Abstract
Background
Endometrial carcinoma (EC) is one of the most common gynecological malignancies among women. Maternal embryonic leucine Zipper Kinase (MELK) is upregulated in a variety of human tumors, where it contributes to malignant phenotype and correlates with a poor prognosis. However, the biological function of MELK in EC progression remains largely unknown.
Methods
We explored the MELK expression in EC using TCGA and GEO databases and verified it using clinical samples by IHC methods. CCK-8 assay, colony formation assay, cell cycle assay, wound healing assay and subcutaneous xenograft mouse model were generated to estimate the functions of MELK and its inhibitor OTSSP167. qRT-PCR, western blotting, co-immunoprecipitation, chromatin immunoprecipitation and luciferase reporter assay were performed to uncover the underlying mechanism concerning MELK during the progression of EC.
Findings
MELK was significantly elevated in patients with EC, and high expression of MELK was associated with serous EC, high histological grade, advanced clinical stage and reduced overall survival and disease-free survival. MELK knockdown decreased the ability of cell proliferation and migration in vitro and subcutaneous tumorigenesis in vivo. In addition, high expression of MELK could be regulated by transcription factor E2F1. Moreover, we found that MELK had a direct interaction with MLST8 and then activated mTORC1 and mTORC2 signaling pathway for EC progression. Furthermore, OTSSP167, an effective inhibitor, could inhibit cell proliferation driven by MELK in vivo and vitro assays.
Interpretation
We have explored the crucial role of the E2F1/MELK/mTORC1/2 axis in the progression of EC, which could be served as potential therapeutic targets for treatment of EC.
Funding
This research was supported by National Natural Science Foundation of China (No:81672565), the Natural Science Foundation of Shanghai (Grant NO:17ZR1421400 to Dr. Zhihong Ai) and the fundamental research funds for central universities (No: 22120180595).
Keywords: Endometrial carcinoma, MELK, E2F1, mTOR, MLST8, OTSSP167
Research in context.
Evidence before this study
Profound upregulation of maternal embryonic leucine zipper kinase (MELK) has been reported in many types of carcinomas and is associated with the malignant phenotype and poor prognosis. In the progression of Endometrial carcinoma (EC), the role of MELK remains unclear and needs to be investigated.
Added value of this study
This study demonstrated that MELK was significantly overexpressed in EC and high-expressed MELK predicted unfavorable overall survival and disease-free survival. MELK promoted EC cell proliferation and migration and regulated cell cycle. MELK can be transactivated by E2F1. Mechanistically, we uncovered that MELK promotes EC progression by activating MELK/mTOR signaling pathway. In addition, we also demonstrated that OTSSP167, one effective MELK inhibitor, may be a candidate drug for EC therapy.
Implications of the available evidence
This research revealed the role of MELK in the progression of EC and uncovered the underlying mechanisms. As the specific MELK inhibitor, OTSSP167 might be a potential targeted drug for EC therapy.
Alt-text: Unlabelled box
1. Introduction
Corpus uteri carcinoma, mainly endometrial carcinoma (EC), is the sixth most prevalent carcinoma among women, accounting for 4.4% of cases in 2018 [1]. The incidence rate of EC is increasing steadily over time especially in the countries undergoing socioeconomic transition [2]. With the increase in the incidence of obesity in younger age groups, the incidence of EC is increasing in the premenopausal population [3]. Modern medicine has made great progress on the therapy for EC, but there is still no effective treatment of advanced recurrent EC. In addition, conservative treatment to preserve fertility is becoming especially important for young women with early-stage EC. Therefore, investigation of the pathogenesis of EC and identification of the potential therapeutic targets need to be addressed in the current scientific research.
It is well known that kinases, as a class of proteins that play vital roles in tumor cells, are easily suppressed by specific inhibitors for antitumor therapy. A variety of kinase-related inhibitors have been used in preclinical or clinical studies on EC therapy, such as inhibitors of the PI3K–AKT–mTOR pathway–associated kinases [4] and HER-2 [5]. Metformin can inhibit EC cell growth via the AMPK pathway [6]. Nonetheless, there are still no effective targeted drugs for EC. AMPK-related kinases (ARKs) are closely related by sequence homology to the catalytic domain of AMPK [7] and remain largely uncharacterized as compared with the depth of research on AMPK. The ARK family contains 12 kinases: BRSK1, BRSK2, NUAK1, NUAK2, QIK, QSK, SIK, MARK1, MARK2, MARK3, MARK4, and maternal embryonic leucine zipper kinase (MELK). We attempted to study one ARK, MELK, which is overexpressed in many tumors.
MELK contains an N-terminal catalytic domain, a ubiquitin-associated domain (UBA domain) adjacent to the catalytic domain, and a kinase-associated 1 domain at the C terminus. MELK is complicatedly regulated by autophosphorylation, autoinhibition, reducing agents, and free Ca2+[8]. Among the 12 ARKs, MELK is the only kinase that cannot be phosphorylated and activated by LKB1 [9]. MELK expression is mainly involved in mesenchymal–epithelial transitions during embryogenesis stages, and there is little or no expression of this protein in adult tissues [10]. MELK is commonly expressed in some tissue-specific progenitor cells [11], is overexpressed in many cancers, and may be a novel biomarker of some malignant tumors [12,13]. Mouse development and physiology are not affected in MELK knockout mice [14], indicating low toxicity of MELK-targeted cancer therapy. OTSSP167, a specific MELK inhibitor, has been tested against many cancer types [15], [16], [17]. Nevertheless, the biological effects of MELK in EC remain poorly understood and are expected to be explored.
Here, we show that MELK is necessary for EC growth. We found that MELK was overexpressed in EC, and the high expression of MELK was caused by a transcription factor (TF) called E2F1. MELK-promoted EC progression may involve the mTORC1 and mTORC2 pathway.
2. Materials and methods
2.1. Clinical samples and database analysis
Two tissue microarrays containing 360 tumor tissues and 40 non-tumor tissues were constructed. All paraffin-embedded samples were obtained from Shanghai Jiao Tong University Affiliated Sixth People's Hospital in China and the First People's Hospital of Huai'an City in Jiangsu, China, between April 2003 and March 2013. Noncancerous samples were obtained from patients with nonmalignant diseases like endometrial polyps, uterine leiomyoma, and endometrial hyperplasia. All of them provided written informed consent, and all the experiments were approved by the Research Ethics Committee of Shanghai Jiao Tong University Affiliated Sixth People's Hospital and complied with the Declaration of Helsinki. A sample was excluded from the analysis if the stripping area was more than 50%.
The gene expression profile results were downloaded from the Gene Expression Omnibus database (GSE17025) at the NCBI. The dataset of UCEC (Uterus Corpus Endometrial Carcinoma) was retrieved from TCGA on January 1, 2017. We subsequently used these databases to perform gene set enrichment analysis (GSEA).
GSEA was performed in the GSEA software provided by the Broad Institute (http://www.broadinstitute.org/gsea/index.jsp). GSEA was performed for comparing the expression profiles in gene sets named h.all.v6.1.symbols.gmt [Hall-marks] from the Molecular Signature Database between a high-MELK-expression group and a low-MELK-expression group based on the median of TCGA or GEO MELK profiles and assesses the enrichment score. We ranked the EC patients according to the mRNA expression level of MELK. The first 30% of the patients were chosen as the low-expression group and the last 30% of the patients were assigned to the high-expression group. Kaplan–Meier analysis and GSEA were conducted by the above grouping method.
2.2. Culture of cell lines and transfection
Human EC cell lines were all stored at Shanghai Cancer Institute. All these cells were cultured in a suggested medium (Invitrogen, Carlsbad, USA) according to ATCC protocols.
SiRNAs targeting different genes were purchased from GenePharma (Shanghai, China), and the sequences are listed in supplementary Table 1 (Table S1). SiRNA transfection was performed using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA). HEC1A and ISK cells were infected with LV3-puro-shMELK (sh1 and sh2) and GV492-puro-MELK lentiviruses and cell clones were selected for puromycin resistance. MELK wild-type and kinase-dead mutant (D150A) plasmids were constructed previously [18] and then were transfected into wild-type ISK cells for further study.
2.3. Quantitative real-time PCR (qRT-PCR)
RNA extracted from EC cell lines was subjected to reverse transcription and subsequently underwent quantitative real-time PCR utilizing a 7500 Real-time PCR system (Applied Biosystems, USA). Relative mRNA expression was calculated by the 2−ΔΔCt method and normalized to 18S mRNA levels. Primer sequences used in this study are listed in Table S2. Experiments were conducted independently in at least triplicate.
2.4. Western blotting
Cell lysis was performed and protein concentration was measured. The cell lysates were separated by SDS-PAGE in a 6–15% gel and transferred to a nitrocellulose membrane. The membrane was probed with primary antibodies and species-specific secondary antibodies. The antibodies are listed in Table S4. Bound secondary antibodies were detected by means of an Odyssey imaging system (LI-COR Biosciences, Lincoln, NE) or Bio-Rad ChemiDoc MP Imaging System (Bio-Rad, USA).
2.5. Immunohistochemistry (IHC) analysis
Briefly, the primary antibodies anti-MELK (1:200, Sigma), anti-E2F1 (1:100, Abcam) and anti-Ki67 (1:1000, Abcam) were incubated with the slides first. Each tumor was assigned a score according to the intensity of the nuclear or cytoplasmic staining (0 = no staining, 1 = weak staining, 2 = moderate staining, and 3 = strong staining) and the proportion of stained tumor cells: (0) <5%, (1) 5–25%, (2) 25–50%, (3) 50–75%, and (4) 75–100%. The scoring was judged independently by two pathologists in a blinded manner. The final immunoreactive score was determined by multiplying the intensity scores by the proportions of stained cells, resulting in “−” for a score of 0, “+” for a score of 1–4, “++” for a score of 5–8, and “+++” for a score of 9–12. Tumors with scores ≥5 were defined as tumors with high expression of MELK (or E2F1), whereas the others were defined as tumors with low MELK (or E2F1) expression.
2.6. Immunofluorescence assay
Briefly, cells were probed with the anti-MELK antibody (1:200, Abcam) and anti-MLST8 antibody (1:50, Abcam) overnight at 4 °C, followed by an Alexa Fluor 594 conjugated anti-rabbit antibody (1:200 Jackson) and Fluor 488–conjugated anti-mouse antibody (1:200, Jackson). The nuclei were stained with 4′,6-diamidine-2′-phenylindole (DAPI; Sigma), and the immunofluorescence signals were captured by confocal microscopy (Carl Zeiss).
2.7. Cell proliferation assay and cell viability assay
Cell proliferation was measured by the Cell Counting Kit-8 (CCK-8) assay (Dojindo, Kumamoto, Japan). The cells (3 × 103 cells/well) were cultured in a 96-well plate and OD450 was measured 1 h after addition of CCK-8 at 0, 1, 2, 3, and 4 days. For the drug sensitivity assessment, AN3CA, ECC-1, HEC1A, HEC1B, ISK and KLE cell lines were treated with different concentrations of OTSSP167 (0, 1, 10, 100, or 1000 nM, Selleck). According to the IC50 curve, the said cell lines (HEC1A) were treated with other concentrations of OTSSP167 (0, 1, 10, 20, 30, 40, or 50 nM). Subsequently, the optimal concentrations of OTSSP167 (10 and 20 nM) were used to treat the cell lines to measure cell proliferation.
2.8. Colony formation assay and Cell cycle analysis
Colony formation assay and cell cycle analysis were performed as previously described [19].
2.9. Wound-healing assay
AN3CA or HEC1A cells were seeded and scratch wounds were made when the cell confluence reached ~80% at ~48 h post-transfection. The resultant cultures were incubated in a serum-free medium for 24 h, and then three visual fields were randomly picked from each scratch wound and visualized by microscopy. The experiments were performed in triplicate.
2.10. Mouse xenograft model
Six-week-old athymic nude mice (BALB/c, females) were housed under standard conditions, and all the animal experiments were carried out according to the animal experimental protocols approved by the Research Ethics Committee of East China Normal University. For subcutaneous xenograft study, 2 × 106 HEC1A cells infected with virus expressing either LV3-puro-shMELK (sh1 and sh2) or empty vector (n = 5 per group) were subcutaneously injected. For OTSSP167 effects on EC, 2 × 106 HEC1A cells were injected subcutaneously into the mice. Mice bearing similar sized tumors (two weeks after injection) were randomly divided into two groups and were treated with either DMSO or OTSSP167 (10 mg/kg, intraperitoneal injection once a week) for 4 weeks [20]. The tumor diameters were measured every 7 days. Tumor volume was estimated via the formula V = 1/2 (a × b × b), where a represents the major tumor axis, and b denotes the minor tumor axis. After 5 or 6 weeks, the mice were euthanized to dissect and weigh tumors. Then the tumor samples were fixed and prepared for subsequent histological analysis.
2.11. Chromatin immunoprecipitation (ChIP)
HEC1A and AN3CA cells were fixed in 1% formaldehyde solution at 37 °C for 10 min. ChIP assay was performed using the Pierce™ agarose ChIP kit (Thermo Fisher Scientific). Premix Taq (CST, USA) was used to quantify the DNA-protein complexes formed by immunoprecipitating DNA with control IgG (CST), anti-E2F1 antibody (1:100, CST) from the sonicated cell lysates. The specific primers used in the process of ChIP were listed in Table S3.
2.12. Luciferase reporter assay
Briefly, wild-type or E2F1 overexpressed HEC1A and AN3CA cells were co-transfected with pGL4.10-promoter vectors and pRL-TK Renilla plasmids. A Dual-Luciferase Reporter Assay System (Promega,USA) was used to analyze the luciferase activity. The data are presented as the fold change relative to the control group. The pRL-TK Renilla luciferase expression plasmid served as an internal control.
2.13. Co-immunoprecipitation (Co-IP) assay
Total protein from AN3CA and HEC1A cells was extracted using total protein extraction buffer (Beyotime, China). Protein A/G Sepharose (Santa Cruz Biotechnology) was preincubated with an anti-MELK (1:50, CST) or anti-MLST8 (1:50, CST) antibody for 60 min on a spinning wheel at 4 °C, followed by two washes. All IPs were performed overnight on a spinning wheel at 4 °C. The beads were collected by centrifugation at 3000 × g, followed by three washes with lysis buffer. The immunoprecipitates were subjected to western blotting.
2.14. Statistical analysis
These analyses were conducted in the SPSS 19.0 software (SPSS, USA). The results are presented as means ± SD, and the comparisons were conducted by two-tailed Student's t-test. The relation between MELK expression and clinical parameters was tested by the χ2 test and Fisher's exact test. For survival analysis, the Kaplan–Meier method was employed in the GraphPad Prism 7 software. Survival curves were analyzed by the logrank test. For all tests, data with p < 0.05 were considered statistically significant.
3. Results
3.1. MELK is overexpressed and correlates with poor prognosis in EC
The physiological roles of ARKs include the regulation of cell polarity, cell migration, and metabolism at cell and organismal levels [21]. ARKs are dysregulated in many tumors and may play vital roles in tumor progression. To explore the potential relations between ARKs and EC, we compared the expression of the 12 ARKs (BRSK1, BRSK2, MARK1, MARK2, MARK3, MARK4, NUAK1, NUAK2, SIK1, SIK2, SIK3, and MELK) between a normal group of patients (NC) and a group of EC patients in the UCEC datasets of TCGA. MELK expression showed the highest fold difference (Tumor/NC) and the best p value (Student's t-test; Supplementary Fig. 1a). What most interested us is that the function of MELK has not been studied deeply in EC. Thus, we focused our research on the roles of MELK in EC progression.
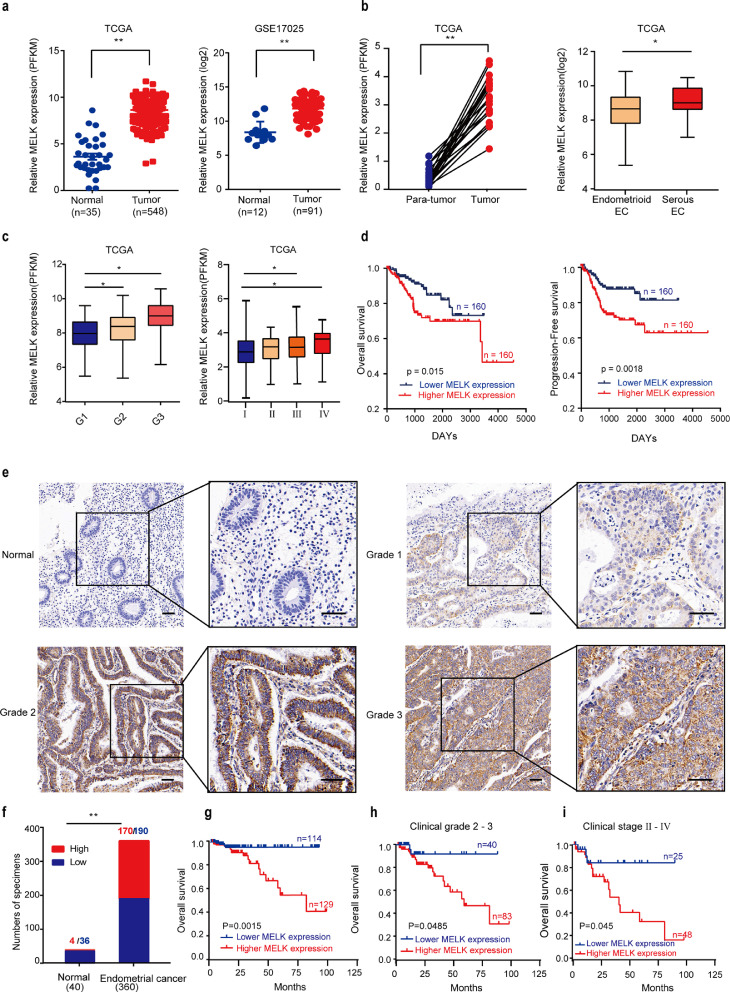
The mRNA expression levels of MELK were analyzed in TCGA and GEO databases. MELK was found to be significantly overexpressed in EC tissue samples compared with the normal counterparts in both TCGA and GEO datasets (Fig. 1a). Moreover, we found that higher MELK expression was related to higher grade (grade 3), later stage (III and IV), and serous EC histological types (Fig. 1b-c and Supplementary Fig. 1b-c). Furthermore, Kaplan–Meier analysis of the patients’ follow-up data from TCGA revealed that the overall survival and disease-free survival of patients with EC overexpressing MELK were significantly shorter than those in patients with low expression of MELK (p < 0.05; Fig. 1d).
Fig. 1.
MELK is overexpressed and correlates with poor prognosis in EC. a. mRNA expression levels of MELK in 548 EC tissue samples and 35 non-tumor tissue samples in TCGA database (left panel); the expression levels of MELK in 91 EC and 12 nontumor tissue samples in the GEO database (middle); the MELK levels in the 22 paired samples in TCGA (right). Values are means ± SD, **p < 0.01 (student's t-test). b. MELK expression in histological types. The median, upper and lower quartiles were plotted, and the whiskers that extend from each box indicate the range values that were outside of the intra-quartile range, *p < 0.05, **p < 0.01(student's t-test). c. Differential mRNA expression of MELK by the histological grade (left), and FIGO stages (right) in TCGA database. The median, upper and lower quartiles were plotted, and the whiskers that extend from each box indicate the range values that were outside of the intra-quartile range, *p < 0.05(student's t-test). d. Overall survival Kaplan–Meier (left) and disease-free survival Kaplan–Meier (right) estimates in relation to MELK alterations in TCGA; the patients were ordered by MELK expression, the first 30% patients as the low-expression group (n = 160), and the last 30% as the high-expression group (n = 160). e. Representative cases of MELK expression in normal tissues and at different histological grades in the tissue microarray of EC. Scale bar, 50 μm. f. The proportion of MELK-expressing cells assessed by blinded IHC analysis in normal tissue samples (n = 40) and EC samples (n = 360), **p < 0.01 (chi-square test). g. Comparisons of overall survival (OS) between MELK low- (n = 114) and high- expression (n = 129) groups of patients with EC. Some staining was omitted because of the stripping of the slide. h. Comparisons of the OS between MELK low- and high-expression groups in the histological grade 2 and 3 cohort. i. Comparisons of the OS between MELK low- and high-expression groups in the FIGO stage Ⅱ-Ⅳ cohort.
To verify the protein expression level of MELK in patients, tissue microarrays (360 EC patients’ samples and 40 control patients’ samples; 250 EC patients had complete follow-up information) were subjected to IHC analysis. Expectedly, MELK turned out to be upregulated in EC relative to the normal controls (Fig. 1e and f). With the increase of histological grade, the positivity rate of MELK expression increased (Fig. 1e). Next, we investigated the relation between the protein levels of MELK and clinical parameters of patients with EC (Table 1). Our results showed that high MELK expression was associated with histological grade (p < 0.001), clinical stage (FIGO, p = 0.001), and lymph node metastasis (p = 0.03). No significant correlation was observed between MELK expression levels and age, menopause status, family history, and pathological type (p > 0.05). Univariate and multivariate analyses of EC prognostic parameters were also conducted among our patients (Table 2). Our results indicated that high expression level of MELK is one of risk factors of EC.
Table. 1.
Correlation between MELK expression and clinicopathological parameters in 250 patients with endometrial cancer.
| Expression of MELK | ||||
| Total | Low (%) | High (%) | P value | |
| Age | ||||
| < 45 years old | 25 | 9 (36.0) | 16 (64.0) | 0.293 |
| ≥45 years old | 225 | 109 (48.4) | 116 (51.6) | |
| Menopause | ||||
| Yes | 163 | 82 (50.3) | 81 (49.7) | 0.186 |
| No | 87 | 36 (41.4) | 51 (58.6) | |
| Pregnancy | ||||
| Yes | 239 | 113 (47.3) | 126 (52.7) | 1 |
| No | 11 | 5 (45.5) | 6 (54.5) | |
| Family history | ||||
| Yes | 15 | 6 (40.0) | 9(60.0) | 0.605 |
| No | 235 | 112(47.7) | 123 (52.3) | |
| Pathological type | ||||
| Ademocarcinoma | 229 | 108(47.2) | 121 (52.8) | 1 |
| Non-ademocarcainoma | 21 | 10(38.5) | 11 (61.5) | |
| Clinical Stage (FIGO) | ||||
| Ⅰ | 174 | 93 (53.4) | 81 (46.6) | 0.001* |
| Ⅱ | 32 | 15 (46.9) | 17 (53.1) | |
| Ⅲ-Ⅳ | 44 | 10 (22.7) | 34 (77.3) | |
| Grade | ||||
| 1 | 123 | 77 (62.6) | 46 (37.4) | 0.000* |
| 2 | 85 | 28 (32.9) | 57 (67.1) | |
| 3 | 42 | 13 (31.0) | 29 (69.0) | |
| LN metastasis | ||||
| Yes | 24 | 6 (25.0) | 18 (75.0) | 0.03* |
| No | 226 | 113 (50.0) | 113 (50.0) | |
FIGO, International Federation of Gynecology and Obstetrics; LN, Lymph node.
P < 0.05, Statistically significant.
Table. 2.
Univariate and multivariate analysis of EC prognostic parameters in Sixth's People Hospital cohort.
| Prognostic parameter HR |
Univariate analysis |
Multivariate analysis |
||||
| HR | 95% CI | P value | HR | 95% CI | P value | |
| Expression of MELK (low vs. high) | 4.288 | 1.692–10.867 | 0.002* | 3.206 | 1.164–8.829 | 0.024* |
| Age (<45 vs.≥45) | 0.537 | 0.158–1.827 | 0.320 | – | – | – |
| Menopause (No vs. Yes) | 1.374 | 0.547–3.452 | 0.499 | – | – | – |
| Pregnancy (No vs. Yes) | 0.731 | 0.098–5.448 | 0.760 | – | – | – |
| Family history (No vs. Yes) | 1.720 | 0.584–5.062 | 0.325 | – | – | – |
| Pathological type (Ademocarcinoma vs non ademocarcinoma) | 1.913 | 0.567–6.446 | 0.296 | – | – | – |
| TNM stage (I vs. II vs III, Ⅳ) | 3.462 | 2.159–5.553 | 0.000* | 2.354 | 1.381–4.014 | 0.002* |
| Grade | 2.835 | 1.697–4.738 | 0.000* | 1.465 | 0.801–2.677 | 0.215 |
| Lymph node metastasis (absent vs. Present) | 12.543 | 3.873–40.618 | 0.000* | 9.775 | 2.615–36.540 | 0.001* |
P<0.05.
To determine the prognostic value of MELK in EC, the correlation between MELK expression and clinical follow-up information was assessed by the Kaplan–Meier analysis. High MELK expression was associated with poor overall survival (p = 0.0015; Fig. 1g). Then, we analyzed the correlation of overall survival with MELK expression among patients with advanced EC that was classified as grade 2 and 3 or stage Ⅱ, Ⅲ, or Ⅳ. These results showed that patients with advanced EC and high MELK expression in the tumor experienced significantly shorter overall survival than those with low MELK expression (Fig. 1h and i).
3.2. MELK promotes cell proliferation and migration of EC cells
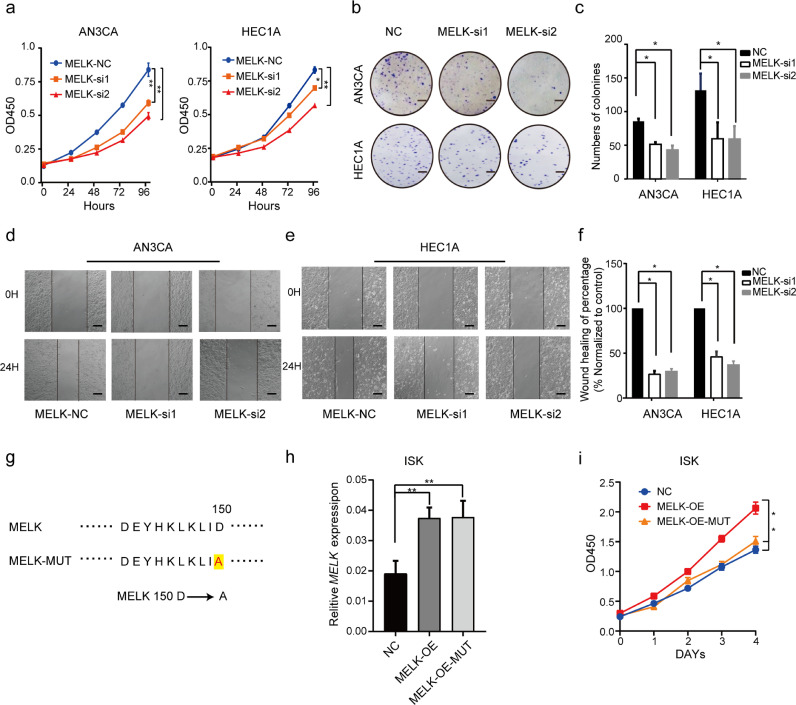
To further investigate the potential biological function of MELK in EC progression, we knocked down MELK in EC cell lines. MELK was found to be highly expressed in AN3CA and HEC1A cells compared with other EC cell lines we have (Supplementary Fig. 2a and c). The interference efficiency of two small interfering RNAs (siRNAs, MELK-si1 and MELK-si2) was measured by qRT-PCR and western blotting (Supplementary Fig. 2b and d). Next, we evaluated the proliferation and migration of AN3CA and HEC1A cells after the knockdown of MELK. A CCK-8 assay and colony formation assay revealed that downregulation of MELK significantly inhibited proliferation of both AN3CA and HEC1A cells (p < 0.05; Fig. 2a and b). The silencing of MELK significantly suppressed the migration of AN3CA and HEC1A cells in a wound-healing assay (Fig. 2d and e). Quantitative and statistical analyses of cell colonies and the percentages of wound-healing uncovered significant differences (p < 0.05) between the siRNA control group and siMELK groups after the experiments were repeated three times (Fig. 2c and f). To investigate whether the kinase activity was required for the functions of MELK in EC, two plasmids MELK-OE (MELK-wide type) and MELK-MUT (D150A-MELK [22], kinase dead, Fig. 2g) were transfected into ISK cells. The results showed that when the kinase activity of MELK was deprived, the ISK cell lost its proliferation ability given by MELK (Fig. 2h and i). Hence, kinase activity was needed when MELK played its oncogenic roles in EC. Recent studies indicate that MELK may be involved in cell cycle regulation [23]. Our results showed that the knockdown of MELK induced cell cycle arrest at the G2–M transition in HEC1A cells (Supplementary Fig. 2e and f). The expression of cyclin B1 was low, further proving the arrest at the G2–M transition after MELK was silenced (Supplementary Fig. 2g).
Fig. 2.
Effects of MELK knockdown on EC cell growth in vitro. a. Cell proliferation after the knockdown of MELK in HEC1A and AN3CA cell lines was measured by the CCK-8 assay. Values are means ± SD, n = 3, *p < 0.05, **p < 0.01(student's t-test). b. The colony formation assay was performed to assess the cell proliferation capability after the MELK knockdown in AN3CA and HEC1A cells. The representative photographs of the colony formation assay are shown. Scale bar, 5 mm. c. Quantitative analysis of the colony formation assay from panel (b), the assay was repeated three times. Values are means ± SD, *p < 0.05(student's t-test). d and e. The wound healing assay was performed to assess the effect of MELK siRNA on the migration of AN3CA and HEC1A cells. The representative images are presented. Black lines indicate the wound edge. Scale bar, 50 μm. f. Quantitative analysis of the wound healing assay, which was repeated three times, and wound healing assay from panels (d and e). Values are means ± SD, *p < 0.05(student's t-test). g-i. The carcinogenic effect of MELK was dependent on its kinase activity. g. Kinase dead MELK (MELK-Mut) refers that the 150th aspartic acid (D) was mutated to alanine (A) in the MELK amino acid sequence. h. RT-PCR was used to detect the expression of MELK after ISK cells transfected with MELK-widetype and MELK-MUT plasmids. Values are means ± SD, **p < 0.01(student's t-test). i. CCK-8 assay was used to detect ISK cell proliferation after the cells transfected with MELK-widetype and MELK- MUT plasmids. Values are means ± SD, *p < 0.05 (Student's t-test).
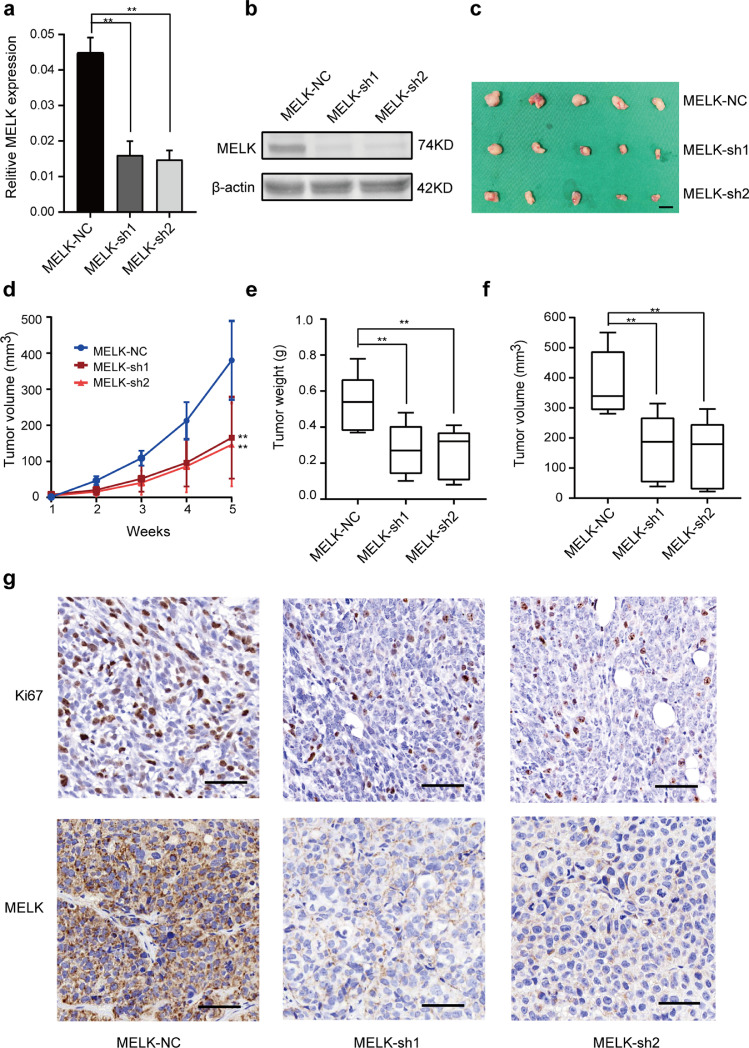
Besides, we analyzed the proliferation-promoting properties of MELK in vivo. To validate the importance of MELK in EC growth regulation, we generated two stable MELK knockdown (MELK-sh1 and MELK-sh2) cells and control HEC1A cells (MELK-NC). The knock-down efficiency of MELK was detected by qPCR and Western blotting (Fig. 3a and b). MELK-sh1, MELK-sh2 and MELK-NC cells were subcutaneously injected into nude mice. The MELK sh1 and sh2 groups manifested a significant delay in the growth of xenografted tumor and a reduced tumor burden as compared with MELK-NC group (Fig. 3c - f). IHC analysis pointed to lower expression of the proliferation marker Ki67 in the MELK-knock-down xenografts (Fig. 3g). This result indicated that MELK promoted the proliferation of HEC1A cells.
Fig. 3.
Effects of MELK knockdown on EC cell growth in vivo. a. and b. HEC1A cells were infected with either the lentivirus expressing MELK short hairpin RNA (MELK-sh1 and MELK-sh2) or control vector (MELK-NC), and cell clones were selected for puromycin resistance. The knock-down efficiency of MELK (sh1 and sh2) were detected by qPCR(a) and western blotting(b). c. Stable MELK knockdown cells and MELK-NC HEC1A cells were injected into female nude mice in each group (n = 5). The mice were euthanized at 5 weeks after the injection. Tumors were excised and weighed. Scale bar, 1 cm. d. Time course of xenograft growth. The tumor volumes of mice described in (c) were measured every week. Each point represents the mean ± SD for the tumors. e. Statistical analysis of tumor weight growth over 5 weeks. The median, upper and lower quartiles were plotted, and the whiskers that extend from each box indicate the range values that were outside of the intra-quartile range; n = 5, **p < 0.01 (Student's t-test). f. Statistical analysis of tumor volumes growth over 5 weeks. The median, upper and lower quartiles were plotted, and the whiskers that extend from each box indicate the range values that were outside of the intra-quartile range; n = 5, **p < 0.01 (Student's t-test). g. Representative images of Ki67 and MELK staining in xenograft tumors from ShMELK (MELK-sh1 and sh2) and MELK-NC mice. Scale bar, 50 μm.
3.3. E2F1 promotes transcriptional activation of MELK in EC
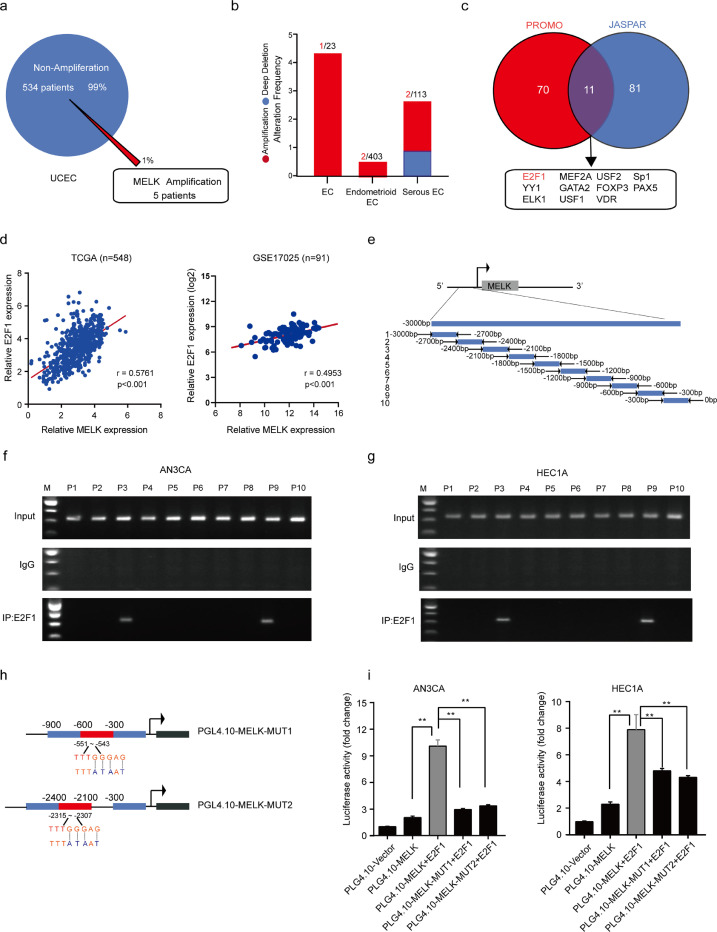
DNA copy number gains are important for gene overexpression. To test whether gene amplification contributes to the higher expression of MELK in EC, MELK alterations were analyzed in the genomic-scale sequencing data on EC in TCGA. The results showed that MELK gene amplification was present in only 5 of 539 samples (1.0%; Fig. 4a and b): insufficient evidence to explain the overexpression of MELK. Several studies and public databases have revealed that there is no genetic alteration such as gene amplification or epigenetic dysregulation affecting MELK expression [24,25]. Thus, we attempt to find the possible TFs that can transactivate MELK expression.
Fig. 4.
MELK is the direct target of a TF called E2F1 in EC. a. MELK DNA copy number amplification findings from the genomic-scale sequencing data on EC in TCGA. b. The proportion of patients with different histological types of EC; the types were unclear in 23 EC patients. c. Two websites, PROMO and JASPAR, were employed to predict the potential TFs. Eleven TFs were possible candidates. d. A correlation between MELK expression and E2F1 expression was demonstrated in both TCGA and GEO databases. The relations between the two variables were determined via Pearson's correlation coefficients. e. A sketch map of primers for MELK promoter sequences. Ten primer sets for 300 bp partitions were designed for PCR to evaluate the direct binding of E2F1 to the MELK promoter, and the primer pairs produced 10 fragments of 300 bp. f and g. The ChIP assay was carried out to verify the potential E2F1-binding site in the MELK promoter region in cell lines HEC1A and AN3CA. Input fractions and IgG served as controls. h. A diagram of MELK mutant plasmids. i. Verification of MELK as an E2F1 target via luciferase reporter assays. Constructs with an intact MELK promoter yielded enhanced luciferase activities in E2F1-expressing HEC1A and AN3CA cells, whereas those carrying mutant sites showed strong repression of luciferase activities. Values are means ± SD, **p < 0.01(student's t-test).
Websites PROMO (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3) and JASPAR (http://jaspar.genereg.net/) were used to explore possible TFs for a primary prediction. Eleven TFs were screened out (Fig. 4c), among which E2F1 attracted our attention. The correlation between E2F1 and MELK expression levels was much higher as compared with the 10 other TFs (R = 0.5761 in TCGA; R = 0.5091 in GEO; p < 0.001; Fig. 4d, Supplementary Fig. 3a).
A ChIP assay was performed to confirm the binding of E2F1 to the MELK promoter (Fig. 4e–g). We designed 10 primer sets covering the individual E2F1-binding motifs in the MELK promoter for PCR. The results clearly indicated E2F1 binding at the MELK promoter sites 3 and 9 in both AN3CA and HEC1A cells. To obtain more definitive evidence, we cloned the intact MELK promoter into luciferase constructs. When introduced into AN3CA and HEC1A cells, the constructs with an intact MELK promoter yielded increased luciferase activity, indicating direct transcriptional modulation of the MELK promoter by E2F1 (Fig. 4i). Moreover, reporter assays revealed that mutations of predicted binding site 3 or 9 abrogated the responsiveness to E2F1 in AN3CA and HEC1A cells, thus pointing to the specificity of regions −551 to −543 and −2315 to −2307 as the E2F1-binding sites in the MELK promoter (Fig. 4h and i).
Next, we detected the regulatory effect of E2F1 on MELK expression by means of an siRNA targeting E2F1. As depicted in Supplementary (Fig. 3b and c), the protein and mRNA expression levels of MELK decreased after E2F1 was knocked down in the two EC cell lines. IHC analysis of the same tissue microarray revealed that MELK expression strongly correlated with E2F1 at the protein level (Supplementary Fig. 3d and e). In addition, we found that E2F1 expression was significantly higher in patients with EC and high expression of E2F1 was associated with shorter overall survival (Supplementary Fig. 4a and b). CCK-8 assay and colony formation assay demonstrated that knockdown of E2F1 could decrease the capacity for cell proliferation in EC (Supplementary Fig. 4c and d). E2F1 may promote EC progression via regulating the expression of MELK. Collectively, these results suggested that high expression of MELK in EC was due to transcriptional activation by E2F1.
3.4. MELK can activate mTORC1 and −2 signaling by interacting with MLST8
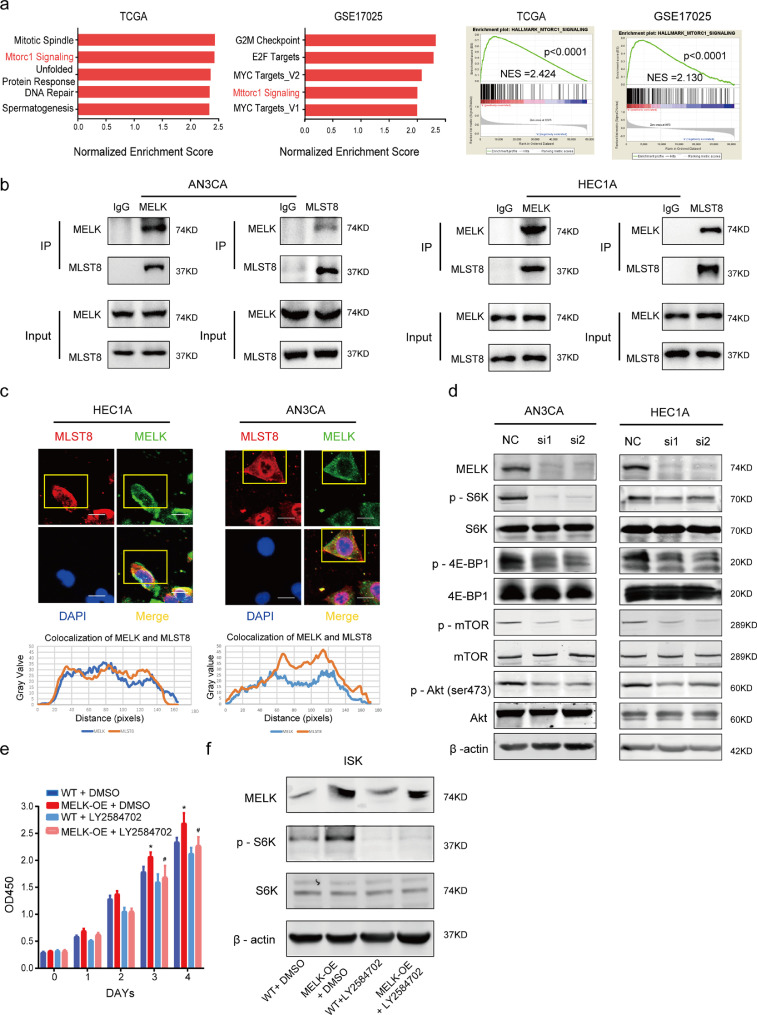
In tumor progression, highly expressed genes frequently activate multiple cell growth–related pathways. To further investigate the potential stimulatory action of MELK on EC progression, GSEA of Hallmarks gene sets was conducted based on the mRNA expression level of MELK in the databases. Our data implied that genes involved in the mTORC1 signaling pathway were particularly enriched in the MELK overexpression group (Fig. 5a). As one of the mTOR complexes (mTORC1 and mTORC2), mTORC1 mainly regulates cell growth and metabolism. It is well known that mTOR complexes primarily function as kinases. How does MELK regulate mTORC1 signaling? Studies indicate that MELK phosphorylates and activates some signaling cascades by directly binding other proteins [26], [27], [28]. We hypothesized that MELK interacts directly with mTORC1. On a protein interaction analysis website [29], we found that MELK can interact with MLST8, one of mTORC1 subunits.
Fig. 5.
MELK activates mTORC1 and −2 by interacting with MLST8. a. GSEA using a hallmark gene set was performed to compare the high MELK expression group and low MELK expression group in both TCGA and GEO databases. MELK expression magnitude in the databases was ordered from low to high: the first 30% of samples were chosen as the low-expression group, and the last 30% of samples were regarded as the high-expression group. Top significant pathways are listed in the left panel, and the mTORC1 signaling pathway was enriched (right panel). b. Co-IP assays were conducted to detect the interactions between MELK and MLST8 in both HEC1A and AN3CA cell lines. c. Immunofluorescence assays were conducted to verify the colocalization of MELK and MLST8. The curves below represent the colocalization of MELK with MLST8, the curves were constructed in the ImageJ software. d. Western blotting analysis of MELK and phosphorylated and total S6K, 4E-BP1, mTOR, and AKT during treatment with MELK-siRNA in both cell lines. β-Actin served as the loading control. e. MELK was artificially overexpressed (MELK-OE) in ISK cells and LY2584702, the S6K inhibitor, was used to treat the MELK-OE and wild type (WT) groups to evaluate the cell proliferation. DMSO is a control. Values are means ± SD, *p < 0.05 (student's t-test) as compared with group “WT+DMSO”; #p < 0.05 (student's t-test) as compared with group MELK-OE+DMSO, n = 3. f. Western blotting analysis of MELK, phosphorylated P70S6K, and total P70S6K according to (e). On the fourth day of treatment with LY2584702, the protein was collected.
Co-IP and immunofluorescence assays confirmed that MELK and MLST8 engage in a direct interaction (Fig. 5b and c). MLST8 is a subunit necessary for mTOR complex activation [30,31]. The interaction between MELK and MLST8 may promote the activation of mTORC1 signaling. Therefore, western blotting was carried out to detect the phosphorylation of mTOR and of important substrates of mTORC1 (P70S6K and 4E-BP1) to determine the participation of MELK in the activation of mTORC1. The results showed that the phosphorylation levels of mTOR, P70S6K, and 4E-BP1 significantly decreased after the knockdown of MELK in HEC1A and AN3CA cells, while the total expression of mTOR, P70S6K, and 4E-BP1 underwent no obvious changes. This finding indicated that the MELK knockdown affected the activation of mTORC1 (Fig. 5d). AKT phosphorylated at Ser473 has been demonstrated to be a reliable indicator of mTORC2 activity [32]. Of note, the phosphorylation of AKT (Ser473) also decreased when MELK was knocked down (Fig. 5d), suggesting that MELK may promote mTORC2 activation as well. Accordingly, MELK seems to activate both mTORC1 and mTORC2.
To further confirm that MELK activates mTORC1, we overexpressed MELK in ISK cells. The wild-type ISK cells (WT) and MELK-overexpressing cells (MELK-OE) were treated with an S6K inhibitor: LY2584702 (1 μM [33,34]). The CCK-8 assay indicated that cell viability in the MELK-OE group significantly increased compared with MELK-WT group. When treated with LY2584702, the cell viability in MELK-OE group was partially decreased compared with MELK-OE cells treated with DMSO. (Fig. 5e). Western blotting revealed that the level of phosphorylated P70S6K increased when MELK was overexpressed and decreased during treatment with LY2584702 (Fig. 5f).
3.5. OTSSP167 can inhibit EC cell growth by targeting MELK
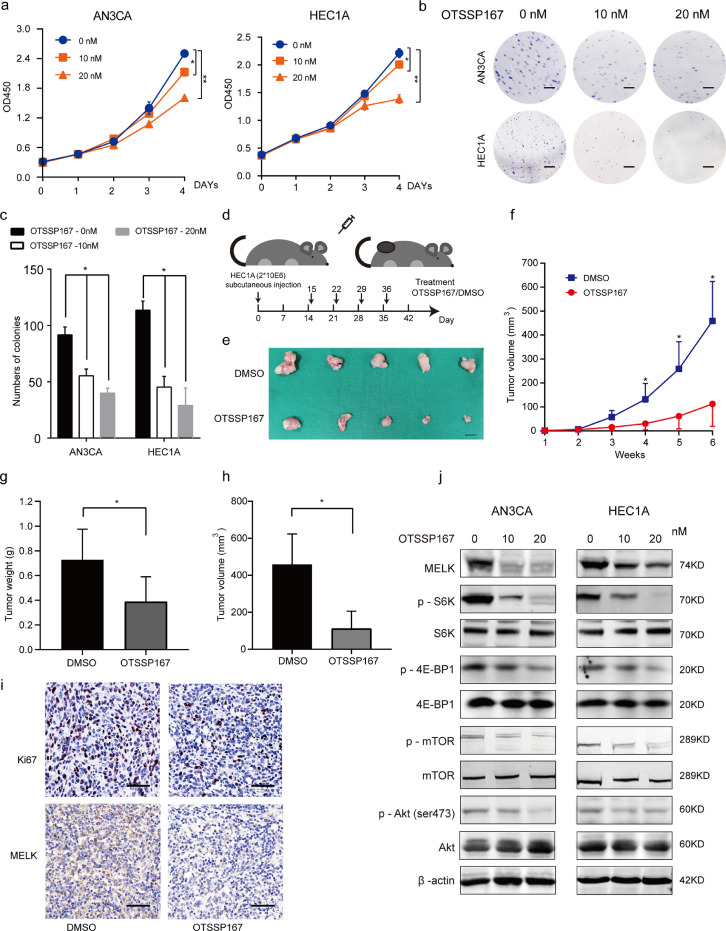
OTSSP167 (OTS167) is a specific MELK inhibitor that has been tested in preclinical trials (https://clinicaltrials.gov/; two studies have been completed and the other two trials are at the recruitment stage). We attempted to evaluate the inhibitory action of OTSSP167 on the progression of EC. First, the toxicity of OTSSP167 to cell lines AN3CA, ECC-1, HEC1A, HEC1B, ISK and KLE were determined, and an IC50 (half-maximal inhibitory concentration) curves were built and the IC50 values were listed (Supplementary Fig. 5a). HEC1A and AN3CA cell lines were continued used for further study.
Different concentrations (0, 1, 10, 20, 30, 40, or 50 nM) of OTSSP167 were incubated with the HEC1A cells, and finally we chose 10 and 20 nM as the optimal concentrations for the subsequent experiments (Supplementary Fig. 5b). Our results revealed that OTSSP167 can significantly inhibit proliferation of both AN3CA and HEC1A cells (Fig. 6a-c). To determine the in vivo effects of OTSSP167, we used the subcutaneous tumor model in nude mice. DMSO or OTSSP167 (10 mg/ml) was administered intraperitoneally once a week for 4 weeks (Fig. 6d). Tumor growth (tumor weight and volume) was significantly suppressed in the OTSSP167-treated group compared with the DMSO group (Fig. 6e-i).
Fig. 6.
OTSSP167 inhibits cell proliferation by inhibiting MELK and thus mTORC1 and −2 signaling. a. CCK-8 assays were conducted to detect cell proliferation after treatment with 10 and 20 nM OTSSP167. Values are means ± SD, n = 3, *p < 0.05, **p < 0.01(student's t-test). b. The colony formation assay was performed to assess the cell proliferation capability after treatment with 10 and 20 nM OTSSP167 in EC cells (AN3CA and HEC1A). Scale bar, 5 mm. c. The representative photographs of the colony formation assay are shown. Values are means ± SD, n = 3, *p < 0.05, **p < 0.01(student's t-test). d. Schematic presentation of the animal study treated with OTSSP167. e. The representative images of tumors on day 42 after treatment of HEC1A xenograft mice. Scale bar, 50 mm. f. Growth curve of tumors treated with OTSSP167. Values are means ± SD, *p < 0.05, **p < 0.01(student's t-test). g. statistical analysis of tumor weight growth over 6 weeks. Values are means ± SD, *p < 0.05, **p < 0.01(student's t-test). h. statistical analysis of tumor volume over 6 weeks. Values are means ± SD, *p < 0.05, **p < 0.01(student's t-test). i. Representative images of Ki67 staining and MELK staining in xenograft tumors from OTSSP167-treated groups and DMSO control groups. Scale bar, 50 μm. j. Western blotting analysis of MELK and phosphorylated and total S6K, 4E-BP1, mTOR, and AKT during treatment with 10 or 20 nM OTSSP167. β-actin served as the loading control.
Western blotting was carried out to quantify the expression of related proteins when the cells were treated with different concentrations (10 and 20 nM) of OTSSP167 for 24 h. The results showed that OTSSP167 inhibited the expression of MELK. OTSSP167 inhibited the phosphorylation of mTOR, p70S6K, 4E-BP1 and AKT (Ser473) in a concentration-dependent manner. The protein expression of total mTOR, p70S6K, 4E-BP1, and AKT did not significantly change (Fig. 6j). These data were consistent with the results of the MELK knockdown, further indicating that MELK can promote cell growth by activating the mTORC1 and −2 signaling. Fig. 7.
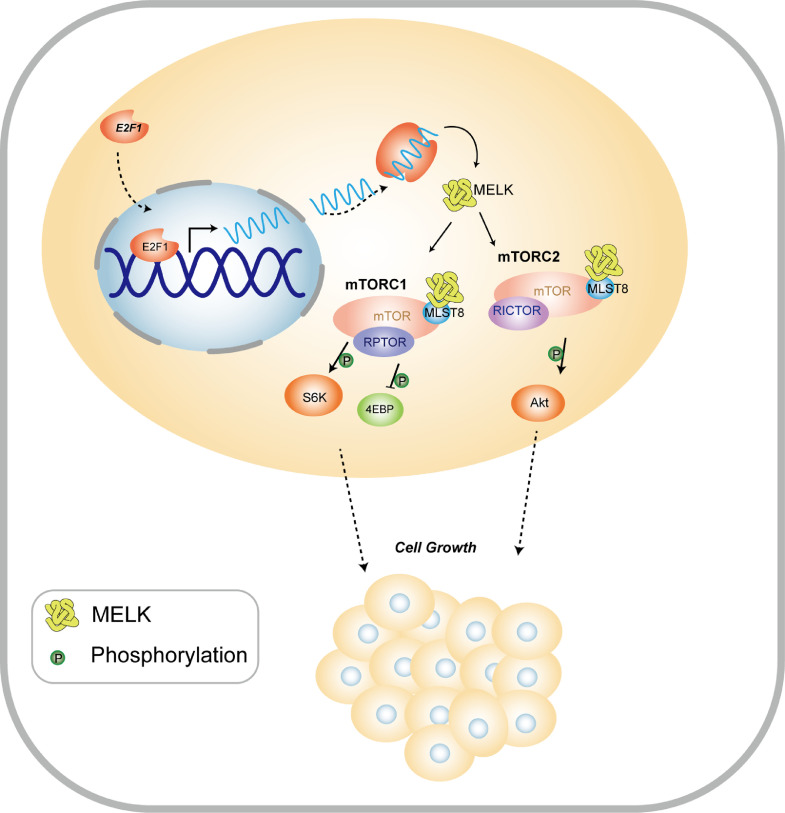
Fig. 7.
The proposed model of the stimulatory influence of MELK on EC progression via the E2F1–MELK–mTORC1/2 pathway.
4. Discussion
Profound upregulation of MELK in many types of carcinomas has been reported and is associated with the malignant phenotype and poor prognosis [35], [36], [37]. Our study revealed that MELK is overexpressed in EC tissues at both mRNA and protein levels. Moreover, high MELK expression correlated with high grade, late stage, lymph node metastasis, and the serous type of EC. Patients with overexpressed MELK tended to have a poor prognosis. Cox regression analysis suggested that MELK overexpression may be one of risk factors of EC. Therefore, MELK may play vital roles in EC progression. In fact, MELK is overexpressed in most carcinomas of TCGA database (http://gdac.broadinstitute.org/). It is essential to study the involvement of MELK in EC development.
MELK participates in many biological processes including the cell cycle, apoptosis, and RNA processing and phosphorylates some specific substrates [8,23,38]. Our results revealed that overexpressed MELK in EC cells is mainly involved in cell proliferation and migration. The cell cycle was significantly arrested at the G2–M transition when MELK was knocked down. MELK upregulated subcutaneous tumorigenesis in vivo. The increased capacity for cell proliferation and migration and an accelerated cell cycle are hallmarks of cancer. We have reasons to believe that MELK is a bona-fide oncogene. MELK shows the largest fold change among ARKs in EC relative to the healthy tissue control. Significant differences between EC and normal tissues are also seen in the expression of some other ARKs: other ARKs can be phosphorylated and activated by LKB1 [9], but they are either upregulated or downregulated in EC. Further studies are needed to investigate other ARKs in EC.
MELK is overexpressed in tumors, but little is known about how the expression of MELK is regulated. Wang et al. [14]have found that FoxM1 can promote the expression of MELK in basal-like breast cancer. MicroRNA-214–3 downregulates MELK, thereby inhibiting cell proliferation and cell cycle procession [39]. We found that high expression of MELK in EC is not caused by DNA copy number amplification. Therefore, we focused on some TFs and confirmed that E2F1 transactivates MELK expression by binding to the promoter of MELK. E2F1 is the most widely studied member of the E2F family in human malignant tumors and is often referred to as an activator because it transcriptionally activates multiple target genes [40]. E2F1 was found to not only highly correlate with the expression of MELK in EC but also to have some similarities in function with MELK. E2F1 can regulate cell proliferation, the cell cycle, cell migration [41], and apoptosis [42]. E2F1 increases MELK expression in EC; this may be one of important mechanisms of EC progression.
High expression of MELK is caused by E2F1, and how exactly MELK promotes EC cell growth remains to be investigated. In this study, GSEA revealed that MELK regulates mTORC1 signaling. Mammalian target of rapamycin (mTOR) is a conserved serine/threonine protein kinase belonging to the phosphatidylinositol 3-kinase (PI3K)-associated kinase family. The mTOR pathway controls cell growth and is often dysfunctional in cancer [43]. The function of mTOR depends on two complexes: mTORC1 and mTORC2. mTORC1 is thought to be primarily involved in the regulation of cellular metabolism, cell growth, autophagy, translation, lipid biosynthesis, and ribosome biosynthesis [44]. Affected by growth factors, mTORC2 regulates survival, metabolism, proliferation, and actin cytoskeletal polarization [45,46].
Our results indicate that MELK can interact with MLST8 and then activates mTORC1 (phosphorylates mTOR, S6K, and 4E-BP1) and mTORC2 (phosphorylates mTOR and AKT at Ser473). MLST8 binds to the catalytic domain of mTOR for stability and is a crucial subunit of both mTORC1 and mTORC2 [30,47]. Structurally, MLST8 contains seven WD40 domain repeats. Most of the proteins containing this domain function as protein–protein interaction adaptors in diverse biological processes [48]. MLST8 overexpression contributes to tumor progression by constitutively activating both mTORC1 and mTORC2 signaling pathways [49]. Our study shows that S6K inhibitor LY2584702 can partially suppress proliferation of MELK-overexpressing ISK cells; this is another example of mTORC1 activation by MELK. Although it has been reported that activation of mTORC2 inactivates mTORC1 [50], this notion does not contradict our findings because of tumor heterogeneity and different growth conditions.
Furthermore, our study indicates that OTSSP167 can suppress the proliferation and colony formation of AN3CA and HEC1A cells. OTSSP167 can suppress EC tumor growth in vivo by inhibiting MELK expression. OTSSP167 inhibited MELK expression and decreased the phosphorylation of mTOR, P70S6K, 4E-BP1, and AKT (Ser473), and this finding is consistent with the results of the MELK knockdown using siRNAs. OTSSP167, a novel quinolone-based compound, potently inhibits MELK expression. OTSSP167 represents one of the most potent MELK inhibitors in most studies and the treatment with OTSSP167 significantly decreases phosphorylation of two proteins—debrin-like (DBNL) and proteasome a subunit 1 (PSMA1)—by MELK [51]. OTSSP167 can inhibit MELK autophosphorylation and break the stability of MELK [52]. OTSSP167 has high target specificity and minimal toxicity and is relatively easy to administer [53].
Notably, recent studies have reported that MELK mutagenization mediated by CRISPR/Cas9 had no effects on 13 cancer cell lines [54]. Meanwhile, some studies raised the concern that the therapeutic effect of the MELK inhibitor, OTSSP167, in multiple cancer models is independent of MELK and due to off-target effects [54], [55], [56], [57]. Zhang et al. and Wang et al. used different genetic and chemical tools (RNA interference, CRISPR, or small -molecule inhibitors) to demonstrate the dependency of MELK in tumor progression [16,58]. Nonetheless, more researchers still tend to verify the roles of MELK in tumor by using RNA interferences and small molecule inhibitors [59,60]. Wang et al. also demonstrate that both tools (RNA interference and CRISPR) can be used to illuminate the requirement of MELK in clonogenic cell growth [58]. In the current study, MELK was knockdown by shRNA (siRNA) and OTSSP167, which strongly reduced the proliferation and colony formation of EC cells in vitro and in vivo. Although related off-target effects are yet to be elucidated, OTSSP167 is still capable in directly reducing MELK levels in our studies. It also indicates that the antitumor effect is unlikely to be unrelated to MELK, and MELK is required for EC progression. Furthermore, these MELK knockdown cells (by RNA interference and OTSSP167) showed the same effects on mTORC1 and mTORC2 activating, proving that this mechanism is indeed MELK dependent, and not due to off-target effects. Comprehensive and in-depth analysis are required to give novel insights on how to use OTSSP167 for anti-EC treatment.
In conclusion, our study uncovered a brand-new signaling axis, E2F1–MELK–mTORC1/2, which may promote the progression of EC. Our results highlight the involvement of a potential therapeutic target, gene MELK, in the development of EC. This gene can be transactivated by E2F1, and the MELK protein interacts with MLST8 to then activate the mTORC1 and −2 pathway (Fig. 6). Furthermore, our data suggest that a MELK-specific inhibitor, OTSSP167, can suppress EC cell proliferation. Thus, OTSSP167 may be a candidate drug for EC treatment.
Funding sources
This research was supported by following foundations: (a). The National Natural Science Foundation of China (No: 81672565) hosted by Yincheng Teng who contributed to experiment design. (b). The Natural Science Foundation of Shanghai (Grant No: 17ZR1421400) hosted by Zhihong Ai who contributed to data collection. (c). The Fundamental Research Funds for central universities (No: 22120180595) hosted by Qiulin Ge who contributed to data analysis.
Author contributions
Z.G.Z. and Y.C.T. designed and supervised this study. Q.Y.X. and Q.L.G. performed the data analysis, statistical analysis and finished the manuscript writing. Q.Y.X., Y.Z., and B.K.Y. performed the whole experiments. Q.Y., S.H.J., Z.H.A. and R.Z.J. provided data collection and technical support. Q.Y. contributed to bioinformatic analysis. Q.Y.X. and Q.L.G contributed equally to this work.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgements
We are grateful to Dr. Jun Li, Dr. Ya-hui Wang, Dr. Qing Li, Dr. Xiao-xin Zhang, (State Key Laboratory of Oncogenes and Related Genes, Shanghai Cancer Institute, Renji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, China) for assistance with our experiments.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2019.102609.
Contributor Information
Zhigang Zhang, Email: zzhang@shsci.org.
Yincheng Teng, Email: ycteng@sjtu.edu.cn.
Appendix. Supplementary materials
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Lortet-Tieulent J., Ferlay J., Bray F., Jemal A. International patterns and trends in endometrial cancer incidence, 1978–2013. J Natl Cancer Inst. 2018;110(4):354–361. doi: 10.1093/jnci/djx214. [DOI] [PubMed] [Google Scholar]
- 3.Moore K., Brewer M.A. Vol. 37. Am Soc Clin Oncol Educ Book; 2017. pp. 435–442. (Endometrial cancer: is this a new disease?). [DOI] [PubMed] [Google Scholar]
- 4.Slomovitz B.M., Coleman R.L. The PI3K/AKT/mTOR pathway as a therapeutic target in endometrial cancer. Clin Cancer Res. 2012;18(21):5856–5864. doi: 10.1158/1078-0432.CCR-12-0662. [DOI] [PubMed] [Google Scholar]
- 5.Buza N., Roque D.M., Santin A.D. HER2/neu in endometrial cancer: a promising therapeutic target with diagnostic challenges. Arch Pathol Lab Med. 2014;138(3):343–350. doi: 10.5858/arpa.2012-0416-RA. [DOI] [PubMed] [Google Scholar]
- 6.Zou J., Hong L., Luo C., Li Z., Zhu Y., Huang T. Metformin inhibits estrogen-dependent endometrial cancer cell growth by activating the AMPK-FOXO1 signal pathway. Cancer Sci. 2016;107(12):1806–1817. doi: 10.1111/cas.13083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manning G., Whyte D.B., Martinez R., Hunter T., Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 8.Beullens M., Vancauwenbergh S., Morrice N., Derua R., Ceulemans H., Waelkens E. Substrate specificity and activity regulation of protein kinase MELK. J Biol Chem. 2005;280(48):40003–40011. doi: 10.1074/jbc.M507274200. [DOI] [PubMed] [Google Scholar]
- 9.Lizcano J.M., Goransson O., Toth R., Deak M., Morrice N.A., Boudeau J. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 2004;23(4):833–843. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heyer B.S., Kochanowski H., Solter D. Expression of MELK, a new protein kinase, during early mouse development. Dev Dyn. 1999;215(4):344–351. doi: 10.1002/(SICI)1097-0177(199908)215:4<344::AID-AJA6>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 11.Nakano I., Paucar A.A., Bajpai R., Dougherty J.D., Zewail A., Kelly T.K. Maternal embryonic leucine zipper kinase (MELK) regulates multipotent neural progenitor proliferation. J Cell Biol. 2005;170(3):413–427. doi: 10.1083/jcb.200412115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pitner M.K., Taliaferro J.M., Dalby K.N., Bartholomeusz C. MELK: a potential novel therapeutic target for tnbc and other aggressive malignancies. Expert Opin Ther Targets. 2017;21(9):849–859. doi: 10.1080/14728222.2017.1363183. [DOI] [PubMed] [Google Scholar]
- 13.Wang J., Wang Y., Shen F., Xu Y., Zhang Y., Zou X. Maternal embryonic leucine zipper kinase: a novel biomarker and a potential therapeutic target of cervical cancer. Cancer Med. 2018;7(11):5665–5678. doi: 10.1002/cam4.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y., Lee Y.M., Baitsch L., Huang A., Xiang Y., Tong H. MELK is an oncogenic kinase essential for mitotic progression in basal-like breast cancer cells. eLife. 2014;3:e01763. doi: 10.7554/eLife.01763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kohler R.S., Kettelhack H., Knipprath-Meszaros A.M., Fedier A., Schoetzau A., Jacob F. MELK expression in ovarian cancer correlates with poor outcome and its inhibition by OTSSP167 abrogates proliferation and viability of ovarian cancer cells. Gynecol Oncol. 2017;145(1):159–166. doi: 10.1016/j.ygyno.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y., Zhou X., Li Y., Xu Y., Lu K., Li P. Inhibition of maternal embryonic leucine zipper kinase with OTSSP167 displays potent anti-leukemic effects in chronic lymphocytic leukemia. Oncogene. 2018;37(41):5520–5533. doi: 10.1038/s41388-018-0333-x. [DOI] [PubMed] [Google Scholar]
- 17.Meel M.H., de Gooijer M.C., Guillen Navarro M., Waranecki P., Breur M., Buil L.C.M. MELK inhibition in diffuse intrinsic pontine glioma. Clin Cancer Res. 2018;24(22):5645–5657. doi: 10.1158/1078-0432.CCR-18-0924. [DOI] [PubMed] [Google Scholar]
- 18.Lin M.L., Park J.H., Nishidate T., Nakamura Y., Katagiri T. Involvement of maternal embryonic leucine zipper kinase (MELK) in mammary carcinogenesis through interaction with Bcl-G, a pro-apoptotic member of the Bcl-2 family. Breast Cancer Res. 2007;9(1):R17. doi: 10.1186/bcr1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang X.-.M., Cao X.-.Y., He P., Li J., Feng M.-.X., Zhang Y.-.L. Overexpression of Rac GTPase activating protein 1 contributes to proliferation of cancer cells by reducing hippo signaling to promote cytokinesis. Gastroenterology. 2018;155(4) doi: 10.1053/j.gastro.2018.07.010. 1233-49.e22. [DOI] [PubMed] [Google Scholar]
- 20.Chlenski A., Park C., Dobratic M., Salwen H.R., Budke B., Park J.H. Maternal embryonic leucine zipper kinase (MELK), a potential therapeutic target for neuroblastoma. Mol Cancer Ther. 2019;18(3):507–516. doi: 10.1158/1535-7163.MCT-18-0819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monteverde T., Muthalagu N., Port J., Murphy D.J. Evidence of cancer-promoting roles for AMPK and related kinases. FEBS J. 2015;282(24):4658–4671. doi: 10.1111/febs.13534. [DOI] [PubMed] [Google Scholar]
- 22.Vulsteke V., Beullens M., Boudrez A., Keppens S., Van Eynde A., Rider M.H. Inhibition of spliceosome assembly by the cell cycle-regulated protein kinase MELK and involvement of splicing factor NIPP1. J Biol Chem. 2004;279(10):8642–8647. doi: 10.1074/jbc.M311466200. [DOI] [PubMed] [Google Scholar]
- 23.Davezac N., Baldin V., Blot J., Ducommun B., Tassan J.P. Human pEg3 kinase associates with and phosphorylates CDC25B phosphatase: a potential role for pEg3 in cell cycle regulation. Oncogene. 2002;21(50):7630–7641. doi: 10.1038/sj.onc.1205870. [DOI] [PubMed] [Google Scholar]
- 24.Sarhadi V.K., Lahti L., Scheinin I., Ellonen P., Kettunen E., Serra M. Copy number alterations and neoplasia-specific mutations in MELK, PDCD1LG2, TLN1, and PAX5 at 9p in different neoplasias. Genes Chromosomes Cancer. 2014;53(7):579–588. doi: 10.1002/gcc.22168. [DOI] [PubMed] [Google Scholar]
- 25.Marie S.K., Okamoto O.K., Uno M., Hasegawa A.P., Oba-Shinjo S.M., Cohen T. Maternal embryonic leucine zipper kinase transcript abundance correlates with malignancy grade in human astrocytomas. Int J Cancer. 2008;122(4):807–815. doi: 10.1002/ijc.23189. [DOI] [PubMed] [Google Scholar]
- 26.Gong X., Chen Z., Han Q., Chen C., Jing L., Liu Y. Sanguinarine triggers intrinsic apoptosis to suppress colorectal cancer growth through disassociation between STRAP and MELK. BMC Cancer. 2018;18(1):578. doi: 10.1186/s12885-018-4463-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H., Sun Q., Sun Y., Zhang J., Yuan H., Pang S. MELK and EZH2 cooperate to regulate medulloblastoma cancer stem-like cell proliferation and differentiation. Molecular cancer research: MCR. 2017;15(9):1275–1286. doi: 10.1158/1541-7786.MCR-17-0105. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y., Begley M., Li Q., Huang H.T., Lako A., Eck M.J. Mitotic MELK-eiF4B signaling controls protein synthesis and tumor cell survival. Proc Natl Acad Sci USA. 2016;113(35):9810–9815. doi: 10.1073/pnas.1606862113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chatr-Aryamontri A., Oughtred R., Boucher L., Rust J., Chang C., Kolas N.K. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 2017;45(D1):D369–DD79. doi: 10.1093/nar/gkw1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim D.H., Sarbassov D.D., Ali S.M., Latek R.R., Guntur K.V., Erdjument-Bromage H. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell. 2003;11(4):895–904. doi: 10.1016/s1097-2765(03)00114-x. [DOI] [PubMed] [Google Scholar]
- 31.Chen E.J., Kaiser C.A. LST8 negatively regulates amino acid biosynthesis as a component of the TOR pathway. J Cell Biol. 2003;161(2):333–347. doi: 10.1083/jcb.200210141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarbassov D.D., Guertin Da Fau - Ali S.M., Ali Sm Fau - Sabatini D.M., Sabatini D.M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307(5712):1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 33.Gu X., Fu X., Lu J., Saijilafu Li B, Luo Z.P. Pharmacological inhibition of S6K1 impairs self-renewal and osteogenic differentiation of bone marrow stromal cells. J Cell Biochem. 2018;119(1):1041–1049. doi: 10.1002/jcb.26272. [DOI] [PubMed] [Google Scholar]
- 34.Madala S.K., Thomas G., Edukulla R., Davidson C., Schmidt S., Schehr A. p70 ribosomal S6 kinase regulates subpleural fibrosis following transforming growth factor-alpha expression in the lung. Am J Physiol Lung Cell Mol Physiol. 2016;310(2):L175–L186. doi: 10.1152/ajplung.00063.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bolomsky A., Heusschen R., Schlangen K., Stangelberger K., Muller J., Schreiner W. Maternal embryonic leucine zipper kinase is a novel target for proliferation-associated high-risk myeloma. Haematologica. 2018;103(2):325–335. doi: 10.3324/haematol.2017.172973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia H., Kong S.N., Chen J., Shi M., Sekar K., Seshachalam V.P. MELK is an oncogenic kinase essential for early hepatocellular carcinoma recurrence. Cancer Lett. 2016;383(1):85–93. doi: 10.1016/j.canlet.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 37.Speers C., Zhao S.G., Kothari V., Santola A., Liu M., Wilder-Romans K. Maternal embryonic leucine zipper kinase (MELK) as a novel mediator and biomarker of radioresistance in human breast cancer. Clin Cancer Res. 2016;22(23):5864–5875. doi: 10.1158/1078-0432.CCR-15-2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Badouel C., Chartrain I., Blot J., Tassan J.P. Maternal embryonic leucine zipper kinase is stabilized in mitosis by phosphorylation and is partially degraded upon mitotic exit. Exp Cell Res. 2010;316(13):2166–2173. doi: 10.1016/j.yexcr.2010.04.019. [DOI] [PubMed] [Google Scholar]
- 39.Li Y., Li Y., Chen Y., Xie Q., Dong N., Gao Y. MicroRNA-214-3p inhibits proliferation and cell cycle progression by targeting MELK in hepatocellular carcinoma and correlates cancer prognosis. Cancer Cell Int. 2017;17:102. doi: 10.1186/s12935-017-0471-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xanthoulis A., Tiniakos D.G. E2F transcription factors and digestive system malignancies: how much do we know? World J Gastroenterol. 2013;19(21):3189–3198. doi: 10.3748/wjg.v19.i21.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang Y.X., Lu J.M., Mo R.J., He H.C., Xie J., Jiang F.N. E2F1 promotes tumor cell invasion and migration through regulating CD147 in prostate cancer. Int J Oncol. 2016;48(4):1650–1658. doi: 10.3892/ijo.2016.3364. [DOI] [PubMed] [Google Scholar]
- 42.Stanelle J., Putzer BM. E2F1-induced apoptosis: turning killers into therapeutics. Trends Mol Med. 2006;12(4):177–185. doi: 10.1016/j.molmed.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 43.Saxton R.A., Sabatini D.M. MTOR signaling in growth, metabolism, and disease. Cell. 2017;168(6):960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim L.C., Cook R.S., Chen J. MTORC1 and mTORC2 in cancer and the tumor microenvironment. Oncogene. 2017;36(16):2191–2201. doi: 10.1038/onc.2016.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jacinto E., Loewith R., Schmidt A., Lin S., Ruegg M.A., Hall A. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6(11):1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 46.Gaubitz C., Prouteau M., Kusmider B., Loewith R. TORC2 structure and function. Trends Biochem Sci. 2016;41(6):532–545. doi: 10.1016/j.tibs.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 47.Yang H., Rudge D.G., Koos J.D., Vaidialingam B., Yang H.J., Pavletich N.P. mTOR kinase structure, mechanism and regulation. Nature. 2013;497(7448):217–223. doi: 10.1038/nature12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu C., Min J. Structure and function of WD40 domain proteins. Protein Cell. 2011;2(3):202–214. doi: 10.1007/s13238-011-1018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kakumoto K., Ikeda J., Okada M., Morii E., Oneyama C. mLST8 promotes mTOR-Mediated Tumor progression. PLoS ONE. 2015;10(4) doi: 10.1371/journal.pone.0119015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mao Z., Zhang W. Role of mTOR in glucose and lipid metabolism. Int J Mol Sci. 2018;19(7) doi: 10.3390/ijms19072043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chung S., Suzuki H., Fau - Miyamoto T., Miyamoto T., Fau - Takamatsu N., Takamatsu N., Fau - Tatsuguchi A., Tatsuguchi A., Fau - Ueda K., Ueda K., Fau - Kijima K. Development of an orally-administrative MELK-targeting inhibitor that suppresses the growth of various types of human cancer. Oncotarget. 2012;3(12):1629–1640. doi: 10.18632/oncotarget.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chung S., Kijima K., Kudo A., Fujisawa Y., Harada Y., Taira A. Preclinical evaluation of biomarkers associated with antitumor activity of MELK inhibitor. Oncotarget. 2016;7(14):18171–18182. doi: 10.18632/oncotarget.7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ganguly R., Hong C.S., Smith L.G., Kornblum H.I., Nakano I. Maternal embryonic leucine zipper kinase: key kinase for stem cell phenotype in glioma and other cancers. Mol. Cancer Ther. 2014;13(6):1393–1398. doi: 10.1158/1535-7163.MCT-13-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin A., Giuliano C.J., Sayles N.M., Sheltzer J.M. CRISPR/Cas9 mutagenesis invalidates a putative cancer dependency targeted in on-going clinical trials. eLife. 2017;6:e24179. doi: 10.7554/eLife.24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giuliano C.J., Lin A., Smith J.C., Palladino A.C., Sheltzer J.M. MELK expression correlates with tumor mitotic activity but is not required for cancer growth. eLife. 2018;7 doi: 10.7554/eLife.32838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Settleman J., Sawyers C.L., Hunter T. Challenges in validating candidate therapeutic targets in cancer. eLife. 2018;7 doi: 10.7554/eLife.32402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ji W., Arnst C., Tipton A.R., Bekier M.E., 2nd Taylor WR, Yen T.J. OTSSP167 abrogates mitotic checkpoint through inhibiting multiple mitotic kinases. PLoS ONE. 2016;11(4) doi: 10.1371/journal.pone.0153518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y., Li B.B., Li J., Roberts T.M., Zhao J.J. A conditional dependency on MELK for the proliferation of triple-negative breast cancer cells. iScience. 2018;9:149–160. doi: 10.1016/j.isci.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meel M.H., Guillen Navarro M., de Gooijer M.C., Metselaar D.S., Waranecki P., Breur M. MEK/MELK inhibition and blood-brain barrier-deficiencies in atypical teratoid/rhabdoid tumors. Neuro-oncology. 2019 doi: 10.1093/neuonc/noz151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li B., Yan J., Phyu T., Fan S., Chung T.H., Mustafa N.B. MELK mediates the stability of EZH2 through site-specific phosphorylation in extranodal natural killer/T-cell lymphoma. Blood. 2019 doi: 10.1182/blood.2019000381. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.