Abstract
Background
An early-life anxious temperament (AT) is a risk factor for the development of anxiety, depression, and comorbid substance abuse. We validated a nonhuman primate (NHP) model of early-life AT and identified the dorsal amygdala as a core component of AT’s neural circuit. Here, we combine RNA sequencing, viral-vector gene manipulation, functional brain imaging, and behavioral phenotyping to uncover AT’s molecular substrates.
Methods
In response to potential threat, AT and brain metabolism were assessed in forty-six young rhesus monkeys. We identified AT-related transcripts using RNA-seq data from dorsal amygdala tissue (including central nucleus of the amygdala [Ce] and dorsal regions of the basal nucleus). Based on the results, we overexpressed the neurotrophin 3 gene (NTF3) in the dorsal amygdala using intraoperative MRI guided surgery (n=5/group).
Results
This discovery-based approach identified AT-related alterations in the expression of well-established and novel genes, including an inverse association between neurotrophin receptor kinase 3 (NTRK3) expression and AT. NTRK3 is an interesting target because it is a relatively unexplored neurotrophic factor that modulates intracellular neuroplasticity pathways. Overexpression of the transcript for NTRK3’s endogenous ligand, NTF3, in the dorsal amygdala resulted in reduced AT and altered function in AT’s neural circuit.
Discussion
Together, these data implicate NT-3/NTRK3 signaling in the dorsal amygdala in mediating primate anxiety. More generally, this approach provides an important step towards understanding the molecular underpinnings of early-life AT and will be useful in guiding the development of treatments to prevent the development of stress-related psychopathology.
Keywords: anxiety, anxious temperament, behavioral inhibition, primate, rna-seq, fdg-pet, aav, ntf3, ntrk3, neurotrophic, central nucleus of the amygdala, amygdala, extended amygdala
Introduction
Anxious temperament (AT) early in life is a major risk factor for the later development of anxiety, and depressive disorders along with comorbid substance abuse (1–3). Understanding the molecular alterations that give rise to extreme AT is an important step toward developing targeted behavioral and pharmacological treatments for early-life anxiety. Given our current understanding of these processes, it is critical to combine discovery-based approaches with interventions targeted to test novel molecular substrates to understand the relevance of the many molecular pathways associated with increased childhood anxiety (4). The rhesus monkey is an ideal species for translational research focused on the molecular underpinnings of AT. In addition to the remarkable similarities between young monkeys and children in the expression of AT, studies in rhesus monkeys allow for the combination of targeted mechanistic techniques with neuroimaging techniques commonly used in humans. Here, we take advantage of the recent evolutionary divergence between humans and rhesus monkeys to identify putative molecular underpinnings of AT in the central nucleus (Ce)-containing dorsal amygdala region. These efforts combine in-depth phenotyping, including brain imaging and behavioral assessments, with post-mortem RNA sequencing along with targeted viral-vector mediated gene expression to test causality.
Neuroimaging studies of anxiety disorders and anxious dispositions performed in children (5), adults (6), and rhesus monkeys (7, 8) reveal a brain-wide network of AT-related regions that encompasses portions of the extended amygdala, including the bed nucleus of the stria terminalis (BST) and the Ce (9). In primates, the Ce-containing dorsal amygdala strongly projects to the BST (10, 11), is functionally connected with the rest of the extended amygdala (12, 13), and is hypothesized to play a critical role in threat learning and processing (14, 15). The dorsal amygdala receives direct and indirect projections from regulatory and evaluative cortical regions, and within the dorsal amygdala, the Ce can initiate defensive behavioral and physiological responses via projections to downstream targets (14, 15). The primate Ce has been causally implicated in AT (16, 17) and dorsal amygdala metabolism is largely nonheritable, suggesting that the environmental factors affecting AT may be mediated by the dorsal amygdala (7, 18). Here, we focused our efforts on understanding molecular alterations in the environmentally sensitive dorsal amygdala region that mediates AT.
Forty-six nonhuman primates were longitudinally assessed for behavioral inhibition, cortisol, and brain metabolism during a 30-minute exposure to a potentially threatening human intruder who made no eye contact (NEC) with the monkey. The NEC context elicits behavioral inhibition, which in children is a prominent risk factor for developing stress-related psychopathology. During NEC, we measured behavioral inhibition (freezing and vocal reductions) as well as plasma cortisol levels and combined them to create a composite measure of AT (7, 8, 18). To assess regional brain metabolism during NEC, animals were injected with 18-FDG immediately prior to the exposure to the NEC context, and integrated brain metabolism occurring during NEC was assessed using positron emission tomography (PET). The phenotyping and imaging data from 22 of the animals was previously presented, and included initial gene expression studies using microarray techniques. Consistent with our previous work (19), AT was stable across repeated assessments (Figure 1a), and metabolism in the AT-network, including the dorsal amygdala, was associated with increased AT (Figure 1b, Table S1 and Figure S1).
Figure 1. AT is associated with a brain-wide AT network, and altered gene expression in the dorsal amygdala.
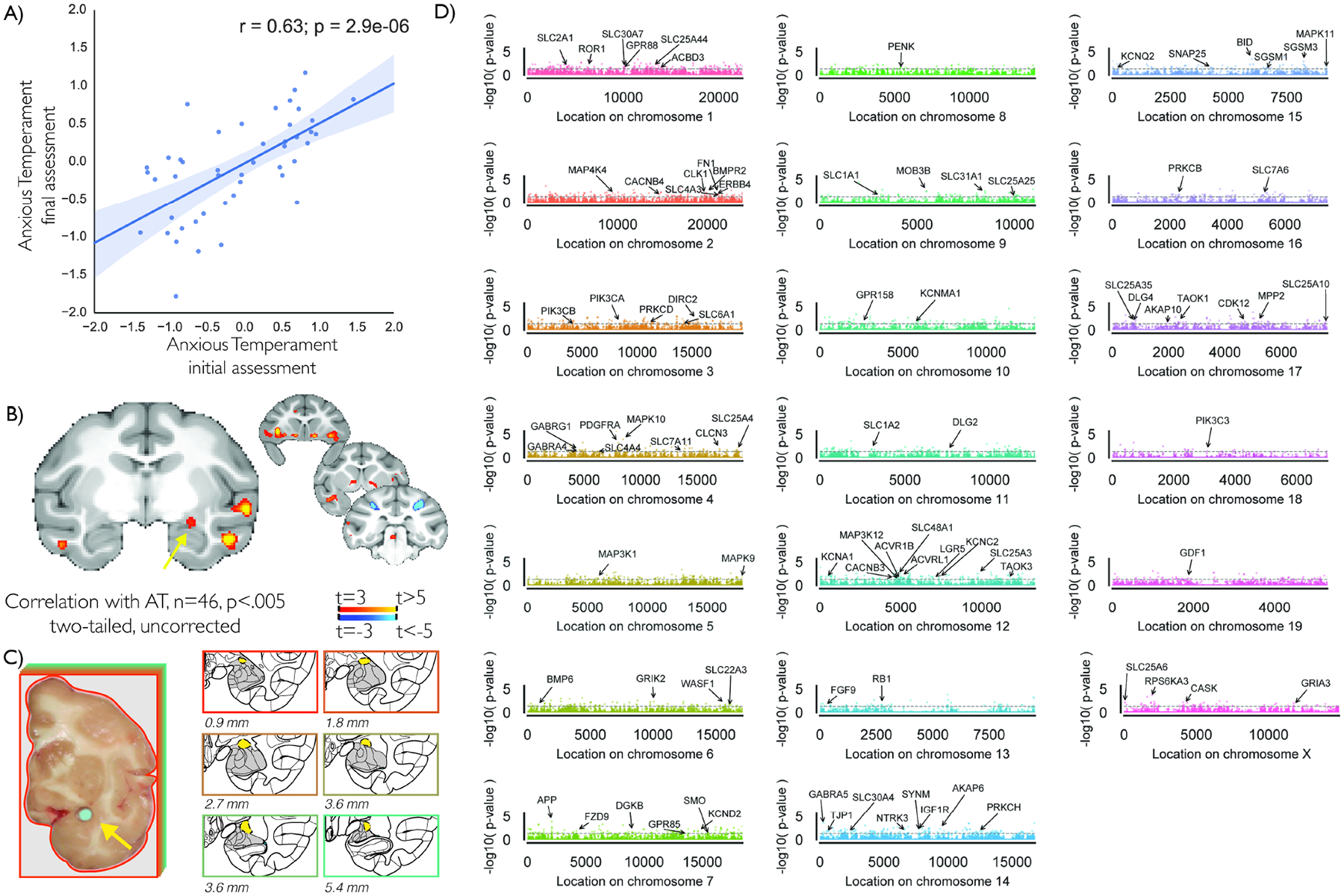
Assessment of AT in 46 young male rhesus monkeys revealed anxious temperament to be stable over time (r=.63, p<.001) (a). Average AT across assessments was associated with metabolic changes in an AT-related brain network (p<.005, two-tailed uncorrected), including the dorsal amygdala region (yellow arrow, also see Figure S1) (b). Dorsal amygdala tissue was harvested from these same animals, yellow arrow (c), RNA was extracted and mapped to the rhesus genome (MaSuRCA v7; UNMC v7.6.8), and a multiple regression was run for each gene with AT as the dependent variable and each exon’s expression levels as the independent measure (see Methods for details). Results demonstrated distributed associations between dorsal amygdala gene expression and AT across chromosomes, as can be seen in this Manhattan plot depicting the log-p value for the F-test of all exons regressed against AT (d). Genes reaching p<.05 significance were annotated if they were a part of the HUGO gene families that included one of the following in its title, “G protein”, “endogenous ligands”, “Kinases”, “aminobutyric”, “Glutamate”, “Mitogen-activated”, “channels”, “SNAREs”, “solute carrier”.
Tissue for RNA-seq was harvested from the dorsal amygdala region from the 46 animals that completed behavioral, endocrine, and brain metabolism assessments (Figure 1a–c). RNA-seq was performed using NuGEN Ovation RNA-seq v2 libraries on Illumina DNA sequencers with ~30 million 100bp reads per animal. Reads were mapped and quantified using an updated version of the RSeqFlow pipeline (20) designed specifically for the rhesus monkey genome and transcriptome (UNMC Rhesus v7.6.8) (21), and resulted in estimates of expression-levels for each annotated exon, intron, and junction of each gene. Performing RNA-seq in these 46 animals allowed for the opportunity to replicate and extend earlier microarray-based findings generated from half of these animals using a more in-depth approach (see SI; findings will be discussed separately when relevant).
Methods
A summary of the methods and procedures most relevant to understanding the RNA-seq and AAV-NTF3 overexpression studies are provided in below. Complete Detailed Methods can be found in the Supplementary Information, including FDG-PET and surgical details.
RNA-seq: Subjects
In 46 young male peri-adolescent rhesus monkeys (mean age=3.3 years old), we examined Ce gene expression using RNA-seq in combination with assessments of behavior, physiology, and functional brain imaging. AT was assessed in response to the potentially threatening no-eye-contact (NEC) condition of the human intruder paradigm, using a composite of increased freezing, decreased vocalizations, and increased cortisol. Brain function was assessed using NEC-related 18FDG-PET. RNA-seq was performed using NuGEN Ovation RNA-seq v2 libraries on Illumina DNA sequencers with ~30 million 100bp reads per animal. Using regression techniques, we examined variation in Ce mRNA expression in relation to individual differences in AT, as well as structural and functional imaging measures. Because of the unique nature and difficulty of this approach, we sequenced RNA from a number of subjects which had previously been examined using a microarray approach (n=22, “RNA sequencing cohort 1”; (19)). The 22 animals that were a part of RNA sequencing cohort 1, represent all animals discussed in Fox et al., 2015, with sufficient RNA remaining to be sequenced. When relevant, we discuss these data separately from the 24 additional subjects. The second cohort of 24 animals were completely new to this study (“RNA sequencing cohort 2”); when relevant we discuss these subjects separately. All procedures were approved by and in accordance with the guidelines established by the Institutional Animal Care and Use Committee at the University of Wisconsin-Madison.
RNA-seq: RNA Purification and Quantification
RNA-Seq was performed using a modification of the SPIA reaction of the NuGEN Ovation RNAseq V2 kit for cDNA Synthesis, followed by library construction using the NuGEN Rapid no-PCR protocol in a NuGEN Mondrian microfluidics instrument. RNA-Seq libraries were then sequenced to a minimum depth of ~30 million single-end 101 base-pair reads. RNA-seq reads were aligned to the Rhesus genome (MaSuRCA v7) using PerM as described in RseqFlow (20), and annotated using the rhesus transcriptome (UNMC v7.6.8). Reads which aligned to coding exons and known junctions in each gene model were summed and normalized (quantile & rpkm) to provide a raw proxy for gene expression.
RNA-seq: Statistical Analyses
Building on our previous work, we examined transcript “features” of each gene model, such as exons, introns, and splice junctions in relation to AT in python using statsmodels (https://github.com/statsmodels/statsmodels/). Because annotation of the rhesus genome is ongoing, and our understanding of splice variation still developing, we performed gene-level multiple regression analyses to predict AT. Each multiple regression analysis was performed in two steps, 1) nuisance variable age was entered into the model to predict AT, 2) estimated expression levels for each exon were simultaneously entered. The test of interest was the significance of the F-change between step 1 and step 2, which accounts for the variance explained by the exonic expression levels. The degrees of freedom for this model vary depending on the number of exons expressed for each gene. Analyses were restricted to genes where we mapped an average of at least 10 reads across subjects, and at least one read in each animal. Additional follow-up analyses can be found in the supplementary information.
AAV5-NTF3: Subjects
Thirty-five potential subjects were behaviorally screened for participation in the NT3 viral vector study with 10 minutes of the NEC condition, and 10 peri-adolescent male animals were selected (mean age at surgery = 2.43 ± 0.19 years). Selected subjects displayed freezing in response to the NEC that ranged from 133.4 seconds to 377.5 seconds (out of 600 seconds total). These subjects were selected because they were in the mid-high range of freezing to maximize the likelihood of observing the hypothesized NT3-induced changes in AT.
All 10 subjects were scanned with MRI and FDG-PET both before and after surgery (AAV5-NTF3 group) or rest (unoperated cage-mate controls). Animals were first assessed using FDG-PET imaging an average of 48.9 (±4.1) days prior to surgery. MRI data were collected roughly two weeks after the PET scan, averaging 33.6 (±1.4) days prior to surgery. As in the RNA-seq subjects, AT was assessed in response to the potentially threatening no-eye-contact (NEC) condition of the human intruder paradigm, using a composite of increased freezing, decreased vocalizations, and increased cortisol, and brain function was assessed using NEC-related 18FDG-PET. An AAV5 viral-vector designed to overexpress NTF3, the primary ligand for NTRK3, was injected into the dorsal amygdala region of 5 animals using real-time intraoperative surgeries. Animals were pair-housed, and one animal from each pair was randomly assigned to receive dorsal amygdala AAV5-NTF3 injections. Post-surgical FDG-PET scans were collected an average of 65.1 (±6.4) days after surgery, allowing sufficient time for recovery. Post-surgical MRI scans were collected an average of 75.8 (±5.1) days after the surgery. All statistical tests compared post-pre change between dorsal amygdala AAV5-NTF3 animals and their cage-mate control. All procedures were approved by and in accordance with the guidelines established by the Institutional Animal Care and Use Committee.
AAV-NTF3: Viral Vector
The DNA sequence corresponding to the entire open reading frame of the rhesus NTF3 (GenBank accession #XM_001118191, bases 8 to 1033) was inserted into the viral vector pAAV-MCS (Vector Biolabs, Malvern, PA). NTF3 protein expression was confirmed (see supplementary methods for details) and the plasmid was then packaged into rAAV5 (Vector Biolabs) with a titer of 1.2 × 1013 genome copies/ml.
AAV-NTF3: Statistical Analyses
Changes between pre- and post-surgical measures of AT were computed for the dorsal amygdala AAV5-NTF3 animals and compared to similarly-spaced assessments of the control animals. Corresponding comparisons were also performed to assess the effects of dorsal amygdala AAV5-NTF3 on the components of AT, i.e. freezing, cooing, and cortisol levels. The effects of NTF3 overexpression were assessed using the statsmodels package in python (https://github.com/statsmodels/statsmodels/). We also performed targeted and voxelwise analyses to examine the effects of NTF3 overexpression on brain metabolism (see supplementary methods for details). Briefly, we first computed the changes in metabolism pre- and post-surgery, and similarly timed assessments in control animals. We then performed group t-tests to compare changes in metabolism between groups. Exploratory voxelwise neuroimaging analysis results were thresholded at a liberal p<.05 two tailed, uncorrected.
Additional detailed methods can be found in the Supplementary Information.
Results
RNA-seq of dorsal amygdala tissue reveals many genes with AT-related expression levels
We first identified AT-related transcripts based on exon expression levels. Because annotation of the rhesus genome is ongoing, and our understanding of splice variation still developing, we performed gene-level multiple regression analyses to predict AT, where expression levels for each exon within a gene were simultaneously entered into a regression model to predict AT, while controlling for age and sex. This approach is well-suited for analysis of genomes with incomplete annotations that preclude a full splice-variant quantification. Additionally, this approach is not biased toward identifying well-annotated genes or genes with many exons (though it is limited by degrees-of-freedom in genes with >40 exons). Results demonstrated 67 genes to have AT-related exonic expression at p<.005 (two-tailed uncorrected; Table S2), and 618 genes at a threshold p<.05 (two-tailed uncorrected; Figure 1d; Table S2).
In addition to our primary analyses, we performed complementary analyses to identify AT-related dorsal amygdala transcripts, including examining various aspects of each gene’s expression profile (e.g. quantifying each intron, exon, and junction independently, averaging expression across the whole gene, and mapping each gene to the human genome), performing gene enrichment analyses, as well as independently examining each component of AT at each assessment (i.e. freezing, cooing, cortisol, at first and last assessment) in relation to gene expression (see Methods). To provide discovery opportunities to interested readers, all AT-related analyses, including post-hoc complementary analyses can be accessed via our web resource (http://at.psychiatry.wisc.edu; https://github.com/asfox/AT_DorsalAmygdala_RNAseq_FoxEtAl).
Results of our gene-level multiple regression approach demonstrated that a number of neuroplasticity-related molecules were inversely associated with AT, including the neurotrophic receptor, neurotrophin receptor kinase 3 (NTRK3, Figure 2a–b), and one of its downstream modulators ribosomal protein S6 kinase alpha-3 (RPS6KA3; Figure S2) (19). Other AT-related transcripts included the inhibitory neurotransmitter receptor subunit, gamma-aminobutyric acid (GABA) A receptor, alpha 5 (GABRA5; see Supplementary Information and Figure S3), GABA B receptor 1 (GABBR1) and amyloid precursor protein, (APP, Figure S4).
Figure 2. NTRK3 expression is associated with AT.
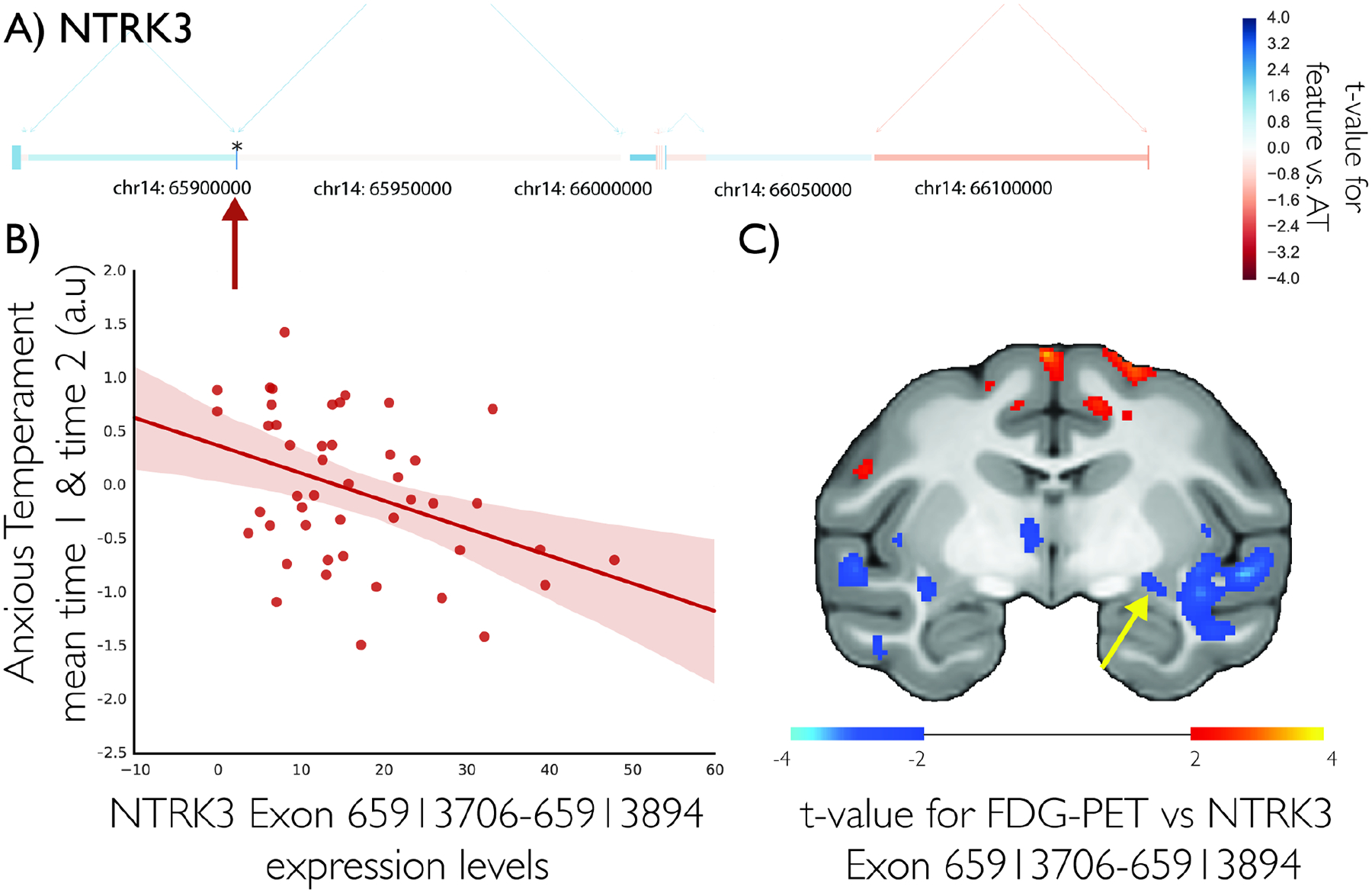
NTRK3 gene-model with each exon colored by the t-value of the association between that exon and AT in 46 young rhesus monkeys (a) shows exon between 65913706–65913894 to be inversely associated with AT (b). NTRK3 gene expression is associated with a distributed brain metabolic network (c; p<.05, two-tailed uncorrected) including an inverse association with the dorsal amygdala region (yellow arrow).
Pathway and ontology analyses underscore the importance of neuroplasticity-related processes in the dorsal amygdala region as important for AT
Consistent with our neurodevelopmental hypothesis (4), gene ontology and KEGG pathway enrichment analyses of the 618 nominally significant AT-related genes revealed significant overrepresentation of genes in the Neurotrophin signaling pathway (Kegg: hsa04722; z=−1.73, p=0.01884; additional significant pathways can be seen in Table S5), which includes one of the stronger hits in our dataset, the ribosomal protein RPS6KA3, a downstream kinase that can be modulated by Trk-receptor activation, which was associated with AT and each of its components (Figure S3). Moreover, we found overexpression in numerous plasticity-related categories, mTOR signaling pathway (Kegg: hsa04150), negative regulation of apoptotic process (GO:0043066), regulation of apoptotic process (GO:0042981), regulation of TOR signaling (GO:0032006), positive regulation of long-term synaptic potentiation (GO:1900273), protein serine/threonine kinase activator activity (GO:0043539). Full tables of overexpression analyses can be seen in Tables S5 and S6. Additionally, we also found significant overrepresentation within other potentially AT-related ontology categories, including behavioral fear response (GO:0001662), regulation of translation in response to stress (GO:0043555), and Wnt signaling pathway (GO:0016055). Although the informatic tools for making these comparisons are still developing alongside our actual knowledge about these pathways, these results continue to support the relevance of multiple molecular contributors to AT, and suggest neuroplasticity-related factors may play an important role.
Post-hoc analyses of RNA-seq of dorsal amygdala tissue support NTRK3 as a reliable target for AT-related alterations
Because of our interest in neuroplasticity as a protective factor for the development of anxiety disorders (4), and in the NTRK3 pathway specifically, we present complementary post-hoc analyses related to NTRK3. Importantly, the negative relation between NTRK3 and AT was significant (n=24, t=−2.08, p=0.0494, r=−0.41 95% CI=[−0.58,−0.22]) when excluding the initial cohort, in which, using microarray technology, we previously identified a relationship between AT and NTRK3. This demonstrates independent replication of the inverse AT-NTRK3 relationship (19). Next, we performed non-parametric analyses in the entire sample, encompassing both cohorts (n=46), which revealed that AT was associated with whole-gene NTRK3 expression levels (n=46, rho=−.35, p=0.019, 95% CI=[−0.47,−0.22]), Post-hoc examination of individual NTRK3 exon expression levels in relation to AT revealed only one exon to be significantly associated with AT on its own (Exon coordinates: 65913706–65913894; n=46, t=−2.76, p=0.009, r=−0.38 95% CI=[−0.5,−0.25]), suggesting specific NTRK3 isoforms may be uniquely associated with AT. Nonparametric spearman’s correlations across the entire sample revealed expression levels of additional NTRK3 gene features along the full length of the gene to be significantly correlated with AT (3/7 exons, 0/8 introns, and 4/9 splice-junctions; p’s<.05).
We then correlated expression levels at the most significant NTRK3 exon with brain metabolism to determine if NTRK3 expression was associated with dorsal amygdala metabolism. More specifically, we performed a voxelwise search for regions where expression levels of this AT-related NTRK3 exon was associated with FDG-uptake during the NEC context. Results demonstrated dorsal amygdala expression of the most AT-related NTRK3 exon to be inversely associated with metabolism in the dorsal amygdala region during NEC (Figure 2c; n=46, p’s<.05, uncorrected).
NT-3 overexpression in dorsal amygdala decreased AT and altered extended amygdala metabolism
Based on these data we hypothesized that increased NTRK3 signaling in dorsal amygdala would decrease AT. To test this hypothesis, we used viral-vector techniques to increase activation of the NTRK3 pathway via overexpression of its endogenous ligand neurotophin-3 (NT-3). Using real-time intraoperative MRI, we infused an AAV5-CMV-NTF3 viral-vector into the dorsal amygdala of 5 young rhesus monkeys and compared them to 5 unoperated control animals (Figure 3a–d; see Methods). Overexpression of NTF3 in dorsal amygdala neurons resulted in a significant decrease in AT compared to control animals (n=5/group; non-parametric Mann-Whitney=4.0, p=0.047; parametric test was two-tailed trend-level significant in the predicted direction both unpaired, t=−2.176, p=0.061, Cohen’s d=1.54 with 95% CI=[−1.1,0.66], and with paired groups, as the study was designed, t= −2.515, p=0.066, Cohen’s d=−1.03 with 95% CI=[−2.16,0.11]; Figure 3f). Further inspection of the components of AT revealed a significant effect of dorsal amygdala NTF3 overexpression in the predicted direction reducing threat-related freezing (t=−3.013, p=0.039, Cohen’s d=−1.23 with 95% CI=[−2.36,−0.1]; Figure 3f, right top). Though in the predicted direction, the post-pre changes were not significant for cooing (though only one animal emitted a coo call during this experiment, t=1.000, p=0.374, Cohen’s d=0.41 with 95% CI=[−0.72,1.54]; Figure 3f, right middle) or cortisol (t= −0.693, p=0.527, Cohen’s d=−0.28 with 95% CI=[−1.42,0.85]; Figure 3f, right bottom). Because of the relatively large confidence interval for point-estimates and the relationships between NTRK3 expression and the components of AT in the RNA-seq study, we choose not to interpret these differences in the overexpression study. Nevertheless, it appears to produce maximal alterations of AT that affect all of its components may require multiple genetic targets. Although we do not interpret these null effects, follow-up analyses focused on freezing.
Figure 3. AAV5-NTF3 overexpression in the dorsal amygdala alters regional metabolism and decreases AT.
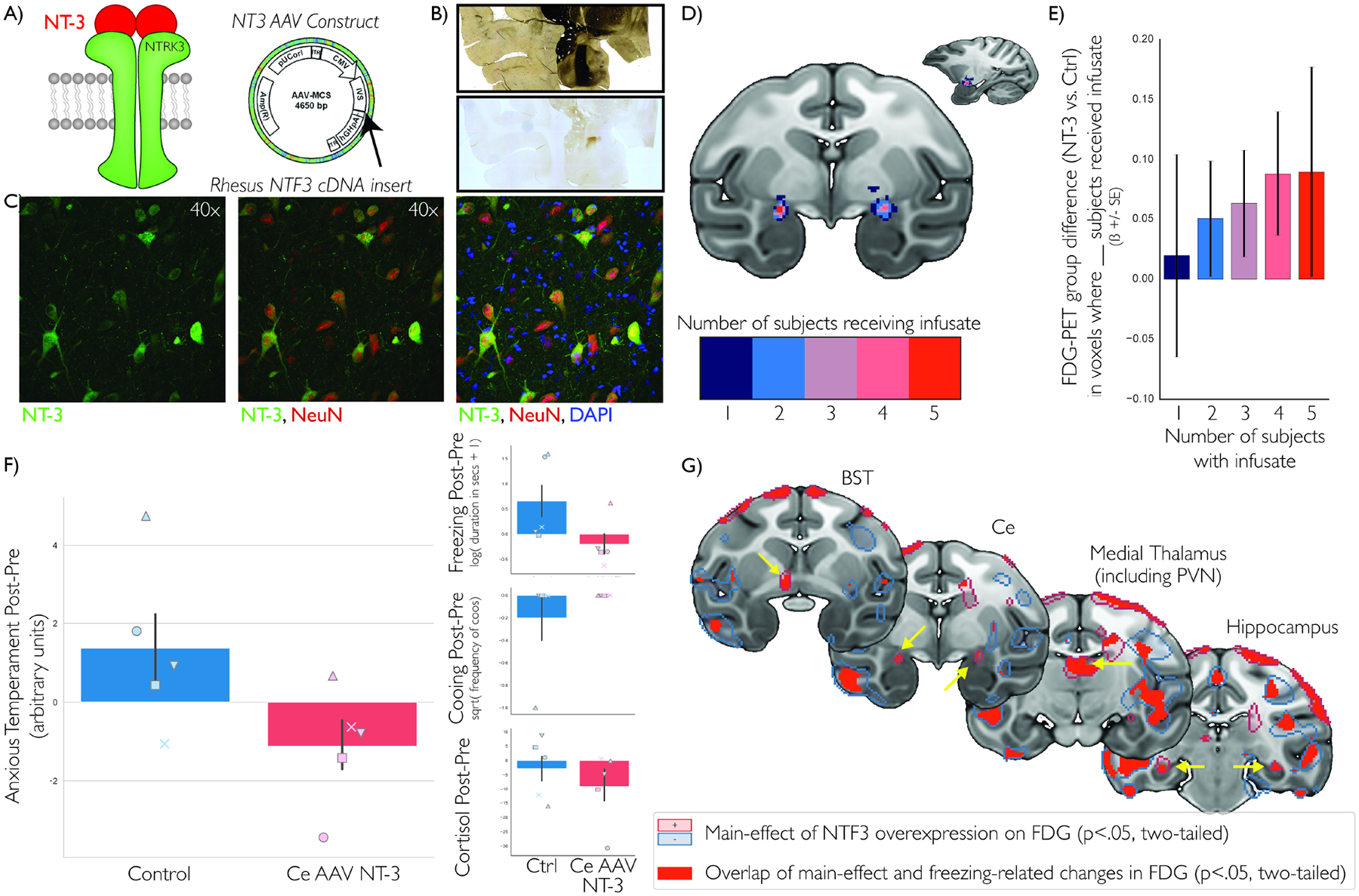
Because NT-3 is the primary ligand for NTRK3 (a, left), we infused AAV5 containing the NTF3 construct (a, right) to overexpress NT-3 in the dorsal amygdala of 5 young rhesus monkeys, using convection enhanced delivery and intraoperative MRI-guided surgical techniques (30, 32). Expression of NT-3 was verified using precise post-mortem dorsal amygdala localization using corresponding acetylcholinesterase (AChE) staining (b, top) with an NT-3 antibody for visualization of overexpression (b, bottom), and high-magnification co-staining demonstrating selective neuronal expression: NT-3 (green), NeuN (red; neurons), and DAPI (blue; cell nuclei) (c). We demonstrated accurate targeting of the infusate into bilateral dorsal amygdala region in all five animals after transformation to standard space (d), using pre-mortem real-time T1-weighted MRI imaging of the viral-vector mixed with radio-opaque Gd infusate. Results demonstrated infusion-overlap related group-differences in NEC-context metabolism, such that the infusion-induced metabolic changes were larger in those voxels in which more subjects received infusate (n=5) compared to uninfused control animals (n=5) (e). AAV-NTF3 infusion was associated with decreases in AT (n=5/group) (f, left). Dorsal amygdala AAV-NTF3 overexpression was significantly associated with decreased freezing, but did not reach significance with cooing and cortisol (though in the predicted direction, suggesting these changes could be specifically associated with freezing (f, right inset). Each Ce-AAV-NTF3 animal has their own marker, which is shared by their matched control (f). Finally, we identified brain-wide metabolic changes that demonstrated a main effect of AAV-NTF3 overexpression (g; p<.05, two-tailed, uncorrected; n=5/group), where changes in metabolism were correlated with changes in freezing across groups (g, red; n=10) in regions that included the BST, dorsal amygdala, medial thalamus, and hippocampus (yellow arrows). These data suggest that the behavioral alterations resulting from dorsal amygdala NTF3 overexpression may be mediated by a distributed network of metabolic changes.
We predicted that dorsal amygdala NTF3 overexpression would alter brain metabolism. We found a post-surgical increase in metabolism compared to controls within the dorsal amygdala intraoperative MRI defined infusion region (Figure 3e). Voxelwise analyses identified NTF3-induced freezing-related metabolic changes within the AT-network (Figure 3g and Tables S3 and S4). These regions, which are likely to mediate the effects of NTF3 on AT, included the Ce-region, BST-region, as well as regions of the medial thalamus and hippocampus. Interestingly, we found that the metabolic alterations in these regions, which are normally positively associated with AT and freezing behavior, were inversely associated with the NTF3-related change in freezing. This unexpected finding highlights the important and interesting disconnect between unitary measures of brain activity and the complex molecular systems that give rise to variation in brain function. Taken together, these findings suggest that neuroplasticity in the dorsal amygdala modulates the function of the distributed neural circuit underlying anxiety.
Discussion
Here, in a highly relevant AT-NHP model, we found variation in dorsal amygdala NTRK3 expression levels to be inversely associated with AT. Importantly, overexpression of NT-3, the major NTKR3-activating ligand, was sufficient to decrease AT. NTRK3, also known as TrkC, is a growth factor receptor located on the surface of the cell, having the potential to alter neuron growth and synaptic plasticity via the intracellular signaling pathways it shares with other Trk receptors (22, 23). The current results implicate a novel translationally-relevant neurotrophic pathway within the primate dorsal amygdala, which complements studies implicating other neurotrophic factors such as BDNF and FGF2 in rodent models of anxiety (24–26). The data presented here support our hypothesis implicating altered dorsal amygdala neuroplasticity as a molecular substrate for the early-life risk to develop anxiety and depressive disorders.
Additional in-depth in vitro and in vivo studies will be important to elucidate the effects of NTRK3 pathway activation at micro-circuit and molecular levels. For example, we found that NTF3 overexpression resulted in both a decrease in AT and an increase in metabolism throughout AT-related regions, which included the Ce-containing dorsal amygdala region and the Ce-projecting BST region. This finding contrasts with our previous work in a large sample of 592 rhesus monkeys that identified AT-related metabolism in these same regions to be positively correlated with AT. It is interesting that we did not observe a positive correlation between AT and aggregate Ce metabolism after NTF3 overexpression. Perhaps this is not surprising, as these data lend important insight into our previous observation that only a small amount of the variability in AT could be accounted for by regional metabolism during NEC (7). Studies in rodents suggest that within regions as small as a typical neuroimaging voxel there often are neurons that produce opposing effects, as is the case with regard to mutually inhibitory populations within intra-Ce circuits (27). This emphasizes the importance of using preclinical models in conjunction with in vivo neuroimaging to understand the relationship between function at a microcircuit level with that represented in a single imaging voxel (14, 27, 28). The disparity in the direction of the effect provides potential insights into why many neuoimaging-phynotype associations typically explain relatively small amounts of variation. We believe, however, that these relatively weak associations do not detract from the importance of identifying regions using neuroimaging. In fact, it is likely that systems- and molecular-level studies can complement clinical neuroimaging and explain the variance that is unaccounted for by the aggregate signal in voxelwise neuroimaging measures (29). Here, at the molecular level, the precise mechanisms by which NT-3 or NTRK3 variation leads to alterations in metabolism and AT remains to be explored. For example, NT-3 can also bind, like BDNF, to NTRK2 (also known as TrkB). Although not observed in the RNA-seq analyses, this raises the possibility that the NTKR2 pathway may also modulate AT in primates (23). We also emphasize that further studies focused on NTRK3 receptor isoforms are warranted, since in vitro studies demonstrate that NT-3 differentially interacts with NTRK3 receptor variants (23).
While we causally implicate the NT-3/NTRK3 system in AT, we emphasize that this target was but one of the many discovery-based associations. We have focused on neuroplasticity-related signaling and the NT-3 system, but this is only one of many potential hypotheses that can be derived from the discovery-based RNA-seq data. Our RNA-seq analyses didn’t reveal results that survive strict multiple comparison correction, and accordingly should be interpreted as moderate evidence in support of a particular molecule’s involvement in anxiety. Nevertheless, there is much to be gleaned from further examination of AT-related transcripts on our online resource (http://at.psychiatry.wisc.edu, or https://github.com/asfox/AT_DorsalAmygdala_RNAseq_FoxEtAl), as there are many other dorsal amygdala transcripts that are associated with AT and/or distributed brain metabolism (e.g. Figures S2, S3, & S4). In addition to the findings presented here, it is likely that other anxiety-related gene expression relationships are being masked by incomplete and ongoing annotation of the rhesus genome. In addition to implicating the NT-3/NTRK3 system in the risk to develop anxiety and depressive disorders, these data underscore the importance of understanding the interactions among multiple molecular mechanisms that likely converge to influence the distributed neural network that underlies anxiety.
The demonstration that AT is decreased by dorsal amygdala NTF3-overexpression complements our previous report of increased AT after overexpression of a gene that is associated with anxiety, corticotropin releasing hormone (CRH), in the dorsal amygdala (30). The NTF3 and CRH overexpression studies were performed in independent samples confirming that alterations in dorsal amygdala gene expression can bidirectionally change AT. From a methodological perspective, concerns regarding possible non-specific effects of the surgery or infusions are mitigated by the demonstration that nearly identical methods were used in these two experiments that produced opposite but predicted behavioral effects. The mechanisms by which NT-3 and CRH overexpression result in opposing effects remains unknown. Rodent studies using Ce-CRH knockouts and optogenetic manipulation of Ce CRH-expressing cells suggest that CRH may provide “gain control” on potentially threatening inputs (14, 27, 31). Previous research on growth-factors suggests that the influence of NT-3 is unlikely to be that specific. Instead, it is more likely that NT-3 is exerting its influence on Ce function by altering the structural properties of Ce cells and altering synapses, including those to and from Ce inhibitory interneurons, some of which are CRH positive. Additional research will be required to understand the overlap between CRH and NTRK3 expressing cells, but it would be reasonable to hypothesize that effects of NT-3 increase local inhibition of CRH and other sets of peptide-expressing cells.
We identified potential molecular targets for the treatment and prevention of anxiety and depressive disorder, and demonstrated causal manipulation of the NT-3/NTRK3 system in early-life AT. Identifying AT-related molecular alterations will provide new insights into the cell-states and physiological features of the dorsal amygdala that are altered in highly anxious individuals. Such work promises to guide the development of new treatments for preventing and alleviating the life-long suffering associated with stress-related psychopathology.
Supplementary Material
| Resource Type | Specific Reagent or Resource | Additional Information | ||
|---|---|---|---|---|
| Add additional rows as needed for each resource type | Include species and sex when applicable. | Include name of manufacturer, company, repository, individual, or research lab. Include PMID or DOI for references; use “this paper” if new. | Include catalog numbers, stock numbers, database IDs or accession numbers, and/or RRIDs. RRIDs are highly encouraged; search for RRIDs at https://scicrunch.org/resources. | Include any additional information or notes if necessary. |
| Antibodies | Mouse monoclonal anti-NeuN (clone A60) | MilliporeSigma | Cat# MAB377; RRID_AB_2314889; Lot# 2806074 | |
| Antibodies | Goat polyclonal anti-human NTF3 | R&D Systems | Cat# AF-267-NA; RRID_AB_354434;Lot# UY0614011 | |
| Virus Strains | AAV5-CMV-NTF3 | Vector Biolabs | ||
| Chemicals, Peptides, and Recombinant Proteins | Gadobenate dimeglumine (Multihance) | Bracco Diagnostics | Cat# 516413; NDC# 0270-5164-13 | |
| Commercial Assay Or Kit | DPC Coat-a-count cortisol radioimmunoassay | Siemens | Cat# TK01; Lot# TKC01 1625;Cat# TK02; Lot# TKC02 1531; Cat# TK05; Lot# TKC05 1530 | |
| Commercial Assay Or Kit | ImmuChem Cortisol Coated Tube radioimmunoassay kit | MP Biomedicals | Cat# 07–221105; Lot# FK1601,02,03 | |
| Commercial Assay Or Kit | RNeasy Plus Mini kit | Qiagen | Cat# 74134 | |
| Commercial Assay Or Kit | NuGEN Ovation RNAseq System v2 kit | NuGEN | Cat# 7102–32 | |
| Experimental Models: Organisms/Strains | Macaca mulatta (RNA-seq; n=46 male) | Wisconsin National Primate Research Center; Harlow Center for Biological Psychology | ||
| Experimental Models: Organisms/Strains | Macaca mulatta (AAV5-NTF3; n=10 male) | Wisconsin National Primate Research Center; Harlow Center for Biological Psychology | ||
| Recombinant DNA | pAAV-MCS | Vector Biolabs | ||
| Recombinant DNA | Coding sequence of rhesus NTF3 | This paper | GenBank accession #XM_001118191, bases 8 to 1033 | |
| Software; Algorithm | RTHawk scanner interface | HeartVista, Palo Alto, CA | N/A | |
| Software; Algorithm | VURTIGO toolkit | Sunnybrook Health Sciences Centre; Toronto, Canada | N/A | |
| Software; Algorithm | Spamalize | http://brainimaging.waisman.wisc.edu/õakes/spam/spam_frames.htm | N/A | |
| Software; Algorithm | ANTs | http://stnava.github.io/ANTs/ | N/A | |
| Software; Algorithm | fmristat | http://www.math.mcgill.ca/keith/fmristat/ | N/A | |
| Software; Algorithm | PerM | Chen and Souaiaia, 2009 | https://code.google.com/archive/p/perm/ | |
| Software; Algorithm | RSeqFlow | Wang et al., 2009 | https://genomics.isi.edu/rnaseq | |
| Software; Algorithm | SageRage – Exon Specific Alignment for Rhesus | Souaiaia et al, 2018 | https://github.com/tadesouaiaia/sage | |
| Software; Algorithm | SageRage – Exon Quantficaton for Rhesus | Souaiaia et al, 2018 | https://github.com/tadesouaiaia/rage | |
| Other | Custom Algorithms For Exon Specific Expression | This paper | http://aml31.genome.wustl.edu | |
| Other | Web-resource for interrogating ancillary analyses. | This Paper | https://github.com/asfox/AT_DorsalAmygdala_RNAseq_FoxEtAl | |
| Deposited Data | Statistical Parametric Maps for voxelwise FDG-PET analyses | This Paper | https://www.neurovault.org/collections/AYJGSCJH/ |
Acknowledgements
We thank the personnel of the Harlow Center for Biological Psychology, the HealthEmotions Research Institute, the Waisman Laboratory for Brain Imaging and Behavior, the Wisconsin National Primate Center, the Wisconsin Institutes for Medical Research, A. Shackman, L. Williams, M. Jesson, D.P.M. Tromp, D. French, S. Shelton and H. Van Valkenberg. We’d also like to thank the University of California, Davis and the California National Primate Research Center for their support. This work was supported by: R01-MH046729 and R01-MH081884, as well as grants to National Primate Center Research Centers (P51-OD011106, P51-RR000167, and P51 OD011107), and the Wiasman Center (P30-HD003352).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial Disclosures
At the time of writing, Dr. Kalin had received honoraria from CME Outfitters, Elsevier, and the Pritzker Consortium; served on scientific advisory boards for Actify Neurotherapies, Neuronetics, and currently serves as an advisor to the Pritzker Neuroscience Consortium and consults to Corcept Therapeutics; served as co-editor of Psychoneuroendocrinology, and currently serves as Editor-in-Chief of The American Journal of Psychiatry; and has patents on promoter sequences for corticotropin-releasing factor CRF2alpha and a method of identifying agents that alter the activity of the promoter sequences (7,071,323; 7,531,356), promoter sequences for urocortin II and the use thereof (7,087,385), and promoter sequences for corticotropin-releasing factor binding protein and the use thereof (7,122,650). All other authors report no biomedical financial interests or potential conflicts of interest.
References
- 1.Biederman J, Hirshfeld-Becker DR, Rosenbaum JF, Herot C, Friedman D, Snidman N, et al. (2001): Further evidence of association between behavioral inhibition and social anxiety in children. The American journal of psychiatry. 158:1673–1679. [DOI] [PubMed] [Google Scholar]
- 2.Clauss JA, Blackford JU (2012): Behavioral inhibition and risk for developing social anxiety disorder: a meta-analytic study. J Am Acad Child Adolesc Psychiatry. 51:1066–1075 e1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Essex MJ, Klein MH, Slattery MJ, Goldsmith HH, Kalin NH (2010): Early risk factors and developmental pathways to chronic high inhibition and social anxiety disorder in adolescence. The American journal of psychiatry. 167:40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fox AS, Kalin NH (2014): A Translational Neuroscience Approach to Understanding the Development of Social Anxiety Disorder and Its Pathophysiology. The American journal of psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams LE, Oler JA, Fox AS, McFarlin DR, Rogers GM, Jesson MA, et al. (2015): Fear of the unknown: uncertain anticipation reveals amygdala alterations in childhood anxiety disorders. Neuropsychopharmacology. 40:1428–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Etkin A, Wager TD (2007): Functional neuroimaging of anxiety: a meta-analysis of emotional processing in PTSD, social anxiety disorder, and specific phobia. The American journal of psychiatry. 164:1476–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fox AS, Oler JA, Shackman AJ, Shelton SE, Raveendran M, McKay DR, et al. (2015): Intergenerational neural mediators of early-life anxious temperament. Proc Natl Acad Sci U S A. 112:9118–9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fox AS, Shelton SE, Oakes TR, Davidson RJ, Kalin NH (2008): Trait-like brain activity during adolescence predicts anxious temperament in primates. PLoS ONE. 3:e2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shackman AJ, Tromp DPM, Stockbridge MD, Kaplan CM, Tillman RM, Fox AS (2016): Dispositional negativity: An integrative psychological and neurobiological perspective. Psychological Bulletin. 142:1275–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Olmos JS, Heimer L (1999): The concepts of the ventral striatopallidal system and extended amygdala In: McGinty JF, editor. Advancing from the ventral striatum to the extended amygdala: Implications for neuropsychiatry and drug abuse. New York: The New York Academy of Sciences, pp 1–32. [DOI] [PubMed] [Google Scholar]
- 11.Oler JA, Tromp DP, Fox AS, Kovner R, Davidson RJ, Alexander AL, et al. (2017): Connectivity between the central nucleus of the amygdala and the bed nucleus of the stria terminalis in the non-human primate: neuronal tract tracing and developmental neuroimaging studies. Brain Struct Funct. 222:21–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oler JA, Birn RM, Patriat R, Fox AS, Shelton SE, Burghy CA, et al. (2012): Evidence for coordinated functional activity within the extended amygdala of non-human and human primates. NeuroImage. 61:1059–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tillman RM, Stockbridge MD, Nacewicz BM, Torrisi S, Fox AS, Smith JF, et al. (2018): Intrinsic functional connectivity of the central extended amygdala. Hum Brain Mapp. 39:1291–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fadok JP, Markovic M, Tovote P, Luthi A (2018): New perspectives on central amygdala function. Curr Opin Neurobiol. 49:141–147. [DOI] [PubMed] [Google Scholar]
- 15.Fox AS, Oler JA, Tromp do PM, Fudge JL, Kalin NH (2015): Extending the amygdala in theories of threat processing. Trends Neurosci. 38:319–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalin NH, Shelton SE, Davidson RJ (2004): The role of the central nucleus of the amygdala in mediating fear and anxiety in the primate. J Neurosci. 24:5506–5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oler JA, Fox AS, Shackman AJ, Kalin NH (2016): The central nucleus of the amygdala is a critical substrate for individual differences in anxiety In: Amaral DG, Adolphs R, editors. Living without an Amygdala. New York, NY: Guilford Press. [Google Scholar]
- 18.Oler JA, Fox AS, Shelton SE, Rogers J, Dyer TD, Davidson RJ, et al. (2010): Amygdalar and hippocampal substrates of anxious temperament differ in their heritability. Nature. 466:864–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fox AS, Oler JA, Shelton SE, Nanda SA, Davidson RJ, Roseboom PH, et al. (2012): Central amygdala nucleus (Ce) gene expression linked to increased trait-like Ce metabolism and anxious temperament in young primates. Proc Natl Acad Sci U S A. 109:18108–18113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Mehta G, Mayani R, Lu J, Souaiaia T, Chen Y, et al. (2011): RseqFlow: workflows for RNA-Seq data analysis. Bioinformatics. 27:2598–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zimin AV, Cornish AS, Maudhoo MD, Gibbs RM, Zhang X, Pandey S, et al. (2014): A new rhesus macaque assembly and annotation for next-generation sequencing analyses. Biol Direct. 9:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamballe F, Klein R, Barbacid M (1991): trkC, a new member of the trk family of tyrosine protein kinases, is a receptor for neurotrophin-3. Cell. 66:967–979. [DOI] [PubMed] [Google Scholar]
- 23.Martinowich K, Manji H, Lu B (2007): New insights into BDNF function in depression and anxiety. Nat Neurosci. 10:1089–1093. [DOI] [PubMed] [Google Scholar]
- 24.Perez JA, Clinton SM, Turner CA, Watson SJ, Akil H (2009): A new role for FGF2 as an endogenous inhibitor of anxiety. J Neurosci. 29:6379–6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song M, Martinowich K, Lee FS (2017): BDNF at the synapse: why location matters. Mol Psychiatry. 22:1370–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turner CA, Clinton SM, Thompson RC, Watson SJ Jr., Akil H (2011): Fibroblast growth factor-2 (FGF2) augmentation early in life alters hippocampal development and rescues the anxiety phenotype in vulnerable animals. Proc Natl Acad Sci U S A. 108:8021–8025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fadok JP, Krabbe S, Markovic M, Courtin J, Xu C, Massi L, et al. (2017): A competitive inhibitory circuit for selection of active and passive fear responses. Nature. 542:96–100. [DOI] [PubMed] [Google Scholar]
- 28.Tye KM, Prakash R, Kim SY, Fenno LE, Grosenick L, Zarabi H, et al. (2011): Amygdala circuitry mediating reversible and bidirectional control of anxiety. Nature. 471:358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fox AS, Shackman AJ (2019): The central extended amygdala in fear and anxiety: Closing the gap between mechanistic and neuroimaging research. Neurosci Lett. 693:58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalin NH, Fox AS, Kovner R, Riedel MK, Fekete EM, Roseboom PH, et al. (2016): Overexpressing Corticotropin-Releasing Hormone in the Primate Amygdala Increases Anxious Temperament and Alters Its Neural Circuit. Biol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanford CA, Soden ME, Baird MA, Miller SM, Schulkin J, Palmiter RD, et al. (2017): A Central Amygdala CRF Circuit Facilitates Learning about Weak Threats. Neuron. 93:164–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emborg ME, Joers V, Fisher R, Brunner K, Carter V, Ross C, et al. (2010): Intraoperative intracerebral MRI-guided navigation for accurate targeting in nonhuman primates. Cell transplantation. 19:1587–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


