Abstract
High-throughput data collection for electron microscopy (EM) demands appropriate software tools. We present a combination of such tools that enable automated acquisition guided by image analysis for a variety of transmission EM acquisition schemes. Py-EM interfaces with the microscope control software SerialEM and expands its updated flexible automation features by image analysis to enable feedback microscopy. We demonstrate dose-reduction in cryo-EM experiments, fully automated acquisition of every cell in a plastic section, and automated targeting on serial sections for volumetric imaging across multiple grids.
Introduction
As quantitative observations become standard in biological research, the need for increased data acquisition throughput grows. New technologies such as improved detectors, microscope hard- and software and the increase in computing performance have led to substantial improvements in speed and image quality in Transmission Electron Microscopy (TEM) imaging1–3. The major bottleneck during the acquisition procedure remains to be the selection of features of interest, namely identifying their exact coordinates for acquisition, which is largely done manually. In image analysis, current developments enable automatic detection of specific features4–6. In high-throughput fluorescence microscopy, feedback from image analysis pipelines is applied to define subsequent acquisition targets7,8. For TEM, existing acquisition software solutions integrate automation for specific tasks such as single particle acquisition or tomography9. In cryo-TEM, feedback from image analysis is employed to identify and judge the quality of particles for acquisition of imaging10–12 or diffraction data13–15. In material science TEM, similar approaches exist that enable ultra-stable imaging16–18. There is, however, no solution that incorporates advanced image analysis procedures into the various kinds of TEM data acquisition workflows in a generic way.
Here we present two software tools that, when combined, allow the user to incorporate established image analysis tools and thus enable specimen-specific automated feedback TEM workflows.
1) SerialEM19 offers control of the microscope and its imaging detectors in a very flexible manner. In the last years of developing this software, new features for automation have been introduced, that we describe in this article. The Navigator functionality is the key element for imaging multiple regions of a specimen with pre-defined acquisition parameters in an automated manner. By using the built-in scripting functionality, users can create and perform highly customized acquisition routines.
2) To provide SerialEM with the necessary coordinate and image information for automated acquisition, we have developed py-EM, a Python module that integrates common image analysis applications. Identifying features of interest automatically from micrographs using specimen-specific image analysis pipelines consequently enables feedback microscopy.
Software
SerialEM
SerialEM19 is a versatile software for TEM data collection originally intended mainly for electron tomography acquisition. While its core is made available as open source, it can communicate with microscope and camera hardware of various manufacturers using a plugin scheme. Over the years, the software has received significant additional functionality to increase automation. Users can control and modify most of its functionality using SerialEM’s built-in scripting feature and thus highly individualize their acquisition procedures. External software tools such as py-EM can be triggered and run through SerialEM’s graphical user interface.
The glossary in Box 2 describes the nomenclature of SerialEM’s key functionality for automated acquisition.
A detailed introduction to the main components of SerialEM required for automated acquisition is provided in Box 3. These functions have been developed over the past since the initial description of the software19, but have not been described in a subsequent scientific publication.
The py-EM module
Py-EM is a collection of Python functions that both interpret and modify SerialEM’s Navigator files and the items within and perform or trigger image analysis tasks on maps. It interfaces between SerialEM – hence the microscope – and any available software tool for image analysis and processing enabling feedback pipelines. We have put a strong focus on making this interface as modular as possible to leave maximum freedom in the choice of external tools for a specific experiment.
Py-EM contains functions that make use of stitching routines available in the IMOD20 software package for processing maps consisting of multiple tiles (i.e. montages). It then makes the resulting single stitched image available for image processing. At the same time, py-EM preserves the Navigator’s coordinate system for that map. As a result of the processing, py-EM generates a Navigator file to be used for further acquisition.
The key concept of our automation approach is to provide SerialEM with a Virtual Map that is generated for each acquisition item. With the information about the visual features of the target object in the associated image, an accurately positioned acquisition can be guaranteed through the Realign To Item procedure. After acquiring a single map with the desired acquisition settings anywhere on the specimen, its Navigator entry will be used as a template containing all important parameters other than position and template image.
Due to SerialEM handling the storage location of map images using relative paths, it is usually possible to process data directly at the microscope for immediate feedback or to transfer the entire data directory including the Navigator file to another computer for processing and back again for acquisition.
We have applied py-EM workflows to automate TEM acquisition either running them as standalone Python scripts or embedded into a KNIME workflow. This generic data analysis software21 includes image processing capabilities22 but also incorporates modules from different other platforms (ImageJ23,24, CellProfiler25,26, Ilastik5, R27 or MATLAB) into a single workflow.
The modular approach of the py-EM functions combined with the KNIME framework thus allows incorporating any type of already existing image processing routine identifying features of interest (Fig. S1). Together with KNIME’s graphical representation of the process, this makes the image analysis procedure very user-friendly.
Applications
High yield automated cryo-EM data acquisition of large particles
Structural determination of protein complexes at high resolution is made possible by averaging of multiple thousands of cryo-EM images. To achieve the necessary throughput, systematic acquisition strategies follow a regular array of acquisition positions10. SerialEM offers tools to create these acquisition patterns and its batch acquisition and scripting routines are optimized for such experiments. Image analysis can be used to target holes on holey carbon grids with good ice conditions9. Both these strategies however cannot capture quality or concentration of the target particles and thus produce large amounts of unusable data when the sample is heterogeneous.
If target particles are large enough to be identified in overview images, these can be manually picked. In this case additional maps at higher magnification need to be acquired at each point in order to successfully relocate and acquire precisely at the selected positions. This exposes the specimen to extra dose before high-resolution imaging and takes a considerable amount of time.
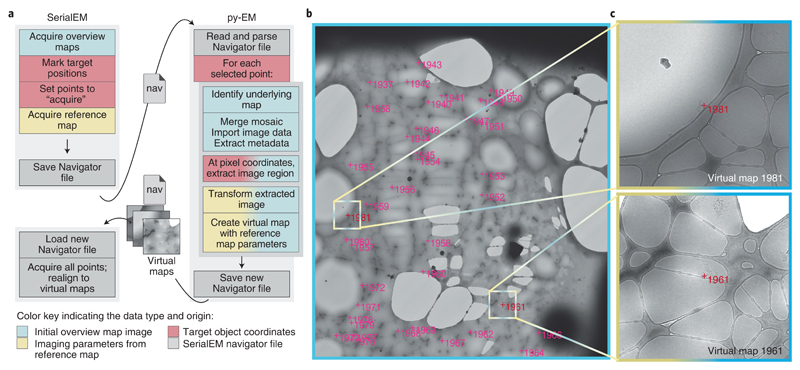
We have developed a workflow that allows high-magnification acquisition of manually selected target items without having to acquire additional maps. Figure 1 shows the workflow and resulting Virtual Maps for a cryo-EM observation of filamentous TORC128 particles. The procedure illustrated in Figure 1a uses py-EM functionality to generate the maps needed for realigning from the already acquired medium magnification overview maps. The py-EM script crops the respective image areas from the grid square maps (Fig. 1b, 2×2 montage at 2250×, pixel size: 6.2 nm) for each selected Navigator item and transforms the extracted images, accounting for both the relative scaling and the rotation to match a template map at medium-high magnification (Fig. 1c). Thus the extracted image that is provided as a new Virtual Map is suitable for direct correlation with an acquired image for realigning. A detailed description of the procedure is provided in Supplementary Protocol 1. The script maps_acquire.py is provided in the applications directory of the py-EM code repository.
Figure 1.
Example application of the automatic generation of Virtual Maps to acquire TORC1 filamentous particles with cryo-EM. The color scheme illustrates the data type and origin as indicated in the bottom legend. a – The application workflow depicting the individual steps in the procedure and the communication and data transfer between SerialEM and py-EM using Navigator files and Virtual Map images. b – Overview map covering a single grid square. The magenta crosses indicate the target positions that were manually clicked within this map. Colored boxes and marks indicate the example regions where the corresponding Virtual Maps were extracted. c – Two example Virtual Maps that the script extracted from the map shown in c. The image data is extracted from the overview map while the microscope parameters are copied from the reference map. Scale: FOV of b: 47 μm; colored boxes in b, FOV of c: 3.8 μm. The presented experiment comprised 2500 points from 125 maps on one grid. The identical procedure was so far successfully applied in 9 experiments on different specimens.
Cryo-EM data collection is typically divided into two phases: selection of acquisition areas and automated data collection. For large particles, the first 24 hours of a 72 hours acquisition slot are spent loading grids, identifying good grid squares and acquiring maps of those. In parallel, the manual picking of particles as well as running the py-EM scripts can happen offline. Instead of acquiring additional maps for realigning, which would take around eight hours, the data acquisition can start directly thereafter. This increases the time available for high-resolution imaging by 20% from 40 to 48 hours.
By avoiding the acquisition of additional maps, the specimen gets irradiated with about 0.1 - 0.2 e-/Å2 less at each position. When maps of close particles overlap, this extra dose would apply multiple times, significantly compromising resolution.
The published result reached 27 Å resolution from 50 micrographs of TORC1 filaments acquired entirely manually28. Using automation and realigning to Virtual Maps, the presented experiment generated 2660 micrographs that allow processing to a resolution where secondary structure elements can be identified.
Systematic acquisition of individual cells on resin sections using feedback TEM
A few organelles, such as centrosomes, are present only as single copies in a given cell. In a typical TEM analysis, the probability of observing a centriole on a 200 nm section of resin-embedded cells ranges between 3 to 5%. When trying to identify a sufficient number of cells that contain such an event, it is necessary to screen the entire population of a section at an intermediate magnification. An alternative would be to implement targeting strategies based on tagging the event of interest, such as in CLEM approaches 29,30. Adapting labeling protocols to primary samples often implies a loss of ultrastructural detail. Systematic screens are extremely challenging by conventional TEM, as it is hard to ensure each cell is imaged only once. This makes the task very tedious and time consuming.
The application presented here is a study of centriole morphology in leukemia cells collected from individual patients. To statistically assess ultrastructural diversity, it was necessary to acquire a large number of centrioles in different samples. We developed a workflow to automatically acquire each individual cell on serial plastic sections. With dimensions of about 500×200 μm, each section would typically display from several hundred up to 1500 cell cross-sections.
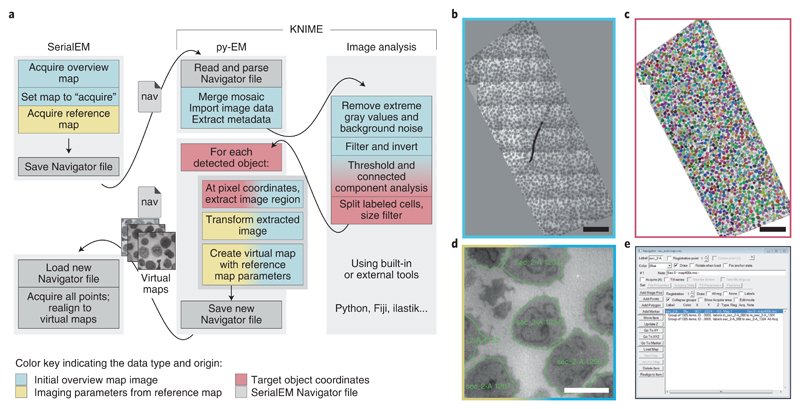
A schematic of the procedure, the input map, the output from the automated cell detection, the resulting Navigator file within SerialEM and an example Virtual Map are shown in Figure 2. To successfully identify each cell, we use the KNIME platform to link and incorporate various independent image analysis tools (in our case Python and ImageJ/Fiji) (Fig. 2a). The user can easily control image analysis parameters through a graphical dialog without needing any knowledge in programming. Figure S1a shows the layout of the KNIME workflow and the graphical controls where users can modify a few selected parameters. The image analysis procedure is illustrated in Figure S1b.
Figure 2.
Illustration of the workflow to automatically acquire all cells on a resin section. The color scheme illustrates the data type and origin as indicated in the bottom legend. a – The application workflow depicting the individual steps in the procedure and the communication and data transfer using Navigator files and Virtual Map images. The KNIME platform embeds py-EM functionality and the image analysis procedure. Interaction with SerialEM happens by exchanging Navigator files and Virtual Maps. b – Overview map, a montage of the entire section. The image limits are defined manually during the merging procedure. c – The result of the automated cell detection. Each cell is represented by a distinct label value, displayed as different colors. d – An example Virtual Map that has been extracted from the overview map for a single cell. The green outline depicts the polygon that determines the acquisition area. e – The resulting Navigator file contains a Virtual Map and the polygon outline for each of the 1325 detected cells. Scale bars: 50 μm (overviews), 5 μm (Virtual Map). The automated identification of cells presented in this figure has been successfully applied to 26 sections with about 1000 cells each for this experiment and with modified image analysis pipelines for two different specimens.
In our example, the workflow will employ py-EM to merge the low magnification mosaic map(s) and to load the map image(s) into the KNIME analysis pipeline for identifying the single cells. It then extracts Virtual Maps at each detected position, generates polygons matching the outline of each cell linked to the corresponding Virtual Maps, and writes a Navigator file ready for acquisition.
We acquire an image stack containing one image per cell that can then be screened easily to categorize cells according to cell type, phenotype or by identifying specific morphological features. In the presented example, the software automatically detected 1325 cell profiles out of which we selected 44 cells that displayed parts of a centriole (see application 3). This manual curation step takes about one hour. The entire image stack is presented in Supplementary Video 1. The necessary files to run the workflow in KNIME are provided as part of the py-EM code.
Previous studies illustrating centriole morphology31,32 that employed manual screening, have required several weeks to months at the microscope resulting in only a few dozens of individual observations, rendering a comparative phenotypic analysis impossible. In the presented experiment, we were able to identify several hundreds of centrioles in leukocytes from different patients for further high-resolution studies requiring only a few days of screening for each. The resulting Virtual Maps enable SerialEM to accurately return to any chosen cell for further acquisition.
Acquiring the entire section at sufficient resolution would require around 70×30 tiles, resulting in a 2D image whose size easily exceeds the memory of most current workstations. Therefore, identifying interesting cells by systematically scanning through such a large montage is challenging either manually or using image analysis. Using automated targeted acquisition, the user can screen a stack of images centered on every individual cell. This makes it easy to focus on morphological features and avoids redundant acquisitions. Also, since every single detected cell is imaged, statistics and classification based on observed phenotype are readily available.
Automated serial-section TEM
In a serial-section TEM experiment, one wants to follow a feature across a series of sections that lie adjacent on the TEM grid33. For reconstructing full cell volumes, series of sections can easily span across dozens of grids. In order to acquire high-resolution data, such as tomograms, positional information needs to be propagated for each location on each section. While doing this manually is feasible when only navigating to a handful of features per section, it quickly becomes a tedious and very time-consuming task when targeting more objects.
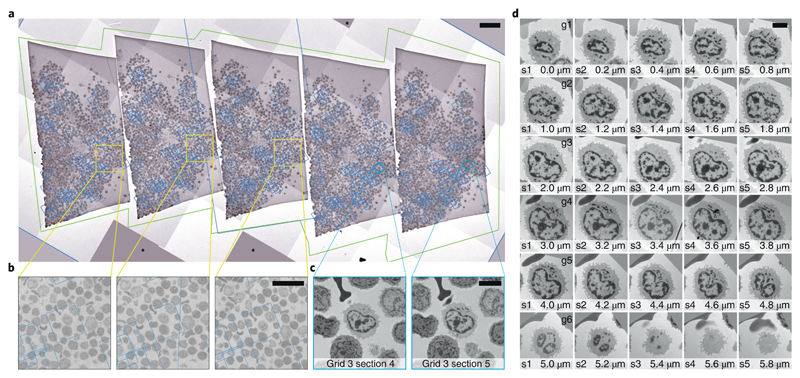
In our example, we selected 60 human leukocyte cells on one of the serial sections (thickness 200 nm) as described above. We wanted to acquire the exact same cells on the 4 other sections on this grid (Fig. 3a) as well as on the consecutive grids and thus follow the selected cells in 3D. We use py-EM scripts to duplicate the list of target items and then employ SerialEM’s registration and realigning functionality to successfully locate cells on neighboring sections based on the maps from the preceding section (Fig. 3b). For each cell of interest, these maps at moderate magnification cover a sufficiently large field of view for realigning (Fig. 3c). We then automatically acquire new maps at every target location wherein we then define the exact position for high-magnification acquisition. The SerialEM script that we use to run the acquisition takes care of keeping consistent file names and Navigator labels throughout the procedure.
Figure 3.
Automated acquisition of cells on serial sections. a – Overview of a ribbon of sections placed on a slot grid. The green outline marks the polygon used to acquire this Montage map. The blue boxes denote the location of the maps used to realign to each cell. b – Magnified regions in a showing the location of one cell of interest. The fact that the blue box that denotes the location of the corresponding maps is not precisely centered on the cell is not crucial for repositioning. The image information used for realigning is taken from the map itself. c – Two maps of a single cell on neighboring sections. These images are used during Realign to Item. d – Gallery of images of an individual cell across 30 sections (thickness: 200 nm) spread across 6 grids. The images were automatically aligned using TrakEM239. Another cell that spans cross 9 sections is shown in Supplementary Video 3. Scales: a: 50 μm, b: 20 μm, C/D: 5 μm. For the presented experiment we have followed a total of 120 cells across 100 sections on 20 grids for 3 different specimens.
We used this approach in the presented example to successfully follow each of 60 cells through a series of 45 serial sections across 9 grids. Figure 3d shows a cell acquired from 30 sections on six grids. An example of aligned images of a cell across 42 of these sections is shown in Supplementary Video 2. A detailed description of the procedure is provided in Supplementary Protocol 2. Supplementary Video 3 provides a step-by-step tutorial on the use of SerialEM to register maps from two adjacent sections.
To resolve the structure of the respective centrosomes at higher resolutions, we acquired tilt series at positions manually defined in the respective maps on the consecutive sections. We use a custom script for fast tomography acquisition. We automatically processed the large amount of acquired tilt series using IMOD’s batch tomogram reconstruction34 on a high-performance compute cluster resulting in full volumes of the desired objects. An example of such a reconstructed tomographic volume is shown in Supplementary Video 4. We have made available all SerialEM scripts used in this experiment at the public Script Repository (https://serialemscripts.nexperion.net/).
Using conventional approaches, manually selecting target cells on consecutive grids is only feasible for a small number of targets and strongly depends on the presence of easily recognizable features on the specimen. Without these, it can take an expert a day to locate just a few target positions across serial sections35. Our automated approach enables parallel tracking and acquisition of hundreds of features.
Transferring maps of 60 cells across five sections by hand took one full day before introducing the scripts. With these, setting up a transition from section to section requires about 10 minutes, independent of the number of objects. Due to the usually high number of acquisition points, the necessary imaging time for the maps strongly depends on secondary tasks such as the speed of the microscope stage or the desired autofocus procedure. For 60 cells, the map acquisition takes between 15 and 30 minutes per section.
The main criterion of the success rate, when trying to automatically relocate cells, is the quality of serial sectioning. Failures to register maps to adjacent sections occur with the presence of large cracks or folds. In our experiments, transferring maps from one section to the next worked almost perfectly for every cell. Problems in identifying the correct feature only occurred when the edge of a section, a grid bar, a crack or fold or large contaminations were blocking the field of view.
When features of cells or tissue do not change dramatically from one section to the following and the quality of the serial sections is good, an alignment can even be done for all sections of a grid in one procedure. The reference maps would then be taken from the last section of the previous grid. There no longer is a need for user interaction after each individual section and the procedure can run fully automated.
The maps cover not only the cell of interest but also the constellation of neighboring cells. Even when a given cell ends in z, the region is usually correctly found. We then manually screen the maps for ending cells and misalignments when setting up the tomography acquisition.
The throughput in tomography acquisition has also increased dramatically, thanks to the availability of fast SCMOS-based detectors. Combined with a sufficiently stable microscopy stage, the built-in drift correction and short exposure times allow skipping otherwise necessary tracking steps. Compared to a traditional tilt series acquisition, this leads to another speed-up by two and reduces the necessary time for a single axis tomogram with 121 tilt images to around 7 minutes.
In the presented experiment we have followed a total of 120 cells across 100 sections on 20 grids for 3 different specimens. In these cells we have acquired a total of 992 tomograms in series on consecutive sections.
Discussion
We have developed both software packages described in this article with a strong focus on flexibility. SerialEM is capable of controlling transmission electron microscopes, accessories and detectors from a range of different vendors and covers acquisition strategies like automated single particle acquisition, cryo-electron tomography, large 2D mosaic tiling as well as batch tomography on serial sections. Data presented in this article was acquired using the following microscopes: JEM 2100Plus (JEOL Ltd., Akishima, Japan), Tecnai F30 and Titan Krios G3 (Thermo Fischer Scientific, Waltham, MA, USA).
The py-EM module aims at universally linking SerialEM with a variety of software options linking image analysis and TEM acquisitions in a feedback logic. This means that the main task in setting up an automated TEM workflow for a specific application is now reduced to developing a suitable image analysis routine. These routines are obviously not limited to the examples described in this article, but could also include advanced feature detection using machine learning5,6 including convoluted neuronal networks36,37. Such approaches will reduce the need for manual identification of particles in cryo-EM by automatically picking holes or large particles. Especially due to the inherent low signal-to-noise in cryo-EM or even worse cryo-tomography, machine learning approaches might enable acquisition of objects that would not be recognizable by eye38.
Being able to image every entity of a specimen in a controlled manner offers entirely new possibilities for TEM. Automated screening and classification into different cell types and phenotypes or detection of aberrant morphology can be performed by computational analysis when having large amounts of comparable data at hand. Automation of serial-section TEM significantly reduces the workload required for this technique and opens new possibilities to perform large-scale 3D-EM while maintaining a high lateral resolution.
A main goal of future developments for both software packages is to make the described automated acquisition approaches easily accessible to users. Demanding applications for the combination of Py-EM and SerialEM will further expand and improve their functionality and we expect encouraging and challenging suggestions from the community. Incorporating image data from multiple modalities, such as in a CLEM experiment, to guide targeted acquisition and their subsequent registration into a common coordinate frame is a challenge itself. Combining such approaches with automation strategies to achieve high throughput would be one of these potential applications.
The increased throughput by automating acquisition schemes and thus the resulting generation of large quantities of data that is subsequently fed into automated analysis pipelines, will enable entirely new types of quantitative TEM experiments.
Supplementary Material
Box 1. Software downloads, documentation and development.
Downloads of SerialEM, all documentation and detailed tutorials are available at http://bio3d.colorado.edu/SerialEM/.
The features presented in this paper are available in version 3.7. SerialEM can be installed on any Windows PC and run without connection to the microscope. As it handles the storage location of map images using relative paths, this enables processing data at a separate workstation while continuing the acquisition of maps or data at the microscope.
External software such as py-EM scripts can be incorporated into the SerialEM GUI via the Tools menu (http://bio3d.colorado.edu/SerialEM/hlp/html/menu_tools.htm).
A collection of SerialEM scripts for various purposes, including all scripts used in the experiments described in this paper, is hosted at the public Script Repository (https://serialemscripts.nexperion.net/).
Both SerialEM (MIT license) and the SerialEMCCD plugin to DigitalMicrograph (GPL v.2) are open source and available at http://bio3d.colorado.edu/SerialEM/OpenSerialEM and http://bio3d.colorado.edu/SerialEM/SerialEMCCD.
The py-EM python module is available open source (GPL v.3) for download and further collaborative development at https://git.embl.de/schorb/pyem. This repository also contains detailed installation instruction and various step-by-step tutorials to introduce the functionality.
Py-EM was tested using both Python 3.6 and 2.7. The module depends on the following Python packages: numpy40, scipy40, scikit-image41, mrcfile42,43, and pandas44 for KNIME integration. Most of these are included in the Anaconda framework (https://anaconda.org/). An interactive Jupyter notebook (https://git.embl.de/schorb/pyem/raw/master/pyEM.ipynb?inline=false) guides the user through the installation procedure.
The exact versions of SerialEM and py-EM described in this paper are provided as supplementary software.
Box 2. Glossary of SerialEM key features.
Map – Navigator item that links an image with coordinates, microscope parameters and imaging state
Montage – image of overlapping single tiles, often used as Map
Navigator – main tool for positioning and targeting in SerialEM; information stored in Navigator files.
Realign to Item – procedure to position the acquisition area precisely to an image feature from a Map.
Registration – independent coordinate systems of objects
Virtual Map – Map item created as a combination of an externally generated image and microscope parameters from a reference Map template. Used to enable Realign to Item.
Box 3. SerialEM key functionality for automated TEM.
Navigator
The Navigator module is the main tool for finding, keeping track of, and positioning at targets of interest. Positions of single points can be recorded either by storing stage coordinates while browsing the specimen, or by marking points on an acquired image.
Maps
A map is a specific image for which the Navigator remembers its storage location, the corresponding acquisition parameters for microscope and camera, and how to display the image to the user. Maps are generally used to provide an overview for picking acquisition targets, but also play an important role in allowing repositioning at a target during batch acquisition.
Polygons
Polygons are connected points drawn on an image to outline an area. Their main use is to define the area to be acquired with a montage at a higher magnification.
Montages
When the area of interest is larger than the field of view of a single, SerialEM can acquire a montage of overlapping tiles. Montages are always taken at regular spacing in a rectangular pattern, but acquisition at tiles that are not needed to fill a polygon can be skipped (Fig. 3a). Very large areas, such as a ribbon of sections, can be defined by moving the stage to each corner of the area and record a corner point there.
Tile positions are reached with electronic image shift, as during tilt series acquisition, or with stage movement for larger areas. Periodic focusing and other features support acquisition of high-quality images from arbitrarily large areas45. Montages are commonly used as overview images for finding targets for data collection.
Realign to Item
The user marks target positions on a map (either a single image, or a montage). Drift will change the exact specimen position over time. Thus, the target feature will no longer be centered when returning to the recorded stage position for acquisition. To accurately reposition the acquisition area to the marked position, SerialEM has a procedure, Realign to Item, that takes images under the same conditions as those used to acquire a map and cross-correlates those images with the map. After aligning to an overview map, the realignment routine can also align to a second map acquired at higher magnification, such as one to define the target area for tilt series acquisition.
Anchor map
For taking a pair of maps, the user indicates the lower-magnification map at the first target as the anchor map template. Thereafter, the user can take images of target areas at the acquisition magnification by pressing the “Anchor Map” button; the program will save that image as a map, will then go to the lower magnification and take and save the anchor map.
Batch acquisition
Batch acquisition by the Navigator is initiated with the Acquire at Points dialog, which can acquire an image, a map, or a tilt series or alternatively run a script at each item marked for acquisition. The dialog offers a choice of operations to be run before acquisition, such as focusing or centering the beam. The Realign to Item routine will typically be chosen to guarantee accurate repositioning.
Registrations
Navigator items that belong to the same coordinate system share the same Registration number. When a grid is rotated, or removed and reinserted, the coordinate system has changed, and any new Navigator items need to be assigned a different registration number. The program then determines the transformation from one registration to another, using a set of points that correspond between the registrations (e.g., features that the user marks in both an imported Light Microscopy image and an EM map). Alternatively, and the program can find the rotation and shift that relate an image before and after rotating the grid for dual-axis tomography by cross-correlation. This procedure is called Align with Rotation. SerialEM also transforms map images of a registration accordingly before using them with Realign to Item.
“Virtual” map templates
Each map item contains the necessary operating conditions for the electron microscope that enables SerialEM to align to features that resemble the map image on the current specimen using “Realign to Item”. Linking a Navigator map item to an image while adding microscope parameters duplicated from a template map will create a “virtual” map entry. We use this approach to precisely realign to a specific cell in neighboring serial sections duplicating maps of target cells from a previous section to identify and target the same cell on the following sections (Application 3, Fig. 3b & c).
Editor’s Summary.
Py-EM and SerialEM enable automated microscope control for high-throughput data acquisition in diverse transmission electron microscopy imaging experiments.
Acknowledgements
We thank Alwin Krämer for initiating and coordinating the scientific project on leukocytes. We thank Ambroise Desfosses and Manoel Prouteau for providing specimens to test and apply the cryo-EM workflow. We acknowledge support from Colin Palmer for mrcfile, Christian Dietz and Clemens von Schwerin for KNIME Image Processing and Python bindings. We like to thank all staff of the EM Core Facility at EMBL for helpful discussions and ideas. We thank Rachel Mellwig for critically reading and commenting the manuscript. We also acknowledge support from the EMBL Center of Bioimage Analysis (CBA). Work on SerialEM was supported originally by grants from NIH and more recently by contributions from users and from JEOL USA, Inc, and by payments for specific projects by Hitachi High Technologies America, Inc, JEOL USA, Inc., and Direct Electron, LP. I.H. is the recipient of a HRCMM (Heidelberg Research Center for Molecular Medicine) Career Development Fellowship.
Footnotes
Author contributions
M.S., I.H., W.J.H.H and Y.S. designed the experimental applications. M.S. and D.N.M. did the software development. I.H., W.J.H.H. and M.S. collected the data. All authors contributed valuable suggestions to the necessary software functionality and edited the article. M.S. and D.N.M wrote the initial manuscript.
Data and Software availability statement
All raw data presented in the applications section of this manuscript is available from the corresponding authors upon request.
The software SerialEM, all documentation and detailed tutorials are available at http://bio3d.colorado.edu/SerialEM/.
Py-EM, step-by-step tutorials and installation instructions are available at https://git.embl.de/schorb/pyem.
Experimental details
Specific details of the experiments presented as applications and exact information on each software component used in the applications is provided in the Life Sciences Reporting Summary.
Competing interest statement
The authors declare no competing interests.
Contributor Information
Martin Schorb, Electron Microscopy Core Facility, EMBL, Heidelberg.
Wim JH Hagen, Structural and Computational Biology Unit and Cryo-Electron Microscopy Service Platform, EMBL, Heidelberg.
Yannick Schwab, Cell Biology and Biophysics Unit and Electron Microscopy Core Facility, EMBL, Heidelberg.
David N Mastronarde, Dept. of Molecular, Cellular & Developmental Biology, University of Colorado, Boulder.
References
- 1.Winey M, Meehl JB, O’Toole ET, Giddings TH. Conventional transmission electron microscopy. Mol Biol Cell. 2014;25:319–323. doi: 10.1091/mbc.E12-12-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McMullan G, Faruqi AR, Henderson R. Direct Electron Detectors. Methods Enzymol. 2016;579:1–17. doi: 10.1016/bs.mie.2016.05.056. [DOI] [PubMed] [Google Scholar]
- 3.Frank J. Advances in the field of single-particle cryo-electron microscopy over the last decade. Nat Protoc. 2017;12:209–212. doi: 10.1038/nprot.2017.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meijering E, Carpenter AE, Peng H, Hamprecht FA, Olivo-Marin J-C. Imagining the future of bioimage analysis. Nat Biotechnol. 2016;34:1250–1255. doi: 10.1038/nbt.3722. [DOI] [PubMed] [Google Scholar]
- 5.Kreshuk A, Koethe U, Pax E, Bock DD, Hamprecht FA. Automated detection of synapses in serial section transmission electron microscopy image stacks. PloS One. 2014;9:e87351. doi: 10.1371/journal.pone.0087351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arganda-Carreras I, et al. Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification. Bioinforma Oxf Engl. 2017;33:2424–2426. doi: 10.1093/bioinformatics/btx180. [DOI] [PubMed] [Google Scholar]
- 7.Conrad C, et al. Micropilot: automation of fluorescence microscopy-based imaging for systems biology. Nat Methods. 2011;8:246–249. doi: 10.1038/nmeth.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neumann B, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan YZ, Cheng A, Potter CS, Carragher B. Automated data collection in single particle electron microscopy. Microsc Oxf Engl. 2016;65:43–56. doi: 10.1093/jmicro/dfv369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suloway C, et al. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 11.Rice WJ, et al. Routine determination of ice thickness for cryo-EM grids. J Struct Biol. 2018;204:38–44. doi: 10.1016/j.jsb.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicholson WV, White H, Trinick J. An approach to automated acquisition of cryoEM images from lacey carbon grids. J Struct Biol. 2010;172:395–399. doi: 10.1016/j.jsb.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coudray N, et al. Automated screening of 2D crystallization trials using transmission electron microscopy: A high-throughput tool-chain for sample preparation and microscopic analysis. J Struct Biol. 2011;173:365–374. doi: 10.1016/j.jsb.2010.09.019. [DOI] [PubMed] [Google Scholar]
- 14.Hu M, et al. Automated electron microscopy for evaluating two-dimensional crystallization of membrane proteins. J Struct Biol. 2010;171:102–110. doi: 10.1016/j.jsb.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng A. Automation of data acquisition in electron crystallography. Methods Mol Biol Clifton NJ. 2013;955:307–312. doi: 10.1007/978-1-62703-176-9_17. [DOI] [PubMed] [Google Scholar]
- 16.Gatel C, Dupuy J, Houdellier F, Hÿtch MJ. Unlimited acquisition time in electron holography by automated feedback control of transmission electron microscope. Appl Phys Lett. 2018;113 133102. [Google Scholar]
- 17.Tejada A, den Dekker AJ, Van den Broek W. Introducing measure-by-wire, the systematic use of systems and control theory in transmission electron microscopy. Ultramicroscopy. 2011;111:1581–1591. doi: 10.1016/j.ultramic.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 18.Liu J, et al. Fully Mechanically Controlled Automated Electron Microscopic Tomography. Sci Rep. 2016;6 doi: 10.1038/srep29231. 29231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 21.Berthold MR, et al. Data Analysis, Machine Learning and Applications. Springer; Berlin, Heidelberg: 2008. KNIME: The Konstanz Information Miner; pp. 319–326. [DOI] [Google Scholar]
- 22.Dietz C, Berthold MR. KNIME for Open-Source Bioimage Analysis: A Tutorial. In: Vos WHD, Munck S, Timmermans J-P, editors. Focus on Bio-Image Informatics. Springer International Publishing; 2016. pp. 179–197. [DOI] [Google Scholar]
- 23.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carpenter AE, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McQuin C, et al. CellProfiler 3.0: Next-generation image processing for biology. PLOS Biol. 2018;16:e2005970. doi: 10.1371/journal.pbio.2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pau G, Fuchs F, Sklyar O, Boutros M, Huber W. EBImage--an R package for image processing with applications to cellular phenotypes. Bioinforma Oxf Engl. 2010;26:979–981. doi: 10.1093/bioinformatics/btq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prouteau M, et al. TORC1 organized in inhibited domains (TOROIDs) regulate TORC1 activity. Nature. 2017;550:265–269. doi: 10.1038/nature24021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Boer P, Hoogenboom JP, Giepmans BNG. Correlated light and electron microscopy: ultrastructure lights up! Nat Methods. 2015;12:503–513. doi: 10.1038/nmeth.3400. [DOI] [PubMed] [Google Scholar]
- 30.Kukulski W, et al. Correlated fluorescence and 3D electron microscopy with high sensitivity and spatial precision. J Cell Biol. 2011;192:111–119. doi: 10.1083/jcb.201009037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elserafy M, et al. Molecular mechanisms that restrict yeast centrosome duplication to one event per cell cycle. Curr Biol CB. 2014;24:1456–1466. doi: 10.1016/j.cub.2014.05.032. [DOI] [PubMed] [Google Scholar]
- 32.Marteil G, et al. Over-elongation of centrioles in cancer promotes centriole amplification and chromosome missegregation. Nat Commun. 2018:9. doi: 10.1038/s41467-018-03641-x. 1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bron C, et al. Three-dimensional electron microscopy of entire cells. J Microsc. 1990;157:115–126. doi: 10.1111/j.1365-2818.1990.tb02952.x. [DOI] [PubMed] [Google Scholar]
- 34.Mastronarde DN, Held SR. Automated tilt series alignment and tomographic reconstruction in IMOD. J Struct Biol. 2016 doi: 10.1016/j.jsb.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quinn TA, et al. Electrotonic coupling of excitable and nonexcitable cells in the heart revealed by optogenetics. Proc Natl Acad Sci U S A. 2016;113:14852–14857. doi: 10.1073/pnas.1611184114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cruz-Roa A, et al. High-throughput adaptive sampling for whole-slide histopathology image analysis (HASHI) via convolutional neural networks: Application to invasive breast cancer detection. PloS One. 2018;13:e0196828. doi: 10.1371/journal.pone.0196828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tajbakhsh N, et al. Convolutional Neural Networks for Medical Image Analysis: Full Training or Fine Tuning? IEEE Trans Med Imaging. 2016;35:1299–1312. doi: 10.1109/TMI.2016.2535302. [DOI] [PubMed] [Google Scholar]
- 38.Buchholz T-O, Jordan M, Pigino G, Jug F. Cryo-CARE: Content-Aware Image Restoration for Cryo-Transmission Electron Microscopy Data. ArXiv181005420 Cs. 2018 [Google Scholar]
- 39.Cardona A, et al. TrakEM2 software for neural circuit reconstruction. PloS One. 2012;7:e38011. doi: 10.1371/journal.pone.0038011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oliphant TE. Python for Scientific Computing. Comput Sci Eng. 2007;9:10–20. [Google Scholar]
- 41.van der Walt S, et al. scikit-image: image processing in Python. PeerJ. 2014;2:e453. doi: 10.7717/peerj.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wood C, et al. Collaborative computational project for electron cryo-microscopy. Acta Crystallogr D Biol Crystallogr. 2015;71:123–126. doi: 10.1107/S1399004714018070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burnley T, Palmer CM, Winn M. Recent developments in the CCP-EM software suite. Acta Crystallogr Sect Struct Biol. 2017;73:469–477. doi: 10.1107/S2059798317007859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKinney W. Data Structures for Statistical Computing in Python. Proceedings of the 9th Python in Science Conference; 2010. pp. 51–56. [Google Scholar]
- 45.Anderson JR, et al. Exploring the retinal connectome. Mol Vis. 2011;17:355–379. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.