Abstract
Fast and accurate detection of viral RNA pathogens is important in apiculture. A polymerase chain reaction (PCR)-based detection method has been developed, which is simple, specific, and sensitive. In this study, we rapidly (in 1 min) synthesized cDNA from the RNA of deformed wing virus (DWV)-infected bees (Apis mellifera), and then, within 10 min, amplified the target cDNA by ultra-rapid qPCR. The PCR products were hybridized to a DNA-chip for confirmation of target gene specificity. The results of this study suggest that our method might be a useful tool for detecting DWV, as well as for the diagnosis of RNA virus-mediated diseases on-site.
Keywords: Ultra-rapid reverse transcription-qPCR, DNA-chip, deformed wing virus, field detection
INTRODUCTION
Deformed wing virus (DWV) is one of the most common and widespread of the 22 known honeybee pathogens, including viruses [1]. DWV has also been documented in other bee species, such as the bumblebee (Bombus terrestris) [2,3], and thus, DWV may have a wide host specificity. Like many other honeybee viral pathogens, DWV generally persists as a latent infection with no apparent symptoms. Infected bees may exhibit damaged appendages, particularly stubby, useless wings, shortened, rounded abdomens, discoloration, and paralysis of the legs and wings [4]. In addition, symptomatic bees are typically expelled from the hive and have severely reduced life-spans, usually less than 48 hours [5]. Therefore, rapid and accurate detection of DWV RNA is very important in apiculture.
Several techniques for the detection of pathogens have been employed in the detection of bee viruses, including indirect fluorescent antibody analysis, agarose gel immunodiffusion, enzyme-linked immunosorbent assay (ELISA), and reverse transcription-polymerase chain reaction (RT-PCR). However, these methods are traditional techniques that are low in sensitivity and specificity [6,7].
Since the development of PCR, it has been used in medical, and clinical laboratory research in a wide range of applications due to its simplicity, specificity, and sensitivity. Reverse transcription-quantitative PCR (RT-qPCR), a variant of real-time quantitative PCR (qPCR), has been a technique commonly used in molecular biology to detect RNA transcript levels. With the technological development of qPCR, it became possible to monitor the amplification process in real-time and to consider quantitative assays for the detection of RNA levels. The use of two-step qPCR and one-step qPCR methods for the detection of viral RNA pathogens has been reported [6,8,9,10]. However, these methods are not suitable for rapid, on-site pathogen detection. PCR-based detection methods have been developed for enhancement of sensitivity when detecting pathogens in environmental samples [11,12,13]. In addition, further development of PCR platforms could lead to enhanced rapid detection of pathogens, and this could enable detection on-site. Recently, real-time micro-scale chip-based PCR systems, which can supply high-speed temperature homogeneity inside the reaction chip, have been developed. In this study, we suggest a method for the rapid detection and confirmation of infection that involves assessment of viral RNA through a real-time micro-scale chip-based PCR system [7,11,14,15,16]. With such a detection system, reliability is an important issue that cannot be ignored. Generally, melting point analysis can distinguish between specific and non-specific amplified products. However, when non-specific products have a similar melting temperature, it is impossible to distinguish between them. For reliability, we explored the use of DNA-chips, a technique that can more easily determine a specific nucleotide sequence via DNA hybridization [17]. This study concentrated on establishing significant time-savings for isolated specific pathogen detection and confirmation. The analysis was not only performed with collected bees that were crippled, but also with asymptomatic bees. Recombinant DNA was used as a positive control.
MATERIALS AND METHODS
Preparation of honeybee (Apis mellifera) samples, DWV-specific recombinant DNA and PCR primers
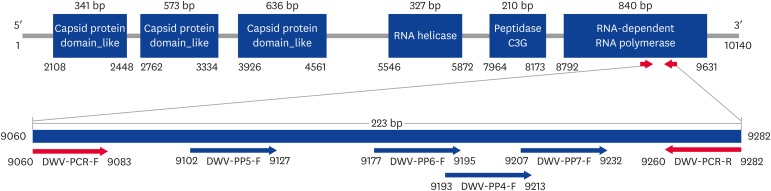
Adult Apis mellifera honeybees were collected from different sites (Suwon and Yeosu) in South Korea. Among them, only the samples from Yeosu exhibited visible signs of DWV infection. We designed primer pairs for the target gene and specific DNA-chip probes for hybridization with PCR products (Table 1). Samples with and without visible DWV signs or symptoms underwent preliminary investigation for the detection of viral RNA pathogens and DWV infection. We ascertained the presence of DWV by detecting a specific amplification product of 223 base pairs (bp). The specific recombinant DNA of DWV, which contains a target gene segment encoding the RNA-dependent RNA polymerase gene (GenBank accession No. HM067437.1), was cloned into the pLUG-Prime TA-cloning vector (Fig. 1). This cloned recombinant DNA was designated as pDWV-RdRp861. It was purified using the FastDNA spin plasmid DNA purification kit (iNtRON Biotechnology, Korea) and used as a template in ultra-rapid qPCR (UR-qPCR).
Table 1. List of nucleotide sequences of the primers and probes used in this study.
| Experiments | Name of oligonucleotide | Sequence (5′→3′) | Reference |
|---|---|---|---|
| RT-qPCR | DWV-SF | ATCAGCGCTTAGTGGAGGAA | [6] |
| DWV-SR | TCGACAATTTTCGGACATCA | ||
| URRT-qPCR | DWV_PCR-F | ACTATAAGAATTTTGGTCCTGGGT | This study |
| DWV-PCR-R | ATGTCCGTTATCGGAGGACCTGA | ||
| Probe for DNA-chip | DWV-PP4-F(D1) | GCAAGAGATCTTAGCGCCTAG | This study |
| DWV-PP5-F(D2) | CTTCAGCGTTCGAAATTATTATCGAC | ||
| DWV-PP6-F(D3) | TAATGTGGACCATGGCGCA | ||
| DWV-PP7-F(D4) | CGCCTAGTCATCTGTGTCGCGATTT |
Fig. 1. Location of primers for URRT-qPCR and probes for DNA-chip. The amplified target region is located at 9060–9282 bp on DWV RNA-dependent RNA polymerase. The size of the amplified target was 223 bp. The four DWV-specific probes represent the PCR product applied to the DNA-chip.
DWV, deformed wing virus; PCR, polymerase chain reaction; URRT-qPCR, ultra-rapid reverse transcription quantitative polymerase chain reaction.
Extraction of total RNA
All samples were quick-frozen using liquid nitrogen and pulverized using a mortar and pestle. The pulverized samples were transferred to tubes and lysed in RNAiso Plus (Takara, Japan) solution, according to the manufacturer's instructions for total RNA extraction. The resultant RNA pellets were resuspended in RNase-free water. Total RNA was used in subsequent experiments after determining its concentration level using a Biophotometer (Eppendorf, Germany).
Determination of optimal conditions for ultra-rapid reverse transcription-quantitative PCR
In the rapid detection of viral RNA, rapid cDNA synthesis is an important factor that allows for a shortening of the overall detection time. The optimal time for reverse transcription that can be applied to ultra-rapid qPCR was determined by performing the reaction at 50°C at different time intervals: 10, 7, 5, 4, 3, 2, 1, 0.5, and 0 min. At different time intervals, the values of Ct and Ct time were compared and analyzed. For comparison with the conventional two-step RT-qPCR method, 1 μg of total RNA isolated from honeybees was converted to cDNA using Superscript III First-strand synthesis system for RT-qPCR (Invitrogen, USA), according to the manufacturer's instructions. The synthesized cDNA was used at a 50 ng/μL concentration and was immediately subjected to qPCR amplification. Ultra-rapid qPCR was performed using GENECHECER (Genesystem Co., Ltd., Korea) with the 2 × Rapi master mix (Genesystem Co., Ltd.). The DWV-specific primer pair and the hybridization control (HC)-specific primer (0.2 pmol/μL) were added to the PCR mixture, and the total volume was adjusted to 10 μL. The PCR was performed with denaturation at 95°C for 1 sec, annealing at 55°C for 3 sec, and polymerization at 72°C for 1 sec for 50 cycles. To determine the optimal conditions for ultra-rapid reverse transcription-quantitative PCR (URRT-qPCR), the annealing temperature was adjusted from 52°C to 55°C in 1°C intervals. The annealing and polymerization times were adjusted from 1 sec to 3 sec, after which we compared Ct values and Ct times; that is, determined the actual minimum time to detect results. URRT-qPCR was performed with serial dilutions of recombinant DNA of pDWV-PnRp861 from 1.3 × 108 to 1.3 × 100 for confirmation of detection sensitivity and to function as a positive control.
DWV-specific probes and detection of samples on a DNA-chip
K-CAP, manufactured by Sugentech Co. (Korea), is a small DNA-chip shaped like a glass rod combined with a 200 μL centrifuge tube. The cross-section of the glass rod was capable of supporting a microarray with a total of 49 oligonucleotides. The glass rod-shaped DNA-chip was soaked in the PCR amplification solution in 200 μL tubes, after which the signal was confirmed. The probes used on the DNA-chip are shown in Table 1. Each probe was spotted on the surface of a DNA-chip and hybridized in a 200 μL centrifuge tube. To ascertain the sensitivity for DWV on the DNA-chip, the specific PCR amplification product was serially diluted from 1.3 × 108 to 1.3 × 100 molecules of the template. The amplified PCR products were transferred to 200 μL PCR tubes and hybridized with the DNA-chip for 1 h at 55°C. After washing using 2 × saline-sodium citrate (SSC) buffer for 5 min, DNA-chips were washed in distilled water for 5 min. The evaluation of hybridization results used calculated spot/background ratio (SBR) values that were determined using a K-SCAN-CAP scanner (Sugentech, Korea) and its related software program. SBR values of the DWV-specific probes were determined.
RESULTS
Optimization for the rapid detection of viral RNA from samples
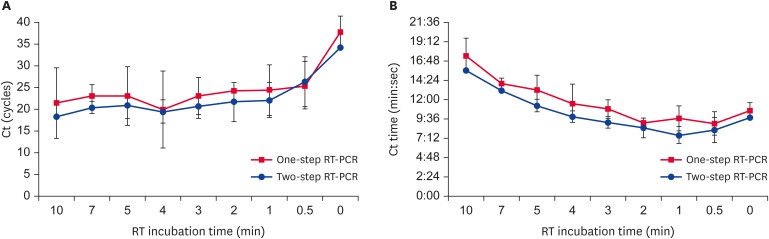
URRT-qPCR-based detection of viral RNA required rapid and effective reverse transcriptase. The cDNA synthesis reactions were performed by comparing one-step RT-qPCR and two-step RT-qPCR under standard conditions. Total RNA was amplified by RT-qPCR and analyzed based on the obtained Ct values and Ct times. The Ct value of the reverse transcription reaction time was 19.24 cycles (Ct time: 6 min 29 sec) at 1 min in the two-step RT-qPCR, while in one-step RT-qPCR, the Ct was 22.19 cycles (Ct time: 8 min 59 sec) with a 0.5 min RT reaction time. The Ct values were inversely proportional to the number of cDNA molecules at the end of the reverse transcription reaction. For the 1 min reaction time, the estimated cDNA amount was approximately 4.03 × 106 molecules. These results indicate that 1 min is sufficient to perform cDNA synthesis. In the two-step RT-qPCR method, the Ct times were less than those in one-step RT-qPCR at the various time intervals tested (Fig. 2A and B). However, total reaction time for the detection of viral RNA in one-step RT-qPCR was shorter than that for two-step RT-qPCR. Thus, we concluded that 1 min was the most suitable duration for accomplishing reverse transcription.
Fig. 2. Comparison of the effectiveness of different reverse transcription reaction times. (A) Cts were compared to determine the efficiencies of cDNA synthesis with different RT reaction times in one-step or two-step RT-qPCR. (B) In one-step or two-step RT-qPCR, the Ct time was analyzed depending on the cDNA synthesis reaction time. Data are presented as mean ± strandard deviation values of three independent experiments.
RT-PCR, reverse transcription-polymerase chain reaction; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Determination of optimal conditions for DWV-specific ultra-rapid RT-qPCR
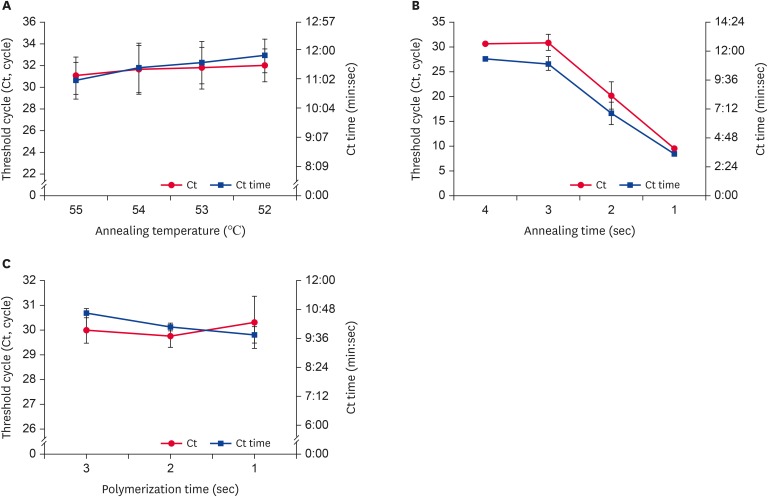
To improve the detection of DWV-specific target genes, URRT-qPCR was performed with recombinant DNA, as prepared above. As the detection process was intended for field applications, we preferred the more rapid method. As shown in Fig. 3A, Ct values and Ct times were highest and fastest, respectively, at 55°C (31.06 ± 1.71 cycles; 11 min 1 sec). When using an annealing time of 3 sec, Ct values and Ct times were significantly greater than those at other times (Fig. 3B). However, with 1 or 2 sec annealing times, we identified amplification graph problems during the reaction (data not shown). Considering the stability of the detection system, we decided on an optimal time of annealing of 3 sec. We fixed the time for polymerization at 1 sec. The Ct value (30.31 cycles) was slightly increased at 1 sec, but the Ct time (9 min 44 sec) decreased (Fig. 3C).
Fig. 3. Optimal conditions for each step of DWV-specific URRT-qPCR. URRT-qPCR conditions were evaluated by examining Ct time values versus (A) annealing temperature, (B) annealing time, and (C) polymerization time. The annealing time was determined based on the stability of the detection system. Data are presented as mean ± strandard deviation values of three independent experiments.
DWV, deformed wing virus; URRT-qPCR, ultra-rapid reverse transcription quantitative polymerase chain reaction.
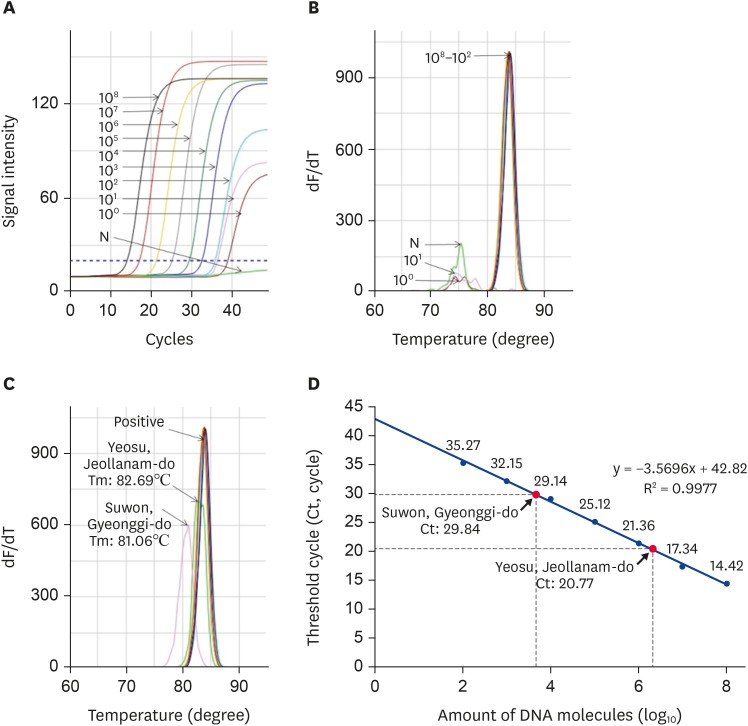
The UR-qPCR was performed with recombinant DNA under the following optimized conditions: pre-denaturation at 95°C for 30 sec, denaturation at 95°C for 1 sec, hybridization at 55°C for 3 sec, and polymerization at 72°C for 1 sec. The detection limit was confirmed, depending on the template concentration. The target gene was significantly amplified from 1.3 × 102 molecules (Fig. 4A and B). The DWV-specific genes in the samples from Suwon and Yeosu were amplified within 29.84 and 20.77 cycles, respectively. The abundance of DWV molecules present was different for each sample and sample source. The Suwon samples contained 4.32 × 103 molecules while those from Yeosu contained 1.5 × 106 molecules (Fig. 4D).
Fig. 4. Detection of DWV-specific target gene using URRT-qPCR. (A) DWV-specific URRT-qPCR was performed using recombinant DNA. The fluorescence signals of the amplified target gene were calculated to determine detection limits. Construction of a standard curve and (B) melting point analysis show that it differed the Tm valuese between specific and nonspecific amplicon. (C) The DWV-specific URRT-qPCR was performed with local samples and recombinant DNA, and it shows that Yeosu and Suwon sample have different values of Tm by melting point analysis. (D) For DWV-infected honeybee samples, we calculated the number of DWV molecules via regression analysis of the amplified target gene product.
DWV, deformed wing virus; URRT-qPCR, ultra-rapid reverse transcription quantitative polymerase chain reaction.
Confirmation of amplification of specific RNA using a DNA-chip
The URRT-PCR method sometimes produces confusion regarding melting temperature (Tm) values when using contaminated nucleic acid samples. In our case, the amplification graph exhibited different Tm values for Suwon samples (Tm 81.06°C) and Yeosu samples (82.69°C), as shown in Fig. 4C.
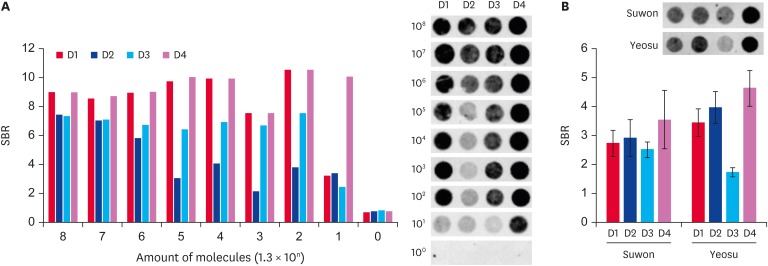
We confirmed the detection of DWV-specific target genes using a DNA-chip. The amplified target gene was hybridized with 1.3 × 108 to 1.3 × 100 molecules on a DNA-chip. An SBR cutoff value of 2 was used. When the SBR value was less than 2, we considered the result false. The SBR values of four DWV-specific probes were detected. With URRT-qPCR, the minimal positive signal was detected when using 1.3 × 102 molecules of the amplified target gene (Fig. 4A and B). The results of hybridization on a DNA-chip showed that the minimal positive signal was detected when using 1.3 × 101 molecules of the target gene as the probe (Fig. 5A). For the samples from Suwon and Yeosu, the SBR values were 2.73, 2.92, 2.51, and 3.55 and 3.44, 3.97, 1.73, and 4.63, respectively, when using probes D1, D2, D3, and D4, respectively. The Suwon sample was detected at above the cutoff value for each probe, but the Yeosu sample was under the cutoff value with the D3 probe (Fig. 5B).
Fig. 5. Confirmation of amplified target gene presence on a DNA-chip. (A) Amplification of the target gene was confirmed on a DNA-chip by using four deformed wing virus-specific probes. The bar graph shows the SBR values, according to the number of recombinant DNA molecules. The SBR in each probe hybridized with deformed wing virus specific gene are shown. (B) Each sample was hybridized to each of the specific probes. The SBR values are shown by the graph. SBR, spot/background ratio.
DISCUSSION
The honeybee (A. mellifera) is an important pollinator with huge ecological and agricultural importance worldwide. However, one of the serious problems that beekeepers frequently encounter is mortality of honeybees, and viral infections are a major cause of honeybee death [9,18,19]. In recent years, the importance of obtaining a rapid diagnosis of such pathogens has been emphasized. PCR is the foremost method for the direct identification of pathogens from field samples. Over the past years, qPCR has demonstrated advantages over conventional PCR in terms of speed, as well as being more sensitive and quantitative [20]; in addition, it is possible to monitor the amplification process in real time. For many years, we have worked on developing a viral pathogen detection method for honeybees that has sensitivity and rapidity [7,14,16]. As previously mentioned, the major goal of this study was to reduce the detection time of the pathogen diagnostic system. For rapid detection of viral RNA, rapid cDNA synthesis is important as it allows for target gene amplification in the PCR assay. We analyzed Ct values and Ct times according to reverse transcription reaction time. Even with a reverse transcription reaction time of 1 min or less, it was possible to synthesize a sufficient amount of cDNA for detection (Fig. 2). In this study, our method could detect 1.3 × 102 molecules of infecting viral RNA in only 11 min when using our specific primer set. These results demonstrate the possibility that our method will be able to provide rapid, simple, and accurate on-site DWV detection. When using the optimized DWV-specific URRT-qPCR, the numbers of molecules of infecting viral RNA in the Suwon and Yeosu samples were calculated as 1.73 × 107 (per bee) and 6.0 × 109 (per bee), respectively. In addition, these samples exhibited differences in Tm values, despite the presence of DWV infection. Sometimes, these Tm differences make it difficult to determine the specific target genes when using URRT-qPCR. Therefore, DNA-chip analysis was performed to provide confirmatory detection of the pathogenic RNA after URRT-qPCR. Generally, DNA-chip-based methods are suitable for a high degree of multiplexing, can be used to detect multiple pathogens, can simultaneously subtype the target agents, and can be easily adapted to accommodate viral variants [17]. Thus far, 11 major pathogens of honeybee have been detected by DNA-chip-based methods, using specific probes for each pathogen [21]. In this study, we used a DNA-chip on which DWV-specific probes had been spotted onto the chip surface and confirmed that Suwon and Yeosu honeybee samples contained DWV RNA molecules. A large initial quantity of viral molecules generated by the URRT-qPCR method yielded a high efficiency of hybridization on the DNA-chip. The results of the DNA-chip analysis were 10 times more sensitive than those obtained by URRT-qPCR.
In conclusion, our results show, for the first time, that it is possible to detect DWV-specific target genes within 11 min when using 1.3 × 102 molecules of the template and under optimized conditions. In addition, our findings suggest that the use of a DNA-chip provides convenience and accuracy, and the development of diagnostic methods combining URRT-qPCR and DNA-chip technology could be applied for the diagnosis of other viral RNA-mediated diseases, as well as honeybee diseases.
Footnotes
Funding: This work (Grants No. 318093-03) was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry(IPET) through Agri-Bio industry Technology Development Program, Export Promotion Technology Development Program (115067-02). Also this work was supported by Korea IPET through Advanced Production Technology Development Program (115102-03) and this work was supported by Rural Development Administration through Agriculture Science and Technology Development (Project No. PJ01408002).
Conflict of Interest: The authors declare no conflicts of interest.
- Conceptualization: Yoon BS.
- Data curation: Kim JM, Lim SJ.
- Formal analysis: Yoon BS, Kim SM.
- Funding acquisition: Yoon BS.
- Investigation: Kim JM, Lim SJ.
- Methodology: Kim JM, Kim SM, Kim MJ, Kim BH.
- Project administration: Yoon BS.
- Resources: Yoon BS.
- Software: Kim JM, Tai TA, Kim MJ.
- Supervision: Kim JM, Yoon BS.
- Validation: Yoon BS, Kim SM.
- Visualization: Kim JM, Lim SJ.
- Writing - original draft: Kim JM.
- Writing - review & editing: Kim SM.
References
- 1.McMenamin AJ, Genersch E. Honey bee colony losses and associated viruses. Curr Opin Insect Sci. 2015;8:121–129. doi: 10.1016/j.cois.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 2.Genersch E, Yue C, Fries I, de Miranda JR. Detection of Deformed wing virus, a honey bee viral pathogen, in bumble bees (Bombus terrestris and Bombus pascuorum) with wing deformities. J Invertebr Pathol. 2006;91:61–63. doi: 10.1016/j.jip.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Tehel A, Brown MJ, Paxton RJ. Impact of managed honey bee viruses on wild bees. Curr Opin Virol. 2016;19:16–22. doi: 10.1016/j.coviro.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 4.Reddy KE, Noh JH, Yoo MS, Kim YH, Kim NH, Doan HT, Ramya M, Jung SC, Van Quyen D, Kang SW. Molecular characterization and phylogenetic analysis of deformed wing viruses isolated from South Korea. Vet Microbiol. 2013;167:272–279. doi: 10.1016/j.vetmic.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 5.Tentcheva D, Gauthier L, Jouve S, Canabady-Rochelle L, Dainat B, Cousserans F, Colina ME, Ballb BV, Bergoin M. Polymerase chain reaction detection of deformed wing virus (DWV) in Apis mellifera and Varroa destructor . Apidologie (Celle) 2004;35:431–439. [Google Scholar]
- 6.Chen YP, Higgins JA, Feldlaufer MF. Quantitative real-time reverse transcription-PCR analysis of deformed wing virus infection in the honeybee (Apis mellifera L.) Appl Environ Microbiol. 2005;71:436–441. doi: 10.1128/AEM.71.1.436-441.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoo MS, Thi KC, Van Nguyen P, Han SH, Kwon SH, Yoon BS. Rapid detection of sacbrood virus in honeybee using ultra-rapid real-time polymerase chain reaction. J Virol Methods. 2012;179:195–200. doi: 10.1016/j.jviromet.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 8.Jamnikar Ciglenečki U, Toplak I. Development of a real-time RT-PCR assay with TaqMan probe for specific detection of acute bee paralysis virus. J Virol Methods. 2012;184:63–68. doi: 10.1016/j.jviromet.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 9.Haddad NJ, Noureddine A, Al-Shagour B, Loucif-Ayad W, El-Niweiri MA, Anaswah E, Hammour WA, El-Obeid D, Imad A, Shebl MA, Almaleky AS, Nasher A, Walid N, Bergigui MF, Yañez O, de Miranda JR. Distribution and variability of deformed wing virus of honeybees (Apis mellifera) in the Middle East and North Africa. Insect Sci. 2017;24:103–113. doi: 10.1111/1744-7917.12277. [DOI] [PubMed] [Google Scholar]
- 10.Kukielka D, Esperón F, Higes M, Sánchez-Vizcaíno JM. A sensitive one-step real-time RT-PCR method for detection of deformed wing virus and black queen cell virus in honeybee Apis mellifera. J Virol Methods. 2008;147:275–281. doi: 10.1016/j.jviromet.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 11.Lim HY. Development of novel rapid detection method for deformed wing virus (DWV) using ultra-fast high-performance PCR (UF-HP PCR) J Apic. 2013;28:237–244. [Google Scholar]
- 12.Ma M, Ma C, Li M, Wang S, Yang S, Wang S. Loop-mediated isothermal amplification for rapid detection of Chinese sacbrood virus. J Virol Methods. 2011;176:115–119. doi: 10.1016/j.jviromet.2011.05.028. [DOI] [PubMed] [Google Scholar]
- 13.Yoo MS, Noh JH, Yoon BS, Reddy KE, Kweon CH, Jung SC, Kang SW. Reverse transcription loop-mediated isothermal amplification for sensitive and rapid detection of Korean sacbrood virus. J Virol Methods. 2012;186:147–151. doi: 10.1016/j.jviromet.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 14.Han SH, Lee DB, Lee DW, Kim EH, Yoon BS. Ultra-rapid real-time PCR for the detection of Paenibacillus larvae, the causative agent of American Foulbrood (AFB) J Invertebr Pathol. 2008;99:8–13. doi: 10.1016/j.jip.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 15.Lee DW, Kim EH, Yoo MS, Han SH. Ultra-rapid real-time PCR for the detection of human immunodeficiency virus (HIV) Korean J Microbiol. 2007;43:91–99. [Google Scholar]
- 16.Lim SJ, Kim JM, Lee CW, Yoon BS. Development of ultra-rapid multiplex PCR detection against 6 major pathogens in honeybee. J Apic. 2017;32:27–39. [Google Scholar]
- 17.Lung O, Fisher M, Beeston A, Hughes KB, Clavijo A, Goolia M, Pasick J, Mauro W, Deregt D. Multiplex RT-PCR detection and microarray typing of vesicular disease viruses. J Virol Methods. 2011;175:236–245. doi: 10.1016/j.jviromet.2011.05.023. [DOI] [PubMed] [Google Scholar]
- 18.Choe SE, Nguyen LT, Noh JH, Koh HB, Jean YH, Kweon CH, Kang SW. Prevalence and distribution of six bee viruses in Korean Apis cerana populations. J Invertebr Pathol. 2012;109:330–333. doi: 10.1016/j.jip.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Tantillo G, Bottaro M, Di Pinto A, Martella V, Di Pinto P, Terio V. Virus infections of honeybees Apis Mellifera . Ital J Food Saf. 2015;4:5364. doi: 10.4081/ijfs.2015.5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai HY, Caswell JL, Prescott JF. Nonculture molecular techniques for diagnosis of bacterial disease in animals: a diagnostic laboratory perspective. Vet Pathol. 2014;51:341–350. doi: 10.1177/0300985813511132. [DOI] [PubMed] [Google Scholar]
- 21.Wang JH, Lee DB, Ku SJ, Peak MC, Min SH, Lim SJ, Lee CW, Yoon BS. Development of a detection method against 11 major pathogens of honey bee using amplification of multiplex PCR and specific DNA-chip. J Apic. 2016;31:133–146. [Google Scholar]