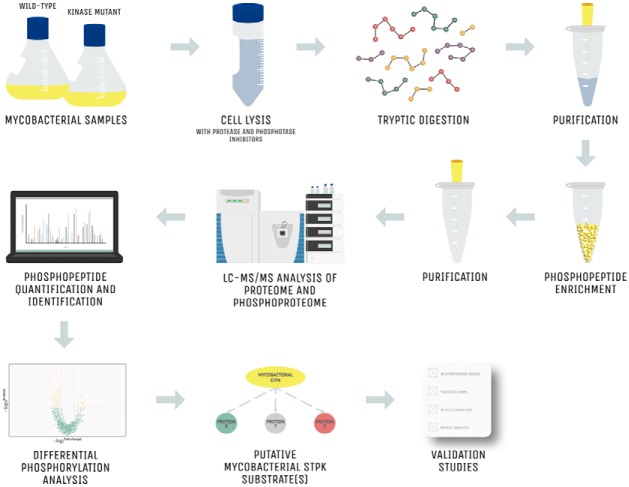
Mycobacterial STPKs are responsible for orchestrating phosphorylation-dependent signaling cascades that mediate bacterial growth and environmental adaptation. Recent advances in MS-based phosphoproteomics have significantly expanded the candidate substrate lists for individual mycobacterial STPKs. Integration of the available phosphoproteomic datasets provide new insights into the functional roles of specific STPKs in cell physiology. Future research should focus on in vivo phosphorylation network reconstruction to expose the fundamental signaling pathways in mycobacteria. Linking STPKs with their physiological substrates may reveal novel antimycobacterial agents.
Keywords: Mass Spectrometry, Serine/Threonine Kinases, Phosphoproteins, Substrate Identification, Phosphorylation, Mycobacterium tuberculosis, Phosphoproteomics
Graphical Abstract
Highlights
Mapping kinase-substrate relationships is vital in discovering new tuberculosis drug targets.
LC-MS/MS-based phosphoproteomics expand mycobacterial STPK substrate catalogues.
We review and integrate MS-generated datasets on novel candidate substrates.
Validation studies are necessary to confirm true physiological substrates of STPKs.
Abstract
Mycobacterial Ser/Thr protein kinases (STPKs) play a critical role in signal transduction pathways that ultimately determine mycobacterial growth and metabolic adaptation. Identification of key physiological substrates of these protein kinases is, therefore, crucial to better understand how Ser/Thr phosphorylation contributes to mycobacterial environmental adaptation, including response to stress, cell division, and host-pathogen interactions. Various substrate detection methods have been employed with limited success, with direct targets of STPKs remaining elusive. Recently developed mass spectrometry (MS)-based phosphoproteomic approaches have expanded the list of potential STPK substrate identifications, yet further investigation is required to define the most functionally significant phosphosites and their physiological importance. Prior to the application of MS workflows, for instance, GarA was the only known and validated physiological substrate for protein kinase G (PknG) from pathogenic mycobacteria. A subsequent list of at least 28 candidate PknG substrates has since been reported with the use of MS-based analyses. Herein, we integrate and critically review MS-generated datasets available on novel STPK substrates and report new functional and subcellular localization enrichment analyses on novel candidate protein kinase A (PknA), protein kinase B (PknB) and PknG substrates to deduce the possible physiological roles of these kinases. In addition, we assess substrate specificity patterns across different mycobacterial STPKs by analyzing reported sets of phosphopeptides, in order to determine whether novel motifs or consensus regions exist for mycobacterial Ser/Thr phosphorylation sites. This review focuses on MS-based techniques employed for STPK substrate identification in mycobacteria, while highlighting the advantages and challenges of the various applications.
PTMs are essential for the proper functioning of certain proteins and create heterogeneity and temporal complexity in mycobacterial proteomes (1–3). Recent developments in MS-based proteomics have highlighted the occurrence of numerous types of PTMs in mycobacteria (2, 3). Phosphorylation is a reversible PTM responsible for diversifying protein functions and orchestrating various signaling networks in mycobacteria, mediated by protein kinases and phosphatases (1, 4–6). Other well-studied PTMs in mycobacteria include glycosylation, lipoylation, acetylation, and pupylation, which have a wide array of physiological consequences in mycobacteria, ranging from the regulation of cell functions to modulation of host-pathogen interactions, as reviewed elsewhere (2, 3).
Being the most dynamic of all common PTMs, Ser/Thr protein phosphorylation is a central control mechanism associated with the growth, virulence, and persistence of mycobacterial species, including Mycobacterium tuberculosis, the etiological agent of tuberculosis (7–11). Through phosphorylation, mycobacterial STPKs and two-component systems coordinate signal transduction cascades that regulate physiological states, environmental adaptation, and pathogenesis (10, 12, 13).
Two-component systems consist of a sensor histidine kinase and a cognate response regulator, which are typically genetically linked and transcriptionally coupled (14, 15). The histidine kinase spans the cytoplasmic membrane and activates the response regulator when prompted by specific external stimuli (14). Communication between the proteins occurs via phosphoryl-group transfer from a conserved histidine residue of the sensor kinase to a conserved aspartate residue of the response regulator (16). The response regulator is usually a transcription factor localized in the cytoplasm that mediates environmental adaptative responses (14).
In contrast to two-component systems, STPKs are pleiotropic signal transducers that can phosphorylate multiple substrates to elicit branched signaling (14). The majority of mycobacterial STPKs are transmembrane proteins with an extracellular sensor domain and an intracellular kinase domain. The extracellular sensor domain transduces extracytoplasmic signals to the intracellular kinase domain, resulting in kinase activation and phosphorylation of Ser/Thr residues on substrate proteins. This phosphorylation may directly affect protein function or influence protein-protein interactions. Notably, Ser/Thr phosphorylation rarely results in direct regulation of transcription, which is often facilitated by two-component signal transduction (10).
Genomic studies have revealed that M. tuberculosis strains are equipped with 11 conserved STPKs (PknA, PknB, PknD, PknE, PknF, PknG, PknH, PknI, PknJ, PknK, and PknL) (17, 18). Furthermore, in mycobacteria with larger genomes, such as M. marinum or M. smegmatis, the number of STPKs outnumber that of two-component system kinases. This suggests that the majority of the signal transduction pathways in these species is via Ser/Thr phosphorylation/dephosphorylation events (19, 20). Hence, mycobacterial STPKs form the focus of this review.
A phylogenetic analysis clustered the 11 mycobacterial STPKs into five groups, Clades I-V (Table I) (20). The group of transmembrane sensor kinases, PknA, PknB, and PknL belong to Clade I. PknD, PknE, and PknH belong to Clade II, representing the group of integral membrane receptor and cytoplasmic kinases. Clade III STPKs comprise PknF, PknI, and PknJ. Finally, Clades IV and V represent the group of soluble kinases that cluster along with PknK and PknG, respectively (20).
Table I. Classification of the 11 mycobacterial STPKs according to their sequence similarity.
| Clade | Mycobacterial STPK |
|---|---|
| I | PknA, PknB, PknL |
| II | PknD, PknE, PknH |
| III | PknF, PknI, PknJ |
| IV | PknK |
| V | PknG |
PknA and PknB are essential for sustaining the growth and survival in vitro and in vivo (11, 19, 21) while modulating cell morphology (22). PknG, a well-studied mycobacterial STPK, is involved in metabolism regulation (7, 23) and promotes the intracellular survival of mycobacteria within macrophages by blocking phagosome-lysosome fusion (24). The remaining kinases have proposed roles in various biological processes, including the regulation of growth and cell division, cell wall synthesis, environmental sensing, and adaptive responses to different abiotic stresses (25–29).
These STPKs, along with their cognate phosphatases, interact functionally to generate a complex signaling architecture (6, 13, 30). Notably, kinases are distinguished from one another based on their substrate repertoires and their modes of regulation (31). Mapping kinase-substrate relationships is, therefore, key to understanding fundamental biochemical pathways and signaling networks in mycobacteria and identifying new, plausible pharmaceutical targets for therapeutic intervention (1).
However, despite the increase in the definition of the functional roles of these individual STPKs, the physiologically relevant substrates and networks responsible for integrating cellular signals remain obscure. Various techniques, including genetic screening, in vitro kinase assays, protein microarrays, two hybrid-based studies and proteomics, have been described for the screening of kinase substrates (32). These approaches often attempt to mimic in vivo contexts to provide useful insights, however, validation is still required. Consequently, improved methods and novel strategies for mycobacterial STPK substrate detection are essential.
The sensitivity and high-throughput capacity of MS have improved significantly in recent years and proved to be of considerable benefit to the detection of phosphorylation events. MS-based methods now surpass classical approaches, especially since MS is able to confidently assign the localization of phosphosites (7–9). Coupled with the use of kinase knock-out mutants or chemical inhibitors, MS also allows for the quantitative analysis of phosphorylation changes in the presence or absence of a specific kinase.
Quantitative phosphoproteomic strategies have been successfully applied to identify novel substrates of protein kinases in bacteria such as Bacillus subtilis (33, 34) and Escherichia coli (35), but also in higher organisms, including yeast (36) and human cells (37, 38). More recently, high-resolution MS has been applied for the identification of STPK physiological substrates and interacting partners in mycobacteria. Accordingly, this review will hereafter focus on quantitative MS-based mycobacterial STPK substrate identification.
MS-Based Ser/Thr Phosphoproteomic Analysis
Classical Methods for STPK Substrate Identification
Technical developments in MS, together with its increasing accessibility, has enabled the refinement of proteomic strategies for the identification of novel physiological substrates of mycobacterial STPKs (4, 7–9, 39). Commonly utilized methods combine in vitro kinase assays with MS to detect kinase substrates. This often entails the use of purified, active protein kinases to phosphorylate proteins or cell lysates in vitro, followed by protein purification and MS analysis for phosphopeptide identification. Applying this strategy, the forkhead-associated (FHA) domain-containing protein, GarA, was identified as a substrate of PknG in vitro (40). Employing a similar proteomic approach, in combination with 2D-PAGE1 and autoradiography, Rv2175c and GarA were found to be substrates of PknL and PknB, respectively (41, 42). This method allowed for the differentiation of several protein species and highlighted the complementarity of 2D-PAGE and MS for increasing proteome coverage.
An alternative approach, termed KESTREL, uses fractions of cell lysates to separate endogenous protein kinases from their various substrates, thereby minimizing background phosphorylation and sample complexity for subsequent MS analysis (43). Mueller and Pieters (23) presented a KESTREL-based approach for mycobacterial STPK substrate identification combining proteome fractionation, in vitro kinase assays and reversed-phase chromatography with 2D-PAGE and tandem MS protein identification (23). Using this method, GarA was, again, identified as a substrate for mycobacterial PknG. However, as demonstrated in previous studies, high enzyme-to-substrate ratios are required to detect phosphorylation using in vitro assays (1, 23, 40). This limitation suggests that these assays may not capture and reflect the true biological selectivity of protein kinases, which are also known to display promiscuity in vitro. The physiologically relevant specificity of these kinases is often lost because of the elevated, nonphysiological kinase and/or substrate concentrations used in kinase assays. Thus, mycobacterial phosphoproteomics research should ideally aim toward in vivo phosphorylation network reconstruction in order to link STPKs with their physiological substrates and binding partners.
Recent Advances in Mycobacterial STPK Substrate Identification
Various quantitative methods for ex vivo LC-MS/MS analysis have been successfully applied to generate phosphoproteomic datasets for mycobacterial species (12, 39, 44–46). For example, Prisic et al. reported global Ser/Thr phosphorylation profiles of the M. tuberculosis laboratory strain H37Rv grown under conditions of hypoxia, nitric oxide stress, oxidative stress, and using glucose or acetate as a carbon source (39). MS analysis of these samples enabled the detection of 506 phosphosites on 301 proteins involved in a vast array of functions. Similar numbers of phosphoproteins were reported in other M. tuberculosis strains (12, 47) and other mycobacterial species (45, 46). However, the high-throughput capability of MS and phosphoproteome profiling is often unable to determine an exact relationship between protein kinases and their substrates due to the presence of multiple different active kinases in a given biological system. Consequently, more laborious methods are required to associate these substrates with their cognate protein kinases (4).
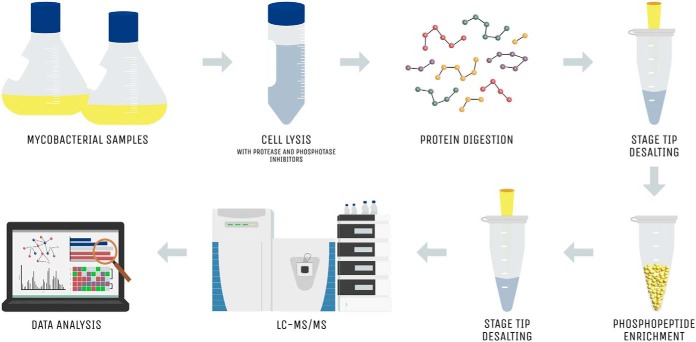
Recently, Nakedi et al. employed label-free quantitative phosphoproteomics to screen for novel physiological substrates of M. bovis BCG PknG in vivo (7). The study compared the phosphoproteomic dynamics of the batch culture growth of M. bovis BCG wild-type against the respective PknG knock-out mutant strain. Titanium dioxide (TiO2) beads were used for phosphopeptide enrichment, followed by LC-MS/MS (Fig. 1). This ex vivo workflow facilitated the identification of 55 differentially phosphorylated phosphopeptides, of which 21 phosphopeptides were phosphorylated only in the M. bovis BCG wild-type and not in the PknG knock-out mutant. Phosphopeptides were identified by mapping acquired peptide MS/MS spectra against the M. bovis BCG Pasteur 1172 reference proteome using the MaxQuant suite, Andromeda (48). Modified peptides underwent score-based filtering, including an Andromeda search score >40, delta score >8, false discovery rate <0.05 and a localization probability ≥0.75 (Fig. 2). A selected number of these novel candidate PknG substrates were further validated through targeted parallel reaction monitoring MS assays. Notably, this study exemplified the utility of parallel reaction monitoring as a new means to validate phosphorylation events in phosphoproteomic experiments (49).
Fig. 1.
General phosphoproteomic workflow. Sample preparation for a standard mycobacterial phosphoproteomic experiment begins with the mycobacterial cells being lysed in the presence of protease and phosphatase inhibitors to preserve all phosphorylation events. Samples then undergo tryptic digestion and subsequent peptide desalting using C18 STAGE (stop-and-go-extraction) tips. Phosphopeptides are then enriched using immobilized metal affinity chromatography, TiO2 or antibody-based immunoaffinity methods, followed by another desalting step. LC-MS/MS data acquisition is then performed, and the generated data are analyzed using various software packages and/or bioinformatic pipelines.
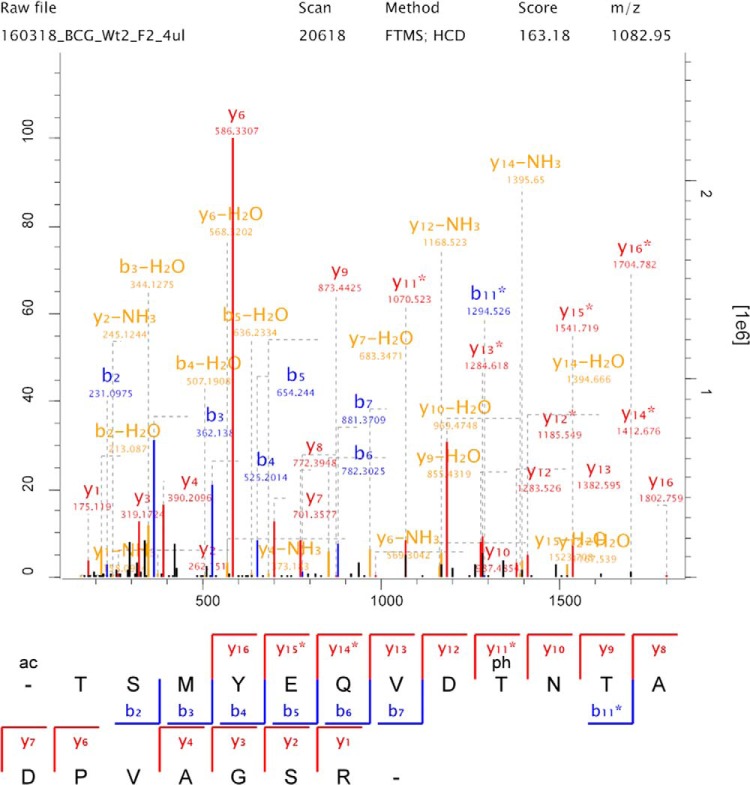
Fig. 2.
Typical phosphopeptide MS/MS fragmentation spectrum. The phosphopeptide MS/MS spectrum shows both b- and y-ions of a peptide (TSMYEQVDTNTADPVAGSR) phosphorylated at a threonine residue derived from the candidate PknG substrate, HpcH_HpaI domain-containing protein.
Following this publication, several other studies employed similar approaches in the search of mycobacterial protein kinase substrates (6, 8, 9, 50, 51). Gil et al. developed an affinity purification-MS strategy to stepwise recover PknG substrates and interactors (8). Using this tailored interactome approach, seven novel candidate PknG substrates were identified, of which six substrates overlapped with those previously reported by Nakedi et al. (7). In addition, this study recovered the only two previously well-characterized PknG substrates, namely GarA and the 50 ribosomal protein L13, thereby substantiating the notion that these proteins represent plausible physiological substrates or interactors of PknG.
In order to identify potential PknA and PknB targets, Carette et al. opted for a phosphoproteomic approach using a small-molecule inhibitor of PknA and PknB (52). They observed differential levels of protein phosphorylation and identified 46 candidate substrates of PknA and PknB. Kaur et al. identified 73 potential targets of PknB using a tandem mass tag-based quantitative phosphoproteomics approach with a PknB tetra mutant, wherein all four putative ligand interacting residues were modified. Simultaneous mutation of these penicillin-binding proteins and Ser/Thr kinase-associated 3–4 linker region residues appeared to abrogate ligand binding (9). Nine of these 73 targets overlapped with those reported previously by Carette et al. (52), providing partial independent verification of this significantly expanded list of candidate PknB substrates. However, the general lack of overlap between the candidate PknB substrates reported by these two studies is notable—even when accepting that the data presented by Carette et al. did not distinguish between PknA and PknB substrates. This suggests that further research is required to verify the full sets of candidate substrates to minimize overreporting and to identify the true physiological PknB substrates.
Notwithstanding caveats concerning the need for robust validation of new candidate substrates for individual kinases, LC-MS/MS phosphoproteomic-based workflows have proven to be the most promising strategy for the ex vivo identification of novel STPK substrates in mycobacteria thus far. Moreover, the cost demand of these workflows can be minimized using label-free quantitation, allowing for the investigation of multiple time points and/or conditions. Above all, these approaches allow for substrate discovery under more biologically relevant conditions. In contrast, in vitro strategies ideally need to factor in subcellular compartmentalization of proteins, as well as physiologically relevant enzyme/substrate concentrations, which ultimately determine whether a protein kinase can interact with, and subsequently phosphorylate, a putative substrate. In vitro kinase assays can, however, serve as a validation technique for novel putative substrates recovered during in vivo phosphoproteomic studies. As part of this review, we have created a manually curated list of all currently reported mycobacterial kinase substrates and their specific phosphosites (Table S1). Table I presents a truncated table of known mycobacterial PknG substrates.
Integration of Novel Substrates Identified by LC-MS/MS: Functional and Localization Enrichment Analyses
The roles of PknA, PknB, and PknG in the cell—Novel candidate PknA, PknB and PknG substrates were analyzed for GO functional and localization enrichment. The putative PknG substrates reported by Nakedi et al. (7) and Gil et al. (8) were converted to M. tuberculosis (strain ATCC 25618/H37Rv) orthologs using Orthologous Matrix Browser (https://omabrowser.org) (53). Potential targets of PknA and PknB identified in Carette et al. (52) and Kaur et al. (9) were classified using Mycobrowser (https://mycobrowser.epfl.ch/) (54). The corresponding protein identifiers (Table S2) were used to perform statistical overrepresentation tests of GO biological processes and GO cellular components using the PANTHER Classification System (http://pantherdb.org) version 14.1 (55) and GO database version 1.2 (released 2019-07–03). The M. tuberculosis (strain ATCC 25618/H37Rv) proteome was used as a reference list.
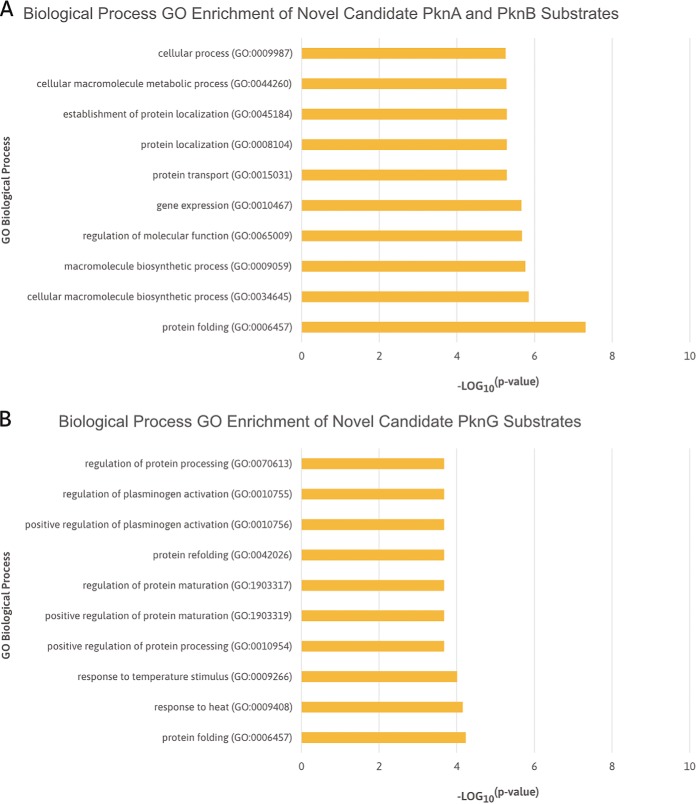
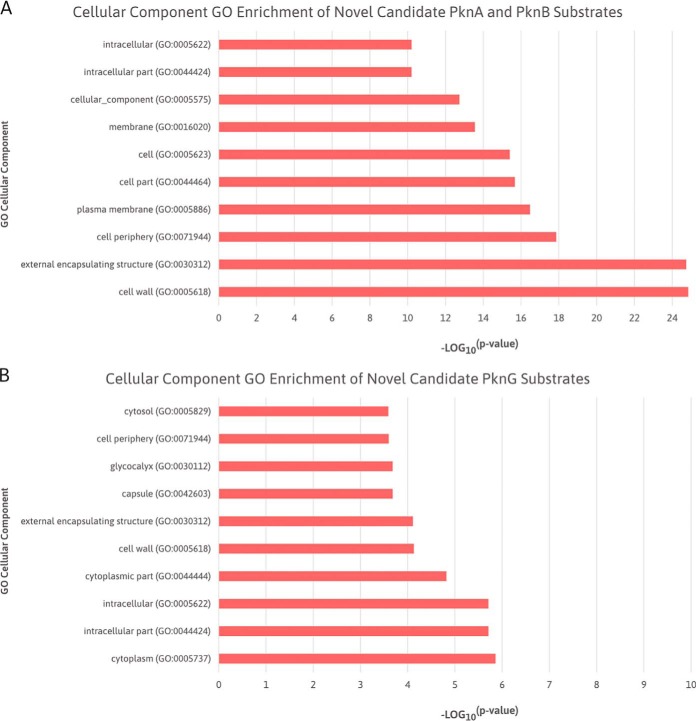
Fig. 3A shows the functions that were enriched in the PknA and PknB candidate substrate list, with a clear enrichment of protein folding, cellular macromolecule biosynthetic process, regulation of molecular function, and gene expression. Moreover, the cellular component enrichment suggests that these substrates are localized in the cell wall, external encapsulation structure, cell periphery, and plasma membrane (Fig. 4A). Both of these results reaffirm the role of PknA and PknB in modulating cell division, cell wall synthesis, cell morphology, and metabolic processes (19, 21, 22, 56–58), while corroborating the localization of these kinases to the cell wall and cell membrane (21, 59).
Fig. 3.
Top 10 significantly enriched biological process GO terms of novel candidate PknA, PknB, and PknG substrates. (A) Bar chart ranking of the significantly enriched biological process GO terms of novel candidate PknA and PknB substrates identified by Carette et al. (52) and Kaur et al. (9). (B) Bar chart ranking of the significantly enriched biological process GO terms of novel candidate PknG substrates identified by Nakedi et al. (7) and Gil et al. (8). Details of the biological process GO enrichments are presented in Table S3.
Fig. 4.
Top 10 significantly enriched cellular component GO terms of novel candidate PknA, PknB, and PknG substrates. (A) Bar chart ranking of the significantly enriched cellular component GO terms of novel candidate PknA and PknB substrates identified by Carette et al. (52) and Kaur et al. (9). (B) Bar chart ranking of the significantly enriched cellular component GO terms of novel candidate PknG substrates identified by Nakedi et al. (7) and Gil et al. (8). Details of the cellular component GO enrichments are presented in Table S4.
The biological processes that were highly enriched in the PknG candidate substrate list were protein folding, response to temperature stimulus, and the regulation of various processes, including protein maturation, plasminogen activation, and protein processing (Fig. 3B). These findings agree with the conclusions drawn in previous studies that suggest PknG to be involved in the regulation of protein folding and translation (7, 8). The cellular component enrichment shows these substrates to be considerably localized in the cytosol, cell wall, and capsule (Fig. 4B). This agrees with PknG being a soluble STPK and cytosolic protein, which is hypothesized to be translocated under specific environmental conditions (60, 61). Furthermore, these results also support the localization of PknG to the cell wall (61–63) and its proposed regulatory role in cell wall biogenesis and integrity (62, 64).
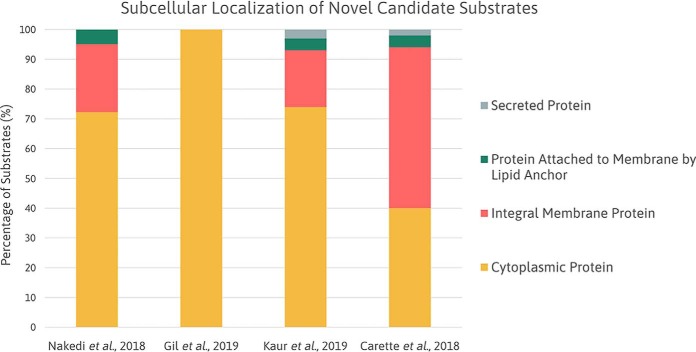
Baer et al. suggested that mycobacterial STPKs should phosphorylate substrates localized in the same subcellular compartment in vivo (30). Therefore, we classified the candidate novel substrates of PknA, PknB (9, 52), and PknG (1, 2) according to UniProt subcellular locations. In cases where no annotation was recorded or ambiguity existed, the tuberculosis-pred online tool (www.tbpred.com) was used. Fig. 5 illustrates the subcellular localization of the MS identified novel candidate PknA, PknB (9, 52), and PknG (7, 8) substrates.
Fig. 5.
Subcellular localization of novel candidate substrates. Column chart illustrating the predicted subcellular localization annotations of novel candidate substrates identified by Nakedi et al. (PknG), Gil et al. (PknG), Kaur et al. (PknB), and Carette et al. (PknA/PknB).
The subcellular localization of a substrate influences its ability to access protein kinases. PknA and PknB are both transmembrane proteins comprising an intracellular kinase domain (18), thereby accounting for the higher percentage of cytosolic protein substrates than membrane protein substrates. Nevertheless, it remains important to determine the circumstances by which a substrate becomes available to interact with a kinase anchored to the cell membrane.
PknG demonstrated preferential phosphorylation of cytoplasmic proteins, as anticipated. However, to understand the recovery of membrane and membrane-associated proteins by Nakedi et al. (7), we analyzed these substrates using the transmembrane topology predicter, Phobius (http://phobius.cgb.ki.se) (65). Intriguingly, none of the detected phosphosites appear to be localized in the cytoplasm, further supporting a possible role of PknG beyond the cytoplasm of the cell. However, further investigation and evidence are required to substantiate the presence and role of PknG in the cell wall.
Characterizing Substrate Specificity Patterns in Mycobacteria
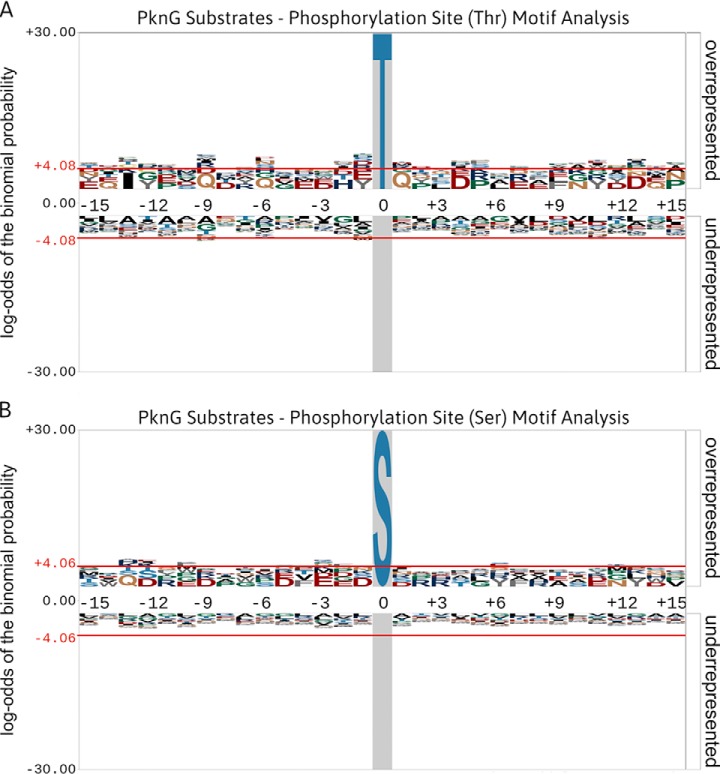
Mycobacterial STPKs have long been suspected of promiscuity toward various substrates, a notion supported in part by the aggregated data on reported mycobacterial substrates (Table S1), which suggests an unexpectedly large number of substrates for each mycobacterial kinase. This contrasts with the tight specificity typically observed for eukaryotic STPKs that underpins the integrity of eukaryotic signaling networks. The apparent promiscuity of mycobacterial STPKs is further supported by the absence of any obvious sequence motifs surrounding preferential Ser/Thr phosphorylation sites for individual STPKs (Fig. 6). However, recent phosphoproteomic-based studies show that a subset of substrates is specifically phosphorylated in vivo only when in the presence of STPK activity (7–9, 52), arguing that these STPKs demonstrate a preference for certain substrates. Furthermore, it is equally clear from the GO enrichment analyses previously presented that the substrates for the individual mycobacterial STPKs are nonrandom at the functional level. Future attempts to determine possible specificity across mycobacterial STPK substrates may, therefore, require the use of more sophisticated Hidden Markov model-type approaches to identify potentially distal structural motifs that can account for the observed substrate selectivity of the mycobacterial STPKs.
Fig. 6.
Phosphorylation site motif analyses of PknG substrates generated using pLogo (68). (A) An in silico motif analysis of Thr-phosphorylated peptides (n = 26) derived from PknG substrates. (B) An in silico motif analysis of Ser-phosphorylated peptides (n = 12) derived from PknG substrates. Phosphorylation site motifs were analyzed using a foreground composed of available sequence windows extracted from the studies cited in Table II. The M. tuberculosis (strain ATCC 25618/H37Rv) proteome was used as the background database. The red horizontal lines (± 4.06) illustrate the relative statistical significance (p value ≤ 0.05, after Bonferroni correction) of residues flanking the central phosphorylation site. Overrepresented residues are above the midline, whereas underrepresented residues are below the midline. No distinct motifs were observed for phosphorylation on either Thr or Ser sites.
Table II. List of known mycobacterial PknG substrates.
| Protein Name | Gene | UniProt ID | P-site | Species | Reference |
|---|---|---|---|---|---|
| 30S ribosomal protein S16 | rpsP | A1KMQ3 | S162 | M. bovis BCG | (7) |
| 4HBT domain-containing protein | BCG_1584c | A0A0H3M6A1 | S2 | M. bovis BCG | (7) |
| 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase (flavodoxin) | ispG | A1KML3 | S387 | M. bovis BCG | (7) |
| 50S ribosomal protein L2 | rplB | A1KGI5 | S32 | M. bovis BCG | (7) |
| 5-methyltetrahydropteroyltriglutamate–homocysteine methyltransferase | metE | A1KHS4 | S713 | M. bovis BCG | (7) |
| 60 kDa chaperonin 1 (GroL1) | groL1/MSMEG_0880 | A0QQU5 | – | M. smegmatis | (8) |
| AAA domain-containing protein | BCG_3931 | A0A0H3MC79 | S277 | M. bovis BCG | (7) |
| Acetyl-/propionyl-coenzyme A carboxylase alpha chain | MSMEG_1807 | A0QTE1 | – | M. smegmatis | (8) |
| Alcohol dehydrogenase, iron-containing | MSMEG_6242 | A0R5M3 | – | M. smegmatis | (8) |
| Antitoxin | BCG_0306c | A0A0H3M1S6 | T152 | M. bovis BCG | (7) |
| Antitoxin | BCG_3207c | A0A0H3MAL0 | T109 | M. bovis BCG | (7) |
| ATP synthase subunit beta | atpD | A1KI98 | S16 | M. bovis BCG | (7) |
| CbiA domain-containing protein | BCG_0573 | A0A0H3M2H1 | T25 | M. bovis BCG | (7) |
| Chaperone protein ClpB | clpB | A0A0H3M7W9 | T79 | M. bovis BCG | (7) |
| Chaperone protein DnaK | dnaK | A1KFH2 | T391 | M. bovis BCG | (7) |
| Chaperone protein DnaK | MSMEG_0709 | A0QQC8 | – | M. smegmatis | (8) |
| Conserved hypothetical proline and threonine-rich protein | BCG_0352 | A0A0H3MAA7 | S403 | M. bovis BCG | (7) |
| DNA gyrase subunit A | gyrA1 | A0A0G2Q9F8 | S263 | M. bovis BCG | (7) |
| Ferredoxin | MSMEG_5122 | A0R2I1 | – | M. smegmatis | (8) |
| FHA domain protein | MSMEG_0035 | A0QNG7 | T18, T116, T377 | M. tuberculosis, M. smegmatis | (8) |
| FHA domain-containing protein | TB39.8 | A0A0H3M751 | T371, Y382 | M. bovis BCG | (7) |
| Glutamine synthetase | MSMEG_4290 | A0R079 | T421, T77, S57 | M. tuberculosis, M. smegmatis | (8) |
| Glycogen accumulation regulator GarA | BQ2027_MB1858, garA, MSMEG_3647 | P64898, P9WJA9, A0QYG2 | T21 | M. bovis BCG, M. tuberculosis, M. smegmatis | (8, 23, 40) |
| HpcH_HpaI domain-containing protein | BCG_3100c | A0A0H3M8P9 | T12 | M. bovis BCG | (7) |
| Inorganic pyrophosphatase | MSMEG_6114 | A0R597 | – | M. smegmatis | (8) |
| Malate dehydrogenase | mdh | A1KI28, P9WK13 | S238, S239 | M. bovis BCG, M. tuberculosis | (7, 67) |
| Probable conserved membrane protein | BCG_0236 | A0A0H3M0X2 | S19 | M. bovis BCG | (7) |
| Protein kinase domain-containing protein | BCG_0696c | A0A0H3MAV4 | S117 | M. bovis BCG | (7) |
| RNA polymerase-binding protein RbpA | rbpA | A0A0H3M6B6 | T18 | M. bovis BCG | (7) |
| UDP-N-acetylmuramate–l-alanine ligase (MurC) | murC | P9WJL7 | – | M. tuberculosis | (64) |
| Uncharacterized protein | BCG_1803c | A0A0H3M4P0 | S11 | M. bovis BCG | (7) |
| Uncharacterized protein | BCG_2052c | A0A0H3M7J9 | T7 | M. bovis BCG | (7) |
Future Perspectives
Quantitative phosphoproteomics is now established as a powerful and reliable strategy for mapping connections between phosphorylated substrates and their respective kinases or phosphatases. As discussed in the preceding, the employment of LC-MS/MS-based strategies has dramatically increased the number of potential substrates for mycobacterial STPKs, specifically for PknB and PknG. While this represents a unique opportunity to better understand the regulatory role of STPKs, it is important to note potential pitfalls associated with such approaches. For instance, it could be argued that differentially phosphorylated proteins may not be direct substrates of the studied kinase but, rather, phosphorylated by another kinase whose activity was affected by the absence of the kinase and/or the presence of a kinase-specific inhibitor. It is, therefore, imperative to further validate these novel substrates. This may include the use of in vitro assays to confirm that the newly identified phosphosites are, in fact, phosphorylated by the corresponding STPK. Additionally, in cases where a new role and/or phenotype is associated with the activity of a protein kinase, it is important to determine the phosphorylation site occupancy of the novel substrate(s). Researchers should then, ideally, verify whether the phenotype is altered by the replacement of the substrate phosphorylation site with an alanine or glutamate residue via site-directed mutagenesis (i.e. knock-out or knock-in of a constitutive phosphorylation phenotype).
It is equally important that future studies investigate and validate the biological significance of these findings in order to increase understanding of the regulatory function(s) of newly identified substrate phosphorylation sites. For example, until very recently, the physiological role of PknG was largely associated with the regulation of mycobacterial metabolism, but newer datasets now suggest that PknG phosphorylation activity likely plays an important role in protein processing, translation, and folding machinery (7, 8). Moreover, it is also important to develop phosphoproteomic strategies to investigate the role of mycobacterial STPKs in host cells in order to further reveal the complexity of host-pathogen interactions. In a recent study, 31 proteins were reported as exclusive interactors of PknG using a human proteome microarray (66). This study provides a valuable foundation for further research on host-pathogen protein-protein interactions to reveal the role of PknG during pathogenetic conditions. However, the biological relevance of these findings remains to be verified by phosphoproteomic studies that account for the subcellular compartmentalization of proteins and the accessibility of a host substrate to a mycobacterial protein kinase in vivo. We, therefore, propose that the next goal in MS-based mycobacterial research should be the identification of host substrates for mycobacterial STPKs during live infections. Work toward this objective is currently underway in our laboratory.
Concluding Remarks
We anticipate that a comprehensive integration of the large phosphoproteomic datasets, aimed at complete identification of novel STPKs substrates, will provide the scientific community with significant new insight into the network mechanisms by which STPKs regulate mycobacterial environmental responses. In addition, this should allow researchers to pinpoint potential sites for new pharmacological interventions. The phosphorylation status of novel substrates can be utilized as biomarkers of the corresponding STPK activity. This, in turn, may aid in determining the efficiency and mode of action of potential inhibitors targeting specific mycobacterial STPKs. Adopting such strategies could provide a new platform to guide further development of next-generation drugs for clinical applications.
Supplementary Material
Acknowledgments
S.S.B. thanks the NRF for a Master's bursary. JMB thanks the NRF for a South African Research Chair grant. NCS thanks the South African Medical Research Council for a Junior Research Fellowship.
Footnotes
* This work is based on the research supported in part by the National Research Foundation (NRF) of South Africa (Grant Numbers: 467126 and 95984). The authors declare that they have no conflicts of interest with the contents of this article.
 This article contains supplemental material Tables S1–S4.
This article contains supplemental material Tables S1–S4.
1 The abbreviations used are:
- 2D-PAGE
- two-dimensional polyacrylamide gel electrophoresis
- FDR
- false discovery rate
- FHA
- fork-head association
- GO
- gene ontology
- IMAC
- immobilized metal affinity chromatography
- KESTREL
- kinase substrate tracking and elucidation
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- MS
- mass spectrometry
- M. bovis BCG
- Mycobacterium bovis Bacillus Calmette-Guérin
- M. smegmatis
- Mycobacterium smegmatis
- M. tuberculosis
- Mycobacterium tuberculosis
- PANTHER
- protein annotation through evolutionary relationship
- P-site
- phosphosite
- PTM
- post-translational modification
- Ser/Thr
- serine/threonine
- STPK
- serine/threonine protein kinase
- TB
- tuberculosis
- TiO2
- titanium dioxide.
REFERENCES
- 1. Wehenkel A., Bellinzoni M., Graña M., Duran R., Villarino A., Fernandez P., Andre-Leroux G., England P., Takiff H., Cerveñansky C., Cole S. T., and Alzari P. M. (2008) Mycobacterial Ser/Thr protein kinases and phosphatases: Physiological roles and therapeutic potential. Biochim. Biophys. Acta 1784, 193–202 [DOI] [PubMed] [Google Scholar]
- 2. van Els C. A., Corbière V., Smits K., van Gaans-van den Brink J. A., Poelen M. C., Mascart F., Meiring H. D., and Locht C. (2014) Toward understanding the essence of post-translational modifications for the Mycobacterium tuberculosis immunoproteome. Front. Immunol. 5, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mehaffy C., Belisle J. T., and Dobos K. M. (2019) Mycobacteria and their sweet proteins: An overview of protein glycosylation and lipoglycosylation in M. tuberculosis. Tuberculosis (Edinb) 115, 1–13 [DOI] [PubMed] [Google Scholar]
- 4. Calder B., Albeldas C., Blackburn J. M., and Soares N. C. (2016) Mass spectrometry offers insight into the role of ser/thr/tyr phosphorylation in the mycobacteria. Front. Microbiol. 7, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chao J., Wong D., Zheng X., Poirier V., Bach H., Hmama Z., and Av-Gay Y. (2010) Protein kinase and phosphatase signaling in Mycobacterium tuberculosis physiology and pathogenesis. Biochim. Biophys. Acta 1804, 620–627 [DOI] [PubMed] [Google Scholar]
- 6. Iswahyudi, Mukamolova G. V., Straatman-Iwanowska A. A., Allcock N., Ajuh P., Turapov O., and O'Hare H. M. (2019) Mycobacterial phosphatase PstP regulates global serine threonine phosphorylation and cell division. Sci. Rep. 9, 8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nakedi K. C., Calder B., Banerjee M., Giddey A., Nel A. J. M., Garnett S., Blackburn J. M., and Soares N. C. (2018) Identification of novel physiological substrates of Mycobacterium bovis BCG protein kinase G (PknG) by label-free quantitative phosphoproteomics. Mol. Cell. Proteomics 17, 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gil M., Lima A., Rivera B., Rossello J., Urdániz E., Cascioferro A., Carrión F., Wehenkel A., Bellinzoni M., Batthyány C., Pritsch O., Denicola A., Alvarez M. N., Carvalho P. C., Lisa M. N., Brosch R., Piuri M., Alzari P. M., and Durán R. (2019) New substrates and interactors of the mycobacterial serine/threonine protein kinase PknG identified by a tailored interactomic approach. J. Proteomics 192, 321–333 [DOI] [PubMed] [Google Scholar]
- 9. Kaur P., Rausch M., Malakar B., Watson U., Damle N. P., Chawla Y., Srinivasan S., Sharma K., Schneider T., Jhingan G. D., Saini D., Mohanty D., Grein F., and Nandicoori V. K. (2019) LipidII interaction with specific residues of Mycobacterium tuberculosis PknB extracytoplasmic domain governs its optimal activation. Nat. Commun. 10, 1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Prisic S., and Husson R. N. (2014) Mycobacterium tuberculosis serine/threonine protein kinases. Microbiol. Spectr. 2, 1–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chawla Y., Upadhyay S., Khan S., Nagarajan S. N., Forti F., and Nandicoori V. K. (2014) Protein kinase B (PknB) of Mycobacterium tuberculosis is essential for growth of the pathogen in vitro as well as for survival within the host. J. Biol. Chem. 289, 13858–13875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fortuin S., Tomazella G. G., Nagaraj N., Sampson S. L., Gey van Pittius N. C., Soares N. C., Wiker H. G., de Souza G. A., and Warren R. M. (2015) Phosphoproteomics analysis of a clinical Mycobacterium tuberculosis Beijing isolate: Expanding the mycobacterial phosphoproteome catalog. Front. Microbiol. 6, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sherman D. R., and Grundner C. (2014) Agents of change—Concepts in Mycobacterium tuberculosis Ser/Thr/Tyr phosphosignalling. Mol. Microbiol. 94, 231–241 [DOI] [PubMed] [Google Scholar]
- 14. Parish T. (2014) Two-component regulatory systems of mycobacteria. Microbiol. Spectr. 2, MGM2-0010-2013 [DOI] [PubMed] [Google Scholar]
- 15. Bretl D. J., Demetriadou C., and Zahrt T. C. (2011) Adaptation to environmental stimuli within the host: Two-component signal transduction systems of Mycobacterium tuberculosis. Microbiol. Mol. Biol. Rev. 75, 566–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zschiedrich C. P., Keidel V., and Szurmant H. (2016) Molecular mechanisms of two-component signal transduction. J. Mol. Biol. 428, 3752–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chakraborti P. K., Matange N., Nandicoori V. K., Singh Y., Tyagi J. S., and Visweswariah S. S. (2011) Signalling mechanisms in mycobacteria. Tuberculosis 91, 432–440 [DOI] [PubMed] [Google Scholar]
- 18. Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E. 3rd, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., Connor R., Davies R., Devlin K., Feltwell T., Gentles S., Hamlin N., Holroyd S., Hornsby T., Jagels K., Krogh A., McLean J., Moule S., Murphy L., Oliver K., Osborne J., Quail M. A., Rajandream M. A., Rogers J., Rutter S., Seeger K., Skelton J., Squares R., Squares S., Sulston J. E., Taylor K., Whitehead S., and Barrell B. G. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 [DOI] [PubMed] [Google Scholar]
- 19. Fernandez P., Saint-Joanis B., Barilone N., Jackson M., Gicquel B., Cole S. T., and Alzari P. M. (2006) The Ser/Thr protein kinase PknB is essential for sustaining mycobacterial growth. J. Bacteriol. 188, 7778–7784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Narayan A., Sachdeva P., Sharma K., Saini A. K., Tyagi A. K., and Singh Y. (2007) Serine threonine protein kinases of mycobacterial genus: Phylogeny to function. Physiol. Genomics 29, 66–75 [DOI] [PubMed] [Google Scholar]
- 21. Nagarajan S. N., Upadhyay S., Chawla Y., Khan S., Naz S., Subramanian J., Gandotra S., and Nandicoori V. K. (2015) Protein kinase a (PknA) of Mycobacterium tuberculosis is independently activated and is critical for growth in vitro and survival of the pathogen in the host. J. Biol. Chem. 290, 9626–9645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kang C. M., Abbott D. W., Park S. T., Dascher C. C., Cantley L. C., and Husson R. N. (2005) The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: Substrate identification and regulation of cell shape. Genes Dev. 19, 1692–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mueller P., and Pieters J. (2017) Identification of mycobacterial GarA as a substrate of protein kinase G from M. tuberculosis using a KESTREL-based proteome wide approach. J. Microbiol. Methods 136, 34–39 [DOI] [PubMed] [Google Scholar]
- 24. Walburger A., Koul A., Ferrari G., Nguyen L., Prescianotto-Baschong C., Huygen K., Klebl B., Thompson C., Bacher G., and Pieters J. (2004) Protein kinase G from pathogenic mycobacteria promotes survival within macrophages. Science. 304, 1800–1804 [DOI] [PubMed] [Google Scholar]
- 25. Papavinasasundaram K. G., Chan B., Chung J. H., Colston M. J., Davis E. O., and Av-Gay Y. (2005) Deletion of the Mycobacterium tuberculosis pknH gene confers a higher bacillary load during the chronic phase of infection in BALB/c mice. J. Bacteriol. 187, 5751–5760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sharma K., Chandra H., Gupta P. K., Pathak M., Narayan A., Meena L. S., D'Souza R. C., Chopra P., Ramachandran S., and Singh Y. (2004) PknH, a transmembrane Hank's type serine/threonine kinase from Mycobacterium tuberculosis is differentially expressed under stress conditions. FEMS Microbiol. Lett. 233, 107–113 [DOI] [PubMed] [Google Scholar]
- 27. Lakshminarayan H., and Rajaram A. (2009) Involvement of serine threonine protein kinase, PknL, from Mycobacterium tuberculosis H37Rv in starvation response of mycobacteria. J. Microb. Biochem. Technol. 1, [Google Scholar]
- 28. Hatzios S. K., Baer C. E., Rustad T. R., Siegrist M. S., Pang J. M., Ortega C., Alber T., Grundner C., Sherman D. R., and Bertozzi C. R. (2013) Osmosensory signaling in Mycobacterium tuberculosis mediated by a eukaryotic-like Ser/Thr protein kinase. Proc. Natl. Acad. Sci. 110, E5069–E5077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kumar D., and Narayanan S. (2012) PknE, a serine/threonine kinase of Mycobacterium tuberculosis modulates multiple apoptotic paradigms. Infect. Genet. Evol. 12, 737–747 [DOI] [PubMed] [Google Scholar]
- 30. Baer C. E., Iavarone A. T., Alber T., and Sassetti C. M. (2014) Biochemical and spatial coincidence in the provisional Ser/Thr protein kinase interaction network of Mycobacterium tuberculosis. J. Biol. Chem. 289, 20422–20433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miller C. J., and Turk B. E. (2018) Homing in: Mechanisms of substrate targeting by protein kinases. Trends Biochem. Sci. 43, 380–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xue L., and Tao W. A. (2013) Current technologies to identify protein kinase substrates in high throughput. Front. Biol. (Beijing). 8, 216–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pompeo F., Rismondo J., Gründling A., and Galinier A. (2018) Investigation of the phosphorylation of Bacillus subtilis LTA synthases by the serine/threonine kinase PrkC. Sci. Rep. 8, 17344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ravikumar V., Shi L., Krug K., Derouiche A., Jers C., Cousin C., Kobir A., Mijakovic I., and Macek B. (2014) Quantitative phosphoproteome analysis of Bacillus subtilis reveals novel substrates of the kinase PrkC and phosphatase PrpC. Mol. Cell. Proteomics 13, 1965–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Semanjski M., Germain E., Bratl K., Kiessling A., Gerdes K., and Macek B. (2018) The kinases HipA and HipA7 phosphorylate different substrate pools in Escherichia coli to promote multidrug tolerance. Sci. Signal. 11, eaat5750. [DOI] [PubMed] [Google Scholar]
- 36. Romanov N., Hollenstein D. M., Janschitz M., Ammerer G., Anrather D., and Reiter W. (2017) Identifying protein kinase-specific effectors of the osmostress response in yeast. Sci. Signal. 10, eaag2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Poss Z. C., Ebmeier C. C., Odell A. T., Tangpeerachaikul A., Lee T., Pelish H. E., Shair M. D., Dowell R. D., Old W. M., and Taatjes D. J. (2016) Identification of mediator kinase substrates in human cells using cortistatin A and quantitative phosphoproteomics. Cell Rep. 15, 436–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xia Q., Cheng D., Duong D. M., Gearing M., Lah J. J., Levey A. I., and Peng J. (2008) Phosphoproteomic analysis of human brain by calcium phosphate precipitation and mass spectrometry. J. Proteome Res. 7, 2845–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Prisic S., Dankwa S., Schwartz D., Chou M. F., Locasale J. W., Kang C.-M., Bemis G., Church G. M., Steen H., and Husson R. N. (2010) Extensive phosphorylation with overlapping specificity by Mycobacterium tuberculosis serine/threonine protein kinases. Proc. Natl. Acad. Sci. 107, 7521–7526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. O'Hare H. M., Durán R., Cerveñansky C., Bellinzoni M., Wehenkel A. M., Pritsch O., Obal G., Baumgartner J., Vialaret J., Johnsson K., and Alzari P. M. (2008) Regulation of glutamate metabolism by protein kinases in mycobacteria. Mol. Microbiol. 70, 1408–1423 [DOI] [PubMed] [Google Scholar]
- 41. Canova M. J., Veyron-Churlet R., Zanella-Cleon I., Cohen-Gonsaud M., Cozzone A. J., Becchi M., Kremer L., and Molle V. (2008) The Mycobacterium tuberculosis serine/threonine kinase PknL phosphorylates Rv2175c: Mass spectrometric profiling of the activation loop phosphorylation sites and their role in the recruitment of Rv2175c. Proteomics 8, 521–533 [DOI] [PubMed] [Google Scholar]
- 42. Villarino A., Duran R., Wehenkel A., Fernandez P., England P., Brodin P., Cole S. T., Zimny-Arndt U., Jungblut P. R., Cerveñansky C., and Alzari P. M. (2005) Proteomic identification of M. tuberculosis protein kinase substrates: PknB recruits GarA, a FHA domain-containing protein, through activation loop-mediated interactions. J. Mol. Biol. 350, 953–963 [DOI] [PubMed] [Google Scholar]
- 43. Knebel A., Morrice N., and Cohen P. (2001) A novel method to identify protein kinase substrates: eEF2 kinase is phosphorylated and inhibited by SAPK4/p38δ. EMBO J. 20, 4360–4369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chao J. D., Papavinasasundaram K. G., Zheng X., Chávez-Steenbock A., Wang X., Lee G. Q., and Av-Gay Y. (2010) Convergence of Ser/Thr and two-component signaling to coordinate expression of the dormancy regulon in Mycobacterium tuberculosis. J. Biol. Chem. 285, 29239–29246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zheng J., Liu L., Liu B., and Jin Q. (2015) Phosphoproteomic analysis of bacillus Calmette-Guérin using gel-based and gel-free approaches. J. Proteomics 126, 189–199 [DOI] [PubMed] [Google Scholar]
- 46. Nakedi K. C., Nel A. J., Garnett S., Blackburn J. M., and Soares N. C. (2015) Comparative Ser/Thr/Tyr phosphoproteomics between two mycobacterial species: The fast growing Mycobacterium smegmatis and the slow growing Mycobacterium bovis BCG. Front. Microbiol. 8, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. de Keijzer J., Mulder A., de Beer J., de Ru A. H., van Veelen P. A., and van Soolingen D. (2016) Mechanisms of phenotypic rifampicin tolerance in Mycobacterium tuberculosis Beijing genotype strain B0/W148 revealed by proteomics. J. Proteome Res. 15, 1194–1204 [DOI] [PubMed] [Google Scholar]
- 48. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 49. Taumer C., Griesbaum L., Kovacevic A., Soufi B., Nalpas N. C., and Macek B. (2018) Parallel reaction monitoring on a Q Exactive mass spectrometer increases reproducibility of phosphopeptide detection in bacterial phosphoproteomics measurements. J. Proteomics 189, 60–66 [DOI] [PubMed] [Google Scholar]
- 50. Turapov O., Forti F., Kadhim B., Ghisotti D., Sassine J., Straatman-Iwanowska A., Bottrill A. R., Moynihan P. J., Wallis R., Barthe P., Cohen-Gonsaud M., Ajuh P., Vollmer W., and Mukamolova G. V. (2018) Two faces of CwlM, an essential PknB substrate, in Mycobacterium tuberculosis. Cell Rep. 25, 57–67.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wong D., Li W., Chao J. D., Zhou P., Narula G., Tsui C., Ko M., Xie J., Martinez-Frailes C., and Av-Gay Y. (2018) Protein tyrosine kinase, PtkA, is required for Mycobacterium tuberculosis growth in macrophages. Sci. Rep. 8, 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Carette X., Platig J., Young D. C., Helmel M., Young A. T., Wang Z., Potluri L. P., Moody C. S., Zeng J., Prisic S., Paulson J. N., Muntel J., Madduri A. V. R., Velarde J., Mayfield J. A., Locher C., Wang T., Quackenbush J., Rhee K. Y., Moody D. B., Steen H., and Husson R. N. (2018) Multisystem analysis of Mycobacterium tuberculosis reveals kinase-dependent remodeling of the pathogen-environment interface. MBio 9, e02333–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Altenhoff A. M., Glover N. M., Train C. M., Kaleb K., Warwick Vesztrocy A., Dylus D., De Farias T. M., Zile K., Stevenson C., Long J., Redestig H., Gonnet G. H., and Dessimoz C. (2018) The OMA orthology database in 2018: Retrieving evolutionary relationships among all domains of life through richer web and programmatic interfaces. Nucleic Acids Res. 46, D477–D485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kapopoulou A., Lew J. M., and Cole S. T. (2011) The MycoBrowser portal: A comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis (Edinb) 91, 8–13 [DOI] [PubMed] [Google Scholar]
- 55. Mi H., Muruganujan A., Ebert D., Huang X., and Thomas P. D. (2019) PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47, D419–D426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dasgupta A., Datta P., Kundu M., and Basu J. (2006) The serine/threonine kinase PknB of Mycobacterium tuberculosis phosphorylates PBPA, a penicillin-binding protein required for cell division. Microbiology 152, 493–504 [DOI] [PubMed] [Google Scholar]
- 57. Thakur M., and Chakraborti P. K. (2006) GTPase activity of mycobacterial FtsZ is impaired due to its transphosphorylation by the eukaryotic-type Ser/Thr kinase, PknA. J. Biol. Chem. 281, 40107–40113 [DOI] [PubMed] [Google Scholar]
- 58. Grundner C., Gay L. M., and Alber T. (2005) Mycobacterium tuberculosis serine/threonine kinases PknB, PknD, PknE, and PknF phosphorylate multiple FHA domains. Protein Sci. 14, 1918–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mir M., Asong J., Li X., Cardot J., Boons G. J., and Husson R. N. (2011) The extracytoplasmic domain of the Mycobacterium tuberculosis ser/thr kinase PknB binds specific muropeptides and is required for PknB localization. PLoS Pathog. 7, e1002182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Koul A., Choidas A., Tyagi A. K., Drlica K., Singh Y., and Ullrich A. (2001) Serine/threonine protein kinases PknF and PknG of Mycobacterium tuberculosis: Characterization and localization. Microbiology 147, 2307–2314 [DOI] [PubMed] [Google Scholar]
- 61. Cowley S., Ko M., Pick N., Chow R., Downing K. J., Gordhan B. G., Betts J. C., Mizrahi V., Smith D. A., Stokes R. W., and Av-Gay Y. (2004) The Mycobacterium tuberculosis protein serine/threonine kinase PknG is linked to cellular glutamate/glutamine levels and is important for growth in vivo. Mol. Microbiol. 52, 1691–1702 [DOI] [PubMed] [Google Scholar]
- 62. van der Woude A. D., Stoop E. J., Stiess M., Wang S., Ummels R., van Stempvoort G., Piersma S. R., Cascioferro A., Jiménez C. R., Houben E. N., Luirink J., Pieters J., van der Sar A. M., and Bitter W. (2014) Analysis of secA2-dependent substrates in Mycobacterium marinum identifies protein kinase G (PknG) as a virulence effector. Cell. Microbiol. 16, 280–295 [DOI] [PubMed] [Google Scholar]
- 63. Hermann C., Giddey A. D., Nel A. J. M., Soares N. C., and Blackburn J. M. (2019) Cell wall enrichment unveils proteomic changes in the cell wall during treatment of Mycobacterium smegmatis with sub-lethal concentrations of rifampicin. J. Proteomics 191, 166–179 [DOI] [PubMed] [Google Scholar]
- 64. Wu F. L., Liu Y., Jiang H. W., Luan Y. Z., Zhang H. N., He X., Xu Z. W., Hou J. L., Ji L. Y., Xie Z., Czajkowsky D. M., Yan W., Deng J. Y., Bi L. J., Zhang X. E., and Tao S. C. (2017) The Ser/Thr protein kinase protein-protein interaction map of M. tuberculosis. Mol. Cell. Proteomics 16, 1491–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Käll L., Krogh A., and Sonnhammer E. L. L. (2007) Advantages of combined transmembrane topology and signal peptide prediction-the Phobius web server. Nucleic Acids Res. 35, W429–W432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wu F. L., Liu Y., Zhang H. N., Jiang H. W., Cheng L., Guo S. J., Deng J. Y., Bi L. J., Zhang X. E., Gao H. F., and Tao S. C. (2018) Global profiling of PknG interactions using a human proteome microarray reveals novel connections with CypA. Proteomics 18, e1800265. [DOI] [PubMed] [Google Scholar]
- 67. Wang X. M., Soetaert K., Peirs P., Kalai M., Fontaine V., Dehaye J. P., and Lefèvre P. (2015) Biochemical analysis of the NAD+-dependent malate dehydrogenase, a substrate of several serine/threonine protein kinases of Mycobacterium tuberculosis. PLoS One 10, e0123327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. O'Shea J. P., Chou M. F., Quader S. A., Ryan J. K., Church G. M., and Schwartz D. (2013) pLogo: A probabilistic approach to visualizing sequence motifs. Nat. Methods 10, 1211–1212 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.