Abstract
The gut microbiome analysis, with specific interest on their direct impact towards the human health, is currently revolutionizing the unexplored frontiers of the pathogenesis and wellness. Although in-depth investigations of gut microbiome, ‘the Black Boxes’, complexities and functionalities are yet at its infancy, profound evidences are being reported for their concurrent involvement in disease etiology and its treatment. Interestingly, studies from the ‘minimal murine’ (Oligo-MM12), ‘humanized’ microbiota gnotobiotic mice models and patient samples, combined with multi-omics and cell biology approaches, have been revealing the implications of these findings in the treatment of gut dysbiosis associated diseases. Nonetheless, due to the inherent heterogeneity of the gut commensals and their unified co-existence with opportunistic pathobionts, it is utmost essential to highlight their functionalities in ‘good or bad’ gut in human wellness. We have specifically reviewed dietary lifestyle and infectious diseases linked with the gut bacterial consortia to delineate the ecobiotic approaches towards their treatment. This notably includes gut mucosal immunity mediated diseases such as Tuberculosis, IBD, CDI, Type 2 Diabetes, etc. Alongside of each dysbiosis, we have described the current therapeutic advancements of the pre- and probiotics derived from human microbiome studies to restore gut microbial homeostasis. With a continuous running debate on the role of microbiota in above mentioned diseases, we have collected numerous scientific evidences highlighting a previously unanticipated complex involvement of gut microbiome in the potential of human health.
Keywords: Gut microbiome, Infectious diseases, Intestinal immune-mediate diseases, Lifestyle disorders, Dysbiosis, Colonizing opportunistic pathogens, Pathobionts
Introduction
Human beings are primate-microbe hybrids and have co-evolved along with the gut microbial world such as bacteria, bacteriophages, viruses, yeasts and fungi. The gut microbiome harbors almost 10–100 trillion of microbial cells, which are largely composed of diverse bacterial commensals, alongside with Colonizing Opportunistic Pathogens (COPs) and Simple Opportunistic Pathogens (SOPs) in a homeostatic ratio and plays the fundamental role in human health [1–5]. The COPs and SOPs are hidden opportunistic pathobionts that continuously invade, colonize and persist asymptomatically in the gut together with the commensals and co-exist with commensals in a mutualistic manner as a ‘good gut’. However, upon any perturbation in the proportional cohabitation of COPs or SOPs residing along with the commensals leads to the dysbiosis, a ‘leaky gut’, and when these opportunistic microbes attain a particular level can cause severe acute, chronic, or latent human diseases [3, 6, 7]. Nonetheless, gut microbiome remarkably serves as a primary immune defense system at the gut mucosal surface, which constantly buffers the effect against COPs and SOPs [3, 8]. The benign coexistence of microbial consortia inside the gut has led to the development of ‘war and peace’ at the gut mucosal surface and always in ‘tug of war’ situation inside human body. Earlier COPs have been primarily reported from many reservoirs like water, soil, air, etc., which typically includes various pathobionts namely Vibrio vulnificus, Mycobacterium marinum, and Legionella pneumophila, etc. Therefore, in-depth understandings on the plausible link between alterations by COPs and disease may lead to more focused and effective treatment strategies.
Recent studies unveil that management of lifestyle can change dysbiosis to restore normal health conditions and gut consortial integrity using prebiotics, probiotics or synbiotics encompassing Lactobacillus, Clostridium, Bifidobacterium, Faecalibacterium and Streptococcus, etc. [4]. Notably, COPs effects over microbiome modulations have also been observed in a range of infectious diseases like Tuberculosis, Inflammatory Bowel Disorder (IBD) and Clostridium difficile Infection (CDI) along with lifestyle disorders like obesity and type-2 diabetes. However, now these COPs have been reported as “normal” part of the human microbiome and can lead to diseases progression at a slow pace due to the latency effect. Despite distinct species of microbes that vary in different individuals, each habitat holds signature species that governs many metabolic pathways [9].
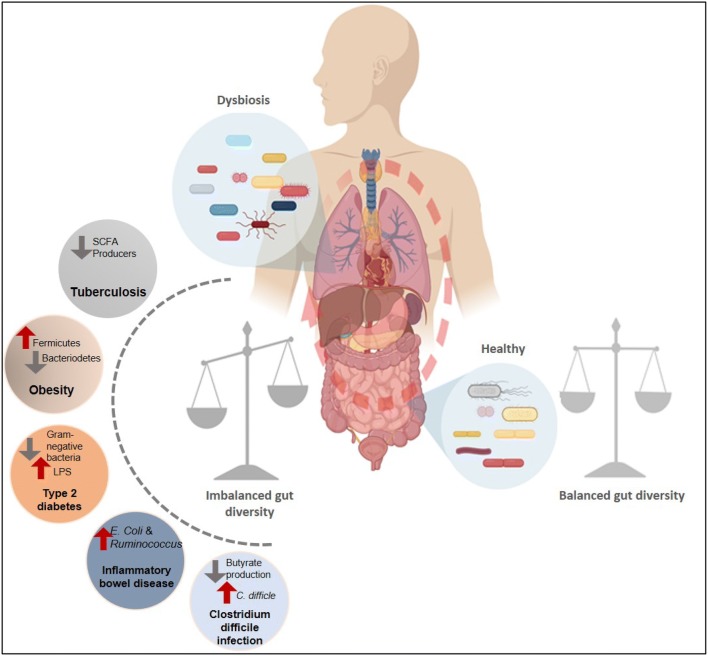
While in the previous articles of the special issue on microbiome, normal and changed microbiome in human beings has already been described [4, 10], here we discuss the modulations of infectious diseases by COPs and their direct link with alterations in the gut commensal bacterial diversity and their effect on pathogenesis (Fig. 1). We have mainly focused on predominant infectious and lifestyle diseases mentioned in Table 1, along with highlighting the applications of prebiotics and probiotics for these diseases.
Fig. 1.
A schematic illustration the cross talk between human microbiome and diseases. Shown in left demonstrates significant alterations of the gut bacterial diversity in various diseases (Tuberculosis, Obesity, Type-2 diabetes, IBD and CDI) and are highlighted by red and grey arrows, respectively
Table 1.
Colonizing opportunistic pathogens (COPs) and triggered diseases
| Disease | COPs/Signature organism | References |
|---|---|---|
| Tuberculosis | Mycobacterium tuberculosis, Faecalibacterium, Roseburia, Eubacterium and Phascolarctobacterium | [14, 17] |
| Inflammatory Bowel Disorder (IBD) | Escherichia coli, Ruminococcus gnavus (CD), Faecalibacterium prausnitzii and Roseburia (UC) | [21, 22] |
| Clostridium difficile Infection (CDI) | Clostridium difficile | [23, 24] |
| Obesity | Fermicutes, Lactobacillus reuteri, Methanobrevibacter smithii | [25–27] |
| Type 2 diabetes | Bifidobacterium spp., Faecalibacterium prausnitzii, Bacteroides and Eubacterium | [28, 29] |
| Sepsis | Staphyllococcus aureus | [30] |
| Pneumonia | Staphyllococcus pneumoniae | [31] |
| Pharyngitis | Streptococcus | [32] |
| Tinea Pedia | Staphylococcus, Trichophyton rubrum, Corynebacterium minutissimum | [33] |
Tuberculosis and Gut Microbiome
The imprecations of Mycobacterium tuberculosis (MTB) caused Tuberculosis (TB), an infectious disease has been recognized since early human antiquity, whereby susceptibility to infections through MTB is marked by the host immune status, nutrition factors, geology and notably the gut diversity responses [11]. However, it has been only recently established that composition of gut microbiota indeed exert major impact on the TB pathology and severity [12–14].
Infection of TB occurs through inhalation of Mycobacterium bacilli which are then phagocytized by resident lung alveolar macrophages and replicate by circumventing host defences. There continuous persistence and co-evolution of these bacilli with the host are predominantly influenced by the gut commensal microbiota that populates our mucosal surfaces [15, 16]. The association of these commensal bacteria in disease severity has not been fully understood, but they most likely contribute to disease outcome by modulating immune status either through dynamics of the gut microbiome or directly through the lung bacteria. Indeed, it has been demonstrated that gut microbiota influences anti-TB therapy (ATT) response. The recent reports on the effect of ATT on gut bacterial diversity showed that there was a persistent microbial imbalance or dysbiosis, in terms of species richness and distribution [17]. This dysbiosis of intestinal microbiota could not be cured and can last up to several years after treatment completion [18]. In addition, variations in the lung microbiome during TB has also been reported in humans as well as in mice system [19].
Effect of Microbiome on Disease Progression: From Latent to Active TB
The infection of MTB in humans is commonly thought to have a binary distribution: active and latent infection. Many of the risk factors involved in the progression from this latent to active state includes HIV infection, exposure to antibiotics, immunodeficiency, air pollution alcohol and smoking addictions. These factors severely affect the gut bacterial consortia, thus playing crucial roles in causing tuberculosis by activation of latent mycobacterial population along with dynamic interaction between the host systems and microbial products [20].
Gut bacteria directly modulate the host immunity, which initiates the innate and adaptive immune responses and regulates the transition from persistent to active TB infection. For instance, the CD4+ and CD8+ cells are key modulators of MTB infection by producing Th-1 mediated pro-inflammatory cytokines which are critical for lysis of infected macrophages [34]. Notably, not only these Th-1 mediated immune responses are important for clearing the pathogens but are also responsible for the host cell damages. Hence, in order to balance this inflammatory response, it is ideal to have Th-2 and regulatory T (T-reg) cell responses thus maintaining a Th-1/Th-2 equilibrium in the body. Gut bacteria play a significant role in maintaining this T cell homeostasis through producing acetate, propionate and butyrate, etc. Short Chain Fatty Acids (SCFAs) by fermentation of dietary fibres [35]. These SCFAs serve as primary precursor substrates in the host metabolism and immunomodulatory proinflammatory cytokines and T-reg cells activation. Current reports on human PBMCs infected with MTB reveal that acetate and butyrate downregulates the production of pro-inflammatory cytokines [36]. Thus, in cell-mediated response against MTB infection, SCFA production is one of the potential mechanisms and link between the microbiome and progression of TB. Recently, Maji et al. [14] reported that patients suffering from pulmonary TB from Indian subcontinents also showed higher Firmicutes to Bacteroidetes ratio, that disturb SCFA and GM-derived bacterial metabolites concentration. This significant increase in the number of bacteria that produces butyrate and propionate including Faecalibacterium, Roseburia, Eubacterium and Phascolarctobacterium develops an inflammatory milieu in host contributing to the pathophysiology of TB [14]. While there are few specific examples as to how these changes in microbial diversity contribute to TB pathology by affecting the anti-inflammatory response in the host, in-depth investigations are essential to delineate their functional roles [19, 37].
Anti-tuberculosis Therapy and Changing Microbiome
Antibiotics therapy has a profound effect on the human microbial diversity, especially the usage of broad-spectrum antibiotics dramatically alters the gut ecology. As of now an effective TB treatment involves use of orally administered antibiotics for a period of at least 6 months [38], which is known to be the longest duration of antibiotics treatment utilized for the patients. The ATT treatment includes a typical combinatorial four-line drug therapeutics that involves the use of rifampicin (R), a drug with broad spectrum antibacterial activity, and three antibiotics (isoniazid, H; pyrazinamide, Z and ethambutol, E) targeting specifically mycobacterial species (2 months), followed by 4 month of rifampicin and isoniazid (HRZ) [38]. Although ATT administration has been the only prescribed cure for millions of patients suffering with MTB infection, its adverse impact on the host microbiome, specifically gut diversity is only recently has become the focus of the comprehensive investigation. The effect of ATT in relation to changes in the intestinal microbial population shows that it adversely affects the gut bacterial composition leading to the dysbiosis, which enables the dominance of opportunistic pathobionts in the gut community (Fig. 1). Emerging studies on the murine models of TB indicate dramatic alterations in the bacterial composition with the start of HRZ treatment and persist for 3 months after cessation of therapy [17]. Furthermore, chances of re-infection increase after a patient has been treated once with the cocktail of TB antibiotics, suggesting the effect dysbacteriosis in TB recurrence [17, 39]. It has studied that during ATT treatment, the members of the Bacteroidetes are decreased while Prevotella are shown to increase, suggesting the shift in gut immune function associated with the host immunity. These studies strongly indicate the effect of ATT on comprehensive microbial ecology and suggest consequences impaired microbiota on human health by increasing risk for resilient TB reinfection and COPs as well as SOPs [3, 6].
Inflammatory Bowel Diseases (IBD) and Gut Microbiome
An autoimmune disease by class, inflammatory bowel disorder is another widespread gut related disorder, where self-immune system attacks the digestive tract components [40]. IBD patients have inflammation in their colon and intestine region. There are two types of IBDs, Crohn’s disease (CD) and Ulcerative colitis (UC) [41]. They differ from each other in location and nature of inflammatory disorder:
Crohn’s disease targets gastrointestinal tract, from mouth to anus, covering full thickness of the bowel wall [42]. Out of all the sites, the three most prominently affected sites include ileal, ileocolic, colonic epithelium [43]. Abdominal pain, diarrhea, fever along with weight loss, underline some of the symptoms experienced under Crohn’s disease including bowel obstructions [44].
Ulcerative colitis (UC) is marked by ulcers or open sores in the colon region of the large intestine. As the name suggests, colitis is inflammation of the colon. Additionally, other symptom includes diarrhea mixed with blood or pus, abdominal and rectal pain [45].
There are various factors that may result in the onset of these disorders such as dysbiosis, immunity, environmental factors and genetic factor [46]. Recent studies have also shown a direct link of oral microbiome dysbiosis along with gut in the commencement of these diseases.
Dysbiosis
The pathogenesis of IBD comprises the chronic and persistent gut response to the commensal gut microbiome, including genetically susceptible host [47]. The remarkable correlation between IBD and impaired gut microbiota can be proved with the fact that the location, pH and anaerobiosis in the gut, e.g. colon and ileum, are shown to harbor highest percentage of total gut microbiota. Of the 4 main divisions of bacteria colonized in these regions, primarily Firmicutes, Bacteroidetes, Proteobacteria and Actinobacteria, the levels of Firmicutes and Bacteroidetes have been found disrupted severely [48]. On the contrary, Proteobacteria and Actinobacteria are comparatively abundant in IBD patients [41, 48, 49]. Faecalibacterium prausnitzii and Roseburia, most predominant species under Firmicutes are known to produce SCFA especially butyrate and acetate, playing a significant role in the protection of the mucosal immunity [49, 50]. Moreover, F. prausnitzii is reported as anti-inflammatory commensal due to the large production of anti-inflammatory factor IL-10. But, in patients suffering from ileal CD, the dynamics of these bacteria are significantly reduced, therefore increasing the risk of early reactivation of the disease. Roseburia is suggested to increase the production of T-regulatory cells, required for the anti-inflammatory purpose, however, with the decrease in their cohorts, the anti-inflammatory agents tend to decrease [48, 51].
In the CD patients, another group of bacteria whose levels elevated were Enterobacteriaceae, including Escherichia coli and Ruminococcus gnavus [22, 52]. E. coli levels as observed in ileal Crohn’s disease were restricted to the region of inflamed mucosa [43]. They promote inflammation by adhering and subsequently degrading the ileal mucosal layers to reach the crypt and attach with the epithelium. Upon attaching with the gut immune cells, this bacterium can replicate within the macrophages and promote the release of tumor necrosis factor. On contrary, R. gnavus is involved in mucolytic activity as well, i.e. breaking down thick mucus layer that is meant for the protection of the intestinal wall [49]. It has been found in several studies that the levels of Bacteroidetes are reduced in IBD patients. However, a subset of Bacteroides fragilis i.e. enterotoxigenic B. fragilis, (ETBF) is also found in high numbers in UC. It secretes proinflammatory zinc-dependent metalloprotease toxin and produces IL-7 in high amounts that cause severe gut inflammation [53, 54].
Studies have shown that oral microecological dysbiosis also a plays role in the compromised gut microbiome causing IBD. During normal body physiology, gut resists colonization of the non-native bacteria thus controlling the expansion of pathobionts. But involvement and colonization of oral dysbacteriosis in the gut tend to cause persistence of inflammatory disease [55, 56]. Atarashi and coworker in 2017 reported strains of Klebsiella spp. from oral region colonized the gut [57]. Streptococcus, Prevotella, Neisseria, Haemophilus, Veillonella and Gemella, were major cause of dysbiosis in the salivary microbiota of IBD patients [58]. Thus, IBD also affects oral cavity of the patients with symptoms like ulcers, dry mouth, aphthous stomatitis, etc. [59]. Intestinal mycobiome such as species of Candida, Saccharomyces, and Aspergillus are another critical component of the microbiota, and modulate immune homeostasis and inflammatory disease. Leonardi et al. [60], reported CX3CR1 (CX3C chemokine receptor 1) and mononuclear phagocytes (MNPs) is significant part in initiation of immune responses (innate and adaptive) to intestinal fungi. They activate antifungal responses by expressing adhesions and antifungal receptors in a Syk-dependent manner. This help in mediating intestinal microbiota and host immune system interactions during inflammatory disease [60].
The dysbiosis is also related to the host genetics. This can be seen with the polymorphism in about more than 75 genes especially in Crohn’s disease that leads abnormalities in mucosal barriers and innate immunity, etc. [21, 61]. A typical example is linked with the IL-23-Th17 pathway, which has an important role in microbial defense and intestine immune homeostasis. Nonetheless, IL-23R receptor in this pathway contains gene that can be affected by environmental factors [61, 62]. Mutation in IL-23R results in overproduction and accumulation of granulocyte-monocyte progenitor cells in the intestine that causes colitis phenotype. High levels of IL-23 are reported in the epithelial mucosal lining of IBD patients [63].
Prebiotic and Probiotics Treatment
Probiotics and prebiotics have prominent roles in resorting commensal gut microbiota and thus are directly involved in the treatment of IBDs. The commonly used probiotics for the treatment have microorganisms like Lactobacillus, Bifidobacterium, and E. coli. They act by reestablishing the mucosal barrier by producing higher amounts of SCFA and metabolites. A similar effect is shown by the intake of prebiotics that is known to increase the levels of Lactobacillus, Bifidobacterium in the gut thus increasing SCFA that is an anti-inflammatory factor [64]. Synbiosis factors produced by microbes can also be used in treatment, for e.g. PSA (Polysaccharide A) made by Bacteroides fragilis suppresses the release of pro-inflammatory IL-17 from intestinal immune cells [65].
Clostridium difficile Infection (CDI) and Gut Microbiome
Another nosocomial infectious disease, spreading worldwide, due to disruptive changes in the gut microbiome is Clostridium difficile infection belonging to firmicutes. CDI, also known as pseudomembranous colitis, is marked with intestinal sores like IBD, whereby inflammation is largely due to elevated levels of C. difficile in the gut [66]. Symptoms include flu like condition, along with bloating, diarrhea, and abdominal pain caused by the toxin released from the bacteria. The main cause behind the infection is the lack of colonization resistance towards C. difficile and commensals [67]. C. difficile is considered as a good gut colonizer, but its rate of colonization varies with ethnicity, malnutrition and age groups. For instance, infants have the highest rate of colonization and with age, this rate tends to decrease. Under stressed conditions such as pH and anaerobiosis, C. difficile can produce dormant spores that are resistant to those conditions and undergo vegetative propagation and colonize upon favorable gut conditions. The main risk factor(s) involved in CDI pathogenesis develops due to antibiotics induced dysbiosis given to patients for other diseases such as diarrhea and TB, which results in collateral disruption of gut bacterial communities [68]. This disruption leads to suppression of dominant species and increased colonization of C. difficile.
Dysbiosis and Colonization of C. difficile
The antibiotic induced, environmental, dietary and age-related changes shift the healthy microbiota of the gut [69, 70]. The changes reported till date majorly include increase in the Bacteroides species like, Enterobacteriaceae, Erysipelotrichaceae, and Bacteroidetes, and decrease in the members of Ruminococcaceae and Lachnospiraceae, which further promote the colonization of C. difficile [68–71]. Nonetheless, the foremost cause of this colonization underlies within the levels of production of butyrate, which decreases in the gut when the numbers of butyrate producers decrease. Butyrate along with SCFAs is associated with C. difficile colonization resistance [67]. With the decline in their number, resistance levels lower, thereby promoting growth during the infection. Further, Bacteroides uniformis, which is particularly involved in improving the malfunctions caused by obesity, also protects the inflammations from C. difficile outgrowth in the gut. Reduction in its numbers leads to declination in the C. difficile colonization resistance [24].
Treatment
The most effective treatments for CDI to restore the normal microbiome is by RePOOPulating the gut with multispecies bacterial community [72]. The classical approach involves fecal bacteriotherapy or stool transplantation, where stool of a healthy individual is infused into patient to re-establish the healthy microbiota of gut [73]. This treatment can cure antibiotic resistant C. difficile colitis; however, practice is limited by inadequacies like pathogen transmission from donor to patients, lack of natural stool stability and its handling. These defects are being overcome by new approaches of synthetic stool substitution known as fecal microbiota transplantation (FMT). In this technique, the synthetic stool is developed by in vitro culturing, isolating microbial diversity and then enriching commensals isolated from the stool of a healthy individual [73, 74]. In addition, current therapy also involves adjoining FMT with synergistic combination of pro- and prebiotics with synbiotics [75]. Together, these combined approaches further enhance the pure commensal, stable stool preparation and reduces the risk of pathogen transmissions. The patient undergoes colon cleansing followed by colonoscopy. Thus, patients become capable to soon restore the normal microbiota composition, and levels of Clostridium reduce to negligible number over time. With rapid advancement in microbiome research, the development of ecobiotic drugs have paved novel ways for quicker treatment, which are detailed in Kumar et al., 2019. In addition, diet also plays crucial roles in restoring the microbiota. It is recommended to consume fiber rich diet and products without probiotics [76]. Age factors also do not adversely impact in the FMT method, such that even if the donor is quite young as compared to the aged patient, the effect of the therapy does not change and is equally effective.
Obesity and Gut Microbiome
While obesity is governed by many factors including environmental exposure, malnutrition, sleep deprivation, and chronic inflammation [77]. However, emerging reports suggest that the gut microbial composition and diet energy plays a significant role in structuring this medical condition [77, 78]. The metabolic capacity that the gut microbiota holds not only includes the degradation of indigestible components, but also includes microbial fermentation of dietary polysaccharides. This results in increased absorption of monosaccharides, fatty acids and their conversion into various biochemical pathways [79]. The first direct association of gut microbiota with obesity has been in-depth demonstrated by J. I. Gordon and co-workers on about 5000 genetically obese mice models (ob/ob). A clear reduction in the number of Bacteroidetes was observed along with increased number of Firmicutes, whereas a completely contrast ratio was observed in genetically lean mice It is currently being considered that Bacteroidetes to Firmicutes ratio largely determines the obese and lean condition in an individual [80, 81].
Bacteroidetes-to-Firmicutes Dysbiosis
Firmicutes possess genes belonging to Gram-positive polysaccharide utilization loci (gpPULs) for the breakdown of the complex dietary polysaccharides into di- and mono- saccharides. However, these enzymes are not yet deciphered in Bacteroidetes [1, 25, 82]. Thus, individuals having ‘obese genetic microbiota makeup’, and consuming carbohydrates and fats enriched westernized diet, harbor a relatively higher proportion of gut Firmicutes and tend to put on weight easily. While, individuals with a genetic makeup of lean microbiota, having higher levels of Bacteroidetes, do not put on excess fat even after consuming the same westernized diet [26, 83]. Lactobacillus reuteri was associated with obesity, whereas, Methanobrevibacter smithii was depleted in obese individuals [25, 27]. With the breakdown of dietary polysaccharides by Firmicutes in a genetically obese individual, a high amount of energy extraction from the diet takes place and the energy is stored in adipose tissue. Thus, relatively low amount of energy remains in their fecal matter, However, due to lower levels of Firmicutes in genetically lean individuals, a relatively less amount of energy is extracted from the diet and higher amounts undigested diet remains in their fecal matter [81, 83].
Probiotic and Prebiotic Therapy as Treatment
As noted earlier that the increased levels of Firmicutes to Bacteroidetes ratio results in obesity. Thus, in probiotic therapy by live microorganisms, like Lactobacillus species is administered to the obese individuals and after a couple of weeks, reduction in weight is observed without reduction in energy intake [4, 84]. Few strains of Bifidobacterium, Enterococcus, Bacteroides, Pediococcus and even Saccharomyces as a formula-fed probiotic, etc. are also included in this therapy. On the contrary, prebiotics are non-digestible carbohydrates that passes small intestine and finally reaches colon where they promote the growth of commensal microorganisms. It is also known to reduce hunger and boost the growth and colonization of Bifidobacterium and Lactobacillus spp. [85]. A comprehensive description of probiotics and prebiotics for the treatment of obese patients has been discussed by Kumar and co-authors (2019).
Type 2 Diabetes and Gut Microbiome
The occurrence of type 2 diabetes is characterized by numerous factors that include individuals’ circadian lifestyle, obesity, age, genetics, and gender, etc. However, along with these factors, upcoming studies evidence the dramatic disproportions in microbial communities residing in the GI tracts, which are directly linked to the insulin resistance [86, 87]. The most important triggering factor that results in the onset of diabetes is the bacterial lipopolysaccharide (LPS) [88]. With the increased intake of high fat diet having higher levels of saturated fats, the microbial dysbiosis is observed and levels of Gram-negative bacteria like Bifidobacterium spp., Faecalibacterium prausnitzii, Bacteroides and Eubacterium decreases [28, 89]. LPS being component of cell wall increases with the increasing mortality of these bacteria [88].
In healthy gut, intestinal epithelium, crypts and mucosal layers along with commensal bacteria act as a barrier to avoid the LPS translocation to the blood stream [88, 90, 91]. Thus, any agent that disrupts these fence(s) is responsible for the promotion of gut associated disorders. Fat rich diet alters the intestinal barrier, leading to increased permeability and LPS absorption by two to three folds resulting in the disruption of the gut barrier [92]. This subsequently enables in the increase of LPS concentration in the serum, leading to a situation called metabolic endotoxemia [93]. The intestinal fence disruption occurs not only due to dysbiosis, but also due to alterations in the tight junction proteins namely, occluding and zonal occluding that are responsible for maintaining the barriers, thus facilitating the permeability of LPS [88, 94]. The involvement of microbiota in these metabolic disorders was proved by a set of experiments performed by Cani and co-workers on genetically obese mice ob/ob. In the first set, LPS quencher like polymyxin B was administered into the mice that function as a blocker for the receptor. And the second set of obese mice lacked CD14, the receptor for LPS. Of note, high levels of LPS promote insulin resistance. In both the conditions, improvement in insulin resistance was observed, clearly demonstrating the contribution of LPS derived from the gut microbiota to metabolic endotoxemia [88].
Treatment
As in obesity, probiotics and prebiotics has crucial role in the treatment of type 2 diabetes. One of the therapies includes use of Bifidobacterium as a component of probiotic that helps to stabilize the gut microbiota of a diabetic patient. They tend to enhance the gut barrier function thus, reducing intestinal endotoxin absorptions [84]. The effects of prebiotics were analogous to the regulation of intestinal mucosal surface with increased mucus layer thickness, and hence inhibition to LPS permeability [95]. Prebiotic like fructo-oligosaccharides helps in regulating the gut-brain axis for glucose metabolism [95]. Drugs used as antidiabetic are also known to normalize the composition of gut microbiota, for example, Metformin acts in the similar manner. One of the species that increase in number after its treatment is Akkermansia, which helps in the stabilization of glucose homeostasis and insulin sensitivity [96].
Conclusions
With the known fact of microbial colonization to almost every specific niche ‘in-and-on’ human body has suggested the symbiosis with not only physiological, but also with clinical connections. Their balanced co-existence is an outcome of several adaptation mechanisms, failure of which results in human health fatalities. The ultimate reason to study microbiome is to improve human health status by all means. Any break down in microbial balances due to stress in lifestyle can be detectible markers for predicting the onset of a disease. The early physiological detection of IBD, obesity and type 2 diabetes using microbial composition before clinical onset is a clear advancement of microbiome studies over other non-clinical analysis. Since these diseases are becoming one of the most prominent health threats in our country, thus their treatment should be taken as a top priority. We have highlighted and presented the recent attempt made to overcome these challenges by using probiotics consumptions as a medicinal alternate or supplement. The latest microbial therapeutic methods could involve prebiotics along with drug- or food-incorporating microbes as a substitute to cure the diseases. Nonetheless, we are only at the beginning to understand their in-depth molecular mechanism of interaction. It is yet to be investigated how gut commensals and COPs interact together. What are the molecular apparatus allowing them to communicate along with the host gut? One of the greatest hurdles is to culture these the gut bacteria in vitro by mimicking host gut conditions. In summary, these upcoming evidences along with current challenges demands further advanced clinical investigations to be translated into clinical practices for FMT and applications of the gut microbial compositions in the wellness human health.
Acknowledgements
NS, MG and VS gratefully acknowledge the Council of Scientific and Industrial Research (CSIR), New Delhi for providing doctoral fellowship. AP acknowledge University Grants Commission (UGC), New Delhi for doctoral fellowship. RL thanks The National Academy of Sciences, India, for support under the NASI‐Senior Scientist Platinum Jubilee Fellowship Scheme.
Compliance with Ethical Standards
Conflict of interest
Authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Nirjara Singhvi and Vipin Gupta have contributed equally.
Contributor Information
Yogendra Singh, Email: ysinghdu@gmail.com.
Gyanendra P. Dubey, Email: gyanendrap.dubey@gmail.com
Rup Lal, Email: ruplal@gmail.com.
References
- 1.Turnbaugh PJ, Ley E, Hamady M, Fraser-Liggett C, Knight R, Gordon J. The human microbiome project: exploring the microbial part of ourselves in a changing world. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ursell LK, Metcalf JL, Parfrey LW, Knight R. Defining the human microbiome. Nutr Rev. 2012;70:S38–S44. doi: 10.1111/j.1753-4887.2012.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Price LB, Hungate BA, Koch BJ, Davis GS, Liu CM. Colonizing opportunistic pathogens (COPs): the beasts in all of us. PLoS Pathog. 2017;13:e1006369. doi: 10.1371/journal.ppat.1006369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar R, Sood U, Gupta V, Singh M, Scaria J, Lal R. Recent advancements in the development of modern probiotics for restoring human gut microbiome dysbiosis. Indian J Microbiol. 2019 doi: 10.1007/s12088-019-00808-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kulkarni AS, Kumbhare SV, Dhotre DP, Shouche YS. Mining the core gut microbiome from a sample Indian population. Indian J Microbiol. 2019;59:90–95. doi: 10.1007/s12088-018-0742-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durack J, Lynch SV. The gut microbiome: relationships with disease and opportunities for therapy. J Exp Med. 2019;216:20–40. doi: 10.1084/jem.20180448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buret AG, Motta JP, Allain T, Ferraz J, Wallace JL. Pathobiont release from dysbiotic gut microbiota biofilms in intestinal inflammatory diseases: a role for iron? J Biomed Sci. 2019;26:1. doi: 10.1186/s12929-018-0495-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sansonetti PJ. War and peace at mucosal surfaces. Nat Rev Immunol. 2004;4:953–964. doi: 10.1038/nri1499. [DOI] [PubMed] [Google Scholar]
- 9.Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, Huttenhower C, Izard J. Composition of the adult digestive tract microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012;13:R42. doi: 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sood U, Bajaj A, Kumar R, Khurana S, Kalia VC. Infection and microbiome: impact of tuberculosis on human gut microbiome of indian cohort. Indian J Microbiol. 2018;58:123–125. doi: 10.1007/s12088-018-0706-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Narasimhan P, Wood J, Macintyre CR, Mathai D. Risk factors for tuberculosis. Pulm Med. 2013;2013:828939. doi: 10.1155/2013/828939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong BY, Maulén NP, Adami AJ, Granados H, Balcells ME, Cervantes J. Microbiome changes during tuberculosis and antituberculous. Therapy Clin Microbiol Rev. 2016;29:915–926. doi: 10.1128/CMR.00096-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Namasivayam S, Sher A, Glickman MS, Wipperman MF. The microbiome and tuberculosis: early evidence for cross talk. MBio. 2018;9:e01420-18. doi: 10.1128/mBio.01420-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maji A, Misra R, Dhakan DB, Gupta V, Mahato NK, Saxena R, Mittal P, Thukral N, Sharma E, Singh A, Virmani R, Gaur M, Singh H, Hasija Y, Arora G, Agrawal A, Chaudhry A, Khurana JP, Sharma VK, Lal R, Singh Y (2018) Gut microbiome contributes to impairment of immunity in pulmonary tuberculosis patients by alteration of butyrate and propionate producers. Environ Microbiol 20:402–419. 10.1111/1462-2920 [DOI] [PubMed]
- 15.Guirado Evelyn, Schlesinger Larry S., Kaplan Gilla. Macrophages in tuberculosis: friend or foe. Seminars in Immunopathology. 2013;35(5):563–583. doi: 10.1007/s00281-013-0388-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balcells ME, Yokobori N, Hong B, Corbett J, Cervantes J. The lung microbiome, vitamin D, and the tuberculous granuloma: a balance triangle. Microb Pathog. 2019;131:158–163. doi: 10.1016/j.micpath.2019.03.041. [DOI] [PubMed] [Google Scholar]
- 17.Namasivayam S, Maiga M, Yuan W, Thovarai V, Costa DL, Mittereder LR, Wipperman MF, Glickman MS, Dzutsev A, Trinchieri G, Sher A. Longitudinal profiling reveals a persistent intestinal dysbiosis triggered by conventional anti-tuberculosis therapy. Microbiome. 2017;5:71–88. doi: 10.1186/s40168-017-0286-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wipperman MF, Fitzgerald DW, Juste MAJ, Taur Y, Namasivayam S, Sher A, Bean JM, Bucci V, Glickman MS. Antibiotic treatment for Tuberculosis induces a profound dysbiosis of the microbiome that persists long after therapy is completed. Sci Rep. 2017;7:10767. doi: 10.1038/s41598-017-10346-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan N, Vidyarthi A, Nadeem S, Negi S, Nair G, Agrewala JN. Alteration in the gut microbiota provokes susceptibility to tuberculosis. Front Immunol. 2016;7:529. doi: 10.3389/fimmu.2016.00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horsburgh CR, Jr, Rubin EJ. Latent tuberculosis infection in the United States. N Engl J Med. 2011;364:1441–1448. doi: 10.1056/NEJMcp1005750. [DOI] [PubMed] [Google Scholar]
- 21.Sartor RB. Genetics and environmental interactions shape the intestinal microbiota to promote inflammatory bowel disease versus mucosal homeostasis. Gastroenterology. 2010;139:1816–1819. doi: 10.1053/j.gastro.2010.10.036. [DOI] [PubMed] [Google Scholar]
- 22.Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P, Vandamme P, Vermeire S. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut. 2011;60:631–637. doi: 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- 23.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crobach MJT, Vernon JJ, Loo VG, Kong LY, Pechine S, Wilcox MH, Kuijper EJ. Understanding clostridium difficile colonization. Clin Microbiol Rev. 2018;31:e00021-17. doi: 10.1128/CMR.00021-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castaner O, Goday A, Park YM, Lee SH, Magkos F, Shiow SATE, Schröder H. The gut microbiome profile in obesity: a systematic review. Int J Endocrinol. 2018;2018:4095789. doi: 10.1155/2018/4095789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turnbaugh PJ, Backhed F, Fulton L, Gordon J. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Million M, Maraninchi M, Henry M, Armougom F, Richet H, Carrieri P, Valero R, Raccah D, Vialettes B, Raoult D. Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. Int J Obes. 2012;36:817–825. doi: 10.1038/ijo.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, Waget A, Delmée E, Cousin B, Sulpice T, Chamontin B, Ferrières J, Tanti JF, Gibson GR, Casteilla L, Delzenne NM, Alessi MC, Burcelin R. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 29.Ganesan K, Chung SK, Vanamala J, Xu B. Causal relationship between diet-induced gut microbiota changes and diabetes: a novel strategy to transplant faecalibacterium prausnitzii in preventing diabetes. Int J Mol Sci. 2018;19:3720. doi: 10.3390/ijms19123720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Oliveira TH, Amorin AT, Rezende IS, Santos Barbosa M, Martins HB, Brito AK, Andrade EF, Gonçalves GK, Campos GB, Silva RA, Timenetsky J, Marques LM. Sepsis induced by Staphylococcus aureus: participation of biomarkers in a murine model. Med Sci Monit. 2015;21:345–355. doi: 10.12659/MSM.892528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ragle BE, Karginov VA, Bubeck Wardenburg J. Prevention and treatment of Staphylococcus aureus pneumonia with a beta-cyclodextrin derivative. Antimicrob Agents Chemother. 2010;54:298–304. doi: 10.1128/AAC.00973-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nobbs AH, Lamont RJ, Jenkinson HF. Streptococcus adherence and colonization. Microbiol Mol Biol Rev. 2009;73:407–450. doi: 10.1128/MMBR.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X, Tan J, Yang H, Gao Z, Cai Q, Meng L, Yang L. Characterization of skin microbiome in tinea pedis. Indian J Microbiol. 2019 doi: 10.1007/s12088-019-00816-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwander S, Dheda K. Human lung immunity against Mycobacterium tuberculosis: insights into pathogenesis and protection. Am J Respir Crit Care Med. 2011;183:696–707. doi: 10.1164/rccm.201006-0963PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lachmandas E, van den Heuvel CN, Damen MS, Cleophas MC, Netea MG, van Crevel R. Diabetes mellitus and increased tuberculosis susceptibility: the role of short-chain fatty acids. J Diabetes Res. 2016;2016:6014631. doi: 10.1155/2016/6014631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui Z, Zhou Y, Li H, Zhang Y, Zhang S, Tang S, Guo X. Complex sputum microbial composition in patients with pulmonary tuberculosis. BMC Microbiol. 2012;12:276. doi: 10.1186/1471-2180-12-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.World Health Organization (2018) Global tuberculosis report 2018. World Health Organization. https://apps.who.int/iris/handle/10665/274453
- 39.Van Rie A, Warren R, Richardson M, Victor TC, Gie RP, Enarson DA, Beyers N, van Helden PD. Exogenous reinfection as a cause of recurrent tuberculosis after curative treatment. N Engl J Med. 1999;341:1174–1179. doi: 10.1056/NEJM199910143411602. [DOI] [PubMed] [Google Scholar]
- 40.Fakhoury M, Negrulj R, Mooranian A, Al-Salami H. Inflammatory bowel disease: clinical aspects and treatments. J Inflamm Res. 2014;7:113–120. doi: 10.2147/JIR.S65979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, Bousvaros A, Korzenik J, Sands BE, Xavier RJ, Huttenhower C. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:79–97. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cosnes J, Gower-Rousseau C, Seksik P, Cortot A. Epidemiology and natural history of inflammatory bowel diseases. Gastroenterology. 2011;140:1785–1794. doi: 10.1053/j.gastro.2011.01.055. [DOI] [PubMed] [Google Scholar]
- 43.Baumgart DC, Sandborn WJ. Crohn’s disease. Lancet. 2012;380:1590–1605. doi: 10.1016/S0140-6736(12)60026-9. [DOI] [PubMed] [Google Scholar]
- 44.Shah J, Etienne D, Reddy M, Kothadia JP, Shahidullah A, Baqui AAMA. Crohn’s disease manifesting as a duodenal obstruction: an unusual case. Gastroenterology Res. 2018;11:436–440. doi: 10.14740/gr1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ito A, Iizuka B, Omori T, Nakamura S, Tokushige K. Development and improvement of simple colonic mucosal ulcer during treatment of severe ulcerative colitis with tacrolimus. Case Rep Gastroenterol. 2017;11:168–177. doi: 10.1159/000456605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaistha A, Levine J. Inflammatory bowel disease: the classic gastrointestinal autoimmune disease. Curr Probl Pediatr Adolesc Health Care. 2014;44:328–334. doi: 10.1016/j.cppeds.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 48.Zuo T, Ng SC. The gut microbiota in the pathogenesis and therapeutics of inflammatory bowel disease. Front Microbiol. 2018;9:2247. doi: 10.3389/fmicb.2018.02247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schroeder BO. Fight them or feed them: how the intestinal mucus layer manages the gut microbiota. Gastroenterol Rep (Oxf) 2019;7:3–12. doi: 10.1093/gastro/goy052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran L, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, Grangette C, Vasquez N, Pochart P, Trugnan G, Thomas G, Blottière HM, Doré J, Marteau P, Seksik P, Langella P. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sokol H, Lay C, Seksik P, Tannock GW. Analysis of bacterial bowel communities of IBD patients: what has it revealed? Inflamm Bowel Dis. 2008;14:858–867. doi: 10.1002/ibd.20392. [DOI] [PubMed] [Google Scholar]
- 52.Hall AB, Yassour M, Sauk J, Garner A, Jiang X, Arthur T, Lagoudas GK, Vatanen T, Fornelos N, Wilson R, Bertha M, Cohen M, Garber J, Khalili H, Gevers D, Ananthakrishnan AN, Kugathasan S, Lander ES, Blainey P, Vlamakis H, Xavier RJ, Huttenhower C. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 2017;9:103. doi: 10.1186/s13073-017-0490-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rabizadeh S, Rhee KJ, Wu S, Huso D, Gan CM, Golub JE, Wu X, Zhang M, Sears CL. Enterotoxigenic bacteroides fragilis: a potential instigator of colitis. Inflamm Bowel Dis. 2007;13:83–1475. doi: 10.1002/ibd.20265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ahmed I, Roy BC, Khan SA, Septer S, Umar S. Microbiome, metabolome and inflammatory bowel disease. Microorganisms. 2016;4:20. doi: 10.3390/microorganisms4020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pickard JM, Zeng MY, Caruso R, Núñez G. Gut microbiota: role in pathogen colonization, immune responses and inflammatory disease. Immunol Rev. 2017;279:70–89. doi: 10.1111/imr.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caballero S, Pamer EG. Microbiota-mediated inflammation and antimicrobial defense in the intestine. Annu Rev Immunol. 2015;33:227–256. doi: 10.1146/annurev-immunol-032713-120238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, Kiguchi Y, Yasuma K, Watanabe E, Tanoue T. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science. 2017;358:359–365. doi: 10.1126/science.aan4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Said HS, Suda W, Nakagome S, Chinen H, Oshima K, Kim S, Kimura R, Iraha A, Ishida H, Fujita J, Mano S, Morita H, Dohi T, Oota H, Hattori M. Dysbiosis of salivary microbiota in inflammatory bowel disease and its association with oral immunological biomarkers. DNA Res. 2014;21:15–25. doi: 10.1093/dnares/dst037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Veloso FT. Extraintestinal manifestations of inflammatory bowel disease: do they influence treatment and outcome? World J Gastroenterol. 2011;17:2702–2707. doi: 10.3748/wjg.v17.i22.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leonardi I, Li X, Semon A, Li D, Doron I, Putzel G, Bar A, Prieto D, Rescigno M, McGovern DPB, Pla J, Iliev ID. CX3CR6+ mononuclear phagocytes control immunity to intestinal fungi. Science. 2018;359:232–236. doi: 10.1126/science.aao1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou M, He J, Shen Y, Zhang C, Wang J, Chen Y. New frontiers in genetics, gut microbiota, and immunity: a rosetta stone for the pathogenesis of inflammatory bowel disease. Biomed Res Int. 2017;2017:8201672. doi: 10.1155/2017/8201672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jostins Luke, Ripke Stephan, Weersma Rinse K., Duerr Richard H., McGovern Dermot P., Hui Ken Y., Lee James C., Philip Schumm L., Sharma Yashoda, Anderson Carl A., Essers Jonah, Mitrovic Mitja, Ning Kaida, Cleynen Isabelle, Theatre Emilie, Spain Sarah L., Raychaudhuri Soumya, Goyette Philippe, Wei Zhi, Abraham Clara, Achkar Jean-Paul, Ahmad Tariq, Amininejad Leila, Ananthakrishnan Ashwin N., Andersen Vibeke, Andrews Jane M., Baidoo Leonard, Balschun Tobias, Bampton Peter A., Bitton Alain, Boucher Gabrielle, Brand Stephan, Büning Carsten, Cohain Ariella, Cichon Sven, D’Amato Mauro, De Jong Dirk, Devaney Kathy L., Dubinsky Marla, Edwards Cathryn, Ellinghaus David, Ferguson Lynnette R., Franchimont Denis, Fransen Karin, Gearry Richard, Georges Michel, Gieger Christian, Glas Jürgen, Haritunians Talin, Hart Ailsa, Hawkey Chris, Hedl Matija, Hu Xinli, Karlsen Tom H., Kupcinskas Limas, Kugathasan Subra, Latiano Anna, Laukens Debby, Lawrance Ian C., Lees Charlie W., Louis Edouard, Mahy Gillian, Mansfield John, Morgan Angharad R., Mowat Craig, Newman William, Palmieri Orazio, Ponsioen Cyriel Y., Potocnik Uros, Prescott Natalie J., Regueiro Miguel, Rotter Jerome I., Russell Richard K., Sanderson Jeremy D., Sans Miquel, Satsangi Jack, Schreiber Stefan, Simms Lisa A., Sventoraityte Jurgita, Targan Stephan R., Taylor Kent D., Tremelling Mark, Verspaget Hein W., De Vos Martine, Wijmenga Cisca, Wilson David C., Winkelmann Juliane, Xavier Ramnik J., Zeissig Sebastian, Zhang Bin, Zhang Clarence K., Zhao Hongyu, Silverberg Mark S., Annese Vito, Hakonarson Hakon, Brant Steven R., Radford-Smith Graham, Mathew Christopher G., Rioux John D., Schadt Eric E., Daly Mark J., Franke Andre, Parkes Miles, Vermeire Severine, Barrett Jeffrey C., Cho Judy H. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Knights D, Lassen KG, Xavier RJ. Advances in inflammatory bowel disease pathogenesis: linking host genetics and the microbiome. Gut. 2013;62:1505–1510. doi: 10.1136/gutjnl-2012-303954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hold GL, Smith M, Grange C, Watt ER, El-Omar EM, Mukhopadhya I. Role of the gut microbiota in inflammatory bowel disease pathogenesis: what have we learnt in the past 10 years? World J Gastroenterol. 2014;20:1192–1210. doi: 10.3748/wjg.v20.i5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nagao-Kitamoto H, Kamada N. Host-microbial cross-talk in inflammatory bowel disease. Immune Netw. 2017;17:1–12. doi: 10.4110/in.2017.17.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Farooq PD, Urrunaga NH, Tang DM, von Rosenvinge EC. Pseudomembranous colitis. Dis Mon. 2015;61:181–206. doi: 10.1016/j.disamonth.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fachi JL, Felipe JS, Pral LP, da Silva BK, Corrêa RO, de Andrade MCP, da Fonseca DM, Basso PJ, Câmara NOS, de Sales E, Souza ÉL, Dos Santos Martins F, Guima SES, Thomas AM, Setubal JC, Magalhães YT, Forti FL, Candreva T, Rodrigues HG, de Jesus MB, Consonni SR, Farias ADS, Varga-Weisz P, Vinolo MAR. Butyrate protects mice from clostridium difficile-induced colitis through an HIF-1-dependent mechanism. Cell Rep. 2019 doi: 10.1016/j.celrep.2019.03.054. [DOI] [PubMed] [Google Scholar]
- 68.Schubert AM, Rogers MA, Ring C, Mogle J, Petrosino JP, Young VB, Aronoff DM, Schloss PD. Microbiome data distinguish patients with Clostridium difficile infection and non-C. difficile-associated diarrhea from healthy controls. MBio. 2014;5:e01021-14. doi: 10.1128/mBio.01021-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Milanović V, Cardinali F, Aquilanti L, Garofalo C, Roncolini A, Sabbatini R, Clementi F, Osimani A. A glimpse into the microbiota of marketed ready-to-eat crickets (Acheta domesticus) Indian J Microbiol. 2019 doi: 10.1007/s12088-019-00817-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sood U, Gupta V, Kumar R, Lal S, Fawcett D, Rattan S, Poinern GEJ, LaL R. Chicken gut microbiome and human health: past scenarios, current perspectives, and futuristic applications. Indian J Microbiol. 2019 doi: 10.1007/s12088-019-00785-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leslie JL, Vendrov KC, Jenior ML, Young VB (2019) The gut microbiota is associated with clearance of clostridium difficile infection independent of adaptive immunity. mSphere 4:e00698-18. 10.1128/mspheredirect.00698-18 [DOI] [PMC free article] [PubMed]
- 72.Petrof EO, Gloor GB, Vanner SJ, Weese SJ, Carter D, Daigneault MC, Brown EM, Schroeter K, Allen-Vercoe E. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: ‘RePOOPulating’ the gut. Microbiome. 2013;1:3–15. doi: 10.1186/2049-2618-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bakken JS. Fecal bacteriotherapy for recurrent Clostridium difficile infection. Anaerobe. 2009;15:285–289. doi: 10.1016/j.anaerobe.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 74.Gupta S, Allen-Vercoe E, Petrof EO. Fecal microbiota transplantation: in perspective. Therap Adv Gastroenterol. 2016;9:229–239. doi: 10.1177/1756283X15607414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.de Vrese M, Schrezenmeir J. Probiotics, prebiotics, and synbiotics. Adv Biochem Eng Biotechnol. 2008;111:1–66. doi: 10.1007/10_2008_097. [DOI] [PubMed] [Google Scholar]
- 76.Choi HH, Cho YS. Fecal microbiota transplantation: current applications, effectiveness, and future perspectives. Clin Endos. 2016;49:257–265. doi: 10.5946/ce.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sun L, Ma L, Ma Y, Zhang F, Zhao C, Nie Y. Insights into the role of gut microbiota in obesity: pathogenesis, mechanisms, and therapeutic perspectives. Protein Cell. 2018;9:397–403. doi: 10.1007/s13238-018-0546-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol. 2009;587:4153–4158. doi: 10.1113/jphysiol.2009.174136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Williams BA, Grant LJ, Gidley MJ, Mikkelsen D. Gut fermentation of dietary fibres: physico-chemistry of plant cell walls and implications for health. Int J Mol Sci. 2017;18:2203. doi: 10.3390/ijms18102203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Baothman OA, Zamzami MA, Taher I, Abubaker J, Abu-Farha M. The role of gut microbiota in the development of obesity and diabetes. Lipids Health Dis. 2016;15:108–116. doi: 10.1186/s12944-016-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ndeh D, Gilbert HJ. Biochemistry of complex glycan depolymerisation by the human gut microbiota. FEMS Microbiol Rev. 2018;42:146–164. doi: 10.1093/femsre/fuy002. [DOI] [PubMed] [Google Scholar]
- 83.Davis CD. The gut microbiome and its role in obesity. Nutr Today. 2016;51(4):167–174. doi: 10.1097/NT.0000000000000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shen J, Obin M, Zhao L. The gut microbiota, obesity, and insulin resistance. Mol Aspects Med. 2013;34:39–58. doi: 10.1016/j.mam.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 85.Guarner F, Khan AG, Garisch J, Eliakim R, Gangl A, Thomson A, Krabshuis J, Lemair T, Kaufmann P, De Paula JA, Fedorak R, Shanahan F, Sanders ME, Szajewska H, Ramakrishna BS, Karakan T, Kim N. World gastroenterology organisation global guidelines: probiotics and prebiotics. J Clin Gastroenterol. 2011;46:468–481. doi: 10.1097/MCG.0b013e3182549092. [DOI] [PubMed] [Google Scholar]
- 86.Sharma D, Goel NK, Sharma MK, Walia DK, Thakare MM, Khaneja R. Prevalence of diabetes mellitus and its predictors among tuberculosis patients currently on treatment. Indian J Community Med. 2018;43:302–306. doi: 10.4103/ijcm.IJCM_230_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chan YK, Estaki M, Gibson DL. Clinical consequences of diet-induced dysbiosis. Ann Nutr Metab. 2013;63:28–40. doi: 10.1159/000354902. [DOI] [PubMed] [Google Scholar]
- 88.Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, Burcelin R. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 89.Singh RK, Chang HW, Yan D, Lee KM, Ucmak D, Wong K, Abrouk M, Farahnik B, Nakamura M, Zhu TH, Bhutani T, Liao W. Influence of diet on the gut microbiome and implications for human health. J Transl Med. 2017;15:73. doi: 10.1186/s12967-017-1175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chassaing B, Koren O, Goodrich JK, Poole AC, Srinivasan S, Ley RE, Gewirtz AT. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature. 2015;519:92–96. doi: 10.1038/nature14232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vancamelbeke M, Vermeire S. The intestinal barrier: a fundamental role in health and disease. Expert Rev Gastroenterol Hepatol. 2017;11:821–834. doi: 10.1080/17474124.2017.1343143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.König J, Wells J, Cani PD, García-Ródenas CL, MacDonald T, Mercenier A, Whyte J, Troost F, Brummer RJ. Human intestinal barrier function in health and disease. Clin Transl Gastroenterol. 2016 doi: 10.1038/ctg.2016.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Neves AL, Coelho J, Couto L, Leite-Moreira A, Roncon-Albuquerque R. Metabolic endotoxemia: a molecular link between obesity and cardiovascular risk. J Mol Endocrinol. 2013;51:51–64. doi: 10.1530/JME-13-0079. [DOI] [PubMed] [Google Scholar]
- 94.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 95.Burcelin R, Serino M, Chabo C, Blasco-Baque V, Amar J. Gut microbiota and diabetes: from pathogenesis to therapeutic perspective. Acta Diabetol. 2011;48:257–273. doi: 10.1007/s00592-011-0333-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Maratos-Flier E. Metabolic disease puts up a fight: microbes, metabolism, and medication. Nat Med. 2013;19:1218–1219. doi: 10.1038/nm.3373. [DOI] [PubMed] [Google Scholar]