Lectins and glycan-binding antibodies are powerful tools in biological research, provided detailed information is available about their glycan-binding specificities. Glycan-arrays, in combination with bioinformatics tools to mine the data, offer the ability to obtain such information. This review focuses on the bioinformatics tools and resources that are available for the analysis of glycan-array data. The tools are enabling new insights into protein-glycan interactions and enhancing the value of glycan-binding proteins in research.
Keywords: Glycoproteins, glycosylation, bioinformatics, glycomics, micro arrays, glycan arrays, lectins
Graphical Abstract
Highlights
Lectins and glycan-binding antibodies are valuable as probe of glycans.
Advanced bioinformatics tools enable the mining of glycan-array data.
New insights into protein-glycan interactions have value in biological research.
Abstract
Proteins that bind carbohydrate structures can serve as tools to quantify or localize specific glycans in biological specimens. Such proteins, including lectins and glycan-binding antibodies, are particularly valuable if accurate information is available about the glycans that a protein binds. Glycan arrays have been transformational for uncovering rich information about the nuances and complexities of glycan-binding specificity. A challenge, however, has been the analysis of the data. Because protein-glycan interactions are so complex, simplistic modes of analyzing the data and describing glycan-binding specificities have proven inadequate in many cases. This review surveys the methods for handling high-content data on protein-glycan interactions. We contrast the approaches that have been demonstrated and provide an overview of the resources that are available. We also give an outlook on the promising experimental technologies for generating new insights into protein-glycan interactions, as well as a perspective on the limitations that currently face the field.
Glycans are a fundamental part of biology. They cover cell surfaces, decorate most secreted proteins, control access to cells, and modify protein-protein and inter-cellular interactions. Glycans form the first-line mode of communication between the microbial world and the human, animal, and plant systems, and they form a main component of innate immune recognition. The adaptive immune system also relies heavily on glycan recognition, contrary to previous predictions, as shown by the large percentage of antibodies in the circulation that recognize glycan epitopes (1). Thus, researchers from diverse fields find glycans a fascinating topic of study.
A common feature among the many fields of study is this: obtaining information about glycan functions and structures is challenging. Researchers do not have the types of well-developed tools that are available for studies of nucleic acids and proteins. Automated synthesis of glycans structures, or the amplification of sequences using biotechnology, are not available. Sequencers to conveniently determine the monosaccharide backbone and linkages of a glycan are not available. And methods to induce or knock-out glycan structures on a specific protein are not available. The tools that are available are the domain of specialists, for the most part, excepting the basic methods to obtain rudimentary information. This situation is improving, owing to efforts on the part of funding agencies and tool developers to bring accessible tools to researchers, but gradually.
In this review, we focus on an approach that has long-standing use in the biological research community and that has the potential for increased and broader value. This approach is the use of affinity reagents, or glycan-binding proteins. Glycan-binding proteins, which include native proteins that bind specific glycan structures, termed lectins, and antibodies that recognize glycans, are used in the same way that antibodies are widely used to study proteins. They can be used to quantify specific features in biological samples or to identify the locations of the features in tissue or on cell surfaces, for example. Lectins have been used in this way for decades (2). The advantages of using lectins and antibodies for studying glycans is that they are easy to use in many types of experiments, they are inexpensive, the assays can be quantitative and high-throughput, and they give measurements about specific glycan motifs or features.
Owing to these advantages, lectins remain the primary method for identifying and quantifying glycan structures in biological samples or on proteins. But this approach also has limitations. The experiments do not give information about complete monosaccharide compositions, or about the heterogeneity of glycosylation between or within proteins, or about the locations of glycosylation on protein backbones. Such information can be accessed through increasingly sophisticated mass spectrometry methods.
Nevertheless, researchers are advancing the use of lectins in biological research. One of the most important advances is improved information about the binding-specificities of lectins. This progress results from improvements in both the experimental methods and the bioinformatics tools. Here we focus primarily on the bioinformatics tools that enhance the value of the experimental data. Developments in the experimental methods are too numerous to be covered in this review. For the researcher who is not a developer of technology, we provide an overview of the experimental and data resources that are currently available and a broad survey of experimental innovations that could eventually provide value to the research community.
Available Experimental and Data Resources
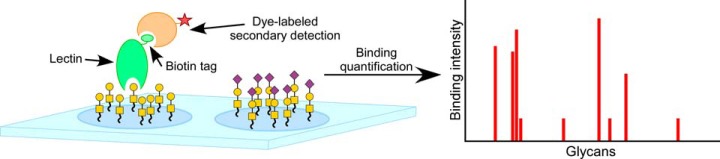
The driving technology in the study of lectin-glycan interactions has been the glycan array. Prior to the introduction of glycan arrays in 2002 (7–9), studies of lectin-glycan binding interactions required serial analyses of individual interactions, for example using elutions from affinity gels or competitive inhibitions of binding (3). These methods required large amounts of each glycan and had limited ability to test many interactions. Subsequent methods providing increased throughput and precision include frontal affinity chromatography (4) and surface-plasmon resonance (5, 6). But the glycan array opened the possibility of probing in parallel dozens of glycans, using tiny amounts of each glycan (Fig. 1).
Fig. 1.
The acquisition of glycan-array data. The typical experiment involves incubating a lectin or glycan-binding antibody on a microarray of diverse glycans, followed by quantifying the amount of binding to each glycan. The protein usually is labeled with a fluorescent tag or another tag that allows fluorescence detection by a secondary agent.
Researchers have produced a huge amount of glycan-array data since the introduction of the technology. The glycan arrays produced by the Consortium for Functional Glycomics (CFG)—a project funded by the National Institutes of Health—have been particularly popular (10), and the data and have been extensively accessed. The public funding was valuable for initiating developments, but several companies are now providing longer-term options for access to glycan-analysis. Core services in academic settings also are offering options as the methods become further standardized. Table I provides a summary of the academic and commercial arrays and datasets that are available as resources. The Table is not exhaustive list of arrays produced by any group, but rather includes those that are provided as a general service; additional arrays produced by academic groups are referenced in the Developments in the Experimental Methods section. In addition, data from glycan arrays other than the CFG array are available through the supplementary data corresponding to publications.
Table I. Experimental and data resources for glycan arrays. The criterion for inclusion was any array that advertised itself as a service or resource and that had appropriate web-accessible information and request forms.
| Source Type | Source | Content Type | Number of Glycans | Data Available |
|---|---|---|---|---|
| Academic | National Center for Functional Glycomics (NCFG) | Previous Versions of the CFG | 250–600+ | Yes |
| Current Version of the CFG | 600 | Yes | ||
| Mannose-6P Array | 26 | No | ||
| Modified Sialic Acid Array | 80 | No | ||
| NCFG General Glycan Array | 100 | No | ||
| NCFG SBA Array | 106 | No | ||
| Asparagine-Linked Array | 38 | Yes | ||
| Imperial College | Custom Arrays | 796 | Yes | |
| Commercial | Z Biotech | General Array | 100 | No |
| N-Glycan Array | 100 | No | ||
| O-Glycan Array | 94 | No | ||
| Heparan Sulfate | 24 | No | ||
| Neu5Gc/Neu5Ac | 80 | No | ||
| Human Milk Oligosaccharide | 46 | No | ||
| Glycosphingolipid | 58 | No | ||
| Glycosaminoglycan | 34 | No | ||
| Chemily | Blood Group Antigen | 21 | No | |
| General Array | 100 | No | ||
| General Array | 300 | No | ||
| Glycosphingolipid | 58 | No | ||
| Human Milk Oligosaccharide | 46 | No | ||
| N-Glycan Array | 100 | No | ||
| Neu5Gc/Neu5Ac | 80 | No | ||
| RayBiotech | General Array | 100 | No | |
| General Array | 300 | No |
Software for Determining Glycan-Binding Specificities
The analysis of glycan-array data has the goal of uncovering the rules that govern the binding of a protein to glycans. One can ask the question, what are the features of a glycan that determine whether a lectin binds or does not bind, or that tune the level of binding? Sometimes the rules appear relatively straightforward. In the case of Vicea villosa lectin (VVL), the presence of a terminal, alpha-linked N-acetyl-galactosamine is necessary and enough for binding to all glycans tested so far. For the complex cases, the rules may involve longer-range interactions with neighboring monosaccharides or separate branches (11, 12). Recent studies of human intelectin-1 (13) and DC-SIGN (14) provide examples of complex rules governing glycan recognition.
Visual inspection of the data can provide qualitative assessments of glycan-binding specificity. This system can function sufficiently well in many cases, such as in studies of changes in influenza specificities (15, 16). But manual analyses have disadvantages. They require expert knowledge; they are subject to the bias of the interpreter; the specificities of proteins are often too complex to be accurately discerned by visual inspection and described by qualitative terms; and they are not amenable to high-throughput processing. Therefore, algorithms for computer analyses are necessary.
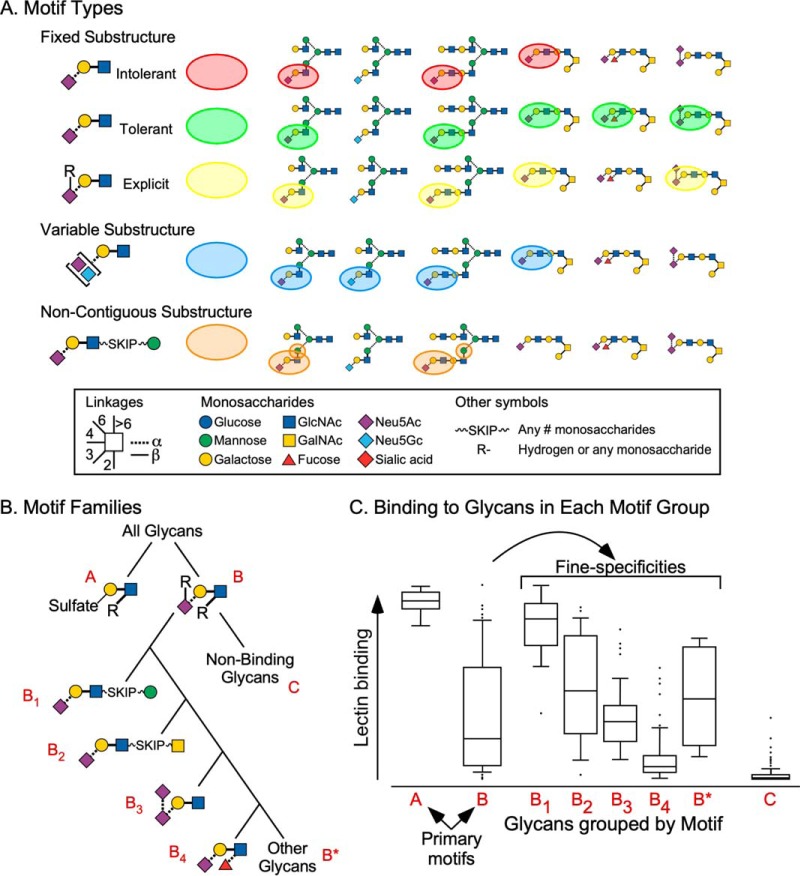
To develop an algorithm for glycan-array analysis, one needs a method of describing the potential binding-determinants of a protein, or the glycan motifs. The glycan motifs are the substructures or patterns of monosaccharides that potentially are bound by a lectin (Fig. 2A). A method of describing motifs enables glycan-array analyses using the basic approach of (1) determining the presence or absence of the motifs on the glycans of an array, and (2) identifying the relationships between the motifs and the binding of the lectin. This approach was demonstrated by Porter and coworkers in 2010 (17). It is analogous to identifying the DNA motifs bound by a transcription factor, but with added complexity. A lectin does not bind a static substructure, but rather a family of substructures, some members stronger and others weaker. The contact points between a lectin and a glycan could involve monosaccharides that are non-contiguous or on different branches. Thus, some monosaccharides could be interchangeable, and the distances between contacts could be variable. Developing a notation to define the glycan motifs that accurately portray lectin binding has been the ongoing challenge for glycan-array analyses (Fig. 2A).
Fig. 2.
Defining motifs and families of motifs. A, Motif types. Fixed substructures are continuous units of defined monosaccharides. Intolerant definitions require the unit to be unsubstituted, and tolerant definitions allow substitutions. Explicit definitions define the locations where substitutions are optional, which gives the highest level of precision in the definition. Variable substructures allow for options in the monosaccharides, providing another level of flexibility in the definition. Non-contiguous substructures allow the components of a motif to be physically separated. This feature is useful when a lectin contacts separate branches of a glycan. B, Motif families. The tree shows the relationships between the groups of glycans with the indicated motifs, using a simulated analysis. The first split represents primary motifs (A and B) to which a protein binds. Motif B can be split into sub-motifs that represent fine specificities. C, The simulated data show the ranges of lectin binding to the glycans in each of the motif groups. For example, the glycans in group B contain motif B but not motif A. The B1-B4 sub-motifs define fine-specificities with differing ranges of binding, potentially explaining the broad range of the parent motif B.
The Porter work used motif definitions based on patterns that are common in mammalian biology. This method had the advantage of incorporating expert knowledge, and it proved accurate in identifying the main specificities of 90 different lectins using data in the CFG database. The user can add new motif definitions, based on additional analyses, to more-accurately describe binding (18). Automated processing of glycan-array data provided global analyses of over 3000 datasets in the CFG database (19).
Another system had the goal of computer-based motif discovery (20), as opposed to user-defined motifs, based on the rationale that an algorithm could pick out unusual specificities that might be missed by a user. The GlycanMotifMiner algorithm identifies the glycans with high binding and uses an iterative search for a subtree that is enriched in the high-binding glycans. The method tests monosaccharide additions to a starting monosaccharide, and then grows the subtree until any addition results in too few binders or too many non-binders. This method has the advantage of not requiring pre-defined or user-refined motifs, but it also has limited ability to find complex specificities, owing to the use of contiguous subtrees as motifs, which do not allow substitutions or gaps between monosaccharides and other complexities. It also requires dichotomizing the glycans into binders and non-binders, which is not a clear distinction in many datasets. A web platform provided convenient access to the GlycoPattern program, which serviced the CFG-array data (21). Related methods have been developed with variations including the use of kernel methods (22) and the use of alpha-closed subtrees (23, 24). These methods showed value in the identification of non-sialylated motifs bound by the influenza virus (25).
A method that incorporates more flexibility into the motifs was demonstrated by Hosada et al. (26). The Multiple Carbohydrate Alignment with Weights (MCAW) algorithm adapted a sequence-alignment algorithm commonly used for DNA alignments, called ClustalW (27). The method aligns the glycans that are strongly-bound by a protein to find a consensus sequence. The consensus sequences are scored by the similarity of the monomers and penalized for gaps. The authors demonstrated the method's effectiveness by analyzing over 1000 CFG data sets and distributing the results in a web-accessible database (28). The method can identify the locations where variability is allowed or disallowed, but disadvantages are that it does not narrow in on the minimal features required for binding, and that it provides little information on lower-affinity motifs. The method could provide interesting insights that are not apparent from other methods, however, and it demonstrates a novel adaptation of DNA-oriented bioinformatics for glyco-bioinformatics.
We previously introduced a method that has the potential of accounting for the complexities of lectin and antibody binding. The method is built on two primary features: (1) flexibility in individual motifs; and (2) families of motifs. The first feature accounts for the variability in the binding-site of a protein. We developed a new syntax, or motif language (29), that uses wildcards and logical operators (AND, OR, etc.) to describe variability in monosaccharides or linkages, and that uses other modifiers to allow for gaps of any length. The monosaccharide carbons can be defined either as “free” (cannot be substituted) or as tolerating substitution, which then distinguishes a terminating monosaccharide from an internal monosaccharide. The result is that motifs of nearly any variability or complexity can be represented (Fig. 2A).
The second feature, using families of motifs (Fig. 2B), accounts for the fact that not all the allowed binding partners of a protein are alike; some modifications to a glycan tune the binding to make it stronger or weaker. These sub-motifs are the fine specificities of a glycan-binding protein. In addition, proteins can have alternate specificities. Unlike fine specificities, primary specificities are different motifs altogether. For example, Concanavalin A primarily binds mannose as found in N-linked glycans, but it also binds terminal, alpha-linked glucose. Another example is given by wheat-germ agglutinin (WGA)1, which has separate preferences for N-acetyl-lactosamine, terminal GlcNAc or GalNAc, or sialic acid. We therefore represent protein binding not as a single motif, but as a family of motifs, with the relationships between motifs organized as primary motifs and fine specificities. The relationships can be visualized graphically (Fig. 2B).
These features are the foundations of the MotifFinder software (29) for automated analyses of glycan-array data. The program searches for the individual motifs that best describe specific fine-specificities or primary-specificities, and it searches for the set of motifs and the relationships between them that account for the overall binding pattern of the protein (Fig. 2C). The flexible motif syntax enables the automated generation of novel motif definitions that can account for unforeseen fine-specificities. This capability could be useful for characterizing protein binding to unusual glycans, as in a previous application of the method (29) to glycans with unequal extensions and substituents on each branch (30). The method identified features that would not have been practically analyzable using manual analyses or previous modes of representing motifs.
Applying the Information
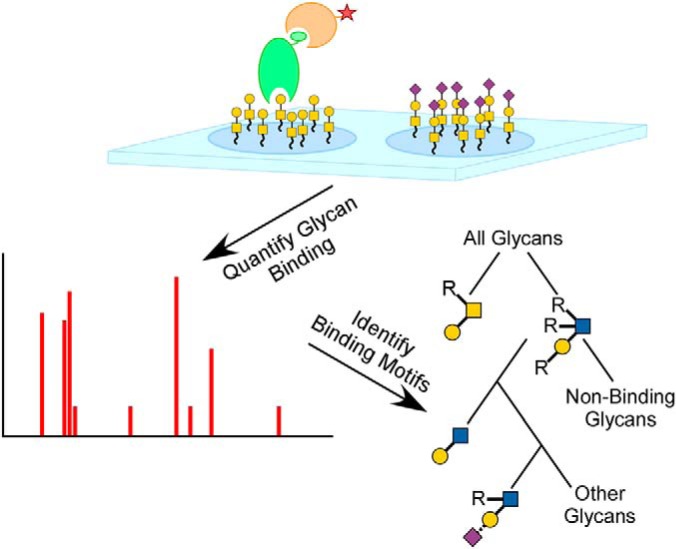
One benefit of the high-precision characterization of glycan-binding specificity is insight into the biology of protein-glycan interactions. Another benefit is the improved use of lectins and antibodies to analyze glycans in biological material. For example, instead of using manual analysis to give a simple, qualitative interpretation of the presence of a motif, a software tool could provide quantitative estimates of glycan motifs. This approach could account for complex and nuanced aspects of protein specificity, and it opens the possibility of integrating information from multiple proteins. A researcher could probe a sample with multiple, different lectins, and the software could use the profile of binding levels across the lectins, combined with the detailed determinations of motif preferences from glycan-array analyses, to provide estimates of the amounts of multiple motifs (Fig. 3). Such datasets are frequently collected using lectin arrays (31, 32).
Fig. 3.
Using the results from glycan-array analyses to interpret experimental data. A lectin (or a glycan-binding antibody) can be applied to glycan arrays and experimental samples in separate experiments. Analysis software is applied to the glycan-array data to determine the lectin specificities, and the amount of lectin binding to the sample with unknown glycans is quantified. The output from the glycan-array analysis is combined with the data from the sample to produce an estimation of the glycans that are present in the sample. This scheme also could be used with integrated data from multiple lectins, and with data acquired after treatments with glycosidases.
Further, one could apply lectin profiling after rounds of exoglycosidase cleavage in order to obtain additional information about the structures. Lectin probing in the presence or absence of glycosidase digestion has been used in affinity-electrophoresis, lectin-blotting, and histochemical analysis (33–36), and we have extended the approach to a micro-scale format in combination with algorithms for the automated interpretation of the data (37, 38). We proposed that this method could provide complementary information to mass-spectrometry and be practical for researchers who are not experts in glycobiology.
Available Software Resources
Table II provides an overview of software resources that are available for glycan-array analysis.
Table II. Software resources for glycan array analyses.
| Software | Method | Summary | Reference |
|---|---|---|---|
| MotifFinder | Motif Binding Association | Uses multiple statistics to test the associations of motifs with lectin binding, using an advanced motif syntax that allows flexible motif definitions. Recent versions include automated motif optimization and modeling families of motifs. | Klamer 2017 (29) |
| GlycoPattern | Frequent Subtree Mining | Uses a graph theory approach to mine new glycan substructures that are frequent in the bound glycans and infrequent in the unbound glycans. | Cholleti 2012 (20) Agravat 2014 (21) |
| Multiple Carbohydrate Alignment with Weights (MCAW) | Weighted Structure Alignment | Adapts traditional sequence alignment algorithms to align the strongest glycan binders for a lectin. Offers a database of analyzed CFG datasets. | Hosoda 2017 (26) Hosoda 2018 (28) |
| GLycan Array Dashboard (GLAD) | Graphical Visualization | Enables researchers to explore trends in glycan array data through graphic visualization and the manual exploration of simple motifs. | Mehta 2019[1] |
Developments in the Experimental Methods
Recent publications (39–43) give good reviews of the many developments in glycan array technology. The approaches differ in fundamental areas such as the production of the glycans, the presentation of the glycans, and the quantification of lectin-glycan interactions. In the end, no single platform gives a complete picture. No platform has all the glycans necessary for such a picture, and each platform has constraints that could influence binding patterns. Direct comparisons between platforms showed that the results can be divergent (44, 45). Many groups have pursued sophisticated enhancements to the experimental systems, and the field has grown beyond what can be reviewed here. Below we provide a sampling of the important work that eventually could be useful for researchers in biology.
A theme that has engaged many technology-developers is the better modeling of the biological environment. Most lectins occur as multimers of repeating subunits (46, 47). For example, the Aurelia aurantia lectin (AAL) has a 6-fold beta-propeller structure with five fucose binding sites on the edges (48, 49). The repeated glycan-binding sites are thought to increase avidity to glycans presented in corresponding units on cell surfaces, where they can change densities (50, 51), or in closely spaced arrangements on a protein. The experimental investigation of this effect using conventional arrays is limited. To produce glycans facilitate studies of multivalency, researchers have synthesized glycopolymers or glycodendrimers, in which glycans decorate a polymer backbone at controlled intervals (52–55). This method offers unique insights into the avidities of hetero-multivalent binding, although it is limited in breadth by the significant synthetic hurdle. Another approach is to measure the agglutination of emulsions containing mixtures of two glycans (56), which enables studies of the kinetics of hetero-multivalent binding and more accurately models a membrane environment. Glycans attached to quantum dots (57) also could be useful for studying multivalent binding, because the glycans can be kept in proximity in the solution phase. An approach that is easier to implement and higher-throughput is to vary the numbers of glycans attached to a protein carrier (58). This method can reveal density-dependent effects but has less control over the molecular details. Bead-based formats (59, 60) could give increased flexibility in experimental design and solution-phase interactions that are not available using planar arrays.
Label-free methods also could provide improved measurements of certain glycan-protein interactions, because the chemical labeling of a protein could affect binding. Surface plasmon resonance offers measurements of binding affinities as well as the ability to identify low-affinity interactions (61, 62). Mass spectrometry could provide solution-phase detection of the glycans bound by a protein and potentially more accurate measurements of binding strengths relative to solid-phase methods (63). A demonstration of this approach using catch-and-release system allowed the assay of glycans bound by various glycan-binding proteins (64, 65). A related method utilized a universal proxy-protein receptor to allow the quantitative screening of glycan binding and carbohydrate-active enzyme activity (66). An inherent challenge with mass-spectrometry is distinguishing between glycans that have the same mass but differences in linkage or sequence, which occur frequently and are functionally important.
Expanding the range of glycans available for the experiments is another significant goal. In contrast to proteins and nucleic acids, glycans need to be synthesized individually or purified from natural sources. In addition, they cannot be amplified through biotechnology. Purification from natural sources is an attractive option for glycans that are not amenable to synthesis and that directly relate to biological samples. The purified glycans could be attached to a linker (67, 68) or fluorescent tag (69, 70) for further analysis. Disadvantages are the difficulty in achieving full purity and the need for structural characterization after purification. Synthetic strategies address these limitations and are making good progress. New synthetic methods have provided structures that previously were difficult to synthesize, such as asymmetrically-branched N-glycans (29, 71–73), which were useful for producing arrays of human milk oligosaccharides (30). Several groups have developed arrays for additional classes such as sialylated structures (74–76), plant cell-wall glycans (77), and microbial glycans (8, 78, 79). The synthesis of these structures currently is limited by the requirement for high expertise and customization, but automated synthesis, which was shown to be feasible selected glycans using enzyme-mediated methods (80, 81), could alleviate that bottleneck.
Though tangential to the in vitro methods, it is worth noting the complementarity of structural analysis and simulation. Grant et al. used computational grafting to generate putative structures for lectin-glycan complexes, giving justification for the observed patterns of binding on a glycan array (12). Anomalous experimental data was explained by the hindered access of bulky, multi-subunit lectins to glycans that are too close to the surface (82). Sood et al. used molecular-dynamics simulations to quantify the contributions made by the functional groups of monosaccharides and to identify the minimum features required for lectin-glycan binding (83). A tool that could facilitate these types of analyses is a curated database called Unilectin3D, which lists the structures in the Protein Database (PDB) of lectins in complex with a glycan (84). The in-silico methods could provide valuable context to the experimental results and produce accurate predictions of binding to glycans that are not represented on the arrays.
Outlook
The resources and tools available to researchers have greatly increased in recent years, with more in development. In addition to the developments covered here, mass-spectrometry methods for glycan analysis are advancing in capabilities and availability, and molecular biology methods involving genetic and chemical manipulations are increasingly powerful. Researchers will generally need to draw upon several approaches to thoroughly study questions. Thus, each of the methods need to be standardized and accessible to researchers who are not specialists in the technologies. In addition, software will be needed to integrate information from multiple, disparate sources.
For glycan-array methods and analyses, researchers would benefit from the calibration of the experiments through standardized material. Well-characterized glycoproteins or manufactured glycoproteins could be used to calibrate data on quantitative scales. Standards also could provide a means to link information between experiments. The published guidelines for glycomics experiments (85) are helpful but do not address standards for calibration. Another need is data repositories. Many groups are producing data that could be useful for others, especially from glycan-array and mass-spectrometry experiments, but databases for deposition are not available. The data are either not available or are spread across the supplementary data of hundreds of papers. In the analogous domains of protein and nucleic-acid research, several databases are accessible for the raw or processed data, such as GEO for gene expression, PDB for structural biology, and many more. Although some glycan-binding datasets have been submitted to the GEO database, this repository is ill-suited for glycan-binding data and is not an intuitive location to find such data.
Another major need is improved information about glycosidase specificity. Glycosidases are standard tools in glycan sequencing by chromatography and electrophoresis, and they are increasingly used in mass-spectrometry experiments. They also can be used to uncover motifs to be probed by lectins (37, 38, 86). In many cases, only basic information is known about the specificity of a glycosidase. Glycan arrays could help by enabling measurements of activity over a huge number of glycans, as demonstrated in a study of influenza neuraminidase (87). Software to interpret data from glycan arrays that were treated by glycosidases and then probed by lectins (38) could provide detailed insights into glycosidase specificity and a means to apply the information.
Footnotes
* This work was funded by National Cancer Institute (Alliance of Glycobiologists for Cancer Detection, U01CA168896; Innovative Molecular Analysis Technology Program, R21CA225474) and the National Institute for General Medical Sciences (STTR/SBIR Program, R43GM131430 and R41GM112750). The authors declare that they have no conflicts of interest with the contents of this article.
1 The abbreviations used are:
- WGA
- wheat-germ agglutinin
- PDB
- protein database.
REFERENCES
- 1. Schneider C., Smith D. F., Cummings R. D., Boligan K. F., Hamilton R. G., Bochner B. S., Miescher S., Simon H. U., Pashov A., Vassilev T., and von Gunten S. (2015) The human IgG anti-carbohydrate repertoire exhibits a universal architecture and contains specificity for microbial attachment sites. Sci. Transl. Med. 7, 269ra261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sharon N. (2007) Lectins: carbohydrate-specific reagents and biological recognition molecules. J. Biol. Chem. 282, 2753–2764 [DOI] [PubMed] [Google Scholar]
- 3. Hayes C. E., and Goldstein I. J. (1974) An alpha-D-galactosyl-binding lectin from Bandeiraea simplicifolia seeds. Isolation by affinity chromatography and characterization. J. Biol. Chem. 249, 1904–1914 [PubMed] [Google Scholar]
- 4. Hirabayashi J., Arata Y., and Kasai K. (2003) Frontal affinity chromatography as a tool for elucidation of sugar recognition properties of lectins. Methods Enzymol. 362, 353–368 [DOI] [PubMed] [Google Scholar]
- 5. Haseley S. R., Talaga P., Kamerling J. P., and Vliegenthart J. F. (1999) Characterization of the carbohydrate binding specificity and kinetic parameters of lectins by using surface plasmon resonance. Anal. Biochem. 274, 203–210 [DOI] [PubMed] [Google Scholar]
- 6. Shinohara Y., Kim F., Shimizu M., Goto M., Tosu M., and Hasegawa Y. (1994) Kinetic measurement of the interaction between an oligosaccharide and lectins by a biosensor based on surface plasmon resonance. Eur. J. Biochem. 223, 189–194 [DOI] [PubMed] [Google Scholar]
- 7. Drickamer K., and Taylor M. E. (2002) Glycan arrays for functional glycomics. Genome Biol. 3, REVIEWS1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang D., Liu S., Trummer B. J., Deng C., and Wang A. (2002) Carbohydrate microarrays for the recognition of cross-reactive molecular markers of microbes and host cells. Nat. Biotechnol. 20, 275–281 [DOI] [PubMed] [Google Scholar]
- 9. Fukui S., Feizi T., Galustian C., Lawson A. M., and Chai W. (2002) Oligosaccharide microarrays for high-throughput detection and specificity assignments of carbohydrate-protein interactions. Nat. Biotechnol. 20, 1011–1017 [DOI] [PubMed] [Google Scholar]
- 10. Blixt O., Head S., Mondala T., Scanlan C., Huflejt M. E., Alvarez R., Bryan M. C., Fazio F., Calarese D., Stevens J., Razi N., Stevens D. J., Skehel J. J., van Die I., Burton D. R., Wilson I. A., Cummings R., Bovin N., Wong C. H., and Paulson J. C. (2004) Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 17033–17038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Taylor M. E., and Drickamer K. (2009) Structural insights into what glycan arrays tell us about how glycan-binding proteins interact with their ligands. Glycobiology 19, 1155–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grant O. C., Tessier M. B., Meche L., Mahal L. K., Foley B. L., and Woods R. J. (2016) Combining 3D structure with glycan array data provides insight into the origin of glycan specificity. Glycobiology 26, 772–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wesener D. A., Wangkanont K., McBride R., Song X., Kraft M. B., Hodges H. L., Zarling L. C., Splain R. A., Smith D. F., Cummings R. D., Paulson J. C., Forest K. T., and Kiessling L. L. (2015) Recognition of microbial glycans by human intelectin-1. Nat. Struct. Mol. Biol. 22, 603–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Coombs P. J., Harrison R., Pemberton S., Quintero-Martinez A., Parry S., Haslam S. M., Dell A., Taylor M. E., and Drickamer K. (2010) Identification of novel contributions to high-affinity glycoprotein-receptor interactions using engineered ligands. J. Mol. Biol. 396, 685–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kumari K., Gulati S., Smith D. F., Gulati U., Cummings R. D., and Air G. M. (2007) Receptor binding specificity of recent human H3N2 influenza viruses. Virol. J. 4, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Childs R. A., Palma A. S., Wharton S., Matrosovich T., Liu Y., Chai W., Campanero-Rhodes M. A., Zhang Y., Eickmann M., Kiso M., Hay A., Matrosovich M., and Feizi T. (2009) Receptor-binding specificity of pandemic influenza A (H1N1) 2009 virus determined by carbohydrate microarray. Nat. Biotechnol. 27, 797–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Porter A., Yue T., Heeringa L., Day S., Suh E., and Haab B. B. (2010) A motif-based analysis of glycan array data to determine the specificities of glycan-binding proteins. Glycobiology 20, 369–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maupin K. A., Liden D., and Haab B. B. (2011) The fine specificity of mannose-binding and galactose-binding lectins revealed using outlier-motif analysis of glycan array data. Glycobiology 22, 160–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kletter D., Singh S., Bern M., and Haab B. B. (2013) Global comparisons of lectin-glycan interactions using a database of analyzed glycan array data. Mol. Cell Proteomics 12, 1026–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cholleti S. R., Agravat S., Morris T., Saltz J. H., Song X., Cummings R. D., and Smith D. F. (2012) Automated motif discovery from glycan array data. Omics 16, 497–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Agravat S. B., Saltz J. H., Cummings R. D., and Smith D. F. (2014) GlycoPattern: a web platform for glycan array mining. Bioinformatics 30, 3417–3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yamanishi Y., Bach F., and Vert J. P. (2007) Glycan classification with tree kernels. Bioinformatics 23, 1211–1216 [DOI] [PubMed] [Google Scholar]
- 23. Aoki-Kinoshita K. F. (2013) Mining frequent subtrees in glycan data using the RINGS glycan miner tool. Methods Mol. Biol. 939, 87–95 [DOI] [PubMed] [Google Scholar]
- 24. Hashimoto K., Takigawa I., Shiga M., Kanehisa M., and Mamitsuka H. (2008) Mining significant tree patterns in carbohydrate sugar chains. Bioinformatics 24, i167–i173 [DOI] [PubMed] [Google Scholar]
- 25. Ichimiya T., Nishihara S., Takase-Yoden S., Kida H., and Aoki-Kinoshita K. (2014) Frequent glycan structure mining of influenza virus data revealed a sulfated glycan motif that increased viral infection. Bioinformatics 30, 706–711 [DOI] [PubMed] [Google Scholar]
- 26. Hosoda M., Akune Y., and Aoki-Kinoshita K. F. (2017) Development and application of an algorithm to compute weighted multiple glycan alignments. Bioinformatics 33, 1317–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thompson J. D., Higgins D. G., and Gibson T. J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hosoda M., Takahashi Y., Shiota M., Shinmachi D., Inomoto R., Higashimoto S., and Aoki-Kinoshita K. F. (2018) MCAW-DB: A glycan profile database capturing the ambiguity of glycan recognition patterns. Carbohydr. Res. 464, 44–56 [DOI] [PubMed] [Google Scholar]
- 29. Klamer Z., Staal B., Prudden A. R., Liu L., Smith D. F., Boons G. J., and Haab B. B. (2017) Mining high-complexity motifs in glycans: a new language to uncover the fine-specificities of lectins and glycosidases. Anal. Chem. 89, 12342–12350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Prudden A. R., Liu L., Capicciotti C. J., Wolfert M. A., Wang S., Gao Z., Meng L., Moremen K. W., and Boons G. J. (2017) Synthesis of asymmetrical multiantennary human milk oligosaccharides. Proc. Natl. Acad. Sci. U.S.A. 114, 6954–6959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pilobello K. T., Krishnamoorthy L., Slawek D., and Mahal L. K. (2005) Development of a lectin microarray for the rapid analysis of protein glycopatterns. Chembiochem. 6, 985–989 [DOI] [PubMed] [Google Scholar]
- 32. Kuno A., Uchiyama N., Koseki-Kuno S., Ebe Y., Takashima S., Yamada M., and Hirabayashi J. (2005) Evanescent-field fluorescence-assisted lectin microarray: a new strategy for glycan profiling. Nat. Methods 2, 851–856 [DOI] [PubMed] [Google Scholar]
- 33. Shimizu K., Katoh H., Yamashita F., Tanaka M., Tanikawa K., Taketa K., Satomura S., and Matsuura S. (1996) Comparison of carbohydrate structures of serum alpha-fetoprotein by sequential glycosidase digestion and lectin affinity electrophoresis. Clin. Chim. Acta 254, 23–40 [DOI] [PubMed] [Google Scholar]
- 34. Evjen G., and Huseby N. E. (1992) Characterization of the carbohydrate moiety of human gamma-glutamyltransferases using lectin-blotting and glycosidase treatment. Clin. Chim. Acta 209, 27–34 [DOI] [PubMed] [Google Scholar]
- 35. Ito N., Nishi K., Nakajima M., Okamura Y., and Hirota T. (1989) Histochemical demonstration of O-glycosidically linked, type 3 based ABH antigens in human pancreas using lectin staining and glycosidase digestion procedures. Histochemistry 92, 307–312 [DOI] [PubMed] [Google Scholar]
- 36. Ito N., Nishi K., Nakajima M., Okamura Y., and Hirota T. (1989) Histochemical analysis of the chemical structure of blood group-related carbohydrate chains in serous cells of human submandibular glands using lectin staining and glycosidase digestion. J. Histochem. Cytochem. 37, 1115–1124 [DOI] [PubMed] [Google Scholar]
- 37. Reatini B. S., Ensink E., Liau B., Sinha J. Y., Powers T. W., Partyka K., Bern M., Brand R. E., Rudd P. M., Kletter D., Drake R., and Haab B. B. (2016) Characterizing protein glycosylation through on-chip glycan modification and probing. Anal. Chem. 88, 11584–11592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Klamer Z., Hsueh P., Ayala-Talavera D., and Haab B. (2019) Deciphering protein glycosylation by computational integration of on-chip profiling, glycan-array data, and mass spectrometry. Mol. Cell Proteomics 18, 29–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rillahan C. D., and Paulson J. C. (2011) Glycan microarrays for decoding the glycome. Annu. Rev. Biochem. 80, 797–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Narla S. N., Nie H., Li Y., and Sun X. L. (2015) Multi-dimensional glycan microarrays with glyco-macroligands. Glycoconj. J. 32, 483–495 [DOI] [PubMed] [Google Scholar]
- 41. Hyun J. Y., Pai J., and Shin I. (2017) The glycan microarray story from construction to applications. Acc. Chem. Res. 50, 1069–1078 [DOI] [PubMed] [Google Scholar]
- 42. Muthana S. M., and Gildersleeve J. C. (2014) Glycan microarrays: powerful tools for biomarker discovery. Cancer Biomark 14, 29–41 [DOI] [PubMed] [Google Scholar]
- 43. McQuillan A. M., Byrd-Leotis L., Heimburg-Molinaro J., and Cummings R. D. (2019) Natural and synthetic sialylated glycan microarrays and their applications. Front. Mol. Biosci. 6, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Temme J. S., Campbell C. T., and Gildersleeve J. C. (2019) Factors contributing to variability of glycan microarray binding profiles. Faraday Discuss. 219, 90–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang L., Cummings R. D., Smith D. F., Huflejt M., Campbell C. T., Gildersleeve J. C., Gerlach J. Q., Kilcoyne M., Joshi L., Serna S., Reichardt N. C., Parera Pera N., Pieters R. J., Eng W., and Mahal L. K. (2014) Cross-platform comparison of glycan microarray formats. Glycobiology 24, 507–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Collins B. E., and Paulson J. C. (2004) Cell surface biology mediated by low affinity multivalent protein-glycan interactions. Curr. Opin. Chem. Biol. 8, 617–625 [DOI] [PubMed] [Google Scholar]
- 47. Dam T. K., and Brewer C. F. (2010) Multivalent lectin-carbohydrate interactions energetics and mechanisms of binding. Adv. Carbohydrate Chem. Biochem. 63, 139–164 [DOI] [PubMed] [Google Scholar]
- 48. Fujihashi M., Peapus D. H., Kamiya N., Nagata Y., and Miki K. (2003) Crystal structure of fucose-specific lectin from Aleuria aurantia binding ligands at three of its five sugar recognition sites. Biochemistry 42, 11093–11099 [DOI] [PubMed] [Google Scholar]
- 49. Wimmerova M., Mitchell E., Sanchez J. F., Gautier C., and Imberty A. (2003) Crystal structure of fungal lectin: six-bladed beta-propeller fold and novel fucose recognition mode for Aleuria aurantia lectin. J. Biol. Chem. 278, 27059–27067 [DOI] [PubMed] [Google Scholar]
- 50. Krishnan P., Singla A., Lee C. A., Weatherston J. D., Worstell N. C., and Wu H. J. (2017) Hetero-multivalent binding of cholera toxin subunit B with glycolipid mixtures. Colloids Surf. B Biointerfaces 160, 281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Worstell N. C., Singla A., Saenkham P., Galbadage T., Sule P., Lee D., Mohr A., Kwon J. S.-I., Cirillo J. D., and Wu H.-J. (2018) Hetero-multivalency of Pseudomonas aeruginosa lectin LecA binding to model membranes. Sci. Rep. 8, 8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gade M., Alex C., Leviatan Ben-Arye S., Monteiro J. T., Yehuda S., Lepenies B., Padler-Karavani V., and Kikkeri R. (2018) Microarray analysis of oligosaccharide-mediated multivalent carbohydrate-protein interactions and their heterogeneity. Chembiochem. 10.1002/cbic.201800037 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Godula K., and Bertozzi C. R. (2012) Density variant glycan microarray for evaluating cross-linking of mucin-like glycoconjugates by lectins. J. Am. Chem. Soc. 134, 15732–15742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Parera Pera N., Branderhorst H. M., Kooij R., Maierhofer C., van der Kaaden M., Liskamp R. M., Wittmann V., Ruijtenbeek R., and Pieters R. J. (2010) Rapid screening of lectins for multivalency effects with a glycodendrimer microarray. Chembiochem. 11, 1896–1904 [DOI] [PubMed] [Google Scholar]
- 55. Narla S. N., and Sun X. L. (2012) Glyco-macroligand microarray with controlled orientation and glycan density. Lab. Chip. 12, 1656–1663 [DOI] [PubMed] [Google Scholar]
- 56. Worstell N. C., Singla A., and Wu H. J. (2019) Evaluation of hetero-multivalent lectin binding using a turbidity-based emulsion agglutination assay. Colloids Surf B Biointerfaces 175, 84–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Guo Y., Nehlmeier I., Poole E., Sakonsinsiri C., Hondow N., Brown A., Li Q., Li S., Whitworth J., Li Z., Yu A., Brydson R., Turnbull W. B., Pohlmann S., and Zhou D. (2017) Dissecting multivalent lectin-carbohydrate recognition using polyvalent multifunctional glycan-quantum dots. J. Am. Chem. Soc. 139, 11833–11844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Oyelaran O., Li Q., Farnsworth D. W., and Gildersleeve J. C. (2009) Microarrays with varying carbohydrate density reveal distinct subpopulations of serum antibodies. J. Proteome Res. 8, 3529–3538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Purohit S., Li T., Guan W., Song X., Song J., Tian Y., Li L., Sharma A., Dun B., Mysona D., Ghamande S., Rungruang B., Cummings R. D., Wang P. G., and She J. X. (2018) Multiplex glycan bead array for high throughput and high content analyses of glycan binding proteins. Nat. Commun. 9, 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yan M., Zhu Y., Liu X., Lasanajak Y., Xiong J., Lu J., Lin X., Ashline D., Reinhold V., Smith D. F., and Song X. (2019) Next-generation glycan microarray enabled by DNA-coded glycan library and next-generation sequencing technology. Anal. Chem. 91, 9221–9228 [DOI] [PubMed] [Google Scholar]
- 61. Gray C. J., Sanchez-Ruiz A., Sardzikova I., Ahmed Y. A., Miller R. L., Reyes Martinez J. E., Pallister E., Huang K., Both P., Hartmann M., Roberts H. N., Sardzik R., Mandal S., Turnbull J. E., Eyers C. E., and Flitsch S. L. (2017) Label-free discovery array platform for the characterization of glycan binding proteins and glycoproteins. Anal. Chem. 89, 4444–4451 [DOI] [PubMed] [Google Scholar]
- 62. Rosencrantz R. R., Nguyen V. H., Park H., Schulte C., Böker A., Schnakenberg U., and Elling L. (2016) Lectin binding studies on a glycopolymer brush flow-through biosensor by localized surface plasmon resonance. Anal. Bioanal. Chem. 408, 5633–5640 [DOI] [PubMed] [Google Scholar]
- 63. Shams-Ud-Doha K., Kitova E. N., Kitov P. I., St-Pierre Y., and Klassen J. S. (2017) Human milk oligosaccharide specificities of human galectins. comparison of electrospray ionization mass spectrometry and glycan microarray screening results. Anal. Chem. 89, 4914–4921 [DOI] [PubMed] [Google Scholar]
- 64. El-Hawiet A., Kitova E. N., and Klassen J. S. (2012) Quantifying carbohydrate-protein interactions by electrospray ionization mass spectrometry analysis. Biochemistry 51, 4244–4253 [DOI] [PubMed] [Google Scholar]
- 65. El-Hawiet A., Shoemaker G. K., Daneshfar R., Kitova E. N., and Klassen J. S. (2011) Applications of a catch and release electrospray ionization mass spectrometry assay for carbohydrate library screening. Anal. Chem. 84, 50–58 [DOI] [PubMed] [Google Scholar]
- 66. Kitov P. I., Kitova E. N., Han L., Li Z., Jung J., Rodrigues E., Hunter C. D., Cairo C. W., Macauley M. S., and Klassen J. S. (2019) A quantitative, high-throughput method identifies protein-glycan interactions via mass spectrometry. Commun. Biol. 2, 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Palma A. S., Liu Y., Zhang H., Zhang Y., McCleary B. V., Yu G., Huang Q., Guidolin L. S., Ciocchini A. E., Torosantucci A., Wang D., Carvalho A. L., Fontes C. M., Mulloy B., Childs R. A., Feizi T., and Chai W. (2015) Unravelling glucan recognition systems by glycome microarrays using the designer approach and mass spectrometry. Mol. Cell Proteomics 14, 974–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Palma A. S., Feizi T., Childs R. A., Chai W., and Liu Y. (2014) The neoglycolipid (NGL)-based oligosaccharide microarray system poised to decipher the meta-glycome. Curr. Opin. Chem. Biol. 18, 87–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yu Y., Lasanajak Y., Song X., Hu L., Ramani S., Mickum M. L., Ashline D. J., Prasad B. V., Estes M. K., Reinhold V. N., Cummings R. D., and Smith D. F. (2014) Human milk contains novel glycans that are potential decoy receptors for neonatal rotaviruses. Mol. Cell Proteomics 13, 2944–2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Song X., Ju H., Lasanajak Y., Kudelka M. R., Smith D. F., and Cummings R. D. (2016) Oxidative release of natural glycans for functional glycomics. Nat. Methods 13, 528–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang Z., Chinoy Z. S., Ambre S. G., Peng W., McBride R., de Vries R. P., Glushka J., Paulson J. C., and Boons G. J. (2013) A general strategy for the chemoenzymatic synthesis of asymmetrically branched N-glycans. Science 341, 379–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wu Z., Liu Y., Li L., Wan X. F., Zhu H., Guo Y., Wei M., Guan W., and Wang P. G. (2017) Decoding glycan protein interactions by a new class of asymmetric N-glycans. Org Biomol. Chem. 15, 8946–8951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Liu L., Prudden A. R., Capicciotti C. J., Bosman G. P., Yang J. Y., Chapla D. G., Moremen K. W., and Boons G. J. (2019) Streamlining the chemoenzymatic synthesis of complex N-glycans by a stop and go strategy. Nat. Chem. 11, 161–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Nycholat C. M., McBride R., Ekiert D. C., Xu R., Rangarajan J., Peng W., Razi N., Gilbert M., Wakarchuk W., Wilson I. A., and Paulson J. C. (2012) Recognition of sialylated poly-N-acetyllactosamine chains on N- and O-linked glycans by human and avian influenza A virus hemagglutinins. Angew Chem. Int. Ed Engl. 51, 4860–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Song X., Yu H., Chen X., Lasanajak Y., Tappert M. M., Air G. M., Tiwari V. K., Cao H., Chokhawala H. A., Zheng H., Cummings R. D., and Smith D. F. (2011) A sialylated glycan microarray reveals novel interactions of modified sialic acids with proteins and viruses. J. Biol. Chem. 286, 31610–31622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Padler-Karavani V., Song X., Yu H., Hurtado-Ziola N., Huang S., Muthana S., Chokhawala H. A., Cheng J., Verhagen A., Langereis M. A., Kleene R., Schachner M., de Groot R. J., Lasanajak Y., Matsuda H., Schwab R., Chen X., Smith D. F., Cummings R. D., and Varki A. (2012) Cross-comparison of protein recognition of sialic acid diversity on two novel sialoglycan microarrays. J. Biol. Chem. 287, 22593–22608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ruprecht C., Bartetzko M. P., Senf D., Dallabernadina P., Boos I., Andersen M. C. F., Kotake T., Knox J. P., Hahn M. G., Clausen M. H., and Pfrengle F. (2017) A synthetic glycan microarray enables epitope mapping of plant cell wall glycan-directed antibodies. Plant Physiol. 175, 1094–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Stowell S. R., Arthur C. M., McBride R., Berger O., Razi N., Heimburg-Molinaro J., Rodrigues L. C., Gourdine J. P., Noll A. J., von Gunten S., Smith D. F., Knirel Y. A., Paulson J. C., and Cummings R. D. (2014) Microbial glycan microarrays define key features of host-microbial interactions. Nat. Chem. Biol. 10, 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Geissner A., Reinhardt A., Rademacher C., Johannssen T., Monteiro J., Lepenies B., Thepaut M., Fieschi F., Mrazkova J., Wimmerova M., Schuhmacher F., Gotze S., Grunstein D., Guo X., Hahm H. S., Kandasamy J., Leonori D., Martin C. E., Parameswarappa S. G., Pasari S., Schlegel M. K., Tanaka H., Xiao G., Yang Y., Pereira C. L., Anish C., and Seeberger P. H. (2019) Microbe-focused glycan array screening platform. Proc. Natl. Acad. Sci. U.S.A. 116, 1958–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhang J., Chen C., Gadi M. R., Gibbons C., Guo Y., Cao X., Edmunds G., Wang S., Liu D., Yu J., Wen L., and Wang P. G. (2018) Machine-driven enzymatic oligosaccharide synthesis by using a peptide synthesizer. Angew Chem. Int. Ed Engl. 57, 16638–16642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Li T., Liu L., Wei N., Yang J. Y., Chapla D. G., Moremen K. W., and Boons G. J. (2019) An automated platform for the enzyme-mediated assembly of complex oligosaccharides. Nat. Chem. 11, 229–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Grant O. C., Smith H. M., Firsova D., Fadda E., and Woods R. J. (2014) Presentation, presentation, presentation! Molecular-level insight into linker effects on glycan array screening data. Glycobiology 24, 17–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sood A., Gerlits O. O., Ji Y., Bovin N. V., Coates L., and Woods R. J. (2018) Defining the specificity of carbohydrate-protein interactions by quantifying functional group contributions. J. Chem. Inf. Model 58, 1889–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bonnardel F., Mariethoz J., Salentin S., Robin X., Schroeder M., Perez S., Lisacek F., and Imberty A. (2019) UniLectin3D, a database of carbohydrate binding proteins with curated information on 3D structures and interacting ligands. Nucleic Acids Res. 47, D1236–D1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Liu Y., McBride R., Stoll M., Palma A. S., Silva L., Agravat S., Aoki-Kinoshita K. F., Campbell M. P., Costello C. E., Dell A., Haslam S. M., Karlsson N. G., Khoo K. H., Kolarich D., Novotny M. V., Packer N. H., Ranzinger R., Rapp E., Rudd P. M., Struwe W. B., Tiemeyer M., Wells L., York W. S., Zaia J., Kettner C., Paulson J. C., Feizi T., and Smith D. F. (2016) The minimum information required for a glycomics experiment (MIRAGE) project: improving the standards for reporting glycan microarray-based data. Glycobiology 27, 280–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ashline D. J., Yu Y., Lasanajak Y., Song X., Hu L., Ramani S., Prasad V., Estes M. K., Cummings R. D., Smith D. F., and Reinhold V. N. (2014) Structural characterization by multistage mass spectrometry (MSn) of human milk glycans recognized by human rotaviruses. Mol. Cell Proteomics 13, 2961–2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Tappert M. M., Smith D. F., and Air G. M. (2011) Fixation of oligosaccharides to a surface may increase the susceptibility to human parainfluenza virus 1, 2, or 3 hemagglutinin-neuraminidase. J. Virol. 85, 12146–12159 [DOI] [PMC free article] [PubMed] [Google Scholar]