Abstract
Outer membrane porin F (OprF) is a major structural membrane protein of Pseudomonas aeruginosa, a recognised human opportunistic pathogen which is correlated with severe hospital-acquired infections. This study investigating a multiphenotypic approach, based on the comparative study of a wild type strain of P. aeruginosa, its isogenic OprF mutant. Both P. aeruginosa PAO1 and OprF mutant strains were grown in same condition and cultures were subjected to further analysis by SDS PAGE, pyocyanin production and biofilm formation that was analyse using scanning electron microscopy. Based on biofilm formation essay and pyocyanin production, the study showed that OprF plays a dynamic role in P. aeruginosa virulence. The absence of OprF results in slow growth rate corresponded to elongated lag phase and reduced biofilm production also a significance reduction in the production of the quorum-sensing-dependent virulence factors pyocyanin. Accordingly, in the OprF mutant scanning electron microscope “SEM” images showed impaired cellular niche and detached cells when compared to regular attached P. aeruginosa wild type cells in the niche. Taken together, this study shows the contribution of OprF in P. aeruginosa virulence, at least partly through impairment of biofilm, cell to cell attachment in niche and pyocyanin production. This study show a vital link between OprF and virulence factor production, providing novel insights for its role in pathogenicity and future could provide the basis for the development of novel drug targets for antibiotics and vaccines.
Keywords: Pseudomonas aeruginosa, OprF mutant, Biofilm production, Pyocyanin, SEM-biofilm
1. Introduction
Pseudomonas aeruginosa is an ubiquitous opportunistic Gram-negative pathogenic bacteria that has a large genome (6.3 megabytes, Mb), which confers environmental versatility (Stover et al., 2000). P. aeruginosa is adapted to cause diseases in different hosts and additionally shows antibiotic resistance by variety of virulence factors including motility, biofilm formation and production of pyocyanin and exo-proteins such as protease, phospholipases and elastase that allowed its tissue invasion and systemic dissemination (Gómez and Prince, 2007). P. aeruginosa is able to grow under a wide range of oxygen tensions. A well-known example of anaerobic conditions developing under infection occurs when the bacterium colonizes the lungs of cystic fibrosis (CF) patients (Worlitzsch et al., 2002, Govan and Deretic, 1996, Hassett et al., 2002). In addition to infecting the CF lung, anaerobic or microaerobic conditions have been found in wound injuries, otitis, and in infections within the upper respiratory tract (Portier et al., 1999, Rumbaugh et al., 1999, Brook, 2002, Carenfelt and Lundberg, 1977, Lyczak et al., 2000). In addition to naturally anaerobic environments, the bacterium often changes its environment directly to bring about anaerobic conditions. One example of this occurring is the generation of biofilm structures. Biofilms are complex bacterial communities formed on attachment to surfaces to allow increased resistance to various environmental insults (Costerton et al., 1999, O'Toole and Kolter, 1998, Xu et al., 1998). It appears that anaerobiosis is the preferred environment for biofilm bound P. aeruginosa as well as having other regulatory mechanisms (Yoon et al., 2002, Filiatrault et al., 2006, Eschbach et al., 2004, Sawers, 1991, Kuchma et al., 2005). Bacteria living in biofilms can be up to 1000 times more resistant to antibacterial compounds than planktonic bacteria because of the biofilm nature and structural composition (Nickel et al., 1985). Biofilm production is a major virulence that has been linked to quorum sensing (QS) system (De Kievit, 2009) were other studies relates P. aeruginosa virulence and QS and to some structural component as Outer membrane proteins (OM) proteins (Fito-Boncompte et al., 2011). OprF is among the very few general porins of P. aeruginosa exposed on the external surface of the bacteria, functioning as a host-pathogen interactions facilitator and involved in various virulence abilities (Fito-Boncompte et al., 2011). The aim of this study is to understand the association of OprF membrane protein in biofilm formation and other virulence factors. However, this could provide the basis for development of novel drug targets for antibiotics and vaccines.
2. Methods
2.1. Bacterial strains, plasmids and media conditions
The strains and plasmids used in this project are P. aeruginosa PAO1 wild type, and P. aeruginosa OprF mutant were obtained from the Pseudomonas aeruginosa PA01 Transposon Mutant Library (Manoil Lab, University of Washington). Strains were cultured routinely at 37 °C for 24 h in Luria Bertani (LB) broth or on LB agar plates or cetrimide agar, (Thermo Scientific Oxoid, UK). LB with tetracycline 50 µg/ml was used to selectively culture the mutant strain.
2.2. Growth curve analysis
From an overnight culture of PAO1 and OprF mutant strains, were inoculated to an OD600, using spectrophotometer (biochrom libra s22), of 0.01 in 250 ml conical flask containing 25 ml of LB broth. OD600 was measured each hour for 24 h.
2.3. SDS PAGE
Total protein was extracted from P. aeruginosa PAO1 and OprF mutant strains, this was achieved by growing the strains in LB broth and the pellet was centrifuged then resuspended in 20 µL of 2 × SDS loading buffer, it was incubated at 50 °C for 5 min before being sonicated for a few seconds, 10 µL of each sample and protein standards (Invitrogen, USA), were electrophoresed on 3% polyacrylamide SDS stacking gel and 12% polyacrylamide SDS resolving gel at 40 mA per gel (Bio-Rad, USA) in SDS running buffer. Proteins were stained with coomassie brilliant blue. The size of the protein bands was determined by comparison with protein standard marker (Invitrogen, USA).
2.4. Biofilm formation assays
Biofilm formation assay were performed in 96- wells polystyrene microtiter plates as previously described with some modifications (Fletcher, 1977). Strains were grown overnight in LB broth at 37 °C. Next day the cultures were diluted in LB to 107 cfu/ml and dispensed in 96-well microtiter plate (Thomas science). The microtiter plates were incubated at 37 °C for 24 h. After that the cell suspension was removed and the plates were washed twice with 0.9% NaCl and inverted to dry at room temperature for 1 h. Following this 150 µL of crystal violet solution (CV; Prolab Diagnostics) was added to the wells and was allowed to stain for 15 min. After staining, CV was removed and the wells were washed 3 times with 0.9% NaCl. The bound CV was then solubilized by adding 200 µL of ethanol-acetone (80:20 v/v). The absorbance of CV was read at 595 nm on the microplate reader (BioTek Instruments, Winooski, VT).
2.5. Scanning electron microscopy (SEM)
To visualize pseudomonas strains biofilm formation, bacteria were grown overnight in LB at 37 °C. Next day the cultures were diluted in LB to 107 cfu/ml. Sterile coverslips (polyvinyl; Fisher Scientific) were placed in each well of a 6-well plate (Thomas science) and then 2 ml of diluted culture plus 2 ml LB medium was added. Biofilms were then grown on the coverslips at 37 °C for 24 h. biofilm samples were fixed with using 3% glutaraldehyde in phosphate buffer pH 7.2 for 24 h. after three washes with phosphate buffer, samples were postfix with 1% osmium tetroxide (in H2O) for 1 hr then the samples were applied in to an ethanol dehydration series of 50, 60, 70, 80, 90, and 2 × 100% (v/v) ethanol, for 5 min at each concentration (Marrie and Costerton, 1984). All samples were then dried for 1 day and sputter-coated with a palladium-gold thin film. The produced biofilm were viewed with a SEM/EDS system (FEI Quanta 400FEG ESEM/EDAX Genesis X4M, FEI Company, USA) in high-vacuum mode at 20 kV.
2.6. Pyocyanin production
Pyocyanin produced by P. aeruginosa was determined as previously described. Briefly, cultures were inoculated with shaking, 200 rpm using (Cheimika, Italy). Starting with OD600 of 0.02 were grown in LB medium at 37 °C, and a humidity of 75% for 16 h. For pyocyanin determination, cultures were extracted with chloroform and re-extracted with 0.2 M HCl. The OD520 was determined using microplate reader and normalized to cell growth measured as OD600. For each sample, cultivation and extraction were performed at least in triplicate and 0.2 M HCl used as a control.
3. Results and discussion
3.1. Growth rate of P. aeruginosa PAO1 wild type, and P. aeruginosa PAO1 OprF mutant
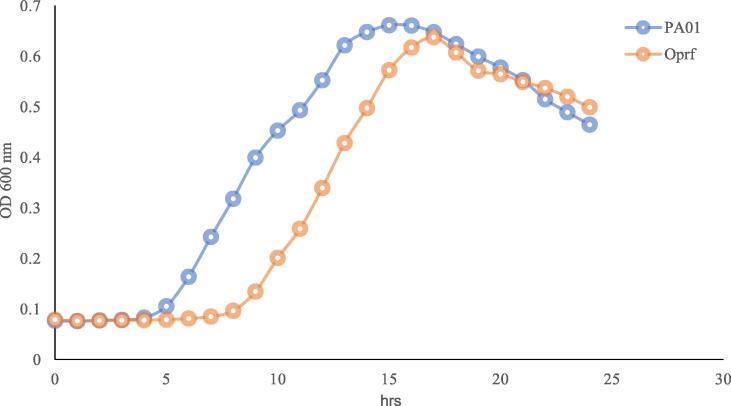
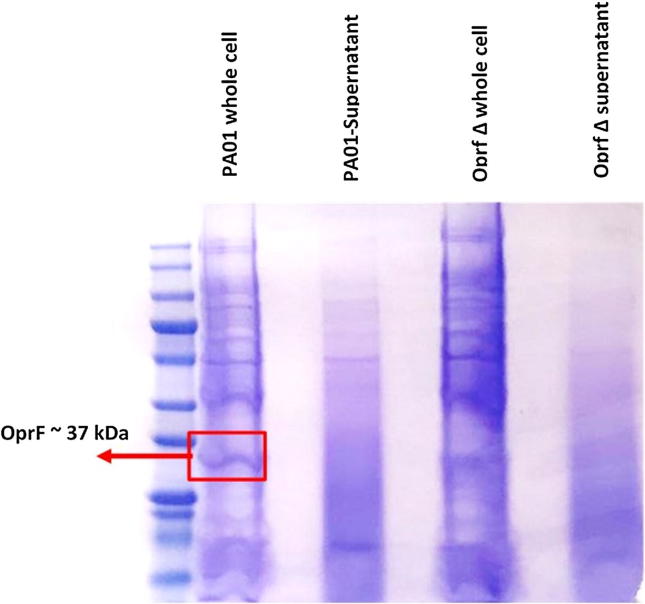
To recognize the effect of mutating OprF gene on the growth rate of P. aeruginosa, the growth of the wild type and OprF deficient strain were compared. Observation of colonies that grown for the same period of time in LB broth, both strains had the same viability by the end of 24 h; time of colonization, but OprF mutant strain was significantly slower in growth rate of colonizing demonstrated by elongated lag phase and slower generation time compared to the wild type P. aeruginosa as shown in Fig. 1. The protein content of both strains were analyzed using SDS page in which the wild type P. aeruginosa showed the presence of OprF protein size around 37 kDa which are missing in the OprF mutant strains as shown in Fig. 2.
Fig. 1.
Comparing bacterial growth of P. aeruginosa PAO1 and OprF mutant strain on LB Broth. It is demonstrated that generation time of the OprF deficient strain was slower and the elongated lag time of inoculation, than the P. aeruginosa wild strain, however, decline phase showed similar pattern for both strains.
Fig. 2.
SDS PAGE Coomassie blue stained comparing P. aeruginosa PAO1 and OprF mutant strain for OprF detection. OprF porin protein size 37 kDa shown in P. aeruginosa wild strain whole cell content and missing in the OprF mutant strain.
3.2. Biofilm formation assay
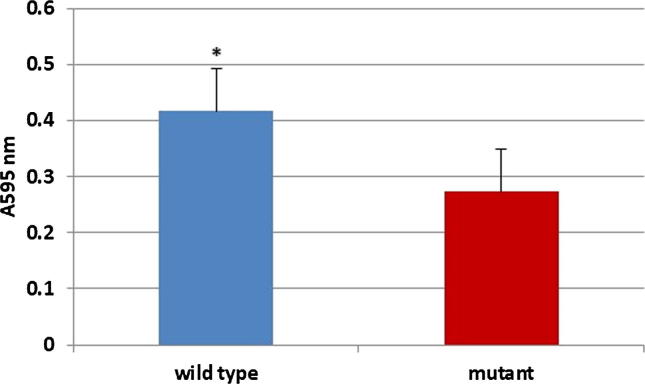
In this study we investigate the association between OprF protein and the biofilm production comparing P. aeruginosa PAO1 wild type, and P. aeruginosa PAO1 mutant, deficient in OprF gene. Fig. 3 showed the significant decline in biofilm production (with p ≤ 0.01) between P. aeruginosa PAO1 and OprF mutant indicating the important role of OprF in the biofilm virulence.
Fig. 3.
Biofilm detection assay of P. aeruginosa shows significant decline in biofilm formation from P. aeruginosa PAO1 wild type strain and the OprF mutant strain. The error bars represent the mean ± standard error. (n = 6, *P < 0.01).
3.3. SEM of P. aeruginosa biofilm
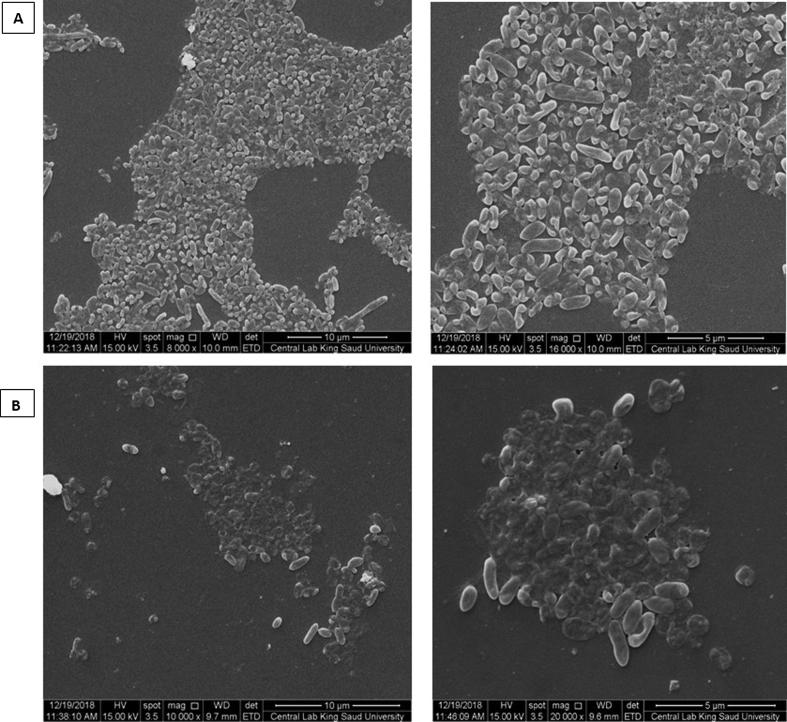
A scanning electron microscope used to visualize both strains biofilm and cells morphology in the niche. With both strains as shown in Fig. 4, biofilm was recognized at 24 h of incubation on glass coverslips as mentioned previously (Marrie and Costerton, 1984). SEM images displayed regular morphology and intense distribution of P. aeruginosa wild type cells in the niche and attached to biofilm, whereas, in OprF mutant the cells were detached in an irregular niche. These images supported the results of biofilm impairment by the absence of OprF.
Fig. 4.
SEM images at different magnification for (A) P. aeruginosa PAO1 wild type, and (B) P. aeruginosa OprF mutant detecting biofilm and cells attachment.
3.4. Pyocyanin production
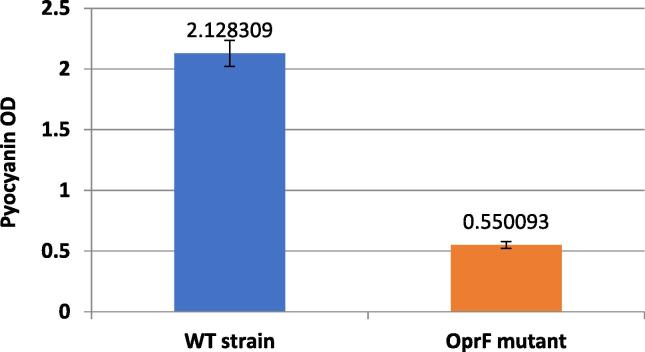
Major virulence factor of P. aeruginosa is the quorum-sensing-dependent virulence factors pyocyanin production. As it was noticed the effect of OprF on culture growth rate and biofilm formation, it was still not yet established if inactivation of the OprF have a direct effect on other virulence factors production. Bacterial culture strains were also checked for the effect of OprF on major virulence factors; exclusively studied the pyocyanin production as a marker of virulence. Measurement of pyocyanin production by P. aeruginosa wild type strain and OprF mutant strain were gathered and compared showing a significant decline (with p = 0.00069) in pyocyanin production among the mutated strains in comparison to wild strain. This clearly magnifies the direct role of OprF gene on the production of pyocyanin as virulence factor.
4. Discussion
OprF is a cornerstone gene in the expression of biological functions of Pseudomonas aeuginosa (Stover et al., 2000, Fito-Boncompte et al., 2011). In this study we demonstrate the association of OprF and its role in obtaining full virulence in P. aeruginosa using a multiphenotypic approach, based on the comparative study of a wild-type strain of P. aeruginosa, its isogenic oprF mutant. Growth rates, biofilm formation and pyocyanin production of different P. aeruginosa strains are followed up and obtained, their affection by presence or absence of OprF was investigated.
The growth of P. aeruginosa PAO1 wild strain and P. aeruginosa PAO1 mutant strain with deficient OprF monitored in a culture for 24 h were both strains verified similar viability for the culturing condition but demonstrated slower growth rate in OprF mutant strain (Fig. 1). This derives in agreement with previous study studying the link of OM proteins in virulence of P. aeruginosa, they hypothesized that OprF is involved in cellular adhesion and/or production of virulence-related factors and the study results showed that the decreased toxicity of the oprF mutant is at least partially due to its lowered ability to adhere to cells. This confirms the role of OprF as a cellular adhesin (Fito-Boncompte et al., 2011). On the other hand biofilm production was not studied or associated to the OprF porin which in this study was directly investigated and our results showed significant reduction in the biofilm formation and these results were supported by SEM images that showed detached in colonizing niche in absence of OprF. SEM images showed the morphology of P. aeruginosa PAO1 wild bacterial cells and their adhesion to each other in a steady niche and intese biofilm in contrast to the OprF mutant were irregular niche biofilm and detached cells were noticed. This may attributed not only to reduced biofilm production in OprF mutants but also to the altered biofilm component which is a future item to be investigated.
The bacterial culture supernatants were also investigated for the presence of major virulence factors, namely, pyocyanin. Altered pyocyanin production by OprF mutant (Fig. 5). Our result comes in agreement with the reduced virulence phenotype of the oprF mutant from previous study (Fito-Boncompte et al., 2011). Inactivation of OprF strongly affected all these virulence factors and revealed vital role in the observed phenotypes. This could be due to the lack of OprF can change the OM composition on the cell surface, transporting a stress signal, which sequentially could be responsible for the observed phenotypes, all leading to less pathogenic strain. Targeting the OprF porin or gene in P. aeruginosa pathogenic strains would render them less pathogenic and more susceptible strains so this could be a novel drug targets for antibiotics and vaccines.
Fig. 5.
OprF mutant strains showing altered pyocyanin production. Compared to the relative amounts of pyocyanin extracted from strain P. aeruginosa PAO1 after the bacteria were grown in LB broth. Experiments were repeated three times and pyocyanin measurements at OD520 was obtained using microplate reader and normalized to cell growth measured as OD600.
Declaration of Competing Interest
We declare that we have no conflict of interest.
Acknowledgement
The authors extend their appreciation to the Deanship of Scientific Research at King Saud University for funding this work through the Research Project No R-17–02-37.
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Sarah I. Bukhari, Email: sbukhari@ksu.edu.sa.
Fadilah Sfouq Aleanizy, Email: faleanizy@ksu.edu.sa.
References
- Brook I. Anaerobic bacteria in upper respiratory tract and other head and neck infections. Ann. Otol., Rhinol. Laryngol. 2002;111:430–440. doi: 10.1177/000348940211100508. [DOI] [PubMed] [Google Scholar]
- Carenfelt C., Lundberg C. Purulent and non-purulent maxillary sinus secretions with respect to pO2, pCO2 and pH. Acta Otolaryngol. 1977;84:138–144. doi: 10.3109/00016487709123952. [DOI] [PubMed] [Google Scholar]
- Costerton J.W., Stewart P.S., Greenberg E. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- de Kievit T. Quorum sensing in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 2009;11:279–288. doi: 10.1111/j.1462-2920.2008.01792.x. [DOI] [PubMed] [Google Scholar]
- Eschbach M., Schreiber K., Trunk K., Buer J., Jahn D., Schobert M. Long-term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. J. Bacteriol. 2004;186:4596–4604. doi: 10.1128/JB.186.14.4596-4604.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filiatrault M.J., Picardo K.F., Ngai H., Passador L., Iglewski B.H. Identification of Pseudomonas aeruginosa genes involved in virulence and anaerobic growth. Infect. Immunity. 2006;74:4237–4245. doi: 10.1128/IAI.02014-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fito-Boncompte L., Chapalain A., Bouffartigues E., Chaker H., Lesouhaitier O., Gicquel G., Bazire A., Madi A., Connil N., Véron W. Full virulence of Pseudomonas aeruginosa requires OprF. Infect. Immunity. 2011;79:1176–1186. doi: 10.1128/IAI.00850-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher M. The effects of culture concentration and age, time, and temperature on bacterial attachment to polystyrene. Canadian J. Microbiol. 1977;23:1–6. [Google Scholar]
- Gómez M.I., Prince A. Opportunistic infections in lung disease: Pseudomonas infections in cystic fibrosis. Curr. Opin. Pharmacol. 2007;7:244–251. doi: 10.1016/j.coph.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Govan J.R., Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassett D.J., Cuppoletti J., Trapnell B., Lymar S.V., Rowe J.J., Yoon S.S., Hilliard G.M., Parvatiyar K., Kamani M.C., Wozniak D.J. Anaerobic metabolism and quorum sensing by Pseudomonas aeruginosa biofilms in chronically infected cystic fibrosis airways: rethinking antibiotic treatment strategies and drug targets. Adv. Drug Deliv. Rev. 2002;54:1425–1443. doi: 10.1016/s0169-409x(02)00152-7. [DOI] [PubMed] [Google Scholar]
- Kuchma S.L., Connolly J.P., O'Toole G.A. A three-component regulatory system regulates biofilm maturation and type III secretion in Pseudomonas aeruginosa. J. Bacteriol. 2005;187:1441–1454. doi: 10.1128/JB.187.4.1441-1454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyczak J.B., Cannon C.L., Pier G.B. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2000;2:1051–1060. doi: 10.1016/s1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- Marrie T., Costerton J. Scanning and transmission electron microscopy of in situ bacterial colonization of intravenous and intraarterial catheters. J. Clin. Microbiol. 1984;19:687–693. doi: 10.1128/jcm.19.5.687-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel J., Ruseska I., Wright J., Costerton J. Tobramycin resistance of Pseudomonas aeruginosa cells growing as a biofilm on urinary catheter material. Antimicrob. Agents Chemother. 1985;27:619–624. doi: 10.1128/aac.27.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole G.A., Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- Portier F., van den Abbeele T., Lecain E., Sauvaget E., Escoubet B., Huy P.T.B., Herman P. Oxygen modulates Na+ absorption in middle ear epithelium. Am. J. Physiol.-Cell Physiol. 1999;276:C312–C317. doi: 10.1152/ajpcell.1999.276.2.C312. [DOI] [PubMed] [Google Scholar]
- Rumbaugh K.P., Griswold J.A., Iglewski B.H., Hamood A.N. Contribution of quorum sensing to the virulence ofpseudomonas aeruginosa in burn wound infections. Infect. Immunity. 1999;67:5854–5862. doi: 10.1128/iai.67.11.5854-5862.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawers R. Identification and molecular characterization of a transcriptional regulator from Pseudomonas aeruginosa PAO1 exhibiting structural and functional similarity to the FNR protein of Escherichia coli. Mol. Microbiol. 1991;5:1469–1481. doi: 10.1111/j.1365-2958.1991.tb00793.x. [DOI] [PubMed] [Google Scholar]
- Stover C.K., Pham X.Q., Erwin A.L., Mizoguchi S.D., Warrener P., Hickey M.J., Brinkman F.S.L., Hufnagle W.O., Kowalik D.J., Lagrou M., Garber R.L., Goltry L., Tolentino E., Westbrock-Wadman S., Yuan Y., Brody L.L., Coulter S.N., Folger K.R., Kas A., Larbig K., Lim R., Smith K., Spencer D., Wong G.K.S., Wu Z., Paulsen I.T., Reizer J., Saier M.H., Hancock R.E.W., Lory S., Olson M.V. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- Worlitzsch D., Tarran R., Ulrich M., Schwab U., Cekici A., Meyer K.C., Birrer P., Bellon G., Berger J., Weiss T. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Invest. 2002;109:317–325. doi: 10.1172/JCI13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K.D., Stewart P.S., Xia F., Huang C.-T., McFeters G.A. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl. Environ. Microbiol. 1998;64:4035–4039. doi: 10.1128/aem.64.10.4035-4039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S.S., Hennigan R.F., Hilliard G.M., Ochsner U.A., Parvatiyar K., Kamani M.C., Allen H.L., Dekievit T.R., Gardner P.R., Schwab U. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Develop. Cell. 2002;3:593–603. doi: 10.1016/s1534-5807(02)00295-2. [DOI] [PubMed] [Google Scholar]