ABSTRACT
Colibacillosis caused by avian pathogenic E. coli (APEC), is an economically important bacterial disease of poultry. APEC are a subgroup of extra intestinal pathogenic E. coli (ExPEC) and poultry are considered potential sources of foodborne ExPEC to humans. Currently, APEC infections in poultry are controlled by antibiotics and/or vaccination; however, their effect is limited due to emergence of antibiotic resistant strains and infections with heterologous serotypes. Therefore, novel approaches are needed. Here, using the bioluminescent quorum sensing (QS) autoinducer 2 (AI-2) indicator Vibrio harveyi BB170, we screened the cell free culture supernatant of APEC O78 prepared from cultures grown in the presence of 4,182 small molecules (SMs; 100 μM). A total of 69 SMs inhibited > 75% of APEC O78 AI-2 activity in the indicator bacteria. Ten SMs that showed highest AI-2 inhibition were selected for further studies. Most of these SMs inhibited the AI-2 activity of other APEC serotypes and significantly reduced APEC O78 biofilm formation and motility. Most compounds showed minimal toxicity on human intestinal cells (Caco-2), chicken macrophage (HD-11), and chicken and sheep red blood cells, and reduced APEC survival in HD-11 and THP-1 macrophages. The SMs induced no or minimal toxicity and conferred protection against APEC in wax moth larval model. SMs affected the expression of APEC O78 QS, virulence, biofilm and motility associated genes providing insight on their potential mode(s) of action. Further testing in chickens will facilitate development of these SMs as novel therapeutics to control APEC in poultry and thereby also reduce zoonotic transmission.
KEYWORDS: APEC, autoinducer-2, quorum sensing inhibitors, virulence, biofilm, motility, infection, chicken, macrophages, wax moth model, gene expression
Introduction
Colibacillosis, caused by avian pathogenic E. coli (APEC), is a significant bacterial disease of poultry worldwide [1,2]. APEC belongs to a subgroup of extra-intestinal pathogenic Escherichia coli (ExPEC). APEC- like bacteria can be transmitted to humans through consumption of contaminated poultry and fresh produce contaminated with poultry litter [3]. Additionally, poultry ExPEC share many important traits with human ExPEC including antimicrobial resistance patterns, resistance genes, and virulence factors; thus, APEC pose a potential zoonotic risk for humans [4]. Even though there are several APEC serotypes implicated in avian colibacillosis, the most predominant serotypes associated with avian colibacillosis are O1: K1, O2: K1, and O78: K80 [5,6].
Avian colibacillosis is characterized by yolk sac infection, swollen-head syndrome, septicemia, and inflammation of different organs such as pericarditis, perihepatitis, airsacculitis, salpingitis, arthritis, and peritonitis [6,7]. APEC infects all ages of commercial poultry and can also negatively affect weight gain and feed conversion. Additionally, APEC infection is associated with high morbidity and mortality and carcass condemnation, leading to significant economic losses to the poultry industry [6].
Currently, APEC infections in poultry are controlled by a commercially available modified-live vaccine (Poulvac E. coli) [8]. However, the vaccination does not provide complete protection against all APEC serotypes, and high mortality in vaccinated broilers due to virulent APEC infections has been reported [8]. In addition, antimicrobials such as cephalosporins, tetracyclines, and quinolones that are used for treating APEC infections have limited effects due to the emergence of antimicrobial resistant strains [7]. Thus, there is a need for identifying novel approaches to enhance the control of APEC infections in poultry.
APEC possesses several virulence factors that have been determined to be involved in different stages of pathogenesis, including P fimbriae, type IV pili, curli, invasins, iron acquisition systems, serum complement resistance factors, anti-phagocytic factors, hemolysin E, outer membrane proteins A, and vacuolating autotransporter toxin [2,9,10]. Pathogenicity of APEC is also regulated by quorum sensing (QS) systems [2,11], which are mechanisms of bacterial cell-to-cell communication that involve density-dependent production, release and detection of extracellular signaling molecules called autoinducers (AIs) [12,13]. The QS autoinducer-2 signal molecule (AI-2) allows interspecies communication [14] and regulates expression of genes involved in various processes, including secretion of virulence factors, biofilm formation, motility, genetic competence, sporulation, and antibiotic production [12,15,16]. Furthermore, luxS which mediates the synthesis of AI-2 has also been shown to regulate motility, biofilm formation, virulence and pathogenesis of many bacterial pathogens, including APEC [2]. Therefore, inhibition of luxS and/or AI-2 activity using novel intervention such as QS small molecule inhibitors (QSI) can be a potential strategy for novel antibacterial development. As the QSI do not interfere with the metabolic processes of a bacterial cell such as protein synthesis, DNA metabolism, cell wall formation which are the targets for the development of drug resistance [17], they do not exert selection pressure on the bacteria during treatment, thus bacteria are less likely to develop resistance [17].
Vibrio harveyi, a marine Gram-negative bioluminescent bacterium, regulates its luminescence through QS [18]. V. harveyi regulates bioluminescence via two-component signaling systems, AI-1 and AI-2, each is composed of a sensor–autoinducer pair [18,19]. V. harveyi AI-2 indicator is capable of detecting the AI-2 of many bacteria that produce similar AI-2 molecules and stimulate light production following the addition of cell-free culture supernatants from these bacteria [20]. For example, AI-2 producing bacteria such as E. coli, Salmonella typhimurium, Pseudomonas aeruginosa, and Vibrio cholerae have been reported to produce signaling molecules that stimulate bioluminescence production in V. harveyi [19,20].
Here, we used V. harveyi BB170 AI-2 indicator bacteria (AI-1+ and AI-2-) [18] to identify small molecule AI-2 inhibitors of APEC. The selected AI-2 inhibitors were tested in vitro to evaluate their toxicity in human intestinal cells, chicken macrophages, chicken and sheep red blood cells (RBCs) and determine their efficacy against APEC infection in chicken and human macrophages and in wax moth (Galleria mellonella) larva model. Further, the expression of several QS, virulence, biofilm and motility associated genes were assessed to provide insight on their potential mode(s) of action on APEC O78. We identified 10 small molecules (SMs) that modulated APEC infection in vitro and in vivo. These SMs can serve as leads for future application in APEC control in poultry and thereby also reduce potential zoonotic transmission of APEC to humans.
Results
Primary screening identified 69 compounds that significantly inhibited AI-2 activity of APEC
A library of 4,182 small molecules (SMs) was screened for APEC O78 growth inhibition. The results showed that 4,122 molecules did not impact the growth of APEC O78 (no elevated OD) (Figure 1A). Supernatants from cultures grown in the presence of 4,122 compounds were screened for their ability to inhibit bioluminescence of the V. harveyi AI-2 indicator. Inhibition of bioluminescence by SM treated APEC O78 culture supernatants was compared to that of the DMSO treated control. A total of 69 compounds inhibited ≥ 75% of the bioluminescence activity. These 69 compounds were further tested for AI-2 mediated bioluminescence inhibition in four independent experiments and 10 compounds (referred hereafter as AI-2 inhibitors) that showed highest AI-2 inhibition (75–98%) were selected for further studies (Figure 1B). However, our analysis revealed that only 25% of AI-2- associated bioluminescence inhibition was achieved with some AI-2 inhibitors (C3- C5, C7, C9 and C10) at 50 µM and no inhibition was observed at lower concentrations (data not shown).
Figure 1.
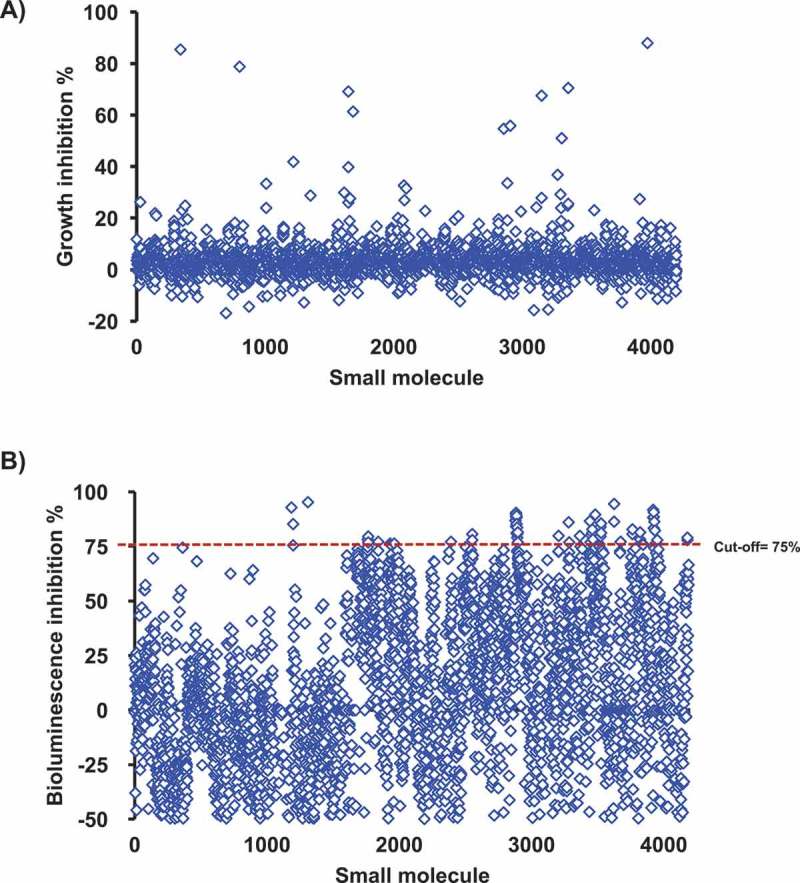
A) High-throughput screening for growth inhibition against APEC O78. 4,122 compounds did not impact the growth of APEC O78 (no elevated OD). B) Screening of the SMs for their effect on the AI-2 activity of APEC O78. The Vibrio harveyi BB170 AI-2 bioluminescent indicator bacteria was used to screen the cell-free culture supernatant of APEC O78. APEC cell-free culture supernatant was prepared from cultures grown in the presence of 100 μM of small molecules. Sixty nine compounds inhibited ≥ 75% of the AI-2 activity of APEC O78. AI-2 bioluminescence indicator assay was repeated four times for these 69 compounds and 10 compounds that showed highest AI-2 inhibition were selected for further studies.
Further, the selected AI-2 inhibitors displayed differential effect on the inhibition of AI-2 activity of multiple APEC serotypes. Most compounds resulted in ≥ 75% inhibition of AI-2 activity of APEC O2 with the exception of C3 and C9, while only C1 and C6 resulted in ≥ 75% inhibition of AI-2 activity of APEC O1. Only C6 exhibited ≥ 75% inhibition of AI-2 activity of O8, O15, O18, and O35. Whereas C8 exhibited ≥ 75% inhibition of AI-2 activity of O109, while C6 and C10 exhibited ≥ 75% inhibition of AI-2 activity of O115 (Table 1).
Table 1.
Effect of the selected SMs on the AI-2 activity of different APEC serotypes.
| % Inhibtion |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Serotypes | C1 | C2 | C3 | C4 | C5 | C6 | C7 | C8 | C9 | C10 | Sources |
| O78 | 84 | 92 | 78 | 75 | 83 | 92 | 94 | 98 | 79 | 84 | [62] |
| O2 | 94 | 96 | 54 | 76 | 89 | 93 | 83 | 94 | 51 | 89 | [63] |
| O1 | 76 | 64 | 65 | 63 | 46 | 93 | 47 | 41 | 19 | 64 | [4] |
| O8 | 20 | 20 | 39 | 0 | 18 | 79 | 2 | 18 | 20 | 0 | [63] |
| O15 | 13 | 27 | 0 | 46 | 45 | 97 | 36 | 25 | 0 | 22 | [63] |
| O18 | 37 | 43 | 8 | 54 | 61 | 95 | 48 | 54 | 14 | 35 | [63] |
| O35 | 48 | 55 | 37 | 60 | 68 | 96 | 62 | 57 | 36 | 47 | [63] |
| O109 | 65 | 57 | 0 | 67 | 69 | 60 | 67 | 75 | 66 | 57 | [63] |
| O115 | 64 | 52 | 35 | 58 | 63 | 99 | 45 | 44 | 44 | 78 | [63] |
Inhibition of AI-2 activity was calculated by comparing bioluminescence inhibition from SM treated APEC O78 culture with that of the DMSO treated control
The selected AI-2 inhibitors affected biofilm formation and motility of APEC O78
Biofilms play a crucial role in APEC virulence and enhance the bacterial resistance to antimicrobials and immune clearance, leading to failure of antimicrobial therapy [21,22]. Therefore, quorum sensing inhibitors have been proposed as promising anti-biofilm agents [21,22]. The effect of the AI-2 inhibitors on APEC O78 biofilm formation was determined using the crystal violet staining (CV) assay. Biofilm formation was assessed after 48 h of incubation in the presence of 100 µM of each compound. We noted that C3 – C7 resulted in 63–66% reduction of APEC O78 biofilm formation, while C2, and C8 – C10 resulted in 52–58% reduction of APEC O78 biofilm formation (P < 0.05) (Figure 2A).
Figure 2.
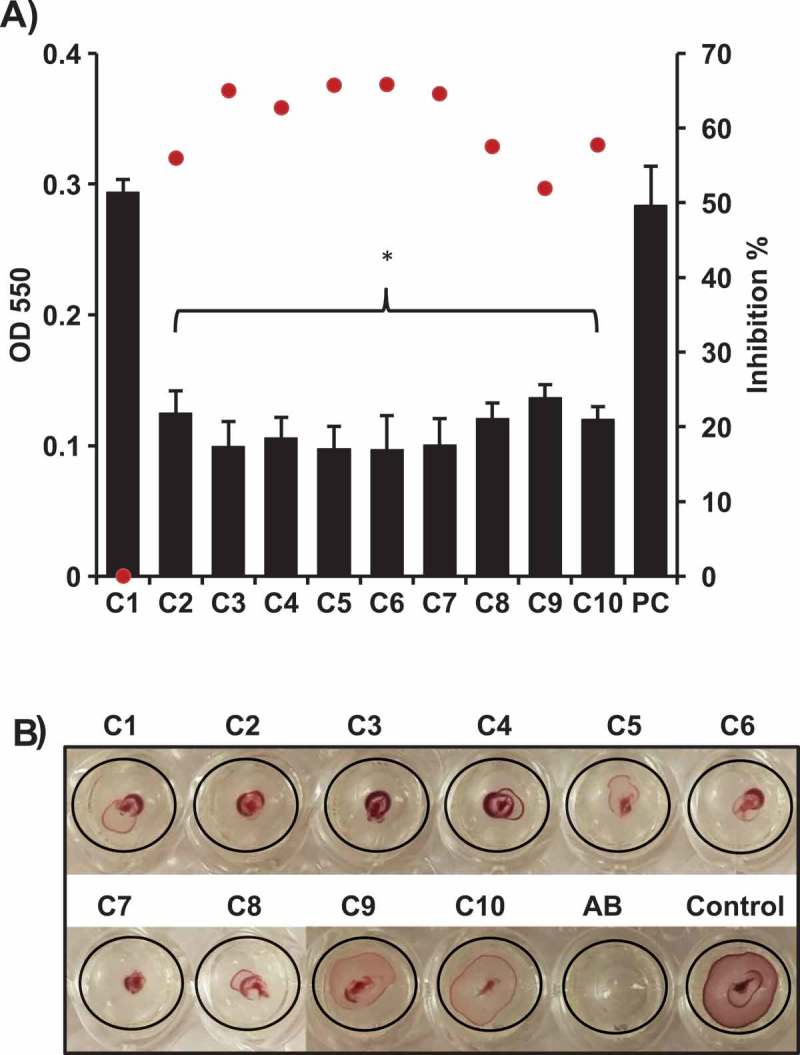
Effect of the selected AI-2 inhibitors on; A) biofilm formation of APEC O78. Biofilm formation was assessed using crystal violet assay by measuring the OD550. All of the compounds significantly inhibited the biofilm formation except C1 in comparison to the DMSO treated control (PC). Two independent experiments were conducted with triplicate wells in each experiment and the average OD550 (black bars) and inhibition % (red dots) are shown. B) motility of APEC O78. Motility was assessed by measuring the halo on a semisolid agar. Except C9 and C10, all compounds resulted in complete inhibition (did not form detectable motility halos) in comparison to the DMSO treated control. *Significant difference between AI-2 inhibitors treated compared to DMSO treated control (P < 0.05).
Motility and chemotaxis allow bacteria to migrate towards or away from favorable environments in response to stimulus and thus contribute to bacterial fitness and virulence [23]. The effect of the AI-2- inhibitors on motility of APEC O78 was determined using 100 µM of each compound. With the exception of C9 and C10, all compounds resulted in inhibition (did not form detectable zones of motility) of APEC O78 motility after 6 h of incubation in comparison to the DMSO treated control (Figure 2B). Notably, the diameter of the zone of motility was reduced up to 75% in comparison to DMSO treated control (from 8 mm in DMSO treated control to 2 mm on an average in AI-2 inhibitors treated wells).
The AI-2 inhibitors showed low toxicity on human colonic adenocarcinoma epithelial cells (Caco-2) and chicken macrophage cells (HD-11) and no hemolytic activity on sheep and chickens red blood cells (RBCs)
The toxicity of the selected AI-2 inhibitors was assessed on Caco-2 and HD-11 cells using lactate dehydrogenase (LDH) assay to quantitatively measure the LDH released into the media from damaged cells after exposure to AI-2 inhibitors as an indicator for cellular cytotoxicity and cytolysis. Our results showed that, when treated with 100 µM of SMs, C8 and C4 exhibited 2% and 12% cytotoxicity, respectively; while C6 and C10 exhibited 27% cytotoxicity and C1-C3, C7, and C9 displayed 36% to 42% cytotoxicity on Caco-2 cells. Further, in HD-11 cells, C5 and C8 exhibited less than 10% cytotoxicity, while C2, C4, C6, and C7 exhibited cytotoxicity between 12% and 18%, and C1, C3, C9 and C10 exhibited toxicity between 22% and 27% (Figure 3A).
Figure 3.
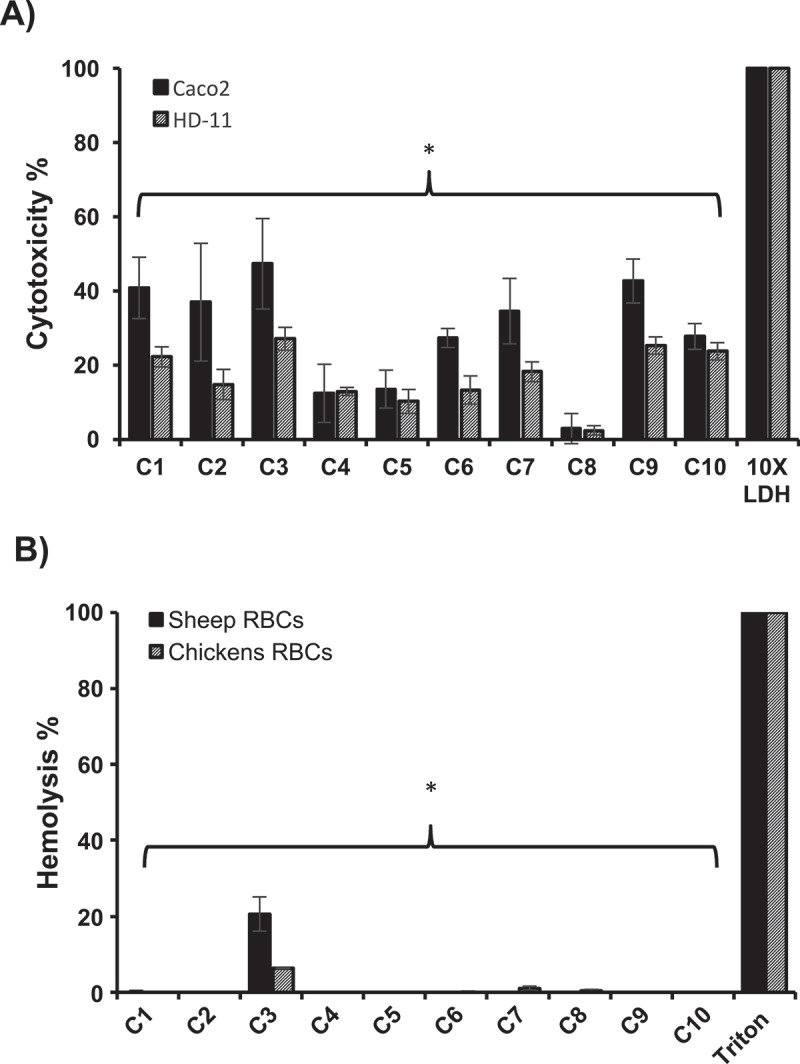
Cytotoxicity (A) and hemolytic activity (B) of the selected AI-2 inhibitors. Cytotoxicity was assessed using Caco-2 and HD-11 cells and hemolytic activity was determined using sheep and chicken RBCs. 100 μM of each compound was used in both assays. Most of the compounds showed significantly less cytotoxicity and hemolytic activity (P ≤ 0.05) compared to DMSO treated control. Two independent experiments were conducted with triplicate wells in each experiment and the average is shown. *Significant difference between AI-2 inhibitors treated wells compared to 10X LDH (cytotoxicity assay control) or Triton X-100 (hemolytic activity assay control).
Additionally, toxicity of the selected AI-2 inhibitors to sheep and chicken RBCs was also tested using 100 µM of each compound. Notably, all compounds exhibited no hemolytic activity against sheep or chicken RBCs with the exception of C3, which exhibited 20.6 % and 6.4% hemolytic activity in sheep and chicken RBCs, respectively (Figure 3B).
The selected AI-2 inhibitors reduced the survival of APEC O78, O2, and O1 in chicken and human macrophage cells
The invasion of APEC into phagocytic cells facilitates its intracellular survival and systemic spread to different organs which is critical for APEC pathogenesis [24]. Therefore, the impact of the AI-2 inhibitors on APEC O78, O1 and O2 (the most predominant serotypes associated with colibacillosis) survival in HD-11 and acute human leukemia macrophage (THP-1 cells) was assessed using 100 µM of each compound. In HD-11 cells, treatment with C1-C3, C5-7, C9 and C10 resulted in 100% clearance of APEC O78 (~ 4 logs reduction; P < 0.001), while C4 and C8 significantly reduced its intracellular survival (up to 1 log; P < 0.05) when compared to DMSO treated control. Additionally, C1-C3, C5, C6, C9 and C10 resulted in 100% clearance of APEC O2 (~ 3.3 logs reduction; P < 0.001), and C4, C7 and C8 significantly reduced its intracellular survival (up to 0.8 log; P < 0.05). Only C3 exhibited 100% clearance of APEC O1 (~ 4 logs reduction; P < 0.001), while C5- C8 and C10 significantly reduced its intracellular survival (up to 0.9 log; P < 0.05) (Figure 4A).
Figure 4.
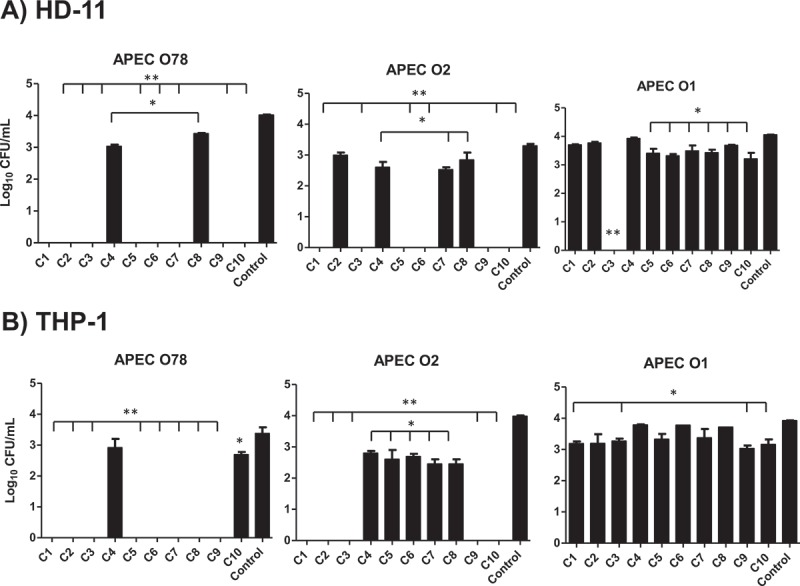
Effect of the AI-2 inhibitors on the intracellular survival of different APEC O78, O2, and O1 in HD-11 (A) and THP-1 (B) cells. Cells were infected with APEC strains at MOI = 100, treated for 6 h with 1 µL (100 µM) of each compound and the internalized bacteria were determined. Two independent experiments were conducted with triplicate wells in each experiment and the average is shown. *Significant difference between AI-2 inhibitors treated cells (P < 0.05) compared to DMSO treated control. **Significant difference between AI-2 inhibitors treated cells (P < 0.001) compared to DMSO treated control.
Similarly, in THP-1 cells, C1- C3, and C5- C9 exhibited 100% clearance of APEC O78 (~ 3.4 logs reduction; P < 0.001), while C10 significantly reduced its intracellular survival (up to 0.7 log; P < 0.05). C1- C3, C9 and C10 exhibited 100% clearance of APEC O2 (4 logs reduction; P < 0.001), and C4- C8 significantly reduced its intracellular survival (up to 1.5 logs; P < 0.05). Similarly, C1, C3, C9 and C10 significantly reduced the intracellular survival of APEC O1 (up to 0.9 log; P < 0.05) (Figure 4B). Interestingly, we observed varying effects of the AI-2 inhibitors on different internalized APEC serotypes in HD-11 and THP-1 cells. Specifically, some AI-2 inhibitors were less effective against O1 and O2. This variation might be due to differences in their capsule composition. Both the O1 and O2 possess K1 (sialic acid) containing capsule, which confers protection against host immunity and increase resistant to the bactericidal effect of serum as compared to O78 which expresses different capsular antigens [25,26]. In addition, O1 also contains pathogenicity island (PAI IAPEC-O1) that carries pap operon (putative virulence genes of APEC), invasion determinant protein encoding gene tia and iron-regulated outer membrane protein encoding gene ireA which are likely to contribute to increased virulence of APEC O1; thus, the lower efficacy of SMs [27].
The AI-2 inhibitors increased the survival of APEC infected wax moth (Galleria mellonella) larvae
The in vivo efficacy of the AI-2 inhibitors was evaluated using the wax moth larva model, which has been previously used to evaluate drug efficacy and bacterial pathogenesis [28]. Treatment of the infected larvae with C1 and C2 resulted in 23% and 30% increase in larval survival, respectively; while compounds C3-10 resulted in 40–60% increase in larval survival in comparison to DMSO treated controls (Figure 5A).
Figure 5.
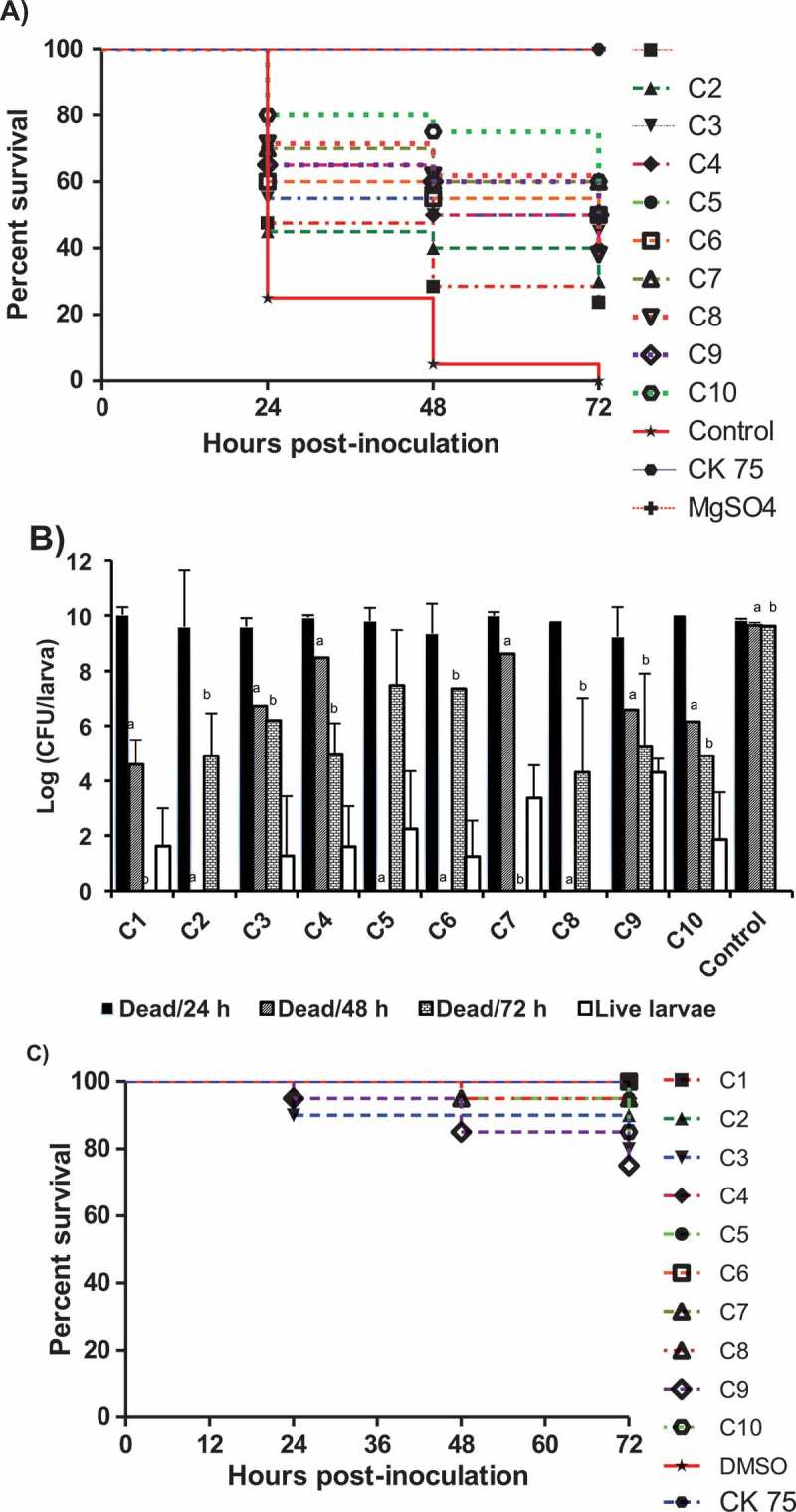
Effect of the AI-2 inhibitors on G. mellonella larvae infected with APEC O78; A) survival rate of the treated larvae, B) bacterial load in dead larvae at different time points (24, 48, 72 h) and in live larvae at 72 h, and C) toxic effect of each compound on G. mellonella larvae. The larvae were treated with the AI-2 inhibitors (12.5 µg/larva) and inoculated with APEC O78 (8.5 µL ~ 4.25 × 104 CFU), survival monitored for 72 h. The results of two independent experiments were averaged. Significant difference between AI-2 inhibitors treated compared to DMSO treated control at 48 ha and 72 hb (P < 0.05).
Treatment of the wax moth larvae with the AI-2 inhibitors also reduced APEC O78 colony forming units (CFU) in both dead and live larvae in comparison to DMSO control. At 72 h post treatment, the bacterial load in dead larvae in all treated groups, except those treated with C1 and C7 that showed no larval death, were significantly reduced (~ 4.3–7.5 logs; P < 0.05) in comparison to DMSO treated control. However, in surviving larvae, C2 and C8 resulted in 100% clearance of APEC (up to 5 logs; P < 0.001), while C4-C6 and C10 significantly reduced the bacterial load (up to 3.5–6 logs; P < 0.05) in comparison to dead larvae. Further, at 48 h post treatment, C2, C5, C6 and C8 treated groups showed no larval death while the rest of the compounds resulted in significant reduction of the APEC O78 load (1.2–3.5 logs; P < 0.05) in comparison to DMSO treated controls (Figure 5B). However, the larvae died at 24 h post treatment possessed APEC load similar to DMSO treated controls. These results indicated that the bacterial load in all AI-2 inhibitors treated groups, except C9, significantly decreased over time (r ≤ −0.87; p ≤ 0.001) in comparison to DMSO treated control.
Notably, all AI-2 inhibitors showed no or low toxicity in larvae. The survival of larvae treated with C1 and C6 was 100%, whereas the survival of larvae was more than 75% for the rest of the AI-2 inhibitors (Figure 5C). These results suggest that the larval death is not due to the toxic effect of AI-2 inhibitors but rather due to APEC O78 infection. Thus, the wax moth model may serve as rapid screening tool to assess the efficacy of potential antimicrobials against APEC and for selection in in vivo studies in the natural host.
The QS AI-2 inhibitors affected the expression of virulence, biofilm formation and motility- associated genes of APEC
Quorum sensing inhibitors have been proposed to affect the expression of QS regulated and virulence- associated genes [17]. Therefore, to elucidate how the AI-2 inhibitors might attenuate APEC pathogenicity [2,13], we assessed the expression of genes representing multiple physiological processes regulated by QS (Table 2) such as AI-2 synthesis (2 genes), small molecules metabolism (5 genes), virulence factors (6 genes), biofilm formation, cell motility and exopolysaccharide formation (11 genes), cell division, DNA processing, and morphology (5 genes).
Table 2.
Real Time PCR Primers used in this study.
| Function | Genes | Gene product | Oligonucleotide sequence (5’ to 3’) | product size (bp) |
|---|---|---|---|---|
| AI-2 synthesis, biofilm formation, cell motility and exopolysaccharide formation | luxS | S-ribosylhomocysteinase | F: ACGAGTGCATCTGGTAAGTG R: CAATGGAAGACGTGCTGAAAG |
87 |
| pfs | 5’-methylthioadenosine/S-adenosylhomocysteine nucleosidase | F: CGGCAGAACCGGTGTTAATAAT R: TGAAATCGGGCATCGGTAAAG |
96 | |
| hha | Hemolysin expression-modulating protein | F: GTGATCTGCGGCTGAGTAAA R: ACGTCGTTGCCAGACAAT |
103 | |
| wzb | Protein-tyrosine-phosphatase | F: TCGTTATCCCAGTGACCAAAC R: GTGTCGCAACTATGACCTGAT |
114 | |
| ompG | Outer membrane protein G precursor | F: CGGTTGGCTGTCGATGTATAA R: AGGTATATTGCAGACCCGTTTC |
95 | |
| rcsB | Transcriptional regulator RcsB | F: CAAGTACATCAAGCGCCATTTC R: CCCTTCGATATCCAGATCCAATAC |
103 | |
| motB | Flagellar motor protein B | F: AGGCTAATACGGTTGGGAATAC R: AGAATCGCCCGATGTTTAGAA |
109 | |
| flgN | Flagella synthesis chaperone protein FlgN | F: CCAGTAACCAGCCGTTATGTT R: TACGCAGGAAAGAACCCAATAC |
117 | |
| fliP | Flagellar biosynthetic protein FliP | F: AGCCATTCAGCGAAGAGAAA R: GTCTGGCAAACAACCCTAAATC |
117 | |
| cheW | Purine-binding chemotaxis protein CheW | F: CATCCACCTGGCTGAACTTA R: GTAACACGGATTGCGAACAC |
106 | |
| nlpC | Probable lipoprotein NlpC precursor | F: GCATCGTCACAACCACAAATC R: GGTTTGAACGACCAGCTACA |
103 | |
| Virulence- associated genes | ompA | Membrane protein | F: TGACCGAAACGGTAGGAAAC R: GGAATACCAGTGGACCAACAA |
99 |
| fimC | Fimbrial chaperone protein | F: GCTGGCAGGTATCCTGATATTC R: TGCCCTGCCGGATAAATTAC |
99 | |
| IucD | IucD protein | F: GTCCGGAGAAGCCTGAAATA R: GAGAAGCGGCGGAAATAAAC |
114 | |
| fyuA | Ferric yersiniabactin uptake A | F: CTTCCCTTCCGGTTCGTTAATC R: GGTACAGCCCAAACACCATATC |
119 | |
| iss | Serum survival protein | F: CGCTCTGGCAATGCTTATTAC R: GAAATGATGGGTGATGGTTTCC |
100 | |
| vat | Vacuolating autotransporter toxin | F: CTGAACCGCGTCCAGATTAT R: ACTCCACGGCAGGAAATATG |
104 | |
| Cell division, DNA processing, and morphology | bolA | BolA protein | F: CAGTACTTTAGCGGAGGAACTC R: CAACCCTTCCCACTCCTTAAT |
82 |
| mreD | Rod shape-determining protein MreD | F: CCGGTCTGAAAGAGACGTTAAT R: TTCCGCAACCTCGCATTAT |
115 | |
| csrA | Carbon storage regulator A | F: GTAACTGGACTGCTGGGATTT R: CCAGGTACGTATTGGCGTAAA |
100 | |
| murD | UDP-N-acetylmuramoylalanine- D-glutamate ligase | F: GCGTGGTTAATGCTGATGATG R: CCTGCTGATGATTCAGGTGATA |
108 | |
| lpp | Major outer membrane lipoprotein precursor | F: CGGTAATCCTGGGTTCTACTCT R: TGCTCAGCTGGTCAACTTTAG |
108 | |
| Small molecules metabolism | frwC | Fructose-like-2 IIC component | F: AGCAGGGCAGCATTGTTAT R: GCAAATCAGCATGAAGGCATAG |
104 |
| Fpr | Ferredoxin-NADP reductase | F: GTTCCTGCATCAGAGGTAGATAG R: TACAGCGATTGGCCCTTATT |
128 | |
| mtlR | Mannitol operon repressor | F: CGTCATTAACCGCCAGGAATA R: GTTCACCAAAGGGTCCAAGTA |
122 | |
| ugpC | SN-glycerol-3-phosphate transport ATP-binding protein UgpC | F: CTGATCGTGGGTAACGTAGAG R: GCAGTGTTCCTGTTTGATGAG |
126 | |
| thiH | Thiazole biosynthesis protein ThiH | F: GCATAAGTCGCCTCGTGATATG R: GACGGAATACGCCGAGTTAAAG |
81 | |
| Housekeeping gene | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | F: CGGTACCGTTGAAGTGAAAGA R: ACTTCGTCCCATTTCAGGTTAG |
99 |
Interestingly, all compounds, with the exception of C4, down-regulated the expression of cytosolic S-ribosylhomocysteine lyase gene (luxS), which mediates AI-2 synthesis. Notably, C4 up-regulated the expression of 5’-methylthioadenosine/S-adenosylhomocysteine nucleosidase gene (pfs), which also mediates AI-2 synthesis (Figure 6A). Further, AI-2 inhibitors affected group of genes that influence AI-2 production and degradation and small- molecules metabolism. These genes have not been directly implicated in cell- cell communication but provide a link between QS and central metabolism [13]. The AI-2 inhibitors down-regulated genes encoding inner membrane sugar transport fructose-like-2 IIC component (frwC; in response to C5- C8 and C10), carbohydrate transport sn-glycerol-3-phosphate import ATP-binding protein (ugpC; C1, C2, C4- C6, C8 and C9), cytoplasmic ferredoxin-NADP reductase (fpr; C3, C5 and C6), and thiazole biosynthesis protein (thiH; C1 and C3- C10) (Figure 6A).
Figure 6.
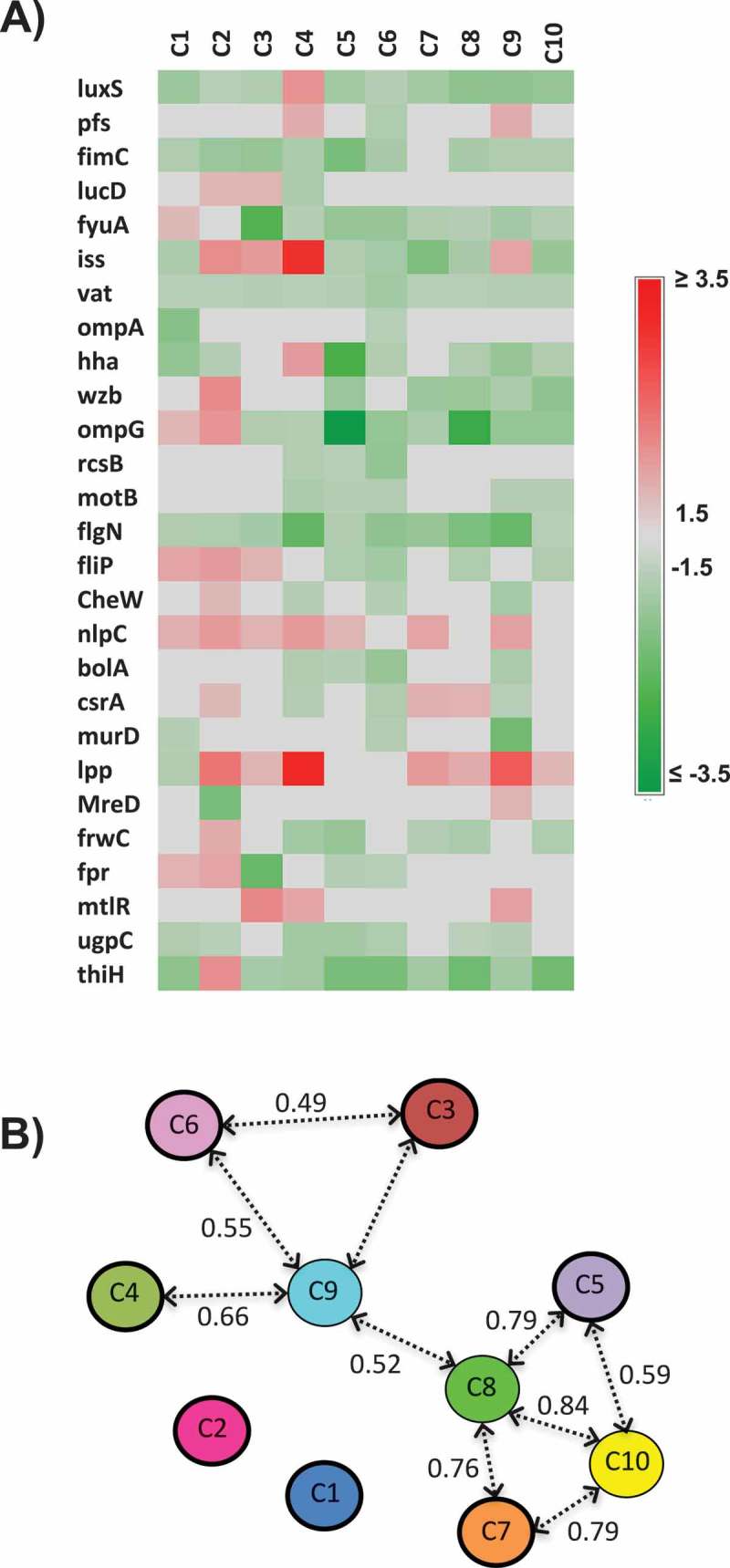
Effect of the AI-2 inhibitors on expression of QS and virulence-associated genes; A) Heat map showing the impact of the selected AI-2 inhibitors on gene expression of QS and virulence associated genes. Effect on gene expression was assessed using 100 µM of AI-2 inhibitors. Three independent experiments were conducted and the average fold change was calculated using ∆∆ct. Fold changes ± 1.5 ≥ or ≤ 1.5 were considered differentially expressed. B) Principal component analysis (PCA) of the qRT-PCR data. Numbers above the arrows indicate the correlation (r) between the AI-2 inhibitors based on the gene expression data. The PCA analysis is based on the fold change of gene expression data.
Since the expression of virulence factors by APEC is among many traits controlled by QS [29], inhibition of these virulence factors by QSI might attenuate APEC virulence. Notably, AI-2 inhibitors down-regulated the expression of genes encoding periplasm fimbrial chaperone protein fimC; C1- C6, and C8- C10), iron uptake chelate protein D (iucD; by C4), ferric yersiniabactin uptake A (fyuA; by C3- C10), serum survival protein (iss; by C1, C5- C8, and C10), and vacuolating autotransporter toxin (vat; by C1-C10). However, C2- C4 and C9 up-regulated the expression of iss (Figure 6A).
AI-2 inhibitors also affected the expression of biofilm formation, motility and exopolysaccharide formation associated genes. The AI-2 inhibitors down-regulated the expression of genes encoding the hemolysin-expression modulating protein (hha; C1, C2, C5, C6 and C8- C10), transcriptional regulator protein (rcsB; C4- C6), outer membrane protein G precursor (ompG; by C2- C10), and tyrosine-phosphatase protein (wzb; by C5, and C7- C10). Both hha and rcsB are located in the cytosol and were shown to be involved in biofilm formation, while ompG and wzb contributes to exopolysacchride formation. Likewise, genes associated with chemotaxis and motility, such as cytoplasmic flagellar synthesis chaperone protein (flgN; C1-10), chemotaxis protein (cheW; C4, C6, and C9), flagellar motor protein B (motB; by C4- C6, and C9- C10) and flagellar biosynthesis protein (fliP; by C5, C6, C8, and C10) were also down-regulated by AI-2 inhibitors (Figure 6A). However, C1-C5, C7, and C9 up-regulated the expression of cell membrane probable endopeptidase (nlpC).
Quorum sensing has also been implicated in the regulation of multiple physiological processes such as DNA replication, cell division and cell morphology [30]. The AI-2 inhibitors down-regulated the expression of genes encoding transcriptional regulators such as cytosolic DNA-binding transcriptional regulator (bolA; C4- C6, and C9), and carbon storage regulator A (csrA; C4, C6, and C9), and UDP-N-acetylmuramoylalanine- D-glutamate ligase (murD; C1, C6, and C9). However, C2-C4, and C7- C10 up-regulated the expression of major outer membrane lipoprotein precursor gene (lpp) (Figure 6A).
Principal component and multivariate analysis were performed to determine the gene expression profiles of APEC treated with the AI-2 inhibitors. The expression profiling divided the AI-2 inhibitors into 2 groups (r > 0.49; P < 0.01). The first group composed of C5, C7, C8 and C10 (0.59 ≤ r ≤ 0.84), while the second group composed of C3, C4, C6 and C9 (0.49 ≤ r ≤ 0.66). The C9 and C8 showed high correlation with other AI-2 inhibitors (r = 0.52; P = 0.005) and they connected these two groups together (Figure 6B). Further, C5 to C10 displayed similarity in their expression profile (r > 0.4; P < 0.05), while C1 and C2 showed no significant expression profiles similarity with the other AI-2 inhibitors (r < 0.33) (Figure 6B). Despite the high similarity scores between certain AI-2 inhibitors, the gene expression profiles differed between AI-2 inhibitors, suggesting that each AI-2 inhibitors might interact with different target(s) that potentially share common biological pathways within APEC.
Discussion
The increase in antibiotic resistant APEC [7], the limited effect of current vaccine [8], and the ability of APEC to establish persistent infections through the formation of biofilms [2] emphasize the need for alternate control strategies for APEC. A great effort has been made to develop anti-pathogenic drugs by reducing the bacterial virulence through QSIs, which proves to be an intriguing approach for antimicrobial chemotherapy [31]. In this study, we identified, using V. harveyi BB170 AI-2 indicator bacteria, 10 novel QS AI-2 inhibitors that did not inhibit APEC growth but interfered with QS- regulated processes including virulence factors, biofilm formation, motility, exopolysaccharide synthesis, stress survival, cell division and pathogenesis in APEC [13]. Use of QSI to attenuate APEC pathogenicity is attractive as this approach is less likely to result in the development of resistant APEC.
All 10 AI-2 inhibitors except C4, attenuated the AI-2–production and down-regulated the expression of luxS. Further, most of the SMs also down-regulated the expression of QS- regulated and virulence-associated genes such as pfs, iucD, fyuA, vat, ompA, iss, and fimC (Figure 6A). Similarly, APEC luxS deletion mutant has been reported to affect AI-2 activity and down-regulate virulence-associated genes [2,32]. In addition, deletion of luxS also resulted in reduced adherence and invasion abilities of APEC O78 and, consequently, its survival in cultured cells [2,5]. Adherence and invasion are important for APEC pathogenesis and mediate colonization, survival and spread in the host [33]. Our results suggest that the selected AI-2 inhibitors, with the exception of C4, might interfere with synthesis, secretion, and/ or transport of AI-2 via their effect on the luxS; affecting QS and cognate pathogenicity of APEC [34]. However, C4 also inhibited AI-2 production, this suggests that C4 might intervene at various points in the AI-2 production cycle without having a direct effect on luxS and/ or luxS dependent mechanisms.
In this study, we found that C2- C9 reduced biofilm formation of APEC O78 (Figure 2A). This result was supported by the inhibitory effect of these compounds on biofilm formation- associated genes (Figure 6A) such as 1) hha, which also influences persister cell formation and bacterial resistance to antibiotics [35]; 2) rcsB, ompG and wzb which are involved in colanic acid capsule biosynthesis that protects the cells against contact-dependent growth inhibition and cell division [36,37]; 3) bolA and murD which are involved in the production of fimbria-like adhesins, curli and colanic acid and regulate cell morphology, cell wall formation, permeability, cell growth and division, motility and peptidoglycan biosynthesis [23,38]; and 4) csrA which regulates central carbohydrate metabolism, glycogen synthesis, gluconeogenesis, cell size and surface properties, motility and flagellum biosynthesis [39]. These findings suggest that the identified AI-2 inhibitors possess anti-biofilm effect against APEC O78 likely through the down-regulation of genes associated with adhesion, motility, and capsule synthesis among others [40]. Our results corroborate previous findings that show other QSIs such as furanone C-30 which inhibited biofilm formation in Streptococcus mutans through the inhibition of QS-regulated genes [41]. Additionally, bacterial motility plays an important role in virulence, biofilm formation and pathogenicity of bacteria [21]. Therefore, motility is considered as an attractive target for AI-2 inhibitors for preventing and/or blocking the infection process. In our study, C1- C8 inhibited APEC O78 motility (Figure 2B). This result was supported by the inhibitory effect of these compounds on motility-associated genes (Figure 6A) such as flgN, motB and fliP. Previously, motility inhibitors have been used to control V. cholerae and S. typhimurium which acted through the down-regulation of flagellar synthesis genes [42,43].
It is known that APEC pathogenesis is controlled by a number of virulence factors and inhibition of these virulence factors by AI-2 inhibitors can render APEC non-virulent and attenuate its pathogenicity. Previously, QSIs such as: furanone C-30 has been reported to regulate virulence of P. aeruginosa through the reduction of QS-regulated virulence factors such as protease, pyoverdin and chitinase and their associated proteins [44; 44]. Hamamelitannin, QSI acts through the inhibition of RNAIII, part of the agr QS system in S. aureus [45], while virstatin was reported to inhibit V. cholerae virulence factors such as cholera toxin and toxin coregulated pilus [46]. Interestingly, AI-2 inhibitors in our study also down-regulated a group of the virulence associated genes (Figure 6B) such as 1) fimC, which plays an important role in APEC adherence to the cells through the regulation of adhesion of type 1 fimbriae [47], 2) iron uptake receptors (fyuA and lucD) which are involved in biofilm formation [48], 3) iss which plays a role in resistance against serum complements [49], and 4) vat which regulates the motility, agglutination, and biofilm formation of APEC [50]. This indicated that the AI-2 inhibitors might interfere with bacterial iron acquisition, metabolism, adhesion and invasion, serum survival; thus, modulate APEC infection of host cells. Notably, all AI-2 inhibitors, except C4, reduced intracellular APEC O78 in chicken and/or human macrophages (Figure 4 A,B), and enhanced G. mellonella survival and reduced APEC loads in surviving larvae (Figure 5(b)).
The two-dimensional structure of the 10 selected AI-2 inhibitors was analyzed using a 2D Tanimoto scoring method [51]. The 10 AI-2 inhibitors were divided into two clusters; the large cluster composed of nine compounds (C1- C3 and C5- C10), while the small cluster was formed only by C4 (Figure 7). All nine compounds within in the larger cluster inhibited the luxS expression, while C4 did not, suggesting a potential association between the chemical structure and the mode of action of the compounds in APEC. Four compounds (C2, C3, C5, C10) possess high structural similarities between each other (chemical structure similarity score = 0.71). C5 and C10 are both composed of a phenylpropyl piperazinyl phenyl groups and showed similarity in their effect on expression profile (r > 0.4; P < 0.05, Figure 6A, B) of QS and virulence associated genes. Similarly, both C2 and C10 are composed of a sequence of piperidinyl phenyl piperazinyl groups; however, C2 displayed better in vitro effect compared to C10, while C10 displayed better in vivo efficacy in wax moths compared to C2 (Figure 5(a)). These findings suggest that the sequence of phenyl piperazinyl functional groups might be key contributors to the AI-2 inhibitory activity of C2, C5 and C10. However, this conclusion requires further experimental analysis.
Figure 7.
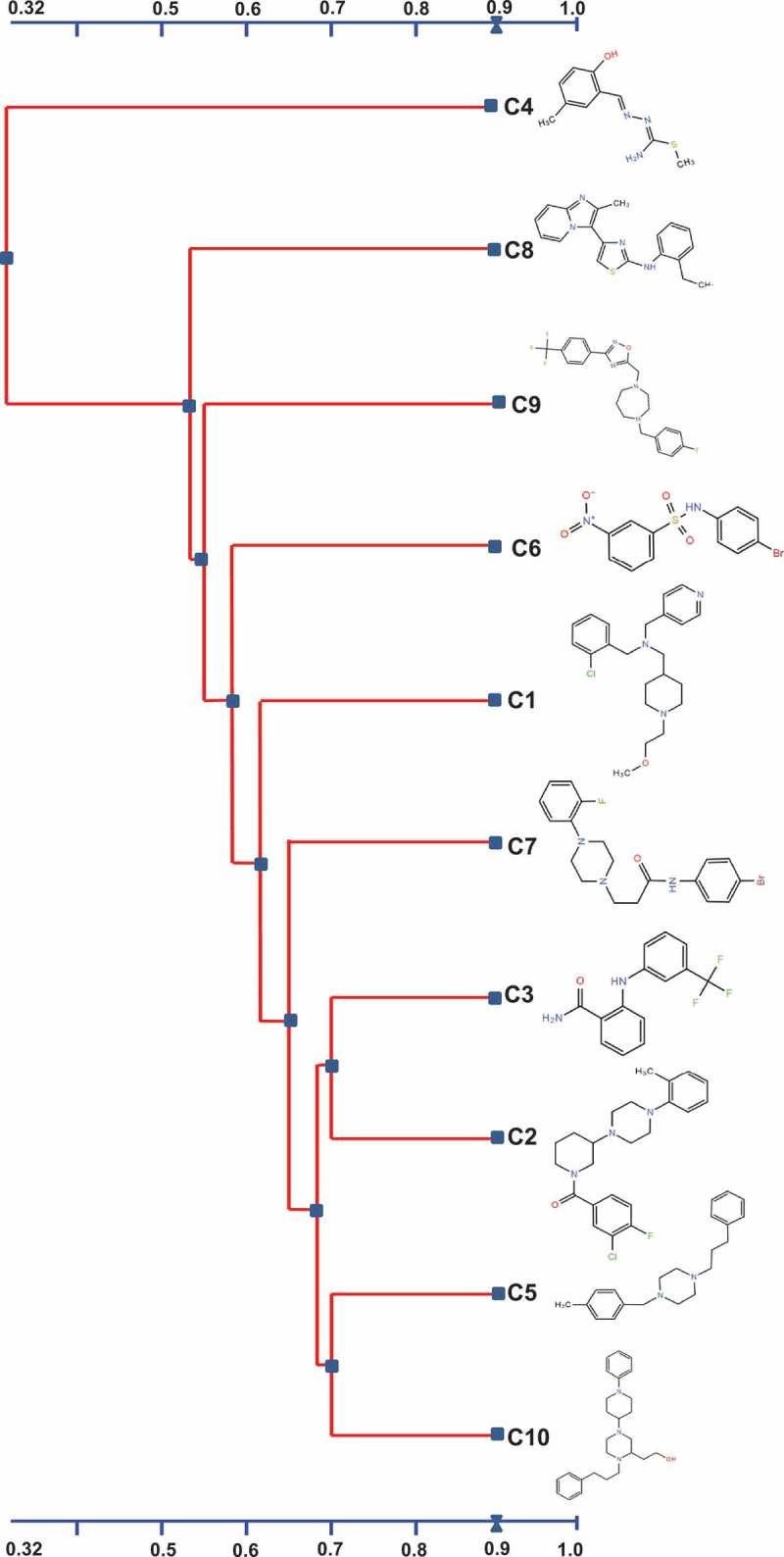
Alignment and the chemical structures of the top 10 AI-2 inhibitors using 2D Tanimoto similarity scoring method. A chemical structure similarity score of 0.68 is statistically significant at the 95% confidence interval.
Although, both C5 and C7 contain piperazinyl functional group (score = 0.58), C6 contains nitrobenzene sulfonamide functional group. However, C6 and C7 have some structural similarity (contain 4- bromophenyl group; score = 0.59), which might explain their similar effect both in vitro and in vivo. C1, C2 and C10 contain piperidine functional group (score = 0.61) but C1 displayed no effect on biofilm formation (Figure 2A) and lower efficacy in vivo compared to C2 and C10 (Figure 5A). This might be due to the presence of piperazine functional group in addition to piperidine functional group in both C2 and C10 suggesting that presence of piperazine functional group might be the key groups contributing to the activity of these compounds (Figure 7). Previously, piperazine based drivatives have been reported to inhibit V. cholerae and P. aeruginosa through down-regulation of RND efflux virulence and AHL- LasR QS-dependent factors, respectively [52,53].
Conclusions
The potential AI-2 inhibitors identified in this study differentially affected the pathobiology of APEC and the expression of QS regulated virulence, biofilm, and motility genes. C5, C6, C7 displayed highest efficacy both in vitro and in vivo and the inhibitory activity seems to be related to piperazine based functional group. Discovery of novel antimicrobials that do not affect bacterial growth is less likely to impose a selective pressure for resistance development by the bacteria and represents an ideal antibiotic independent approach. Further in vivo studies in chickens and specific target identification of these potential leads will facilitate development of these AI-2 inhibitors as novel therapeutics to augment APEC control in poultry and prevent its potential zoonotic transmission. Because of their small size and convenient synthesis scheme, we expect the cost of large scale synthesis of these inhibitors to be equivalent or less than the cost associated with synthesis of current antibiotics used in the industry to treat APEC infections. In addition, since human pathogenic ExPECs genetically resemble APEC, our future studies will also test the effect of these inhibitors against human pathogenic ExPECs particularly those that are multidrug resistant.
Material and methods
Small molecules library
A library of 4,182 compounds (yactives) were selected via pre-screening of 81,320 compounds and were obtained from Chembridge, Inc. (San Diego, CA, USA) [54]. These compounds were dissolved in 100% dimethyl sulfoxide (DMSO) to a concentration of 10 mM in a 96-well plate and stored at −80 °C for further use.
Bacterial strains and culture conditions
APEC serotypes O1 (GenBank accession no. NC_008563), O2 (GenBank accession no. NZ_CP006834), and O78 (GenBank accession no. CP004009) were kindly provided by Dr. Tim Johnson (University of Minnesota, Saint Paul, MN). Other APEC serotypes such as O8, O15, O18, O35, O109, and O115 were kindly provided by Drs. Lisa K. Nolan and Catherine M. Logue (University of Georgia, Athens, GA). Luria-Bertani broth (LB; BD Difco) was used for routine propagation of APEC serotypes. APEC serotypes were routinely grown in LB broth overnight at 37 °C with shaking at 200 rpm. Rifampicin resistant APEC O78 (RifR) was isolated by plating the APEC O78 on LB agar containing 50 µg/mL of rifampicin [55], and one spontaneous resistant mutant was used for the wax moth studies. E. coli DH5α (Invitrogen) was grown overnight in LB broth at 37 °C with shaking at 200 rpm. V. harveyi BB170 (AI-1+; AI-2-) and V. harveyi BB120 (AI-1+; AI-2-) were kindly provided by Dr. Bonnie L. Bassler (Princeton University) and were grown in AB medium at 30 °C aerobically with shaking as described previously [19].
Primary screening for non-growth inhibitors
Overnight culture of APEC O78 prepared in LB broth was adjusted to an optical density (OD600) of 0.05 (7 × 107 CFU/mL) in fresh LB broth. One hundred micro-liters of the culture was transferred to a 96-well plate and 1µL (100 μM final concentration) of the compound was added to each well using a pin tool [54]. Chloramphenicol (40 μg/mL) or kanamycin (30 μg/mL) and 1μL of 100% DMSO were included as controls in each plate. Plates were incubated at 37 °C for 10 h with shaking in a SunriseTM Tecan plate reader (Tecan Group Ltd. San Jose, CA, USA) and the growth was kinetically monitored every 30 min by measuring the OD at 600 nm. Compounds that resulted in no growth inhibition (no elevated OD) were chosen for screening in AI-2 bioluminescence inhibition assay.
AI-2 bioluminescence indicator assay
The AI-2 bioluminescence assay was performed as described previously [2,18,19]. Briefly, APEC O78 cultures grown in the presence of SMs (see above) were centrifuged at 5000 × g for 10 min, and cell-free culture supernatants were prepared by using a 0.22 µm filter 96 well plate (Millipore). The bioluminescence reporter V. harveyi BB170 (AI-1+ and AI-2-) was grown overnight in AB medium at 30 °C aerobically with shaking [19]. The overnight culture was diluted to 1:5000 in fresh AB medium and incubated at 30 °C for 3 h. Following incubation, 180 μL of the V. harveyi BB170 culture was distributed into each well of a 96-well plate and mixed with 20 µL of the cell free culture supernatant, the plate was then incubated at 30 °C for 2.5 h in the dark and the bioluminescence was measured using in vivo imaging system (IVIS Lumina Series III, PerkinElmer, USA). The incubation time was determined based on the preliminary studies which showed optimal induction of bioluminescence at 2.5 h (data not shown). Cell-free culture supernatants collected from overnight cultures of V. harveyi BB120 (AI-1+andAI-2+) and E. coli DH5α (a strain lacks AI-2) were used as controls [2,19]. Bioluminescence of SMs treated culture supernatants were compared to DMSO treated control. The Z-score was calculated to evaluate the quality of the bioluminescence screening [56]. Four independent experiments were conducted for the 69 compounds that inhibited ≥ 75% of the AI-2 mediated bioluminescence and the average inhibition percentage was calculated. Ten compounds that showed highest AI-2 inhibition were selected for further studies. The details of the selected SMs are listed in Table S1.
The selected compounds were also tested for inhibition of AI-2 production in other APEC serotypes such as O1, O2, O8, O15, O18, O35, O109, and O115 that are commonly implicated in colibacillosis using the same procedure described above.
Biofilm assay
Effect of the selected AI-2 inhibitors on biofilm formation was assessed using the crystal violet (CV) assay [57]. Briefly, APEC O78 was grown in LB broth in the presence of 100 µM of each compound in a 96-well plate at 37 °C for 10 h. The culture was then diluted 1:100 in fresh LB broth. The biofilm assay was performed in a 96-well plate containing 150 µL of the diluted culture and 100 µM of each compound. The plate was then incubated aerobically without shaking at 37 °C for 48 h, washed twice with PBS to remove the non-adherent cells and stained with 200 µL of 0.1% CV in water at room temperature for 10 min. The plate was washed with PBS and biofilm was quantified by measuring the absorbance at 550 nm after solubilizing the CV in 200 μL of 30% acetic acid in water for 15 min. Two independent experiments were conducted in triplicate wells in each experiment.
Motility assay
The effect of selected AI-2 inhibitors on APEC motility was performed as described previously [2]. Briefly, overnight culture of APEC O78 was adjusted to an OD600 of 0.05 and 100 µL was transferred to each well of a 96 well plate and grown in the presence of 100 µM of each compound at 37 °C for 10 h. The culture was then adjusted to an OD600 of 0.05 and used for the motility assay. The motility assay was performed in a 48-well plate using semisolid agar media (0.4% LB agar) containing 0.01% tetrazolium chloride and 100 µM of each compound. One microliter of the OD adjusted culture was stabbed onto the middle of the agar and the plate was incubated at 37 °C for 6 h. The motility was assessed by measuring the diameter of the halo zone in comparison to DMSO treated control. Two independent experiments were conducted with duplicate wells in each experiment.
Lactate dehydrogenase assay
The selected 10 AI-2 inhibitors were evaluated for their toxicity to Caco-2 (ATCC® HTB-37™) using LDH assay [56]. Briefly, Caco-2 cells (1.4 × 105 cells/well) were grown in a 96-well plate in minimal essential medium (MEM) supplemented with 20% fetal bovine serum (FBS; Gibco), 1% non-essential amino acid (NEAA, Invitrogen Life Technologies) and 1mM sodium pyruvate, at 37 °C in a humidified 5% CO2 incubator for 48 h until a complete monolayer was formed. For LDH assay, cells were washed twice with media containing no FBS/no antibiotics and incubated with 150 µL of fresh media containing 100 μM of each compound at 37 °C for 24 h in a humidified 5% CO2 incubator. Fifty microliters of supernatant was analyzed using LDH Cytotoxicity Assay Kit (Thermo Scientific). Two independent experiments were conducted in triplicate wells in each experiment.
Toxicity was also determined on HD-11 cells (CVCL-4685). The HD-11 cells (1.4 × 105 cells/well) were grown in Iscove’s modified Dulbecco’s medium (IMEM; Gibco) supplemented with 2 mM glutamine and 10% FBS at 37 °C in a humidified 5% CO2 for 48 h until a monolayer was completely formed. LDH assay was conducted as described above. Two independent experiments were conducted in triplicate wells in each experiment.
Hemolysis assay
The hemolytic activity of the selected AI-2 inhibitors was determined as described before [56]. Briefly, 200 µL of the 10% RBCs (LAMPIRE Biological Laboratories) suspension in PBS was incubated with 100 μM of each compound for 1 h in a 96- well plate. The plate was then centrifuged at 5000 × g for 5 min, placed on ice for 5 min and the absorbance of the supernatant was measured at 540 nm. PBS, 1% DMSO, and 0.1% Triton X-100 were used as controls. Two independent experiments were conducted in triplicate wells in each experiment.
Intracellular survival assay
The effect of the selected AI-2 inhibitors on the survival of APEC in macrophage cells was tested as described before [58]. Briefly, HD-11 cells was grown as described above and infected with 1 × 107 CFU (MOI = 100) of mid-log phase APEC strains (O78, O1 and O2) at 37 °C for 1 h. Cells were washed with PBS and treated with gentamicin (150 μg/mL) for 1 h to kill the extracellular bacteria [59]. The cells were then washed and incubated with 100 µM of each compound in media containing 10 μg/mL of gentamicin at 37 °C for 6 h. Following treatment, cells were washed, lysed with 0.1% Triton X-100 for 5 min, ten-fold serially diluted in PBS and plated on LB agar plates to determine CFUs. Cells treated with chloramphenicol (40 μg/mL) and 1 μL of 100% DMSO were used as controls. The experiment was conducted two times with four replicates in each experiment. Intracellular survival was expressed as the log change of APEC CFUs in the AI-2 inhibitors treated cells.
Similarly, the intracellular survival assay was also performed in THP-1 cells (ATCC® TIB-202™) as described above. The THP-1 cells were grown in RPMI 1640 medium (Gibco) supplemented with 10% FBS and 2 mM glutamine. In order to differentiate the THP-1 monocyte to macrophage, 100 nM phorbol myristate acetate (PMA; Sigma-Aldrich) was added to the media. The cells (1.4 × 105 cells/well) were grown in a 96-well plate in a 5% CO2 incubator at 37 °C for 48 h, infected with APEC (MOI = 100), treated with each compound and the intracellular bacteria was determined as above.
Wax moth (G. mellonella) larva infection model
G. mellonella caterpillars (larvae) in the final instar stage (fifth instar) were obtained from Vanderhorst, Inc. (St. Mary’s, Ohio, USA), stored in wood shavings in a petri dish in the dark and used within 7 days of receiving. Larvae with 15–25 mm length, 250–350 mg weight, having a creamy color with minimal speckling and no grey markings were used in this study. The infection was performed as described previously [28,60]. For the inoculation, AI-2 inhibitors were diluted in a buffer mix (30% DMSO plus 10 mM MgSO4) as described previously and each larva (n = 10) was inoculated with 8.5 µL (50 mg/kg; 12.5 µg/larva) of the AI-2 inhibitors into the hemocoel via the last left proleg using PB600-1 repeating dispenser (Hamilton) attached to insulin syringe (31 gauge, 8 mm needle length; ReliOn). Larvae were placed inside sterile petri dishes and incubated for 2 h in the dark at 37 °C. Then, larvae were infected with 8.5 µL of (4.25 × 104 CFU) of RifR APEC O78 in 10 mM MgSO4 on the right hind proleg. RifR APEC O78 was generated as described above by plating APEC on LB agar plate containing 50 µg/ mL rifampicin for specific monitoring of APEC population inside the larvae. The AI-2 inhibitors exhibited similar AI-2 inhibition against RifR APEC O78 as that of parent wild-type APEC O78 (data not shown). Infection dose of RifR APEC O78 to larvae was identified based on preliminary study (Figure S1). Larvae were incubated at 37°C in the dark and the survival was monitored daily for 72 h. Infected larvae treated with buffer mix and 75 mg/kg of chloramphenicol were used as controls.
For the quantification of APEC inside the larvae, dead and live larvae were surface sterilized with 70% ethanol, homogenized in PBS. The suspension was tenfold serially diluted and plated on MacConkey agar plates supplemented with 50 µg/mL of rifampicin. The plates were then incubated overnight at 37 °C and APEC load was enumerated. Two independent experiments were conducted using larvae (n = 10) obtained in different batches.
Toxicity of SMs in wax moth larvae
To confirm that the death of the larvae was not due to the compounds, intrinsic toxicity the AI-2 inhibitors were assessed in a separate experiment. G. mellonella larvae (fifth instar; n = 10) were inoculated with 8.5 µL of SMs (50 mg/kg body weight; 12.5 µg/ larva) into the hemocoel via the last left proleg. Post inoculation, larvae were placed inside sterile petri dishes and incubated in the dark at 37 °C for 72 h and larval survival was monitored every 24 h. Two independent experiments were conducted
Quantitative real-time reverse transcription PCR (qRT-PCR)
The effect of the selected AI-2 inhibitors on the expression of QS-regulated and virulence factors genes of APEC was determined using qRT-PCR. APEC O78 was grown in the presence of 100 µM of each compound in a 96-well plate at 37 °C for 10 h. APEC O78 culture treated with 1µL of 100% DMSO was used as control. Total RNA was extracted from duplicate wells for each compound (200 µL) using a miRNeasy Mini Kit (Qiagen). RNA quality and quantity was determined by nanodrop 2000 C spectrophotometer (Thermo scientific). Traces of DNA were removed using Genomic DNA removal mix (Qiagen). Approximately, 5 μg of purified RNA was used to synthesize cDNA using the Qiagen RT2 First Strand Kit (Qiagen). The qRT-PCR was performed using SensiMixTM SYBR® Hi-ROX qPCR Master Mix (Bioline) according to manufacturer instructions in a realplex2 mastercycler (Eppendorf) with 55 oC annealing temperature. Gene-specific primers were designed using PrimerQuest Tool and obtained from integrated DNA technologies (IDT). The primers used with target genes’ description are listed in Table 2. The data were normalized to the house-keeping gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) [2]. The relative fold change was calculated using the ∆∆Ct method [61]. Three independent experiments were conducted.
Statistical analysis
Data from biofilm formation, motility, qRT-PCR, toxicity, hemolytic activity, intracellular survival, and wax moth infection assays were expressed as the mean ± standard deviation. ANOVA followed by Tukey test was used to analyze these data and a P-value < 0.05 was used to determine statistically significant differences between means. A fold change of ± 1.5 ≥ or ≤ 1.5 and a P-value ≤ 0.05 were used to determine statistically significant differences in gene expression. The differences between the gene expression profiles were analyzed using a principal component analysis (PCA) on JMP PRO 13 software (SAS Institute). Statistical analyses of the wax moth infection studies were performed using GraphPad Prism 5 software (GraphPad, Inc.) and plotted using the Kaplan-Meier graph.
Funding Statement
Research in Dr. Rajashekara’s laboratory is funded by Agriculture and Food Research Initiative Competitive Grant # 2015-68004-23131 from the National Institute for Food and Agriculture (NIFA), U.S. Department of Agriculture (USDA) and Federal and State funds appropriated to the Ohio Agricultural Research and Development Center, The Ohio State University.
Acknowledgments
We thank Dr. Pierce A. Paul for his assistance with statistical analysis.
Disclosure statement
The authors declare no conflict of interest.
Supplementary material
Supplemental data can be accessed here.
References
- [1].Wang S, Niu C, Shi Z, et al. Effects of ibeA deletion on virulence and biofilm formation of avian pathogenic Escherichia coli. Infect Immun. 2011. January;79(1):279–287. PubMed PMID: 20974831; PubMed Central PMCID: PMC3019902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Han X, Bai H, Liu L, et al. The luxS gene functions in the pathogenesis of avian pathogenic Escherichia coli. Microb Pathog. 2013. February;55:21–27. PubMed PMID: 23046700. [DOI] [PubMed] [Google Scholar]
- [3].Markland SM, LeStrange KJ, Sharma M, et al. Old friends in new places: exploring the role of extraintestinal E. coli in intestinal disease and foodborne illness. Zoonoses Public Health. 2015;62(7):491–496. [DOI] [PubMed] [Google Scholar]
- [4].Johnson TJ, Kariyawasam S, Wannemuehler Y, et al. The genome sequence of avian pathogenic Escherichia coli strain O1: K1:h7shares strong similarities with human extraintestinal pathogenic E. coli genomes. J Bacteriol. 2007. April;189(8):3228–3236. PubMed PMID: 17293413; PubMed Central PMCID: PMC1855855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Palaniyandi S, Mitra A, Herren CD, et al. LuxS contributes to virulence in avian pathogenic Escherichia coli O78: K80:H9. Vet Microbiol. 2013. October 25;166(3–4):567–575. PubMed PMID: 23958403. [DOI] [PubMed] [Google Scholar]
- [6].Barnes HJ, Nolan LK, Vaillancourt JF.. Colibacillosis In: Saif YM, Fadly AM, Glisson JR, et al, editors. Diseases of Poultry. 12th ed. Ames: Blackwell Publishing; 2008. p. 691–732. [Google Scholar]
- [7].Zhao S, Maurer JJ, Hubert S, et al. Antimicrobial susceptibility and molecular characterization of avian pathogenic Escherichia coli isolates. Vet Microbiol. 2005. May 20;107(3–4):215–224. [DOI] [PubMed] [Google Scholar]
- [8].Ghunaim H, MA Abu-Madi, Kariyawasam S. Advances in vaccination against avian pathogenic Escherichia coli respiratory disease: potentials and limitations. Vet Microbiol. 2014. August 06;172(1–2):13–22. PubMed PMID: 24878325. [DOI] [PubMed] [Google Scholar]
- [9].Stacy AK, Mitchell NM, Maddux JT, et al. Evaluation of the prevalence and production of Escherichia coli common pilus among avian pathogenic E. coli and its role in virulence. PloS one. 2014;9(1):e86565 PubMed PMID: 24466152; PubMed Central PMCID: PMC3900561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Moulin-Schouleur M, Reperant M, Laurent S, et al. Extraintestinal pathogenic Escherichia coli strains of avian and human origin: link between phylogenetic relationships and common virulence patterns. J Clin Microbiol. 2007. October;45(10):3366–3376. PubMed PMID: 17652485; PubMed Central PMCID: PMC2045314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wang S, Liu X, Xu X, et al. Escherichia coli Type III Secretion System 2 ATPase EivC Is Involved in the Motility and Virulence of Avian Pathogenic Escherichia coli. Front Microbiol. 2016;7:1387 PubMed PMID: 27630634; PubMed Central PMCID: PMC5005338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med. 2012. November;2(11). PubMed PMID: 23125205; PubMed Central PMCID: PMC3543102 DOI: 10.1101/cshperspect.a012427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].DeLisa MP, Wu CF, Wang L, et al. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J Bacteriol. 2001. September;183(18):5239–5247. PubMed PMID: 11514505; PubMed Central PMCID: PMC95404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Surette MG, Bassler BL. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc Natl Acad Sci U S A. 1998. June 9;95(12):7046–7050. PubMed PMID: 9618536; PubMed Central PMCID: PMC22733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sperandio V, Torres AG, Jarvis B, et al. Bacteria-host communication: the language of hormones. Proc Natl Acad Sci U S A. 2003. July 22;100(15):8951–8956. PubMed PMID: 12847292; PubMed Central PMCID: PMC166419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Schauder S, Bassler BL. The languages of bacteria. Genes Dev. 2001. June 15;15(12):1468–1480. PubMed PMID: 11410527. [DOI] [PubMed] [Google Scholar]
- [17].Rasmussen TB, Givskov M. Quorum sensing inhibitors: a bargain of effects. Microbiology. 2006. April;152(Pt 4):895–904. PubMed PMID: 16549654. [DOI] [PubMed] [Google Scholar]
- [18].Bassler BL, Wright M, Silverman MR. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol Microbiol. 1994. July;13(2):273–286. PubMed PMID: 7984107. [DOI] [PubMed] [Google Scholar]
- [19].Surette MG, Miller MB, Bassler BL. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Natl Acad Sci U S A. 1999. February 16;96(4):1639–1644. PubMed PMID: 9990077; PubMed Central PMCID: PMC15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bassler BL, Greenberg EP, Stevens AM. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J Bacteriol. 1997. June;179(12):4043–4045. PubMed PMID: 9190823; PubMed Central PMCID: PMC179216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Barrios AF, Zuo R, Ren D, et al. Hha, YbaJ, and OmpA regulate Escherichia coli K12 biofilm formation and conjugation plasmids abolish motility. Biotechnol Bioeng. 2006. January 5;93(1):188–200. PubMed PMID: 16317765. [DOI] [PubMed] [Google Scholar]
- [22].Brackman G, Coenye T. Quorum sensing inhibitors as anti-biofilm agents. Curr Pharm Des. 2015;21(1):5–11. PubMed PMID: 25189863. [DOI] [PubMed] [Google Scholar]
- [23].Dressaire C, Moreira RN, Barahona S, et al. BolA is a transcriptional switch that turns off motility and turns on biofilm development. mBio. 2015. February 17;6(1):e02352–14. PubMed PMID: 25691594; PubMed Central PMCID: PMC4337573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chanteloup NK, Porcheron G, Delaleu B, et al. The extra-intestinal avian pathogenic Escherichia coli strain BEN2908 invades avian and human epithelial cells and survives intracellularly. Vet Microbiol. 2011. January 27;147(3):435–439. [DOI] [PubMed] [Google Scholar]
- [25].Huja S, Oren Y, Trost E, et al. Genomic avenue to avian colisepticemia. mBio. 2015. January 13;6(1). PubMed PMID: 25587010; PubMed Central PMCID: PMC4313913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Pourbakhsh SA, Boulianne M, Martineau-Doize B, et al. Virulence mechanisms of avian fimbriated Escherichia coli in experimentally inoculated chickens. Vet Microbiol. 1997. November;58(2–4):195–213. PubMed PMID: 9453131. [DOI] [PubMed] [Google Scholar]
- [27].Kariyawasam S, Johnson TJ, Nolan LK. The pap operon of avian pathogenic Escherichia coli strain O1: K1is located on a novel pathogenicity island. Infect Immun. 2006. January;74(1):744–749. PubMed PMID: 16369033; PubMed Central PMCID: PMC1346673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hill L, Veli N, Coote PJ. Evaluation of Galleria mellonella larvae for measuring the efficacy and pharmacokinetics of antibiotic therapies against Pseudomonas aeruginosa infection. Int J Antimicrob Agents. 2014. March;43(3):254–261. PubMed PMID: 24361354. [DOI] [PubMed] [Google Scholar]
- [29].Cui ZQ, Wu ZM, Fu YX, et al. Autoinducer-2 of quorum sensing is involved in cell damage caused by avian pathogenic Escherichia coli. Microb Pathog. 2016. October;99:247–252. PubMed PMID: 27569532. [DOI] [PubMed] [Google Scholar]
- [30].Sperandio V, Torres AG, Giron JA, et al. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157: H7. J Bacteriol. 2001. September;183(17):5187–5197. PubMed PMID: 11489873; PubMed Central PMCID: PMC95396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Rasmussen TB, Givskov M. Quorum-sensing inhibitors as anti-pathogenic drugs. Int J Med Microbiol. 2006. April;296(2–3):149–161. PubMed PMID: 16503194. [DOI] [PubMed] [Google Scholar]
- [32].Han X, Bai H, Tu J, et al. Deletion of luxS further attenuates the virulence of the avian pathogenic Escherichia coli aroA mutant. Microb Pathog. 2015. November;88:39–47. PubMed PMID: 26271577. [DOI] [PubMed] [Google Scholar]
- [33].Pilatti L, Boldrin de Paiva J, Rojas TC, et al. The virulence factor ychO has a pleiotropic action in an Avian Pathogenic Escherichia coli (APEC) strain. BMC Microbiol. 2016. March 10;16:35 10.1186/s12866-016-0654-2. PubMed PMID: 26965678; PubMed Central PMCID: PMC4787180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gao Q, Zheng D, Yuan Z. Substrate preference of 5’-methylthioadenosine/S-adenosylhomocysteine nucleosidase in Burkholderia thailandensis. FEMS Microbiol Lett. 2013. February;339(2):110–116. PubMed PMID: 23198993. [DOI] [PubMed] [Google Scholar]
- [35].Kim Y, Wood TK. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem Biophys Res Commun. 2010. January 1 PubMed PMID: WOS:000273624500038;391(1):209–213. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Takeda S, Fujisawa Y, Matsubara M, et al. A novel feature of the multistep phosphorelay in Escherichia coli: a revised model of the RcsC YojN RcsB signalling pathway implicated in capsular synthesis and swarming behaviour. Mol Microbiol. 2001. April;40(2):440–450. PubMed PMID: 11309126. [DOI] [PubMed] [Google Scholar]
- [37].Aoki SK, Malinverni JC, Jacoby K, et al. Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB. Mol Microbiol. 2008. October;70(2):323–340. PubMed PMID: 18761695; PubMed Central PMCID: PMC2579741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Vieira HL, Freire P, Arraiano CM. Effect of Escherichia coli morphogene bolA on biofilms. Appl Environ Microbiol. 2004. September;70(9):5682–5684. PubMed PMID: 15345459; PubMed Central PMCID: PMC520865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Romeo T, Gong M, Liu MY, et al. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J Bacteriol. 1993. August;175(15):4744–4755. PubMed PMID: 8393005; PubMed Central PMCID: PMC204926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Roberts IS. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu Rev Microbiol PubMed PMID: WOS:A1996VL56000010 1996;50:285–315. English. [DOI] [PubMed] [Google Scholar]
- [41].He Z, Wang Q, Hu Y, et al. Use of the quorum sensing inhibitor furanone C-30 to interfere with biofilm formation by Streptococcus mutans and its luxS mutant strain. Int J Antimicrob Agents. 2012. July;40(1):30–35. PubMed PMID: 22578766. [DOI] [PubMed] [Google Scholar]
- [42].Rasmussen L, White EL, Pathak A, et al. A high-throughput screening assay for inhibitors of bacterial motility identifies a novel inhibitor of the Na+-driven flagellar motor and virulence gene expression in Vibrio cholerae. Antimicrob Agents Chemother. 2011. September;55(9):4134–4143. PubMed PMID: 21709090; PubMed Central PMCID: PMC3165335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Malapaka VR, Barrese AA, Tripp BC, et al. High-throughput screening for antimicrobial compounds using a 96-well format bacterial motility absorbance assay. J Biomol Screen. 2007. September;12(6):849–854. PubMed PMID: 17644774. [DOI] [PubMed] [Google Scholar]
- [44].Hentzer M, Wu H, Andersen JB, et al. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. Embo J. 2003. August 1;22(15):3803–3815. PubMed PMID: 12881415; PubMed Central PMCID: PMC169039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kiran MD, Adikesavan NV, Cirioni O, et al. Discovery of a quorum-sensing inhibitor of drug-resistant staphylococcal infections by structure-based virtual screening. Mol Pharmacol. 2008. May;73(5):1578–1586. PubMed PMID: 18314496. [DOI] [PubMed] [Google Scholar]
- [46].Hung DT, Shakhnovich EA, Pierson E, et al. Small-molecule inhibitor of Vibrio cholerae virulence and intestinal colonization. Science. 2005. October 28;310(5748):670–674. PubMed PMID: 16223984. [DOI] [PubMed] [Google Scholar]
- [47].Krogfelt KA, Bergmans H, Klemm P. Direct evidence that the fimh protein is the mannose-specific adhesin of escherichia-coli type-1 fimbriae. Infect Immun. 1990. June;58(6):1995–1998. PubMed PMID: WOS:A1990DF20800078 English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hancock V, Ferrieres L, Klemm P. The ferric yersiniabactin uptake receptor FyuA is required for efficient biofilm formation by urinary tract infectious Escherichia coli in human urine. Microbiology. 2008. January;154(Pt 1):167–175. PubMed PMID: 18174135. [DOI] [PubMed] [Google Scholar]
- [49].Nolan LK, Horne SM, Giddings CW, et al. Resistance to serum complement, iss, and virulence of avian Escherichia coli. Vet Res Commun. 2003. February;27(2):101–110. PubMed PMID: 12718504. [DOI] [PubMed] [Google Scholar]
- [50].Zhao Y, Wang S, Yang D, et al. [Vacuolating autotransporter toxin affects biological characteristics and pathogenicity of avian pathogenic Escherichia coli]. Wei Sheng Wu Xue Bao. 2015. September 4;55(9):1208–1214. PubMed PMID: 26762034. [PubMed] [Google Scholar]
- [51].Bajusz D, Racz A, Heberger K. Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations? J Cheminform. 2015;7:20 PubMed PMID: 26052348; PubMed Central PMCID: PMC4456712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Musthafa KS, Balamurugan K, Pandian SK, et al. 2,5-Piperazinedione inhibits quorum sensing-dependent factor production in Pseudomonas aeruginosa PAO1. J Basic Microbiol. 2012. December;52(6):679–686. PubMed PMID: 22359266. [DOI] [PubMed] [Google Scholar]
- [53].Bina XR, Philippart JA, Bina JE. Effect of the efflux inhibitors 1-(1-naphthylmethyl)-piperazine and phenyl-arginine-beta-naphthylamide on antimicrobial susceptibility and virulence factor production in Vibrio cholerae. J Antimicrob Chemother. 2009. January;63(1):103–108. PubMed PMID: 19010827. [DOI] [PubMed] [Google Scholar]
- [54].Wallace IM, Urbanus ML, Luciani GM, et al. Compound prioritization methods increase rates of chemical probe discovery in model organisms. Chem Biol. 2011. October 28;18(10):1273–1283. PubMed PMID: 22035796; PubMed Central PMCID: PMC4193902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Cai XC, Xi H, Liang L, et al. Rifampicin-resistance mutations in the rpoB gene in bacillus velezensis CC09 have pleiotropic effects. Front Microbiol. 2017;8:178 PubMed PMID: 28243227; PubMed Central PMCID: PMC5303731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Kumar A, Drozd M, Pina-Mimbela R, et al. Novel anti-campylobacter compounds identified using high throughput screening of a pre-selected enriched small molecules library. Front Microbiol. 2016;7:405 PubMed PMID: 27092106; PubMed Central PMCID: PMC4821856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].O’Toole GA. Microtiter dish biofilm formation assay. J Vis Exp. 2011;(47). PubMed PMID: 21307833; PubMed Central PMCID: PMC3182663 DOI: 10.3791/2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Gao Q, Xu H, Wang X, et al. RfaH promotes the ability of the avian pathogenic Escherichia coli O2 strain E058 to cause avian colibacillosis. J Bacteriol. 2013. June;195(11):2474–2480. PubMed PMID: 23504015; PubMed Central PMCID: PMC3676060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Helmy YA, Kassem II, Kumar A, et al. In vitro evaluation of the impact of the probiotic E. coli Nissle 1917 on campylobacter jejuni’s invasion and intracellular survival in human colonic cells. Front Microbiol. 2017;8:1588 PubMed PMID: 28878749; PubMed Central PMCID: PMC5572226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Desbois AP, Coote PJ. Wax moth larva (Galleria mellonella): an in vivo model for assessing the efficacy of antistaphylococcal agents. J Antimicrob Chemother. 2011. August;66(8):1785–1790. PubMed PMID: 21622972. [DOI] [PubMed] [Google Scholar]
- [61].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001. December;25(4):402–408. PubMed PMID: 11846609. [DOI] [PubMed] [Google Scholar]
- [62].Mangiamele P, Nicholson B, Wannemuehler Y, et al. Complete genome sequence of the avian pathogenic Escherichia coli strain APEC O78. Genome Announc. 2013. March 21;1(2):e0002613 PubMed PMID: 23516182; PubMed Central PMCID: PMC3622999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Rodriguez-Siek KE, Giddings CW, Doetkott C, et al. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology. 2005. June;151(Pt 6):2097–2110. PubMed PMID: 15942016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


