Abstract
Purpose
To investigate the pathogenesis of the recurrent preimplantation embryonic arrest characterized by direct cleavage.
Methods
Two affected individuals underwent time-lapse imaging to observe the cleavage behaviors in their final ICSI attempts. In addition, both patients were subjected to whole-exome sequencing. After the identification of possible causative genes, molecular modeling analyses were used to evaluate the possible effects of candidate mutations on protein secondary structure.
Results
All the bipronucleated (2PN) zygotes from both individuals presented multiple abnormal cleavage behaviors, particularly direct cleavage (DC) and subsequent cleavage arrest. Mutation analysis identified one new frameshift mutation c.1521dupC (p.S508Qfs*5) and two missense mutations c.A1117C and c.C1708T (p.T373P and p.R570C, respectively) of the PADI6 gene, which were in the protein-arginine deiminase (PAD) domain and highly conserved.
Conclusion
This study expands the mutation spectrum of PADI6 and is the first to propose that the preimplantation embryonic arrest with concomitant abnormal cleavage behaviors, especially DC, maybe associated with PADI6 mutations.
Electronic supplementary material
The online version of this article (10.1007/s10815-019-01606-7) contains supplementary material, which is available to authorized users.
Keywords: PADI6, Preimplantation embryonic arrest, Direct cleavage, Mutation
Introduction
Normal preimplantation embryonic development is a key step in establishing a successful pregnancy. The zygote undergoes three consecutive occurrences of cytokinesis and produces six to eight blastomeres on day 3 of cultivation. Only approximately 40–70% of the human embryos produced during in vitro fertilization (IVF) are viable embryos, and the others arrest at different stages [1]. If all her embryos undergo developmental arrest, the patient’s IVF/ intracytoplasmic sperm injection (ICSI) cycle fails.
The quality of the embryo is usually evaluated according to cell number, cell symmetry, and fragmentation by means of morphological assessments at several predefined points in time during embryo development [2, 3]. With the widespread application of time-lapse imaging, opportunities to generate a complete and continuous picture of the kinetics of embryo development have arisen [1, 4–6]. Numerous recent studies have identified maternal-effect factors that play essential roles in preimplantation embryonic development. Mutations in TUBB8 (tubulin beta 8 class VIII) [7, 8], PADI6 (peptidyl arginine deiminase, type VI) [9], and TLE6 (transducin-like enhancer of split 6) [10, 11] cause embryonic arrest before the 8-cell stage, with concomitant fragmentation. However, mutations in these genes just account for a small number of patients, and the genetic basis of preimplantation embryonic arrest remains poorly understood.
In this study, we observed all the embryos have abnormal cleavage patterns, mainly direct cleavage (DC), and identified a new compound-heterozygous missense mutation and a new homozygous frameshift mutation in PADI6 (OMIM ID: 617234; NM_207421) in two infertile individuals by time-lapse imaging and whole-exome sequencing (WES). These data are the first to suggest the possible pathogenesis of bipronucleated (2PN) zygote DC and extend the spectrum of phenotypes for mutations in PADI6.
Materials and methods
Study subjects
All the preimplantation embryonic arrest and control subjects examined in this study were from the Reproductive and Genetic Hospital of CITIC-Xiangya. All blood samples were donated for the investigation after informed consent was obtained. This study was approved by the Ethics Committee of the Reproductive and Genetic Hospital of CITIC-Xiangya (reference LL-SC-2017-009).
Time-lapse imaging
The 2PN zygotes were placed individually and cultured in G1 plus medium for time-lapse imaging with the Primo Vision system (Vitrolife, Goteborg, Sweden). Images of each embryo were recorded every 5 min.
Whole-exome sequencing (WES) and variant analysis
The probands and control individuals were subjected to WES. Then, combined the time-lapse data and focused on patients of the DC occurred. Sequencing, variant calling, and annotation were previously described [12]. Variants were filtered using the following criteria: (1) variations with minor allele frequencies less than 1% in the following databases (Genome AD, 1000 Genomes Project and ExAC), (2) exonic nonsynonymous or splice site variants or coding INDELs, (3) homozygous/compound-heterozygous mutations in the proband, (4) mRNA/proteins that were highly expressed or specifically expressed in the human oocyte, (5) coexistence in both probands, and (6) absence in both controls. In addition, additional infertile individuals with recurrent preimplantation embryonic arrest (n = 98), poor fertilization (n = 50), and oocyte mature disorder (n = 30) were also recruited to estimate the prevalence of PADI6 mutation in different infertile populations.
Sanger sequencing
Specific primers flanking the mutation in the PAID6 gene were used for amplification by PCR with an ABI 3100 DNA analyzer (Applied Biosystems, Foster City, CA, US). The variants were validated by Sanger sequencing in the affected individuals, other family members and 100 women with normal fertility.
Molecular modeling and evolutionary conservation analysis
WT and mutated PADI6 (NP_997304.3, p.T373P, and p.R570C) were assessed using SWISS-MODEL software (https://swissmodel.expasy.org) based on the template of 4dkt.1.A.pdb. Mutated PADI6 was mapped onto the atomic model using PyMol (http://www. pymol.org). Evolutionary conservation analysis was performed with MultiAlin (http:// multalin.toulouse.inra.fr/multalin/multalin.html) software.
Results
Clinical characteristics and phenotypes of patients and control individuals
We recruited two patients and two control individuals from independent and hitherto uncharacterized families with primary female infertility. Both control subjects underwent 1 IVF/ICSI cycle. The 11 embryos were cleaved at the 8-cell stage on day 3 and further cultured to form blastocysts on day 5, and one blastocyst was implanted, and one healthy baby was born. Both patients underwent 2–3 failed IVF/ICSI attempts at our center. Patient II-1 in family 1 underwent two different COH protocols for separate IVF/ICSI attempts. The number of retrieved oocytes (24 and 20 in attempts 1 and 2, respectively) and frequency of oocyte maturity (83.3% and 80%, respectively) were not clearly abnormal, but the normal fertilization (2PN) rate was low (20% and 50%, respectively); however, some 0PN fertilized eggs also cleaved. After cultivation, no viable embryos were produced, all zygotes were cleaved at the 2–4-cell stage on day 3, and all were arrested during further blastocyte culture. Patient II-1 in family 2 underwent three COH protocols and different IVF/ICSI attempts, which also resulted in many retrieved oocytes (14, 17, and 18 in attempts 1, 2, and 3, respectively), high oocyte maturity (100%, 76.4%, and 77.8%, respectively), and an especially low 2PN rate (21.4%, 23.1%, and 14.3, respectively). In addition, all the zygotes were cleaved at the 2–5-cell stage on day 3 and were arrested during further blastocyte culture. Table 1 summarizes the detailed information of the two patients.
Table 1.
Oocyte and embryo characteristics of two patients undergoing IVF and ICSI
| Patients | Age | IVF and ICSI attempts | COH protocol | Total oocytes | MII Oocytes | Fertilized oocytes | Outcomes |
|---|---|---|---|---|---|---|---|
| II-1 in family 1 | 31 | First IVF | Ultra-long | 24 | 20 | 6 (4 2PN/2 1PN) | The 2PN zygotes were cleaved to 1 grade II 4-cell stage, 2 grade II 2-cell stage; 0PN zygotes were cleaved to 3 grade II 2-cell stage; and the remaining 14 embryos showed cleavage failure on day 3. |
| 33 | Secord ICSI/time-lapse | Long | 20 | 16 | 11 (8 2PN/3 1PN) | The 2PN zygotes were cleaved to 2 grade II 5-cell stage, 1 grade I 4-cell stage, 2 grade II 4-cell stage, and 2 grade II 3-cell stage; 1PN zygotes were cleaved to 2 grade II 3- to 5-cell stage; 0PN zygotes were cleaved to 3 2- to 4-cell stage; and the remaining 4 embryos showed cleavage failure on day 3. All embryos arrested during subsequent blastocyst culture. | |
| II-1 in family 2 | 26 | First IVF | Long | 14 | 14 | 11 (3 2PN/5 1PN/3 3PN) | The 2PN zygotes were cleaved to 2 grade II 4-cell stage; 1PN zygotes were cleaved to 4 2- to 4-cell stage; 3PN embryos were cleaved to 3 2- to 4-cell stage; 0PN zygotes were cleaved to 3 2- to 3-cell stage; and the remaining 2 embryos showed cleavage failure on day 3. |
| 26 | Secord half ICSI | Ultra-long | 17 | 13 | 11 (3 2PN/7 1PN/1 3PN) | The 2PN zygotes were cleaved to 2 grade II 4-cell stage and 1 grade III 2-cell stage; 1PN zygotes were cleaved to 4 2- to 3-cell stage; 3PN embryos were cleaved to 3-cell stage; 0PN zygotes were cleaved to 2 2- to 5-cell stage; and the remaining 3 embryos showed cleavage failure on day 3. All embryos arrested during subsequent blastocyst culture. | |
| 27 | Third ICSI/time-lapse | GnRH-a | 18 | 14 (4 degeneration) | 5 (3 2PN/2 1PN) | The 2PN zygotes were cleaved to 1 grade II/1 grade III/1 grade IV 2-cell stage, and the remaining 7 embryos showed cleavage failure on day 3. |
Time-lapse imaging of abnormal division behaviors
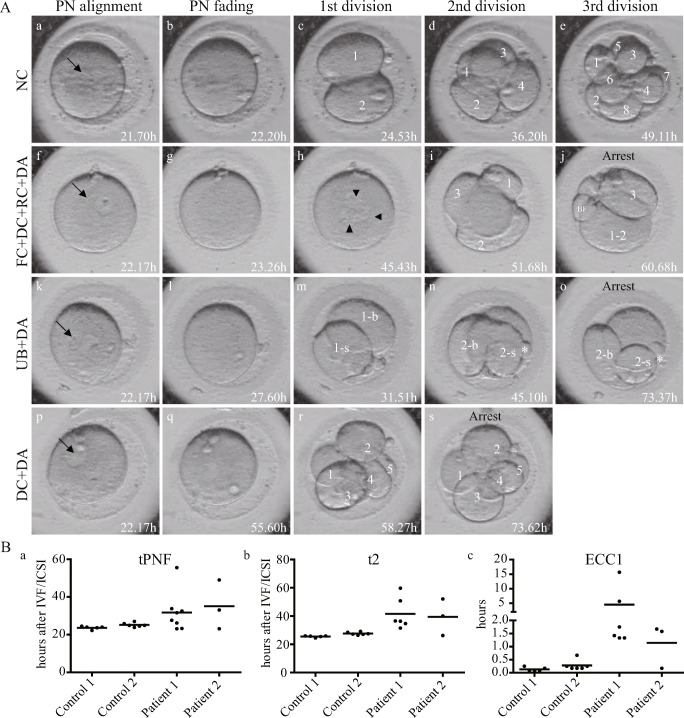
We adopted time-lapse imaging in the final attempt of both patients and control individuals. The kinetic parameters focused on the tPNF (timing of the fading of both pronuclei), t2 (timing of division to two completely separated blastomeres, for the patients, the t2 indicated the timing of the finish of the first division), and ECC1 (the duration of the first division). Additionally, morphological dynamic parameters were analyzed. In the 11 control 2PN zygotes, both pronuclei had faded at 22.2 to 27.0 h after IVF/ICSI, and the length of time was nearly the same for different zygotes. However, the tPNF was clearly longer for the 2PN embryos of both patients (23.1–55.6 h and 23.2–49.0 h, respectively), and the lengths of time were dispersed from each other, and the t2 and ECC1 showed the similar change tendency with tPNF (Fig. 1B a–c). The normal cleavage pattern (NC) and typical abnormal cleavage patterns are shown in Fig. 1A, and detailed morphological dynamic parameter analyses are described in Table 2. The control zygotes underwent three continuous cleavages of each blastomere to two blastomeres. Finally, an 8-cell embryo was observed on day 3 of cultivation (Fig. 1A a–e and Supplemental Video 1). For patient 1, multiple abnormal division behaviors were observed. Regarding embryo 5, the zygote cleaved into two uneven blastomeres (UBs) (Fig. 1A m), and then, only one blastomere entered the next cell cycle, cleaved into two UBs again, and finally arrested at the 3-cell stage (Fig. 1A n–o). Embryos 4 and 6 underwent the first failed cytokinesis (FC), and then DC in the second cell cycle (Fig. 1A h–j). The other five zygotes directly divided into three to five blastomeres during the first cell cycle, and then developmentally arrested (DA) on day 3 (Fig. 1A p–s and Supplemental Video 2). However, in patient 2, all three zygotes were directly divided into three or four blastomeres in the first division, and then developmental arrest occurred. Meanwhile, embryo 2 also exhibited reverse cleavage (RC) behavior (blastomere fusion from 4 cells to 3 cells during the first cleavage), and scattered fragments emerged after division in embryos.
Fig. 1.
Morphological characteristics and kinetic parameter abnormities observed by time-lapse imaging. A Representative images show three consecutive occurrences of division and the production of eight blastomeres in the control zygote (a–e), as well as three abnormal cleavage patterns during embryonic development of the patients’ zygotes. NC, normal cleavage; PN, pronucleus; DA, developmental arrest; DC, direct cleavage; FC, failed cytokinesis; RC, reverse cleavage; BF, big fragment; UB, uneven blastomeres; -b, the larger blastomeres of the UB; -s, the smaller blastomeres of the UB. The arrow indicates the PN, and the arrowhead indicates the three nuclei produced despite the first cytokinesis failure. B Kinetic parameters such as the tPNF(a), t2(b) and ECC1(c) delays in the patients’ zygotes. Dates are shown as the average
Table 2.
Summary of kinetic and morphological dynamic parameters
| tPNF (hours) | t2 (hours) | ECC1 (hours) | First cleavage | Secord cleavage | Third cleavage | ||
|---|---|---|---|---|---|---|---|
| Control 1 | Embryo 1 | 23.6 | 25.8 | 0.25 | Normal, cleaved to 2-cell | Normal, 2-cell cleaved to 4-cell | Normal, 4-cell cleaved to 8-cell |
| Embryo 2 | 24.0 | 25.9 | 0.17 | ||||
| Embryo 3 | 24.0 | 25.5 | 0.07 | ||||
| Embryo 4 | 24.5 | 26.0 | 0.08 | ||||
| Embryo 5 | 22.2 | 24.5 | 0.08 | ||||
| Control 2 | Embryo 1 | 20.5 | 26.4 | 0.17 | Normal, cleaved to 2-cell | Normal, 2-cell cleaved to 4-cell | Normal, 4-cell cleaved to 8-cell |
| Embryo 2 | 24.5 | 27.5 | 0.25 | ||||
| Embryo 3 | 25.9 | 27.1 | 0.67 | ||||
| Embryo 4 | 24.9 | 29.2 | 0.17 | ||||
| Embryo 5 | 21.1 | 28.5 | 0.25 | ||||
| Embryo 6 | 20.9 | 27.3 | 0.17 | ||||
| Patient 1 | Embryo 1 | 55.6 | 59.7 | 1.42 | DC to 5-cell, FR | DA | |
| Embryo 2 | 26.2 | 34.7 | 5.67 | DC to 3-cell, BF | 1/3 DA to 5-cell, FR | DA | |
| Embryo 3 | 32.5 | 36.4 | 1.33 | DC to 4-cell, BF | DA | ||
| Embryo 4 | 23.1 | / | / | FC | DC to 4-cell | DA | |
| Embryo 5 | 27.6 | 31.5 | 1.33 | UB to 2-cell | 1/2 DA to 3-cell | DA | |
| Embryo 6 | 23.3 | / | / | FC | DC to 3-cell and RC to 2-cell, FR | DA | |
| Embryo 7 | 31.6 | 50.8 | 15.67 | DC to 3-cell and RC to 2-cell, FR | 1/2 DA to 3-cell | 1/3 DA to 5-cell | |
| Embryo 8 | 33.8 | 36.6 | 1.75 | DC to 3-cell | DA | ||
| Patient 2 | Embryo 1 | 49.0 | 52.1 | 1.67 | DC to 3-cell, BF | DA | |
| Embryo 2 | 33.8 | 40.0 | 1.58 | DC to 4-cell and RC to 3-cell, FR | DA | ||
| Embryo 3 | 23.2 | 26.2 | 0.17 | DC to 4-cell | DA |
tPNF, timing of the fading of both pronuclei; t2, timing of division to two completely separated blastomeres, for the patients, the t2 indicated the timing of the finish of the first division; ECC1, the duration of the first division; DC, direct cleavage; DA, developmental arrest; FC, failed cytokinesis; RC, reverse cleavage; UB, uneven blastomeres; BF, big fragment; FR, scattered fragments after division
Identification of PADI6 pathogenic variants
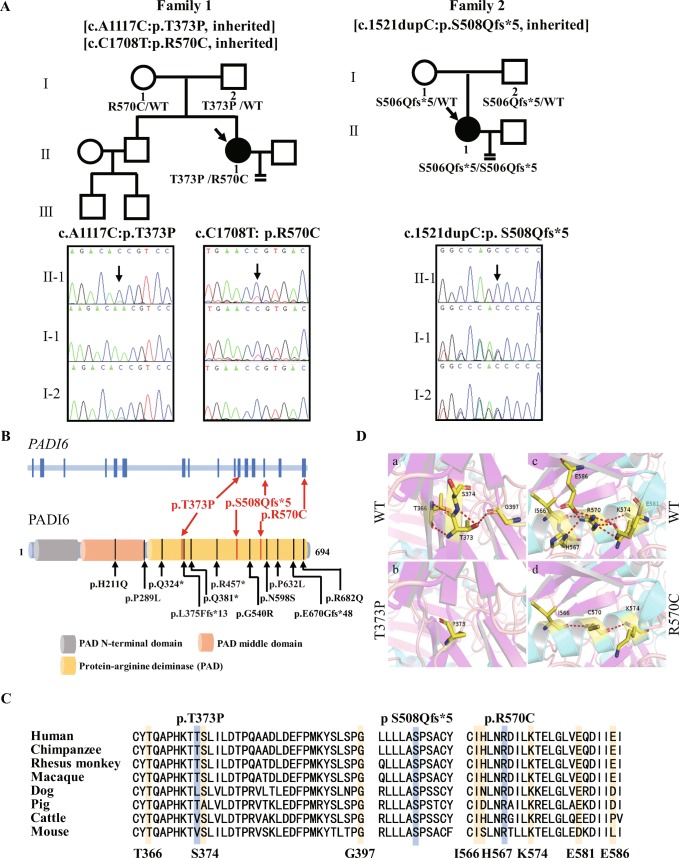
We performed WES in two patients and control individuals. After filtering was performed according to the criteria described in “Materials and Methods,” only PADI6 (OMIM ID: 617234; NM_207421) variants remained in both patients (Supplemental Table 1). Patient 1 had a compound-heterozygous missense mutation, namely c.A1117C and c.C1708T (p.T373P and p.R570C, respectively). Sanger sequencing confirmed that p.T373P and p.R570C were from her father and mother, respectively (Fig. 2A left). Patient 2 had a homozygous frameshift mutation c.1521dupC (p.S508Qfs*5), although her parents were not consanguineous according to the patient’s statement. However, Sanger sequencing confirmed that both her parents had a heterozygous mutation (Fig. 2A right). These corresponding mutations were not found in the East Asian population of the Genome AD exome, 1000 Genome, or exome aggregation consortium (ExAC) database, and were also not found in 100 healthy Chinese women with normal fertility.
Fig. 2.
Patient genotypic features. A Pedigree analysis of two cases of recurrent preimplantation embryonic arrest. Black squares with arrows indicate affected individuals. The black arrows in the chromatograms show the locations of the PADI6 mutations. B Localization of variants in the genomic and protein structure. Red arrows represent 3 new mutations in our study, and black arrows represent 11 mutations reported in previous studies. C Conservation of mutated amino acids in 7 different species. The residues S508 and R570 and nearby residues T366, G397, I566, and K574 are highly conserved across species; however, others are conserved in primates. D Protein conformation predictions of mutations in PADI6. The magnified views show the comparison of missense mutant residues P373 and C570 with views of wild-type residues T373 and R570. Red-dashed lines represent hydrogen bonds. The number of hydrogen bonds is decreased for both mutant residues
All three mutations were located in the protein-arginine deiminase (PAD) domain, and the hotspot region included the most (9/11) reported mutant residues (Fig. 2B). However, biallelic mutations in PADI6 were absent in 98 additional infertile individuals with recurrent preimplantation embryonic arrest, 50 individuals with poor fertilization and 30 individuals with oocyte mature disorder.
Impact of PADI6 mutations
The amino acids at positions p.S508 and p.R570 were highly conserved across species, and p.T373 was conserved in primates (Fig. 2C). According to a three-dimensional (3D) structure of PADI6, the hydrogen bonds of mutated residues T373 and R570 with nearby residues were predicted to have significant changes. The wild-type residue T373 is capable of hydrogen bonds and connects with residues T366, S374, and G397 (Fig. 2D a), but all three connections are absent in the mutated residue P373 (Fig. 2D b). In addition, the wild-type residue R570 is capable of hydrogen bonds with residues I566, H577, K574, E581, and E586 (Fig. 2D c); however, the mutated residues only have hydrogen bonds with the I566 and K574 residues (Fig. 2D d). All the obliterated hydrogen bond-connected residues are conserved in, minimally, primates (Fig. 2C).
Discussion
PADI6 is a member of the subcortical maternal complex (SCMC) and is critical to the formation of the oocyte-restricted fibrous structure, namely the cytoplasmic lattice (CPL). An increasing number of studies with mouse models have indicated that PADI6 plays a role in de novo protein synthesis prior to the maternal-to-embryonic transition and in microtubule-mediated organelle positioning and movement, and PADI6 null embryos do not develop past the two-cell stage [13, 14].
Four previous studies identified 11 different mutations in PADI6, including 8 compound-heterozygous mutations (p.H211Q and p.E670Gfs*48, p.Q324* and p.G540R, p.P289L and p.P632L, and p.N598s and p.R582Q) and 3 homozygous mutations (p.Q381*, p.L375Ffs*13, and p.R457*) (Fig. 2B and Supplemental Table 2). Here, we report three additional novel PADI6 mutations, which further expand the mutation spectrum of PADI6.
Previous investigations have indicated that mutations in PADI6 cause early embryonic arrest, underlying almost all cases of embryo arrest between the 2- and 5-cell stage [9]. Nevertheless, some viable embryos were also produced on day 3; however, these embryos failed to form blastocysts on day 5 or were transferred into the uterus but failed to establish a pregnancy [9]. Recent research has shown that PADI6 missense variants also cause miscarriages and molar pregnancies. The embryos are permissive for blastocyst implantation and the differentiation of some embryonic tissues, but the pregnancy is retained by the mother only until 8–12 weeks [15]. Our result is consistent with the aforementioned results, in which preimplantation embryonic arrest occurs. Meanwhile, previous studies have indicated that biallelic mutations in PADI6 may just cause embryonic arrest (Supplemental Table 2). Patients in our study demonstrated poor 2PN fertilization (Table 1), especially for patient 2 (21.4%, 23.1%, and 14.3%); however, some 0PN fertilized eggs also cleaved, combine with these fertilized eggs, the average fertilization rates were 63.9% (patient 1) and 86.5% (patient 2). Meanwhile, in our 50 poor fertilization patients WES data, we did not find any PADI6 mutations. Due to the sample size limitation, the relationship between poor fertilization and PADI6 mutation requires further research.
We have highlighted the previously reported mutation type, the location of all variants in the sequence of PADI6 and the phenotype (Fig. 2B and Supplemental Table 2). Most (9/11) of the mutations were located in the PAD domain, and most (5/7) were one- or two-protein-truncating variants in one patient [9, 11, 15, 16]. The compound-heterozygous missense mutation p.P289L and p.P632L in the patient caused the same phenotype as the previously described patients [11]; however, the p.N598S and p.R682Q compound-heterozygous missense mutation has a milder effect, namely hydatidiform moles [15], which may be because the two mutations are both near the end of the PAD domain mutation. In our study, we identified one compound front/middle PAD domain mutation and one PAD domain-truncating variant, resulting in preimplantation embryonic arrest.
There are growing concerns about the relationship between abnormal cleavage patterns and embryonic developmental potential. With the detailed observations of our patients’ cleavage anomalies, we noticed that almost all the tPNF, t2, and ECC1 were delayed and that all embryos had DC. The membranes of the two pronuclei disappeared, which corresponded to the M phase in the cell cycle [17, 18]. In 2PN embryos, the pronuclei usually disappear 20–24 h after insemination or ICSI [6, 19]. For our patients, all the pronuclei disappeared within 23.1–55.6 h. The delayed disappearance of the pronuclei may imply an obstacle to M phase entry.
Meanwhile, cleavage abnormalities have been shown to occur quite frequently, and DC accounts for a large proportion of these abnormalities [20, 21]. The cause of the DC pattern is not clear. Some studies regarded it as the formation of tripolar spindles, which cause abnormal distribution of chromosomes to the blastomeres during cell division in polyspermic human oocytes division [22, 23]. However, for properly fertilized 2PN zygotes, the pathogenesis of DC is poorly understood. These DC embryos have been confirmed to have a markedly decreased blastocyst formation rate, and the ability of these embryos to establish a pregnancy has been shown to be significantly reduced [2, 4]. Almost all (8/11) 2PN zygotes in our patients underwent direct division during the first cleavage, which may explain the abovementioned continuous pregnancy failure by morphological analysis on day 3 in the PADI6 mutation patients [9].
Recent researches indicated the CPL can regulate mouse oocyte microtubules, and PADI6 knockout causes altered spindle microtubule assembly [13]. We speculate that DC may be associated with to an abnormal spindle during cell division because of the obstacle to microtubule assembly. However, because of scarcity of human oocytes and embryos, the exact molecular mechanism is largely unknown.
In conclusion, our study expands the mutation spectrum of PADI6 and is the first to propose that the preimplantation embryonic arrest may be associated with PADI6 mutations is due to abnormal cleavage, mainly DC.
Electronic supplementary material
(AVI 4696 kb)
(AVI 3633 kb)
(DOCX 21 kb)
Acknowledgments
We thank all families that participated in our study.
Funding information
This study is supported by grants from National Key R&D Program of China (grant 2018YFC1003100, to L.H.) and Natural Science Foundation of Hunan Province (2018JJ3893).
Compliance with ethical standards
This study was approved by the Ethics Committee of the Reproductive and Genetic Hospital of CITIC-Xiangya (reference LL-SC-2017-009).
Conflicts of interest
The authors declare that they have no conflicts of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gardner DK, Lane M. Culture and selection of viable blastocysts: a feasible proposition for human IVF? Hum Reprod Update. 1997;3(4):367–382. doi: 10.1093/humupd/3.4.367. [DOI] [PubMed] [Google Scholar]
- 2.Rubio I, Kuhlmann R, Agerholm I, Kirk J, Herrero J, Escribá M-J, Bellver J, Meseguer M. Limited implantation success of direct-cleaved human zygotes: a time-lapse study. Fertil Steril. 2012;98(6):1458–1463. doi: 10.1016/j.fertnstert.2012.07.1135. [DOI] [PubMed] [Google Scholar]
- 3.Meseguer M, Herrero J, Tejera A, Hilligsøe KM, Ramsing NB, Remohí J. The use of morphokinetics as a predictor of embryo implantation. Hum Reprod. 2011;26(10):2658–2671. doi: 10.1093/humrep/der256. [DOI] [PubMed] [Google Scholar]
- 4.Yang S, Shi J, Gong F, Zhang S, Lu C, Tan K, Leng LZ, Hao M, He H, Gu YF, Lu GX, Lin G. Cleavage pattern predicts developmental potential of day 3 human embryos produced by IVF. Reprod BioMed Online. 2015;30(6):625–634. doi: 10.1016/j.rbmo.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Yang S-H, Wu C-H, Chen Y-C, Yang C-K, Wu T-H, Chen P-C, et al. Effect of morphokinetics and morphological dynamics of cleavage stage on embryo developmental potential: a time-lapse study. Taiwan J Obstet Gynecol. 2018;57(1):76–82. doi: 10.1016/j.tjog.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 6.Lemmen J, Agerholm I, Ziebe S. Kinetic markers of human embryo quality using time-lapse recordings of IVF/ICSI-fertilized oocytes. Reprod BioMed Online. 2008;17(3):385–391. doi: 10.1016/S1472-6483(10)60222-2. [DOI] [PubMed] [Google Scholar]
- 7.Chen B, Li B, Li D, Yan Z, Mao X, Xu Y, Mu J, Li Q, Jin L, He L, Kuang Y, Sang Q, Wang L. Novel mutations and structural deletions in TUBB8: expanding mutational and phenotypic spectrum of patients with arrest in oocyte maturation, fertilization or early embryonic development. Hum Reprod. 2017;32(2):457–464. doi: 10.1093/humrep/dew322. [DOI] [PubMed] [Google Scholar]
- 8.Feng R, Yan Z, Li B, Yu M, Sang Q, Tian G, Xu Y, Chen B, Qu R, Sun Z, Sun X, Jin L, He L, Kuang Y, Cowan NJ, Wang L. Mutations in TUBB8 cause a multiplicity of phenotypes in human oocytes and early embryos. J Med Genet. 2016;53(10):662–671. doi: 10.1136/jmedgenet-2016-103891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Y, Shi Y, Fu J, Yu M, Feng R, Sang Q, Liang B, Chen B, Qu R, Li B, Yan Z, Mao X, Kuang Y, Jin L, He L, Sun X, Wang L. Mutations in PADI6 cause female infertility characterized by early embryonic arrest. Am J Hum Genet. 2016;99(3):744–752. doi: 10.1016/j.ajhg.2016.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alazami AM, Awad SM, Coskun S, Al-Hassan S, Hijazi H, Abdulwahab FM, et al. TLE6 mutation causes the earliest known human embryonic lethality. Genome Biol. 2015;16(1):240. doi: 10.1186/s13059-015-0792-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Song D, Mykytenko D, Kuang Y, Lv Q, Li B, Chen B, Mao X, Xu Y, Zukin V, Mazur P, Mu J, Yan Z, Zhou Z, Li Q, Liu S, Jin L, He L, Sang Q, Sun Z, Dong X, Wang L. Novel mutations in genes encoding subcortical maternal complex proteins may cause human embryonic developmental arrest. Reprod BioMed Online. 2018;36(6):698–704. doi: 10.1016/j.rbmo.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 12.Dai J, Zheng W, Dai C, Guo J, Lu C, Gong F, Li Y, Zhou Q, Lu G, Lin G. New biallelic mutations in WEE2: expanding the spectrum of mutations that cause fertilization failure or poor fertilization. Fertil Steril. 2019;111(3):510–518. doi: 10.1016/j.fertnstert.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 13.Kan R, Yurttas P, Kim B, Jin M, Wo L, Lee B, Gosden R, Coonrod SA. Regulation of mouse oocyte microtubule and organelle dynamics by PADI6 and the cytoplasmic lattices. Dev Biol. 2011;350(2):311–322. doi: 10.1016/j.ydbio.2010.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yurttas P, Vitale AM, Fitzhenry RJ, Cohen-Gould L, Wu W, Gossen JA, Coonrod SA. Role for PADI6 and the cytoplasmic lattices in ribosomal storage in oocytes and translational control in the early mouse embryo. Development. 2008;135(15):2627–2636. doi: 10.1242/dev.016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qian J, Nguyen NMP, Rezaei M, Huang B, Tao Y, Zhang X, et al. Biallelic PADI6 variants linking infertility, miscarriages, and hydatidiform moles. Eur J Hum Genet. 2018;26(7):1007–1013. doi: 10.1038/s41431-018-0141-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maddirevula S, Coskun S, Awartani K, Alsaif H, Abdulwahab F, Alkuraya F. The human knockout phenotype of PADI6 is female sterility caused by cleavage failure of their fertilized eggs. Clin Genet. 2017;91(2):344–345. doi: 10.1111/cge.12866. [DOI] [PubMed] [Google Scholar]
- 17.Azzarello A, Hoest T, Mikkelsen A. The impact of pronuclei morphology and dynamicity on live birth outcome after time-lapse culture. Hum Reprod. 2012;27(9):2649–2657. doi: 10.1093/humrep/des210. [DOI] [PubMed] [Google Scholar]
- 18.Ezoe K, Ohata K, Morita H, Ueno S, Miki T, Okimura T, et al. Prolonged blastomere movement induced by the delay of pronuclear fading and first cell division adversely affects pregnancy outcomes after fresh embryo transfer on Day 2: a time-lapse study. Reprod BioMed Online. 2018. [DOI] [PubMed]
- 19.Verlinsky Y, Kuliev A. An atlas of preimplantation genetic diagnosis: an illustrated textbook & reference for clinicians: CRC Press; 2004.
- 20.Barrie A, Homburg R, McDowell G, Brown J, Kingsland C, Troup S. Preliminary investigation of the prevalence and implantation potential of abnormal embryonic phenotypes assessed using time-lapse imaging. Reprod BioMed Online. 2017;34(5):455–462. doi: 10.1016/j.rbmo.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 21.Ciray HN, Campbell A, Agerholm IE, Aguilar J, Chamayou S, Esbert M, Sayed S, Time-Lapse User Group Proposed guidelines on the nomenclature and annotation of dynamic human embryo monitoring by a time-lapse user group. Hum Reprod. 2014;29(12):2650–2660. doi: 10.1093/humrep/deu278. [DOI] [PubMed] [Google Scholar]
- 22.Kola I, Trounson A, Dawson G, Rogers P. Tripronuclear human oocytes: altered cleavage patterns and subsequent karyotypic analysis of embryos. Biol Reprod. 1987;37(2):395–401. doi: 10.1095/biolreprod37.2.395. [DOI] [PubMed] [Google Scholar]
- 23.Somfai T, Inaba Y, Aikawa Y, Ohtake M, Kobayashi S, Konishi K et al. Relationship between the length of cell cycles, cleavage pattern and developmental competence in bovine embryos generated by in vitro fertilization or parthenogenesis. J Reprod Dev. 2010:0912210236-. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(AVI 4696 kb)
(AVI 3633 kb)
(DOCX 21 kb)