Abstract
Research question
Using RNA-sequencing analysis, we investigated the relationship between ovarian stimulation and endometrial transcriptome profiles during the window of implantation (WOI) to identify candidate predictive factors for the WOI and to optimize timing for embryo transfer.
Methods
Twelve women with normal basal hormone levels and regular ovulation were randomly assigned into three groups based on sampling time: late-proliferate phase (P group), and receptive phase in natural cycles (LH+7, N group) and stimulated cycles (hCG+7, S group). Transcriptome profiles of mRNAs and long non-coding RNAs (lncRNAs) were then compared among the three groups. Validation was performed using real-time qPCR.
Results
Comparison of transcriptome profiles between the natural and stimulated endometrium revealed 173 differentially expressed genes (DEGs), with a > 2-fold change (FC) and p < 0.05, under the influence of supraphysiological estradiol (E2) induced by ovarian stimulation. By clustering and KEGG pathway analysis, molecules and pathways associated with endometrial receptivity were identified. Of the 39 DEGs common to the three groups, eight genes were validated using real-time PCR. ESR1, MMP10, and HPSE were previously reported to be associated with endometrial receptivity. In addition, three novel genes (IL13RA2, ZCCHC12, SRARP) and two lncRNAs (LINC01060, LINC01104) were new potential endometrial receptivity-related markers.
Conclusion
Using mRNA and lncRNA sequencing, we found that supraphysiological E2 levels from ovarian stimulation had a marked impact upon endometrial transcriptome profiles and may result in a shift of the WOI. The precise mechanisms underlying the supraphysiological hormone-induced shift of the WOI require further research.
Registration number
ChiCTR180001453
Electronic supplementary material
The online version of this article (10.1007/s10815-019-01616-5) contains supplementary material, which is available to authorized users.
Keywords: Ovarian stimulation, Window of implantation, Endometrial receptivity, mRNAs, lncRNAs
Introduction
A viable embryo, receptive endometrium, and the synchronization of these two factors provide three essential elements for a successful pregnancy [1]. The endometrium is a dynamic tissue which grows and sheds monthly under the control of estradiol (E2) and progesterone. The window of implantation (WOI) is a period of time usually 5–10 days after ovulation that lasts 12 to 48 h, when the endometrium has transformed for the implantation of embryos [2]. During this time-specified and hormone-regulated phase, the endometrium gains the optimal ability to accept embryos, termed receptivity [3]. The acquisition of endometrial receptivity is the result of the sequential effect of E2 and progesterone [4]. Implantation failure makes up 30% of all pregnancy loses when excluding embryonic factors [5].
Controlled ovarian stimulation (COH) allows the simultaneous development of multiple follicles, but it induces supraphysiologic levels of E2, which increases the risk of ovarian hyperstimulation syndrome (OHSS) and defective endometrial receptivity [6]. In the 1990s, it was first noticed that high responders to COH had lower implantation rates compared with normal and low responders [7]. However, this phenomenon was not found in frozen-thawed embryo transfer (FET) cycles and donor cycles, indicating that endometrial receptivity was likely impaired by COH treatment [8]. Progesterone and estrogen were both found to be essential for the preparation of receptive endometrium, while the degree of endometrial estrogen exposure was related to impairment of endometrial receptivity [9]. However, the mechanism remains unclear.
Differences in endometrial morphology between natural and stimulated cycles have been extensively reported. The endometrium of stimulated endometrium was advanced, with delayed gland and advanced stroma development, sometimes accompanying stromal edema [10]. The asynchronization of endometrial development may lead to the failure of implantation. It was also found that greater than 3 days difference between the endometrial maturity and menstrual period resulted no pregnancy [11]. During stimulated cycles, pinopodes developed in advance and exhibited morphology different from those in natural cycles [12]. Although the morphological assessment of endometrium has benefits, this kind of evaluation is observer-dependent and thus biased. Moreover, there is no consensus regarding the morphological evaluation of endometrium.
The altered expression of molecular markers related to endometrial receptivity has been reported. Estrogen and progesterone receptors [13], leukemia inhibitory factors, cell adhesion molecules, such as integrins, abnormal expression of growth factors, and some immune cells, [14–18] were shown to be related to the process of embryo implantation or the establishment of endometrial receptivity. However, whether a certain molecular change will influence the outcome of pregnancy remains unknown [11].
One of the major challenges in this field is to find new potential markers of endometrial receptivity. Early and intensive endeavors explored gene profiles during the peri-implantation phase in a natural cycle [19–27]. In 2004, Mirkin et al. [28] reported for the first time that COH leads to differential gene expression. Subsequent work using DNA microarrays found more transcriptome changes induced by supraphysiological estrogens [29–33]. However, due to various factors, including inter-patient variability, limited sample sizes, and different timing of biopsy, the results of these studies were highly heterogeneous and few genetic overlaps were found [34]. It has to be pointed out that gene array techniques have limitations. The recent and rapid emergence of next-generation sequencing (NGS) technology is overcoming some of the array analysis shortcomings [35]. Unlike traditional Sanger sequencing and microarray, RNA-seq is a deep and high-throughput sequencing method. Instead of specific probe hybridization, NGS can detect and quantify the entire transcriptome, including encoding RNA (mRNA) and long non-coding RNA (lncRNA), providing more comprehensive transcriptomic information. NGS is very suitable for detecting rare transcripts in complex and unknown tissues, such as the endometrium, and for revealing important pathways for further analysis. To our knowledge, this is the first report to compare the differences in gene expression between natural and stimulated cycles using NGS.
In this study, we used NGS to investigate the effects of ovarian stimulation on functional genomic profiles of the endometrium, and to discover potential regulatory genes and involved pathways to provide a better understanding of embryo implantation and the WOI.
Material and methods
Human subjects
Twelve women with normal ovulation and basal hormonal levels undergoing IVF/ICSI-ET at the Reproductive Department of Beijing Chaoyang Hospital were recruited. Inclusion criteria were as follows: (1) age 20–35 years; (2) body mass index (BMI) 18–23 kg/m2; (3) regular menstrual cycle with confirmed ovulation, basal follicle stimulating hormone (FSH) or LH < 8 IU/L; (4) gynecological ultrasound showed uterus, uterine cavity, and bilateral attachment had no abnormalities, endometrial thickness on ovulation day ≥ 6 mm; (5) COH protocol: GnRH long-agonist or GnRH antagonist protocols. Exclusion criteria were as follows: (1) uterine malformations; (2) endometrial lesions: endometritis, polyps, submucosal fibroids, uterine adhesion; (3) endometriosis; (4) uterine surgery that may damage the endometrium: curettage, polyp removal, submucosal myomectomy; (5) hydrosalpinx. Subjects were evenly and randomly allocated to three groups according to the sampling time.
This study was registered in the Chinese Clinical Trials Registration Center with registration number: ChiCTR180001453. The study was approved by the ethics committee of Chaoyang Hospital and informed consent was signed before the biopsy. Patients were required to use condoms for contraception during the study.
Controlled ovarian stimulation and tissue collection
In natural cycles, we assigned the day of the urinary LH peak (hemtrue®, Shanghai, China) as LH+0. Samples were collected in late-proliferative phase (P group) and on the day of LH+7 (N group). Patients in P group were biopsied on the cycle day 11–14 in their regular 28–30 periods. In the meantime, ultrasound was used to monitor ovulation. Histological analysis was performed to reconfirm the biopsy time.
Biopsies were performed at the fundal and upper part of the uterus under sterile condition using Pipelle (Yajie®, Jiangxi, China). Biopsies were performed by the same surgeon. After washing to remove mucus and blood, the endometrium samples were divided into two parts. One was placed in 4% paraformaldehyde (Cat no. KGIHC016CS, KGI Bio, China) for histological assessment, and the other was snap-frozen and stored in liquid nitrogen for further processing.
In stimulation cycles, ovarian stimulation was performed using standard long GnRH-agonist or GnRH-antagonist protocol. Long GnRH-agonist protocol: 7 days after ovulation, a subcutaneous injection of 0.05 mg triptorelin acetate (Decapeptyl®, Ferring Pharmaceuticals, Switzerland) was administered daily for 14 days until criteria of downregulation was met (E2 < 50 pg/ml, Luteinizing hormone (LH) and FSH < 5 IU/ml, endometrial thickness < 5 mm, and the maximal follicle diameter was less than 10 mm). Then individualized injections of 150–225 IU rFSH (Gonal-f®; Serono, Geneva, Switzerland) were administered daily for ovarian stimulation. Triptorelin acetate was continued until trigger day. For the GnRH-antagonist protocol, rFSH was started on the 2nd day of menstruation, and 0.25 mg cetrorelix acetate (Cetrotide®; Serono, Geneva, Switzerland) was added daily until trigger day and serum E2 had reached 400 pg/ml or when a follicle > 14-mm diameter was confirmed by ultrasound. When the leading follicle was 18–20 mm in diameter and there were more than three follicles at 16 mm, 5000–10000 IU HCG (Livzon Pharmaceutical, Guangdong, China) was injected to trigger final oocyte maturation. Oocyte retrieval was performed 34–36 h after hCG injection. Serum E2 levels were measured on the same day with an electro-chemiluminescence immunoassay kit (Roche Diagnostics GmbH, Mannheim, Germany). Endometrial biopsies were done 7 days after hCG administration in four patients with a high risk of developing OHSS.
RNA extraction and RNA-Seq analysis
RNA extraction and RNA-sequencing
Total RNA was extracted using the TRIZOL method according to the protocol recommended by the manufacturer (Life Technologies, Inc., Gaithersburg, MD, USA). The total RNA quantity was measured with a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies Inc. DE, USA) and RNA integrity was assessed with an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA, USA). RNA samples with an RNA integrity number (RIN) ≥ 7 and 28S:18S ≥ 1.5:1 in electrophoresis were chosen for mRNA Library Construction. Samples were sequenced according to the product protocol using the mRNA-Seq Sample Preparation Kit (Cat no. RS-930-1001 Illumina containing 1 box, part no. 1004814 and 1 bag, part no. 1004815). The libraries were sequenced on the Illumina HiSeq X Ten generating 150 bp paired end reads.
Detection of differentially expressed genes
Raw reads were mapped to the human genome (hg19) using hista2 software. Gene expression quantity was performed using feature-count with the ensemble gene Annotation file by the t Strand-Specific params. The differentially expressed genes (DEGs) were identified by edgeR R package. RPKM (Reads Per Kilobase per Million mapped reads) was used to calculate gene expression. Qualified gene expression was defined as gene count-per-million (CPM) > 2 in more than 2 samples within one group. Reads with low quality were discarded. The DEGs were identified with the p value < 0.05 and fold change (FC) > 2. P-values were corrected using false discovery rate (FDR) (5%).
Gene annotation: gene ontology and pathway analysis
DEGs were used for gene ontology (GO) and KEGG (Kyoto Encyclopedia of Genes and Genomes database) pathway analysis. GO and KEGG pathway enrichment analysis was performed using the ClusterProfiler package from BioConductor (http://www.bioconductor.org/). GO was classified as cellular component (CC), biological process (BP), and molecular function (MF). Hypergeometric exact test was used to classify enrichment GO categories and KEGG pathway, and only the terms with p < 0.05 were considered enriched.
Quantitative RT-PCR
The remaining RNA of each sample after sequencing was used for quantitative RT-PCR. Genomic DNA elimination reaction was first performed. 5× gDNA Eraser Buffer (2 μl, TAKARA) and gDNA Eraser (1 μl, TAKARA) were added to 2 µg of total RNA. RNase Free dH2O was then added to make the total reaction volume to 10 μl. The mixture was heated at 42 °C for 2 min and chilled on ice. Reverse-transcription reaction was then performed. cDNA was synthesized in a total volume of 20 μl containing 10 μl reaction solution from step 1 and 10 μl Master mix (4 μl 5× PrimeScript Buffer2 for Real Time, 1 μl PrimeScript RT Enzyme Mix1, 1 μl RT Primer Mix, 4 μl RNase Free dH2O, all from TAKARA). The subsequent incubation process was 15 min at 37 °C, then 5 sec at 85 °C. The cDNA was stored at 4 °C for further use.
RT-PCR was performed using cDNA in a total of 20 ul PCR mix. The total mix contained 1 µl cDNA, 300 nM forward primer, 300 nM reverse primer, and 1× SYBRgreen PCR Master Mix (Applied Biosystems). The Q-PCR was performed in a MicroAmp® Optical 96-well Reaction Plate (Applied Biosystems) with an ABI Prism Optical Adhesive Cover (Applied Biosystems) in the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). The selected protocol consisted of 10 min at 95 °C, then 40 cycles of 15 s at 95 °C and 1 min at 60 °C, followed by a dissociation curve step of 15 s at 95 °C, 1 min at 60 °C, 30 s at 95 °C, and 15 s at 60 °C. Standard curve material consisted of pooled endometrial RNA from all three groups. All reactions were performed in triplicate. Supplemental Table S1 shows the primer sequences, forward (F) and reverse (R), for each gene in RT-PCR.
Relative gene expression in the P and S groups compared with the N group was calculated by delta delta Ct. Validation data and presented as mean ± standard error of mean (SEM).
Results
Demographic data
There were no significant differences in age, duration of infertility, and basal hormone levels among the three groups. The serum E2 levels in the stimulation cycle group were significantly higher than that in the natural cycle, and the histology dating was consistent with the time of sampling. The results are shown in Table 1.
Table 1.
Demographic data of the 12 subjects
| Stimulated cycle (hCG+7) n = 4 |
Natural cycle | p value | ||
|---|---|---|---|---|
| Proliferative phase n = 4 |
LH+7 n = 4 |
|||
| Age (years) | 32.5 ± 2.5 | 31.0 ± 3.7 | 34.3 ± 2.1 | NS |
| Cause of infertility | Tubal | Tubal | Tubal | NS |
| Duration (years) | 3.0 ± 1.8 | 2.8 ± 1.0 | 2.3 ± 1.3 | NS |
| BMI (kg/m2) | 21.2 ± 1.7 | 19.8 ± 0.90 | 20.6 ± 2.4 | NS |
| Period (days) | 31.75 ± 3.3 | 27.5 ± 1.0 | 29.0 ± 1.2 | NS |
| Basal FSH (mIU/ml) | 5.2 ± 1.7 | 7.6 ± 1.6 | 6.5 ± 1.5 | NS |
| AFC | 14 ± 5.2 | 16 ± 2.9 | 13.2 ± 5.4 | NS |
| Histology | Secretory phase | Proliferative phase | Secretory phase | — |
| rFSH dosage (IU) | 2362.5 ± 256.17 | — | — | — |
| rFSH duration (days) | 10.75 ± 1.89 | — | — | — |
| E2 on the hCG day (pg/ml) | 3121.5 ± 1445.63 | — | — | — |
| No. of retrieved oocytes | 15.5 ± 6.76 | — | — | — |
All the data are presented as mean ± standard deviation (SD), NS indicates no significant difference between groups (p > 0.05)
BMI, body mass index; AFC, antral follicle count; COH, controlled ovarian stimulation; rFSH, recombinant follicle stimulating hormone; hCG, human chorionic gonadotropin; E2, estradiol
Different gene expression profiles between natural and stimulated cycles
Twelve samples were analyzed using RNA-seq. Four triplicate libraries were generated and sequenced to a depth of 55 million to 82 million reads (Supplemental Table S2). The reads were aligned to a human reference genome (hg19) using hisat2 software with the --rna-strandness RF. On average, 91.70% of sequences were aligned to the genome, with an average of 78.13% of the reads being uniquely aligned; 93.39% of the uniquely aligned reads were aligned to mRNA, with 58.18% of reads aligned to exons.
The reads of each gene were counted using the feature count. Reads aligned to multiple genes were discarded, and the reads of genes were normalized, and an average of 19702 genes were detected (Supplemental Table S3). The edger was used to identify differentially expressed genes. Genes with a > 2-fold change with a p value < 0.05 were identified as differentially expressed genes (DEGs).
Among the 18487 genes, there were 1063 DEGs in P group compared with the N group, of which 513 (48.3%) were upregulated and 550 (51.7%) were downregulated. Compared with the S group, there were 173 DEGs in the natural cycle group, of which 130 (75.1%) were upregulated and 43 (24.9%) were downregulated. Thirty-nine overlapping genes were in common between the two comparisons.
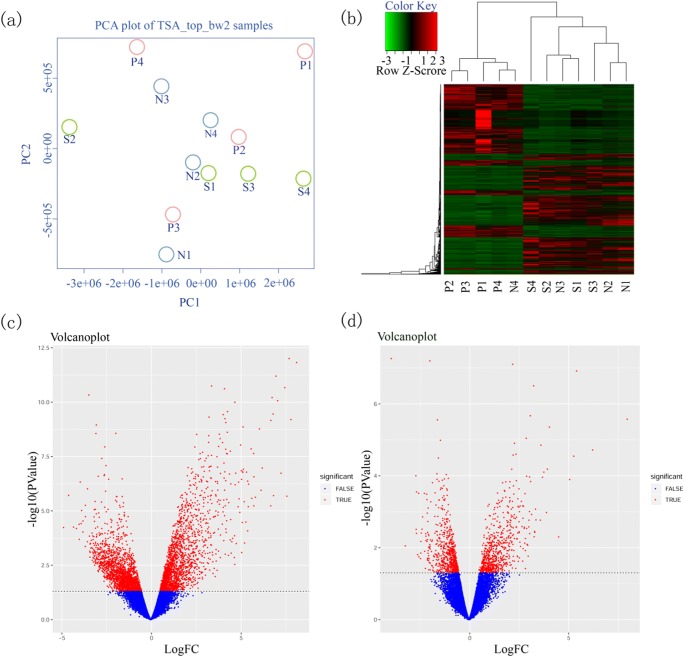
Clustering and principal component analyses (PCA) of the gene expression patterns for the three groups are shown in Fig. 1a. The samples were labeled with three groups in the PCA analysis. The 4 samples in P group were randomly distributed, while samples in N and S group were well clustered. Plus, the two groups can also be separated, indicating the difference between N and S group was originating from inter-group variability rather individual heterogeneity. The heat map showed significantly different expression patterns between the N, S, and P groups (Fig. 1b), and genes representing the secretory phase were more similar in groups N and S compared with group P. These DEGs could be almost clustered together according to the abundance of gene expression. Volcano plots representing the distribution of the different genomic profiles from the compared groups are shown in Fig. 1c, d.
Fig. 1.
a Principal component analysis (PCA) of DEGs. b Unsupervised hierarchical clustering of gene list of 12 samples. x-axis represents samples and y-axis represents the expression abundance of different genes. The color from green to red represents the gene expression abundance from low to high. Volcano plot of differentially expressed genes. c N vs P. d N vs S. The x-axis represented the log2 of fold change (FC) and y-axis represented log10 of p values. Genes with FC > 2 and p < 0.05 were painted as red dots. Red dots on the left were downregulated genes and on the right were upregulated genes. The leftover blue dots were the genes without significant difference
Gene ontologies clustering and pathway analysis
Gene ontology and pathway analysis of genes in the natural cycle and stimulation cycle were performed to find possible biological processes related to endometrial receptivity (Fig. S1). The involved DEGs belong to almost all the signaling pathways and biological functions. Interestingly, of the many involved pathways, immune system and cancer-specific cell types, proliferation, and immune responses exhibited the most significant differences.
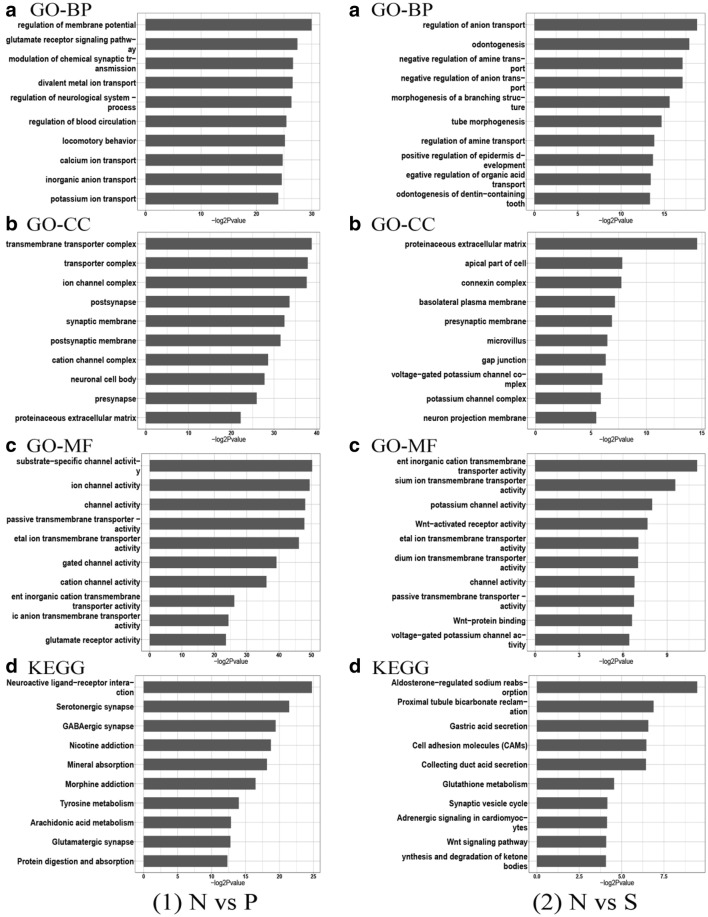
Using the enrichment score, we selected the top ten items from GO in biological processes (BP), molecular functions (MF), and cellular components (CC) (Fig. 2). In the natural cycle, the most significant biological processes included ion transport channels, membrane potential regulation, calcium ion pathways, and synaptic connections. According to KEGG analysis, signal pathways including signal transduction of cells, such as ligand-receptor interaction and metabolism, showed the most significant differences. In the stimulation cycle, cell adhesion molecules (CAMs) and Wnt signaling pathways showed significant differences. Many members of the CAM family were previously shown to be associated with endometrial receptivity [16, 17, 36–38]. The Wnt signaling pathway was also found to play an important role in embryo implantation [39].
Fig. 2.
The top 10 enrichment GO (gene ontology) and KEGG pathway terms in (1) N vs P and (2) N vs S. x-axis represents negative log2 p values.
A GO analysis for biological process (BP); B GO analysis for molecular function (MF); C GO analysis for cellular components; (CC) D 10 most significant KEGG pathways
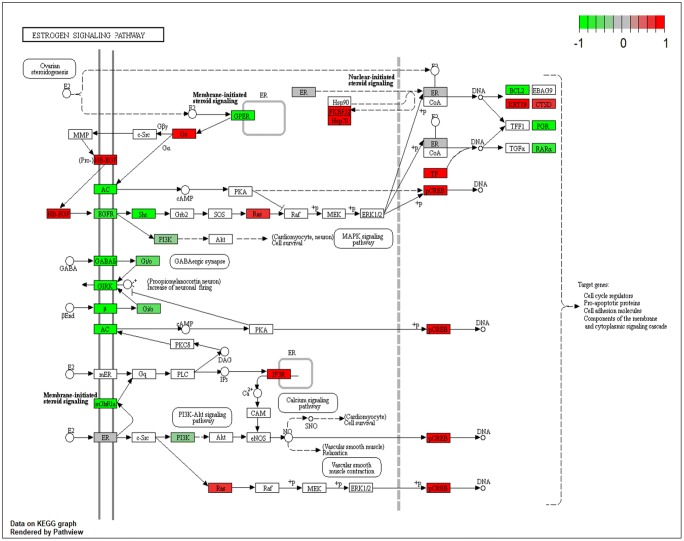
Notably, analysis of KEGG pathways of the DEGs showed that genes associated with the estrogen signaling pathway exhibited significant differential expression in the stimulation cycle (Fig. 3). Under the influence of excessive estrogen, genes involved in the estrogen receptor pathway were almost universally differentially expressed. Upstream genes were generally down-regulated, while the downstream genes were generally up-regulated. In the E2-mediated pathways, the CAMs and proapoptotic proteins were both involved. In addition, ERK 1/2 on the estrogen receptor pathway was homologous to the target gene of the LINC01060 we later predicted. The Ras–Raf–MEK–ERK signaling cascade was found to regulate cell proliferation and apoptosis, but the mechanism remained unknown [40]. It was also reported to induce differentiation in endometrial angioblast [41] and death receptor-mediated apoptosis in endometrial cells [42].
Fig. 3.
The DEGs in the estrogen receptor pathway. The red boxes represented upregulated genes, and the green ones represented downregulated genes, and color shade represented fold change value. The darker the color, the more significant the difference
Potential biomarkers of endometrial receptivity in stimulated cycles and validation of data by RT-PCR
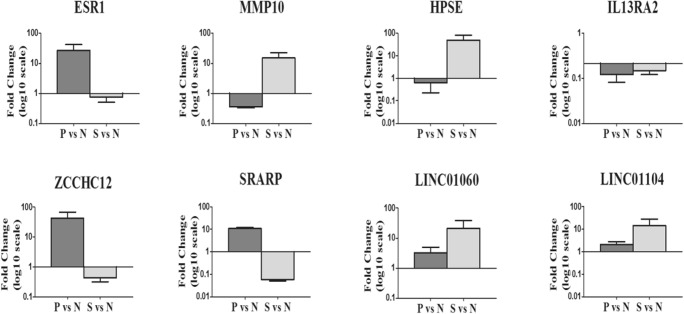
Known genes and their biological functions that were differentially expressed in both the natural and stimulated cycles are shown in Supplemental Table S4. Based on biological function and relevance to reproduction, and by comparisons with DEGs in related studies, we identified three DEGs, ESR1, MMP10, and HPSE that were related to endometrial receptivity. In addition, ZCCHC12, IL13RA2, SRARP, and two lncRNAs, LINC01060 and LINC01104, were selected as potential novel markers or regulators of endometrial receptivity.
Expression of the eight selected transcripts was examined in the 3 groups by real-time PCR, and the results were consistent with the sequencing data (Fig. 4).
Fig. 4.
Validation data of the selected genes using quantitative RT-PCR. x-axis showed the gene names, and the fold changes on a log10 scale comparing P vs N (dark grey) and S vs N (light grey) were displayed on the y-axis. The data are presented as the mean ± standard error of mean (SEM)
Analysis of lncRNAs
The two differentially expressed lncRNAs, LINC01060 and LINC01104, are intergenic non-coding RNAs, and their functions remain unclear. The website (http://rtools.cbrc.jp/cgi-bin/RNARNA/index.pl) was used to predict their target genes for LINC01060 (http://rtools.cbrc.jp/cgi-bin/RNARNA/createRankedHTML_bin.pl?gene=ENST00000513313&fm=1&to=100&ngenes=100&rmeth=min) and LINC01104 (http://rtools.cbrc.jp/cgi-bin/RNARNA/_createRankedHTML_bin.pl?gene=ENST00000452354&to=100&rmeth=min&fm=1&submit=submit). The top 100 target genes were selected for function analysis using toppgene (https://toppgene.cchmc.org/). We found that the potential target genes of LINC01060, such as MAPK7 and MAP3K2, were enriched in the Erk5 MAPK signaling pathway (https://rgd.mcw.edu/rgdweb/Ontology/view.html?acc_id=PW:0000603, p=0.0001279, fdr=0.04273), as shown in Fig. S2. However, the target genes of this pathway were not differentially expressed among the N, P, and S groups. Enriched GO and pathways of LINC01104 were not found (https://toppgene.cchmc.org/output.jsp?userdata_id=1b13f39b-d23c-4fac-9ad9-679b0a9b6466).
Discussion
To our knowledge, this is the first study using NGS to compare the differential expression of genomic profiles between stimulated and natural endometrium. Compared with traditional commercial array methods, the NGS provides more comprehensive transcriptome information, including coding and non-coding mRNA, underlying the transition of endometrium [43]. We selected the late-proliferative phase as the non-receptive control group. As shown in Table 2, previous studies generally used LH+2 (i.e., the early secretory phase) as the pre-receptive endometrium model. However, the progesterone-induced transition from the proliferative to secretory phase is more essential for endometrial receptivity. It can be deduced that genes playing a key role in the transition process also have a role in endometrial receptivity or embryo implantation. To date, we have found only one study exploring changes in genomic expression of endometrium between the proliferative and mid-secretory phases in natural cycles [44].
Table 2.
Overview of studies on the differentially expressed genes of endometrium under ovarian stimulation
| Study | Comparator | Type of GnRH-a/ant | Sample size | Type of array | Fold change | Gene ontology (GO) | Differentially expressed genes (DEGs) |
|---|---|---|---|---|---|---|---|
| Stimulation protocol | KEGG pathways | ||||||
| Mirkin et al. (2004) | LH+8 VS hCG+9 | Leuprolide acetate | 15 | Affymetrix HG-U95Av2 | ≥ 1.2 |
Up: agonist—5 antagonist—6 Down: agonist—1 antagonist—6 |
|
| GnRH agonist VS GnRH antagonist | Ganirelix or Cetrorelix | Up: 13 Down: 0 | |||||
| Simon et al. (2005) | LH+2/+7 VS hCG+2/+7 | Buserelin | 12 | Affymetrix HG-U133A | ≥ 2 | No relevant alteration was Observed. | |
| GnRH agonist VS GnRH antagonist | Ganirelix | ||||||
| Horcajads et al. (2005) | LH+2/LH+7 VS hCG+7 | Leuprolide acetate | 24 |
Affymetrix HG-U133A |
≥ 3 |
Up: 281 Down: 277 |
|
| GnRH agonist | |||||||
| Macklon et al. (2008) | 5 days after ovulation VS 5 days after ovum pick up | Ganirelix | 8 |
Affymetrix HGU133plus2 |
≥ 1.5 | Cell adhesion, T-cell receptor signaling, regulation of signal transduction, cell growth, proliferation, and programmed cell death |
Up: 142 Down: 98 |
| GnRH antagonist | |||||||
| Horcajads et al. (2008) | LH+1/+3/+5/+7/+9 VS hCG+1/+3/+5/+7/+9 | Leuprolide acetate | 49 |
Affymetrix HGU133A |
— | Biological processes including cell proliferation, angiogenesis, cellular physiological process, cell death, and apoptosis were listed. |
Up: 203 Down :97(2-day delay of genomic profile in COH) |
| GnRH agonist | |||||||
| Liu et al. (2008) |
LH+7 VS hCG+7 Moderate responders VS High responders |
Buserelin (nasal spray) |
13 |
Affymetrix HG-U133A |
≥ 2 | enzyme, signal transducer, ion-binding protein, transporter, receptor, transcription factor, cell adhesion molecule, regulatory protein | Moderate responders: Up: 5 Down: 2 |
| GnRH agonist | Wnt-signaling pathway, MAPK signaling pathway, nicotinamide metabolism | High responders: Up: 244 Down: 159 | |||||
| Haouzi et al. (2009) | LH+2/LH+7 VS hCG+2/hCG+7 | Not mention | 84 |
Affymetrix HGU133plus2 |
> 2 | cell division cycle (CDC) members, cyclin-dependent kinases (CDK), and members of the E2F family of transcription factors |
Up: 777 Down: 221 |
| Not mention | TGFβ signaling pathway, the complement and coagulation cascades and leukocyte transendothelial migration | ||||||
| Haouzi et al. (2009) | LH+2/LH+7 VS hCG+2/hCG+5 | Not mention | 84 |
Affymetrix HGU133plus2 |
> 2 | Chemokines in inflammatory processes, growth factors related to endometrium | GnRH agonist: Up: 731 Down: 451 |
| GnRH agonist VS GnRH antagonist | G1/S checkpoint regulation | GnRH antagonist: Up: 634 Down: 210 | |||||
| Senapati et al. (2018) | LH+12 VS hCG+13 | Leuprolide acetate | 23 | Affymetrix Human Gene 2.0 ST array+ Infinium DNA methylation 450 K BeadChip |
> 1.5 > 2 |
FC > 2: 165 DEGs, FC > 1.5: 641 DEGs 3322 genes with differential methylation, HPSE2, SERPINA5, MMP2, and MMP26 were similar to our common DEGs. |
|
| GnRH antagonist (7) and GnRH agonist (4) | Type of GnRH-anta not mention |
Gene enrichment in endometrial remodeling Proteolytic pathway involved in extracellular matrix degradation |
Our results indicated that hyperphysiological E2 levels resulted in the alteration of endometrial transcriptomic profiles. The hundreds of identified DEGs, whether new or familiar, were related to different aspects of endometrial receptivity. Some DEGs were shown in many studies to be associated with WOI or endometrial receptivity. For example, ESR1 is mainly expressed on endometrial cells and mediates the proliferation and differentiation of endometrial cells by E2 [45]. Polymorphisms of ESR1 were associated with infertility, endometriosis, recurrent spontaneous abortion, and the transportation of embryos in the oviduct [46–49]. Types of ESR1 polymorphism were found to provide a predictive value on the ovarian response to exogenous FSH and the outcomes of assisted reproduction [50, 51]. MMP10, a family member of lytic enzymes that cleave protein components of the extracellular matrix, plays a role in uterine matrix degradation and embryo implantation [52]. Mutations in MMP10 were detected by whole exon sequencing in patients with recurrent spontaneous abortion, suggesting this mutation affected MMP10 function and may lead to miscarriage [53]. A significant decrease in MMP10 expression was found in the placentas of women with pre-eclampsia, demonstrating a role for MMP10 in maternal-fetal communication and maintenance of pregnancy [54]. Similarly, changes in HPSE expression have been found in the placentas of women with pre-eclampsia [55].
We also revealed some new potential markers. For instance, ZCCHC12, encoding a downstream effector of bone morphogenetic protein (BMP) signaling, was previously found by RNA-Seq to be an important DEG in the regulation of endometrial receptivity in natural cycles [35]. Despite this consistent finding, the mechanism has not been elucidated. There is limited available data on SRARP and IL13RA2. SRARP is involved in steroid hormone synthesis, and IL13RA2 belongs to the family, in which many members were found to be closely associated with endometrial receptivity [56].
Two long non-coding RNAs, LINC01060 and LINC011104, were found in our study, and to date, no studies have been reported relating either LncRNA to reproduction. LINC011104 is a non-coding RNA mostly found in fat tissue, lymph nodes, and testes. Enriched GO and pathways of LINC011104 were not found by enrichment analysis, and its function and interaction with other genes require further research. LINC01060 was found to be significantly downregulated in pancreatic cancer tissues and was closely related to tumor invasion and metastasis [57]. Reported functional studies of its target genes were mainly focused on its role in regulating neurodevelopment. It is noteworthy that LINC01060 was found to have a link with the ERK5 MAPK pathway, which was previously found to participate in embryonic angiogenesis by regulating the expression of vascular endothelia growth factor, essential for embryo implantation [58]. Nevertheless, the target genes of LINC01060 in this signaling pathway were not differentially expressed between the current study groups. However, it is possible that genes with low expression levels may be indispensable for some essential pathways. The current results could also reflect limited sample size or the analytical methods used in our genetic analysis. Information about these lncRNAs and related genes was rather limited, and further in vitro or in vivo experiments are needed to investigate their functions.
Mirkin et al. [28] first reported transcriptome differences in stimulated endometrium using microarrays, but identified less than 20 DEGs. Simon [33] and Horcajads [32] took a step forward by finding a delay in changes of genomic expression for about 2 days, a feature more significant in the GnRH agonist protocol compared with GnRH antagonist protocol [29]. Liu grouped patients as high and moderate responders based on the level of E2 and found more DEGs in the high responders [31]. Interestingly, there were few overlapping DEGs among these studies. The most recent study found DEGs that were critical in the remodeling of endometrium and placenta formation, in which the identified MMP2 and HPSE2 were similar to our findings [43]. The variation of enrolled patients, techniques of data processing, and analysis may account for the heterogenicity of DEGs.
Successful embryo implantation depends on the balance of endometrial receptivity and selectivity under the precise control of functional and regulatory genes [59]. Despite many investigations of embryo implantation in vivo or in vitro, little is known of the underlying molecular mechanisms. There are hundreds of candidate genes or pathways with ambiguous function, which require further analysis. To improve our knowledge of the underlying mechanisms of embryo implantation, the signaling pathways of WOI or endometrial receptivity may provide valuable opportunities in the near future.
The current study has shed light on the special role of E2 during the establishment of endometrial receptivity. For the first time, we identified two lncRNAs that may participate in the regulation of WOI under excessive E2 levels, providing a new avenue for future research. However, there were some limitations of this study. For example, the sample size of each group was limited. Also, samples in natural and stimulated cycles were not paired due to the difficulties of multiple biopsies on the same patients in one period, possibly causing intra-individual bias. Fortunately, by the time the manuscript was completed, all 12 patients enrolled in the current study were getting a live birth or ongoing pregnancy after standard FET protocol, suggesting that the endometrium was healthy or receptive.
Conclusion
The phenomenon of lower implantation rates in high responders has puzzled reproductive endocrinologists for years. Side effects of excessive E2 produced by COH have drawn much attention. Supraphysiologic hormonal levels lead to a high risk of OHSS and impaired endometrium function. Our study suggested that high E2 levels in COH alter the expression of endometrial receptivity-related mRNA and LncRNA. Further research studying on their pathways in vitro or functional identification in endometrial cell lines is needed to validate the biological function of these identified genes. Our findings may provide new genetic markers or pathways that will contribute to a better understanding of the WOI. Current methods for endometrial sampling and testing require further improvement, and a less invasive and more efficient method for endometrial biopsy is needed for clinical practice.
Electronic supplementary material
(RAR 2715 kb)
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lingxiu Li, Email: lilingxiumsf@163.com.
Peng Wang, Email: wangpengdoc@hotmail.com.
Shan Liu, Email: liushan821009@126.com.
Xueyan Bai, Email: xueyan_0000@163.com.
Binbin Zou, Email: zoubinbin100@163.com.
Yuan Li, Email: cyliyuan@126.com.
References
- 1.Zhang S, Lin H, Kong S, Wang S, Wang H, Wang H, et al. Physiological and molecular determinants of embryo implantation. Mol Asp Med. 2014;34:939–980. doi: 10.1016/j.mam.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Norwitz ER, Schust DJ, Fisher JS. Implantation and the survival of early pregnancy. N Engl J Med. 2001;345:1400–1408. doi: 10.1056/NEJMra000763. [DOI] [PubMed] [Google Scholar]
- 3.Rachel SMD. The receptivity of the endometrium to implementation. Journal of Obstetrics & Gynaecology of the British Empire. 2010;69(1):107–109. doi: 10.1111/j.1471-0528.1962.tb00015.x. [DOI] [PubMed] [Google Scholar]
- 4.Paulson Richard J. Hormonal induction of endometrial receptivity. Fertility and Sterility. 2011;96(3):530–535. doi: 10.1016/j.fertnstert.2011.07.1097. [DOI] [PubMed] [Google Scholar]
- 5.Macklon NS, Geraedts JPM, Fauser BCJM. Conception to ongoing pregnancy: The “black box” of early pregnancy loss. Hum Reprod Update. 2002;8:333–343. doi: 10.1093/humupd/8.4.333. [DOI] [PubMed] [Google Scholar]
- 6.Kumar Pratap, Sharma Alok. Understanding implantation window, a crucial phenomenon. Journal of Human Reproductive Sciences. 2012;5(1):2. doi: 10.4103/0974-1208.97777. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Pellicer A, Valbuena D, Cano F, Remohi J, Simon C. Lower implantation rates in high responders: Evidence for an altered endocrine milieu during the preimplantation period. Fertil Steril. 1996;65:1190–1195. doi: 10.1016/s0015-0282(16)58337-x. [DOI] [PubMed] [Google Scholar]
- 8.Evans J, Hannan NJ, Edgell TA, Vollenhoven BJ, Lutjen PJ, Osianlis T, et al. Fresh versus frozen embryo transfer: Backing clinical decisions with scientific and clinical evidence. Hum Reprod Update. 2014;20:808–821. doi: 10.1093/humupd/dmu027. [DOI] [PubMed] [Google Scholar]
- 9.Ozkaya E, Kutlu T, Yayla CA, et al. Area under the curve of temporal estrogen and progesterone measurements during assisted reproductive technology: Which hormone is the main determinant of cycle outcome? Journal of Obstetrics and Gynaecology Research. 2018;44(2):263–269. doi: 10.1111/jog.13492. [DOI] [PubMed] [Google Scholar]
- 10.Basir GS, Wai-sum O, Ng EHY, Ho PC. Morphometric analysis of peri-implantation endometrium in patients having excessively high oestradiol concentrations after ovarian stimulation. Hum Reprod. 2001;16:435–440. doi: 10.1093/humrep/16.3.435. [DOI] [PubMed] [Google Scholar]
- 11.Bourgain C, Devroey P. The endometrium in stimulated cycles for IVF. Hum Reprod Update. 2003;9:515–522. doi: 10.1093/humupd/dmg045. [DOI] [PubMed] [Google Scholar]
- 12.Nikas G. Endometrial receptivity: Changes in cell-surface morphology. Semin Reprod Med. 2000;18:229–235. doi: 10.1055/s-2000-12561. [DOI] [PubMed] [Google Scholar]
- 13.Hsiu JG, Toner JP, Oehninger S, Jones HW. Endometrial estrogen and progesterone receptor and pinopode expression in stimulated cycles of oocyte donors. Fertil Steril. 1999;71:1040–1047. doi: 10.1016/s0015-0282(99)00137-5. [DOI] [PubMed] [Google Scholar]
- 14.Valdez-Morales FJ, Domà nguez AG, Vital-Reyes VS, Hinojosa Cruz JC, Chimal-Monroy J, Franco-Murillo Y, et al. Changes in receptivity epithelial cell markers of endometrium after ovarian stimulation treatments: Its role during implantation window. Reprod Health. 2015;12:1–11. doi: 10.1186/s12978-015-0034-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boomsma Carolien M., Kavelaars Annemieke, Eijkemans Marinus J.C., Fauser Bart C.J.M., Heijnen Cobi J., Macklon Nick S. Ovarian stimulation for in vitro fertilization alters the intrauterine cytokine, chemokine, and growth factor milieu encountered by the embryo. Fertility and Sterility. 2010;94(5):1764–1768. doi: 10.1016/j.fertnstert.2009.10.044. [DOI] [PubMed] [Google Scholar]
- 16.Chen Q j, Sun X x, Li L, Gao X h, Gemzell-Danielsson K, Cheng L n. Effects of ovarian stimulation on endometrial integrin β3 and leukemia inhibitory factor expression in the peri-implantation phase. Fertil Steril. 2008;89:1357–1363. doi: 10.1016/j.fertnstert.2007.03.073. [DOI] [PubMed] [Google Scholar]
- 17.Thomas K, Thomson AJ, Sephton V, Cowan C, Wood S, Vince G, et al. The effect of gonadotrophic stimulation on integrin expression in the endometrium. Hum Reprod 2002;17:63–68. Available from: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=11756363&retmode=ref&cmd=prlinks%5Cnpapers2://publication/uuid/D41C33EE-8D6D-4CFC-86A6-FAD09F5B5F19 (Accessed: 15 June 2017) [DOI] [PubMed]
- 18.Bourgain C, Ubaldi F, Tavaniotou A, Smitz J, Van Steirteghem AC, Devroey P. Endometrial hormone receptors and proliferation index in the periovulatory phase of stimulated embryo transfer cycles in comparison with natural cycles and relation to clinical pregnancy outcome. Fertil Steril. 2002;78:237–244. doi: 10.1016/s0015-0282(02)03228-4. [DOI] [PubMed] [Google Scholar]
- 19.Altmae S., Martinez-Conejero J.A., Salumets A., Simon C., Horcajadas J.A., Stavreus-Evers A. Endometrial gene expression analysis at the time of embryo implantation in women with unexplained infertility. Molecular Human Reproduction. 2009;16(3):178–187. doi: 10.1093/molehr/gap102. [DOI] [PubMed] [Google Scholar]
- 20.Kuokkanen Satu, Chen Bo, Ojalvo Laureen, Benard Lumie, Santoro Nanette, Pollard Jeffrey W. Genomic Profiling of MicroRNAs and Messenger RNAs Reveals Hormonal Regulation in MicroRNA Expression in Human Endometrium1. Biology of Reproduction. 2010;82(4):791–801. doi: 10.1095/biolreprod.109.081059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.PONNAMPALAM Anna P., WESTON Gareth C., SUSIL Beatrice, ROGERS Peter A. W. Molecular profiling of human endometrium during the menstrual cycle. The Australian and New Zealand Journal of Obstetrics and Gynaecology. 2006;46(2):154–158. doi: 10.1111/j.1479-828X.2006.00547.x. [DOI] [PubMed] [Google Scholar]
- 22.Ponnampalam AP, Weston GC, Trajstman AC, Susil B, Rogers PAW. Molecular classification of human endometrial cycle stages by transcriptional profiling. Mol Hum Reprod. 2004;10:879–893. doi: 10.1093/molehr/gah121. [DOI] [PubMed] [Google Scholar]
- 23.Horcajadas JA, Riesewijk A, Martin J, Cervero A, Mosselman S, Pellicer A, et al. Global gene expression profiling of human endometrial receptivity. J Reprod Immunol. 2004;63:41–49. doi: 10.1016/j.jri.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 24.Borthwick JM, Charnock-jones DS, Tom BD, Hull ML, Teirney R, Phillips SC, et al. Determination of the transcript pro ® le of human endometrium. Mol Hum Reprod. 2003;9:19–33. Available from: http://molehr.oxfordjournals.org/content/9/1/19.long . [DOI] [PubMed]
- 25.Riesewijk A. Gene expression profiling of human endometrial receptivity on days LH+2 versus LH+7 by microarray technology. Molecular Human Reproduction. 2003;9(5):253–264. doi: 10.1093/molehr/gag037. [DOI] [PubMed] [Google Scholar]
- 26.Kao L. C., Tulac S., Lobo S., Imani B., Yang J. P., Germeyer A., Osteen K., Taylor R. N., Lessey B. A., Giudice L. C. Global Gene Profiling in Human Endometrium during the Window of Implantation. Endocrinology. 2002;143(6):2119–2138. doi: 10.1210/endo.143.6.8885. [DOI] [PubMed] [Google Scholar]
- 27.Carson D. D. Changes in gene expression during the early to mid-luteal (receptive phase) transition in human endometrium detected by high-density microarray screening. Molecular Human Reproduction. 2002;8(9):871–879. doi: 10.1093/molehr/8.9.871. [DOI] [PubMed] [Google Scholar]
- 28.Mirkin S, Nikas G, Hsiu JG, Díaz J, Oehninger S. Gene expression profiles and structural/functional features of the peri-implantation endometrium in natural and gonadotropin-stimulated cycles. J Clin Endocrinol Metab. 2004;89:5742–5752. doi: 10.1210/jc.2004-0605. [DOI] [PubMed] [Google Scholar]
- 29.Macklon NS, Van Der Gaast MH, Hamilton A, Fauser BCJM, Giudice LC. The Impact of Ovarian Stimulation With Recombinant. :357–65. [DOI] [PubMed]
- 30.Haouzi D, Assou S, Mahmoud K, Tondeur S, Rème T, Hedon B, et al. Gene expression profile of human endometrial receptivity: Comparison between natural and stimulated cycles for the same patients. Hum Reprod. 2009;24:1436–1445. doi: 10.1093/humrep/dep039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Yunao, Lee Kai-Fai, Ng Ernest H.Y., Yeung William S.B., Ho Pak-Chung. Gene expression profiling of human peri-implantation endometria between natural and stimulated cycles. Fertility and Sterility. 2008;90(6):2152–2164. doi: 10.1016/j.fertnstert.2007.10.020. [DOI] [PubMed] [Google Scholar]
- 32.Horcajadas José A., Mínguez Pablo, Dopazo Joaquín, Esteban Francisco J., Domínguez Francisco, Giudice Linda C., Pellicer Antonio, Simón Carlos. Controlled Ovarian Stimulation Induces a Functional Genomic Delay of the Endometrium with Potential Clinical Implications. The Journal of Clinical Endocrinology & Metabolism. 2008;93(11):4500–4510. doi: 10.1210/jc.2008-0588. [DOI] [PubMed] [Google Scholar]
- 33.Simon C, Oberye J, Bellver J, Vidal C, Bosch E, Horcajadas JA, et al. Similar endometrial development in oocyte donors treated with either high- or standard-dose GnRH antagonist compared to treatment with a GnRH agonist or in natural cycles. Hum Reprod. 2005;20:3318–3327. doi: 10.1093/humrep/dei243. [DOI] [PubMed] [Google Scholar]
- 34.Lessey BA, Salamonsen LA, Simón C, Altmäe S, Macklon NS, Campoy C, et al. Guidelines for the design, analysis and interpretation of ‘omics’ data: focus on human endometrium. Hum Reprod Update. 2013;20:12–28. doi: 10.1093/humupd/dmt048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu S, Yao G, Wang Y, Xu H, Ji X, He Y, et al. Transcriptomic changes during the pre-receptive to receptive transition in human endometrium detected by RNA-Seq. J Clin Endocrinol Metab. 2014;99:E2744–E2753. doi: 10.1210/jc.2014-2155. [DOI] [PubMed] [Google Scholar]
- 36.Ganesh A, Chauhan N, Das S, Chakravarty B, Chaudhury K. Endometrial receptivity markers in infertile women stimulated with letrozole compared with clomiphene citrate and natural cycles. Syst Biol Reprod Med. 2014;60:105–111. doi: 10.3109/19396368.2013.862316. [DOI] [PubMed] [Google Scholar]
- 37.Miller PB, Parnell BA, Bushnell G, Tallman N, Forstein DA, Higdon HL, et al. Endometrial receptivity defects during IVF cycles with and without letrozole. Hum Reprod. 2012;27:881–888. doi: 10.1093/humrep/der452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He Z, Ma Y, Li L, Liu J, Yang H, Chen C, et al. Osteopontin and Integrin αvβ3 Expression during the Implantation Window in IVF Patients with Elevated Serum Progesterone and Oestradiol Level. Geburtshilfe Frauenheilkd. 2016;76:709–717. doi: 10.1055/s-0041-111697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y, Kodithuwakku SP, Ng PY, Chai J, Ng EHY, Yeung WSB, et al. Excessive ovarian stimulation up-regulates the Wnt-signaling molecule DKK1 in human endometrium and may affect implantation: An in vitro co-culture study. Hum Reprod. 2010;25:479–490. doi: 10.1093/humrep/dep429. [DOI] [PubMed] [Google Scholar]
- 40.Mebratu Y, Tesfaigzi Y. How ERK1/2 Activation Controls Cell Proliferation and Cell Death Is Subcellular Localization the Answer? Cell Cycle. 2010;8:1168–1175. doi: 10.4161/cc.8.8.8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arlier S, Murk W, Guzeloglu-Kayisli O, Semerci N, Larsen K, Tabak MS, et al. The extracellular signal-regulated kinase 1/2 triggers angiogenesis in human ectopic endometrial implants by inducing angioblast differentiation and proliferation. Am J Reprod Immunol. 2017;78:1–11. doi: 10.1111/aji.12760. [DOI] [PubMed] [Google Scholar]
- 42.Fluhr Herbert, Spratte Julia, Bredow Marike, Heidrich Stephanie, Zygmunt Marek. Constitutive activity of Erk1/2 and NF-κB protects human endometrial stromal cells from death receptor-mediated apoptosis. Reproductive Biology. 2013;13(2):113–121. doi: 10.1016/j.repbio.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 43.Wang Z, Gerstein M, Snyder M. RNA-Seq : a revolutionary tool for transcriptomics. Nature Reviews Genetics. 2010;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jemt A, Sigurgeirsson B, Hanna A, Ujvari D, Westgren M, Lundeberg J. Comprehensive RNA sequencing of healthy human endometrium at two time points of the menstrual cycle. Biol Reprod. 2016;96:24–33. Available from: https://academic.oup.com/biolreprod/article/96/1/24/2725477/Comprehensive-RNA-sequencing-of-healthy-human . [DOI] [PubMed]
- 45.Ganesh Vijaya, Venkatesan Vettriselvi, Koshy Teena, Reddy Sanjeeva Nellapalli, Muthumuthiah Suruli, Paul Solomon Franklin Durairaj. Association of estrogen, progesterone and follicle stimulating hormone receptor polymorphisms with in vitro fertilization outcomes. Systems Biology in Reproductive Medicine. 2018;64(4):260–265. doi: 10.1080/19396368.2018.1482030. [DOI] [PubMed] [Google Scholar]
- 46.Paskulin DD, Cunha-filho JS, Paskulin LD, Augusto C, Souza B, Ashton-prolla P. ESR1 rs9340799 Is Associated with Endometriosis-Related Infertility and In Vitro Fertilization Failure. 2013;35:907–13. [DOI] [PMC free article] [PubMed]
- 47.Swaminathan M, Ganesh V, Koshy T, et al. A Study on the Role of Estrogen Receptor Gene Polymorphisms in Female Infertility. Genetic Testing and Molecular Biomarkers. 2016;20(11):692–695. doi: 10.1089/gtmb.2016.0097. [DOI] [PubMed] [Google Scholar]
- 48.Biosciences M, Medicine V, Li S, Neill SRSO, Zhang Y, Holtzman MJ, et al. Estrogen receptor a is required for oviductal transport of embryos. :1595–607. [DOI] [PMC free article] [PubMed]
- 49.Pan H, Suo P, Liu C, Wang J, Zhou S, Ma X, et al. The ESR1 gene in unexplained recurrent spontaneous abortion. Syst Biol Reprod Med. 2014;60:161–164. doi: 10.3109/19396368.2013.877540. [DOI] [PubMed] [Google Scholar]
- 50.Boudjenah R, Molina-gomes D, Torre A, Bergere M, Bailly M, Wainer R, et al. Genetic Polymorphisms Influence the Ovarian Response to rFSH Stimulation in Patients Undergoing In Vitro Fertilization Programs with ICSI. 2012;7. [DOI] [PMC free article] [PubMed]
- 51.De Mattos CS, Trevisan CM, Peluso C, Adami F, Cordts EB, Christofolini DM, et al. ESR1 and ESR2 gene polymorphisms are associated with human reproduction outcomes in Brazilian women. J Ovarian Res. 2014;7:1–9. doi: 10.1186/s13048-014-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu X, Pan Y. Molecular characterization, mapping, and haplotype analysis of porcine matrix metalloproteinase genes MMP1 and MMP10. Biochem Genet. 2009;47:763–774. doi: 10.1007/s10528-009-9275-x. [DOI] [PubMed] [Google Scholar]
- 53.Quintero-Ronderos P, Mercier E, Fukuda M, Suarez C, Gonzalez R, Patarroyo M, et al. Novel genes and mutations in patients affected by recurrent spontaneous abortion. PLoS One. 2017;Submitted:1–14. [DOI] [PMC free article] [PubMed]
- 54.Kim MS, Yu JH, Lee MY, Kim AL, Jo MH, Kim MG, et al. Differential expression of extracellular matrix and adhesion molecules in fetalorigin amniotic epithelial cells of preeclamptic pregnancy. PLoS One. 2016;11:1–16. doi: 10.1371/journal.pone.0156038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaartokallio T, Cervera A, Kyllönen A, Laivuori K, Kere J, Laivuori H. Gene expression profiling of pre-eclamptic placentae by RNA sequencing. Sci Rep [Internet]. Nature Publishing Group; 2015;5:1–15. Available from: 10.1038/srep14107 [DOI] [PMC free article] [PubMed]
- 56.Davidson LM, Coward K. Molecular mechanisms of membrane interaction at implantation [J] Birth Defects Research Part C: Embryo Today: Reviews. 2016;108(1):19–32. doi: 10.1002/bdrc.21122. [DOI] [PubMed] [Google Scholar]
- 57.Shi X, Guo X, Li X, Wang M, Qin R. Loss of Linc01060 induces pancreatic cancer progression through vinculin-mediated focal adhesion turnover. Cancer Lett. 2018;433:76–85. doi: 10.1016/j.canlet.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 58.Sohn SJ, Sarvis BK, Cado D, Winoto A. ERK5 MAPK regulates embryonic angiogenesis and acts as a hypoxia-sensitive repressor of vascular endothelial growth factor expression. J Biol Chem. 2002;277:43344–43351. doi: 10.1074/jbc.M207573200. [DOI] [PubMed] [Google Scholar]
- 59.Evans Jemma, Salamonsen Lois A., Winship Amy, Menkhorst Ellen, Nie Guiying, Gargett Caroline E., Dimitriadis Eva. Fertile ground: human endometrial programming and lessons in health and disease. Nature Reviews Endocrinology. 2016;12(11):654–667. doi: 10.1038/nrendo.2016.116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(RAR 2715 kb)