SUMMARY
The hippocampus formation, although prominently implicated in schizophrenia pathogenesis, has been overlooked in large-scale genomics efforts in the schizophrenic brain. We performed RNA-seq in hippocampi and dorsolateral prefrontal cortices (DLPFCs) from 551 individuals (286 with schizophrenia). We identified substantial regional differences in gene expression and found widespread developmental differences that were independent of cellular composition. We identified 48 and 245 differentially expressed genes (DEGs) associated with schizophrenia within the hippocampus and DLPFC, with little overlap between the brain regions. 124 of 163 (76.6%) of schizophrenia GWAS risk loci contained eQTLs in any region. Transcriptome-wide association studies in each region identified many novel schizophrenia risk features that were brain region-specific. Last, we identified potential molecular correlates of in vivo evidence of altered prefrontal-hippocampal functional coherence in schizophrenia. These results underscore the complexity and regional heterogeneity of the transcriptional correlates of schizophrenia and offer new insights into potentially causative biology.
In Brief
Collado-Torres et al. describe the BrainSeq Phase II gene expression resource encompassing two brain regions from 551 genotyped individuals spanning the entire human lifespan (286 with schizophrenia). This resource can answer region-specific questions about development and schizophrenia and its genetic risk.
INTRODUCTION
Schizophrenia is a psychiatric disorder that affects ~1% of the population worldwide and is linked to major socio-economic costs (Gore et al., 2011; Millan et al., 2016). As a highly heritable disorder (Gejman et al., 2010; Gottesman and Shields, 1982), it results from deficits in brain development and maturation, and it is typically diagnosed in young adults (Birnbaum and Weinberger, 2017; Messias et al., 2007). Different treatments exist that have various degrees of efficacy, mainly against so-called positive symptoms, although improving the treatment of these and other symptoms remains a crucial goal (Leucht et al., 2013; Millan et al., 2016).
Genetic studies play a critical role in understanding the etiology of neurodevelopmental disorders and informing the development of novel therapeutics to treat these diseases. Recent efforts by the Psychiatric Genomics Consortium (PGC) have identified well over 100 loci with a significant genome-wide association with risk for schizophrenia (Pardiñas et al., 2018; Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). However, translating an association between a locus and a disease into a therapeutic intervention is a challenging task; the biological importance of associated genetic variation must first be understood.
BrainSeq, a human brain genomics consortium consisting of seven pharmaceutical companies working pre-competitively with the Lieber Institute for Brain Development, was initiated with the goal of generating publicly available archival neurogenomic datasets (RNA sequence, genotype, and DNA methylation) in post-mortem brain tissue to enhance the understanding of psychiatric disorders (BrainSeq: A Human Brain Genomics Consortium, 2015). In the first phase of BrainSeq, we identified widespread genetic, developmental, and schizophrenia-associated changes in polyadenylated RNAs in the dorsolateral prefrontal cortex (DLPFC) (Jaffe et al., 2018) using poly(A)+ RNA sequencing (RNA-seq) to prioritize protein-coding changes in gene expression. Other research groups, in particular the CommonMind Consortium, have also focused on understanding gene expression and regulation in the schizophrenic brain (Fromer et al., 2016). However, in general, the transcriptional landscape of the hippocampus (HIPPO),another region prominently implicated in the pathogenesis of schizophrenia (Callicott et al., 1998; Rasetti et al., 2014; Weinberger, 1999), is much less explored because current large consortia have prioritized neocortical brain regions like the DLPFC (Fromer et al., 2016; Akbarian et al., 2015) despite differences in neuronal clonal organization (Xu et al., 2014) and the timing of their formation (Rice and Barone, 2000; Weinberger, 1999).
Here, in the second phase of the BrainSeq Consortium, we studied the expression differences between the DLPFC and HIPPO by performing RNA-seq using RiboZero libraries on 900 tissue samples across 551 individuals (286 with schizophrenia) in both the DLPFC (n = 453) and HIPPO (n = 447; Figures S1 and S2A). We quantified the expression of multiple feature summarizations of the Gencode v.25 reference transcriptome, including genes, exons, and splice junctions. These summarization methods are consistent with earlier work (Jaffe et al., 2018) and allow exploration of both the annotated and unannotated transcriptome. We compared expression within and across brain regions, modeled age-related changes in controls using linear splines, integrated genetic data to perform expression quantitative trait locus (eQTL) analyses, and performed differential expression analyses controlling for observed and latent confounders. By extending the experiment-based quality surrogate variable (qSVA) framework (Jaffe et al., 2017), we adjusted for RNA degradation across multiple brain regions and reduced false positives. We further leveraged the eQTL data in a transcriptome-wide association analysis (TWAS) to refine the summary statistics from the current largest genome-wide association study (GWAS) of schizophrenia (Pardiñas et al., 2018). We also explored, at the molecular level, evidence from in vivo imaging studies showing that the pattern of functional coherence between the DLPFC and HIPPO is altered in patients with schizophrenia. Together, these results shed light on the complex regional and molecular heterogeneity of schizophrenia-associated gene expression in the human brain.
RESULTS
The BrainSeq Phase II dataset was created to explore the differences in the regulation of brain gene expression during development and the possibility of dysregulation in schizophrenia (Figure S1; Experimental Model and Subject Details; RNA Sequencing). After processing the RNA-seq data, we applied extensive quality control procedures to identify low-quality samples and resolve potential sample swaps prior to filtering features with low expression across both brain regions (Figures S3 and S4; Method Details; RNA-Seq Processing Pipeline; Feature Filtering by Expression Levels; Genotype Data Processing; and Quality Control). We analyzed 24,652 genes, 396,583 exons, and 297,181 exon-exon junctions that were expressed across experiment-specific subsets of the 900 samples for all subsequent analyses.
Regional Differences in Expression between Brain Regions across Development
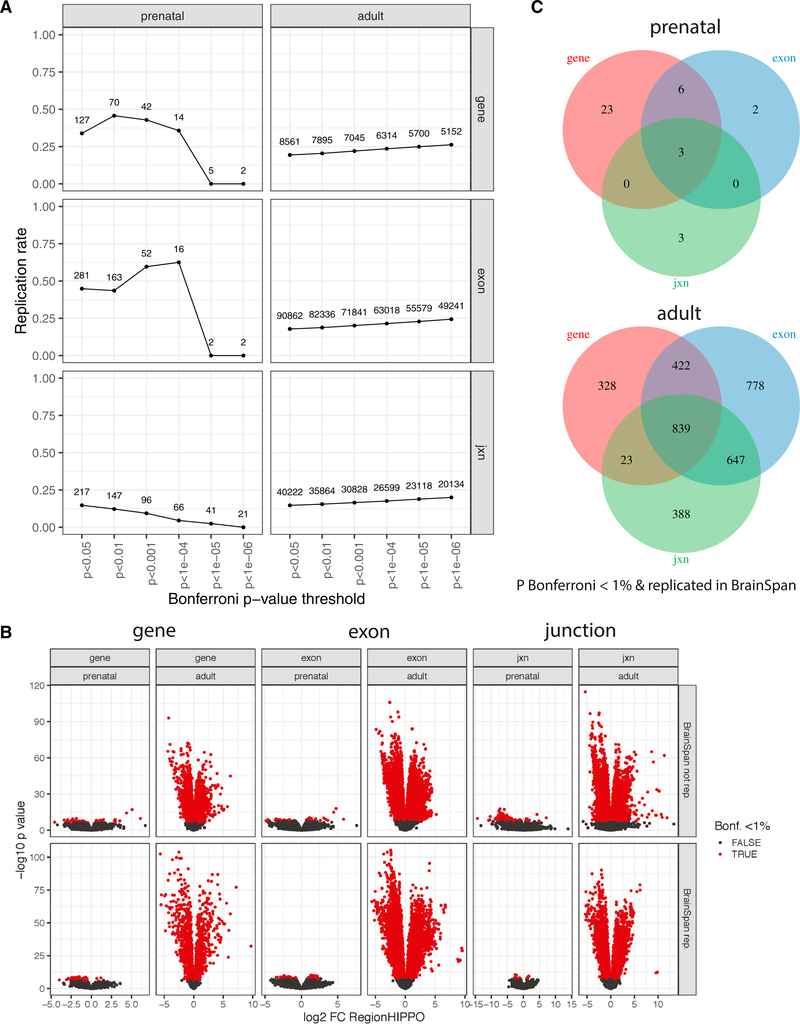
We first explored differences in expression between the DLPFC and HIPPO among neurotypical controls (Method Details; Differential Expression by Brain Region in Prenatal and Adult Samples). We compared the regions using both adult (age ≥ 18 years; range, 18 to 96 years) and prenatal (age < 0; range, 14 to 22 post-conception weeks) samples separately (Figure S2B; total adult, n = 460; prenatal, n = 57). We also used 26 subjects (adult, n = 8; prenatal, n = 18) across 52 samples from the same two regions available in the smaller BrainSpan project as a potential replication dataset (BrainSpan, 2011; Figure S2C). The replication rate of differentially expressed features was determined across several Bonferroni-adjusted p value thresholds while further requiring a consistent directionality of the expression differences (Figure 1A; Method Details; Replication Analysis with BrainSpan). Overall, there were large regional expression differences, particularly among adults by effect size, as shown in Figure 1. However, we noted that many of the strongest effects did not replicate in BrainSpan, which could be due to the smaller sample size in that dataset. Furthermore, the differences in dissection for the prenatal HIPPO between our study and BrainSpan may explain the lower concordance we observed in the prenatal age group compared with the adults (Figure S5). We did observe, however, higher replication rates when only considering directionality (Figure S6). This conservative analysis identified 1,612 and 32 genes differentially expressed between the DLPFC and HIPPO among the adult and prenatal age groups, respectively (Figure 1C) at Bonferroni-adjusted p < 0.01 and replicating in BrainSpan that were robust to differences in sample size, replication potential, and statistical power (Figure S7). Moreover, individual exons from a total of 2,686 genes and exon-exon junctions from 1,897 genes were differentially expressed between these two regions in adults (Figure 1C). Transcript-level results were mostly complementary in adults because only one differentially expressed transcript was detected in the prenatal age group (Figure S8). These differential expression results in adults reflect fewer reads spanning exon-exon junctions and differences in complexity between gene and exon read assignments and coverage, particularly using ribosomal depletion. The modest differential expression results for the prenatal age group could, at least in part, be due to the relatively small sample sizes in both BrainSeq Phase II and BrainSpan and also potentially due to less complete cortical maturation and differentiation in the fetal samples.
Figure 1. Adult and Prenatal DLPFC and HIPPO Expression Differences.
(A) Replication rate with consistent fold change for the adult and prenatal differences between brain regions using the BrainSpan dataset (n = 16 for adult, n = 36 for prenatal) across Bonferroni p value thresholds for the exon, gene, and exon-exon junction features. The labels for each point denote the number of features considered at the given Bonferroni p value threshold.
(B) Volcano plots of the differential expression signal between brain regions for each feature type and age group stratified by whether the differential expression replicated in BrainSpan (p < 0.05 and consistent fold change).
(C) Differentially expressed features grouped by gene ID for the adult and prenatal age groups.
See also Figures S5–S10 and Tables S11 and S14.
We then explored gene ontology and pathway analyses in both adult and prenatal samples. In the adult samples, enriched Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways for differentially expressed genes (DEGs) include biological processes such as axonogenesis and neurogenesis that are consistently observed across the three main expression features (Figure S9A) with higher expression in the DLPFC than in the HIPPO. We also observed DEG enrichment for ion channel activity and binding molecular functions (Figure S9B), synaptic membrane and transporter complex cellular components (Figure S9C), and GABAergic and cholinergic synapse pathways (Figure S9D; exons and junctions only). In the prenatal age group, although overall there were fewer enriched GO terms and KEGG pathways (Figure S10) for the regional DEGs, likely because there were fewer DEGs, increased positive regulation of neurogenesis (Figure S10A) as well as relatively increased basal transcription machinery binding (Figure S10B), nuclear chromatin (Figure S10C), and biosynthesis of amino acids (Figure S10D) were found in the HIPPO compared with the DLPFC. These findings are likely consistent with the earlier neuronal maturation of HIPPO compared with the DLPFC (Rice and Barone, 2000). These analyses suggest that expression landscapes in the HIPPO and DLPFC begin to diverge in prenatal life and diverge further across brain development and aging.
RNA Deconvolution and Developmental Differences between Brain Regions
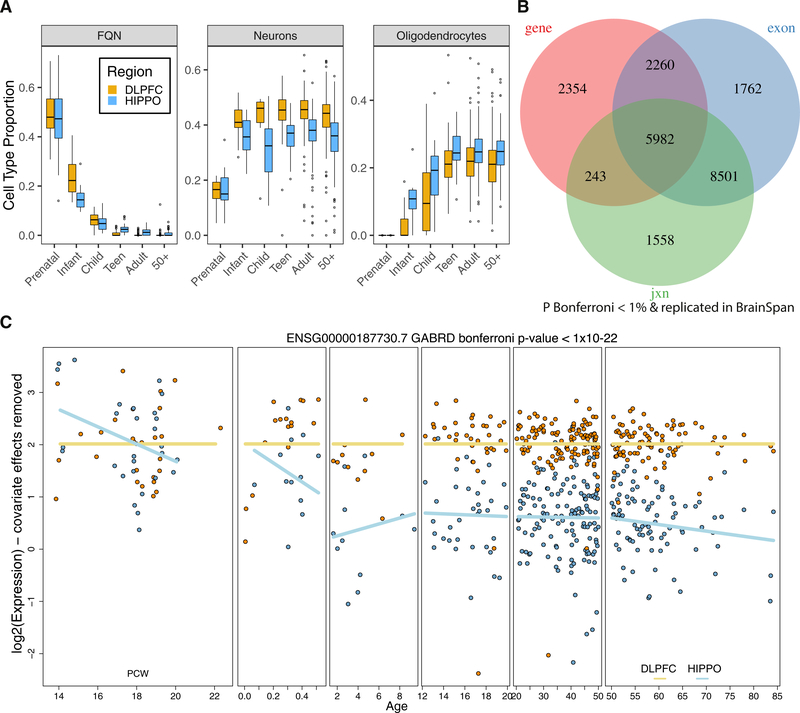
We hypothesized that these differences in expression between brain regions could arise from differences in the cellular components of these regions. We therefore used independent single-cell RNA-seq data (Darmanis et al., 2015) as well as DNA methylation data to estimate the RNA and cellular proportions for various main cell types, including fetal quiescent neurons, neurons, and oligodendrocytes (Method Details; RNA Deconvolution; DNA Methylation). We identified no difference in RNA fractions between the DLPFC and HIPPO within the prenatal age group. However, the RNA fractions associated with neurons and oligodendrocytes became significantly different in postnatal life, involving relatively decreased oligodendrocytes and increased neurons in the DLPFC compared with the HIPPO (Figure 2A; Figure S11). These findings support the results from the above DEGs between the two brain regions when comparing the prenatal and adult age groups (Figure 1C). We further hypothesized that the oligodendrocyte RNA fraction could serve as a proxy of maturation reflected in myelinogenic activity and, indeed, found increased fractions in the HIPPO compared with the DLPFC between the ages of 0 and 20 (Figure 2A; p = 5.48e–4), providing additional evidence for the earlier maturation of the HIPPO in comparison with the DLPFC. These results also echo archival data about myelogenic cycles in the postnatal cortex based on tissue myelin staining (Yakovlev and Lecours, 1967).
Figure 2. Differences across Development between the DLPFC and HIPPO.
(A) Estimated cell type RNA fractions for fetal quiescent neurons (FQNs), neurons, and oligodendrocytes. See Figure S11A for the other five cell type RNA fractions estimated with RNA deconvolution.
(B) Differentially expressed features between brain regions across development grouped by gene ID.
(C) GABRD is differentially expressed between the DLPFC and HIPPO across development (Bonferroni p value < 1 × 10−22). The y axis shows the residualized expression after removing modeled covariates. PCW, post-conception week.
See also Figures S11–S19 and Tables S1, S11, and S14.
Although there are large effects of cellular composition on the developing and aging brain within a brain region (Jaffe et al., 2016, 2018), it was unclear how these RNA fractions associate with expression trajectories contrasting the two brain regions. Sensitivity analyses within the regional gene expression differences within adult life suggested potentially independent effects of regional differences and cellular composition; 93.4% of genes that were different by brain region (that also replicated in BrainSpan) retained significance when further adjusting for the above RNA fractions. We therefore partitioned age into six age groups and used a linear spline model to identify changes in expression between the DLPFC and HIPPO across development and aging among all neurotypical controls (Figure S2B; DLPFC, n = 300; HIPPO, n = 314). We Bonferroni-adjusted the p values and used BrainSpan (Figure S2C; n = 79) to survey the features that replicated in this external dataset (p < 0.05). Exploration of the top 8 principal components (PCs) showed that brain region, pre- versus post-natal age (Figure S12; PC1 mean, 20.5%; PC2 mean, 10.9%) and sex (Figure S13) represented the main sources of variation across all feature types, whereas race was not associated with the top 8 PCs (Figure S14). The BrainSpan dataset presented similar patterns (age, Figure S15; sex, Figure S16; race, Figure S17), and we included these and other covariates as adjustment variables in the model (Method Details; Differential Expression over Development).
We identified widespread differences in transcriptional regulation between the DLPFC and HIPPO across development, with 10,839 genes differentially expressed between these regions, dependent on age period (Bonferroni < 0.01), that are nominally replicated in BrainSpan (p < 0.05; Figure 2B). Of these genes, 5,982 (55%) contained differentially expressed exons and splice junctions that replicated in BrainSpan (Method Details; Replication Analysis with BrainSpan). For example, GABRD (Figure 2C), which encodes a subunit of the major inhibitory neurotransmitter receptor in the mammalian brain, shows decreased expression in the HIPPO compared with the DLPFC in several of the age groups. We evaluated whether these age trajectory differences were sensitive to differences in RNA composition across the two regions. We found that the majority of genes still had different expression trajectories when further adjusting for composition (93.1%), showing no strong evidence for confounding by RNA composition, just like we found for the regional differences. We hypothesized that these contrasts would be more confounded by differences in trajectories of RNA fractions across age (i.e., slopes) rather than by differences by region alone (i.e., intercepts), and this hypothesis was largely supported by the data (Table S1; Figure S18). Enriched GO terms among DEGs dependent on development at the gene, exon, or exon-exon junction levels include dendrite development biological processes as well as synaptic membrane and postsynaptic density cellular components (Figure S19). These results suggest unique developmental profiles of expression in the HIPPO and DLPFC and likely further associate with the shifting cellular landscapes during development in each brain region and echo earlier reports (Sousa et al., 2017), suggesting that cortical gene expression during fetal life reflects less differentiation of cell types and connectivity patterns compared with the adult cortex.
Unique Schizophrenia-Associated Expression Differences in the HIPPO Compared with the DLPFC
Given the different expression levels and developmental regulation between the DLPFC and HIPPO, we next asked whether different genes were associated with schizophrenia diagnosis in the two regions. As we have shown previously (Jaffe et al., 2017), RNA degradation is a major, if not universal, confounder in schizophrenia disorder (SCZD) cases versus neurotypical control comparisons, in part because of different ante- and postmortem conditions that are not fully addressed by adjusting for observable quality metrics (e.g., RNA integrity number [RIN], mitochondrial mapping rate, pH, etc.) (Figures S20A and S20B). A more effective paradigm to adjust for postmortem RNA degradation is use of the qSVA framework, which is based on an ex vivo RNA degradation experiment (Jaffe et al., 2017). We modified the qSVA framework so that it could be applied to more than one brain region by identifying a common set of expressed genomic regions (Collado-Torres et al., 2017) that are associated with degradation across both brain structures (Figure S21; Method Details; Degradation Data Generation; Determining Multi-region Quality Surrogate Variables). Exploratory data analysis led us to exclude 43 HIPPO samples prepared with a different sequencing kit as well as age <17 samples, given that there are no SCZD cases in that age range in the BrainSeq Phase II dataset (Figures S20C–S20F and S2D). We identified 15 and 16 quality surrogate variables (qSVs) for the DLPFC and HIPPO, respectively, and 22 when using both regions together. The top qSV for each brain region was similarly associated with RIN and SCZD diagnosis (Figure S22) as well as with other technical covariates. Although the endothelial RNA fraction was significantly different by SCZD diagnosis in both brain regions (Bonferroni-adjusted-p value [p-bonf] < 5%; Figure S23), the quality surrogate variables are associated (p-bonf < 5%) with multiple RNA fractions (Figure S24) and thus adjust for the cell fractions. Degradation quality plots for each brain region suggest that the adapted qSVA workflow is effective in substantially reducing the confounding effects of RNA degradation on the SCZD case control analysis (Figure S25; Method Details; DEqual Plots). For example, a univariate model generated 6,429 DEGs by SCZD status in the HIPPO at a false discovery rate (FDR) of less than 5% while a model that does not adequately control for degradation—i.e., one based only on observed quality control metrics (e.g., RIN, pH, mitochondrial [mito] mapping rate)—still showed positive correlation between SCZD and degradation susceptibility.
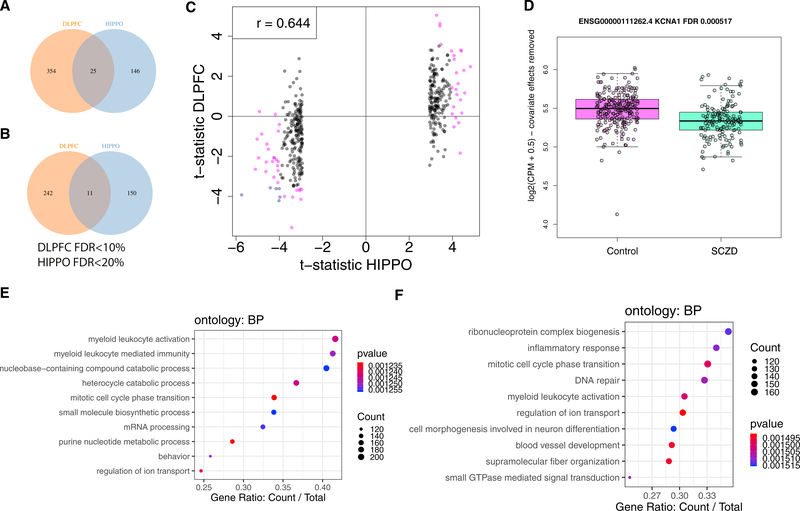
In contrast, when using qSVA, in the HIPPO, we identified only 48 significantly DEGs between patients and controls (27 control > SCZD, down; 21 control < SCZD, up), whereas, in the DLPFC, we identified 245 genes significantly differentially expressed (142 control > SCZD, down; 103 control < SCZD, up) at FDR < 5%. The number of these genes scaled with the FDR because there were 101 genes (52 down, 49 up) in the HIPPO and 632 in the DLPFC (379 down, 253 up) at FDR < 10%. We considered a more liberal set of DEGs in the HIPPO (FDR < 20%, n = 332, 171 down, 161 up) than in the DLPFC (FDR <10%, n = 632) for gene set enrichment analyses to ensure a sufficient number of input genes for inference. Interestingly, among the two sets of genes associated with differential expression based on diagnosis in the two regions, there was remarkably little overlap at different feature levels (Figures 3A and 3B for genes; Figure S26 for other expression features) and also little overlap across expression features (Figure S27) when grouped by Gencode gene ID.
Figure 3. SCZD Differential Expression in the DLPFC and HIPPO.
(A) Venn diagram of differentially expressed genes with higher expression in neurotypical controls than SCZD-affected individuals by brain region (DLPFC, FDR < 10%; HIPPO, FDR < 20%).
(B) Similar to (A) but for genes with higher expression in SCZD than in neurotypical controls.
(C) t-statistics for the top 400 differentially expressed genes in the HIPPO compared with their t-statistics in the DLPFC. Pink points, FDR < 5% in the DLPFC or HIPPO; blue points, FDR < 5% in both brain regions.
(D) KCNA1 is differentially expressed in the DLPFC (FDR = 0.000517).
(E and F) Gene set enrichment analysis (GSEA) on the DLPFC (E) with biological process ontology terms and HIPPO (F). Count, number of enriched genes for the given ontology term; gene ratio, ratio of genes enriched for the ontology term among the number of genes that are contained in the given ontology term. Terms are colored by their p value of enrichment.
See also Figures S24 and S26–S35 and Tables S2, S4, S5, S11, and S14.
We used previously published data from BrainSeq Phase I (BrainSeq P1) and the CommonMind Consortium (CMC) to examine replication of these differences in the DLPFC, which could specifically assess differences in library preparation protocols (poly(A)+ versus RiboZero) for a largely overlapping sample set (BrainSeq P1) and independently sequenced subjects in different labs (CMC), respectively. At the gene level, the DLPFC data significantly correlate (ρ = 0.81) with the DLPFC BrainSeq P1 results based on poly(A)+ (Figure S28) as well as the CMC dataset based on RiboZero (ρ = 0.53; Figure S28B), which we had previously re-processed for a more appropriate comparison (Fromer et al., 2016; Jaffe et al., 2018). In contrast with the lack of regional overlap among “significantly” DEGs, the t-statistics for the top 400 DEGs by SCZD status in the HIPPO correlate with the t-statistics in the DLPFC (Figure 3C; ρ = 0.64). Similar comparisons against BrainSeq P1 (ρ = 0.70) and CMC (ρ = 0.18; Figure S28) show greater concordance with our DLPFC results than HIPPO results. Across all genes, the correlation is reduced between brain regions (ρ = 0.28, DLPFC versus HIPPO) and datasets (Figure S29). For example, among the genes identified in BrainSeq P1 at FDR < 10% that replicated in the CMC, KCNA1 is differentially expressed in the DLPFC but not in the HIPPO (Figure 3 D), despite BrainSeq P1 and the DLPFC having different RNA-seq library preparation protocols. The correlation between the DLPFC and HIPPO as well as the correlation of each brain region with the CMC increased when not adjusting for quality surrogate variables (Figure S30), further highlighting the initial confounding by degradation.
We performed gene set enrichment analyses at the gene level for DEGs based on diagnosis in each brain region, which did show more overlap in biological processes than individual DEGs had suggested. In both the DLPFC and HIPPO, we found that myeloid leukocyte activation and regulation of ion transport processes (Figures 3E and 3F; Figures S31A and S32A; Table S2) and peptidase and ATPase activity molecular functions (Figures S31B and S32B) were enriched. We also performed GO enrichment analyses (Figure S33) and primarily found enrichment across genes with decreased expression among SCZD cases for both brain regions. We further performed a series of sensitivity analyses to better identify biological correlates of these expression differences. First, we tested for differences in annotated gene classes to potentially better leverage the RiboZero libraries and found strong depletion of non-protein-coding RNAs (odds ratio, 0.13; p < 2.2e–16), in line with the relatively lower expression of these classes of RNAs (Table S3). Next, we identified enrichment of more prenatal-like expression in patients with schizophrenia compared with unaffected controls in these RiboZero-based data, in line with our previous observations from DLPFC poly(A)+ data (Jaffe et al., 2018), with stronger effects in the DLPFC compared with the HIPPO (Table S4]; Figure S34). Given the differences in prevalence and symptoms of schizophrenia in males versus females (McGrath et al., 2008), we tested for enrichment of sex differences among these schizophrenia-associated expressed features and found no association (Table S5).
We identified 111 region-dependent DEGs (FDR < 5%) using interaction modeling between SCZD diagnosis status and brain region (Figure S35A; 424 with support at the gene, exon, or junction expression level), such as SCLO2A1 (Figure S35B), that are enriched for axonogenesis, axon development, postsynaptic density, and neuron-to-neuron synapse biological processes and cellular components (Figures S35C and S35D). These interaction DEGs, which showed differential effects across the two brain regions, had minimal overlap with either the SCZD DEGs in the HIPPO (4 of 48) or DLPFC (5 of 245). These results, in total, suggest largely unique DEGs and processes associated with schizophrenia in the HIPPO compared with the DLPFC and underscore the incompleteness of the molecular pathology associated with schizophrenia based on only the DLPFC or HIPPO. The enrichment of immune processes in those genes relatively decreased in expression in schizophrenia echoes several recent studies that have not confirmed earlier proposals of an upregulated immune response in the brain of patients with this illness (Birnbaum et al., 2018; Plavén-Sigray et al., 2018).
Decreased Regional Coherence in Schizophrenia via Co-expression Analysis
The clinical imaging research literature contains a number of reports of altered patterns of correlated measures of the HIPPO and DLPFC in SCZD subjects compared with neurotypical controls (Meyer-Lindenberg et al., 2001; Weinberger et al., 1992). Evidence of altered coherence of gene expression across these regions in schizophrenia has not been explored previously. Using the subset of individuals with data in both brain regions (n = 265 subjects, 530 RNA-seq samples), we first computed correlations across individuals for each gene to determine how genes were co-expressed across these brain regions. Genes most consistently co-expressed (at family-wise error rate [FWER] < 5%) across these regions were enriched for GO terms related to the immune response, response to virus, cytokine activity (Figures S36A–S36C), and cytokine-cytokine receptor interaction KEGG pathways (Figure S36D), suggesting that immune processes are particularly coherent across these brain regions within individuals and depleted for housekeeping gene status (odds ratio, 0.4; p = 0.011; Table S6). Interestingly, there was no association between gene co-expression across brain regions and subsequent differential expression within either region in schizophrenia, supporting the regional heterogeneity across brain region by diagnosis described above (Figure S37).
We next assessed decreased coherence at the subject rather than gene level across the entire transcriptome, which would be more comparable with identifying decreased connectivities via neuroimaging and other studies (Friston et al., 2016). We found that SCZD-affected individuals had significantly lower transcriptome-wide correlation across the DLPFC and HIPPO for all expression feature types we assessed (Figure S38; gene-level p = 0.0164), which was more robust than differences by sex (Figure S39; gene-level p = 0.15). Interestingly, although the 48 SCZD DEGs in the HIPPO were associated with decreased correlation across brain region (Figure S40A; p = 1.89 × 10−9), no other differentially expressed features in our data or previous datasets (DLPFC, BrainSeq P1, and CMC) showed decreased correlation (Figures S40B–S40D). Furthermore, leaving out those 48 DEGs in the HIPPO did not alter the decreased correlation across all expressed features (p = 0.0181), suggesting that the global decreased correlations were not driven by DEGs. We further refined these analyses by computing more specific individual-level correlations for sets of genes defined by biological functions (grouped by GO term and KEGG pathways). We identified 10 cellular component, 14 biological process, and 3 molecular function GO terms as well as 13 KEGG pathways with significant correlation differences (at FDR < 5%) between SCZD-affected individuals and neurotypical controls (Table S7). Consistent with the dysconnection hypothesis and prior evidence from neuroimaging studies (Friston et al., 2016; Meyer-Lindenberg et al., 2001; Weinberger et al., 1992), 35 of the 40 (87.5%) enriched terms and pathways had decreased correlation in SCZD-affected individuals (p = 4.53 × 10−6, χ12 = 20.0; Table S7). We highlight gene sets involving the dendritic spine neck (Figure S41A) and positive regulation of long-term synaptic depression (Figure S41B) having decreased coherence in SCZD-affected individuals, which have been previously linked to schizophrenia (Crabtree and Gogos, 2014; Hasan et al., 2012; Penzes et al., 2011). We also found that coherent activation of the immune response (Figure S41C) across regions was also decreased in SCZD individuals, in agreement with our differential expression analyses among individual features. At the pathway level, the lysosome pathway (Figure S41D) is among the pathways with decreased correlation, which is in agreement with previous studies that identified the lysosome as one of the components with dysregulated function in schizophrenia (Zhao et al., 2015). These findings provide novel insight into the potential molecular mechanisms underlying decreased functional coherences between the hippocampus and frontal cortex in schizophrenia.
Differences in Genetic Regulation of Expression between Brain Regions
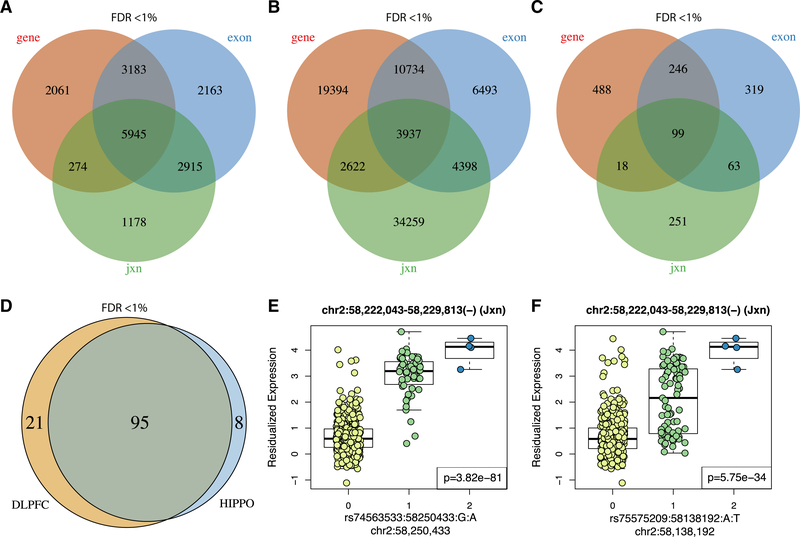
To explore the transcriptional effect of genetic risk alleles found in GWASs (Pardiñas et al., 2018; Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014), we combined genotype data with the RNA-seq data to assess the overlap in genetic regulation by brain region. We identified eQTLs for each of the expression features and brain regions (Method Details;eQTLs) using individuals with age > 13 (n = 477). In the HIPPO, we found 11,237,357 eQTL associations (SNP-feature pairs at FDR < 1%) across genes, exons, and junctions, corresponding to 17,719 genes (Figure 4A; Figure S42A). In contrast, in the DLPFC, we found 15,766,398 eQTLs for 19,482 genes across the same spectrum of expression features, which were largely replicated in an analysis of previous DLPFC poly(A)+ data in BrainSeq P1 (Table S8). We then performed joint analyses in the DLPFC and HIPPO to identify 205,618 (at FDR < 1%) brain region-dependent eQTLs (i.e., statistical interaction between genotype and region; Figure 4B; Figure S42B), corresponding to 1,484 genes (Figure 4C; Figure S42C) across all three expression feature types. These results show substantial regional specificity in genetic regulation of expression. Furthermore, the brain region-dependent eQTL associations involve genes that are enriched among DEGs between neurons and non-neurons in the brain (p = 3.601 × 10~−86; Method Details; Neuronal DEG Enrichment in Brain Region-Dependent eQTLs). The eQTL associations we identified showed low replication rates in GTEx v.6 (GTEx Consortium, 2015) when considering directionality and p < 0.05 but higher rates when only considering directionality (HIPPO, 31.9% versus 69.9%; DLPFC, 32.5% versus 70%; brain region-dependent, 24.6% versus 67.4% at the gene level; Table S8), likely because of the much smaller sample sizes (GTEx, 92 and 82; BrainSeq P2, 397 and 395 for the DLPFC and HIPPO, respectively). We further verified the statistical power gains by using a mixed-population eQTL analysis compared with subsetting to a single self-reported race (Method Details; CAUC-Only Sensitivity Analysis; Table S8).
Figure 4. DLPFC and HIPPO eQTLs and Schizophrenia Risk.
(A) HIPPO eQTLs grouped by gene ID.
(B) Region-dependent eQTLs grouped by SNP ID.
(C) Region-dependent eQTLs group by gene ID.
(D) Unique risk schizophrenia GWAS index SNPs from PCG2 (Pardiñas et al., 2018) by brain region that are in eQTLs.
(E) Top HIPPO eQTLs among schizophrenia risk SNPs. This exon-exon junction skips exon 4/14 of the FANCL gene.
(F) Index schizophrenia risk SNP corresponding to the proxy SNP from (E).
eQTL p values are shown in (E) and (F).
See also Figures S42–S44 and Tables S8, S11, S14, S15, and S16.
We explored whether any of these eQTLs were previously identified schizophrenia GWAS risk variants (Pardiñas et al., 2018). Among the HIPPO eQTLs, we found eQTL associations to 60 risk SNPs such as rs42945 and expression features from the NDRG4 gene (Figures S43A and S43B), which has been implicated previously in sudden cardiac death among users of antipsychotics in patients with schizophrenia (Watanabe et al., 2017). Similarly, 3 risk SNPs (rs4144797, rs12293670, and rs324015) were brain region-dependent eQTLs that involved features from genes LRP1, NRGN, and NGEF (Figures S44A–S44D) and have been identified previously as methylation QTL SNPs in the DLPFC (Jaffe et al., 2016). Although risk SNPs rs12293670 and rs4144797 were specific to the DLPFC, with a weak to no eQTL signal in the HIPPO, rs324015 showed opposing eQTL directionality by brain region in features in LRP1 (Figures S44A–S44D).
To more fully characterize region-specific eQTLs in the context of schizophrenia risk SNPs, we carried out a targeted eQTL analysis by using samples age >13 (Figure S42D, n = 477; Method Details; Risk Loci) and GWAS variants. Using rAggr (Edlund et al., 2017), we identified proxy SNPs that have a linkage disequilibrium (LD) R2 score of greater than 0.8 with the 179 index GWAS (Pardiñas et al., 2018) risk SNPs and used 163 risk loci that were common (minor allele frequency [MAF] > 5%) and well imputed in our genotype data (Figure S42E; Method Details; Risk Loci). Of these 163 risk loci, 124 (76%) were either eQTLs (FDR < 1%) in the HIPPO or DLPFC, with 95 variants (76.6% of the eQTLs) shared across both regions (Figure 4D). This percentage of GWAS-significant SNPs showing association with gene expression is more than 50% greater than in previous reports, again illustrating the incompleteness of eQTL knowledge when based on only limited surveys of brain regions. Overall, 5,510 and 6,780 of the 9,692 proxy and index SNPs were eQTLs (FDR < 1%) in the HIPPO and DLPFC, spanning 1,731 and 2,525 different expression features, respectively (Figure S42F). The top eQTL in the HIPPO corresponds to an exon-skipping event in the FANCL gene (Figure 4E) and involves proxy SNP rs74563533. The corresponding index SNP rs75575209 is also an eQTL (Figure 4F) with the same exon-exon junction (chr2:58,222,043–58,229,813 in hg38 coordinates). The eQTL involving this junction and proxy SNP rs74563533 was also the top result in the DLPFC in BrainSeq P1 (Jaffe et al., 2018) (p = 2.796e–85, chr2:58,449,178–58,456,948 in hg19 coordinates). FANCL has been linked to Fanconi anemia disease (Meetei et al., 2003) and is involved in the DNA repair pathway (Machida et al., 2006). From the 103 and 116 risk loci in these eQTLs, only 38 (36.9%) and 37 (31.9%) of the loci pair to a single gene in the HIPPO and DLPFC (Figure S42G). This finding demonstrates that, in both brain regions, roughly two-thirds of the risk loci (63.1% and 68.1%, respectively) are ambiguous to resolve. Through our eQTL browser resource (http://eqtl.brainseq.org/phase2/), we made all eQTLs sets available for further exploration: global HIPPO and DLPFC eQTLs, brain-region dependent eQTLs, and schizophrenia risk HIPPO and DLPFC eQTLs.
TWASs Identify Novel Schizophrenia Genetic Risk Associations
We next more formally integrated GWAS and eQTL statistics by performing TWAS (Gusev et al., 2016). We constructed SNP weights across the four feature summarizations (gene, exon, junction, and transcript) using both brain regions (DLPFC and HIPPO). We then applied these weights to the summary statistics from the entire collection of schizophrenia GWAS summary statistics from PGC2 and the Clozapine Clinic in the UK (CLOZUK), as described by Pardiñas et al. (2018). We identified widespread TWAS associations with genome-wide significant and marginally significant GWAS risk loci, including 8,185 features (538 genes, 4,258 exons, 2,297 junctions, and 1,092 transcripts) significantly associated with schizophrenia risk (at TWAS FDR < 5%) that were annotated to 2,044 unique gene IDs. At more stringent Bonferroni significance, there were 1,140 features (110 genes, 530 exons, 302 junctions, and 198 transcripts) in 333 gene IDs significantly associated with schizophrenia genetic risk (Table S9A). Because the TWAS approach combines GWAS and eQTL information, it is possible for a GWAS signal that does not reach GWAS genome-wide significance (p < 5e–8) to still achieve TWAS transcriptome-wide significance. We therefore annotated the strongest GWAS variant for each significant TWAS feature back to the clumped GWAS risk loci and found that 77.7% of the TWAS Bonferroni-significant features (n = 931) mapped back to the published GWAS risk loci. Although a sizable fraction of Bonferroni-significant TWAS features were outside of GWAS risk loci (n = 209 features, 18.3%), a much larger fraction of FDR-significant TWAS features identified potentially novel genes and corresponding GWAS loci implicated in schizophrenia (n = 5,789 features, 70.7% corresponding to 1,576 genes; 77.1%).
When comparing DLPFC and HIPPO, more features were heritable and had TWAS weights in the DLPFC (74,327 features spanning 11,421 genes) compared with the HIPPO (52,924 features spanning 9,949 genes; Table S9A). There were also more TWAS genome-wide significant (FDR < 5%) features associated with schizophrenia for the DLPFC (5,760 features spanning 1,513 genes) compared with the HIPPO (4,081 features spanning 1,254 genes). Only 1,656 features (20.2%) spanning 624 genes (30.5%) were significantly associated with schizophrenia in both brain regions. Although some features were region-specific in these TWAS analyses, the TWAS Z scores were highly correlated (0.86 to 0.93 by brain region, feature, and risk loci status) and concordant (only 2 were discordant) among the DLPFC and HIPPO for the features that did have TWAS weights in both brain regions (Figure S45).
Although becoming increasingly popular, we view the TWAS approach as complementary to the eQTL analyses we carried out on risk loci above. Of the 124 risk loci we identified with eQTLs, only 92 had SNP weights in TWASs (74.2%), resulting in 32 loci not even being considered in TWASs (Table S9B). Of these 92 risk loci with TWAS weights, 82 (89.1%) across both brain regions had a feature with a genome-wide significant TWAS signal at FDR < 5% (n = 61 for Bonferroni < 5%, 66.3%), suggesting concordance between TWAS and direct eQTL analysis. To assess the robustness and potential replication of our TWAS findings, we compared our TWAS weights applied to the PGC2 GWAS statistics (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014) with the TWAS published by Gusev et al. (2018). Gusev et al. (2018) identified 83 unique genes with TWAS Bonferroni < 5% across the CMC gene level and “CMC splicing” datasets in the DLPFC. Using our weights applied to the PGC2 GWAS, we replicated 67 (80.7%) with TWAS FDR < 5% and 36 (43.4%) with TWAS Bonferroni < 5% across the expression feature levels we assessed. Furthermore, of the 406 DLPFC genes with significant TWAS (FDR < 5%) associations we identified in this study, 336 (82.8%) were recently tested by Gandal et al. (2018) using the PGC2+CLOZUK GWAS (Pardiñas et al., 2018). Finally, we used these TWAS results to more formally test for association between schizophrenia genetic risk and illness state. We compared the TWAS Z scores across all heritable features with the corresponding differential expression t-statistic for schizophrenia in either brain region and found no correlation (Figure S47). These results suggest that genetic risk and illness state impart an orthogonal signal on the transcriptome, where the latter likely reflects the consequences and not causes of illness, inline with our previous findings (Jaffe et al., 2018). Nevertheless, these TWAS analyses across two brain regions identified specific expressed features that robustly associate with genetic risk for schizophrenia.
DISCUSSION
In this second phase of the BrainSeq consortium project, we generated and processed 900 RNA-seq samples from human postmortem brains from 551 individuals, 286 (52%) of whom were given the diagnosis of schizophrenia (Figure S1). We used the RiboZero library preparation method to preserve non-coding RNA and examined two brain regions, the HIPPO and DLPFC, expanding our previous analysis of 495 (175 with schizophrenia) DLPFC samples analyzed using poly(A)+ RNA-seq (Jaffe et al., 2018). This second phase of the BrainSeq consortium project expands the variety of analyses that can be carried out to further investigate differences among the two brain regions most consistently implicated in schizophrenia pathogenesis. This phase of the project also permits an examination of the trajectory of gene expression across development (Figure S1). Our results underscore the limitations of findings related to brain development and schizophrenia based on earlier gene expression data in the DLPFC alone. In particular, we find minimal overlap in DEGs between SCZD cases and controls common to both regions. We also find many eQTLS with unique regional patterns. Last, we find that TWAS identify unique schizophrenia genes uncovered by the gene expression differences between regions. By analyzing the expression data with multiple complementary annotation methods (gene, exon, exon-exon junction, transcript), we gleaned a more complete understanding of differences in the transcriptome (annotated and unannotated) between regions and cases versus neurotypical controls (Figure S1).
Perhaps not surprisingly, we found that expression in the HIPPO and DLPFC is more similar at a prenatal age than at an adult age. Using estimates from RNA deconvolution, we found widespread differences across development and cell RNA fraction. These differences were supported by cell type composition estimates derived from DNA methylation data from neurotypical controls. These results are important to understand whether expression differences associated with a given disorder are intrinsic to a specific brain region or related to underlying cell types. Given that SCZD cases generally have a longer postmortem interval and lower pH than neurotypical controls (Figures S20A and S20B) and that RNA degradation is a major and virtually universal confounder in SCZD case control analyses, we adapted the experiment-based qSVA framework (Jaffe et al., 2017) for more than one brain region (Figure S21). Using the qSVs for each brain region, we were able to substantially reduce the effect of RNA degradation in our SCZD case control analyses (Figure S25). By expanding the qSVA framework for two regions, we have laid the groundwork for analyzing multiple brain regions with this framework. This will require degradation experiments for additional brain regions.
Our analytical approach yielded 48 and 245 DEGs among SCZD cases and neurotypical controls in the HIPPO and DLPFC (FDR < 5%), respectively, with little overlap among these genes, even when studied with higher FDR thresholds or examining other expressed features. This finding, by itself, highlights the molecular heterogeneity of this disorder across these regions. We determined that genes that are significantly co-expressed between the HIPPO and DLPFC (FWER < 5%) independent of diagnosis status were enriched for immune processes but showed no relation with the SCZD case versus neurotypical control differential expression signal. This adds to the growing evidence that immune activation is not a characteristic of the schizophrenia brain (Birnbaum et al., 2018). Interestingly, at the individual subject level, we found that the HIPPO and DLPFC have significantly (p < 0.05) lower transcriptome-wide correlation at the gene, exon, exon-exon junction, and transcript levels (Figure S38) in SCZD cases compared with neurotypical controls, which may echo and could provide molecular insights into in vivo evidence of altered connectivity between these regions (Friston et al., 2016; Meyer-Lindenberg et al., 2001; Weinberger et al., 1992). To our knowledge, this is the first evidence showing that gene expression is less coherent between these regions in schizophrenia. Among the GO terms with significant correlation differences, we identified dendritic spine neck and positive regulation of long-term synaptic depression (Figures S41A–S41C), which had significantly decreased correlation in SCZD cases and have been linked previously to schizophrenia (Crabtree and Gogos, 2014; Hasan et al., 2012; Penzes et al., 2011). We further found more supporting evidence linking the lysosome pathway (Figure S41D) to SCZD, complementary to previous findings (Zhao et al., 2015).
Last, we characterized extensive genetic regulation of gene expression with significantly different effects across the two brain regions: we identified 205,618 brain region-dependent eQTLs (at FDR < 1%), corresponding to 1,484 genes, at the gene, exon, or exon-exon junction level. These brain region-dependent eQTLs included clinically relevant risk variants because five schizophrenia risk loci showed significant differential regional regulation. We also identified millions of eQTLs in both the HIPPO and DLPFC (FDR < 1%) across genes, exons, and exon-exon junctions. A sub-analysis focusing on schizophrenia GWAS risk loci showed that 124 of the 163 risk loci observed in our dataset contain eQTLs, with 95 (76% of the eQTLs) being shared across the HIPPO and DLPFC. Furthermore, we found that 6,970 risk SNPs (GWAS index and their proxies) are eQTLs in either brain region. In each brain region, over 60% of the risk loci are associated with more than one gene, highlighting the complexity of SNP association for this disorder and the potential difficulty in interpreting the eQTL results to identify risk genes.
Given the complexity and richness of these eQTL findings, we made all five sets of eQTL results (global and risk-focused for each brain region plus regional-dependent ones) publicly available via the user-friendly Lieber Institute for Brain Development (LIBD) eQTL browser at http://eqtl.brainseq.org/phase2/ to facilitate independent analyses. These genotype and expression datasets were more formally integrated to form TWAS weights for all expressed features, which extended previous analyses that were only performed on genes and splicing events. The use of all expressed features (exons, junctions, and transcripts), rather than just gene-level summaries, resulted in far more schizophrenia risk associations. Features in over 1,500 genes (73.7%) showed genome-wide significant associations that would have been missed doing only gene-level analyses. These TWAS weights can be downloaded and applied to any genetic dataset to impute expression of the DLPFC- and HIPPO-expressed features for integration into studies of living subjects, particularly when combined with clinical and neuroimaging data.
Overall, we showed extensive regional specificity of developmental and genetic regulation and schizophrenia-associated expression differences between the HIPPO and DLPFC. These findings suggest that some components of the transcriptional correlates of development and genetic risk for schizophrenia are brain region-specific. Thus, it may be necessary to incorporate regionally targeted therapies for schizophrenia or develop therapies directed at molecular pathology that is shared across brain regions.
Our resource across multiple brain regions further complements the recent publications from the first wave of the PsychENCODE consortium (Akbarian et al., 2015), particularly by Wang et al. (2018) and Gandal et al. (2018). These two capstone papers combined DLPFC data from over 1,800 brains, including ~500 from the first phase of our BrainSeq Consortium (Jaffe et al., 2018), and found widespread differential expression of genes associated with schizophrenia (4,821 genes) and other neuropsychiatric disorders as well as eQTLs to nearly every expressed gene. These landmark resources have created the new standard for interrogation of cortical RNA-seq data, and, indeed, our independently generated DLPFC samples here (which partially overlap the second phase of the CMC National Institute of Mental Health [NIHM] Human Brain Collection Core [HBCC] cohort) replicate many of the findings from these previous analyses. Our analytical framework of interrogating feature-level expression beyond genes and transcripts, like exons and splice junctions, and controlling for latent expression heterogeneity via qSVA (Jaffe et al., 2017) further complements these PsychENCODE efforts. Our new analyses of the HIPPO provides novel insights into the etiology and neurobiology of schizophrenia. These data and analysis methods, generated as part of the BrainSeq Phase II project, strengthen the foundation of our understanding of schizophrenia and enhance our ability to find new ways to improve the lives of individuals affected by this disease.
STAR⋆METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact: Andrew E. Jaffe (andrew.jaffe@libd.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Table S10 includes summary demographic information for the samples used in this study. Detailed individual level demographic information (including age and sex) is further provided in the RangedSummarizedExperiment objects (colData slot) listed under “Data and Software Availability.” See “Method Details: Postmortem brain tissue acquisition and processing” for information on where the human brain samples were collected from and how the brain tissue dissections were performed.
As previously described in Jaffe et al. (2016), postmortem human brain tissue was obtained by autopsy primarily from the Offices of the Chief Medical Examiner of the District of Columbia, and of the Commonwealth of Virginia, Northern District, all with informed consent from the legal next of kin (protocol 90-M-0142 approved by the NIMH/NIH Institutional Review Board). Additional post-mortem prenatal, infant, child, and adolescent brain tissue samples were provided by the National Institute of Child Health and Human Development Brain and Tissue Bank for Developmental Disorders (http://medschool.umaryland.edu/BTBank) under contracts NO1-HD-4–3368 and NO1-HD-4–3383. Postmortem human brain tissue was also provided by donation with informed consent of next of kin from the Office of the Chief Medical Examiner for the State of Maryland (under Protocol No. 12–24 from the State of Maryland Department of Health and Mental Hygiene) and from the Office of the Medical Examiner, Department of Pathology, Homer Stryker, M.D. School of Medicine (under Protocol No. 20111080 from the Western Institute Review Board). The Institutional Review Board of the University of Maryland at Baltimore and the State of Maryland approved the protocol, and the tissue was donated to the Lieber Institute for Brain Development under the terms of a Material Transfer Agreement. Clinical characterization, diagnoses, and macro- and microscopic neuropathological examinations were performed on all samples using a standardized paradigm, and subjects with evidence of macro- or microscopic neuropathology were excluded, as were all subjects with any psychiatric diagnoses. Details of tissue acquisition, handling, processing, dissection, clinical characterization, diagnoses, neuropathological examinations, and quality control measures were further described previously (Lipska et al., 2006). Postmortem tissue homogenates of the prefrontal cortex (dorsolateral prefrontal cortex, DLPFC, BA46/9) were obtained from all subjects.
All hippocampus dissections were performed by the same neuroanatomist (Thomas M. Hyde, M.D., Ph.D.) on all samples in this study. The hippocampus was dissected from the anterior tip posteriorly through to the mid body of the hippocampus at the level of the lateral geniculate nucleus. The dissection included the hippocampus proper (i.e., Ammons Horn and CA1–3) plus the subicular complex. The dissections were performed as uniformly as possible using anatomical landmarks and well as visual inspection of the hippocampus itself to guide dissections from the anterior pole of the hippocampus back through the mid body. The hippocampal formation in the fetal brain was dissected under visual guidance using a hand held dental drill. Briefly, the dorsal-medial aspect of the temporal lobe was removed, including the adjacent cortex, medial to the hippocampal sulcus. In the rostral-caudal axis, the dissection was performed at the mid-point of the Sylvian Assure.
METHOD DETAILS
RNA sequencing
Total RNA was extracted from samples using the RNeasy Lipid Tissue Mini Kit (QIAGEN). Paired-end strand-specific sequencing libraries were prepared from 300ng total RNA using the TruSeq Stranded Total RNA Library Preparation kit with Ribo-Zero Gold ribosomal RNA depletion for all DLPFC 453 samples and 43 adult HIPPO samples (https://www.illumina.com/products/selection-tools/rrna-depletion-selection-guide.html) which removes rRNA and mtRNA and 404 HIPPO samples were run with the Illumina TruSeq Stranded Total RNA Library Prep Human/Mouse/Rat (HMR) kit (https://www.illumina.com/products/selection-tools/rrna-depletion-selection-guide.html) which just removes rRNA (and not mtRNA). An equivalent amount of synthetic External RNA Controls Consortium (ERCC) RNA Mix 1 (Thermo Fisher Scientific) was spiked into each sample for quality control purposes. The libraries were sequenced on an Illumina HiSeq 3000 at the LIBD Sequencing Facility, after which the Illumina Real Time Analysis (RTA) module was used to perform image analysis and base calling and the BCL converter (CASAVA v1.8.2) was used to generate sequence reads, producing a mean of 125.2 million 100-bp paired-end reads per sample.
RNA-seq processing pipeline
Raw sequencing reads were quality checked with FastQC (Babraham Bioinformatics, 2016), and where needed leading bases were trimmed from the reads using Trimmomatic (Bolger et al., 2014) as appropriate. Quality checked reads were mapped to the hg38/GRCh38 human reference genome with splice-aware aligner HISAT2 version 2.0.4 (Kim et al., 2015). Feature-level quantification based on GENCODE release 25 (GRCh38.p7) annotation was run on aligned reads using featureCounts (subread version 1.5.0-p3) (Liao et al., 2014) with a mean 45.3% (SD = 7.4%) of mapped reads assigned to genes. Exon-exon junction counts were extracted from the BAM files using regtools (Feng et al., 2018) v.0.1.0 and the bed_to_juncs program from TopHat2 (Kim et al., 2013) to retain the number of supporting reads (in addition to returning the coordinates of the spliced sequence, rather than the maximum fragment range) as described in Jaffe et al. (2018). Annotated transcripts were quantified with Salmon version 0.7.2 (Patroet al., 2017) and the synthetic ERCC transcripts were quantified with Kallisto version 0.43.0 (Bray et al., 2016). For an additional QC check of sample labeling, variant calling on 740 common missense SNVs was performed on each sample using bcftools version 1.2. We generated strand-specific base-pair coverage BigWig files for each sample using bam2wig.py version 2.6.4 from RSeQC (Wang et al., 2012) and wigToBigWig version 4 from UCSC tools (Kent et al., 2010). Table S10 includes demographics for different subsets of samples and differences (if any) among technical covariates.
Genotype data processing
Genotype data were processed and imputed as previously described (Jaffe et al., 2018). Briefly, genotype imputation was performed on high-quality observed genotypes (removing low quality and rare variants) using the prephasing/imputation stepwise approach implemented in IMPUTE2 (Howie et al., 2009) and Shape-IT (Delaneau et al., 2008), with the imputation reference set from the full 1000 Human Genomes Project Phase 3 dataset (Auton et al., 2015), separately by Illumina platform using genome build hg19. We retained common variants (MAF >5%) that were present in the majority of samples (missingness < 10%) that were in Hardy Weinberg equilibrium (at p > 1×10−6) using the Plink tool kit version 1.90b3a (Purcell et al., 2007). Multidimensional scaling (MDS) was performed on the autosomal LD-independent construct genomic ancestry components on each sample, which can be interpreted as quantitative levels of ethnicity – the first component separated the Caucasian and African American samples. This processing and quality control steps resulted in 7,023,860 common variants in this dataset of 551 unique subjects. We remapped variants to hg38 first using the dbSNP database (Sherry et al., 2001) (from v142 on hg19 to v149 on hg38) and then the liftOver tool (Hinrichs et al., 2006) for unmapped variants (that were dropped in dbSNP v149). eQTL analyses were performed using the 7,023,286 variants with hg38 coordinates (574 variants did not liftOver).
Quality control
After completing the preprocessing pipeline, samples were checked for quality control measures. All samples had expected ERCC concentrations. Forty-two samples with poor alignment rates (< 70%), gene assignment rates (< 20%), and mitochondrial mapping rates (> 6%, DLPFC only) were dropped. Next, for each sample we correlated the genotypes of missense single nucleotide variants (SNVs) found in the processing pipeline to those same SNVs based on genotyping described above, and dropped ten samples based on possible incorrect sample labeling, where the genotypes were lowly correlated to the same sample number, or highly correlated do a different sample number. Ultimately 900 samples passed quality control checks (HIPPO n = 447, DLPFC n = 453).
Feature filtering by expression levels
We filtered lowly expressed features using the expression_cutoff() function from the jaffelab (Collado-Torres and Jaffe, 2017) package v0.99.18 resulting in cutoffs based on the mean expression across all 900 samples: RPKM 0.25 for genes, RPKM 0.30 for exons, RP10M 0.46 for exon-exon junctions and 0.32 TPM for transcripts (Figure S3). 24,652 genes (42.5%), 396,583 exons (69.4%), 297,181 exon-exon junctions (35.5%) and 92,732 transcripts (46.8%) passed the expression filters. Figure S4 shows the number of expressed features passing the expression cutoffs grouped by gene ids.
Differential expression by brain region in prenatal and adult samples
Using the neuropsychiatric control samples (Figure S2B; Table S10) for the adult age group (age > = 18, HIPPO n = 238, DLPFC n = 222) or the prenatal group (age < 0, HIPPO and DLPFC n = 28) we identified differentially expressed features by brain region while adjusting for age, technical covariates (mitochondrial mapping rate, total assigned gene rate, RIN) and ethnicity (first five principal components based on the genotype data) as shown in Equation 1. We used the voom method (Law et al., 2014) for genes, exons, and exon-exon junctions and calculated the t-statistics for β1 = 0 using limma (Ritchie et al., 2015) v3.34.5 while adjusting for repeated-measures using duplicateCorrelation(). Resulting p values were Bonferroni-adjusted within each feature type.
| (Equation 1) |
Equation 1. Full model for the differential expression analysis by brain region subsetted to specific age groups.
The full list and statistics for the differentially expressed features are available in Table S11.
DNA methylation
DNA methylation was assessed using the Illumina HumanMethylation450 (“450k”) microarray as previously described (Jaffe et al., 2016). Samples from both brain regions were jointly normalized using stratified quantile normalization in the minfi Bioconductor package (Aryee et al., 2014). We retained a single array in the case of duplicates by choosing the sample that had the closest quality profile (via Methylated and Unmethylated signal intensity) to all other arrays. We implemented in silico estimation of the relative proportions of five cell types (ESCs, ES-derived NPCs, and derived dopamine neurons from culture, and adult cortex neuronal and non-neuronal cells from adult tissue) using the reference profile described in Jaffe et al. (2016) with the deconvolution algorithm described by Houseman et al. (2012).
RNA deconvolution
We used normalized expression data (log2(RPKM+1)) from 25 replicating and 110 quiescent fetal neurons, 18 oligodendrocyte progenitor cells (OPCs), 131 neurons, 62 astrocytes, 38 oligodendrocytes,16 microglia and 20 endothelial cells from Fluidigm single cell sequencing data from Darmanis et al. (2015). We picked the 20 most cell-type specific genes for each of the 8 cell types using t-statistics analogous to Jaffe and Irizarry (2014) after scaling gene expression to the standard normal distribution. This process resulted in 158 unique genes (since some genes could be markers for multiple cell types, Table S12) which were used with the deconvolution method (Houseman et al., 2012) using minfi (Aryee et al., 2014). This approach was described in more detail in Burke et al. (2018) but only involved data from Darmanis et al. (2015) here.
Differential expression over development
Using the neuropsychiatric control samples (Figure S2B; Table S10, n = 614) we fit a linear spline model with break points at birth/0,1, 10, 20, and 50 years of age while adjusting for the brain region (DLPFC was set as the reference, n = 300, HIPPO n = 314), sex, technical covariates (mitochondrial mapping rate, total assigned gene proportion and RIN) and ethnicity (first five principal components based on the genotype data). We computed F-statistics based on the null (Equation 2) and full (Equation 3) models shown below where we tested for an interaction between the age splines and the region indicator. For genes, exons, and exon-exon junctions we used the voom method (Law et al., 2014) for normalizing the expression. To take into account the correlation induced by measuring two brain regions from the same individuals (in most cases) we used the duplicateCorrelation() function in limma where the block and design arguments were set to the individual identifier and the intercept and region indicator, respectively. The resulting estimated correlation was passed to the lmFit() function in limma (Ritchie et al., 2015) version 3.34.5. Statistics were then calculated using eBayes() and topTable() with the coef argument set to the additional terms in the full model (Equation 3). Resulting p values were Bonferroni-adjusted within each feature type.
| (Equation 2) |
Equation 2. Null model for differential expression over development where x+ = max(0, x).
| (Equation 3) |
Equation 3. Additional terms present in the full model absent from the null model (Equation 2) that are tested in the F-statistic.
The full list and statistics for the differentially expressed features are available in Table S11.
Replication analysis with BrainSpan
To assess replication, we downloaded the fastq files from BrainSpan (BrainSpan, 2011) and processed the samples using the same RNA-seq processing pipeline and software versions. We retained only the samples with region code DFC and HIP and subsetted the measured features to those retained in our expression filtering cutoffs for BrainSeq Phase II (Figure S2C). We then used the same models and procedure as for Equation 1 for the differential expression by brain region analysis, and Equation 2 and Equation 3 for determining differential expression over development: p value in BrainSpan had to be < 0.05 to be considered as replicating. For the differential expression by brain region analysis we required the log fold change to be consistent by directionality (same sign across datasets). For the concordance analysis across the top 5,000 features with BrainSpan for the differences between DLPFC and HIPPO in prenatal and adult age groups we used ffpe version 1.26.0 (Waldron et al., 2012).
Degradation data generation
The qSVA algorithm begins with measuring brain tissue degradation in a separate RNA-seq dataset to determine regions most-associated with degradation as illustrated in Figure S21. In this study, degradation was tested at four different time points for tissue from two brain regions, HIPPO and DLPFC, from five individuals (Jaffe et al., 2017). An aliquot of ~100 mg of pulverized tissue for each brain region from each donor was left on dry ice, and placed at room temperature until reaching the respective time interval, at which point the tissue was placed back onto dry ice (Jaffe et al., 2017). The four time intervals tested were 0,15, 30, and 60 min, with the 0-minute aliquot remaining on dry ice for the entirety of the experiment, and RNA extraction began immediately after the end of the final time interval (Jaffe et al., 2017).
Determining multi-region quality surrogate variables (qSVs)
Raw fastq files were processed with the same RNA-seq processing pipeline as the BrainSeq Phase II data using the same software versions. The base-pair coverage RNA-seq expression data were normalized to 40 million reads using recount.bwtool (Ellis et al., 2018) version 0.99.28, and a cutoff of 5 reads was used to identify the expressed regions (ERs) of interest using derfinder version 1.12.6 (Collado-Torres et al., 2017). Transcript features most susceptible to RNA degradation were then identified using a regression model for degradation time adjusting for the brain region and individuals as shown in Equation 4. Individuals were adjusted for to account for variability due to subject effect, since brain tissue degradation was measured in the same brain at 4 different time points. The top 1000 ERs most associated with degradation (by FDR) were then quantified in the BrainSeq Phase II data using recount.bwtool.
| (Equation 4) |
Equation 4. Linear model for identifying degradation-associated expressed regions.
We also used a model (shown in Equation 5) that included an interaction term between degradation time and brain region and found 620 ERs with a significant interaction term (FDR < 5%) of which 603 had a significant degradation time term in the initial model. The genomic coordinates of the degradation-associated ERs used are available in Table S13.
| (Equation 5) |
Equation 5. Same as Equation 4 but also including an interaction term between degradation time and brain region.
From our exploratory data analysis of all 900 BrainSeq Phase II samples (Figure S20) we determined that it was best to drop the HIPPO Gold samples (n = 43) and samples age <17 due to the lack of early SCZD cases (Table S10) before determining the actual qSVs (right side of Figure S21). We found three sets of qSVs with the BE algorithm (Buja and Eyuboglu, 1992) using sva (Leek and Storey, 2007) version 3.26.0: one for DLPFC samples (k = 15, n = 379), one for HIPPO samples (k = 16, n = 333) and one for both regions combined (k = 22, n = 712). The DLPFC and HIPPO top qSVs have similar relationships to RIN and SCZD diagnosis status as shown in Figure S22.
DEqual plots
To assess the performance of the qSVA approach we used three different linear regression models for each brain region: a naive model with only the SCZD diagnosis term (Equation 6), a model with all covariates we adjust for excluding qSVs (Equation 7), and a full model with all adjustment covariates and qSVs (Equation 8). We then compared the log2 fold change by SCZD diagnosis against the log2 fold change by degradation time for each brain region as shown in Figure S25.
| (Equation 6) |
Equation 6. Linear regression model for the case-control analysis with a naive model.
| (Equation 7) |
Equation 7. Linear regression model for the case-control analysis using all adjustment covariates except the quality surrogate variables (qSVs).
| (Equation 8) |
Equation 8. Linear regression model for the case-control analysis where k was determined by the BE algorithm (Buja and Eyuboglu, 1992) using sva (Leek and Storey, 2007) version 3.26.0 for each brain region separately (Table S10; DLPFC k = 15, n = 379; HIPPO k= 16, n = 333).
Differential expression between SCZD cases and neurotypical controls
Using voom (Lawet al., 2014) from limma (Ritchie et al., 2015) version 3.34.9 we identified differentially expressed features using the model described in Equation 8 for each brain region for genes, exons, and exon-exon junctions. For transcripts we skipped the voom step. For comparisons with BrainSeq Phase 1 (Jaffe et al., 2018) and the CommonMind Consortium dataset (Fromer et al., 2016) we used the log fold changes we determined previously (Jaffe et al., 2018) and matched the sets by gene ids. The full list and statistics for the differentially expressed features are available in Table S11.
To identify differentially expressed features by the interaction between SCZD diagnosis status and brain region, we expanded Equation 8 to include the interaction term and used the same methodology as previously described. The qSVs used in this analysis were determined across both the DLPFC and HIPPO degradation matrices (n = 712) resulting in k = 22 qSVs.
Gene ontology and gene set enrichment analyses
Unless otherwise noted, we used the compareCluster() function from clusterProfiler (Yu et al., 2012) version 3.6.0 for gene ontology (Ashburner et al., 2000; The Gene Ontology Consortium, 2017) and KEGG (Kanehisa et al., 2017) enrichment analyses with parameters pvalueCutoff = 0.1 and qvalueCutoff = 0.05 with the set of Ensembl gene ids expressed in genes, exons, and exon-exon junctions as the background universe (Figure S4). For gene set enrichment analysis in the SCZD case-control model, we used the gseGO() function from clusterProfiler with default parameters using Ensembl gene ids.
Visualization of differential expression results
We used the cleaningY() function from the jaffelab package (Collado-Torres and Jaffe, 2017) version 0.99.20 to regress out adjustment covariates from our main differential expression models (Equations 1,3, and 8) to visualize the expression as shown in Figure 2C and Figure 3D. Doing so helps visualize what the model is observing.
Venn diagrams
Colored venn diagrams were made using the VennDiagram R package version 1.6.18 while non-colored venn diagrams were made using gplots version 3.0.1.
Brain region co-expression analyses
Using the age >17 RNA-seq samples from the SCZD case-control analysis, we identified the individuals with measured expression in both DLPFC and HIPPO (n = 265 subjects, 530 RNA-seq samples). We computed the log2(RPKM + 0.5) for gene and exon expression levels, log2(RP10M + 0.5) for exon-exon junctions and log(TPM + 0.5) for transcripts. For DLPFC and HIPPO separately, we then used the cleaningY() function from the jaffelab package (Collado-Torres and Jaffe, 2017) version 0.99.21 to remove all the covariates from Equation 8 while keeping the intercept and SCZD diagnosis effects.
At the gene level, we computed the Pearson correlation for each gene across all 265 individuals producing one correlation value per gene. We then permuted the individual labels 1000 times to compute a null distribution of the gene correlation values and calculated FWER p values using derfinder:::.calcFVal() function (Collado-Torres et al., 2017). With the genes that had a significant correlation among brain regions (FWER < 5%) we identified enriched GO terms and KEGG pathways using the same parameters previously described. We did this process for both the raw expression (say log2(RPKM + 0.5) at the gene level) and the “cleaned” expression (post use of the cleaningY()function).
We computed correlations at the individual level for all 4 expression summarizations (gene, exon, exon-exon junction, transcript) across all features, resulting in one correlation value per individual and expression level. We performed two-sided t tests comparing the mean correlation among SCZD cases and neurotypical controls for each expression level. We grouped all the genes expressed in our dataset by their GO level 3 category for the biological process, cellular component, and molecular function ontologies as well as by their KEGG pathway id. For each of these 4 gene sets, we calculated the individual level correlation between DLPFC and HIPPO, their two-sides t test p value for a difference in mean correlation, and corrected the p values using the FDR method. GO terms and KEGG pathways with a significant difference are listed in Table S7.
We further computed correlations across brain regions using the sets of genes with significant differential expression by SCZD status identified in our previous analysis for both brain regions as well as in either the BSP1 and CMC datasets.
Identifying eQTLs
MatrixEQTL (Shabalin, 2012) version 2.2 was used to identify eQTLs for each brain region using samples age > 13 (Figure S42D, DLPFC n = 397, HIPPO n = 395) with either the log2(RPKM+1) for genes and exons, log2(RP10M+1) for exon-exon junctions or log2(TPM+1) for transcripts. For the eQTL analysis we adjusted for SCZD diagnosis status, sex, SNP PCs as well as expression PCs as shown in Equation 9. To determine the number of expression PCs to adjust (p) for we used the num.sv() function from the sva package with the model in Equation 10 and the vfilter = 50000 parameter. Then the Matrix_eQTL_main() was used to identify the eQTLs using the parameters pvOutputThreshold.cis = 0.001, pvOutputThreshold = 0, useModel = modelLINEAR, cisDist = 5e5.
| (Equation 9) |
Equation 9. Main eQTL model for DLPFC and HIPPO.
| (Equation 10) |
Equation 10. Model used for determining the number of expression PCs to adjust for in the eQTL analysis.
To identify eQTLs that have different effects by brain region, we ran asimilar analysis using all the age > 13 samples (n = 792) using the model from Equation 11 and by changing some parameters of Matrix_eQTL_main() to useModel = modelLINEAR_CROSS, cisDist = 2.5e5.
| (Equation 11) |
Equation 11. Model used for the brain region interaction eQTL analysis between DLPFC and HIPPO. num.sv() from the sva package was used to determine the number of expression PCs to adjust for.
The eQTL snp-feature pairs with FDR < 1% and their replication statistics are available in Table S15.
Risk loci
We downloaded the list of 179 schizophrenia GWAS risk SNPs determined by the Psychiatric Genomics Consortium (Pardiñas et al., 2018). We used this list as input to the rAggr tool available at http://biostats.usc.edu/software.html (Edlund et al., 2017) to identify proxy markers in linkage disequilibrium (LD > 0.8) with the list of the 179 index SNPs based on the 1000 Genomes Project Phase 3 database. A maximum distance of 500kb and minumum MAF of 0.001 were used as cutoffs, and reference populations used were African, Americas, East Asian, and European, based on the races present in our samples. Of the 44 index SNPs absent in our genotype data, 28 (64%) had at least one proxy present, so we considered 163 out of 179 risk loci (91%) and 135 index SNPs we directly analyzed. The rAggr software returned a set of 10,981 markers that included both the index and proxy SNPs, of which we found 9,736 of them present in our genotype data (Figure S42D). We then used those 9,736 SNPs to identify eQTLs for each brain region by repeating a similar analysis as described above using Equations 9 and 10, with the exception of now using pvOutputThreshold.cis = 1 when running Matrix_eQTL_main() for the set of risk SNPs.
The significant (FDR < 1%) snp-feature eQTL pairs from this analysis focused on risk loci for DLPFC and HIPPO are listed in Table S16.
GTEx eQTL replication
We downloaded the GTEx v6 (GTEx Consortium, 2015) HIPPO (“Brain - Hippocampus”) and DLPFC [“Brain - Frontal Cortex (BA9)”] fastq files from SRA (Leinonen et al., 2011) using fastq-dump as well as their corresponding genotype data for samples passing the GTEx quality controls (SMAFRZE labeled as “USE ME”). RNA-seq data were processed using the same pipeline and expression features were subsetted to those observed in BrainSeq Phase II. Genotype data were processed as previously described and variantes were subsetted to those observed in BrainSeq Phase II. eQTL analyses were run using matrixEQTL (Shabalin, 2012) for all the expression features and all the variants that had a significant eQTL association (FDR < 0.01) in any of the previous 5 eQTL analyses. Replication was assessed for the DLPFC, HIPPO and the brain region interaction eQTLs by directionality or by nominal p value < 0.05.
BrainSeq Phase I replication
BrainSeq Phase I DLPFC polyA+ data were re-processed using the same pipeline used for this analysis and the expression features were subsetted to those observed in BrainSeq Phase II. Replication eQTL analyses were carried out similar to those for the GTEx eQTL replication.
CAUC-only sensitivity analysis
We ran a sensitivity analysis by subsetting each of our eQTL models (DLPFC, HIPPO and interaction between the two brain regions) to just the self-reported CAUC samples (43.8 to 46.1% of the original sample size, Table S8B) at the gene expression feature level. We still adjusted for quantitative genomic ancestry components to account for differences between self-reported and genomic ancestry, as well as to further control for samples being genotyped across multiple microarray platforms.
Each of the three eQTL models showed high directionality agreement between the genotype regression coefficient from the significant mixed ethnicity eQTL SNP-gene pairs and those from the CAUC-only analysis: interaction: 94.5%, DLPFC: 94.1%, HIPPO: 94.5% (Table S8). While allelic directionally was highly concordant between ethnicities among those eQTLs identified in combined analysis, many of these eQTLs are much less significant within CAUCs only. For example, while between 82%–87% of mixed-ethnicity eQTLs were concordant and at least marginally significant in CAUC at p < 0.05 (interaction 87.2%, DLPFC: 82.3%, HIPPO: 83.1%), many fewer eQTLs remain significant at more stringent p values, like p < 0.001 (interaction: 57.9%, DLPFC: 57.5%, HIPPO: 59.1%, Table S8). Given the high directional consistency (~95%), we believe this decrease in identified eQTLs in CAUC-only is likely due to the decrease in power from using a smaller number of individuals. This is further supported by the vast majority of the SNP-gene pairs in CAUCs at FDR < 0.01 were themselves genome-wide significant (FDR < 0.01) in our original mixed ancestry analyses (interaction 78.4%, DLPFC: 85.2%, HIPPO: 84.8%, Table S8C) with identical directionality for those observed in both analyses (interaction 100%, DLPFC: 99.99%, HIPPO: 99.99%).
Neuronal DEG enrichment in brain-region dependent eQTLs
We used the RNA-seq data obtained from sorted cell populations (Price et al., 2018) to identify differentially expressed genes between neurons and non-neurons (NeuN+ versus NeuN−). Among the 44,338 genes from this cell sorted RNA-seq dataset that match our dataset based on Ensembl gene ids (95.6% were matched), there are 9,848 (22.21%) differentially genes between neurons and non-neurons (neuron > non-neuron: 3,439, non-neuron > neuron: 6,409) at FDR <5%. We observed that 41.49% of the unique genes in our brain-region dependent eQTLs across all features (1,798 based on Ensembl IDs) are differentially expressed between neurons and non-neurons, which is significantly higher than the 22.21% (p = 3.601×10−86, one sided proportion test) observed in the sorted RNA-seq data. Comparable results are obtained if we focus on the 205,618 SNP-feature pairs obtained excluding the transcript level eQTLs (proportion = 41.91%, p = 9.124×10−82).
TWAS
We adapted the FUSION TWAS software (Gusev et al., 2016, 2018) from hg19 to hg38 as described in our step-by-step guide at https://github.com/LieberInstitute/brainseq_phase2/tree/master/twas. First, three sets of SNPs were homogenized in terms of coordinates, names, and reference alleles: 1) the LD reference set from: gusevlab.org/projects/fusion/, 2) the SNPs used to calculate the TWAS weights, i.e., the same set of SNPs used in the eQTL analyses, and 3) the GWAS summary statistic SNPs from PGC2 and the Walters Group Data Repository (Pardiñas et al., 2018). Feature weights were then computed for DLPFC and HIPPO at all four expression summarization features separately by running the weight computing TWAS-FUSION R script based off of Gusev’s work. After the functional weight was computed for each feature, two more TWAS-FUSION scripts were run (adapted from FUSION.assoc_test.R and FUSION.post_process.R) which applied the weights to the GWAS summary statistic SNPs and calculated the functional-GWAS association statistics. We reclassified variants as proxies if they have GWAS p values < 5e-8 even if they were not in strong LD (R^2 <0.8) with the index SNP. We computed TWAS FDR- and Bonferroni-adjusted p values within each feature type separately.
We repeated this TWAS processing for the PGC2 GWAS (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014) for both DLPFC and HIPPO across all four expression summarization features. We then assessed which of the genes identified by the TWAS analysis Gusev et al. (2018) either in CMC or the CMC splicing data were observed in our TWAS analyses (PGC2+CLOZUK and PGC2). See Table S17 for the genome-wide significant TWAS results (FDR < 5%) in either DLPFC or HIPPO with either the PGC2+CLOZUK or the PGC2 GWAS as well as the comparison with the Gusev et al. (2018) TWAS results.
QUANTIFICATION AND STATISTICAL ANALYSIS
Table S10 describes the sample sizes for the differential expression analyses we performed. More specifically, for the DLPFC versus HIPPO analysis with prenatal samples, DLPFC versus HIPPO with adult neurotypical control samples, the DLPFC versus HIPPO developmental analysis with neurotypical control samples, and the schizophrenia versus neurotypical controls analysis per brain region. The different subsections of the “Method Details” further specify the statistical models and tests used as well as the versions of the specific software used. Overall, statistical tests were performed using R versions 3.3, 3.4 and 3.5 with detailed R session information provided in the code GitHub repositories listed under “Data and Software Availability.” The threshold and method used for statistical significance is listed in the main text along the description of the results.
DATA AND SOFTWARE AVAILABILITY
Raw and processed data are available from http://eqtl.brainseq.org/phase2/. Code is available through GitHub at https://github.com/LieberInstitute/brainseq_phase2 and https://github.com/LieberInstitute/qsva_brain, both of which are described in their README.md files. The FUSION TWAS code modified for hg38 is available from https://github.com/LieberInstitute/fusion_twas with modifications described in detail at https://github.com/LieberInstitute/brainseq_phase2/tree/master/twas. Supplementary figures and tables are available via Mendeley Data (https://doi.org/10.17632/3j93ybf4md.1).
ADDITIONAL RESOURCES
As part of BrainSeq Phase II, we created an eQTL browser available at http://eqtl.brainseq.org/phase2/ that enables exploring the millions of eQTL snp-feature pairs for DLPFC, HIPPO and the brain region dependent results.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| DLPFC and HIPPO human brain dissections. | This paper. Lieber Institute for Brain Development. The collection of human brains and dissections are described in further detail under “STAR Methods” section of the manuscript. | Unique brain identifies (BrNum) and RNA extraction identifies (RNum) are provided in the RangedSummarizedExperiment R objects (colData slot) listed under the “Data and Software Availability” section of the manuscript. |
| Critical Commercial Assays | ||
| RNeasy Lipid Tissue Mini Kit for RNA extraction | QIAGEN | Cat#: 74804 |
| TruSeq Stranded Total RNA Library Prep Human/Mouse/Rat | Illumina: https://www.illumina.com/products/selection-tools/rrna-depletion-selection-guide.html | Cat#: 20020599 |
| TruSeq Stranded Total RNA Library Prep Gold | Illumina: https://www.illumina.com/products/selection-tools/rrna-depletion-selection-guide.html | Cat#: 20020597 |
| Deposited Data | ||
| Raw and processed data | This paper | http://eqtl.brainseq.org/phase2/ |
| Supplementary Figures and Tables at Mendeley Data | This paper | https://doi.org/10.17632/3j93ybf4md.1 |
| DLPFC RNA-seq FASTQ raw data | This paper | Globus collection jhpce#bsp2-dlpfc https://app.globus.org/file-manager?origin_id=0dd03924-6853-11e9-bf44-0e4a062367b8&origin_path=%2F |
| HIPPO RNA-seq FASTQ raw data | This paper | Globus collection jhpce#bsp2-hippo https://app.globus.org/file-manager?origin_id=96be20a2-6853-11e9-bf44-0e4a062367b8&origin_path=%2F |
| Software and Algorithms | ||
| Analysis code | This paper | https://github.com/LieberInstitute/brainseq_phase2 |
| Analysis code for SCZD case-control DEG analysis | This paper | https://github.com/LieberInstitute/qsva_brain |
| FUSION TWAS software modified for hg38 | This paper | https://github.com/LieberInstitute/fusion_twas |
| Guide for using performing a TWAS analysis with hg38 | This paper | https://github.com/LieberInstitute/brainseq_phase2/tree/master/twas |
| Other | ||
| BrainSeq eQTL browser | This paper | http://eqtl.brainseq.org/phase2/eqtl/ |
Highlights.
Dorsolateral prefrontal cortex and hippocampus gene expression across development
Novel region-specific schizophrenia genetic risk features
Decreased regional functional coherence in schizophrenia
Public brain gene expression and eQTL resource at http://eqtl.brainseq.org/phase2
ACKNOWLEDGMENTS
The authors would like to express their gratitude to our colleagues whose tireless efforts have led to the donation of postmortem tissue to advance these studies: the Office of the Chief Medical Examiner of the District of Columbia; the Office of the Chief Medical Examiner for Northern Virginia, Fairfax Virginia; and the Office of the Chief Medical Examiner of the State of Maryland, Baltimore, Maryland. We would also like to acknowledge Llewellyn B. Bigelow, MD, for his diagnostic expertise. We also thank R. Zielke, R.D. Vigorito, and R.M. Johnson of the National Institute of Child Health and Human Development Brain and Tissue Bank for Developmental Disorders at the University of Maryland for providing fetal, child, and adolescent brain specimens. Finally, we are indebted to the generosity of the families of the decedents, who donated the brain tissue used in these studies. This project was supported by the Lieber Institute for Brain Development, the BrainSeq Consortium, and partially by NIH R21-MH109956-01 (to A.E.J.). We would like to thank the GTEx consortium. The data used for the analyses described in this manuscript were obtained from dbGaP accession number phs000424.v6.p1 on October 6, 2015.
Footnotes
DECLARATION OF INTERESTS
The following BrainSeq Consortium members have competing interests. M.M., T.S., K.T., and D.J.H. are employees of Astellas Pharma. N.J.B. and A.J.C. are employees of AstraZeneca. D.A.C., J.N.C., C.L.A.R., B.J.E., P.J.E., D.C.A., Y. Li, Y. Liu, K.M., B.B.M., J.E.S., and H.W. are employees of Eli Lilly and Company. M.F., D.H., and H.K. are employees of Janssen Research & Development LLC and Johnson and Johnson. M.D. and L.F. are employees of H. Lundbeck A/S. T.K.-T. and D.M. are employees of F. Hoffmann-La Roche. P.O., S.X., and J.Q. are former employees of Pfizer.
CONSORTIA
Members of the BrainSeq Consortium include Mitsuyuki Matsumoto, Takeshi Saito, Katsunori Tajinda, Daniel J. Hoeppner, Nicholas J. Brandon, Alan J. Cross, David A. Collier, John N. Calley, Cara Lee A. Ruble, Brian J. Eastwood, Philip J. Ebert, David Charles Airey, Yupeng Li, Yushi Liu, Karim Malki, Bradley B. Miller, James E. Scherschel, Hong Wang, Maura Furey, Derrek Hibar, Hartmuth Kolb, Michael Didriksen, Lasse Folkersen, Tony Kam-Thong, Dheeraj Malhotra, Patricio O’Donnell, Simon (Hualin) Xi, Jie Quan, JooHeon Shin, Andrew E. Jaffe, Rujuta Narurkar, Richard E. Straub, Amy Deep-Soboslay, Thomas M. Hyde, Joel E. Kleinman, and Daniel R. Weinberger.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.neuron.2019.05.013.
REFERENCES
- Babraham Bioinformatics (2016). FastQC (Babraham Institute; ). https://www.bioinformatics.babraham.ac.uk/projects/fastqc/. [Google Scholar]
- Akbarian S, Liu C, Knowles JA, Vaccarino FM, Farnham PJ, Crawford GE, Jaffe AE, Pinto D, Dracheva S, Geschwind DH, et al. ; PsychENCODE Consortium (2015). The PsychENCODE project. Nat. Neurosci 18, 1707–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, and Irizarry RA (2014). Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30, 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. ; The Gene Ontology Consortium (2000). Gene ontology: tool for the unification of biology. Nat. Genet 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, and Abecasis GR; 1000 Genomes Project Consortium (2015). A global reference for human genetic variation. Nature 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum R, and Weinberger DR (2017). Genetic insights in to the neurodevelopmental origins of schizophrenia. Nat. Rev. Neurosci 18, 727–740. [DOI] [PubMed] [Google Scholar]
- Birnbaum R, Jaffe AE, Chen Q, Shin JH, Kleinman JE, Hyde TM, and Weinberger DR; BrainSeq Consortium (2018). Investigating the neuroimmunogenic architecture of schizophrenia. Mol. Psychiatry 23, 1251–1260. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BrainSeq: A Human Brain Genomics Consortium. BrainSeq: A Human Brain Genomics Consortium (2015). Brainseq: neurogenomics to drive novel target discovery for neuropsychiatric disorders. Neuron 88, 1078–1083. [DOI] [PubMed] [Google Scholar]
- BrainSpan (2011). Atlas of the Developing Human Brain. http://www.brainspan.org.
- Bray NL, Pimentel H, Melsted P, and Pachter L (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Buja A, and Eyuboglu N (1992). Remarks on parallel analysis. Multivariate Behav. Res 27, 509–540. [DOI] [PubMed] [Google Scholar]
- Burke EE, Chenoweth JG, Shin JH, Collado-Torres L, Kim SK, Micali N, Wang Y, Straub RE, Hoeppner DJ, Chen H-Y, et al. (2018). Dissecting transcriptomic signatures of neuronal differentiation and maturation using iPSCs. bioRxiv. 10.1101/380758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callicott JH, Egan MF, Bertolino A, Mattay VS, Langheim FJ, Frank JA, and Weinberger DR (1998). Hippocampal N-acetyl aspartate in unaffected siblings of patients with schizophrenia: a possible intermediate neurobiological phenotype. Biol. Psychiatry 44, 941–950. [DOI] [PubMed] [Google Scholar]
- Collado-Torres L, and Jaffe AE (2017). jaffelab-package: commonly used functions by the Jaffe lab (Lieber Institute; ). https://rdrr.io/github/LieberInstitute/jaffelab/. [Google Scholar]
- Collado-Torres L, Nellore A, Frazee AC, Wilks C, Love MI, Langmead B, Irizarry RA, Leek JT, and Jaffe AE (2017). Flexible expressed region analysis for RNA-seq with derfinder. Nucleic Acids Res. 45, e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree GW, and Gogos JA (2014). Synaptic plasticity, neural circuits, and the emerging role of altered short-term information processing in schizophrenia. Front. Synaptic Neurosci 6, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, Hayden Gephart MG, Barres BA, and Quake SR (2015). A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. USA 112, 7285–7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O, Coulonges C, and Zagury J-F (2008). Shape-IT: new rapid and accurate algorithm for haplotype inference. BMC Bioinformatics 9, 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund CK, Conti DV, and Van Den Berg DJ (2017). rAggr. http://biostats.usc.edu/software.html.
- Ellis SE, Collado-Torres L, Jaffe A, and Leek JT (2018). Improving the value of public RNA-seq expression data by phenotype prediction. Nucleic Acids Res. 46, e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y-Y, Ramu A, Cotto KC, Skidmore ZL, Kunisak J, Conrad DF, Lin Y, Chapman W, Uppaulri R, Govindan R, et al. (2018). RegTools: Integrated analysis of genomic and transcriptomic data for discovery of splicing variants in cancer. bioRxiv. 10.1101/336634v2. [DOI] [Google Scholar]
- Friston K, Brown HR, Siemerkus J, and Stephan KE (2016). The dysconnection hypothesis (2016). Schizophr. Res. 176, 83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, Ruderfer DM, Oh EC, Topol A, Shah HR, et al. (2016). Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci 19, 1442–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandal MJ, Zhang P, Hadjimichael E, Walker RL, Chen C, Liu S, Won H, van Bakel H, Varghese M, Wang Y, et al. ; PsychENCODE Consortium (2018). Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 362, eaat8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gejman PV, Sanders AR, and Duan J (2010). The role of genetics in the etiology of schizophrenia. Psychiatr. Clin. North Am 33, 35–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore FM, Bloem PJN, Patton GC, Ferguson J, Joseph V, Coffey C, Sawyer SM, and Mathers CD (2011). Global burden of disease in young people aged 10–24 years: a systematic analysis. Lancet 377, 2093–2102. [DOI] [PubMed] [Google Scholar]
- Gottesman II, and Shields J (1982). Schizophrenia (CUP Archive).
- Consortium GTEx (2015). Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BWJH, Jansen R, de Geus EJC, Boomsma DI, Wright FA, et al. (2016). Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet 48, 245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A, Mancuso N, Won H, Kousi M, Finucane HK, Reshef Y, Song L, Safi A, McCarroll S, Neale BM, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium (2018).Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat. Genet 50, 538–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A, Nitsche MA, Herrmann M, Schneider-Axmann T, Marshall L, Gruber O, Falkai P, and Wobrock T (2012). Impaired long-term depression in schizophrenia: a cathodal tDCS pilot study. Brain Stimul. 5, 475–483. [DOI] [PubMed] [Google Scholar]
- Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, Furey TS, Harte RA, Hsu F, et al. (2006). The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 34, D590–D598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, and Kelsey KT (2012). DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, and Marchini J (2009).A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AE, and Irizarry RA (2014). Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 15, R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AE, Gao Y, Deep-Soboslay A, Tao R, Hyde TM, Weinberger DR, and Kleinman JE (2016). Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat. Neurosci 19, 40–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AE, Tao R, Norris AL, Kealhofer M, Nellore A, Shin JH, Kim D, Jia Y, Hyde TM, Kleinman JE, et al. (2017). qSVA framework for RNA quality correction in differential expression analysis. Proc. Natl. Acad. Sci. USA 114,7130–7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AE, Straub RE, Shin JH, Tao R, Gao Y, Collado-Torres L, Kam-Thong T, Xi HS, Quan J, Chen Q, et al. ; BrainSeq Consortium (2018). Developmental and genetic regulation of the human cortex transcriptome illuminate schizophrenia pathogenesis. Nat. Neurosci 21, 1117–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Tanabe M, Sato Y, and Morishima K (2017). KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45 (D1), D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Zweig AS, Barber G, Hinrichs AS, and Karolchik D (2010). BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics 26, 2204–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, and Smyth GK (2014). voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, and Storey JD (2007). Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 3, 1724–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen R, Sugawara H, and Shumway M; International Nucleotide Sequence Database Collaboration (2011). The sequence read archive. Nucleic Acids Res. 39, D19–D21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucht S, Cipriani A, Spineli L, Mavridis D, Orey D, Richter F, Samara M, Barbui C, Engel RR, Geddes JR, et al. (2013). Comparative efficacy and tolerability of 15 antipsychotic drugs in schizophrenia: a multiple-treatments meta-analysis. Lancet 382, 951–962. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Lipska BK, Deep-Soboslay A, Weickert CS, Hyde TM, Martin CE, Herman MM, and Kleinman JE (2006). Critical factors in gene expression in postmortem human brain: focus on studies in schizophrenia. Biol. Psychiatry 60, 650–658. [DOI] [PubMed] [Google Scholar]
- Machida YJ, Machida Y, Chen Y, Gurtan AM, Kupfer GM, D’Andrea AD, and Dutta A (2006). UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol. Cell 23, 589–596. [DOI] [PubMed] [Google Scholar]
- McGrath J, Saha S, Chant D, and Welham J (2008). Schizophrenia: a concise overview of incidence, prevalence, and mortality. Epidemiol. Rev 30, 67–76. [DOI] [PubMed] [Google Scholar]
- Meetei AR, de Winter JP, Medhurst AL, Wallisch M, Waisfisz Q, van de Vrugt HJ, Oostra AB, Yan Z, Ling C, Bishop CE, et al. (2003). A novel ubiquitin ligase is deficient in Fanconi anemia. Nat. Genet 35, 165–170. [DOI] [PubMed] [Google Scholar]
- Messias EL, Chen C-Y, and Eaton WW (2007). Epidemiology of schizophrenia: review of findings and myths. Psychiatr. Clin. North Am 30,323–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Poline JB, Kohn PD, Holt JL, Egan MF, Weinberger DR, and Berman KF (2001). Evidence for abnormal cortical functional connectivity during working memory in schizophrenia. Am. J. Psychiatry 158, 1809–1817. [DOI] [PubMed] [Google Scholar]
- Millan MJ, Andrieux A, Bartzokis G, Cadenhead K, Dazzan P, Fusar-Poli P, Gallinat J, Giedd J, Grayson DR, Heinrichs M, et al. (2016). Altering the course of schizophrenia: progress and perspectives. Nat. Rev. Drug Discov 15, 485–515. [DOI] [PubMed] [Google Scholar]
- Pardiñas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, Legge SE, Bishop S, Cameron D, Hamshere ML, et al. ; GERAD1 Consortium; CRESTAR Consortium; GERAD1 Consortium; CRESTAR Consortium; GERAD1 Consortium; CRESTAR Consortium (2018). Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet 50, 381–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, and Kingsford C (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Cahill ME, Jones KA, VanLeeuwen J-E, and Woolfrey KM (2011). Dendritic spine pathology in neuropsychiatric disorders. Nat. Neurosci 14, 285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plavén-Sigray P, Matheson GJ, Collste K, Ashok AH, Coughlin JM, Howes OD, Mizrahi R, Pomper MG, Rusjan P, Veronese M, et al. (2018). Positron emission tomography studies of the glial cell marker translocator protein in patients with psychosis: a meta-analysis using individual participant data. Biol. Psychiatry 84, 433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AJ, Collado-Torres L, Ivanov NA, Xia W, Burke EE, Shin JH, Tao R, Ma L, Jia Y, Hyde TM, et al. (2018). Divergent neuronal DNA methylation patterns across human cortical development: critical periods and a unique role of CpH methylation. bioRxiv. 10.1101/328391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, and Sham PC (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasetti R, Mattay VS, White MG, Sambataro F, Podell JE, Zoltick B, Chen Q, Berman KF, Callicott JH, and Weinberger DR (2014). Altered hippocampal-parahippocampal function during stimulus encoding: a potential indicator of genetic liability for schizophrenia. JAMA Psychiatry 71, 236–247. [DOI] [PubMed] [Google Scholar]
- Rice D, and Barone S Jr. (2000). Critical periods of vulnerability for the developing nervous system: evidence from humans and animal models. Environ. Health Perspect 108 (Suppl 3), 511–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalin AA (2012). Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics 28, 1353–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, and Sirotkin K (2001). dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa AMM, Zhu Y, Raghanti MA, Kitchen RR, Onorati M, Tebbenkamp ATN, Stutz B, Meyer KA, Li M, Kawasawa YI, et al. (2017). Molecular and cellular reorganization of neural circuits in the human lineage. Science 358, 1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium (2017). Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 45 (D1), D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron L, Ogino S, Hoshida Y, Shima K, McCart Reed AE, Simpson PT, Baba Y, Nosho K, Segata N, Vargas AC, et al. (2012). Expression profiling of archival tumors for long-term health studies. Clin. Cancer Res 18, 6136–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang S, and Li W (2012). RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185. [DOI] [PubMed] [Google Scholar]
- Wang D, Liu S, Warrell J, Won H, Shi X, Navarro FCP, Clarke D, Gu M, Emani P, Yang YT, et al. ; PsychENCODE Consortium (2018). Comprehensive functional genomic resource and integrative model for the human brain. Science 362, eaat8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe J, Fukui N, Suzuki Y, Sugai T, Ono S, Tsuneyama N, Saito M, Tajiri M, and Someya T (2017). Effect of GWAS-identified genetic variants on maximum QT interval in patients with schizophrenia receiving antipsychotic agents: a 24-hour holter ECG study. J. Clin. Psychopharmacol 37, 452–455. [DOI] [PubMed] [Google Scholar]
- Weinberger DR (1999). Cell biology of the hippocampal formation in schizophrenia. Biol. Psychiatry 45, 395–402. [DOI] [PubMed] [Google Scholar]
- Weinberger DR, Berman KF, Suddath R, and Torrey EF (1992). Evidence of dysfunction of a prefrontal-limbic network in schizophrenia: a magnetic resonance imaging and regional cerebral blood flow study of discordant monozygotic twins. Am. J. Psychiatry 149, 890–897. [DOI] [PubMed] [Google Scholar]
- Xu H-T, Han Z, Gao P, He S, Li Z, Shi W, Kodish O, Shao W, Brown KN, Huang K, and Shi SH (2014). Distinct lineage-dependent structural and functional organization of the hippocampus. Cell 157, 1552–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovlev PI, and Lecours AR (1967). The myelogenetic cycles of regional maturation of the brain In Regional Development of the Brain in Early Life, Minkowski A, ed. (Blackwell Scientific Publications; ), pp. 3–70. [Google Scholar]
- Yu G, Wang L-G, Han Y, and He Q-Y (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Xu J, Chen J, Kim S, Reimers M, Bacanu S-A, Yu H, Liu C, Sun J, Wang Q, et al. (2015). Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. Mol. Psychiatry 20, 563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed data are available from http://eqtl.brainseq.org/phase2/. Code is available through GitHub at https://github.com/LieberInstitute/brainseq_phase2 and https://github.com/LieberInstitute/qsva_brain, both of which are described in their README.md files. The FUSION TWAS code modified for hg38 is available from https://github.com/LieberInstitute/fusion_twas with modifications described in detail at https://github.com/LieberInstitute/brainseq_phase2/tree/master/twas. Supplementary figures and tables are available via Mendeley Data (https://doi.org/10.17632/3j93ybf4md.1).