma cells. Leukemia 21, 1580–1584 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Fowler JA, Lwin ST, Drake MT, Edwards JR, Kyle RA, Mundy GR, Edwards CM, Host-derived adiponectin is tumor-suppressive and a novel therapeutic targetfor multiple myeloma and the associated bone disease. Blood 118, 5872–5882 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khandekar MJ, Cohen P, Spiegelman BM, Molecular mechanisms of cancer development in obesity. Nat. Rev. Cancer 11, 886–895 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Chen Y, Jacamo R, Shi Y.-x., Wang R.-y., Battula VL, Konoplev S, Strunk D, Hofmann NA, Reinisch A, Konopleva M, Andreeff M, Human extramedullary bone marrow in mice: A novel in vivo model of genetically controlled hematopoietic microenvironment. Blood 119, 4971–4980 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tontonoz P, Spiegelman BM, Fat and beyond: The diverse biology of PPARγ. Annu. Rev. Biochem 77, 289–312 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Attwood JT, Yung RL, Richardson BC, DNA methylation and the regulation of gene transcription. Cell. Mol. Life Sci 59, 241–257 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greer EL, Shi Y, Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet 13, 343–357 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz YB, Pirrotta V, A new world of Polycombs: Unexpected partnerships and emerging functions. Nat. Rev. Genet 14, 853–864 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G, Genome regulation by polycomb and trithorax proteins. Cell 128, 735–745 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Guillerey C, Ferrari de Andrade L, Vuckovic S, Miles K, Ngiow SF, Yong MC, Teng MW, Colonna M, Ritchie DS, Chesi M, Bergsagel PL, Hill GR, Smyth MJ, Martinet L, Immunosurveillance and therapy of multiple myeloma are CD226 dependent. J. Clin. Invest 125, 2904 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sugii S, Olson P, Sears DD, Saberi M, Atkins AR, Barish GD, Hong S-H, Castro GL, Yin Y-Q, Nelson MC, Hsiao G, Greaves DR, Downes M, Yu RT, Olefsky JM, Evans RM, PPARγ activation in adipocytes is sufficient for systemic insulin sensitization. Proc. Natl. Acad. Sci. U.S.A 106, 22504–22509 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vidal-Puig AJ, Considine RV, Jimenez-Liñan M, Werman A, Pories WJ, Caro JF, Flier JS, Peroxisome proliferator-activated receptor gene expression in human tissues. Effects of obesity, weight loss, and regulation by insulin and glucocorticoids. J. Clin. Invest 99, 2416–2422 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J-E, Wang C, Xu S, Cho Y-W, Wang L, Feng X, Baldridge A, Sartorelli V, Zhuang L, Peng W, Ge K, H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. eLife 2, e01503 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L, Xu S, Lee JE, Baldridge A, Grullon S, Peng W, Ge K, Histone H3K9 methyltransferase G9a represses PPARγ expression and adipogenesis. EMBO J 32, 45–59 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ravazzolo R, Cappato S, Bocciardi R, Hints on transcriptional control of essential players in heterotopic ossification of Fibrodysplasia Ossificans Progressiva. Bone 109, 187–191 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Nieman KM, Romero IL, Van Houten B, Lengyel E, Adipose tissue and adipocytes support tumorigenesis and metastasis. Biochim. Biophys. Acta 1831, 1533–1541 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y-C, Sosnoski DM, Mastro AM, Breast cancer metastasis to the bone: Mechanisms of bone loss. Breast Cancer Res 12, 215 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lipton A, Uzzo R, Amato RJ, Ellis GK, Hakimian B, Roodman GD, Smith MR, The science and practice of bone health in oncology: Managing bone loss and metastasis in patients with solid tumors. J. Natl. Compr. Cancer Netw 7 (Suppl 7), S1–S29 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chesi M, Matthews GM, Garbitt VM, Palmer SE, Shortt J, Lefebure M, Stewart AK, Johnstone RW, Bergsagel PL, Drug response in a genetically engineered mouse model of multiple myeloma is predictive of clinical efficacy. Blood 120, 376–385 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berry R, Jeffery E, Rodeheffer MS, Weighing in on adipocyte precursors. Cell Metab 19, 8–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X, Ling W, Pennisi A, Wang Y, Khan S, Heidaran M, Pal A, Zhang X, He S, Zeitlin A, Abbot S, Faleck H, Hariri R, Shaughnessy JD Jr., F. van Rhee, B. Nair, B. Barlogie, J. Epstein, S. Yaccoby, Human placenta-derived adherent cells prevent bone loss, stimulate bone formation, and suppress growth of multiple myeloma in bone. Stem Cells 29, 263–273 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, Romero IL, Carey MS, Mills GB, Hotamisligil GS, Yamada SD, Peter ME, Gwin K, Lengyel E, Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat. Med 17, 1498–1503 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bancroft JD, Stevens A, Theory and Practice of Histological Techniques (Churchill Livingstone, ed. 3, 1990), pp. xiv, 726 p. [Google Scholar]
- 38.Ma W, Wang M, Wang Z-Q, Sun L, Graber D, Matthews J, Champlin R, Yi Q, Orlowski RZ, Kwak LW, Weber DM, Thomas SK, Shah J, Kornblau S, Davis RE, Effect of long-term storage in TRIzol on microarray-based gene expression profiling. Cancer Epidemiol. Biomarkers Prev 19, 2445–2452 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spicer PP, Kretlow JD, Young S, Jansen JA, Kasper FK, Mikos AG, Evaluation of bone regeneration using the rat critical size calvarial defect. Nat. Protoc 7, 1918–1929 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H, A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Gating/sorting strategy for adipocytes and characterization of primary adipocytes by fluorescent staining.
Fig. S2. Characterization of marrow adipocytes.
Fig. S3. Myeloma-associated adipocytes enhance osteoclastogenesis in vitro.
Fig. S4. Myeloma-associated adipocytes inhibit osteoblastogenesis in vitro.
Fig. S5. Myeloma-associated adipocytes inhibit in vivo extramedullary bone formation.
Fig. S6. Expression of adipokines in myeloma-associated adipocytes.
Fig. S7. Adipokines secreted from myeloma-associated adipocytes enhance osteoclast differentiation.
Fig. S8. Adipokines secreted from myeloma-associated adipocytes suppress osteoblast differentiation.
Fig. S9. Reduced expression of PPARγ in myeloma-associated adipocytes.
Fig. S10. Elevated expression of the PRC2 proteins EZH2 and SUZ12 in myeloma-associated adipocytes.
Fig. S11. Characterization of adipocyte-targeted Ezh2-knockout mice.
Fig. S12. Distribution of myeloma cells in the Vk*MYC myeloma cell–bearing mice.
Table S1. Identification of SP1 that binds to the PRC2 complex in adipocytes using mass spectrometry.
Table S2. Characteristics of patients with myeloma and donors with normal BM.
Table S3. Primers used in real-time reverse transcription PCR analysis.
Table S4. Primers used in ChIP-PCR.
Data file S1. Primary data (Excel file).