Abstract
Purpose
High progesterone is associated with low implantation rate. Our previous study demonstrated that DNA methylation in endometrium was increased in women with high progesterone in IVF cycles. However, the DNA methylation status is still not yet confirmed, and how it affects endometrial receptivity in high progesterone is still unknown. Current study investigated the effects of high progesterone on DNA methylation and gene expression of adhesion molecules on endometrium during implantation window.
Methods
A cohort study included 20 women with high progesterone (HP) and 20 with normal progesterone (NP) on the day of human chorionic gonadotropin (hCG) administration after controlled ovarian hyperstimulation in IVF cycle. Endometrial tissues were collected on the 7th day after hCG administration. Immunohistochemical staining of DNA methyltransferases (DNMT1 and DNMT3B) and adhesion molecules (MUC1, CDH1 and CTNNB1) were performed. Methylation of MUC1, CDH1, and CTNNB1 promoter regions was detected by Sequenom MassARRAY or bisulfite sequencing PCR. RT-qPCR was used to quantify mRNA expression levels, and correlation of methylation and gene expression level of the adhesion molecules were determined.
Results
DNMT3B, but not DNMT1, in nucleus of luminal and glandular epithelial cells in HP group was significantly higher than that in NP group. Promoter regions of CDH1 and CTNNB1, but not MUC1, in endometrium of HP group were hypermethylated. Protein and mRNA expression of MUC1, CDH1, and CTNNB1 in endometrium of HP group was significantly lower than that in NP group. Level of DNA methylation was negatively correlated with the gene expression of CDH1 and CTNNB1, but not MUC1.
Conclusions
DNA hypermethylation and low expression of adhesion molecules on endometrium were associated with high progesterone during implantation window, which may contribute to the underlying epigenetic mechanism in the failure of IVF treatment.
Electronic supplementary material
The online version of this article (10.1007/s10815-019-01623-6) contains supplementary material, which is available to authorized users.
Keywords: DNA methylation, Adhesion molecules, Endometrium, In vitro fertilization, Progesterone
Introduction
In 1991, Schoolcraft et al. is the first group to report high progesterone on day of human chorionic gonadotropin (hCG) administration in IVF cycles [1]. Incidence of high progesterone varies from different protocols used in controlled ovarian hyperstimulation (COH), which is as high as 35% (5–35%) in women that used gonadotropin-releasing hormone (GnRH) agonists and 38% (20–38%) in women that used GnRH antagonists [2, 3]. Many studies demonstrated that high serum progesterone on the day of hCG administration was associated with implantation failure [2–7]. However, no detrimental effect of high progesterone on the quality of oocyte or embryo was found. Pregnancy rate of women with the embryo transferred from oocyte donation with high progesterone on the day of hCG administration was not deleteriously affected [8]. It indicated that high progesterone may rather influence endometrium and result in poor IVF outcomes.
Gene expression profile of endometrium is also altered in women with high progesterone on the day of hCG administration [9–12]. Genes involved in cell adhesion, developmental processes, and immune system were affected, which may determine the detrimental effects of high progesterone on endometrial receptivity and thereby implantation failure. Adhesion molecules on endometrium, such as Mucin 1 (MUC1), cadherin 1 (CDH1), and β catenin (CTNNB1), are considered as endometrium receptivity markers which play an essential role for embryo attachment and subsequent invasion during the window of implantation [13–16]. How the high progesterone level on the day of hCG administration regulate expression level of adhesion molecules on the endometrium is still not fully understood; one of the possible mechanisms is epigenetics. In our previous study, we demonstrated that high progesterone level on the day of hCG administration in IVF cycle was associated with epigenetic modification in endometrium [17], including 5-methylcytosine (5-mc) was upregulated. 5-mc was formed when a methyl group was transferred to the 5th position of cytosine residues in a CpG dinucleotide context. It refers as a golden marker of DNA methylation [18, 19], whereas DNA methyltransferases (DNMTs), including DNMT1 and DNMT3B, are responsible for the maintenance and deposition of DNA methylation [20, 21]. However, the DNA methylation status is still not yet confirmed, and how it affects endometrial receptivity in high progesterone is still largely unknown.
In present study, we aimed to investigate the effect of high progesterone on the day of hCG administration in IVF cycle on DNA methylation and correlate with gene expression of adhesion molecules on endometrium during implantation window.
Materials and methods
The study was conducted in the Department of Obstetrics and Gynaecology, Prince of Wales Hospital, teaching hospital of The Chinese University of Hong Kong, Hong Kong. The samples were collected from the Reproductive Medical Centre, the First Affiliated Hospital of Zhengzhou University, Henan, China, between February 2017 and July 2017. The study was approved by the ethics committee of both universities concerned. Written informed consents were obtained from all participating women.
Subjects and tissue collection
In total, 40 women were recruited in our study and underwent the first cycle of IVF treatment. Twenty women had high serum progesterone level (HP) ≥ 1.7 ng/ml, and the other 20 women had normal serum progesterone level (NP) < 1.7 ng/ml on the day of hCG administration. The cutoff level of progesterone levels at 1.7 ng/mL was based on our previously published data [22], in which 1.7 ng/mL on the day of hCG administration represented the 90th percentile of the serum progesterone level derived from the data of 1457 fresh embryo transfer (ET) cycles. Inclusion criteria included (1) age between 25 and 40 years; (2) body mass index (BMI) between 18 and 25 kg/m2; (3) regular menstrual cycle (25–35 days); (4) normal basal serum hormone levels on day 2–4 of the menstrual cycle (FSH <10 IU/L and E2 < 60 pg/mL); (5) infertility because of tubal or male factor; and (6) normal karyotype in both couples. Exclusion criteria included (1) polycystic ovarian syndrome, (2) hydrosalpinx, (3) uterine abnormalities, (4) thyroid dysfunction, and (5) recurrent miscarriage. Among all women, 38 underwent controlled ovarian hyperstimulation (COH) using super GnRH-a long protocol, while 2 underwent COH using GnRH-a long protocol. hCG at dose 6500–10,000 IU (Merck-Serono) was injected for ovulation induction after COH when more than 3 follicles reached a mean diameter of 16–18 mm.
Serum progesterone levels were measured by Electrochemiluminescence Immunoassay Kit Progesterone II (Cobas 12145383) using immunoanalyzer (Roche Cobas e411; Roche Diagnostics, Mannheim, Germany) during COH on every 2–4 days, during the late follicular phase and on the day of hCG administration in each cycle. Detection limit of the assay was 0.03 ng/ml, and the sensitivity was 0.15 ng/ml. Intra-assay and inter-assay coefficients of variation were 3.0 and 5.5 %, respectively.
All 40 women cancelled their fresh ET due to the risk of ovarian hyperstimulation syndrome (OHSS), as part of the preimplantation genetic testing or personal reasons. Endometrial tissues were collected from the fundus of uterus using Pipelle sampler (Prodimed) under sterile conditions on the 7th day after hCG administration. Part of the endometrial tissue was immediately placed into 10% neutral buffered formalin overnight after biopsy and embedded into paraffin before experiment. Other parts of the endometrial tissue were snap frozen for DNA and RNA studies.
Immunohistochemistry
Spatial protein expression of DNMTs in the endometrium was examined by IHC staining using anti-DNMT1 rabbit polyclonal antibody (HPA002694, Sigma, diluted 1:300) and anti-DNMT3B rabbit polyclonal antibody (HPA001595, Sigma, diluted 1:250). Spatial protein expression of endometrium adhesion molecules was studied by immunohistochemical (IHC) staining using anti-MUC1 rabbit polyclonal antibody (HPA004179, Sigma, diluted 1:200), anti-CDH1 rabbit polyclonal antibody (HPA004812, Sigma, diluted 1:200), and anti-CTNNB1 rabbit polyclonal antibody (HPA029160, Sigma, diluted 1:300).
Paraffin-embedded endometrial tissue sections (5 μm) were dewaxed in xylene, rehydrated through descending ethanol to double distilled water (ddH2O), and then followed with permeabilization in Triton X-100 for 25 minutes. Then antigen retrieval was performed in a microwave oven with 10 mmol/L citrate buffer with pH = 6 for 20 minutes. Endogenous peroxidase activities and nonspecific binding were blocked with 0.3% hydrogen peroxide in methanol for 20 minutes and 1% rabbit serum at room temperature for 1 hour. The sections were then incubated with each primary antibody overnight at 4 °C. Serial control sections were incubated with normal rabbit or mouse serum without specific primary antibodies. After incubation, the slides were washed thrice with 0.2% Tween in PBS and once with ddH2O and then incubated 1 hour in secondary goat anti-rabbit antibody (ab205718, Abcam). After washing thrice with 0.2% Tween in PBS and once with ddH2O, the specific antibody binding was visualized by incubation with peroxidase substrate 3,3'-diamino-benzidene tetrahydrochloride (DAB, Dako) and counterstained with hematoxylin for less than 3 minutes. Dark brown nuclear (for DNMTs) or cytoplasmic (for MUC1, CDH1, and CTNNB1) staining indicated a positive reaction.
For semiquantitative IHC analysis, 10 random views with luminal epithelium and glandular epithelium at 400 magnification for each section were captured by using Leica digital camera (DM6000B, Leica, Germany). DNMT1 and DNMT3B expression was evaluated by counting the number of positively stained nuclei over the total number of cells in luminal and glandular epithelium. Two independent observers (X.Y., W.M.Y.) counted the cells by using Image J software (1.49v, Wayne Rasband, National Institutes of Health, Bethesda, MD). MUC1, CDH1, and CTNNB1 were evaluated by histochemical score (H-score) on the positively stained cytoplasm according to an equation: H score = ∑ Pi (i + 1), in which i was the intensity of staining (1 = weak; 2 = moderate; 3 = strong; and 4 = very strong) and Pi was the percentage of cells positively stained at each intensity (0%–100%). Two independent observers (X.Y., W.M.Y.) scored each section. The final scores for all the adhesion molecules in luminal epithelium and glandular epithelium were obtained by taking the mean of the H-scores in triplicates.
RNA extraction and reverse transcription and quantitative real-time PCR (RT-qPCR)
Total RNA was extracted from each sample using RNeasy Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer. The concentration and quality of all RNA samples were evaluated by a NanoDrop 2000 apparatus (Thermo Fisher Scientific, Wilmington, DE, USA).
Total RNA was reverse transcribed using PrimeScriptTM 1st strand cDNA Synthesis Kit (TaKaRa Bio, USA). Quantitative PCR was performed on a LightCycler 480 (Roche Diagnostics, Mannheim, Germany) using TaqMan™ Universal Master Mix II (Thermo Scientific). The conditions for RT-qPCR were as follows: 50 °C for 2 min and 95 °C for 10 min followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. ACTB was used as internal controls for normalization. Primers are purchased from Thermo Fisher Scientific: ACTB, Hs01060665; MUC1, Hs00159357_m1; CDH1, Hs01023895_m1; and CTNNB1, Hs00355045_m1. ∆∆CT method was used to calculate the CT-value for each sample, and the results were expressed as 2−∆∆CT to analyze the fold change (HP vs. NP): ∆∆CT = (CTtarget gene−CTactin) HP − (CTtarget gene−CTactin) NP.
DNA extraction and sequenom MALDI-TOF mass spectrometry or bisulfite sequencing PCR
Genomic DNA from frozen endometrial samples was extracted using the QIAamp DNA Mini Kit (Qiagen, Valencia, CA, USA). The concentration and quality of all DNA samples were evaluated by a NanoDrop 2000 apparatus (Thermo Fisher Scientific, Wilmington, DE, USA) and gel electrophoresis. The DNA samples with concentration more than 20 ng/ul, OD260/OD280 ratio between 1.5 and 2.0, OD260/OD230 ratio more than 0.6, and clear smear on gel were considered qualified.
One microgram of genomic DNA from each sample was bisulfite treated using NaHSO3 (Zymo Research, Irvine, CA, USA). The bisulfite-treated genomic DNA was further processed by polymerase chain reaction (PCR) and in vitro transcription (IVT). PCR was performed in a total volume of 8 ul by using 2 pmol of each primer/200 μM dNTP/0.5 units of PCR enzyme (primers of CDH1 and CTNNB1 are showed in Table 1). The reaction mixture was preactivated for 4 min at 94 °C. The reactions were amplified in 45 cycles of 94 °C for 20 s, 56 °C for 30 s, and 72 °C for 1 min, followed by 72 °C for 3 min. Then 1.7 μl 0.3 units of shrimp alkaline phosphatase (SAP) enzyme in H2O were added in order to dephosphorylate unincorporated dNTPs. The reaction was incubated at 37 °C for 20 min, and SAP was then heat inactivated for 5 min at 85 °C. IVT was performed by using 2 μl PCR reaction mix as template in a 7-μl transcription reaction with the addition of RNase A to cleave the in vitro transcript and kept at 37 °C for 3 hours. The samples were then cleaned by Resin. At last, the mixture from each sample was dispensed to a 384-well SpectroCHIP® bioarray by MassARRAY NanodispenserRS1000. The chip was read by a MassARRAY Analyzer 4.0 mass spectrometer system. Data were collected and analyzed by using MassARRAY EpiTyper v1.0 software (Agena, Inc).
Table 1.
Primers of MUC1 for BSP and primers of CDH1, CTNNB1 and PAEP for Sequenom MALDI-TOF Mass Spectrometry
| Genes | Label | Start | Primer sets | Product size | No of CpG’s covered | |
|---|---|---|---|---|---|---|
| MUC1 | 1 | 578 | Forward | GAGGTTTGGGGTYGTTTATTTAGT | 498 | 48 |
| 1075 | Reverse | ACAAAATAATATTAAAAACAAATAAACAC | ||||
| 2 | 1148 | Forward | GTTTTGAGAYGGAGTTTTGTTTT | 337 | 20 | |
| 1484 | Reverse | AAAAAATTAAACACRTTCTATATAAACC | ||||
| 3 | 1439 | Forward | GTTTTTTAAGTATATAAYGGTTTATATAGAA | 370 | 25 | |
| 1808 | Reverse | CTCTAATAATCACCRAAAACCTTAC | ||||
| CDH1 | 1 | 5142 | Forward | aggaagagagGGTTTTAAGGGTTTATGGTTGGT | 428 | 26 |
| 4715 | Reverse | cagtaatacgactcactatagggagaaggctTAAAAACCCTTTCTAATCCCAAATC | ||||
| 2 | 5600 | Forward | aggaagagagTTTGGTATGGTAGGTGTTTTTATTTTT | 481 | 36 | |
| 5120 | Reverse | cagtaatacgactcactatagggagaaggctACCAACCATAAACCCTTAAAACC | ||||
| CTNNB1 | 1 | 4196 | Forward | aggaagagagGGTATTTTTAAGGATTTGTTGAATTG | 542 | 38 |
| 4737 | Reverse | cagtaatacgactcactatagggagaaggctATAACCCTAATATCCTCCCCTATCC | ||||
| 2 | 4713 | Forward | aggaagagagGGATAGGGGAGGATATTAGGGTTAT | 514 | 38 | |
| 5226 | Reverse | cagtaatacgactcactatagggagaaggctAAACACCTCAAAAAAACAAACTCCT | ||||
| PAEP | 1 | 595 | Forward | aggaagagagTTTGGTAGTTTTATTTTTGGGTATTT | 417 | 14 |
| 179 | Reverse | cagtaatacgactcactatagggagaaggctCCAAAATAATCTCAATCTCCTAACC | ||||
| 2 | 4616 | Forward | aggaagagagTTTGTGGTATTTTGGGGTAGTTATG | 532 | 11 | |
| 5147 | Reverse | cagtaatacgactcactatagggagaaggctCAAACCTTTAAAAACTCCAAATCCT | ||||
| 3 | 3094 | Forward | aggaagagagATGGGAAATGATTTTGTGTTAGAGT | 566 | 15 | |
| 2529 | Reverse | cagtaatacgactcactatagggagaaggctACAACTAACTTCCTATCCACCTCAA |
Since there was no suitable primer of MUC1 for Sequenom MALDI-TOF mass spectrometry, bisulfite sequencing PCR (BSP) was employed. Two microgram of genomic DNA from each sample was denatured by adding NaOH. Then saturated metabisulfite and Quinol were added to sulfonate and hydrolytic deaminate the denatured DNA. By passing through a desalting column, the free bisulfite ions in DNA were removed. At last, freshly prepared NaOH was added for DNA desulfonation. Then the converted genomic DNA was processed into PCR amplification. PCR reaction was performed in a total volume of 50 ul by using 1 ul of each primer/1 ul dNTP/4 units of PCR enzyme (primers of MUC1 is showed in Table 1). The reaction mix was preactivated for 4 min at 98 °C. The reactions were amplified in 20 cycles of 94 °C for 45 s, 66 °C for 45 s, and 72 °C for 1 min, followed by another 20 cycles of 94 °C for 45 s, 56 °C for 45 s, and 72 °C for 1 min, and then followed by 72 °C for 8 min. The PCR results were verified by gel-based electrophoresis. A single, bright, and specific band was considered as successful PCR amplification. For cloning purposes, purified PCR products were ligated to pUC18-T vector (Sangon Biotech Co., Ltd, Shanghai, China) and transformed into competent JM109 cells. Then white colonies which represented vectors inserted with target PCR product were selected on IPTG/X-gal plate. For each sample, we selected at least 10 clones for bisulfite conversion and then DNA sequencing. Plasmids containing the target DNA were extracted by using the QIAprep Spin Miniprep Kit (Qiagen) and subjected to standard sequencing analysis by using 3730xl DNA analyzer (Applied Biosystems Inc., Foster City, USA).
Statistical analysis
Statistical analysis was performed by using SPSS statistical software package, version 20.0 (IBM, New York, USA). Student’s t-test was used to compare numerical data, including age, BMI, duration of infertility, number of IVF cycle, basal FSH level, basal E2 level, duration of gonadotropin (Gn), dose of Gn, and level of E2 and P on day of hCG administration. Categorical data were compared using chi-square test. mRNA expression level of receptivity markers and semiquantitative IHC analysis of DNMTs were also analyzed by using student’s t-test. DNA methylation level of MUC1, CDH1, and CTNNB1 was analyzed by using student’s t-test. Correlation analysis was performed by Pearson correlation. A P value < 0.05 was considered statistically significant.
Results
Patient demographic characteristics
Table 2 showed the demographic characteristics of women in HP and NP groups. There was no significant difference between the two groups, except the dose of GnRH-a used, and the progesterone level on day of hCG administration in NP group was significantly lower than that in HP group (1890 ± 698.3 vs 2507.5 ± 771.1, P = 0.012; 0.9 ± 0.4 ng/ml vs 3.1 ± 0.9 ng/ml, P < 0.001, respectively).
Table 2.
Demographic characteristics of women from HP and NP group
| NP (n = 20) | HP (n = 20) | P value | |
|---|---|---|---|
| Age (years) | 28.8 ± 3.1 | 29.3 ± 3.7 | 0.619 |
| BMI(Kg/m2) | 21.7 ± 3.2 | 23.3 ± 2.5 | 0.092 |
| Year of infertility (Years) | 3.9 ± 3.5 | 4.0 ± 3.1 | 0.925 |
| No. of IVF cycle | 1 (50%) | 1 (50%) | – |
| Cause of infertility | |||
| Tubal | 30% | 20% | 0.206 |
| Male factor | 12.5% | 7.5% | 0.429 |
| Anovulation | 0 | 2.5% | 0.311 |
| Unexplained | 7.5% | 20% | 0.077 |
| Basal FSH (IU/L) | 5.9 ± 1.2 | 5.8 ± 1.5 | 0.793 |
| Basal E2 (pg/ml) | 29.3 ± 16.5 | 37.9 ± 19.4 | 0.121 |
| Stimulation protocol | |||
| Super GnRH-a long protocol | 45% | 50% | 0.147 |
| GnRH-a long protocol | 5% | 0% | 0.147 |
| Duration of ovarian stimulation (days) | 13.0 ± 1.5 | 14.1 ± 1.9 | 0.053 |
| Dose of Gn | 1890.0 ± 698.3 | 2507.5 ± 771.1 | 0.012 |
| E2 on day of hCG administration (pg/ml) | 5561 ± 2409 | 4601 ± 1894 | 0.169 |
| P on day of hCG administration (ng/ml) | 0.9 ± 0.4 | 3.1 ± 0.9 | <0.001 |
Expression level of DNMTs
Figure 1 shows the protein expression levels of DNMT1 and DNMT3B in endometrium in NP and HP groups. We found that DNMT1 and DNMT3B were mainly expressed in the nucleus of luminal and glandular epithelial cells for both DNMT1 and DNMT3B and in the nucleus of some stromal cells for DNMT1 only. There was no significant difference in the expression level of DNMT1 in both luminal and glandular epithelium between NP and HP groups. However, the expression level of DNMT3B in HP group was significantly higher than that in NP group in both luminal and glandular epithelium.
Fig. 1.
Expression level of DNA methyltransferases in NP and HP group. Expression and comparison of (A–D) DNMT1 and (E–H) DNMT3B in two compartments of endometrium from women with normal progesterone level and high progesterone level. 20 samples in each group were tested. There was no significant difference in the expression of DNMT1 between endometrium from HP and NP group. The expression of DNMT3B in endometrium from HP group was significantly higher than that from NP group. Red arrow indicates positive staining. Scale bar = 50 μm. GE glandular epithelia; LE luminal epithelia; SC stromal cell. Values expressed are mean + SEM. n.s., not significant
DNA methylation level of adhesion molecules
To quantify the DNA methylation status of adhesion molecules on endometrium, we quantified the methylation of CpG sites in the promoter regions of MUC1, CDH1, and CTNNB1 and compared between HP and NP groups (Fig. 2). The results showed that there was no significant difference in the DNA methylation in MUC1 between the two groups in which the DNA methylation level in three regions of promoter was measured by BSP (P = 0.841, P = 0.151, and P = 0.690), in which 5 samples for each group were tested. For CDH1 and CTNNB1, the overall methylation level on the CpG sites at their promoter regions in HP group was more (11 vs 9 sites/clusters for CDH1; 12 vs 11 sites/clusters for CTNNB1) and higher (0.069 ± 0.052 vs 0.063 ± 0.067 for CDH1; 0.078 ± 0.055 vs 0.074 ± 0.049 for CTNNB1) than that in NP group, but hypermethylation was mainly clustered in the 9th to 13th CpG sites with significance only in the 12th to 13th CpG sites in CDH1 and in the 36th to 41st CpG sites with significance only in the 41st CpG site in CTNNB1 (P = 0.006 and P = 0.018, respectively; Supplemental Tables 1 and 2).
Fig. 2.
DNA methylation level of adhesion molecules in NP and HP group. Comparison of DNA methylation level of (A–B) MUC1, (C–D) CDH1, and (E–F) CTNNB1 on endometrium from HP and NP group. 5 samples in each group were used for testing methylation level of MUC1 by bisulfite sequencing methods, and 20 samples in each group were used for testing methylation level of CDH1 and CTNNB1 by Sequenom MassARRAY. There was no significant difference in the methylation level of MUC1 between endometria from HP and NP group. CDH1 and CTNNB1 were significantly hypermethylated in endometrium from HP group than that from NP group. Profiling of DNA methylation level of CpG sites in (A) MUC1, (C) CDH1, and (E) CTNNB1 promoter region was presented as an epigram
Protein and mRNA expression level of receptivity markers
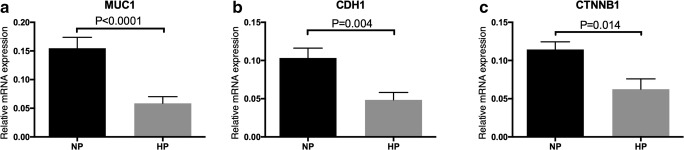
To verify whether the hypermethylation affects the gene expression of the adhesion molecules, we detected the protein and mRNA expression of MUC1, CDH1, and CTNNB1 on endometrium. Figure 3 shows the IHC staining results of MUC1, CDH1, and CTNNB1. All proteins were expressed in luminal and glandular epithelium only, but not stroma. The protein expression levels of MUC1, CDH1, and CTNNB1 in both cytoplasm of the luminal and glandular epithelial cells in NP group were significantly higher than that in HP group. Figure 4 shows the mRNA expression level of MUC1, CDH1, and CTNNB1 quantified by RT-qPCR. mRNA expression level of MUC1, CDH1, and CTNNB1 in HP group was also significantly lower than that in NP group.
Fig. 3.
Protein expression level of adhesion molecules in NP and HP group. Expression and comparison of (A–D) MUC1, (E–H) CDH1, and (I–L) CTNNB1 in two compartments of endometrium from women with normal progesterone level and high progesterone level. 20 samples in each group were tested. The expression of MUC1, CDH1, and CTNNB1 in endometrium from HP group was significantly higher than that from NP group. Red arrow indicates positive staining. Scale bar = 50 μm. GE glandular epithelia; LE luminal epithelia; SC stromal cell. Values expressed are mean + SEM
Fig. 4.
mRNA expression level of adhesion molecules in NP and HP group. Comparison of mRNA expression level of (A) MUC1, (B) CDH1, and (C) CTNNB1 in endometrium from HP and NP group. 20 samples in each group were tested. The mRNA expression level of MUC1, CDH1, and CTNNB1 in endometrium from HP group was significantly lower than that from NP group. Values expressed are mean + SEM. n.s., not significant
Correlation analysis of mRNA expression level and DNA methylation of receptivity markers
Figure 5 showed the correlation analysis of mRNA expression level and DNA methylation level of the MUC1, CDH1, and CTNNB1. We found that the mRNA expression level of CDH1 and CTNNB1 were negatively correlated with the DNA methylation levels in promoter region (P = 0.016, P = 0.003, respectively). However, no significant correlation was found between mRNA expression level and DNA methylation of MUC1 (P = 0.437).
Fig. 5.
Correlation analysis between methylation level and mRNA expression level of adhesion molecules in NP and HP group. Correlation analysis between mean methylation levels of the promoter region of (A) MUC1, (B) CDH1, and (C) CTNNB1 with corresponding mRNA expression level. There was no significant correlation in MUC1 between the DNA methylation level and mRNA expression level. The DNA methylation level of CDH1 and CTNNB1 was significantly negatively correlated with mRNA expression level. Red dots, NP group; green dots, HP group
Discussion
To our best knowledge, this is the first study to investigate the association between high progesterone on the day of hCG administration and aberrant methylation and gene expression of adhesion molecules on endometrium during the implantation window. In our study, we found that DNMT3B was upregulated in endometrium from women with high progesterone level. While the protein and mRNA expression of MUC1, CDH1, and CTNNB1 was downregulated. In addition, CDH1 and CTNNB1, but not MUC1, were hypermethylated in endometrium in HP group, and the expression level of CDH1 and CTNNB1 was negatively correlated with their mRNA expression levels.
Methylation of the fifth position of cytosine is one of the best studied epigenetic modifications [23]. Among the 3 common DNA methyltransferases, DNMT1 plays a role in maintaining methylation at hemimethylated CpG sites, which is responsible for the preservation of the methylation patterns, while DNMT3B are responsible for de novo methylation [24]. In our study, we detected expression level of DNMTs by using IHC staining and found higher expression level of DNMT3B in HP group than that in NP group but no difference of DNMT1 expression level between the two groups. It suggested that high progesterone level may alter the de novo methylation through DNMT3B expression level on endometrium. Aberrant DNA methylation and gene expression of adhesion molecules emerge subsequently; it is possible that the altered DNMT3B expression level may also affect other molecules related to endometrial receptivity, more than the selected markers examined in our study.
CpG hypermethylation of CDH1 promoter is an important molecular mechanism in the transcriptional inactivation of CDH1 which has been reported in tumors of epithelial origin [25]. Some studies demonstrated that there was aberrant methylation of the CDH1 promoter region in the endometrium of women with uterine fibroids [26], whereas the methylation status of CTNNB1 in endometrium has not been addressed before. In our study, CpG hypermethylation of both CDH1and CTNNB1 promoter was found in endometrium from women with high progesterone on the day of hCG administration, and the mRNA expression level of CDH1 and CTNNB1 was negatively correlated with DNA methylation, suggesting that high progesterone may regulate CDH1 and CTNNB1 expression through DNA methylation. It has been reported that MUC1 gene expression is regulated by DNA methylation and histone H3 lysine 9 (H3-K9) modification of the promoter region in cancer cell line [27]. In our study, the expression level of MUC1 in HP group was significantly lower than that in NP group, while no significant difference was found upon the DNA methylation of MUC1 in HP group and NP group. It suggested that the altered MUC1 gene expression under high progesterone may not be regulated by DNA methylation but may be due to other epigenetic mechanism, such as histone modification. In our previous study, we demonstrated that the expression level of H3K9me2 in luminal and glandular epithelium in HP group was higher than that in NP group [17], which may be associated with the altered expression level of MUC1 through histone modification. Further studies, such as ChIP assays and Western blot, are needed to confirm whether H3K9me2 binds to MUC1 gene and thus regulates its expression level alternatively.
Interestingly, our study found that the expression level of MUC1 was high in endometrium during implantation window. MUC1 is the first molecule that blastocyst encounters when attaching to endometrium [16]. On luminal epithelium, MUC1 forms a barrier protecting the mucosa from infection and, at the same time, protecting blastocyst from attaching until the appropriate time and location for implantation [15]. Based on this mechanism, MUC1 was expected to be downregulated during window of implantation. However, MUC1 in both mRNA and protein levels is significantly upregulated from the proliferative to the mid-secretory phase of menstrual cycle [28]. Therefore, a locally acting mechanism of MUC1 in endometrium was put forward [29, 30], in which human blastocysts produce a specific, highly localized downregulation/cleavage of MUC1 at attachment sites, which can explain the paradoxical phenomenon of MUC1. This mechanism was also confirmed by an in vitro study, in which MUC1 was lost locally at the site of implantation while no change was found in non-implantation site [31].
Successful implantation requires a receptive endometrium, an embryo with implantation competency, and a synchronized dialogue between maternal and embryonic tissues [32], in which the endometrium adhesion molecules play a key role. In our study, the aberrant methylation and downregulation of adhesion molecules on endometrium by high progesterone during implantation window may explain the impaired endometrial receptivity and implantation failure in women affected by high progesterone level on the day of hCG administration in IVF cycles. Although the role of epigenetic alterations on pregnancy outcome in IVF cycles remains to be defined, our present data shed light on the association between epigenetic regulation and the expression of adhesion molecules on endometrium. In the future, method to correct the epigenetic status in endometrium may be able to improve the implantation rate in those women with high progesterone on the day of hCG administration.
Strengths and limitations
One of the strengths of our study was that we obtained all the endometrial samples on a precisely defined day; that was the 7th day after hCG administration. Since genes and functions of endometrium change differently in even 1 or 2 days around implantation window, collecting all samples on precise time would be very essential to reduce the variation in our results. One limitation of our study was that the whole endometrium biopsy was used for detecting the mRNA expression level and DNA methylation level of endometrium receptivity markers. Previous study and our IHC staining confirmed that MUC1, CDH1, and CTNNB1 were only expressed on epithelial cells. Laser capture microdissection to isolate the epithelial cells should provide a more precise and accurate expression and methylation data though. However, the cell preparation will affect the quality of the results, and large amount of cells is required for the assay; therefore both sorting and microdissection may not be very practical for the purpose. Before knowing which cells were hypermethylated or hypomethylated, other more advanced technology such as single cell methylation sequencing is still under development. Due to limitations of current technology, in this study, we first tested the overall methylation status in the tissue level and then check the expression in both tissue and cell levels. Another limitation of our study was the small sample size for detecting DNA methylation, especially MUC1 in which the number was 5 for NP and HP group separately.
Conclusions
In conclusion, our study demonstrated a significantly altered DNA methylation and gene expression of adhesion molecules on endometrium under high serum progesterone level on the day of hCG administration in IVF cycle. The findings provide an insight into an epigenetic mechanism of high progesterone detrimentally influence on the endometrium during implantation window in IVF cycles.
Electronic supplementary material
(XLSX 14.2 kb)
(XLSX 15.2 kb)
Acknowledgements
This work was supported by the Direct Grant for Research 2016/2017 (2016.058) to Y.X., H.X., and C C.W and Shenzhen Science Project Funding JCYJ20160427113429186 to T.Z. and Postdoctoral International Exchange Program to Y.X.
Compliance with ethical standards
Conflicts of interest
The authors declare that they have no conflict(s) of interest.
Footnotes
The paper was selected to present at the 34th annual meeting, European Society of Human Reproduction and Embryology, Barcelona, Spain, July 1–4, 2018.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yingpu Sun, Email: syp2008@vip.sina.com.
Chi Chiu Wang, Email: ccwang@cuhk.edu.hk.
References
- 1.Schoolcraft W, Sinton E, Schlenker T, Huynh D, Hamilton F, Meldrum DR. Lower pregnancy rate with premature luteinization during pituitary suppression with leuprolide acetate. Fertil Steril. 1991;55(3):563–566. doi: 10.1016/S0015-0282(16)54186-7. [DOI] [PubMed] [Google Scholar]
- 2.Venetis CA, Kolibianakis EM, Papanikolaou E, Bontis J, Devroey P, Tarlatzis BC. Is progesterone elevation on the day of human chorionic gonadotropin administration associated with the probability of pregnancy in in vitro fertilization? A systematic review and meta-analysis. Hum Reprod Update. 2007;13(4):343–355. doi: 10.1093/humupd/dmm007. [DOI] [PubMed] [Google Scholar]
- 3.Bosch E, Labarta E, Crespo J, Simon C, Remohi J, Jenkins J, et al. Circulating progesterone levels and ongoing pregnancy rates in controlled ovarian stimulation cycles for in vitro fertilization: analysis of over 4000 cycles. Hum Reprod. 2010;25(8):2092–2100. doi: 10.1093/humrep/deq125. [DOI] [PubMed] [Google Scholar]
- 4.Venetis CA, Kolibianakis EM, Bosdou JK, Tarlatzis BC. Progesterone elevation and probability of pregnancy after IVF: a systematic review and meta-analysis of over 60 000 cycles. Hum Reprod Update. 2013;19(5):433–457. doi: 10.1093/humupd/dmt014. [DOI] [PubMed] [Google Scholar]
- 5.Kolibianakis EM, Venetis CA, Bontis J, Tarlatzis BC. Significantly lower pregnancy rates in the presence of progesterone elevation in patients treated with GnRH antagonists and gonadotropins: a systematic review and meta-analysis. Curr Pharm Biotechnol. 2012;13(3):464–470. doi: 10.2174/138920112799361927. [DOI] [PubMed] [Google Scholar]
- 6.Huang CC, Lien YR, Chen HF, Chen MJ, Shieh CJ, Yao YL, et al. The duration of pre-ovulatory serum progesterone elevation before hCG administration affects the outcome of IVF/ICSI cycles. Hum Reprod. 2012;27(7):2036–2045. doi: 10.1093/humrep/des141. [DOI] [PubMed] [Google Scholar]
- 7.Kilicdag EB, Haydardedeoglu B, Cok T, Hacivelioglu SO, Bagis T. Premature progesterone elevation impairs implantation and live birth rates in GnRH-agonist IVF/ICSI cycles. Arch Gynecol Obstet. 2010;281(4):747–752. doi: 10.1007/s00404-009-1248-0. [DOI] [PubMed] [Google Scholar]
- 8.Legro RS, Ary BA, Paulson RJ, Stanczyk FZ, Sauer MV. Premature luteinization as detected by elevated serum progesterone is associated with a higher pregnancy rate in donor oocyte in-vitro fertilization. Hum Reprod. 1993;8(9):1506–1511. doi: 10.1093/oxfordjournals.humrep.a138288. [DOI] [PubMed] [Google Scholar]
- 9.Labarta E, Martinez-Conejero JA, Alama P, Horcajadas JA, Pellicer A, Simon C, et al. Endometrial receptivity is affected in women with high circulating progesterone levels at the end of the follicular phase: a functional genomics analysis. Hum Reprod. 2011;26(7):1813–1825. doi: 10.1093/humrep/der126. [DOI] [PubMed] [Google Scholar]
- 10.Van Vaerenbergh I, Fatemi HM, Blockeel C, Van Lommel L, In't Veld P, Schuit F, et al. Progesterone rise on HCG day in GnRH antagonist/rFSH stimulated cycles affects endometrial gene expression. Reprod BioMed Online. 2011;22(3):263–271. doi: 10.1016/j.rbmo.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 11.Haouzi D, Bissonnette L, Gala A, Assou S, Entezami F, Perrochia H, Dechaud H, Hugues JN, Hamamah S. Endometrial receptivity profile in patients with premature progesterone elevation on the day of HCG administration. Biomed Res Int. 2014;2014:951937. doi: 10.1155/2014/951937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li R, Qiao J, Wang L, Li L, Zhen X, Liu P, Zheng X. MicroRNA array and microarray evaluation of endometrial receptivity in patients with high serum progesterone levels on the day of hCG administration. Reprod Biol Endocrinol. 2011;9:29. doi: 10.1186/1477-7827-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seppala M, Taylor RN, Koistinen H, Koistinen R, Milgrom E. Glycodelin: a major lipocalin protein of the reproductive axis with diverse actions in cell recognition and differentiation. Endocr Rev. 2002;23(4):401–430. doi: 10.1210/er.2001-0026. [DOI] [PubMed] [Google Scholar]
- 14.Lessey BA. Assessment of endometrial receptivity. Fertil Steril. 2011;96(3):522–529. doi: 10.1016/j.fertnstert.2011.07.1095. [DOI] [PubMed] [Google Scholar]
- 15.Brayman M, Thathiah A, Carson DD. MUC1: a multifunctional cell surface component of reproductive tissue epithelia. Reprod Biol Endocrinol. 2004;2:4. doi: 10.1186/1477-7827-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Achache H, Revel A. Endometrial receptivity markers, the journey to successful embryo implantation. Hum Reprod Update. 2006;12(6):731–746. doi: 10.1093/humupd/dml004. [DOI] [PubMed] [Google Scholar]
- 17.Xiong Y, Wang J, Liu L, Chen X, Xu H, Li TC, et al. Effects of high progesterone level on the day of human chorionic gonadotropin administration in in vitro fertilization cycles on epigenetic modification of endometrium in the peri-implantation period. Fertil Steril. 2017;108(2):269–76.e1. doi: 10.1016/j.fertnstert.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Ziller MJ, Muller F, Liao J, Zhang Y, Gu H, Bock C, et al. Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLoS Genet. 2011;7(12):e1002389. doi: 10.1371/journal.pgen.1002389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14(3):204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 20.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/S0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 21.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69(6):915–926. doi: 10.1016/0092-8674(92)90611-F. [DOI] [PubMed] [Google Scholar]
- 22.Liu L, Zhao L, Li TC, Zhu H, Lin X, Jin X, Tong X, Zhang S. Comparison of progesterone measurement on day of, and day after, HCG administration in IVF-embryo transfer cycles. Reprod BioMed Online. 2015;30(2):157–165. doi: 10.1016/j.rbmo.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 23.Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330(6004):622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeltsch A, Jurkowska RZ. New concepts in DNA methylation. Trends Biochem Sci. 2014;39(7):310–318. doi: 10.1016/j.tibs.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 25.Grady WM, Willis J, Guilford PJ, Dunbier AK, Toro TT, Lynch H, Wiesner G, Ferguson K, Eng C, Park JG, Kim SJ, Markowitz S. Methylation of the CDH1 promoter as the second genetic hit in hereditary diffuse gastric cancer. Nat Genet. 2000;26(1):16–17. doi: 10.1038/79120. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Ran R, Guan Y, Zhu X, Kang S. Aberrant methylation of the E-cadherin gene promoter region in the endometrium of women with uterine fibroids. Reprod Sci. 2016;23(8):1096–1102. doi: 10.1177/1933719116630415. [DOI] [PubMed] [Google Scholar]
- 27.Yamada N, Nishida Y, Tsutsumida H, Hamada T, Goto M, Higashi M, Nomoto M, Yonezawa S. MUC1 expression is regulated by DNA methylation and histone H3 lysine 9 modification in cancer cells. Cancer Res. 2008;68(8):2708–2716. doi: 10.1158/0008-5472.can-07-6844. [DOI] [PubMed] [Google Scholar]
- 28.Hey NA, Graham RA, Seif MW, Aplin JD. The polymorphic epithelial mucin MUC1 in human endometrium is regulated with maximal expression in the implantation phase. J Clin Endocrinol Metab. 1994;78(2):337–342. doi: 10.1210/jcem.78.2.8106621. [DOI] [PubMed] [Google Scholar]
- 29.Thathiah A, Carson DD. MT1-MMP mediates MUC1 shedding independent of TACE/ADAM17. Biochem J. 2004;382(Pt 1):363–373. doi: 10.1042/bj20040513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meseguer M, Aplin JD, Caballero-Campo P, O'Connor JE, Martin JC, Remohi J, et al. Human endometrial mucin MUC1 is up-regulated by progesterone and down-regulated in vitro by the human blastocyst. Biol Reprod. 2001;64(2):590–601. doi: 10.1095/biolreprod64.2.590. [DOI] [PubMed] [Google Scholar]
- 31.Hoffman LH, Olson GE, Carson DD, Chilton BS. Progesterone and implanting blastocysts regulate Muc1 expression in rabbit uterine epithelium. Endocrinology. 1998;139(1):266–271. doi: 10.1210/endo.139.1.5750. [DOI] [PubMed] [Google Scholar]
- 32.Simon C, Martin JC, Pellicer A. Paracrine regulators of implantation. Baillieres Best Pract Res Clin Obstet Gynaecol. 2000;14(5):815–826. doi: 10.1053/beog.2000.0121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX 14.2 kb)
(XLSX 15.2 kb)