Abstract
Background
HER2 plays a critical role in tumourigenesis and is associated with poor prognosis of patients with HER2-positive breast cancers. Although anti-HER2 drugs are beneficial for treating breast cancer, de novo, or acquired resistance often develops. Epigenetic factors are increasingly targeted for therapy; however, such mechanisms that interact with HER2 signalling are poorly understood.
Methods
RNA sequencing was performed to identify PHF8 targets downstream of HER2 signalling. CHIP-qPCR were used to investigate how PHF8 regulates HER2 transcription. ELISA determined cytokine secretion. Cell-based assay revealed a feed forward loop in HER2 signalling and then evaluated in vivo.
Findings
We report the synergistic interplay between histone demethylase PHF8 and HER2 signalling. Specifically, PHF8 levels were elevated in HER2-positive breast cancers and upregulated by HER2. PHF8 functioned as a coactivator that regulated the expression of HER2, markers of the HER2-driven epithelial-to-mesenchymal transition and cytokines. The HER2-PHF8-IL-6 regulatory axis was active in cell lines and in newly established MMTV-Her2/MMTV-Cre/Phf8fl°x/fl°x mouse models, which revealed the oncogenic function of Phf8 in breast cancer for the first time. Further, the PHF8-IL-6 axis contributed to the resistance to trastuzumab in vitro and may play a critical role in the infiltration of T cells in HER2-driven breast cancers.
Interpretation
These findings provided informative mechanistic insight into the potential application of PHF8 inhibitors to overcome resistance to anti-HER2 therapies.
Funding
This work was supported by Carver Trust Young Investigator Award (01-224 to H.H.Q); and a Breast Cancer Research Award (to H.H.Q.).
Keywords: PHF8, HER2, IL-6, Breast cancer, Drug resistance
Research in context.
Evidence before this study
HER2 plays critical roles in tumourigenesis and is associated with poor prognosis of breast cancers. Although some tumours respond well to anti-HER2 therapies, many are resistant de novo or following therapy. Epigenetic mechanisms influencing HER2-driven cancers and drug resistance are largely unknown.
Added value of this study
This study is novel in revealing a feed forward loop in HER2 signalling and discovering novel epigenetic mechanisms in HER2 gene expression and HER2 signalling in breast cancers. This study screened secretion of cytokines affected by histone demethylase PHF8 in HER2 positive breast cells. The HER2-PHF8-IL-6 regulatory axis verified here contributes to the resistance to Trastuzumab in vitro and may play a critical role in the infiltration of T-cells in HER2-driven breast cancers.
Implications of all the available evidence
Elevated PHF8 in HER2 positive breast cancer may play an important role in the immune response by altering the tumour microenvironment and influencing T cell trafficking to tumour sites by regulating cytokine production. This study gains mechanistic insights into the potential application of PHF8 inhibitors in the resistance of anti-HER2 therapies.
Alt-text: Unlabelled box
1. Introduction
Breast cancer is the most commonly diagnosed cancer and the second leading cause of cancer death of American women. Thus, approximately 268,600 new cases of breast cancer will be diagnosed, and approximately 41,760 women will die from breast cancer in 2019 in the United States [1]. Breast cancers include the following (not mutually exclusive) categories: oestrogen receptor (ER)-positive; ERBB2/HER2/NEU (HER2)-positive (HER2+), and triple-negative. HER2+ breast cancers represent 20%–30% of breast cancers and are often associated with poor prognosis [2]. HER2 is a transmembrane receptor protein tyrosine kinase that plays critical roles in the development of cancer and resistance to therapy of patients with HER2+ [2,3] and HER2-negative (HER2-) [2,3] and HER2-negative (HER2-) [4], [5], [6] breast cancers. In the later cases, such as luminal or triple-negative breast cancer, HER2 expression is elevated within a defined group of cancer stem cells that are believed to be the true oncogenic population in the heterogeneous breast cancer and to confer resistance to both hormone and radiation therapies [4], [5], [6]. Trastuzumab, a humanised anti-HER2 antibody, and lapatinib, a HER2 kinase inhibitor, dramatically improve the efficacy of treatment of patients with HER2+ breast cancer or gastric cancer [7]. Notably, these anti-HER2 therapies achieve beneficial outcomes when administered to HER2+ patients with cancer [8]. However, drug resistance often develops de novo, which hinders therapy [2]. Thus, to identify novel therapeutic targets that are critical for HER2-driving tumour development and resistance to therapy is still needed.
Investigations of the importance of epigenetic mechanisms in oncogenesis have shifted focus on developing cancer therapeutics that target chromatin regulators [9,10]. For example, targeting bromodomain and extra terminal domain proteins (BETs) using the inhibitor JQ1 antagonises the proliferation of multiple myeloma cells by repressing MYC and its downstream effectors [11]. Similarly, targeting the histone demethylase KDM4 family member NCDM-32B effectively inhibits the proliferation and malignant transformation of breast cancer cells [12]. In the context of HER2, the association of epigenetic changes including DNA methylation, histone modifications, and ncRNAs/miRNAs associated with HER2+ breast cancer susceptibility are the focus of a detailed review [13]. Importantly, histone deacetylase (HDAC) and DNA methylation inhibitors upregulate HER2 expression [13,14]. Moreover, methylation of histone-3 lysine 4 (H3K4me3) and that of histone-3 lysine 9 (H3K9me2) are associated with the induction or downregulation of HER2 expression, respectively [13]. Thus, WDR5, a core component of H3K4me3 methyltransferase and G9a, the H3K9me2 methyltransferase, may be responsible for the changes in these modifications [13]. However, whether and how histone demethylase, another major contributor to epigenetic mechanisms, influences HER2 expression, and HER2-driven tumour development and resistance to therapy are unknown.
Our team recently reported that histone demethylase PHD finger protein 8 (PHF8) promotes the epithelial-to-mesenchymal transition (EMT) and contributes to breast tumourigenesis [15]. Further, PHF8 is expressed at relatively higher levels of HER2+ breast cancer cell lines, and PHF8 is required for their anchorage-independent growth. PHF8 demethylates histones H3K9me2 and H3K27me2 [16], [17], [18], [19] and H4K20me1 [20,21]. These studies discovered the general transcriptional coactivator function of PHF8. Further, PHF8 is overexpressed and associated with the malignant phenotypes of diverse cancers such as prostate cancer [22,23], oesophageal squamous cell carcinoma [24], lung cancer [25], and hepatocellular carcinoma [26]. Our team further identified the MYC-miR-22-PHF8 regulatory axis upregulates MYC expression, which in turn indirectly upregulates PHF8 expression through repression of microRNA-22 (miR-22) that represses PHF8 [15,23]. Moreover, a USP7-PHF8-positive feedback loop was discovered in which deubiquitinase USP7 stabilises PHF8, and PHF8 transcriptionally upregulates USP7 in breast cancer cells [27]. Through this mechanism, stabilised PHF8 upregulates CCNA2 to augment the proliferation of breast cancer cells [27]. These data support the conclusion that elevated expression of PHF8 contributes to its oncogenic activity. However, the epigenetic regulatory role that PHF8 plays in HER2-driven tumour development and resistance to anti-HER2 therapy is unknown.
We report here that PHF8 expression was elevated in HER2+ breast cancers and in cells that overexpressed HER2. The upregulation of PHF8 played a coactivator role in HER2 expression and that of genes upregulated by activated HER2 signalling. Moreover, we demonstrate that PHF8 contributed to the upregulation of IL-6 expression in vitro and in vivo and that the PHF8-IL-6 axis mediated resistance to anti-HER2 drugs. This study illuminates the potential for drug development aimed at inhibiting histone demethylase in HER2-driven tumour development and in the resistance to therapy.
2. Materials and methods
2.1. Cell lines and treatments
All cell lines used in this study were obtained from the American Type Culture Collection (Rockville, MD, USA). MCF-7, MDA-MB-231, K562 and HEK293T cells were grown in Dulbecco's Modified Eagle Medium (DMEM) containing 10% FBS (Gibco). HCC1954, SKBR3 and BT474 cells were cultured in RPMI1640 medium containing 10% FBS. MCF10A cells were cultured in DMEM-F12 supplemented with 20 ng/ml Epidermal Growth Factor (EGF) (Sigma), 100 ng/ml cholera toxin (Sigma), 10 g/ml insulin (Sigma), 500 ng/ml hydrocortisone (Sigma), and 5% horse serum. All cell lines used in this study were maintained in the specified medium supplemented with 1 × Penicillin–Streptomycin (Gibco) and incubated in 5% CO2 at 37 °C. MCF7-HER2, MCF10A-HER2 (overexpressing HER2), PHF8 inducible knocking down SKBR3, BT474 and HCC1954 stable cell lines were obtained as described earlier [15]. Briefly, cDNA of HER2 were cloned into the XhoI and NotI sites of the pOZ retroviral vector, PHF8 shRNAs were cloned into the AgeI and EcoRI sites of a TetON-pLKO-puro lentivirus vector, which was a gift from Dmitri Wiederschain (Addgene plasmid #21915, RRID:Addgene_98398) [28]. Retrovirus and lentivirus packaging of the pOZ and TetON-pLKO-puro vectors, infection, and stable selection were performed as described previously [20]. Briefly, Cells were infected with pOZ virus for 72 h and selected with anti-IL-2α receptor, which was conjugated with magnetic beads. Cell infected with control or PHF8 shRNAs were subjected to puromycin selection. Doxycycline at 1 μg/ml and 0.5 μg/ml was applied to induce the expression of target shRNAs for experiments lasting < 72 h and 6 days, respectively. Knockdown efficiency was verified by quantitative RT-PCR and western blotting. Two different shRNAs per target gene were tested to reduce off-target effects. PHF8 in house rabbit-anti-PHF8 [20] for western blotting, HER2 (29D8, RRID:AB_2799587), PARP (46D11, RRID:AB_659884), CDH2 (13A9, RRID:AB_2798427), pAKT (Ser473) (193H12, RRID:AB_331168) from Cell signalling Technology, γ-TUBULIN (MA1-850, RRID:AB_2211249) from Thermo Fisher; TFAP2C (sc-12762, RRID:AB_667770), c-MYC (SC-40, RRID:AB_627268), ZEB1 (sc-10572, RRID:AB_2273177), pSTAT3 (SC-7993, RRID:AB_656682), STAT3 (sc-482, RRID:AB_632440) from Santa Cruz, β-ACTIN (Ab8227, RRID:AB_2305186) were used for western blotting.
T-DM1[29] (Kadcyla, Genentech) was used to treat cells for 24 h. Dimethyl sulfoxide (equal volume to that of treated cells) was added to the culture media of the control cells. Human Cytokine Antibody Array blots probed with the cell cultured media. Serum free media were added after doxycycline induction for 72 h of each cell line and collected after 24 h culturing. Recombinant human IL-6 was purchased from PeproTech (Rocky Hill, NJ). Enzyme-linked immunosorbent array (ELISA) 10,000 cells were seeded in 6-well plates with complete medium of each cell line with FBS for 24 h. Then the medium was completely replaced to serum-free medium for another 24 h, and the supernatant was tested using the Quantikine human IL-6 (sensitivity < 5 pg/mL) ELISA kit (R&D Systems, Inc., Minneapolis, MN) according to manufacturer's recommended conditions. For some experiments, cells were induced with doxycycline for 72 h first before serum-free medium replacement, and the cytokine in the media was analysed by ELISA.
2.2. RNA-seq analysis
Gene expression analysis by next generation RNA sequencing (RNA-seq) was performed on RNA isolated from overexpression of HER2 MCF10A cells with two PHF8 inducible shRNAs versus double mock vectors control cells, which were marked as mock/ctlshRNA, HER2/ctlshRNA, HER2/PHF8shRNA1 and HER2/PHF8shRNA2 groups. Expression values were calculated as FPKM (fragment per kilobase of exon per million of mapped fragments) and were used to determine differential expression of mRNAs in four groups of samples.
Transcripts were called expressed if FPKM values in overexpression of HER2 with shRNA control samples were ≥1.0 for mRNAs. The mean expression level and differences in expression between four groups were calculated, and from these statistically significant differences in expression between each group was determined using a paired t-test. Fold change (HER2/shNC versus mock/shNC) was calculated to identify differentially expressed transcripts. Transcripts were deemed differentially expressed by OE-HER2 if the fold change was ≥1.5 or ≤0.5, with p value less than or equal to 0.05. Additionally, transcripts were deemed as PHF8 conserved regulated if mean fold change of HER2/ PHF8shRNA1 or 2 versus HER2/shNC was ≥1.3 or ≤0.7 (p ≤ 0.05) and regulation by two shRNAs were of same trend. Differentially expressed mRNAs that significantly regulated by PHF8 are represented by heatmaps by Graphpad prism 7, and Z scores were scaled by row using standard Z score calculation of log 10 absolute FPKM values.
2.3. ChIP and ChIP-qPCR
Chromatin immunoprecipitations (ChIPs) were performed as described previously [15]. Briefly, formaldehyde crosslinked cells were lysed and sonicated to shear the DNA. The sonicated DNA–Protein complexes were immunoprecipitated with the following antibodies: control IgG (A01008, RRID:AB_1108307), anti-TFAP2C (sc-12762, RRID:AB_667770), anti-PHF8 (ab36068, RRID:AB_2050177), anti-H3K4me3 (ab8580, RRID:AB_306649), anti-H3K27ac (ab4729, RRID:AB_2118291)). The immuno complexes were collected using protein A/G agarose beads. The eluted DNA and 1% of respective input DNA were reverse cross-linked at 65 °C overnight and used for the qPCR using SYBR Green qPCR mix and a CFX96 instrument (BioRad).
2.4. Cell proliferation assay (MTT)
Cells were seeded at a density of 3 × 103 cells/well in a 96-well plate with outer wells left empty for addition of PBS. After 24 h of culture, the media was changed and vehicle, drugs, or IL-6 (100 ng/mL) [30] were added. The cells were incubated with inhibitors or drugs for the time specified; then 0.5 mg/mL MTT dye was added and the cells were incubated for an additional 4 hr. Formazan crystals were dissolved in dimethyl sulfoxide (DMSO) for 15 min and the plates were read spectrophotometrically at 590 nm with a reference of 650 nm. Each assay was performed at least three times with five wells replication.
2.5. Mouse works
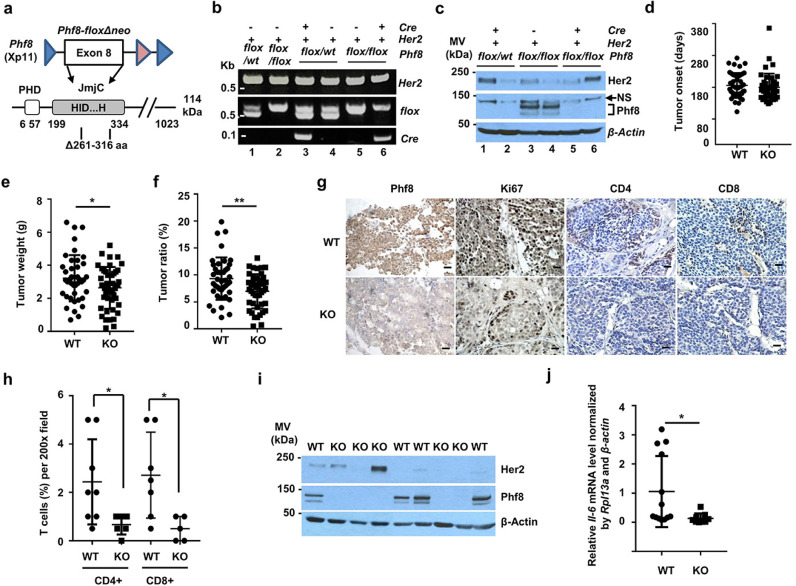
Phf8 knockout mice were established by Dr. Yang Shi's lab at Harvard Medical School. Briefly, the Phf8fl°x/fl°x allele was generated by flanking exon 8 with two loxp cassettes. Exon 8 of Phf8 encodes amino acid residues 261–316 of the C-terminal JmjC domain containing a 2-oxoglutarate (2-OG)-binding residue (K264) (Fig. 5A). Deletion of this region truncates PHF8 and abolishes its demethylase activity. We used MMTV-Cre to knockout Phf8 (KO) from mammary epithelial cells.
Fig. 5.
Functional impact of PHF8 on HER2-induced tumourigeneses and inflammatory response in vivo. a.Schematic of floxed Phf8 and the corresponding amino acid sequence. b. Genotyping of Her2, flox, and Cre using tail DNA from the indicated mouse. kbp: Kilobase pair. c. Western blotting analysis of PHF8 and HER2 levels of mice with the indicated genotypes. d–f. Tumour onset date (d), tumour weight (e), and tumour ratio (f) of WT to Phf8 KO mice. Each point represents an individual animal. The Student t-test was used to calculate the significance of differences between the KO and WT groups (n = 86). g. IHC analysis of PHF8, Ki67, CD4, and CD8 levels in tumour tissues from Phf8 KO and WT mice (magnification, 200 ×, bar = 10 μm. h. Comparison of infiltrating T cells in tumours from WT and Phf8 KO mice. The percentages of intraepithelial and stromal tumour-infiltrating CD8+ and CD4+ T cells were quantified in 200x fields. I. Western blotting analysis of HER2 and PHF8 levels in primary tumour cell lines from Phf8 KO and WT mice. J. RT-qPCR mRNA levels of Il-6 in primary tumour cell lines. Il-6 expression was normalised by both Rpl13a and β-actin. * p ≤ 0.05.
MMTV-Her2 mice were provided by Dr. Weizhou Zhang. MMTV-Her2, MMTV-Cre, and Phf8fl°x/fl°x mice were crossed to generate wild-type (WT) PHF8 mice: MMTV-Her2/MMTV-Cre, MMTV-Her2/Phf8fl°x/fl°x, and PHF8 KO mice: MMTV-Her2/MMTV-Cre/Phf8fl°x/fl°x. Notably, because Phf8 resides on the X chromosome, a segment of MMTV-HER2/MMTV-Cre/Phf8fl°x/wt is completely deleted because of inactivation of the X chromosome as shown in Fig. 5C. All mice have been fully backcrossed to FVB/N mice (from the Jackson Laboratory) for 8 generations. Her2-driven mammary tumours were monitored every 3 days. At experimental endpoint when largest tumour reaches 2 cm in diameter, animals were sacrificed, and all mammary tumours were removed and weighed. After the mice were sacrificed, tumour weight was directly measured, and tumour ratio was calculated as percentage of body weight. Tumours were fixed, embedded in paraffin, and serially sectioned at a thickness of 6–8 μm, and IHC staining was performed as described previously [15].
2.6. Immunohistochemical staining (IHC)
Histological tissue arrays (BR10010c, BR1503d, BR1504a, BR244 and BR082a) purchased from USBIOMAX were routinely processed as previously described [15]. Both protease-induced epitope retrieval and heat-induced epitope retrieval were carried out. Endogenous peroxidase activity was blocked with 3% H2O2. After blocking by reagent (Immunoperoxidase Secondary Detection System, Millipore), immunohistochemical blotting was performed using PHF8 antibody (PHF8 IHC-00343, RRID: AB_1264338) 100x diluted in PBST with 1% goat serum. Biotinylated secondary goat anti-mouse IgG/goat anti-rabbit IgG (Millipore) antibody was used in labelling with IHC Select® Immunoperoxidase Secondary Detection System (Millipore) for PHF8 detection. Pictures were taken by a Nikon digital camera through Nikon Eclipse 80i microscope.
2.7. Ethics statement
The mouse work in this study was conducted according to the procedures approved by the Ethics Committee (the Institutional Animal Care and Use Committee (IACUC)) at The University of Iowa and complied with animal use guidelines. Human tissue microarrays were purchased from USBIOMAX from which the clinical information on patient cohorts for expression analysis is openly available for research use and has been de-identified.
2.8. Statistical analysis
GraphPad Prism software (v7) was used to conduct statistical analysis. Results are expressed as the mean ± SD. Differences between experimental groups were compared using an unpaired two-tailed Student's t-test (for two conditions). A *p value <=0.05 was considered statistically significant. ** p value <=0.01 was considered highly significant.
2.9. Oligonucleotides
All oligonucleotides were synthesized by IDT and sequence of each was listed below.
| Oligonucleotides names | Sequence |
|---|---|
| PHF8 RT-qPCR F | GCAAACCGCAGCACCACACCT |
| PHF8 RT-qPCR R | CGAGTCTCTGCTTTGCTGTG |
| HER2 RT-qPCR F | TGTGTGGACCTGGATGACAAGG |
| HER2 RT-qPCR R | CTCCGTTTCCTGCAGCAGTCT |
| IL-6 RT-qPCR F | AGCCAGAGCTGTGCAGATGAGTA |
| IL-6 RT-qPCR R | TGACCAGAAGAAGGAATGCCCAT |
| RPL13A RT-qPCR F | CCTGGAGGAGAAGAGGAAAGAGA |
| RPL13A RT-qPCR R | TTGAGGACCTCTGTGTATTTGTCAA |
| Mouse Rpl13a RT-qPCR F | GGTGGAAGTACCAGGCAGTGACA |
| Mouse Rpl13a RT-qPCR R | GAGGACCTCTGTGAACTTGCAGAT |
| Mouse actinb RT-qPCR F | GGCTGTATTCCCCTCCATCG |
| Mouse actinb RT-qPCR R | CCAGTTGGTAACAATGCCATGT |
| Mouse il-6 RT-qPCR F | TCCAGTTGCCTTCTTGGGAC |
| Mouse il-6 RT-qPCR R | GTACTCCAGAAGACCAGAGG |
| Mouse Phf8 RT-qPCR F | TGACTCCAACCCTACCCAAG |
| Mouse Phf8 RT-qPCR R | CGGCTGTTCTACCTCCTTCA |
| IL-6 ChIP-qPCR TSS F | ACATCCTCGACGGCATCTCA |
| IL-6 ChIP-qPCR TSS R | GAACCCAGCAAAGACCTCCTA |
| IL-6 ChIP-qPCR UTR F | AAAGGCTTTGGCCACCAGTA |
| IL-6 ChIP-qPCR UTR R | CAGGATCAGCACCAAGGGTT |
| Human HER2 cDNA XhoI F | GAC GAC CTC GAG ATG GAG CTG GCG GCCTTG TGC CGC T |
| Human HER2 cDNA NotI R | GAC GAC GCG GCC GCC ACT GGC ACG TCC AGACCC AGG TA |
| HER2 ChIP promoter 1 F | CCCTGCTGTGTCCATATATCGAG |
| HER2 ChIP promoter 1 R | GGATAGTTACAGGTACGTTTAGGAA |
| HER2 ChIP promoter 2 F | CGAAGAGAGGGAGAAAGTGAAGCT |
| HER2 ChIP promoter 2 R | GGAATCTCAGCTTCACAACTTCAT |
| HER2 ChIP CTCF F | CCCCGACTTGAGGTATCCTT |
| HER2 ChIP CTCF R | GGGGCATACAAAAGAGGGCT |
| HER2 ChIP NRE F | CCCTCTGACGTCCATCATCTCT |
| HER2 ChIP NRE R | CTCCGTTTCCTGCAGCAGTCT |
| MMTVHER2genotyping F | TTT CCT GCA GCA GCC TAC GC |
| MMTVHER2genotyping R | CGG AAC CCA CAT CAG GCC |
| MMTVCre genotyping F | GCG GTC TGG CAG TAA AAA CTA TC |
| MMTVCre genotyping R | GTG AAA CAG CAT TGC TGT CAC TT |
| Flox genotyping F | CAGTAGGTAGCATGGTTTTGTGTGGA |
| Flox genotyping R | TTCAATAAGAGTATTACCCTATACATTTC |
3. Results
3.1. PHF8 expression is elevated in HER2+ breast cancers and upregulated by HER2
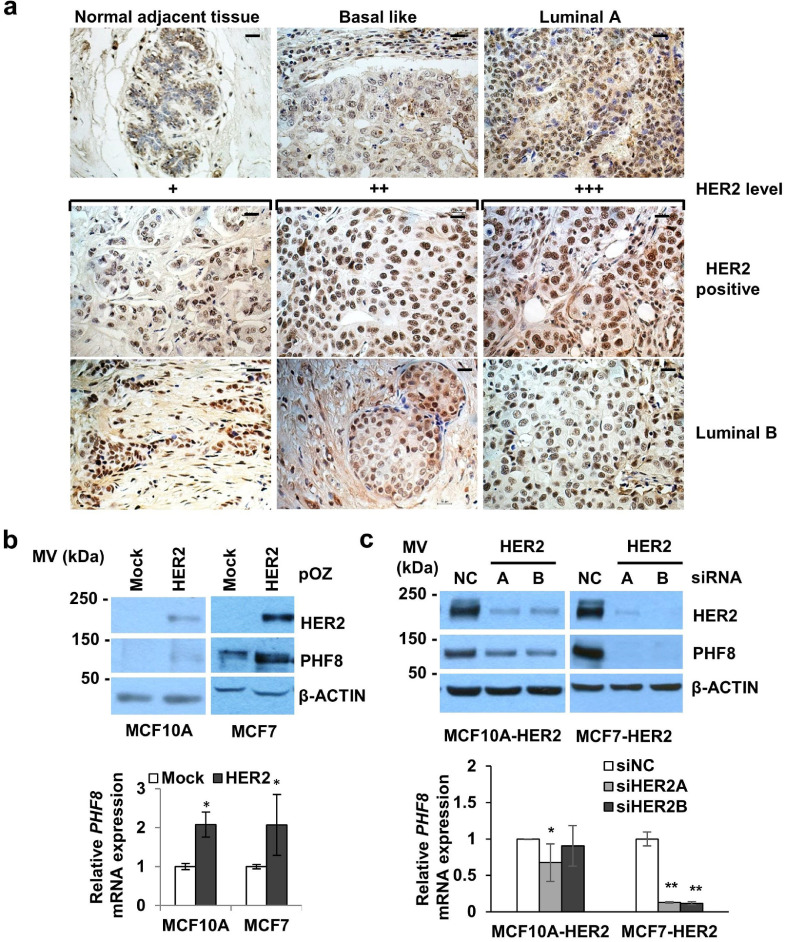
Prompted by findings that higher expression of PHF8 in HER2+ breast cancer cells is required for their anchorage-independent growth [15], we performed Gene Expression Profiling Interactive Analysis [31] to initially evaluate PHF8 mRNA levels in breast cancers. Intriguingly, we found that PHF8 mRNA levels were only slightly elevated amongst breast cancers and their subtypes (n = 1085) compared to those of normal tissues (n = 291) (Supplementary Figure 1A and 1B). PHF8 is subject to the activities of post-transcriptional and post-translational regulators such as MYC-miR-22-PHF8 [15,23] and USP7-PHF8 [27]; consequently, the actual PHF8 protein levels in cancers can differ from those of its mRNA. Therefore, we used immunohistochemistry (IHC) to analyse cancer tissue arrays with 486 breast cancers, 20 normal breast tissues, and 50 metastatic lymph nodes samples (US BIOMAX). Significant increases were detected in PHF8 protein levels (strong nuclear staining) across breast cancers and all subtypes, including HER2+ breast cancers (Supplementary Table 1 and Fig. 1A). These data, together with higher PHF8 levels in the HER2+ breast cancer cell lines SKBR3 and BT474 [15], suggested the regulation of PHF8 by HER2.
Fig. 1.
PHF8 expression is elevated in HER2+ breast cancers and is upregulated by HER2.a. Immunohistochemistry (IHC) was used to measure PHF8 levels in breast cancer tissue arrays. Representative PHF8 staining of breast cancer and normal adjacent tissues: Basal like, Luminal A, Luminal B, and HER2+ samples. Magnification: 200 ×, bar = 10 μm. b. Western blot and RT-PCR analyses of the levels of PHF8 (upper panel) and its mRNA (lower panel) in MCF10A and MCF7 cells with or without overexpression of HER2. c. Western blotting and RT-PCR analyses of PHF8 protein (upper panel) and mRNA (lower panel) levels in MCF10A-HER2 and MCF7-HER2 cells with or without HER2 knockdown. * p ≤ 0.05; ** p ≤ 0.01.
Moreover, we found that the overexpression of HER2 in cells infected using the pOZ retroviral system upregulated the levels of PHF8 and its mRNA levels in MCF10A and MCF7 (Fig. 1B). Conversely, HER2 knockdown using siRNAs specific for the HER2 gene body inhibited the upregulation of PHF8 in these cells but significantly increased PHF8 mRNA levels in MCF7 cells (Fig. 1C). Further, HER2 knockdown in SKBR3, BT474 and HCC1954 cells downregulated PHF8 levels (Supplementary Figure 2), such decrement on PHF8 levels were also observed by anti-HER2 drugs (Supplementary Figure 2 and Supplementary Figure 8a). Moreover, a significant positive correlation between HER2 and PHF8 mRNAs was found through analyses of the databases as follows: Cancer Genome Atlas Breast Invasive Carcinoma (TCGA-BRCA), TCGA normal breast tissue, and Genotype-Tissue Expression (GTEx) Program mammary tissue (Supplementary Figure 1C). Together, these findings support the hypothesis that elevated expression of HER2 upregulates that of PHF8.
3.2. PHF8 is a transcriptional coactivator of HER2
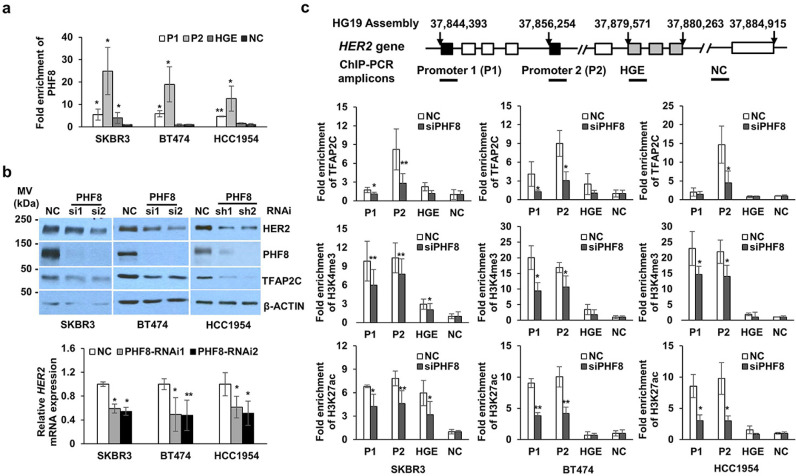
The coactivator role of PHF8 caused us to hypothesise that PHF8 participates in the transcriptional regulation of HER2. PHF8 ChIP-seq data acquired from human embryonic H1 cells and K562 cells [32] indicate enrichment of PHF8 bound to the two promoter regions of HER2 (Supplementary Figure 3). It is not surprising, therefore, that PHF8 colocalized with H3K4me3, because PHF8 binds to H3K4me3 through its PHD domain [20]. Importantly, we identified similar enrichment of PHF8 on HER2 promoters in SKBR3, BT474, and HCC1954 cells (Fig. 2A). Loss of function of PHF8 in cells expressing a PHF8-specific siRNA or shRNA in SKBR3, BT474, and HCC1954 cells uniformly downregulated the levels of HER2 and those of its mRNA (Fig. 2B), supporting the conclusion that PHF8 functions as a coactivator in the transcriptional regulation of HER2. We recently identified a novel HER2 gene body enhancer (HGE), which recruits transcription factor TFAP2C [33], a positive regulator of HER2 expression [34], [35], [36], [37], [38]. Notably, TFAP2C is a direct target of PHF8 [20]. Thus, when we were searching for if PHF8 regulated TFAP2C in HER2+ cells, we found that PHF8 knockdown downregulated levels of TFAP2C and those of its mRNA (Figs. 2B and Supplementary Figure 4). Moreover, the enrichment of TFAP2C at HER2 promoters was reduced in PHF8-RNAi cells (Fig. 2C), indicating that the regulation of TFAP2C by PHF8 contributed to HER2 expression.
Fig. 2.
PHF8 functions as a transcriptional coactivator that regulates HER2 expression. a.Chromatin immunoprecipitation (ChIP) analysis of PHF8 bound to HER2 in HER2+ cells. Enrichment of PHF8 on the HER2 promoter 1 (P1), promoter 2 (P2), HER2 gene body regulatory element (HGE), and an unmodified region (NC) were analysed using ChIP-qPCR. Relative enrichment represents the average fold-enrichment of PHF8 vs. the input, normalised to the NC region. b. Western blotting and RT-PCR analyses of the levels of HER2 and its mRNA in the indicated cells with and without PHF8 knockdown. NC: control scrambled siRNA or shRNA; PHF8-siRNAs, PHF8-si1 and PHF8-si2 and PHF8 shRNAs, PHF8-sh1 and PHF8-sh2, are described in the methods section. Lower panel: RT-qPCR analysis of HER2 mRNA expression normalised to that of RPL13A. c. ChIP-qPCR analysis of the recruitment of H3K27ac, H3K4me3, and TFAP2C to HER2 in HER2+ cells with PHF8 knockdown. The Student t-test was performed to evaluate the significance of differences between variables. * p ≤ 0.05, ** p ≤ 0.01.
HER2 is constitutively active in HER2+ breast cancer cells, therefore, possesses an active chromatin state. Thus, PHF8 demethylation substrates such as H3K9me2, H4K20me1, and H3K27me2 may not be highly enriched at HER2 promoters. By contrast, H3K4 me3 is required for the transcriptional regulation of HER2 in breast cancer cells [39]. Moreover, PHF8 plays a critical role in sustaining the levels of H3K4me3 in diverse cell types [20,40]. ChIP experiments reinforced this role of PHF8. Thus, PHF8 knockdown reduced H3K4me3 levels at HER2 promoters in the three cell lines tested (Fig. 2C). These data support the conclusion that PHF8 participated in the transcriptional regulation of HER2 by sustaining H3K4 me3 levels. Moreover, H3K27ac, a general activation marker, was downregulated on HER2 promoter regions in PHF8 knockdown cells (Fig. 2C). Thus, PHF8 may demethylate H3K27 to prime the acetylation of, or indirectly regulate, H3K27ac through its acetyltransferase. Together, our data reveal the role of PHF8 in the transcriptional regulation of HER2, in which the underlying mechanisms may involve direct and indirect regulation of multiple factors.
3.3. PHF8 functions as a dominant coactivator downstream in the HER2 signalling pathway
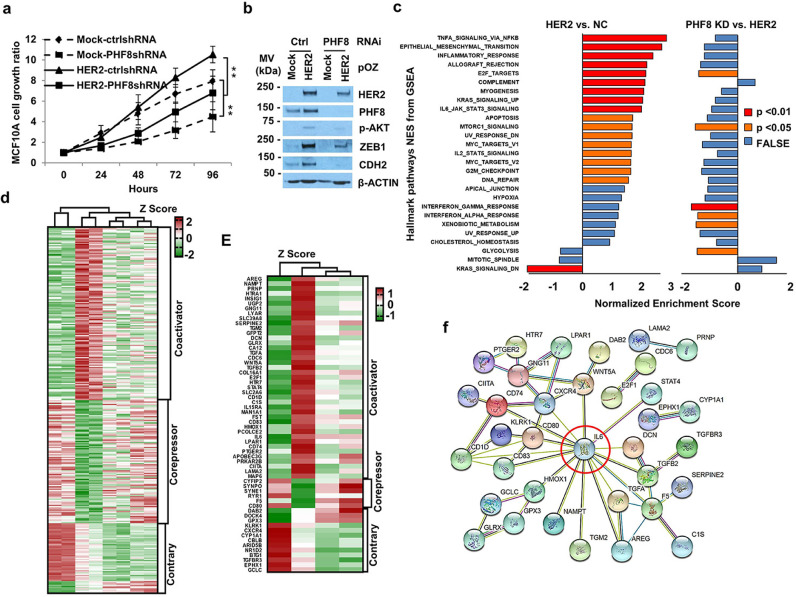
Although PHF8 directly participates in the transcriptional regulation of HER2, PHF8 knockdown reduced HER2 mRNA levels by approximately 30% (Fig. 2B). Thus, we aimed to further identify the genome-wide effects of PHF8 on HER2-regulated genes. MCF10A cells are extensively used to study HER2 function [41], [42], [43]. Thus, we established MCF10A cells that stably expressed HER2 and under doxycycline-inducible PHF8 shRNAs. HER2 can induce genomic instability [44], therefore, we used early-passage cell lines to maintain their isogenic status. Consistent with previous reports [42,45], HER2 overexpression induced proliferation, AKT phosphorylation (p-AKT), and expression of EMT markers (N-cadherin [CDH2] and zinc finger E-box binding homeobox 1 [ZEB1]) (Fig. 3A and B). Notably, silencing PHF8 attenuated these effects (Fig. 3A and B). Moreover, PHF8 knockdown downregulated overexpressed HER2, indicating that PHF8 may indirectly regulate HER2.
Fig. 3.
PHF8 facilitates HER2 signalling through its tumour-promoter activity. a. The proliferation of MCF10A cells overexpressing HER2, with PHF8 knockdown, or both was assessed using the MTT assay. Data are presented as mean ± SD of three independent experiments. **p ≤ 0.01. b. Western blotting analysis of the expression of HER2, PHF8, and other indicated proteins shown in panel a in MCF10A cells. The data represent three independent experiments. A and B, Mock: overexpression control; HER2: HER2 overexpression; shNC: scrambled shRNA; shPHF8: PHF8 shRNAs. c. PHF8 knockdown counteracted the activity of most pathways induced by HER2 overexpression. Gene set Enrichment Analysis (GSEA) of Hallmark pathways in HER2-overexpressing cells vs. control cells compared with HER2-overexpressing cells vs. PHF8 knocking down. The p values are coloured; normalised enrichment scores (NES) are shown on the x-axis. d. Heat map of RNA sequencing Z-score results of 298 PHF8 differentially regulated genes (DRG) that were significantly regulated by HER2 overexpression. e. Heat map of the Z-scores of 60 PHF8-associated DRGs contributing most to enriched pathways. f. Protein–protein association networks of 60 PHF8-associated DRGs were analysed using STRING. Edges represent protein-protein associations such as direct interactions, gene neighbourhood, co-expression, and co-occurrence.
We next used RNA-seq to analyse cell lines expressing mock/control shRNA, HER2/control shRNA, and HER2/PHF8shRNAs 1 and 2. We identified 838 upregulated and 536 downregulated genes in cells overexpressing HER2 (cut-off of actual fold change = 1.5 [FC ≥1.5 or ≤ −1.5] and adjusted p value < 0.05) (Supplementary Table 2). Gene Set Enrichment Analysis (GSEA) [46] of hallmark gene sets revealed that HER2-regulated genes were significantly enriched in 18 pathways such as TNFα signalling, the EMT transition, the inflammatory response, and mTOR signalling (Fig. 3C and Supplementary Table 5), consistent with previous reports using similar parameters [47], [48], [49]. PHF8 knockdown attenuated most of the pathways induced by HER2 overexpression (Fig. 3C and Supplementary Table 6). Importantly, the pathways of E2F targets, mTOR signalling, and interferon responses elevated by HER2 overexpression were significantly counteracted by PHF8 loss of function (Fig. 3C). These data strongly suggest that the general coactivator functions of PHF8 were associated with an HER2-induced transcriptome.
We next defined 298 genes that were differentially regulated (DRGs) by PHF8 by subtraction from HER2-regulated genes using the criteria as follows: same trend of regulation by two PHF8 shRNAs, where at least one shRNA accounted for >30% of DRGs, and p ≤ 0.05. The 30% cut-off was selected according to the published data for the general regulatory function of PHF8 [20,21,27]. These 298 DRGs clustered into a coactivator (PHF8 knockdown attenuated the expression of genes upregulated by HER2 or enhanced the expression of genes downregulated by HER2) or a corepressor (PHF8 knockdown counteracted the expression of genes downregulated by HER2 or enhanced the expression of genes upregulated by HER2) groups (Figs. 3D, Supplementary Table 3, and 4). These analyses led us to the general conclusion as follows: The transcriptional coactivator function of PHF8 was dominant compared to its corepressor function. GO biological processes indicate that PHF8 coactivator genes (upregulated by HER2) were enriched in cell proliferation and cytokine production, whereas PHF8 corepressor genes (downregulated by HER2) were enriched in axon and neuron regeneration (Supplementary Figure 5).
To further investigate the biological effects of PHF8 on HER2-regulated genes, we identified genes that contributed to significantly enriched pathways regulated by HER2 overexpression or PHF8 knockdown from the 298. The result was a 60-gene signature (Figs. 3E and Supplementary Table 7). The identification of these genes further demonstrated the dominant coactivator functions of PHF8 downstream of HER2 signalling. Analysis of protein–protein association networks of these 60 genes using STRING [50] revealed that interleukin-6 (IL-6) is a hub (Fig. 3F) that contributes to nine pathways, including the interferon response, TNFa signalling, the EMT, and inflammation (Supplementary Table 7). These data suggest that PHF8 contributed to the tumourigenic functions of HER2 through IL-6.
3.4. PHF8 promotes resistance to the anti-HER2 effects of breast cancer cells through IL-6
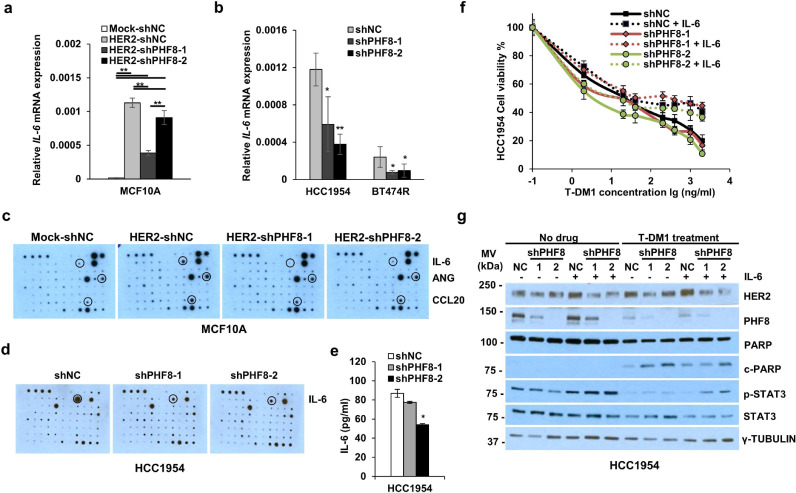
We next investigated the regulation of IL-6 by PHF8 and its contribution to HER2 signalling and the resistance of breast cancers to anti-HER2 drugs. Our rationale was as follows: the regulation of IL-6 by HER2 overexpression that ranked higher (Supplementary Table 7), the central position of IL-6 in the protein–protein association network of PHF8-associated DRGs in the context of HER2 (Fig. 3F), and the functional importance of IL-6 in HER2 signalling [51] and in drug resistance [52,53] especially the resistance to trastuzumab [51,54,55] and lapatinib [56]. First, we confirmed the regulation of IL-6 by HER2 and PHF8 in MCF10A cell lines (Fig. 4A) and in HCC1954 and BT474 lapatinib-resistant (-R) cells [57] cells (Fig. 4B), which possess higher IL-6 mRNA and protein levels compared with the other cell lines tested (Supplementary Figure 6). We next analysed a human cytokine antibody array and obtained similar results. PHF8 knockdown attenuated the upregulation of IL-6 by HER2 overexpression in MCF10A cells (Fig. 4C) and downregulated IL-6 expression in HCC1954 cells (Fig. 4D). The regulation of IL-6 by PHF8 in HCC1954 cells was further validated using an ELISA assay (Fig. 4E). Notably, angiogenin (ANG) and CCL20 were regulated by HER2 and PHF8 in a pattern similar to that of IL-6 (Fig. 4C). However, similar data for PHF8 were not acquired using HCC1954 cells (Fig. 4D).
Fig. 4.
PHF8 promotes resistance to anti-HER2 therapies in breast cancer cells through IL-6.a and b. RT-qPCR analysis of IL-6 expression. RPL13A mRNA served as the control. c and d. Human Cytokine Antibody Array blots probed with the media from the indicated cells. Western blotting was performed in duplicate, an ELISA was used to measure expression, and the regulated cytokines are marked. e. An ELISA was used to measure IL-6 secreted from HCC1954 cells with or without PHF8 knockdown; * p ≤ 0.05, ** p ≤ 0.01. f. MTT analysis of the viability of HCC1954 cells with or without PHF8 knockdown in the presence or absence of IL-6. Cells were treated doxycycline for 72 h and then reseeded into the wells of 96-well plates. IL-6 (100 ng/mL) was added to the cells 24 h before the addition of T-DM1 (48 h) (n = 5). g. Western blotting analysis of the levels of HER2, PHF8, PARP/c-PARP (cleaved PARP), and STAT3/PSTAT3 in HCC1954 cells treated as in f, γ-Tubulin served as the loading control. The data represents three independent experiments.
HCC1954 cells are resistant to trastuzumab [58] and express PHF8 (Supplementary Figure 7). Therefore, we investigated whether PHF8 and the PHF8-IL-6 axis contributed to such resistance. The IC50 of trastuzumab (T-DM1) of HCC1954 cells transfected with a control shRNA was 92.44 ng/ml (Fig. 4F). Knockdown of PHF8 expression by the two shRNAs reduced the IC50 values to 33.72 ng/ml and 29.68 ng/ml (Fig. 4F), suggesting a positive role for PHF8 in the resistance of HCC1954 cells to trastuzumab. When exposed to IL-6 (Fig. 4F), inhibition of viability of the HCC1954 cells infected with control shRNA stayed static with increasing T-DM1 concentrations (IC50 value = 106.43 ng/ml) (Fig. 4F). However, exposure of IL-6 increased the IC50 to 104.57 ng/ml and 64.47 ng/ml, in two PHF8-knockdown cell lines, supporting the conclusion that the PHF8-IL6 axis contributed to the resistance of HCC1954 cells to trastuzumab.
Fig. 4G illustrates the results of the western blotting analysis of these cells in the presence and absence of T-DM1 (2 ng/ml). PHF8 knockdown slightly reduced the levels of activated STAT3 (p-STAT3), and the addition of IL-6 restored p-STAT3 levels (Fig. 4G), supporting the role of PHF8 in regulating IL-6 signalling. T-DM1 treatment of the control cells significantly reduced p-STAT3 levels and induced apoptosis, reflected by the detection of cleaved PARP (c-PARP) (Fig. 4G). PHF8 knockdown increased T-DM1-induced apoptosis, and the addition of IL-6 counteracted the induction of apoptosis (Fig. 4G), supporting the contribution of the PHF8-IL-6 axis to the resistance of HCC1954 cells to trastuzumab. Experiments with the same setup of lapatinib plus trastuzumab treatment were performed in HCC1954 cells (Supplementary Figure 8a). The results are of the same trend; however, apoptosis cannot be detected obviously since HCC1954 is highly resistant to drugs. Moreover, overexpressing PHF8 in drug-sensitive parental SKBR3 cells showed PHF8 gain-of-function contributes to lapatinib plus trastuzumab resistance of these cells (Supplementary Figure 8b).
3.5. Phf8 contributes to HER2-driven breast tumour development in vivo
PHF8 is required for the anchorage-independent growth of HER2+ breast cancer cells [15]. Our current data support synergism between PHF8 and HER2. Thus, we sought to further study the role of PHF8 in HER2-driven tumour development in vivo: For this purpose, we used a Phf8 knockout mouse model (illustrated in Fig. 5A) in which Her2 was overexpressed under the control of the mouse mammary tumour virus (MMTV) long terminal repeat. WT mice (n = 42) and Phf8 KO mice (n = 44) with an MMTV-Her2 background were genotyped (tail) (Fig. 5B), and PHF8 levels of tumour samples were determined using western blotting (Fig. 5C). There was no significant difference between the numbers and time to detection of mammary tumours that developed in the WT and Phf8 KO mice (185.8 ± 42.8 [mean ± SD] and 180 ± 39.2 days, respectively) (Fig. 5D). However, the average weight (2.64 ± 1.26 g) of tumours of Phf8 KO mice was significantly lower compared with that of the WT mice (3.22 ± 1.40 g) (Fig. 5E). Moreover, the ratio of the relative tumour weight as a percentage of total body weight of Phf8 KO mice was significantly reduced (6.97% ± 3.17% from 9.31% ± 1.26% for WT mice) (Fig. 5F). These data indicate that PHF8 played critical roles in tumour growth rather than in tumour initiation. This conclusion is further supported by the significantly reduced proliferative index of Phf8 KO mice (n = 7) compared to that of WT mice (n = 6) (12.47% ± 4.56% vs. 3.57% ± 2.18%; p = 0.002).
PHF8 contributes to the regulation of IL-6 and the IL-6-immune response network associated with tumour growth [59], [60], [61]. Therefore, we next analysed infiltrating T cells. Compared with widespread T cells in WT mice (n ≥ 7), the tumours from Phf8 KO mice (n ≥ 5) had few intratumoural T cells and few T cells peripheral to the tumour mass and in collagen bundles (Fig. 5G). Analysis of CD4 and CD8 expression revealed a significant reduction of intratumoural and peritumoural infiltrating T cells in Phf8 KO mice compared with those in WT mice (Fig. 5H). These data are consistent with those of studies indicating that in the tumour microenvironment, IL-6 promotes inflammation-induced CD8+ T cell trafficking in tumours [62] and in stromal cells such as CD4+ regulatory T cells (Treg) that support tumourigenesis. Next, we asked whether the regulation of IL-6 by PHF8 is conserved. For this purpose, we established 13 WT and 9 KO primary cultures from WT (n = 8) and KO (n = 6) mice, respectively. HER2 and PHF8 levels were determined using western blotting (Fig. 5I). Importantly, the Il-6 mRNA levels in cells of KO mice were significantly lower than those of WT mice (Fig. 5J), consistent with the cell line data. Together, these data indicate that the PHF8-IL-6 axis mediated T cell infiltration in HER2-driven tumour development.
4. Discussion
The resistance to anti-HER2 drugs, such as to lapatinib or trastuzumab, remains a hurdle to achieving successful therapy of HER2+ breast cancers [2]. Thus, it is critically important to identify novel therapeutic targets. Specific inhibitors of epigenetic factors may serve this purpose. Here, we used in vitro and in vivo approaches to demonstrate synergy between the histone demethylase PHF8 and HER2 and the oncogenic functions of PHF8 in HER2-driven tumour development. Our data have significant therapeutic implications for targeting PHF8 in HER2+ breast cancers.
The oncogenic functions of PHF8 contribute to diverse cancers [22], [23], [24], [25], [26]. We [15] and Wang et al. [27] discovered such functions of PHF8 in several subtypes of breast cancers that are associated with a significant increase of PHF8 mRNA levels. However, our previous [15] and current approaches using an updated dataset that includes more samples [29] normal and 1085 breast cancer tissues did not demonstrate significant upregulation of PHF8 mRNA levels in HER2+ breast cancers. By contrast, our IHC analysis presented here from a pool of >500 samples revealed a significant increase in PHF8 levels in all subtypes of breast cancers. Although the mRNA data do not agree with those acquired using breast cancer arrays, the discrepant mRNA and protein levels of PHF8 in breast cancers indicate the roles of the post-transcriptional [15] and post-translational [27] regulation of PHF8 expression. Although our previous study [15] demonstrated the transcriptional regulation of PHF8 by ectopically expressed HER2, the MYC-miR-22-PHF8 regulatory axis may contribute to the regulation of PHF8 by HER2 signalling for the following reasons: 1) MYC is a key player in HER2-mediated oncogenesis [63,64]; 2) The expression of miR-22 expression is lower in HER2+ breast cancers [65]; 3) miR-22 downregulates PHF8 levels in SKBR3 cells [15]. Moreover, LET-7 targets PHF8, which implicates LET-7 as a component of a putative HER2–LET-7–PHF8 axis for the following reasons: 1) LET-7 levels are lower in HER2+ breast cancers [65]; 2) HER2 represses LET-7 expression through ERK signalling [66] by activating LIN28, which inhibits the synthesis of LET-7 family members [67]; 3) HER2 inhibits LET-7 through a pathway involving AKT-MYC [68]. Together, these findings indicate that HER2 regulates PHF8 through multiple mechanisms. Further studies are required to prove the existence of an HER2-microRNAs–PHF8 axis.
RNA-seq and pathway analyses reported here indicate that the transcriptional coactivator role of PHF8 downstream of HER2 signalling is dominant over its corepressor role, consistent with the results of other studies [15,20,21,27]. HER2 is expressed at a higher rate as a function of its copy number in HER2-amplified breast cancer cells [39,69,70]. Thus, including PHF8 as a component of the MLL complex that influences the activity of H3K9 acetyltransferase [39] helps to decipher the epigenetic regulatory machinery of HER2. We reasoned, therefore, that constitutive transcription of HER2 may lower H3K9me2 levels, and it is possible that PHF8 plays a role of sustaining the low occupancy of H3K9me2 in the HER2 genomic region. Further, HER2 is transcriptionally upregulated by tamoxifen, an ER antagonist, in ER+ breast cancers and by radiation therapy of triple-negative breast cancers [4], [5], [6]. Thus, HER2 signalling contributes to resistance to endocrine treatment or radiotherapy, and PHF8 may serve as a transcriptional coactivator in breast cancer cells with HER2 amplification that overexpresses HER2.
HER2 signalling-specific DRGs regulated by PHF8 were enriched in the GO category “positive regulation of cell proliferation” followed by chemokine/cytokine biosynthetic pathways, implicating PHF8 in the immune response to tumours and its established functions in the regulation of the cell cycle. IL-6, CD74, TGFB2, WNT5A, and HMOX1 represent major components for cytokine-associated pathways.
IL-6 signalling is considered a “malevolent player” in tumour progression [59]. Analysis on IL-6 levels in subgroups of HER2/PHF8 using TCGA breast cancer data revealed no significant correlation between HER2/PHF8 and IL6 mRNA levels (data not shown). It is possibly due to two reasons: First, PHF8 is heavily subject to post-transcriptional and post-translational regulations, therefore, its mRNA levels may not truthfully reflect its actual protein levels; Secondly, tumour tissue from patients include immune, neutrophils and stromal cells from which IL-6 is mainly produced [59,71]. Importantly, our data using primary tumour cell lines from the mouse model are consistent with our data from the cell lines (Fig. 5j). Notably, IL-6 has significant tumour-inhibitory effects, because it influences tissue recruitment of T cells [72] and serves as a key player in the activation, proliferation, and survival of lymphocytes during immune responses that promote antitumour adaptive immunity [59]. Thus, our IHC data on CD4 and CD8 expressions on tumour tissues for the infiltration T cells can indirectly interpret the consequence of Il-6 secretion by tumour cells.
TGF-β and WNT signalling pathways play key roles in the EMT and in the acquisition of the metastatic potential of cancer stem cells and their resistance to therapy, and they are immunosuppressive [73]. The coactivator functions of PHF8 that influence the expression of cytokines may lead to opposing outcomes. However, the reduction of infiltrating T cells in the tumours of Phf8 KO mice demonstrates a positive role for PHF8 in the infiltration of T cells into a tumour. Thus, PHF8 may play an important role in the immune response by altering the tumour microenvironment and influencing T cell trafficking to tumour sites by regulating cytokine production.
In addition to its dominant coactivator function, PHF8 acts as a corepressor [74]. Specifically, PHF8 is phosphorylated by ERK2 upon IFNꝩ treatment and dissociates from repressive promoters where PHF8 forms a complex with HDAC1 and SIN3A in the static state [74]. This mechanism may apply to the HER2 function, because ERK can be activated by HER2 signalling [2,3]. Genome-wide PHF8 ChIP-seq in the MCF10A cell system is sought to decipher how PHF8 plays its corepressor function on the genes identified in this study. Genome-wide PHF8 ChIP-seq analyses of MCF10A cells may help decipher the mechanism through which PHF8 acts as a corepressor to regulate the genes identified here.
Declaration of competing interests
The authors claim no conflict of interest.
Acknowledgments
Acknowledgements
We thank Dr. Yang Shi and Dr. Hui-Jun Lim for providing Phf8 knockout mice. We also thank Dr. Songhai Chen for technical assistance on the mouse work and the IHC data analysis, Dr. Brad Amendt and his lab for helpful discussions.
Funding Sources
This work was supported by lab start-up funds to H.H.Q from the Department of Anatomy and Cell Biology, the Carver College of Medicine, University of Iowa; Carver Trust Young Investigator Award (01-224 to H.H.Q) from the Roy J. Carver Charitable Trust; a Breast Cancer Research Award (to H.H.Q.) by the Holden Comprehensive Cancer Centre at University of Iowa; The NIH grant (P30 CA086862) to the Genomics facility at the University of Iowa. N.B. was supported by NIH M.D./Ph.D. fellowship (F30 CA206255); W.Z was supported by NIH grants CA200673, and CA203834, the V Scholar award, a Breast Cancer Research Award and an Oberley Award (National Cancer Institute Award P30CA086862) from Holden Comprehensive Cancer Centre at the University of Iowa. The funders have no roles in study design, data collection, data analysis, interpretation, writing of the report.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2019.102612.
Appendix. Supplementary materials
Data for reference
RNA sequencing data were submitted to GEO database with accession number GSE140373.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Roskoski R., Jr The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol. Res. 2014;79:34–74. doi: 10.1016/j.phrs.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Baselga J., Swain S.M. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nature reviews Cancer. 2009;9(7):463–475. doi: 10.1038/nrc2656. [DOI] [PubMed] [Google Scholar]
- 4.Hurtado A., Holmes K.A., Geistlinger T.R., Hutcheson I.R., Nicholson R.I., Brown M. Regulation of ERBB2 by oestrogen receptor-PAX2 determines response to tamoxifen. Nature. 2008;456(7222):663–666. doi: 10.1038/nature07483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duru N., Fan M., Candas D., Menaa C., Liu H.C., Nantajit D. HER2-associated radioresistance of breast cancer stem cells isolated from HER2-negative breast cancer cells. Clin Cancer Res. 2012;18(24):6634–6647. doi: 10.1158/1078-0432.CCR-12-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao N., Li S., Wang Z., Ahmed K.M., Degnan M.E., Fan M. NF-kappaB-mediated HER2 overexpression in radiation-adaptive resistance. Radiat. Res. 2009;171(1):9–21. doi: 10.1667/RR1472.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iqbal N. Iqbal N. human epidermal growth factor receptor 2 (HER2) in cancers: overexpression and therapeutic implications. Mol Biol Int. 2014;2014 doi: 10.1155/2014/852748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paik S., Kim C., Wolmark N. HER2 status and benefit from adjuvant trastuzumab in breast cancer. N Engl J Med. 2008;358(13):1409–1411. doi: 10.1056/NEJMc0801440. [DOI] [PubMed] [Google Scholar]
- 9.Verma M., Banerjee H.N. Epigenetic inhibitors. Method Mol Biol. 2015;1238:469–485. doi: 10.1007/978-1-4939-1804-1_24. [DOI] [PubMed] [Google Scholar]
- 10.Greer E.L., Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13(5):343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delmore J.E., Issa G.C., Lemieux M.E., Rahl P.B., Shi J., Jacobs H.M. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye Q., Holowatyj A., Wu J., Liu H., Zhang L., Suzuki T. Genetic alterations of KDM4 subfamily and therapeutic effect of novel demethylase inhibitor in breast cancer. Am J Cancer Res. 2015;5(4):1519–1530. [PMC free article] [PubMed] [Google Scholar]
- 13.Singla H., Ludhiadch A., Kaur R.P., Chander H., Kumar V., Munshi A. Recent advances in HER2 positive breast cancer epigenetics: susceptibility and therapeutic strategies. Eur J Med Chem. 2017;142:316–327. doi: 10.1016/j.ejmech.2017.07.075. [DOI] [PubMed] [Google Scholar]
- 14.Ramadan W.S., Vazhappilly C.G., Saleh E.M., Menon V., AlAzawi A.M., El-Serafi A.T. Interplay between epigenetics, expression of estrogen Receptor- alpha, HER2/ERBB2 and sensitivity of triple negative breast cancer cells to hormonal therapy. Cancers. 2018;11(1) doi: 10.3390/cancers11010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shao P., Liu Q., Maina P.K., Cui J., Bair T.B., Li T. Histone demethylase PHF8 promotes epithelial to mesenchymal transition and breast tumorigenesis. Nucleic Acids Res. 2017;45(4):1687–1702. doi: 10.1093/nar/gkw1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loenarz C., Ge W., Coleman M.L., Rose N.R., Cooper C.D., Klose R.J. PHF8, a gene associated with cleft lip/palate and mental retardation, encodes for an Nepsilon-dimethyl lysine demethylase. Hum Mol Genet. 2010;19(2):217–222. doi: 10.1093/hmg/ddp480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fortschegger K., de Graaf P., Outchkourov N.S., van Schaik F.M., Timmers H.T., Shiekhattar R. PHF8 targets histone methylation and RNA polymerase II to activate transcription. Mol Cell Biol. 2010;30(13):3286–3298. doi: 10.1128/MCB.01520-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleine-Kohlbrecher D., Christensen J., Vandamme J., Abarrategui I., Bak M., Tommerup N. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol Cell. 2010;38(2):165–178. doi: 10.1016/j.molcel.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng W., Yonezawa M., Ye J., Jenuwein T., Grummt I. PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me1/2 demethylation. Nat Struct Mol Biol. 2010;17(4):445–450. doi: 10.1038/nsmb.1778. [DOI] [PubMed] [Google Scholar]
- 20.Qi H.H., Sarkissian M., Hu G.Q., Wang Z., Bhattacharjee A., Gordon D.B. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature. 2010;466(7305):503–507. doi: 10.1038/nature09261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu W., Tanasa B., Tyurina O.V., Zhou T.Y., Gassmann R., Liu W.T. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010;466(7305):508–512. doi: 10.1038/nature09272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bjorkman M., Ostling P., Harma V., Virtanen J., Mpindi J.P., Rantala J. Systematic knockdown of epigenetic enzymes identifies a novel histone demethylase PHF8 overexpressed in prostate cancer with an impact on cell proliferation, migration and invasion. Oncogene. 2012;31(29):3444–3456. doi: 10.1038/onc.2011.512. [DOI] [PubMed] [Google Scholar]
- 23.Maina P.K., Shao P., Liu Q., Fazli L., Tyler S., Nasir M. c-MYC drives histone demethylase PHF8 during neuroendocrine differentiation and in castration-resistant prostate cancer. Oncotarget. 2016;7(46):75585–75602. doi: 10.18632/oncotarget.12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun X., Qiu J.J., Zhu S., Cao B., Sun L., Li S. Oncogenic features of PHF8 histone demethylase in esophageal squamous cell carcinoma. PLoS ONE. 2013;8(10):e77353. doi: 10.1371/journal.pone.0077353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen Y., Pan X., Zhao H. The histone demethylase PHF8 is an oncogenic protein in human non-small cell lung cancer. Biochem Biophys Res Commun. 2014 doi: 10.1016/j.bbrc.2014.07.076. [DOI] [PubMed] [Google Scholar]
- 26.Zhou W., Gong L., Wu Q., Xing C., Wei B., Chen T. PHF8 upregulation contributes to autophagic degradation of E-cadherin, epithelial-mesenchymal transition and metastasis in hepatocellular carcinoma. J Exper Clin Cancer Res. 2018;37(1):215. doi: 10.1186/s13046-018-0890-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Q., Ma S., Song N., Li X., Liu L., Yang S. Stabilization of histone demethylase PHF8 by USP7 promotes breast carcinogenesis. J Clin Invest. 2016;126(6):2205–2220. doi: 10.1172/JCI85747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiederschain D., Wee S., Chen L., Loo A., Yang G., Huang A. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle. 2009;8(3):498–504. doi: 10.4161/cc.8.3.7701. [DOI] [PubMed] [Google Scholar]
- 29.Lewis Phillips G.D., Li G., Dugger D.L., Crocker L.M., Parsons K.L., Mai E. Targeting HER2-positive breast cancer with trastuzumab-DM1, an antibody-cytotoxic drug conjugate. Cancer Res. 2008;68(22):9280–9290. doi: 10.1158/0008-5472.CAN-08-1776. [DOI] [PubMed] [Google Scholar]
- 30.Block K.M., Hanke N.T., Maine E.A., Baker A.F. IL-6 stimulates STAT3 and Pim-1 kinase in pancreatic cancer cell lines. Pancreas. 2012;41(5):773–781. doi: 10.1097/MPA.0b013e31823cdd10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ram O., Goren A., Amit I., Shoresh N., Yosef N., Ernst J. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell. 2011;147(7):1628–1639. doi: 10.1016/j.cell.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Q., Kulak M.V., Borcherding N., Maina P.K., Zhang W., Weigel R.J. A novel HER2 gene body enhancer contributes to HER2 expression. Oncogene. 2018;37(5):687–694. doi: 10.1038/onc.2017.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kulak M.V., Cyr A.R., Woodfield G.W., Bogachek M., Spanheimer P.M., Li T. Transcriptional regulation of the GPX1 gene by TFAP2c and aberrant CpG methylation in human breast cancer. Oncogene. 2013;32(34):4043–4051. doi: 10.1038/onc.2012.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vernimmen D., Begon D., Salvador C., Gofflot S., Grooteclaes M., Winkler R. Identification of HTF (HER2 transcription factor) as an AP-2 (activator protein-2) transcription factor and contribution of the HTF binding site to ERBB2 gene overexpression. Biochem J. 2003;370(Pt 1):323–329. doi: 10.1042/BJ20021238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ailan H., Xiangwen X., Daolong R., Lu G., Xiaofeng D., Xi Q. Identification of target genes of transcription factor activator protein 2 gamma in breast cancer cells. BMC Cancer. 2009;9:279. doi: 10.1186/1471-2407-9-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bosher J.M., Totty N.F., Hsuan J.J., Williams T., Hurst H.C. A family of AP-2 proteins regulates c-erbB-2 expression in mammary carcinoma. Oncogene. 1996;13(8):1701–1707. [PubMed] [Google Scholar]
- 38.Perissi V., Menini N., Cottone E., Capello D., Sacco M., Montaldo F. AP-2 transcription factors in the regulation of ERBB2 gene transcription by oestrogen. Oncogene. 2000;19(2):280–288. doi: 10.1038/sj.onc.1203303. [DOI] [PubMed] [Google Scholar]
- 39.Mungamuri S.K., Murk W., Grumolato L., Bernstein E., Aaronson S.A. Chromatin modifications sequentially enhance ErbB2 expression in ErbB2-positive breast cancers. Cell Rep. 2013;5(2):302–313. doi: 10.1016/j.celrep.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maina P.K., Shao P., Jia X., Liu Q., Umesalma S., Marin M. Histone demethylase PHF8 regulates hypoxia signaling through HIF1alpha and H3K4me3. Biochimica et biophysica acta Gene regulatory mechanisms. 2017;1860(9):1002–1012. doi: 10.1016/j.bbagrm.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yong H.Y., Kim I.Y., Kim J.S., Moon A. ErbB2-enhanced invasiveness of H-Ras MCF10a breast cells requires MMP-13 and uPA upregulation via p38 MAPK signaling. Int J Oncol. 2010;36(2):501–507. [PubMed] [Google Scholar]
- 42.Kim I.Y., Yong H.Y., Kang K.W., Moon A. Overexpression of ErbB2 induces invasion of MCF10a human breast epithelial cells via MMP-9. Cancer Lett. 2009;275(2):227–233. doi: 10.1016/j.canlet.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 43.Bollig-Fischer A., Dziubinski M., Boyer A., Haddad R., Giroux C.N., Ethier S.P. HER-2 signaling, acquisition of growth factor independence, and regulation of biological networks associated with cell transformation. Cancer Res. 2010;70(20):7862–7873. doi: 10.1158/0008-5472.CAN-10-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burrell R.A., Juul N., Johnston S.R., Reis-Filho J.S., Szallasi Z., Swanton C. Targeting chromosomal instability and tumour heterogeneity in HER2-positive breast cancer. J Cell Biochem. 2010;111(4):782–790. doi: 10.1002/jcb.22781. [DOI] [PubMed] [Google Scholar]
- 45.Ingthorsson S., Andersen K., Hilmarsdottir B., Maelandsmo G.M., Magnusson M.K., Gudjonsson T. HER2 induced EMT and tumorigenicity in breast epithelial progenitor cells is inhibited by coexpression of EGFR. Oncogene. 2015 doi: 10.1038/onc.2015.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pradeep C.R., Zeisel A., Kostler W.J., Lauriola M., Jacob-Hirsch J., Haibe-Kains B. Modeling invasive breast cancer: growth factors propel progression of HER2-positive premalignant lesions. Oncogene. 2012;31(31):3569–3583. doi: 10.1038/onc.2011.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong L., Meng F., Wu L., Mitchell A.V., Block C.J., Zhang B. Cooperative oncogenic effect and cell signaling crosstalk of cooccurring HER2 and mutant PIK3CA in mammary epithelial cells. Int J Oncol. 2017;51(4):1320–1330. doi: 10.3892/ijo.2017.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu S., Lee J.S., Jie C., Park M.H., Iwakura Y., Patel Y. HER2 overexpression triggers an IL1alpha proinflammatory circuit to drive tumorigenesis and promote chemotherapy resistance. Cancer Res. 2018;78(8):2040–2051. doi: 10.1158/0008-5472.CAN-17-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–DD13. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chung S.S., Giehl N., Wu Y., Vadgama J.V. STAT3 activation in HER2-overexpressing breast cancer promotes epithelial-mesenchymal transition and cancer stem cell traits. Int J Oncol. 2014;44(2):403–411. doi: 10.3892/ijo.2013.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Conze D., Weiss L., Regen P.S., Bhushan A., Weaver D., Johnson P. Autocrine production of interleukin 6 causes multidrug resistance in breast cancer cells. Cancer Res. 2001;61(24):8851–8858. [PubMed] [Google Scholar]
- 53.Ghandadi M., Sahebkar A. Interleukin-6: a critical cytokine in cancer multidrug resistance. Curr Pharm Des. 2016;22(5):518–526. doi: 10.2174/1381612822666151124234417. [DOI] [PubMed] [Google Scholar]
- 54.Korkaya H., Kim G.I., Davis A., Malik F., Henry N.L., Ithimakin S. Activation of an IL6 inflammatory loop mediates trastuzumab resistance in HER2+ breast cancer by expanding the cancer stem cell population. Mol Cell. 2012;47(4):570–584. doi: 10.1016/j.molcel.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhong H., Davis A., Ouzounova M., Carrasco R.A., Chen C., Breen S. A novel IL6 antibody sensitizes multiple tumor types to chemotherapy including trastuzumab-resistant tumors. Cancer Res. 2016;76(2):480–490. doi: 10.1158/0008-5472.CAN-15-0883. [DOI] [PubMed] [Google Scholar]
- 56.Huang W.C., Hung C.M., Wei C.T., Chen T.M., Chien P.H., Pan H.L. Interleukin-6 expression contributes to lapatinib resistance through maintenance of stemness property in HER2-positive breast cancer cells. Oncotarget. 2016;7(38):62352–62363. doi: 10.18632/oncotarget.11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stuhlmiller T.J., Miller S.M., Zawistowski J.S., Nakamura K., Beltran A.S., Duncan J.S. Inhibition of lapatinib-induced kinome reprogramming in ERBB2-Positive breast cancer by targeting bet family bromodomains. Cell Rep. 2015;11(3):390–404. doi: 10.1016/j.celrep.2015.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sahin O., Frohlich H., Lobke C., Korf U., Burmester S., Majety M. Modeling erbb receptor-regulated G1/S transition to find novel targets for de novo trastuzumab resistance. BMC Syst Biol. 2009;3:1. doi: 10.1186/1752-0509-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fisher D.T., Appenheimer M.M., Evans S.S. The two faces of IL-6 in the tumor microenvironment. Semin Immunol. 2014;26(1):38–47. doi: 10.1016/j.smim.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mauer J., Denson J.L., Bruning J.C. Versatile functions for IL-6 in metabolism and cancer. Trends Immunol. 2015;36(2):92–101. doi: 10.1016/j.it.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 61.Tsukamoto H., Senju S., Matsumura K., Swain S.L., Nishimura Y. IL-6-mediated environmental conditioning of defective Th1 differentiation dampens antitumour immune responses in old age. Nat Commun. 2015;6:6702. doi: 10.1038/ncomms7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fisher D.T., Chen Q., Skitzki J.J., Muhitch J.B., Zhou L., Appenheimer M.M. IL-6 trans-signaling licenses mouse and human tumor microvascular gateways for trafficking of cytotoxic t cells. J Clin Invest. 2011;121(10):3846–3859. doi: 10.1172/JCI44952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hynes N.E., Lane H.A. Myc and mammary cancer: Myc is a downstream effector of the ErbB2 receptor tyrosine kinase. J Mammary Gland Biol Neoplasia. 2001;6(1):141–150. doi: 10.1023/a:1009528918064. [DOI] [PubMed] [Google Scholar]
- 64.Nair R., Roden D.L., Teo W.S., McFarland A., Junankar S., Ye S. c-Myc and Her2 cooperate to drive a stem-like phenotype with poor prognosis in breast cancer. Oncogene. 2014;33(30):3992–4002. doi: 10.1038/onc.2013.368. [DOI] [PubMed] [Google Scholar]
- 65.Mattie M.D., Benz C.C., Bowers J., Sensinger K., Wong L., Scott G.K. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol. Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu D., Deng Q., Sun L., Wang T., Yang Z., Chen H. A Her2-let-7-beta2-AR circuit affects prognosis in patients with Her2-positive breast cancer. BMC Cancer. 2015;15:832. doi: 10.1186/s12885-015-1869-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paroo Z., Ye X., Chen S., Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139(1):112–122. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chang T.C., Zeitels L.R., Hwang H.W., Chivukula R.R., Wentzel E.A., Dews M. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci USA. 2009;106(9):3384–3389. doi: 10.1073/pnas.0808300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kraus M.H., Popescu N.C., Amsbaugh S.C., King C.R. Overexpression of the EGF receptor-related proto-oncogene erbB-2 in human mammary tumor cell lines by different molecular mechanisms. EMBO J. 1987;6(3):605–610. doi: 10.1002/j.1460-2075.1987.tb04797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bofin A.M., Ytterhus B., Martin C., O'Leary J.J., Hagmar B.M. Detection and quantitation of HER-2 gene amplification and protein expression in breast carcinoma. Am J Clin Pathol. 2004;122(1):110–119. doi: 10.1309/8A2D-JFT0-7NE6-EWHE. [DOI] [PubMed] [Google Scholar]
- 71.Xing Z., Jordana M., Kirpalani H., Driscoll K.E., Schall T.J., Gauldie J. Cytokine expression by neutrophils and macrophages in vivo: endotoxin induces tumor necrosis factor-alpha, macrophage inflammatory protein-2, interleukin-1 beta, and interleukin-6 but not RANTES or transforming growth factor-beta 1 mRNA expression in acute lung inflammation. Am J Respir Cell Mol Biol. 1994;10(2):148–153. doi: 10.1165/ajrcmb.10.2.8110470. [DOI] [PubMed] [Google Scholar]
- 72.Hunter C.A., Jones S.A. IL-6 as a keystone cytokine in health and disease. Nat Immunol. 2015;16(5):448–457. doi: 10.1038/ni.3153. [DOI] [PubMed] [Google Scholar]
- 73.Esquivel-Velazquez M., Ostoa-Saloma P., Palacios-Arreola M.I., Nava-Castro K.E., Castro J.I., Morales-Montor J. The role of cytokines in breast cancer development and progression. J interf Cytok Res. 2015;35(1):1–16. doi: 10.1089/jir.2014.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Asensio-Juan E., Fueyo R., Pappa S., Iacobucci S., Badosa C., Lois S. The histone demethylase PHF8 is a molecular safeguard of the IFNgamma response. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkw1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data were submitted to GEO database with accession number GSE140373.