Abstract
Classical human leukocyte antigen (HLA) molecules of the major histocompatibility class II (MHCII) complex present peptides for the development, surveillance and activation of CD4+ T cells. The nonclassical MHCII-like protein HLA-DM (DM) catalyzes the exchange and loading of peptides onto MHCII molecules, thereby shaping MHCII immunopeptidomes. Natural variations of DM in both chains of the protein (DMA and DMB) have been hypothesized to impact peptide presentation, but no evidence for altered function has been reported. Here we define the presence of DM allotypes in human populations covered by the 1000 Genomes Project and probe their activity. The functional properties of several allotypes are investigated and show strong enhancement of peptide-induced T cell activation for a particular combination of DMA and DMB. Biochemical evidence suggests a broader pH activity profile for the new variant relative to that of the most commonly expressed DM allotype. Immunopeptidome analysis indicates that the compartmental activity of the new DM heterodimer extends beyond the late endosome and suggests that the natural variation of DM has profound effects on adaptive immunity when antigens bypass the canonical processing pathway.
Keywords: MHC class II; Limited polymorphism; HLA-DM, Peptide editing; Epitope selection; Immunopeptidome
Subject terms: Antigen presentation, Immunogenetics
Introduction
Classical major histocompatibility class II (MHCII) molecules (HLA-DP, -DQ and -DR) are heterodimeric proteins that display peptides on the cell surface of antigen-presenting cells (APCs) for CD4+ T cell surveillance. The key players and basic mechanisms resulting in the selection and display of certain peptides at the cell surface by MHCII molecules are well understood.1 In this context, the nonclassical MHCII protein HLA-DM (DM) assumes a dominant role in epitope selection. First identified as an essential antigen presentation factor and facilitator of peptide dissociation,2,3 DM was demonstrated to couple its catalytic exchange properties with chaperoning empty MHCII molecules to bind peptides.4,5 DM exerts its function by sensing rare conformations of the peptide–MHCII complex5,6 which are more often visited by suboptimal complexes. As a result of DM function, highly kinetically stable peptide–MHCII complexes, more resistant to DM-catalyzed exchange, are selected and are likely to be transported to the cell surface. This fundamental function of DM is modulated in specific cell subsets (B cells and thymic epithelial cells) by another nonclassical MHCII molecule, HLA-DO (DO), that acts as a competitive inhibitor of the DM-MHCII interaction.7 While classical MHCII molecules are highly polymorphic and restrict the primary sequence of peptides displayed at the cell surface to a considerable extent, the nonclassical MHCII genes are oligomorphic and encode for proteins that define the exact composition of the cellular immunopeptidome. Therefore, DM function, further modulated in specific cell types by DO, defines to a great extent the epitopes that are encountered by the T cell receptor of CD4+ T cells.
Variations in MHCII antigen processing and presentation, and in particular DM function, have been implicated in health and disease.8 Several studies have demonstrated that DM activity affects the presentation of autoimmune-relevant peptides,9–12 endogenous peptides relevant for allogeneic stem cell transplantation,13 and epitopes of major allergens.14 Indeed, insights from different knockout murine models for DM have shown that T cell development, the ability to mount effective immune responses to pathogens, and the outcome of allograft transplantation are all compromised in the absence of DM to an extent that is modulated by the classical MHCII background (reviewed in ref. 15). DM peptide exchange activity is favored at the low pH of late endosomal compartments in the so-called canonical antigen processing pathway.1,3 Although alternative routes of antigen processing and presentation have important consequences affecting T cell responses in vivo,8 the role of DM in these pathways has often been disregarded owing to the observation that its catalytic peptide exchange activity is largely compromised at non-acidic pH levels.1,3 However, DM is widely distributed in different APC types,16,17 and its presence on the surface of B cells and immature dendritic cells facilitates antigen internalization.17 Similarly, the specific activity of different allotypes of DM, as well as its natural regulator DO, have traditionally been neglected. Limited polymorphisms of these molecules have been assigned to different protein regions, but their potential impact on protein function has only recently been investigated. We were previously able to show that the in vitro peptide exchange activity of DM differs for allotypes bearing distinct DMA allelic variants.18 More recently, Denzin and collaborators have provided evidence that natural variations of DO also impact its functional properties at the cellular level.19
Here we present an experimental approach to assess functional differences due to natural variations of DM. Hypothesizing that DM’s function will depend on the specific combination of DMA and DMB gene products, we first analyzed the presence of particular DM haplotypes in human populations covered by the 1000 Genomes Project (1000GP). Our analysis reveals eight major DM haplotypes at the human global population level. These haplotypes are differentially distributed among the world’s subpopulations and, in most cases, include the DMA*0101 allelic variant. Moreover, the DMA*0103 allele, which was previously shown to impair DM peptide exchange function,18 is strongly linked to DMB*0107. The functional investigations reported here identify changes in immunopeptidomes and T cell activation in the presence of DMA*0103-DMB*0107 relative to DMA*0101-DMB*0101. Interestingly, the DMA*0103 and DMB*0107 combination encodes for an allotype that is more effective in synthetic peptide loading, which may take place in alternative subcellular compartments. This effect coincides with an altered pH activity profile of this particular heterodimer that is present in approximately 2% of the human population. Such distinct editing properties will have to be taken into account when considering T cell-mediated immunity in the context of autoimmunity, pathogen defense, or tumor immunotherapy.
Results
HLA-DM genes associate in eight major haplotypes at the global human population level
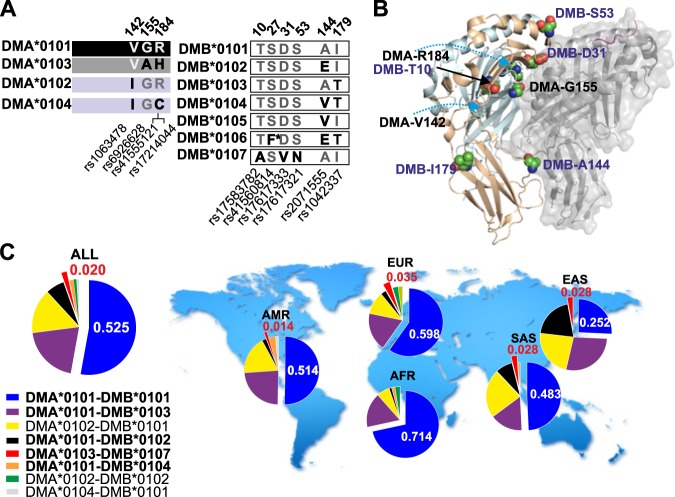
The HLA locus exhibits strong linkage disequilibrium (LD), and it is likely that specific DMA and DMB alleles associate with haplotypes. We therefore investigated the presence of specific DM haplotypes at a global scale using the reference single-nucleotide polymorphisms (rsSNPs)20 (Fig. 1a, Table S1) present in each DMA and DMB allelic variant21 in conjunction with the publicly available sequence data from the 1000GP.22 Noteworthy, many of the residues affected by natural variations lie at or close to the interface between DM and HLA-DR (DR), as inferred from the three-dimensional structure of the DM-DR1-HA complex23 (Fig. 1b), and could modulate the interaction of these two molecules.
Fig. 1.
Overview of DMA and DMB natural variation. a DMA and DMB allelic variants described in the HLA/IMGT database. Polymorphic residues in DMA and DMB are shown with single-letter code. Substitutions resulting from each SNP are indicated, and the positions affected in the mature protein sequence and the rsSNP codes are shown above and below the boxes, respectively. b Cartoon representation of the DM heterodimer (DMA in yellow and DMB in cyan) in complex with HLA-DR1-HA (gray, transparent surface for HLA-DR1, PDB 4fqx). Residues affected by rsSNPs are shown as spheres. c DM haplotype frequencies shown as pie charts for the whole dataset from the 1000 Genomes Project (ALL, left). Haplotypes in bold letters were functionally characterized. The haplotype distribution is also shown for different subpopulations studied by the 1000 Genomes Project. AFR African, AMR American, EAS East Asian, EUR European, SAS South Asian. Only haplotypes with frequencies > 0.01 are shown
The 1000GP dataset used comprises 2504 genomes (ALL) grouped in five superpopulations. Notably, the previously reported SNP rs41560814 (resulting in DMB-S45F; found in DMB*0106) is not identified in this dataset, indicating that, if present at all, this substitution occurs only at a very low frequency in the populations covered by the 1000GP. We probed the association between rsSNPs and defined the presence and frequency of DMA and DMB allelic variants (Fig. S1A) as well as their association with haplotypes24 (Fig. S1B). This analysis yields eight DMA-DMB haplotypes (hereafter referred to as the main haplotypes or allotypes, indicated as DMxy, where “x” refers to the DMA allelic variant and “y” to the corresponding DMB variant) with frequencies >0.01 on a global scale. The most widespread haplotype is DM11, which, together with DM13 and DM21, accounts for >75% of the haplotypes detected (Fig. 1c). All DMA alleles are represented within the 8 major haplotypes, but neither DMB*0105 nor DMB*0106 reaches the 0.01 threshold. Finally, the haplotypes described are unequally distributed across the world’s populations covered by this dataset (Fig. 1c). Thus, for instance, DMA*0103- and DMA*0104-bearing haplotypes are absent in some of the populations covered (AFR and EAS, respectively).
HLA-DM haplotypes feature specific cellular functions and differentially affect T cell activation
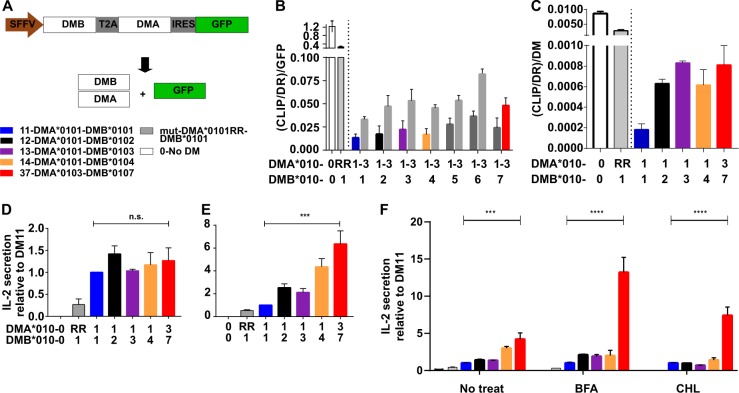
We have previously observed lower peptide exchange activity for DM31 relative to DM11 in vitro18 and hypothesized that the specific features of HLA-DM heterodimers depend upon the combination of DMA and DMB.15 We sought to test this hypothesis with all DM1y and DM3y heterodimers—particularly those resulting in any of the main haplotypes—in the presence of transmembrane domains due to their potential influence on DM function.25 DM peptide exchange function can be determined at the cellular level by measuring the cell surface accumulation of Class-II associated invariant chain peptide (CLIP) bound to MHCII26 as well as the ability to affect T cell activation using model antigens.2 The T2.DR3 cell line was transduced with all potential DM1y and DM3y allotypes as well as a DMA mutant (DMA*0101-R98A-R194A, also referred to as RR) that is known to reduce peptide exchange activity23 (DMmut). The expression cassette used here features a green fluorescent protein (GFP) as a surrogate marker of DM expression27 (Fig. 2a). Upon transduction and cell sorting (GFP), the different cell lines were stained for surface expression of CLIP and for DR (Fig. S2A), allowing us to determine the surface CLIP/DR ratio. Surface DR is slightly decreased upon DM expression, but there was no significant difference between DR or CLIP levels for any of the DM-expressing cell lines. Since DM expression levels affect the outcome of CLIP/DR ratios,28 these values were further normalized to the DM expression level based on GFP27 (Fig. 2b, DM confirmed as in S2A). CLIP/DR ratios for different DM-expressing subpopulations confirmed the differences measured at the global level and the dose-dependent behavior of the ratios and GFP (DM) expression levels (Fig. S2B). All of the main DM allotypes tested displayed higher catalytic activity than that of DMmut (ten-fold less active), but the differences in CLIP/DR ratios normalized to DM expression between DM1y and DM3y heterodimers were not statistically significant. We performed a more detailed analysis using single-cell clone-derived cell lines bearing DM allotypic combinations representative of the main haplotypes and with similar expression of DM (Fig. S2C). In this case, direct detection of DM by intracellular staining confirmed our previous findings (Fig. 2c, S2C). We used the geometric mean fluorescent intensity (gMFI) of MaP.DM1-PE to normalize (CLIP/DR) ratios and directly quantified the presence of DM for the cell lines subsequently used in T cell activation experiments. The main conclusion from these experiments is that the dynamic range of the differences between DM allotypes tested was rather small under the DM overexpression conditions used. Similarly, the analysis of sodium dodecyl sulfate-stable dimer formation using clonally expanded cell lines did not reveal substantial differences for any of the main haplotypes, indicating a similar rate of high/low affinity peptides bound to HLA-DR3 (Fig. S2D). Analysis of CLIP/DR ratios using T2.DR1 transfectants confirmed these findings for a second MHCII allotype (Fig. S2E and S2F).
Fig. 2.
Characterization of the cellular function of HLA-DM allotypes. a Construct design for the expression of selected DM allotypes. A bicistronic cassette consisting of the cDNAs of DMB and DMA spaced by a self-cleavable T2A peptide was placed under the control of the SFFV promoter. An IRES sequence allows the expression of GFP as a surrogate marker of DM. The color legend shown at the bottom applies to those DM allotypes analyzed in detail. b Quantification of (CLIP/DR)/GFP ratios for all cell lines generated on the T2.DR3 background using flow cytometry (gMFI of each marker was used). DMA and DMB genes combined for each cell line are indicated along the “x” axes. Each pair of bars after the controls shows DMA1 (left) and DMA3 (right), and DMB is indicated below. Mean and SD values are shown for n = 3 independent experiments measured in duplicate. Gray symbols above the bars indicating statistical significance refer to the analysis of differences between DM allotypes only. c (CLIP/DR)/DM ratios determined for single-cell clone cell lines of the indicated selected DM allotypes as indicated in (b). In this case, the values are normalized to the gMFI of DM intracellular staining. The mean and SD are for n = 4 independent experiments measured in duplicate. d Antigen presentation assays using T2.DR3 cell lines stably expressing the indicated DM allotypes. Human reactive CD4+ T cells for an immunodominant MTB-A85B epitope (MTB-A85B53–67: VPSPSMGRDIKVQFQ) were used as responder cells. T2 APCs were fed with 50 mg/ml MTB-85B antigen (protein) for 16 h and then fixed. One hundred and five of these APCs were incubated together with 105 responder cells for 48 h. IL-2 was measured by ELISA. The bar chart shows the mean and SD of n = 3 independent experiments measured in duplicate. All values are relative to the most abundant DM allotype (average IL-2 secretion by DM11 = 120 ± 9 pg/ml). e The same experiment was repeated with the T2.DR3 cell lines supplied with 50 mg/ml MTB-A85B53–67 peptide for 6 h. Downstream processing was performed as described in (d). Mean and SD of n = 3 independent experiments measured in duplicate (average IL-2 secretion by DM11 = 373 ± 25 pg/ml). f Same as (d) using the corresponding peptide added to the cells at a concentration of 25 mg/ml for 4.5 h. APCs were either not treated (No treat) or treated either with 25 mM brefeldin A (BFA) or 100 mM chloroquine (CHL). Mean and SD values from n = 3 independent experiments measured in duplicate (average IL-2 secretion by DM11 No treat = 75 ± 8 pg/ml; BFA = 40 ± 5 pg/ml; CHL = 53 ± 5 pg/ml)
DM activity facilitates the presentation of known epitopes originating from antigenic proteins that require intracellular processing,2 and it can edit the peptides bound to MHCII molecules in different subcellular compartments.16,17 Although editing of MHCII-CLIP does not vary significantly for different DM allotypes, more subtle shifts in the presented peptidome can still lead to substantial differences in the stimulation of specific T cell clones. Thus we performed T cell activation experiments in which the clonally expanded T2.DR3-DM cell lines bearing the main haplotypes were tested for their ability to present a DR3-restricted epitope derived from Mycobacterium tuberculosis antigen 85B (MTB-A85B), either using whole antigen or the corresponding restricted A85B53–67 peptide. CD4+ T cells reactive to the MTB-A85B53–67 peptide were used as responder cells, and the amount of interleukin-2 (IL-2) secreted was interpreted as a direct consequence of the extent of antigen presented (normalized to DM11-expressing cells). T cell assays using whole-protein antigen showed no significant difference for the presentation of this peptide between DM allotypes (Fig. 2d). However, using synthetic peptide, we measured a six-fold increase in IL-2 production in the case of the T2.DR3-DM37 cell line and a four-fold increase for T2.DR3-DM14 (Fig. 2e). The lower T cell activation levels obtained using the DMmut control for both peptides and protein (Fig. 2d) indicated that the presentation of this epitope is dependent on DM activity.
The distinct antigen loading behavior of each DM allotype was further evaluated in the presence of brefeldin A (BFA) and chloroquine (CHL). BFA blocks anterograde protein transport from the Golgi, preventing nascent MHCII molecules from reaching endosomal compartments. CHL prevents the acidification of endosomes, attenuating antigen degradation and compromising DM activity. As expected, treatment with these drugs essentially blocked antigen presentation in experiments using the whole protein (Fig. S2G). In the case of peptide delivery, BFA treatment resulted in substantially increased T cell responses when using T2.DR3-DM37 cells as APCs relative to T2.DR3-DM11 cells (12-fold), confirming the distinct behavior of DM37 vs. DM11 (Fig. 2f). CHL treatment in this case revealed a six-fold increase in IL-2 release, which was slightly higher than that seen without drug using the same peptide concentration, indicating that peptide loading does indeed take place in organelles with a less acidic pH. The corresponding T2.DR1 transfectants showed a similar trend for increased presentation of synthetic peptide in the presence of DM37 relative to DM11 under the same conditions (Fig. S2H). Together, these results indicate that the synthetic peptide is not required to be proteolytically degraded and is preferentially loaded onto MHCII molecules in the presence of DM37, suggesting that this event occurs in nonlate endosomal compartments.
The DM37 allotype (DMA*0103-DMB*0107) features a pH-broadened peptide-exchange activity
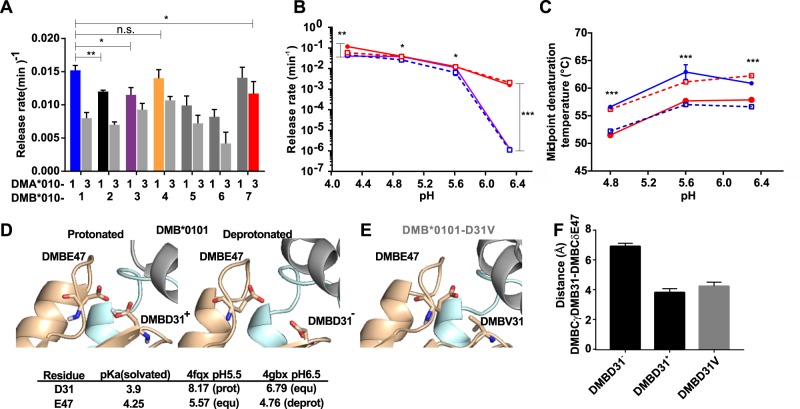
To shed light on the distinct peptide-loading behavior of these DM heterodimers, we characterized the peptide exchange function of these proteins under experimentally defined conditions in vitro (details regarding expression and purification in Fig. S3A and S3B). We confirmed that, at the pH of the late endosomal compartment (5.3), all DM3y heterodimers had reduced catalytic activity relative to DM1y but to differing degrees for the individual DMB variants (Fig. 3a and S3C). Although DM37 is less active than DM11 in peptide release experiments at a fixed pH (5.3), its activity is substantially higher than that of any other DM3y allotype, indicating partial compensation of the reduced peptide exchange activity of DMA*0103-bearing allotypes. Interestingly, peptide release rates measured at different pH values showed an atypical profile for DM37 and DM17. Thus dissociation rates catalyzed by DM37 and DM17 were significantly higher at low (4.2) and high (6.4) pH relative to any other DM allotype (Fig. 3b and S3D). This suggests that such distinct pH-insensitive behavior is provided by the substitutions found in the DMB*0107 allele, consisting of DMBT10A, DMBD31V, and DMBS53N. Additional assays showed that those substitutions do not have a significant effect on the thermal stability of DM heterodimers, hence protein folding is not compromised (Fig. 3c and S3E). Thus DM37 is the only main allotype comprising DMA*0103 that features distinct biochemical behavior with more pH-independent activity.
Fig. 3.
Biochemical characterization of HLA-DM allotypes. a Dissociation rates of MBP-FITC catalyzed by different DM allotypes measured by fluorescence polarization. DR2/MBP-FITC (250 nM) complexes were incubated in the presence of different DM allotypes (75 nM) and an excess of unlabeled MBP (50 µM). Reactions were performed in citrate phosphate buffer at pH 5.3 at 25 °C, and FP was determined over 200 min. Mean and SD of n = 3 independent experiments measured in triplicate. b Representative examples of release rates determined as in (a) under different pH conditions. Values show the mean and SD of n = 3 experiments measured in triplicate. DM11 is shown as filled circles (blue) and DM31 in open circles (blue). DM37 is shown as filled circles (red) and DM17 as open circles (red). c Representative examples of the thermal stability of DMA-DMB allotypes at different pH values. The midpoint denaturation temperature of each protein is represented over the pH range of the assay. The results show the average and SD of n = 6 independent experiments. Symbols are the same as in (b). d The 3D structures of the DM-DR1 (PDB accession codes 4fqx and 4gbx) complex are used to model the implications of protonation and deprotonation of DMBD31 and DMBE47. DM-DR structures are depicted as cartoons, and the side chains of the corresponding amino acids are shown as sticks. DR is shown in gray, DMB in cyan, and DMA in yellow. Protonation of these two acidic amino acids results in a structured loop with their side chains close to each other and to the DR1 alpha chain (left). Equilibration of the structure after deprotonation of these two residues yields an open loop conformation (right) in which the side chain of DMBE47 is moved away from the DM-DR interface. The predicted pKa values for the protonatable groups of residues D31 and E47, as well as their predicted protonation states in each solved 3D structure, are indicated; prot protonated, deprot deprotonated, equ equilibrium protonated–deprotonated. e DMBD31V substitution found in the DMB*0107 results is also modeled similar to the substitutions studied in d. In this latter case, the Val side chain is not affected by protonation–deprotonation, resulting in nonrepulsion of the residues due to electrostatic interaction. f Bar chart showing the average and SD of the distance (in Å) between the Cγ of the side chain of residue DMB31 and the corresponding Cδ DMB47 for the three scenarios shown in (d) and (e) during the energy minimization procedure
To understand the distinct pH profile of DM37, we rationalized the structural properties of this variant based on the known crystal structure of DM11 when interacting with HLA-DR1.23 In silico predictions of pKa values of the side chains of DMBD31 and DMBE47 in complex with HLA-DR1 indicated a protonated side chain of DMBD31 (Fig. 3d) that forms a hydrogen bond with DMBE47. To understand the pH dependency of this interaction, we generated two models of the complex with either a protonated (DMBD31+) or deprotonated (DMBD31−) side chain of DMBD31. We also generated a model of the DMBD31V mutant that was shown experimentally to be more pH independent (Fig. 3e). All models were subjected to energy minimization and equilibration by short molecular dynamics simulations. Interestingly, DMBD31+ showed a similar conformation to that of the crystal structure (Fig. 3d left) with placement of Cδ(E31) and Cγ(E47) in close proximity (3.8 Å, Fig. 3f). However, deprotonation of DMBD31 resulted in electrostatic repulsion between the side chains of DMBD31 and DMBE47 that led to a loop rearrangement and a Cδ(E31)−Cγ(E47) distance of ~6.8 Å, associated with a loosened DM-HLA-DR1 interface (Fig. 3d and Supplemental video1). Introducing valine at position DMB31 was compatible with the loop conformation seen in the crystal structure at pH 5.5 (Fig. 3e, f) and should not be subject to pH-dependent deprotonation. Overall, our data suggest that the protonation state of DMB-31 is responsible for the structural rearrangements that are the basis for the pH dependency of DM.
HLA-DM11 and DM37 imprint distinct specific signatures on the HLA-DR3 immunopeptidome
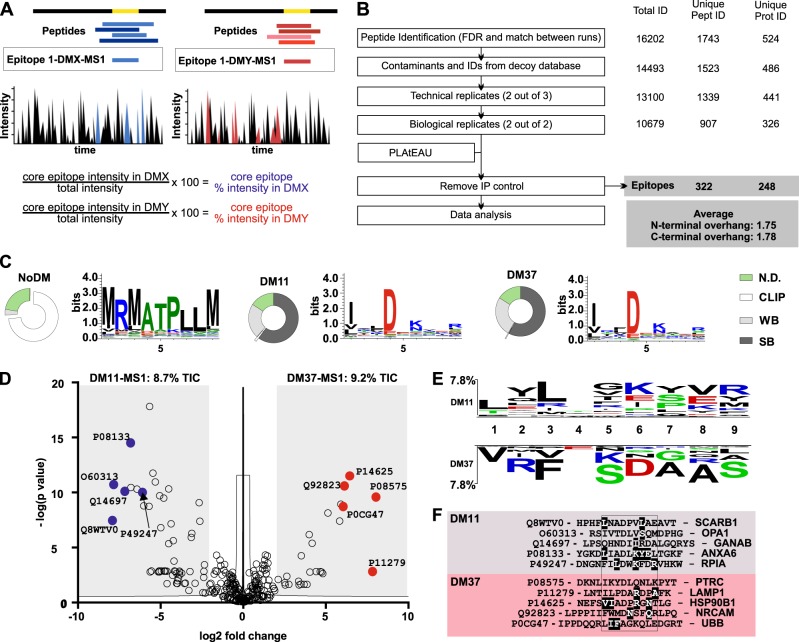
Since our cellular and biochemical experiments demonstrated that DM11 and DM37 have clearly distinct editing functions, we hypothesized that the composition of the MHCII immunopeptidome should also reflect such differences. We therefore analyzed the pool of peptides presented by the above-described T2.DR3 cell lines expressing the two selected DM haplotypes, DM11 and DM37. To this end, we used a validated and published in-house-developed algorithm termed PLAtEAU (Peptide Landscape Antigenic Alignment Utility),27 which allows us to perform label-free quantification of sets of nested peptides isolated from MHCII molecules. The algorithm aligns the identified peptides to their protein sources and thus defines the longest overlapping amino acid sequence from all peptides of a consensus epitope. Additionally, the MS1 intensity of all peptides used to define this epitope is summed up on a per residue basis, yielding a total MS1 intensity that can be normalized to the total ion current of the run, producing a relative abundance value for each epitope (Fig. 4a). The performance of our analysis is shown in Fig. 4b. The initial 1523 unique peptide identifications for the complete dataset (all conditions pooled) yielded a total of 322 consensus epitopes (Fig. S4A) from 248 protein entries (Fig. S4B) with their corresponding relative intensities (Table S2). The performance of our replication scheme for each DM condition was similar to the overlap between DM11 and DM37 (85%, Figures S4C and S4D), indicating that the two epitope pools were very similar qualitatively.
Fig. 4.
Influence of DM on the immunopeptidome presented by HLA-DR molecules. a Schematic representation of MHCII immunopeptidome analysis using the PLAtEAU algorithm. PLAtEAU aligns the peptides eluted from MHCII molecules under each condition to their protein sources (black and gray) and defines a consensus epitope that spans the maximum amino acid sequence from any series of nested peptides. During this process, the algorithm also takes into account the MS1 intensities of individual peptides (color intensity) to derive a defined relative intensity for each consensus epitope. The relative intensity of each epitope is calculated on a per residue basis using the MS1 intensity of each peptide and the Total Ion Current (TIC) of each condition. For two specific conditions, DMX and DMY conventional analysis focuses on the peptides, while using PLAtEAU yields epitopes with associated relative intensities (boxed) that can be used for LFQ. b Overview of the performance of the immunopeptidome analysis and the criteria applied for peptide identification and quantification. Known contaminants (included in the MaxQuant platform) and identifications from the decoy database are initially filtered out. Two extra filters are applied, one for technical replication and another for biological replication. The resulting dataset is subsequently used as the input for our recently described algorithm, PLAtEAU56. Epitopes found in the IP control samples are removed. Number of identifications, peptides, and protein sources are shown on the right side. c Features of the immunopeptidome isolated from the T2.DR3-NoDM (left), -DM11 (middle) and -DM37 (right) cell lines. For each dataset, we retrieved the consensus epitopes and their corresponding relative intensities. We used these lists of consensus epitopes to predict binding affinities and cores (NetMHCIIpan). Weighing the presence of each consensus epitope (relative intensity) allows us to define the relative abundances of strong binders (SB), weak binders (WB, including CLIP, highlighted), and those for which no affinity is predicted (N.D. not defined). The results are shown as donut charts. The same weighted consensus epitope list is used to define Seqlogos for each condition. d Volcano plot depicting the log2-fold intensity difference between the epitopes identified for DM37 and DM11 vs. the –log(P) of the differences. Representative examples of epitopes enriched for each condition are shown, indicating the UniProt accession code. e Two-sample logo analysis of the weighted binding cores for each condition (DM11 or DM37). The predicted position for each residue within the binding groove of DR3 molecules is shown, ranging from P1(1) to P9.(9) Only significant differences (p < 0.05) are shown. Residues shown on the upper or the lower part indicate those that were preferentially found in the pool of peptides eluted from DM11 or DM37, respectively. f Representative examples of epitopes enriched for each condition (d). Predicted binding cores are aligned and contained within a box. UniProt accession codes for the protein sources are indicated, as are the gene names. Highlighted residues are also found in the two-sample logo analysis
Identified epitopes (PLAtEAU) were analyzed quantitatively to generate Seqlogos and the distribution of binding affinities (NetMHCIIpan) for each condition (Fig. 4c). Such analysis revealed that the expression of either DM enriched a very similar proportion of high- and low-affinity binders (considerably higher than the NoDM condition) and that they selected essentially the same anchor residues for the binding groove. More importantly, relative quantification by PLAtEAU, which assigns MS1 intensities to core epitopes, enabled a robust comparison of the significantly but relatively small differences in the peptidomes for the two DM haplotypes. Differences in the log2 relative intensities of all epitopes, identified across the 6 replicates measured, indicated that there were 110 epitopes differentially selected by one or the other DM allotype (Table S3). Those epitopes represented approximately 9% of the total ion current of each condition (Fig. 4d), and a two-sample logo analysis of the predicted binding cores (based on the epitope abundance, Fig. 4c) revealed subjacent differences in each of the pools (DM11 vs. DM37, Fig. 4e). Importantly, these differences at the amino acid level are not necessarily conservative but also affect the charge (K to D substitution) or bulkiness (V to A alteration) of nonmain anchor residues. Representative examples enriched for each DM condition already indicated the contribution of certain residues to these observed compositional differences (Fig. 4f). A conventional gene ontology (GO) annotation analysis yielded basically the same GO term distribution for both DM conditions (Figure S4E). However, we rationalized that given the quantitative nature of the generated data, a one-dimensional (1D) annotation enrichment could be applied to determine the preferential sampling of protein sources in the immunopeptidomes analyzed.29 This approach takes into account the summed relative intensities for each protein entry and compares the distribution of log2 intensities for each GO term to the total log2 intensity measured at the corresponding condition. In our case, epitopes originating from proteins annotated with the GO terms “vesicles” and “extracellular space” were specifically enriched (relative to the total log2 intensity distribution) only in the presence of DM37 (Figure S4F-S4I). In sum, epitope-based quantification by PLAtEAU demonstrates that, under the same HLA-DR and DM expression levels, the DM allotype has a considerable impact in shaping the specific composition of the immunopeptidome.
Discussion
The presence of DM haplotypes has been reported for a global subpopulation (Han Chinese, included in the EAS group in this study (Fig. 1)). A study by Feng et al.30 analyzed 1000 individuals of this subpopulation and found a distribution of DMA-DMB haplotypes similar to that seen in our analysis (DM13: 27% vs. 31%; DM21: 22% vs. 27%; DM12: 20% vs. 14% in our report and in ref. 30, respectively). Thus it can be assumed that the even more limited genomic 1000GP dataset for this subpopulation bears statistical significance for all major haplotypes. The eight haplotypes of DM with frequencies > 1% at the global human population level clearly contrast with the several thousands of alleles and hundreds of haplotypes described for classical MHCII genes. Differences in genetic variations between classical and nonclassical MHCII genes are thought to arise from the fact that classical MHCII genes are subject to functional diversification, while nonclassical MHCII genes undergo purifying selection.31,32 Thus, from an evolutionary and clinical perspective, one should wonder whether these nonclassical DM haplotypes are neutral or if they carry functional features for which they have been selected. The selection of such specific properties could have consequences shaping adaptive immune responses and eventually modulate the ability to fight pathogens or predispose to MHCII-related diseases. While limited to only two MHCII allotypes and a particular antigen, our study represents the first report in which altered T cell function may be linked to genetic variations in HLA-DM genes. Future studies should focus on clarifying the implications of DM natural variations under physiologically relevant conditions.
We only detected distinct T cell activation profiles when antigen was provided as a synthetic peptide in the presence of defined DM allotypes. Since we could not detect DM at the cell surface, similar to previous reports using the same cell line,33 we suggest that peptides are loaded in intracellular organelles. The substantial enhancement of T cell activity observed for DM37 relative to DM11 and the fact that this activity was not abolished in the presence of CHL indicates that the peptides are not loaded onto MHCII molecules in late endosomes but rather encounter DM earlier in the secretory pathway. In the presence of BFA, which hinders transport of newly synthesized proteins from the Golgi along the secretory pathway, the difference in T cell activation between DM37 and DM11 became even more pronounced. In this case, the amount of DM and MHCII present in endosomes might become limiting over time, and the catalytic difference between the two haplotypes may become more apparent under such conditions. However, while still speculative at this point, the observed differences in the pH profile of DM37 strongly argue for more effective MTB-peptide loading in cellular compartments with higher pH for this haplotype. Interestingly, low DM editing activity in alternative endosomal compartments, in which physico-chemical conditions are expected to deviate from those found in late endosomal compartments, is responsible for the presentation of specific pMHCII isomers34 recognized by autoimmune-related T cells.12,35 At least two types of conformers, Type A and B (representing register shifts of the same peptide), arise from different antigen processing routes. Type A results from endosomal degradation of an antigen and reactive T cells, while Type B arises from the loading of peptides or antigens in alternative compartments. In the latter example, low or no DM editing is believed to be responsible for the presentation of this conformer and is extremely capable of triggering autoreactive T cells. In this context, it is therefore plausible that DM37 could promote the presence of Type B conformers.
Molecular insights into DM function first postulated and later demonstrated the importance of the amino acids DMBE8, DMBD31, and DMBE47 for pH-dependent peptide exchange activity.36 Global changes in the secondary structure of DM upon pH increase had already been noted37 and described in the two HLA-DR-DM structures solved at two different pH values (5.5 and 6.5).23 In these structures, residues DMBE47 and DMBD31 move apart from each other at higher pH,23 likely as a result of the electrostatic repulsion between their side chains upon deprotonation, as previously proposed.38,39 As a consequence of such repulsion, a loop in DM required for peptide exchange contacts the MHCII molecules. The side chain of Val in position DMB-31 (DMB*0107), which is not subject to protonation/deprotonation, would allow the placement of E47 close to the DM–DR interface, even at higher pH values. Moreover, the presence of the DMBV31 substitution in DMB*0107 affects one of the most prominent regions for the interaction between DM and MHCII.5,23 This region is particularly variable for DQ alleles,40 and such variation has been shown to be responsible for the specific DM requirements of different DQ allotypes.11,41 Thus, while it has already been noted that DM’s function depends on a particular MHCII allele,42 our study shows that the DM allotype also impinges on MHCII function. A daunting question in this context is whether these DM allotypes also show distinct editing behavior toward different MHCII allotypes. Our cellular and in vitro experiments capitalize mostly on DR3 and DR15, two DM-susceptible allotypes, but it will be extremely interesting to test DM editing for other classical MHCII allotypes with presumably different DM susceptibilities. Furthermore, considering that the interaction surfaces for DM-MHCII23 and DM-DO7 are largely overlapping, it is likely that the DMB*0107 polymorphisms described here also lead to an altered DM–DO interaction, and further studies should seek to answer this question.
The functional and biochemical differences observed between DM11 and DM37 could have a significant impact on physiologically relevant processes such as epitope selection upon infection or display of the internal immunopeptidome during the acquisition of central tolerance. We applied a recently developed experimental pipeline, which minimizes the impact of inherent changes in peptidomes, for example, by the fortuitous cleavage of proteins by cathepsins, on data analysis.27 Our algorithm, PLAtEAU, combines information on biological and technical replicates at the peptide level and assigns MS1 spectral intensities to consensus epitopes, thereby yielding a reduced but more robust set of epitopes recognized by MHCII molecules. We find that one third of the epitopes displayed by DR3 are preferentially loaded in the presence of one of the two DM allotypes investigated. This is reflected in the differences observed when comparing the sequence logos of the binding cores for each condition, taking into account the relative intensities (approximately 8% of the total pool of peptides). Interestingly, such differences include amino acids pointing toward the TcR recognition site, which could have a considerable impact on TcR engagement.43 One of the examples found to be enriched in the presence of DM37 is an epitope of lysosome-associated membrane protein 1 (LAMP1, P11279), which has been identified in samples isolated from individuals suffering from multiple sclerosis and arthritis.44,45
The recent report of altered DO functionality due to natural variations in murine strains19 and this current work describing distinct DM editing functions for human DM allotypes add another layer of complexity to the field of antigen presentation. The DM37 allotype described here features atypical peptide exchange properties, probably related to a single amino acid substitution (DMB-D31V), resulting in a catalytic mechanism with an atypical pH activity profile. This leads to changes in the immunopeptidome that we anticipate will have consequences for T cell reactivity in individuals expressing this particular DM haplotype. Thereby, carriers of the DMA0103–DMB0107 allelic combination present an interesting subpopulation for which functional, immunopeptidomic, and genetic association studies should help probe its contribution to HLA-linked diseases.
Materials and methods
Bioinformatics
The association between the DMA and DMB genes was analyzed using the Haploview software version 4.2.24 All rsSNPs20 found in DMA and DMB alleles (HLA/IMGT database21) were used to estimate all pairwise combinations within an approximately 20-kb region of chromosome 6 spanning the DMA and DMB genes (positions 32934628 to 32953121). As input for our analysis, we used the 1000GP dataset GRCh37p13. Notably, for haplotype definition and LD calculation, we forced the inclusion of all rsSNPs, even though in some cases their distribution did not follow the Hardy–Weinberg equilibrium.
Cloning and cDNA constructs
The cDNA constructs encoding HLA-DR2-CLIP as well as DMA*0101/DMA*0103 and DMB*0101, as cloned in pFastBacDual (protein luminal domains for insect cell expression) and pCDNA3.1 (full length proteins for mammalian expression), have been previously described.27 In the case of recombinant proteins expressed in baculovirus, the original DMA*0101-DMB*0101 and DMA*0103-DMB*0101 were used to generate all different allotypes. For experiments using mammalian cells, we followed the same approach using cDNAs encoding full-length proteins within pCDNA3.1 as templates.18 DMB and DMA cDNAs were first fused by PCR spacing the two starting codons with a cDNA encoding the T2A peptide (in frame). The resulting PCR products were subsequently cloned into the pLeGO-iG2 vector (Addgene plasmid #27341 viaBamHI and NotI).
Lentiviral transduction
HEK293T cells in Dulbecco’s modified Eagle’s medium supplemented with pen/strep (50 U/ml each) were used as lentiviral packaging cells (10% fetal calf serum (FCS)). The T2-DR3 cell line was kindly provided by Peter Cresswell. These cells, as well as the stably transduced cells generated from them, were grown in Iscove’s Modified Dulbecco’s Medium (IMDM) supplemented with 10% FCS and pen/strep (50 U/ml each).
For lentivirus packing, HEK293T cells were transiently transfected using PEI with the pRSV, pMDL, and pVSV vectors and corresponding pLeGO-iG2 constructs.46 Supernatants were harvested after 24 and 48 h and concentrated. Titered viruses (5 multiplicity of infection (MOI)) were used for spinoculation (1 h at 1200 × g at 32 °C) of T2 cells (2 × 105) using polybrene (8 μg/ml). Subsequently, the cells were resuspended in medium in a 3.5-cm dish. After 72 h, GFP-positive cells were sorted using a FACS Aria (BD) and allowed to grow under normal conditions. Each cell line was checked for stable GFP expression on a regular basis. Single-cell clones were isolated as previously described.27
Generation of MTB-reactive CD4+ T cells
Expansion of MTB-specific T cell lines was performed as previously described.47 In brief, DR3+-typed isolated peripheral blood mononuclear cells were incubated with the corresponding MTB-A85B peptide (50 µg/ml) in the presence of CD40 (1 µg/ml) and CD28 (1 µg/ml) antibodies for 7 h followed by magnetic separation of peptide-reactive (CD154+) cells using the CD154 MicroBead Kit (Miltenyi Biotec). MTB-reactive, isolated CD154+ cells were cultured with autologous feeder cells treated with mitomycin C (Sigma-Aldrich) at a ratio of 1:100 in X-VIVO™ 15 (Lonza) supplemented with 5% human AB serum (PAN-Biotech), 100 U/ml penicillin, 100 μg/ml streptomycin (PAN-Biotech), and 50 ng/ml IL-2 (PeproTech). Cells were expanded for 14 days, and the culture medium was replenished when needed. Before experiments, MTB-A85B-reactive CD4+ T cell lines were allowed to rest for 48 h in IL-2-free medium.
Antibodies and flow cytometry
The following antibodies were used for flow cytometric measurements: L243-Alexa647 Mouse IgG2a, κ (Biolegend), VB500-CerCLIP Mouse IgG1 κ (BD), MaP.DM1-PE Mouse IgG1, κ (Biolegend), Alexa647-Mouse IgG2a κ (Biolegend), isotype control-(MOPC-173) (Biolegend), BV500-Mouse IgG1, κ isotype Control (X40) (BD), PE-Mouse IgG1, and κ isotype control (MOPC-21) (Biolegend).
Cells were split the day before measurement was performed to a cell density of 3 × 105 cells per ml. The day after, 5 × 105 cells per sample were collected in mid-logarithmic growth phase, then washed and stained as described. For T2.DR3 cells, we used a 1:50 dilution of CerCLIP-BV500 and 1:50 dilution of L243. MaP.DM1-PE was applied after fixation and permeabilization of the cells using Cytofix/Cytoperm (BD 55474) at a 1:40 dilution. Isotype controls were used at the same concentration as the corresponding staining antibody for each experiment. Measurements were performed in a Canto II flow cytometer (BD) and analyzed using FCS Express 6.0 (De Novo Software). The gMFI of each gated population was used to define CLIP and DR surface expression and the intracellular levels of DM. CLIP/DR ratios were further normalized to the GFP gMFI for bulk-sorted DM-positive cell lines or the gMFI of DM for clonally expanded cell lines.
Antigen presentation assays
For antigen presentation assays, we used M. tuberculosis antigen 85B (recombinant protein; Lionex) as well as the peptide MTB-A85B53–66 (SPSMGRDIKVQFQSG). For all experiments, T2.DR cell lines were fed the corresponding antigens (25 or 50 μg/ml for protein or peptide, respectively) for a defined period of time in IMDM medium with 1% FCS (4.5 h or 16 h), and subsequently cells were washed in cold phosphate-buffered saline (PBS). Following fixation (paraformaldehyde), the cells were washed using first PBS and then T cell medium. Finally, 5 × 104 fixed APCs and 5 × 104 responder cells were added to each well (96-well plate), and the culture supernatant was harvested after 36 h and assayed for cytokine production. For DR3-restricted antigen presentation, we measured IL-2 secretion using an Enzyme-linked Immunosorbent Assay Kit (hIL-2; Biolegend). The assays were performed following the manufacturers’ specifications, and each condition was measured in duplicate. CHL (Sigma-Aldrich) and BFA (Biolegend) were used at concentrations of 25 and 100 μM, respectively. Drugs were added 1 h prior to adding the antigen and were maintained throughout antigen processing.
Recombinant protein expression
Constructs based on the pFastBacDual vector and verified by sequencing were used to transform DH10Bac cells. After blue-white screening following the manufacturer specifications, three clones per construct were used to isolate bacmids and generate recombinant baculovirus. Recombinant baculoviruses were generated upon transfection of the bacmids into Sf9 cells using Cellfectin. One out of the three clones was selected for expression based on the secretion levels after the first round of virus expansion. Viruses expanded for two generations were used for protein expression at an MOI = 5 in suspended Sf9 cell cultures. Supernatants were harvested after 72 h, and proteins were purified as described.
Biochemical characterization of DM allotypes
All recombinant HLA proteins (and the constructs used) were produced in an insect cell-baculovirus expression system as previously described.18 Secreted proteins were purified by affinity chromatography from the cell culture supernatants using the L243 antibody covalently bound to a CNBr matrix for HLA-DR2-CLIP (DR2-C) and an M2 affinity gel for DMs (Sigma-Aldrich). DR2-C was further dialyzed vs. PBS pH 7.4 and treated with thrombin (20 U/mg in PBS; Sigma-Aldrich), yielding DR2/C. CLIP was then replaced by MBP-FITC—DENPVVHFFKNIVTPRT (K is used for conjugation of fluorescein isothiocyanate (FITC)) from these complexes. To this end, DR2/C complexes at 20 μM were incubated with 100 μM MBP-FITC in the dark for 48 h (PBS pH 5.8 at 37 °C). Affinity-purified DM and DR2/MBP-FITC after the peptide-exchange reaction were gel-filtered using a Sephadex S200 column. Fractions containing the protein of interest were pooled and concentrated using a Vivaspin 10-kDa MWCO spin filter. The homogeneity and purity of the samples were checked by SDS–polyacrylamide gel electrophoresis.
Fluorescence polarization was used to assess the effect of DM in peptide dissociation experiments. DR2/MBP-FITC (250 nM) was incubated with an excess of competing, non-labeled peptide (MBP83–99 50 μM) in the presence of titrated amounts of DM (75 and 250 nM). Reactions were performed in 50 mM citrate phosphate buffer with 150 mM NaCl at the indicated pH. Samples were prepared by premixing DM, unlabeled MBP peptide, and buffer, to which we added purified DR2/MBP-FITC (predialyzed vs. the buffer used in the assay). Reactions were prepared on ice and measured at 25 °C with a Victor 3V plate reader (PerkinElmer).
Thermofluor assays were used to estimate the thermal stability of all recombinant DM allotypes as previously described.18 Monomeric protein (0.3–0.5 mg/ml) in 50 mM citrate phosphate buffer containing 150 mM NaCl and adjusted to the desired pH was mixed with 5× SyPro Orange (Life Technologies) in a 96-well plate. Samples were heated from 25 to 95 °C with a temperature increase of 2 °C/min. Excitation was carried out at 490 nm, and the fluorescent signal was detected at 575 nm. The change in fluorescence intensity was plotted vs. the temperature, and a sigmoidal function was fit to determine the midpoint denaturation temperature (Tm). All measurements were performed in triplicate in at least three independent experiments.
Modeling the DMBD31V substitution
The crystal structure of the HLA-DRB1*01:01/DM complex (PDB-ID:4FQX, pH 5.5) served as the starting structure to build the models of DMBD31−, DMBD31+, and the D31V mutant. The unresolved atoms in the structure were modeled and refined with the MODELLER software48 using the corresponding sequence. The mutation in the starting structure of the D31V mutant was modeled by substituting the aspartate (ASP31) side chain with valine (VAL). The pKa values of the titratable side chains of D31 and E47 were estimated with the program propKa3.49 Then protonation states were assigned for pH 5.5 with the Gromacs program pdb2gmx.50 With the Amber 12 simulation package, each complex was initially placed in an octahedral TiP3 water box and neutralized with 33 counterions. Then each complex was energy-minimized, positionally restrained (25 kcal/mol/Å2), and heated from 100 to 300 K. The restraints were resolved in five steps, and then each complex was equilibrated for 2 ns.51 Short-range nonbonded interactions were taken into account up to a cutoff value of 9 Å.
Proteomics
For each of the two biological replicates stated per condition, 2 × 108 cells in complete culture medium were washed twice with medium lacking FCS and finally resuspended in medium with 1% FCS. After 6 h, the cells were harvested, and pMHCII complexes were purified as previously described.27 Briefly, cells from each culture were divided into three aliquots at harvesting, which were used as technical replicates. These individual cell pellets were kept on rotation for 1 h in cell lysis buffer (Tris HCl 50 mM, pH 8.0, NaCl 150 mM, plus 1% CHAPS, 1:20 complete protease inhibitor) and spun for 30 min at 15,000 × g. Supernatants of each technical replicate were transferred to columns loaded with FF-Sepharose-L243 beads and kept on rotation for 2 h. Subsequently, beads were washed with 10 volumes of Tris HCl 50 mM, pH 8.0, alternating high- and low-salt concentrations. A final wash with H2O was followed by acidic elution of the peptides from the MHCII complexes using 5 volumes of 0.02% trifluoroacetic acid. The resulting peptide mixtures were fractionated using a microspin filter device with a 10-kDa cutoff and washed using C18 zip-tips. Peptides were analyzed by a reversed-phase capillary system (Ultimate 3000 nanoLC) connected to an Orbitrap Velos (Thermo Fischer). Liquid chromatography was performed on a reversed-phase C18 column (Thermo Scientific) and used for technical replicates from the moment of cell lysis up to measurement.
MaxQuant software (version 1.5.2.8)52 was used for peptide identification. A customized database featuring reviewed and nonredundant UniProt human proteins (accessed March 2017) was used for peptide identification. No enzyme specificity was used for the search, and a tolerance of 10 ppm was allowed for the main ion search and 50 ppm for MSMS identification. The “match between runs” feature was enabled. False Discovery Rate (FDR) was set at 0.01. The mass spectrometric (MS) proteomics data have been deposited into the ProteomeXchange Consortium via the PRIDE53 partner repository with the dataset identifier PXD009791. Subsequent data processing using the PLAtEAU algorithm has been recently described in ref. 27 and is available viahttps://github.com/e-morrison/plateau.
Perseus software54 was used to analyze the MS data. The identified epitopes and their corresponding percentage of intensity from the Total Ion Current (TIC) determined by PLAtEAU were initially loaded as matrices. Protein sources were annotated using the Homo sapiens annotation file (mainAnnot.homo_sapiens.txt.gz), removing specific terms related to the specific functions of the entries (cell part; cell body: apical part of cell: cell leading edge; bleb; cytoskeleton; cell projection; macromolecular complex; lipid particle; chromosome; chromatin remodeling complex; cilium; transcription elongation factor complex; ubiquitin ligase complex; phosphatidylinositol 3-kinase complex; pseudopodium; site of polarized growth; filopodium; proton-transporting two-sector ATPase complex; MHC class I peptide loading complex; polysome; small ribosomal subunit; transcription factor complex; midbody; receptor complex; protein complex; NADH dehydrogenase complex; neuron projection; respiratory chain complex IV; respiratory chain complex III; proteasome complex; DNA helicase complex; histone methyltransferase complex; microtubule organizing center; synapse; nucleoplasm; nucleosome; nucleolus; microbody; apical part of cell; cell cortex; ribbon; intracellular membrane-bounded organelle; centrosome; intracellular nonmembrane-bounded organelle; site of pol. growth). All data were log2-transformed, and missing values were imputed as the minimum observed value.
Peptide MHCII affinity and determination of the core epitopes was determined using the NetMHCIIpan 3.1 software.55 We set the epitope size to a minimum size of nine amino acids using the standard configuration. In cases where multiple registers were allowed, we considered the register with the strongest affinity.
Quantification and statistical analysis
GraphPad Prism 7.0 software (GraphPad Software, San Diego CA, USA) was used for statistical analysis. Variance was calculated with the two-way analysis of variance method. The null hypothesis was rejected when the p value was < 0.05. The corresponding p values for each test are indicated as follows: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
For the proteomics analysis approach, we used the Perseus software.54 To evaluate whether there were quantitative differences between the epitopes eluted from each DM condition, all measurements were used to define mean intensity values for each epitope. p Values were calculated based on the observed intensities using a t test, setting the FDR to 0.01 and the S0 value to 0.2. 1D annotation enrichment is based on Wilcoxon–Mann–Whitney test, which assumes independence of the values. In this case, an FDR of 0.02 was applied.
Electronic supplementary material
Table S1. Haploview statistics for the rsSNPs found in coding regions of DMA and DMB
Table S2. Relative intensities of the epitopes defined by PLAtEAU
Table S3. Epitope enrichment in samples isolated from T2.DR3 cell lines expressing DM11 or DM37
Acknowledgements
We thank Peter Cresswell for providing the cell lines used in this study. We also would like to thank the group of Petra Knaus for the S2 working space and David H. Canaday for the 9AF6 T cell hybridoma. For mass spectrometry, we would like to acknowledge the assistance of the Core Facility BioSupraMol supported by the Deutsche Forschungsgemeinschaft (DFG). C.F. is thankful for funding by the DFG (FR-1325/17–1, SFB958 (project Z03) and TRR186 (projects A05, A11)). M.A.-B. is thankful for funding from the Freie Universität Berlin Forschungskommision.
Author contributions
M.A.-B. designed and performed the research. E.M. contributed to the MS experiments and bioinformatics analysis. E.T.A. analyzed the influence of pH on DM structure in silico. M.U. and M.W. performed the biochemical experiments. F.E. contributed to the antigen presentation assays. C.F. initiated and supervised the project. M.A.-B. wrote the paper with support from E.M. and major input from C.F.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Miguel Álvaro-Benito, Email: malvaro@zedat.fu-berlin.de.
Christian Freund, Email: chfreund@zedat.fu-berlin.de.
Electronic supplementary material
The online version of this article (10.1038/s41423-018-0181-1) contains supplementary material.
References
- 1.Blum JS, Wearsch PA, Cresswell P. Pathways of antigen processing. Annu. Rev. Immunol. 2013;31:443–473. doi: 10.1146/annurev-immunol-032712-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mellins E, et al. Defective processing and presentation of exogenous antigens in mutants with normal HLA class II genes. Nature. 1990;343:71–74. doi: 10.1038/343071a0. [DOI] [PubMed] [Google Scholar]
- 3.Sloan VS, et al. Mediation by HLA-DM of dissociation of peptides from HLA-DR. Nature. 1995;375:802–806. doi: 10.1038/375802a0. [DOI] [PubMed] [Google Scholar]
- 4.Grotenbreg GM, et al. Empty class II major histocompatibility complex created by peptide photolysis establishes the role of DM in peptide association. J. Biol. Chem. 2007;282:21425–21436. doi: 10.1074/jbc.M702844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wieczorek M, et al. MHC class II complexes sample intermediate states along the peptide exchange pathway. Nat. Commun. 2016;7:13224. doi: 10.1038/ncomms13224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wieczorek M, Tolba ET, Stich J, Álvaro-Benito M, Stolzenberg S, Noe F, Freund C. Conformational plasticity of MHC proteins. Frontiers Immunol. 8, 292 (2017). [DOI] [PMC free article] [PubMed]
- 7.Guce AI, et al. HLA-DO acts as a substrate mimic to inhibit HLA-DM by a competitive mechanism. Nat. Struct. Mol. Biol. 2013;20:90–98. doi: 10.1038/nsmb.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Unanue ER, Turk V, Neefjes J. Variations in MHC class II antigen processing and presentation in health and disease. Annu. Rev. Immunol. 2016;34:265–300. doi: 10.1146/annurev-immunol-041015-055420. [DOI] [PubMed] [Google Scholar]
- 9.Lindner R, Unanue ER. Distinct antigen MHC class II complexes generated by separate processing pathways. EMBO J. 1996;15:6910–6920. doi: 10.1002/j.1460-2075.1996.tb01083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amria S, et al. HLA‐DM negatively regulates HLA-DR4-restricted collagen pathogenic peptide presentation and T cell recognition. Eur. J. Immunol. 2008;38:1961–1970. doi: 10.1002/eji.200738100. [DOI] [PubMed] [Google Scholar]
- 11.Hou T, et al. An insertion mutant in DQA1*0501 restores susceptibility to HLA-DM: implications for disease associations. J. Immunol. 2011;187:2442–2452. doi: 10.4049/jimmunol.1100255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohan JF, Petzold SJ, Unanue ER. Register shifting of an insulin peptide-MHC complex allows diabetogenic T cells to escape thymic deletion. J. Exp. Med. 2011;208:2375–2383. doi: 10.1084/jem.20111502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kremer AN, et al. Endogenous HLA class II epitopes that are immunogenic in vivo show distinct behavior toward HLA-DM and its natural inhibitor HLA-DO. Blood. 2012;120:3246–3255. doi: 10.1182/blood-2011-12-399311. [DOI] [PubMed] [Google Scholar]
- 14.Rosskopf S, et al. Creation of an engineered APC system to explore and optimize the presentation of immunodominant peptides of major allergens. Sci. Rep. 2016;6:31580. doi: 10.1038/srep31580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Álvaro-Benito M, Morrison E, Wieczorek M, Sticht J, Freund C. Human leukocyte antigen-DM polymorphisms in autoimmune diseases. Open Biol. 2016;6:160165. doi: 10.1098/rsob.160165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pierre P, et al. HLA-DM is localized to conventional and unconventional MHC class II–containing endocytic compartments. Immunity. 1996;4:229–239. doi: 10.1016/S1074-7613(00)80431-8. [DOI] [PubMed] [Google Scholar]
- 17.Arndt SO, et al. Functional HLA‐DM on the surface of B cells and immature dendritic cells. EMBO J. 2000;19:1241–1251. doi: 10.1093/emboj/19.6.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Álvaro-Benito M, Wieczorek M, Sticht J, Kipar C, Freund C. HLA-DMA polymorphisms differentially affect MHC class II peptide loading. J. Immunol. 2014;194:803–816. doi: 10.4049/jimmunol.1401389. [DOI] [PubMed] [Google Scholar]
- 19.Denzin LK, et al. Neutralizing antibody responses to viral infections are linked to the non-classical MHC class II gene H2-Ob. Immunity. 2017;47:310–322.e7. doi: 10.1016/j.immuni.2017.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson J, et al. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 2015;43:D423–D431. doi: 10.1093/nar/gku1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibbs RA, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pos W, et al. Crystal structure of the HLA-DM–HLA-DR1 complex defines mechanisms for rapid peptide selection. Cell. 2012;151:1557–1568. doi: 10.1016/j.cell.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 25.Weber DA, Dao CT, Jun J, Wigal JL, Jensen PE. Transmembrane domain-mediated colocalization of HLA-DM and HLA-DR is required for optimal HLA-DM catalytic activity. J. Immunol. 2001;167:5167–5174. doi: 10.4049/jimmunol.167.9.5167. [DOI] [PubMed] [Google Scholar]
- 26.Denzin LK, Robbins NF, Carboy-Newcomb C, Cresswell P. Assembly and intracellular transport of HLA-DM and correction of the class II antigen-processing defect in T2 cells. Immunity. 1994;1:595–606. doi: 10.1016/1074-7613(94)90049-3. [DOI] [PubMed] [Google Scholar]
- 27.Álvaro-Benito M, Morrison E, Abualrous ET, Kuropka B, Freund C. Quantification of HLA-DM-dependent major histocompatibility complex of class II immunopeptidomes by the peptide landscape antigenic epitope alignment utility. Front. Immunol. 2018;9:872. doi: 10.3389/fimmu.2018.00872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramachandra L, Kovats S, Eastman S, Rudensky AY. Variation in HLA-DM expression influences conversion of MHC class II alpha beta:class II-associated invariant chain peptide complexes to mature peptide-bound class II alpha beta dimers in a normal B cell line. J. Immunol. 1996;156:2196–2204. [PubMed] [Google Scholar]
- 29.Cox J, Mann M. 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics. 2012;13(Suppl 16):S12. doi: 10.1186/1471-2105-13-S16-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng ML, et al. Analysis of HLA-DM polymorphisms in the Chinese Han population. Tissue Antigens. 2012;79:157–164. doi: 10.1111/j.1399-0039.2012.01838.x. [DOI] [PubMed] [Google Scholar]
- 31.Hughes AL, Nei M. Nucleotide substitution at major histocompatibility complex class II loci: evidence for overdominant selection. Proc. Natl. Acad. Sci. USA. 1989;86:958–962. doi: 10.1073/pnas.86.3.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughes AL, Hughes MK, Howell CY, Nei M. Natural selection at the class II major histocompatibility complex loci of mammals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1994;346:359–366. doi: 10.1098/rstb.1994.0153. [DOI] [PubMed] [Google Scholar]
- 33.Denzin LK, Cresswell P. HLA-DM induces clip dissociation from MHC class II αβ dimers and facilitates peptide loading. Cell. 1995;82:155–165. doi: 10.1016/0092-8674(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 34.Pu Z, Lovitch SB, Bikoff EK, Unanue ER. T cells distinguish MHC-peptide complexes formed in separate vesicles and edited by H2-DM. Immunity. 2004;20:467–476. doi: 10.1016/S1074-7613(04)00073-1. [DOI] [PubMed] [Google Scholar]
- 35.Stadinski BD, et al. Diabetogenic T cells recognize insulin bound to IAg7 in an unexpected, weakly binding register. Proc. Natl. Acad. Sci. USA. 2010;107:10978–10983. doi: 10.1073/pnas.1006545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fremont DH, Crawford F, Marrack P, Hendrickson WA, Kappler J. Crystal structure of mouse H2-M. Immunity. 1998;9:385–393. doi: 10.1016/S1074-7613(00)80621-4. [DOI] [PubMed] [Google Scholar]
- 37.Busch R, Reich Z, Zaller DM, Sloan V, Mellins ED. Secondary structure composition and pH-dependent conformational changes of soluble recombinant HLA-DM. J. Biol. Chem. 1998;273:27557–27564. doi: 10.1074/jbc.273.42.27557. [DOI] [PubMed] [Google Scholar]
- 38.Pashine A, et al. Interaction of HLA-DR with an acidic face of HLA-DM disrupts sequence-dependent interactions with peptides. Immunity. 2003;19:183–192. doi: 10.1016/S1074-7613(03)00200-0. [DOI] [PubMed] [Google Scholar]
- 39.Nicholson MJ, et al. Small molecules that enhance the catalytic efficiency of HLA-DM. J. Immunol. 2006;176:4208–4220. doi: 10.4049/jimmunol.176.7.4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Z, Jensen PE. Structural characteristics of HLA-DQ that may impact DM editing and susceptibility to type-1 diabetes. Front. Immunol. 2013;4:262. doi: 10.3389/fimmu.2013.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Z, et al. Type 1 diabetes associated HLA-DQ2 and DQ8 molecules are relatively resistant to HLA-DM mediated release of invariant chain-derived CLIP peptides. Eur. J. Immunol. 2016;46:834–845. doi: 10.1002/eji.201545942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Lith M, McEwen-Smith RM, Benham AMHLADP. HLA-DQ, and HLA-DR have different requirements for invariant chain and HLA-DM. J. Biol. Chem. 2010;285:40800–40808. doi: 10.1074/jbc.M110.148155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcia KC, Adams EJ. How the T cell receptor sees antigen—a structural view. Cell. 2005;122:333–336. doi: 10.1016/j.cell.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 44.Fissolo N, et al. Naturally presented peptides on major histocompatibility complex I and II molecules eluted from central nervous system of multiple sclerosis patients. Mol. Cell. Proteomics. 2009;8:2090–2101. doi: 10.1074/mcp.M900001-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heyder T, et al. Approach for identifying human leukocyte antigen (HLA)-DR bound peptides from scarce clinical samples. Mol. Cell. Proteomics. 2016;15:3017–3029. doi: 10.1074/mcp.M116.060764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weber K, Bartsch U, Stocking C, Fehse B. A multicolor panel of novel lentiviral "gene ontology" (LeGO) vectors for functional gene analysis. Mol. Ther. 2008;16:698–706. doi: 10.1038/mt.2008.6. [DOI] [PubMed] [Google Scholar]
- 47.Bacher P, et al. Antigen-reactive T cell enrichment for direct, high-resolution analysis of the human naive and memory Th cell repertoire. J. Immunol. 2013;190:3967–3976. doi: 10.4049/jimmunol.1202221. [DOI] [PubMed] [Google Scholar]
- 48.Webb B, Sali A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics. 2016;54:5.6.1–5.6.37. doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olsson MHM, Søndergaard CR, Rostkowski M, Jensen JH. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 50.Pronk S, et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Case DA, et al. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 53.Vizcaíno JA, et al. update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;2016:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tyanova S, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 55.Andreatta M, et al. Accurate pan-specific prediction of peptide-MHC class II binding affinity with improved binding core identification. Immunogenetics. 2015;67:641–650. doi: 10.1007/s00251-015-0873-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Álvaro-Benito M, Morrison E, Abualrous ET, Kuropka B, Freund C. Quantification of HLA-DM-dependent major histocompatibility complex of class II immunopeptidomes by the peptide landscape antigenic epitope alignment utility. Front Immunol. 9:872 (2018). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Haploview statistics for the rsSNPs found in coding regions of DMA and DMB
Table S2. Relative intensities of the epitopes defined by PLAtEAU
Table S3. Epitope enrichment in samples isolated from T2.DR3 cell lines expressing DM11 or DM37