Abstract
Polycomb repressive complex 2 (PRC2) is a key epigenetic multiprotein complex involved in the regulation of gene expression in metazoans. PRC2 is formed by a tetrameric core that endows the complex with histone methyltransferase activity, allowing it to mono-, di- and tri-methylate histone H3 on lysine 27 (H3K27me1/2/3); H3K27me3 is a hallmark of facultative heterochromatin. The core complex of PRC2 is bound by several associated factors that are responsible for modulating its targeting specificity and enzymatic activity. Depletion and/or mutation of the subunits of this complex can result in severe developmental defects, or even lethality. Furthermore, mutations of these proteins in somatic cells can be drivers of tumorigenesis, by altering the transcriptional regulation of key tumour suppressors or oncogenes. In this review, we present the latest results from structural studies that have characterised PRC2 composition and function. We compare this information with data and literature for both gain-of function and loss-of-function missense mutations in cancers to provide an overview of the impact of these mutations on PRC2 activity.
Subject terms: Epigenetics, Cancer
Background
Transcriptional diversity is one of the hallmarks of cellular identity. It is largely regulated at the level of chromatin, where different protein complexes act as initiators, enhancers and/or repressors of transcription. Among these complexes, are epigenetic modifiers, which are able to catalyse post-translational modifications (PTMs)—such as methylation, acetylation, phosphorylation or ubiquitination—of histone proteins. These modifications can influence gene expression by modulating chromatin accessibility, its interaction with other proteins and its three-dimensional organisation. The Polycomb group (PcG) proteins form histone-modifying complexes whose activity is associated with transcriptional silencing of facultative heterochromatin.1–4 Two catalytically distinct complexes can be distinguished: the Polycomb repressive complex (PRC) 1, and PRC2. PRC1 catalyses the mono-ubiquitination of lysine 119 on histone H2A (H2AK119ub),5,6 whereas PRC2 catalyses the mono-, di- and tri-methylation of lysine 27 on the histone H3 tail (H3K27me1/2/3).7 In mice and humans, PRC2 is essential for proper embryonic stem cell (ESC) fate specification, as it regulates the expression of key developmental genes.8–10 Indeed, depletion of PRC2 subunits leads to severe developmental defects with early embryonic or perinatal lethality.10–17 Mutations and/or dysregulation of PcG genes are found in several cancer types, especially haematological ones,18,19 as well as in rare genetic diseases associated with overgrowth, such as Weaver syndrome.20 These alterations can affect PRC2 recruitment and enzymatic activity, leading to changes in the expression of tumour suppressors or oncogenes.
The PRC2 core comprises three stoichiometric factors: enhancer of Zeste (EZH)1 or EZH2, which has a SET domain and is the catalytic subunit of the complex;7,21 suppressor of Zeste (SUZ) 12; and embryonic ectoderm development (EED) (Table 1). These three proteins form the minimal core that confers histone methyltransferase (HMT) activity. A fourth factor, retinoblastoma-binding protein (RBBP)4/7 (also known as RBAP48/46), has a slightly lower stoichiometry and is dispensable for the enzymatic activity of the complex.22,23 Although there is very little diversity in PRC2 core components, a large number of facultative subunits have been shown to bind PRC2 in a sub-stoichiometric and cell-type specific manner,24–26 adding both to the complexity of recruitment of this complex to chromatin and to additional possibilities of regulation of its enzymatic activity.27 Studies carried out over the past 5 years have shown that many of these facultative subunits bind in a mutually exclusive manner, giving rise to two versions of the PRC2 complex. The first variant (PRC2.1) comprises one of three Polycomb-like (PCL) proteins (PCL1/2/3, also named PHF1, MTF2 and PHF19, respectively) as well as Elongin BC and Polycomb repressive complex 2-associated protein (EPOP) or PRC2-associated LCOR isoform 1 (PALI1/2), while the other variant (PRC2.2) comprises Jumonji and AT-rich interaction domain 2 (JARID2) and adipocyte enhancer-binding protein 2 (AEBP2),26,28–30 in addition to the core components.
Table 1.
Domain composition of PRC2 subunits
| Protein | Name | Acronym |
|---|---|---|
| SUZ12 | Zn finger binding domain | ZnB |
| WD-domain binding 1 | WDB1 | |
| C2 domain | C2 | |
| Zn Finger | Zn | |
| WD-domain binding 2 | WDB2 | |
| VRN2-EMF2-FIS2-Su(z)12 box | VEFS | |
| EZH2 | SANT1L-binding domain | SBD |
| EED-binding domain | EBD | |
| β-addition motif | BAM | |
| SET activation loop | SAL | |
| stimulation-responsive motif | SRM | |
| Swi3, Ada2, N-CoR and TFIIIB DNA-binding domain 1 like | SANT1 | |
| Motif connecting SANT1 and SANT2 | MCSS | |
| SANT2-like | SANT2 | |
| CXC domain | CXC | |
| Su(var)3-9, E(z) and Trx domain | SET | |
| Post-SET | Post-SET | |
| EED | WD-repeat region | WD1 |
| WD2 | ||
| WD3 | ||
| WD4 | ||
| WD5 | ||
| WD6 | ||
| WD7 | ||
| PALI1 | Nuclear receptor binding box | NR |
| CTBP binding motifs (x2) | CTBP | |
| G9A interaction region | ||
| Pali interaction with PRC2 domain | PIP | |
| RBBP4 | WD-repeat region | WD1 |
| WD2 | ||
| WD3 | ||
| WD4 | ||
| WD5 | ||
| WD6 | ||
| WD7 | ||
| RBBP7 | WD-repeat region | WD1 |
| WD2 | ||
| WD3 | ||
| WD4 | ||
| WD5 | ||
| WD6 | ||
| WD7 | ||
| PHF1/PCL1 | Tudor domain | Tudor |
| PHD Domain | PHD1 | |
| PHD Domain | PHD2 | |
| Extended Homology domain | EH | |
| Chromo domain | Chromo | |
| MTF2/PCL2 | Tudor domain | Tudor |
| PHD Domain | PHD1 | |
| PHD Domain | PHD2 | |
| Extended Homology domain | EH | |
| Chromo domain | Chromo | |
| PHF19/PCL3 | Tudor domain | Tudor |
| PHD Domain | PHD1 | |
| PHD Domain | PHD2 | |
| Extended Homology domain | EH | |
| Chromo domain | Chromo | |
| EPOP | ELOBC binding box | BC box |
| C-terminal region | CTR | |
| AEBP2 | Zn finger | Zn1 |
| Zn finger | Zn2 | |
| Zn finger | Zn3 | |
| Lysine/Arginine-rich domain | KR | |
| C2 binding domain | C2B | |
| H3K4 displacement domain | H3K4D | |
| JARID2 | Transrepression domain | TR |
| Ezh1/2-binding domain | ||
| Nucleosome interaction domain | ||
| Jumonji N-term | JmjN | |
| AT-rich interaction domain | ARID | |
| Jumonji C-term | JmjC | |
| Zinc finger | ZF |
Along with the interest in characterising the functional role of accessory factors in regulating PRC2 activity, effort has also been put into trying to gain structural insights into the complexity of PRC2 and its subtypes. In this review, we discuss the latest findings regarding the PRC2 structure, focusing on the aspects that define the formation of different complex subtypes, its chromatin targeting and its enzymatic activity. Finally, building on all the current structural knowledge, we highlight the potential effect of PRC2 mutations on complex integrity and activity, and the role of mutations of PRC2 components in cancer.
Structural basis for PRC2 complex formation and function
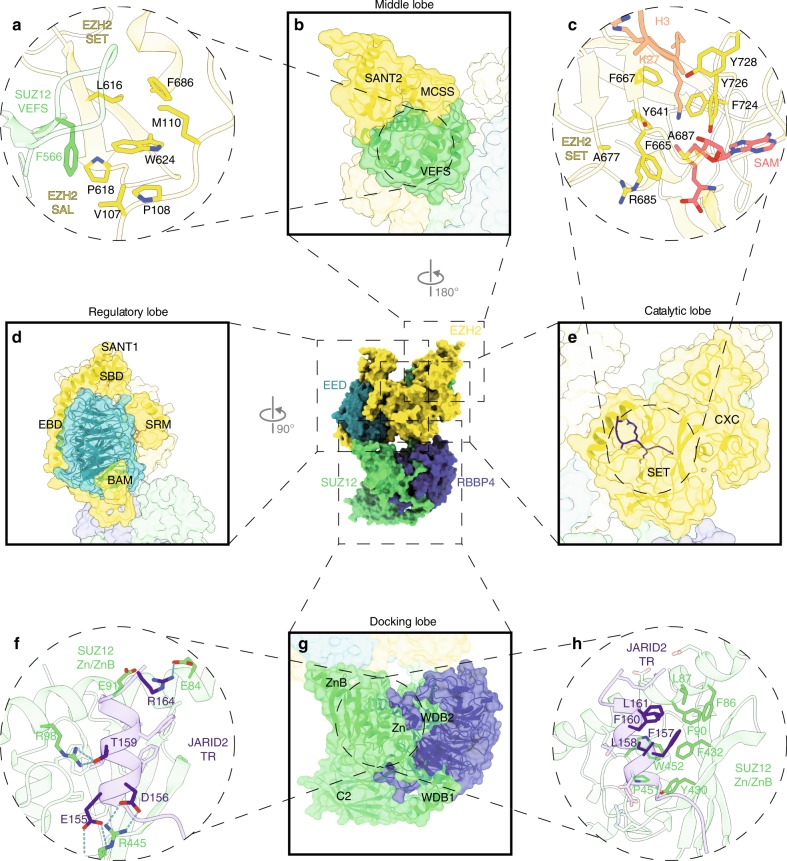
The association of the trimeric core (EZH2, SUZ12 and EED) with RBBP4 results in a stable, four-lobed structure (Fig. 1)27,28 that comes together to mediate HMT activity.
Fig. 1.
Four-lobed structure of the PRC2 core, comprising EZH2, SUZ12, EED and RBBP4 (PDB: 5WAI and 6C23). a The middle lobe is essential for PRC2 histone methyltransferase activity. The GWG motif of EZH2 (W624) is stabilised by a hydrophobic pocket at the interface between EZH2 (SAL/SET) and SUZ12 (VEFS) (PDB: 6C23). b The middle lobe extends to the back of the catalytic lobe (e), bridging it with the regulatory lobe (d). c PRC2’s HMT activity resides in the SET domain of EZH2. The lysine substrate (K27) is accommodated in a hydrophobic pocket that goes into the cofactor (SAM) binding pocket (PDB: 5TQR and 6C23). d EZH2 wraps around the EED WD propeller. Regulatory contacts occur at the open end of the propeller and are transmitted to the catalytic lobe (e) by the SRM domain of EZH2. e The catalytic lobe is formed by the SET and the CXC domains of EZH2. f–h The docking lobe is formed by the association of RBBP4/7 with SUZ12 N-terminus (g) and serves as a platform for the association of accessory factors. Binding of JARID2 TR domain to the SUZ12 Zn/ZnB pocket involves hydrogen bonds (f) as well as hydrophobic interactions (h) (PDB: 5WAI)
The catalytic lobe
The C-terminal region of EZH2, comprising the CXC domain (a cysteine-rich region) and the SET domain, forms the catalytic lobe, in which the HMT activity of PRC2 resides (Fig. 1e). The active site presents two pockets in the SET domain: the first one is a highly hydrophobic channel (Y641, F667, F724, Y726 and Y728), which accommodates the long aliphatic chain of the lysine substrate (Fig. 1c). The end of this channel is connected to a second pocket, in which the cofactor S-adenosyl methionine (SAM) is positioned in an orientation that brings its methyl group in close proximity to the ε-amino group of the lysine. Residues that lie at the interface of these two pockets (e.g. Y641, A677 and A687) are crucial for catalysis, and their mutation results in changes in affinity for the substrate that are associated with gain-of-function phenotypes (as discussed below). The assembly of the trimeric core is essential for HMT activity: in isolation, EZH2 adopts an autoinhibited conformation, with the post-SET domain (the region C-terminal to the SET domain) folded upwards into the lysine-binding cleft, blocking the substrate from engaging the active site.31–33 This mechanism, which seems to be conserved in the H3K9 methyltransferase Suv39h2,34 might provide a ‘safety catch’ against spurious histone methylation.
The regulatory lobe
The catalytic lobe is in close contact with the regulatory lobe, which is formed by the association of EED with the N-terminal domain of EZH2 (Fig. 1d). The long α-helix of the EED-binding domain (EBD) and the β-addition motif (BAM) of EZH2 wrap around the bottom (or closed end) and side, respectively, of the WD-repeat seven-bladed β-propeller of EED.35,36 This conformation is necessary to maintain EED in a stable position while leaving the opposite side of the propeller open for stimulatory ligand binding.37 The open end of the propeller faces the rest of the EZH2 N-terminal region. The SET activation loop (SAL) and the stimulation-responsive motif (SRM) of the EZH2 N-terminal region are responsible for the allosteric changes in the catalytic lobe upon trimethyl-peptide binding in the EED pocket. Indeed, although the trimeric core retains basal HMT activity, the recognition of the H3K27me3 mark further activates the complex resulting in an enhanced catalysis (discussed below). Finally, the loop around EED is closed by the association of the SANT1 domain of EZH2 with the distal part of EBD, named SANT1-binding domain (SBD).
The middle lobe
The central part of EZH2 (comprising the MCSS and SANT2 domains) and the globular VEFS domain of SUZ12 are part of a bridging middle lobe (Fig. 1b). In particular, VEFS (which is packed between the regulatory and the catalytic modules) seals together the SAL domain and the GWG loop (W624) of the SET domain via a hydrophobic pocket (SUZ12 F566; EZH2 V107, P108, M110, L616, P618 and F686) (Fig. 1a), thereby stabilising the active site.35
The docking lobe
Finally, the SUZ12 N-terminal region protrudes away from VEFS to form an extended fourth lobe that serves as a docking platform for factors that associate with PRC2 (Fig. 1g).38,39 RBBP4/7 is inserted into this lobe, its WD propeller tied by multiple interactions with the SUZ12 C2, WDB1 and WDB2 domains. The ‘cage’ built by SUZ12 around RBBP4/7 prevents the latter from binding many of its known nuclear interactors, including nucleosomes,39 thereby inhibiting its ability to sense active chromatin states. The functional relevance of incorporating an ‘inactive’ WD propeller into the PRC2 complex is still unclear.
Overall, spatial segregation of the docking lobe with respect to the rest of the complex reflects the functional separation between recruitment and catalysis, with the former mediated by accessory proteins, and the latter endowed into the other three lobes.
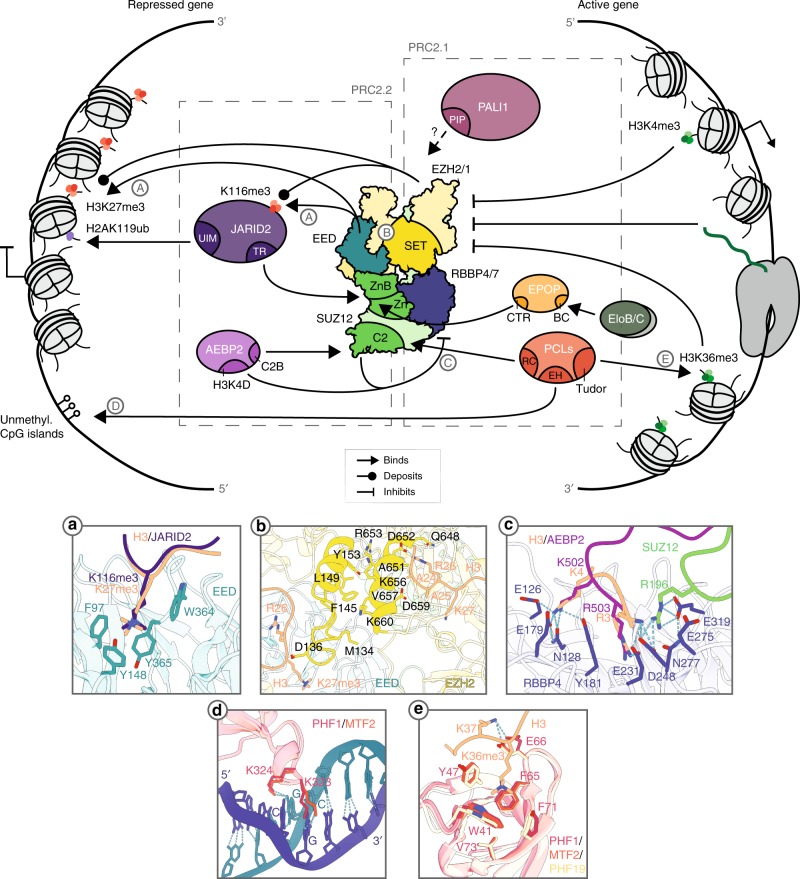
PRC2-associated factors
The interaction of facultative subunits with the PRC2 core, as well as the occurrence of mutual exclusivity, was based only on biochemical evidence until 2018, when two studies resolved the structure of the human PRC2 core complex in association with facultative subunits, in a first attempt at elucidating the structural basis for differential usage of associated factors, as well as their role in regulation of enzymatic activity39,40 These structures show that a non-canonical C2 domain in SUZ12 provides a binding platform for either the C2B domain of AEBP2 or the RC domain of PHF19 (Fig. 2). In parallel, the ZnB-Zn domain of SUZ12 functions as a docking site for the C-terminal region of EPOP as well as the transrepression (TR) domain of JARID2 (Fig. 1f, h; Fig. 2).39 These findings provided the first structural evidence for the previously observed mutual exclusiveness of the binding to PRC2 of EPOP/PCL proteins and AEBP2/JARID2. However, they still did not rule out the possibility that hybrid complexes might form containing either AEBP2–EPOP or PCL–JARID2. Indeed, the appearance of a hybrid MTF2–JARID2 complex was observed upon AEBP2 depletion in mouse ESCs, suggesting that further protein–protein interactions help dictate which pair of subunits interacts with the core complex.14
Fig. 2.
PRC2’s activity and recruitment depend on the chromatin context. a The EED aromatic cage of PRC2 is able to recognise both H3K27me3 and JARID2K116me3 (PDB: 3IIW and 6C23). b Recognition of the stimulatory ligands results in extensive interactions between the SAL, SRM and iSET domains, which results in the opening of the active site and enhanced histone methyltransferase activity (PDB: 5HYN). c Binding of the RBBP4 acidic pocket of unmodified H3K4 is inhibited in the context of the PRC2 complex (PDB: 2YBA and 5WAK). d PCL proteins specifically bind unmethylated CpG dinucleotides through their EH domain (PDB: 5XFQ and 5XFR). e The Tudor domain of PCL proteins is able to recognise the H3K36me3 mark (PDB: 4BD3, 5XFQ and 5XFR). Residues numbering in (c) and (e) is relative to PHF1
Although these structures provide crucial insight into PRC2 holocomplex formation, many PRC2-associated factors have yet to be characterised in such detail, such as the ELOB and ELOC heterodimer (also termed TCEB2 and TCEB1, respectively; hereafter referred to as ELOBC), and the PALI1/2 proteins. However, some biochemical evidence provides preliminary insights into how these factors associate both with the core complex and other accessory proteins. In mouse ESCs, ELOBC associates with PRC2 on chromatin28,29 in an interaction mediated by EPOP (depletion of EPOP disrupts the association of ELOBC with PRC2 and its subsequent binding on chromatin). Binding of ELOBC to EPOP (and, consequently, to PRC2) is mediated by the BC box motif, a classical ELOBC binding motif, on the N-terminus of EPOP. Mutation of a crucial leucine residue (L40) in this motif impairs the association of ELOBC with EPOP and PRC2.28,29 The interaction of EPOP with ELOBC is crucial for proliferation of several human cancer cell lines. Indeed, depletion of EPOP leads to a decrease in their proliferative capacity. This phenotype is dependent on EPOP interaction with ELOBC as only the wild-type version of the EPOP protein (but not the L40 mutant) is able to rescue cancer cells proliferation.29
PALI1 and 2 are vertebrate-specific subunits of the PRC2.1 complex, which associate with PRC2 through their highly conserved C-terminal PALI-interaction with PRC2 (PIP) domain (Fig. 2). Structural information about these proteins is still lacking, but two tryptophan residues (W1125 and W1186 in PALI1) conserved in both PALI proteins have been found to play an essential role in PRC2 binding, as the concomitant mutation of both residues abolishes the association of the PIP domain with PRC2 in vitro.30 Although this biochemical evidence provides crucial information about how these factors associate with PRC2, the exact molecular functions of EPOP, ELOBC or PALI1/2 within PRC2 so far remain unknown.
Functional interplay between PRC2 and chromatin
PRC2 mediates gene repression through direct interaction with its target genes and deposition of the repressive H3K27me3 mark. The ability of PRC2 to identify its target genes and correctly perform its enzymatic activity relies on a complex but tightly regulated interaction with different elements of the chromatin environment. This is mainly facilitated by the many sub-stoichiometric associated factors that provide PRC2 with the capability to bind DNA, nucleosomes, histones and RNA.
DNA and nucleosomes
In Drosophila melanogaster, PcG proteins are recruited to their target genes via Polycomb response elements (PREs), which are cis-regulatory DNA sequences that provide a platform for Polycomb subunit binding and subsequent gene repression.41,42 Although Polycomb proteins are well conserved between flies and mammals (reviewed in 43), no PRE-like specific DNA sequences have been identified to date in mammals.44
Of the core components of PRC2, both EZH2 and SUZ12 possess zinc finger domains, which could potentially mediate an interaction with DNA. However, rather than playing a major role in targeting PRC2 to chromatin, binding of the core components of the complex to DNA seems to stabilise the interaction of PRC2 with the nucleosome template and thereby facilitate catalysis; specifically, the EZH2 CXC Zn-binding clusters contact the DNA exiting from the substrate nucleosome. Additionally, the EZH2 SBD domain contacts the DNA minor groove in the H3K27me3-marked nucleosome, after it has been bound by EED. Concurrently, EED might also contact the DNA of the modified nucleosome, through an unstructured, positively charged N-terminal stretch.45 Stable interaction with the chromatin is indeed key for PRC2 stimulation: HMT activity is enhanced on di-/oligonucleosomes as compared with mononucleosomes, especially for linker DNA with lengths of 35–40 bp, allowing for engagement of both the stimulating and the substrate lysine residues.45,46 Interestingly, this feature seems to be specific for EZH2–PRC2, as EZH1–PRC2 does not display such a preference for its substrate.46
AEBP2 and JARID2 contain three and one zinc finger domains, respectively, with JARID2 also harbouring an AT-rich-interacting domain (ARID). Both proteins display DNA-binding ability in vitro47,48 and were therefore the first candidates to be considered for facilitating DNA-mediated recruitment of PRC2. AEBP2 was shown to have a binding preference for a highly degenerate consensus sequence with a bipartite structure (CTT(N)15–23cagGCC),49 suggesting that the recognition of this motif could provide a targeting mechanism for PRC2. In addition, in vitro biochemical assays showed that the presence of AEBP2 increases the capacity of PRC2 to bind nucleosomes, through its KR motif (a lysine/arginine-rich positively charged patch), resulting in enhanced HMT activity.46 Nonetheless, AEBP2 does not seem to be essential for PRC2 recruitment in vivo, as depletion of AEBP2 in mouse ESCs does not affect PRC2 occupancy on chromatin and even leads to a slight increase in H3K27me3 levels.14 For JARID2, in vitro experiments have demonstrated its ability to bind DNA, with a slight bias for GC-rich elements. This binding is mediated by the C-terminal region of JARID2, potentially through its C5HC2 zinc finger.50 Independent of its DNA-binding ability, JARID2 can also mediate the interaction with nucleosomes, resulting in enhanced PRC2 activity. This interaction is mediated by a region with no annotated structure, which spans residues 349–450.51 In line with these observations, evidence has shown that PRC2.2—but not PRC2.1—is able to bind nucleosomes in vitro.39 In agreement with a role for JARID2 in mediating PRC2 recruitment to chromatin loci, depletion of JARID2 reduces PRC2 levels at specific target genes.50,52–54
Notably, JARID2 is mutated and/or deleted in various types of leukaemia where PRC2 plays an onco-suppressor role.55,56 This suggests that JARID2-mediated PRC2 recruitment is essential for its tumour-suppressive functions.
PCL proteins are essential for PRC2 recruitment, as their depletion in mouse ESCs leads to a strong reduction in the levels of PRC2 on chromatin.57–59 In 2017, Li and colleagues59 resolved the crystal structure of human N-terminal PHF1, which contains Tudor, PHD1 and PHD2 domains as well as a newly identified extended homologous (EH) domain. The EH domain folds into three α-helices and a curved three-stranded β-sheet. The W1 loop of the winged helix structure within the EH domain (EHWH) binds to unmethylated CpG DNA motifs, but not to GpC or AT-rich elements. Binding specificity is provided by the first two lysine residues of the W1 loop (K323 and K324 in PHF1), which form extensive contacts with both cytosine and guanine nucleotides (Fig. 2d), and their mutation to alanine abolishes the DNA-binding ability of all three PCL proteins.59 This interaction with DNA increases the residence time of PRC2 on nucleosomes and on naked DNA, leading to its increased catalytic activity.60 These results provide a potential explanation for the observed correlation between PRC2-occupied sites and unmethylated CpGs observed in vivo.8,61 However, a large fraction of unmethylated CpG islands remains unbound by PRC2 in vivo, suggesting that additional features might be required for target specificity. Consistent with this, a 2018 report showed that specific DNA helical shape characteristics (a wider minor groove and a decreased propeller and helix twist) are required for MTF2 to bind to unmethylated CpG motifs,62 therefore narrowing down the potential target regions. These observations point to a more degenerate definition of vertebrate PREs that would be defined by DNA helical shape rather than specific primary sequence.
Histone post-translational modifications
Another crucial aspect of PRC2 regulation and recruitment on chromatin comes from its direct interaction with many post-translation modifications that are present on histone tails. Indeed, PRC2 activity strongly depends on the epigenetic state of its chromatin environment.
On the one hand, recognition of PRC2-deposited H3K27me3 products by EED results in catalytic stimulation of the complex: the open end of the EED WD-propeller is oriented towards the side of the catalytic lobe, thereby exposing its aromatic cage in this direction. Following the engagement of H3K27me3 with the aromatic cage (F97, Y148, Y365 and W364; Fig. 2a), the SRM domain of EZH2 makes extensive interactions with both EED and the trimethyl-peptide, passing from a disordered unstructured conformation to a fully structured α-helix shape. This conformation can now contact the iSET domain, which undergoes a counterclockwise rotation of 20°, leading to the opening of the SET substrate-binding cleft (Fig. 2b).35,37,63,64 These dynamics result in 4–6-fold enhanced substrate binding35 and 3–7-fold increased catalytic activity.37,65 Interestingly, H3K27me3 is not the only epigenetic mark that can be recognised by EED, as other repressive chromatin marks (e.g. H1K26me3, H3K9me3, H4K20me3)—but not active marks (e.g. H3K4me3, H3K36me3, H3K79me3)—are bound with comparable affinity. However, H3K27me3 is the only mark that is able to elicit PRC2 catalytic stimulation.37,65 Although it is tempting to speculate that EED-mediated recognition of repressive histone marks could serve as a mechanism for recruitment of PRC2 to silenced loci, a large body of evidence has shown that stimulation of PRC2 catalytic activity and its recruitment on chromatin are completely uncoupled.64,66–71 Rather, this mechanism accounts for a positive-feedback loop: upon PRC2 deposition of the H3K27me3 mark through its basal level of activity, methylation of the neighbouring nucleosome is favoured, resulting in spreading of the histone mark along the linear chromatin fibre as well as along the regions that are in spatial proximity.72
In addition to this, PRC2 is also able to self-stimulate via trimethylation of JARID2 at lysine 116 (JARID2K116me3) (Fig. 2a). This modification mimics that of H3K27me3 and is able to elicit the same structural rearrangement,73 but the physiological relevance of this mechanism is not yet clear. However, as JARID2K116me3 activates PRC2 catalysis regardless of the presence of the H3K27me3 mark, this mechanism could account for the deposition of the mark on unmodified nucleosomes. In other words, the incorporation of JARID2 in the complex could serve as a mechanism to confer PRC2 with a pioneering function that can initiate silencing.
Besides its own marks, PRC2 also recognises marks set by PRC1. Indeed, although H3K27me3 deposited by PRC2 is known to provide a docking site for PRC1 through its recognition by CBX proteins,74 this inter-complex crosstalk appears to be bidirectional, as PRC2 can bind the H2AK119ub mark deposited by PRC1.75 This binding is specifically mediated by an ubiquitin-interaction motif (UIM) on the N-terminus of JARID2 and is essential for PRC2 recruitment to H2Aub119-marked regions (Fig. 2).76 This creates a positive-feedback loop between PRC2 and PRC1 that keeps genes from exiting this repressed state.
On the other hand, however, histone post-translation modifications associated with an active transcriptional state (e.g. H3K4me3 and H3K36me2/3) have the opposite effect, strongly inhibiting PRC2 di- and tri-methylation activity.77,78 Initial studies focusing on the role of the nucleosome-binding proteins RBBP4/779–81 revealed that the ability of these proteins to bind histone tails is inhibited by the presence of H3K4me3,82 leading to speculation that these proteins could serve as a sensor for active chromatin states. Although appealing, this model was soon rejected, as H3K4me3-dependent PRC2 inhibition is independent of RBBP4 inclusion in the complex: a minimal EZH2–EED–SUZ12(VEFS) complex retains both HMT activity and inhibition by H3K4me3/H3K36me2/3.77 Two studies published in 2018 have helped to explain how RBBP4 progressively loses its nucleosome-binding capacity in the context of the human PRC2 complex: distinct residues from SUZ12 (C2 R196) and AEBP2 (H3K4D, K502 and R503) directly compete with the H3 tail for binding to the RBBP4/7 acidic pocket (comprising E126, N128, E179, Y181, E231, D248, E275, N277 and E319) (Fig. 2c). Moreover, PRC2 appears to engage dinucleosomes in an orientation that is incompatible with the interaction of RBBP4/7 with the H3 tail.45 Finally, SUZ12 (WDB1, WDB2) wrapping around RBBP4/7 eventually imposes steric hindrance to the interactions of RBBP4/7 with histone H4.39
An alternative hypothesis has therefore been put forward by two reports showing that EZH2 is able to sense the modification state of both H3K36 and H4K16/20 via two independent pockets located in its CXC/SET and SANT1 domains, respectively.83,84 Although the functional relevance of H4 binding is still unclear, Jani and colleagues83 have shown that unmodified H3K36 (but not H3K36me2/3) can increase the catalytic activity of EZH2, providing a rationale for the lower PRC2 activity in active chromatin environments.
As well as the PRC2 core components, the PCL family of proteins has long been known to interact directly with the H3K36me2/3 mark. PHF1, PHF19 and MTF2 can all bind the H3K36me2/3 mark through their aromatic cage (W41, Y47, F65, F71 and V73 in human PHF1) located in their highly conserved Tudor domain (Fig. 2e).57–59,85–87 Interactions with these histone marks are abolished when any of these residues is mutated to an alanine.57,85 The significance of this interaction for PRC2 recruitment remains unclear, considering the limited overlap between the genomic localisation of H3K36me3 and H3K27me3 on chromatin in mouse ESCs.57 One of the proposed mechanisms is that PCL-containing PRC2 transiently binds to H3K36me3, together with a H3K36me2/3-specific demethylase (such as KDM2B or NO66), in order to establish de novo H3K27me3 domains and to impose gene silencing during differentiation.57,58 Thus, while histone post-translation modifications associated with active transcription could mediate negative feedback on EZH2 resulting in inhibition of PRC2 activity, an intriguing possibility is that incorporation of PCL proteins into PRC2.1 confers on the complex the ability to initiate the silencing of actively transcribed regions.
However, many genomic regions in mouse ESCs show co-localisation of the H3K36me2 and H3K27me2 marks: in this scenario, the H3K36me2 mark, deposited by NSD1, prevents the spreading of H3K27me3.88 Notably, H3K36me2/3 decorated regions seem to be devoid of PRC2,88,89 suggesting that H3K27me2 deposition at these loci is the result of a transient interaction. Whether PCL proteins play a role in this mechanism is not clear yet.
RNA
Along with the many chromatin features that regulate PRC2 recruitment and activity, other studies suggest that RNA might play a role in both of these two processes: despite the lack of a known RNA-binding domain, PRC2 binds RNA in vivo and in vitro in a non-sequence-specific manner, although it does show a preference for GC-rich sequences and G-quadruplexes.90–92 Moreover, direct binding to RNA results in inhibition of PRC2 HMT activity both in vitro90,93 and in vivo,94–96 probably due to competition with chromatin binding (Fig. 2)95,97 (for a comprehensive discussion please refer to98,99).
However, the molecular mechanisms of binding and of the resulting inhibition are not yet clear. To address these questions, Zhang and colleagues96 used a targeted proteomic identification of RNA-binding regions (RBR-ID) approach to map, at high resolution, the interactions within the nucleus between PRC2 components and RNA. Their results showed that binding involves neither a single subunit of the complex nor discrete RNA-binding domains within them but, rather, dispersed amino acid patches on the exposed surface of the complex;96 this is in line with previous in vitro observations by hydrogen deuterium exchange mass spectrometry.100 Interestingly, RNA interactions were observed for all PRC2 subunits (that is, all core and accessory factors belonging to PRC2.1 and PRC2.2). However, the most consistent points of interaction were at regions of EZH2 at the interface with EED (namely, the SBD, EBD, SRM and iSET domains), leading the authors to speculate that inhibition of PRC2 HMT activity by RNA could be, at least partially, due to interference with transduction of the stimulatory signal.96 These observations suggest that the inhibition of PRC2 activity in active chromatin environments is mediated not only at the epigenetic level (through negative regulation by H3K4me3 and H3K36me2/3) but also by RNA itself, as the final output of actively transcribed regions.
Mutations in PRC2 components and their roles in cancer
Early observations demonstrated that EZH2 was overexpressed in prostate tumours and played a role in the progression of this cancer type.101 Since then, many reports have shown that other PRC2 factors are subject to deletions, mutations, translocations and/or dysregulation in different cancer types. In the majority of cancer types, high expression of PRC2 factors correlates with sustained proliferation of cancer cells.102,103 However, whether this reflects a causative role for PRC2 in tumorigenesis is difficult to assess. In some cases, PRC2 has been shown to repress key tumour-suppressor genes, therefore directly promoting tumour progression.18,104,105 Accordingly, a lot of effort is being put in developing targeting strategies to inhibit the activity of PRC2 in order to treat various cancer types (for a comprehensive review on PRC2 inhibition strategies see 19,106,107).
PRC2 genes, both core and associated factors, are mostly mutated in haematological malignancies. This is in line with their role in haematopoiesis, where they regulate crucial processes such as hematopoietic stem cells self-renewal and B-cells germinal centre formation.19 In haematological malignancies, contrary to most other cancer types, PRC2 can exhibit both oncogenic and tumour-suppressor functions, depending on the cellular context.19,105
SET domain loss-of-function mutations
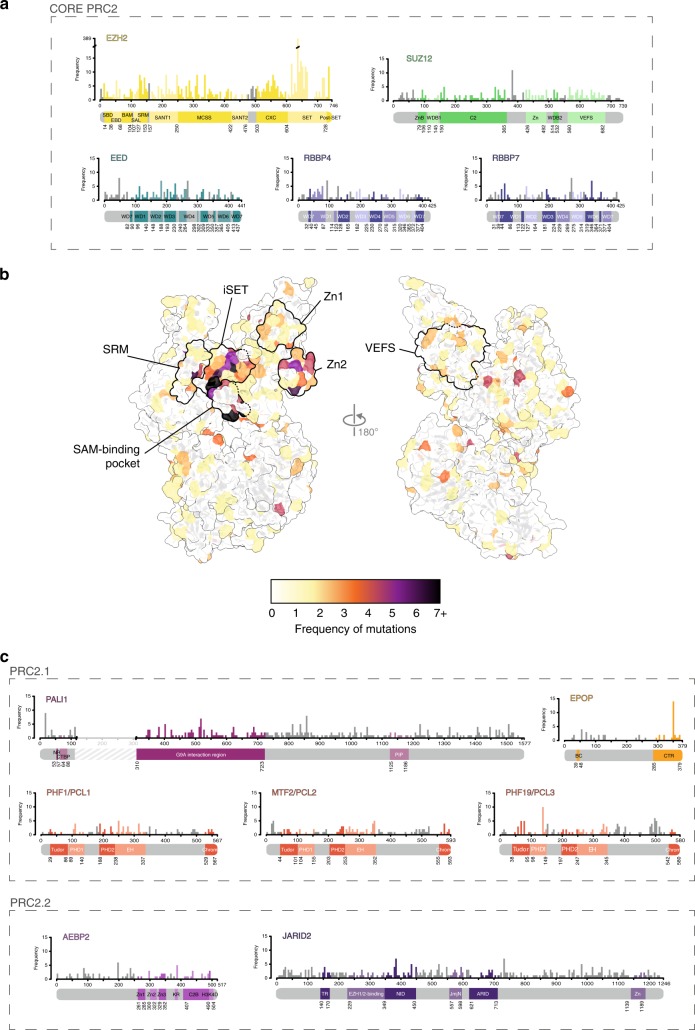
Among all PRC2 factors, EZH2 displays the highest rate of mutations (data from the COSMIC database)108 (Fig. 3a–c), in line with its essential role as the catalytic subunit of the complex. Indeed, most of the mutations in this protein occur in the CXC and SET domains, which are required for HMT activity. In particular, in the SET domain, hotspots of mutations are localised on residues that form part of the active site, binding both the substrate and the SAM cofactor (Fig. 3a, b).
Fig. 3.
The PRC2 cancer mutational landscape. a Missense mutation frequency mapped on the linear structure of PRC2 core components (bin = 5 residues). b Structural mapping of missense mutations of PRC2 genes in cancer. Numbers were obtained from the Catalogue Of Somatic Mutations In Cancer (COSMIC) database108 and mapped on a composite structural model of PRC2, comprising core components with the addition of JARID2 TR domain and AEBP2 C-terminus (PDB: 5WAI and 6C23). c Missense mutation frequency mapped on the linear structure of PRC2-associated factors (bin = 5 residues)
As previously discussed, the lysine substrate is accommodated in an aromatic cage (Fig. 1c), and mutations of the hydrophobic residues involved in this structure (F665C/L/S, F667L, Y726C/D/N/H and Y728H) are mostly found in myeloid malignancies, mainly in acute myeloid leukaemia (AML), chronic myeloid leukaemia (CML) and myelodysplastic syndrome (MDS).109–116 Disruption of one of these residues could result in impaired substrate recognition and, possibly, loss of methyltransferase activity, as demonstrated for the Y726D mutant.113 The R685 residue, which lies just below the bottom of the hydrophobic pocket, seems to be essential for the correct orientation of F665, and its mutation to cytosine abolishes methyltransferase activity in vitro.113,117 Indeed, this amino acid is found mutated (R685C/H) in AML, early T-cell precursor acute lymphoblastic leukaemia (ETP-ALL), MDS and myelofibrosis.113,115,117–124
iSET is also responsible for stabilisation of the H3 substrate, as it is sensitive to the activating signal coming from the EED–trimethyl-peptide–SRM axis. Specifically, the α-helix of iSET shapes the channel in which the H3 tail lies and is essential for enhancing PRC2 activity upon stimulus (Fig. 2b). Mutations in many of the residues that face the H3 tail (e.g. Q648, A651, D652, G655 and D659) are found in AML, MDS and T-cell acute lymphoblastic leukaemia (T-ALL).110,111,113,115,116,125–130 These mutations can either cause steric hindrance to the H3 tail (e.g. G655R) or abolish interactions between the H3 backbone and the iSET (for instance, D659V/A/G impairs the stabilisation of the two alanine residues preceding the lysine substrate). However, to our knowledge, the impact of these mutations on the HMT activity or stimulation of PRC2 has not been assessed.
With respect to the SAM-binding cleft, residues forming the walls of this channel (V621, N688, H689 and S690) are essential, and their relevance is underscored by their relatively high frequency of mutation in AML, CML, ETP-ALL and MDS110–113,115,117,122,130–134 (Fig. 3b). In particular, the mutations V621M and H689R abolish the HMT activity of PRC2, probably by impairing SAM binding and abrogating the deposition of H3K27me3 on chromatin.117,135
SET domain gain-of-function mutations
Mutations in Y641 of EZH2 have been observed to occur at a high frequency (in around 25% of patients) in patients with follicular lymphoma (FL) and diffuse large B-cell lymphoma (DLBCLs),113,136–149 as well as in those with melanoma151–152 and Ewing sarcoma.153,154 Mutations of this residue are thought to be an early driving event in cancer progression, as expression of the EZH2 Y641F mutant in mouse B-cells and melanocytes leads to the formation of lymphoma and melanoma, respectively.137,155 At the biochemical level, the EZH2 Y641F mutant shows an increased ability to modify dimethyl substrates yet is almost unable to act on nucleosomes with unmethylated or monomethylated H3K27.64,156–158 Molecularly, Y641 is positioned at the interface between the substrate lysine and the SAM cofactor, with the potential to form hydrogen bonds with the unmethylated lysine (Fig. 1c). Mutation of Y641 is thought to enlarge the binding channel, rendering the mutant form more permissive to H3K27me2 engagement but preventing this residue from interacting with the unmethylated substrate.144,159 Although initially interpreted as a loss-of-function mutation, it was later discovered that this mutation can act synergistically with the wild-type form of EZH2, as the latter is more proficient on H3K27me0/1 substrates and can provide the perfect substrate (H3K27me2) for the Y641 mutant form, resulting in overall hyper-trimethylation.137,157,158,160,161 Indeed, mutations in Y641 are exclusively found in heterozygosity. How this excess of H3K27me3 confers tumorigenicity is not well understood, but evidence points to a role for this mark in mediating the silencing of multiple tumour suppressors through changes in promoter-enhancer contacts within chromatin compartments.162
Two other gain-of-function mutations, A677G and A687V, are present in around 1–2% of patients with FL or DLBCL.142–144,156,163 The A677G mutation enlarges the lysine-binding pocket of EZH2 without affecting the interaction of Y641 with the unmodified substrate, therefore resulting in highly efficient catalysis on all methylation states and eventually leading to aberrantly high levels of H3K27me3 and lower levels of H3K27me2.35,156 On the other hand, A687V increases the efficiency of EZH2 for monomethyl substrates, by weakening the interaction with a water molecule that needs to be displaced for demethylation to happen. This results in a more balanced substrate preference with respect to wild-type EZH2, thereby increasing overall levels of H3K27me3 without affecting those of H3K27me2.164 Presumably, the resulting aberrantly high levels of H3K27me3 are responsible for the tumorigenicity of these EZH2 mutants.
CXC domain mutations
The two zinc-binding clusters inside the CXC domain (Zn3C9 and Zn3C8H1) are placed right on top of the active site and are involved in contacting DNA in the substrate nucleosome.45 These clusters accumulate a high frequency of mutations, probably leading to loss-of-function mutant isoforms that are not able to stabilise the interaction of PRC2 with the nucleosome. So far, only the C571W mutation has been found in homozygosis in myelodysplasia patients. EZH2 proteins bearing this mutation show complete loss of PRC2 HMT activity and can partially destabilise the integrity of the PRC2 complex.113
Stimulatory axis mutations (EED–SAL–SRM)
The SAL and SRM domains of EZH2 are responsible for transducing the stimulatory signal from the recognition of the H3K27me3 or JARID2K116me3 mark by EED to the iSET motif (Fig. 2b). Almost all residues involved in this process can be mutated in different types of tumour. Among these, major hotspots are represented by P132 and F145, which have been found to be mutated in patients with myelofibrosis, MDS or T-ALL111,113,116,120,121,126,165 (Fig. 3a, b). Interestingly, mutation of these residues specifically affects PRC2 stimulation but not its basal activity, complex stability or recruitment to chromatin. Moreover, mouse ESCs expressing these EZH2 mutant forms show a dramatic reduction of H3K27me2/3 levels and an impaired pluripotency capacity,64 demonstrating that stimulation of PRC2 HMT activity is key for its correct functioning in the nucleus. In line with these results, mutations in EED that affect its interaction with the SAL–SRM module (namely, S259F and R302G) lead to the very same stimulation-deficient phenotype.64 S259F and R302G mutations have been observed in T-ALL and myelofibrosis patients, respectively.123,132
VEFS domain mutations
The VEFS domain has a moderately high frequency of mutations, with a diffuse mutational pattern rather than specific hotspots (Fig. 3a, b). Many of these mutations have been experimentally validated in vitro, and indicate that even single amino acid substitutions in the VEFS domain can result in a dramatic decrease in the HMT activity of PRC2. This is the case for F603L, D605V and E610G, which have been found in patients with chronic myelomonocytic leukaemia (CMML), myelofibrosis or B-ALL.166,167 In addition to these mutations, other mutations such as W591C/R and N618Y have been observed in osteosarcoma and T-ALL130,168 and could have deleterious effects: mutations of the homologous residues in D. melanogaster Su(z)12 result in decreased HMT activity and impaired complex assembly.169 Interestingly, N618 forms H bonds with the amine and carboxyl groups of Y292 in the CXC domain, suggesting that mutation to a tyrosine residue would disrupt this interaction and thereby destabilise SUZ12’s association with EZH2.
Other hotspots
Apart from the aforementioned hotspots, only a few other residues in most of the PRC2 proteins show a higher frequency of mutation with respect to the rest. However, the functional impact of most of these mutations has not yet been addressed, in some cases due to the lack of a resolved structure. This is the case for EZH2 E740K/Q in both solid tumours and leukaemias108,115,126,146,170,171 (Fig. 3a). This residue lies in the post-SET region and its mutation could potentially affect the autoinhibition mechanism of EZH2 (see above), resulting in aberrant H3K27me3 deposition and consequently transcriptional deregulation. EPOP A350V/P mutations, which have been observed in lung and thyroid tumours,172 reside in the C-terminal region of EPOP, which is essential for its binding to PRC2 (Fig. 3c). AEBP2 A198E and R388Q have been observed in kidney and oesophagus tumours, respectively.173,174 The latter mutation, located in the positively charged patch of the KR domain, potentially affects the interaction of AEBP2 with nucleosomes and, consequently, PRC2 stimulation46 (Fig. 3c). Finally, the SUZ12 mutations E383G/K/V have been observed in oesophageal squamous cell carcinomas (SCC);174 the EED mutations R52C/H/L/P in a variety of solid tumours108,146 (Fig. 3a); and the PHF19 mutation D136A in thyroid neoplasms and upper aerodigestive tract SCC108,174 (Fig. 3c). The consequences of these latter mutations on PRC2 assembly and activity are difficult to predict as they do not involve residues of known function.
Conclusion
The PRC2 complex is a tightly regulated machine that governs the correct spatial and temporal expression of key developmental regulators via its function as a transcriptional repressor. Its activity is regulated via interactions with both facultative subunits and its chromatin environment. In general, the interaction of PRC2 with chromatin appears to be a way to safeguard the different chromatin states. Indeed, PRC2 is able to perpetuate a repressive state in a number of ways: first, by binding its own H3K27me3 mark; second, by binding the H2A119ub mark deposited by PRC1; and, third, by trimethylating and binding to JARID2K116. All of these interactions increase PRC2 activity, either by increasing residence time on chromatin (by PHF1, for example60) or stabilising its interaction with nucleosomes through AEBP2 and JARID2.46,51 By contrast, an active chromatin state is maintained by inhibition of PRC2 activity through the inhibition of the core complex by H3K4me3 and H3K36me3, and by PRC2 binding to RNA, which impedes chromatin binding and/or transduction of the stimulatory signal. In other words, both chromatin states are able to maintain a positive-feedback loop through, in part, inhibition or activation of the PRC2 complex. However, in some cases, maintenance of a chromatin state can be challenged. For instance, both AEBP2 and JARID2 can mimic histone tails through their binding to PRC2. This competition between PRC2 subunits and histone tails could be a mechanism by which PRC2 autoregulates its activity to override the epigenetic context of the targeted chromatin.
Evidence points towards the idea that the catalytic modulation of human PRC2 is structurally uncoupled from its recruitment to chromatin. The minimal complex of EZH2–EED–VEFS(SUZ12) is sufficient to mediate enzymatic activity and modulate it by sensing the chromatin state, as is reflected by the higher incidence of missense mutations in the domains that mediate the catalysis and stimulation of methyltransferase activity.
Interestingly, the ability of core components to interact with chromatin features seems to be limited to modulation of the catalysis rather than recruitment to target regions. This observation is in line with the emerging view that differential recruitment of PRC2 during distinct developmental stages is achieved by incorporating accessory factors that are dynamically expressed during development. Cell-type-specific expression and partial redundancy of accessory subunits could therefore account for their lower mutation rates observed in cancer In line with this, it is worth noting that depletion of PRC2 accessory subunits only slightly affects genome-wide levels of H3K27me3. This suggests that, once the pattern is established, the core complex alone (without accessory factors) is able to maintain it, potentially through the positive-feedback mediated by the EED–SRM–iSET axis. Therefore, mutations/dysregulation of core components might be necessary to induce changes in the methylation pattern, as observed during tumorigenesis. While structural studies have greatly deepened our understanding of the function and regulation of PRC2, many unanswered questions relating to PRC2 and its role in controlling gene expression remain to be addressed. For instance, what is the role of the recently characterised associated subunits (e.g. EPOP, PALI1/2, ELONGIN B/C), for which structures with the complete core complex are still lacking? How much of our knowledge about EZH2 function and structure applies to EZH1, considering their differential response to stimulatory elements as well as their distinct mutational landscape? Furthermore, what is the impact of those mutation hotspots for which we have neither structural nor functional details? Answering these questions will help us to better understand both the physiological and the cancer-associated functions of PRC2.
Acknowledgements
We thank Claudia Vivori for figure editing, Veronica A. Raker for scientific editing of the manuscript, and Pedro Vizán for critical reading of the manuscript. We apologise to any colleagues whose work has not been cited due to space limitations.
Author contributions
All authors participated in writing, generation of figures and literature survey for this review article.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Not applicable.
Funding
The work in the Di Croce laboratory is supported by grants from the Spanish Ministry of Economy, Industry and Competitiveness (MEIC) (BFU2016-75008-P), Fundacion Vencer El Cancer (VEC), the European Regional Development Fund (FEDER), and from AGAUR. I. Mocavini is supported by an FPI fellowship. We acknowledge support of the Spanish Ministry of Economy, Industry and Competitiveness (MEIC) through the Instituto de Salud Carlos III and to the EMBL partnership; Centro de Excelencia Severo Ochoa; CERCA Programme / Generalitat de Catalunya.
Consent for publish
Not applicable.
Data availability
Not applicable.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Paul Chammas, Ivano Mocavini.
References
- 1.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science. 2002;298(Nov):1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 2.Kirmizis A. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 2004;18(Jul):1592–1605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431(Oct):873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 4.Cao R, Tsukada Y-I, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20(Dec):845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu C, Bender W, et al. Stabilization of chromatin structure by PRC1, a polycomb complex. Cell. 1999;98(Jul):37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 6.Saurin AJ, Shao Z, Erdjument-Bromage H, Tempst P, Kingston RE. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature. 2001;412(Aug):655–660. doi: 10.1038/35088096. [DOI] [PubMed] [Google Scholar]
- 7.Kuzmichev A. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16(Nov):2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441(May):349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 9.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, et al. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;125(Apr):301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell Biol. 2007;27(May):3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faust C, Schumacher A, Holdener B, Magnuson T. The eed mutation disrupts anterior mesoderm production in mice. Development. 1995;121(Feb):273–285. doi: 10.1242/dev.121.2.273. [DOI] [PubMed] [Google Scholar]
- 12.O’Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell Biol. 2001;21(Jul):4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takeuchi T, Yamazaki Y, Katoh-Fukui Y, Tsuchiya R, Kondo S, Motoyama J, et al. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev. 1995;9(May):1211–1222. doi: 10.1101/gad.9.10.1211. [DOI] [PubMed] [Google Scholar]
- 14.Grijzenhout A, Godwin J, Koseki H, Gdula MR, Szumska D, McGouran JF, et al. Functional analysis of AEBP2, a PRC2 Polycomb protein, reveals a Trithorax phenotype in embryonic development and in ESCs. Development. 2016;143(Aug):2716–2723. doi: 10.1242/dev.123935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pasini D, Bracken AP, Jensen MR, Denchi EL, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23(Oct):4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S, He F, Xiong W, Gu S, Liu H, Zhang T, et al. Polycomblike-2-deficient mice exhibit normal left–right asymmetry. Dev. Dyn. 2007;236(Mar):853–861. doi: 10.1002/dvdy.21070. [DOI] [PubMed] [Google Scholar]
- 17.Rothberg JLM, Maganti HB, Jrade H, Porter CJ, Palidwor GA, Cafariello C, et al. Mtf2-PRC2 control of canonical Wnt signaling is required for definitive erythropoiesis. Cell Discov. 2018;4(Dec):21. doi: 10.1038/s41421-018-0022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piunti A, Pasini D. Epigenetic factors in cancer development: polycomb group proteins. Futur. Oncology. 2011;7(Jan):57–75. doi: 10.2217/fon.10.157. [DOI] [PubMed] [Google Scholar]
- 19.Di Carlo, V., Mocavini, I., Di Croce, L. Polycomb complexes in normal and malignant hematopoiesis. J. Cell Biol. 218, 55–69 (2019). [DOI] [PMC free article] [PubMed]
- 20.Gibson WT, Hood RL, Zhan SH, Bulman DE, Fejes AP, Moore R, et al. Mutations in EZH2 cause weaver syndrome. Am. J. Hum. Genet. 2012;90(Jan):110–118. doi: 10.1016/j.ajhg.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal polycomb sites. Cell. 2002;111(Oct):185–196. doi: 10.1016/S0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 22.Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 23.Ketel CS, Andersen EF, Vargas ML, Suh J, Strome S, Simon JA. Subunit contributions to histone methyltransferase activities of fly and worm polycomb group complexes. Mol. Cell Biol. 2005;25(Aug):6857–6868. doi: 10.1128/MCB.25.16.6857-6868.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smits AH, Jansen PWTC, Poser I, Hyman AA, Vermeulen M. Stoichiometry of chromatin-associated protein complexes revealed by label-free quantitative mass spectrometry-based proteomics. Nucleic Acids Res. 2013;41(Jan):e28–e28. doi: 10.1093/nar/gks941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alekseyenko AA, Gorchakov AA, Kharchenko PV, Kuroda MI. Reciprocal interactions of human C10orf12 and C17orf96 with PRC2 revealed by BioTAP-XL cross-linking and affinity purification. Proc. Natl Acad. Sci. USA. 2014;111(Feb):2488–2493. doi: 10.1073/pnas.1400648111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kloet SL, Makowski MM, Baymaz HI, van Voorthuijsen L, Karemaker ID, Santanach A, et al. The dynamic interactome and genomic targets of Polycomb complexes during stem-cell differentiation. Nat. Struct. Mol. Biol. 2016;23(Jul):682–690. doi: 10.1038/nsmb.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holoch D, Margueron R. Mechanisms regulating PRC2 recruitment and enzymatic activity. Trends Biochem. Sci. 2017;42(Jul):531–542. doi: 10.1016/j.tibs.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 28.Beringer M, Pisano P, Di Carlo V, Blanco E, Chammas P, Vizán P, et al. EPOP functionally links elongin and polycomb in pluripotent stem cells. Mol. Cell. 2016;64(Nov):645–658. doi: 10.1016/j.molcel.2016.10.018. [DOI] [PubMed] [Google Scholar]
- 29.Liefke R, Karwacki-Neisius V, Shi Y. EPOP interacts with elongin BC and USP7 to modulate the chromatin landscape. Mol. Cell. 2016;64(Nov):659–672. doi: 10.1016/j.molcel.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conway E, Jerman E, Healy E, Ito S, Holoch D, Oliviero G, et al. A family of vertebrate-specific polycombs encoded by the LCOR/LCORL genes balance PRC2 subtype activities. Mol Cell. 2018;70(May):408–421.e8. doi: 10.1016/j.molcel.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 31.Antonysamy S, Condon B, Druzina Z, Bonanno JB, Gheyi T, Zhang F, et al. Structural context of disease-associated mutations and putative mechanism of autoinhibition revealed by X-Ray crystallographic analysis of the EZH2-SET domain. PLoS One. 2013;8(Dec):e84147. doi: 10.1371/journal.pone.0084147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu H, Zeng H, Dong A, Li F, He H, Senisterra G, et al. Structure of the catalytic domain of EZH2 reveals conformational plasticity in cofactor and substrate binding sites and explains oncogenic mutations. PLoS One. 2013;8(Dec):e83737. doi: 10.1371/journal.pone.0083737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bratkowski M, Yang X, Liu X. Polycomb repressive complex 2 in an autoinhibited state. J. Biol. Chem. 2017;292(Aug):13323–13332. doi: 10.1074/jbc.M117.787572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu H, Min J, Lunin VV, Antoshenko T, Dombrovski L, Zeng H, et al. Structural biology of human H3K9 methyltransferases. PLoS One. 2010;5(Jan):e8570. doi: 10.1371/journal.pone.0008570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Justin N, Zhang Y, Tarricone C, Martin SR, Chen S, Underwood E, et al. Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat. Commun. 2016;7:11316. doi: 10.1038/ncomms11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han Z, Xing X, Hu M, Zhang Y, Liu P, Chai J. Structural basis of EZH2 recognition by EED. Structure. 2007;15(Oct):1306–1315. doi: 10.1016/j.str.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 37.Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, III, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461(Oct):762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ciferri C, Lander GC, Maiolica A, Herzog F, Aebersold R, Nogales E. Molecular architecture of human polycomb repressive complex 2. elife. 2012;1(Oct):e00005. doi: 10.7554/eLife.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen S, Jiao L, Shubbar M, Yang X, Liu X. Unique structural platforms of Suz12 dictate distinct classes of PRC2 for chromatin binding. Mol. Cell. 2018;69(Mar):840–852.e5. doi: 10.1016/j.molcel.2018.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kasinath V, Faini M, Poepsel S, Reif D, Feng XA, Stjepanovic G, et al. Structures of human PRC2 with its cofactors AEBP2 and JARID2. Science. 2018;359(Feb):940–944. doi: 10.1126/science.aar5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horard B, Tatout C, Poux S, Pirrotta V. Structure of a polycomb response element and in vitro binding of polycomb group complexes containing GAGA factor. Mol. Cell Biol. 2000;20(May):3187–3197. doi: 10.1128/MCB.20.9.3187-3197.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hodgson JW, Argiropoulos B, Brock HW. Site-specific recognition of a 70-base-pair element containing d(GA)n repeats mediates bithoraxoid polycomb group response element-dependent silencing. Mol. Cell Biol. 2001;21(Jul):4528–4543. doi: 10.1128/MCB.21.14.4528-4543.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuettengruber B, Bourbon H-M, Di Croce L, Cavalli G. Genome regulation by polycomb and trithorax: 70 years and counting. Cell. 2017;171(Sep):34–57. doi: 10.1016/j.cell.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 44.Bauer M, Trupke J, Ringrose L. The quest for mammalian Polycomb response elements: are we there yet? Chromosoma. 2016;125(Jun):471–496. doi: 10.1007/s00412-015-0539-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poepsel S, Kasinath V, Nogales E. Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. Nat. Struct. Mol. Biol. 2018;25(Feb):154–162. doi: 10.1038/s41594-018-0023-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee C-H, Holder M, Grau D, Saldaña-Meyer R, Yu J-R, Ganai RA, et al. Distinct stimulatory mechanisms regulate the catalytic activity of polycomb repressive complex 2. Mol. Cell. 2018;70(May):435–448.e5. doi: 10.1016/j.molcel.2018.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patsialou A. DNA-binding properties of ARID family proteins. Nucleic Acids Res. 2005;33(Jan):66–80. doi: 10.1093/nar/gki145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He G-P, Kim S, Ro H-S. Cloning and characterization of a novel zinc finger transcriptional repressor. J. Biol. Chem. 1999;274(May):14678–14684. doi: 10.1074/jbc.274.21.14678. [DOI] [PubMed] [Google Scholar]
- 49.Kim H, Kang K, Kim J. AEBP2 as a potential targeting protein for Polycomb Repression Complex PRC2. Nucleic Acids Res. 2009;37(May):2940–2950. doi: 10.1093/nar/gkp149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24(Feb):368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Son J, Shen SS, Margueron R, Reinberg D. Nucleosome-binding activities within JARID2 and EZH1 regulate the function of PRC2 on chromatin. Genes Dev. 2013;27(Dec):2663–2677. doi: 10.1101/gad.225888.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pasini D, Cloos PAC, Walfridsson J, Olsson L, Bukowski J-P, Johansen JV, et al. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464(Mar):306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- 53.Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, et al. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139(Dec):1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, et al. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139(Dec):1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Su C-L, Deng T-R, Shang Z, Xiao Y. JARID2 inhibits leukemia cell proliferation by regulating CCND1 expression. Int. J. Hematol. 2015;102(Jul):76–85. doi: 10.1007/s12185-015-1797-x. [DOI] [PubMed] [Google Scholar]
- 56.Puda A, Milosevic JD, Berg T, Klampfl T, Harutyunyan AS, Gisslinger B, et al. Frequent deletions of JARID2 in leukemic transformation of chronic myeloid malignancies. Am. J. Hematol. 2012;87(Mar):245–250. doi: 10.1002/ajh.22257. [DOI] [PubMed] [Google Scholar]
- 57.Ballaré C, Lange M, Lapinaite A, Martin GM, Morey L, Pascual G, et al. Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nat. Struct. Mol. Biol. 2012;19(Dec):1257–1265. doi: 10.1038/nsmb.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brien GL, Gambero G, O’Connell DJ, Jerman E, Turner SA, Egan CM, et al. Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 to embryonic stem cell genes during differentiation. Nat. Struct. Mol. Biol. 2012;19(Dec):1273–1281. doi: 10.1038/nsmb.2449. [DOI] [PubMed] [Google Scholar]
- 59.Li H, Liefke R, Jiang J, Kurland JV, Tian W, Deng P, et al. Polycomb-like proteins link the PRC2 complex to CpG islands. Nature. 2017;549:287–291. doi: 10.1038/nature23881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi J, Bachmann AL, Tauscher K, Benda C, Fierz B, Müller J. DNA binding by PHF1 prolongs PRC2 residence time on chromatin and thereby promotes H3K27 methylation. Nat. Struct. Mol. Biol. 2017;24(Dec):1039–1047. doi: 10.1038/nsmb.3488. [DOI] [PubMed] [Google Scholar]
- 61.Tanay A, O’Donnell AH, Damelin M, Bestor TH. Hyperconserved CpG domains underlie Polycomb-binding sites. Proc. Natl Acad. Sci. USA. 2007;104(Mar):5521–5526. doi: 10.1073/pnas.0609746104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perino M, van Mierlo G, Karemaker ID, van Genesen S, Vermeulen M, Marks H, et al. MTF2 recruits Polycomb Repressive Complex 2 by helical-shape-selective DNA binding. Nat. Genet. 2018;50(Jul):1002–1010. doi: 10.1038/s41588-018-0134-8. [DOI] [PubMed] [Google Scholar]
- 63.Jiao L, Liu X. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science. 2015;350(Oct):aac4383. doi: 10.1126/science.aac4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee C-H, Yu J-R, Kumar S, Jin Y, LeRoy G, Bhanu N, et al. Allosteric activation dictates PRC2 activity independent of its recruitment to chromatin. Mol. Cell. 2018;70(May):422–434.e6. doi: 10.1016/j.molcel.2018.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu C, Bian C, Yang W, Galka M, Ouyang H, Chen C, et al. Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2) Proc. Natl Acad. Sci. USA. 2010;107:19266–19271. doi: 10.1073/pnas.1008937107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Curtin ML, Pliushchev MA, Li H-Q, Torrent M, Dietrich JD, Jakob CG, et al. SAR of amino pyrrolidines as potent and novel protein-protein interaction inhibitors of the PRC2 complex through EED binding. Bioorg. Med. Chem. Lett. 2017;27:1576–1583. doi: 10.1016/j.bmcl.2017.02.030. [DOI] [PubMed] [Google Scholar]
- 67.He Y, Selvaraju S, Curtin ML, Jakob CG, Zhu H, Comess KM, et al. The EED protein–protein interaction inhibitor A-395 inactivates the PRC2 complex. Nat Chem Biol. 2017;13(Apr):389–395. doi: 10.1038/nchembio.2306. [DOI] [PubMed] [Google Scholar]
- 68.Qi W, Zhao K, Gu J, Huang Y, Wang Y, Zhang H, et al. An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of EED. Nat. Chem. Biol. 2017;13(Apr):381–388. doi: 10.1038/nchembio.2304. [DOI] [PubMed] [Google Scholar]
- 69.Youmans DT, Schmidt JC, Cech TR. Live-cell imaging reveals the dynamics of PRC2 and recruitment to chromatin by SUZ12-associated subunits. Genes Dev. 2018;32(Jun):794–805. doi: 10.1101/gad.311936.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li L, Zhang H, Zhang M, Zhao M, Feng L, Luo X, et al. Discovery and molecular basis of a diverse set of polycomb repressive complex 2 inhibitors recognition by EED. PLoS One. 2017;12(Jan):e0169855. doi: 10.1371/journal.pone.0169855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barnash KD, The J, Norris-Drouin JL, Cholensky SH, Worley BM, Li F, et al. Discovery of peptidomimetic ligands of EED as allosteric inhibitors of PRC2. ACS Comb. Sci. 2017;19(Mar):161–172. doi: 10.1021/acscombsci.6b00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oksuz O, Narendra V, Lee C-H, Descostes N, LeRoy G, Raviram R, et al. Capturing the onset of PRC2-mediated repressive domain formation. Mol. Cell. 2018;70(Jun):1149–1162.e5. doi: 10.1016/j.molcel.2018.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sanulli S, Justin N, Teissandier A, Ancelin K, Portoso M, Caron M, et al. Jarid2 methylation via the PRC2 complex regulates H3K27me3 deposition during cell differentiation. Mol. Cell. 2015;57(Mar):769–783. doi: 10.1016/j.molcel.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol. Cell Biol. 2006;26(Apr):2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kalb R, Latwiel S, Baymaz HI, PWTC Jansen, Müller CW, Vermeulen M, et al. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat. Struct. Mol. Biol. 2014;21(Jun):569–571. doi: 10.1038/nsmb.2833. [DOI] [PubMed] [Google Scholar]
- 76.Cooper S, Grijzenhout A, Underwood E, Ancelin K, Zhang T, Nesterova TB, et al. Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat. Commun. 2016;7(Dec):13661. doi: 10.1038/ncomms13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schmitges FW, Prusty AB, Faty M, Stützer A, Lingaraju GM, Aiwazian J, et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell. 2011;42(May):330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 78.Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J. Biol. Chem. 2011;286(Mar):7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Murzina NV, Pei X-Y, Zhang W, Sparkes M, Vicente-Garcia J, Pratap JV, et al. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure. 2008;16(Jul):1077–1085. doi: 10.1016/j.str.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87(Oct):95–104. doi: 10.1016/S0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 81.Vermaak D, Wade PA, Jones PL, Shi Y-B, Wolffe AP. Functional analysis of the SIN3-histone deacetylase RPD3-RbAp48-histone H4 connection in the Xenopus oocyte. Mol. Cell Biol. 1999;19:5847–5860. doi: 10.1128/MCB.19.9.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nowak AJ, Alfieri C, Stirnimann CU, Rybin V, Baudin F, Ly-Hartig N, et al. Chromatin-modifying complex component Nurf55/p55 associates with histones H3 and H4 and polycomb repressive complex 2 subunit Su(z)12 through partially overlapping binding sites. J. Biol. Chem. 2011;286(Jul):23388–23396. doi: 10.1074/jbc.M110.207407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jani KS, Jain SU, Ge EJ, Diehl KL, Lundgren SM, Müller MM, et al. Histone H3 tail binds a unique sensing pocket in EZH2 to activate the PRC2 methyltransferase. Proc. Natl Acad. Sci. USA. 2019;116(Apr):8295–8300. doi: 10.1073/pnas.1819029116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weaver TM, Liu J, Connelly KE, Coble C, Varzavand K, Dykhuizen EC, et al. The EZH2 SANT1 domain is a histone reader providing sensitivity to the modification state of the H4 tail. Sci. Rep. 2019;9(Dec):987. doi: 10.1038/s41598-018-37699-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cai L, Rothbart SB, Lu R, Xu B, Chen W-Y, Tripathy A, et al. An H3K36 methylation-engaging Tudor motif of polycomb-like proteins mediates PRC2 complex targeting. Mol. Cell. 2013;49(Feb):571–582. doi: 10.1016/j.molcel.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kycia I, Kudithipudi S, Tamas R, Kungulovski G, Dhayalan A, Jeltsch A. The Tudor Domain of the PHD finger protein 1 is a dual reader of lysine trimethylation at lysine 36 of histone H3 and lysine 27 of histone variant H3t. J. Mol. Biol. 2014;426(Apr):1651–1660. doi: 10.1016/j.jmb.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 87.Musselman CA, Avvakumov N, Watanabe R, Abraham CG, Lalonde M-E, Hong Z, et al. Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1. Nat. Struct. Mol. Biol. 2012;19(Dec):1266–1272. doi: 10.1038/nsmb.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Streubel G, Watson A, Jammula SG, Scelfo A, Fitzpatrick DJ, Oliviero G, et al. The H3K36me2 methyltransferase Nsd1 demarcates PRC2-mediated H3K27me2 and H3K27me3 domains in embryonic stem cells. Mol. Cell. 2018;70(Apr):371–379.e5. doi: 10.1016/j.molcel.2018.02.027. [DOI] [PubMed] [Google Scholar]
- 89.Ferrari KJ, Scelfo A, Jammula S, Cuomo A, Barozzi I, Stützer A, et al. Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol. Cell. 2014;53(Jan):49–62. doi: 10.1016/j.molcel.2013.10.030. [DOI] [PubMed] [Google Scholar]
- 90.Cifuentes-Rojas C, Hernandez AJ, Sarma K, Lee JT. Regulatory interactions between RNA and polycomb repressive complex 2. Mol. Cell. 2014;55(Jul):171–185. doi: 10.1016/j.molcel.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Davidovich C, Zheng L, Goodrich KJ, Cech TR, Promiscuous RNA. binding by Polycomb repressive complex 2. Nat. Struct. Mol. Biol. 2013;20(Nov):1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang X, Goodrich KJ, Gooding AR, Naeem H, Archer S, Paucek RD, et al. Targeting of polycomb repressive complex 2 to RNA by short repeats of consecutive guanines. Mol. Cell. 2017;65(Mar):1056–1067.e5. doi: 10.1016/j.molcel.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 93.Kaneko S, Son J, Bonasio R, Shen SS, Reinberg D. Nascent RNA interaction keeps PRC2 activity poised and in check. Genes Dev. 2014;28(Sep):1983–1988. doi: 10.1101/gad.247940.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat. Struct. Mol. Biol. 2013;20(Nov):1258–1264. doi: 10.1038/nsmb.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Beltran M, Yates CM, Skalska L, Dawson M, Reis FP, Viiri K, et al. The interaction of PRC2 with RNA or chromatin is mutually antagonistic. Genome Res. 2016;26(Jul):896–907. doi: 10.1101/gr.197632.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang Q, McKenzie NJ, Warneford-Thomson R, Gail EH, Flanigan SF, Owen BM, et al. RNA exploits an exposed regulatory site to inhibit the enzymatic activity of PRC2. Nat. Struct. Mol. Biol. 2019;26(Mar):237–247. doi: 10.1038/s41594-019-0197-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang X, Paucek RD, Gooding AR, Brown ZZ, Ge EJ, Muir TW, et al. Molecular analysis of PRC2 recruitment to DNA in chromatin and its inhibition by RNA. Nat. Struct. Mol. Biol. 2017;24(Dec):1028–1038. doi: 10.1038/nsmb.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Davidovich C, Wang X, Cifuentes-Rojas C, Goodrich KJ, Gooding AR, Lee JT, et al. Toward a consensus on the binding specificity and promiscuity of PRC2 for RNA. Mol. Cell. 2015;57(Feb):552–558. doi: 10.1016/j.molcel.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Betancur JG. Pervasive lncRNA binding by epigenetic modifying complexes — The challenges ahead. Biochim. Biophys. Acta Gene Regul. Mech. 2016;1859(Jan):93–101. doi: 10.1016/j.bbagrm.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 100.Long, Y., Bolanos, B., Gong, L., Liu, W., Goodrich, K. J., Yang, X., et al. Conserved RNA-binding specificity of polycomb repressive complex 2 is achieved by dispersed amino acid patches in EZH2. elife. 6, e31558 (2017). [DOI] [PMC free article] [PubMed]
- 101.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419(Oct):624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 102.Deb G, Singh AK, Gupta S. EZH2: not EZHY (Easy) to deal. Mol Cancer Res. 2014;12(May):639–653. doi: 10.1158/1541-7786.MCR-13-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jiang T, Wang Y, Zhou F, Gao G, Ren S, Zhou C. Prognostic value of high EZH2 expression in patients with different types of cancer: a systematic review with meta-analysis. Oncotarget. 2016;7(Jan):4584–4597. doi: 10.18632/oncotarget.6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Laugesen A, Højfeldt JW, Helin K. Role of the polycomb repressive complex 2 (PRC2) in transcriptional regulation and cancer. Cold Spring Harb. Perspect. Med. 2016;6(Sep):a026575. doi: 10.1101/cshperspect.a026575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Comet I, Riising EM, Leblanc B, Helin K. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat. Rev. Cancer. 2016;16(Dec):803–810. doi: 10.1038/nrc.2016.83. [DOI] [PubMed] [Google Scholar]
- 106.Xu B, Konze KD, Jin J, Wang GG. Targeting EZH2 and PRC2 dependence as novel anticancer therapy. Exp. Hematol. 2015;43(Aug):698–712. doi: 10.1016/j.exphem.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Herviou, L., Cavalli, G., Cartron, G., Klein, B., Moreaux, J. EZH2 in normal hematopoiesis and hematological malignancies. Oncotarget. 7, 2284–2296 (2016). [DOI] [PMC free article] [PubMed]
- 108.Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019;47(Jan):D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schwaab J, Ernst T, Erben P, Rinke J, Schnittger S, Ströbel P, et al. Activating CBL mutations are associated with a distinct MDS/MPN phenotype. Ann. Hematol. 2012;91(Nov):1713–1720. doi: 10.1007/s00277-012-1521-3. [DOI] [PubMed] [Google Scholar]
- 110.Xu C, Bian C, Yang W, Galka M, Ouyang H, Chen C, et al. Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2) Proc. Natl Acad. Sci. USA. 2010;107(Nov):19266–19271. doi: 10.1073/pnas.1008937107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122(Nov):3616–3627. doi: 10.1182/blood-2013-08-518886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gambacorti-Passerini CB, Donadoni C, Parmiani A, Pirola A, Redaelli S, Signore G, et al. Recurrent ETNK1 mutations in atypical chronic myeloid leukemia. Blood. 2015;125(Jan):499–503. doi: 10.1182/blood-2014-06-579466. [DOI] [PubMed] [Google Scholar]
- 113.Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat. Genet. 2010;42(Aug):722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 114.Palomo, L., Garcia, O., Arnan, M., Xicoy, B., Fuster, F., Cabezón, M., et al. Targeted deep sequencing improves outcome stratification in chronic myelomonocytic leukemia with low risk cytogenetic features. Oncotarget. 7, 57021–57035 (2016). [DOI] [PMC free article] [PubMed]
- 115.Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016;374(Jun):2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bartels, S., Schipper, E., Hasemeier, B., Kreipe, H., Lehmann, U. Routine clinical mutation profiling using next generation sequencing and a customized gene panel improves diagnostic precision in myeloid neoplasms. Oncotarget. 7, 30084–30093 (2016). [DOI] [PMC free article] [PubMed]
- 117.Makishima H, Jankowska AM, Tiu RV, Szpurka H, Sugimoto Y, Hu Z, et al. Novel homo- and hemizygous mutations in EZH2 in myeloid malignancies. Leukemia. 2010;24(Oct):1799–1804. doi: 10.1038/leu.2010.167. [DOI] [PubMed] [Google Scholar]
- 118.Nikoloski G, Langemeijer SMC, Kuiper RP, Knops R, Massop M. Tönnissen ERLTM, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat. Genet. 2010;42(Aug):665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 119.Jankowska AM, Makishima H, Tiu RV, Szpurka H, Huang Y, Traina F, et al. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, and DNMT3A. Blood. 2011;118(Oct):3932–3941. doi: 10.1182/blood-2010-10-311019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Guglielmelli P, Biamonte F, Score J, Hidalgo-Curtis C, Cervantes F, Maffioli M, et al. EZH2 mutational status predicts poor survival in myelofibrosis. Blood. 2011;118(Nov):5227–5234. doi: 10.1182/blood-2011-06-363424. [DOI] [PubMed] [Google Scholar]
- 121.Grossmann V, Kohlmann A, Eder C, Haferlach C, Kern W, Cross NCP, et al. Molecular profiling of chronic myelomonocytic leukemia reveals diverse mutations in >80% of patients with TET2 and EZH2 being of high prognostic relevance. Leukemia. 2011;25(May):877–879. doi: 10.1038/leu.2011.10. [DOI] [PubMed] [Google Scholar]
- 122.Lundberg P, Karow A, Nienhold R, Looser R, Hao-Shen H, Nissen I, et al. Clonal evolution and clinical correlates of somatic mutations in myeloproliferative neoplasms. Blood. 2014;123(Apr):2220–2228. doi: 10.1182/blood-2013-11-537167. [DOI] [PubMed] [Google Scholar]
- 123.Nangalia J, Massie CE, Baxter EJ, Nice FL, Gundem G, Wedge DC, et al. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N. Engl. J. Med. 2013;369(Dec):2391–2405. doi: 10.1056/NEJMoa1312542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rampal R, Ahn J, Abdel-Wahab O, Nahas M, Wang K, Lipson D, et al. Genomic and functional analysis of leukemic transformation of myeloproliferative neoplasms. Proc. Natl Acad. Sci. USA. 2014;111(Dec):E5401–E5410. doi: 10.1073/pnas.1407792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.López C, Bergmann AK, Paul U, Murga Penas EM, Nagel I, Betts MJ, et al. Genes encoding members of the JAK-STAT pathway or epigenetic regulators are recurrently mutated in T-cell prolymphocytic leukaemia. Br. J. Haematol. 2016;173(Apr):265–273. doi: 10.1111/bjh.13952. [DOI] [PubMed] [Google Scholar]
- 126.Liu Y, Easton J, Shao Y, Maciaszek J, Wang Z, Wilkinson MR, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat. Genet. 2017;49(Aug):1211–1218. doi: 10.1038/ng.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Thol F, Klesse S, Köhler L, Gabdoulline R, Kloos A, Liebich A, et al. Acute myeloid leukemia derived from lympho-myeloid clonal hematopoiesis. Leukemia. 2017;31(Jun):1286–1295. doi: 10.1038/leu.2016.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yoshida K, Toki T, Okuno Y, Kanezaki R, Shiraishi Y, Sato-Otsubo A, et al. The landscape of somatic mutations in Down syndrome–related myeloid disorders. Nat. Genet. 2013;45(Nov):1293–1299. doi: 10.1038/ng.2759. [DOI] [PubMed] [Google Scholar]
- 129.Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(Oct):64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 130.Perry JA, Kiezun A, Tonzi P, Van Allen EM, Carter SL, Baca SC, et al. Complementary genomic approaches highlight the PI3K/mTOR pathway as a common vulnerability in osteosarcoma. Proc. Natl Acad. Sci. USA. 2014;111(Dec):E5564–E5573. doi: 10.1073/pnas.1419260111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hirsch P, Zhang Y, Tang R, Joulin V, Boutroux H, Pronier E, et al. Genetic hierarchy and temporal variegation in the clonal history of acute myeloid leukaemia. Nat. Commun. 2016;7(Nov):12475. doi: 10.1038/ncomms12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481(Jan):157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Metzeler KH, Herold T, Rothenberg-Thurley M, Amler S, Sauerland MC, Gorlich D, et al. Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood. 2016;128(Aug):686–698. doi: 10.1182/blood-2016-01-693879. [DOI] [PubMed] [Google Scholar]
- 134.Garg M, Nagata Y, Kanojia D, Mayakonda A, Yoshida K, Haridas Keloth S, et al. Profiling of somatic mutations in acute myeloid leukemia with FLT3-ITD at diagnosis and relapse. Blood. 2015;126(Nov):2491–2501. doi: 10.1182/blood-2015-05-646240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lui JC, Barnes KM, Dong L, Yue S, Graber E, Rapaport R, et al. Ezh2 mutations found in the weaver overgrowth syndrome cause a partial loss of H3K27 histone methyltransferase activity. J. Clin. Endocrinol. Metab. 2018;103(Apr):1470–1478. doi: 10.1210/jc.2017-01948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.McCabe MT, Graves AP, Ganji G, Diaz E, Halsey WS, Jiang Y, et al. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27) Proc. Natl Acad. Sci. USA. 2012;109(Feb):2989–2994. doi: 10.1073/pnas.1116418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bodor C, Grossmann V, Popov N, Okosun J, O’Riain C, Tan K, et al. EZH2 mutations are frequent and represent an early event in follicular lymphoma. Blood. 2013;122(Oct):3165–3168. doi: 10.1182/blood-2013-04-496893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bohers E, Mareschal S, Bouzelfen A, Marchand V, Ruminy P, Maingonnat C, et al. Targetable activating mutations are very frequent in GCB and ABC diffuse large B-cell lymphoma. Genes Chromosom Cancer. 2014;53(Feb):144–153. doi: 10.1002/gcc.22126. [DOI] [PubMed] [Google Scholar]
- 139.Pasqualucci L, Khiabanian H, Fangazio M, Vasishtha M, Messina M, Holmes AB, et al. Genetics of follicular lymphoma transformation. Cell Rep. 2014;6(Jan):130–140. doi: 10.1016/j.celrep.2013.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Morin RD, Assouline S, Alcaide M, Mohajeri A, Johnston RL, Chong L, et al. Genetic landscapes of relapsed and refractory diffuse large B-cell lymphomas. Clin. Cancer Res. 2016;22(May):2290–2300. doi: 10.1158/1078-0432.CCR-15-2123. [DOI] [PubMed] [Google Scholar]
- 141.Juskevicius D, Jucker D, Klingbiel D, Mamot C, Dirnhofer S, Tzankov A. Mutations of CREBBP and SOCS1 are independent prognostic factors in diffuse large B cell lymphoma: mutational analysis of the SAKK 38/07 prospective clinical trial cohort. J. Hematol. Oncol. 2017;10(Dec):70. doi: 10.1186/s13045-017-0438-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(Aug):298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl Acad. Sci. USA. 2012;109(Mar):3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Majer CR, Jin L, Scott MP, Knutson SK, Kuntz KW, Keilhack H, et al. A687V EZH2 is a gain-of-function mutation found in lymphoma patients. FEBS Lett. 2012;586(Sep):3448–3451. doi: 10.1016/j.febslet.2012.07.066. [DOI] [PubMed] [Google Scholar]
- 145.Bödör, C., O’Riain, C., Wrench, D., Matthews, J., Iyengar, S., Tayyib, H., et al. EZH2 Y641 mutations in follicular lymphoma. Leukemia25, 726–729 (2011). [DOI] [PubMed]
- 146.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017;23(Jun):703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ryan RJH, Nitta M, Borger D, Zukerberg LR, Ferry JA, Harris NL, et al. EZH2 codon 641 mutations are common in BCL2-rearranged germinal center B cell lymphomas. PLoS One. 2011;6(Dec):e28585. doi: 10.1371/journal.pone.0028585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang J, Grubor V, Love CL, Banerjee A, Richards KL, Mieczkowski PA, et al. Genetic heterogeneity of diffuse large B-cell lymphoma. Proc. Natl Acad. Sci. USA. 2013;110(Jan):1398–1403. doi: 10.1073/pnas.1205299110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Saieg MA, Geddie WR, Boerner SL, Bailey D, Crump M, da Cunha Santos G. EZH2 and CD79B mutational status over time in B-cell non-Hodgkin lymphomas detected by high-throughput sequencing using minimal samples. Cancer Cytopathol. 2013;121(Jul):377–386. doi: 10.1002/cncy.21262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat J-P, et al. A landscape of driver mutations in melanoma. Cell. 2012;150(Jul):251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat. Genet. 2012;44(Sep):1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Durinck S, Ho C, Wang NJ, Liao W, Jakkula LR, Collisson EA, et al. Temporal dissection of tumorigenesis in primary cancers. Cancer Discov. 2011;1(Jul):137–143. doi: 10.1158/2159-8290.CD-11-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tirode F, Surdez D, Ma X, Parker M, Le Deley MC, Bahrami A, et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov. 2014;4(Nov):1342–1353. doi: 10.1158/2159-8290.CD-14-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Crompton BD, Stewart C, Taylor-Weiner A, Alexe G, Kurek KC, Calicchio ML, et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov. 2014;4(Nov):1326–1341. doi: 10.1158/2159-8290.CD-13-1037. [DOI] [PubMed] [Google Scholar]
- 155.Souroullas GP, Jeck WR, Parker JS, Simon JM, Liu J-Y, Paulk J, et al. An oncogenic Ezh2 mutation induces tumors through global redistribution of histone 3 lysine 27 trimethylation. Nat. Med. 2016;22(Jun):632–640. doi: 10.1038/nm.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.McCabe MT, Graves AP, Ganji G, Diaz E, Halsey WS, Jiang Y, et al. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27) Proc. Natl Acad. Sci. USA. 2012;109(Feb):2989–2994. doi: 10.1073/pnas.1116418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, Richon VM, et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc. Natl Acad. Sci. USA. 2010;107(Dec):20980–20985. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wigle TJ, Knutson SK, Jin L, Kuntz KW, Pollock RM, Richon VM, et al. The Y641C mutation of EZH2 alters substrate specificity for histone H3 lysine 27 methylation states. FEBS Lett. 2011;585(Oct):3011–3014. doi: 10.1016/j.febslet.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 159.Brooun A, Gajiwala KS, Deng Y-L, Liu W, Bolaños B, Bingham P, et al. Polycomb repressive complex 2 structure with inhibitor reveals a mechanism of activation and drug resistance. Nat. Commun. 2016;7(Sep):11384. doi: 10.1038/ncomms11384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yap DB, Chu J, Berg T, Schapira M, Cheng S-WG, Moradian A, et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117(Feb):2451–2459. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Swalm BM, Knutson SK, Warholic NM, Jin L, Kuntz KW, Keilhack H, et al. Reaction coupling between wild-type and disease-associated mutant EZH2. ACS Chem. Biol. 2014;9(Nov):2459–2464. doi: 10.1021/cb500548b. [DOI] [PubMed] [Google Scholar]
- 162.Donaldson-Collier MC, Sungalee S, Zufferey M, Tavernari D, Katanayeva N, Battistello E, et al. EZH2 oncogenic mutations drive epigenetic, transcriptional, and structural changes within chromatin domains. Nat. Genet. 2019;51(Mar):517–528. doi: 10.1038/s41588-018-0338-y. [DOI] [PubMed] [Google Scholar]
- 163.Okosun J, Bödör C, Wang J, Araf S, Yang C-Y, Pan C, et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat. Genet. 2014;46(Feb):176–181. doi: 10.1038/ng.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ott HM, Graves AP, Pappalardi MB, Huddleston M, Halsey WS, Hughes AM, et al. A687V EZH2 is a driver of histone H3 lysine 27 (H3K27) hypertrimethylation. Mol. Cancer Ther. 2014;13:3062–3073. doi: 10.1158/1535-7163.MCT-13-0876. [DOI] [PubMed] [Google Scholar]
- 165.Patel KP, Newberry KJ, Luthra R, Jabbour E, Pierce S, Cortes J, et al. Correlation of mutation profile and response in patients with myelofibrosis treated with ruxolitinib. Blood. 2015;126(Aug):790–797. doi: 10.1182/blood-2015-03-633404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Score J, Hidalgo-Curtis C, Jones AV, Winkelmann N, Skinner A, Ward D, et al. Inactivation of polycomb repressive complex 2 components in myeloproliferative and myelodysplastic/myeloproliferative neoplasms. Blood. 2012;119(Feb):1208–1213. doi: 10.1182/blood-2011-07-367243. [DOI] [PubMed] [Google Scholar]
- 167.Nikolaev SI, Garieri M, Santoni F, Falconnet E, Ribaux P, Guipponi M, et al. Frequent cases of RAS-mutated Down syndrome acute lymphoblastic leukaemia lack JAK2 mutations. Nat. Commun. 2014;5(Dec):4654. doi: 10.1038/ncomms5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ntziachristos P, Tsirigos A, Van VlierbergheP, Nedjic J, Trimarchi T, Flaherty MS, et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat. Med. 2012;18(Feb):298–302. doi: 10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Rai AN, Vargas ML, Wang L, Andersen EF, Miller EL, Simon JA. Elements of the polycomb repressor SU(Z)12 needed for histone H3-K27 methylation, the interface with E(Z), and in vivo function. Mol. Cell Biol. 2013;33(Dec):4844–4856. doi: 10.1128/MCB.00307-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Mason CC, Khorashad JS, Tantravahi SK, Kelley TW, Zabriskie MS, Yan D, et al. Age-related mutations and chronic myelomonocytic leukemia. Leukemia. 2016;30(Apr):906–913. doi: 10.1038/leu.2015.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Duployez N, Marceau-Renaut A, Boissel N, Petit A, Bucci M, Geffroy S, et al. Comprehensive mutational profiling of core binding factor acute myeloid leukemia. Blood. 2016;127(May):2451–2459. doi: 10.1182/blood-2015-12-688705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Fernandez-Cuesta L, Peifer M, Lu X, Sun R, Ozretić L, Seidel D, et al. Frequent mutations in chromatin-remodelling genes in pulmonary carcinoids. Nat. Commun. 2014;5(May):3518. doi: 10.1038/ncomms4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat. Genet. 2014;46(Mar):225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hao J-J, Lin D-C, Dinh HQ, Mayakonda A, Jiang Y-Y, Chang C, et al. Spatial intratumoral heterogeneity and temporal clonal evolution in esophageal squamous cell carcinoma. Nat. Genet. 2016;48(Dec):1500–1507. doi: 10.1038/ng.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.