Abstract
We have encountered pancreatic tumors with unique histologic features, which do not conform to any of the known tumors of the pancreas or other anatomical sites. We aimed to define their clinicopathologic features and whether they are characterized by recurrent molecular signatures.
Eight cases were identified; studied histologically and by immunohistochemistry. Selected cases were also subjected to whole-exome sequencing (WES; n=4), RNA-sequencing (n=6), Archer FusionPlex assay (n=5), methylation profiling using the Illumina MethylationEPIC (850k) array platform (n=6) and TERT promoter sequencing (n=5).
Six neoplasms occurred in females. The mean age was 43 years (range, 26–75). Five occurred in the head/neck of the pancreas. All patients were treated surgically; none received neoadjuvant/adjuvant therapy. All patients are free of disease after 53 months of median follow-up (range, 8–94). The tumors were well-circumscribed, and the median size was 1.8 cm (range, 1.3–5.8). Microscopically, the unencapsulated tumors had a geographic pattern of epithelioid cell nests alternating with spindle cell fascicles. Some areas showed dense fibrosis, in which enmeshed tumor cells imparted a slit-like pattern. The predominant epithelioid cells had scant cytoplasm and round-oval nuclei with open chromatin. The spindle cells displayed irregular, hyperchromatic nuclei. Mitoses were rare. No lymph node metastases were identified. All tumors were positive for vimentin, CD99 and cytokeratin (patchy), while negative for markers of solid pseudopapillary neoplasm, neuroendocrine, acinar, myogenic/rhabdoid, vascular, melanocytic, or lymphoid differentiation, gastrointestinal stromal tumor as well as MUC4. Whole exome sequencing revealed no recurrent somatic mutations or amplifications/homozygous deletions in any known oncogenes or tumor suppressor genes. RNA-sequencing and the Archer FusionPlex assay did not detect any recurrent likely pathogenic gene fusions. Single sample gene set enrichment analysis revealed that these tumors display a likely mesenchymal transcriptomic program. Unsupervised analysis (t-SNE) of their methylation profiles against a set of different mesenchymal neoplasms demonstrated a distinct methylation pattern.
Here, we describe pancreatic neoplasms with distinctive morphologic/immunophenotypic features and a distinct methylation pattern, along with a lack of abnormalities in any of key genetic drivers, supporting that these neoplasms represent a novel entity with an indolent clinical course. Given their mesenchymal transcriptomic features, we propose the designation of “sclerosing epithelioid mesenchymal neoplasm” of the pancreas.
INTRODUCTION
The pancreas gives rise to a range of epithelial neoplasms with ductal, acinar, and neuroendocrine differentiation, as well as the enigmatic solid pseudopapillary neoplasm that has no defined cell lineage. Additionally, most mesenchymal neoplasms that have been well characterized in other anatomic locations also rarely arise primarily within the pancreas. Over the last several years, we have encountered pancreatic tumors with distinctive histologic features, which do not conform to any of the known types of pancreatic epithelial neoplasms. In fact, the tumors also appear different from both epithelial and mesenchymal neoplasms described in other anatomical sites. They are characterized by well-demarcated nests of epithelioid and spindle cells in a densely sclerotic stroma, a histologic pattern we have termed “sclerosing epithelioid mesenchymal neoplasm” of the pancreas.
In the current study, we analyzed eight such cases and sought to define their diagnostic histologic features, clinical behavior, and molecular underpinning. After thoroughly assessing a wide range of defined epithelial and mesenchymal entities that could be considered in the differential diagnosis, we believe that, based on their striking and distinctive morphologic, immunohistochemical and molecular features, these cases represent a novel entity.
MATERIALS AND METHODS
Cases
Eight cases of pancreatic neoplasms with the distinctive morphologic features described above were identified among the consultation cases of the authors (D.S.K.−5 cases, V.A.−2 cases, G.Z.−1 case). The study was approved by the institutional review boards of the respective institutions. Available gross photographs and descriptions as well as all histologic sections were evaluated to characterize the spectrum of gross and histologic findings. Available medical records, including pathology reports, were reviewed to obtain clinical data including age, sex, presenting symptoms, and treatment. Outcome information was obtained when possible.
Immunohistochemical Analysis
Representative formalin-fixed paraffin-embedded tissue sections of each case, for which a paraffin block or unstained sections were available, were immunolabeled using the standard avidin-biotin peroxidase method. The antibodies used along with their sources, dilutions, and pretreatment conditions are listed in Supplementary Table 1.
Molecular Analyses
To detect possible recurrent somatic genetic alterations, whole-exome sequencing (n=4), RNA-sequencing (n=6), the Archer FusionPlex assay (n=5), methylation profiling using the Illumina MethylationEPIC (850k) platform (n=6), and targeted Sanger resequencing of the TERT promoter region (n=5) were performed (Supplementary Table 2). The choice of sequencing platforms for each case was based on tissue availability and nucleic acid quantity and quality.
Whole exome sequencing analysis
Microdissected tumor and normal DNA samples extracted from formalin-fixed paraffin-embedded tissue sections of four cases were subjected to whole exome sequencing analysis at Memorial Sloan Kettering Cancer Center Integrated Genomics Operation [with a median depth of 200x (range, 180x-216x) and 169x (range, 100x-195x) for tumor and matched normal DNA samples, respectively (Supplementary Table 3)]. Sequencing data were analyzed as previously described [14]. Somatic single nucleotide variants were identified using MuTect (v1.0) [5]; small insertions and deletions (indels) using Strelka (v2.0.15), VarScan2 (v2.3.7), Lancet (v1.0.0) and Scalpel (v0.5.3) [69]. Somatic copy number alterations and loss of heterozygosity (LOH) were obtained using FACETS [10] as described [2; 11]. The cancer cell fractions (CCFs) of all mutations were computed using ABSOLUTE (v1.0.6) [12] as described [1; 2]. A combination of mutation function predictors was employed to define the potential functional impact of each mutation as described [13], and mutational hotspots were assigned [14]. For the quantification of microsatellite instability based on the whole exome sequencing data, microsatellite instability sensor was employed as described [2; 15].
RNA-Sequencing and Archer FusionPlex Assay for Fusion Detection
Since the majority of the neoplasms in the differential diagnosis (See Discussion section) are driven by gene-fusions, both RNA-sequencing and Archer FusionPlex assay were performed to detect possible fusions in these cases. Total RNA was extracted from formalin-fixed paraffin-embedded tissue sections of seven cases. Samples from six cases of sufficient quality and quantity were subjected to paired-end RNA sequencing (2× 50bp cycles; Supplementary Table 2) at Memorial Sloan Kettering Cancer Center’s Integrated Genomics Operation as previously described [1]. Sequence read pairs for each case were aligned to the reference genome GRCh37 using STAR [16], Bowtie2 [17] and the bwa (v0.7.10) [3]. Aligned read pairs supporting fusion transcripts from each case were identified using INTEGRATE [18], STAR-Fusion, [19] MAPSplice [20], and FusionCatcher [21]. The oncogenic potential of putative fusion genes was annotated using OncoFuse [22]. To account for alignment artifacts, normal transcriptional variants and formalin-fixed paraffin-embedded tissue preservation artifacts, we excluded all fusion genes and read-throughs if present i) in a set of 297 normal samples from The Cancer Genome Atlas (TCGA) [23; 24], ii) in six normal formalin-fixed paraffin-embedded tissue samples subjected to RNA-sequencing or iii) in FusionFilter [19]. After filtering, fusion genes were considered as candidates if they had two or more chimeric junction reads and were in-frame. Out-of-frame fusions were also considered as candidates if a partner gene was a known cancer gene [25]. For gene expression analysis, the raw read counts were calculated using HTSeq, converted to normalized Reads Per Kilobase Million (RPKM) counts per gene, and the genes overexpressed per sample (Z-score >10) identified. The overexpressed genes were then subjected to single sample gene set enrichment analysis (GSEA) as described [26].
In addition, total RNA from five of these six cases was subjected to the Memorial Sloan Kettering Cancer Center-Solid Fusion assay (Supplementary Table 2), a custom-targeted, RNA-based panel that utilizes Archer Anchored Multiplex PCR (AMP) technology and next-generation sequencing to detect gene fusions [27; 28]. Unidirectional gene-specific primers were designed to target specific exons in 62 genes known to be involved in chromosomal rearrangements (Supplementary Table 4). Gene-specific primers, in combination with adaptor-specific primers, enriched for known and novel fusion transcripts. Final targeted amplicons were sequenced on an Illumina MiSeq. Data were analyzed using Archer Software (v4.0.10). This custom assay has been validated and approved for clinical use at Memorial Sloan Kettering Cancer Center by the New York State Department of Health Clinical Laboratory Evaluation Program.
Genome-wide methylation profiling using the Illumina MethylationEPIC (850k) array platform
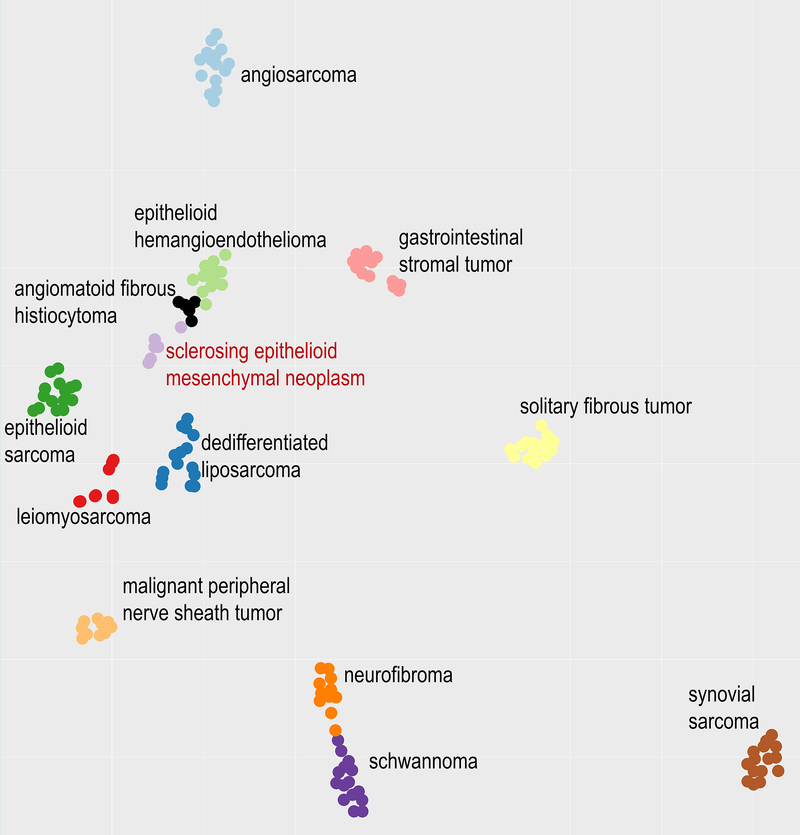
Genome-wide methylation profiles were obtained for six cases (Supplementary Table 2). For each case, 500 ng of DNA were subjected to bisulfite conversion, formalin-fixed paraffin-embedded tissue restoration, and processing on the Illumina MethylationEPIC array according to the manufacturer’s protocol. The array interrogates the methylation status of approximately 850,000 CpG sites across the genome. DNA-methylation data were normalized by performing background correction and dye bias correction (shifting of negative control probe mean intensity to zero and scaling of normalization control probe mean intensity to 20,000, respectively). Probes targeting sex chromosomes, probes containing multiple single nucleotide polymorphisms and those that could not be uniquely mapped were removed. Probes were excluded if the predecessor Illumina Infinium 450k BeadChip did not cover them, thereby making data generated by both 450k and EPIC comparable for subsequent analyses. In total, 438,370 probes were kept for analysis. Unsupervised analysis was performed against a reference set of different mesenchymal neoplasms using the t-distributed stochastic neighbor embedding (t-SNE) algorithm that has the most variable 20,000 non-XY CpG sites (by standard deviation). The set of mesenchymal neoplasms, which have been previously published in part [29; 30], included solitary fibrous tumors, gastrointestinal stromal tumors, angiomatoid fibrous histiocytomas, epithelioid hemangioendotheliomas, angiosarcomas, epithelioid sarcomas, synovial sarcomas, leiomyosarcomas, dedifferentiated liposarcomas, malignant peripheral nerve sheath tumors, neurofibromas and schwannomas.
TERT promoter mutation analysis
Recurrent hotspot mutations in the promoter of TERT, leading to upregulated telomerase expression and decreased cell death, have been documented in many cancer types [28]. Since the TERT promoter is typically not covered by whole exome sequencing analysis, Sanger sequencing of the TERT promoter region was performed in five cases. Primer sets that amplify the −124C>T and −146C>T hotspot sites of the TERT promoter were employed for Sanger sequencing as previously described [31]. Sequences of the forward and reverse strands were analyzed using MacVector software (MacVector, Inc). All analyses were performed in triplicate.
RESULTS
Clinicopathologic Features
The clinicopathologic findings of each case, including age, sex, location of tumor and other pertinent features are summarized in Table 1 and Supplementary Table 2.
Table 1.
Clinicopathologic Features of the Cases Analyzed
| n (%) | |
|---|---|
| Mean age (range), years | 43 (26–75) |
| Female:Male | 6:2 |
| Tumor location | |
| Head/Neck | 5 (62) |
| Body/Tail | 3 (38) |
| Median tumor size (range), cm | 1.8 (1.3–5.8) |
| Margin status | |
| R0 | 8 (100) |
| Lymph node status (n=6) | |
| N0 | 6 (100) |
| Follow-up | |
| No evidence of disease | 8 (100) |
| Median (range), months | 53 (8–94) |
Of the eight cases, six occurred in females and two in males. The mean age was 43 years (range, 26–75). One patient had a history of renal cell carcinoma diagnosed eight years before the pancreatic lesion. Five of the lesions were detected in the head/neck of the pancreas, one in the body and two in the tail. None of three cases with available data had elevated serum tumor markers (CA19.9 and CEA). All patients were treated surgically (six resections, two excisions); none received neoadjuvant or adjuvant chemotherapy. At a median follow-up of 53 months (range, 8–94), all patients were alive with no evidence of disease recurrence or a primary neoplasm at another anatomic site.
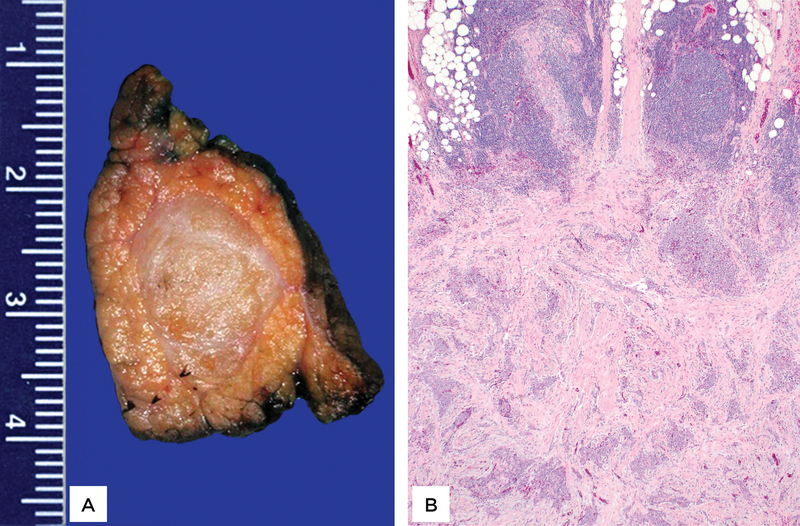
Macroscopically, six tumors were limited to the pancreas. Of the remaining two tumors, one invaded into the peripancreatic adipose tissue, the other into the duodenum. The median size of the tumors was 1.8 cm (range, 1.3–5.8). The tumors did not have a capsule but were well-circumscribed and solid; cut surfaces were described to be tan-white with firm or even sclerotic consistency (Figure 1A).
Figure 1:
The tumors were well-circumscribed and solid. Cut surfaces were tan-white with firm/ sclerotic consistency (A). Sections revealed epithelioid and spindled cell neoplasms associated with extensive fibrosis as well as dense lymphoid aggregates at the periphery (B).
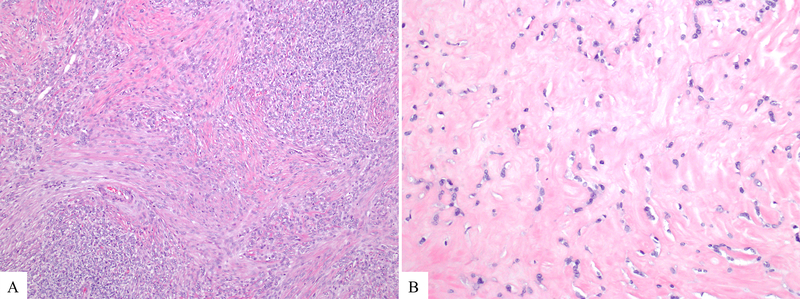
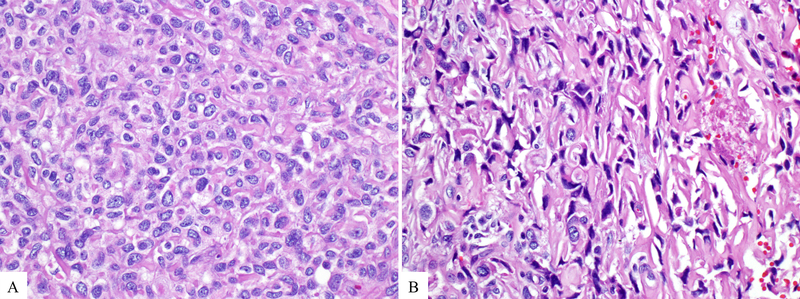
Microscopic sections revealed solid neoplasms associated with extensive fibrosis as well as dense lymphoid aggregates at the periphery (Figure 1B). The density of tumor cells varied significantly not only among the tumors and but also throughout each tumor, creating a geographic appearance of hypercellular and hypocellular areas (Figure 2). In some areas the tumor cells formed cellular sheets; other areas showed dense hyaline fibrosis with only rare nests of neoplastic cells. There were also foci in which linear arrays of tumor cells infiltrated around hyalinized collagen deposits (Congo-Red stain negative), imparting a slit-like pattern (Figure 3). No true epithelial structures such as glands, papillae or pseudopapillae were identified. Similarly, there was no hemorrhage, necrosis, clusters of foamy macrophages or eosinophilic globules. The tumor cells exhibited variable morphology as well. Most tumor cells were epithelioid to spindled and contained scant cytoplasm lacking glycogen or mucin (PAS stain negative) and round to oval nuclei with open chromatin, some with conspicuous nucleoli but no grooves or indentations. These cells intermingled with cells that displayed more irregular, hyperchromatic nuclei (Figure 3). Occasional (range, 0–3 per 10 HPF) mitotic figures were also noted. There was no lymphovascular invasion, but two cases revealed perineural invasion. At the periphery, the fibrosis and neoplastic cells entrapped the surrounding pancreatic parenchyma. In one case, the tumor extended into the peripancreatic adipose tissue and in another also into the duodenum. None of the six resections revealed lymph node metastasis.
Figure 2.
The density of tumor cells varied significantly among the tumors and throughout each tumor. Although in some areas the tumor cells formed cellular sheets (A); other areas showed dense hyaline fibrosis with only rare nests of neoplastic cells (B).
Figure 3:
The tumor cells exhibited variable morphology. Most cells were epithelioid and contained scant cytoplasm and round to oval nuclei with open chromatin, some with conspicuous nucleoli (A). Spindled cells displayed more irregular, hyperchromatic nuclei (B).
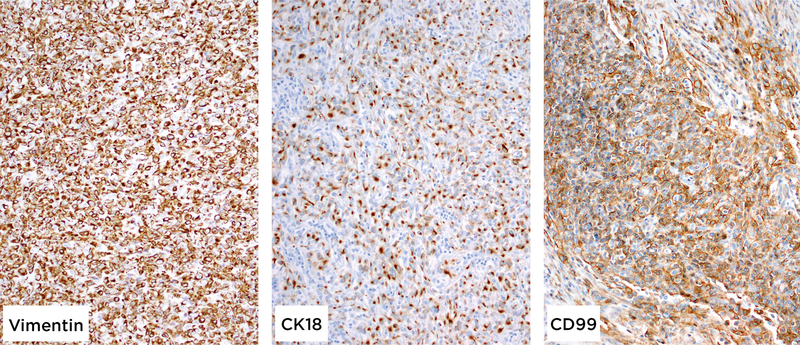
Immunohistochemically, all tumors were positive for vimentin (diffuse), CD99 (diffuse, membranous) and cytokeratin (AE1/3 and CK18, both patchy; Figure 4). One tumor (Case 3) expressed patchy and weak synaptophysin but all tumors were negative for chromogranin. S100 was also focally and weakly positive in Case 3. Markers of solid pseudopapillary neoplasm (abnormal nuclear β-catenin, progesterone receptor, and CD10 expression), acinar differentiation (trypsin, chymotrypsin), myogenic/rhabdoid differentiation (desmin, myogenin, INI1), vascular differentiation (ERG, CD31, and CD34), melanocytic differentiation (S100, HMB45, Melan-A), gastrointestinal stromal tumor (CD117, DOG1), as well as TTF1, HepPar-1, MUC4, BCL2, ALK, STAT6, CD21, CD35, and CD45 were negative in all tumors (Table 2).
Figure 4:
All tumors were positive for vimentin, CD99 and cytokeratins (AE1:AE3 and CK18, both patchy), while negative for markers of solid pseudopapillary neoplasm, acinar- myogenic/rhabdoid-, vascular-, melanocytic-, lymphocytic differentiation as well as MUC4.
Table 2.
Results of the Immunohistochemical Analysis
| Antibody | Positive (%) |
|---|---|
| AE1:AE3 & CK 18 | 8/8 (100) |
| CD99 | 8/8 (100) |
| Vimentin | 8/8 (100) |
| Chromogranin | 0/8 (0) |
| Synaptophysin | 1/8 (13), patchy and weak |
| PR | 0/8 (0) |
| CD10 | 0/8 (0) |
| β-Catenin (nuclear) | 0/8 (0) |
| Trypsin & Chymotrypsin | 0/8 (0) |
| TTF1 | 0/8 (0) |
| HepPar-1 | 0/8 (0) |
| MUC4 | 0/8 (0) |
| Desmin & Myogenin | 0/8 (0) |
| INI1 (BAF-47) | 3/3 (100), retained |
| CD117 & DOG1 | 0/8 (0) |
| S100 | 1/8 (13), very focal |
| HMB45 & Melan-A | 0/8 (0) |
| CD31 & ERG | 0/8 (0) |
| CD21 & CD35 | 0/8 (0) |
| CD45 | 0/8 (0) |
| BCL2 | 0/8 (0) |
| ALK | 0/8 (0) |
| CD34 | 0/8 (0) |
| STAT6 | 0/3 (0) |
Lastly, electron microscopy performed on one case (Case 1) showed non-specific findings including occasional desmosomes, perinuclear tonofilaments and abundant rough endoplasmic reticulum.
Molecular Features
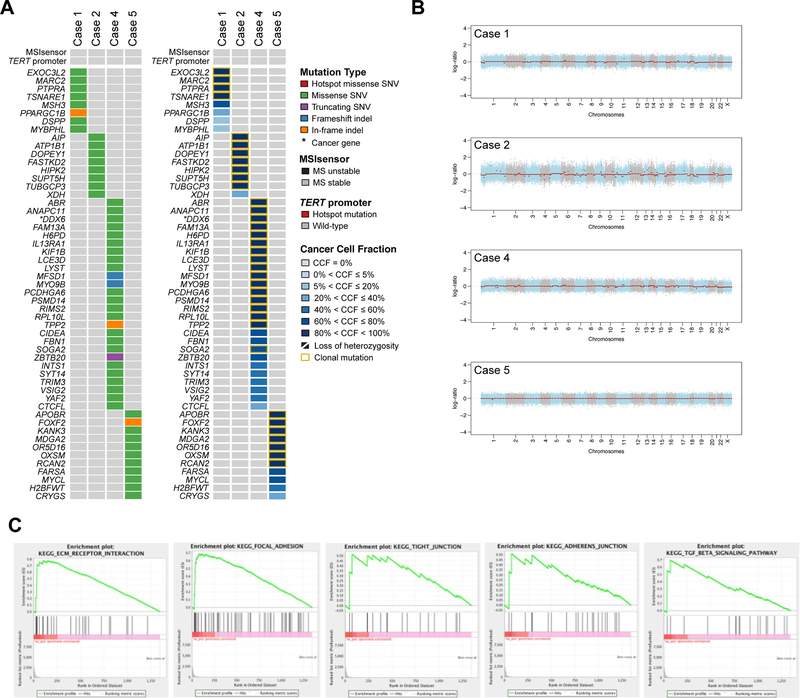
Whole exome sequencing analysis revealed a low mutation burden in these tumors, with a median of 15 (range 9–35) somatic mutations and a median of 9.5 (range 8–26) non-synonymous somatic mutations (Figure 5; Supplementary Table 5). No recurrent somatic mutation or recurrently mutated genes were identified (Figure 5A). Only one of the non-synonymous somatic mutations found in one case (Case 4) affected a known cancer gene (DDX6), and most of the mutations (89%) were predicted to be passenger mutations (Supplementary Table 5) [13]. Similarly, copy number analysis did not reveal any recurrent amplifications or homozygous deletions. In fact, the tumors displayed low levels of genetic instability with a paucity of gene copy number alterations (Figure 5B). All tumors tested were microsatellite stable as defined by microsatellite instability sensor (Figure 5A) [2; 15]. Finally, Sanger sequencing of the TERT promoter region did not identify any of the known TERT gene promoter hotspot positions (Figure 5A; Supplementary Figure 1).
Figure 5. Genomic and transcriptomic characterization of sclerosing epithelioid mesenchymal neoplasm of the pancreas.
A) Non-synonymous somatic mutations in sclerosing epithelioid mesenchymal neoplasm of the pancreas detected by whole-exome sequencing. The mutation types (left) and cancer cell fractions of each mutation (right) are shown, color-coded according to the legend. The phenobar (top) provides information about the presence of TERT promoter hotspot mutations and the microsatellite instability sensor score (microsatellite instability). Indel, small insertion and deletion; MS, microsatellite; SNV, single nucleotide variant. B) Chromosome plots of the four sclerosing epithelioid mesenchymal neoplasm of the pancreas subjected to whole-exome sequencing. Log2-ratios plotted on the y‐axis according to their genomic coordinates on the x‐axis. C) Results of the Gene Set Enrichment Analysis of genes overexpressed in six sclerosing epithelioid mesenchymal neoplasm of the pancreas subjected to RNA-sequencing. The pathways found to be enriched are shown in the title of each plot.
RNA-sequencing analysis did not reveal any recurrent or functionally recurrent likely pathogenic fusion gene (i.e., fusion genes that are in-frame, have intact functional domains and high driver probabilities; Supplementary Table 6). A customized Archer FusionPlex assay confirmed the absence of gene fusions involving 62 known target genes.
To assess the transcriptional program of the sclerosing epithelioid mesenchymal neoplasm of the pancreas, we subjected the transcriptomic data to Gene Set Enrichment Analysis (GSEA). This analysis revealed that pathways related to the extracellular matrix, focal adhesion, tight junctions, adherens junctions and TGFβ-signaling were enriched in these lesions (Figure 5C). Previous studies reported the activation/ enrichment of these pathways in cells of mesenchymal origin [32; 33].
Unsupervised analysis of the methylation profiles demonstrated a distinct methylation signature as evidenced by cluster separation and t-distributed stochastic neighbor embedding analysis. The sclerosing epithelioid mesenchymal neoplasms formed a unique cluster when compared to the most variable 20,000 non-XY CpG sites against the reference set of other mesenchymal neoplasms (Figure 6).
Figure 6.
Unsupervised analysis (t-SNE dimensionality reduction algorithm) of the most variable 20,000 CpG sites demonstrates a distinct clustering of these pancreatic neoplasms as compared to other mesenchymal neoplasms in the differential diagnosis.
DISCUSSION
This manuscript documents our experience with eight hitherto undescribed pancreatic neoplasms that predominantly occur in the head and neck region of the pancreas of middle-aged female patients and have an indolent, if not fully benign, clinical course. The neoplasms appear as solid, relatively well circumscribed tumors composed of epithelioid to spindled cells with moderately atypical nuclei and occasional mitotic figures. There is a variable degree of collagenization, including cellular fibrous bands as well as small ropey collagen deposits between strands of cells. Dense lymphoid aggregates are present at the tumor periphery. To investigate the histogenesis and facilitate the diagnosis of these unusual neoplasms, we studied the cases immunohistochemically using a panel of antibodies directed against a wide variety of proteins. The results showed that the neoplasms only express vimentin, CD99 and cytokeratin – non-specific findings that do not support a specific relationship to any of the defined types of primary epithelial neoplasms of the pancreas. In addition, to determine whether these neoplasms could be driven by a highly recurrent somatic genetic alteration, we performed extensive molecular analyses including whole exome sequencing, RNA-sequencing, and targeted sanger sequencing. These studies also revealed very low prevalence of alterations in common oncogenic signaling pathways as the neoplasms did not display any recurrent mutations, amplifications, deletions or likely pathogenic gene fusions.
Given the lack of an identifiable cell lineage, the differential diagnosis of these neoplasms is challenging. A wide variety of tumors with significant sclerotic and epithelioid components, including those not known to occur in the pancreas, were considered (Table 3). Immunohistochemical and molecular analyses are essential to fully explore the differential diagnosis, in this case largely by arguing against other diagnostic possibilities under consideration and thus supporting that sclerosing epithelioid mesenchymal neoplasm is a novel tumor entity in the pancreas.
TABLE 3:
Differential Diagnoses of Pancreatic Sclerosing Epithelioid Mesenchymal Neoplasm
| Sclerosing Epithelioid Mesenchymal Neoplasm | Solid Pseudopapillary Neoplasms | Sarcomatoid Carcinomas of the Pancreas | Sclerosing Epithelioid Fibrosarcomas | Myoepithelial Neoplasms of Soft Tissue | Angiomatoid Fibrous Histiocytoma | Solitary Fibrous Tumor | Perivascular Epithelioid Cell Neoplasm (PEComa) | Leiomyosarcoma | Gastrointestinal Stromal Tumor | |
|---|---|---|---|---|---|---|---|---|---|---|
| Median Age | 40s | 20–30s | 50–60s | 50s | 40s | 10s-20s | 50s | 30s-40s | 60s | 60s |
| Gender | Female predominance | Female predominance | Female predominance | No gender predilection | No gender predilection | Slight female predominance | Female predominance | Female predominance | No gender predilection | Slight male predominance |
| Microscopic Features | Geographic apparence of hypo and hypercellular areas with collagenous stroma ("ropey collagen"); epithelioid to spindle cells with scant cytoplasm,round-oval nuclei and open chromatin; mild to moderate nuclear atypia | Variety of patterns (solid, pseudopapillary, cystic, trabecular, etc); small to medium-sized and polygonal to elongated cells with clear to eosinophilic cytoplasm and ovoid nuclei (often have grooves) | Sheets of spindle cells, sometimes with pleomorphic giant cells and/or heterologous elements | Hypo or hypercellular with large areas of hyalinized fibrous stroma; small to medium sized, uniform epithelioid cells with clear to scant eosinophilic cytoplasm and bland nuclei with vesicular or finely stipple chromatin | Often infiltrative with heterogenous morphology and myxoid or hyalinized stroma; epithelioid, spindled or plasmacytoid cells arranged in small nests, strands or file | Often highly cellular with sheets, short fascicles or whorls of bland spindle to ovoid eosinophilic cells | Spindle cells in a collagenous stroma ("ropey collagen") with irregularly distributed thick walled "staghorn" vessels | Nested, trabecular, or sheet-like architecture, sometime with a dense collagenous stroma; epithelioid and/or spindle cells with granular eosinophilic to clear cytoplasm | Interlacing spindle cells with varying degree of pleomorphism and atypia, necrosis; mitotic figures are common | Spindle cell, epithelioid or mixed morphology; extracellular collagen globules ("skeinoid fibres") may be seen; variable mitotic activity |
| Immunohistochemical Features | Vimentin, CD99 and cytokeratins (weak) | PR and abnormal B-catenin | Vimentin and cytokeratin (weak) | MUC4 | S100, cytokeratins, EMA | Desmin, SMA, CD68, CD99, EMA | CD34, Bcl2, nuclear STAT6 | Smooth muscle (SMA, desmin - less common) and melanocytic (HMB-45, Melan-A) markers | HHF35, SMA and desmin | CD117, DOG1, CD34 |
| Molecular Features | No recurrent somatic mutations, amplifications or deletions in any known oncogenes or suppressor genes; distinct methylation pattern | Somatic activating mutation in exon 3 of CTNNB1 | Oncogenic mutations of KRAS, and loss of function mutations and/or deletions of the tumor suppressor genes TP53, SMAD4/DPC4, CDKN2A/p16 | EWSR1-CREB3L1 or FUS-CREB3L21, FUS-CREB3L2 fusions | EWSR1-PBX1, EWSR1-POU5F1, EWSR1-ZNF44, FUS-KLF17 or EWSR1-KLF17 fusions | EWSR1-CREB1, EWSR1-ATF1, or FUS-ATF1 fusions | NAB2-STAT6 fusion | TSC1/ TSC2 mutations, SFPQ-TFE3, RAD51B-OPHN1, RAD51B -BRAGB fusions | Complex genotype; TP53 mutations common | Mutually exclusive activating mutations of KIT or PDGFRA receptor tyrosine kinases |
Technically, because of the well demarcated nature and sclerotic consistency of the lesions, pancreatic hamartoma could be considered in differential diagnosis. However, microscopically, pancreatic hamartomas reveal a disordered arrangement of variably cystic ductal structures lined by cuboidal or flattened epithelium, surrounded by well-differentiated acini embedded in inflammatory or paucicellular stroma [34]. We believe the pathologic findings also essentially exclude all pancreatic epithelial neoplasms with well-defined cell lineages. However, the pancreatic tumor type of undetermined lineage, solid pseudopapillary neoplasm, could theoretically be considered. Although solid pseudopapillary neoplasms can be sclerotic, the typical pseudopapillary structures and characteristic nuclear features were not found in sclerosing epithelioid mesenchymal neoplasms; also, solid pseudopapillary neoplasms are typically either negative or only very focally positive for cytokeratin but consistently express PR and CD10. Furthermore, solid pseudopapillary neoplasms have activating gene mutations in CTNNB1 and consequently reveal diffuse nuclear β-catenin staining [35]; none of these findings were encountered in sclerosing epithelioid mesenchymal neoplasms.
Due to the mesenchymal morphology as well as vimentin and weak cytokeratin expression, the default differential diagnoses include sarcomatoid carcinoma and true sarcomas that express keratin, such as Ewing sarcoma/primitive neuroectodermal tumor, sclerosing epithelioid fibrosarcoma and myoepithelial neoplasms of soft tissue or salivary glands. Not surprisingly, one of the cases was originally diagnosed as “anaplastic carcinoma”. It should be noted, however, that most sarcomatoid carcinomas of the pancreas, which are considered variants of ductal adenocarcinoma, are larger infiltrative tumors and reveal considerable pleomorphism, necrosis and proliferative activity, as well as high mutation burden, with KRAS hotspot mutations and mutations in TP53, SMAD4, and CDKN2A, which typify most pancreatic carcinomas of ductal lineage. Also, sarcomatoid carcinomas are rapidly lethal malignancies [36 38]. None of these features were present in sclerosing epithelioid mesenchymal neoplasms. Likewise, lack of the characteristic EWSR1 gene rearrangements argues against the possibility of Ewing sarcoma/primitive neuroectodermal tumor, despite the finding of CD99 immunolabeling [39], as well as sclerosing epithelioid fibrosarcoma [40] or myoepithelial neoplasms of soft tissue [41].
In fact, given the morphologic features, including epithelioid cells with clear or eosinophilic cytoplasm embedded within a densely sclerotic stroma, and vimentin expression, sclerosing epithelioid fibrosarcoma was a proposed diagnosis for several of our cases, despite its occurrence being exceedingly rare in visceral organs [42; 43]. Unlike sclerosing epithelioid fibrosarcomas, however, the neoplasms analyzed here lacked S100 and MUC4 expression [44], the EWSR1-CREB3L1 or FUS-CREB3L2 fusion genes, or any other recurrent expressed chimeric genes [40], based on RNA-sequencing and the Archer FusionPlex assay.
The widespread but weak membranous expression of CD99, along with some of the histologic features, raised the possibility of an angiomatoid fibrous histiocytoma. However, lack of cystic, hemorrhagic spaces, desmin expression and, EWSR1-CREB1, EWSR1-ATF1, or FUS-ATF1 fusions, combined with cytokeratin labeling found in our cases, would be unusual for this entity [45]. We also performed BCL2, CD34, ALK and STAT6 immunohistochemistry to explore the possibility of an intrapancreatic solitary fibrous tumor [46–49] or inflammatory myofibroblastic tumor [48; 50; 51], but these studies were negative. Similarly, lack of S100 labeling makes the possibility of a myoepithelial neoplasms of soft tissue or salivary glands, a melanocytic lesion, or a nerve sheath tumor unlikely [41; 52]. Perivascular epithelioid cell neoplasms would reveal smooth muscle (SMA, desmin) and melanocytic (HMB-45, Melan-A) markers as well as TSC1/TSC2 mutations [53; 54]. Absence of CD117, DOG1, CD34 expression and KIT or PDGFRA receptor tyrosine kinase mutations argues against gastrointestinal stromal tumor [55]. Hematolymphoid neoplasms such as Langerhans cell histiocytosis [56–59] and follicular dendritic cell sarcoma would be positive for CD1a, CD21 or CD35 [60; 61].
Thus, studies failed to point to a specific cell lineage and we have been unable to equate these neoplasms with any other previously-defined entity. It should be noted, however, that Gene Set Enrichment Analysis based on RNA-sequencing provided evidence suggesting that the neoplasms analyzed here either display a mesenchymal origin or have adopted mesenchymal differentiation. Interestingly, the neoplasms also displayed a distinct methylation signature forming a unique cluster when compared against the reference set of other mesenchymal neoplasms (Figure 6).
This study has several limitations. First, given the rarity of this entity, our sample size is small. However, we and others have demonstrated that with 4 samples, we have 80% statistical power to detect a pathognomonic fusion gene or somatic mutation, if this pathognomonic mutation is present in >70% of cases [62; 63]. Second, we have only surveyed the protein coding genes by whole exome sequencing analysis and the RNA-sequencing analysis was performed with RNA extracted from formalin-fixed paraffin-embedded samples; it is possible that whole-genome sequencing may result in the identification of a pathognomonic genetic alteration affecting non-coding elements. Further studies with optimally accrued samples of this entity are warranted.
In summary, although the histologic features are distinctive and bring to mind a number of specific diagnostic considerations, a wide array of immunohistochemical studies have failed to align these tumors with well-recognized epithelial or mesenchymal entities, and molecular studies to date have not disclosed specific genetic alterations except activation/enrichment of certain pathways including extracellular matrix, tight junctions, adherens junctions and TGFβ-signaling by Gene Set Enrichment Analysis, pathways that typify cells of mesenchymal origin [32; 33]. Methylation profiling also demonstrates a distinct methylation signature. Therefore, this entity appears to be unique to the pancreas and has not been previously described or named. We propose the descriptive term “Sclerosing Epithelioid Mesenchymal Neoplasm” given the inability to specifically define cell lineage. The follow-up information from these cases failed to demonstrate malignant behavior; however, since the number of cases we present was small and the follow-up was limited, it is premature to assure fully benign biology. Awareness of the occurrence of sclerosing epithelioid mesenchymal neoplasm in the pancreas may help to identify additional cases leading to more information on the clinicopathologic and prognostic features of this unusual tumor.
Supplementary Material
Supplementary Table 1: List of antibodies.
Supplementary Table 2: Summary of cases included in this study.
Supplementary Table 3: Whole-exome sequencing statistics.
Supplementary Table 4: List of the genes in Memorial Sloan Kettering Cancer Center-Solid Fusion Panel.
Supplementary Table 5: Non-synonymous somatic mutations identified in sclerosing epithelioid mesenchymal neoplasm of the pancreas using whole-exome sequencing.
Supplementary Table 6: Fusion candidate genes and/or readthroughs identified in sclerosing epithelioid mesenchymal neoplasm of the pancreas by RNA-sequencing analysis.
Supplementary Figure 1: TERT promoter mutation assessment using Sanger sequencing. Representative Sanger sequence electropherograms of the TERT promoter region encompassing the −124C>T and −146C>T hotspot sites in both tumor and matched normal samples.
ACKNOWLEDGMENTS
We are indebted to Drs. Günter Klöppel, Ralph H. Hruban, Meera Hameed, Christopher D. Fletcher, and Elizabeth A. Montgomery for their assessment of selected cases and concurrence regarding the novelty of this entity.
The authors also thank Ms. Marina Asher for her assistance with the immunohistochemical stains.
FUNDING
This work has been supported in part by the NCI/NIH Cancer Center Support Grant P30 CA008748 and by the German Cancer Aid Grant 70112499. J.S. Reis-Filho is supported by the Breast Cancer Research Foundation.
REFERENCES
- 1.Pareja F, Lee JY, Brown DN, Piscuoglio S, Gularte-Merida R, Selenica P, et al. The genomic landscape of mucinous breast cancer. J Natl Cancer Inst 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashley CW, Da Cruz Paula A, Kumar R, Mandelker D, Pei X, Riaz N, et al. Analysis of mutational signatures in primary and metastatic endometrial cancer reveals distinct patterns of DNA repair defects and shifts during tumor progression. Gynecol Oncol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mckenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013; 31:213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koboldt DC, Zhang Q, Larson DE, Shen D, Mclellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012; 22:568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 2012; 28:1811–7. [DOI] [PubMed] [Google Scholar]
- 8.Narzisi G, O’rawe JA, Iossifov I, Fang H, Lee YH, Wang Z, et al. Accurate de novo and transmitted indel detection in exome-capture data using microassembly. Nat Methods 2014; 11:1033–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Narzisi G, Corvelo A, Arora K, Bergmann EA, Shah M, Musunuri R, et al. Genome-wide somatic variant calling using localized colored de Bruijn graphs. Commun Biol 2018; 1:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 2016; 44:e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weigelt B, Bi R, Kumar R, Blecua P, Mandelker DL, Geyer FC, et al. The landscape of somatic genetic alterations in breast cancers from ATM germline mutation carriers. J Natl Cancer Inst 2018; 110:1030–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carter SL, Cibulskis K, Helman E, Mckenna A, Shen H, Zack T, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 2012; 30:413–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martelotto LG, Ng CK, De Filippo MR, Zhang Y, Piscuoglio S, Lim RS, et al. Benchmarking mutation effect prediction algorithms using functionally validated cancer-related missense mutations. Genome Biol 2014; 15:484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang MT, Asthana S, Gao SP, Lee BH, Chapman JS, Kandoth C, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol 2016; 34:155–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niu B, Ye K, Zhang Q, Lu C, Xie M, Mclellan MD, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics 2014; 30:1015–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012; 9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, White NM, Schmidt HK, Fulton RS, Tomlinson C, Warren WC, et al. INTEGRATE: gene fusion discovery using whole genome and transcriptome data. Genome Res 2016; 26:108–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haas B, Dobin A, Stransky N, Li B, Yang X, Tickle T, et al. STAR-Fusion: fast and accurate fusion transcript detection from RNA-Seq. 2017; Available from: https://www.biorxiv.org/content/early/2017/03/24/120295.
- 20.Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res 2010; 38:e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicorici D, Satalan M, Edgren H, Kangaspeska S, Murumagi A, Kallioniemi O, et al. FusionCatcher - a tool for finding somatic fusion genes in paired-end RNA-sequencing data. 2014; Available from: https://www.biorxiv.org/content/early/2014/11/19/011650.
- 22.Shugay M, Ortiz De Mendibil I, Vizmanos JL, Novo FJ. Oncofuse: a computational framework for the prediction of the oncogenic potential of gene fusions. Bioinformatics 2013; 29:2539–46. [DOI] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012; 490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 2018; 173:291–304 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, et al. Comprehensive characterization of cancer driver genes and mutations. Cell 2018; 174:1034–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005; 102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng Z, Liebers M, Zhelyazkova B, Cao Y, Panditi D, Lynch KD, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med 2014; 20:1479–84. [DOI] [PubMed] [Google Scholar]
- 28.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017; 23:703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koelsche C, Hartmann W, Schrimpf D, Stichel D, Jabar S, Ranft A, et al. Array-based DNA-methylation profiling in sarcomas with small blue round cell histology provides valuable diagnostic information. Mod Pathol 2018; 31:1246–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koelsche C, Stichel D, Griewank KG, Schrimpf D, Reuss DE, Bewerunge-Hudler M, et al. Genome-wide methylation profiling and copy number analysis in atypical fibroxanthomas and pleomorphic dermal sarcomas indicate a similar molecular phenotype. Clin Sarcoma Res 2019; 9:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piscuoglio S, Ng CK, Murray M, Burke KA, Edelweiss M, Geyer FC, et al. Massively parallel sequencing of phyllodes tumours of the breast reveals actionable mutations, and TERT promoter hotspot mutations and TERT gene amplification as likely drivers of progression. J Pathol 2016; 238:508–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silva WA Jr., Covas DT, Panepucci RA, Proto-Siqueira R, Siufi JL, Zanette DL, et al. The profile of gene expression of human marrow mesenchymal stem cells. Stem Cells 2003; 21:661–9. [DOI] [PubMed] [Google Scholar]
- 33.Groger CJ, Grubinger M, Waldhor T, Vierlinger K, Mikulits W. Meta-analysis of gene expression signatures defining the epithelial to mesenchymal transition during cancer progression. PLoS One 2012; 7:e51136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pauser U, Kosmahl M, Kruslin B, Klimstra DS, Kloppel G. Pancreatic solid and cystic hamartoma in adults: characterization of a new tumorous lesion. Am J Surg Pathol 2005; 29:797–800. [DOI] [PubMed] [Google Scholar]
- 35.Terris B, Cavard C. Diagnosis and molecular aspects of solid-pseudopapillary neoplasms of the pancreas. Semin Diagn Pathol 2014; 31:484–90. [DOI] [PubMed] [Google Scholar]
- 36.Hruban R, Pitman MB, Klimstra DS. Tumors of the Pancreas. AFIP Atlas of Tumor Pathology. Washington, DC: American Registry of Pathology, 2007. [Google Scholar]
- 37.Bosman FT, Carneiro F, Hruban RH, Theise ND. WHO Classification of Tumors of the Digestive System. Lyon: WHO Press, 2010. [Google Scholar]
- 38.Thompson LDR, Basturk O, Adsay V. Pancreas In: Mills SE, Editor. Sternberg’s Diagnostic Surgical Pathology. Wolters Kluwer Health, 2015. [Google Scholar]
- 39.Ladanyi M, Lewis R, Garin-Chesa P, Rettig WJ, Huvos AG, Healey JH, et al. EWS rearrangement in Ewing’s sarcoma and peripheral neuroectodermal tumor. Molecular detection and correlation with cytogenetic analysis and MIC2 expression. Diagn Mol Pathol 1993; 2:141–6. [PubMed] [Google Scholar]
- 40.Arbajian E, Puls F, Antonescu CR, Amary F, Sciot R, Debiec-Rychter M, et al. In-depth genetic analysis of sclerosing epithelioid fibrosarcoma reveals recurrent genomic alterations and potential treatment targets. Clin Cancer Res 2017; 23:7426–7434. [DOI] [PubMed] [Google Scholar]
- 41.Jo VY, Fletcher CD. Myoepithelial neoplasms of soft tissue: an updated review of the clinicopathologic, immunophenotypic, and genetic features. Head Neck Pathol 2015; 9:32–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frattini JC, Sosa JA, Carmack S, Robert ME. Sclerosing epithelioid fibrosarcoma of the cecum: a radiation-associated tumor in a previously unreported site. Arch Pathol Lab Med 2007; 131:1825–8. [DOI] [PubMed] [Google Scholar]
- 43.Tomimaru Y, Nagano H, Marubashi S, Kobayashi S, Eguchi H, Takeda Y, et al. Sclerosing epithelioid fibrosarcoma of the liver infiltrating the inferior vena cava. World J Gastroenterol 2009; 15:4204–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doyle LA, Wang WL, Dal Cin P, Lopez-Terrada D, Mertens F, Lazar AJ, et al. MUC4 is a sensitive and extremely useful marker for sclerosing epithelioid fibrosarcoma: association with FUS gene rearrangement. Am J Surg Pathol 2012; 36:1444–51. [DOI] [PubMed] [Google Scholar]
- 45.Thway K, Fisher C. Angiomatoid fibrous histiocytoma: the current status of pathology and genetics. Arch Pathol Lab Med 2015; 139:674–82. [DOI] [PubMed] [Google Scholar]
- 46.Luttges J, Mentzel T, Hubner G, Kloppel G. Solitary fibrous tumour of the pancreas: a new member of the small group of mesenchymal pancreatic tumours. Virchows Arch 1999; 435:37–42. [DOI] [PubMed] [Google Scholar]
- 47.Chetty R, Jain R, Serra S. Solitary fibrous tumor of the pancreas. Ann Diagn Pathol 2009; 13:339–43. [DOI] [PubMed] [Google Scholar]
- 48.Kim JY, Song JS, Park H, Byun JH, Song KB, Kim KP, et al. Primary mesenchymal tumors of the pancreas: single-center experience over 16 years. Pancreas 2014; 43:959–68. [DOI] [PubMed] [Google Scholar]
- 49.Huang SC, Li CF, Kao YC, Chuang IC, Tai HC, Tsai JW, et al. The clinicopathological significance of NAB2-STAT6 gene fusions in 52 cases of intrathoracic solitary fibrous tumors. Cancer Med 2016; 5:159–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamamoto H, Yamaguchi H, Aishima S, Oda Y, Kohashi K, Oshiro Y, et al. Inflammatory myofibroblastic tumor versus IgG4-related sclerosing disease and inflammatory pseudotumor: a comparative clinicopathologic study. Am J Surg Pathol 2009; 33:1330–40. [DOI] [PubMed] [Google Scholar]
- 51.Basturk O, Askan G. Benign tumors and tumor-like lesions of the pancreas. Surg Pathol Clin 2016; 9:619–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kane SV, Bagwan IN. Myoepithelial carcinoma of the salivary glands: a clinicopathologic study of 51 cases in a tertiary cancer center. Arch Otolaryngol Head Neck Surg 2010; 136:702–12. [DOI] [PubMed] [Google Scholar]
- 53.Zamboni G, Pea M, Martignoni G, Zancanaro C, Faccioli G, Gilioli E, et al. Clear cell “sugar” tumor of the pancreas. A novel member of the family of lesions characterized by the presence of perivascular epithelioid cells. Am J Surg Pathol 1996; 20:722–30. [DOI] [PubMed] [Google Scholar]
- 54.Pan CC, Jong YJ, Chai CY, Huang SH, Chen YJ. Comparative genomic hybridization study of perivascular epithelioid cell tumor: molecular genetic evidence of perivascular epithelioid cell tumor as a distinctive neoplasm. Hum Pathol 2006; 37:606–12. [DOI] [PubMed] [Google Scholar]
- 55.Schaefer IM, Marino-Enriquez A, Fletcher JA. What is new in gastrointestinal stromal tumor? Adv Anat Pathol 2017; 24:259–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goyal R, Das A, Nijhawan R, Bansal D, Marwaha RK. Langerhans cell histiocytosis infiltration into pancreas and kidney. Pediatr Blood Cancer 2007; 15:748–50. [DOI] [PubMed] [Google Scholar]
- 57.El Demellawy D, Young JL, De Nanassy J, Chernetsova E, Nasr A. Langerhans cell histiocytosis: a comprehensive review. Pathology 2015; 47:294–301. [DOI] [PubMed] [Google Scholar]
- 58.Rollins BJ. Genomic alterations in langerhans cell histiocytosis. Hematol Oncol Clin North Am 2015; 29:839–51. [DOI] [PubMed] [Google Scholar]
- 59.Ozkaya N, Dogan A, Abdel-Wahab O. Identification and targeting of kinase alterations in histiocytic neoplasms. Hematol Oncol Clin North Am 2017; 31:705–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saygin C, Uzunaslan D, Ozguroglu M, Senocak M, Tuzuner N. Dendritic cell sarcoma: a pooled analysis including 462 cases with presentation of our case series. Crit Rev Oncol Hematol 2013; 88:253–71. [DOI] [PubMed] [Google Scholar]
- 61.Griffin GK, Sholl LM, Lindeman NI, Fletcher CD, Hornick JL. Targeted genomic sequencing of follicular dendritic cell sarcoma reveals recurrent alterations in NF-kappaB regulatory genes. Mod Pathol 2016; 29:67–74. [DOI] [PubMed] [Google Scholar]
- 62.Weinreb I, Piscuoglio S, Martelotto LG, Waggott D, Ng CK, Perez-Ordonez B, et al. Hotspot activating PRKD1 somatic mutations in polymorphous low-grade adenocarcinomas of the salivary glands. Nat Genet 2014; 46:1166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pareja F, Brandes AH, Basili T, Selenica P, Geyer FC, Fan D, et al. Loss-of-function mutations in ATP6AP1 and ATP6AP2 in granular cell tumors. Nat Commun 2018; 9:3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: List of antibodies.
Supplementary Table 2: Summary of cases included in this study.
Supplementary Table 3: Whole-exome sequencing statistics.
Supplementary Table 4: List of the genes in Memorial Sloan Kettering Cancer Center-Solid Fusion Panel.
Supplementary Table 5: Non-synonymous somatic mutations identified in sclerosing epithelioid mesenchymal neoplasm of the pancreas using whole-exome sequencing.
Supplementary Table 6: Fusion candidate genes and/or readthroughs identified in sclerosing epithelioid mesenchymal neoplasm of the pancreas by RNA-sequencing analysis.
Supplementary Figure 1: TERT promoter mutation assessment using Sanger sequencing. Representative Sanger sequence electropherograms of the TERT promoter region encompassing the −124C>T and −146C>T hotspot sites in both tumor and matched normal samples.