Abstract
Introduction
To develop effective therapies and identify novel early biomarkers for chronic kidney disease, an understanding of the molecular mechanisms orchestrating it is essential. We here set out to understand how differences in chronic kidney disease (CKD) origin are reflected in gene expression. To this end, we integrated publicly available human glomerular microarray gene expression data for 9 kidney disease entities that account for most of CKD worldwide. Our primary goal was to demonstrate the possibilities and potential on data analysis and integration to the nephrology community.
Methods
We integrated data from 5 publicly available studies and compared glomerular gene expression profiles of disease with that of controls from nontumor parts of kidney cancer nephrectomy tissues. A major challenge was the integration of the data from different sources, platforms, and conditions that we mitigated with a bespoke stringent procedure.
Results
We performed a global transcriptome-based delineation of different kidney disease entities, obtaining a transcriptomic diffusion map of their similarities and differences based on the genes that acquire a consistent differential expression between each kidney disease entity and nephrectomy tissue. We derived functional insights by inferring the activity of signaling pathways and transcription factors from the collected gene expression data and identified potential drug candidates based on expression signature matching. We validated representative findings by immunostaining in human kidney biopsies indicating, for example, that the transcription factor FOXM1 is significantly and specifically expressed in parietal epithelial cells in rapidly progressive glomerulonephritis (RPGN) whereas not expressed in control kidney tissue. Furthermore, we found drug candidates by matching the signature on expression of drugs to that of the CKD entities, in particular, the Food and Drug Administration–approved drug nilotinib.
Conclusion
These results provide a foundation to comprehend the specific molecular mechanisms underlying different kidney disease entities that can pave the way to identify biomarkers and potential therapeutic targets. To facilitate further use, we provide our results as a free interactive Web application: https://saezlab.shinyapps.io/ckd_landscape/. However, because of the limitations of the data and the difficulties in its integration, any specific result should be considered with caution. Indeed, we consider this study rather an illustration of the value of functional genomics and integration of existing data.
Keywords: CKD, drug repositioning, signaling pathway, transcription factor
Graphical abstract
CKD is a significant public health burden affecting more than 10% of the population globally.1 There is no specific therapy, and the associated costs are enormous.2 The origin of CKD is heterogeneous and has slowly changed in recent years due to an aging population with increased number of patients with hypertension and diabetes. Major contributors to worldwide CKD include diabetic nephropathy (DN) and hypertensive nephropathy (HN). Other contributors are immune diseases, such as lupus nephritis (LN), and glomerulonephritides, including IgA nephropathy (IgAN), membranous glomerulonephritis (MGN), and minimal change disease (MCD), as well as focal segmental glomerulosclerosis (FSGS), and RPGN.
Regardless of the type of initial injury to the kidney, the stereotypic response to chronic repetitive injury is scar formation with subsequent kidney functional decline. Scars form in the tubulointerstitium as tubulointerstitial fibrosis and in the glomerulus as glomerulosclerosis. Despite this stereotypic response, the initiating stimuli are quite heterogeneous, ranging from an auto-immunological process in LN to poorly controlled blood glucose levels in DN. A better understanding of similarities and differences in the complex molecular processes orchestrating disease initiation and progression will guide the development of novel targeted therapeutics.
A powerful tool to understand and model the molecular basis of diseases is the analysis of genome-wide gene expression data. This has been applied in the context of various kidney diseases contributing to CKD,3, 4, 5, 6, 7 and many studies are available in the online resource NephroSeq.8,9 However, to the best of our knowledge, no study so far has combined these data sets to build a comprehensive landscape of the molecular alterations underlying different kidney diseases that account for most CKD cases. We collected from 5 extensive studies microarray gene expression data from kidney biopsies of patients of 8 separate glomerular disease entities leading to CKD (from here on referred to as CKD entities), FSGS, MCD, IgAN, LN, MGN, DN, HN, and RPGN. We normalized the data with a bespoke stringent procedure, which allowed us to study the similarities and differences among these entities in terms of deregulated genes, pathways, and transcription factors, as well as to identify drugs that revert their expression signatures and thereby might be useful to treat them.
Methods
Data Collection
Raw data CEL files of each microarray dataset, GSE20602,10 GSE32591,11 GSE37460,11 GSE47183,12,13 and GSE50469,14 were downloaded and imported to R (R version 3.3.2). For more information see the Supplementary Methods.
Normalization
Cyclic loess normalization was applied using the limma package.15, 16, 17 YuGene transformation was carried out using the YuGene R package.18
Batch Effect Mitigation: Method
First, we structured the data in a platform-specific manner. Then, we conducted differential gene expression analysis between the identical biological conditions from distinct study sources after cyclic loess normalization. We subsequently removed the genes that are significantly differentially expressed between them, as it indicated differences mainly due to the data source, rather than the biological difference. We applied this procedure for the data fragments coming from Affymetrix (Santa Clara, CA) Human Genome U133 Plus 2.0 Array and Affymetrix Human Genome U133A Array. Next, we merged the data sets between the 2 platforms using the overlapping genes, followed by a process to mitigate the platform-induced batch effect. This latter procedure is similar to the one used for the data source–specific batch effect mitigation.
Detection of Genes With Consistently Small P Values Across All Studies
Based on the assumption that common mechanisms might contribute to all CKD entities, we performed a Maximum P value (maxP) method,19 which uses the maximum P value as the test statistic on the output of the differential expression analysis of the hypothetically separate studies. For more information see the Supplementary Methods.
Diffusion Map
The batch-mitigated data containing merely the maxP identified 1790 genes (Supplementary Material and Supplementary Data and Code) (false discovery rate < 0.01), were YuGene transformed,18 and the destiny R package20 was used to produce the diffusion maps.
Functional Analysis
Transcription Factor Activity Analysis
We estimated transcription factor activities in the glomerular CKD entities using DoRothEA,21 which is a pipeline that tries to estimate transcription factor activity via the expression level of its target genes using a curated database of transcription factor (TF)–target gene interactions (TF Regulon). For more information see the Supplementary Methods.
Inferring Signaling Pathway Activity Fusing PROGENy
We used the cyclic loess normalized and batch effect mitigated expression values for PROGENy,22 a method that uses downstream gene expression changes due to pathway perturbation to infer the upstream signaling pathway activity. For more information see the Supplementary Methods.
Pathway Analysis With Piano
Pathway analysis was performed using the Piano package from R.23 For more information see the Supplementary Methods.
Drug Repositioning
For each CKD entity, the signature of cosine distances computed by characteristic direction was applied to a signature search engine, L1000CDS2,24 with the mode of reverse in configuration.
Immunofluorescent Staining of Human Kidney Biopsies and Analysis
Validation involving human kidney biopsies was approved by the local ethics committee at Karolinska Institutet (Dnr 2017/1991-32). Stainings were performed on 2-μm paraffin-embedded sections as previously described.25 For more information see the Supplementary Methods.
Results
Assembly of a pan-CKD Collection of Patient Gene Expression Profiles
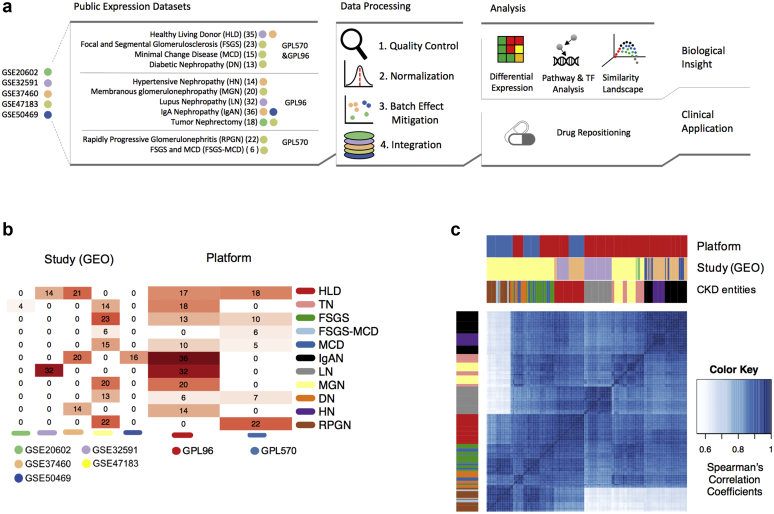
We searched in NephroSeq (www.nephroseq.org) and the Gene Expression Omnibus26,27 and identified 5 studies, GSE20602,10 GSE32591,11 GSE37460,11 GSE47183,12,13 GSE5046914, with human microarray gene expression data for 9 different glomerular disease entities: FSGS, FSGS-MCD, MCD, IgAN, LN, MGN, DN, HN, and RPGN, as well as healthy tissue and nontumor part of kidney cancer nephrectomy tissues as controls (Figure 1a and b). In addition, in one dataset, patients were labeled as an overlap of FSGS and MCD (FSGS-MCD) and we left it as such. These studies were generated in 2 different microarray platforms. To jointly analyze and compare the different CKD entities, we performed a stringent preprocessing and normalization procedure involving quality control, either cyclic loess normalization or YuGene transformation, and a batch effect mitigation procedure (see the Methods section and the Supplementary Material). At the end, we kept 6289 genes from 199 samples in total. From the 2 potential controls, healthy tissue, and nephrectomies, we chose the latter for further analysis as the batch mitigation removed a large number of genes from the healthy tissue samples.
Figure 1.
Visual abstract and descriptive analysis. (a) Flow of analysis followed in this study. (b) Heatmap of the distribution of samples across studies and microarray platforms. (c) Hierarchical clustering of the arrays based on gene expression Spearman’s correlation coefficients. DN, diabetic nephropathy; FSGS, focal segmental glomerulosclerosis; GEO, Gene Expression Omnibus; HLD, healthy living donor; HN, hypertensive nephropathy; IgAN, IgA nephropathy; LN, lupus nephritis; MCD, minimal change disease; MGN, membranous glomerulonephritis; RPGN, rapidly progressive glomerulonephritis; TN, tumor nephrectomy.
Technical Heterogeneity Across Samples
We first examined the similarities between the samples to assess further batch effects. Data did not primarily cluster by study source or platform, which can be attributed to our batch mitigation procedure (Figure 1c; Supplementary Figure S1), although some technical sources of variance potentially remained (Supplementary Figure S1). Samples from RPGN and FSGS-MCD conditions seemed to be more affected by platform-specific batch effects than samples from other conditions, due to the unbalanced distribution of samples: RPGN and FSGS-MCD samples were exclusively represented in 1 study and in 1 of the 2 platforms (Affymetrix Human Genome U133 Plus 2.0 Array [GPL570]). Therefore, the batch effect mitigation procedure could not be conducted on them.
Biological Heterogeneity of CKD Entities
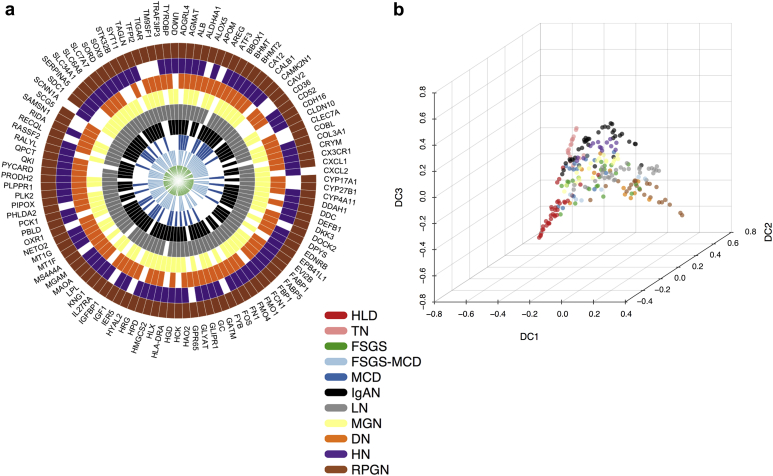
We set out to find molecular differences among glomerular CKD entities. First, we calculated the differential expression of individual genes between the different CKD entities and tumor nephrectomy (TN) samples using limma.17,28 From the 6289 genes included in the integrated dataset, 1791 showed significant differential expression (|logFC| > 1, P < 0.05) in at least 1 CKD entity. RPGN was the CKD entity with the largest number of significantly differentially expressed genes (885), and MCD was the one with the least (75). Twelve genes showed significant differential expression across all the CKD entities (AGMAT, ALB, BHMT2, CALB1, CYP4A11, FOS, HAO2, HMGCS2, MT1F, MT1G, PCK1, SLC6A8). Interestingly, all these genes were underexpressed across all the CKD entities compared with TN. In contrast, QKI and LYZ genes were significantly overexpressed in HN, IgAN, and LN, and significantly underexpressed in FSGS-MCD and RPGN (and DN for QKI); 107 different genes were significantly differentially expressed relative to TN in at least 6 CKD entities (Figure 2a). Of note, several of the previously mentioned genes are considered to be expressed mainly in tubule. This could be explained by contamination of the glomerular samples with tubular cells during the microdissection procedure. Future studies using single-cell RNA sequencing (scRNA-seq) will dissect which genes are specifically expressed in glomerular cells during homeostasis and disease.
Figure 2.
Transcription-based map of chronic kidney disease (CKD) entities. (a) Radial heatmap of consistently differentially expressed genes across 6 or more disease entities (upregulation or downregulation). (b) Diffusion map of CKD entities reveals the underpinning geometric structure of the glomerular CKD transcriptomics data. DN, diabetic nephropathy; FSGS, focal segmental glomerulosclerosis; HLD, healthy living donor; HN, hypertensive nephropathy; IgAN, IgA nephropathy; LN, lupus nephritis; MCD, minimal change disease; MGN, membranous glomerulonephritis; RPGN, rapidly progressive glomerulonephritis; TN, tumor nephrectomy.
To better comprehend the divergence and similarities of the CKD samples, we asked how the distinct CKD entities localized with respect to each other using a common set of differentially expressed genes with regard to the nontumorous part of tumor nephrectomies using diffusion maps (Figure 2b). For illustrative purposes, we included the healthy tissue samples in the diffusion map; we did not use the healthy samples for differential expression analysis. The diffusion distances of each given CKD entity sample relative to TN samples reflect a nonlinear lower dimensional representation of the differences in gene expression profiles between those samples. The diffusion map orders the patients along a “pseudo-temporal” order, which we interpret here as an indicator of disease progression severity in glomeruli.29
The most distant condition from nephrectomy samples was RPGN, which is arguably the most drastic kidney disease condition with the most rapid functional decline among the entities included (Figure 2b). Healthy donor samples were distinct from TN samples even though the latter were resected distantly from the tumors. This might be explained by either minor contamination with cancer cells or paraneoplastic effects on the nonaffected kidney tissue, such as immune cell infiltration or solely that the nephrectomy tissue was exposed to short ischemia whereas the biopsy tissue from healthy donors was not. DN and LN were in close proximity to RPGN, whereas HN localized near IgAN. Differences were harder to assess in the middle of the diffusion map, but were visible when plotting the dimension components pair-wise (Supplementary Figure S2). For instance, MCD samples spanned from a point proximal to TN to near FSGS, but some MCD samples were in close proximity to MGN or even HN. Although it makes sense that MCD, as a relatively mild disease with normal light microscopy, is relatively close to the control groups of TN and healthy living donor, it remains unclear why other disease entities such as LN and DN spread widely in the diffusion map. Unfortunately, the data we used did not include information about disease severity, which might help to explain this heterogeneity, with early-stage disease possibly closer to the control groups and late-stage disease closer to RPGN. Dimension component 1 (DC1) seems to focus on the dissimilarity between the 2 reference healthy conditions, TN and healthy living donor from the CKD entities. Dimension component 2 (DC2) provides more insight into the disparity of the reference conditions. Dimension component 3 (DC3) discerns the subtle geometrical manifestation of the distinct CKD entities with regard to each other. In summary, using diffusion maps, we could visualize the intertwinement of the CKD entities that are present in our studies.
Transcription Factor Activity in CKD Entities
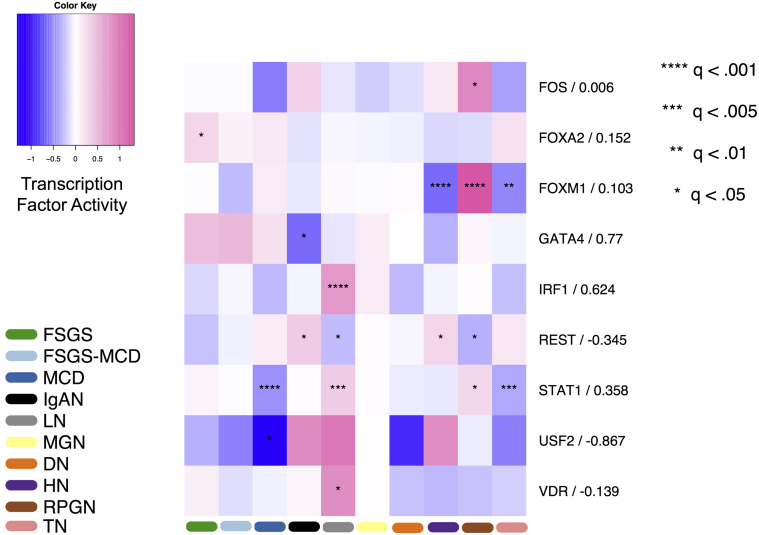
To further characterize the differences among the CKD entities, we performed various functional analyses. First, we assessed the activity of TFs based on the levels of expression of their known putative targets (see the Methods). These changes provide superior estimates of the TF activity than the expression level of the transcription factor itself21,30 (Figure 3). We found 10 TFs differentially regulated in at least one CKD entity. Furthermore, we correlated the activities of the identified TFs with their expression. Those TFs with no correlation indicate factors whose activity may be significantly modulated using posttranslational modifications or factors whose regulation or expression measurements are unconfident. For instance, interferon regulatory factor-1 (IRF1) is significantly enriched in LN and moderately correlated (Spearman’s rho, rs = 0.624) with the expression level of the gene encoding for IRF1. This might imply an as of yet undiscovered potential role of IRF1 as a transcriptional activator in LN. In addition, the transcriptional activity of IRF1 was elevated in LN compared with the other disease entities. The activity of the upstream stimulatory factor 2 (USF-2)31 was estimated to be significantly decreased in MCD compared with the rest of the conditions. Interestingly, the estimated activity of USF-2 across the CKD entities was inversely correlated, Spearman’s rho (rs = −0.867), with the expression level of the gene USF-2 itself.
Figure 3.
Transcription factor activity in glomerular chronic kidney disease (CKD) entities. Heatmap depicting transcription factor activity (color) for each CKD entity and tumor nephrectomy (TN) in glomerular tissue. Negative numbers (blue) signify decreased transcription factor activity, positive numbers (pink) indicate increased transcription factor activity. The significance according to the corresponding q-value of each transcription factor in each disease entity is represented by asterisk(s). The numbers to the right of factor names are Spearman’s rank-based correlation coefficients of factor activity and factor expression across different CKD entities. DN, diabetic nephropathy; FSGS, focal segmental glomerulosclerosis; HLD, healthy living donor; HN, hypertensive nephropathy; IgAN, IgA nephropathy; LN, lupus nephritis; MCD, minimal change disease; MGN, membranous glomerulonephritis; RPGN, rapidly progressive glomerulonephritis.
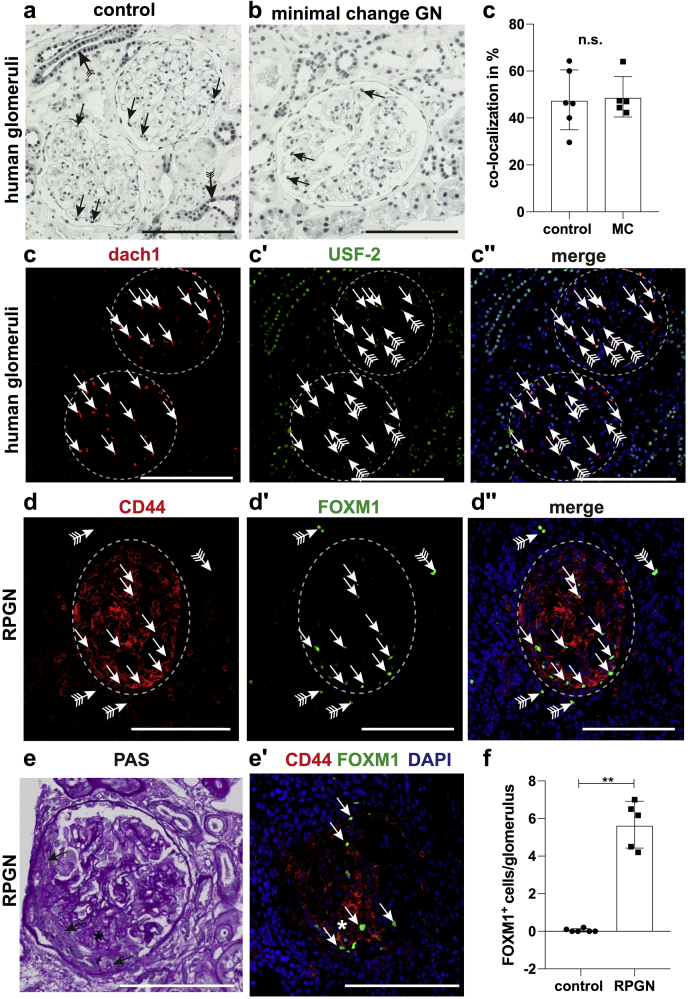
We next sought to validate the expression of the 2 previously identified TFs USF-2 and FOXM1 in human tissue by immunostaining. We choose these 2 TFs because the activity of USF2 was predicted to be the lowest in MCD, and FOXM1 to be highest in RPGN (Figure 3). We stained for USF2 in human kidney biopsies from healthy controls and patients with MCD. In glomeruli, USF2 was expressed in podocytes, the mainly affected glomerular cell-type in MCD (Figure 4a and b). However, when compared with controls, USF2 expression in podocytes showed no significant difference detectable by immunofluorescence (Figure 4c–c′′). This does not exclude reduced activity of USF-2, as this might be regulated not only by its abundance in the nucleus but also by its DNA binding capability, influenced in turn by, for example, posttranslational modifications and the interaction with other proteins. We then stained for FOXM1, a transcription factor of the forkhead box family. Our analysis suggests a highly increased activity of FOXM1 in RPGN (Figure 3). We next validated this observation in human biopsy samples from patients with RPGN and healthy controls. FOXM1 showed a unique expression in CD44-positive glomerular parietal epithelial cells in RPGN lesions, whereas we did not find any expression of FOXM1 in healthy human glomeruli (Figure 4d–f). Consistent with our TF activity analysis, quantification of this finding in 5 RPGN biopsies versus 6 controls yielded a highly significant difference (Figure 4f), indicating that FOXM1 could play a role in RPGN progression. These data suggest that our computational method might be useful to identify novel regulators in CKD.
Figure 4.
Validation of upstream stimulatory factor 2 (USF-2) and FOXM1 in human kidney biopsies. (a–c) Histological validation of USF-2 expression in human biopsies from patients with minimal change disease (n = 5) and controls (n = 6). (a) Immunohistochemical staining of USF-2 showed expression in nuclei of many cell types of the kidney including tubular cells (strongest in collecting duct, arrow with tails). In the glomeruli, USF-2 expression could be detected in podocytes (arrows). (b) USF-2 staining in biopsies from patients with minimal change disease demonstrated a similar staining pattern compared with controls including expression in podocytes (arrows). (c–c′′) Colocalized Ddach1 (podocyte marker in red) and USF-2 (in green). Arrows mark Dach1-USF-2 double-positive podocytes. (d–f) Histological validation of FoxM1 expression in human biopsies from patients with rapidly progressive glomerulonephritis (RPGN) (n = 5) and controls (n = 6). FoxM1 expression was detected most abundantly in glomeruli with crescentic CD44+ lesions (arrows in d–d′′). Rarely expression could be noted in the tubular compartment (arrows with tails). (e–e′) Serial sections revealed that FoxM1 expression was mainly detected in CD44+ cells in the glomerular crescentic lesions (arrows in e and e′). (f) Quantification of number of glomerular FoxM1+ cells control versus RPGN (P = 0.0043). **P < 0.01 by unpaired Mann-Whitney t test (c and f and bar plot data represent mean ± SD. Bars = 100 μm. DAPI, 4′,6-diamidino-2-phenylindole; n.s., not significant; PAS, periodic acid–Schiff.
Signaling Pathway Analysis
We complemented the functional characterization of transcription factor activities with an estimation of pathway activities with the tools PROGENy22 and Piano.23
Pathway Activity of CKD Entities Using PROGENy
PROGENy infers pathway activity by looking at the changes in levels of the genes downstream of the corresponding pathways, rather than to the genes that constitute the pathway. This provides a better proxy of pathway activity than assessing the genes in the actual pathway.22 We used PROGENy to estimate pathway activity in the different disease entities from the gene expression data (Figure 5a). Essentially, the degree of pathway deregulation was associated with the degree of disease severity, and present rather divergent activities across the CKD entities. For example, vascular endothelial growth factor pathway was estimated to be significantly influential in 5 CKD entities: RPGN, HN, DN, LN, and IgAN, of which it is predicted to be deactivated in RPGN and DN, but more activated in HN, LN, and IgAN. Ten of 11 pathways were predicted to be significantly deregulated in RPGN with respect to TN, in accordance with the diffusion map (Figure 2b) outcome; the divergence of RPGN from TN (control) was considerably more prominent both at a global transcriptome landscape and signaling pathway level. Intriguingly, the Janus kinase–signal transducers and activators of transcription (JAK-STAT) pathway did not appear to be affected in RPGN, but was considerably activated in LN and markedly deactivated in DN in comparison with TN. Overall, the CKD entities were characterized by distinct combinations, magnitudes, and directions of signaling pathway activities according to PROGENy.
Figure 5.
Pathway activity alterations in chronic kidney disease (CKD) entities. (a) Heatmap depicting pathway activity (color) for each CKD entity relative to tumor nephrectomy in glomerular tissue, according to PROGENy.22 The magnitude and direction (positive or negative) of PROGENy scores indicate the degree of pathway deregulation in a given CKD entity with regard to the reference condition, tumor nephrectomy. Permutation q-values are used to indicate statistical significance of each pathway in each disease entity, indicated by asterisks. (b) Radial heatmap of consensually enriched pathways across 3 or more disease entities (upregulated, downregulated, or nondirectional regulation) according to Piano23 using MSigDB-C2-CP gene sets. DN, diabetic nephropathy; FSGS, focal segmental glomerulosclerosis; HN, hypertensive nephropathy; IgAN, IgA nephropathy; LN, lupus nephritis; MCD, minimal change disease; MGN, membranous glomerulonephritis; RPGN, rapidly progressive glomerulonephritis.
Pathway Enrichment With Piano
Although PROGENy can give accurate estimates of pathway activity, it is limited to 11 pathways for which robust signatures could be generated.22 To get a more global picture, we complemented that analysis with a gene set enrichment analysis using Piano.23 A total of 160 pathways of 1329 were significantly enriched (upregulated/downregulated, corrected P < 0.05) in at least 1 CKD entity. HN was the entity with the largest number of differentially enriched pathways (81; 25 downregulated, 56 upregulated), whereas FSGS-MCD did not show significant enrichment for any pathway. Cell-cycle and immune-system–related pathways were significantly upregulated in 7 of 9 CKD entities (FSGS, HN, IgAN, LN, MGN, and RPGN in both cases, DN for immune system, and MCD for cell-cycle); in contrast, the vascular endothelial growth factor pathway was differentially enriched in LN only. Interestingly, the tumor necrosis factor receptor 2 pathway was differentially enriched in IgAN, HN, and LN, in line with the results from PROGENy, in which the vascular endothelial growth factor pathway was significantly deregulated not only in IgAN, HN, and LN, but also in RPGN and DN. A total of 59 different pathways showed significant enrichment in at least 3 CKD entities (Figure 5b). Figure 4b also shows that HN (52), MGN (45), and IgAN (37) are the CKD entities with more pathways differentially enriched in at least 3 entities, a result that agrees with Figure 2b showing these entities in the center of the diffusion map.
Prediction of Potential Novel Drugs That Might Affect the Identified Disease Signature in Different Kidney Diseases
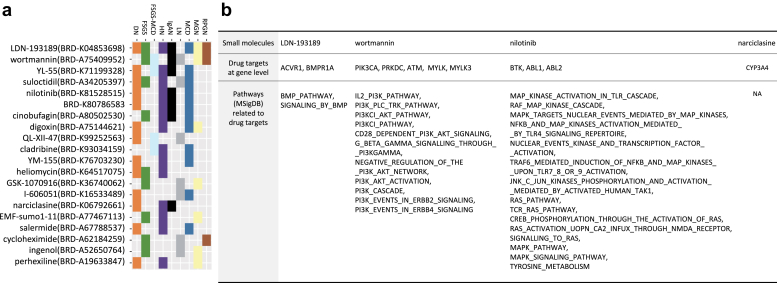
Finally, we applied a signature search engine, L1000CDS2,24 that prioritizes drugs that are expected to have a reverse signature compared with the disease signature. This engine is based on computing the distance between the signature of the disease and the signature of the LINCS-L1000 data, a large collection of changes in gene expression driven by drugs. We performed this analysis separately for the 9 CKD entities and identified 220 small molecules across the CKD entitles (Supplementary Figure S5). To narrow down the list of 220 small molecules, we focused on 20 small molecules observed in the L1000CDS2 output of at least 3 subtypes (Figure 6a).
Figure 6.
Top 20 drug candidates from drug repositioning. (a) Distribution of 20 small molecules reversely correlated with at least 3 chronic kidney disease entities. (b) Table of 4 small molecules of the 20 of (a) supported by manual curation. Table shows drugs (first row), protein coding genes targeted by these 4 drugs (second row), and pathways (MSigDB) related to the biological functions these drugs affect (third row).
By curation of scientific publications, we found that 4 small molecules have experimental evidence to support their clinical relevance in CKD or renal disease animal model testing (Supplementary Figure S7). BRD-K04853698 (LDN-193189), which is known as a selective bone morphogenic protein signaling inhibitor, has been shown to suppress endothelial damage in mice with CKD.32 Wortmannin, a cell-permeable PI3K inhibitor, decreased albuminuria and podocyte damage in early DN in rats.33 The tyrosine kinase inhibitor nilotinib is used to treat chronic myelogenous leukemia in humans.34 Nilotinib treatment resulted in stabilized kidney function and prolonged survival after subtotal nephrectomy in rats when compared with vehicle.35 Finally, narciclasine was identified, and it has been reported to reduce macrophage infiltration and inflammation in the mouse unilateral ureteral obstruction model of kidney fibrosis.36
To further explore the association of these drugs with CKD and its progression, we analyzed the expression data for the targets of the drug candidates. First, each drug candidate was mapped to genes that encode the proteins targeted by these drugs (Figure 6b). For each gene, its differential expression of any CKD entity against TN was evaluated. Of the 11 mapped genes, MYLK3, a target of narciclasine, was significantly differentially expressed (underexpressed, logFC <−1, P < 0.05) in 2 CKD entities (IgAN and LN) (Supplementary Figure S6). Complementarily, screened drugs were mapped to the pathways they affect based on their functional information. The enrichment of the subset of pathways was evaluated using the previous results from the gene set analysis algorithm (Piano). This time, only the PI3KCI pathway appeared to be enriched in HN (upregulated, P < 0.05), and as the pathway affected by the candidate repositioned drug (Wortmannin, PI3K inhibitor). Taken together, these data suggest that kidney transcriptomics might be useful to predict potential drug candidates novel for CKD.
Conclusion
We have aimed to shed light on the commonalities and differences among glomerular transcriptomes of major kidney diseases contributing to the CKD epidemic affecting >10% of the population worldwide. Multiple pathologies are covered under the broad umbrella of CKD and, although they share a physiological manifestation (i.e., loss of kidney function), the driving molecular processes can be different. In this study, we explored these processes by analyzing glomerular gene expression from kidney biopsies obtained via microdissection. We observed expression data of many genes that are considered to be tubule-specific in the glomerular data set (e.g., ALB and CALB1). This might be due to contamination of the glomerular samples with surrounding tubular cells as a consequence of imperfect microdissection. Current technologies, including scRNA-seq will help to dissect expression in particular cell types of the glomerulus.
Genes such as Quaking (QKI) or Lysozyme C (LYZ), were significantly overexpressed, underexpressed, or not altered depending on the underlying kidney disease. It is known that QKI is associated with angiogenic growth factor release and plays a pathological role in the kidney,37 whereas LYZ is known to be related to the extent of vascular damage and heart failure38 and has recently been found to be increased in plasma during CKD progression.39 These data support the fact that despite a stereotypic response of the kidney to injury with glomerulosclerosis, interstitial fibrosis, and nephron loss, there are various disease-specific differences that are important to understand so as to develop novel personalized therapies.
CKD is a complex disease that can be acquired through a variety of biological mechanisms. Our pathway analysis reflects this heterogeneity. There was little to no overlap in significantly enriched pathways between the different kidney disease entities. We found 59 different pathways that showed significant enrichment in at least 3 disease entities (Figure 5b), indicating that different disease entities share some general mechanisms but their underlying pathophysiology differs from one to another. Besides increasing the interpretability, the pathway analysis identified many more differences among disease identities than the gene-level analysis (Figures 2a and 5b). For example, pathway analysis identified pathways related to the metabolism of lipids and lipoproteins significantly downregulated in MCD, MGN, and HN; and pathways related to fatty acid metabolism significantly downregulated in MCD, IgAN, MGN, and HN, results similar to those reported by Kang et al.6
PROGENy (Figure 5a) yielded JAK-STAT, a major cytokine signal transduction regulator,40 to be significantly activated in LN with respect to TN, and DoRothEA (Figure 3) predicted the TFs IRF1 and STAT1 to be significantly enriched in LN and downregulated in DN. A pathogenic role of JAK-STAT/STAT1/interferon signaling in LN is supported by various studies.41, 42, 43 Indeed, different human and mouse studies have shown an upregulation of JAK-STAT signaling in DN,44,45 in contrast to our results showing decrease in JAK-STAT. However, the study of Berthier et al.45 also revealed a downregulation of JAK2 mRNA in glomeruli of advanced/progressive diabetic kidney disease in humans. Pathway activities could vary depending on, among many other factors, the state of pathology of the cohort. Such a difference or other confounding factors could explain this discrepancy.
We next aimed to compare some of the predicted pathway activity to the literature. Interestingly, our analysis predicted increased nuclear factor-κB pathway activity in LN and it has been shown that selective inhibition of nuclear factor-κB inducing kinase reduces disease severity in an LN mouse model.46 Furthermore, we predicted increased phosphoinositide 3-kinase activity in FSGS and a human causing FSGS mutation in the anilin gene was shown to increase phosphoinositide 3-kinase activity in podocytes.47
We also used a signature-matching algorithm to explore potential drugs that could revert the disease phenotype. We found that 4 drugs hold promise in different CKD entities. Even though more experimental and clinical validation is required, our approach suggests that it is possible to find promising treatments for CKD via drug repositioning. In particular, for one of the identified drugs, nilotinib, use in humans has already been granted in leukemia and there are supporting data of its valuable insight at indications for CKD.35
Analysis of expression of the drug targets found that MYLK3, a gene encoding for one of the targets of narciclasine, was significantly underexpressed in IgAN and LN when compared with TN. Similarly, the PI3KCI pathway, the target of Wortmannin was enriched in HN (upregulated, P < 0.05). This analysis attempted to refine the outcome of the repositioning analysis and at the same time helped to connect it to the disease mechanism at both the gene and pathway levels.
The analysis of TF activity revealed significantly higher FOXM1 activity in RPGN over all other kidney diseases analyzed. RPGN is characterized by a rapid decline in kidney function due to proliferation of parietal epithelial cells in the glomerulus, which leads to deterioration of the associated nephron.48 FOXM1, a TF crucially involved in proliferation,49 could represent a potential therapeutic target in glomerular parietal epithelial cells in RPGN. The fact that protein level of USF-2 was not significantly changed does not exclude reduced activity of USF-2, as the activity of transcription factors is very often influenced not only by their expression but also by posttranslational modifications and binding to other proteins.
We view our analysis as a first, preliminary step toward a characterization of the similarities and differences of the various pathologies that lead to CKD. As more data become available, either from micro-arrays or RNA sequencing, these can be integrated in our pipeline. Furthermore, the burgeoning field of scRNA-seq has just started to produce data from kidneys50,51 and can revolutionize our understanding of the functioning of the kidney and its pathologies.52,53 In particular, scRNA-seq data can provide signatures of the many cell types in the kidney, which in turn can be used to deconvolute the composition of cell types12 in the more abundant and cost-effective bulk expression data sets.53 Other data sets, such as (phospho)proteomics54 and metabolomics,55 may complement gene expression toward a more complete picture of the CKD entities.56 Ideally, all these data sets will be collected in a standardized manner to facilitate integration, which was a major hurdle in our study. Such a comprehensive analysis across large cohorts, akin to what has happened for the different tumor types thanks to initiatives such as the International Cancer Genome Consortium, can lead to major improvements in our understanding and treatment of CKD.57
Looking forward, our aim is to extend our collection and subsequent analyses as more datasets are available. Our methods complement those already available elsewhere, in particular in NephroSeq. While we provide our own user-friendly interface to mine the results, we could envision to embed them within NephroSeq and/or other resources.
Limitations
Our results should be interpreted with caution because of the limitations coming from the data, which required an aggressive batch effect mitigation procedure. Nonetheless, our analysis could provide pointers to mechanisms in CKD entities to be further studied. Furthermore, our work provides an example of the potential usefulness of integrating publicly available data. Further, the limitations observed in this study shed light into the lack of standardization of basic experimental designs in the CKD community. To learn the most of CKD, the community should work collectively to create fundamental experimental and data handling guidelines. This should result in more comparable and robust data across research laboratories.
The integration of the data from different sources and platforms requires batch effect management, which should be customized to the data at hand. The current data were heavily affected by platform- and study-specific batch effects, because the outcome categories (CKD entities and their samples) were unevenly distributed across studies and microarray platforms. The commonly used algorithms for correcting batch effects assume a balanced distribution of outcome categories across batches and are vulnerable to the group-batch imbalance.58, 59, 60, 61 We conducted a stringent batch effect mitigation process to minimize the influence of technical heterogeneity. Note that this is a more stringent approach than other batch correction approaches that seek to “model-away” batch-related variance but retain all the data. In our case, we opted to remove genes that are most affected by batch effects. For the illustration of this procedure, see Supplementary Figure S1.
Further, we had to omit crucial and pertinent studies, such as Woroniecka et al.62 or Beckerman et al.3 Adding Woroniecka et al.62 would have required introducing a third microarray platform, further complicating the batch mitigation procedure. We did not include Beckerman et al.,3 as we focused on microdissected glomerular fractions, whereas this study provides data from tubuli.
Because of the heterogeneity of the included samples and strong batch effects, we identified only a small number of genes that were differentially expressed, and thus one has to be cautious when drawing conclusions from this analysis. However, our aim is primarily to demonstrate several computational tools that have been developed mainly in cancer settings and will be of major interest in future analyses when more kidney transcriptomic data is openly available.
One important limitation of the study is the lack of detailed individual clinical data that were not deposited together with the raw data and also not available on request. Further, public freely available data were included into this work and thus we did not have any influence on the data generation, quality, and standardization. The limited number of available samples also made use of pool data from patients, only differentiating based on the reported disease entities. Unfortunately, data on glomerular filtration rate and histological scores are highly sparse, and given the limited number of samples, further stratification would have left us with a considerably diminished statistical power. Our analysis resulted in major differences between disease subtypes, although there are likely confounding effects due to different degrees of disease/clinical phenotype. Thus, our results should be taken with caution and rather considered as hypotheses requiring further studies to be validated.
Furthermore, microdissection often results in cross-contamination of tubule or glomerular fragments, and thus the presented glomerular data do contain potentially various tubule-specific genes. Future scRNA-seq experiments will demonstrate the cell-specific and compartment-specific expression of genes and overcome the current issues with microdissection. In addition, for the drug-matching approach, we had to rely on cell lines that are not necessarily originating from kidney tissues. We plan to revisit this method when a new kidney-specific data set will be available. This will likely improve the prediction accuracy.
In summary, with this article, we do not claim to derive specific precise insights, given the clear limitations in the quality of the public kidney data available so far. Rather, we wish to demonstrate what is possible to achieve with computational functional genomics tools that can be used with high-quality omics and clinical data that will hopefully be available soon.
Disclosure
All the authors declared no competing interests.
Acknowledgments
This work was supported by the JRC for Computational Biomedicine, which was partially funded by Bayer AG, the European Union Horizon 2020 grant SyMBioSys MSCA-ITN-2015-ETN #675585, which provided financial support for AA, and by grants of the German Research Foundation (SFB/TRR57 and P30), a grant of the European Research Council (ERC-StG 677448) to RK, and a grant of the German Society of Internal Medicine to CK. We thank Nicolas Palacio for feedback on the manuscript.
Footnotes
Figure S1. Batch effect mitigation procedure. (A) Principal component analysis (PCA) of gene expression measurements corresponding of IgAN samples from the 2 distinct studies prior batch effect mitigation. The second principal component separates samples by study. (B) MA plot visualizing the difference in gene expression between the GSE37460 and GSE50469 IgAN samples. (C) Percentage of variance explained by principal component 2 (PC2) as a function of the gradual removal of the most affected genes (−log10 adjusted P value of a particular removed affected gene). (D) PCA of gene expression corresponding to the IgAN samples from the 2 distinct studies after batch effect mitigation. (E) Depiction of variance for each gene that is explained by group (CKD entity), study, and platform after batch effect mitigation. CKD entity-related variation explains most of the variance in the data. The scater R package1 was used for producing the batch effect management-related plots.
Figure S2. Two-dimensional diffusion maps of CKD entities unravel the geometric trajectory of CKD entities based on their comparative transcriptome profile. (A) Dimension component 1 (DC1) is depicted against dimension component 2 (DC2), so that the divergence between the controls and the CKD entities are apparent. (B) DC1 is visualized against dimension component 3 (DC3), revealing the fine distinctions between CKD entities.
Figure S3. Transcriptional regulation in CKD entities. Heatmap of consistently differentially expressed genes across 6 or more disease entities (upregulation or downregulation).
Figure S4. Hierarchical clustering of CKD entities based on a common set of differentially expressed genes with regard to the nontumorous part of tumor nephrectomies. The figure is a complementary representation of Figure 2b.
Figure S5. Heatmap depicting the expression of the genes encoding for the transcription factors shown in Figure 3. The expression values were averaged within each condition, then scaled and centered across the conditions. The numbers to the right of factor names are Spearman’s rank-based correlation coefficients of factor activity and factor expression across different CKD entities.
Figure S6. Enrichment of metabolic pathways after gene set analysis. Pathway analysis result in metabolic pathways (“METABOL”): and their corresponding enrichment: upregulation (green), downregulation (red), and nonsignificant (white). Metabolic pathways are listed on the y-axis and disease entities in the x-axis. Only pathways enriched in at least 1 disease are shown. Note that FSGS, FSGS-MCD, and RPGN do not have any metabolic pathway significantly affected.
Figure S7. Bar graph (count of CKD entities) and heatmap of the distribution of 220 small molecules reversely correlated with 9 CKD entities. Colored bars on both the bar graph and heat map correspond to the subtype of CKD entities and 220 small molecules are represented on the x-axis of both graphs.
Figure S8. Volcano plot of differential expression of CKD entities versus TN for glomerular samples for the drug-targeted genes. The x-axis indicates the log2 of the fold change (FC) and the y-axis the −log10 of the P value after differential expression analysis using limma.
Figure S9. Manual curation of 4 small molecules. The figure includes drug names corresponding to 4 small molecules, biological function, Food and Drug Administration (FDA) approval status, and publications describing the clinical relevance of the particular small molecule in CKD.
Figure S10. Cell lines used in the drug-matching paradigm. The number of significant perturbations of a given cell line per condition.
Data and Code.
Supplementary Methods. Data collection, preprocessing and mapping, correlation of arrays, batch effect mitigation, detection of genes with consistently small p values across all studies, transcription factor activity analysis with DoRothEA, inferring signaling pathway activity using PROGENy, pathway analysis with piano, drug repositioning, immunofluorescent staining of human kidney biopsies and analysis, and supplementary references.
Contributor Information
Rafael Kramann, Email: rkramann@gmx.net.
Julio Saez-Rodriguez, Email: julio.saez@bioquant.uni-heidelberg.de.
Translational Statement
Different etiologies cause chronic kidney disease. We integrate and analyze transcriptomic analysis of glomerular and tubular compartments from different entities to dissect their different pathophysiology, what might help to identify novel entity-specific therapeutic targets.
Supplementary Material
References
- 1.Hill N.R., Fatoba S.T., Oke J.L. Global prevalence of chronic kidney disease - a systematic review and meta-analysis. PLoS One. 2016;11 doi: 10.1371/journal.pone.0158765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamer R.A., El Nahas A.M. The burden of chronic kidney disease. BMJ. 2006;332:563–564. doi: 10.1136/bmj.332.7541.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beckerman P., Qiu C., Park J. Human kidney tubule-specific gene expression based dissection of chronic kidney disease traits. EBioMedicine. 2017;24:267–276. doi: 10.1016/j.ebiom.2017.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nair V., Komorowsky C.V., Weil E.J. A molecular morphometric approach to diabetic kidney disease can link structure to function and outcome. Kidney Int. 2018;93:439–449. doi: 10.1016/j.kint.2017.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schena F.P., Nistor I., Curci C. Transcriptomics in kidney biopsy is an untapped resource for precision therapy in nephrology: a systematic review. Nephrol Dial Transplant. 2017;32:1776. doi: 10.1093/ndt/gfx268. [DOI] [PubMed] [Google Scholar]
- 6.Kang H.M., Ahn S.H., Choi P. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat Med. 2015;21:37–46. doi: 10.1038/nm.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ju W., Nair V., Smith S. Tissue transcriptome-driven identification of epidermal growth factor as a chronic kidney disease biomarker. Sci Transl Med. 2015;7:316ra193. doi: 10.1126/scitranslmed.aac7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Athey B.D., Cavalcoli J.D., Jagadish H.V. The NIH National Center for Integrative Biomedical Informatics (NCIBI) J Am Med Inform Assoc. 2012;19:166–170. doi: 10.1136/amiajnl-2011-000552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martini S., Eichinger F., Nair V. Defining human diabetic nephropathy on the molecular level:integration of transcriptomic profiles with biological knowledge. Rev Endocr Metab Disord. 2008;9:267–274. doi: 10.1007/s11154-008-9103-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neusser M.A., Lindenmeyer M.T., Moll A.G. Human nephrosclerosis triggers a hypoxia-related glomerulopathy. Am J Pathol. 2010;176:594–607. doi: 10.2353/ajpath.2010.090268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berthier C.C., Bethunaickan R., Gonzalez-Rivera T. Cross-species transcriptional network analysis defines shared inflammatory responses in murine and human lupus nephritis. J Immunol. 2012;189:988–1001. doi: 10.4049/jimmunol.1103031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ju W., Greene C.S., Eichinger F. Defining cell-type specificity at the transcriptional level in human disease. Genome Res. 2013;23:1862–1873. doi: 10.1101/gr.155697.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martini S., Nair V., Keller B.J. Integrative biology identifies shared transcriptional networks in CKD. J Am Soc Nephrol. 2014;25:2559–2572. doi: 10.1681/ASN.2013080906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hodgin J.B., Berthier C.C., John R. The molecular phenotype of endocapillary proliferation: novel therapeutic targets for IgA nephropathy. PLoS One. 2014;9 doi: 10.1371/journal.pone.0103413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smyth G.K. limma: linear models for microarray data. In: Gentleman R., Carey V.J., Huber W., editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Statistics for Biology and Health. Springer; New York, NY: 2005. pp. 397–420. [Google Scholar]
- 16.Smyth G.K., Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 17.Ritchie M.E., Phipson B., Wu D. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lê Cao K.-A., Rohart F., McHugh L. YuGene: a simple approach to scale gene expression data derived from different platforms for integrated analyses. Genomics. 2014;103:239–251. doi: 10.1016/j.ygeno.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Wilkinson B. A statistical consideration in psychological research. Psychol Bull. 1951;48:156–158. doi: 10.1037/h0059111. [DOI] [PubMed] [Google Scholar]
- 20.Angerer P., Haghverdi L., Büttner M. destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics. 2016;32:1241–1243. doi: 10.1093/bioinformatics/btv715. [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Alonso L.M., Iorio F., Matchan A. Transcription factor activities enhance markers of drug sensitivity in cancer. Cancer Res. 2018;78:769–780. doi: 10.1158/0008-5472.CAN-17-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schubert M., Klinger B., Klünemann M. Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat Commun. 2018;9:20. doi: 10.1038/s41467-017-02391-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Väremo L., Nielsen J., Nookaew I. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 2013;41:4378–4391. doi: 10.1093/nar/gkt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duan Q., Reid S.P., Clark N.R. L1000CDS2:LINCS L1000 characteristic direction signatures search engine. NPJ Syst Biol Appl. 2016;2 doi: 10.1038/npjsba.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuppe C., Gröne H.-J., Ostendorf T. Common histological patterns in glomerular epithelial cells in secondary focal segmental glomerulosclerosis. Kidney Int. 2015;88:990–998. doi: 10.1038/ki.2015.116. [DOI] [PubMed] [Google Scholar]
- 26.Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrett T., Wilhite S.E., Ledoux P. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phipson B., Lee S., Majewski I.J. Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann Appl Stat. 2016;10:946–963. doi: 10.1214/16-AOAS920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haghverdi L., Buettner F., Theis F.J. Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics. 2015;31:2989–2998. doi: 10.1093/bioinformatics/btv325. [DOI] [PubMed] [Google Scholar]
- 30.Alvarez M.J., Shen Y., Giorgi F.M. Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat Genet. 2016;48:838–847. doi: 10.1038/ng.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuda M., Tamura K., Wakui H. Upstream stimulatory factors 1 and 2 mediate the transcription of angiotensin II binding and inhibitory protein. J Biol Chem. 2013;288:19238–19249. doi: 10.1074/jbc.M113.451054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kajimoto H., Kai H., Aoki H. BMP type I receptor inhibition attenuates endothelial dysfunction in mice with chronic kidney disease. Kidney Int. 2015;87:128–136. doi: 10.1038/ki.2014.223. [DOI] [PubMed] [Google Scholar]
- 33.Kim S.H., Jang Y.W., Hwang P. The reno-protective effect of a phosphoinositide 3-kinase inhibitor wortmannin on streptozotocin-induced proteinuric renal disease rats. Exp Mol Med. 2012;44:45–51. doi: 10.3858/emm.2012.44.1.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blay J.-Y., von Mehren M. Nilotinib: a novel, selective tyrosine kinase inhibitor. Semin Oncol. 2011;38(Suppl 1):S3–S9. doi: 10.1053/j.seminoncol.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iyoda M., Shibata T., Hirai Y. Nilotinib attenuates renal injury and prolongs survival in chronic kidney disease. J Am Soc Nephrol. 2011;22:1486–1496. doi: 10.1681/ASN.2010111158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fuchs S., Hsieh L.T., Saarberg W. Haemanthus coccineus extract and its main bioactive component narciclasine display profound anti-inflammatory activities in vitro and in vivo. J Cell Mol Med. 2015;19:1021–1032. doi: 10.1111/jcmm.12493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gomez I.G., Nakagawa N., Duffield J.S. MicroRNAs as novel therapeutic targets to treat kidney injury and fibrosis. Am J Physiol Renal Physiol. 2016;310:F931–F944. doi: 10.1152/ajprenal.00523.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abdul-Salam V.B., Ramrakha P., Krishnan U. Identification and assessment of plasma lysozyme as a putative biomarker of atherosclerosis. Arterioscler Thromb Vasc Biol. 2010;30:1027–1033. doi: 10.1161/ATVBAHA.109.199810. [DOI] [PubMed] [Google Scholar]
- 39.Glorieux G., Mullen W., Duranton F. New insights in molecular mechanisms involved in chronic kidney disease using high-resolution plasma proteome analysis. Nephrol Dial Transplant. 2015;30:1842–1852. doi: 10.1093/ndt/gfv254. [DOI] [PubMed] [Google Scholar]
- 40.Rawlings J.S., Rosler K.M., Harrison D.A. The JAK/STAT signaling pathway. J Cell Sci. 2004;117:1281–1283. doi: 10.1242/jcs.00963. [DOI] [PubMed] [Google Scholar]
- 41.Dong J., Wang Q.X., Zhou C.Y. Activation of the STAT1 signalling pathway in lupus nephritis in MRL/lpr mice. Lupus. 2007;16:101–109. doi: 10.1177/0961203306075383. [DOI] [PubMed] [Google Scholar]
- 42.Wang S., Yang N., Zhang L. Jak/STAT signaling is involved in the inflammatory infiltration of the kidneys in MRL/lpr mice. Lupus. 2010;19:1171–1180. doi: 10.1177/0961203310367660. [DOI] [PubMed] [Google Scholar]
- 43.Ripoll È., de Ramon L., Draibe Bordignon J. JAK3-STAT pathway blocking benefits in experimental lupus nephritis. Arthritis Res Ther. 2016;18:134. doi: 10.1186/s13075-016-1034-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hodgin J.B., Nair V., Zhang H. Identification of cross-species shared transcriptional networks of diabetic nephropathy in human and mouse glomeruli. Diabetes. 2013;62:299–308. doi: 10.2337/db11-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berthier C.C., Zhang H., Schin M. Enhanced expression of Janus kinase–signal transducer and activator of transcription pathway members in human diabetic nephropathy. Diabetes. 2009;58:469–477. doi: 10.2337/db08-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brightbill H.D., Suto E., Blaquiere N. NF-κB inducing kinase is a therapeutic target for systemic lupus erythematosus. Nat Commun. 2018;9:179. doi: 10.1038/s41467-017-02672-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hall G., Lane B.M., Khan K. The human FSGS-causing ANLN R431C mutation induces dysregulated PI3K/AKT/mTOR/Rac1 signaling in podocytes. J Am Soc Nephrol. 2018;29:2110–2122. doi: 10.1681/ASN.2017121338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smeets B., Uhlig S., Fuss A. Tracing the origin of glomerular extracapillary lesions from parietal epithelial cells. J Am Soc Nephrol. 2009;20:2604–2615. doi: 10.1681/ASN.2009010122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wierstra I., Alves J. FOXM1, a typical proliferation-associated transcription factor. Biol Chem. 2007;388:1257–1274. doi: 10.1515/BC.2007.159. [DOI] [PubMed] [Google Scholar]
- 50.Sivakamasundari V., Bolisetty M., Sivajothi S. Comprehensive cell type specific transcriptomics of the human kidney. bioRxiv. 2017 [Google Scholar]
- 51.Wu H., Uchimura K., Donnelly E. Comparative analysis and refinement of human PSC-derived kidney organoid differentiation with single-cell transcriptomics. Cell Stem Cell. 2018;23:869–881.e8. doi: 10.1016/j.stem.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu H., Humphreys B.D. The promise of single-cell RNA sequencing for kidney disease investigation. Kidney Int. 2017;92:1334–1342. doi: 10.1016/j.kint.2017.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kiryluk K., Bomback A.S., Cheng Y.-L. Precision medicine for acute kidney injury (AKI): redefining AKI by agnostic kidney tissue interrogation and genetics. Semin Nephrol. 2018;38:40–51. doi: 10.1016/j.semnephrol.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu P., Lassén E., Nair V. Transcriptomic and proteomic profiling provides insight into mesangial cell function in IgA nephropathy. J Am Soc Nephrol. 2017;28:2961–2972. doi: 10.1681/ASN.2016101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hocher B., Adamski J. Metabolomics for clinical use and research in chronic kidney disease. Nat Rev Nephrol. 2017;13:269–284. doi: 10.1038/nrneph.2017.30. [DOI] [PubMed] [Google Scholar]
- 56.Saez-Rodriguez J., Rinschen M.M., Floege J. Big science and big data in nephrology. Kidney Int. 2019;95:1326–1337. doi: 10.1016/j.kint.2018.11.048. [DOI] [PubMed] [Google Scholar]
- 57.Mariani L.H., Pendergraft W.F., 3rd, Kretzler M. Defining glomerular disease in mechanistic terms: implementing an integrative biology approach in nephrology. Clin J Am Soc Nephrol. 2016;11:2054–2060. doi: 10.2215/CJN.13651215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 59.Goh W.W.B., Wang W., Wong L. Why batch effects matter in omics data, and how to avoid them. Trends Biotechnol. 2017;35:498–507. doi: 10.1016/j.tibtech.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 60.Nygaard V., Rødland E.A., Hovig E. Methods that remove batch effects while retaining group differences may lead to exaggerated confidence in downstream analyses. Biostatistics. 2016;17:29–39. doi: 10.1093/biostatistics/kxv027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leek J.T., Scharpf R.B., Bravo H.C. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet. 2010;11:733. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Woroniecka K.I., Park A.S.D., Mohtat D. Transcriptome analysis of human diabetic kidney disease. Diabetes. 2011;60:2354–2369. doi: 10.2337/db10-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.