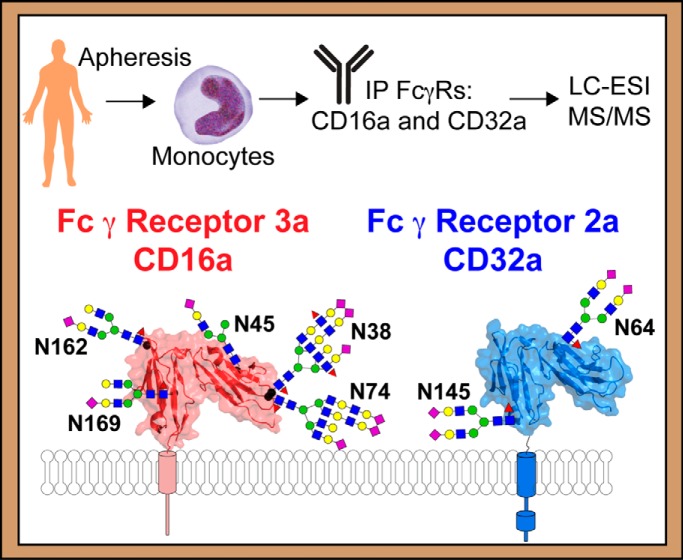
The composition of post-translational modifications, particularly carbohydrate chains attached to specific asparagine residue (N-glycans), that are attached to antibodies are well described in the human body and modulate the binding to Fc γ receptors. The N-glycan composition of these receptors likewise impacts antibody binding affinity, however receptor composition in the human body is unknown because of challenges associated with obtaining sufficient receptor from a single donor and limited analytical techniques. Here we characterize all N-glycosylation sites of CD16a and CD32a isolated from primary human monocytes that are isolated from a normally discarded filter following platelet and plasma donation.
Keywords: Antibodies, therapeutic antibodies, glycoproteomics, n-glycosylation, glycoproteins
Graphical Abstract
Highlights
Monocytes are isolated from single donors after apheresis.
Monocytes process CD16a and CD32a N-glycosylation differently by site and donor.
CD16a with hybrid and oligomannose type N-glycans bind IgG1 Fc with stronger affinity than complex type.
Abstract
FcγRIIIa (CD16a) and FcγRIIa (CD32a) on monocytes are essential for proper effector functions including antibody dependent cellular cytotoxicity (ADCC) and phagocytosis (ADCP). Indeed, therapeutic monoclonal antibodies (mAbs) that bind FcγRs with greater affinity exhibit greater efficacy. Furthermore, post-translational modification impacts antibody binding affinity, most notably the composition of the asparagine(N)-linked glycan at N162 of CD16a. CD16a is widely recognized as the key receptor for the monocyte response, however the post-translational modifications of CD16a from endogenous monocytes are not described. Here we isolated monocytes from individual donors and characterized the composition of CD16a and CD32a N-glycans from all modified sites. The composition of CD16a N-glycans varied by glycosylation site and donor. CD16a displayed primarily complex-type biantennary N-glycans at N162, however some individuals expressed CD16a V158 with ∼20% hybrid and oligomannose types which increased affinity for IgG1 Fc according to surface plasmon resonance binding analyses. The CD16a N45-glycans contain markedly less processing than other sites with >75% hybrid and oligomannose forms. N38 and N74 of CD16a both contain highly processed complex-type N-glycans with N-acetyllactosamine repeats and complex-type biantennary N-glycans dominate at N169. The composition of CD16a N-glycans isolated from monocytes included a higher proportion of oligomannose-type N-glycans at N45 and less sialylation plus greater branch fucosylation than we observed in a recent analysis of NK cell CD16a. The additional analysis of CD32a from monocytes revealed different features than observed for CD16a including the presence of a predominantly biantennary complex-type N-glycans with two sialic acids at both sites (N64 and N145).
Circulating leukocytes express Fc γ receptors (FcγRs)1 that trigger a protective response after encountering an antibody-coated target cell. Therapeutic monoclonal antibodies (mAbs) are a rapidly growing class of highly specific and well tolerated drugs with a predicted global market of $140B by 2024 with many new variants in the development pipeline (1). Many mAbs treat diseases by eliciting effector functions including antibody dependent cellular cytotoxicity (ADCC) and antibody dependent cellular phagocytosis (ADCP) that are triggered by FcγRs.
FcγRIII (CD16) binds to the IgG1 crystallizable fragment (Fc) and is likely the predominant receptor contributing to the therapeutic efficacy of mAbs that require effector functions, thus the affinity of Fc binding to CD16a is an important therapeutic consideration (2–6). NK cells, macrophages and a subset of monocytes express CD16a as an integral membrane protein whereas neutrophils express the related glycosylphosphatidylinositol-anchored receptor CD16b (7). Though stimulating NK cell-mediated effector functions represents a goal for mAb development, other CD16+ cell types including monocytes respond to antibody-coated targets, contributing to clearance and potentially therapeutic efficacy (8, 9). Indeed, monocytes appeared as the dominant effector cell for B cell removal and platelet phagocytosis with little contribution from NK or T cells (10, 11). Furthermore, increasing the strength of IgG binding by monocyte CD16a increased ADCC, indicating that increasing the strength of the monocyte CD16a-IgG interaction may lead to greater clinical efficacy (12). These results indicate that enhancing mAb affinity for CD16a expressed on monocytes/macrophage may lead to enhanced clinical efficacy.
Monocytes display classical, non-classical and intermediate phenotypes (13, 14). The classical CD14++,CD16- phenotype represents >80% of peripheral monocytes with non-classical (CD14+, CD16++) and intermediate (CD14++, CD16+) monocytes accounting for the remainder. Monocytes also express the FcγRs CD32a and CD64. Monocytes can differentiate into macrophages, dendritic cells and osteoclasts and adopt critical roles in tissue specific immune responses as well as wound healing (8, 15). A recent study demonstrated that monocyte ADCC activity is exclusive to CD16+ non-classical/intermediate monocytes (16). This manuscript showed that CD16 is indispensable for proper ADCC activity whereas CD32a contributes to a lesser extent.
The affinity between IgG and CD16a is dependent on polymorphisms and post-translational modification. The CD16a V158 allele encodes a receptor with 4–5-fold greater affinity for IgG1 than the CD16a F158 allele (17). Interestingly, clinical efficacy of glycoengineered mAbs designed to bind CD16a with higher affinity is greater in V158 homozygous individuals than F158 homozygotes, suggesting the affinity between IgG and CD16a is directly correlated to clinical response (18, 19). Furthermore, composition of the asparagine(N)-linked glycans also impacts the affinity of the CD16a-IgG interaction, though most studies focused on IgG N-glycan composition (2–6, 20–23). Recent in vitro studies indicated that CD16a N-glycan composition likewise affects binding to IgG (24–26). CD16a with an oligomannose-type N-glycan bound with greater affinity to IgG1 Fc than CD16a with complex-type N-glycans (21, 23, 27). Of the five CD16a N-glycans, the effect of N-glycan composition is primarily because of the modification at N162. Prior studies from our group identified the presence of minimally-processed endogenous CD16a glycoforms from primary NK cells and substantial donor-to-donor variability in processing at select sites (27, 28). Thus, post-translational modification plays a critical role in antibody-mediated functions, though little is known about how monocytes process CD16a.
A description of FcγR processing from monocytes is expected to define what receptor forms and affinities are present at the cell surface. Isolating receptors from the surface of primary human cells represents an enormous challenge because of the small amounts of receptor in the body. We recently described the analysis of CD16a N-glycans from NK cells (28), however, we expect that monocytes represent a greater challenge. Despite the greater abundance of monocytes in the body compared with NK cells, CD16a+/non-classical monocytes represent ∼17% of the total and it is unclear if monocytes express as much CD16a as NK cells.
CD16a and CD32a are highly modified molecules with five and two conserved N-glycan sites, respectively. N-glycans are commonly found on secreted proteins, particularly serum proteins that exhibit variable composition resulting from differential processing. In general, N-glycans are attached immediately following translation and processed in the ER and Golgi into one of three main N-glycosylation types: oligomannose, hybrid or complex-type (reviewed in (29)). Processing enzymes in the medial and trans Golgi modify N-glycans with “caps” including sialic acid or introduce branches to generate highly-processed complex-type N-glycans. Some N-glycans escape the majority of the processing enzymes; these N-glycan types include oligomannose-type which are minimally processed and hybrid-type which experience extensive processing on only part of the glycan. N-glycosylation heterogeneity is likely dictated by abundance of the remodeling enzymes as well as enzyme localization in the ER and Golgi network (30, 31). Cell-specific glycosylation differences have been described to be important in many biological processes, especially in development and the immune system (30, 32). Here we characterize the N-glycosylation profile of CD16a and CD32a from monocytes retained during plasma and platelet donation.
EXPERIMENTAL PROCEDURES
Materials
All materials were purchased from Millipore-Sigma (Burlington, MA) unless otherwise noted.
Experimental Design
This study was judged by Iowa State University's institutional review to be exempt because the donors were de-identified, the only available donor information is shown in Table I. Biological samples are represented by four single donors and six donors combined into two separate pools (MoA and MoB). Lysates from certain donors were pooled and processed as a single sample because of low monocyte yields or unavailability of basic information typically available for other donors. Technical replicates were not possible because cell isolation from a single donor or donor pool only provided enough material to perform a single CD16a characterization.
Table I. Donor information.
| Donor | Gender | Age | ABO group | Rh factor | Date of collection | Monocytes isolated × 106 | CD16a genotype |
|---|---|---|---|---|---|---|---|
| MoA (pool) | |||||||
| Mo01 | n.d. | n.d. | n.d. | n.d. | 6/9/2017 | 200 | V/F |
| Mo03 | n.d. | n.d. | n.d. | n.d. | 6/9/2017 | 329 | V/F |
| Mo05 | n.d. | n.d. | n.d. | n.d. | 6/9/2017 | 351 | V/V |
| MoB (pool) | |||||||
| Mo06-i | n.d. | n.d. | n.d. | n.d. | 6/9/2017 | 580 | V/F |
| Mo06-ii | F | 73 | AB | + | 3/16/2017 | 280 | F/F |
| Mo06-iii | F | 35 | AB | + | 4/6/2017 | 139 | V/F |
| Mo02 | F | 17 | B | + | 6/9/2017 | 200 | V/V |
| Mo11 | M | 61 | A | + | 6/8/2018 | 406 | V/F |
| Mo13 | F | 40 | O | + | 8/28/2018 | 303 | F/F |
| Mo15 | M | 27 | O | + | 8/28/2018 | 588 | V/F |
n.d. = not determined.
Protein Expression
Anti-CD16 (humanized Fc) 3g8, Anti-CD16 mouse 3g8, or Anti-CD16 mouse 3g8 N297Q (heavy chain), which caused removal of N-glycosylation and was used for most of the immunoprecipitations in this study. Recombinant CD16a in HEK293F and HEK293S were transfected and purified as previously described (33). CD16a-Man9 and CD16a-Hybrid recombinant proteins were made by adding kifunensine and swainsonine to the culture, respectively (5 μm).
Surface Plasmon Resonance
Experiments were performed and IgG1 Fc was glycoengineered as described previously (33).
Cell Isolation
Leukocyte reduction system (LRS) filters were obtained from either LifeServe (Ames, Iowa) or the DeGowin Blood Center (University of Iowa, Iowa City) and processed within three hours of plasma or platelet donation. Cell isolations for NK cells were performed as described (27) and then frozen at −80 °C except the RosetteSep Human Monocyte Enrichment Mixture (Stem Cell Technologies, Vancouver, British Columbia, Canada) was used to isolate purified monocytes through negative selection using Lymphoprep (Stem Cell Technologies).
Flow Cytometry
Nonspecific antibody binding sites on 5 × 105 monocytes in 0.2 ml were blocked with 1 μl human serum. Then, anti-human CD14 (a mouse IgG2a, clone M5E2, BioLegend, San Diego, California) and/or anti-human CD16 (3G8, mouse IgG1) was added to a final concentration of 2 μg/ml and cells incubated on ice for 30 min. Cells were then washed 2 × 1 ml of phosphate-buffered saline plus 0.05% sodium azide; cells were pelleted during each wash by centrifugation at 600 × g for 8 min. The secondary antibodies anti-mIgG1 (RMG1–1 conjugated to allophycocyanin, BioLegend) and anti-mIgG2a (RMG2a-62 conjugated to phycoerythin, BioLegend) were then added to a final concentration of 2 μg/ml, mixed and cells incubated on ice for 30 min. Cells were then washed again as described above and fixed in 1% paraformaldehyde before flow cytometry analysis on the BD FACSanto (BD Biosciences, San Jose, California) instrument. Gating of the monocyte population based on forward and side scattering was determined by excluding cell debris and smaller contaminating cells such as platelets and erythrocytes. CD14 and CD16 gating was decided based on negative controls lacking the primary antibody.
Immunoprecipitation
Frozen cell pellets were thawed and immediately lysed using 40 μl of lysis buffer (100 mm tris(hydroxymethyl)aminomethane, 100 mm sodium chloride, 5 mm ethylenediamenetetraacetic acid, 5 mm oxidized glutathione, 10 μm potassium ferricyanide, 1 mm 4-(2-aminoethyl) benzenesulfonyl fluoride, 10 mg/ml dodecylmaltoside, pH 8.0) per 1 × 106 cells by repeated pipetting. Lysed cells were centrifuged for 20 min at 4400 × g in 4 °C to clarify the supernatant, which was saved separately, frozen overnight, thawed on ice and centrifuged again at 4400 × g for 20 min at 4 °C. Supernatant was sonicated for 1 min at full power in a bath sonicator, 100 μl protein G Sepharose (GE healthcare, Chicago, Illinois) was added to the supernatant and incubated for 2 h with end-over-end mixing. Resin was removed by centrifugation at 500 × g for 5 min, supernatant was moved to a new tube. Either 200 μl Anti-CD16 (humanized Fc) 3g8, Anti-CD16 mouse 3g8, or Anti-CD16 mouse 3g8 N317Q covalently coupled to aldehyde-functionalized agarose beads (Thermo Fisher Scientific, Waltham, Massachusetts) was added, and incubated overnight with end-over-end mixing at 4 °C. Resin was pelleted by centrifugation at 500 × g for 5 min and supernatant was removed and stored as the “flow through” fraction. Resin was then washed 2 × 0.5 ml with lysis buffer plus 0.5 m KCl, 2 × 0.5 ml lysis buffer, 2 × 0.5 ml wash buffer (25 mm 3-(N-morpholino)propanesulfonic acid, 100 mm sodium chloride) and 2 × 0.5 ml 50 mm ammonium bicarbonate, pH 8.0. Resin was then moved to a micro biospin column (BioRad, Philadelphia, Pennsylvania) and eluted with 5 × 0.2 ml of (50:49.9:0.1 acetonitrile/water/trifluoroacetic acid) into 20 μl 1 m ammonium bicarbonate. Samples were then pooled, frozen and lyophilized. Immunoprecipitation of CD16a from NK cells was performed the same as described (27).
Western Blots
The amount of recovered CD16a was estimated by Western blotting using recombinant CD16a as a standard. Western blots were performed as described (27).
CD16a Glycosidase Treatment
CD16a from primary NK cell and monocyte was immunoprecipitated as mentioned above except CD16a was not eluted after the wash steps and digestions were performed on CD16a bound to 3G8 agarose resin. Endo F1 was expressed using a plasmid provided by Dr. Kelley Moremen (University of Georgia) and isolated as mentioned previously (34). CD16a-loaded 3G8 agarose resin was diluted using 250 mm sodium phosphate buffer, pH 5.5. Endo F1 was added and incubated at 37 °C for 1.5 h. The reaction was stopped by boiling for 5 min in the presence of SDS-PAGE loading buffer (BioRad). PNGase F digestion was performed by following the manufacturer's protocol (New England Biolab, Ipswich, Massachusetts). Briefly, resin was boiled in presence of denaturing buffer before adding NP-40, reaction buffer and PNGaseF. The reaction was incubated at 37 °C for 4 h and was stopped by boiling the samples in presence of SDS-PAGE gel loading buffer. Soluble CD16a (sCD16a) from NK cell and monocytes was obtained by incubating isolated NK cell and monocytes at 37 °C for 1 h at a cell density of 10 × 106 cells/ml in RPMI 1640 (Thermo Scientific) medium supplemented with 1% fetal bovine serum (VWR, Suwanee, Georgia), 1 μg/ml phorbol 12-myristate 13-acetate and 1 mm 4-(2-aminoethyl) benzenesulfonyl fluoride (Millipore Sigma). The cells were removed by centrifugation and the supernatant frozen until sCD16a was immunoprecipitated using the method described above. Negative controls for each reaction were treated as digested samples except enzymes were not added. Recombinant CD16a (250 ng) expressed in HEK293F or HEK293S served as a positive control. Boiled samples were directly loaded on 12% SDS-PAGE gels and analyzed using antiCD16 Western blotting.
CD16a Proteolysis
CD16a was resuspended in 100 μl of 50 mm ammonium bicarbonate, pH 8.0 and heat denatured at 90 °C for 5 min and cooled on ice. A 1:40 (w:w) ratio of chymotrypsin and 1:30 (w:w) of Glu-C (Promega, Madison, Wisconsin) was added and the solution incubated at 25 °C overnight. Fresh chymotrypsin and Glu-C was added and the solutions incubated for 3 h. Chymotrypsin is specific for FYW with lower specificity for ML whereas Glu-C is specific for E. Both enzymes cut at the C-terminal of the mentioned amino acid residue. Following proteolysis, dithiothreitol was added to a final concentration of 5 mm and the solution incubated for 1 h at 37 °C. Iodacetamide was then added to a final concentration of 15 mm and the solution incubated in the dark at RT for 1 h. The sample was then centrifuged for 2 min at 9600 × g, the supernatant transferred to a new tube, frozen at −80 °C and lyophilized. Lyophilized samples were resuspended in 10 μl of water and then glycopeptides were enriched using the ProteoExtract Glycopeptide Enrichment Kit following the manufacturer's guidelines to separate peptides from glycopeptides. Isolated glycopeptides were then frozen, lyophilized, and resuspended in a solution containing 5% acetonitrile and 0.1% formic acid before MS analysis.
Detergent present in the peptide sample following glycopeptide depletion was removed using ethyl acetate. First, an empty glass tube was washed with 200 μl ethyl acetate. Water saturated ethyl acetate was made by mixing 5 ml ethyl acetate with 625 μl water in the washed glass tube. 1 ml of the upper phase was added per 100 μl sample. Samples were vortexed for 1 min, centrifuged for 12,500 × g and the upper layer was removed and discarded. This washing procedure was repeated 3 times. Residual ethyl acetate was removed from the top under a nitrogen stream. Isolated peptides were then frozen, lyophilized, resuspended in 20 μl of a solution containing 5% acetonitrile and 0.1% formic acid in water prior to MS analysis (35).
Mass Spectrometry
Complete methods were implemented as described (28). Additional to the cited study, AGC values were set to 5 × 105, the resolving power was set to 17500 and NCE was stepped at 17, 27 and 37 eV for glycopeptides and 37 eV for peptides. The instrument automatically determined a mass rage for the MS2 spectra based on the parent ion mass. The lower mass cutoff was often 200–250 m/z, excluding important fragments corresponding to the most abundant carbohydrate decomposition product ions.
Data Analysis
Raw data files were initially analyzed with Byonic (Protein Metrics Version 2.12.0, Cupertino, California) against FASTA files including CD32a, CD16a, IgG1, FcRγ and CD3ζ. The search parameters included Cleavage site (FYWEL), Cleavage side (C-terminal), Digestion specificity (Semi specific (slow)), Missed cleavages (2), Precursor mass tolerance (5 ppm), Fragment mass tolerance (0.05 Dalton), Recalibration (lock mass) (none), maximum precursor mass (5000), Precursor and mass charge assignments (Compute from MS1), Maximum # of precursors per scan (10), smoothing width (0.01 m/z) and the following modifications included (in addition to N- and O-glycan databases): Carbamidomethyl/+57.021464 @ C | fixed; Deamidated/+0.984016 @ N | common1; Acetyl/+42.010565 @ Protein N-term | rare1; Phospho/+79.966331 @ C, D, H, K, R, S, T,Y | common2; HexNAc/+203.079373 @ N,S,T | common1. The combyne cutoff threshold scores was set to “auto” in Byonic. Hits were then manually validated and additional peaks not identified by Byonic were found. Glycopeptide annotations were made in Glycoworkbench Version 2.1 (36). Annotated MS2s for each species are shown in a Supplemental PDF (glycopeptide_annotations.pdf) and a recent characterization of this purification and analysis strategy using NK cells (28). The 600 m/z peak in MS2 spectra is because of carryover from a calibration standard and is present in spectra taken following water injection that were collected between each sample analysis. MS2 fragmentation was used to confirm oxonium ions expected from the glycan composition and the glycan fragmentation attached to the intact peptide is annotated with the peptide + GlcNAc peak being the most intense, peptide diagnostic ion. Species without clear MS2 spectra were assigned if they showed retention times and predictable differences from known species (for example those with a mass difference of 162.05 Da or 365.15 Da). The supplemental MCP_glycopeptides.xls sheet scores each glycopeptide based on mass error (ppm), retention time, and whether the MS2 included glycan specific-oxonium ions and/or a peak corresponding to the peptide + GlcNAc (a diagnostic ion). A glycopeptide with mass error <1 ppm, <5 ppm, <10 ppm, <20 ppm, and >20 ppm were given a respective score of 2, 1, 0.5, 0.25, or −5. Those glycopeptides receiving a mass score of −5 differed by 1 Da and likely represent an inability to resolve the monoisotopic peak in MS spectra. Glycopeptides with a retention time (min) of <1, <2 or >2 standard deviations of the mean were given a score of 1, 0.5 or 0. If the MS2 contained oxonium ions or the peptide + GlcNAc ion it would get a score of 1 or 0 if not. If the glycopeptide was clearly identified through MS spectra in another dataset and elutes at a comparable retention time it was given a score of 1 or 0 if not. This system adds individual scores for a range between −5 to 6 with a 6 indicating the highest probability of correct identification. In the glycan column, the fifth number is a phosphate/sulfate modification considered an adduct in this analysis (see (28) for a complete description of these species). The analyses of MS1 and MS2 spectra provided assignments of moderate confidence. A mascot search of ESI-MS/MS data for the peptide samples searched the human database for matches. N-glycan structures indicate composition only without linkage information elucidated. Analyte intensities were measured from MS1 spectra using scripts in R based on retention time windows defined for each species (28). This approach measured the intensity of as many as seven isotopolog peaks for each glycopeptide. Isobaric species were not differentiated.
Donor Genotyping
Blood retained in the tubing of the apheresis filters (∼50 μl) was centrifuged at 10,000 × g for 10 min and the cell pellet and serum fractions were stored separately at −80 °C. The RNA from the cell pellet was isolated using the Trizol Reagent as directed by manufacturer (Thermo Fisher Scientific). cDNA was prepared from the isolated RNA using the High Capacity RNA to cDNA as directed by the manufacturer (Thermo Fisher Scientific). The full FCGR3a ORF including region coding for amino acid 158 in the final protein was then amplified using AccuPrime PFX with forward (5′-CAGTGTGGCATCATGTGGCAG-3′) and reverse (5′-TTTGTCTTGAGGGTCCTTTCT-3′) primers. Resulting DNA was purified with a Gel Extraction Kit (Qiagen, Germantown, Maryland) and submitted for Sanger Sequencing at the Iowa State Sequencing Facility.
RESULTS
Apheresis Filters Entrap Monocytes
We isolated primary human monocytes from apheresis filters following plasma or platelet donation to define specific features of CD16a expressed by monocytes. Apheresis filters provided an average of 4 × 108 monocytes per donor following negative selection (Table I). Sequencing cDNA from each donor's leukocytes revealed that most donors expressed both the V158 and F158 alleles, as expected (Table I and supplemental Fig. S1). Flow cytometry indicated a high percentage of cells (82.3–97.6%) expressed CD14 (supplemental Fig. S2) in the expected proportion of “classical” CD14+ CD16dim to “non-classical” CD14dim CD16+ monocytes (83–85% and 15–17%, respectively; (13, 14, 37)). Individual donors also displayed variation in the level of CD16 staining for the CD16+ monocytes (supplemental Fig. S2).
Striking Similarity of the Monocyte and NK cell CD16a Extracellular Domains
CD16a enrichment is essential to study processing. CD16a purified from monocyte lysate through immunoprecipitation with typical yields of ∼0.5 μg CD16a/400 × 106 monocytes (supplemental Fig. S3). By comparison, immunoprecipitation yields from NK cells that express higher levels of CD16a are more than 10× greater on a per-cell basis (13, 14, 27, 28, 37).
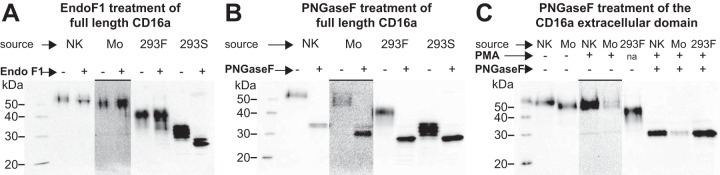
Though CD16a is expressed from the same gene by NK cells and monocytes, mobility differences appeared in SDS-PAGE, a valuable technique to probe CD16 processing using intact protein (23, 27). CD16a from NK cells migrates slower than the same protein from monocytes (Fig. 1A and 1C). The relative mobility difference is maintained following PNGaseF treatment, indicating that N-glycans likely restrict CD16a mobility from NK cells or monocytes to a similar extent (Fig. 1B). Consistent with this conclusion, the mobility of CD16a from NK cells and monocytes, following proteolytic shedding induced with phorbol 12-myristate 13-acetate (PMA), is comparable before and after PNGaseF treatment (Fig. 1C; (38–41)). Furthermore, the extracellular domains migrate at the same position as the extracellular domain of CD16a expressed in HEK293F cells following PNGaseF digestion. These data suggest the polypeptide backbone of the extracellular domains are similarly mobile and that the N-glycans of CD16a from NK cell and monocytes impact mobility of the extracellular domains to a similar extent. However, there is a difference in CD16a processing that causes restricted mobility of the full length CD16a expressed in NK cells, and this modification is most likely present in the portion of the protein retained on the cell following proteolytic shedding. Thus, contrary to previous reports, the mobility differences between CD16a on monocytes and NK cells are not because of PNGaseF-cleavable N-glycans (42, 43).
Fig. 1.
The CD16a extracellular domain analyzed from the NK cells and monocytes of a single donor. A, CD16a immunoprecipitated from the NK cells and monocytes from a single donor were incubated with (+) or without (−) Endo F1, an enzyme that cleaves afucosylated oligomannose and hybrid-type N-glycans. B, CD16a immunoprecipitated from the NK cells and monocytes from a different single donor were incubated with (+) or without (−) PNGase F, an enzyme which cuts complex, hybrid and oligomannose-type N-glycans. C, Treating monocytes and NK cells from two different donors with PMA activates the cells, causing the proteolytic shedding of the CD16a extracellular domain. Areas with an overbar are adjusted to have greater contrast because of low abundance of CD16a from monocytes in these experiments. “293F” and “293S” refer to protein expressed in HEK293F or HEK293S cells with complex type or oligomannose-type N-glycans, respectively.
It is also worthwhile to consider the effect of Endoglycosidase (Endo)F1 treatment on CD16a. Endo F1 treatment cleaves oligomannose and certain hybrid N-glycans and increased the mobility of NK cell CD16a but had little noticeable impact on monocyte CD16a (Fig. 1A). This result is consistent with previous reports that indicated NK cell CD16a contains hybrid and oligomannose-type N-glycans (27, 28). Based on this result, we expect less EndoF1-sensitive hybrid and oligomannose forms in monocyte CD16a. Soluble CD16a expressed in HEK293S cells displays oligomannose forms at each site and shows a substantial mobility increase.
CD16a N-glycan Modifications
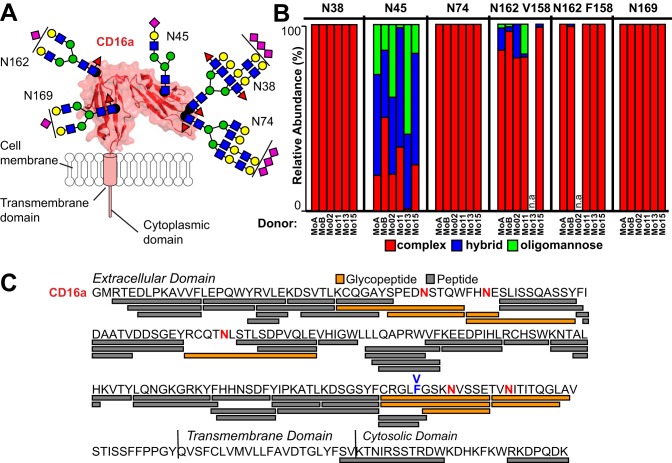
Previous studies indicated CD16a N-glycosylation composition affects binding to IgG1, however it was unknown if forms that bind with high affinity in vitro are present on circulating monocytes (23, 27). We analyzed six monocyte CD16a samples, four isolated from single donors and two as pools of three donors (Table I). Monocyte lysates from six donors were pooled into groups of three donors (MoA and MoB) to increase CD16a yield and improve the depth of coverage at a single site. The most abundant N-glycan for each glycosite varied and is illustrated in Fig. 2A with N-glycan types shown by individual donor or pool in Fig. 2B. Data from these donors revealed between 13 to 39 unique species at each N-glycosylation site and a total of 77 unique N-glycan compositions (Table II; Supplemental glycopeptide_annotations.pdf). Our analysis revealed only N-glycan modifications. We also characterized non-modified peptides for a total of 84% coverage of the peptide backbone (Fig. 2C). An analysis of the peptide spectra indicated that IgG and CD16a were the predominant glycoproteins in each dataset (supplemental MCP_Proteomics.xls). The MoA and MoB datasets also contained CD32a. Byonic identified fibrinogen glycopeptides in only the MoB dataset that compromised 5 out of 183 total spectral counts.
Fig. 2.
CD16a N-glycosylation composition varies by site and donor on monocytes. A, CD16a contains 5 N-glycosylation sites on the extracellular domain. These sites are modeled from the most abundant forms found on monocytes; the ribbon diagram and surface rendering are based on known structures of CD16a determined by X-ray crystallography. Carbohydrate residues are modeled as cartoons using the SNFG convention and roughly scaled to size (46). B, The relative abundance of each N-glycan type for the four individual donors and two donor pools as specified in Table I. n.a. - not applicable. C, ESI-MS/MS identified peptides and glycopeptides covering 84% of the CD16a sequence. Glycosylated Asn residues are shown in red. The V/F158 amino acid residue is labeled in blue.
Table II. Glycopeptides identified in this study.
| site | # | Peptide | Mo02 (V/V) | MoA (V/F) | MoB (V/F) | Mo11 (V/F) | Mo13 (F/F) | Mo15 (V/F) | glycopeptide glycoforms |
|---|---|---|---|---|---|---|---|---|---|
| N38 | 1 | KCQGAYSPEDNSTQW | 6 | 2 | 5 | 7 | n.d. | 2 | 11 |
| 2 | SPEDNSTQW | 3 | 3 | n.d. | 7 | n.d. | n.d. | 9 | |
| N45 | 1 | FHNE | 17 | 16 | 10 | 14 | 11 | 15 | 21 |
| 2 | FHNESLISSQASSY | 5 | 18 | 18 | 8 | 3 | 10 | 28 | |
| N74 | 1 | RCQTNLSTLSDPVQLE | 16 | 7 | 5 | 3 | 4 | 3 | 24 |
| N162 (V158) | 1 | CRGLVGSKNVSSE | 10 | 20 | 8 | 15 | n.a. | 6 | 21 |
| N162 (F158) | 2 | GSKNVSSE | n.a. | 1 | 6 | 2 | 2 | 4 | 6 |
| N169 | 1 | TVNITITQGL | 5 | 7 | 3 | 2 | 2 | 2 | 9 |
| 2 | TVNITITQGLA | n.d. | 1 | 2 | 3 | 2 | n.d. | 4 | |
| # of total glycan compositions: | 77 | ||||||||
n.a. = not applicable; n.d. = not detected.
Subtle Donor-dependent Variation in N162-Glycosylation
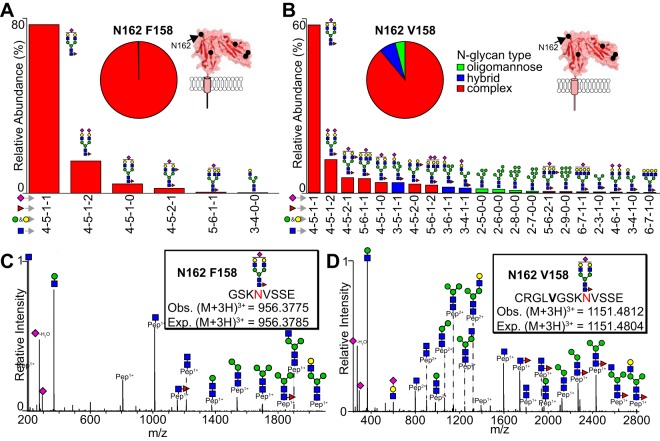
The N162 glycopeptides also contain F158 or V158, linking the CD16a allele with processing at N162. The glycopeptides identified for each donor matched those expected based on cDNA sequencing (Table II and supplemental Fig. S1). F158 introduces a chymotrypsin cut site resulting in a shorter N162 glycopeptide (Peptide 2) (Fig. 3A and 3C). The N162 glycopeptides produced ions with relatively high intensity and high quality MS2s for most species as shown in Fig. 3A–3D. The identification of more peptide 1 (V158) species could result from increased ionization efficiency as compared with peptide 2 (F158), increased proteolysis or differences in V158 and F158 expression cannot be distinguished with these methods. HCD fragmented ions confirm composition of the parent ion, in particular through monosaccharide species specific for sialic acid (292.10 Da or 274.09 Da (-H2O)) and HexNAc (204.09 Da), disaccharide species including HexNAc-Hexose (366.14 Da), and trisaccharide species including HexNAc-Hexose-Sialic acid (657.24 Da). A peak corresponding to the peptide + HexNAc is a common diagnostic glycopeptide and one of the most intense as noted previously (28, 44). Most of these ions contain a deoxyhexose residue and fragments corresponding to the peptide + HexNAc + deoxyhexose and are likely core fucosylated, though we cannot eliminate the possibility of branch fucosylation because isomeric species were likely not separated during chromatography.
Fig. 3.
The CD16a N162-glycan is predominantly a monosialylated biantennary complex-type with some hybrid/oligomannose. The relative abundance represents the averages of each species from datasets originating from individual donors and donor pools and the pie charts represent the average abundance of each N-glycan type. A, Relative abundance of F158 glycopeptides (1 and 7). B, Relative abundance of glycopeptides for a single V158-specific peptide. C, A representative MS2 spectrum for most abundant N162 F158 glycopeptide. D, A representative MS2 spectrum for most abundant N162 V158 glycopeptide. Cartoons indicate one possible glycan configuration consistent with the composition, isobaric ions are not distinguished.
The predominant N162 glycans, averaged across all donors and allotypes, are summarized in Fig. 3A–3B with a core-fucosylated monosialylated complex-type N-glycan representing the most abundant species, though other forms also appear including complex-type biantennary, triantennary and hybrid-types. Other low abundant species include tetrantennery complex-type as well as oligomannose-type N-glycans. Interestingly, four datasets revealed small amounts of oligomannose-type N162-glycans (<10%). Overall, we determined that the N162-(F158)-glycan from these six data sets was 99.7% complex-type and 0.3% hybrid-type whereas the N162-(V158)-glycan was 88.9% complex-type, 7.1% hybrid-type and 3.9% oligomannose-type N-glycans (Fig. 3A–3B).
CD16a V158 with Hybrid or Oligomannose-type N162-glycans Exhibits Greater Affinity for IgG1 Fc
The appearance of under-processed N162 glycoforms from some donors is noteworthy because CD16a V158 with oligomannose-type N162 glycans binds IgG1 Fc with greater affinity in vitro, suggesting the monocytes from some donors express CD16a with enhanced affinity (20, 21, 23, 27). However, it is unclear how hybrid-type N-glycans impact affinity or if the CD16a F158 allotype displays a similar enhancement. SPR binding studies between IgG1 Fc and CD16a expressing complex-type (CT), Man9 or hybrid-type N-glycans indicate an affinity enhancement when CD16a has Man9 or hybrid-type as compared with complex-type (Table III). These results are comparable to previous measurements from our lab, though the absolute difference in enhancement because of the presence of a oligomannose-type N-glycans is reduced (20, 21, 23, 27). Likewise, these are consistent with previous data showing the negative impact of Fc fucosylation on affinity for all three V158 glycoforms. In this experiment, the hybrid-type N-glycans increased CD16a V158 binding to IgG1 Fc by 1.4–2.3-fold, whereas oligomannose N-glycans increased affinity by 2.5–2.9-fold as compared with CD16a V158-CT. As expected, CD16a F158 bound IgG1 Fc with lower affinity than the V158 allotype (17). Surprisingly, receptor glycan composition did not appear to impact binding affinities of the CD16a F158 allotype.
Table III. Equilibrium dissociation constants (KD) from SPR analysis of the CD16a and IgG1 Fc interaction (Errors represent errors of fit to the Langmuir binding isotherm; this experiment is representative of three independent experiments).
| IgG1 Fc G0F | IgG1 Fc G0 | |||
|---|---|---|---|---|
| Receptor | KD (nM) | error | KD (nM) | error |
| CD16a V158-CT | 500 | 30 | 100 | 10 |
| CD16a V158-Man9 | 170 | 20 | 40 | 10 |
| CD16a V158-Hybrid | 220 | 20 | 70 | 10 |
| CD16a F158-CT | 1500 | 100 | 240 | 40 |
| CD16a F158-Man9 | 1300 | 200 | 200 | 70 |
| CD16a F158-Hybrid | 1000 | 100 | 200 | 10 |
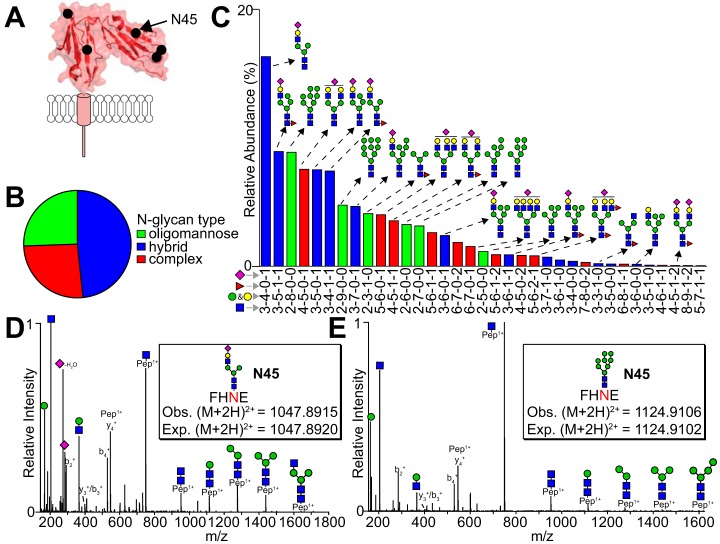
Restricted Processing and Variability at N45
N45 glycosylation shows the most restricted processing and greatest variability compared with the other four CD16a N-glycosylation sites (Fig. 4A–4C). Two N45 peptides contained the N45-glycan (Table II), N45 peptide 1 is the expected degradation product and peptide 2 is the product of a missed Glu-C cleavage. A hybrid-type N-glycan with a sialic acid residue, three mannose residues that lacks fucosylation is the most predominant species found in all six datasets (Fig. 4C and 4D). The N45 glycan exhibited substantial variability between donors with a range of 1.0–50.2% complex types, 36.0–64.0% hybrid types and 1.7–58.8% for oligomannose types. MS2 fragmentation of hybrid-type N-glycan with one fucose and one sialic acid as well as an oligomannose-type glycan are illustrated in Fig. 4D and 4E. Diagnostic fragments in the MS2 spectra include hexose (163.06 Da), HexNAc (204.09 Da) and HexNAc + hexose (366.14 Da) appeared in many MS2 spectra along with peptide fragmentation including y- and b-series ions.
Fig. 4.
N-glycosylation at N45 shows restricted processing with mostly hybrid and oligomannose N-glycans. A, Schematic showing the location of N45. B, Charts showing the average abundance of each N-glycan type. C, The most abundant N45-glycan species identified, ranked according to average abundance. The intensities of N45 glycopeptides 1 and 2 were combined for this analysis. MS2 fragmentation for the most abundant hybrid (D) and oligomannose-type N-glycans (E).
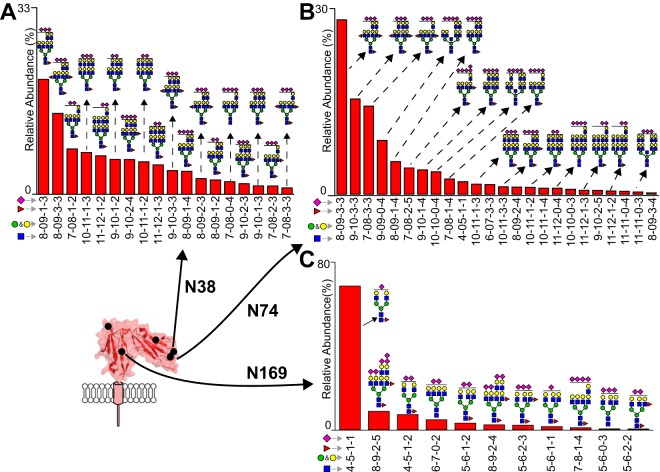
Extensive Modification with LacNAc Units at N38 and N74
N-glycans at the N38 and N74 sites displayed exclusively highly processed complex-type N-glycans (Fig. 5A and 5B). Repeats of a hexose-HexNAc moiety define the predominant features of these species, though sialic acids and fucosylation are also present. The most intense ions differed by the number of hexose-HexNAc units and contained 2–5 sialic acids. Analysis of the monoisotopic peaks from the parent ions in the MS1 dimension provided differentiation of species to determine the presence of two fucose residues versus one sialic. Because of the large size of these ions and overall lower intensity compared with the N45 and N162 glycopeptides, MS2 fragmentation proved much weaker for these species and only showed the diagnostic peptide + GlcNAc ion for N38 and N74 (supplemental glycopeptide_annotations.pdf).
Fig. 5.
Processing of the N38, N74 and N169 N-glycans. N-glycans at N38 (A) and N74 (B) are exclusively highly processed forms with LacNAc repeats. C, N-glycosylation at N169 is predominantly a complex-type biantennary form with some tetra-antennary complex-types as determined by combining intensities of peptide 1 and 2.
N169-glycans are Predominantly Complex-type
Complex-type N-glycans exclusively occupied N169 in all donors (Fig. 5C). A complex-type biantennary with one fucose and one sialic acid was the most predominant N169-glycan, the same predominant form found at N162 (Fig. 3). The ADAM17 cleaved peptide (N169 peptide 2) was identified but at a much lower intensity when compared with the expected Glu-C and chymotrypsin peptide (N169 peptide 1) (Table II and supplemental MCP_glycopeptides.xls), though this may result from nonspecific proteolysis during peptide purification.
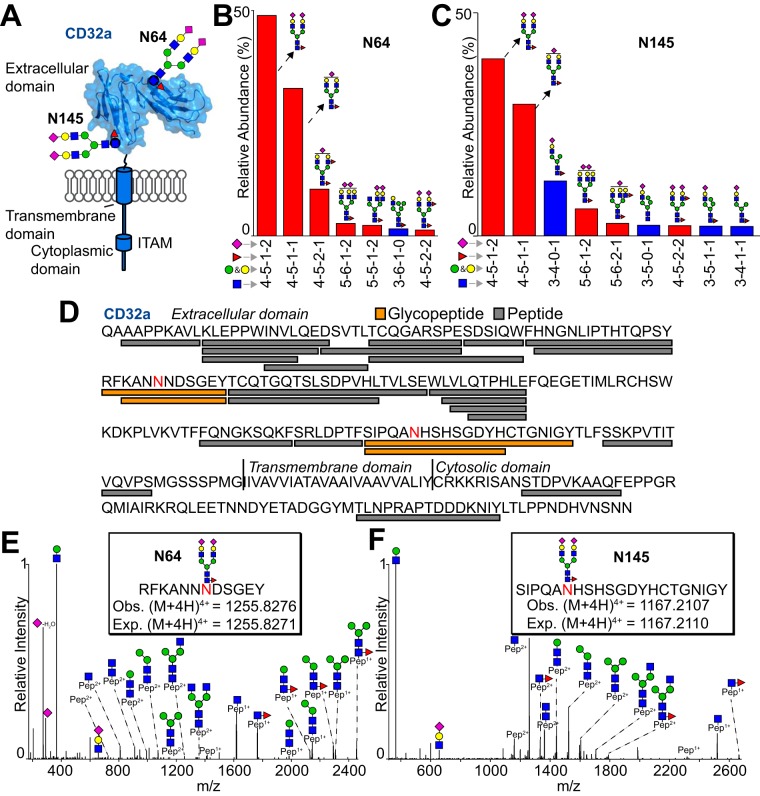
Monocytes Process CD32a Differently than CD16a
Monocytes also express CD32a that contains two N-glycosylation sites and glycan profiles that are unlike CD16a (Fig. 6A). Two pooled datasets (MoA and MoB) included CD32a peptides because we purified CD16a from these samples using 3G8 (a mouse IgG1) that contained an Fc N297-glycan that captured CD32a, unlike the single-donor data sets purified over immobilized 3G8 without Fc N-glycosylation that didn't bind CD32a (45). Analysis of the peptides from these datasets provided 61% sequence coverage of CD32a (Fig. 6C and supplemental MCP_glycopeptides.xls). Of these two datasets, MoB produced N64 and N145 N-glycopeptide ions with high intensity in the MS1 dimension and high quality MS2 spectra (Fig. 6B–6F). N-glycans at these sites were predominantly complex-type biantennary species with a core fucose residue and two sialic acids. MS2 fragmentation confirmed these assignments (Fig. 6E and 6F). MoA provided only a single N64 glycopeptide that matched the predominant N64 species from MoB. Similarly, the most abundant N145 species from both datasets proved identical. The MoA dataset included 7% oligomannose forms and 2% hybrid forms unlike the MoB dataset that revealed no oligomannose forms and 17% hybrid forms (supplemental MCP_glycopeptides.xls). These data indicate the possibility for a surprising degree of variability at the N145 site and stark differences in processing compared with CD16a from the same donor pools.
Fig. 6.
Purified CD32a contains predominantly complex-type biantennary N-glycans. A, CD32a contains 2 N-glycosylation sites on the extracellular domain. These sites are modeled from the most abundant forms found on monocytes; the ribbon diagram and surface rendering are based on known structures of CD32a determined by X-ray crystallography. The most abundant N-glycans at N64 (B) and N145 (C). D, ESI-MS/MS identified peptides and glycopeptides covering 61% of the CD32a sequence. Glycosylated Asn residues are shown in red. Representative MS2 spectra for the most abundant N-glycoforms at N64 (D) and N145 (E). Data in B, C, E and F are from the MoB pool.
DISCUSSION
We defined the composition of CD16a N-glycans isolated using primary human monocytes recovered from apheresis filters. These data indicated that the predominant N-glycoforms at a single location shared greater similarity to N-glycoforms at that site from a different donor than they did to glycoforms at a different site from the same donor. This is evident in many ways, but is most clearly shown with glycan type in Fig. 2B. In general, the CD16a N38 and N74-glycans are exclusively of a highly-processed complex-type with N-acetyllactosamine repeats, N169 are complex-type, N162 are predominantly complex-type with some less-processed forms present in some donors, and N45 contains a mixture of complex, hybrid and oligomannose types. This analysis reports similar glycopeptide coverage with less purified CD16a compared with the previous analysis of CD16a from detergent-lysed NK cells of single donors reported by our group (28). Though monocytes are more abundant in the peripheral blood of most donors, there is less monocyte CD16a than NK cell CD16a because only a subset of monocytes express CD16a (∼17%) and those that do usually express less CD16a compared with NK cells as judged by flow cytometry (supplemental Fig. S2).
The initial comparison of CD16a from monocytes and NK cells revealed differences that are attributable to N-glycan composition (Fig. 1A–1C). Surprisingly, EndoF1 digested NK cell CD16a but not monocyte CD16a. This is likely because of differences in N45-glycan composition; monocyte N45 contains a much higher proportion of smaller hybrid types that lack a fourth mannose residue (likely the α1–3Man on the α1–6Man branch) that is required for EndoF1 activity (46) and is found on the NK cell CD16a N45 N-glycans (28).
Restricted processing at N45 is not unique to monocyte CD16a (Fig. 4B and 4C). NK cell CD16a N-glycosylation displays more restricted processing than recombinant CD16a expressed in HEK293F cells (27). A recent study found CD16a N45 on NK cells displays predominantly hybrid-type (28) and hardly any oligomannose forms unlike CD16a from monocytes reported here which has mostly hybrid and oligomannose-type. Furthermore, CD16b, the highly related FcγR expressed by neutrophils contains predominantly oligomannose and minimal hybrid-type N45-glycans unlike CD16a from both NK cells and monocytes (44, 47). CD16 N45 N-glycosylation processing can be summarized as NK cell > monocyte > neutrophil. NMR and molecular dynamics simulations indicate the CD16 N45 N-glycan makes intramolecular contacts with the polypeptide leading to decreased mobility of the N-glycan (21, 22). Thus, restricted processing of N45 likely results from carbohydrate-polypeptide contacts that reduce accessibility to glycosyl transferases and glycosidases.
CD16 N162 processing on NK cells varied by donor though neutrophils or monocytes expressed receptor with much less variability at this site (Fig. 3) (28, 44, 47). Processing at the remaining three sites is more directly comparable between the three cells types. Gene microarray data indicate that monocytes express the biosynthetic machinery for multiantennary branching of N-glycans including N-acetyllactosamine structures (48). MGAT5 catalyzes addition of the fourth branch GlcNAc residue to the 6-OH of the α1–6Man residue of a triantennary complex-type N-glycan, providing the likely site for LacNAc polymerization (49). Interestingly, Galectin 3 binds poly-LacNAc motifs on N-glycans and regulates T cell receptor clustering and cell activation (50, 51). It remains unclear if CD16a LacNAc motifs on NK cells, monocytes or neutrophils adopt similar roles.
This cell isolation and purification strategy proved sufficient to determine the unique distribution of N-glycoforms at each of the five CD16a N-glycan sites and provided a comparable analysis of CD32a from the same cells. Interestingly, the two N-glycosylation sites on CD32a, N64, and N145, were predominantly complex-type biantennary with two sialic acids and one fucose, unlike any of the major forms on the five CD16a sites. Thus, these monocytes processed CD16a and CD32a N-glycosylation differently. Because the purification targeted CD16a, it is possible that stronger binding CD32a glycoforms preferentially bound the mIgG1 and are thus enriched in these datasets, however this appears unlikely because binding differences of CD32a glycoforms for human IgG1 Fc were not identified (23). Saggu and coworkers recently characterized CD32a N-glycans purified from neutrophils and identified the identical predominant N64-glycan but a high percentage of under-processed N145 glycans including oligomannose and hybrid-types (45). These authors found sialylated CD32a interacted with Mac1 which downregulated CD32a activity implicating CD32a N-glycosylation as functionally relevant mediators of neutrophil activation. Interestingly, monocytes reportedly downregulate Mac-1 expression after incubation with IgG; CD32a N-glycans may perform a comparable role on monocytes (52).
This analysis clearly identifies the predominant CD16a glycoforms expressed by primary human monocytes, though because of the relatively low sample size we are unable to define the distribution of modifications at each site in the broader population. We observed variable CD16a surface expression between donors that, when combined with different cell counts from each donor, led to recovery yields that varied by an order of magnitude (100 ng – 1000 ng) (supplemental Fig. S2–S3). This variability in recovery diminished the number of species identified from some donors that is evident from extracted ion currents for characteristic oxonium ions (supplemental Figs. S4–S9). Despite the low yields from certain donors, the predominant species identified in these cases matched the most abundant species from the datasets with deeper coverage, supporting the identification of the predominant N-glycoforms.
In summary, these data provide insight into how an individual donor modifies individual N-glycosylation sites on two different FcγRs recovered from a single cell type. Differential modifications potentially provide an individual with the ability to modify the activation threshold by controlling receptor affinity, and this approach affords the ability to characterize FcγRs from circulating monocytes using cells isolated from a single donor.
DATA AVAILABILITY
Raw data available at: https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp MassIVE accession number: (MSV000084012).
Supplementary Material
Acknowledgments
We thank donors and staff at the DeGowin Blood Center (Iowa City, IA) and LifeServe Blood Center (Ames and Des Moines, IA) for providing materials. We also thank Joel Nott for the MS ananlysis (ISU Protein Facility) and Dr. Sean Rigby (ISU Flow Cytometry Facility) for performing the flow cytometry measurements. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Institutes of Health.
Footnotes
* This material is based upon work supported by the National Institutes of Health under Award No. R01 GM115489 (NIGMS) and R21 AI142122 (NIAID). The authors declare that they have no conflicts of interest with the contents of this article.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- FcγRs
- Fc γ receptors
- ADCC
- antibody-dependent cell mediated cytotoxicity
- ADCP
- antibody-dependent cell mediated phagocytosis
- NK
- natural killer
- ER
- endoplasmic reticulum
- ESI-MS/MS
- electrospray ionization-tandem mass spectrometry
- PTMs
- post-translational modifications
- GlcNAc
- N-acetylglucosamine
- HexNAc
- N-acetylhexosamine
- PMA
- phorbol 12-myristate 13-acetate
- DDM
- dodecyl maltoside
- LacNAc
- N-acetyllactosamine.
REFERENCES
- 1. Mastrangeli R., Palinsky W., and Bierau H. (2019) Glycoengineered antibodies: towards the next-generation of immunotherapeutics. Glycobiology 29, 199–210 [DOI] [PubMed] [Google Scholar]
- 2. Mossner E., Brunker P., Moser S., Puntener U., Schmidt C., Herter S., Grau R., Gerdes C., Nopora A., van Puijenbroek E., Ferrara C., Sondermann P., Jager C., Strein P., Fertig G., Friess T., Schull C., Bauer S., Dal Porto J., Del Nagro C., Dabbagh K., Dyer M. J., Poppema S., Klein C., and Umana P. (2010) Increasing the efficacy of CD20 antibody therapy through the engineering of a new type II anti-CD20 antibody with enhanced direct and immune effector cell-mediated B-cell cytotoxicity. Blood 115, 4393–4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nimmerjahn F., and Ravetch J. V. (2005) Divergent immunoglobulin g subclass activity through selective Fc receptor binding. Science 310, 1510–1512 [DOI] [PubMed] [Google Scholar]
- 4. Gerdes C. A., Nicolini V. G., Herter S., van Puijenbroek E., Lang S., Roemmele M., Moessner E., Freytag O., Friess T., Ries C. H., Bossenmaier B., Mueller H. J., and Umana P. (2013) GA201 (RG7160): a novel, humanized, glycoengineered anti-EGFR antibody with enhanced ADCC and superior in vivo efficacy compared with cetuximab. Clin. Cancer Res. 19, 1126–1138 [DOI] [PubMed] [Google Scholar]
- 5. Sehn L. H., Assouline S. E., Stewart D. A., Mangel J., Gascoyne R. D., Fine G., Frances-Lasserre S., Carlile D. J., and Crump M. (2012) A phase 1 study of obinutuzumab induction followed by 2 years of maintenance in patients with relapsed CD20-positive B-cell malignancies. Blood 119, 5118–5125 [DOI] [PubMed] [Google Scholar]
- 6. Illidge T. M. (2012) Obinutuzumab (GA101)–a different anti-CD20 antibody with great expectations. Expert Opin. Biol. The.r 12, 543–545 [DOI] [PubMed] [Google Scholar]
- 7. Patel K. R., Roberts J. T., and Barb A. W. (2019) Multiple variables at the leukocyte cell surface impact Fc gamma receptor-dependent mechanisms. Front. Immunol. 10, 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gordan S., Biburger M., and Nimmerjahn F. (2015) bIgG time for large eaters: monocytes and macrophages as effector and target cells of antibody-mediated immune activation and repression. Immunol. Rev. 268, 52–65 [DOI] [PubMed] [Google Scholar]
- 9. Wang W., Erbe A. K., Hank J. A., Morris Z. S., and Sondel P. M. (2015) NK Cell-mediated antibody-dependent cellular cytotoxicity in cancer immunotherapy. Front. Immunol. 6, 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Uchida J., Hamaguchi Y., Oliver J. A., Ravetch J. V., Poe J. C., Haas K. M., and Tedder T. F. (2004) The innate mononuclear phagocyte network depletes B lymphocytes through Fc receptor-dependent mechanisms during anti-CD20 antibody immunotherapy. J. Exp. Med. 199, 1659–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Biburger M., Aschermann S., Schwab I., Lux A., Albert H., Danzer H., Woigk M., Dudziak D., and Nimmerjahn F. (2011) Monocyte subsets responsible for immunoglobulin G-dependent effector functions in vivo. Immunity 35, 932–944 [DOI] [PubMed] [Google Scholar]
- 12. Herter S., Birk M. C., Klein C., Gerdes C., Umana P., and Bacac M. (2014) Glycoengineering of therapeutic antibodies enhances monocyte/macrophage-mediated phagocytosis and cytotoxicity. J. Immunol. 192, 2252–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Passlick B., Flieger D., and Ziegler-Heitbrock H. W. (1989) Identification and characterization of a novel monocyte subpopulation in human peripheral blood. Blood 74, 2527–2534 [PubMed] [Google Scholar]
- 14. Wong K. L., Tai J. J., Wong W. C., Han H., Sem X., Yeap W. H., Kourilsky P., and Wong S. C. (2011) Gene expression profiling reveals the defining features of the classical, intermediate, and nonclassical human monocyte subsets. Blood 118, e16–e31 [DOI] [PubMed] [Google Scholar]
- 15. Crawford K., Gabuzda D., Pantazopoulos V., Xu J., Clement C., Reinherz E., and Alper C. A. (1999) Circulating CD2+ monocytes are dendritic cells. J. Immunol. 163, 5920–5928 [PubMed] [Google Scholar]
- 16. Yeap W. H., Wong K. L., Shimasaki N., Teo E. C., Quek J. K., Yong H. X., Diong C. P., Bertoletti A., Linn Y. C., and Wong S. C. (2016) CD16 is indispensable for antibody-dependent cellular cytotoxicity by human monocytes. Sci. Rep. 6, 34310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bruhns P., Iannascoli B., England P., Mancardi D. A., Fernandez N., Jorieux S., and Daeron M. (2009) Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood 113, 3716–3725 [DOI] [PubMed] [Google Scholar]
- 18. Musolino A., Naldi N., Bortesi B., Pezzuolo D., Capelletti M., Missale G., Laccabue D., Zerbini A., Camisa R., Bisagni G., Neri T. M., and Ardizzoni A. (2008) Immunoglobulin G fragment C receptor polymorphisms and clinical efficacy of trastuzumab-based therapy in patients with HER-2/neu-positive metastatic breast cancer. J. Clin. Oncol. 26, 1789–1796 [DOI] [PubMed] [Google Scholar]
- 19. Weng W. K., and Levy R. (2003) Two immunoglobulin G fragment C receptor polymorphisms independently predict response to rituximab in patients with follicular lymphoma. J. Clin. Oncol. 21, 3940–3947 [DOI] [PubMed] [Google Scholar]
- 20. Roberts J. T., and Barb A. W. (2018) A single amino acid distorts the Fc gamma receptor IIIb/CD16b structure upon binding immunoglobulin G1 and reduces affinity relative to CD16a. J. Biol. Chem. 293, 19899–19908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Falconer D. J., Subedi G. P., Marcella A. M., and Barb A. W. (2018) Antibody fucosylation lowers the FcgammaRIIIa/CD16a affinity by limiting the conformations sampled by the N162-Glycan. ACS Chem. Biol. 13, 2179–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Subedi G. P., Falconer D. J., and Barb A. W. (2017) Carbohydrate-polypeptide contacts in the antibody receptor CD16A identified through solution NMR spectroscopy. Biochemistry 56, 3174–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Subedi G. P., and Barb A. W. (2018) CD16a with oligomannose-type N-glycans is the only “low-affinity” Fc gamma receptor that binds the IgG crystallizable fragment with high affinity in vitro. J. Biol. Chem. 293, 16842–16850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hayes J. M., Frostell A., Cosgrave E. F., Struwe W. B., Potter O., Davey G. P., Karlsson R., Anneren C., and Rudd P. M. (2014) Fc gamma receptor glycosylation modulates the binding of IgG glycoforms: a requirement for stable antibody interactions. J. Proteome Res. 13, 5471–5485 [DOI] [PubMed] [Google Scholar]
- 25. Hayes J. M., Frostell A., Karlsson R., Muller S., Martin S. M., Pauers M., Reuss F., Cosgrave E. F., Anneren C., Davey G. P., and Rudd P. M. (2017) Identification of Fc Gamma Receptor Glycoforms That Produce Differential Binding Kinetics for Rituximab. Mol. Cell. Proteomics 16, 1770–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zeck A., Pohlentz G., Schlothauer T., Peter-Katalinic J., and Regula J. T. (2011) Cell type-specific and site directed N-glycosylation pattern of FcgammaRIIIa. J. Proteome Res. 10, 3031–3039 [DOI] [PubMed] [Google Scholar]
- 27. Patel K. R., Roberts J. T., Subedi G. P., and Barb A. W. (2018) Restricted processing of CD16a/Fc gamma receptor IIIa N-glycans from primary human NK cells impacts structure and function. J. Biol. Chem. 293, 3477–3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Patel K. R., Nott J. D., and Barb A. W. (2019) Primary human natural killer cells retain proinflammatory IgG1 at the cell surface and express CD16a glycoforms with donor-dependent variability. Mol. Cell. Proteomics 18, 2178–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moremen K. W., Tiemeyer M., and Nairn A. V. (2012) Vertebrate protein glycosylation: diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 13, 448–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nairn A. V., Aoki K., dela Rosa M., Porterfield M., Lim J. M., Kulik M., Pierce J. M., Wells L., Dalton S., Tiemeyer M., and Moremen K. W. (2012) Regulation of glycan structures in murine embryonic stem cells: combined transcript profiling of glycan-related genes and glycan structural analysis. J. Biol. Chem. 287, 37835–37856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fisher P., Spencer H., Thomas-Oates J., Wood A. J., and Ungar D. (2019) Modeling glycan processing reveals golgi-enzyme homeostasis upon trafficking defects and cellular differentiation. Cell Rep. 27, 1231–1243.e1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lyons J. J., Milner J. D., and Rosenzweig S. D. (2015) Glycans instructing immunity: the emerging role of altered glycosylation in clinical immunology. Front. Pediatr. 3, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Subedi G. P., and Barb A. W. (2016) The immunoglobulin G1 N-glycan composition affects binding to each low affinity Fc gamma receptor. MAbs 8, 1512–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barb A. W. (2015) Intramolecular N-glycan/polypeptide interactions observed at multiple N-glycan remodeling steps through [(13)C,(15)N]-N-acetylglucosamine labeling of immunoglobulin G1. Biochemistry 54, 313–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yeung Y. G., and Stanley E. R. (2010) Rapid detergent removal from peptide samples with ethyl acetate for mass spectrometry analysis. Curr. Protoc. Protein Sci. Chapter 16, Unit 16 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Damerell D., Ceroni A., Maass K., Ranzinger R., Dell A., and Haslam S. M. (2012) The GlycanBuilder and GlycoWorkbench glycoinformatics tools: updates and new developments. Biol. Chem. 393, 1357–1362 [DOI] [PubMed] [Google Scholar]
- 37. Andersen M. N., Al-Karradi S. N., Kragstrup T. W., and Hokland M. (2016) Elimination of erroneous results in flow cytometry caused by antibody binding to Fc receptors on human monocytes and macrophages. Cytometry A 89, 1001–1009 [DOI] [PubMed] [Google Scholar]
- 38. Wang Y., Wu J., Newton R., Bahaie N. S., Long C., and Walcheck B. (2013) ADAM17 cleaves CD16b (FcgammaRIIIb) in human neutrophils. Biochim. Biophys. Acta 1833, 680–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lajoie L., Congy-Jolivet N., Bolzec A., Gouilleux-Gruart V., Sicard E., Sung H. C., Peiretti F., Moreau T., Vie H., Clemenceau B., and Thibault G. (2014) ADAM17-mediated shedding of FcgammaRIIIA on human NK cells: identification of the cleavage site and relationship with activation. J. Immunol. 192, 741–751 [DOI] [PubMed] [Google Scholar]
- 40. Romee R., Foley B., Lenvik T., Wang Y., Zhang B., Ankarlo D., Luo X., Cooley S., Verneris M., Walcheck B., and Miller J. (2013) NK cell CD16 surface expression and function is regulated by a disintegrin and metalloprotease-17 (ADAM17). Blood 121, 3599–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jing Y., Ni Z., Wu J., Higgins L., Markowski T. W., Kaufman D. S., and Walcheck B. (2015) Identification of an ADAM17 cleavage region in human CD16 (FcgammaRIII) and the engineering of a non-cleavable version of the receptor in NK cells. PLoS ONE 10, e0121788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Edberg J. C., Barinsky M., Redecha P. B., Salmon J. E., and Kimberly R. P. (1990) Fc gamma RIII expressed on cultured monocytes is a N-glycosylated transmembrane protein distinct from Fc gamma RIII expressed on natural killer cells. J. Immunol. 144, 4729–4734 [PubMed] [Google Scholar]
- 43. Edberg J. C., and Kimberly R. P. (1997) Cell type-specific glycoforms of Fc gamma RIIIa (CD16): differential ligand binding. J. Immunol. 159, 3849–3857 [PubMed] [Google Scholar]
- 44. Washburn N., Meccariello R., Duffner J., Getchell K., Holte K., Prod'homme T., Srinivasan K., Prenovitz R., Lansing J., Capila I., Kaundinya G., Manning A. M., and Bosques C. J. (2019) Characterization of endogenous human FcgammaRIII by mass spectrometry reveals site, allele and sequence specific glycosylation. Mol. Cell. Proteomics 18, 534–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Saggu G., Okubo K., Chen Y., Vattepu R., Tsuboi N., Rosetti F., Cullere X., Washburn N., Tahir S., Rosado A. M., Holland S. M., Anthony R. M., Sen M., Zhu C., and Mayadas T. N. (2018) Cis interaction between sialylated FcgammaRIIA and the alphaI-domain of Mac-1 limits antibody-mediated neutrophil recruitment. Nat. Commun. 9, 5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Varki A. (2017) Biological roles of glycans. Glycobiology 27, 3–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yagi H., Takakura D., Roumenina L. T., Fridman W. H., Sautes-Fridman C., Kawasaki N., and Kato K. (2018) Site-specific N-glycosylation analysis of soluble Fcgamma receptor IIIb in human serum. Sci. Rep. 8, 2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Trottein F., Schaffer L., Ivanov S., Paget C., Vendeville C., Cazet A., Groux-Degroote S., Lee S., Krzewinski-Recchi M. A., Faveeuw C., Head S. R., Gosset P., and Delannoy P. (2009) Glycosyltransferase and sulfotransferase gene expression profiles in human monocytes, dendritic cells and macrophages. Glycoconj. J. 26, 1259–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sasai K., Ikeda Y., Fujii T., Tsuda T., and Taniguchi N. (2002) UDP-GlcNAc concentration is an important factor in the biosynthesis of beta1,6-branched oligosaccharides: regulation based on the kinetic properties of N-acetylglucosaminyltransferase V. Glycobiology 12, 119–127 [DOI] [PubMed] [Google Scholar]
- 50. Nabi I. R., Shankar J., and Dennis J. W. (2015) The galectin lattice at a glance. J. Cell Sci. 128, 2213–2219 [DOI] [PubMed] [Google Scholar]
- 51. Demetriou M., Granovsky M., Quaggin S., and Dennis J. W. (2001) Negative regulation of T-cell activation and autoimmunity by Mgat5 N-glycosylation. Nature 409, 733–739 [DOI] [PubMed] [Google Scholar]
- 52. Fernandez-Calotti P. X., Salamone G., Gamberale R., Trevani A., Vermeulen M., Geffner J., and Giordano M. (2003) Downregulation of mac-1 expression in monocytes by surface-bound IgG. Scand J. Immunol. 57, 35–44 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data available at: https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp MassIVE accession number: (MSV000084012).