In this study, Zhang et al. set out to describe the molecular mechanisms underlying meiotic chromatin remodeling and meiotic resumption during oocyte development. Using a combination of in vivo and genomic approaches, the authors demonstrate that Snf2h, the catalytic subunit of ISWI family complexes, is critical in driving meiotic progression and acts by regulating the expression of genes important for maturation-promoting factor (MPF) activation.
Keywords: chromatin remodeling, Snf2h, transcriptional regulation, germ cell development, meiotic resumption, MPF activity
Abstract
Oocytes are indispensable for mammalian life. Thus, it is important to understand how mature oocytes are generated. As a critical stage of oocytes development, meiosis has been extensively studied, yet how chromatin remodeling contributes to this process is largely unknown. Here, we demonstrate that the ATP-dependent chromatin remodeling factor Snf2h (also known as Smarca5) plays a critical role in regulating meiotic cell cycle progression. Females with oocyte-specific depletion of Snf2h are infertile and oocytes lacking Snf2h fail to undergo meiotic resumption. Mechanistically, depletion of Snf2h results in dysregulation of meiosis-related genes, which causes failure of maturation-promoting factor (MPF) activation. ATAC-seq analysis in oocytes revealed that Snf2h regulates transcription of key meiotic genes, such as Prkar2b, by increasing its promoter chromatin accessibility. Thus, our studies not only demonstrate the importance of Snf2h in oocyte meiotic resumption, but also reveal the mechanism underlying how a chromatin remodeling factor can regulate oocyte meiosis.
Germ cell meiotic defect is one of the major causes of female reproductive disorders including aneuploidy, miscarriage, and infertility. Oocytes initiate meiosis during fetal development and then remain arrested at the prophase I stage until puberty (Holt et al. 2013). After puberty, the luteinizing hormone (LH) surge triggers oocytes to re-enter the meiotic cell cycle, which is termed as meiotic resumption. The first visible sign of meiotic resumption is breakdown of the nuclear membranes, which is referred to as germinal vesicle (GV) breakdown (GVBD). Following GVBD, oocytes undergo metaphase I (MI) spindle formation, microtubule-to-kinetochore attachments, anaphase-to-telophase transition, and first polar body extrusion. After completion of meiosis I, oocytes enter meiosis II and stay arrested at the metaphase II (MII) until fertilization. Upon fertilization, oocytes complete meiosis II with the extrusion of second polar body (Clift and Schuh 2013).
Meiotic progression is controlled by the maturation-promoting factor (MPF), a protein complex composed of cyclin-dependent kinase 1 (Cdk1) and its coactivator cyclin B1 (Cyclin B1). MPF is inactive when GV oocytes are arrested at prophase I and its activation is a perquisite for meiotic resumption (Holt et al. 2013). In GV oocytes, Cdk1 is inactivated by Wee1b-mediated phosphorylation at Thr14 and Tyr15 (Han et al. 2005) and Cyclin B1 is maintained at minimal level by anaphase-promoting complex/cyclosome (APC/C)-mediated degradation (Marangos et al. 2007). To trigger meiotic resumption, LH surge decreases oocyte cAMP level and its downstream protein kinase A (PKA) activity, which activates Cdk1 by reducing the phosphorylation at Thr14/Tyr15. Active Cdk1 then represses APC/C activity to allow accumulation of Cyclin B1 and leads to MPF activation (Holt et al. 2013). Besides the cAMP-PKA signaling cascade, additional factors such as phosphatases (e.g., Dusp7 and Ppp2cb) (Su et al. 2012; Pfender et al. 2015; Tischer and Schuh 2016) and kinases (e.g., Fyn) (Zheng et al. 2007; Levi et al. 2010) have been reported to control meiotic resumption through regulating the MPF activity. Thus, the role of posttranslational regulation of the cAMP–PKA–MPF pathway in meiosis has been well established.
In addition to regulation of MPF activity, meiotic progression is also accompanied with dramatic chromatin remodeling (Sasaki and Matsui 2008) and epigenetic changes. For example, depletion of the CxxC zinc finger protein Cfp1, a major mediator of H3K4me3, delays meiotic resumption and causes defective spindle assembly (Sha et al. 2018). In addition, histone lysine methyltransferase Setdb1 and demethylase Lsd1 have been shown to be required for chromatin condensation and meiotic resumption in mouse oocytes (Kim et al. 2015, 2016; Eymery et al. 2016). However, depletion of these epigenetic regulators only delays meiotic resumption, but not completely blocks GVBD, suggesting that either functional redundancy between these epigenetic factors or the key regulator(s) of meiotic resumption functioning at chromatin level is yet to be identified.
ATP-dependent chromatin remodeling complexes play important roles in regulating gene transcription and consist of four subfamilies: the switching/sucrose nonfermenting (SWI–SNF) family, the imitation switch (ISWI) family, the chromodomain helicase DNA-binding protein (CHD) family, and the inositol requiring 80 (INO80) family (Clapier and Cairns 2009). All chromatin remodeling complexes use ATP hydrolysis to alter histone–DNA contacts, and the catalytic subunits share a similar ATPase-containing domain. Since ATP-dependent chromatin remodeling complexes are mainly characterized in biochemical assays or cell lines (Clapier et al. 2017), their physiological roles remain less understood. With regard to meiosis, ISWI has been shown to be required for germline stem cell (GSC) self-renewal and early oogenesis in Drosophila (Deuring et al. 2000; Xi and Xie 2005). Consistently, depletion of Acf1, a subunit of an ISWI complex, CHRAC, leads to defects in oogenesis in Drosophila (Börner et al. 2016). In addition, Snf2h, the catalytic subunit of ISWI family complexes, has been postulated to play a role in meiotic resumption given its transient presence in bovine GV oocytes (Wee et al. 2010). However, whether ISWI complex is functionally important for mammalian meiotic progression remains unknown.
Through oocyte-specific depletion, we demonstrate here that Snf2h (also known as Smarca5) plays an essential role in oocyte meiotic resumption in mouse. Loss of function of Snf2h results in dysregulation of a number of genes involved in MPF activation and oocyte maturation, including Ccnb2 (Li et al. 2018), Prkaca (Bornslaeger et al. 1986), Prkar2b (Yoon et al. 2018), and Ndc80 (Gui and Homer 2013). In addition, ATAC-seq analysis revealed a global alteration in chromatin accessibility in Snf2h-deficient oocytes and a significant decrease of chromatin accessibility at the Prkar2b promoter. Our study supports the notion that Snf2h deficiency-mediated chromatin remodeling defects of the key meiotic genes in growing oocytes manifest to cause gene expression changes in fully grown GV oocytes underlying the meiotic resumption defects in CKO Snf2h female mice.
Results
Snf2h is required for MII oocyte generation
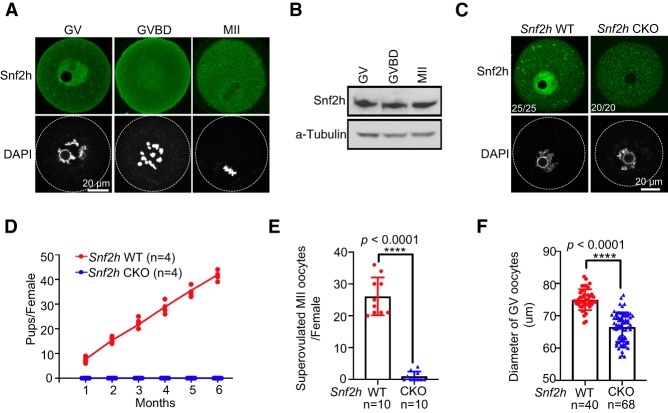
A previous study in bovine indicated that Snf2h protein is transiently detectable in GV but not GVBD oocytes, implying that Snf2h may have a specific function for oocyte maturation (Wee et al. 2010). In this study, we used mouse oocyte as a model system to test this hypothesis. We first assessed Snf2h level by immunostaining during mouse oocyte maturation and found Snf2h signal is readily detectable in the nucleus of GV oocytes but appears dispersed in GVBD and MII oocytes (Fig. 1A). Western blot analysis confirmed that Snf2h protein is present with a similar level at all three stages (Fig. 1B).
Figure 1.
Snf2h is essential for female fertility in mouse. (A) Immunostaining of Snf2h in oocytes at germinal vesicle (GV), germinal vesicle breakdown (GVBD), and meiosis metaphase II (MII) stages. Oocytes were collected from adult ovaries (6–8 wk). Scale bar, 20 µm. (B) Western blotting of Snf2h at GV, GVBD, and MII stages. a-Tubulin was used as a loading control. (C) Immunostaining of Snf2h in WT and CKO GV oocytes. Oocytes were collected from adult WT and CKO ovaries (6–8 wk). The numbers represent the number of oocytes showing the staining pattern and the total number of oocytes analyzed, respectively. Scale bar, 20 µm. (D) Fertility test of WT and CKO female mice. The WT (n = 6) and CKO (n = 6) female mice were co-caged with WT fertile male mice for 6 mo and the total numbers of pups per female are shown. (E) The numbers of MII oocytes per adult female mice (6–8 wk) after superovulation. The WT (n = 10 from three independent biological replicates) and CKO (n = 10 from three independent biological replicates) female mice were used for the analysis. Data are presented as mean ± SEM. Each dot represents a single female mouse analyzed. (****) P < 0.0001 by two-tailed Student's t-tests. (F) Quantification of oocyte size by measuring the diameters of GV oocytes (n = 40 from WT and n = 68 from CKO adult female mice, N = 3 independent biological replicates). Each dot represents a single oocyte analyzed. Data are presented as mean ± SEM. (****) P < 0.0001 by two-tailed Student's t-tests.
To assess the Snf2h function in oogenesis, an oocyte-specific knockout of Snf2h (CKO) was generated by crossing the Zp3Cre mice (Lewandoski et al. 1997) with the Snf2hfloxed/floxed mice (Snf2hfl/fl) (Alvarez-Saavedra et al. 2014). The Zp3-Cre is an oocyte-specific Cre line, in which Cre is expressed in early growing oocytes of mice at postnatal day five (P5) (Lan et al. 2004). In this study, Snf2hfl/fl and Zp3cre-Snf2hfl/fl female mice served as wild type (WT) and CKO, respectively. In CKO oocytes, exon 5 of Snf2h is depleted and a truncated protein without the ATPase domain is expected to be produced (Supplemental Fig. S1A). Immunostaining with an antibody against the full length Snf2h protein confirmed that Snf2h was undetectable in the CKO GV oocytes (Fig. 1C).
The Snf2h CKO female mice are infertile as no pups were obtained when they were crossed with WT fertile male mice for 6 mo (Fig. 1D). Remarkably, almost no MII oocytes can be collected from CKO female mice (Fig. 1E), indicating a severe defect in meiotic maturation. The sizes of WT and CKO ovaries are similar (Supplemental Fig. S1B) and comparable number of GV oocytes can be retrieved from both groups (Supplemental Fig. S1C). The ratio of GV oocytes with nonsurrounded nucleolus (NSN) and surrounded nucleolus (SN) chromatin configuration showed no difference between WT and CKO (Supplemental Fig. S1D). Interestingly, the average diameter of CKO GV oocytes (66 µm) is significantly smaller than that of the WT GV oocytes (75 µm) (Fig. 1F). Since the meiotic and developmental competences are acquired gradually during oocyte growth and the competences correlate with oocyte size, chromatin conformation, and cytoplasmic maturation (Wickramasinghe et al. 1991; Eppig et al. 1994), the above data suggests that the CKO GV oocytes may have impaired meiotic and/or developmental competence.
Oocyte-specific depletion of Snf2h blocks meiotic resumption
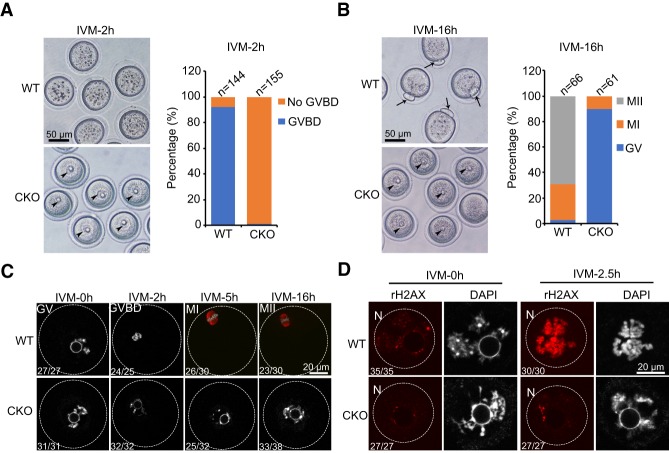
To determine when Snf2h CKO oocytes arrest during meiotic maturation, we monitored the meiotic progression of GV oocytes collected from 6- to 8-wk female mice by in vitro maturation (IVM). After 2 h, most (92.2%) WT GV oocytes underwent GVBD, but almost all (98.6%) CKO oocytes were still arrested at prophase I (Fig. 2A). At 16 h, while 69% of the WT oocytes developed to MII stage, as evident by the extrusion of second polar body, only a few CKO oocytes entered into MI stage and most (90%) CKO oocytes still maintain the intact nuclear envelope (Fig. 2B). Similar meiotic resumption defects were also observed in oocytes collected from CKO mice without prior hormone (pregnant mare serum gonadotrophin [PMSG]) administration (Supplemental Fig. S2A), ruling out the possibility that such defects are artifacts caused by hormone stimulation. Interestingly, CKO oocytes from 3-wk-old female mice showed a weaker phenotype, in which ∼35% of oocytes can develop to MII stage after 16 h of in vitro culture (Supplemental Fig. S2B,C). This observation suggests that the longer oocyte growth phase in adult female mice may lead to accumulation of more severe defects caused by Snf2h deficiency, which further impair the meiotic competence. To better characterize the Snf2h deficiency caused oogenesis defects, we used the GV oocytes from 6- to 8-wk-old female mice in the subsequent experiments.
Figure 2.
Oocyte-specific depletion of Snf2h blocks meiotic resumption. (A) GVBD ratio of WT and Snf2h CKO oocytes after in vitro maturation (IVM) for 2 h. The left panel shows the representative bright field images. Arrowheads point to germinal vesicles. Scale bar, 50 µm. The right panel shows the quantification of GVBD ratio for WT and CKO oocytes. One-hundred-forty-four WT and 155 CKO GV oocytes from adult ovaries (6–8 wk) were used for the analysis. N = 6 independent biological replicates. (B) Oocyte maturation ratio of WT and CKO oocytes after IVM for 16 h. Left panel shows the representative bright field images. Arrows point to first polar bodies and arrowheads point to germinal vesicles. Scale bar, 50 µm. Right panel shows the quantification of maturation ratio for GV oocytes. Sixty-six WT and 61 CKO GV oocytes from adult ovaries (6–8 wk) were used for the analysis. N = 3 independent biological replicates. (C) Immunostaining showing the maturation process of WT and CKO oocytes. Stages are determined based on DAPI and α-Tubulin staining. The numbers represent the combined number of oocytes exhibiting the staining pattern and the total number of oocytes analyzed in three independent biological replicates. Scale bar, 20 µm. (D) Immunostaining showing the level of γH2AX in WT and CKO oocytes during IVM. The numbers represent the combined number of oocytes exhibiting the staining pattern and the total number of oocytes analyzed in three independent biological replicates. Scale bar, 20 µm.
We next performed tubulin and DNA immunostaining at different time points of oocyte maturation to monitor the chromatin configuration changes. Contrary to WT oocytes, which underwent chromatin condensation, spindle formation and first polar body extrusion during IVM, most CKO oocytes were arrested at GV stage without any apparent chromatin configuration changes (Fig. 2C). Furthermore, CKO oocytes did not exhibit increased γH2AX signals at GVBD as observed in WT oocytes (Fig. 2D), indicating a defect in chromatin remodeling at meiotic resumption.
Snf2h is required for cytoplasmic MPF activation in oocytes
At the initial stages of meiotic resumption, oocytes undergo chromatin condensation and nuclear membrane breakdown. As a chromatin remodeling factor, Snf2h can participate in this process by directly shaping the chromatin or indirectly through regulating the expression of genes important for meiotic resumption. To differentiate between these two possibilities, we first analyzed the chromatin organizations by immunofluorescence in WT and CKO GV oocytes. During the NSN-to-SN transition in GV oocytes, chromatin becomes condensed and the number of chromocenters (aggregation of heterochromatin regions, indicated by the immunostaining of HP1β) is decreased from approximately seven to approximately three per oocyte (Supplemental Fig. S3A,B; Bonnet-Garnier et al. 2012). However, in CKO GV oocytes, the number of chromocenters was only slightly decreased during NSN-to-SN transition (from approximately seven to approximately six per oocyte). Thus, more chromocenters in CKO SN oocytes were observed than that in WT SN oocytes (Supplemental Fig. S3A,B). Furthermore, as indicated by HP1β staining, while chromocenters in WT SN GV oocytes were mostly scattered away from the nucleolus-like body (NLB), they were still in proximity to the perinucleolar chromatin in CKO SN GV oocytes (Supplemental Fig. S3A,C,D). Therefore, Snf2h depletion affects both the number of chromocenters and their localization during NSN-to-SN transition in GV oocytes.
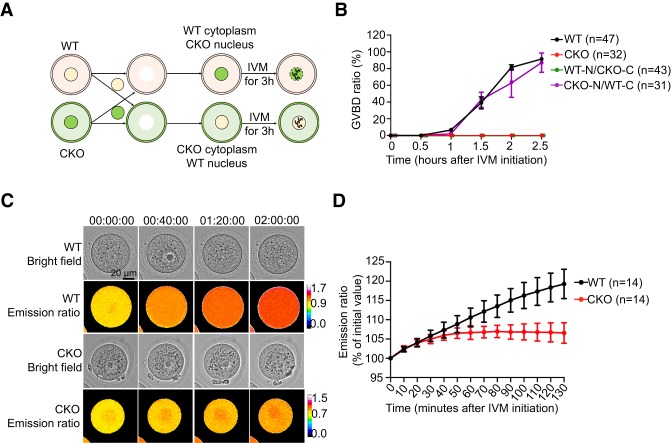
To determine whether the chromatin organization defects are the cause of the meiotic arrest of CKO GV oocytes, we exchanged the nucleus between WT and CKO oocytes and monitored the meiotic progression by IVM (Fig. 3A). Although most GV oocytes reconstructed with WT cytoplasm and CKO nucleus successfully went through meiotic resumption after 2.5-h of IVM, none of the GV oocytes reconstructed with CKO cytoplasm and WT nucleus underwent GVBD (Fig. 3B). This result indicates that cytoplasm defects, but not the chromatin defects, in the CKO oocytes are the major causes of the meiotic arrest. These data also suggest that Snf2h contributes to oocyte maturation, likely through regulating the expression of meiosis-related genes.
Figure 3.
Cytoplasmic defects cause CKO oocytes unable to activate maturation-promoting factor (MPF) for meiotic resumption. (A) Schematic diagram of the experimental strategy for nuclei exchange experiments. The GV nuclei from WT or CKO mice were removed and reconstructed with enucleated CKO or WT oocytes, respectively. (B) GVBD ratio of WT, CKO, and reconstructed oocytes at different time points during IVM. N = 3 independent biological replicates and n > 10 oocytes were used for each replicate. Data are presented as mean ± SEM. (C) Representative images (one image per 40 min) showing the dynamic FRET signal during IVM of WT and CKO oocytes. (D) Quantification of the emission ratio over time in WT (n = 14) and CKO (n = 14) oocytes from N = 3 independent biological replicates. Data are presented as mean ± SEM.
It is well established that meiotic arrest of GV oocytes is maintained by high levels of cAMP. cAMP activates PKA to phosphorylate downstream effector proteins, which in turn suppresses the MPF (Cdk1/Cyclin B1) activity (Holt et al. 2013). To determine whether MPF fails to activate in CKO oocytes and causes the meiotic arrest, we used a fluorescence resonance energy transfer (FRET) assay to monitor the MPF activity during IVM (Levasseur et al. 2019). As expected, the MPF activity of WT GV oocytes increased during the 2-h IVM period and underwent GVBD. In contrast, the MPF activity of CKO oocytes only slightly increased and reached a plateau by 50 min and GVBD did not take place (Fig. 3C,D). These data indicate that Snf2h CKO GV oocytes fail to activate MPF to initiate meiotic resumption.
Depletion of Snf2h leads to dysregulation of genes involved in MPF activation
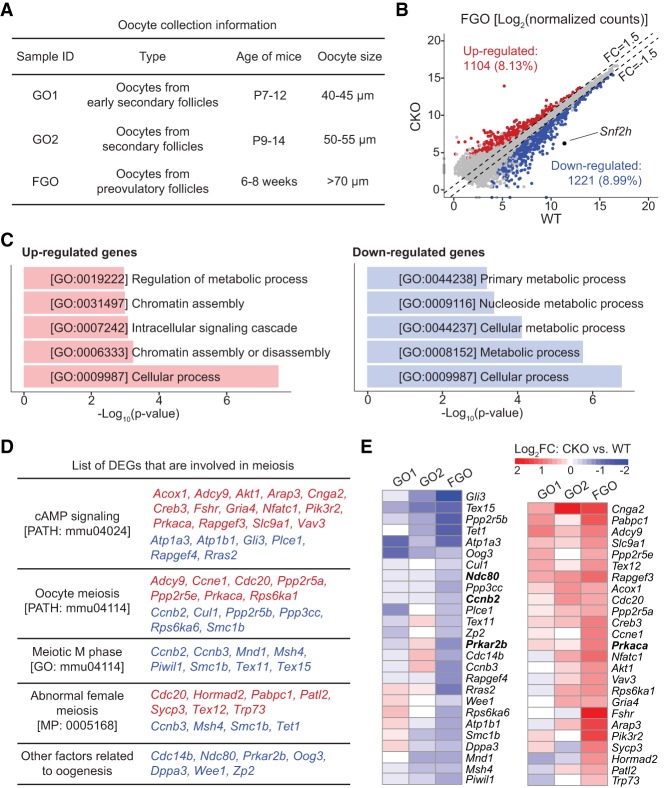
Since no active transcription takes place after GV NSN-to-SN transition, the inability of cytoplasmic products to activate MPF should reflect the cumulative defects caused by Snf2h deficiency during oocyte growth. To understand how Snf2h deficiency impairs MPF activation, we performed RNA sequencing (RNA-seq) analysis of growing oocytes from early secondary follicles (growing oocytes 1 [GO1], 40–45 µm) and secondary follicles (growing oocytes 2 [GO2], 50–55 µm), and fully grown oocytes from preovulatory follicles (FGO, >70 µm) (Fig. 4A). The transcriptomes of two replicates from each group exhibited high correlations (R > 0.975) (Supplemental Fig. S4A,B) and Snf2h was completely depleted at all three stages (Supplemental Fig. S4C). Notably, our transcriptome data sets are highly correlated to a previously published RNA-seq data set (Supplemental Fig. S4D; Zhang et al. 2016), validating our oocyte staging, collection, and RNA-seq analysis procedures.
Figure 4.
Depletion of Snf2h leads to dysregulation of genes related to MPF activation. (A) Schematic illustration of oocytes collection for RNA-seq. Oocytes were collected based on the age of the mice and the size of the oocytes. Twenty oocytes were used for each RNA-seq library and two RNA-seq replicates were performed for WT and CKO groups at each stage. (B) Scatter plots comparing the gene expression profiles of WT and CKO fully grown GV oocytes. Two replicates for WT and CKO FGO oocytes were used for differential gene expression analyses (FC > 1.5, P-value <0.05, RPKM > 1). (C) Gene ontology analysis of the differentially expressed genes in CKO FGO oocytes. (D) Functional classification of differentially expressed genes associated with cAMP signaling, oocyte meiosis, meiotic M phase, abnormal female meiosis, and other reported factors involved in oogenesis. Up-regulated and down-regulated genes are colored with red and blue, respectively. (E) Heat map showing the log2 (fold change) of genes listed in D for GO1, GO2, and FGO oocytes.
Comparative analysis revealed that the most significant transcriptome differences between WT and CKO were observed in fully grown oocytes with 1104 genes up-regulated and 1221 genes down-regulated (FC > 1.5, P < 0.05, and RPKM > 1 as a cutoff) (Fig. 4B; Supplemental Fig. S4E; Supplemental Table S1). This result is consistent with the principle component analysis (PCA) (Supplemental Fig. S4B). One likely explanation for the milder gene expression changes in growing oocytes compared with those in fully grown oocytes is that Snf2h depletion by Zp3-Cre occurs at P5 and the transcriptional changes are accumulated during oocyte growth. Gene Ontology (GO) analysis revealed that the up-regulated genes in fully grown GV oocytes is enriched for the GO term of chromatin assembly (Fig. 4C), consistent with the chromatin remodeling function of Snf2h. Additionally, both up- and down-regulated genes in GV oocytes are enriched for the GO terms of cellular and metabolic processes (Fig. 4C) known to be important for oocyte maturation (Gu et al. 2015).
Although unlikely, these transcriptional changes might be caused by hormone administration. To rule out this possibility, we performed RNA-seq using non-hormone-stimulated fully grown GV oocytes (nFGO). Comparative analysis revealed a good correlation not only between the two replicates of nFGO, but also between the FGO samples collected with or without hormone stimulation (Supplemental Fig. S5A). Similar to the FGO collected under hormone stimulation, a significant transcriptome change was observed between WT and CKO nFGO with 880 genes up-regulated and 1065 genes down-regulated (Supplemental Fig. S5B), and these differentially expressed genes were highly correlated with those identified under hormone stimulated conditions (Supplemental Fig. S5C). These data strongly support the notion that the gene expression change is due to loss of Snf2h function rather than caused by hormone stimulation.
To establish a link between the transcriptome changes and the MPF activation defect, we analyzed expression levels of genes involved in cAMP signaling and female meiotic cell cycle and found a number of meiosis-related genes were indeed dysregulated in both CKO FGO and nFGO (Fig. 4D; Supplemental Fig. S5D). We validated the differential expression of Ccnb2, Ndc80, Prkar2b, and Prkaca in CKO oocytes using quantitative PCR and confirmed that the protein abundance of Prkaca, Prkar2b, and Cyclin B2 were also obviously altered in Snf2h-depleted oocytes (Supplemental Fig. S6A,B). Based on previous studies (Yin et al. 2007; Gui and Homer 2013; Li et al. 2018), a combined up-regulation of Adcy9, Prkaca, and Cdc20, and down-regulation of Ccnb2, Ccnb3, Prkar2b, and Ndc80 is likely to be responsible for the failure of MPF activation in CKO oocytes. Since most genes involved in cAMP signaling and female meiosis was not as severely altered in growing oocytes (i.e., GO1 and GO2) as compared with the fully grown oocytes (Fig. 4E; Supplemental Fig. S6A), it is likely that the overall defects accumulated during oocyte growth are responsible for the MPF activation defect.
The meiotic resumption defect can be rescued by restoring MPF activity
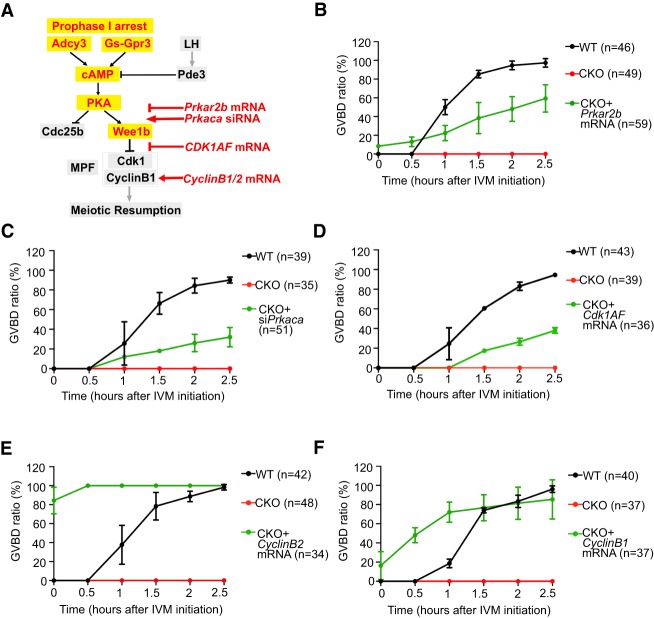
The cAMP-PKA-MPF signaling pathway plays a critical role in regulating meiotic resumption (Fig. 5A). Prkar2b and Prkaca, respectively, encode the regulatory and catalytic subunit of PKA (Kirschner et al. 2009). After binding to cAMP, the PKA catalytic subunit is released from the regulatory subunit and becomes capable of phosphorylating downstream substrates (e.g., Wee1b) to inhibit MPF activity (Fig. 5A). Previous studies have shown that the ratio of catalytic to regulatory subunit (C/R ratio) is important for PKA activity (Nishimura et al. 2012) and that overexpression of Prkaca could inhibit meiotic resumption (Bornslaeger et al. 1986). Thus, simultaneous up-regulation of Prkaca and down-regulation of Prkar2b in CKO GV oocytes (Fig. 4E) may elevate the C/R ratio, leading to the increase in PKA activity and ultimate inhibition of MPF and meiotic resumption. We attempted to rescue the meiotic arrest phenotype by Prkar2b mRNA overexpression or Prkaca siRNA-mediated knockdown (Fig. 5A). Importantly, both manipulations partially rescued the meiotic resumption defects (Fig. 5B,C). Similarly, overexpression of a constitutively active form of Cdk1 (Cdk1AF), which cannot be phosphorylated at Thr14/Tyr15 residues (Adhikari et al. 2016), also partially rescued the phenotype (Fig. 5D).
Figure 5.
Meiotic resumption defects caused by Snf2h deficiency can be rescued by restoring MPF activity. (A) Schematic illustration of the cAMP-PKA-MPF pathway. Red words with yellow background positively regulate cAMP-PKA activity, which leads to meiotic arrest at prophase I stage. Black words with gray background negatively regulate cAMP-PKA activity, which activates MPF and promotes meiotic resumption. Red words with no background are the strategies used for the rescue experiments. (B–F) GVBD ratio at different time points during IVM for oocytes injected with Prkar2b mRNA (B), Prkaca siRNA (C), Cdk1AF mRNA (D), Ccnb2 mRNA (E), or Ccnb1 mRNA (F). N = 3 independent biological replicates for each rescue experiment and n > 10 oocytes were used for each replicate. Data are presented as mean ± SEM.
Cyclin B2 (encoded by Ccnb2), similar to Cyclin B1 (encoded by Ccnb1), can associate with Cdk1 (Satyanarayana and Kaldis 2009) and compensate for Cyclin B1 deficiency in Ccnb1 CKO oocytes (Li et al. 2018). In addition, Ndc80 (also known as Hecl) has been shown to protect Cyclin B2 from destruction by Cdh1-activated anaphase-promoting complex (APCCdh1) and depletion of Ndc80 or Ccnb2 by antisense morpholinos resulted in severe defects in GVBD (Gui and Homer 2013). In Snf2h CKO oocytes, we found that expression levels of both Ccnb2 and Ndc80 were decreased (Fig. 4E), which resulted in a decrease of Cyclin B2 protein abundance (Supplemental Fig. S6B). Consistently, overexpression of Ccnb2 fully rescued the defects of meiotic resumption in CKO oocytes, even when they were cultured in the MEM medium containing cAMP phosphodiesterase inhibitor 3-Isobutyl-1-methylxanthine (IBMX) (Fig. 5E). Since Cyclin B1 and Cyclin B2 have redundant functions in meiotic cell cycle regulation, we also overexpressed Ccnb1 mRNA and achieved almost a complete rescue of the meiotic phenotype (Fig. 5F). Collectively, the above results support the notion that Snf2h regulates meiotic resumption by regulating transcription of genes involved in MPF activation.
Snf2h regulates transcription by altering chromatin accessibility
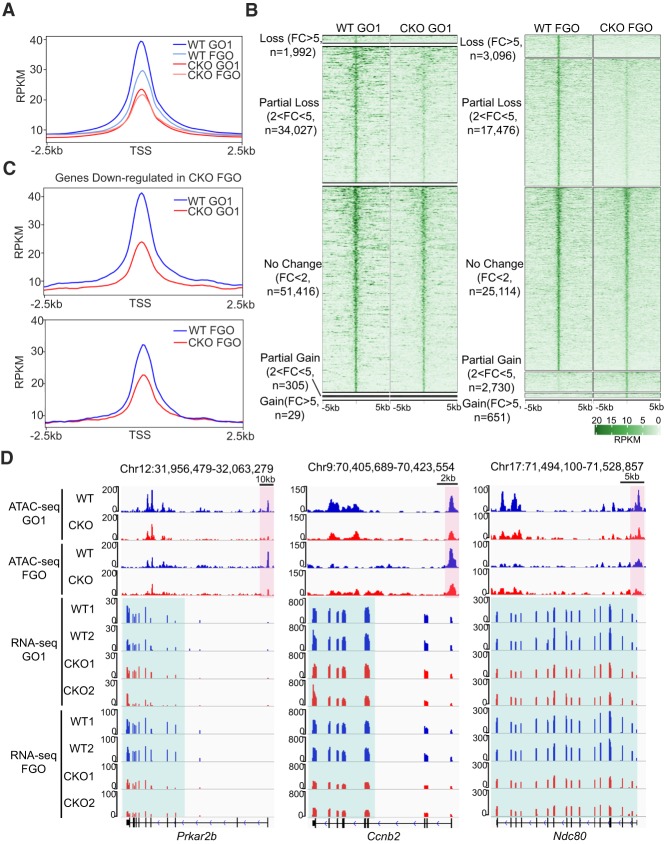
We next set out to understand how Snf2h may regulate transcription of meiosis-related genes during oogenesis. Since chromatin remodeling factors are known to regulate gene expression by altering nucleosome positioning and promoter accessibility (Clapier and Cairns 2009), we hypothesize that dysregulation of meiotic genes may be due to the alternation of promoter chromatin accessibility in the CKO oocytes. To this end, we assessed chromatin accessibility by performing ATAC-seq analysis of growing oocytes (GO1) and fully grown GV oocytes (FGO) from WT and CKO female mice with two replicates (Supplemental Table S2). After confirming the data reproducibility (Supplemental Fig. S7A), we combined the data for subsequent analysis. Consistent with previous reported chromatin “open-to-close” transition at the transcription start sites (TSS) during oocyte growth (Gu et al. 2019), lower level of ATAC-seq signal at TSS was observed in fully grown GV oocytes when compared with that in GO1 oocytes from WT mice (Fig. 6A). Compared with WT oocytes, an overall reduction of ATAC-seq signal was observed in both CKO GO1 and FGO (Fig. 6A,B), indicating a global decrease of chromatin accessibility due to Snf2h depletion.
Figure 6.
Snf2h regulates transcription by altering chromatin accessibility. (A) Metaplot showing the ATAC-seq signals at transcription start sites (TSSs) in WT and CKO growing oocytes (GO1) and fully grown GV oocytes (FGO). (B) Heat map showing the ATAC peaks classified according to their changes in CKO versus WT oocytes. Each row represents a locus (ATAC-seq peak center ± 5 kb), and the green gradient color indicates the ATAC-seq signal intensity. (C) Metaplot showing the ATAC-seq signals at TSS of down-regulated genes in CKO FGO. (D) Genome browser views showing the ATAC-seq and RNA-seq signals at Prkar2b, Ccnb2, and Ndc80 loci. The pink boxes indicate the ATAC peaks at the promoter regions, and the green boxes indicate the RNA-seq signals.
To determine whether the mRNA abundance differences in CKO fully grown GV oocytes correlate with chromatin accessibility changes during oocyte growth, we integrated the ATAC-seq data sets with the transcriptome of WT and CKO oocytes. We found that the down-regulated genes in CKO fully grown GV oocytes, as a whole, exhibited a more dramatic decrease of promoter ATAC signal in CKO GO1 compared with the CKO FGO (Fig. 6C). To analyze this correlation in more detail, we generated a heat map followed by clustering analysis, which revealed that 70% of down-regulated genes were associated with decreased ATAC-seq signals in GO1 and/or FGO (Supplemental Fig. S7B,D). In contrast, only < 25% of up-regulated genes were associated with increased ATAC-seq signals at either stage (Supplemental Fig. S7B). Importantly, some meiotic genes, including those whose dysregulation contribute to the Snf2h CKO meiotic arrest, such as Prkar2b, Ccnb2, and Ndc80, had already reduced or lost their promoter ATAC peaks in CKO GO1 prior to their down-regulation in the fully grown oocytes (Fig. 6D; Supplemental Fig. S7C,D). Collectively, these data suggest that loss function of Snf2h causes decreased promoter accessibility of meiotic genes at GO1 stage, which likely manifest to affect gene expression at fully grown oocytes, resulting in meiotic defects.
Discussion
The precise progression of meiotic cell cycle is essential for female reproduction, and defects in meiotic resumption have been one of the major causes of infertility. Although meiotic progression is known to be accompanied by dramatic chromatin remodeling, the molecular mechanisms underlying meiotic chromatin remodeling and meiotic resumption are poorly understood. Here, we demonstrate that Snf2h, the catalytic subunit of ISWI family complexes, is critical in driving meiotic progression by regulating the expression of genes important for MPF activation. The study represents an important progress towards understanding the molecular mechanism of how chromatin remodeling contributes to meiotic resumption.
Snf2h is an essential regulator of meiotic resumption
It is well known that chromatin remodeling factors use the energy derived from ATP hydrolysis to move, eject, or restructure nucleosomes (Clapier et al. 2017). The ISWI complex has been previously shown to alter DNA and histone contact by nucleosome sliding and histone replacement in vitro (Clapier et al. 2017). However, it is not clear how ISWI-mediated nucleosome positioning or spacing regulates transcription and whether this causes any physiological outcome. As the catalytic subunit of the ISWI complex, Snf2h has been reported to be critical for embryonic development (Stopka and Skoultchi 2003), embryonic lens differentiation (He et al. 2016), hematopoietic stem and progenitor cell (HSPC) proliferation and differentiation (Kokavec et al. 2017), as well as cerebellar morphogenesis and neural maturation (Alvarez-Saavedra et al. 2014). Here we extended these studies by uncovering its essential function in oocyte meiotic resumption.
Unlike WT oocytes, which undergo GVBD and chromatin condensation during meiotic resumption, neither takes place in Snf2h CKO oocytes (Fig. 2). Despite the defects in chromocenter formation in the Snf2h CKO oocytes, nucleus exchange experiments demonstrated that nucleus defects did not contribute to the meiotic arrest phenotype. Rather, it is the defects in CKO cytoplasm that prevented the reconstructed oocytes from entering meiotic resumption (Fig. 3). This unexpected result indicates that Snf2h controls meiotic resumption through regulating meiosis-related gene expression, rather than regulating chromatin configuration changes. Consistent with this notion, RNA-seq analysis revealed dysregulation of a number of genes involved in MPF activation in CKO fully grown GV oocytes (Fig. 4). Importantly, restoring the MPF activity through mRNA/siRNA injection can partially or fully rescue the meiotic arrest phenotype of CKO oocytes (Fig. 5). These data strongly support that transcriptional dysregulation of genes involved in MPF activation is the cause of the meiotic arrest in Snf2h CKO oocytes. Thus, our study demonstrates that Snf2h-mediated chromatin remodeling is indispensable for meiotic gene expression and meiotic resumption during oogenesis.
Dysregulation of CDK1 activation and Cyclin B2 expression prevent MPF activation
In GV oocytes, MPF remains inactive due to Wee1b-mediated inhibitory phosphorylation of Cdk1 and APCCdh1-mediated active degradation of Cyclin B1/B2 (Gui and Homer 2013; Adhikari et al. 2016). During meiotic resumption, MPF activation requires Cdk1 activation and Cyclin B1/B2 accumulation (Fig. 5A). It is known that down-regulation of cAMP-PKA activity is essential for MPF activation as low levels of the cAMP-PKA activity can lead to the decrease of Wee1b phosphorylation, which releases Cdk1 from the inhibitory phosphorylation. Furthermore, Cdk1 activation can lead to accumulation of Cyclin B1/B2 by inhibiting APCCdh1 activity (Holt et al. 2013).
In CKO oocytes, both Cdk1 activation and Cyclin B1/B2 accumulation are impaired due to misregulation of relevant genes. On one hand, down-regulation of Prkar2b and up-regulation of Prkaca in CKO oocytes elevates the catalytic/regulatory ratio of PKA and increases PKA activity, which ultimately impairs Cdk1 activation. On the other hand, both Ccnb2 and Ndc80, a transcript encoding a protector of Cyclin B2 from degradation, were down-regulated in CKO oocytes, which may lead to insufficient levels of Cyclin B2 for MPF activation. Therefore, combined dysregulation of both Cdk1 activation and Cyclin B2 level can explain the meiotic arrest phenotype of the CKO oocytes.
We showed that Ccnb1/2 mRNA overexpression fully rescued the meiotic arrest phenotype of CKO oocytes (Fig. 5). This observation is not surprising considering the fact that in mitotic cells a high level of Cyclin B can lead to a full activation of Cdk1 by activating Cdc25, which can remove the inhibitory phosphorylation of Cdk1 (Deibler and Kirschner 2010). Thus, although the high PKA activity can inhibit Cdk1 activation in CKO oocytes, overexpression of Ccnb1/2 may overcome this inhibitory effect by activating Cdc25/Cdk1 to achieve a full rescue of the meiotic arrest phenotype. In contrast, although Prkar2b and Cdk1AF mRNA overexpression or Prkaca siRNA knockdown can reduce PKA-mediated inhibition of Cdk1, MPF activation may be limited by the insufficient level of Cyclin B2. As mentioned earlier, both Ccnb2 mRNA and a Cyclin B2 degradation protector Ndc80 are down-regulated in CKO oocytes. Thus, insufficient Cyclin B2 cannot be rescued by inhibiting PKA activity or increasing Cdk1 activity, which explains why only partial rescue was achieved under these conditions.
Snf2h regulates transcription of meiotic genes by altering chromatin accessibility
Compared with the well-studied nucleosome-positioning mechanism, whether Snf2h regulates transcription by altering chromatin accessibility remains largely unknown. In fact, only a couple of studies suggested that Snf2h can alter chromatin accessibility in T cells and ES cells. For example, it was reported that Snf2h may regulate cytokine gene expression by altering chromatin accessibility in the murine EL4 T cells (Precht et al. 2010). Recently, Barisic et al. (2019) compared chromatin accessibility landscape in WT and Snf2h KO ESCs and suggested that target binding of a subset of transcription factors may depend on Snf2h-mediated chromatin remodel. Therefore, in vivo evidences are still lacking regarding how Snf2h-mediate chromatin remodeling can control gene expression.
In this study, using ATAC-seq, we showed that depletion of Snf2h resulted in a global decrease in chromatin accessibility during oocyte growth (Fig. 6). Although there is no direct correlation between global RNA abundance and chromatin accessibility in fully grown GV oocytes, we indeed observed a strong correlation between decrease of ATAC-seq signals and transcriptional down-regulation during oogenesis (Fig. 6; Supplemental Fig. S7). These data indicate that Snf2h may function as a chromatin remodeler to facilitate gene expression by increasing chromatin accessibility during oocyte growth. Importantly, the key meiotic genes whose dysregulation contribute to the meiotic arrest of Snf2h CKO oocytes, such as Prkar2b, Ccnb2, and Ndc80, had already exhibited reduced chromatin accessibility at GO1 stage prior to their reduced gene expression in fully grown oocytes. Collectively, these data support the notion that Snf2h-mediated promoter accessibility might be a prior requirement for meiotic gene activation. Failure to establish open chromatin at GO1 in Snf2h CKO mice may prevent activation of key meiotic genes in fully grown oocytes, resulting in meiotic resumption defect.
Materials and methods
Mice
Snf2h CKO mice were generated by crossing Zp3cre mice (Lewandoski et al. 1997) with Snf2hfl/fl mice (Alvarez-Saavedra et al. 2014). All animal studies were performed in accordance with guidelines of the Institutional Animal Care and Use Committee at Harvard Medical School.
Oocyte collection and in vitro maturation
For fully grown GV oocytes collection, ovaries were dissected from 3- or 6- to 8-wk-old female mice 46–48 h after injection with 7.5 international units (IU) of pregnant mare serum gonadotrophin (PMSG) (Millipore). For non-hormone-stimulated fully grown GV oocytes (nFGO), ovaries were dissected from 6- to 8-wk-old female mice without PMSG injection. The ovaries were transferred to M2 medium (Sigma-Aldrich) and punctured with a 27-gauge needle to release cumulus–oocyte complexes. The cumulus cells were gently removed from the cumulus–oocyte complexes using a narrow-bore glass pipette. The oocytes were then transferred into α-MEM (Life technologies) supplemented with 5% FBS (Sigma-Aldrich), 10 ng/mL EGF (Sigma-Aldrich), and 0.2 mM 3-isobutyl-1-methylxanthine (IBMX) (Sigma-Aldrich). About 30–40 GV oocytes can be obtained from each hormonally primed mouse. After culture in MEM + IBMX medium for 1 h, GV oocytes with perivitelline space (PVS) were selected for subsequent experiments as these oocytes are of high meiotic competence (Inoue et al. 2007). For the meiotic resumption and in vitro oocyte maturation (IVM) analyses, GV oocytes were washed at least three times to remove IBMX before being transferred to IBMX-free α-MEM medium containing 5% FBS and 10 ng/mL EGF.
For growing oocytes collection, ovaries were removed and digested in PBS supplemented with 2 mg/mL collagenase (Sigma-Aldrich) and 0.025% trypsin (Sigma-Aldrich) for 30 min at 37°C. M2 medium (Sigma-Aldrich) was then added to neutralize the digestion mix and oocytes were picked up with a drawn-out glass pipette. To avoid potential contamination by somatic cells, oocytes were washed extensively in clean drops of M2 medium. A stage micrometer was used in combination with an eyepiece reticle to measure the sizes of oocytes. Mice of postnatal days p7–12 and p9–14 were used to collect growing oocytes with diameters of 40–45 µm (from early secondary follicles) and 50–55 µm (from secondary follicles), respectively.
mRNA synthesis and microinjection
mRNAs were synthesized by in vitro transcription (IVT) as described previously (Matoba et al. 2014). Briefly, MPF activity sensor (Addgene plasmid #26064) and full-length cDNA of Ccn1b, Ccnb2, Prkar2b, and Cdk1AF were cloned into pcDNA3.1 vector with in-fusion cloning kit (Clontech). The constructed plasmids were linearized by XbaI. After purification, the linearized plasmid DNA were used as templates for IVT using mMESSAGE mMACHINE T7 Ultra kit (Thermo Fisher Scientific). The synthesized mRNA was dissolved in nuclease-free water and quantified by NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies) and stored at −80°C.
GV oocytes with PVS were injected with ∼10 pL of mRNA with specific concentrations (1.5 µg/µL Ccnb1 mRNA, 1.0 µg/µL Ccnb2 mRNA, 2.5 µg/µL Prkar2b mRNA, 1.5 µg/µL Cdk1AF mRNA, 1.5 µg/µL MPF activity sensor) using a Piezo-driven micromanipulator (Eppendorf). Injected GV oocytes were cultured in MEM + IBMX medium for 3 h to allow mRNA translation and then transferred into IBMX-free MEM medium for IVM. For siRNA injection, GV oocytes were injected with 4 µM siPrkaca and arrested in MEM + IBMX medium for 24 h before IVM. Oligonucleotides used in this study are listed in Supplemental Table S3.
Nucleus exchange experiments in fully grown GV oocytes
GV oocytes nuclear transfer was carried out as described previously (Inoue et al. 2008). Briefly, GV oocytes were cultured in M2 medium containing 0.2 mM IBMX, 10 mg/mL cytochalasin B (Calbiochem), and 0.1 mg/mL colcemid (Sigma-Aldrich) for 15 min. The nucleus was removed with minimal cytoplasm carryover and fused with an enucleated oocyte using GenomONE - CF EX Sendai virus Envelope cell fusion kit (Cosmo Bio HVJ-E). The reconstructed oocytes were cultured in MEM + IBMX for 1 h to ensure complete fusion before IVM experiments.
Western blotting
For immunoblot experiments, 120 oocytes were collected and zona pellucida were removed with acid Tyrode's solution (Sigma-Aldrich). After washing three times with prewarmed 1% polyvinylalcohol (PVA) (Sigma-Aldrich)/PBS, oocytes in 5 µL of 1%PVA/PBS were transferred using a calibrated glass pipette to 15 µL of sample buffer containing 5 µL of 4× NuPAGE LDS sample buffer (Thermo Fisher Scientific) and 10 µL of sterile filtered water. The samples were then snap-frozen in dry ice for 5 min and the freeze–thaw procedures were repeated twice before storing them at −80°C. The samples were heated for 5 min at 90°C before loading to SDS-PAGE gels. Western blotting was performed as described previously (Marangos 2016). Primary antibodies against Snf2h (1/1000 dilution; Abcam ab168653), Prkar2b (1/50 dilution; Santa Cruz Biotechnology sc-376778), Prkaca (1/1000 dilution; Cell Signaling CST4782), CDK1(Y15) (1/500 dilution; Cell Signaling CST9111), Cyclin B2 (1/1000 dilution; Abcam ab185622), and a-Tubulin (1/2000 dilution; Sigma-Aldrich T6199) were used in this study.
Reverse transcription and quantitative PCR analysis
Twenty oocytes were collected and zona pellucida were removed with acid Tyrode's solution (Sigma-Aldrich). After being washed three times with prewarmed 0.2% BSA/PBS, oocytes were subjected to reverse transcription with the SuperScript III CellsDirect cDNA synthesis kit according to the manufacturer's instruction. RT-qPCR was performed using Fast SYBR Green Master Mix (Thermo Fisher Scientific 4385612) using primers listed in Supplemental Table S3 for analysis of Prkar2b, Prkaca, Ccnb2, and Ndc80. Relative quantification was performed using comparative CT method and normalized with Gapdh.
Immunofluorescence and imaging
Immunofluorescence assay was carried out as described previously (Matoba et al. 2014). Oocytes were fixed and permeabilized in PBS containing 3.7% PFA and 0.2% Triton X-100 for 20 min, and then washed with PBS containing 10 mg/mL BSA (PBS/BSA). Oocytes were incubated with primary antibodies overnight at 4°C and secondary antibodies for 1 h at room temperature. The oocytes were then mounted on a glass slide in VectaShield antibleaching solution with DAPI (Vector Laboratories). Fluorescence was imaged using Zeiss LSM800. Primary antibodies against Snf2h (1/200 dilution; Abcam ab168653), α-Tubulin (1/2000 dilution; Sigma T6199), and γH2AX (1/1000 dilution; Millipore 05-636) were used in this study.
FRET imaging and analysis
GV oocytes were injected with MPF activity sensor mRNA and cultured in MEM + IBMX medium for 3 h for sufficient mRNA translation. Then oocytes were transferred to IBMX-free MEM medium in a confocal dish (Green BioResearch LLC). FRET imaging was performed as reported (Levasseur et al. 2019) using Screeny (HIM 1023) microscopy with the help of the MicRoN team at Harvard Medical School. All quantifications were performed using ImageJ software.
RNA-seq
RNA-seq libraries were prepared as previously described (Matoba et al. 2014). Twenty oocytes at GO1, GO2, and FGO stages from WT and CKO female mice were used for RNA-seq library preparation. Briefly, reverse transcription and cDNA amplification were performed using SMARTer ultralow input RNA cDNA preparation kit (Clontech) following the manufacturer's instructions. Nextera XT DNA library preparation kit (Illumina) was used for cDNA fragmentation, adaptor ligation, and amplification according to the manufacturer's instructions. Single-end 100-bp sequencing was performed on a HiSeq2500 sequencer (Illumina).
ATAC-seq
ATAC-seq libraries of growing oocytes and fully grown GV oocytes were prepared as previously described with some modifications (Wu et al. 2016). Briefly, 100 fully grown GV nuclei were collected after incubation in M2 medium containing 0.2 mM IBMX, 10 mg/mL cytochalasin B, and 0.1 mg/mL colcemid for 15 min. After washing with 0.2%BSA/PBS, growing oocytes or fully grown GV nuclei were added to 8 µL of tagmentation mix (33 mM Tris-acetate, 66 mM K-acetate, 10 mM Mg-acetate, 16% DMF, 0.02% digitonin) containing 0.5 µL of assembled Tn5/adapter (760 µg/mL unassembled Tn5 enzyme with 0.143 vol of 100 µM Tn5MEDS-A/B oligonucleotides). After tagmentation for 30 min at 37°C, the reaction was stopped with freshly made stop buffer (100 mM Tris at pH 8.0, 100 mM NaCl, 0.4% SDS, 40 µg/mL Proteinase K) and incubated for 16 h at 55°C. After quenching SDS by adding 5 µL of 25% Tween-20, PCR was performed to amplify the library for 16 cycles using the following PCR conditions: 5 min at 72°C and5 min at 98°C, and thermocycling for 20 sec at 98°C, 30 sec at 63°C, and 60 sec at 72°C, and 5 min at 72°C. After the PCR reaction, libraries were purified with the 1.6× SPRI beads (Beckman).
RNA-seq analyses
Methods for RNA-seq read alignments and differential gene expression analysis were similar to previously described (Chen and Zhang 2019). The RNA-seq reads were aligned to the mm10 reference and the gene annotation file was downloaded from GENECODE M20 (https://www.gencodegenes.org/mouse/release_M20.html). Genes were considered differentially expressed if they met the following criteria: RPKM > 1, fold change (FC) > 1.5, and P-value < 0.05. For PCA analyses, raw read counts were normalized by DESeq package, and the “prcomp” function available in R (http://www.r-project.org) was used. Gene ontology analyses were performed using DAVID Bioinformatics Resources 6.8 (Huang da et al. 2009). Bamcoverage from deeptools (version 3.0.2) (Ramírez et al. 2014) was used to generate RNA-seq bigwig tracks with parameters “–skipNonCoveredRegions –binSize 10 –scaleFactor 1/DESeq's size factor”.
ATAC-seq analyses
Low-quality bases and adaptor sequences were trimmed for raw ATAC-seq reads as described for RNA-seq reads. Quality-trimmed ATAC-seq reads were aligned to mm10 reference genome using bowtie2 (version 2.2.9) (Langmead and Salzberg 2012) with default parameters. Following alignment, uniquely mapped reads (reads without “XS” tag and mapping quality > 30) were retrieved, and PCR duplicates were removed using MarkDuplicates command from Picard tools (version 2.8.0; (https://broadinstitute.github.io/picard). ATAC-seq bigwig tracks were generated using bamcoverage from deeptools (version 3.0.2) (Ramírez et al. 2014) with parameters “–binSize 20 -e 250 –minMappingQuality 30 –scaleFactor 1 –normalizeUsing RPKM.”
The remaining read alignments were then subject to peak calling using Macs2 (version 2.2.1.20160309) (Zhang et al. 2008) with parameters “-g mm –bdg –SPMR –nomodel -q 0.01 –nolambda.” The overlapping peaks from all ATAC-seq libraries were merged using “findOverlapsOfPeaks” function of the ChIPpeakAnno Package (version 3.16.1) (Zhu et al. 2010). The reads mapped to each peak were counted using “summarizeOverlaps” function from the GenomicAlignments package (version 1.18.1) (Lawrence et al. 2013) and then normalized to RPKM values. The PCA analyses were performed using RPKM values of all of the merged peaks with the “prcomp” function available in R (http://www.r-project.org). To identify peaks with differential signal between WT and CKO oocytes, the following criteria were applied: average RPKM > 4, RPKM in each library > 2, and FC > 2 for partial gain or partial loss peaks and FC > 5 for gain or loss peaks.
The GENCODE M20 annotation file (https://www.gencodegenes.org/mouse/release_M20.html) was used to annotate ATAC peaks at promoter regions. The “makeTxDbFromGFF” function from the GenomicFeatures package (1.34.6) (Lawrence et al. 2013) was used to load the GTF file, and “annotatePeak” function from ChIPpeakAnno package (version 3.16.1) (Zhu et al. 2010) was used to assign peaks at promoter regions (< ±2 kb from transcription start sites). Genes with multiple ATAC-seq peaks at promoter regions were not included for scatter plots in Supplemental Figure S7C if the peaks show different direction of changes in CKO oocytes.
Statistical analyses and graph generation
The experiments were randomized and no statistical method was used to predetermine sample size. Informed consent was obtained from all subjects. Results are given as means ± SEM. Each experiment included at least three independent samples (n) or experiments (N). Results for WT and CKO were compared by two-tailed unpaired Student's t-tests. Statistically significant values were P < 0.05 (*), P < 0.01 (**), P < 0.001 (***), and P < 0.0001 (****). Statistical analyses and graph generation were performed by Prism 8 and Excel 2018.
Reproducibility between RNA-seq libraries were assessed using Pearson's r coefficient, which was calculated using “cor” function and were visualized using “smoothScatter” function available in R (http://www.r-project.org) (Supplemental Figs. S4A, S5A). Reproducibility between ATAC-seq libraries were assessed by comparing the enrichment at 5-kb bin resolution for the entire genome. Gene expression heat maps were generated using the R function “pheatmap” (Fig. 4E) or the ComplexHeatmap package (Supplemental Figs. S4D, S5C,D, S7B; Gu et al. 2016). ATAC-seq heat maps (Fig. 6B) were generated using EnrichedHeatmap package (version 1.12.0) (Gu et al. 2018). All genome browser tracks were visualized in the Integrative Genomic Viewer genome browser (Fig. 6D; Supplemental Figs. S4C, S7D; Robinson et al. 2011).
Data availability
All RNA-seq and ATAC-seq data sets generated in this study are summarized in Supplemental Table S4 and have been deposited in the Gene Expression Omnibus under accession number GSE134279.
Supplementary Material
Acknowledgments
This project is supported by the National Institutes of Health (R01HD092465 to Y.Z. and GM116143 to A.I.S.). Y.Z. is an Investigator of the Howard Hughes Medical Institute.
Author Contributions: Y.Z. conceived the project. C.Z. designed and performed most of the experiments. Z.C. and Y.L. performed sequencing and data analysis. Q.Y. performed ATAC-seq. X.F. performed Western blot. T.S. and A.I.S. provided the Snf2hfl/fl mice. C.Z. and Y.Z. wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.331157.119.
References
- Adhikari D, Busayavalasa K, Zhang J, Hu M, Risal S, Bayazit MB, Singh M, Diril MK, Kaldis P, Liu K. 2016. Inhibitory phosphorylation of Cdk1 mediates prolonged prophase I arrest in female germ cells and is essential for female reproductive lifespan. Cell Res 26: 1212–1225. 10.1038/cr.2016.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Saavedra M, De Repentigny Y, Lagali PS, Raghu Ram EV, Yan K, Hashem E, Ivanochko D, Huh MS, Yang D, Mears AJ, et al. 2014. Snf2h-mediated chromatin organization and histone H1 dynamics govern cerebellar morphogenesis and neural maturation. Nat Commun 5: 4181 10.1038/ncomms5181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barisic D, Stadler MB, Iurlaro M, Schübeler D. 2019. Mammalian ISWI and SWI/SNF selectively mediate binding of distinct transcription factors. Nature 569: 136–140. 10.1038/s41586-019-1115-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet-Garnier A, Feuerstein P, Chebrout M, Fleurot R, Jan HU, Debey P, Beaujean N. 2012. Genome organization and epigenetic marks in mouse germinal vesicle oocytes. Int J Dev Biol 56: 877–887. 10.1387/ijdb.120149ab [DOI] [PubMed] [Google Scholar]
- Börner K, Jain D, Vazquez-Pianzola P, Vengadasalam S, Steffen N, Fyodorov DV, Tomancak P, Konev A, Suter B, Becker PB. 2016. A role for tuned levels of nucleosome remodeler subunit ACF1 during Drosophila oogenesis. Dev Biol 411: 217–230. 10.1016/j.ydbio.2016.01.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornslaeger EA, Mattei P, Schultz RM. 1986. Involvement of cAMP-dependent protein kinase and protein phosphorylation in regulation of mouse oocyte maturation. Dev Biol 114: 453–462. 10.1016/0012-1606(86)90209-5 [DOI] [PubMed] [Google Scholar]
- Chen Z, Zhang Y. 2019. Loss of DUX causes minor defects in zygotic genome activation and is compatible with mouse development. Nat Genet 51: 947–951. 10.1038/s41588-019-0418-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. 2009. The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304. 10.1146/annurev.biochem.77.062706.153223 [DOI] [PubMed] [Google Scholar]
- Clapier CR, Iwasa J, Cairns BR, Peterson CL. 2017. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Biol 18: 407–422. 10.1038/nrm.2017.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clift D, Schuh M. 2013. Restarting life: fertilization and the transition from meiosis to mitosis. Nat Rev Mol Cell Biol 14: 549–562. 10.1038/nrm3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deibler RW, Kirschner MW. 2010. Quantitative reconstitution of mitotic CDK1 activation in somatic cell extracts. Mol Cell 37: 753–767. 10.1016/j.molcel.2010.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuring R, Fanti L, Armstrong JA, Sarte M, Papoulas O, Prestel M, Daubresse G, Verardo M, Moseley SL, Berloco M, et al. 2000. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol Cell 5: 355–365. 10.1016/S1097-2765(00)80430-X [DOI] [PubMed] [Google Scholar]
- Eppig JJ, Schultz RM, O'Brien M, Chesnel F. 1994. Relationship between the developmental programs controlling nuclear and cytoplasmic maturation of mouse oocytes. Dev Biol 164: 1–9. 10.1006/dbio.1994.1175 [DOI] [PubMed] [Google Scholar]
- Eymery A, Liu Z, Ozonov EA, Stadler MB, Peters AH. 2016. The methyltransferase Setdb1 is essential for meiosis and mitosis in mouse oocytes and early embryos. Development 143: 2767–2779. 10.1242/dev.132746 [DOI] [PubMed] [Google Scholar]
- Gu L, Liu H, Gu X, Boots C, Moley KH, Wang Q. 2015. Metabolic control of oocyte development: linking maternal nutrition and reproductive outcomes. Cell Mol Life Sci 72: 251–271. 10.1007/s00018-014-1739-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Eils R, Schlesner M. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32: 2847–2849. 10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- Gu Z, Eils R, Schlesner M, Ishaque N. 2018. EnrichedHeatmap: an R/Bioconductor package for comprehensive visualization of genomic signal associations. BMC Genomics 19: 234 10.1186/s12864-018-4625-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C, Liu S, Wu Q, Zhang L, Guo F. 2019. Integrative single-cell analysis of transcriptome, DNA methylome and chromatin accessibility in mouse oocytes. Cell Res 29: 110–123. 10.1038/s41422-018-0125-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui L, Homer H. 2013. Hec1-dependent cyclin B2 stabilization regulates the G2-M transition and early prometaphase in mouse oocytes. Dev Cell 25: 43–54. 10.1016/j.devcel.2013.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SJ, Chen R, Paronetto MP, Conti M. 2005. Wee1B is an oocyte-specific kinase involved in the control of meiotic arrest in the mouse. Curr Biol 15: 1670–1676. 10.1016/j.cub.2005.07.056 [DOI] [PubMed] [Google Scholar]
- He S, Limi S, McGreal RS, Xie Q, Brennan LA, Kantorow WL, Kokavec J, Majumdar R, Hou H, Edelmann W, et al. 2016. Chromatin remodeling enzyme Snf2h regulates embryonic lens differentiation and denucleation. Development 143: 1937–1947. 10.1242/dev.135285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt JE, Lane SI, Jones KT. 2013. The control of meiotic maturation in mammalian oocytes. Curr Top Dev Biol 102: 207–226. 10.1016/B978-0-12-416024-8.00007-6 [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Inoue A, Akiyama T, Nagata M, Aoki F. 2007. The perivitelline space-forming capacity of mouse oocytes is associated with meiotic competence. J Reprod Dev 53: 1043–1052. 10.1262/jrd.19064 [DOI] [PubMed] [Google Scholar]
- Inoue A, Nakajima R, Nagata M, Aoki F. 2008. Contribution of the oocyte nucleus and cytoplasm to the determination of meiotic and developmental competence in mice. Hum Reprod 23: 1377–1384. 10.1093/humrep/den096 [DOI] [PubMed] [Google Scholar]
- Kim J, Singh AK, Takata Y, Lin K, Shen J, Lu Y, Kerenyi MA, Orkin SH, Chen T. 2015. LSD1 is essential for oocyte meiotic progression by regulating CDC25B expression in mice. Nat Commun 6: 10116 10.1038/ncomms10116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Zhao H, Dan J, Kim S, Hardikar S, Hollowell D, Lin K, Lu Y, Takata Y, Shen J, et al. 2016. Maternal Setdb1 is required for meiotic progression and preimplantation development in mouse. PLoS Genet 12: e1005970 10.1371/journal.pgen.1005970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner LS, Yin Z, Jones GN, Mahoney E. 2009. Mouse models of altered protein kinase A signaling. Endocr Relat Cancer 16: 773–793. 10.1677/ERC-09-0068 [DOI] [PubMed] [Google Scholar]
- Kokavec J, Zikmund T, Savvulidi F, Kulvait V, Edelmann W, Skoultchi AI, Stopka T. 2017. The ISWI ATPase Smarca5 (Snf2h) is required for proliferation and differentiation of hematopoietic stem and progenitor c. Stem Cells 35: 1614–1623. 10.1002/stem.2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan ZJ, Xu X, Cooney AJ. 2004. Differential oocyte-specific expression of Cre recombinase activity in GDF-9-iCre, Zp3cre, and Msx2Cre transgenic mice. Biol Reprod 71: 1469–1474. 10.1095/biolreprod.104.031757 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. 2013. Software for computing and annotating genomic ranges. PLoS Comput Biol 9: e1003118 10.1371/journal.pcbi.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levasseur MD, Thomas C, Davies OR, Higgins JMG, Madgwick S. 2019. Aneuploidy in oocytes is prevented by sustained CDK1 activity through degron masking in cyclin B1. Dev Cell 48: 672–684.e5. 10.1016/j.devcel.2019.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi M, Maro B, Shalgi R. 2010. The involvement of Fyn kinase in resumption of the first meiotic division in mouse oocytes. Cell Cycle 9: 1577–1589. 10.4161/cc.9.8.11299 [DOI] [PubMed] [Google Scholar]
- Lewandoski M, Wassarman KM, Martin GR. 1997. Zp3-cre, a transgenic mouse line for the activation or inactivation of loxP-flanked target genes specifically in the female germ line. Curr Biol 7: 148–151. 10.1016/S0960-9822(06)00059-5 [DOI] [PubMed] [Google Scholar]
- Li J, Tang JX, Cheng JM, Hu B, Wang YQ, Aalia B, Li XY, Jin C, Wang XX, Deng SL, et al. 2018. Cyclin B2 can compensate for Cyclin B1 in oocyte meiosis I. J Cell Biol 217: 3901–3911. 10.1083/jcb.201802077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marangos P. 2016. Preparation of cell lysate from mouse oocytes for Western blotting analysis. Methods Mol Biol 1457: 209–215. 10.1007/978-1-4939-3795-0_15 [DOI] [PubMed] [Google Scholar]
- Marangos P, Verschuren EW, Chen R, Jackson PK, Carroll J. 2007. Prophase I arrest and progression to metaphase I in mouse oocytes are controlled by Emi1-dependent regulation of APCCdh1. J Cell Biol 176: 65–75. 10.1083/jcb.200607070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S, Liu Y, Lu F, Iwabuchi KA, Shen L, Inoue A, Zhang Y. 2014. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell 159: 884–895. 10.1016/j.cell.2014.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Fujii W, Kano K, Sugiura K, Naito K. 2012. Analyses of the involvement of PKA regulation mechanism in meiotic incompetence of porcine growing oocytes. Biol Reprod 87: 53 10.1095/biolreprod.112.101279 [DOI] [PubMed] [Google Scholar]
- Pfender S, Kuznetsov V, Pasternak M, Tischer T, Santhanam B, Schuh M. 2015. Live imaging RNAi screen reveals genes essential for meiosis in mammalian oocytes. Nature 524: 239–242. 10.1038/nature14568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Precht P, Wurster AL, Pazin MJ. 2010. The SNF2H chromatin remodeling enzyme has opposing effects on cytokine gene expression. Mol Immunol 47: 2038–2046. 10.1016/j.molimm.2010.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. 2014. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42: W187–W191. 10.1093/nar/gku365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Matsui Y. 2008. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet 9: 129–140. 10.1038/nrg2295 [DOI] [PubMed] [Google Scholar]
- Satyanarayana A, Kaldis P. 2009. Mammalian cell-cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene 28: 2925–2939. 10.1038/onc.2009.170 [DOI] [PubMed] [Google Scholar]
- Sha QQ, Dai XX, Jiang JC, Yu C, Jiang Y, Liu J, Ou XH, Zhang SY, Fan HY. 2018. CFP1 coordinates histone H3 lysine-4 trimethylation and meiotic cell cycle progression in mouse oocytes. Nat Commun 9: 3477 10.1038/s41467-018-05930-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopka T, Skoultchi AI. 2003. The ISWI ATPase Snf2h is required for early mouse development. Proc Natl Acad Sci 100: 14097–14102. 10.1073/pnas.2336105100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YQ, Sugiura K, Sun F, Pendola JK, Cox GA, Handel MA, Schimenti JC, Eppig JJ. 2012. MARF1 regulates essential oogenic processes in mice. Science 335: 1496–1499. 10.1126/science.1214680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischer T, Schuh M. 2016. The phosphatase Dusp7 drives meiotic resumption and chromosome alignment in mouse oocytes. Cell Rep 17: 1426–1437. 10.1016/j.celrep.2016.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee G, Shin ST, Koo DB, Han YM. 2010. Behaviors of ATP-dependent chromatin remodeling factors during maturation of bovine oocytes in vitro. Mol Reprod Dev 77: 126–135. [DOI] [PubMed] [Google Scholar]
- Wickramasinghe D, Ebert KM, Albertini DF. 1991. Meiotic competence acquisition is associated with the appearance of M-phase characteristics in growing mouse oocytes. Dev Biol 143: 162–172. 10.1016/0012-1606(91)90063-9 [DOI] [PubMed] [Google Scholar]
- Wu J, Huang B, Chen H, Yin Q, Liu Y, Xiang Y, Zhang B, Liu B, Wang Q, Xia W, et al. 2016. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534: 652–657. 10.1038/nature18606 [DOI] [PubMed] [Google Scholar]
- Xi R, Xie T. 2005. Stem cell self-renewal controlled by chromatin remodeling factors. Science 310: 1487–1489. 10.1126/science.1120140 [DOI] [PubMed] [Google Scholar]
- Yin S, Liu JH, Ai JS, Xiong B, Wang Q, Hou Y, Chen DY, Sun QY. 2007. Cdc20 is required for the anaphase onset of the first meiosis but not the second meiosis in mouse oocytes. Cell Cycle 6: 2990–2992. 10.4161/cc.6.23.4993 [DOI] [PubMed] [Google Scholar]
- Yoon H, Jang H, Kim EY, Moon S, Lee S, Cho M, Cho HJ, Ko JJ, Chang EM, Lee KA, et al. 2018. Knockdown of PRKAR2B results in the failure of oocyte maturation. Cell Physiol Biochem 45: 2009–2020. 10.1159/000487978 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Zheng H, Huang B, Li W, Xiang Y, Peng X, Ming J, Wu X, Zhang Y, Xu Q, et al. 2016. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 537: 553–557. 10.1038/nature19361 [DOI] [PubMed] [Google Scholar]
- Zheng KG, Meng XQ, Yang Y, Yu YS, Liu DC, Li YL. 2007. Requirements of Src family kinase during meiotic maturation in mouse oocyte. Mol Reprod Dev 74: 125–130. 10.1002/mrd.20613 [DOI] [PubMed] [Google Scholar]
- Zhu LJ, Gazin C, Lawson ND, Pagès H, Lin SM, Lapointe DS, Green MR. 2010. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11: 237 10.1186/1471-2105-11-237 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-seq and ATAC-seq data sets generated in this study are summarized in Supplemental Table S4 and have been deposited in the Gene Expression Omnibus under accession number GSE134279.