Abstract
Using different chromatographic methods, four new compounds were isolated from the aerial parts of Suaeda monoica (Chenopodiaceae) along with 2-hydroxy-1-naphthoic acid (SCM-3). The structures of the new compounds were established as 6′-hydroxy-10′-geranilanyl naphtha-1-oate (SMC-1), 4,4,8β,10β-Tetramethyl-9β-isobutanyl decalin-13-ol-13-O-β-D-xylopyranoside (SCM-2), 6′-(2-hydroxynaphthalen-3-yl) hexanoic acid (SCM-4) and 1′-(2-Methoxy-3-naphthyl)-4′-(2′'-methylbenzoyl)-n-butane (SMC-5) by IR, EIMS and NMR (1 & 2D) analyses. All compounds (50 μg/mL) were tested for cell proliferative potential on cultured human liver cell HepG2 cells by MTT assay. The results revealed a marked cell proliferative potential of all compounds (1.42–1.48 fold) as compared to untreated control. The results of molecular docking and binding with specific proteins such as PTEN (Phosphatase and Tensin homolog) and p53 also justify the cell proliferative potential of the isolated compounds. Glide program with Schrodinger suit 2018 was used to evaluate the binding between SMC compounds and proteins (PTEN and p53). The binding affinity of all compounds was in order of 104–105 M−1 towards both PTEN and p53. All the SMC compounds have been found to bind at the active site of PTEN, thereby may prevent the binding of phosphatidylinositiol 3,4,5-triphosphate (PI3P). In the locked position, PTEN would not be able to hydrolyze PI3P and hence the PI3P regulated signaling pathway remains active. Similarly, SMC molecules were found to interact with the amino acid residues (Ser99, Thr170, Gly199, and Asp224) which are critically involved in the formation of tetrameric p53. The blockage of p53 to attain its active conformation thus may prevent the recruitment of p53 on DNA and hence may promote cell proliferation.
Keywords: Suaeda monoica, Cell-proliferation, Molecular docking, β-Naphthol, Caproic acid, Chenopodiaceae, HepG2 cells
1. Introduction
Genus Suaeda belongs to halophytes category from the family Chenopodiaceae, consisting of around 75 species widely distributed around seacoasts, salty marshes and deserts (Boulos, 1991). Suaeda monoica Forssk. ex. J.F. Gmel (SMC) is a bushy dark green succulent leaved shrub with pale green flowers. In Saudi Arabia genus Suaeda is represented by nine species (Boulos, 1999) out of which five are found in Al Jouf area (S. aegyptica, S. vera, S. vermiculata, S. fruiticosa and S. mollis (Al-Hassan, 2006) while S. monoica distributed in the southern lowlands and sometimes forming a tree of approx. 3 m height (Collenette, 1999).
Mangrove plants have already established their identity as one of the useful category of medicinal plants in traditional system of medicine (Premnathan et al., 1992, Premnathan et al., 1996). Antiviral, wound healing and hepatoprotective potential of S. monoica leaves have also been reported (Ravikumar et al., 2010, Ravikumar et al., 2011a). Earlier reports proved the medicinal potential of S. monoica against Plasmodium falciparum, insects and microbes (Chandrasekaran et al., 2008, Kassem et al., 2003, Ravikumar et al., 2011b). The existing literature reports the presence of a broad range of chemical constituents in S. monoica such as proteins, tannins, resins, terpenoids, glycosides and flavonoids (Lakshmanan et al., 2013). From the phytochemical point of view, the major classes of compounds reported in S. monoica are different fatty acids, triterpenoids and sterols (Chandrasekaran et al., 2008, Bandaranayake, 1998, Yezhelyev et al., 2006). A polysaccharide which is active against human immune deficiency virus also reported from S. monoica (Premnathan et al., 1992, Padmakumar and Ayyakkannu, 1997). The recent phytochemical findings suggest the presence of gallic acid, catechin, caffeic acid, syringic acid, rutin, coumaric acid, vanillin and quercetin in methanolic extract of aerial parts of S. monoica and S. pruinosa (Elsharabasy et al., 2019). Elsharabasy et al also reported the presence of several amino acids in both the species. These amino acids include histidine, arginine, threonine, alanine, proline, tyrosine, methionine, cysteine, isoleucine, leucine, lysine, aspartic acid, glutamic acid and glycine in S. monoica and S. pruinosa. Several fatty acids such as lauric acid, myristoleic acid, palmitic acid, palmitoleic acid, stearic acid, oleic acid, linoleic acid and linolenic acid were also identified in S. monoica and S. pruinosa by Elsharabasy et al., 2019. A norsesquaterpenol named as 13,17-octahydropentalene-4,4,10,23-tetramethyl-17,21-diisopropyltetradecahydrocyclo-[a]-phenanthrene-(14), 20(23), 21(30)-trien-5a-ol and a monocyclic triterpenoid named as [1,4,4-trimethylcyclopent- 1(5)-enyl]-9,10,17,21-tetramethyl-9a-ol-16a (17a)-epoxy heptadecan-6,10-dione along with 3-epi-lupeol and 4-cyclopentylpyrocatechol were reported by Al-Said et al., 2017 in ethanolic extract of aerial parts of S. monoica.
Natural products play a vital role in healing, protection and rejuvenation of the tissues probably by enhancing the cell division and ultimately leading to cell proliferation. Natural products have well established history of healing and regenerative potential for various tissues in human body (Hasan and Al Sorkhy, 2014). Several herbal drugs have been utilized in the treatment of disease arise due to decline of stem cell proliferation and utilized for regeneration and rehabilitation of tissues by promoting cell proliferation. The essential obligation of cell proliferation is in tissue regeneration, repair and aging (Al-Said et al., 2017). Herbal extracts also stimulated skin cells growth like hyperforin from Hypericum perforatum stimulated keratinocyte differentiation, polysaccharides in Actinidia chinensis stimulated human keratinocytes and other herbal extracts induce dermal papilla cells proliferation (Ashtiani et al., 2012). Taking into consideration the demand of cell proliferative agents, this study was planned to explore cell proliferative effect of various phytoconstituents isolated from S. monoica because till now S. monoica phytoconstituents have not been extensively explored.
The cell proliferative potential of S. monoica was investigated against HepG2 cells by MTT assay. Furthermore, a bioassay guided fractionation and isolation was carried out to isolate four new compounds (SMC-1, -2, -4, -5) along with a known compound (SMC-3). In addition, molecular docking studies by binding them to PTEN and p53 proteins using AutoDock 4.2 were performed to assure the cell proliferation effect (AlAjmi et al., 2018).
2. Materials and methods
2.1. Instruments
Infrared spectra were recorded from ATI Mattson genesis series Fourier transform (FT-IR) infrared spectrophotometer. Bruker Avance DRX 500 MHz spectrometer (Massachusetts, United States) was used to obtain 1D and 2D NMR spectra at 500 and 125 MHz for 1H and 13C, respectively. Bruker Bioapex FT–MS (Massachusetts, United States) was used to obtain the EI mass spectra.
2.2. Plant material
The collection of S. monoica (aerial parts) was done in March 2017 from Aqabaat Al-Makhwah, after tunnel # 13, Saudi Arabia. The Identification of the plant was done by Field Taxonomist, Pharmacognosy Department, College of Pharmacy, KSU, Riyadh. Voucher Specimen (Voucher # 15235) was deposited in herbarium of Pharmacognosy Department.
2.3. Extraction and isolation of phytoconstituents
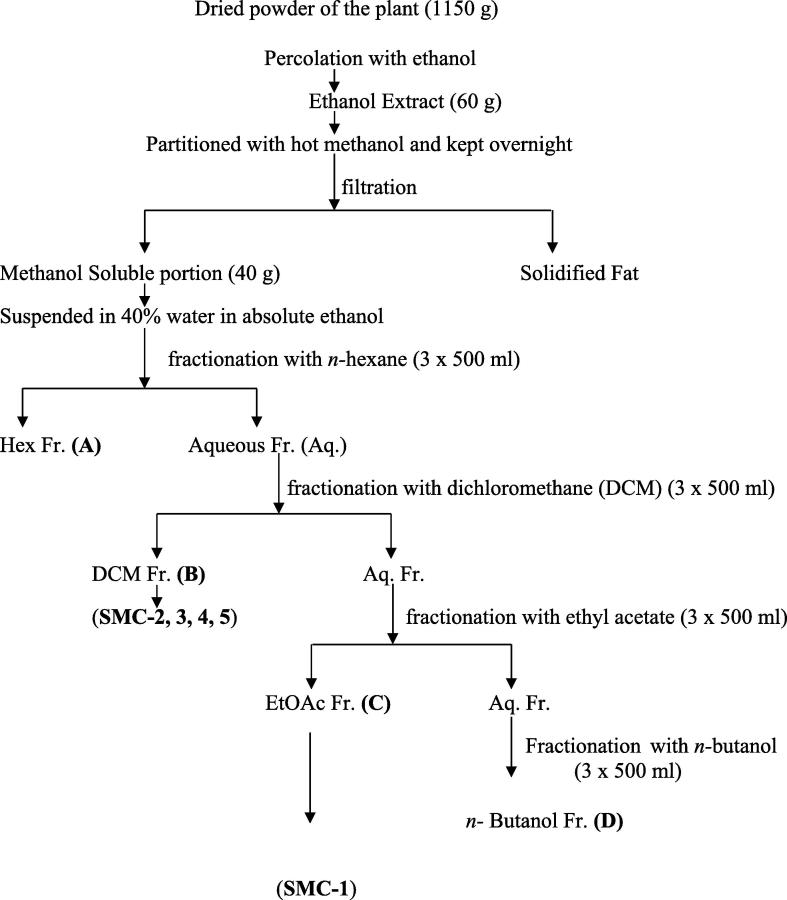
The aerial parts of S. monoica were dried in air and ground to coarse powder (1150 g) and, then extracted by percolation using ethanol (95%) as extractive solvent. Extraction was repeated for three times (3 × 2 L) to complete exhaustion of the plant material, then all cuts were combined and concentrated in rotary vacuum evaporator to get 60 g of a dried semi solid extract. The total amount of the extract was dissolved in hot MeOH and left for overnight, then filtered off to afford a dry soluble portion of 40 g and a fatty-nature solid. This 40 g of the extract was processed as mentioned in the scheme of isolation. After multiple and stepwise partitioning of the total amount of the extract, fractions A (n-hexane), B (dichloromethane), C (ethyl acetate) and D (n-butanol) were produced (Scheme 1). The ethyl acetate-fraction (fraction C, 1.2 g) was separated utilizing column chromatography on sephadex LH-20 column (20 mm i.d. × 400 mm) to give 5 fractions (C1-C5). The elution was performed with methanol: water (9:1). Fraction C4 showed a major compound, which was purified on a sephadex LH-20 column) (15 mm i.d. × 600 mm) with methanol: water (9:1) to afford SMC-1 (25 mg). The dichloromethane-fraction (fraction B, 1.8 g) was subjected to a pre-packed silica gel column (35 mm i.d. × 400 mm) using a gradient of dichloromethane: methanol (100:1) to pure methanol as a mobile phase to afford 10 fractions (B1-B10). Fraction B3 was eluted with dichloromethane and methanol (19:1) using sephadex LH-20 column (10 mm i.d. × 600 mm) to afford SMC-2 (14 mg). The purification of the major compound in fraction B6 utilizing sephadex LH-20 column (10 mm i.d. × 600 mm) with dichloromethane and methanol (9:1) as an eluent, gave SMC-3 (11 mg). The isolation of SMC-4 (6 mg) was achieved from fraction B7 utilizing sephadex LH-20 column (10 mm i.d. × 600 mm) with dichloromethane and methanol (9:1) as a solvent. SMC-5 (8 mg) was isolated from fraction B8 by eluting with dichloromethane and methanol (5:1) using sephadex LH-20 column (10 mm i.d. × 600 mm).
Scheme 1.
Isolation scheme of SMC compounds.
2.3.1. 6′-hydroxy-10′-geranilanyl naphtha-1-oate (SMC-1)
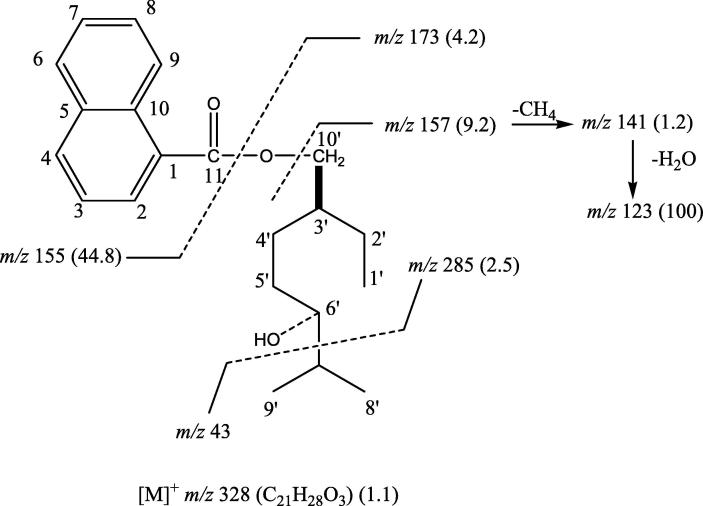
It was a yellow semisolid compound isolated from the ethyl acetate fraction. IR υmax (KBr): 3360, 2964, 1737, 1614, 1519, 1434, 1361, 1197, 1064, 979, 783, 745 cm−1; 1H NMR (CD3OD): δ 6.62 (1H, m, H-2), 6.60 (2H, m, H-6, H-9), 6.58 (1H, m, H-4), 6.45 (1H, m, H-3), 6.41 (1H, m, H-7), 6.38 (1H, m, H-8), 4.01 (1H, d, J = 7.5 Hz, H2-10′a), 3.95 (1H, d, J = 7.0 Hz, H2-10′b), 3.52 (1H, m, w1/2 = 7.5 Hz, H-6′β), 2.63 (2H, m, H2-5′), 2.54 (1H, m, H-3′), 2.50 (1H, m, H-7′), 1.88 (2H, m, H2-4′), 1.38 (2H, m, H-6, H2-2′), 1.25 (3H, d, J = 6.5 Hz, Me-8′), 1.23 (3H, d, J = 6.3 Hz, Me-9′), 0.84 (3H, t, J = 6.5 Hz, Me-1′); 13C NMR (CD3OD): δ 130.40 (C-1), 121.25 (C-2), 121.32 (C-3), 131.80 (C-4), 132.63 (C-5), 126.52 (C-6), 117.01 (C-7), 116.32 (C-8), 130.82 (C-9), 131.16 (C-10), 169.13 (C-11), 9.65 (C-1′), 22.42 (C-2′), 49.21 (C-3′), 30.26 (C-4′),36.21 (C-5′), 70.10 (C-6′), 39.66 (C-7′), 20.85 (C-8′), 21.13 (C-9′), 64.62 (C-10′); EIMS m/z (rel. int.): 328 [M] (C21H28O3) (1.1), 285 (2.5), 173 (4.2), 157 (9.2), 155 (44.8), 141 (1.2), 123 (100).
(C21H28O3) (1.1), 285 (2.5), 173 (4.2), 157 (9.2), 155 (44.8), 141 (1.2), 123 (100).
2.3.2. 4,4,8β,10β-Tetramethyl-9β-isobutanyl-13-ol-13-β-D-xylopyranoside (SMC-2)
It was a colorless solid compound isolated from dichloromethane fraction. IR υmax (KBr): 3415, 3392, 2958, 2867, 1458, 1367, 1164, 1074, 1024 cm−1; 1H NMR (DMSO‑d6): δ 4.23 (1H, d, J = 7.5 Hz, H-1′), 3.51 (1H, dd, J = 6.2, 1.1 Hz, H2-13a), 3.42 (1H, dd, J = 6.3, 1.1 Hz, H2-13b), 3.40 (1H, m, H-4′), 3.23 (1H, dd, H-2′), 3.09 (1H dd, J = 7.1, 1.2 H2-5′a), 3.06 (2H,dd, J = 6.9, 1.2 Hz, H2-5b'), 3.02 (1H, dd, H-3′), 2.89 (1H, m, H-12), 2.21 (1H, m, H-9), 1.89 (2H, m, H2-1), 1.80 (2H, m, H2-7), 1.51 (1H, m, H-5), 1.47 (2H, m, H2-2), 1.20 (2H, m, H2-3), 1.14 (2H, m, H2-6), 0.96 (2H, m, H2-11), 0.90 (3H, s, Me-17), 0.82 (3H, s, Me-15), 0.76 (3H, d, J = 6.1 Hz, Me-18), 0.73 (3H, d, J = 6.5 Hz, Me-16), 0.65 (3H, s, Me-14); 13C NMR (DMSO‑d6): δ 35.91 (C-1), 32.63 (C-2), 30.22 (C-3), 46.22 (C-4), 57.18 (C-5), 29.75 (C-6), 42.35 (C-7), 45.08 (C-8), 56.23 (C-9), 35.18 (C-10), 25.48 (C-11), 49.56 (C-12), 61.04 (C-13), 11.74 (C-14), 18.90 (C-15), 19.06 (C-16), 19.68 (C-17), 18.58 (C-18), 101.25 (C-1′), 73.82 (C-2′), 76.71 (C-3′), 70.04 (C-4′), 66.15 (C-5′); EIMS m/z (rel. int.): 398 [M] (C23H42O5) (100), 383 (22.7), 368 (6.3), 234 (3.2), 205 (8.3), 163 (5.2), 149 (28.5), 133 (30.2).
(C23H42O5) (100), 383 (22.7), 368 (6.3), 234 (3.2), 205 (8.3), 163 (5.2), 149 (28.5), 133 (30.2).
2.3.3. 6′-(2-hydroxynaphthalen-3-yl)hexanoic acid (SMC-4)
It was a yellow amorphous solid compound isolated from dichloromethane fraction. IR υmax (KBr): 3407, 3209, 2971, 2852, 1677, 1521, 1471, 1355, 1296, 1272, 1228, 1108, 775 cm−1; 1H NMR (CD3OD): δ 6.95 (1H, dd, J = 7.3, 2.0 Hz, H-9), 6.73 (1H, s, H-1), 6.69 (1H, d, J = 8.0, 1.3 Hz, H-6), 6.60 (1H, s, H-4), 6.45 (1H, m, H-8), 6.02 (1H, m, H-7), 2.86 (2H, t, J = 7.5 Hz, H2-6′), 2.42 (2H, t, J = 7.5 Hz, H2-2′), 2.29 (2H, m, H2-5′), 1.58 (2H, m, H2-4′), 1.27 (2H, m, H2-3′); 13C NMR (CD3OD): δ 124.80 (C-1), 164.56 (C-2), 131.12 (C-3), 127.10 (C-4), 129.10 (C-5), 120.62 (C-6), 117.02 (C-7), 118.14 (C-8), 127.82 (C-9), 131.70 (C-10), 178.37 (C-1′), 38.66 (C-2′), 26.04 (C-3′), 30.02 (C-4′), 35.10 (C-5′), 52.10 (C-6′); EIMS m/z (rel. int.): 258 [M] (C16H18O3) (2.3).
(C16H18O3) (2.3).
2.3.4. 1′-(2-Methoxy-3-naphthyl)-4′-(2′'-methylbenzoyl)-n-butane (SMC-5)
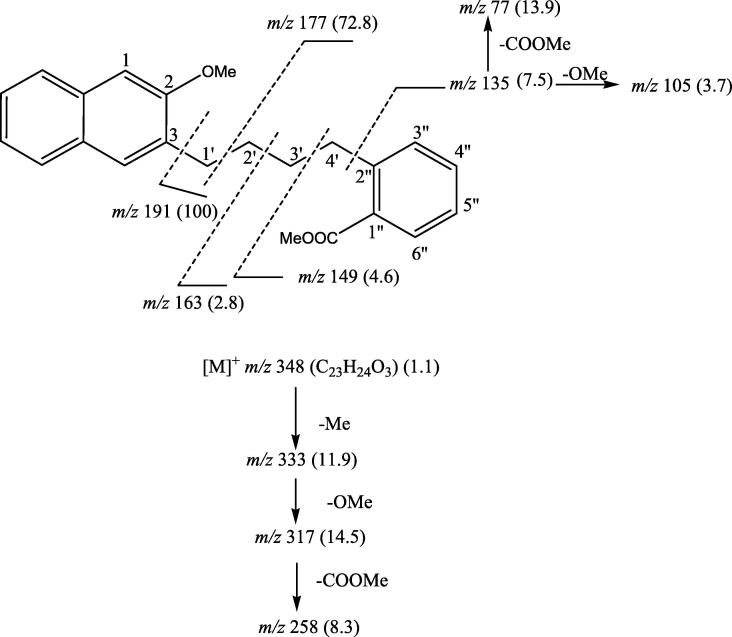
It was a yellow crystalline compound isolated from dichloromethane fraction. IR υmax (KBr): 2927, 2851, 1733, 1637, 1596, 1515, 1269, 1031, 979, 848 cm−1; 1H NMR (CD3OD): δ 7.46 (1H, dd, J = 10.5, 2.3 Hz, H-6′'), 7.12 (1H, s, H-1), 7.06 (1H, dd, J = 9.0, 1.0 Hz, H-3′'), 7.04 (1H, dd, J = 8.5, 1.0 Hz, H-6), 6.82 (1H, dd, J = 8.1, 1.5 Hz, H-9), 6.75 (2H, m, H-7, H-4′′), 6.45 (2H, m, H-8, H-5′′), 6.41 (1H, s, H-4), 3.85 (3H, s, OCOMe), 3.46 (2H, t, J = 7.5 Hz, H2-1′), 3.37 (3H, s, OMe), 3.33 (2H, t, J = 7.3 Hz, CH2-4′), 2.78 (2H, m, H2-3′), 1.26 (2H, m, H2-2′); 13C NMR (CD3OD): δ 123.43 (C-1), 156.61 (C-2), 123.38 (C-3), 128.01 (C-4), 130.32 (C-5), 121.02 (C-6), 116.41 (C-7), 118.81 (C-8), 128.21 (C-9), 130.95 (C-10), 49.67 (C-1′), 35.73 (C-2′), 42.59 (C-3′), 42.71 (C-4′), 132.24 (C-1′'), 130.48 (C-2′'), 128.37 (C-3′'), 118.75 (C-4′'), 118.66 (C-5′'), 132.19 (C-6′'), 169.47 (C-7′'), 56.57 (OCOMe), 50.01 (OMe); EIMS m/z (rel. int.): 348 [M] (C23H24O3) (1.1), 333 (11.9), 317 (14.5), 258 (8.3), 191 (100), 177 (72.8), 163 (2.8), 149 (4.6), 135 (7.5), 120 (43.9), 105 (3.7), 77 (13.9).
(C23H24O3) (1.1), 333 (11.9), 317 (14.5), 258 (8.3), 191 (100), 177 (72.8), 163 (2.8), 149 (4.6), 135 (7.5), 120 (43.9), 105 (3.7), 77 (13.9).
2.3.5. 2-Hydroxy-1-naphthoic acid (SMC-3)
It was a yellow amorphous solid compound isolated from dichloromethane fraction with m. p. 220–222 °C. IR υmax (KBr): 3377, 3221, 2948, 2851, 1670, 1611, 1512, 1448, 1328, 1313, 1244, 1216, 1172, 977, 833 cm−1; 1H NMR (CD3OD): δ 7.59 (1H, d, J = 9.7 Hz, H-3), 7.44 (1H, d, J = 9.0 Hz, H-6), 7.42 (1H, d, J = 9.0 Hz, H-9), 6.84 (1H, m, H-7), 6.82 (1H, m, H-8), 6.60 (1H, s, OH), 6.30 (1H, d, J = 9.7 Hz, H-4); 13C NMR (CD3OD): δ 131.28 (C-1), 160.93 (C-2), 131.85 (C-3), 131.21 (C-4), 131.05 (C-5), 127.33 (C-6), 115.56 (C-7), 116.26 (C-8), 127.53 (C-9), 131.19 (C-10), 171.49 (C-11); EIMS m/z (rel. int.): 188 [M] (C11H8O3) (1.1).
(C11H8O3) (1.1).
2.4. Cell proliferation assay
Human hepatoblastoma cells, HepG2 were maintained in RPMI-1640 medium (Gibco, USA), supplemented with 10% bovine serum (Gibco, USA) and 1 × penicillin–streptomycin mixture (Invitrogen, USA) and incubated at 37 °C with 5% CO2 supply in a humid chamber. HepG2 cells (0.5 × 105/well) were seeded in a 96-well flat-bottom plate (Becton-Dickinson Labware, USA) and incubated overnight. Stock solutions of compounds (SMC-1, SMC-2, SMC-3, SMC-4 and SMC-5; 1 mg, each) were prepared by first dissolving in 50 μL DMSO and then in RPMI medium (1 mg/mL; DMSO > 0.1%, final), followed by reconstitution of four working concentration (50, 25, 12.5 and 6.25, μg/mL) in the medium. DMSO vehicle or untreated control was also included.
For cell proliferation assay (TACS MTT Cell Proliferation Assay Kit, Tervigen, USA), the cells were treated with the four different doses of SMC-compounds, including control in triplicate. After 48 h of incubation, MTT reagent (10 μL/well) was added to each well and incubated at room-temperature for 4 h in dark. Immediately upon appearance of purple color, detergent solution (100 μL/well) was added and the cells further incubated for 1.5 h at 37 °C. The optical density (OD; λ = 570 nm) was measured (Microplate reader ELx800; BioTek, USA). Non-linear regression analysis was performed (Excel software 2010; Microsoft Corp., USA) to determine the cell proliferation fraction using the following equation:
where ODs, ODb and ODc are the absorbance of sample, blank and negative control, respectively. The data were presented as fold-increase in cell proliferation in relation to the untreated control (Al-Said et al., 2017).
2.5. Molecular docking studies
Interaction of the isolated compounds (SMC 1–5) with cell signaling molecules like PTEN and p53 was determined by performing molecular docking with the help of different modules of Schrodinger suite (Schrodinger, LLC, NY, USA) as described previously (AlAjmi et al., 2018). Briefly, the two-dimensional structures of all the compounds were drawn in 2D sketcher and prepared for docking in LigPrep (LigPrep, Schrodinger, LLC, NY, USA) by generating different ionization states of each ligand at pH 7.0 ± 2.0 with the help of Epik-module (Epik, Schrodinger 2018, LCC, NY, USA) followed by desaltation. Further, a maximum of 32 stereoisomers for each ligand was generated and the energy of each stereoisomer was minimized using OPLS3e forefield. The 3D coordinates of PTEN and p53 core domain were retrieved from RCSB database available at https://www.rcsb.org/ with the PDB IDs 5BZX (2.50 Å resolution) and 4HJE (1.91 Å resolution) respectively (Chen et al., 2013, Lee et al., 2015). Protein structures were processed and optimized for docking after removing heterogeneous molecules such as water and any bound ligand molecule using protein preparation wizard of Glide (Glide, Schrodinger, LLC, NY, USA). Moreover, missing hydrogen atoms were added, proper bond orders were assigned, zero bond order was assigned to disulfide bonds. Prime (Prime, Schrodinger, LLC, NY, USA) was employed to add any missing side chains and/or missing loops before optimizing the protein for hydrogen-bond network. Finally, energy of the protein was minimized using OPLS3e forecefield. The grid box of 56 × 56 × 56 Å dimensions was generated for PTEN core domain by selecting the bound ligand (tartaric acid) as the centroid of the grid box. and the Solis and Wets local search methods were employed to perform molecular docking. For p53, SiteMap (SiteMap, Schrodinger, LLC, NY, USA) was employed to identify the most probable binding site(s) of p53. A total number of three binding sites (I-III) have been identified for p53. The grid box was generated by selecting the residues of the respective binding sites as the centroid of the corresponding grid boxes with a dimension of 64 × 64 × 64 Å, 64 × 64 × 64 Å, and 56 × 56 × 56 Å for binding sites I, II and III respectively. The binding affinity (Kd) was calculated from Gibb’s free energy (ΔG) using the following relation (AlAjmi et al., 2018).
where, R (=1.987 cal/mol/K) is the universal gas constant, and T is the temperature (=298 K).
3. Results and discussion
3.1. Isolation and structure elucidation of SMC-1, -2, -3, -4 and -5
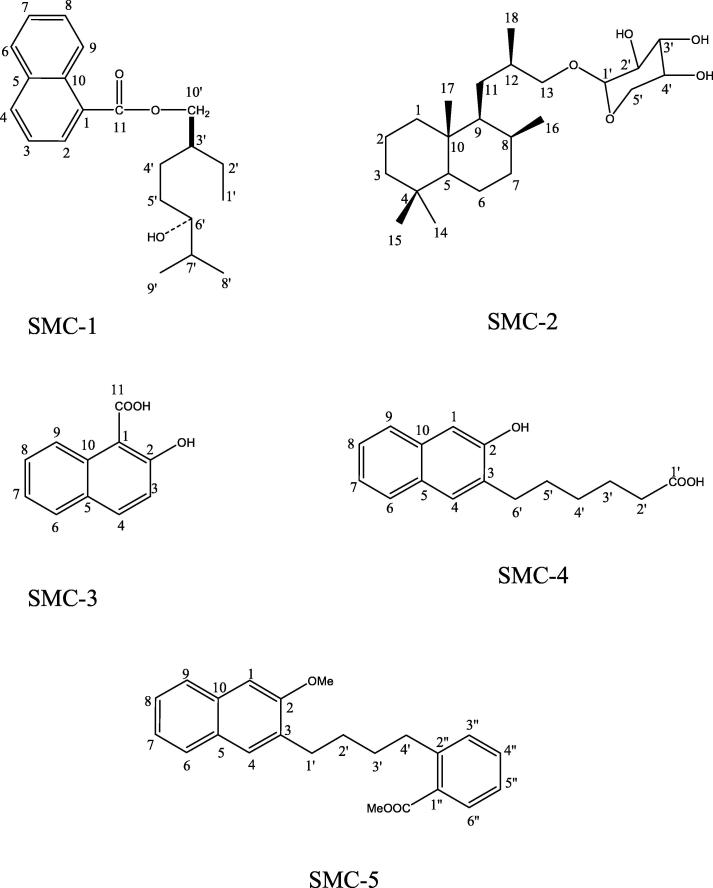
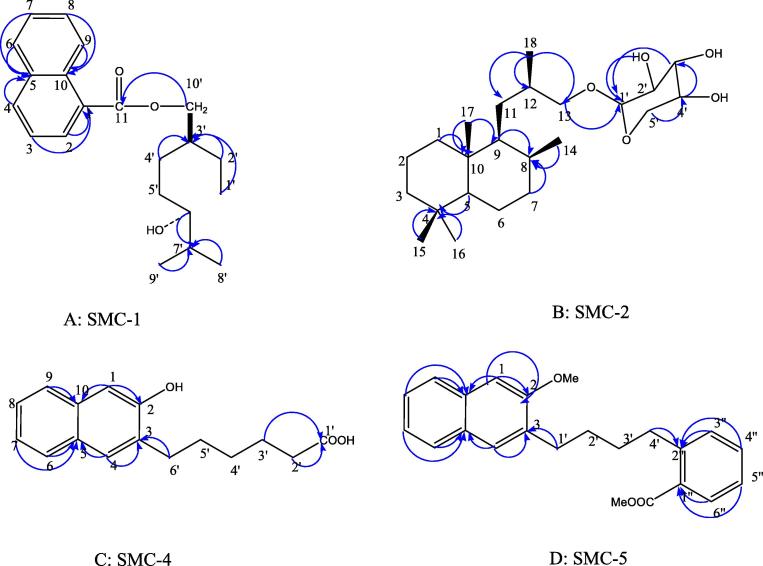
Five compounds of diverse nature have been isolated (Fig. 1) from Suaeda monoica and were coded as SMC-1, SMC-2, SMC-3, SMC-4 and SMC-5. SMC-1 showed IR absorption bands for hydroxyl group (3360 cm−1), ester function (1737 cm−1) and aromatic ring (1614, 1519, 1064 cm−1). The EIMS spectrum showed a molecular ion peak at m/z 328 that was consisting with a molecular formula of C21H28O3. The ion fragments arising at m/z 285 [C6′ - C7′ fission, C18H21O3]+, 155 [CO-O fission, C11H7O]+, 173 [M − 155, C10H21O2]+, 157 [O-CH2 fission, C10H21O]+, 141 [157 – CH4]+ and 123 [141 - H2O]+ indicated that a hydroxyl geranilane-type monoterpene moiety could be esterified with naphthoic acid (Fig. 2). The 1H NMR spectrum of SMC-1 displayed five one proton and a two- proton multiplets from δ 6.62 to 6.38 assigned to naphthyl protons. Two one proton doublets at δ 4.01 (J = 7.5 Hz) and 3.95 (J = 7.0 Hz) were assigned to oxymethylene H2-10′ protons. One proton multiplet at δ 3.52 with half width of 7.5 Hz was accounted to β-oriented carbinol H-6′ proton. Two three proton doublets at δ 1.25 (J = 6.5 Hz) and 1.23 (J = 6.3 Hz) and a three proton triplet at δ 0.84 (J = 6.5 Hz) were associated with the secondary C-8′ and C-9′ and primary C-1′ methyl protons, respectively. The remaining methylene and methine protons resonated at δ 2.63 to 1.38. The 13C NMR spectrum of SMC-1 exhibited signals for ester carbon at δ 169.13 (C-11), aromatic carbons in the range of δ 132.63–116.32, oxymethylene carbon at δ 64.62 (C-10′), carbinol carbon at δ 70.10 (C-6′), methyl carbons at δ 9.65 (C-1′), 20.85 (C-8′) and 21.13 (C-9′), and methylene and methine carbons between δ 49.21–22.42. The DEPT spectrum of SMC-1 showed the presence of three methyl, four methylene and ten methine carbons. The 1H–1H COSY spectrum of SMC-1 exhibited correlations of H-2 with H-3 and H-4; H-7 with H-6, H-8 and H-9; H-3′ with H2-2′, H2-4′, and H2-10′; and H-7′ with H-6′, Me-8′ and H-9′. The HMBC spectrum of SMC-1 showed interactions of H-2, H-3 and H-9 with C-1; H-4, H-6 and H-7 with C-5; H-8 and H-9 with C-10; H2-10′ with C-11; Me-8′, Me-9′ and H-6′ with C-7′; and Me-1′, H2′-2′ and H2-4′ with C-3′ (Fig. 5A). The HSQC spectrum of SMC-1 exhibited correlation of 1H NMR signals of H2-10′ at δ 4.01 and 3.95 with carbon signal at δ 64.62 (C-10′); H-6′ 1H NMR signal at δ 3.52 with carbon signal at δ 70.10 (C-6′), methyl proton signals at δ 1.25 (Me-8′), 1.23 (Me-9′) and 0.84 (Me-1′) with their respective carbons at δ 20.85 (C-8′), 21.13 (C-9′) and 9.65 (C-1′), and aromatic proton 1H NMR signal with their corresponding carbon signals. The 1H and 13C NMR spectral data of the naphthyl unit were compared with the reported values of the naphthylene moieties (Aslam et al., 2012, Ali et al., 2014). The spectral data of the geranilany units were compared with the published data of acyclic monoterpenes (Ali, 2001). On the basis of these spectral data analysis the structure of SMC-1 has been elucidated as 6′-hydroxy-10′-geranilanyl 1-naphthoate, a new aromatic monoterpenic ester.
Fig. 1.
Chemical structures of compounds (SMC- 1, 2, 3, 4, & 5) isolated from S. monoica.
Fig. 2.
Mass fragmentation pattern of SMC-1.
Fig. 5.
HMBC correlations of SMC compounds.
SMC-2 gave positive test of glycosides and showed IR absorption bands for hydroxyl group (3415, 3392 cm−1). Its molecular ion peak was determined at m/z 398 in the EIMS spectrum that was consistent with a molecular formula of a labdane-type norditerpenic glycoside C23H42O5. The ion peaks arising at m/z 133 [C1′-O fission, C5H9O4]+, and 149 [C13-O fission, C5H9O5]+ suggested the presence of a pentose sugar in the molecule. The ion fragments generated at m/z 383 [M−Me]+, 368 [M−2xMe]+, 234 [383–149]+, 163 [C12-C13 fission, C5H9O5-CH2]+, 205 [C9-C11 fission, C5H9O5-CH2-CH-(Me)–CH2]+ supported nor diterpenic nature of the molecule (Fig. 3). The 1H NMR spectrum of SMC-2 exhibited a one- proton doublet at δ 4.23 (J = 7.5 Hz) assigned to the anomeric H-1′ proton. Two one proton double doublets at δ 3.51 (J = 6.2 Hz, 1.1 Hz) and 3.42 (J = 6.3 Hz, 1.1 Hz) were ascribed to oxymethylene H2-13 protons. Two one-proton double doublets at δ 3.09 (J = 7.1, 1.2 Hz) and 3.06 (J = 6.9, 1.2 Hz) were due to oxymethylene H2-5′ protons. Three one-proton multiplets at δ 3.40, 3.23 and 3.02 were accounted to sugar carbinol H-4′, H-2′ and H-3′ protons, respectively. Two doublets at δ 0.76 (J = 6.1 Hz) and 0.73 (J = 6.5 Hz) and three singlets at δ 0.90, 0.82 and 0.65, all integrated for three protons each, were accounted to secondary C-18 and C-16, and tertiary C-17, C-15 and C-14 methyl protons, respectively, all attached to saturated carbons. The remaining methylene and methine protons resonated between δ 2.89–1.14. The 13C NMR spectrum of SMC-2 displayed signals for anomeric carbon at δ 101.25 (C-1′), other sugar carbons from δ 76.71 to 66.15, oxymethylene carbon at δ 61.04 (C-13), methyl carbons at δ 11.74 (C-14), 18.90 (C-15), 19.06 (C-16), 19.68 (C-17), and 18.58 (C-18), and the remaining methine and methylene carbons in the range of δ 57.18–25.48. The absence of any 1H NMR signal beyond δ 4.23 and 13C NMR signal after δ 101.25 supported saturated nature of the molecule. The DEPT spectrum of SMC-2 showed the presence of five methyl and eight each of methylene and methine carbons. The 1H–1H COSY spectrum of SMC-2 exhibited correlations of H-1′ with H-2′, H-3′ with H-4′; H-5 with H2-6, H2-6 with H2-7, and H-9 with H-8 and H2-11. The HMBC spectrum of SMC-2 showed correlations of H2-13, H-3′ and H-2′ with C-1′; H2-11, H-13 and Me-18 with C-12; H-3′ and H2-5′ with C-4′; H2-1, Me-17, H2-11 and H-9 with C-10; and H2-3, H-5, Me-14 and Me-15 with C-4 (Fig. 5B). The HSQC spectrum of SMC-2 displayed interactions of 1H NMR signals at δ 3.51 and 3.42 (H2-13) with 13C NMR at δ 61.04 (C-13), anomeric 1H NMR signal at δ 4.23 (H-1′) with the carbon signal at δ 101.25 (C-1′) and the methyl protons signals with the respective carbon signals. The 1H and 13C NMR data of diterpenic nucleus of SMC-2 were compared with the reported values of the labdane-type and related compounds and it was concluded that the molecule was a norlabdanoyl glycoside possessing two less carbon atoms in the side chain (Ali, 2001, Al-Massarani et al., 2017, Zheng et al., 2012, Demetzos and Dimas, 2001, Sy and Brown, 1997). Acid hydrolysis of SMC-2 yielded norlabdan-13-ol and D-xylose, Rf 0.76 (n-butanol: acetic acid: water, 4:1:1.6). On the basis of foregoing discussion, the structure of compound SMC–2 has been characterized as 4,4,8β,10β-Tetramethyl-9β-isobutanyl-13-ol-13-β-D-xylopyranoside, an unknown labdane-type nor diterpene xyloside.
Fig. 3.
Mass fragmentation pattern of SMC-2.
SMC-4 produced effervescences with sodium bicarbonate solution and showed IR absorption bands for hydroxyl group (3407 cm−1), carboxyl function (3209, 1677 cm−1) and aromatic ring (1521, 1108 cm−1) and aliphatic chain (775 cm−1). Its molecular ion peak was determined at m/z 258 on the basis of mass and 13C NMR spectra corresponding to molecular formula of alkylated naphthol. The 1H NMR spectrum of SMC-4 exhibited two one- proton singlets assigned to p-coupled aromatic H-1 and H-4 protons, respectively. Other aromatic protons as a one- proton double doublet at δ 6.95 (J = 2.0, 7.3 Hz, H-9), as a one- proton doublet at δ 6.69 (J = 8.0 Hz, H-6), and as one- proton multiplets at δ 6.45 (H-8) and 6.02 (H-7). Two two- proton triplets at δ 4.43 (J = 7.5 Hz) and 2.86 (J = 7.5 Hz), and three two- proton multiplets at δ 2.29, 1.58 and 1.27 were ascribed to methylene H2-6′ attached to the aromatic ring and methylene H2-2′ adjacent to the carboxyl function and the other methylene H2-5′, H2-4′ and H2-3′ of the side chain. The 13C NMR spectrum of SMC-4 displayed signals for carboxylic carbon at δ 178.37 (C-1′), aromatic carbons between δ 164.56–117.02 and methylene carbons from δ 52.10 to 26.04. The DEPT spectrum of SMC-4 showed the presence of five methylene and six methine carbons. The 1H–1H COSY spectrum of SMC-4 exhibited correlations of H-7 with H-6, H-8 and H-9; H-4 with H2-6′, and H2-2′ with H2-3′ and H2-4′. The HMBC spectrum of SMC-4 showed interactions of H-1 and H-9 with C-10; H-4 and H2-6′ with C-3; H-1 and H-4 with C-2; H-4 and; H-6 and H-7 with C-5; and H2-2′ with C-1′ (Fig. 5C). The HSQC spectrum of SMC-4 displayed correlation of 1H NMR signals at δ 6.73 (H-1), 6.60 (H-4), 6.95 (H-9) and 6.69 (H-6) with their respective aromatic carbon signals at δ 124.80 (C-1), 127.10 (C-4), 127.82 (C-9) and 120.62 (C-6); and methylene proton signals at δ 2.86 (H2-6′) and 2.42 (H2– 2′) with the carbon signals at δ 52.10 (C-6′) and 38.66 (C-2′). On the basis of these evidences, the structure of SMC-4 was elucidated as 6-(2-hydroxynaphthalen-3-yl)hexanoic acid; a new alkylated β-naphthol.
SMC-5 had IR absorption bands for ester group (1733 cm−1) and aromatic rings (1637, 1596, 1515, 1031 cm−1). On the basis of mass and 13C NMR spectra the molecular ion peak of SMC-5 was established at m/z 348 corresponding to a molecular formula of a naphthyl benzoyl butane derivative, C23H24O3. The ion peaks arising at m/z 135 [C4′-C2′' fission, C6H4-COOMe]+, 105 [135-OMe]+ and 77 [135-COOMe]+ indicated that methyl benzoate group was present in the molecule. The ion fragments generated at m/z 149 [C3′-C4′ fission, CH2-C6H4-COOMe]+ [M−Me]+, 163 [C2′-C3′ fission, CH2-CH2-C6H4-COOMe]+, 177 [C1′-C2′ fission, (CH2)3-C6H4-COOMe]+ and 191 [C3-C1 fission, (CH2)4-C6H4-COOMe]+ suggested that methyl benzoate was attached to one of the end of n-butane and methoxy (Fig. 4). Naphthalene was linked to another end carbon of n-butane chain. The 1H NMR spectrum of SMC-5 exhibited four one- proton double doublets at δ 7.46 (J = 10.5, 2.3 Hz), 7.06 (J = 9.0, 1.0 Hz), 7.04 (J = 8.5, 1.0 Hz) and 6.82 (J = 8.1, 1.5 Hz) were assigned to ortho-, meta- coupled aromatic H-6′', H-3′', H-6 and H-9 protons, respectively. Two one proton singlets at δ 7.12 and 6.41, and two three-proton singlets at δ 3.85 and 3.37 were attributed correspondingly to p-coupled H-1 and H-4 and to methoxy protons. The other aromatic protons resonated as two proton multiplets at δ 6.75 (H-7, H-4′') and 6.45 (H-5′and H-8). Two triplets at δ 3.46 (J = 7.5 Hz) and 3.33 (J = 1.5 Hz) and two-multiplets at δ 2.78 and δ 1.26, all integrating for two-protons each, were associated with the methylene H2-1′, H-4′, H-3′ and H-2′ protons, respectively. The 13C NMR spectrum of SMC-5 displayed carbon signals for ester carbon at δ 169.47 (C-7′'), aromatic carbons between δ 156.61 to 116.41, methoxy carbons at δ 56.57, 50.01 and methylene carbons from δ 49.67 to 35.73. The DEPT spectrum of SMC-5 showed the presence of two methoxy, four methylene and ten methine carbons. The 1H–1H COSY spectrum of SMC-5 exhibited correlations of H-4′' with H-3′' and H-5′'; H-5′' with H-6′'; H-7 with H-6 and H-8; H-8 with H-9; H-4′' with H-3′'; and H-3′ with H2-4′ and H2-2′. The HMBC spectrum of SMC-5 displayed interactions of H-4, H-1 and H2-1′ with C-3; H-1, H-8 and H-9 with C-10; H-4, H-6 and H-7 with C-5; H2-4′ and H-3′' with C-2′'; and H-6′' and H-5′' with C-1′' (Fig. 5D). The HSQC spectrum of SMC-5 showed correlations of 1H NMR signals at δ 7.12 (H-1), 6.41 (H-4) and 3.46 (H2-1′) with the respective carbon signals at δ 123.43 (C-1), 128.01 (C-4) and 49.67 (C-1′); 1H NMR signals at δ 7.04 (H-3′'), 7.46 (H-6′') and 3.33 (H2-4′), with the carbon signals at δ 128.37 (C-3′'), 132.19 (C-6′') and 42.71 (C-4′), respectively. On the basis of these evidences the structure of SMC-5 has been established as 1′-(2-Methoxy-3-naphthyl)-4′-(2′'-methylbenzoyl)-n-butane, a new β-methoxy-naphthalene derivative.
Fig. 4.
Mass fragmentation pattern of SMC-5.
SMC-3 gave effervescence with sodium bicarbonate solution and showed IR absorption bands for hydroxyl group (3477 cm−1), carboxylic function (1670 cm−1) and aromatic ring (1600, 1512, 977 cm−1). Its molecular ion peak was determined at m/z 188 on the basis of mass and 13C NMR spectra consistent with the molecular formula of hydroxyl aromatic acid, C11H8O3. The 1H NMR spectrum of SMC-3 exhibited four one- proton doublets at δ 7.59 (J = 16.0 Hz), 7.44 (J = 9.0 Hz), 7.42 (J = 9.0 Hz) and 6.30 (J = 16.0 Hz) assigned to ortho-coupled H-3, H-6, H-9 and H-4 and two one-proton multiplets at δ 6.84 and 6.82 accounted to H-7 and H-8 protons, respectively. The 13C NMR spectrum of SMC-3 displayed the presence of eleven carbon attributed to carboxylic carbon at δ 171.49 (C-11) and hydroxyl naphthalene carbons from δ 160.93 to 115.56. The DEPT spectrum of SMC-3 showed six methine carbon signals. Its 1H–1H COSY spectrum displayed correlations of H-3 with H-4; and H-7 with H-6, and H-8; and H-8 with H-9. The HMBC spectrum of SMC-3 exhibited interactions of H-3 and H-4 with C-2 and C-5; H-6 with C-5, and H-8 with C-10. The HSQC spectrum showed that 1H NMR signals at δ 7.59 (H-3) and 6.30 (H-4) correlated with carbon signals at δ 131.85 (C-3) and 131.21 (C-4), respectively, and proton signals at δ 7.44 (H-6), 7.42 (H-9), 6.84 (H-7) and 6.82 (H-8) with their corresponding carbon signals at δ 127.33 (C-6), 127.53 (C-9), 115.56 (C-7) and 116.26 (C-8). The chemical structure of SMC-3 has been confirmed from the spectral data of previous research work reported by Elsayed et al., 2013.On the basis of above discussion, the structure of SMC-3 has been elucidated as 2-Hydroxy-1-naphthoic acid. The acid is utilized by Burkholderia species strain BC1 as the sole source of carbon and energy (Chowdhury et al., 2014).
3.2. Cell proliferative potential of SMC compounds
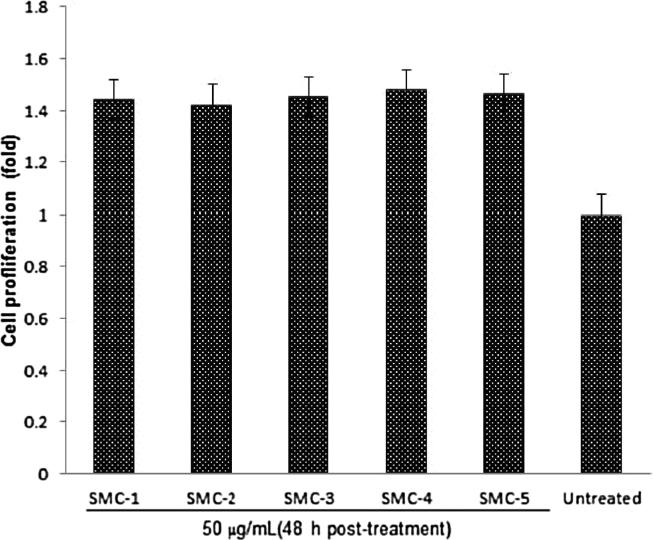
All five compounds were found to be nontoxic towards HepG2 cells (data not shown). Notably, the final concentration of DMSO was < 0.1% in the highest tested dose (50 μg/mL) where the cell viability was comparable to the untreated control. Therefore, the compounds were further evaluated for cell proliferative activities at the 50 μg/mL dose. The MTT assay showed the marked cell proliferative potential of all five compounds (1.42–1.48 fold) as compared to untreated control (Fig. 6).
Fig. 6.
The HepG2 cell proliferative potential of the five isolated compounds from S. monoica.
Many herbs have shown the cell proliferation potential, which is considered as one of the most useful methods for tissue regeneration, repair and aging. Blue berry, green tea, catechin, carnosine, and vitamin D3 were proved to have cell proliferation potential on human bone marrow as compared with human granulocyte macrophage colony-stimulating factor (h GM-CSF). Further studies showed that an interaction of nutrients with stem cell populations can promote rejuvenation (Bickford et al., 2006). There are several other natural compounds such as betulin and betulinic acid, which had previously been reported for cell proliferative potential and exhibited mitogenic activity with the ability to induce and modulate cytokine production in human blood leucocyte cells (Zdzisińska et al., 2003). It is observed that cell proliferation rate of mouse bone marrow mesenchymal stem cells enhanced up to 122.24% compared to untreated cells when treated with roots of Aconiti lateralis (Kim et al., 2013). The findings of this experiment, which showed 1.42- to 1.48-fold increase in cell proliferation as compared to untreated cells may pave a way to explore the removal of blockade on check points or stimulation of immunotherapeutic channels to treat some dreaded diseases.
3.3. Molecular docking analysis: Interaction of SMC compounds with cell signaling molecules (PTEN and p53)
3.3.1. Interaction between SMC compounds and PTEN
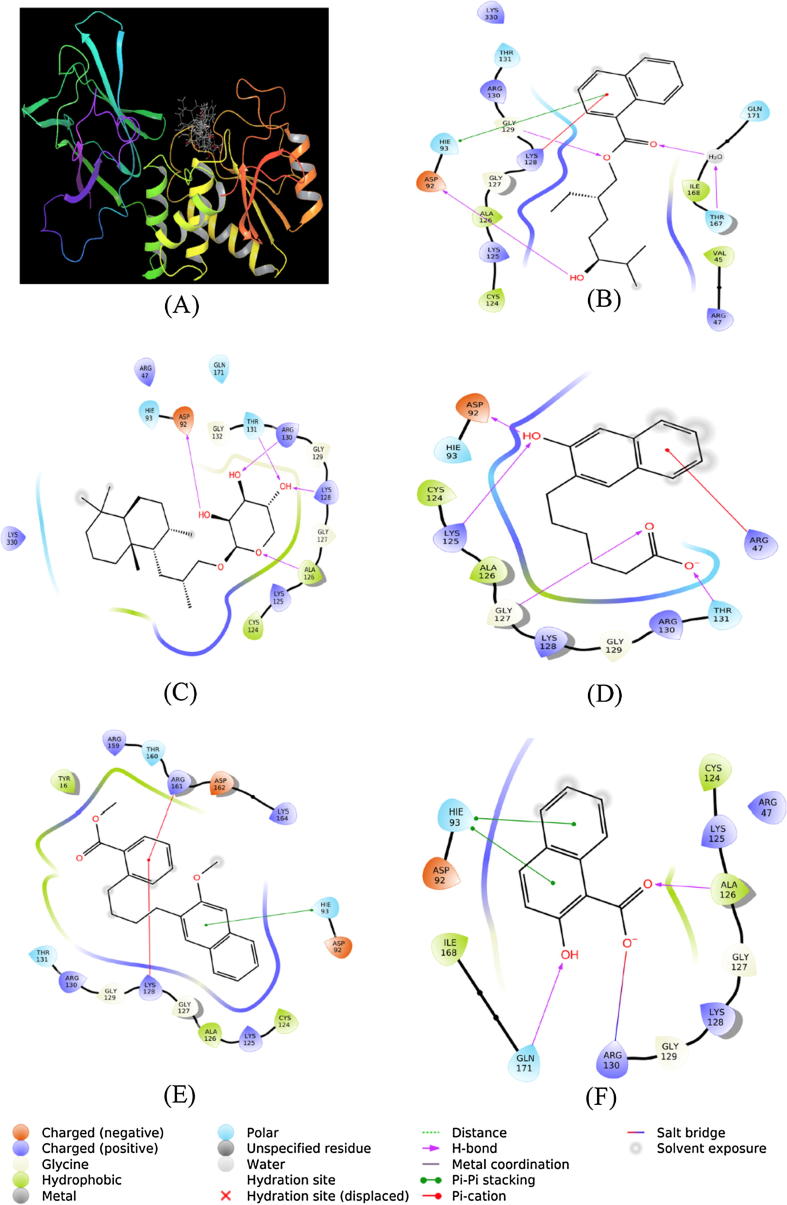
PTEN (phosphatase and tensin homologue deleted on chromosome 10) is a PI (phosphoinositide) 3-phosphatase. It acts as a tumor suppressor protein by inhibiting cellular proliferation, survival, and growth by inactivating PI3K-dependent signaling pathways. It antagonizes PI3K signaling pathway by dephosphorylating phosphatidylinositol 3,4,5-triphosphate into phosphatidylinositol 4,5-diphosphate (Leslie and Downes, 2004). In most of the cases where uncontrolled cell proliferation occurs, PTEN has been found to be inhibited or inactivated by different mutations. Thus, it is interesting to see whether the SMC compounds isolated in this study promote cell proliferation by inhibiting or inactivating PTEN. For this purpose, we employed molecular docking to gain an insight into the mechanism by which SMC compounds bind to PTEN. It is clear from Fig. 7A that all the SMC molecules (1–5) were bound at the same site.
Fig. 7.
Binding of SMC compounds with PTEN. Binding of (A) all SMC compounds, (B) SMC-1, (C) SMC-2, (D) SMC-3, (E) SMC-4, (F) SMC-5.
SMC-1 interacted with PTEN by forming one hydrophobic interaction with His93 in addition to three hydrogen bonds with Asp92, Gly129 and Tyr176 (Fig. 7B). Moreover, a cation-Pi interaction was observed between SMC-1 and Lys128. Other amino acid residues involved in the interaction were Val45, Arg47, Cys124, Lys125, Ala126, Gly127, Ile168, Gln171, Arg130, Thr131, and Lys330. The Gibb’s free energy of binding was estimated to be −6.660 kcal/mol, corresponding to a binding affinity of 7.67 × 104 M−1 (Table 1).
Table 1.
Interaction of SMC compounds with PTEN.
| Compound ID | Hydrophobic interactions | Hydrogen bonding | Cation-Pi interactions | Other residues involved | Binding energy (ΔG), kcal/mol | Binding affinity, Kd (M−1) |
|---|---|---|---|---|---|---|
| SMC-1 | His93 | Asp92, Gly129, Thr167 | Lys128 | Val45, Arg47, Cys124, Lys125, Ala126, Gly127, Ile168, Gln171, Arg130, Thr131, Lys330 | −6.660 | 7.67 × 104 |
| SMC-2 | – | Asp92, Ala126, Lys128, Arg130, Thr131 | – | Arg47, His93, Cys124, Lys125, Gly127, Gly129, Gly132, Gln171, Lys330 | −6.564 | 6.52 × 104 |
| SMC-3 | – | Asp92, Lys125, Gly127, Thr131 | Arg47 | His93, Cys124, Ala126, Lys128, Gly129, Arg130 | −7.149 | 1.75 × 105 |
| SMC-4 | His93 | – | Lys128, Arg161 | Tyr16, Asp92, Cys124, Lys125, Ala126, Gly127, Gly129, Arg130, Thr131, Arg159, Thr160, Asp162, Lys164 | −6.748 | 8.89 × 104 |
| SMC-5 | His93# | Ala126, Gln171 | Arg130* | Arg47, Asp92, Cys124, Lys125, Gly127, Lys128, Gly129, Ile168 | −7.321 | 2.34 × 105 |
Two interactions.
Salt bridge.
The PTEN-SMC-2 complex was stabilized by five hydrogen bonds with Asp92, Ala126, Lys128, Arg130, Thr131 (Table 1). Moreover, Arg47, His93, Cys124, Lys125, Gly127, Gly129, Gly132, Gln171, and Lys330 of PTEN were also involved in the stability of PTEN-SMC-2 complex (Fig. 7C). The PTEN-SMC-2 complex was stabilized by −6.564 kcal/mol of free energy, which is corresponded to a binding affinity of 6.52 × 104 M−1 (Table 1).
The complex between SMC-3 and PTEN was stabilized by one cation-Pi interaction with Arg47 and four hydrogen bonds with Asp92, Lys125, Gly127, and Thr131 (Fig. 7D). Other amino acid residues involved in the interaction between SMC-3 and PTEN were His93, Cys124, Ala126, Lys128, Gly129, and Arg130 (Fig. 7D). The binding free energy and binding affinity of PTEN-SMC-3 complex formation were – 7.149 kcal/mol, and 1.75 × 105 M−1, respectively (Table 1).
SMC-4 formed one hydrophobic interaction with His93 and two cation-Pi interactions with Lys128 and Arg161 of PTEN. Other amino acid residues involved in PTEN-SMC-4 complex formation were Tyr16, Asp92, Cys124, Lys125, Ala126, Gly127, Gly129, Arg130, Thr131, Arg159, Thr160, Asp162, and Lys164 (Fig. 7E). Gibb’s free energy of the interaction between SMC-4 and PTEN was estimated to be −6.748 kcal/mol while the binding affinity was determined to be 8.89 × 104 M−1 (Table 1).
Similarly, SMC-5 interacted with PTEN by forming two hydrophobic interactions with His93 and two hydrogen bonds with Ala126 and Gln171 (Table 1). Moreover, Arg130 formed a salt bridge with PTEN. Several other residues such as Arg47, Asp92, Cys124, Lys125, Gly127, Lys128, Gly129, and Ile168 were also involved in the formation of a stable complex between SMC-5 and PTEN (Fig. 7F). Gibb’s free energy of stabilization and binding affinity of SMC-5 towards PTEN were estimated to be −7.321 kcal/mol, and 2.34 × 105 M−1, respectively (Table 1).
The results of molecular docking could be better interpreted after gaining an insight into the three-dimensional structure of PTEN. It is a 403 amino acid long protein composed of two major domains namely N-terminal phosphatase domain (residues 7–185), and C-terminal C2 domain (residues 186–351). The active site of PTEN is made up of residues 123–130 with a signature HCXXGXXR motif (Leslie and Downes, 2004). In addition, Asp92 acts as a general acid to facilitate protonation of the O-atom of the tyrosyl leaving group. Several other residues such as residues 42–52, residues 91–94, residues 163–166, and Gln171 play significant role in the recognition of the substrate and its hydrolysis (Lee et al., 1999). From our docking results, we observed that the SMC compounds (SMC 1–5) bind at the active site of and thus may sterically hinder the accessibility of phosphatidylinositol 3,4,5-triphosphate to the active site thereby preventing the dephosphorylation of phosphatidylinositol 3,4,5-triphosphate by PTEN. The intact phosphatidylinositol 3,4,5-triphosphate molecule will then be available for PI3K signaling pathway and other downstream pathways involved in the cellular growth and survival.
3.3.2. Interaction between SMC compounds and p53
p53 is a tumor suppressor protein which subdues tumor formation and progression by regulating various transcriptional pathways involved in cell cycle arrest, DNA repair, and apoptosis (Bai and Zhu, 2006). p53 is a tetrameric phosphoprotein comprising 393 amino acids organized into well-structured domains as well as unstructured regions working synergistically. It comprises four domains that perform specific functions such as (i) activation of transcription factors by transactivation domain, (ii) recognition of specific DNA sequences by DNA-binding domain, (iii) tetramerization of the protein by tetramerization domain, and (iv) recognition of damaged DNA by regulatory domain. The loss of p53 tumor suppressor function results in uncontrolled proliferation of cells thereby leading to the formation of cancers. Thus, it is interesting to know whether SMC compounds induce cellular progression by binding to p53 and thus inactivating it.
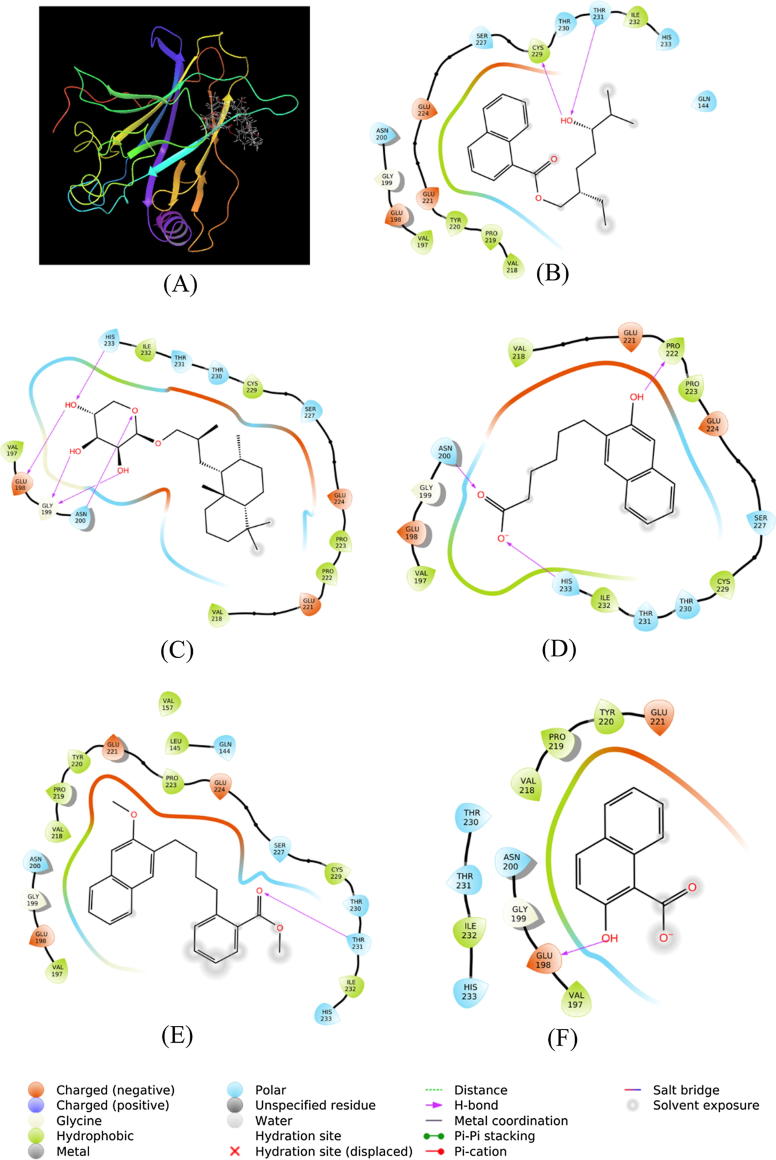
The most probable binding site(s) on p53 was determined using Epik-module (Epik, Schrodinger 2018, LCC, NY, USA). We found that three binding sites (I-III) were present on p53, relative locations of which are shown in supplementary Fig. S1. The analysis of binding site has shown that SMC molecules have relatively higher affinity towards binding sites I and II as compared to binding site III. Thus, the results of binding sites I and II have been discussed here, while the results of binding site III has been reported as supplementary data (Fig. S1 and Table S1). Results representing the nature of interactions and the type of amino acid residues involved in the formation of stable SMC-p53 complex are given in Table 2 and Fig. 8,9. The results presented in Fig. 8A and Fig. 9A confirm that all the SMC molecules (1–5) were bound at the site I and site II, respectively.
Table 2.
Interaction of SMC compounds with p53.
| Compound ID | Hydrophobic interactions | Hydrogen bonding | Cation-Pi interactions | Other residues involved | Binding energy (ΔG), kcal/mol | Binding affinity, Kd (M−1) |
|---|---|---|---|---|---|---|
| Site I | ||||||
| SMC-1 | – | Cys229, Thr231 | – | Gln144, Val197, Glu198, Gly199, Asn200, Val218, Pro219, Tyr220, Glu221, Glu224, Ser227, Thr230, Ile232, His233 | −6.186 | 3.44 × 104 |
| SMC-2 | – | Glu198, Gly199#, Asn200, His233 | – | Val197, Val218, Glu221, Pro222, Pro223, Glu224, Ser227, Cys229, Thr230, Thr231, Ile232 | −6.529 | 6.15 × 104 |
| SMC-3 | – | Asn200, Pro222, His233 | – | Val197, Glu198, Gly199, Val218, Glu221, Pro223, Glu224, Ser227, Cys229, Thr230, Thr231, Ile232 | −6.887 | 1.12 × 105 |
| SMC-4 | – | Thr231 | – | Gln144, Ile145, Val157, Val197, Glu198, Gly199, Asn200, Val218, Pro219, Tyr220, Glu221, Pro223, Glu224, Ser227, Cys229, Thr230, Ile232, His233 | −5.608 | 1.29 × 104 |
| SMC-5 | – | Glu198 | – | Val197, Gly199, Asn200, Val218, Pro219, Tyr220, Glu221, Thr230, Thr231, Ile232, His233 | −5.698 | 1.51 × 104 |
| Site II | ||||||
| SMC-1 | – | Val97, Ser99 | – | Ser96, Pro98, Arg158, Met160, Ile254, Thr256, Glu258, Arg267 | −5.106 | 5.56 × 103 |
| SMC-2 | – | Glu258, Gly262#, Arg267 | – | Ser96, Val97, Pro98, Ser99, Leu206, Asp208, Thr211, Arg213, Arg158, Met160, Ile254, Thr256, Asn263, Leu264 | −5.598 | 1.28 × 104 |
| SMC-3 | – | Ser99, Glu258 | Arg158 | Pro98, Met160, Arg213, Thr211, Asp208, Ile254, Thr256, Gly262, Leu264, Arg267 | −6.010 | 2.56 × 104 |
| SMC-4 | – | – | – | Ser96, Val97, Pro98, Ser99, Asp208, Arg209, Asn210, Thr211, Arg158, Met160, Ile254, Thr256, Glu258, Leu264, Arg267 | −5.711 | 1.54 × 104 |
| SMC-5 | – | Gly262, Arg267 | – | Pro98, Ser99, Arg158, Met160, Ile254, Thr256, Glu258, Asn263, Leu264 | −5.793 | 1.77 × 104 |
* Salt bridge.
Two bonds.
Fig. 8.
Binding of SMC compounds at site 1of P53. Binding of (A) all SMC compounds, (B) SMC-1, (C) SMC-2, (D) SMC-3, (E) SMC-4, (F) SMC-5.
Fig. 9.
Binding of SMC compounds at site 2 of P53. Binding of (A) all SMC compounds, (B) SMC-1, (C) SMC-2, (D) SMC-3, (E) SMC-4, (F) SMC-5.
3.3.2.1. Interaction of SMC molecules at the binding site I
SMC-1 interacted with p53 by forming two hydrogen bonds with Cys229 and Thr231 (Fig. 8B). Other amino acid residues involved in the interaction were Gln144, Val197, Glu198, Gly199, Asn200, Val218, Pro219, Tyr220, Glu221, Glu224, Ser227, Thr230, Ile232, and His233. The Gibb’s free energy of binding was estimated to be −6.186 kcal/mol, corresponding to a binding affinity of 3.44 × 104 M−1 (Table 2).
The p53-SMC-2 complex was stabilized by five hydrogen bonds with Glu198, Gly199, Asn200, and His233 (Fig. 8C). Moreover, Val197, Val218, Glu221, Pro222, Pro223, Glu224, Ser227, Cys229, Thr230, Thr231, and Ile232 of p53 formed multiple van der Waals interactions with SMC-2 (Fig. 8C). The p53-SMC-2 complex was stabilized by −6.529 kcal/mol of free energy, which is corresponded to a binding affinity of 6.15 × 104 M−1 (Table 2).
The complex between SMC-3 and p53 was stabilized by three hydrogen bonds with Asn200, Pro222 and His233 (Fig. 8D). Other amino acid residues involved in the interaction between SMC-3 and p53 were Val197, Glu198, Gly199, Val218, Glu221, Pro223, Glu224, Ser227, Cys229, Thr230, Thr231, and Ile232 (Fig. 8D). The binding free energy and binding affinity of p53-SMC-3 complex formation were −6.887 kcal/mol, and 1.12 × 105 M−1 respectively (Table 2).
SMC-4 formed one hydrogen bond with Thr231 of p53. Other amino acid residues involved in p53-SMC-4 complex formation were Gln144, Ile145, Val157, Val197, Glu198, Gly199, Asn200, Val218, Pro219, Tyr220, Glu221, Pro223, Glu224, Ser227, Cys229, Thr230, Ile232, and His233 (Fig. 8E). Gibb’s free energy of the interaction between SMC-4 and p53 was estimated to be −5.608 kcal/mol while the binding affinity was determined to be 1.29 × 104 M−1 (Table 2).
Similarly, SMC-5 interacted with p53 by forming one hydrogen bond with Glu198 (Table 2). Several other residues such as Val197, Gly199, Asn200, Val218, Pro219, Tyr220, Glu221, Thr230, Thr231, Ile232, and His233 were also involved in the formation of a stable complex between SMC-5 and p53 (Fig. 8F). Gibb’s free energy of stabilization and binding affinity of SMC-5 towards p53 were estimated to be −5.698 kcal/mol, and 1.51 × 104 M−1, respectively (Table 2).
3.3.2.2. Interaction of SMC molecules at the binding site II
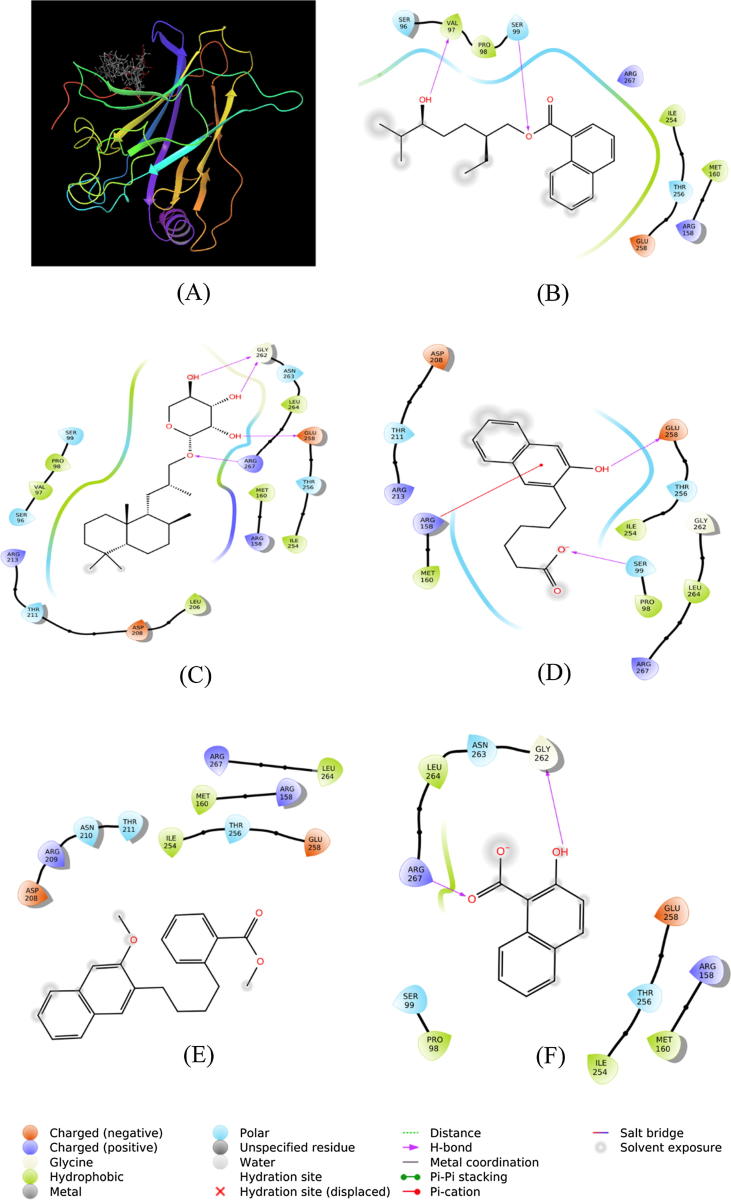
SMC-1 interacted with p53 by forming two hydrogen bonds with Val97 and Ser99. Other amino acid residues involved in the interaction were Ser96, Pro98, Arg158, Met160, Ile254, Thr256, Glu258, and Arg267 (Fig. 9B). The Gibb’s free energy of binding was estimated to be −5.106 kcal/mol, corresponding to a binding affinity of 5.56 × 103 M−1 (Table 2).
The p53-SMC-2 complex was stabilized by four hydrogen bonds with Glu258, Gly262 and Arg267 (Fig. 9C). Moreover, Ser96, Val97, Pro98, Ser99, Leu206, Asp208, Thr211, Arg213, Arg158, Met160, Ile254, Thr256, Asn263, and Leu264 of p53 formed multiple van der Waals interactions with SMC-2 (Fig. 9C). The p53-SMC-2 complex was stabilized by −5.598 kcal/mol of free energy, which is corresponded to a binding affinity of 1.28 × 104 M−1 (Table 2).
The complex between SMC-3 and p53 was stabilized by two hydrogen bonds with Ser99 and Glu258, and one cation-Pi interaction with Arg518 (Fig. 9D). Other amino acid residues involved in the interaction between SMC-3 and p53 were Pro98, Met160, Arg213, Thr211, Asp208, Ile254, Thr256, Gly262, Leu264, and Arg267 (Fig. 9D). The binding free energy and binding affinity of p53-SMC-3 complex formation were −6.010 kcal/mol, and 2.56 × 104 M−1, respectively (Table 2).
SMC-4 formed did not form any hydrogen bond, hydrophobic or electrostatic interactions with p53 at binding site III. Several amino acid residues such as Ser96, Val97, Pro98, Ser99, Asp208, Arg209, Asn210, Thr211, Arg158, Met160, Ile254, Thr256, Glu258, Leu264, and Arg267 were involved in p53-SMC-4 complex formation (Fig. 9E). Gibb’s free energy of the interaction between SMC-4 and p53 was estimated to be −5.711 kcal/mol while the binding affinity was determined to be 1.54 × 104 M−1 (Table 2).
Similarly, SMC-5 interacted with p53 by forming two hydrogen bonds with Gly262 and Arg267 (Table 2). Several other residues such as Pro98, Ser99, Arg158, Met160, Ile254, Thr256, Glu258, Asn263, and Leu264 were also involved in the formation of SMC-5-p53 complex (Fig. 9F). Gibb’s free energy of stabilization and binding affinity of SMC-5 towards p53 were estimated to be −5.793 kcal/mol and 1.77 × 104 M−1 respectively (Table 2).
The results of molecular docking suggest that SMC compounds bind different amino acid residues of distinct domains of p53 with binding affinity in the range of 104-105 M−1 (Table 2). It is noticeable that the amino acid residues of binding sites I and II such as Ser99, Thr170, Gly199 and Asp224 have been reported to participate in the formation of active tetrameric form of p53 (AlAjmi et al., 2018). On the other hand, Arg280, which is located in binding site III of p53, has been shown to participate in the formation of p53-DNA complex. The binding of SMC compounds to p53 thus may prevent it to bind to DNA and thus preclude the recruitment of other factors necessary for cell cycle arrest. Moreover, SMC compounds may change the p53 binding activity towards its downstream targets and thus may induce aberrant cell proliferation (Bai and Zhu, 2006).
4. Conclusion
The DCM fraction of the S. monoica was found to be the most active in cell proliferation. The chromatographic purification of DCM faction led to the isolation of four new compounds along with one known with remarkable cell proliferative potential (1.42–1.48 fold), which supports the use of S. monoica as folklore medicine for wound healing. The in vitro results of this study confirm that SMC compounds (SMC 1–5) have the potential to induce cellular proliferation in HepG2 cells. Moreover, an insight into the possible mechanism of SMCs action through molecular docking revealed that the isolated SMC compounds are able to bind and inactivate or modulate the tumor suppressor proteins such as PTEN and p53. The exact mechanism of cell proliferation by S. monoica need to be explored in further studies but the findings of this research justifies the use of S. monoica in traditional medicines.
Acknowledgments
Acknowledgement
The authors extend their appreciation to the Deanship of Scientific Research at King Saud University, Riyadh, Saudi Arabia for funding the work through the research group project number (RGP-073).
Declaration of Competing Interest
The authors declare that they don’t have any conflict of interest.
Footnotes
Peer review under responsibility of King Saud University.
References
- AlAjmi M.F., Rehman M.T., Hussain A., Rather G.M. Pharmacoinformatics approach for the identification of Polo-like kinase-1 inhibitors from natural sources as anti-cancer agents. Int. J. Biol. Macromol. 2018;116:173–181. doi: 10.1016/j.ijbiomac.2018.05.023. [DOI] [PubMed] [Google Scholar]
- Al-Hassan HO., 2006. Wild Plants of the Northern Region. Ministry of Agriculture, Camel and range research center in collaboration with FAO, Al-Jouf, SA.
- Ali A., Jameel M., Ali M. New naphthyl esters from the bark of Ficus religiosa Linn. The Nat. Prod. J. 2014;4(4):248–253. [Google Scholar]
- Ali, M., 2001. Techniques in Terpenoid Identification, Birlas Publications (Regd.), Shahdara, Delhi, pp. 15–23.
- Al-Massarani S.M., El-Gamal A.A., Parvez M.K., Al-Dosari M.S., Al-Said M.S., Abdel-Kader M.S., Basudan O.A. New Cytotoxic seco-type triterpene and labdane-type diterpenes from Nuxia oppositifolia. Molecules. 2017;22(3):389. doi: 10.3390/molecules22030389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Said M.S., Siddiqui N.A., Mukhair M.A., Parvez M.K., Alam P., Ali M. A novel monocyclic triterpenoid and a norsesquaterpenol from the aerial parts of Suaeda monoica Forssk. ex J.F. Gmel with cell proliferative potential. Saudi Pharm. J. 2017;25(7):1005–1010. doi: 10.1016/j.jsps.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashtiani A., Aghaei M., Ehsani A., Rastegar H., Salout M.Y. Combination of herbal extracts and platelet rich plasma induced dermal papilla cells proliferation Involvement of ERK and AKT pathway. Res. Pharm. Sci. 2012;7(5):23–27. doi: 10.1111/jocd.12033. [DOI] [PubMed] [Google Scholar]
- Aslam M., Ali M., Dayal R., Javed K. Coumarins and a naphthyl labdanoate diarabinoside from the fruits of Peucedanum grande. C. B. Clarke, Z. Naturforsch. 2012;67 c:580–586. doi: 10.1515/znc-2012-11-1208. [DOI] [PubMed] [Google Scholar]
- Bai L., Zhu W.-G. p53: structure, function and therapeutic applications. J. Cancer Mol. 2006;2(4):141–153. [Google Scholar]
- Bandaranayake W.M. Traditional and medicinal uses of mangroves. Mangroves Salt Marshes. 1998;2(3):133–148. [Google Scholar]
- Bickford P.C., Tan J., Shytle R.D., Sanberg C.D., El-Badri N., Sanberg P.R. Nutraceuticals synergistically promote proliferation of human stem cells. Stem Cells Dev. 2006;15(1):118–123. doi: 10.1089/scd.2006.15.118. [DOI] [PubMed] [Google Scholar]
- Boulos L. Notes on Suaeda Forssk. ex Scop. Studies in the Chenopodiaceae of Arabia: 2. Kew Bull. 1991;46(2):291–296. [Google Scholar]
- Boulos L. Flora of Egypt. 1999;vol. 1:417. [Google Scholar]
- Chandrasekaran M., Kannathasan K., Venkatesalu V. Antimicrobial activity of fatty acid methyl esters of some members of Chenopodiaceae. Z. Naturforsch. C. 2008;63(5–6):331–336. doi: 10.1515/znc-2008-5-604. [DOI] [PubMed] [Google Scholar]
- Chowdhury P.P., Sarkar J., Basu S., Dutta T.D. Metabolism of 2-hydroxy-1-naphthoic acid and naphthalene via gentisic acid by distinctly different sets of enzymes in Burkholderia sp. strain BC1. Microbiology. 2014;160(Pt 5):892–902. doi: 10.1099/mic.0.077495-0. [DOI] [PubMed] [Google Scholar]
- Chen Y., Zhang X., Dantas Machado A.C., Ding Y., Chen Z., Qin P.Z. Structure of p53 binding to the BAX response element reveals DNA unwinding and compression to accommodate base-pair insertion. Nucl. Acids Res. 2013;41(17):8368–8376. doi: 10.1093/nar/gkt584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collenette S. Riyadh; SA: 1999. Wildflowers of Saudi Arabia: National Commission for Wildlife Conservation and Development (NCWCD) [Google Scholar]
- Demetzos C., Dimas K. Labdane-type diterpenes: Chemistry and biological activity. Stud. Nat. Prod. Chem. 2001;25:235–292. [Google Scholar]
- Elsayed S.A., El-Hendawy A.M., Butler I.S., Mostafa S.I. New complexes of 2-hydroxy-1-naphthoic acid and X-ray crystal structure of [Pt(hna)(PPh3)2] J. Mol. Struct. 2013;1036:196–202. [Google Scholar]
- Elsharabasy F.S., Metwally N.S., Mahmoud A.H., Soliman M.S., Youness E.R., Farrag A.H., Arafa S. Phytoconstituents and hepatoprotective effect of Suaeda monoica Forssk and Suaeda pruinosa Lang. Biomed. Pharmacol. J. 2019;12(1):117–129. [Google Scholar]
- Hasan N., Al Sorkhy M. Herbs that promote cell proliferation. Int. J. Herb Med. 2014;1(6):18–21. [Google Scholar]
- Kassem F.A., Ibrahim H.A., El-Shazly A.M., Abdallah E.S.A. Plant Derived Pest Control Agents III-Insecticidal and Biochemical studies of different plant extracts against Culex pipiens larvae. J. Agric. Env. Sci. 2003;2(1):56–68. [Google Scholar]
- Kim D.R., Kim H.Y., Park J.K., Park S.K., Chang M.S. Aconiti Lateralis Preparata Radix Activates the Proliferation of Mouse Bone Marrow Mesenchymal Stem Cells and Induces Osteogenic Lineage Differentiation through the Bone Morphogenetic Protein-2/Smad-Dependent Runx2 Pathway. Evid. Based Complement. Alternat. Med. 2013;2013:586741. doi: 10.1155/2013/586741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmanan G., Rajeshkannan C., Kavitha A., Mekala B., Kamaladevi N. Preliminary screening of biologically active constituents of Suaeda monoica and Sesuvium portulacastrum from palayakayal mangrove forest of Tamilnadu. J. Pharmacog. Phytochem. 2013;2(3):149–152. [Google Scholar]
- Lee C.U., Hahne G., Hanske J. Redox Modulation of PTEN Phosphatase Activity by Hydrogen Peroxide and Bisperoxidovanadium Complexes. Angew Chem. 2015;54(46):13796–13800. doi: 10.1002/anie.201506338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-O., Yang H., Georgescu M.-M., Di Cristofano A., Maehama T., Shi Y. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99(3):323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- Leslie N.R., Downes C.P. PTEN function: how normal cells control it and tumor cells lose it. Biochem. J. 2004;382(Pt 1):1–11. doi: 10.1042/BJ20040825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmakumar K., Ayyakkannu K. Antiviral activity of marine plants. Indian J. Virol. 1997;13(1):33–36. [Google Scholar]
- Premnathan M., Chandra K., Bajpai S., Kathiresan K. A survey of some Indian marine plants for antiviral activity. Botanica marina. 1992;35(4):321–324. [Google Scholar]
- Premnathan M., Nakashima H., Kathiresan K., Rajendran N., Yamamoto N. In vitro anti human immunodeficiency virus activity of mangrove plants. Indian J. Med. Res. 1996;103:278–281. [PubMed] [Google Scholar]
- Ravikumar S., Gnanadesigan M., Inbaneson S.J., Kalaiarasi A. Hepatoprotective and antioxidant properties of Suaeda maritima (L.) dumort ethanolic extract on concanavalin-A induced hepatotoxicity in rats. Indian J. Exp. Biol. 2011;49(6):455–460. [PubMed] [Google Scholar]
- Ravikumar S., Gnanadesigan M., Seshserebiah J., Jacob Inbaneson S. Hepatoprotective effect of an Indian salt marsh herb Suaeda monoica Forsk. Ex. Gmel against concanavalin-A induced toxicity in rats. Life Sci. Med. Res. 2010;2010(LSMR-2):1–9. [Google Scholar]
- Ravikumar S., Ramanathan G., Inbaneson S.J., Ramu A. Antiplasmodial activity of two marine polyherbal preparations from Chaetomorpha antennina and Aegiceras corniculatum against Plasmodium falciparum. J. Parasitol. Res. 2011;108(1):107–113. doi: 10.1007/s00436-010-2041-5. [DOI] [PubMed] [Google Scholar]
- Sy L.-K., Brown G.D. Labdane diterpenoids from Alpinia chinensis. J. Nat. Prod. 1997;60(9):904–908. [Google Scholar]
- Yezhelyev M.V., Gao X., Xing Y., Al-Hajj A., Nie S., O'Regan R.M. Emerging use of nanoparticles in diagnosis and treatment of breast cancer. Lancet Oncol. 2006;7(8):657–667. doi: 10.1016/S1470-2045(06)70793-8. [DOI] [PubMed] [Google Scholar]
- Zdzisińska B., Rzeski W., Paduch R., Szuster-Ciesielska A., Kaczor J., Wejksza K., Kandefer-Szerszeń M. Differential effect of betulin and betulinic acid on cytokine production in human whole blood cell cultures. Pol. J. Pharmacol. 2003;55(2):235–238. [PubMed] [Google Scholar]
- Zheng C.-J., Lan X.-P., Wang Y., Huang B.-K., Han T., Zhang Q.-Y., Qin L.-P. A new labdane diterpene from Vitex negundo. Pharm. Biol. 2012;50(6):687–690. doi: 10.3109/13880209.2011.597410. [DOI] [PubMed] [Google Scholar]