In this study, Denholtz et al. set out to investigate chromatin architecture changes that accompany the activation of human neutrophils. Using in situ HIC and other genome-wide analysis, the authors propose a model in which the microbe-induced transcriptional signature of activated neutrophils is driven by activated enhancer repertoires. Activated enhancers marked by elevated levels of H3K27Ac, in turn, rapidly recruit the cohesin machinery to dictate changes in chromatin folding and nuclear positioning of genes associated with an inflammatory gene program.
Keywords: gene expression, inflammation, neutrophils, nuclear architecture
Abstract
Differentiating neutrophils undergo large-scale changes in nuclear morphology. How such alterations in structure are established and modulated upon exposure to microbial agents is largely unknown. Here, we found that prior to encounter with bacteria, an armamentarium of inflammatory genes was positioned in a transcriptionally passive environment suppressing premature transcriptional activation. Upon microbial exposure, however, human neutrophils rapidly (<3 h) repositioned the ensemble of proinflammatory genes toward the transcriptionally permissive compartment. We show that the repositioning of genes was closely associated with the swift recruitment of cohesin across the inflammatory enhancer landscape, permitting an immediate transcriptional response upon bacterial exposure. We found that activated enhancers, marked by increased deposition of H3K27Ac, were highly enriched for cistromic elements associated with PU.1, CEBPB, TFE3, JUN, and FOSL2 occupancy. These data reveal how upon microbial challenge the cohesin machinery is recruited to an activated enhancer repertoire to instruct changes in chromatin folding, nuclear architecture, and to activate an inflammatory gene program.
The organization of the human genome within the nucleus is central to the control of gene expression and thus cell identity and function. At the largest scale, the genome is folded into chromosome territories that, with the exception of nucleoli, rarely intermingle. However, chromosomes are not randomly distributed across the nucleus. Large and gene-poor chromosomes are predominantly positioned at the lamina, whereas small and gene-rich chromosomes concentrate in the nuclear interior (Fritz et al. 2016). Chromosomes themselves fold into loop domains that physically associate to establish the transcriptionally repressive or inert heterochromatic B compartment or transcriptionally permissive euchromatic A compartment (Lieberman-Aiden et al. 2009; Dixon et al. 2012). The heterochromatic compartment is highly enriched at the nuclear lamina, whereas the euchromatic compartment is positioned in the nuclear interior (Kosak et al. 2002).
Loop domains are established in part by the CTCF protein (Dixon et al. 2012; Rao et al. 2014). Convergently oriented pairs of CTCF-bound loci can form CTCF-anchored loops generated by recruitment of the cohesin complex (Nora et al. 2017; Rao et al. 2017). Cohesin is loaded onto transcribed regions located throughout loop bodies (Busslinger et al. 2017). Once sequestered, the cohesin complex extrudes chromatin in a progressive manner until a pair of convergent CTCF-bound sites are reached, a process termed loop extrusion (Fudenberg et al. 2016). Gene promoters connected to transcriptional enhancers by CTCF-mediated loops tend to be highly expressed (Rao et al. 2014), and CTCF occupancy at nearby sites contributes to the maintenance of gene expression and stable chromatin structure (Nora et al. 2017; Rao et al. 2017; Schwarzer et al. 2017; Bintu et al. 2018). Finally, recent studies have demonstrated that cohesin-dependent looping is closely associated with the control of inducible gene expression (Bruno et al. 2018).
Human neutrophils are abundant, short-lived circulating white blood cells that are critical first-responders to infection and tissue damage. Upon injury or infection, neutrophils exit the circulation via extravasation, migrate toward damaged tissues or infectious foci, phagocytose small pathogens, release reactive oxygen and nitrogen species, and extrude their chromatin as cytotoxic granule-laced extracellular traps (NETs). In addition to their direct role in killing invading pathogens, activated neutrophils rapidly induce the expression of a wide range of cytokines and chemokines to orchestrate an immediate inflammatory response (Ley et al. 2018).
The nuclei of polymorphonuclear (PMN) neutrophils are composed of multiple distinct but internally continuous lobes allowing them to swiftly migrate between (paracellular route) or through (transcellular route) endothelial cells that line blood vessels and interstitial spaces of tissues while maintaining their nuclear integrity (Olins et al. 2009; Muller 2013; Rowat et al. 2013). The Lamin B receptor (LBR) is an important determinant for imposing a multi-lobular nuclear architecture on neutrophils (Hoffmann et al. 2002; Shultz et al. 2003). Neutrophils of mice deficient in the Lbr gene fail to adopt a multi-lobular nuclear shape (Shultz et al. 2003), and mouse neutrophilic cell lines lacking Lbr cannot form characteristic toroidal nuclei during differentiation (Zhu et al. 2017). Similarly, humans with LBR mutations manifest the Pelger-Huët anomaly, characterized by a reduction in nuclear lobe number in granulocytes (Hoffmann et al. 2002).
Chromatin folding in murine neutrophils is highly enriched for remote genomic interactions, primarily involving heterochromatic regions. These interactions span vast genomic distances resulting in large-scale chromosome condensation. Terminal differentiation of murine neutrophils is also associated with the relocation of centromeres, pericentromeres, telomeres, LINE elements, and ribosomal DNA from the nuclear interior to the nuclear lamina, a process that requires the Lbr gene (Zhu et al. 2017). As neutrophils differentiate, the LBR deforms the malleable nuclear envelope by wrapping it around the heterochromatic component of the neutrophil genome, resulting in its characteristic lobed shape.
Upon reaching a tissue site of infection, neutrophils neutralize bacteria in multiple ways: (1) engulfment through phagocytosis, (2) degranulation to release microbicidal factors into the extracellular space, (3) release of extracellular traps or NETs that are composed of extruded chromatin fibers and antimicrobial factors, and (4) rapid induction of cytokine gene expression to coordinate a broader immune response (Brinkmann et al. 2004; Ley et al. 2018). To detect and respond appropriately to diverse invading pathogens, neutrophils express a variety of pattern recognition receptors including cell-surface and endolysosomal Toll-like receptors (TLRs), C-type lectin receptors, and formyl peptide receptors, among others. Once activated, a variety of downstream signaling pathways converge on the NF-κB and AP1 transcription factors to induce an inflammatory gene program including the cytokines and chemokines IL-8/CXCL8, TNFα, IL-1β, IL-17, and IFNγ (Garcia-Romo et al. 2011; Thomas and Schroder 2013; Tecchio et al. 2014).
The mechanisms by which pathogen-sensing pathways interface with the neutrophil genome to induce a rapid and stimulant-appropriate inflammatory gene expression program remain unclear. Here we found that human neutrophil genomes display highly segmented compartments and contracted heterochromatin when compared with human embryonic stem cells. Upon microbe encounter, a specific subset of modestly euchromatic subdomains, spatially segregated from the highly euchromatic A compartment, displayed strengthening of their euchromatic character, and relocalized from a perinuclear envelope position toward the nuclear interior. Prominent among the regions that repositioned during human neutrophil activation were gene loci associated with an activated neutrophil-specific gene expression program. Microbial-induced changes in long-range chromatin interactions were globally associated with rapid loss of insulation at euchromatic subdomain boundaries, as well as the formation of de novo chromatin loops linking immune response genes to pre-existing and de novo formed transcriptional enhancers. The loop-mediated juxtaposition of inflammatory genes to transcriptional enhancers upon microbial exposure was closely associated with the deposition of histone 3 lysine 27 acetylation (H3K27ac), an enhancer-associated histone modification and rapid loading (<3 h) of the cohesin complex at the subset of enhancer elements that control an inflammatory gene program. Based on these observations, we propose that the microbe-induced transcriptional signature of activated neutrophils is driven by activated enhancer repertoires. Activated enhancers marked by elevated levels of H3K27Ac, in turn, rapidly recruit the cohesin machinery to dictate changes in chromatin folding and nuclear positioning of genes associated with an inflammatory gene program.
Results
Human neutrophil development is associated with segmented compartments and contracted genomes
Neutrophil nuclei undergo dramatic morphological changes during differentiation from multipotent progenitors, with terminally differentiated neutrophil nuclei having three to five internally continuous but spatially distinct lobes (Supplemental Fig. S1A). To characterize the genomic interactions established during the development of PMN cells, neutrophils were isolated from human peripheral blood, formaldehyde-fixed, and analyzed using in situ HiC (Supplemental Table S1; Rao et al. 2014). The genomes of human neutrophils were slightly enriched for interchromosomal interactions when compared with human embryonic stem cells (hESCs) (Fig. 1A). Chromosome territories remained intact and we found no evidence of individual chromosomes being split across multiple lobes (Supplemental Fig. S1B). Notably, compared with hESCs, human neutrophils were depleted for genomic interactions that spanned <3 Mb but were enriched for interactions that covered >3 Mb (Fig. 1A).
Figure 1.
Heterochromatic super-contraction and segmentation of the neutrophil genome during acquisition of the PMN shape. (A) Neutrophil (blue) and H1 hESC (teal) chromatin interaction frequencies as a function of linear genomic distance. Percent interchromosomal paired end tags (PETs) are indicated. (B) Normalized HiC contact matrices for H1 hESC (left) and neutrophil (right) chromosome 20. First principal component 1 eigenvector (PC1) for each HiC matrix is displayed above its respective matrix. Positive PC1 values correspond to the gene-rich A compartment, negative values to the gene-poor B compartment, based on human genome build hg38. (C) HiC contact matrix showing the log2 fold change in normalized interactions between the H1 hESC and neutrophil matrices in B, illustrating changes in the organization of chromosome 20 during terminal differentiation from the pluripotent state and acquisition of the PMN shape. PC1 values for H1 hESCs and neutrophils are shown above and at the right of the matrix, respectively. (D) Density plot showing the distribution of A (solid lines) and B (dotted lines) compartment domain sizes in H1 hESCs (teal) and neutrophils (blue). Total number of domains for each data set are listed at top. Domains <100,000 bp were not considered. (E) Example of a new TAD and compartment boundary formed during differentiation and hypercompartmentalization of the neutrophil genome. (Top, from top to bottom) IGV tracks showing H1 hESC PC1 values, H1 hESC TADs, neutrophil PC1 values, and neutrophil TADs. (Bottom) Normalized HiC contact matrix of H1 hESC and neutrophil HiC matrices and a log2 fold change difference matrix at a new TAD/compartment boundary on chromosome 14. (F) Percent of TAD and compartment boundaries shared between H1 hESCs and neutrophils. Domain boundaries within 100 kb (< 3 windows) were considered shared. (G) Insulation scores in H1 hESCs and neutrophils calculated for each 40-kb bin genome-wide, at neutrophil-specific, H1 hESC-specific, and shared PC1 compartment/TAD boundaries for both cell types. Grouped pairs are all significantly different by the Wilcoxon rank sum test with log(P) values <1 × 10−11.
We next constructed contact matrices for hESCs cells and human neutrophils (Fig. 1B). We found that a larger fraction of the neutrophil genome was sequestered in the B compartment when compared with hESCs (Supplemental Fig. S1C). The stereotypic plaid pattern, resulting from the spatial segregation of the A and B compartments, was much more pronounced in human neutrophils compared with hESCs (Fig. 1B). Intrachromosomal and interchromosomal interactions between A and B compartments were both less prevalent in neutrophils versus hESCs (Fig. 1C; Supplemental Fig. S1D). Conversely, long-range genomic interactions across the B compartment were significantly more extensive in human neutrophils than hESCs (Fig. 1C; Supplemental Fig. S1D). During differentiation, large genomic regions that exhibited a continuum of either positive or negative PC1 values in hESCs fragmented into smaller genomic regions that switched PC1 values in neutrophils (Fig. 1C,D). Many of the genomic regions that switched from negative to positive PC1 values during development were associated with a neutrophil-specific transcription signature, whereas those regions switching from positive to negative PC1 values were associated with silencing of lineage-inappropriate genes (Supplemental Fig. S1E–G; Supplemental Table S2). Notably, the hypersegmentation of compartment domains in the neutrophil genome established de novo loop domain and compartment boundaries (Fig. 1E). Specifically, although >75% of loop domain boundaries identified in hESCs were conserved in neutrophils, <40% of loop domain boundaries in neutrophils were present in hESCs (Fig. 1F). Overall compartment boundaries were poorly conserved between these two cell types (Fig. 1F). Genome-wide analysis of cell type-specific loop domain and compartment boundary element insulation strength confirmed this finding, indicating the existence of cell type-specific boundaries that were associated specifically with either hESCs or human neutrophils, in addition to shared boundaries (Fig. 1G). Taken together our data reveal that human neutrophils, when compared with hESCs, are characterized by a contracted genome with increased enforcement of compartmentalization and highly segmented A and B compartments.
PMA-induced activation of neutrophils rapidly modulates nuclear architecture
Upon detecting invading microbes, neutrophils rapidly activate an inflammatory-specific transcription signature. As a first approach to examine whether and how the nuclear architecture of neutrophils responds to inflammatory signals, HiC was performed on neutrophils cultured in both the absence and presence of the canonical neutrophil activator phorbol 12-myristate 13-acetate (PMA), a protein kinase C. PMA stimulation of human neutrophils resulted in a global decrease in short-range intrachromosomal interactions and a global increase interchromosomal interactions (Fig. 2A), while exerting minimal effects on A-B compartmentalization and loop domain boundaries (Fig. 2B). Likewise, PMA-induced activation did not trigger large-scale switching of genes or regulatory elements between the A and B compartments (Fig. 2C). However, further scrutiny of chromatin folding across the A compartment revealed a small but significant number of discrete genomic regions that underwent significant PMA-dependent changes from low but positive PC1 values to highly positive PC1 values, indicating an increase in euchromatic character (Fig. 2C). We refer to these regions as PMA ΔPC1 domains (Fig. 2C; Materials and Methods). Notably, PMA ΔPC1 domains were strongly enriched for genes implicated in the neutrophil defense response, including genes downstream from key innate immune receptors such as the complement receptors, FCγ receptor, and dectin-2, as well as genes implicated in cell migration and regulation of lysosomal pH (Fig. 2D; Supplemental Table S2).
Figure 2.
PMA activation increases euchromatic character at distinct genomic loci encoding for neutrophil activation genes. (A) Unstimulated (blue) and PMA-activated neutrophil (red) (3 h) chromatin interaction frequencies as a function of linear genomic distance. Percent interchromosomal PETs are indicated. (B). Percentages of TAD and compartment boundaries shared between unstimulated and PMA-activated neutrophils. (C) Scatter plot comparing 10-kb-windowed PC1 values between unstimulated and PMA-activated neutrophils. PMA ΔPC1 domains are marked in red. (D) Metascape-defined functional groups enriched for genes found within 100 kb of PMA ΔPC1 domains. Complete metascape results can be found in Supplemental Table S2. (E) HiC contact matrices of chromosome 18 for unstimulated (left) and PMA-activated (right) neutrophils, 100-kb resolution. Respective PC1 values are shown above each matrix. (F) Difference matrix showing the log2 fold change in normalized interactions between unstimulated and PMA-activated neutrophils. PC1 differences are shown at top, PMA ΔPC1 domains are marked by red triangles. (G) Black and red points indicate genome-wide interaction correlations for each 40-kb bin in the genome, ranked from most to least differential, left to right. Bins containing PMA ΔPC1 domains are marked with red points. The red line with red shading indicates the proportion of total PMA ΔPC1 domains found at a given rank or lower, showing a preference for PMA ΔPC1 domains to fall in genomic regions with the most differential chromatin interactions upon PMA stimulation. (H) Box plots of genome-wide interaction correlation values for PMA ΔPC1 domains and the remainder of the genome during PMA stimulation. Box plot outliers are not shown. (*) Wilcoxon rank-sum test P-value <2 × 10−16.
Analysis of intrachromosomal HiC contact matrices revealed few significant changes across chromosome 18 following PMA-induced activation (Fig. 2E). PMA ΔPC1 domains, however, often displayed changes in interactions with euchromatin, both in their immediate vicinity, as well as across chromosome 18 (Fig. 2F). Direct measure of genome-wide changes in chromatin organization showed that in activated neutrophils PMA ΔPC1 domains showed large-scale changes in contact frequencies (Fig. 2G). Specifically, ∼25% of PMA ΔPC1 domains fell within the top 10% most differentially interacting genomic regions (Fig. 2G). Likewise, PMA ΔPC1 domains on average displayed significantly lower chromatin interaction correlation with unstimulated neutrophils when compared with the genome as a whole (Fig. 2H). Taken together, these data indicate that in PMA-activated neutrophils genic and intergenic domains associated with innate immune genes increase their euchromatic character and undergo alterations in remote genomic interactions.
Upon microbial exposure, a subset of neutrophil inflammatory genes increase their euchromatic character
To validate the alterations in neutrophil euchromatic character using a physiologically relevant stimulus, human neutrophils were cultured in the presence of live Escherichia coli bacteria for a period of 3 h. E. coli cocultured neutrophils were isolated, formaldehyde cross-linked, and analyzed using HiC. Genomes of human neutrophils cultured in the presence of E. coli only displayed minor alterations in contact frequencies, maintained overall compartment and loop domain structures (Fig. 3A,B), and remained essentially free from detectable A–B compartment switching (Fig. 3C). However, similar to PMA-activated neutrophils, a distinct subset of genomic regions positioned in the A compartment displayed a substantial increase in euchromatic character upon E. coli encounter (Fig. 3C, E. coli ΔPC1 domains). Notably, the E. coli ΔPC1 domains included genes encoding for cytokines and chemokines, genes associated with neutrophil degranulation, and genes linked with the inflammatory response (Fig. 3D; Supplemental Table S2).
Figure 3.
Neutrophil E. coli coculture increases euchromatic character at distinct loci encoding for neutrophil pathogen response genes. (A) Unstimulated (blue) and E. coli cocultured neutrophil (green) chromatin interaction frequencies as a function of linear genomic distance. Percent interchromosomal PETs are indicated. Note: Unstimulated neutrophil data is identical to that shown in Figure 2A and is shown here to illustrate differences between unstimulated and E. coli cocultured neutrophil data. Neutrophils were cultured in the presence of E. coli for a 3-h period. (B) Percent of TAD and compartment boundaries shared between unstimulated and E. coli cocultured neutrophils. (C) Scatter plot comparing 10-kb-windowed PC1 scores between unstimulated and E. coli cocultured neutrophils. E. coli ΔPC1 domains are marked in green. (D) Metascape-defined functional groups enriched for genes found within 100 kb of E. coli ΔPC1 domains. Complete metascape results are described in Supplemental Table S2. (E) HiC contact matrices of chromosome 20 for unstimulated (left) and E. coli cocultured (right) neutrophils, 100-kb resolution. Respective PC1 values are shown above each matrix. (F) Difference matrix showing the log2 fold change in normalized interactions between unstimulated and E. coli cocultured neutrophils. PC1 differences are shown at the top with E. coli ΔPC1 domains marked with green triangles. (G) Black and green points indicate genome-wide interaction correlations for each 40-kb bin in the genome, ranked from most to least differential, left to right. Bins containing E. coli ΔPC1 domains are marked with green points. The green line with green shading indicates the proportion of total E. coli ΔPC1 domains found at a given rank or lower, showing a preference for E. coli ΔPC1 domains to fall within genomic regions with the most differential chromatin interactions upon E. coli encounter. (H) Box plots of genome-wide interaction correlation values for E. coli ΔPC1 domains and the remainder of the genome during E. coli encounter. Box plot outliers are not shown. (*) Wilcoxon rank-sum test P-value <2 × 10−16.
Similar to PMA-activated neutrophils, E. coli cocultured neutrophils showed few large-scale changes in chromatin organization compared with unstimulated neutrophils (Fig. 3E). E. coli ΔPC1 domains, however, showed dramatic increases in genomic interactions involving neighboring euchromatic regions, as well as the remainder of the chromosome upon coculture with E. coli (Fig. 3F). Similar to PMA ΔPC1 domains, E. coli ΔPC1 domains were among the most restructured genomic regions in response to E. coli, with 25% of E. coli ΔPC1 domains assigned to the top 15% of the most differentially interacting regions globally (Fig. 3G). E. coli ΔPC1 domains overall displayed significantly lower correlation with unstimulated neutrophil genome structure than the remainder of the genome (Fig. 3H). These data indicate that upon microbial exposure, a subset of genes associated with an inflammatory response increase their euchromatic character.
We next sought to ascertain to what degree ΔPC1 domains differed between stimuli. E. coli ΔPC1 domains only partially overlapped with PMA ΔPC1 domains (Supplemental Fig. S2A). The identities of genes in ΔPC1 domains also depended on the stimulus that neutrophils encountered. E. coli-specific ΔPC1 domains were highly enriched for chemokine and cytokine genes as well as genes involved in chemotaxis (Supplemental Fig. S2B). In contrast, PMA-specific ΔPC1 regions were enriched for defensin gene clusters (Supplemental Fig. S2B). These data suggest that the changes in euchromatic character regulate stimulant-appropriate inflammatory responses. Supporting this hypothesis, genes residing in stimulus-specific ΔPC1 domains underwent stimulus-specific changes in gene expression. Genes in E. coli ΔPC1 domains were more highly expressed upon E. coli encounter than upon PMA stimulation, whereas genes in PMA ΔPC1 domains were more highly expressed upon PMA stimulation than during E. coli coculture (Supplemental Fig. S2C). Taken together, these data indicate that neutrophil activation enhances the euchromatic character of a subset of inflammatory response gene loci in a stimulus-dependent manner.
Rapid assembly and relocalization of a CXCL transcriptional hub upon E. coli encounter
To determine how euchromatic character is strengthened upon microbial activation, we focused on an archetypal E. coli ΔPC1 domain containing inflammatory-specific genes encoded within the extended CXCL locus. The CXCL locus spans a cluster of genes encoding a class of chemokines that include CXCL8 (IL8), CXCL1, and CXCL2 (MIP2α), each of which is rapidly induced when exposed to microbial agents. We found that in unstimulated neutrophils the CXCL locus exists as a loop domain associated with a modestly positive PC1 score that is insulated from neighboring euchromatin (Fig. 4A). Notably, within 3 h of exposure to E. coli, the euchromatic character of the CXCL locus was significantly strengthened (Fig. 4A), accompanied by large scale changes in chromatin folding, with genomic interactions and transcriptional activation spreading into neighboring regions (Fig. 4B,C).
Figure 4.
E. coli co-culture-induced topological changes at the CXCL sub-domain are associated with noncoding transcription, cohesin recruitment, and locus repositioning. (A) HiC contact maps of the extended CXCL gene cluster in unstimulated (top) and E. coli cocultured (bottom) human neutrophils. PC1 scores are shown above their respective matrices, CXCL E. coli ΔPC1 domain position is noted. (B) Log2 difference matrix comparing HiC contacts between unstimulated and E. coli cocultured neutrophils within the extended CXCL gene cluster. PC1 differential values (E. coli cocultured–unstimulated PC1 values) are shown above matrix, protein-coding genes in the CXCL gene locus are shown below. (C, top) Linear genomic features and significant chromatin interactions at the CXCL E. coli ΔPC1 domain. CTCF and SMC3 ChIP-seq, RNA-seq, and HOMER-defined chromatin interactions with −log(P) values less than −50 are shown for unstimulated (blue, top) and E. coli cocultured neutrophils (green, bottom) with PC1 differential values (E. coli cocultured–unstimulated PC1 values) shown between. The bottom panel displays SMC3 ChIP-seq and RNA-seq tracks at the CXCL8 (IL8) gene, demonstrating transcription-associated recruitment of SMC3 to the CXCL8 locus. Peaks of transcription and associated SMC3 recruitment are highlighted in yellow. (D) Representative FISH image (z-projection) showing the euchromatic CXCL8 locus (green) in unstimulated (top) and E. coli cocultured (bottom) neutrophils. (E) Quantification of the proportion of nuclear volume between the FISH signal and the nuclear periphery (Eroded volume fraction) in unstimulated and E. coli co-cultured neutrophils. Number of alleles analyzed are listed below the box plots. Wilcoxon rank sum test P-value for the data distributions: (**) P < 0.00005. (F) Quantification of DAPI signal intensity at the FISH spots identified in B. Wilcoxon rank sum test P-values: (**) P < 0.00005.
To determine whether the alterations in genome folding were associated with gene expression, activated neutrophils were analyzed for transcript abundance as well as CTCF and SMC3 occupancy (Fig. 4C). As expected, CXCL8, CXCL1, and CXCL2 transcript abundance was significantly elevated upon E. coli encounter (Fig. 4C). Notably, a recently described noncoding genomic region located immediately upstream of CXCL8 was also transcriptionally induced upon exposure to bacteria (Fig. 4C; Fanucchi et al. 2019). While CTCF occupancy was elevated at a site closely linked with the CXCL8 locus, other CTCF-bound sites in the locus were not modulated upon activation (Fig. 4C). In contrast, we found that E. coli encounter substantially enriched cohesin occupancy across the locus (Fig. 4C). Cohesin occupancy was particularly prominent at sites closely associated with de novo loops that linked the CXCL8, CXCL1, and CXCL2 gene bodies, promoter regions, and SMC3-enriched intergenic regions into a shared transcriptional hub (Fig. 4C).
To validate these findings in single cells, we performed fluorescence in situ hybridization (FISH) using a probe corresponding to the E. coli-specific CXCL ΔPC1 domain (Fig. 4D). In unstimulated neutrophils, the CXCL E. coli ΔPC1 domain localized near the nuclear periphery (Fig. 4D). Upon E. coli encounter the CXCL E. coli ΔPC1 domain rapidly relocated away from the heterochromatic nuclear periphery toward the nuclear interior, concomitant with its change in euchromatic character and elevated transcript levels (Fig. 4D). Specifically, the E. coli ΔPC1 domain relocated from the DAPI-dense portion of the nucleus near the nuclear periphery to the DAPI-sparse nuclear interior (Fig. 4E,F). This change in nuclear positioning was not an indirect result of changes in nuclear morphology, nor activation-induced loss of nuclear integrity, as heterochromatic control probes remained tightly associated with the nuclear periphery during E. coli encounter (Supplemental Fig. S3A). Collectively, these observations indicate that upon microbial exposure human neutrophils rapidly remodel nuclear architecture to assemble a CXCL transcriptional hub in the nuclear interior.
Neutrophil activation is associated with global loss of insulation at inflammatory genes
The data described above reveal that when human neutrophils encounter bacteria, a subset of inflammatory genes undergo large-scale changes in chromatin folding that spread into neighboring loop domains. To quantitatively describe this loss of subdomain insulation, we computed the insulation scores for genomic regions that surrounded the boundaries of E. coli ΔPC1 domains and upon microbial exposure gained euchromatic character to merge with surrounding euchromatin. We found that upon E. coli encounter the gain of euchromatic character across E. coli ΔPC1 domains was closely associated with decreased insulation strength at E. coli ΔPC1 domain boundaries (Fig. 5A). Although globally the genomic distances separating chromatin interaction anchor points were significantly decreased in activated versus unstimulated neutrophils, the distance separating anchor points of chromatin interactions with E. coli ΔPC1 domains increased (Fig. 5B). This loss of insulation at E. coli ΔPC1 domain boundaries suggests de novo formation of regulatory interactions with the surrounding area (Fig. 5B). Additionally, chromatin interactions contained entirely within E. coli ΔPC1 domains were found on average to be significantly stronger in unstimulated neutrophils as compared with E. coli cocultured neutrophils, suggesting a loss of subdomain structure and self-association during microbial encounter (Fig. 5C).
Figure 5.
E. coli ΔPC1 domains lose local spatial insulation and are associated with increased cohesin-bound chromatin loop strength. (A) Insulation score meta-analysis. Insulation scores were calculated for genomic regions surrounding E. coli ΔPC1 domain boundaries. Normalized insulation scores for unstimulated (blue) and E. coli cocultured (green) neutrophils are shown. Vertical dotted lines demarcate meta-domain boundaries. Horizontal dotted lines show median normalized insulation scores at PC domain boundaries genome-wide for each condition. Calculations shown are for all E. coli ΔPC1 domains larger than 100 kb. (B) Distribution of linear genomic distances between chromatin interaction anchor points genome-wide, for interactions anchored in ΔPC1 domains, and for interactions fully contained within E. coli ΔPC1 domains. Wilcoxon rank sum test: (**) P < 2.2 × 10−16; (*) P < 1 × 10−5; (+) not significant. (C) Interaction strength bias (see the Materials and Methods) of HOMER-defined chromatin interactions for unstimulated and E. coli cocultured neutrophils genome-wide, for those interactions with a single anchor in an E. coli ΔPC1 domain, and for those interactions contained entirely within E. coli ΔPC1 domains. (D) HICCUPS defined loop strength with respect to E. coli ΔPC1 domains in unstimulated and E. coli cocultured neutrophils. (E) Distance between HICCUPS loops and E. coli ΔPC1 domains. Differences are not significant by the Kolmogorov-Smirnov test. Five out of 45 (11%) E. coli coculture-specific loops are in E. coli ΔPC1 domains, and 19/49 are within 1 Mb of E. coli ΔPC1 domains (45%). E. coli ΔPC1 domains make up ∼0.3% of the human neutrophil genome. (F) Log2 (normalized E. coli cocultured/unstimulated) FPKM values for genes at chromatin interaction anchors shared between unstimulated and E. coli cocultured neutrophils, or at chromatin interactions specific to one condition. Wicoxon rank sum test: (**) P < 1 × 10−4; (*) P < 5×10−3. (G) Change in PC1 values at genes with the given mRNA expression differential during E. coli encounter.
Given the loss of insulation at E. coli ΔPC1 domain boundaries, we next sought to determine the relationship between ΔPC1 domains and gene regulatory chromatin interactions. Although chromatin interactions within E. coli ΔPC1 domains were on average weakened during E. coli encounter (Fig. 5C), E. coli encounter-specific chromatin loops within ΔPC1 domains were significantly stronger than chromatin loops found only in unstimulated neutrophils (Fig. 5D). This finding suggested a gene regulatory role for E. coli encounter-dependent loops, and a tight link between these loops and E. coli ΔPC1 domains. Supporting this finding, E. coli-dependent chromatin loops were generally closer to E. coli ΔPC1 domains than were unstimulated neutrophil-specific chromatin loops, and 11% of E. coli-dependent chromatin loops were identified in E. coli ΔPC1 domains, which make up only 0.3% of the genome (Fig. 5E). Importantly, genes near E. coli coculture-specific chromatin loop anchors were significantly more highly expressed than genes at chromatin loop anchors found only in unstimulated neutrophils (Fig. 5F).
Given the enrichment of neutrophil inflammatory response genes in E. coli ΔPC1 domains (Fig. 3D) and the link between expression levels and an increase in euchromatic character (Supplemental Fig. S2C), we next determined the relationship between euchromatic character (PC1 score) and transcript levels during microbe encounter. Notably, we found a strong correlation between PC1 score dynamics and transcriptional dynamics, with the most highly induced genes also showing the largest increases in PC1 score, and the most repressed genes showing the largest decreases in PC1 score (Fig. 5G). These phenomena are readily visible at a number of inflammatory loci, wherein the tight self-association of E. coli ΔPC1 domains in unstimulated cells is lost in favor of distal regulatory interactions and transcriptional activation during E. coli encounter (Supplemental Fig. S4).
Taken together these data indicate that neutrophil transcriptional state, euchromatic character, and spatial localization of genes are closely linked.
Microbial exposure rapidly recruits cohesin to inflammatory enhancers
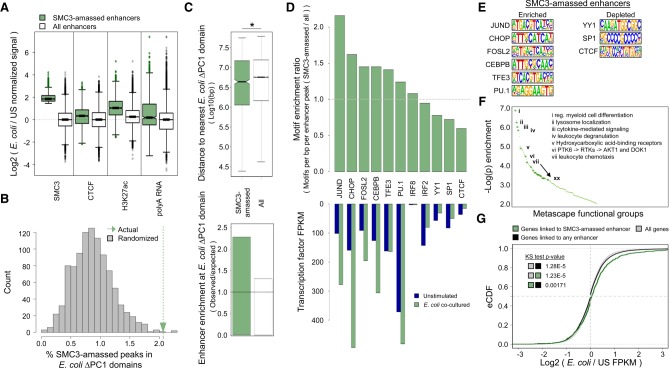
Examination of gene regulatory interactions associated with E. coli ΔPC1 domains (Fig. 4; Supplemental Fig. S4) hinted that a large number of E. coli-dependent interactions were associated with the recruitment of the cohesin complex to cis regulatory elements. To study this phenomenon and understand its role in E. coli-dependent changes in gene expression, we analyzed unstimulated and E. coli-exposed neutrophils for SMC3 and CTCF occupancy, as well as changes in H3K27Ac-marked enhancer repertoires and transcription. We then focused our analysis on a specific subset of SMC3-amassed enhancers: those H3K27ac-defined enhancers present in E. coli cocultured neutrophils that gained substantial SMC3 occupancy during E. coli encounter (Fig. 6A; Materials and Methods).
Figure 6.
E. coli coculture induces cohesin recruitment to a subset of H3K27ac-defined enhancers. (A) Log2 ratio (E. coli cocultured/unstimulated) of the normalized ChIP-seq and RNA-seq signals at H3K27ac-defined enhancers that amass SMC3 as well as all enhancers genome-wide. (B) Histogram showing the percent of SMC3-amassed enhancers falling in E. coli ΔPC1 domains randomly positioned within the A compartment (gray), and the percent of SMC3-amassed enhancers falling within actual E. coli ΔPC1 domains (green arrow). Out of 1000 random permutations, 997 resulted in lower overlap between SMC3-amassed enhancers and ΔPC1 domains than were observed in the empirical data. (C) The top panel shows distance distribution between E. coli ΔPC1 domain boundaries and SMC3-amassed enhancers (green) and all enhancers (white). The bottom panel shows observed enrichment of SMC3-amassed enhancers or all enhancers in ΔPC1 domains divided by the expected enrichment of these enhancers in E. coli ΔPC1 domains based on 1000 random permutations of E. coli ΔPC1 domain positions within the A compartment. (*) Wilcoxon rank sum test P-value < 0.005. (D) The top panel indicates ratio of mean transcription factor motif density (motifs per base pair per peak, SMC3-amassed enhancers/all enhancers) for representative transcription factors. The bottom panel shows gene expression values (FPKM) of representative transcription factors in unstimulated and E. coli cocultured neutrophils. (E) Known transcription factor motifs identified in D. (F) Metascape gene functional analysis for genes interacting with SMC3-amassed enhancers. Full metascape analysis results are shown in Supplemental Table S2. (G) Empirical cumulative distribution of log2(E. coli cocultured/unstimulated FPKM) values for all genes, genes interacting with any enhancer, and genes interacting with E. coli coculture-dependent SMC3-amassed enhancers.
SMC3-amassed enhancers are characterized by modestly increased CTCF binding, and substantially increased H3K27ac deposition and polyadenylated RNA abundance (Fig. 6A). Supporting the importance of SMC3 and H3K27ac deposition at new regulatory interactions, E. coli coculture-specific chromatin interaction anchors were found to be enriched for SMC3 occupancy and H3K27ac deposition, with only modest changes in CTCF occupancy compared with unstimulated neutrophil-specific interactions (Supplemental Fig. S5A–C). Similarly, H3K27ac-defined enhancers enriched for polyadenylated RNA signal were likewise enriched for SMC3 occupancy and H3K27ac deposition (Supplemental Fig. S5D).
Supporting the importance of SMC3-amassed enhancers in E. coli ΔPC1 domain behavior, SMC3-amassed enhancers were enriched at E. coli ΔPC1 domains (Fig. 6B). SMC-amassed enhancers were also, on average, localized closer to ΔPC1 domains when compared with the global enhancer repertoire, and were more enriched in ΔPC1 domains than the entire enhancer repertoire (Fig. 6C).
To understand the mechanism of cohesin targeting to SMC3-amassed enhancers, we identified transcription factor binding motifs enriched within both SMC3-amassed enhancers and non-SMC3-amassed enhancers found in E. coli cocultured neutrophils. We then computed the enrichment of transcription factor motif density in SMC3-amassed enhancers compared with all enhancers found in E. coli cocultured neutrophils. Validating this approach, we found that DNA-binding motifs associated with known inflammatory regulating factor AP1 including JUND and FOSL2, as well as transcriptional regulators that orchestrate neutrophil differentiation and physiology including CEBPB, CEBP homolog CHOP, and PU.1, were significantly enriched across the bacterial-induced SMC3-amassed enhancer repertoire compared with the entire enhancer repertoire (Fig. 6D [top], E). Notably, transcript abundance associated with these factors was elevated in neutrophils exposed to E. coli (Fig. 6D, bottom). Apart from known inflammatory and myeloid regulatory transcription factors, we also found that DNA sequences associated with TFE3 occupancy were enriched at SMC3-amassed enhancers (Fig. 6D,E).
Recent studies revealed that in activated macrophages and microglial cells TFE3 orchestrate an inflammatory program of gene expression (Pastore et al. 2016). Notably, upon activation TFE3 relocated from the cytoplasm to the nuclear interior (Pastore et al. 2016). As a first approach to determine whether likewise TFE3 translocates from the cytoplasm to the nucleus we examined naïve and microbial-exposed neutrophils for TFE3 localization using immunofluorescence. We found that in naive neutrophils TFE3 was predominantly localized in the cytoplasm. Notably, however, we found that within 3 h upon exposure to E. coli TFE3 repositioned from the cytoplasm to the nuclear interior in the vast majority of neutrophils (Supplemental Fig. S6).
Genes interacting with SMC3-amassed enhancers were next analyzed for functional group enrichment. Notably, bacterial-induced SMC3-amassed enhancers were closely associated with genes involved in neutrophil activation, including cytokine signaling and response, chemotaxis, and degranulation (Fig. 6F; Supplemental Fig. S7; Supplemental Table S2). Analysis of RNA-seq data revealed that genes interacting with enhancers in general showed little preference to be induced upon E. coli encounter as compared with any other gene in the genome. In contrast, genes linked to SMC3-amassed enhancers showed a significant increase in gene expression during E. coli encounter compared with genes globally, or to genes linked to enhancers in general (Fig. 6G). This phenomenon appears to depend on both SMC3 occupancy and H3K27ac deposition, as interactions with either SMC3-amassed E. coli-specific enhancers or SMC3-amassed pre-existing enhancers were both associated with increased gene expression, whereas interactions with enhancers only found in unstimulated cells were not associated with increased gene expression, regardless of SMC3 occupancy (Supplemental Fig. S8).
Taken together, these data indicate that upon bacterial exposure human neutrophils rapidly sequester the cohesin machinery at a specific subset of enhancers to modulate chromatin folding and activate an inflammatory gene transcription program.
Discussion
The unique morphology of neutrophils has been an enigma since its discovery more than a century ago (Cavaillon 2011). How neutrophil genomes are folded into three-dimensional space and how neutrophil nuclear architecture is altered upon microbial exposure has remained largely unknown. Here we used a genome-wide chromosome conformation capture approach (HiC) to address these questions. We found that human neutrophil nuclei, when compared with embryonic stem cells, displayed a distinct nuclear architecture: (1) a decline in genomic interactions across loop domains (<3 Mb); (2) a segmentation of large, continuous A and B compartments into numerous small compartments, resulting in the establishment of new compartment and loop domain boundaries; and, (3) an increase in remote chromosomal interactions across loop domains (>3 Mb). This increase in long-range genomic interactions primarily involved heterochromatic regions indicating a key role for heterochromatic interactions in influencing human neutrophil genome topology. Our data are consistent with previous studies involving murine neutrophils that also displayed a highly contracted genome when compared with progenitor cells and show that key features of neutrophil genome structure are conserved between the murine and human genomes (Zhu et al. 2017).
The neutrophil genome undergoes large-scale alterations in morphology upon bacterial encounter. Using genome-wide chromosome conformation capture studies, we found that such changes involve the repositioning of euchromatic E. coli ΔPC1 domains enriched for cytokine and other immune response genes. Upon encountering activating stimuli, these domains gained euchromatic character, repositioning themselves from the nuclear periphery to the more euchromatic nuclear interior. During this process, the boundaries of these domains lost insulation, allowing the domain to merge with neighboring highly euchromatic regions, and further allowing for new chromatin interactions to form and activate an inflammatory gene program. These subdomains resemble a previously identified euchromatic A2 spatial subcompartment positioned between the nuclear periphery and the nuclear interior (Rao et al. 2014; Chen et al. 2018). Based on our observations, we propose that the A2 subcompartment is associated with genes or regulatory elements that need to be transcriptionally repressed, but accessed quickly, precluding both their sequestration to the fully heterochromatic B compartment, as well as their presence in the transcriptionally active A1 compartment.
Our data further provide mechanistic insight as to how neutrophils instruct changes in nuclear positioning and domain insulation upon bacterial encounter. Alterations in chromatin topology both at ΔPC1 domains and across the genome are closely associated with the rapid recruitment of cohesin to a subset of H3K27ac-defined enhancers. While cohesin occupancy is substantially enriched at these enhancers, CTCF binding is only modestly elevated upon bacterial encounter. These observations imply that changes in nuclear architecture are predominantly activated by cohesin-dependent loop extrusion. This finding then raises the question as to how cohesin is being recruited to inflammatory genes upon bacterial encounter. We found that the increase in cohesin occupancy at SMC3-amassed enhancers was closely accompanied by substantial enrichment for the enhancer mark H3K27ac. Hence, we suggest that upon bacterial encounter, human neutrophils activate a signaling response that involves the Toll-like receptor pathway. Motif analysis suggests that Toll-like receptor mediated signaling modulates the expression and/or biochemical activities of key neutrophil-associated transcriptional regulators such as PU.1, CEBP/β, CEBP homolog CHOP, AP1 factors JUN and FOS, as well as TFE3. The activities of such regulators, in turn, would promote the assembly of an active enhancer repertoire as evidenced by the deposition of H3K27Ac, which then rapidly sequesters cohesin at inflammatory response enhancer-gene promoter clusters. Once recruited to SMC3-amassed enhancers, cohesin may act to extrude chromatin until convergent CTCF sites are reached, removing insulation at ΔPC1 domain boundaries by forming de novo loop domains in which activated enhancers are placed within close spatial proximity of gene promoters, altogether facilitating the rapid activation of an inflammatory response gene program (Fig. 7).
Figure 7.

Microbial-induced human neutrophil activation instructs rapid changes in nuclear architecture to orchestrate an inflammatory gene program. Activation-induced transcription factor binding results in H3K27ac deposition, cohesin recruitment, and formation of de novo chromatin loops linking enhancers to inflammatory genes to orchestrate an inflammatory gene program.
Why has such an elaborate mechanism of gene activation, including loop extrusion, evolved in human neutrophils? We suggest that segregating enhancers and promoters in spatially distinct loop domains ensures efficient silencing and prevents stochastic activation of an inflammatory-specific gene program in unstimulated neutrophils. Only upon exposure to activating stimuli are unstimulated neutrophils instructed to juxtapose the inflammatory enhancer repertoire with their target gene promoters, thus facilitating enhancer-promoter communication and the induction of an inflammatory-specific gene program. We hypothesize that the specificity of this response is likely governed by transcription factors downstream from activated receptors that bind their target enhancers, allowing cohesin and histone acetyl transferase recruitment, juxtaposition of target gene promoters, and stabilization of transcription units.
As documented here for human neutrophils during a microbial encounter, enhancers and promoters may be spatially segregated from each other in distinct loop domains until an appropriate environmental signal is received in order to prevent inappropriate or pathological activation of gene expression. Previous studies have documented a related mechanism that orchestrates the developmental progression of lymphoid cells. Specifically, regulatory regions associated with key developmental regulators such as EBF1 and Bcl11b are, in progenitor cells, positioned at the nuclear lamina to suppress premature activation during developmental progression. Upon reaching the appropriate developmental stage, alterations in chromatin folding readily reposition such enhancers away from the transcriptionally repressive environment at the lamina into the euchromatic nuclear interior, leading to assembly of transcriptionally productive enhancer–promoter interactions. The repositioning also directs the enhancer into a single loop domain to facilitate enhancer–promoter communication. Once placed within the euchromatic compartment and within spatial proximity to EBF1 and Bcl11b, enhancers and promoters then act to establish B or T cell identity, respectively (Lin et al. 2012; Isoda et al. 2017). Thus, the inflammatory gene response and activation of a developmental-specific gene expression programs share a common mechanism that assures appropriate timing of gene expression.
In sum, here we demonstrate that in human neutrophils, prior to encounter with bacteria, an armamentarium of inflammatory genes was positioned in a transcriptionally passive environment suppressing premature transcriptional activation. Upon microbial exposure, however, human neutrophils rapidly (<3 h) repositioned the ensemble of proinflammatory genes towards the transcriptionally permissive compartment. We found that the repositioning of genes was closely associated with the swift recruitment of cohesin across the inflammatory enhancer landscape, permitting an immediate transcriptional response upon bacterial exposure. These data reveal at the mechanistic level how upon microbial challenge human neutrophils undergo rapid changes in nuclear architecture to orchestrate an immediate inflammatory gene program.
Materials and methods
Human subject details
Blood for neutrophil isolation was obtained via venopuncture from healthy human volunteers under written informed consent approved by the University of California at San Diego Human Research Protection Program (#131002X).
Blood draws and neutrophil isolation
Whole blood was layered onto Polymorphprep reagent (Accurate Chemical and Scientific Corp. AN1114683), centrifuged for 45 min at 500g, 25°C, and allowed to stop without braking. The granulocyte layer was extracted and contaminating red blood cells were lysed as needed (generally one to three times) with brief resuspensions in sterile H2O followed by immediate flooding with 1× phosphate-buffered saline (PBS) and centrifugation at 500g for 7 min at 25°C. Cells were checked for purity via Wright-Giemsa staining; the final granulocyte fraction was generally >95% neutrophils. For RNA-sequencing experiments, neutrophils were further purified to homogeneity using an EasySep human neutrophil enrichment kit (Stem Cell Technologies 19257) as per the manufacturer's protocol.
For all experiments, neutrophils were cultured in HBSS +Ca/+Mg/−Phenol red (Thermo Fisher 14025092) with the addition of 0.5% endotoxin-free BSA (Akron AK8917-0100) at 37°C in a 5% CO2 humidified incubator.
Wright-Giemsa staining
Neutrophils (1 × 105) were spun onto cover slips using a Cytospin3 (Shandon 74010121 GB) and flooded with Wright stain (Sigma WS16-500ML) for 3 min. Cover slips were then washed with six consecutive dips in water baths. Cover slips were then allowed to air dry and were then flooded with Giemsa stain (Sigma GS500-500ML) and allowed to incubate for 7 min before being washed as above and allowed to air dry.
Neutrophil activation
Neutrophils were plated at the desired cell numbers and treated with 25 nM phorbol 12-myristate 13-acetate (PMA, Promega V1171) or cocultured in the presence of E. coli strain K1 at a multiplicity of infection (MOI) of 5. Stimulations were performed for 3 h and cells were harvested as detailed below.
Chromatin immunoprecipitation with sequencing (ChIP-seq)
Neutrophils were plated at 10 × 106 to 20 × 106 cells/10 mL for each ChIP experiment. At the completion of each experiment cells were washed with fresh media, formaldehyde was added to the culture to a final concentration of 1%, and cells were cross linked with agitation for 10 min at room temperature. Fixation was then quenched for 5 min with glycine at a final concentration of 0.13 M. Fixed cells were scraped from the plate and washed three times in ice cold 1× phosphate buffered saline (PBS) with 0.1 mM EDTA and 1×EDTA-free complete protease inhibitors (Roche 05056489001). Cell pellets were snap frozen in liquid nitrogen and stored at −80°C until processing.
To bind antibody to Protein G Dynabeads (Invitrogen 10004D), beads were washed three times with 1 mL of bead wash buffer (1×PBS, 5 mg/mL BSA, Roche complete EDTA-free protease inhibitor, 0.22 µM filtered) and resuspended in 500 µL of the same. One to five micrograms of antibody was added and allowed to bind beads overnight at 4°C with rotation. The following day, beads were washed three times with 1 mL of bead wash buffer and resuspended in 100 µL of RIPA 150 (50 mM Tris at pH 8.0, 150 mM NaCl, 0.1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100, 1 mM EDTA).
For each ChIP, cells were thawed and lysed for 10 min on ice with inversion in Farnham lysis buffer (5 mM PIPES at pH 8.0, 85 mM KCl, 0.5% NP-40, 10 mM EDTA, protease inhibitors) with or without 20 draws through an 18-gauge needle. Nuclei were spun down at 2000 rpm for 5 min at 10°C in a benchtop microfuge, supernatant was discarded, and nuclei were resuspended in 300 µL of RIPA 150. Chromatin was then sonicated in a Diagenode Bioruptor 300 chilled to 4°C three times for eight cycles of 30 sec on and 30 sec off, set on high with 5 min of cooling time between each set of eight cycles. The insoluble fraction was spun down at maximum speed for 20 min at 4°C in a benchtop microfuge. Input and IP samples were split to separate new tubes, IP volume was adjusted to 900 µL with RIPA 150, and 100 µL of Protein G dynabeads bound to the antibody of interest in RIPA 150 was added to each IP. Chromatin was allowed to bind to antibody-bead conjugates overnight at 4°C while rotating. Following binding, beads were washed twice for 5 min in RIPA 150, twice for 5 min in RIPA 500 (50 mM Tris at pH 8.0, 500 mM NaCl, 0.1% SDS, 01% sodium deoxycholate, 1% Triton X-100, 1 mM EDTA), twice for 3 min in LiCl wash (10 mM Tris at pH 8.0, 250 mM LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA), and once in 1× TE. Beads were transferred to clean tubes at the start of each new wash buffer. DNA was eluted from beads with 200 µL of elution buffer (1 mM sodium carbonate, 1% SDS) for 1 h at 65°C with shaking, at which point beads were removed and cross-links were reversed overnight at 65°C. Eluted DNA was purified using a ChIP DNA clean and concentrator kit (Zymo D5205).
DNA for ChIP and other high-throughput sequencing approaches was processed as follows: End repair was performed using an Epicenter End-It kit (Lucigen ER0720), according to manufacturer's instructions and column purified in a Zymo Minelute column (Zymo D4013). A-tails were added by incubating DNA in 1×NEB buffer 2 (New England Biolabs B7002S) with the addition of 200 µM dATP and 7.5 units of Klenow (exo-) (New England Biolabs M0212L) for 45 min at 37°C. NEB Next adaptors (New England Biolabs E7337A) were ligated using an NEB quick ligation kit (New England Biolabs M2200L) for 30 min at benchtop temperature followed by treatment with 2 µL of USER enzyme (New England Biolabs M5505L) for 15 min at 37°C. DNA was purified using an AmpureXP bead-analogous two-step SPRI bead protocol (Rohland and Reich 2012), resulting in purification of DNA fragments between ∼200 and 800 bp.
PCR amplification of final libraries for sequencing was performed with Phusion hot start polymerase II system (Thermo Fisher F549L) in conjunction with the NEB Next indexing system (New England Biolabs E7335L and E7500S). Final size selection for all high-throughput sequencing libraries was performed using a home-made two-step SPRI bead-based DNA purification system, resulting in final DNA fragment sizes of ∼200–800 bp.
RNA sequencing
At specified time points neutrophils were washed once with PBS and lysed in the RLT buffer component of the Qiagen RNeasy mini kit (Qiagen 74106) with the addition of 10 µL/mL 2-mercaptoethanol, homogenized via Qiashredder (Qiagen 79654), and snap-frozen in liquid nitrogen. Total RNA was purified via RNeasy mini kit (Qiagen 74106) according to the manufacturer's instructions, including the RNase-free DNase (Qiagen 79254) treatment step. RNA was eluted in H2O, Turbo DNase kit buffer (Thermo Fisher/Ambion AM1907) was added to a 1× concentration, and RNA was treated with 4 U of Turbo DNase for 30 min at 37°C. Turbo DNase was then treated with inactivation reagent per manufacturer's specifications. mRNA was purified from total RNA using a Dynabead mRNA purification kit (Life Technologies 61006). First strand synthesis was performed using the SuperScript III first strand synthesis system (Thermo Fisher 18080051) as follows: 100–500 ng of RNA, 0.5 µL of oligo(dT) primer, 0.8 µL of random hexamer, 1 µL of 10 mM dNTP, and H2O to 9.5 µL. The mixture was incubated for 10 min at 70°C and then snap frozen. First strand synthesis mix composed of 2 µL of 10× RT buffer, 4 µL of 25 mM MgCl2, 2 µL of 0.1 M DTT, 0.5 µL of 120 ng/µL ActinomycinD, 40 U of RNaseOUT, and 200 units of SuperScriptIII was added to the mixture, which was then incubated for 10 min at 25°C, 45 min at 42°C, 25 min at 50°C, and15 min at 75°C. Unincorporated nucleotides were removed from the mixture using a ProbeQuant G-50 column (Sigma GE28-9034-08). First strand synthesis reaction was then brought to 51 µL with H2O and cooled on ice. Twenty-four microliters of second strand mixture composed of 1 µL of 10× RT buffer; 2 µL of 25 mM MgCl2; 1 µL of 0.1 M DTT; 2 µL of 10 mM dATP, dGTP, dCTP, and dUTP mix; 15 µL of 5× second strand synthesis buffer (New England Biolabs B6117S); 0.5 µL E. coli ligase (New England Biolabs M0205S); 2 µL of DNA polymerase I (New England Biolabs M0209S); and 0.5 µL of RNase H was added and the mixture was incubated for 2 h at 16°C. DNA was purified using a DNA clean and concentrator kit (Zymo D4013) and sonicated on a Covaris E220 with the following settings; duty cycle 10%; intensity 5; cycle per burst 200; time (seconds) 180. Sonicated DNA was purified using a DNA clean and concentrator kit. DNA was prepared for high-throughput sequencing using the methodology described above for ChIP-seq, with the addition of 1 µL of UNG (Thermo Fisher/Applied Biosystems N8080096) during USER enzyme treatment.
Whole-genome bisulfite sequencing
Neutrophils were washed twice with PBS and genomic DNA was isolated using a DNeasy Blood and Tissue kit (Qiagen 69504). One microgram of genomic DNA mixed with unmethylated λ DNA at a concentration of 0.5% of total DNA was sonicated by Biorupter 300 with 20 cycles (30 sec on/30 sec off at low power). Fragmented DNA was end-repaired and A-tailed as described above. TruSeq adapters (Illumina FC-121-2001) were ligated to fragmented DNA, which was then purified by running on a 2% agarose gel. Bisulfite conversion was performed using the MethylCode kit as described by the manufacturer (Invitrogen MECOV-50). Bisulfite-treated DNA was amplified by using a TruSeq PCR primer mixture and Pfu Turbo Cx Polymerase, agarose gel-purified, and sequenced on an Illumina HiSeq 2500 sequencer with paired-end 150-bp reads.
E. coli culture and MOI determination
E. coli strain K1 was grown in LB at 37°C with shaking overnight, and diluted into a fresh culture and grown to exponential phase the day of each experiment. E. coli was then pelleted at 3000 rpm for 10 min at 10°C on a benchtop centrifuge, washed in cell culture medium, and added to neutrophil cultures at an MOI of ∼5 in HBSS +Ca/+Mg/−Phenol red with 0.5% endotoxin free BSA. Nine 1:10 serial dilutions of E. coli-containing media were plated on LB agar and grown overnight at 37°C. The resulting colonies were counted in order to assess MOI for individual experiments.
In situ HiC
In situ HiC was performed as described (Rao et al. 2014), modifying only the MboI restriction enzyme digest time to assure proper digestion of chromatin. Generally, HiC libraries prepared from activated neutrophils were digested for 2–4 h with 50 U of MboI to avoid overdigesting the chromatin. The remainder of the library preparation adhered to the published protocol and reagents exactly. HiC library DNA was prepared for high throughput sequencing using the NEB Next platform according to manufacturer's instructions, and sequenced using paired-end 100-bp reads.
Fluorescence in situ hybridization (FISH)
Cover slips were incubated overnight in 1%HCl in 70% ethanol, washed 3× with H2O, once in 70% ethanol, and stored in 100% ethanol. Coverslips were allowed to air dry prior to adding cells. Cells were incubated on cover slips in 24-well plates as described above. At the completion of incubation times, cells were washed three times for 3 min in PBS and fixed for 30 min in 6% paraformaldehyde (Electron Microscopy Sciences 15710) in 1× PBS. PFA was flushed out with >5 volumes of PBS/0.05% Tween-20 (PBST), ensuring that cells never contact the air. Residual PFA was quenched via incubation with fresh 20 mM glycine in PBS for 15 min at room temperature. Cells were permeabilized in PBS + 0.5% Triton X-100 for 20 min at room temperature, washed twice with PBST, and incubated in PBS + 100 µg/mL RNase A (Qiagen 19101) for 1 h at 37°C. Cells were then treated with 0.1 N HCl for 5 min at room temperature, washed twice for 3 min with 1× PBS and twice for 5 min with 2× SSC, and then incubated for >48 h in 2× SSC/50% formamide at 4°C. Coverslips were then blotted dry and 5 µL of probe containing 75–200 ng of labeled DNA was added to each coverslip. Cover slips were then sealed on top of glass slides along with probe using rubber cement. Probes and genomic DNA were denatured together for 5 min at 78°C on a heat block and allowed to hybridize for 16–48 h at 37°C. Following hybridization cover slips were washed 1 × 15 min in SSC/50% formamide prewarmed to 37°C, three times for 15 min in 2× SSC prewarmed to 37°C, three times for 7 min in 0.1× SSC prewarmed to 60°C, three times for 7 min in 4× SSC/0.02% Tween-20 prewarmed to 42°C, once for 5 min with 2× SSC prewarmed to 37°C, and twice for 5 min in 1× PBS. Cells were then postfixed in 4% PFA in 1× PBS for 10 min at room temperature, and PFA was flushed out as above. Cells were washed once for 10 min in PBST + DAPI, four times for 5 min in 1× PBS, and mounted in Prolong Gold mounting medium (Thermo Fisher P36930).
FISH probes were prepared from bacterial artificial chromosomes (BACs) using nick/translation (Roche 11745808910). One microgram of BAC DNA was used in each 20-µL nick/translation reaction along with the following fluorophores, as needed: ChromaTide Alexa fluor 488-5-dUTP (Thermo Fisher/Life Technologies C11397), Cy3-dUTP (VRW 42501), or Alexa fluor 647-aha-dUTP (Thermo Fisher/Life Technologies A32763). Nick/Translation was performed for 5–16 h at 15°C and terminated by addition of 1 µL of 0.5 M EDTA. Unincorporated nucleotides were removed with ProbeQuant G-50 columns per manufacturer's instructions. One-hundred nanograms of labeled probe DNA was run on a 1.5% agarose gel following each nick/translation reaction to ensure that the majority of probe fragments were in the 300- to 800-bp range. Up to 200 ng of total probe per cover slip was combined with 10 µg of salmon sperm DNA (Thermo Fisher 15632011), 4 µg of human Cot1 DNA (Thermo Fisher 15279011), 1/10 volume of 3 M sodium acetate (pH 5.2), and 2.5 volumes of 100% ethanol. Probes were allowed to precipitate for 30 min at −20°C, centrifuged at maximum speed for 20 min at 4°C, washed twice with 70% ethanol and once with 100% ethanol, air-dried, and resuspended in 6 µL of 100% formamide at 56°C. Six microliters of 2× hybridization buffer (40% dextran sulfate in 8× SSC [20× SSC: 3 M NaCl, 0.3 M sodium citrate]) was then added to each probe. Probes were denatured for 5 min at 80°C and snap-cooled on ice. Probes were then added to cover slips and denatured and hybridized to genomic DNA as noted above. The CXCL locus FISH probe utilized BAC RP11-243E9, the heterochromatic control probe used BAC RP11-134J16.
Imaging of FISH samples was performed at the Waitt Biophotonics Center at the Salk Institute. FISH samples were imaged on Zeiss Airyscan 880 microscopes using the Airyscan Fast mode (Huff 2016) at a resolution of 40 nm in the x and y axes. Z sections were imaged every 160 nm. Quantification of FISH data was performed using TANGO (Ollion et al. 2013) for FIJI (Schindelin et al. 2012). Nuclei and spot detection were performed with built-in tools in TANGO. Image metrics analyzed in TANGO include “eroded volume fraction” and “signal quantification layer” in Figure 4, and “distances” in Supplemental Figure S3. Metrics were exported from TANGO as text files and statistical analysis and figure generation were performed in R using built-in tools (https://www.R-project.org).
HiC analysis
Raw HiC library read alignment to human genome build hg38, valid read pair filtering, matrix assembly at various resolutions, and ICE normalization of said matrices were performed using HiC-pro with default settings (Servant et al. 2015). Biological replicates were pooled following valid read pair filtering, and pooled data sets were used for analysis except where noted.
For all direct comparisons of HiC data (topological domain boundary location comparisons, insulation scores, plotted contact matrices, log2 differential matrices) ICE normalized sparse matrix files were created containing only the subset of interacting bins that recorded reads in all data sets being compared. Read numbers at these bins were then quantile normalized in R using the normalize.quantiles() function in the preprocessCore package (https://github.com/bmbolstad/preprocessCore), allowing direct comparison of chromatin interactions between libraries with different read distributions and sequencing depths (Hsu et al. 2017).
Topological domain boundaries were called on normalized HiC data at 40-kb resolution using the domain calling software published in Dixon et al. (2012).
HiC-Pro defined valid read pairs were used in conjunction with HOMER (Heinz et al. 2010) to run principal component analysis (PCA, runHiCpca.pl -res 10000), generate distance versus interaction frequency plots (makeTagDirectory), define compartment boundaries (findHiCCompartments.pl), determine interaction correlations (getHiCcorrDiff.pl -res 40000 -superRes 40000), define distance-normalized chromatin interactions (analyzeHiC -res 20000 -superRes 40000 -minDist 100000), and to generate whole-chromosome pairing plots (analyzeHiC -res 400000000). CTCF anchored-type loops were called using HICCUPS (Rao et al. 2014).
Insulation scores were determined as follows: The genome was divided into 40-kb segments. Insulation scores for each segment were defined as the number of normalized (ICE and quantile) (see above) valid read pairs within a 500-kb window centered on the segment of interest whose ends map to opposite sides of the segment of interest divided by the total number of valid read pairs whose ends both map within the 500-kb window.
ΔPC1 domains were identified as follows: PCA was run at 10,000-bp resolution on pooled HiC data using the runHiCpca.pl command in HOMER with the following settings: -res 10000 -superRes 10000 -genome hg38. Visual inspection showed that positive PC1 values corresponded to the gene-rich A compartment, and negative PC1 values corresponded to the gene-poor B compartment on all chromosomes and across all conditions. Genomic regions with PC1 score differentials between conditions greater than three standard deviations above the mean PC1 score differential between conditions were identified as potential ΔPC1 domains. PCA was then run on individual HiC biological replicates and only those potential ΔPC1 domains with a reproducible gain in PC1 value in each biological replicate were retained. Finally, reproducible ΔPC1 domains within 100 kb of each other were merged into single continuous ΔPC1 domains which were used for downstream analysis.
ChIP-seq analysis
Raw fastq files were aligned to the human genome build hg38 using Bowtie (Langmead et al. 2009) with the following parameters: -m1 –best –strata. Downstream processing of ChIP-seq data was performed using HOMER, except where noted. Uniquely mapped reads from high quality biological replicates were pooled for downstream analysis (Landt et al. 2012). Sequencing data was reorganized as a HOMER-formatted tag directory for each replicate and multiple reads mapping to the same base pair were collapsed to a single read using the makeTagDirectory command in HOMER with the following parameters: -tbp 1. ChIP peaks were called using the findPeaks command in HOMER with default parameters. Genes at ChIP peaks were identified using annotatePeaks.pl in HOMER, and the GenomicRanges package (Lawrence et al. 2013) in R.
SMC3-amassed enhancers were defined as follows: Enhancers were defined as H3K27ac peaks called as above. In order to identify enhancers with activation-dependent cohesin recruitment (SMC3-amassed enhancers), total unique SMC3 ChIP-seq reads mapping to enhancers were calculated using annotatePeaks.pl in HOMER. To directly compare binding strength between conditions, read numbers at enhancers were quantile normalized across conditions using the preprocessCore R package. Those reads with a log2(normalized activated/normalized unstimulated read numbers) value >1.5 were defined as SMC3-amassed. The GenomicRanges package in R was used to identify genes in contact with SMC3-amassed enhancers in conjunction with HOMER-defined chromatin interactions (detailed below). Enhancer-gene pairs were called as interacting if the center of one interaction anchor was within 10 kb of an enhancer and the center of the other interaction anchor was within 50 kb of a gene promoter.
RNA-seq analysis
RNA-seq data was analyzed using the Tuxedo tools, except where noted. Raw fastq files were aligned to the human genome build hg38 using tophat2 (Kim et al. 2013) with the following parameters: –library-type fr-firststrand -a 15. Duplicated reads were removed using Picard tools command MarkDuplicates REMOVE_DUPLICATES=T, and RNA-seq quality metrics were assessed using Picard tools command CollectRnaSeqMetrics (http://broadinstitute.github.io/picard). Gene expression values were computed for each replicate across each condition using cuffdiff with an hg38 refflat file as reference with the following parameters: –library-type fr-firststrand. Subsequent analysis of gene expression and integration of gene expression data with other data types was performed in R.
Metascape analysis
Genes associated with various genomic features were identified using the GenomicRanges package in R and were analyzed for functional enrichment in the Metascape Web portal (http://metascape.org) using “Express Analysis” on default settings (Tripathi et al. 2015). Metascape gene set enrichment visualizations were performed in R.
Bisulfite-seq analysis
Bisulfite converted DNA sequencing data was processed using the BSseeker2 software suite (Guo et al. 2013). A bisulfite-sequencing amenable hg38 reference genome was built using the bs_seeker2-build.py command. DNA sequences were aligned to the hg38 bisulfite sequencing-amenable genome build using the bs_seeker2-align.py command with the following options: -m 6 -I 0 -X 800. Cytosine methylation levels were determined using bs_seeker2-call_methylation.py with default settings. Awk was used to convert CGmap files to HOMER-compatible allC formatted files. HOMER-formatted tag directories were built using HOMER's makeTagDirectories command with the following options: -format allC -minCounts 0 -genome hg38. Due to sequencing coverage-induced biases in DNA methylation meta-analysis (data not shown), awk was used to create HOMER-formatted tag directories containing only those cytosine residues covered by both unstimulated and PMA-activated neutrophil data sets. HOMER's annotatePeaks.pl command was used with the -ratio option to determine DNA methylation levels at particular genomic features.
Data visualization
Normalized HiC contact matrices presented in this study were generated using HiCPlotter (Akdemir and Chin 2015). HiC interactions and ChIP-seq data in Figure 4 and Supplemental Figures S4 and S6 were visualized using Sushi (Phanstiel et al. 2014). The remainder of linear genomic data was visualized using the Integrated Genomics Viewer (Robinson et al. 2011; Thorvaldsdóttir et al. 2013). FISH images were processed in FIJI. All other data were visualized using R.
Data availability
Data sets generated in this study are available as a series in the GEO database under accession number GSE126758.
Supplementary Material
Acknowledgments
We thank Alex Bortnick and other members of the Murre laboratory for editing the manuscript, and Yolanda Markaki and Kathrin Plath for assistance with the FISH protocol. This study was supported by funding from the Center for Computational Biology and Bioinformatics (UL1TRR001442), the California Institute for Regenerative Medicine (RB5-07025), and the National Institutes of Health to C.M. (AI082850, AI00880, and AI09599) and V.N. (1 U01 AI124316); from the Frontiers of Innovation Scholars Program to M.D.; and by the Waitt Advanced Biophotonics Core Facility of the Salk Institute with funding from National Institutes of Health-National Cancer Institute Cancer Center Support Grant (P30 014195) and the Waitt Foundation. T.I. was supported by the Uehara Memorial Foundation. High-throughput sequencing was performed at the Institutes for Genomic Medicine Genomics Center, University of California at San Diego.
Author contributions: M.D. performed the majority of the experiments and analysis. Y.Z. processed HiC samples. A.H., H.L., T.I., and S.D. provided technical support and advice. M.D. and C.M. wrote the manuscript. V.N. and C.M. supervised the study.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.333708.119.
References
- Akdemir K, Chin L. 2015. HiCPlotter integrates genomic data with interaction matrices. Genome Biol 16: 198 10.1186/s13059-015-0767-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintu B, Mateo LJ, Su J-HH, Sinnott-Armstrong NA, Parker M, Kinrot S, Yamaya K, Boettiger AN, Zhuang X. 2018. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science (New York, NY) 362: eaau1783 10.1126/science.aau1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. 2004. Neutrophil extracellular traps kill bacteria. Science (New York, NY) 303: 1532–1535. 10.1126/science.1092385 [DOI] [PubMed] [Google Scholar]
- Bruno L, Ramlall V, Studer RA, Sauer S, Bradley D, Dharmalingam G, Carroll T, Ghoneim M, Chopin M, Nutt SL, et al. 2018. Control of inducible gene expression links cohesin to hematopoietic progenitor self-renewal and differentiation. Nat Immunol 9: 932–941. 10.1038/s41590-019-0471-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busslinger GA, Stocsits RR, van der Lelij P, Axelsson E, Tedeschi A, Galjart N, Peters J-MM. 2017. Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature 544: 503–507. 10.1038/nature22063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaillon J-MM. 2011. The historical milestones in the understanding of leukocyte biology initiated by Elie Metchnikoff. J Leuk Biol 90: 413–424. 10.1189/jlb.0211094 [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhang Y, Wang Y, Zhang L, Brinkman EK, Adam SA, Goldman R, van Steensel B, Ma J, Belmont AS. 2018. Mapping 3D genome organization relative to nuclear compartments using TSA-seq as a cytological ruler. J Cell Biol 217: 4025–4048. 10.1083/jcb.201807108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanucchi S, Fok ET, Dalla E, Shibayama Y, Börner K, Chang EY, Stoychev S, Imakaev M, Grimm D, Wang KC, et al. 2019. Immune genes are primed for robust transcription by proximal long noncoding RNAs located in nuclear compartments. Nat Genet 51: 138–150. 10.1038/s41588-018-0298-2 [DOI] [PubMed] [Google Scholar]
- Fritz AJ, Barutcu A, Martin-Buley L, van Wijnen AJ, Zaidi SK, Imbalzano AN, Lian JB, Stein JL, Stein GS. 2016. Chromosomes at Work: organization of chromosome territories in the interphase nucleus. J Cell Biochem 117: 9–19. 10.1002/jcb.25280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. 2016. Formation of chromosomal domains by loop extrusion. Cell Rep 15: 2038–2049. 10.1016/j.celrep.2016.04.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Romo GS, Caielli S, Vega B, Connolly J, Allantaz F, Xu Z, Punaro M, Baisch J, Guiducci C, Coffman RL, et al. 2011. Netting neutrophils are major inducers of type I IFN production in pediatric systemic lupus erythematosus. Sci Transl Med 3: 73ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Fiziev P, Yan W, Cokus S, Sun X, Zhang MQ, Chen PY, Pellegrini M. 2013. BS-Seeker2: a versatile aligning pipeline for bisulfite sequencing data. BMC Genomics 14: 774 10.1186/1471-2164-14-774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. MolCell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann K, Dreger CK, Olins AL, Olins DE, Shultz LD, Lucke B, Karl H, Kaps R, Müller D, Vayá A, et al. 2002. Mutations in the gene encoding the lamin B receptor produce an altered nuclear morphology in granulocytes (Pelger-Huët anomaly). Nat Gen 31: 410–414. 10.1038/ng925 [DOI] [PubMed] [Google Scholar]
- Hsu SC, Gilgenast TG, Bartman CR, Edwards CR, Stonestrom AJ, Huang P, Emerson DJ, Evans P, Werner MT, Keller CA, et al. 2017. The BET protein BRD2 cooperates with CTCF to enforce transcriptional and architectural boundaries. Mol Cell 66: 102–116.e7. 10.1016/j.molcel.2017.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff J. 2016. The Fast mode for ZEISS LSM 880 with Airyscan: high-speed confocal imaging with super-resolution and improved signal-to-noise ratio. Nat Meth 13: i–ii. 10.1038/nmeth.f.398 [DOI] [Google Scholar]
- Isoda T, Moore AJ, He Z, Chandra V, Aida M, Denholtz M, Piet van Hamburg J, Fisch KM, Chang AN, Fahl SP, et al. 2017. Non-coding transcription instructs chromatin folding and compartmentalization to dictate enhancer-promoter communication and T cell fate. Cell 171: 103–119.e18. 10.1016/j.cell.2017.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. 2002. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science (New York, NY) 296: 158–162. 10.1126/science.1068768 [DOI] [PubMed] [Google Scholar]
- Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. 2012. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 22: 1813–1831. 10.1101/gr.136184.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg S. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès P, Aboyoun P, Carlson M, Gentleman R, Morgan M, Carey V. 2013. Software for computing and annotating genomic ranges. PLoS Comp 9: e10003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley K, Hoffman HM, Kubes P, Cassatella MA, Zychlinsky A, Hedrick CC, Catz SD. 2018. Neutrophils: new insights and open questions. Sci Immunol 3: eaat4579 10.1126/sciimmunol.aat4579 [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science (New York, NY) 326: 289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Benner C, Mansson R, Heinz S, Miyazaki K, Miyazaki M, Chandra V, Bossen C, Glass CK, Murre C. 2012. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat Immunol 13: 1196–1204. 10.1038/ni.2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller W. 2013. Getting leukocytes to the site of inflammation. Vet Pathology 50: 7–22. 10.1177/0300985812469883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Goloborodko A, Valton A-LL, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, Bruneau BG. 2017. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169: 930–944.e22. 10.1016/j.cell.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins AL, Hoang TV, Zwerger M, Herrmann H, Zentgraf H, Noegel AA, Karakesisoglou I, Hodzic D, Olins DE. 2009. The LINC-less granulocyte nucleus. European Cell Biol 88: 203–214. 10.1016/j.ejcb.2008.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollion J, Cochennec J, Loll F, Escudé C, Boudier T. 2013. TANGO: a generic tool for high-throughput 3D image analysis for studying nuclear organization. Bioinformatics 29: 1840–1841. 10.1093/bioinformatics/btt276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore N, Brady OA, Diab HI, Martina JA, Sun L, Huynh T, Lim JA, Zare H, Raben N, Ballabio A, et al. 2016. TFEB and TFE3 cooperate in the regulation of the innate immune response in activated macrophages. Autophagy 12: 1240–1258. 10.1080/15548627.2016.1179405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phanstiel D, Boyle A, Araya C, Snyder MP. 2014. Sushi.R: flexible, quantitative and integrative genomic visualizations for publication-quality multi-panel figures. Bioinformatics 30: 2808–2810. 10.1093/bioinformatics/btu379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huang S-CC, Glenn St Hilaire B, Engreitz JM, Perez EM, Kieffer-Kwon K-RR, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID, et al. 2017. Cohesin loss eliminates all loop domains. Cell 171: 305–320.e24. 10.1016/j.cell.2017.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J, Thorvaldsdóttir H, Winckler W, Guttman M, Lander E, Getz G, Mesirov J. 2011. Integrative genomics viewer. Nat Biotech 29: 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohland N, Reich D. 2012. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res 22: 939–946. 10.1101/gr.128124.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowat AC, Jaalouk DE, Zwerger M, Ung W, Eydelnant IA, Olins DE, Olins AL, Herrmann H, Weitz DA, Lammerding J. 2013. Nuclear envelope composition determines the ability of neutrophil-type cells to passage through micron-scale constrictions. J Biol Chem 288: 8610–8618. 10.1074/jbc.M112.441535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer W, Abdennur N, Goloborodko A, Pekowska A, Fudenberg G, Loe-Mie Y, Fonseca NA, Huber W, Haering CH, Mirny L, et al. 2017. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551: 51–56. 10.1038/nature24281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant N, Varoquaux N, Lajoie BR, Viara E, Chen C-JJ, Vert J-PP, Heard E, Dekker J, Barillot E. 2015. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol 16: 259 10.1186/s13059-015-0831-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz LD, Lyons BL, Burzenski LM, Gott B, Samuels R, Schweitzer PA, Dreger C, Herrmann H, Kalscheuer V, Olins AL, et al. 2003. Mutations at the mouse ichthyosis locus are within the lamin B receptor gene: a single gene model for human Pelger-Huët anomaly. Hum Mol Genet 12: 61–69. 10.1093/hmg/ddg003 [DOI] [PubMed] [Google Scholar]
- Tecchio C, Micheletti A, Cassatella MA. 2014. Neutrophil-derived cytokines: facts beyond expression. Front Immunol 5: 508 10.3389/fimmu.2014.00508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CJ, Schroder K. 2013. Pattern recognition receptor function in neutrophils. Trends Immunol 34: 317–328. 10.1016/j.it.2013.02.008 [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov J. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinformatics 14: 178–192. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che J, Mulder LC, et al. 2015. Meta- and orthogonal integration of influenza ‘OMICs’ data defines a role for UBR4 in virus budding. Cell Host Microbe 18: 723–735. 10.1016/j.chom.2015.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Gong K, Denholtz M, Chandra V, Kamps MP, Alber F, Murre C. 2017. Comprehensive characterization of neutrophil genome topology. Genes Devel 31: 141–153. 10.1101/gad.293910.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data sets generated in this study are available as a series in the GEO database under accession number GSE126758.