In this study, Vargas et al. set out to elucidate the downstream effectors of the Rtca-mediated RNA repair/splicing pathway. Using genome-wide transcriptome analysis, the authors demonstrate that the microtubule-associated protein (MAP) tubulin polymerization-promoting protein (TPPP) ringer functions downstream from and is suppressed by Rtca via Xbp1-dependent transcription. Ringer cell-autonomously promotes axon regeneration in the peripheral and central nervous system.
Keywords: axon regeneration, Drosophila, HDAC6, MAP1B, Rtca, TPPP, dendritic arborization neuron, futsch, microtubule, ringer
Abstract
Promoting axon regeneration in the central and peripheral nervous system is of clinical importance in neural injury and neurodegenerative diseases. Both pro- and antiregeneration factors are being identified. We previously reported that the Rtca mediated RNA repair/splicing pathway restricts axon regeneration by inhibiting the nonconventional splicing of Xbp1 mRNA under cellular stress. However, the downstream effectors remain unknown. Here, through transcriptome profiling, we show that the tubulin polymerization-promoting protein (TPPP) ringmaker/ringer is dramatically increased in Rtca-deficient Drosophila sensory neurons, which is dependent on Xbp1. Ringer is expressed in sensory neurons before and after injury, and is cell-autonomously required for axon regeneration. While loss of ringer abolishes the regeneration enhancement in Rtca mutants, its overexpression is sufficient to promote regeneration both in the peripheral and central nervous system. Ringer maintains microtubule stability/dynamics with the microtubule-associated protein futsch/MAP1B, which is also required for axon regeneration. Furthermore, ringer lies downstream from and is negatively regulated by the microtubule-associated deacetylase HDAC6, which functions as a regeneration inhibitor. Taken together, our findings suggest that ringer acts as a hub for microtubule regulators that relays cellular status information, such as cellular stress, to the integrity of microtubules in order to instruct neuroregeneration.
In recent years, several strategies have shown efficacy augmenting nerve regeneration in various experimental models (Massey et al. 2006; Park et al. 2008; Smith et al. 2009; Côté et al. 2010; Hellal et al. 2011; Nakamura and Okano 2013; Wu et al. 2015; Matamoros et al. 2019). Unfortunately, therapeutic interventions to promote nerve regeneration and functional recovery still do not exist. Previous work has also helped shape the approach researchers have taken toward better understanding regeneration and drawing connections between successful paradigms. Here, we report a link between two cellular mechanisms that are essential for regeneration: RNA processing (Anthony and Gallo 2010; Hörnberg and Holt 2013; Kosmaczewski et al. 2015; Song et al. 2015; Klim et al. 2019) and microtubule dynamics (Hellal et al. 2011; Sengottuvel et al. 2011; Chen et al. 2012; Stone et al. 2012; Ruschel and Bradke 2018; Matamoros et al. 2019).
In Drosophila, sensory dendritic arborization (da) neurons show differential regenerative potentials between the periphery and the central nervous system (CNS), resembling that of mammalian neurons. Moreover, distinct subclasses of da neurons also regenerate differently (Song et al. 2012). We previously developed a two-photon-based axon injury model that assays class III (C3da) and class IV (C4da) da neurons to identify and analyze targets that enhance regeneration (Li et al. 2018). Using this model, we identified Rtca (RNA 3′-terminal phosphate cyclase), an RNA-binding protein (RBP), as an inhibitor of axon regeneration (Song et al. 2015). Rtca is involved in stress induced Xbp1 mRNA splicing, and its knockout or neuronal knockdown promotes axon regeneration both in the peripheral nervous system (PNS) and CNS. However, its downstream effectors and signaling mechanisms remain unexplored. RBPs are increasingly shown to regulate complex cellular processes associated with neurodegenerative diseases and regeneration (Anthony and Gallo 2010; Elsaeidi et al. 2014; Klim et al. 2019). Herein, we report the results from transcriptome profiling revealing that a microtubule associated protein, ringer (also known as ringmaker, which is the fly homolog of the mammalian tubulin polymerization-promoting proteins [TPPPs]), is strongly increased following Rtca removal.
Microtubules and the cytoskeletal network are essential for neuronal function and are paramount to an axon's ability to respond to guidance cues, transport proteins and organelles, grow, survive, and regenerate (Baas et al. 1991; Tanaka et al. 1995; Buck and Zheng 2002; Witte et al. 2008; Matamoros and Baas 2016; Hilton and Bradke 2017). Microtubule-binding small molecules and microtubule-associated proteins (MAPs) that regulate microtubule dynamics are attractive therapeutic targets to augment axon regeneration (Blanquie and Bradke 2018). Ringer belongs to the brain-specific protein, p25α, also known as the TPPP protein family. TPPPs regulate tubulin polymerization and are implicated in neurodegenerative disorders such as α-synucleinopathies and Multiple System Atrophy (Lindersson et al. 2005; Kovács et al. 2007; Song et al. 2007). Drosophila has only one TPPP ortholog, ringer, and it directly binds tubulin, promotes microtubule bundling and polymerization in vitro, and is critical for microtubule stabilization and developmental axon growth (Mino et al. 2016). Here we show that transcription of ringer is negatively regulated by Rtca via Xbp1. We found that ringer functions as a neuronal intrinsic promoter of axon regeneration, working in concert with other MAPs, specifically Futsch/MAP1B and HDAC6, which have been previously shown to be integral for axonal health and integrity (Gordon-Weeks and Fischer 2000; Bettencourt da Cruz et al. 2005; Rivieccio et al. 2009; Godena et al. 2011; Li et al. 2011; Lin et al. 2015; Prior et al. 2018). Our results reveal MAPs as important arbiters of axon regeneration and propose ringer (TPPP homologs) as an attractive therapeutic target for promoting axon regeneration.
Results
Rtca loss of function increases the expression of ringer mRNA and protein
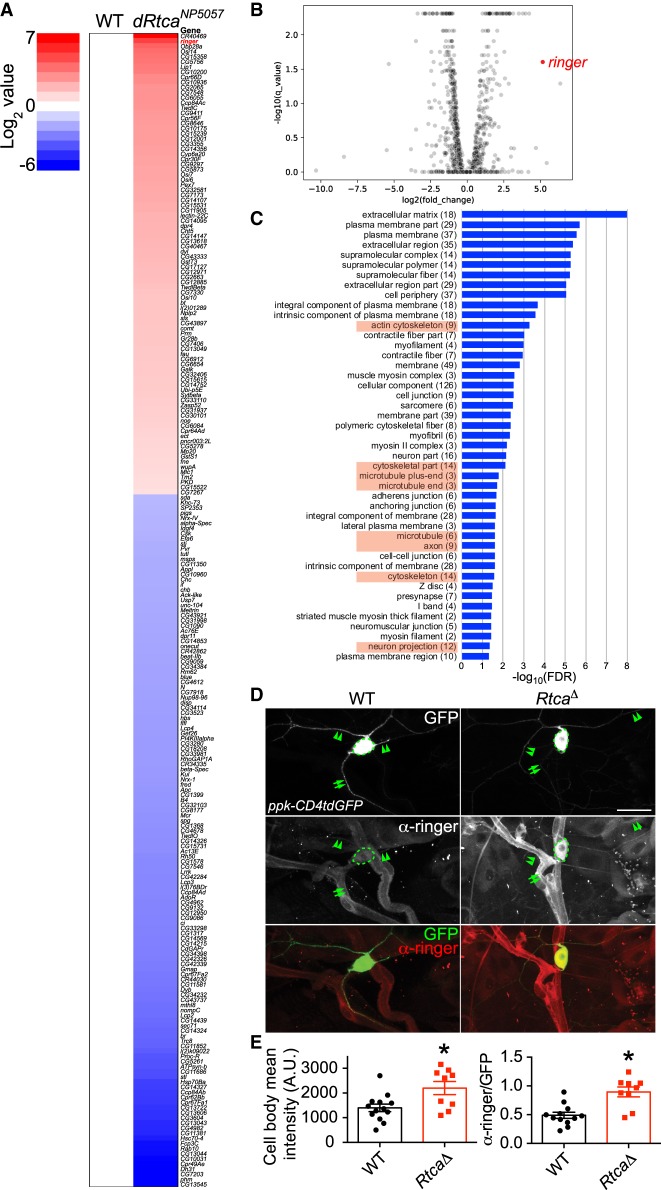
In order to identify the downstream effectors mediating Rtca's inhibitory function on axon regeneration, we performed RNA sequencing (RNA-seq) of wild-type (WT) and Rtca loss-of-function (LOF) mutants: RtcaNP5057 (Song et al. 2015). Specifically, we focused our analyses on the C4da neurons, which were labeled by expressing ppk-CD4tdGFP and enriched with fluorescence-activated cell sorting (FACS). Bioinformatic analyses were performed to determine the altered gene expression and signaling pathways. In total, 225 genes were found to be differentially expressed in Rtca mutant C4da neurons compared with WT (Fig. 1A,B). Interestingly, pathways regulating the cytoskeleton were enriched in both the up- and down-regulated gene sets (Fig. 1C). In particular, Rtca LOF is associated with the gene ontology (GO) terms in actin, microtubule, axon, and neuron projection (Fig. 1C). Subsequently, we decided to focus on the MAP ringer, because it was the most highly up-regulated protein coding gene. To confirm the results from the RNA-seq experiment, we performed immunostaining in third instar larvae using the ringer antibody (Mino et al. 2016). In WT, we found that ringer was expressed in multiple cell types (Fig. 1D). In particular, ringer was detected in the cell body (Fig. 1D, dashed circle), proximal dendrites (arrowheads) and axon (Fig. 1D, arrows) of C4da neurons (Fig. 1D). In accordance with our RNA-seq results, ringer expression was drastically enhanced in a Rtca deletion allele, RtcaΔ (Song et al. 2015), with robust signal throughout C4da neurons (Fig. 1D). The ringer antibody fluorescence intensity was quantified using confocal imaging of C4da neuron cell bodies, confirming an ∼60% increase (Fig. 1E). Given ringer's known function in maintaining microtubule integrity and the importance of microtubule stability in axon growth and regrowth, we went on to determine whether and how manipulation of ringer affects axon regeneration.
Figure 1.
Transcriptome profiling and increased ringer expression in Rtca mutants. (A) Heat map showing the significantly up-regulated (red) or down-regulated (blue) genes in a hypomorphic allele of Rtca: RtcaNP5057. Ringer, highlighted in red, is the second most up-regulated protein-coding gene. n = 3 replicates for each genotype. (B) Volcano plot showing differential gene expression in C4da neurons in RtcaNP505 versus WT. Positive log2(fold change) indicates an increase in mutants relative to WT. ringer is highlighted in red. (C) The GO term analyses (http://geneontology.org) of the differentially regulated gene sets. The pathways in the GO Cellular Component are ranked based on the false discovery rate (FDR). (D) Immunostaining for the ringer protein in larvae shows that it is expressed in various cell types including neurons, glial cells, and epithelial cells. Ringer is present in the soma (dashed circle), axon (arrows), and dendrites (arrowheads) of C4da neurons (labeled with ppk-CD4tdGFP). Ringer protein is up-regulated in a deletion allele of Rtca: RtcaΔ. (E) Ringer expression level in the soma is quantified by measuring the mean fluorescence intensity with and without normalization to GFP, and it is significantly increased in RtcaΔ. n = 9 to 13 neurons from three to five larvae. (*) P < 0.05 by two-tailed unpaired Mann-Whitney test. Scale bar, 20 μm.
Ringer cell-autonomously promotes axon regeneration in the PNS and CNS
To study axon regeneration, we used the previously established Drosophila da sensory neuron injury model (Song et al. 2012, 2015). Briefly, axons of the nociceptive C4da neurons (labeled with ppk-CD4tdGFP) were severed in the PNS of third instar larvae at ∼72 h after egg laying (h AEL) using a two-photon laser. Degeneration of the distal axon was confirmed at 24 h after injury (h AI) and regeneration was assessed at 48 h AI (Fig. 2A). WT C4da neurons robustly regrew their axon from the retracted axon stem and extended along the original trajectory, beyond the injury site (Fig. 2A, arrowheads, axons are marked by the green dashed line). The percentage of axons that regenerated in WT neurons was 73%. The regeneration index was measured to assess the length of regeneration normalized to the growth of the larvae (Materials and Methods), as described previously (Song et al. 2012). WT axons had a regeneration index of 0.35 ± 0.04. In a null mutant of ringer, ringer915 (Mino et al. 2016), a substantial number of injured axons stalled, retracted, or displayed limited regrowth (Fig. 2A, arrow). A significant reduction in the regeneration percentage, the regeneration index and regeneration length was observed (Fig. 2B,C; Supplemental Fig. S1A). RNAi knockdown in C4da neurons (ppk-Gal4 > ringer RNAiBL38287) recapitulated the impaired regeneration observed in the whole-body knockout (Fig. 2A–C, arrow), suggesting that ringer functions cell-autonomously. The RNAi experiment was performed in heterozygotes of Df(3L)BSC649/+, a deficiency allele that covers the ringer locus, to provide a sensitized background. Heterozygotes of Df(3L)BSC649/+ by itself did not display defects in regeneration and thus served as the control. These results indicate ringer as a neuronal arbiter of regeneration.
Figure 2.
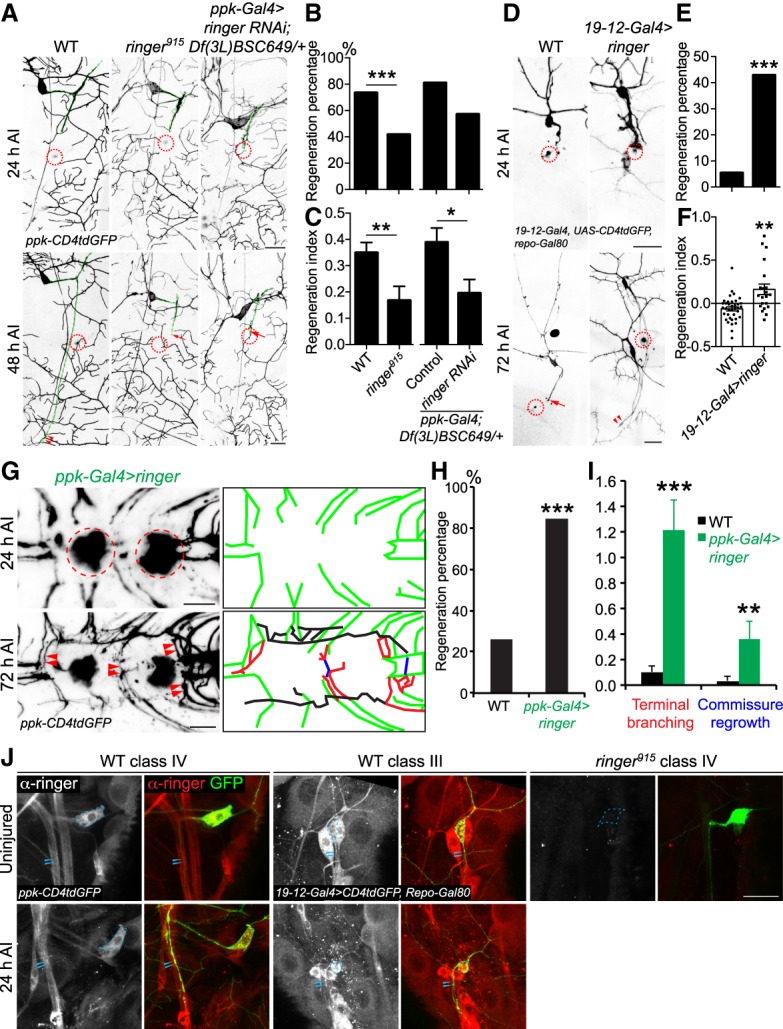
Loss-of-function and gain-of-function analyses for ringer in axon regeneration in the PNS and CNS. (A) C4da neuron axons robustly regenerate in WT. Ringer loss of function in a null allele, ringer915, significantly reduces the regenerative potential. C4da neuron-specific ringer RNAi in a sensitized background—deficiency Df(3L)BSC649/+ heterozygotes—also leads to reduced axon regeneration, indicating that ringer is cell-autonomously required for axon regeneration. The injury site is demarcated by the dashed circle. Arrow marks axon stalling while arrowheads show the regrowing axon tips. Axons are marked with dashed green lines. (B,C) Quantifications of C4da neuron axon regeneration with regeneration percentage (B) and regeneration index (C). N = 21 to 62 neurons from six to 16 larvae. (D) C3da neuron axons fail to regenerate in WT. C3da neuron-specific overexpression of ringer leads to increased axon regeneration. (E,F) Quantifications of C3da neuron axon regeneration. (G) C4da neuron specific overexpression of ringer increases axon regrowth in the VNC. The injury site is demarcated by the dashed circle. The regenerating axons are illustrated in schematic diagrams with terminal branching marked in red, commissure regrowth in blue, and other regrowing axons in black. (H) Compared with WT, which shows limited regrowth, the regeneration percentage is increased after ringer overexpression. n = 13 to 46 injured segments from seven to 29 larvae. (I) Ringer overexpression increases terminal branching and commissure regrowth of C4da neurons after injury in the VNC. N = 7 to 29 larvae. (J) Ringer expression pattern before and axon injury. Ringer protein is present in both C4da and C3da neurons and is detected in the axons before and after injury, as compared with the ringer915 negative control, which is devoid of ringer immunolabeling. Ringer is highly localized to the cell bodies and diffuse throughout the uninjured axon. After injury, ringer immunolabeling remains high in the soma and was concentrated within the proximal and medial portions of the injured axons. Strong immunolabeling of ringer protein is not observed in the growth cones or retraction bulbs of injured axons. The dashed circle marks the cell bodies and arrowheads mark the axons. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001 by Fisher's exact test (B, E, H) or two-tailed unpaired Student's t-test (C, F, I). Scale bar, 20 μm.
Conversely, we determined whether gain-of-function (GOF) of ringer would be sufficient to enhance the regenerative potential of C3da neurons, which are normally incapable of regeneration (Song et al. 2012). We labeled C3da neurons with 19-12-Gal4, UAS-CD4tdGFP, repo-Gal80 and used the following injury paradigm: Axotomy was induced at 48–72 h AEL, degeneration was confirmed at 24 h AI, and regeneration was assayed at 72 h AI (Song et al. 2012). Given the reduced regeneration potential of C3da neurons, axons were given more time to regenerate in comparison with C4da neurons. Compared with WT C3da neurons, which exhibited axon regeneration at only about 5% of the time (Fig. 2D–F, arrow), overexpression of ringer in C3da neurons significantly enhanced the axon regeneration percentage to 43% and increased the regeneration index and regeneration length (Fig. 2D–F, arrowheads; Supplemental Fig. S1B).
To address whether overexpression of ringer is beneficial for axon regeneration in the CNS, we examined C4da neuron axon regeneration within the ventral nerve cord (VNC) after CNS lesion, as previously described (Song et al. 2015). Compared with WT control, which showed limited regrowth, ringer overexpression improved the regeneration percentage, with a significant increase in “terminal branching” and “commissure regrowth” (Fig. 2G–I, arrowheads; Materials and Methods). These LOF and GOF analyses all together demonstrate that ringer is required for proper neuroregeneration cell-autonomously and that it can function as a potent booster for axon regeneration both in the peripheral and central nervous system.
We next examined the expression pattern of ringer in da neurons more closely. We found that ringer was present in both C4da and C3da neurons and was detected in the axons before and after axon injury, as compared with the ringer915 negative control, which was devoid of ringer immunolabeling (Fig. 2J, arrowheads). Ringer was highly localized to the cell bodies and diffuse throughout the uninjured axon, consistent with ringer's role in stabilizing and bundling microtubules throughout the neuronal cytoplasm (Mino et al. 2016). After injury, ringer immunolabeling remained high in the soma and was concentrated within the proximal and medial portions of the injured axons. Strong immunolabeling of ringer protein was not observed in the growth cones or retraction bulbs of injured axons. This result suggests that rather than determining the class-specific regenerative potential, ringer likely acts as an obligatory regeneration factor in all neurons.
Ringer functions downstream from Rtca through Xbp1-dependent transcription
Transcriptional regulation is a prime candidate for the regulation of ringer across cell types given our findings from the Rtca null mutants detailed above. We previously reported that a main function of Rtca is to antagonize the nonconventional splicing of the Xbp1 mRNA during ER stress (Song et al. 2015). The cellular-stress-induced splice variant Xbp1s then enters the nucleus and functions as a transcription factor orchestrating the ER stress response (Yoshida et al. 2001). In Rtca mutants, more Xbp1s is induced and inhibiting Xbp1 in Rtca mutants abolishes the enhanced axon regeneration phenotype seen in Rtca mutants, confirming that Xbp1 lies downstream from Rtca (Song et al. 2015). We hypothesized that the transcriptional increase of ringer in Rtca mutants is also dependent on Xbp1 and subsequently examined the expression pattern of ringer in double mutants of Rtca and Xbp1, compared with Rtca mutants alone. We found that the drastic increase of ringer expression in Rtca mutants, especially in the C4da neurons (soma, axon, and dendrites) was completely abolished in the Rtca and Xbp1 double mutants (Fig. 3A,B), confirming that the transcription factor activity of Xbp1 is necessary for the up-regulation of ringer in Rtca mutants.
Figure 3.
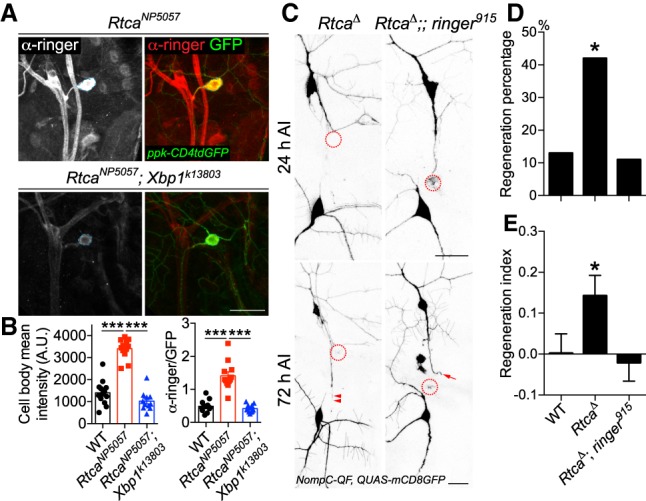
Ringer functions downstream from Rtca, in a Xbp1-dependent manner. (A) Xbp1 mediates the increase of ringer expression in Rtca mutants. The drastic increase of ringer expression in RtcaNP5057 mutants is abolished in Rtca and Xbp1 double mutants. Dashed circle marks the C4da neuron cell body. (B) The ringer immunolabeling is quantified by measuring the mean fluorescence intensity in the C4da neuron soma with and without normalization to GFP, showing that the increased ringer expression in Rtca mutants is eliminated after Xbp1 deficiency. n = 11 to 13 neurons from three to five larvae. (C) While Rtca LOF improves C3da neuron axon regeneration after peripheral injury, the enhancement is completely eliminated in Rtca and ringer double mutants. The injury site is demarcated by the dashed circle. Arrow marks axon stalling while arrowheads show the regrowing axon tips. (D,E) Quantifications of C3da neuron axon regeneration with regeneration percentage (D) and regeneration index (E). n = 19 to 24 neurons from four to six larvae. (*) P < 0.05; (***) P < 0.001 by Fisher's exact test (D), one-way ANOVA followed by Tukey's test (B), or Dunn's test (E). Scale bar, 20 μm.
To further elucidate the connection between Rtca and ringer, we sought to determine whether the increased expression of ringer mediates the regeneration enhancement in Rtca mutants by performing epistasis analysis. While loss of Rtca alone significantly promoted C3da neuron axon regeneration (Fig. 3C–E, arrowheads), double mutants of Rtca and ringer abolished the regeneration enhancement (Fig. 3C–E, arrow). This result, together with the ringer expression analyses, indicates that ringer is a major downstream effector of Rtca and Xbp1 in regulating axon regeneration.
Ringer maintains microtubule integrity during axon regeneration
Given the previous report that ringer directly affects tubulin polymerization and promotes microtubule bundling and stability (Mino et al. 2016), we assessed microtubule properties during axon regeneration in the presence or absence of ringer. We focused on two processes essential for microtubule integrity: α-tubulin lysine 40 acetylation and the interactions between MAPs. Acetylated α-tubulin (Ac-Tub) resides on the lumen of microtubules. While it does not confer stability directly, it is a part of the tubulin code (Yu et al. 2015) and regulates the interaction between MAPs and microtubules (Reed et al. 2006; Sudo and Baas 2010). Ac-Tub plays an important role in the growth, integrity, polarization, and overall health of the neuron (Baas and Black 1990; Dompierre et al. 2007; Witte et al. 2008; Perdiz et al. 2011; Matamoros and Baas 2016). We immunolabeled Ac-Tub and measured its expression both in the cell body (Fig. 4A, dotted line) and proximal axon (Fig. 4A, arrowheads) of injured C4da neurons. While there was a larger standard deviation for the immunofluorescence of Ac-Tub in ringer mutants, we did not find significant differences in Ac-Tub of the cell bodies, which is consistent with tubulin posttranslational modifications being tightly regulated (Fig. 4B). However, we measured a significant 40% reduction in the immunofluorescence of Ac-Tub in the proximal axon of injured neurons in ringer915 mutants, compared with WT axons (Fig. 4C). This result suggests that compromised microtubule stability, as reflected by the reduction of Ac-Tub level within the axon, contributes to the impaired axon regenerative potential.
Figure 4.
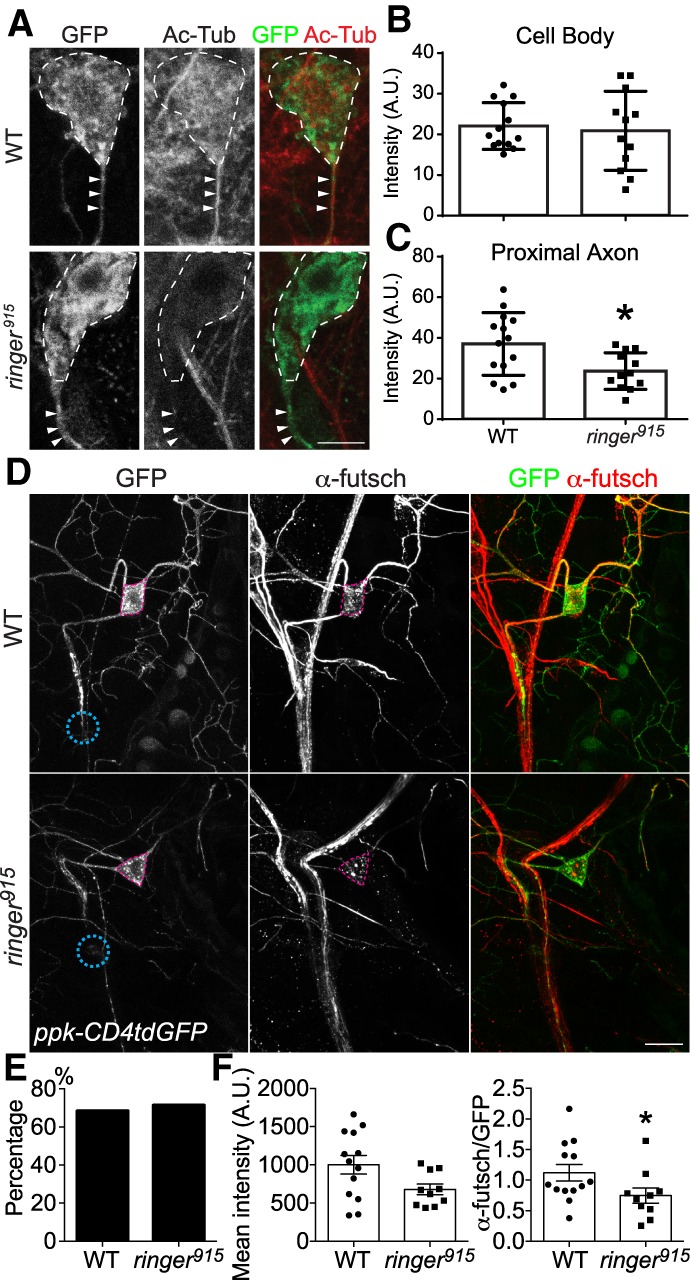
Ringer deficiency impairs microtubule integrity during axon regeneration. (A) Ac-Tub immunofluorescence, as viewed with confocal microscopy, demonstrates a reduction in microtubule acetylation in the cell body (dotted line) and axon (arrowhead) of injured ringer mutant axon. Scale bar, 5 µm. (B) There is no significant decrease in Ac-Tub immunofluorescence between WT (n = 13) and ringer mutants (n = 12), although variance is greater for ringer mutants. (C) Fluorescence intensity is significantly reduced from 37.02 ± 4.114 (n = 14) for WT to 23.73 ± 2.599 (n = 12) for ringer mutants within the proximal axon (5 µm). (D) In WT C4da neurons at 24 h AI, futsch is strongly expressed in both the cell body and the proximal and medial axon. Futsch immunolabeling is reduced in ringer915 mutant C4da neuron cell bodies and axons. The pink dashed circle marks the C4da neuron cell body and the teal dashed circle demarcated the injury site. Scale bar, 20 μm. (E) The percent of injured C4da neurons expressing futsch is comparable between WT and ringer mutants. (F) The fluorescence intensity of futsch staining in the soma of futsch-expressing C4da neurons (with and without normalization to GFP) is significantly reduced in ringer mutants. n = 10 to 13 neurons from five to seven larvae. (*) P < 0.05 by Fisher's exact test (E) or two-tailed unpaired Mann-Whitney test (B,C,F).
In a second line of investigation, we inquired whether MAPs might interact with ringer during axon regeneration. Recently, futsch, also known as MAP1B, was found to associate with ringer to form synapses at neuromuscular junctions (Shi et al. 2019). Futsch is essential to the health of a neuron and its dysregulation leads to neurodegeneration (Bettencourt da Cruz et al. 2005; Godena et al. 2011). Therefore, we sought to determine whether futsch and ringer were working together to promote regeneration. To test whether futsch expression is regulated by ringer, we examined futsch immunolabeling in WT C4da neurons at 24 h AI. Futsch was strongly expressed in both the cell body and the proximal and medial axon (Fig. 4D). In line with our hypothesis, futsch immunolabeling was starkly reduced in ringer915 mutant C4da neuron cell bodies and axons (Fig 4D). The percentage of injured C4da neurons expressing futsch did not change (Fig. 4E); however, the intensity of staining was significantly reduced by > 30% (Fig. 4F). This reduction is at least partially due to transcriptional repression, as futsch mRNA level is significantly down-regulated in ringer mutants (Supplemental Fig. S2A). In addition, posttranscriptional and posttranslational mechanisms are also likely to contribute. Therefore, our result demonstrates that futsch is highly expressed in the injured axons of the regeneration-capable C4da neurons, under positive regulation by ringer.
Ringer and futsch/MAP1B cooperate to promote axon regeneration
Futsch deficiency, as in a hypomorphic allele, FutschN94 (Hummel et al. 2000), resulted in a severe regeneration-deficient phenotype. The axons of futschN94 mutants failed to regenerate beyond the site of injury and axons retracted (Fig. 5A, arrow). FutschN94 mutants showed a lower regeneration percentage, compared with ringer915 mutants and the regeneration index was significantly reduced to nearly zero (Fig. 5B,C). Futsch mutation resulted in a more dramatic loss of regenerative potential in C4da neurons compared with ringer mutants, suggesting that futsch may receive additional input parallel to ringer. We next performed genetic interaction analysis to determine the functional relationship between ringer and futsch. Both ringer915/+ and futschN94/+ heterozygotes exhibited robust axon regeneration (Fig. 5A, arrowheads) and showed no significant reduction in regeneration percentage or regeneration index compared with WT axons (Fig. 5B,C). However, transheterozygotes of ringer915/+ and futschN94/+ were unable to regenerate after injury, with regeneration percentage and regeneration index resembling those in ringer915 mutants (Fig. 5A–C), confirming that ringer and futsch indeed operate in the same genetic pathway required for axon regeneration.
Figure 5.
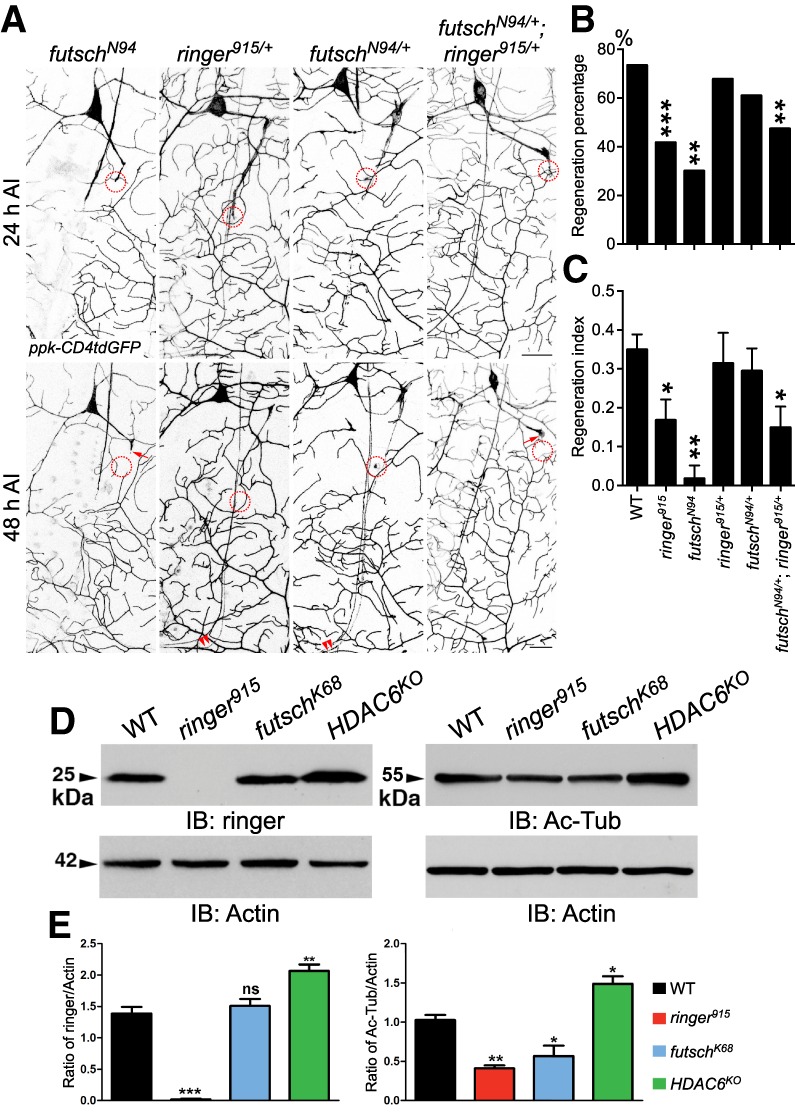
Ringer and futsch operate in the same genetic pathway to promote axon regeneration, and Western blotting of ringer and Ac-Tub in various mutants. (A) Genetic interaction analysis between ringer and futsch. While C4da neuron axons in heterozygotes of futschN94/+ or ringer915/+ behave similarly to WT, significant reduction of regeneration is observed in homozygotes of futschN94 and transheterozygotes of futschN94/+ and ringer915/+ (futschN94/+; ringer915/+). The injury site is demarcated by the dashed circle. Arrow marks axon stalling while arrowheads show the regrowing axon tips. (B,C) Quantifications of C4da neuron axon regeneration with regeneration percentage (B) and regeneration index (C). n = 20 to 64 neurons from five to 16 larvae. (D,E) Ringer and Ac-Tub levels in WT and mutants of ringer, futsch, and HDAC6. (D) Representative immunoblots showing total levels of ringer (left) and Ac-Tub (right) from larval lysates of specified genotypes. Actin was used as the loading control. (E) Quantification of the ratio of band intensities of ringer (left) and Ac-Tub (right) with respect to Actin in represented genotypes. n = 3 independent experiments. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001 by Fisher's exact test (B), one-way ANOVA followed by Holm-Sidak's test (C), or Dunnett's test (E). Scale bar, 20 μm.
Western blotting of the entire larval body wall lysate was then performed for assessing changes in protein expression and Ac-Tub abundance across various mutants (Fig. 5D,E). Ringer knockout was confirmed with its 25-kDa band completely absent in ringer915 mutants. Although futsch mutants, as in a hypomorphic allele, futschK68 (Hummel et al. 2000), did not exhibit any change in ringer protein expression, reduced ringer mRNA level was observed (Supplemental Fig. S2B), suggesting possible posttranscriptional compensation. Ac-Tub levels were reduced by approximately half in mutants of ringer and futsch, consistent with our observation of reduced Ac-Tub staining in ringer915 mutant C4da neurons after injury (Fig. 4A–C). Given the dysregulated Ac-Tub levels in ringer and futsch mutants, we investigated the effect of histone deacetylase 6 (HDAC6), the primary cytoplasmic α-tubulin deacetylase, on Ac-Tub and ringer protein levels. As expected, Ac-Tub level was significantly increased in HDAC6 knockout (HDAC6KO) (Fig. 5D,E). Importantly, HDAC6KO also led to a substantial increase of ringer protein expression (Fig. 5D,E), prompting us to determine the role HDAC6 plays during axon regeneration in vivo.
Futsch binds to microtubule in a ringer-dependent manner and regulates microtubule dynamics
In order to understand the mechanistic link between ringer/futsch and axon regeneration, we first focused on their interaction with the microtubule. It has been established that ringer, futsch, and tubulin exist in a molecular complex (Shi et al. 2019). We thus wanted to determine whether loss of ringer or futsch results in the breakdown of this molecular complex in vivo. We performed immunoprecipitations (IPs) using adult head lysates from WT, ringer915, and futschK68 mutants (Fig. 6A,B). Equal protein was used for each IP experiment from each genotype. Immunoblots from WT and ringer915 mutant lysates (Fig. 6A) showed loss of ringer (Fig. 6A, panel a) in ringer915 mutants and same levels of total tubulin (Fig. 6A, panel c). Immunoblots from WT and futschK68 mutant lysates (Fig. 6B) show loss of futsch (Fig. 6B, panel a) in futschK68 mutants and same levels of ringer (Fig. 6B, panel c). Actin was used as the loading control (Fig. 6A [panel b], B [panel b]). IPs using ringer and tubulin efficiently precipitated ringer and tubulin, respectively, confirming the specificity of the IPs (Shi et al. 2019). While loss of ringer did not completely abolish the binding of futsch to tubulin, tubulin levels in the IP complex were significantly reduced (∼45% reduction) in ringer915 mutants (Fig. 6A, panel d). On the other hand, loss of futsch did not change the complex formation between ringer and tubulin and the levels of ringer remained unchanged (Fig. 6B, panel d). These data suggest that while loss of any one of these proteins does not abolish the complex formation between the two remaining proteins, ringer primes/facilitates futsch–tubulin binding.
Figure 6.
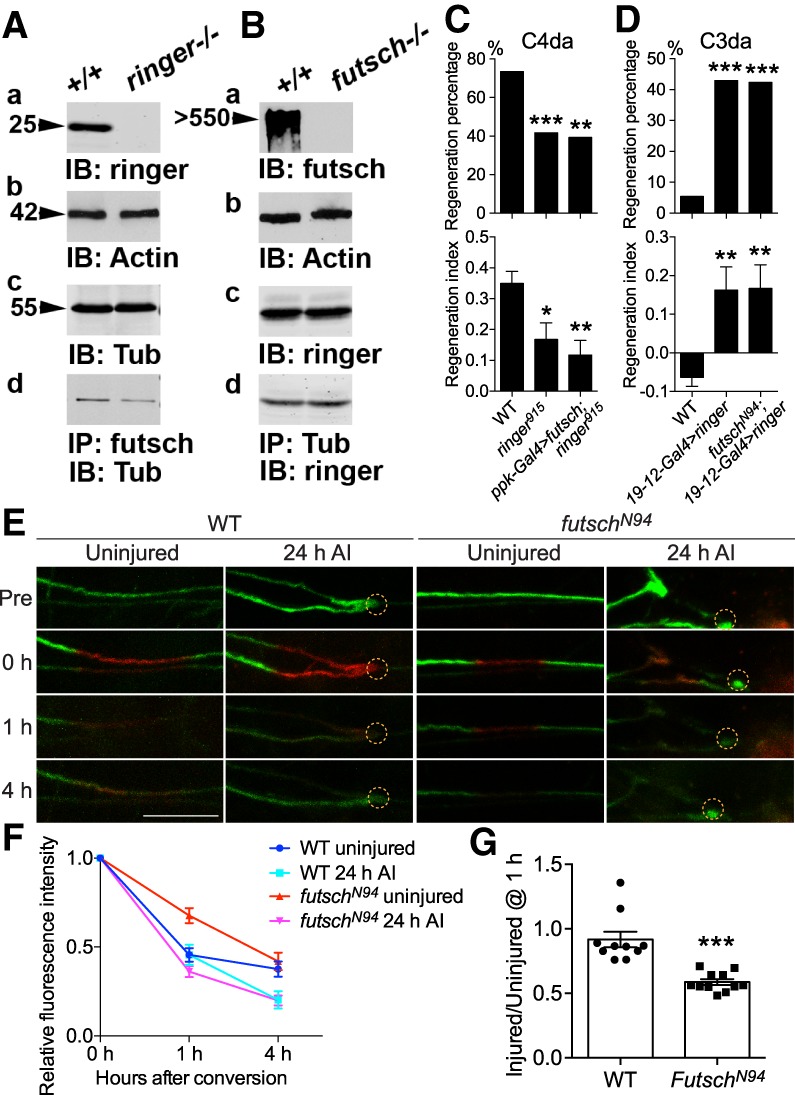
Futsch binds to microtubule in a ringer-dependent manner and regulates microtubule dynamics. (A,B) Immunoblots from equal amounts of adult fly head lysates from WT and ringer mutants (A, panel a), and WT and futsch mutants (B, panel a) were immunoblotted for anti-ringer (A, panel a) and anti-futsch (B, panel a), respectively. Anti-Actin was used for loading control (A [panel b], B [panel b]). The levels of tubulin (A, panel c) and ringer (B, panel c) were unchanged in WT and ringer mutant lysates (A, panel c), and WT and futsch mutant lysates (B, panel c), respectively. IPs from the WT and ringer mutants (A, panel d), and WT and futsch mutant fly heads (B, panel d) show that while loss of any one of these proteins does not abolish the complex formation between the remaining two proteins, ringer is required to facilitate futsch binding to tubulin (A, panel d). (C,D) Quantifications of C4da (C) and C3da (D) neuron axon regeneration with regeneration percentage and regeneration index. N = 21–64 neurons from five to 16 larvae. C4da neuron overexpression of futsch does not rescue the reduced axon regeneration in ringer915 mutants, while futsch mutation does not suppress the enhanced axon regeneration induced by C3da neuron overexpression of ringer. (E–G) The assay for microtubule turnover/dynamics using tdEOS-labeled α-tubulin. A 20-μm region in the proximal axon was photoconverted by using a 405-nm laser. After 0, 1, and 4 h, the converted axon was checked for the remaining red tdEOS. Examples of photoconversion are shown in E. The dashed circle marks the injury site. Photoconverted segments were imaged at different time points and the red fluorescence was measured both in the conversion region and outside it. The difference between the signal in the conversion region and outside regions was normalized to the same measurement at 0 h. The relative fluorescence intensity (FI) over time is plotted (F). In WT, the relative FI is significantly reduced (*) in the injured versus the uninjured conditions at 4 h. At 1 h, the FI in uninjured futschN94 mutants is significantly higher (***) than that of WT, whereas the FI in injured futschN94 mutants is lower than the injured WT. (G) The ratio of the injured versus uninjured FI at 1 h was calculated and shows a significant reduction in futschN94 mutants. N = 7 to 11 neurons from five to 10 larvae. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001 by Fisher's exact test (C,D, top), one-way ANOVA followed by Tukey's test (C,D, bottom), two-way ANOVA followed by Sidak's test (F), or two-tailed unpaired Student's t-test (G). Scale bar, 20 μm.
Second, to determine the genetic relationship between ringer and futsch, we performed epistasis analysis. We found that C4da neuron-specific overexpression of futsch failed to rescue the reduced axon regeneration defect in ringer915 mutants (Fig. 6C), whereas futsch LOF was unable to suppress the enhanced axon regeneration in C3da neurons caused by ringer overexpression (Fig. 6D). Combined with the IP experiments above, we speculate that ringer and futsch operate in the same complex with ringer lying more downstream.
Third, to directly assess microtubule dynamics, we performed imaging of the photoconvertible tdEos-α-tubulin, which has previously been used to analyze microtubule sliding (Barlan et al. 2013; Lu et al. 2013) and turnover (Tao et al. 2016). Unconverted tdEos-α-tubulin emits green fluorescence. After illuminating a 20-μm axon segment with 405-nm light, tdEos–α-tubulin emitted red fluorescence which gradually decayed (Fig. 6E). In WT, while the decay rate between uninjured and injured larvae was similar at 1 h, the rate was significantly higher (P < 0.05) at 4 h in the injured condition, suggesting faster turnover/dynamics during axon regrowth (Fig. 6E,F). In the futschN94 mutants, the turnover/dynamics at 1 h was significantly lower (P < 0.001) in the uninjured condition, whereas the injured larvae exhibited a trend toward faster decay (Fig. 6E,F), indicating dysregulated turnover/dynamics. We then calculated the ratio of injured versus uninjured conditions at 1 h, and found a drastic reduction in futschN94 mutants (Fig. 6G), suggesting aberrantly accelerated turnover/dynamics. These data are also consistent with the reduced Ac-Tub level in futsch mutants (Fig. 5D,E), correlating faster turnover/dynamics with reduced stability. We conclude that ringer and futsch corporately bind to microtubule, maintaining its stability and dynamics, which is essential for axon regeneration.
HDAC6 suppresses axon regeneration by inhibiting ringer
HDAC6 inhibition has been shown to be neuroprotective (Rivieccio et al. 2009; Li et al. 2011; Cho and Cavalli 2014; Hanson et al. 2018; Prior et al. 2018). More specifically, HDAC6 has been implicated in injury-mediated microtubule deacetylation, and through its inhibition dorsal root ganglia (DRG) neurons enhance their neurite outgrowth in vitro (Rivieccio et al. 2009; Li et al. 2011; Cho and Cavalli 2014; Hanson et al. 2018; Prior et al. 2018). We first assessed the regeneration-incompetent C3da neurons and found that HDAC6KO strongly enhanced their axon regeneration, increasing the regeneration percentage to 54% and the regeneration index to 0.34 (Fig. 7A–C). However, this beneficial effect was abolished in HDAC6KO and ringer915 double mutants (Fig. 7A–C), suggesting that ringer functions downstream from HDAC6. Next, we went on to determine whether HDAC6 also regulates the regeneration capabilities of C4da neurons. Overexpression of the WT HDAC6 (HDAC6-WT) significantly reduced the regeneration percentage to 50% (Fig. 7D,E). On the other hand, overexpression of a dominant-negative HDAC6 mutant with the disrupted second catalytic domain (Zhang et al. 2014) HDAC6-H664A significantly increased the regeneration index of C4da neurons (Fig. 7D,E). Furthermore, we pharmacologically inhibited HDAC6 and found that the HDAC6 specific inhibitor tubacin was efficacious in promoting C3da neurons axon regeneration when applied to the fly food (50 µM) (Xiong et al. 2013) compared with the DMSO vehicle control (Supplemental Fig. S3A–C). Therefore, HDAC6 functions as a negative regulator of axon regeneration in both C3da and C4da neurons.
Figure 7.
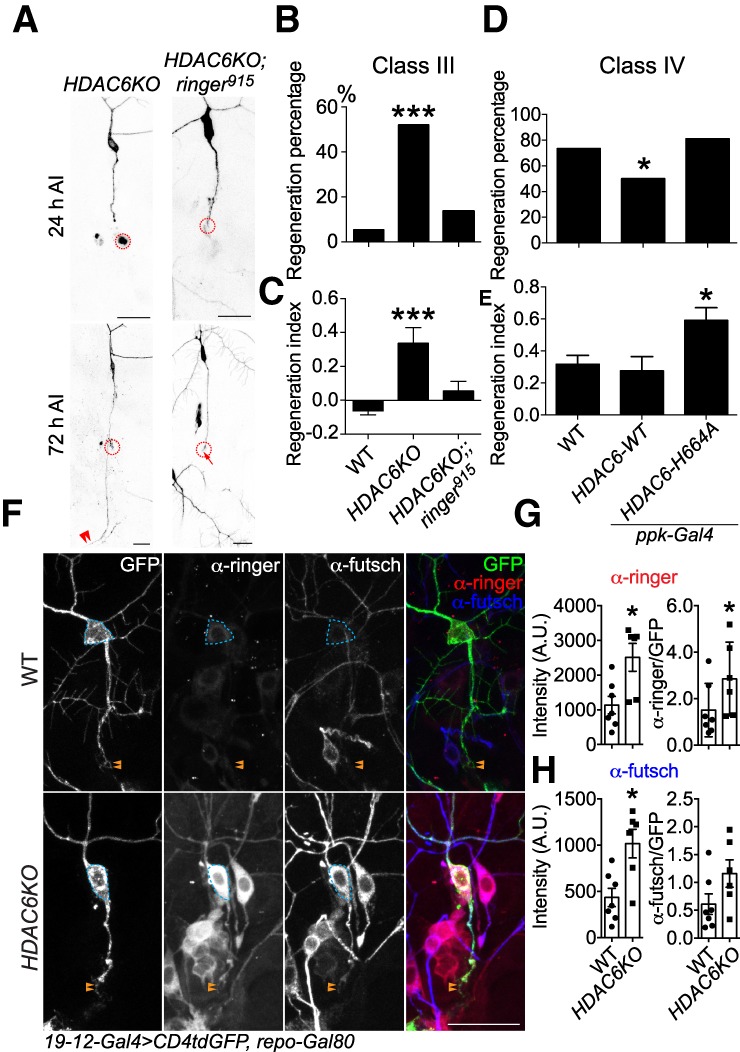
HDAC6 suppresses axon regeneration by inhibiting ringer and futsch. (A) Epistasis analysis of HDAC6 and ringer. Whereas HDAC6KO drastically promotes C3da neuron axon regeneration after peripheral injury, the enhancement is completely eliminated in HDAC6 and ringer double knockouts. The injury site is demarcated by the dashed circle. Arrow marks axon stalling while arrowheads show the regrowing axon tips. (B,C) Quantifications of C3da neuron axon regeneration with regeneration percentage (B) and regeneration index (C). n = 22 to 37 neurons from six to 10 larvae. (D,E) Quantifications of C4da neuron axon regeneration with regeneration percentage (D) and regeneration index (E). C4da neuron-specific overexpression of HDAC6-WT significantly reduces the regeneration percentage, without significantly affecting the regeneration index. On the other hand, overexpression of a dominant-negative HDAC6 mutant with the disrupted second catalytic domain HDAC6-H664A significantly increased the regeneration index. n = 24–64 neurons from six to 16 larvae. (F) Removal of HDAC6 drastically up-regulates ringer and futsch expression in both the soma and injured axon at 24 h AI, with a prominent increase within the growth cone. The dashed circle marks the cell body and arrowheads mark the growth cone. (G,H) Quantifications of the ringer (G) and futsch (H) immunofluorescence intensity in the soma (with and without normalization to GFP) shows significant increase in HDAC6KO, relative to WT. n = 6 to 7 neurons from three to four larvae. (*) P < 0.05; (***) P < 0.001 by Fisher's exact test (B,D), one-way ANOVA followed by Dunnett's test (C,E), or two-tailed unpaired Mann-Whitney test (G,H). Scale bar, 20 μm.
Given the enhancement of axon regeneration in C3da neurons, we went on to determine how ringer and futsch localized after injury in HDAC6KO. We found that removal of HDAC6 drastically up-regulated ringer and futsch protein expression in both the soma and injured axon, with a prominent increase within the growth cone (Fig. 7F, dashed circle and arrowheads). Relative to WT, quantifications of ringer and futsch immunofluorescence intensity in HDAC6KO showed a significant increase in the soma (Fig. 7G,H), corroborating our conclusion that HDAC6 negatively regulates ringer to suppress axon regeneration. Interestingly, this increased protein expression is likely due to posttranscriptional mechanisms, because the mRNA levels of ringer and futsch are either not changed or actually reduced in HDAC6KO (Supplemental Fig. S2A,B). Taken together, our results revealed a signaling pathway that starts with Rtca and Xbp1, involves HDAC6 as a negative regulator of regeneration and converges on both ringer and futsch as positive regulators (Supplemental Fig. S4).
Discussion
RBPs have been shown to be crucial in regulating complex cellular processes such as mRNA editing, transport and local translation (Kanai et al. 2004; Wells 2006; Bramham and Wells 2007; Sahoo et al. 2018a,b). Aberrant processing of RNA is present in neuronal diseases and injury (Kawahara et al. 2004; Kwak and Kawahara 2005; Peng et al. 2006; Aizawa et al. 2010; Anthony and Gallo 2010; Kosmaczewski et al. 2015). How these processes are affected after nervous system trauma and their regulation during neural repair are poorly understood. We previously identified Rtca, an RNA-binding protein regulating RNA repair and splicing, as a potential damage sensor that inhibits axon regeneration. Rtca LOF enhances axon regeneration in both fly and mammalian neurons. To better understand its underlying mechanism, we performed RNA-seq to assess the transcriptome of Rtca mutant neurons and found that ringer transcripts were highly expressed. Ringer is a MAP homologous to the mammalian tubulin polymerization-promoting proteins (TPPPs), in particular TPPP3 or TPPP1, which has been shown to be a regulator of axonal microtubule organization by promoting microtubule polymerization, assembly, and stability both in vitro and in vivo (Mino et al. 2016). We have revealed a connection between the injury-evoked RNA repair/splicing system and the MAP ringer, and propose that Rtca suppresses Xbp1 via nonconventional mRNA splicing, which in turn reduces ringer expression to inhibit axon regeneration. Furthermore, we provide evidence for an association between futsch and HDAC6, additional MAPs capable of regulating microtubule stability and posttranslational modifications. Ringer is also inhibited by HDAC6, and it cooperates with futsch to relay a cellular stress signal to the microtubule network. In addition, our data suggest that Rtca and Xbp1 likely have additional downstream effectors independent of ringer, and that futsch likely receives additional inputs, in parallel to ringer, to support axonal regeneration (Supplemental Fig. S4). Future studies to directly monitor microtubule dynamics in Rtca LOF mutants will help further validate this model and offer clues to the identity of additional players in this pathway.
The capacity of an axon to regenerate depends on both the external environment and cell-intrinsic mechanisms, which ultimately converge onto axonal microtubules. MAPs have become popular targets for augmenting nerve regeneration given the importance of microtubule stability and polymerization in both the nascent axon and the regenerating axon's growth cone. As an axon elongates, microtubules engorge the growth cone to fill it with microtubule mass. As the growth cone advances, microtubules bundle and consolidate within the nascent axon to provide structure and support. Ringer has been shown to be essential for proper microtubule bundling (Mino et al. 2016). Microtubules are inherently polarized because newly added tubulin dimers only assemble and disassemble at the “plus” end of the lattice, whereas the minus end of a microtubule is highly stabilized with special tubulin variants, abundant post translational modifications (e.g., acetylation of α-tubulin), and minus-end associating proteins (Baas et al. 2016). Therefore, a single microtubule can be thought of as having two general domains; a plus-end that is labile (i.e., where dynamic instability occurs) and a minus end that is stable and resists depolymerization (Baas and Black 1990). Microtubule stabilization prevents depolymerization and favors microtubule growth, which is beneficial for the axon's growth cone to advance (Conde and Cáceres 2009). Inducing microtubule stabilization using extremely low doses of the drugs paclitaxel or epithilones has resulted in significant augmentation of nerve regeneration in vivo (Hellal et al. 2011; Ruschel and Bradke 2018). Our results demonstrated a loss of microtubule acetylation in whole-cell lysate and specifically within the proximal axon of injured neurons in ringer mutants. This is in line with the function of ringer, which has been associated with microtubule polymerization and stability. Future experiments to dynamically track ringer proteins in accordance with microtubule polymerization during axon regeneration, and an extensive investigation of microtubule posttranslational modifications following axotomy are warranted.
Futsch, a MAP1B homolog, was recently shown to associate with ringer (Shi et al. 2019). Together, ringer and futsch were found to regulate synapse formation at neuromuscular junctions via a microtubule-based mechanism. It can be inferred that ringer and futsch may help promote the formation of a growth cone rather than a retracting dystrophic end within injured axons, similar to its maintenance of synaptic integrity. Ringer mutation led to a decrease in futsch mRNAs and immunolabeling, suggesting a role in regulating futsch transcription, localization, and protein levels. Both ringer and futsch mutations impaired axon regeneration, albeit futsch had a more dramatic effect, suggesting that futsch may contribute to additional signaling independent of ringer. While heterozygous mutants for futsch and ringer did not have a reduction in regeneration, transheterozygotes of futsch and ringer mutations exhibited a similar reduction in regeneration as ringer mutants alone. Coimmunoprecipitation experiments showed that ringer, futsch, and tubulin physically interact and form a molecular complex, and that ringer facilitates futsch binding to tubulin. Epistasis analysis further demonstrated that overexpression of futsch failed to rescue the reduced axon regeneration in ringer mutants, while overexpression of ringer is sufficient to promote axon regeneration despite the absence of futsch. Importantly, we found that microtubule turnover is faster in injured versus uninjured axons, and that futsch LOF dysregulates microtubule dynamics, accelerating its turnover after injury. Taken together, our data suggest that ringer and futsch cooperate in the same complex with tubulin, to maintain microtubule dynamics/stability, and that both are critical to the ability of sensory neurons to regenerate. Futsch is phosphorylated by GSK3 and sustained GSK3 activity promotes axon regeneration and increases the pool of dynamic microtubule mass (Gögel et al. 2006; Gobrecht et al. 2016; Leibinger et al. 2017), which further lead us to speculate that futsch might be regulated by additional signaling pathways.
Elucidating how microtubule stability properties are altered following an injury and the MAPs responsible for mediating those changes may identify novel therapeutic targets. We found that acetylation properties were altered by ringer mutations and, therefore, we sought to explore the role HDAC6, the primary tubulin deacetylase, may play in instructing regeneration. HDAC6 knockout and pharmacological inhibition increased regeneration in C3da neurons, a subtype of sensory neurons incapable of regeneration in WT flies. Previous studies have shown that HDAC6 inhibition and deletion leads to the hyperacetylation of microtubules (Haggarty et al. 2003; Zilberman et al. 2009). Early studies found that HDAC6 was neuroprotective after a CNS injury and associated these findings with HDAC6's role in transcriptional regulation (Rivieccio et al. 2009). However, more recent studies found that HDAC6 is neuroprotective in a manner that was associated with its deacetylation of microtubules (Li et al. 2011; Hanson et al. 2018; Prior et al. 2018). Other studies have shown that HDAC6 is essential for healthy axonal transport and influences MAP-microtubule interactions (Iwata et al. 2005; Dompierre et al. 2007; Ding et al. 2008; Chen et al. 2010; Simoes-Pires et al. 2013). In our study, we show that HDAC6 LOF leads to increased protein levels of ringer and futsch, likely through posttranscriptional mechanisms. It may also be possible that HDAC6 knockout affects microtubule-binding kinetics and the protein localization of ringer and futsch (i.e., concentrated versus diffuse). Augmented regeneration following HDAC6 knockout was lost with a ringer mutation. These results, along with the changes observed in Ac-Tub levels, suggest an interaction between HDAC6 and ringer, where ringer may function to either directly or indirectly restrict HDAC6 deacetylase activity with respect to α tubulin acetylation. This is likely, given that ringer has been shown to regulate microtubule bundling and stability, which are associated with highly acetylated domains of microtubules. Ringer may be essential to protecting highly acetylated and stable microtubule domains from HDAC6 deacetylation by occluding its interaction with α tubulin or directly blocking deacetylase activity. This would be consistent with in vitro studies suggesting that mammalian TPPP modulates microtubule acetylation by binding to HDAC6 and inhibiting its activity (Tokési et al. 2010). Alternatively, HDAC6 could inhibit TPPP nucleation by binding to TPPP and preventing its association to tubulin. Furthermore, HDAC6 can also physically modify kinases shown to negatively interrupt TPPP function such as ERK2 (Hlavanda et al. 2007). This network hypothesis could help explain an underlying positive feedback loop regulating microtubule stability: Increase of TPPP would inhibit HDAC6 leading to an enhancement of acetylated, potentially stable microtubule; in contrast, modification of kinases by HDAC6 could lead to kinase activation and downstream phosphorylation of TPPP, limiting its microtubule binding activity. We believe that HDAC6 and ringer are involved in a pathway that ultimately affects the stability and dynamics of microtubules. Future studies will explore whether ringer and HDAC6 expression, along with posttranslational modifications of tubulin, can predict the regenerative potential of da sensory neurons. C4da neurons show only ∼75% regeneration and we propose that the other 25% will show differences in the expression of MAPs and microtubule posttranslational modifications, specifically acetylation of α-tubulin.
The future treatments for nerve regeneration will most likely be combinatorial, with a need to address the extrinsic and intrinsic barriers to regeneration. We have identified a link between RNA repair/splicing and microtubule organization via a damage-evoked mechanism involving Rtca and ringer. We present further evidence that therapeutic targets capable of augmenting nerve regeneration ultimately converge on microtubules. Microtubules are a bottleneck to regeneration and identifying intrinsic signaling cascades that regulate microtubule dynamics using fly genetics will reveal pathways critical to microtubule-mediated nerve regeneration. Given the complexity of MAPs and the increasing number of candidate proteins, utilizing our fly injury model system allows us to screen for promising targets that warrant an investigation into their mammalian homologs with in vitro and in vivo mammalian nerve injury models (Kim et al. 2012; Matamoros et al. 2019). Excitingly, the zebrafish homolog of TPPP3 was recently shown to promote axon regeneration in Mauthner cells and is regulated at the transcript level by microRNA 133b (Huang et al. 2017). This corroborates our findings, allowing us to propose that ringer/TPPP is tightly regulated and may function as a relay station at multiple levels. Moreover, HDAC6 was also recently shown to be inhibitory in a regeneration screen performed in C. elegans (Kim et al. 2018). In summary, we identified a RNA repair/splicing pathway that up-regulates the MAP ringer, which interacts with other MAPs associated with microtubule stability/dynamics and tubulin posttranslational modifications, processes that are evolutionarily conserved and promising targets for regenerative therapies.
Materials and methods
Fly stocks
19-12-Gal4 (Xiang et al. 2010), repo-Gal80 (Awasaki et al. 2008), ppk-CD4-tdGFP (Han et al. 2011), ppk-Gal4 (Han et al. 2011), RtcaNP5057, RtcaΔ (Song et al. 2015), ringer915, UAS-ringer RNAiBL38287 (Mino et al. 2016), Xbp1k13803 (Song et al. 2015), NompC-QF (Petersen and Stowers 2011), QUAS-mCD8GFP (Potter et al. 2010), futschN94, futschK68 (Hummel et al. 2000), UAS-tdEos-α-tubulin (UAS-αTub84B.tdEOS) (Tao et al. 2016) and HDAC6KO (Du et al. 2010) have been described previously. To generate the UAS-ringer stock, the entire open reading frame of ringer (from DGRC) was cloned into the pACU2 vector, and the construct was then injected (Rainbow Transgenic Flies, Inc.). Randomly selected male and female larvae were used. Analyses were not performed blind to the conditions of the experiments.
RNA sequencing
To isolate and purify C4da neurons, 50–60 age-matched third instar larvae expressing the C4da neuron-specific ppk-CD4tdGFP were collected, washed, and dissected in cold PBS. The larval body walls were then dissociated using a combination of enzymatic (1 mg/mL collagenase) and mechanical perturbations (10× trituration at room temperature and then incubated for 15 min at 37°C with 10× trituration every 5 min) to yield single-cell suspensions that were then filtered using a 70-μm membrane. The filtrate was then sorted by flow cytometry, typically giving a yield of ∼15% with ∼300–500 cells. The RNA was extracted using the RNAqueous-micro kit (Thermo Fisher AM1931). cDNA was synthesized using the SMARTer Ultra Low RNA Kit (Takara), and then fragmented using the Nextera DNA library preparation kit (Illumina). The samples were sequenced using the single-read sequencing technology from Illumina. A total of six samples (three WT and three Rtca mutants) were sequenced and analyzed.
Sensory axon lesion in Drosophila
Da neuron axon lesion and imaging in the PNS or within the VNC were performed in live fly larvae as previously described (Song et al. 2012, 2015; Stone et al. 2014).
Quantitative analyses of sensory axon regeneration in flies
Quantification was performed as previously described (Song et al. 2012, 2015). Briefly, for axon regeneration in the PNS, we used “regeneration percentage,” which depicts the percent of regenerating axons among all the axons that were lesioned; “regeneration length,” which measures the increase of axon length; and “regeneration index,” which is calculated as an increase of “axon length”/“distance between the cell body and the axon converging point (DCAC).” An axon is defined as regenerating only when it obviously regenerated beyond the retracted axon stem, and this was independently assessed of the other parameters. “Regeneration index” is a more accurate measurement of axon regeneration than “regeneration length,” as it is normalized to larval growth. For axon regeneration in the CNS, we used “regeneration percentage,” which depicts the percent of lesioned commissure segments that showed obvious regrowth; “terminal branching,” which counts the number of axons that reached the commissure bundle and appeared to form elaborate branches; and “commissure regrowth,” which counts the number of axons that regenerated to connect commissure segments, longitudinally or laterally. The regeneration parameters from various genotypes were compared with that of the WT if not noted otherwise, and only those with significant difference were labeled with the asterisks.
Live imaging of Drosophila larvae
Live imaging was performed as described (Emoto et al. 2006; Parrish et al. 2007). Embryos were collected for 2–24 h on yeasted grape juice agar plates and were aged at 25°C or room temperature. At the appropriate time, a single larva was mounted in 90% glycerol under coverslips sealed with grease, imaged using a Zeiss LSM 880 microscope, and returned to grape juice agar plates between imaging sessions.
Immunohistochemistry and quantification
Third instar larvae or cultured neurons were fixed according to standard protocols. The following antibodies were used: guinea pig anti-ringer (1:1000) (Mino et al. 2016), mouse anti-futsch antiserum (1:100; Developmental Studies Hybridoma Bank 22C10), mouse monoclonal antiacetylated α-tubulin (1:100; Sigma T7451), and fluorescence-conjugated secondary antibodies (1:1000; Jackson ImmunoResearch). For microtubule staining, a 1× PHEM (1.8% PIPES, 0.5% HEPES, 0.4% EGTA, 0.04% MgCl2) microtubule stabilization buffer was used for dissections before and after 20 min of methanol fixation at −20°C. Larval body walls were permeabilized with 10% triton x-100 in 5% normal goat serum and 1× PBS overnight at 4°C to improve antibody penetrance for microtubule staining. Images were acquired using a confocal microscope and analyzed in ImageJ software. Z-stacks containing the regions of the cell to be quantified were used to make maximum intensity projections that were then outlined and measured for fluorescence intensity of the respective immunofluorescence in the cell body and axons (proximal and distal).
Western blotting, immunoprecipitations, and quantification
Third instar larval body walls without brain lobes, ventral nerve cord, or any attached imaginal discs were homogenized in ice-cold lysis buffer (Banerjee et al. 2017). The supernatants with equal amounts of proteins from each genotype were separated on SDS-PAGE for immunoblotting with respective antibodies. Each experiment was done independently three times and the most representative blots are shown. Primary antibodies used for immunoblotting were guinea pig anti-ringer (Mino et al. 2016, 1:15,000), anti-Ac-Tub (1:5000; Sigma T7451), and anti-β-Actin (1:15,000; Cell Signaling 4967S). Image J (NIH, USA) was used for quantification of band intensities of Western blots from three independent experiments. The intensity of the bands of interest was divided by their respective Actin loading control.
For immunoprecipitations studies, WT, ringer915 and futschK68 mutant fly heads were processed using identical experimental conditions according to previously described protocols (Banerjee et al. 2010). Each experiment was done independently three times and the most representative blots are shown.
Quantitative RT-PCR
Quantitative RT-PCR was performed for ringer, futsch, and rp49 (Ribosomal protein 49) in fly larvae. Rp49 was used as loading control. Ringer was amplified with primers F 5′-ACAAATTCCTGGACGATCTGG-3′ and R 5′-ttgctcgtatccgtcaatcg-3′; futsch was amplified with primers F 5′-TTAGCAGTTTCACCCGCC-3′ and R 5′-CCCGAATCACCCTTTGTACG-3′; rp49 was amplified with F 5′-CAGTCGGATCGATATGCTAAGCTG-3′ and R 5′-TAACCGATGTTGGGCATCAGATAC-3′.
Tubulin photoconversion assay
This was performed as described previously (Tao et al. 2016). Briefly, ppk-Gal4 was used to drive the expression of UAS-tdEos-α-tubulin in C4da neurons. A segment of ∼20-μm of the proximal axon was illuminated with a 405-nm laser to photoconvert the tdEos-α-tubulin from green to red. Converted neurons were imaged immediately after conversion (0 h) and re-imaged for red fluorescence at 1 and 4 h later using a 543-nm laser. Counter-images of the unconverted tdEos-α-tubulin were acquired with a 488-nm laser. Fluorescence intensity was measured within the 20-μm segment using ImageJ. Red fluorescence intensity was measured in converted segment and in the nonconverted region. Remaining fluorescence intensity (FI) was defined as (FIconverted− FIunconverted)timecourse/(FIconverted− FIunconverted)0h.
HDAC6 inhibitor treatment
For the HDAC6 inhibitor treatment, tubacin (MedChem Express HY-13428) was dissolved in DMSO at a concentration of 10 mM and then added at a final concentration of 50 μM (1:200 dilution) to grape juice agar plates. Control flies were treated with the same volume of DMSO (vehicle). The embryos/larvae were continuously cultured on medium containing the inhibitor or DMSO till imaging. Tubacin led to an overall developmental delay.
Statistical analysis
No statistical methods were used to predetermine sample sizes; however, sample sizes are similar to those reported in previous publications (Song et al. 2012, 2015), and the statistical analyses were done afterward without interim data analysis. Data distribution was assumed to be normal but this was not formally tested. All data were collected and processed randomly. Each experiment was successfully reproduced at least three times and performed on different days. Values of “n” (sample size) are provided in the figure legends. Data are expressed as mean ± SEM in bar graphs. No data points were excluded. Two-tailed unpaired Student's t-test was performed for comparison between two groups of samples. One-way ANOVA followed by multiple comparison test was performed for comparisons among three or more groups of samples. Fisher's exact test was used to compare the percentage. Statistical significance was assigned (P < 0.05 [*], P < 0.01 [**], and P < 0.001 [***]).
Supplementary Material
Acknowledgments
We thank J. Goldshteyn and S. Younger for technical assistance, Bloomington Stock Center and Vienna Drosophila Resource Center for fly stocks, Drosophila Genomics Resource Center for plasmids, and members of the Song laboratory for helpful discussions. Y.S. is a recipient of the National Institute of Neurological Disorders and Stroke (NINDS) Pathway to Independence Award. This work was supported by an Intellectual and Developmental Disabilities Research Center New Program Development Award (CHOP/Penn), an NINDS K99/R00 award (R00NS088211), and a National Institutes of Health grant (1R01NS107392) to Y.S., and a Predoctoral Training Program in Developmental Biology (T32 HD083185) to E.J.M.V.
Author contributions: E.J.M.V., A.J.M., J.Q., Y.N.J., and Y.S. conceived and designed the study. E.J.M.V., A.J.M., J.Q., Q.W., and Y.S. performed the experiments with C.H.J., D.G., T.W.H., and J.S.W. for RNA-seq analysis and with S.B. for the Western blot and IP experiments. E.J.M.V., A.J.M., J.Q., Q.W., and Y.S. analyzed the data. E.J.M.V., A.J.M., and Y.S. wrote the paper. All authors discussed the results and commented on the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.331330.119.
References
- Aizawa H, Sawada J, Hideyama T, Yamashita T, Katayama T, Hasebe N, Kimura T, Yahara O, Kwak S. 2010. TDP-43 pathology in sporadic ALS occurs in motor neurons lacking the RNA editing enzyme ADAR2. Acta Neuropathol 120: 75–84. 10.1007/s00401-010-0678-x [DOI] [PubMed] [Google Scholar]
- Anthony K, Gallo JM. 2010. Aberrant RNA processing events in neurological disorders. Brain Res 1338: 67–77. 10.1016/j.brainres.2010.03.008 [DOI] [PubMed] [Google Scholar]
- Awasaki T, Lai SL, Ito K, Lee T. 2008. Organization and postembryonic development of glial cells in the adult central brain of Drosophila. J Neurosci 28: 13742–13753. 10.1523/JNEUROSCI.4844-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas PW, Black MM. 1990. Individual microtubules in the axon consist of domains that differ in both composition and stability. J Cell Biol 111: 495–509. 10.1083/jcb.111.2.495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas PW, Slaughter T, Brown A, Black MM. 1991. Microtubule dynamics in axons and dendrites. J Neurosci Res 30: 134–153. 10.1002/jnr.490300115 [DOI] [PubMed] [Google Scholar]
- Baas PW, Rao AN, Matamoros AJ, Leo L. 2016. Stability properties of neuronal microtubules. Cytoskeleton (Hoboken) 73: 442–460. 10.1002/cm.21286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Blauth K, Peters K, Rogers SL, Fanning AS, Bhat MA. 2010. Drosophila neurexin IV interacts with Roundabout and is required for repulsive midline axon guidance. J Neurosci 30: 5653–5667. 10.1523/JNEUROSCI.6187-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Venkatesan A, Bhat MA. 2017. Neurexin, Neuroligin and Wishful Thinking coordinate synaptic cytoarchitecture and growth at neuromuscular junctions. Mol Cell Neurosci 78: 9–24. 10.1016/j.mcn.2016.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlan K, Lu W, Gelfand VI. 2013. The microtubule-binding protein ensconsin is an essential cofactor of kinesin-1. Curr Biol 23: 317–322. 10.1016/j.cub.2013.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt da Cruz A, Schwärzel M, Schulze S, Niyyati M, Heisenberg M, Kretzschmar D. 2005. Disruption of the MAP1B-related protein FUTSCH leads to changes in the neuronal cytoskeleton, axonal transport defects, and progressive neurodegeneration in Drosophila. Mol Biol Cell 16: 2433–2442. 10.1091/mbc.e04-11-1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanquie O, Bradke F. 2018. Cytoskeleton dynamics in axon regeneration. Curr Opin Neurobiol 51: 60–69. 10.1016/j.conb.2018.02.024 [DOI] [PubMed] [Google Scholar]
- Bramham CR, Wells DG. 2007. Dendritic mRNA: transport, translation and function. Nat Rev Neurosci 8: 776–789. 10.1038/nrn2150 [DOI] [PubMed] [Google Scholar]
- Buck KB, Zheng JQ. 2002. Growth cone turning induced by direct local modification of microtubule dynamics. J Neurosci 22: 9358–9367. 10.1523/JNEUROSCI.22-21-09358.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Owens GC, Makarenkova H, Edelman DB. 2010. HDAC6 regulates mitochondrial transport in hippocampal neurons. PLoS One 5: e10848 10.1371/journal.pone.0010848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Stone MC, Tao J, Rolls MM. 2012. Axon injury and stress trigger a microtubule-based neuroprotective pathway. Proc Natl Acad Sci 109: 11842–11847. 10.1073/pnas.1121180109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, Cavalli V. 2014. HDAC signaling in neuronal development and axon regeneration. Curr Opin Neurobiol 27: 118–126. 10.1016/j.conb.2014.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde C, Cáceres A. 2009. Microtubule assembly, organization and dynamics in axons and dendrites. Nat Rev Neurosci 10: 319–332. 10.1038/nrn2631 [DOI] [PubMed] [Google Scholar]
- Côté MP, Hanna A, Lemay MA, Ollivier-Lanvin K, Santi L, Miller K, Monaghan R, Houlé JD. 2010. Peripheral nerve grafts after cervical spinal cord injury in adult cats. Exp Neurol 225: 173–182. 10.1016/j.expneurol.2010.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, Dolan PJ, Johnson GV. 2008. Histone deacetylase 6 interacts with the microtubule-associated protein tau. J Neurochem 106: 2119–2130. 10.1111/j.1471-4159.2008.05564.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dompierre JP, Godin JD, Charrin BC, Cordelieres FP, King SJ, Humbert S, Saudou F. 2007. Histone deacetylase 6 inhibition compensates for the transport deficit in Huntington's disease by increasing tubulin acetylation. J Neurosci 27: 3571–3583. 10.1523/JNEUROSCI.0037-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du G, Liu X, Chen X, Song M, Yan Y, Jiao R, Wang CC. 2010. Drosophila histone deacetylase 6 protects dopaminergic neurons against α-synuclein toxicity by promoting inclusion formation. Mol Biol Cell 21: 2128–2137. 10.1091/mbc.e10-03-0200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsaeidi F, Bemben MA, Zhao XF, Goldman D. 2014. Jak/Stat signaling stimulates zebrafish optic nerve regeneration and overcomes the inhibitory actions of Socs3 and Sfpq. J Neurosci 34: 2632–2644. 10.1523/JNEUROSCI.3898-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto K, Parrish JZ, Jan LY, Jan YN. 2006. The tumour suppressor Hippo acts with the NDR kinases in dendritic tiling and maintenance. Nature 443: 210–213. 10.1038/nature05090 [DOI] [PubMed] [Google Scholar]
- Gobrecht P, Andreadaki A, Diekmann H, Heskamp A, Leibinger M, Fischer D. 2016. Promotion of functional nerve regeneration by inhibition of microtubule detyrosination. J Neurosci 36: 3890–3902. 10.1523/JNEUROSCI.4486-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godena VK, Romano G, Romano M, Appocher C, Klima R, Buratti E, Baralle FE, Feiguin F. 2011. TDP-43 regulates Drosophila neuromuscular junctions growth by modulating Futsch/MAP1B levels and synaptic microtubules organization. PLoS One 6: e17808 10.1371/journal.pone.0017808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gögel S, Wakefield S, Tear G, Klämbt C, Gordon-Weeks PR. 2006. The Drosophila microtubule associated protein Futsch is phosphorylated by Shaggy/Zeste-white 3 at an homologous GSK3β phosphorylation site in MAP1B. Mol Cell Neurosci 33: 188–199. 10.1016/j.mcn.2006.07.004 [DOI] [PubMed] [Google Scholar]
- Gordon-Weeks PR, Fischer I. 2000. MAP1B expression and microtubule stability in growing and regenerating axons. Microsc Res Tech 48: 63–74. [DOI] [PubMed] [Google Scholar]
- Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL. 2003. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc Natl Acad Sci 100: 4389–4394. 10.1073/pnas.0430973100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C, Jan LY, Jan YN. 2011. Enhancer-driven membrane markers for analysis of nonautonomous mechanisms reveal neuron-glia interactions in Drosophila. Proc Natl Acad Sci 108: 9673–9678. 10.1073/pnas.1106386108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson K, Tian N, Vickers JC, King AE. 2018. The HDAC6 inhibitor trichostatin A acetylates microtubules and protects axons from excitotoxin-induced degeneration in a compartmented culture model. Front Neurosci 12: 872 10.3389/fnins.2018.00872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellal F, Hurtado A, Ruschel J, Flynn KC, Laskowski CJ, Umlauf M, Kapitein LC, Strikis D, Lemmon V, Bixby J, et al. 2011. Microtubule stabilization reduces scarring and causes axon regeneration after spinal cord injury. Science 331: 928–931. 10.1126/science.1201148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton BJ, Bradke F. 2017. Can injured adult CNS axons regenerate by recapitulating development? Development 144: 3417–3429. 10.1242/dev.148312 [DOI] [PubMed] [Google Scholar]
- Hlavanda E, Klement E, Kókai E, Kovács J, Vincze O, Tőkési N, Orosz F, Medzihradszky KF, Dombrádi V, Ovádi J. 2007. Phosphorylation blocks the activity of tubulin polymerization-promoting protein (TPPP): identification of sites targeted by different kinases. J Biol Chem 282: 29531–29539. 10.1074/jbc.M703466200 [DOI] [PubMed] [Google Scholar]
- Hörnberg H, Holt C. 2013. RNA-binding proteins and translational regulation in axons and growth cones. Front Neurosci 7: 81 10.3389/fnins.2013.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Chen M, Yang L, Wagle M, Guo S, Hu B. 2017. MicroRNA-133b negatively regulates zebrafish single Mauthner-Cell axon regeneration through targeting tppp3 in vivo. Front Mol Neurosci 10: 375 10.3389/fnmol.2017.00375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel T, Krukkert K, Roos J, Davis G, Klämbt C. 2000. Drosophila Futsch/22C10 is a MAP1B-like protein required for dendritic and axonal development. Neuron 26: 357–370. 10.1016/S0896-6273(00)81169-1 [DOI] [PubMed] [Google Scholar]
- Iwata A, Riley BE, Johnston JA, Kopito RR. 2005. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem 280: 40282–40292. 10.1074/jbc.M508786200 [DOI] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. 2004. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43: 513–525. 10.1016/j.neuron.2004.07.022 [DOI] [PubMed] [Google Scholar]
- Kawahara Y, Ito K, Sun H, Aizawa H, Kanazawa I, Kwak S. 2004. Glutamate receptors: RNA editing and death of motor neurons. Nature 427: 801 10.1038/427801a [DOI] [PubMed] [Google Scholar]
- Kim HJ, Park JW, Byun JH, Vahidi B, Rhee SW, Jeon NL. 2012. Integrated microfluidics platforms for investigating injury and regeneration of CNS axons. Ann Biomed Eng 40: 1268–1276. 10.1007/s10439-012-0515-6 [DOI] [PubMed] [Google Scholar]
- Kim KW, Tang NH, Piggott CA, Andrusiak MG, Park S, Zhu M, Kurup N, Cherra SJ 3rd, Wu Z, Chisholm AD, et al. 2018. Expanded genetic screening in Caenorhabditis elegans identifies new regulators and an inhibitory role for NAD+ in axon regeneration. Elife 7: e39756 10.7554/eLife.39756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klim JR, Williams LA, Limone F, Guerra San Juan I, Davis-Dusenbery BN, Mordes DA, Burberry A, Steinbaugh MJ, Gamage KK, Kirchner R, et al. 2019. ALS-implicated protein TDP-43 sustains levels of STMN2, a mediator of motor neuron growth and repair. Nat Neurosci 22: 167–179. 10.1038/s41593-018-0300-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmaczewski SG, Han SM, Han B, Irving Meyer B, Baig HS, Athar W, Lin-Moore AT, Koelle MR, Hammarlund M. 2015. RNA ligation in neurons by RtcB inhibits axon regeneration. Proc Natl Acad Sci 112: 8451–8456. 10.1073/pnas.1502948112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovács GG, Gelpi E, Lehotzky A, Höftberger R, Erdei A, Budka H, Ovádi J. 2007. The brain-specific protein TPPP/p25 in pathological protein deposits of neurodegenerative diseases. Acta Neuropathol 113: 153–161. 10.1007/s00401-006-0167-4 [DOI] [PubMed] [Google Scholar]
- Kwak S, Kawahara Y. 2005. Deficient RNA editing of GluR2 and neuronal death in amyotropic lateral sclerosis. J Mol Med (Berl) 83: 110–120. 10.1007/s00109-004-0599-z [DOI] [PubMed] [Google Scholar]
- Leibinger M, Andreadaki A, Golla R, Levin E, Hilla AM, Diekmann H, Fischer D. 2017. Boosting CNS axon regeneration by harnessing antagonistic effects of GSK3 activity. Proc Natl Acad Sci 114: E5454–E5463. 10.1073/pnas.1621225114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Jiang H, Chang M, Xie H, Hu L. 2011. HDAC6 α-tubulin deacetylase: a potential therapeutic target in neurodegenerative diseases. J Neurol Sci 304: 1–8. 10.1016/j.jns.2011.02.017 [DOI] [PubMed] [Google Scholar]
- Li D, Li F, Guttipatti P, Song Y. 2018. A Drosophila in vivo injury model for studying neuroregeneration in the peripheral and central nervous system. J Vis Exp 10.3791/57557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Nazif K, Smith A, Baas PW, Smith GM. 2015. Histone acetylation inhibitors promote axon growth in adult dorsal root ganglia neurons. J Neurosci Res 93: 1215–1228. 10.1002/jnr.23573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindersson E, Lundvig D, Petersen C, Madsen P, Nyengaard JR, Højrup P, Moos T, Otzen D, Gai WP, Blumbergs PC, et al. 2005. p25α Stimulates α-synuclein aggregation and is co-localized with aggregated α-synuclein in α-synucleinopathies. J Biol Chem 280: 5703–5715. 10.1074/jbc.M410409200 [DOI] [PubMed] [Google Scholar]
- Lu W, Fox P, Lakonishok M, Davidson MW, Gelfand VI. 2013. Initial neurite outgrowth in Drosophila neurons is driven by kinesin-powered microtubule sliding. Curr Biol 23: 1018–1023. 10.1016/j.cub.2013.04.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey JM, Hubscher CH, Wagoner MR, Decker JA, Amps J, Silver J, Onifer SM. 2006. Chondroitinase ABC digestion of the perineuronal net promotes functional collateral sprouting in the cuneate nucleus after cervical spinal cord injury. J Neurosci 26: 4406–4414. 10.1523/JNEUROSCI.5467-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matamoros AJ, Baas PW. 2016. Microtubules in health and degenerative disease of the nervous system. Brain Res Bull 126: 217–225. 10.1016/j.brainresbull.2016.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matamoros AJ, Tom VJ, Wu D, Rao Y, Sharp DJ, Baas PW. 2019. Knockdown of fidgetin improves regeneration of injured axons by a microtubule-based mechanism. J Neurosci 39: 2011–2024. 10.1523/JNEUROSCI.1888-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mino RE, Rogers SL, Risinger AL, Rohena C, Banerjee S, Bhat MA. 2016. Drosophila Ringmaker regulates microtubule stabilization and axonal extension during embryonic development. J Cell Sci 129: 3282–3294. 10.1242/jcs.187294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Okano H. 2013. Cell transplantation therapies for spinal cord injury focusing on induced pluripotent stem cells. Cell Res 23: 70–80. 10.1038/cr.2012.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KK, Liu K, Hu Y, Smith PD, Wang C, Cai B, Xu B, Connolly L, Kramvis I, Sahin M, et al. 2008. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science 322: 963–966. 10.1126/science.1161566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish JZ, Emoto K, Kim MD, Jan YN. 2007. Mechanisms that regulate establishment, maintenance, and remodeling of dendritic fields. Annu Rev Neurosci 30: 399–423. 10.1146/annurev.neuro.29.051605.112907 [DOI] [PubMed] [Google Scholar]
- Peng PL, Zhong X, Tu W, Soundarapandian MM, Molner P, Zhu D, Lau L, Liu S, Liu F, Lu Y. 2006. ADAR2-dependent RNA editing of AMPA receptor subunit GluR2 determines vulnerability of neurons in forebrain ischemia. Neuron 49: 719–733. 10.1016/j.neuron.2006.01.025 [DOI] [PubMed] [Google Scholar]
- Perdiz D, Mackeh R, Poüs C, Baillet A. 2011. The ins and outs of tubulin acetylation: more than just a post-translational modification? Cell Signal 23: 763–771. 10.1016/j.cellsig.2010.10.014 [DOI] [PubMed] [Google Scholar]
- Petersen LK, Stowers RS. 2011. A gateway MultiSite recombination cloning toolkit. PLoS One 6: e24531 10.1371/journal.pone.0024531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CJ, Tasic B, Russler EV, Liang L, Luo L. 2010. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell 141: 536–548. 10.1016/j.cell.2010.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior R, Van Helleputte L, Klingl YE, Van Den Bosch L. 2018. HDAC6 as a potential therapeutic target for peripheral nerve disorders. Expert Opin Ther Targets 22: 993–1007. 10.1080/14728222.2018.1541235 [DOI] [PubMed] [Google Scholar]
- Reed NA, Cai D, Blasius TL, Jih GT, Meyhofer E, Gaertig J, Verhey KJ. 2006. Microtubule acetylation promotes kinesin-1 binding and transport. Current biology : CB 16: 2166–2172. 10.1016/j.cub.2006.09.014 [DOI] [PubMed] [Google Scholar]
- Rivieccio MA, Brochier C, Willis DE, Walker BA, D'Annibale MA, McLaughlin K, Siddiq A, Kozikowski AP, Jaffrey SR, Twiss JL, et al. 2009. HDAC6 is a target for protection and regeneration following injury in the nervous system. Proc Natl Acad Sci 106: 19599–19604. 10.1073/pnas.0907935106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruschel J, Bradke F. 2018. Systemic administration of epothilone D improves functional recovery of walking after rat spinal cord contusion injury. Exp Neurol 306: 243–249. 10.1016/j.expneurol.2017.12.001 [DOI] [PubMed] [Google Scholar]
- Sahoo PK, Lee SJ, Jaiswal PB, Alber S, Kar AN, Miller-Randolph S, Taylor EE, Smith T, Singh B, Ho TS, et al. 2018a. Axonal G3BP1 stress granule protein limits axonal mRNA translation and nerve regeneration. Nat Commun 9: 3358 10.1038/s41467-018-05647-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo PK, Smith DS, Perrone-Bizzozero N, Twiss JL. 2018b. Axonal mRNA transport and translation at a glance. J Cell Sci 131: jcs196808 10.1242/jcs.196808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengottuvel V, Leibinger M, Pfreimer M, Andreadaki A, Fischer D. 2011. Taxol facilitates axon regeneration in the mature CNS. J Neurosci 31: 2688–2699. 10.1523/JNEUROSCI.4885-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q, Lin YQ, Saliba A, Xie J, Neely GG, Banerjee S. 2019. Tubulin polymerization promoting protein, ringmaker, and MAP1B Homolog futsch coordinate microtubule organization and synaptic growth. Front Cell Neurosci 13: 192 10.3389/fncel.2019.00192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes-Pires C, Zwick V, Nurisso A, Schenker E, Carrupt PA, Cuendet M. 2013. HDAC6 as a target for neurodegenerative diseases: what makes it different from the other HDACs? Mol Neurodegener 8: 7 10.1186/1750-1326-8-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PD, Sun F, Park KK, Cai B, Wang C, Kuwako K, Martinez-Carrasco I, Connolly L, He Z. 2009. SOCS3 deletion promotes optic nerve regeneration in vivo. Neuron 64: 617–623. 10.1016/j.neuron.2009.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YJ, Lundvig DM, Huang Y, Gai WP, Blumbergs PC, Højrup P, Otzen D, Halliday GM, Jensen PH. 2007. p25α relocalizes in oligodendroglia from myelin to cytoplasmic inclusions in multiple system atrophy. Am J Pathol 171: 1291–1303. 10.2353/ajpath.2007.070201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Ori-McKenney KM, Zheng Y, Han C, Jan LY, Jan YN. 2012. Regeneration of Drosophila sensory neuron axons and dendrites is regulated by the Akt pathway involving Pten and microRNA bantam. Genes Dev 26: 1612–1625. 10.1101/gad.193243.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Sretavan D, Salegio EA, Berg J, Huang X, Cheng T, Xiong X, Meltzer S, Han C, Nguyen TT, et al. 2015. Regulation of axon regeneration by the RNA repair and splicing pathway. Nat Neurosci 18: 817–825. 10.1038/nn.4019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone MC, Rao K, Gheres KW, Kim S, Tao J, La Rochelle C, Folker CT, Sherwood NT, Rolls MM. 2012. Normal spastin gene dosage is specifically required for axon regeneration. Cell Rep 2: 1340–1350. 10.1016/j.celrep.2012.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone MC, Albertson RM, Chen L, Rolls MM. 2014. Dendrite injury triggers DLK-independent regeneration. Cell Rep 6: 247–253. 10.1016/j.celrep.2013.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo H, Baas PW. 2010. Acetylation of microtubules influences their sensitivity to severing by katanin in neurons and fibroblasts. J Neurosci 30: 7215–7226. 10.1523/JNEUROSCI.0048-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka E, Ho T, Kirschner MW. 1995. The role of microtubule dynamics in growth cone motility and axonal growth. J Cell Biol 128: 139–155. 10.1083/jcb.128.1.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J, Feng C, Rolls MM. 2016. The microtubule-severing protein fidgetin acts after dendrite injury to promote their degeneration. J Cell Sci 129: 3274–3281. 10.1242/jcs.188540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokési N, Lehotzky A, Horvath I, Szabo B, Olah J, Lau P, Ovadi J. 2010. TPPP/p25 promotes tubulin acetylation by inhibiting histone deacetylase 6. J Biol Chem 285: 17896–17906. 10.1074/jbc.M109.096578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells DG. 2006. RNA-binding proteins: a lesson in repression. J Neurosci 26: 7135–7138. 10.1523/JNEUROSCI.1795-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte H, Neukirchen D, Bradke F. 2008. Microtubule stabilization specifies initial neuronal polarization. J Cell Biol 180: 619–632. 10.1083/jcb.200707042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Klaw MC, Connors T, Kholodilov N, Burke RE, Tom VJ. 2015. Expressing constitutively active Rheb in adult neurons after a complete spinal cord injury enhances axonal regeneration beyond a chondroitinase-treated glial scar. J Neurosci 35: 11068–11080. 10.1523/JNEUROSCI.0719-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Yuan Q, Vogt N, Looger LL, Jan LY, Jan YN. 2010. Light-avoidance-mediating photoreceptors tile the Drosophila larval body wall. Nature 468: 921–926. 10.1038/nature09576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Zhao K, Wu J, Xu Z, Jin S, Zhang YQ. 2013. HDAC6 mutations rescue human tau-induced microtubule defects in Drosophila. Proc Natl Acad Sci 110: 4604–4609. 10.1073/pnas.1207586110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107: 881–891. 10.1016/S0092-8674(01)00611-0 [DOI] [PubMed] [Google Scholar]
- Yu I, Garnham CP, Roll-Mecak A. 2015. Writing and reading the tubulin code. J Biol Chem 290: 17163–17172. 10.1074/jbc.R115.637447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Wu CT, Qi X, Meijering RA, Hoogstra-Berends F, Tadevosyan A, Cubukcuoglu Deniz G, Durdu S, Akar AR, Sibon OC, et al. 2014. Activation of histone deacetylase-6 induces contractile dysfunction through derailment of α-tubulin proteostasis in experimental and human atrial fibrillation. Circulation 129: 346–358. 10.1161/CIRCULATIONAHA.113.005300 [DOI] [PubMed] [Google Scholar]
- Zilberman Y, Ballestrem C, Carramusa L, Mazitschek R, Khochbin S, Bershadsky A. 2009. Regulation of microtubule dynamics by inhibition of the tubulin deacetylase HDAC6. J Cell Sci 122: 3531–3541. 10.1242/jcs.046813 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.