Abstract
Background
Whereas the role of neoadjuvant radiotherapy in rectal cancer is well-established, the ability to discriminate between radioresistant and radiosensitive tumors before starting treatment is still a crucial unmet need. Here we aimed to develop an in vivo test to directly challenge living cancer cells to radiotherapy, using zebrafish xenografts.
Methods
We generated zebrafish xenografts using colorectal cancer cell lines and patient biopsies without in vitro passaging, and developed a fast radiotherapy protocol consisting of a single dose of 25 Gy. As readouts of the impact of radiotherapy we analyzed proliferation, apoptosis, tumor size and DNA damage.
Findings
By directly comparing isogenic cells that only differ in the KRASG13D allele, we show that it is possible to distinguish radiosensitive from radioresistant tumors in zebrafish xenografts, even in polyclonal tumors, in just 4 days. Most importantly, we performed proof-of-concept experiments using primary rectum biopsies, where clinical response to neoadjuvant chemoradiotherapy correlates with induction of apoptosis in their matching zebrafish Patient-Derived Xenografts-Avatars.
Interpretation
Our work opens the possibility to predict tumor responses to radiotherapy using the zebrafish Avatar model, sparing valuable therapeutic time and unnecessary toxicity.
Keywords: Chemoradiotherapy, Colorectal cancer, Zebrafish avatars, Personalized medicine, Radiotherapy
Research in Context.
Evidence before this study
To date, there is no robust method of predicting whether a tumor will respond to radiotherapy or not. Neoadjuvant radiotherapy or chemoradiation (nRT), followed by total mesorectal excision surgery represent the standard of care in selected patients with locally advanced rectal cancer (LARC). Clinical and pathological response or TNM down-staging reported after nRT are currently the strongest markers of favourable oncological outcomes in LARC. Poor response with persistently involved lymph nodes (ypN+) is associated with adverse outcomes. Yet there are no effective methods to identify patients who will most likely benefit from nRT. Therefore, a biomarker that predicts response to nRT at an early time point to reduce unnecessary toxicities associated with ineffective treatments is a critical unmet need.
Added value of this study
Here, we developed the zebrafish larvae xenograft model as a cost-effective and fast in vivo biomarker of response to radiotherapy. Our results show that, in just 4 days it is possible to distinguish radiosensitive from radioresistant tumors in zebrafish xenografts, even in polyclonal tumors. We also performed proof-of-concept experiments that demonstrate the feasibility of using fresh rectal cancer biopsies to generate zebrafish Avatars. In two case studies patient clinical response correlated with its matching Avatar.
Implications of all the available evidence
Future work will be aimed at increasing the number of patients to test the predictive power of the zebrafish Avatar model. Moreover, since we use freshly cryopreserved samples, it is possible to receive out-patient’ samples for a multi-center study. Therefore, our work has the potential to revolutionize clinical practice in the future by providing a timely assessment of predicted radio-sensitivity, thus enabling personalized treatment, while sparing valuable therapeutic time and unnecessary toxicities.
Alt-text: Unlabelled box
1. Background
Colorectal cancer (CRC) is the third most common cancer worldwide, with 1.8 million new cases diagnosed in 2018 [1]. Rectal cancer accounts for approximately 30% of CRC and is associated with worse clinical outcome [2]. Neoadjuvant radiotherapy (nRT) is the preferred approach to down-size and down-stage locally/advanced rectal cancer (LARC). Short-Course Preoperative Radiotherapy (SCRT) or combination of long-course radiotherapy with chemotherapy (CRT) are the established approaches for intermediate stage or LARC [3]. SCRT consists of the delivery of 5 Gy over five consecutive days (5 × 5 Gy), while CRT consists of 25–28 fractions of 1.8–2 Gy [4], [5], [6]. After a standard interval of 6–12 weeks after nCRT, tumor response can be assessed and graded as: complete response (8%–20% of patients), partial response (40%−60%) or no response to therapy (20%) [2]. Therefore, ~20% of patients are exposed to unnecessary side effects and to a delay in more effective therapeutic strategy. This 20% may be even underestimated, since some studies point to 40%–45% of patients that do not respond to neoadjuvant CRT [3].
Response to treatment is highly heterogeneous. Therapies that are efficient and successful for some patients, may be relatively ineffective for others [4]. Thus, the ability to discriminate patients that will benefit (responders) from those who will not (non-responders) remains a challenge. With the aim to provide pre-clinical insights and to guide treatment decisions, studies have been conducted with mice Patient-Derived Xenografts (PDX) as a model for drug screening [5]. However, it is a time-consuming and costly model, where the required time for sample engraftment and treatment is typically about 2–4 months, which makes it unfeasible for clinical decision-making [6] (see Table S1). In vitro clonogenic assays have been also a topic of extensive research. However, primary cells are difficult to grow in vitro and due to the clonal selection that these cells are subjected to, phenotypes that no longer represent the initial tumor might become over-represented, impairing the accurate prediction of the patient outcome [7]. More importantly, in vitro clonogenic assays take several weeks to perform, which is not compatible with the time-frame of treatment decision [7].
The use of zebrafish larvae xenografts has developed into a promising in vivo model for human cancer studies [8], [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [41]. Recently, we showed that in just 4 days, several hallmarks of cancer such as proliferation, metastatic and angiogenic potentials can be recapitulated in zebrafish larvae tumor xenografts. As a proof-of-principle, we screened the current standard of care guidelines for CRC from 1st to 3rd lines of treatment, and showed similar drug responses between zebrafish and mouse xenografts. We generated zebrafish Patient-Derived Xenografts (zPDX) and compared response to chemotherapy with actual clinical response in patients, with promising results [11].
Here, we assessed the possibility of using zebrafish xenografts to distinguish between radiosensitive and radioresistant tumors. To this end, we developed a Single Dose Radiotherapy (SDRT, 1 × 25 Gy) protocol to assess in vivo sensitivity in just 4 days. This SDRT protocol is time- and cost-effective. In addition, we compared our SDRT protocol to a hypofractionated short-course radiotherapy (SCRT 5 × 5 Gy) protocol similar to the one applied in the clinic. Our results suggest that our SDRT protocol provides a good proxy of tumor response. Moreover, we generated polyclonal xenografts by mixing radiosensitive cells with their isogenic radioresistant clones and show that we can distinguish both phenotypes in the same xenograft. By directly comparing isogenic cells, we also show that KRAS sensitizes cells to radiotherapy in HCT116 CRC cells. Finally, we performed proof-of-concept experiments using patient biopsies, setting the ground for a future personalized radiotherapy assay.
2. Methods
2.1. Human colorectal cancer cell lines
HCT116 (KRASG13D) and Hke3 (KRASWT) were donated by Dr. Ângela Relógio (Institute for Theoretical Biology, Berlin). Cell lines were tested for mycoplasma and authenticated through Short Tandem Repeat (STR) profiling.
2.2. Cell culture
CRC cell lines were expanded and maintained in Dulbecco's Modified Eagle Medium (DMEM) High Glucose (Biowest) supplemented with 10% (v/v) Fetal Bovine Serum (FBS) (Sigma-Aldrich) and 1% (v/v) Penicillin-Streptomycin 10,000 U/mL (Hyclone) in a humidified atmosphere containing 5% CO2 at 37 °C.
2.3. Zebrafish care and handling
In vivo experiments were performed using zebrafish (Danio rerio), nacre, casper and Tg(Fli1:eGFP), which were handled according to European animal welfare Legislation, Directive 2010/63/EU (European Commission, 2016).
2.4. Cell labelling
CRC cell lines were labelled with Vybrant CM-DiI (Vybrant™ CM-DiI, Thermo Fisher Scientific), at 4 μL/mL concentration, or with Deep Red dye (CellTracker™, Thermo Fisher Scientific), at a concentration of 1 μL/mL. Staining was performed according to manufacturer's instructions. Cells were resuspended to final concentration of 0.25 × 106 cells/μL. The polyclonal CRC xenografts consisted in a mixture of 1:1 of HCT116 and Hke3 cells.
2.5. CRC primary samples processing
The study was approved by the Ethics Committee of the Champalimaud Foundation. CRC patient samples were provided by Champalimaud Clinical Center's (CCC) Digestive Unit, after signed informed consent. Primary tissue from surgical resected rectal cancer samples and biopsies were collected (Table S3) and cryopreserved (90% FBS and 10% DMSO). For microinjection, samples were thawed and minced in Mix1 (Table S3) with subsequent mechanic tissue fragmentation and centrifugation (150xg, 4 min). The remaining tissue fragments were enzymatically digested in Mix2 (Table S3), passed through a 70 μm cell strainer and labeled with DiI or Deep Red for 8 min at 37 °C. Cell suspension was resuspended in Mix3 to a final concentration of ~0.25 × 106 cells/μL and viability was assessed by trypan blue exclusion method. Prior injection, a small aliquot of the processed/dissociated tumor sample was stained with MGG Grunwald-Giemsa (Bio-Optica) method according to the manufacturer's instructions.
2.6. Zebrafish xenograft microinjection
Labelled cells were injected into the perivitelline space (PVS) of anesthetized 2 days post fertilization (dpf) zebrafish larvae. Following microinjection, all larvae were maintained at 34 °C until the end of the experiments in E3 medium.
The tumor implantation percentage was calculated as follows:
2.7. Xenografts irradiation and drug administration
At 1 day post injection (dpi) all zebrafish xenografts were screened regarding the presence of successfully injected tumor mass and grouped according to their tumor size. All non-successful xenografts were discarded and ethically euthanized. At 1dpi zebrafish xenografts were anesthetized and randomly distributed into the several experimental conditions. Single doses of 0, 2, 4, 6, 10, 14, 20 or 25 Gy were delivered to zebrafish xenografts, ending the assay at 4dpi, 6dpi or at 8dpi, according to the experiment. 5FU was administered at 4.2 mM in E3 medium during three successive days. In the end of each experiment, xenografts were sacrificed and fixed in 4% (v/v) formaldehyde (Thermo Scientific) and transferred to 100% methanol for long-term preservation. Irradiation procedures and regimens were adapted for zebrafish xenografts by the Champalimaud Foundation Radiation Oncology Department. The 6MV X-rays beams with the corresponding prescription dose (Gy) were calculated with the same algorithm used in clinical practice (ECLIPSE, Varian Medical System, CA) and was delivered via a linear accelerator (Truebeam, Varian Medical Systems, CA). Irradiation was targeted to the center of a defined area of 30 × 30 cm where the 6-well plates with the anesthetized zebrafish were placed (6 mL of E3 medium per well). The well plates were positioned with a source-to-surface distance of 100 cm. No build up material was needed.
2.8. Xenograft whole-mount immunofluorescence
Primary antibodies: anti-Activated Caspase3 (rabbit, Cell signaling, 1:100, code#9661), anti-Human mitochondria (mouse, Merck Millipore, 1:100, cat#MAB1273), anti-γH2AX, serine 139 (mouse, Merck Millipore, 1:1000, cat#05-636), anti-Ki67 (mouse, Leica-Novocastra, 1:100, cat#NCL-Ki67-MM1), and anti-phospho Histone H3 (rabbit, Merck Milipore, 1:100 cat#06-570). Secondary antibodies: Alexa goat anti-rabbit 488 (Molecular probes, 1:400), anti-mouse 488 (Molecular probes, 1:400), and anti-mouse 647 (Molecular probes 1:400) were applied simultaneously with DAPI. Xenografts were mounted with Mowiol.
2.9. Xenografts imaging and quantification
All xenografts were acquired in Zeiss LSM 710 fluorescence confocal microscope with a 5 μm interval z stacks. Images were analyzed using ImageJ software, using the Cell Counter plugin [19]. Total number of cells / tumor = AVG (3 slices Zfirst, Zmiddle, Zlast) x total n° slices/1.5. Mitosis and activated Caspase 3 were quantified in all slices. Nuclear size was assessed by: AVG ((Zfirst ROI/ Zfirst DAPI) + (Zmiddle ROI/ Zmiddle DAPI) + (Zlast ROI/Zlast DAPI)). zPDX quantifications were performed in all tumor depth.
2.10. Statistical analysis
Statistical analysis was performed using GraphPad Prism software. Data sets were challenged by normality tests (D'Agostino & Pearson and the Shapiro-Wilk). Data with Gaussian distribution were analyzed by unpaired t-test. Datasets that did not pass the normality test were analyzed by the Mann–Whitney-test. All normalised data (fold induction or tumor size normalised) were analyzed by Mann–Whitney test. Differences were considered significant at P < .05 and statistical output was represented by stars as non-significant (NS)> 0.05, *≤ .05, **≤ .01, ***≤ .001. All graphs presented the results as AVG ± standard error of the mean (SEM).
3. Results
3.1. Zebrafish xenograft dose-response curve to ionizing radiation
A dose response curve to test the survival and tumor response to increasing doses of ionizing radiation (IR) was generated. First, we determined larvae survival rates after irradiation under our experimental conditions, i.e., irradiation at 3 days post fertilization (dpf). Our results show no significant differences in larvae survival compared to non-irradiated controls (Fig. 1i). In agreement with previous studies in zebrafish [20], our results show that zebrafish larvae are highly resistant to IR during this stage of development (from 3dpf to 8dpf).
Fig. 1.
Zebrafish xenografts reveal cellular sensitivity to increasingly doses of ionizing radiation. 2dpf zebrafish larvae were injected with fluorescently labelled HCT116 cells (red) in the PVS. At 1dpi, the zebrafish xenografts were randomly divided and submitted to 0, 2, 4, 6, 10, 14, 20 or 25 Gy doses as a single radiation scheme (a–h) and tumor size (total number of tumor cells) was quantified (j). The total number of quantified xenografts is indicated in the images. Survival curve of non-injected larvae at 3dpf, 50 larvae were irradiated in each condition (i). j and i are results from one independent experiment expressed as AVG±SEM. Scale bar (50 µm). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
We next generated human CRC xenografts using the HCT116CRC radiosensitive cell line [21,22]. As in our previous studies, HCT116 tumor cells were fluorescently labeled and injected into the perivitelline space (PVS) of 2dpf zebrafish larvae [11]. At one day post injection (dpi), xenografts were irradiated with single doses of 0, 2, 4, 6, 10, 14, 20 and 25 Gy, and evaluated 5 days later, at 6dpi (Fig. 1a–h’). To study the in vivo tumor response dynamics in the zebrafish xenograft model we analyzed the impact of radiation on tumor size (number of cells per tumor). The tumor size curve of irradiated xenografts showed almost a linear relationship with radiation dose between 2Gy and 6 Gy, with 6 Gy leading to a ~50% reduction of the tumor mass. This curve is consistent with previous studies using the same CRC cell line HCT1116 [21,22]. Between 6 and 14 Gy the effect seems to stabilize, followed by a drop to 63% reduction of tumor size with 20 Gy and 73% in 25 Gy. Our results are consistent with previous studies in spheroids, which show a higher survival rate than the classical adherent 2D in vitro cell cultures [23].
3.2. The SDRT protocol distinguishes sensitive from resistant tumors
Our goal is to develop a fast in vivo protocol to distinguish radiosensitive from radioresistant tumors. This would allow to spare non-responders from unnecessary toxicities and explore alternative treatment strategies. We selected a high dose to maximize the effect. Since SDRT (1 × 25 Gy) leads to a ~73% shrinkage of HCT116 tumors after 5 days (Fig. 1) with no significant impact on xenograft survival, we reduced the duration of the assay to a total of 4 days, to have a practical and quick protocol for future translation into the clinical setting.
Previous studies suggest that KRAS may sensitize tumors to radiotherapy [2]. HCT116 tumor cells harbor a KRASG13D mutation, rendering them highly proliferative in comparison to the isogenic KRASWT Hke3 cell line [11,24] (see Table S2). Since radiosensitivity is highly correlated with proliferation, we hypothesized that Hke3, with a lower proliferative potential, could be considered as a negative control (i.e., a radioresistant isogenic control). We generated HCT116 and Hke3 xenografts and, at 1dpi, xenografts were subjected to a single dose of 25 Gy. Three days after IR (and 4dpi), we assessed the impact of radiation on proliferation, apoptosis, and tumor size. As expected, a 25 Gy radiation leads to a significant reduction of proliferation of HCT116 (Fig. 2i, P < .0001, see Fig. S1 for comparison with pHH3 and Ki-67), a significant increase of apoptosis (Fig. 2j, 7.4-fold, P < .0001, see a’-d’ for maximal Z projection of Activated Caspase3) accompanied by a ~55% reduction of tumor size (Fig. 2k, P < .0001). In contrast, although radiation reduced proliferation of Hke3 cells, it neither induced apoptosis nor did it reduce tumor mass (Fig. 2e, g, i–k).
Fig. 2.
Single Dose Radiotherapy (1 × 25 Gy) regimen is able to discriminate radiosensitive from radioresistant CRC zebrafish xenografts. Human CRC cells HCT116 and Hke3 were injected into the PVS of 2dpf zebrafish larvae. At 1dpi, successfully implanted xenografts were submitted to 5FU chemotherapy for 3 consecutive days (b and f), to a single radiation dose of 25 Gy (c and g), or the combination of both treatments (d and h), and compared to non-irradiated xenograft controls (a and e). Maximum Z projections of Activated Caspase3, HCT116 (a’–d’) and Hke (e’–h’). At 4dpi, cell proliferation (mitotic figures%), apoptosis (% activated Caspase3, green), and tumor size (number of tumor cells, DAPI, blue) were analyzed and quantified (i–k, respectively). i–k results are the average of 3 independent experiments and results are expressed as AVG ± SEM. Each dot represents a xenograft and the total number of quantified xenografts is indicated in the images (a–h). **P ≤ .01, ***P ≤ .001; NS, nonsignificant. Dashed white line is delimitating the tumor of each represented xenograft. Scale bar-50 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Since neoadjuvant therapy often consists of a combination of radiation with 5FU-based chemotherapy (CRT) [25], we tested the impact of its addition to radiation in our model, using 5FU alone as control, on HCT116 and Hke3 xenografts. As expected, CRT induced a significant reduction of proliferation (Fig. 2i) and induction of apoptosis (Fig. 2j, 10.3-fold increase, P < .0001), as well as a significant shrinkage of HCT116 tumors (Fig. 2k, P < .0001). Combination of 5FU and radiotherapy resulted in an additive effect (Fig. 2i,j), clearly observed in the reduction of mitotic figures and induction of cell death, but not in tumor shrinkage. CRT in Hke3 cells had no effect on apoptosis or tumor size (Fig. 2e, g, i–k).
There are several reports from in vivo and in vitro experiments suggesting that ionizing radiation may stimulate the metastatic process. Clinical observations do not directly confirm these data, leaving open the question whether or not radiotherapy enhances metastasis [26].Taking advantage of the zebrafish xenograft model we analyzed whether radiation could alter the metastatic potential of HCT116 and Hke3 tumor xenografts. However, no significant differences were observed (Supplementary Fig. S2).
As a note, despite of being widely used and accepted in radiation studies [10,[27], [28], [29], [30], [31]], HCT116 and Hke3 are colon cancer cells lines and different responses may occur in rectal cancer cells.
Overall, our results show that the zebrafish xenograft model has sufficient resolution to distinguish between CRC HCT116/Hke3 radiosensitive and radioresistant cells in vivo in just 4 days. Also, our results highlight how KRAS mutations may sensitize cells to radiotherapy.
3.3. Zebrafish xenografts show resolution to distinguish radiosensitivity in polyclonal heterogeneous tumors
To confirm the difference in HCT116 and Hke3 clones radiosensitivity profiles, we co-injected the two cell lines in the same host, each labeled with a different lipophilic dye. By co-injecting, we can directly compare response under the same experimental conditions. With this experiment, we intended to reproduce a common characteristic of CRC tumors: their clonal heterogeneity.
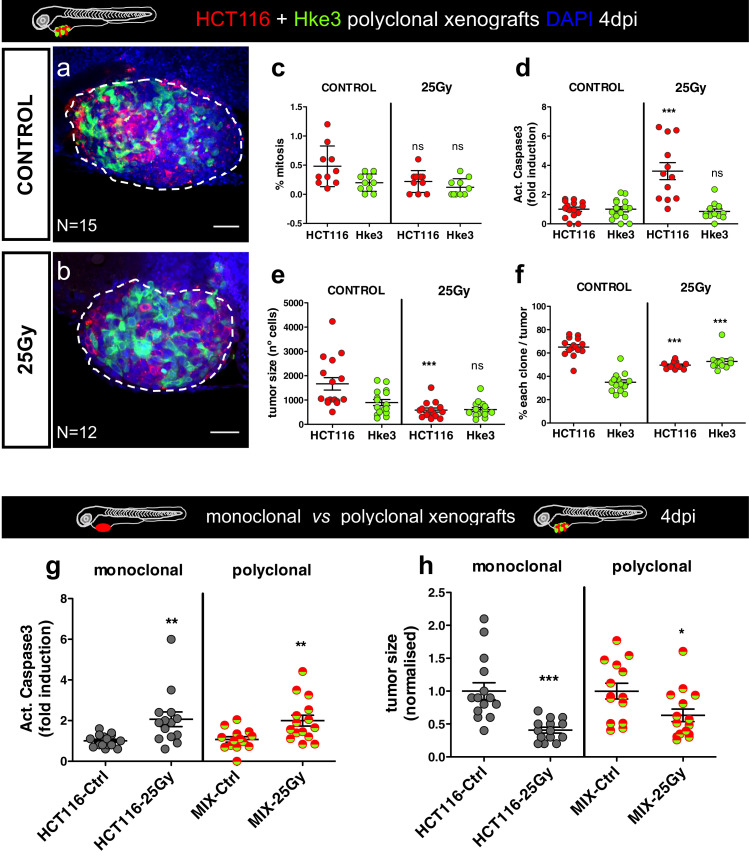
At 1dpi, mixed xenografts (HCT116+Hke3) were exposed to 1 × 25 Gy and compared to non-irradiated controls. The two populations were analyzed separately at 4dpi. Similar to the monoclonal tumors, HCT116 tumors reduced their proliferation index upon radiation (Fig. 3c, P = .017), apoptosis was induced (Fig. 3d, P = .0001), and the number of HCT116 cells per tumor reduced to half (Fig. 3e, P < .0001). In contrast, radiation did not impact on survival of Hke3 cells within the heterogenous tumor mass (Fig. 3c, P = .1; d-P = .5; e-P = .36). Interestingly, in control non-irradiated tumors, HCT116KRASG13D cells become dominant over Hke3KRAS-WT given their higher proliferative capacity, representing ~70% of the tumors (Fig. 3f). However, upon 1 × 25 Gy, HCT116 significantly reduced its frequency (P < .0001) in relation to controls, while Hke3 increased its presence (P < .0001).
Fig. 3.
Zebrafish xenografts have resolution to distinguish radiotherapy sensitivity in polyclonal xenografts. HCT116 (red) and Hke3 (green-fake color) cells were labelled with different lipophilic dyes and mixed in a proportion of 1:1 (a, b). Polyclonal CRC xenografts (1dpi) were submitted in vivo to SDRT (1 × 25 Gy) and compared to their respective controls (non-irradiated xenografts). The cellular response to the ionizing radiation was assessed by the quantification of mitotic figures % (c), apoptosis % (d), tumor size (total number cells, e) and the % of each clone present in each xenograft (f). The total number of analyzed xenografts is indicated in the images. Comparison of response to radiotherapy in monoclonal vs. polyclonal xenografts: apoptosis (g) and tumor size (h). All results, c–h, are expressed in AVG ± SEM and c, d, g and h were normalised to the respective controls. Each dot of c–h graphics represents one xenograft, resulting from one independent experiment. The statistical analysis was performed using Mann–Whitney-test, with P values *P ≤ .05, **P ≤ .01, ***P ≤ .001; NS, nonsignificant. a and b are 25x magnification confocal images, with the tumor surrounded by a white dashed line. Scale bar- 50 µm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Indeed, tumor clonal heterogeneity may represent a potential caveat for our assay, as we could miss response to treatment. To directly test this idea, we compared the response of monoclonal sensitive tumors (HCT116) to the polyclonal tumor overall response (mix HCT116+Hke3) without distinguishing the two populations. Strikingly, even in this hypothetical situation of a 1:1 mixture of both clones, we can detect the response of the sensitive clone in terms of apoptosis and tumor size (Fig. 3g, h, apoptosis P = .0056; tumor size P = .0307), although tumor size response was more heterogeneous.
All together, these results demonstrate that zebrafish xenografts and our new SDRT-25 Gy protocol completed in 4 days, can distinguish radiosensitive from radioresistant tumors even in the same xenograft. In heterogeneous tumors, our data illustrates how KRAS mutations, which provide a proliferative advantage, can be an “Achilles heel”, making cells more sensitive to therapy. On the other hand a minor resistant clone, unaffected by treatment, may become more representative, or even dominant.
3.4. Direct comparison between SDRT and fractionated radiotherapy protocols
SCRT or long-course fractionated (25-28 × 1.8 Gy) are recommended neoadjuvant treatment approaches delivered routinely in the clinic for selected patients with LARC cancer [32]. We therefore, sought to compare our SDRT protocol with one of the protocols given in the clinic, namely the SCRT-5 × 5 Gy, since the fractionated 25–28 × 1.8 Gy is not practical as an assay. HCT116 and Hke3 zebrafish xenografts were generated and randomly distributed in three experimental conditions at 1dpi: control (non-irradiated xenografts), SCRT (5 × 5 Gy), and SDRT (1 × 25 Gy). To directly compare the two protocols, the same time after treatment was given in all conditions: xenografts were analyzed 3 days post treatment (3dpT), i.e., at 4dpi for the SDRT and 8dpi for SCRT (Fig. 4a).
Fig. 4.
Single Dose Radiotherapy (1 × 25 Gy) regimen allows to discriminate responders and non-responders xenografts. The different radiotherapy schemes are illustrated in a. At 1dpi, CRC xenografts (HCT116 and Hke3) were submitted to the SDRT (1 × 25 Gy) in a single radiation session and to the SCRT (5 × 5 Gy), 5 Gy daily dose for five consecutive days, and compared with the controls (non-irradiated xenografts). In order to evaluate the radiobiology of both protocols, the same overall time following treatment ending (3dpT) was given: fixed larvae at 4dpi for SDRT and 8dpi for SCRT. The respective non-irradiated controls were also fixed (4 and 8dpi). Zebrafish xenografts were analyzed (b–q) and quantified for: mitotic figures % (r), apoptosis (activated Caspase3) (s), tumor size (total DAPI number) (t), and nuclear area size (total DAPI number/tumor area size (ROI)) (u). Results in r–u were normalised to the respective control and are expressed in AVG ± SEM. Mann–Whitney test was the statistical analysis performed. P values *P ≤ .05, **P ≤ .01, ***P ≤ .001, **** P ≤ .0001 and NS, nonsignificant. The nuclei dimension for the isogenic pair is represented in f–i and n–q, with white dashed line limitation and mitotic figures indicated by white arrows. The total number of analyzed xenografts are indicated (b–e and j–m) (25x magnification) and are the results from two independent experiments. Scale bar (50 µm).
In HCT116 radiosensitive tumors (Fig. 4b–i), both regimens reduced the number of cells undergoing mitosis (SDRT: 88% reduction, ****P < .0001, SCRT: 61% reduction, *P = .0194, Fig. 4r). However, only the SDRT showed a significant 2-fold induction of apoptosis (**P = 00018, Fig. 4s). Nevertheless, quantification of tumor size revealed an anti-tumor effect of both protocols: SDRT elicited a 59% reduction of tumor size (Fig. 4t, ****P < .0001) whereas SCRT induced 80% reduction (Fig. 4t, ***P < .0001). Therefore, our results suggest that the SDRT regimen exhibits a stronger effect than SCRT regarding proliferation and apoptosis, but not in tumor shrinkage.
IR increases the size of the nuclei due to chromatin modification, and arrest in G2 phase of the cell cycle [33]. Thus in order to investigate this, we analysed the nuclear area size of irradiated cells. Our results show that both RT protocols induced the enlargement of HCT116 cells nuclei (****P < .0001). However, the impact was superior in the fractionated regimen (SCRT AVG = 1.9 vs. SDRT AVG=1.4-fold increase, ****P < .0001, Fig. 4i).
Of note, in the Hke3 xenografts, we could not detect induction of apoptosis or reduction of tumor size, independently of the protocol used (Fig. 4j-m, s, t, note that AVG apoptosis is reducing with IR). Nevertheless, an increase in nuclear area size was detected with the SDRT protocol but not with the SCRT, suggesting that DNA damage and alterations on chromatin structure are being triggered. It also suggests that SDRT protocol is more efficient when used with resistant tumor cells. Similarly to the effect on the proliferation previously observed, there was a reduction of mitotic figures in both RT regimens (SCRT: 61% reduction, *P = .0194; SDRT: 88% reduction, ****P < .0001, Fig. 4r). The impact on cell proliferation and nuclear size shows that radiation damage is being inflicted. However, Hke3 cells are possibly more efficient at repairing or coping with the damage than HCT116. This is also reflected in differential amount of phosphorylation of γH2AX, a histone variant involved in the DNA damage response (Supplementary Fig. S3).
Overall, our results show that both RT protocols can elicit CRC cells proliferation arrest, trigger cell death through apoptosis, and a subsequent reduction of tumor size. Moreover, the nuclei enlargement observed in cancer cells is consistent with the described radiation effect in chromatin [33], which is considered an indicator of radiosensitivity. Thus, our data suggests that the SDRT (1 × 25 Gy) is an appropriate and efficient regimen to discriminate radiosensitive from radioresistant zebrafish xenografts, as well as convenient and less time-consuming.
3.5. Zebrafish Patient-Derived Xenografts respond to the SHD-RT protocol
To optimize and test the feasibility of our protocol, we first used surgical resected rectum cancer samples without in vitro passaging to generate zebrafish Patient-Derived Xenografts (zPDX) (Fig. 5). The average implantation rate obtained for the total of samples tested was ~43% (Fig. 5a, N = 19 patient samples). Next, we tested our radiotherapy protocol in two of these patient samples. Tumor tissue was prepared for injection (see Methods) and at 1dpi zPDX were subjected to the SDRT (1 × 25 Gy) protocol. Activated Caspase 3 and tumor size were analyzed at 4dpi (Fig. 5b–e). Although we were unable to observe a reduction of the tumor size, we detected a significant increase in apoptosis in both zPDX (zPDX#1, 3.6-fold increase, P = .0025; zPDX#2, 2.8-fold induction, P = .0006). These results gave us confidence to proceed to more challenging biopsy samples.
Fig. 5.
(a) Implantation rates of surgically resected rectal cancer samples. (b–e) 2dpf zebrafish were injected with rectum surgical resected samples (fluorescently labelled, magenta). One day after injection zPDX were submitted to SDRT (1 × 25 Gy) radiotherapy regimen. At 4dpi, apoptosis (d) and tumor size (e) were analyzed. **P ≤ .05, **P ≤ .01, ***P ≤ .001 and ns, nonsignificant. Scale bar-50 µm.
3.6. Comparison of patient response to nCRT treatment with their matching zPDX
In order to compare responses to radiotherapy between the zPDX-Avatars and the corresponding patient clinical response, we initiated a prospective observational study in patients subjected to neoadjuvant chemoradiotherapy (nCRT). For this end we started receiving rectal cancer biopsy samples, where a mirror was sent to pathology assessment. From the seven samples we had access in our institute from February to May 2019, three were necrotic, one was diagnosed as an adenoma, one did not implant and two we were able to proceed.
We present the findings from two patients with low rectal cancer (Fig. 6, P#B1 and P#B2) enrolled in the study, both with confirmed colorectal carcinoma and candidates to neoadjuvant treatment with nCRT under Multidisciplinary Board recommendations.
Fig. 6.
zPDX response to nCRT may predict patient clinical response to nCRT. (a) Design of the study. Before starting the nCRT, rectum biopsies were collected for pathology and to generate zPDX. Comparison of MRIs before and after nCRT treatment of Patient#B1 (b–c’’) and Patient#B2 (d–e’’). 2dpf zebrafish were injected with rectum biopsies from Patient#B1 (f–i) and Patient#B2 (j–m) (fluorescently labelled, white). At 1dpi, half of the zPDX were irradiated with SDRT (1 × 25 Gy) and treated with 5FU for 2 consecutive days. The other half was kept as control (non-irradiated and non-treated). Activated Caspase3 (h, l) and the tumor cell mass (i, m) were quantified. **P ≤ .05, **P ≤ .01, ***P ≤ .001 and NS, nonsignificant. Scale bar-50 µm.
A 69-year-old male, herein Patient#B1, presented with a low rectal tumor staged with Magnetic Resonance Imaging (MRI) as mrT4N1, showing involved mesorectal fascia and extramural venous invasion (Fig. 6b–b’’). The tumor was anteriorly invading the prostate gland and inferiorly the anal canal. An abdominal MRI showed four synchronous liver metastases at segments IV, V, transition of segments IV/VII and segment III (M1). Patient#B2, a 50-year-old male, presented also with a low rectal tumor in contact with the internal anal sphincter, staged as an mrT2N1 and no distant metastasis found when full staging was completed (M0) (Fig. 6d–d’’). Patient#B1 was treated with LC-CRT (RT plus Capecitabine) followed with immediate consolidation with CAPOX. An image guided – volumetric modulated arc therapy technique (IGRT-VMAT) was used, boosting the RT dose simultaneously to the areas of gross tumor detectable by planning images (simultaneous integrated boost-SIB). A total dose of 45 Gy was delivered to the pelvis and a SIB dose of 56 Gy to the involved areas with tumor. The RT dose was delivered concomitantly with Capecitabine 1250 mg/m2 (prodrug of 5FU), five days per week during 5 weeks. Two weeks after the end of LC-CRT, patient started with consolidation with CAPOX every three weeks. Patient#B2 was proposed for LC-CRT only and the same RT technique and doses were delivered concomitantly with Capecitabine. Both patients were treated at the Champalimaud Clinical center between February and March 2019.
In order to compare the patient clinical response with their matching zPDX, before starting the nCRT, fresh biopsies were collected for pathology and for the zebrafish clinical study. Samples were processed for injection (see Fig. S4 and Methods) and zPDX generated. One day after, successfully injected xenografts were randomly distributed into the two conditions (control and 25 Gy + 5FU). Importantly, the results from this experiment were obtained and analyzed before knowing the patient´s clinical response to treatment.
Upon CRT of zPDX#B1, we could not detect a statistically significant induction in Caspase3 levels (P = .06) or in the tumor size (Fig. 6f–i). In contrast, zPDX#B2 showed a significant induction of activated Caspase3 (1.6-fold induction, P = .0007), indicating that apoptosis was triggered by CRT (Fig. 6j–m).
Both patients were assessed for clinical response at around 10 weeks after the end of CRT. A pelvic MRI and a rectosigmoidoscopy (RS) were performed. Consistent with the zPDX results, Patient#B1 achieved an overall poor MRI response, with an MRI tumor regression grade (mrTRG) of 4, a Diffusion-Weighted Imaging (mrDWI) grade 3, and a decrease in tumor size (Dimensional Δ) around −32%. Endoluminal response was also poor, classified with a grade 3 (Fig. 6c–c’’). In contrast, Patient#B2 presented a major initial clinical response to CRT achieving an mrTRG1, mrDWI 1 with a Dimensional Δ of −76%, and the tumor was not palpable or visible by RS (Fig. 6e–e’’). Albeit with only two patients our results constitute a proof-of-principle that the zebrafish Avatar assay was able to predict the clinical response to CRT.
4. Discussion
The zebrafish Avatar model coupled with a single radiation exposure of 25 Gy is able to identify radioresistant CRC clones and its predictive accuracy in the clinical setting is being explored in a prospective study. Neoadjuvant radiotherapy followed by total mesorectal excision surgery is the standard of care in selected patients with LARC. Response to treatment varies widely with ~20% of tumors exhibiting resistance. Since pathological complete response (pCR) is associated with improved prognosis, alternative approaches such as the “watch and wait” [34] program or transanal local excision are being considered to avoid the adverse effects of surgery (urinary, bowel, and sexual dysfunction) [2]. In contrast, patients whose tumors are resistant are subjected to potentially unnecessary side effects, with a delay on more effective treatment strategies. Therefore, a biomarker that predicts response to nRT at an early time point has long been a critical need in clinical decision-making to tailor the adequate therapy for each individual patient. Despite its widespread use in cancer treatment, in fact, radiotherapy has not yet entered the era of precision medicine, with limited approaches to predict radiosensitivity and adjust dose based on biological differences between or within tumors. Early efforts to develop assays for measuring human tumor radiosensitivity involved clonogenic assays, such as the surviving fraction at 2 Gy (2SF2) [35]. The usefulness of these assays in prospective clinical trials, however, has proven to be limited due to the inherent tumor heterogeneity, which is inadequately accounted for by these models [36]. More recently, the advent of high-throughput profiling approaches have led to an attempt to associate gene expression signatures with tumor radiosensitivity. A signature of 10 genes related to SF2 was used to build a rank-based linear regression algorithm to predict radiosensitivity (a radiosensitivity index, RSI) [3]. The RSI has been shown to be prognostic in several tumor types cancer [37,38,3] constituting a very promising assay.
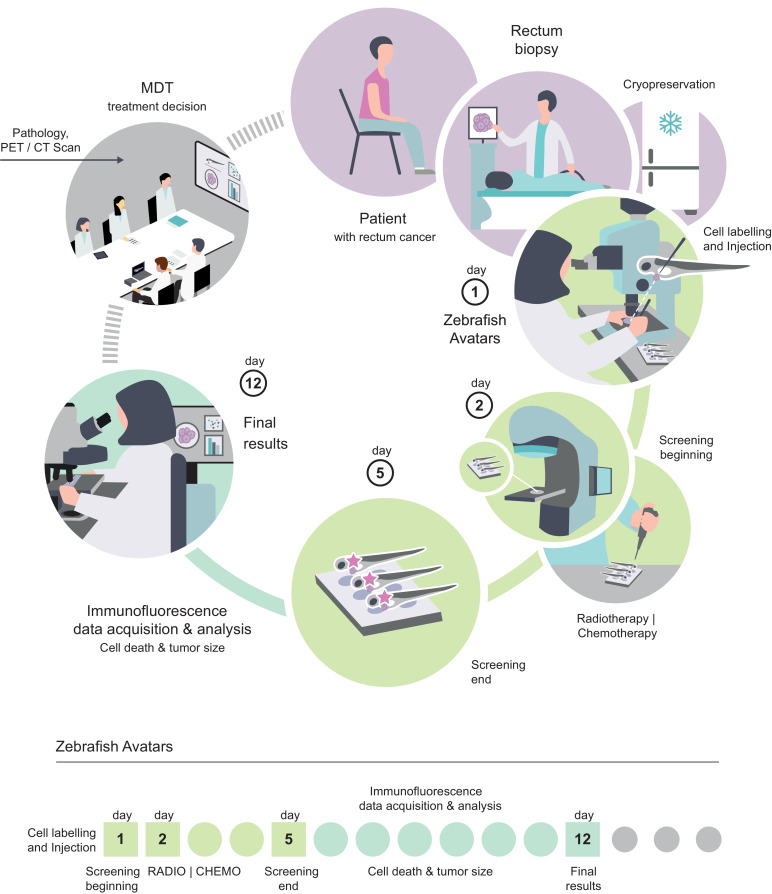
Several other molecular biomarkers, either tissue- or blood-based, have been proposed to predict response to nCRT at an early time point [2]. However, none has reached the clinical use due to lack of robustness or inherent practical limitations. Dayde and colleagues proposed there should be an integration of several biomarkers such as clinicopathological and imaging features, identification of mechanisms of tumor biology, to develop such a robust cost-effective molecular biomarker [2]. However, the vast tumor genetic heterogeneity found in CRC [39,40] may account for the difficulty in identifying a specific molecular biomarker of response. Therefore, a test that directly challenges in vivo the patient-derived tumor cells, independent of their genetic profile, may prove more robust and complement other strategies. Here, we tested whether the fast zebrafish larvae xenograft model could be used as an in vivo biomarker of response to radiotherapy. The zebrafish-larvae model provides a reduction of scale that enables to inject tumor cells directly into multiple zebrafish larvae (statistical analysis), without in vitro passaging and to obtain single cell resolution. This reduction of scale also allows a reduction of the time, enabling us to swiftly evaluate the impact of therapy on tumor cells, in a clinically relevant time-frame, prior to treatment. In addition, since we use freshly cryopreserved samples, it is possible to receive out-patient’ samples for a multi-center study in the future (Fig. 7).
Fig. 7.
Schematic illustration of the workflow and time-frame of the zebrafish Avatar assay. Illustration by Gil Costa.
5. Conclusion
We developed a SDRT (1 × 25 Gy) protocol delivered at one day post-injection to the zebrafish xenografts and analysis at 3 days post-radiation. We analyzed the impact of radiation on mitosis, apoptosis, tumor and nuclear area size, and showed that HCT116KRASG13D cells are sensitive to radiation, whereas their isogenic cells, Hke3KRASwt are resistant. This demonstrates that our assay is able to distinguish radiosensitive from radioresistant tumors, even in polyclonal-heterogeneous tumors. We performed proof-of-concept experiments that demonstrate the feasibility of using rectal cancer biopsies with promising results, suggesting that zebrafish Avatars may indeed predict clinical response to neoadjuvant therapy (see summary Supplementary Fig. S5). Preliminary patient clinical response data appears to correlate with induction of apoptosis upon treatment in their matching zPDX. In summary, this work opens a new avenue to study the predictive power of the zebrafish Avatar model as a radiotherapy sensor for future personalized medicine and potentially change clinical practice.
Declaration of Competing Interest
The authors declare no conflict of interest and nothing to disclose.
Acknowledgments
Funding
Champalimaud Foundation, Howard Hughes Medical Institute (HHMI), Congento (LISBOA-01-0145-FEDER-022170, co-financed by FCT/Lisboa2020) and FCT-PTDC/MEC-ONC/31627/2017.
Acknowledgments
We would like to thank Dr. Tania Gilot Carvalho and Dr. Mireia Castillo for pathology assessment and support. We thank the Surgery and Histopathology Units of Champalimaud Clinical center; the Champalimaud Fish Facility (C. Certal and J. Monteiro) for excellent animal care. We thank Marta Estrada for the critical reading of the manuscript. We thank Gil Costa for Fig. 7 illustration. We thank the Champalimaud Foundation, Howard Hughes Medical Institute (HHMI), Congento (LISBOA-01-0145-FEDER-022170, co-financed by FCT/Lisboa2020) and FCT-PTDC/MEC-ONC/31627/2017 for financial support.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2019.11.039.
Contributor Information
Miguel Godinho Ferreira, Email: miguel-godinho.ferreira@unice.fr.
Rita Fior, Email: rita.fior@research.fchampalimaud.org.
Appendix. Supplementary materials
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. Available from: [DOI] [PubMed] [Google Scholar]
- 2.Dayde D., Tanaka I., Jain R., Tai M.C., Taguchi A. Predictive and prognostic molecular biomarkers for response to neoadjuvant chemoradiation in rectal cancer. Int J Mol Sci. 2017;18(3):573. doi: 10.3390/ijms18030573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torres-Roca J.F. A molecular assay of tumor radiosensitivity: a roadmap towards biology-based personalized radiation therapy. Per Med. 2012;9(5):547–557. doi: 10.2217/pme.12.55. https://www.ncbi.nlm.nih.gov/pubmed/23105945 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagle P.W., Plukker J.T.M., Muijs C.T., van Luijk P., Coppes R.P. Patient-derived tumor organoids for prediction of cancer treatment response. Semin Cancer Biol. 2018;53:258–264. doi: 10.1016/j.semcancer.2018.06.005. [Internet] Available from: http://www.sciencedirect.com/science/article/pii/S1044579×18300464. [DOI] [PubMed] [Google Scholar]
- 5.Williams J.A. Using PDX for preclinical cancer drug discovery: the evolving field. J Clin Med. 2018;7(3):41. doi: 10.3390/jcm7030041. https://www.ncbi.nlm.nih.gov/pubmed/29498669 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pompili L., Porru M., Caruso C., Biroccio A., Leonetti C. Patient-derived xenografts: a relevant preclinical model for drug development. J Exp Clin Cancer Res. 2016;35(1):189. doi: 10.1186/s13046-016-0462-4. https://www.ncbi.nlm.nih.gov/pubmed/27919280 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torres-Roca J.F., Stevens C.W. Predicting response to clinical radiotherapy: past, present, and future directions. Cancer Control. 2008;15(2):151–156. doi: 10.1177/107327480801500207. Available from: [DOI] [PubMed] [Google Scholar]
- 8.Barriuso J., Nagaraju R., Hurlstone A. Zebrafish: a new companion for translational research in oncology. Clin Cancer Res. 2015;21(5):969–975. doi: 10.1158/1078-0432.CCR-14-2921. http://clincancerres.aacrjournals.org/content/21/5/969.abstract Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fazio M., Zon L.I. Fishing for answers in precision cancer medicine. Proc Natl Acad Sci USA. 2017;114(39):10306–10308. doi: 10.1073/pnas.1713769114. Epub 2017 Sep 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gnosa S., Capodanno A., Murthy R.V., Jensen L.D.E., Sun X.-F. AEG-1 knockdown in colon cancer cell lines inhibits radiation-enhanced migration and invasion in vitro and in a novel in vivo zebrafish model. Oncotarget. 2016;7(49):81634–81644. doi: 10.18632/oncotarget.13155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fior R., Póvoa V., Mendes R.V., Carvalho T., Gomes A., Figueiredo N. Single-cell functional and chemosensitive profiling of combinatorial colorectal therapy in zebrafish xenografts. Proc Natl Acad Sci USA. 2017;114(39):E8234–E8243. doi: 10.1073/pnas.1618389114. Epub 2017 Aug 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Letrado P., de Miguel I., Lamberto I., Díez-Martínez R., Oyarzabal J. Zebrafish: speeding up the cancer drug discovery process. Cancer Res. 2018;78(21):6048–6058. doi: 10.1158/0008-5472.CAN-18-1029. http://cancerres.aacrjournals.org/content/78/21/6048.abstract Available from: [DOI] [PubMed] [Google Scholar]
- 13.Haldi M., Ton C., Seng W.L., McGrath P. Human melanoma cells transplanted into zebrafish proliferate, migrate, produce melanin, form masses and stimulate angiogenesis in zebrafish. Angiogenesis. 2006;9:139–151. doi: 10.1007/s10456-006-9040-2. [DOI] [PubMed] [Google Scholar]
- 14.Marques I.J., Weiss F.U., Vlecken D.H., Nitsche C., Bakkers J., Lagendijk A.K. Metastatic behaviour of primary human tumours in a zebrafish xenotransplantation model. BMC Cancer. 2009;9(1):128. doi: 10.1186/1471-2407-9-128. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu J.-Q., Zhai J., Li C.-Y., Tan A.-M., Wei P., Shen L.-Z. Patient-derived xenograft in zebrafish embryos: a new platform for translational research in gastric cancer. J Exp Clin Cancer Res. 2017;36(1):160. doi: 10.1186/s13046-017-0631-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stoletov K., Kato H., Zardouzian E., Kelber J., Yang J., Shattil S. Visualizing extravasation dynamics of metastatic tumor cells. J Cell Sci. 2010;123(13):2332–2341. doi: 10.1242/jcs.069443. http://jcs.biologists.org/content/123/13/2332.abstract Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicoli S., Ribatti D., Cotelli F., Presta M. Mammalian tumor xenografts induce neovascularization in zebrafish embryos. Cancer Res. 2007;67(7):2927–2931. doi: 10.1158/0008-5472.CAN-06-4268. http://cancerres.aacrjournals.org/content/67/7/2927.abstract Available from: [DOI] [PubMed] [Google Scholar]
- 18.Zhao C., Wang X., Zhao Y., Li Z., Lin S., Wei Y. A novel xenograft model in zebrafish for high-resolution investigating dynamics of neovascularization in tumors. PLoS One. 2011;6(7):e21768. doi: 10.1371/journal.pone.0021768. 2011/07/13e21768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676. doi: 10.1038/nmeth.2019. [Internet] Jun 28 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li R., Liao G., Yin G., Wang B., Yan M., Lin X. Ionizing radiation blocks hair cell regeneration in zebrafish lateral line neuromasts by preventing wnt signaling. Mol Neurobiol. 2018;55(2):1639–1651. doi: 10.1007/s12035-017-0430-9. [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 21.Xiao Y., Zheng X., Huang A., Liu T., Zhang T., Ma H. Deficiency of 53BP1 inhibits the radiosensitivity of colorectal cancer. Int J Oncol. 2016;49(4):1600–1608. doi: 10.3892/ijo.2016.3629. [DOI] [PubMed] [Google Scholar]
- 22.Ramzan Z., Nassri A.B., Huerta S. Genotypic characteristics of resistant tumors to pre-operative ionizing radiation in rectal cancer. World J Gastrointest Oncol. 2014;6(7):194–210. doi: 10.4251/wjgo.v6.i7.194. https://www.ncbi.nlm.nih.gov/pubmed/25024812 [Internet] 2014/07/15 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martins-Neves S.R., Lopes Á.O., do Carmo A., Paiva A.A., Simões P.C., Abrunhosa A.J. Therapeutic implications of an enriched cancer stem-like cell population in a human osteosarcoma cell line. BMC Cancer. 2012;12:139. doi: 10.1186/1471-2407-12-139. https://www.ncbi.nlm.nih.gov/pubmed/22475227 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shirasawa S., Furuse M., Yokoyama N., Sasazuki T. Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science. 1993;260:85–88. doi: 10.1126/science.8465203. http://science.sciencemag.org/content/260/5104/85.abstract Available from: [DOI] [PubMed] [Google Scholar]
- 25.Glynne-Jones R., Wyrwicz L., Tiret E., Brown G., Rödel C., Cervantes A. Rectal cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29(Supplement_4):iv263. doi: 10.1093/annonc/mdy161. [DOI] [PubMed] [Google Scholar]
- 26.Sundahl N., Duprez F., Ost P., De Neve W., Mareel M. Effects of radiation on the metastatic process. Mol Med. 2018;24(1):16. doi: 10.1186/s10020-018-0015-8. [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martins I., Raza S.Q., Voisin L., Dakhli H., Allouch A., Law F. Anticancer chemotherapy and radiotherapy trigger both non-cell-autonomous and cell-autonomous death. Cell Death Dis. 2018;9(7):716. doi: 10.1038/s41419-018-0747-y. http://europepmc.org/articles/PMC6006149 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bodo S., Campagne C., Thin T.H., Higginson D.S., Vargas H.A., Hua G. Single-dose radiotherapy disables tumor cell homologous recombination via ischemia/reperfusion injury. J Clin Invest. 2019;129(2):786–801. doi: 10.1172/JCI97631. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marill J., Mohamed Anesary N., Paris S. DNA damage enhancement by radiotherapy-activated hafnium oxide nanoparticles improves cGAS-STING pathway activation in human colorectal cancer cells. Radiother Oncol. 2019 doi: 10.1016/j.radonc.2019.07.029. [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 30.Terranova-Barberio M., Pecori B., Roca M.S., Imbimbo S., Bruzzese F., Leone A. Synergistic antitumor interaction between valproic acid, capecitabine and radiotherapy in colorectal cancer: critical role of p53. J Exp Clin Cancer Res. 2017;36(1):177. doi: 10.1186/s13046-017-0647-5. https://www.ncbi.nlm.nih.gov/pubmed/29212503 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim J.Y., Park S.G., Kim K.-S., Choi Y.H., Kim N.K. The Krüppel-like factor (KLF5) as a predictive biomarker in preoperative chemoradiation therapy for rectal cancer. Ann Surg Treat Res. 2019;97(2):83–92. doi: 10.4174/astr.2019.97.2.83. http://europepmc.org/articles/PMC6669127 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Cutsem E., Cervantes A., Adam R., Sobrero A., Van Krieken J.H., Aderka D. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol. 2016;27(8):1386–1422. doi: 10.1093/annonc/mdw235. Available from: [DOI] [PubMed] [Google Scholar]
- 33.Meyer B., Fabbrizi M.R., Raj S., Zobel C.L., Hallahan D.E., Sharma G.G. Histone H3 lysine 9 acetylation obstructs atm activation and promotes ionizing radiation sensitivity in normal stem cells. Stem Cell Rep. 2016;7(6):1013–1022. doi: 10.1016/j.stemcr.2016.11.004. https://www.ncbi.nlm.nih.gov/pubmed/27974220 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bernier L., Balyasnikova S., Tait D., Brown G. Watch-and-Wait as a therapeutic strategy in rectal cancer. Curr Colorectal Cancer Rep. 2018;14(2):37–55. doi: 10.1007/s11888-018-0398-5. https://www.ncbi.nlm.nih.gov/pubmed/29576755 [Internet] 2018/03/07 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buffa F.M., Davidson S.E., Hunter R.D., Nahum A.E., West C.M.L. Incorporating biologic measurements (SF2, CFE) into a tumor control probability model increases their prognostic significance: a study in cervical carcinoma treated with radiation therapy. Int J Radiat Oncol Biol Phys. 2001;50(5):1113–1122. doi: 10.1016/s0360-3016(01)01584-x. [Internet] Aug 1 Available from: [DOI] [PubMed] [Google Scholar]
- 36.Adamus-Górka M., Mavroidis P., Lind B.K., Brahme A. Comparison of dose response models for predicting normal tissue complications from cancer radiotherapy: application in rat spinal cord. Cancers (Basel) 2011;3(2):2421–2443. doi: 10.3390/cancers3022421. https://www.ncbi.nlm.nih.gov/pubmed/24212817 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eschrich S.A., Fulp W.J., Pawitan Y., Foekens J.A., Smid M., Martens J.W.M. Validation of a radiosensitivity molecular signature in breast cancer. Clin Cancer Res. 2012;18(18):5134–5143. doi: 10.1158/1078-0432.CCR-12-0891. https://www.ncbi.nlm.nih.gov/pubmed/22832933 2012/07/25 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scott J.G., Berglund A., Schell M.J., Mihaylov I., Fulp W.J., Yue B. A genome-based model for adjusting radiotherapy dose (GARD): a retrospective, cohort-based study. Lancet Oncol. 2017;18(2):202–211. doi: 10.1016/S1470-2045(16)30648-9. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A., Kinzler K.W. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. http://science.sciencemag.org/content/339/6127/1546.abstract Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Almendro V., Marusyk A., Polyak K. Cellular heterogeneity and molecular evolution in cancer. Annu Rev Pathol Mech Dis. 2013;8(1):277–302. doi: 10.1146/annurev-pathol-020712-163923. Available from: [DOI] [PubMed] [Google Scholar]
- 41.Taylor A.M., Zon L.I. Zebrafish tumor assays: the state of transplantation. Zebrafish. 2009;6(4):339–346. doi: 10.1089/zeb.2009.0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.