Abstract
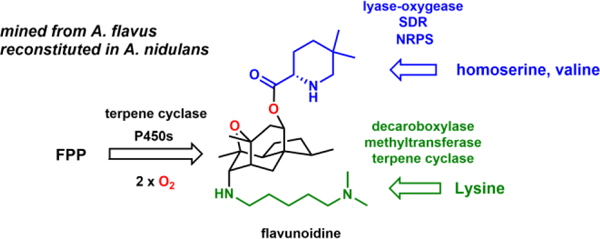
Biosynthetic pathways containing multiple core enzymes have potential to produce structurally complex natural products. Here we mined a fungal gene cluster that contains two predicted terpene cyclases (TCs) and a nonribosomal peptide synthetase (NRPS). We showed the flv pathway produces flavunoidine 1, an alkaloidal terpenoid. The core of 1 is a tetracyclic, cage-like and oxygenated sesquiterpene that is connected to dimethylcadaverine via a C-N bond, and is acylated with 5,5-dimethyl-l-pipecolate. The roles of all flv enzymes are established based on metabolite analysis from heterologous expression.
Graphical Abstract
Structural complexities of natural products (NPs) are generated by enzymes in the biosynthetic pathways.1 Scaffolds assembled by core enzymes such as polyketide synthases (PKSs), nonribosomal peptide synthetases (NRPSs) or terpene cyclase (TCs), etc., can be morphed into complex NPs by accessory enzymes including transferases2 and oxidoreductases,3 etc. In fungi, the combinations of different core enzymes in the same biosynthetic pathway, such as PKS/PKS,4 PKS/NRPS,5 PKS/TC (Figure 1A),6 can result in complex hybrid NPs unachievable with a single core enzyme. In contrast, biosynthetic pathways containing the combination of NRPS and TC have not been well-studied. While many metabolites are derived from prenylation of peptidyl cores via prenyltransferases,7 the use of a TC to generate a terpene core that is decorated by a NRPS is rare. However, bioinformatic scanning of sequenced fungal genomes suggests TC/NRPS hybrid clusters are common (Figure S2). Isolation of fungal aminoacylated terpenoids also suggests such hybrid molecules can be synthesized by fungi (Figure 1B).8 Recent characterization of the aculene A biosynthetic pathway demonstrates such collaboration between the TC and a single module NRPS.9 Based on these evidences, we believe there is significant potential in mining fungal TC/NRPS pathways for new NPs.
Figure 1.
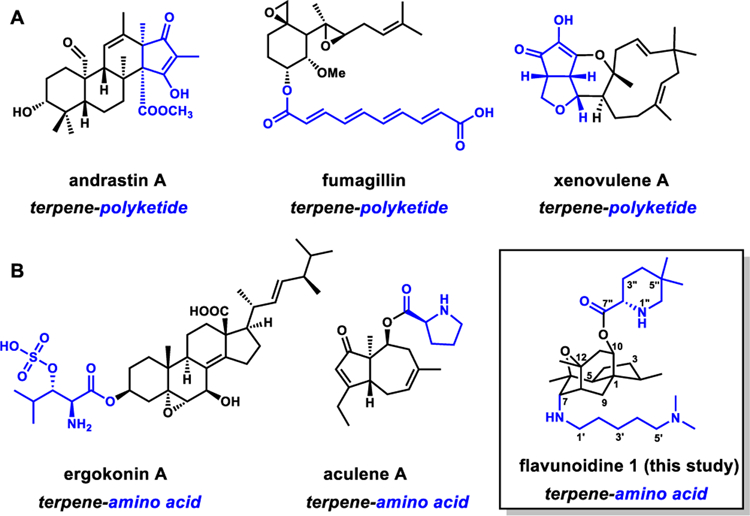
Structures of fungal polyketides synthesized by collaborative efforts of core enzymes. (A) meroterpenoids derived from TC/PKS,6b–d etc.; (B) compounds derived from TC and NRPS enzymes include ergokonin A8a (proposed), aculene A,9 and flavunoidine 1 (this study).
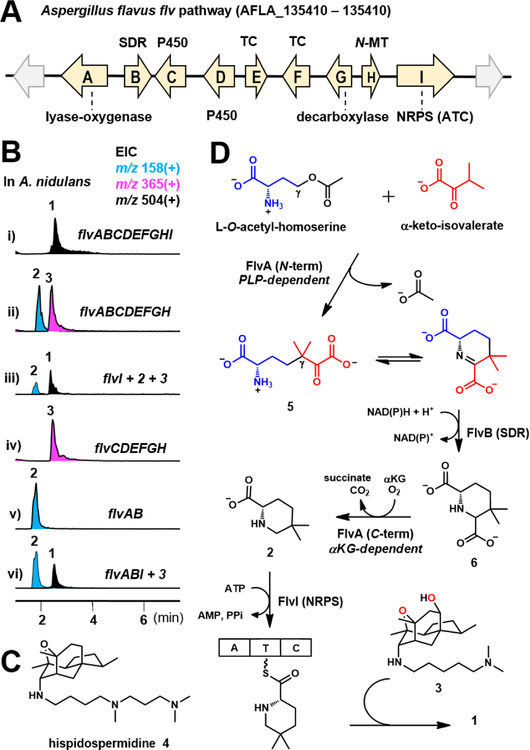
Among the predicted gene clusters containing both TC and NRPS, we selected an uncharacterized nine-gene cluster conserved in several well-studied fungal species (Figure 2A, Figure S3, Table S1). The flv cluster in Aspergillus flavus encodes two TCs: FlvE and FlvF, with sequence similarity to trichodiene synthase10 and ophiobolin synthase,11 respectively; and a single-module NRPS FlvI. This cluster also contains redox enzymes, including FlvB (short chain reductase, SDR), FlvC and FlvD (P450s), and a didomain enzyme FlvA. The N-terminal half of FlvA is predicted to be a PLP-dependent lyase,12 while the C-terminal half is predicted to be a non-heme Fe, α-ketoglutarate dependent oxygenase (α–KG) (Figures S15–16).13 The remaining enzymes encoded in the gene cluster include FlvG, which is a homolog of ornithine decarboxylase; and FlvH, which has sequence similarity to histone lysine N-methyltransferase (N-MT).14 To investigate the metabolite that can be biosynthesized from the flv cluster, we used Aspergillus nidulans as a heterologous host to mine the pathway.
Figure 2.
Heterologous expression of flv pathway. (A) The flv gene cluster. (B) LC/MS analysis of extracts from A. nidulans expressing different combinations of flv genes. (C) The structure of hispidospermidin 4. (D) Proposed biosynthesis of 1 and 2 (For details see Figure S4).
When the entire flv gene cluster was introduced into A. nidulans, we detected and isolated a new metabolite 1 with molecular weight (MW) of 503 (Figure 2B, i) (1.2 mg/L). The structure of 1 was solved to be flavunoidine shown in Figure 1B (Table S5 and Figures S17–22). The tetracyclic cage was only previously found in the phospholipase C inhibitor hispidospermidin 4 (Figure 2C).15 The C7 axial trimethyl-spermidine substituent in 4 is replaced by N,N-dimethylcadaverine in 1, while the C10 position is hydroxylated and acylated with 5,5-dimethyl-l-pipecolate. 4 was the only NP with the same tetracyclic core, and has been the subject of total synthesis by Danishefsky,16a–b Overman16c and Sorenson,16d–e etc. 1 does not display notable cytotoxicity, nor is it antifungal or antibacterial.
We first investigated formation of the dimethylpipecolate in 1. To probe the role of the NRPS FlvI, we expressed the other eight genes FlvA-H. This led to the absence of 1 but the emergence of 2 (MW 157) (0.7 mg/L) and 3 (MW 364) (0.9 mg/L) (Figure 2B, ii). NMR analysis showed that 2 is 5,5-dimethyl-l-pipecolic acid (Table S6 and Figures S23–27), while 3 is the unacylated precursor of 1 (Table S7 and Figures S28–33). The accumulation of these separate building blocks suggests the NRPS FlvI is responsible for esterifying 2 and 3 (Figure 2D). Indeed, feeding of 2 and 3 to A. nidulans expressing FlvI led to the biotransformation to 1 (Figure 2B, iii).
Pipecolate biosynthesis from lysine has been shown to involve a PLP-dependent enzyme and reductase,17 which led us to propose that the didomain enzyme FlvA and SDR FlvB may be involved in biosynthesis of 2. When FlvC-H were expressed, we only observed accumulation of 3 (Figure 2B, iv), while coexpression of FlvA and FlvB separately resulted in formation of 2 (Figure 2B, v). Individual expression of either FlvA or FlvB did not result in formation of 2. In addition, coexpression of FlvA, FlvB and FlvI, accompanied by feeding of 3, led to the production of 1 (Figure 2B, vi). These results implicate FlvA and FlvB in the biosynthesis of 2. We propose the PLP-dependent lyase12 domain of FlvA catalyzes a γ-replacement reaction as shown in Figure 2D (detailed mechanism shown in Figure S4). l-O-acetyl-homoserine can bind and form the aldimine, which can undergo two proton abstraction steps to eliminate acetate and form the vinyl glycine quinonoid. This species can be attacked by α-keto-isovalerate to form a new C-C bond, which upon protonation and transaldimination can lead to release of the ketone 5 that can cyclize intramolecularly to yield the imine. The imine can be reduced by FlvB to yield the 6-carboxylated pipecolate 6. The C-terminal α-KG dependent oxygenase domain of FlvA is then proposed to catalyze the decarboxylation of 6 to yield 2. The mechanism, shown in Figure S4, can involve a radical decarboxylative route facilitated by an active site tyrosyl18, 19 or thiyl20 radical generated from the Fe(IV)=O. Upon decarboxylation to generate the C4 radical, hydrogen delivery from the active residue forms 2, and reductive quenching of the radical carrier regenerates the enzyme.
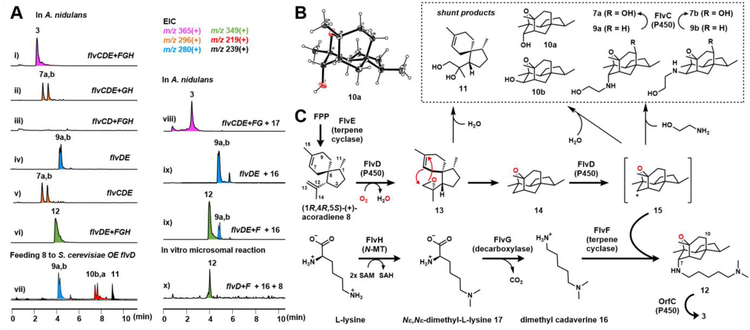
We next investigated formation of the tetracyclic core and the unusual trans diaxial nitrogen and oxygen functionality in 3. We have established that coexpression of six enzymes FlvC-H can synthesize 3 (Figure 2A, iv). To examine the roles of the TCs (FlvE and FlvF), we removed either gene and analyzed the resulting metabolic profiles. Removing flvE abolished all related metabolites (Figure 3A, iii), suggesting its involvement in core synthesis; while removing flvF led to accumulation of a pair of metabolites 7a and 7b with the same MW (Figure 3, ii). 7a was purified and structurally determined (Figure 3C, Tables S8, Figures S34–39) to contain the same core as 3, but substituted at C7 by ethanolamine via an axial C-N bond. Based on data below for the stereoisomer pair 9a and 9b, we propose 7b is the C7 equatorial stereoisomer of 7a. Formation of both axial and equatorial isomers indicates the C-N bonds formed in 7a and 7b may be uncatalyzed. This also hints FlvF may be responsible for stereoselectively forming the C-N bond in 3.
Figure 3.
Biosynthesis of the core of 1. (A) LC-MS analysis of metabolites in vivo and in vitro assays; (B) Crystal structure of 10a; (C) Proposed biosynthetic pathway of 3. Observed shunt products are shown in the dashed box.
To analyze the function of core TC FlvE, we expressed the enzyme in Saccharomyces cerevisiae JHY651.21 GCMS analysis revealed a sesquiterpene product 8 (Figure S5). Purified FlvE also synthesized 8 using farnesyl diphosphate (FPP) (Figures S6 and S14). Isolation and characterization confirmed 8 to be (1R,4R,5S)-(+)-acoradiene.22 The (−)-enantiomer was previously isolated from plants.23 We then coexpressed the P450s, FlvD and FlvC, with FlvE to determine if oxidative modifications of 8 can generate the core in 3. Coexpression of both P450s with FlvE in A. nidulans resulted in formation of 7a and 7b (Figure 3A, v), while coexpression of only FlvD and FlvE led to the C7-stereoisomers 9a (axial) and 9b (equatorial) (Figure 3a, iv) (Table S10 and Figures S42–53), which do not contain the C10 hydroxyl. Coexpression of FlvD-H without FlvC in A. nidulans resulted in the formation of 12 (Figure 3a, vi), which is the C10-deshydroxy version of 3. Collectively, these results implicate FlvC as the C10 hydroxylase (Figure 3C), while FlvD alone can oxidatively convert 8 into the tetracyclic cage, which nonenzymatically forms a C-N bond with ethanolamine in A. nidulans.
Feeding 8 to A. nidulans expressing FlvD-H, but without the TC FlvE, restored the otherwise abolished production of 12 (Figure S7), suggesting 8 is a precursor in the pathway. To analyze the function of FlvD in morphing 8, we overexpressed the enzyme in yeast and fed 8 to analyze biotransformation products. In addition to 9a and 9b, new metabolites 10a, 10b and 11 were detected (Figures 3a, vii and S8). 11 retains the carbon scaffold in 8, but with the C12, C13 diol (Table S12, Figures S60–65). 10a is substituted with an axial hydroxyl group at C7 (Figure 3C) (Table S11 and Figure S54–59). We obtained an X-ray crystal structure of 10a (Figure 3B), which confirmed the tetracyclic structure and allowed assignment of absolute stereochemistry of compounds. Based on the structures of 9a and 9b, we propose 10b is the equatorial stereoisomer at C7. Purified microsomes from yeast expressing FlvD converted 8 to 10a and 10b (Figure S9). No 9a or 9b was detected since no ethanolamine was present in the reaction. Performing the microsomal assay in H218O led to incorporation of labeled 18O into 10a and 10b, indicating the C7 hydroxyl groups are derived from water (Figure S10). We verified 9 and 10 are shunt products, as feeding these compounds to A. nidulans expressing FlvD-H without FlvE did not restore biosynthesis of 12 (Figure S7).
We propose 11 is derived from the nonenzymatic epoxide opening of 13, which can be formed stereoselectively from 8 by FlvD. 13 can undergo intramolecular [3+2]-cycloaddition between the olefin and epoxide to directly forge the tetracyclic core 14. This reaction may be assisted by an active site Lewis acid in FlvD and proceed in a step-wise mechanism.24 This represents a very concise way to morph the terpene 8 into the caged core. The intermediate 14, which was not isolated in vivo nor in vitro, may be further oxidized at C7 by FlvD to yield a carbocation 15. 15 can then be quenched by nucleophiles such as water to yield 10a and 10b. Ethanolamine, which is biosynthesized by both yeast and A. nidulans,25 may enter the active site of FlvD and quench 15 to yield 9a and 9b.
When the TC homolog FlvF is coexpressed, the nonenzymatic quenching is suppressed and dimethylcadaverine 16 can stereoselectively quench 15 to afford 12 (Figure 3A, vi). To test this, we coexpressed FlvD and FlvE and fed the strain with 16. This strain still synthesized only 9a and 9b, and no 12 was formed (Figure 3A, ix). Upon coexpression of FlvF with FlvD and FlvE, feeding of 16 led to formation of 12 (Figure 3, ix). We then performed an in vitro assay using yeast microsomes containing FlvD and purified FlvF (Figure S14) in the presence of 8 and 16. This reaction produced 12 and no shunt products were detected (Figure 3A, x). Excluding FlvF from this assay led to formation of only 10a and 10b, even in the presence of 16 (Figure S11). Directly adding 10 and 16 to FlvDF did not lead to formation of 12, confirming 10 is a shunt product from nonenzymatic quenching. The mechanism by which FlvF can enable the stereoselective C-N bond formation in 12 is unexpected for an enzyme annotated as TC. Moore reported an algal TC that can catalyze N-geranylation of l-Glu.26 However, the mechanism here is different since the terpene substrate is not pyrophosphorylated. We propose FlvF may form a complex with FlvD and deliver 16 to the active site where 15 is generated. The mechanism of this reaction is under investigation.
The two remaining enzymes in the pathway, FlvH (N-MT) and FlvG (decarboxylase) are proposed to synthesize 16 from l-lysine in a two-step reaction, in which FlvH performs methylation to give 17, and is decarboxylated by FlvG to afford 16 (Figure 3C). When flvH was removed from A. nidulans that produces 3, trace amounts of 3 were formed (Figure S12). The titer of 3 can be restored upon feeding of 17 (Figure 3, viii). When 17 was fed to the same strain without flvG, biosynthesis of 3 was abolished and only 7a and 7b were observed. Feeding of 16 restored production of 3, establishing FlvG catalyzes decarboxylation of 17 to 16 (Figure S13).
In summary, we mined a fungal biosynthetic gene cluster that contained both TC and NRPS core enzymes, and discovered a new alkaloidal terpenoid 1. The tetracyclic core of 1 is synthesized by the TC FlvE and P450 FlvD, while a second TC FlvF is required for attachment of the C7 axial dimethylcadaverine. The NRPS acylates the terpenoid core with dimethylpipecolate. The unexpected structural features of 1 highlight potential of fungal genome mining using combinations of core biosynthetic enzymes as a criterion.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the NIH 1R35GM118056 to YT.
Footnotes
ASSOCIATED CONTENT
Supporting Information.
This material is available free of charge via the Internet at http://pubs.acs.org. Experimental procedures, chromatograms, and spectroscopic data.
No competing financial interests have been declared.
REFERENCES
- (1).Lin CI; McCarty RM; Liu HW. The enzymology of organic transformations: a survey of name reactions in biological systems. Angew. Chem., Int. Ed 2017, 56, 3446–3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).(a) Rix U; Fischer C; Remsing LL; Rohr J. Modification of post-PKS tailoring steps through combinatorial biosynthesis. Nat. Prod. Rep 2002, 19, 542–580. [DOI] [PubMed] [Google Scholar]; (b) Olano C; Méndez C; Salas JA. Post-PKS tailoring steps in natural product-producing actinomycetes from the perspective of combinatorial biosynthesis. Nat. Prod. Rep 2010, 27, 571–616. [DOI] [PubMed] [Google Scholar]
- (3).(a) Tang MC; Zou Y; Watanabe K; Walsh CT; Tang Y. Oxidative cyclization in natural product biosynthesis. Chem. Rev 2016, 117, 5226–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Cochrane RV; Vederas JC. Highly selective but multifunctional oxygenases in secondary metabolism. Acc. Chem. Res 2014, 47, 3148–3161. [DOI] [PubMed] [Google Scholar]; (c) Cox R. Oxidative rearrangements during fungal biosynthesis. Nat. Prod. Rep 2014, 31, 1405–1424. [DOI] [PubMed] [Google Scholar]
- (4).(a) Cox RJ. Polyketides, proteins and genes in fungi: programmed nano-machines begin to reveal their secrets. Org. Biomol. Chem 2007, 5, 2010–2026. [DOI] [PubMed] [Google Scholar]; (b) Sato M; Dander JE; Sato C; Hung YS; Gao SS; Tang MC; Hang L; Winter JM; Garg NK; Watanabe K; Tang Y. Collaborative biosynthesis of maleimide-and succinimide-containing natural products by fungal polyketide megasynthases. J. Am. Chem. Soc 2017, 139, 5317–5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Boettger D; Hertweck C. Molecular diversity sculpted by fungal PKS–NRPS hybrids. ChemBioChem 2013, 14, 28–42. [DOI] [PubMed] [Google Scholar]
- (6).(a) Itoh T; Tokunaga K; Matsuda Y; Fujii I; Abe I; Ebizuka Y; Kushiro T. Reconstitution of a fungal meroterpenoid biosynthesis reveals the involvement of a novel family of terpene cyclases. Nat. Chem 2010, 2, 858–864. [DOI] [PubMed] [Google Scholar]; (b) Matsuda Y; Abe I. Biosynthesis of fungal meroterpenoids. Nat. Prod. Rep 2016, 33, 26–53. [DOI] [PubMed] [Google Scholar]; (c) Lin HC; Chooi YH; Dhingra S; Xu W; Calvo AM; Tang Y. The fumagillin biosynthetic gene cluster in Aspergillus fumigatus encodes a cryptic terpene cyclase involved in the formation of β-trans-bergamotene. J. Am. Chem. Soc 2013, 135, 4616–4619. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Schor R; Schotte C; Wibberg D; Kalinowski J; Cox RJ. Three previously unrecognised classes of biosynthetic enzymes revealed during the production of xenovulene A. Nat. Commun 2018, 9, 1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).(a) Li SM. Prenylated indole derivatives from fungi: structure diversity, biological activities, biosynthesis and chemoenzymatic synthesis. Nat. Prod. Rep 2010, 27, 57–78. [DOI] [PubMed] [Google Scholar]; (b) Ding Y; Wet JRD; Cavalcoli J; Li S; Greshock TJ; Miller KA; Finefield JM; Sunderhaus JD; McAfoos TJ; Tsukamoto S; Williams RM. Genome-based characterization of two prenylation steps in the assembly of the stephacidin and notoamide anticancer agents in a marine-derived Aspergillus sp. J. Am. Chem. Soc 2010, 132, 12733–12740. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Grundmann A; Kuznetsova T; Afiyatullov SS; Li SM. FtmPT2, an N-Prenyltransferase from Aspergillus fumigatus, Catalyses the Last Step in the Biosynthesis of Fumitremorgin B. ChemBioChem 2008, 9, 2059–2063. [DOI] [PubMed] [Google Scholar]
- (8).(a) Augustiniak H; Forche E; Reichenbach H; Wray V; Graefe U; Hoefle G. Isolation and structure elucidation of ergokonin A and B; two new antifungal sterol antibiotics from Trichoderma koningii. Liebigs Ann Chem 1991, 4, 361–366. [Google Scholar]; (b) Cóbar OM. Survey of 2, 11-cyclized cembranoids from Caribbean sources. Nat. Prod. Res 2009, 23, 26–43. [DOI] [PubMed] [Google Scholar]
- (9).Lee CF; Chen LX; Chiang CY; Lai CY; Lin HC. Biosynthesis of Norsesquiterpene Aculenes Requires Three Cytochrome P450s to Catalyze a Stepwise Demethylation Process. Angew. Chem., Int. Ed 2019, 58, 1–6. [DOI] [PubMed] [Google Scholar]
- (10).Rynkiewicz MJ; Cane DE; Christianson DW. Structure of trichodiene synthase from Fusarium sporotrichioides provides mechanistic inferences on the terpene cyclization cascade. Proc. Natl. Acad. Sci 2001, 98, 13543–13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Chiba R; Minami A; Gomi K; Oikawa H. Identification of ophiobolin F synthase by a genome mining approach: a sesterterpene synthase from Aspergillus clavatus. Org. Lett 2013, 15, 594–597. [DOI] [PubMed] [Google Scholar]
- (12).(a) Nagasawa T; Kanzaki H; Yamada H. Cystathionine gamma-lyase of Streptomyces phaeochromogenes. The occurrence of cystathionine gamma-lyase in filamentous bacteria and its purification and characterization. J. Biol. Chem 1984, 259, 10393–10403. [PubMed] [Google Scholar]; (b) Wei Y; Perez LJ; Ng WL; Semmelhack MF; Bassler BL. Mechanism of Vibrio cholerae autoinducer-1 biosynthesis. ACS Chem. Biol 2011, 6, 356–365. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Liang J; Han Q; Tan Y; Ding H; Li J. Current Advances on Structure-Function Relationships of Pyridoxal 5’ Phosphate-Dependent Enzymes. Front. Mol. Biosci 2019, 6, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).(a) Gao SS; Naowarojna N; Cheng R; Liu X; Liu P. Recent examples of α-ketoglutarate-dependent mononuclear non-haem iron enzymes in natural product biosyntheses. Nat. Prod. Rep 2018, 35, 792–837. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Herr CQ; Hausinger RP. Amazing diversity in biochemical roles of Fe (II)/2-oxoglutarate oxygenases. Trends Biochem. Sci 2018, 43, 517–532. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Islam MS; Leissing TM; Chowdhury R; Hopkinson RJ; Schofield CJ. 2-Oxoglutarate-dependent oxygenases. Annu. Rev. Biochem 2018, 87, 585–620. [DOI] [PubMed] [Google Scholar]
- (14).Qian C; Zhou MM. SET domain protein lysine methyltransferases: Structure, specificity and catalysis. Cell. Mol. Life Sci 2006, 63, 2755–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).(a) Yanagisawa M; Sakal A; Adachi K; Sano T; Watanabe K; Tanaka Y; Okuda T. Hispidospermidin, a novel phospholipase C inhibitor produced by Chaetosphaeronema hispidulum (Cda) Moesz NR 7127. J. Antibiot 1994, 47, 1–5. [DOI] [PubMed] [Google Scholar]; (b) Ohtsuka T; Itezono Y. Nakayama N; Sakai A; Shimma N; Yokose K; Seto H. Hispidospermidin, a novel Phospholipase c inhibitor produced by Chaetosphaeronema hispidulum (Cda) Moesz NR 7127. J. Antibiot 1994, 47, 6–15. [DOI] [PubMed] [Google Scholar]
- (16).(a) Frontier AJ; Raghavan S; Danishefsky SJ. Stereocontrolled Total Synthesis of Hispidospermidin. J. Am. Chem. Soc 1997, 119, 6686–6687. [Google Scholar]; (b) Frontier AJ; Raghavan S; Danishefsky SJ. A highly stereoselective total synthesis of hispidospermidin: derivation of a pharmacophore model. J. Am. Chem. Soc 2000, 122, 6151–6159. [Google Scholar]; (c) Overman LE; Tomasi AL. Enantioselective total synthesis of hispidospermidin. J. Am. Chem. Soc 1998, 120, 4039–4040. [Google Scholar]; (d) Tamiya J; Sorensen EJ. A Concise Synthesis of (−)-Hispidospermidin Guided by a Postulated Biogenesis. J. Am. Chem. Soc 2000, 122, 9556–9557. [Google Scholar]; (e) Tamiya J; Sorensen EJ. A spontaneous bicyclization facilitates a synthesis of (−)-hispidospermidin. Tetrahedron 2003, 59, 6921–6932. [Google Scholar]
- (17).Hartmann M; Kim D; Bernsdorff F; Ajami-Rashidi Z; Scholten N; Schreiber S; Zeier T; Schuck S; Reichel-Deland V; Zeier J. Biochemical principles and functional aspects of pipecolic acid biosynthesis in plant immunity. Plant Physiol 2017, 174, 124–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Phelan RM; Townsend CA. Mechanistic insights into the bifunctional non-heme iron oxygenase carbapenem synthase by active site saturation mutagenesis. J. Am. Chem. Soc 2013, 135, 7496–7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Yan W; Song H; Song F; Guo Y; Wu CH; Her AS; Pu Y; Wang S; Naowarojna N; Weitz A; Hendrich MP. Endoperoxide formation by an α-ketoglutarate-dependent mononuclear non-haem iron enzyme. Nature 2015, 527, 539. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- (20).Backman LR; Funk MA; Dawson CD; Drennan CL. New tricks for the glycyl radical enzyme family. Crit. Rev. Biochem. Mol. Biol 2017, 52, 674–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Harvey CJ; Tang M; Schlecht U; Horecka J; Fischer CR; Lin HC; Li J; Naughton B; Cherry J; Miranda M; Li YF; Chu AM; Hennessy JR; Vandova GA; Inglis D; Aiyar RS; Steinmetz LM; Davis RW; Medema MH; Sattely E; Khosla C; Onge RP; Tang Y; Hillenmeyer ME. HEx: a heterologous expression platform for the discovery of fungal natural products. Sci. Adv 2018, 4, eaar5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Kurosawa S; Bando M; Mori K. Synthesis of (1R, 4R, 5S)-(+)-Acoradiene, the Structure Proposed for the Aggregation Pheromone of the Broad-Horned Flour Beetle. Eur. J. Org. Chem 2001, 2001, 4395–4399. [Google Scholar]
- (23).Tomita B; Hirose Y. Allo-cedrol: a new tricarbocyclic sesquiterpene alcohol. Phytochemistry 1973, 12, 1409–1414. [Google Scholar]
- (24).Shuler WG; Combee LA; Falk ID; Hilinski MK. Intermolecular Electrophilic Addition of Epoxides to Alkenes: [3+ 2] Cycloadditions Catalyzed by Lewis Acids. Eur. J. Org. Chem 2016, 2016, 3335–3338. [Google Scholar]
- (25).(a) McGee TP; Skinner HB; Bankaitis VA. Functional redundancy of CDP-ethanolamine and CDP-choline pathway enzymes in phospholipid biosynthesis: ethanolamine-dependent effects on steady-state membrane phospholipid composition in Saccharomyces cerevisiae. J. Bacteriol 1994, 176, 6861–6868. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Fillinger S; Ruijter G; Tamás MJ; Visser J; Thevelein JM; D’enfert C. Molecular and physiological characterization of the NAD-dependent glycerol 3-phosphate dehydrogenase in the filamentous fungus Aspergillus nidulans. Mol. Microbiol 2001, 39, 145–157. [DOI] [PubMed] [Google Scholar]; (c) Dijkema C; Rijcken RP; Kester HC; Visser J. 13C-NMR studies on the influence of pH and nitrogen source on polyol pool formation in Aspergillus nidulans. FEMS Microbiol. Lett 1986, 33, 125–131. [Google Scholar]
- (26).Brunson JK; McKinnie SM; Chekan JR; McCrow JP; Miles ZD; Bertrand EM; Bielinski VA; Luhavaya H; Oborník M; Smith GJ; Hutchins DA; Allen AE; Moore BS. Biosynthesis of the neurotoxin domoic acid in a bloom-forming diatom. Science 2018, 361, 1356–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.