Alphaviruses are a genus of positive-stranded RNA viruses and include numerous important human pathogens, such as Chikungunya virus, Ross River virus, Western equine encephalitis virus, etc., which create the emerging and reemerging public health threat worldwide. RNA interference (RNAi) is one of the most important antiviral mechanisms in plants and insects. Accumulating evidence has provided strong support for the existence of antiviral RNAi in mammals. In response to antiviral RNAi, viruses have evolved to encode viral suppressors of RNAi (VSRs) to antagonize the RNAi pathway. It is unclear whether alphaviruses encode VSRs that can suppress antiviral RNAi during their infection in mammals. In this study, we first uncovered that capsid protein encoded by Semliki Forest virus (SFV), a prototypic alphavirus, had a potent VSR activity that can antagonize antiviral RNAi in the context of SFV infection in mammalian cells, and this mechanism is probably used by other alphaviruses.
KEYWORDS: alphavirus, antiviral RNAi, capsid, Semliki Forest virus, viral suppressors of RNAi
ABSTRACT
RNA interference (RNAi) is a conserved antiviral immune defense in eukaryotes, and numerous viruses have been found to encode viral suppressors of RNAi (VSRs) to counteract antiviral RNAi. Alphaviruses are a large group of positive-stranded RNA viruses that maintain their transmission and life cycles in both mosquitoes and mammals. However, there is little knowledge about how alphaviruses antagonize RNAi in both host organisms. In this study, we identified that Semliki Forest virus (SFV) capsid protein can efficiently suppress RNAi in both insect and mammalian cells by sequestrating double-stranded RNA and small interfering RNA. More importantly, when the VSR activity of SFV capsid was inactivated by reverse genetics, the resulting VSR-deficient SFV mutant showed severe replication defects in mammalian cells, which could be rescued by blocking the RNAi pathway. Besides, capsid protein of Sindbis virus also inhibited RNAi in cells. Together, our findings show that SFV uses capsid protein as VSR to antagonize RNAi in infected mammalian cells, and this mechanism is probably used by other alphaviruses, which shed new light on the knowledge of SFV and alphavirus.
IMPORTANCE Alphaviruses are a genus of positive-stranded RNA viruses and include numerous important human pathogens, such as Chikungunya virus, Ross River virus, Western equine encephalitis virus, etc., which create the emerging and reemerging public health threat worldwide. RNA interference (RNAi) is one of the most important antiviral mechanisms in plants and insects. Accumulating evidence has provided strong support for the existence of antiviral RNAi in mammals. In response to antiviral RNAi, viruses have evolved to encode viral suppressors of RNAi (VSRs) to antagonize the RNAi pathway. It is unclear whether alphaviruses encode VSRs that can suppress antiviral RNAi during their infection in mammals. In this study, we first uncovered that capsid protein encoded by Semliki Forest virus (SFV), a prototypic alphavirus, had a potent VSR activity that can antagonize antiviral RNAi in the context of SFV infection in mammalian cells, and this mechanism is probably used by other alphaviruses.
INTRODUCTION
RNA interference (RNAi) is a conserved posttranscriptional gene silencing mechanism that originally evolves as an intrinsic antiviral immune mechanism in a broad range of eukaryotic organisms (1 – 3). In the process of antiviral RNAi, the viral replicative intermediate double-stranded RNAs (vRI-dsRNAs) synthesized during virus replication are sensed and cleaved by host endoribonuclease Dicer into approximately 21- to 23-nucleotide (nt) virus-derived small interfering RNA (vsiRNA). These vsiRNAs are then loaded into the Argonaute (AGO) protein of the RNA-induced silencing complexes (RISCs) to mediate the cleavage of cognate viral genomic RNAs (2, 4, 5). Thus far, RNAi has been widely recognized as the major antiviral response in fungi, plants, and insects (1, 2). For mammals, vRI-dsRNAs and other viral nucleic acids usually induce innate antiviral immunity such as the type I interferon (IFN-I) system (1). Recently, accumulating evidence have provided strong support for the existence of antiviral RNAi in mammals (6 – 10).
In response to antiviral RNAi, viruses have evolved to encode VSRs to antagonize the RNAi pathway through different mechanisms (11). Some VSRs bind to and sequestrate long dsRNAs and/or siRNAs to shield them from Dicer cleavage or to prevent their loading into AGO, while others inhibit certain important components of the RNAi pathway such as Dicer or AGO (11). For instance, Flock house virus (FHV) B2 acts as a VSR by preventing dsRNA from being cleaved by Dicer-2, as well as sequestering the siRNAs produced by Dicer-2 (12 – 14), whereas Wuhan nodavirus B2 can directly bind and inhibit Drosophila Dicer-2 required for vsiRNA production (15, 16). Moreover, cricket paralysis virus 1A directly inhibits the endonuclease activity of AGO2 and simultaneously targets AGO2 for proteasomal degradation in Drosophila (17).
In mammals, a number of viral proteins, such as Ebola virus VP35 (18), HIV-1 Tat (19), hepatitis C virus core (20), dengue virus NS4B (21), Yellow Fever virus (YFV) capsid (22), and coronavirus 7a and nucleocapsid (23, 24), have been shown to suppress ectopic dsRNA/shRNA-induced RNAi in vitro. On the other hand, a few viral proteins encoded by mammalian viruses, including Enterovirus A71 (EV-A71) 3A, influenza A virus NS1, and Nodamura virus (NoV) B2, have been found to act as bona fide VSRs that antagonize antiviral RNAi in the course of viral infections (6, 8, 10).
Alphaviruses are a large group of positive-stranded RNA viruses that belong to the genus Alphavirus in the family Togaviridae (25) and include numerous medically important human pathogens such as Sindbis virus (SINV), Chikungunya virus (CHIKV), Ross River virus, Eastern equine encephalitis virus, Western equine encephalitis virus, Venezuelan equine encephalitis virus, etc. The infections by these viruses are responsible for a broad spectrum of diseases, ranging from mild, undifferentiated, febrile illness to debilitating polyarthralgia, encephalitis and even death in humans and horses (26 – 29). To date, there is no approved antiviral therapy specific for alphaviruses (30). Alphaviruses transmit between mosquito vectors and vertebrate hosts (31, 32) and create an emerging and reemerging public health threat worldwide (33). Although previous studies indicated the critical role of antiviral RNAi in regulating the replication of alphaviruses, such as CHIKV and SINV in mosquitoes (31), it is unclear whether alphavirus encodes a bona fide VSR that can suppress antiviral RNAi during viral infection in mammals.
SFV is a member of the Alphavirus genus. Although SFV infection only causes a mild febrile illness in human, it is highly pathogenic in rodents and serves a model virus to investigate the mechanisms of viral replication, virus-host interaction, and innate immunity (34 – 36). SFV contains a single positive-stranded RNA genome of ∼12 kb, which consists of two open reading frames (ORFs) that encode four nonstructural proteins (nsP1 to nsP4), three structural proteins (capsid, envelope glycoproteins E1 and E2), and two small cleavage products (E3 and 6K) (36). Both ORFs are translated as polyproteins, which undergo cis and trans cleavage to form the mature viral proteins. SFV capsid protein is multifunctional and plays a critical role in the encapsidation of genome and formation of viral nucleocapsid capsid (37 – 39). In this study, we first uncovered that SFV-encoded capsid protein had a potent in vitro VSR activity that suppressed artificially induced RNAi in both insect and mammalian cells. We further demonstrated that SFV capsid can act as bona fide VSR to antagonize RNAi in the context of SFV infection in mammalian cells.
RESULTS
SFV capsid protein is a potential VSR.
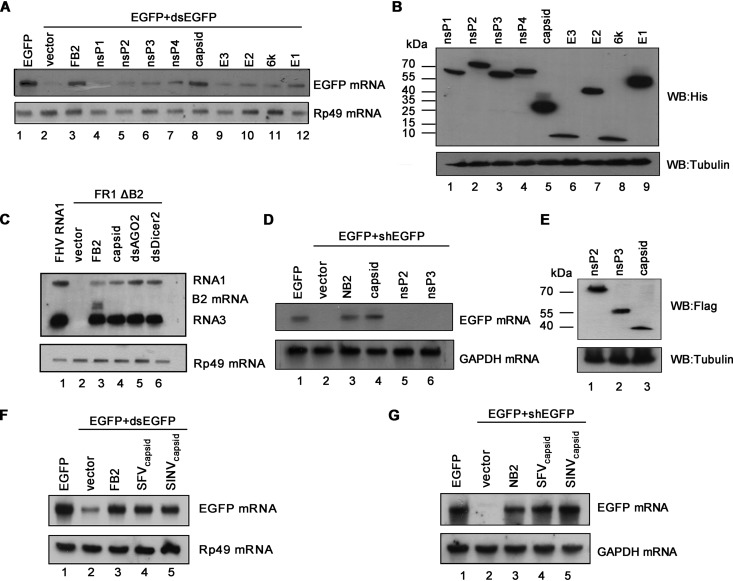
To evaluate whether SFV encodes any protein that works as a potential VSR, we examined all SFV-encoded proteins via a reversal-of-silencing assay in Drosophila S2 cells, which was previously used by us to screen VSRs of other viruses (15). In brief, cultured S2 cells were cotransfected with the plasmid encoding enhanced green fluorescent protein (EGFP) and EGFP-specific dsRNA, which is cleaved by fly Dicer-2 to produce siRNA and induce RNAi, together with the plasmid encoding one of the SFV proteins (Fig. 1A). The expression of the viral proteins was confirmed by Western blotting with anti-His antibody (Fig. 1B). At 48 h posttransfection (hpt), the mRNA levels of EGFP were detected by Northern blotting with a digoxigenin (DIG)-labeled RNA probe targeting 520 to 700 nt of the EGFP ORF. The EGFP-specific dsRNA can induce RNAi to destruct EGFP transcript (Fig. 1A, lane 2). FHV B2 (FB2), a well-characterized VSR, was used as a positive control, which expectedly restored EGFP mRNA levels (Fig. 1A, lane 3). Our data show that the ectopic expression of SFV capsid protein effectively restored the accumulation of EGFP mRNA (Fig. 1A, lane 8), indicating that SFV capsid protein is a potential VSR.
FIG 1.
SFV capsid protein is a potential VSR. (A) S2 cells were cotransfected with a plasmid encoding EGFP (0.1 μg) and dsEGFP (0.3 μg), together with either empty plasmid or a plasmid encoding SFV protein or FHV B2 (FB2) (1 μg for each). At 48 hpt, total RNAs were extracted, and the level of EGFP mRNA was examined by Northern blotting with a DIG-labeled RNA probe targeting nt 500 to 720 of the EGFP ORF region. Rp49 mRNA was used as loading control. (B) The expression of SFV proteins was detected by Western blotting. (C) S2 cells were transfected with pMT FHV RNA1 (FHV RNA1) (0.01 μg) or pMT FHV ΔB2 RNA1 (FR1 ΔB2) (0.3 μg), together with a plasmid encoding SFV capsid or FB2, as indicated above. At 48 hpt, FHV RNA transcription was induced by incubation with CuSO4 (0.5 mM). At 24 h after induction, the total RNAs were harvested for Northern blot analysis. The band between RNA1 and RNA3 was the B2 mRNA transcribed from the expression plasmid. The dsRNAs targeting AGO2 (dsAGO2) and Dicer2 (dsDicer2) were used as positive controls. (D) 293T cells were cotransfected with a plasmid encoding EGFP (0.1 μg) and EGFP-specific shRNA (shEGFP) (0.3 μg), together with either empty plasmid or a plasmid encoding SFV capsid protein, nsP2, nsP3, or NB2 (1 μg for each). At 48 hpt, the total RNAs were extracted, and the level of EGFP mRNA was examined by Northern blotting. (E) The expression of SFV nsP2, nsP3, and capsid proteins in 293T cells was detected by Western blotting. (F) S2 cells were cotransfected with a plasmid encoding EGFP (0.1 μg) and EGFP-dsRNA (dsEGFP) (0.3 μg), together with either empty plasmid or a plasmid encoding SINV capsid protein, SFV capsid protein, or FB2 (1 μg for each). At 48 hpt, the total RNAs were extracted, and the level of EGFP mRNA was examined by Northern blotting. Rp49 mRNA was used as the loading control. (G) 293T cells were cotransfected with a plasmid encoding EGFP (0.1 μg) and EGFP-specific shRNA (shEGFP) (0.3 μg), together with either empty plasmid or a plasmid encoding SINV capsid protein, SFV capsid protein, or NB2 (1 μg for each). At 48 hpt, the total RNAs were extracted, and the level of EGFP mRNA was examined by Northern blotting. GAPDH mRNA was used as the loading control.
To further confirm the VSR activity of SFV capsid, we examined whether capsid could rescue the replication of a B2-deficient FHV RNA1 replicon (FR1 ΔB2) in S2 cells (13). This mutant replicon lost the ability to suppress RNAi, leading to a defective self-replication of FHV RNA1 and subgenomic RNA3 in S2 cells (Fig. 1C, compare lanes 1 and 2). Our data show that the replication defect could be partially rescued by the ectopic expression of either FHV B2 or SFV capsid (Fig. 1C, lanes 3 and 4) or by the knockdown of fly AGO2 or Dicer-2 (Fig. 1C, lanes 5 and 6).
Because that RNAi pathway is conserved from insects to mammals, we sought to determine whether SFV capsid can suppress RNAi in cultured human 293T cells via the reversal-of-silencing assay, in which RNAi is induced by coexpressing EGFP expression vector and EGFP-specific short hairpin RNA (shRNA). Our data show that ectopic expression of SFV capsid effectively suppressed the shRNA-induced RNAi in 293T cells (Fig. 1D and E). Moreover, it was expectedly that ectopically expressing NoV B2 (NB2), another well-established VSR, suppressed RNAi (Fig. 1D, lane 3). Together, our data demonstrate that SFV capsid protein is a potential VSR that can suppress RNAi in both insect and mammalian cells.
Capsid proteins from alphaviruses share high homology in amino acid sequences, suggesting a conserved function (40). Thus, we examined the VSR activity of capsid protein from SINV via the reversal-of-silencing assays in both Drosophila S2 and human 293T cells, respectively. Our data show that SINV capsid inhibited RNAi in both cells (Fig. 1F and G), suggesting that the in vitro VSR activity is a common feature for alphaviral capsid proteins.
SFV capsid suppressed Dicer-mediated siRNA production by sequestrating dsRNA.
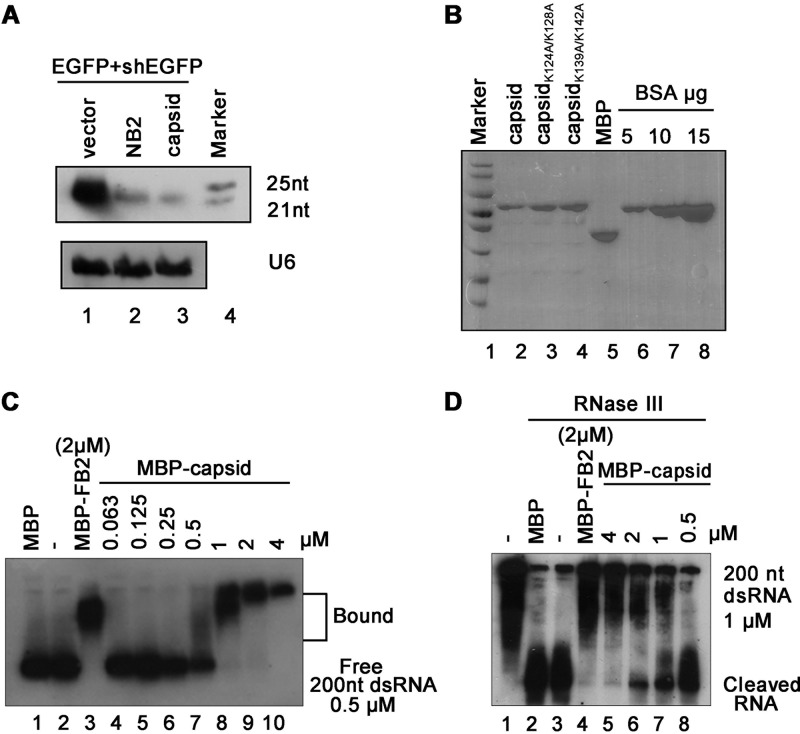
It is well established that dsRNA/shRNA-induced RNAi requires the Dicer-mediated cleavage of dsRNA or shRNA into siRNA (1). To explore whether SFV capsid can suppress this process, RNAs harvested from 293T cells coexpressing EGFP expression vector and EGFP-shRNA together in the presence or absence of SFV capsid were subjected to small RNA Northern blotting by using a DIG-labeled RNA oligonucleotide probe targeting EGFP siRNA. As shown in Fig. 2A, the accumulation of 22-nt Dicer-cleaved siRNA product was reduced in cells overexpressing SFV capsid compared to that in cells overexpressing empty vector, indicating that SFV capsid can inhibit Dicer-mediated siRNA production. These results are consistent with the observation that SFV capsid could effectively restore shRNA-mediated elimination of EGFP transcript in 293T cells (Fig. 1D).
FIG 2.
SFV capsid suppresses Dicer-mediated siRNA production by sequestrating dsRNA. (A) 293T cells were cotransfected with a plasmid encoding EGFP (0.1 μg) and EGFP-specific shRNA (shEGFP) (0.3 μg), together with either empty plasmid or a plasmid encoding SFV capsid protein or NB2 (1 μg for each). At 48 hpt, the total RNAs were extracted for small Northern blotting with a DIG-labeled RNA oligonucleotide probe targeting EGFP siRNA. U6 was used as the loading control. (B) SDS-PAGE of purified recombinant SFV capsid. BSA was used as a quantity control. (C) Increasing amounts (0 to 4 μM) of MBP fusion capsid (MBP-capsid) were incubated with 0.5 μM 200-nt DIG-labeled dsRNA at 25°C for 30 min. Complexes were separated on 1.5% native-TBE agarose gel, transferred to membranes, and then incubated with anti-DIG antibody conjugated to alkaline phosphatase. MBP-FB2 and MBP were used as controls. (D) Increasing amounts (0 to 4 μM) of MBP-capsid were incubated with 1 μM 200-nt DIG-labeled dsRNA at 25°C for 30 min. The protein-dsRNA complexes were then incubated with 1 U of RNase III at 37°C for 30 min. The reaction products were subjected to 7 M urea–15% PAGE analysis.
We sought to examine whether SFV capsid inhibits siRNA production by directly binding to long dsRNA. To this end, we purified the recombinant maltose-binding protein (MBP)-fusion capsid protein (MBP-capsid; Fig. 2B, lane 2) and conducted electrophoretic mobility shift assay (EMSA) by incubating the in vitro transcribed DIG-labeled 200-nt dsRNA together with the MBP-capsid. As shown in Fig. 2C, capsid protein can bind to dsRNA directly, and the shifting amount of labeled dsRNAs was increased with the increasing amounts of MBP-capsid used in the reaction. MBP fusion FHV B2 (MBP-FB2) was used as a positive control.
Subsequently, we sought to examine whether SFV capsid can protect dsRNA from Dicer cleavage by using an in vitro RNase III assay (41). RNase III was widely used as the Dicer substitute to examine the activities of VSRs, as previously described (16). Our data show that the presence of MBP-capsid efficiently protected dsRNA from RNase III digestion in a dose-dependent manner (Fig. 2D, lanes 5 to 8), while dsRNA was cleaved into siRNA in the presence of MBP alone (Fig. 2D, lane 2).
Altogether, our findings suggest that SFV capsid could suppress RNAi by sequestrating dsRNA from Dicer cleavage.
SFV capsid suppressed siRNA-induced RNAi.
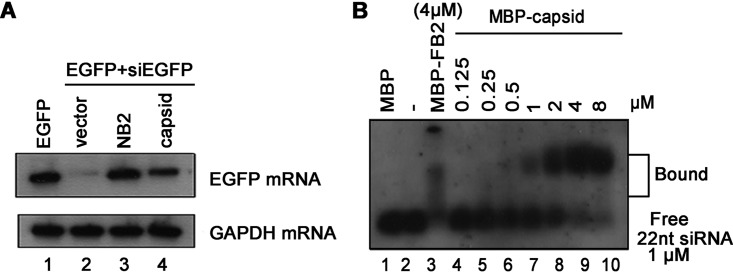
In the RNAi pathway, when being processed from dsRNA, siRNAs are incorporated into RISC to mediate the cleavage of cognate mRNAs (1). Since we have found that SFV capsid can inhibit RNAi by sequestrating dsRNA, it would be intriguing to examine whether SFV capsid could also suppress siRNA-induced RNAi, which occurs after siRNA biogenesis. To this end, 293T cells were cotransfected with a plasmid expressing EGFP and chemically synthesized EGFP-specific siRNA (siEGFP), together with the expression vector for SFV capsid. As shown in Fig. 3A, the chemically synthesized siRNA mediated the silencing of EGFP mRNA, whereas SFV capsid efficiently restored the level of EGFP mRNA, indicating that SFV capsid can suppress siRNA-induced RNAi in cells.
FIG 3.
SFV capsid suppresses siRNA-induced RNAi. (A) 293T cells were cotransfected with a plasmid encoding EGFP (0.1 μg) and EGFP-specific siRNA (siEGFP) (50 nM), together with either empty plasmid or a plasmid encoding SFV capsid protein or NB2 (1 μg for each). At 48 hpt, the total RNAs were extracted for Northern blotting. GAPDH mRNA was used as the loading control. (B) Increasing amounts (0 to 8 μM) of MBP fusion capsid were incubated with 1 μM DIG-labeled synthetic 22-nt siRNA at 25°C for 30 min. Complexes were separated on 1.5% native-TBE agarose gel, transferred to membranes, and then incubated with anti-DIG antibody conjugated with alkaline phosphatase.
Because SFV capsid can suppress siRNA-induced RNAi, we speculate that capsid protein may have siRNA-binding activity. To test this possibility, we conducted EMSA by incubating purified MBP-capsid together with DIG-labeled synthetic 22-nt siRNA. Our data show that SFV capsid protein can directly bind to siRNA in a dose-dependent manner (Fig. 3B).
Taken together, our findings indicate that SFV capsid can suppress RNAi by sequestrating both dsRNA and siRNA.
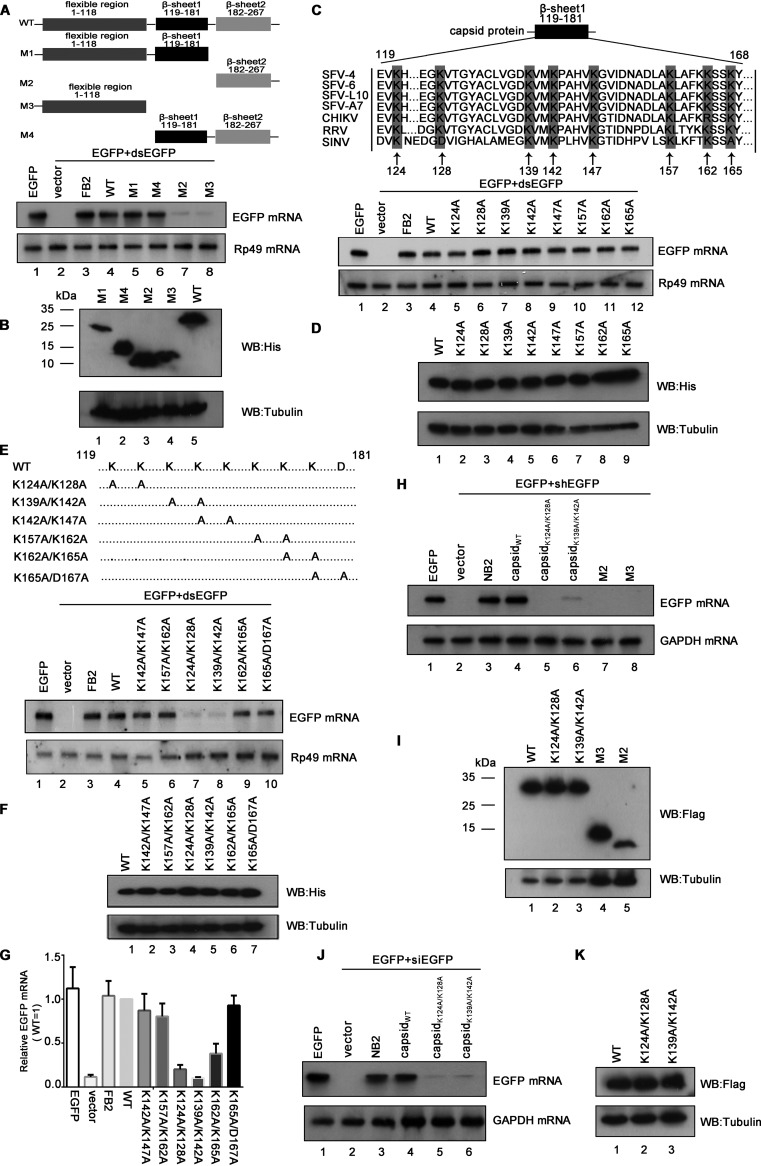
K124/K128 and K139/K142 of SFV capsid are critical for the VSR activity.
After determining the in vitro VSR activity of SFV capsid, we sought to identify the critical domain or amino acids (aa) required for its VSR activity. Previous studies showed that SFV capsid consists of an N-terminal segment (aa 1 to 118) and a C-terminal protease domain (aa 119 to 267) formed by two β-sheet domains (aa 119 to 181 and aa 182 to 267) (40). Accordingly, we constructed a set of capsid protein truncations, as illustrated in Fig. 4A, and examined their activities to suppress RNAi via the reversal-of-silencing assay in S2 cells. Our data show that the region of aa 119 to 181 is critical for the capsid’s VSR activity (Fig. 4A). The expression of capsid protein truncations was confirmed by Western blotting with anti-His antibody (Fig. 4B).
FIG 4.
K124/K128 and K139/K142 of SFV capsid are critical for the VSR activity. (A and B) S2 cells were cotransfected with a plasmid encoding EGFP (0.1 μg) and dsEGFP (0.3 μg), together with either empty plasmid or a plasmid encoding SFV capsid deletion mutant as indicated or FB2 (1 μg for each). At 48 hpt, the total RNAs were extracted for Northern blotting. Rp49 mRNA was used as the loading control. The expression of capsid mutations in S2 cells was detected by Western blotting (B). (C and D) The amino acid sequence alignments of alphaviral capsids are as follows: SFV4 (KP699763.1), SFV6 (KT009012.1), SFV-L10 (KP271965.1), SFV-A7 (Z48163.2), CHIKV (AOT86261.1), RRV (P08491.3), and SINV (AKZ17419.1). S2 cells were cotransfected with a plasmid encoding EGFP (0.1 μg) and dsEGFP (0.3 μg), together with either empty plasmid or the plasmid encoding the indicated single-point mutations of SFV capsid (1 μg for each). At 48 hpt, the total RNAs were extracted for Northern blotting. The expression of capsid mutations in S2 cells was detected by Western blotting (D). (E to G) Schematic illustration of double-point mutations of SFV capsid. S2 cells were cotransfected with a plasmid encoding EGFP (0.1 μg) and dsEGFP (0.3 μg), together with either empty plasmid or the plasmid encoding the indicated double-point mutations of SFV capsid (1 μg for each). At 48 hpt, the total RNAs were extracted for Northern blotting (E) and qRT-PCR (G); the expression of capsid mutations in S2 cells was detected by Western blotting (F). (H and I) 293T cells were cotransfected with a plasmid encoding EGFP (0.1 μg) and EGFP-specific shRNA (shEGFP) (0.3 μg), together with either empty plasmid or a plasmid encoding SFV capsid protein or mutations (NB2; 1 μg for each). (H) At 48 hpt, the total RNAs were extracted, and the level of EGFP mRNA was examined by Northern blotting. GAPDH mRNA was used as the loading control. (I) The expression of capsid mutations in 293T cells was detected by Western blotting. (J and K) 293T cells were cotransfected with a plasmid encoding EGFP (0.1 μg) and EGFP-specific siRNA (siEGFP) (50 nM), together with either empty plasmid or a plasmid encoding capsidWT, capsidK124A/K128A, capsidK139A/K142A, or NB2 (1 μg for each). At 48 hpt, the total RNAs were extracted, and the level of EGFP mRNA was examined by Northern blotting. (K) The expression of capsid mutations in 293T cells was detected by Western blotting.
To identify the critical residue(s) within the region from aa 119 to 181 required for the VSR activity, we performed multiple sequences alignments of capsids encoded by RRV, SFV, SINV, and CHIKV. The conserved positively charged residues, lysine (K) and aspartic acid (D), were subjected to single-point or double-point mutations to alanine (A), and the resulting mutant capsid proteins were then examined via the reversal-of-silencing assay in S2 cells. Although none of the single-point mutations disrupted the VSR activity (Fig. 4C and D), the K124A/K128A or K139A/K142A mutation (capsidK124A/K128A or capsidK139A/K142A) significantly suppressed the activity of SFV capsid to suppress RNAi (Fig. 4E, lanes 7 and 8; Fig. 4F and G) in S2 cells. In addition, the truncation mutations (M2 and M3) and capsidK124A/K128A or capsidK139A/K142A also lost the VSR activity in 293T cells (Fig. 4H and I). We also found that the K124A/K128A or K139A/K142A mutation significantly suppressed the activity of SFV capsid to suppress siRNA-induced RNAi in 293T cells (Fig. 4J and K).
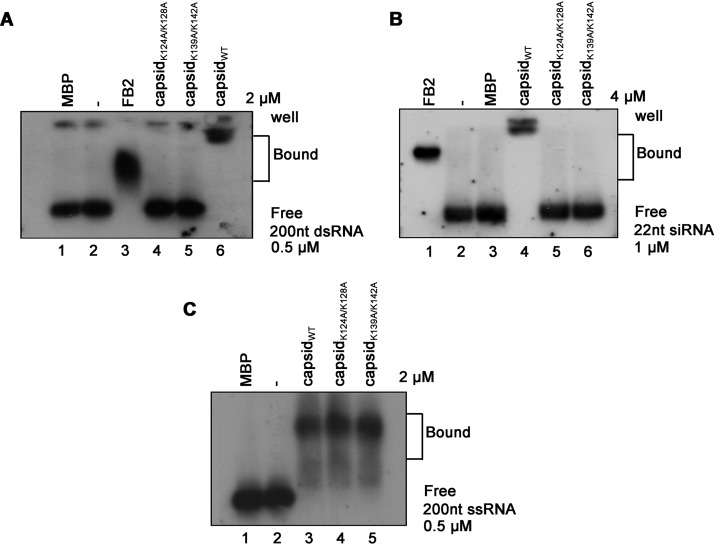
Moreover, we found that either K124A/K128A or K139A/K142A mutation abolished the dsRNA- and siRNA-binding activities of SFV capsid (Fig. 5A and B). Interestingly, although capsidK124A/K128A and capsidK139A/K142A failed to bind to dsRNA or siRNA, their activities to bind to ssRNA were still intact (Fig. 5C).
FIG 5.
K124/K128 and K139/K142 of the SFV capsid are critical for the dsRNA and siRNA binding. (A to C) MBP-capsidWT, MBP-capsidK124A/K128A, or MBP-capsidK139A/K142A was incubated with 0.5 μM 200-nt DIG-labeled dsRNA (A), 1 μM 22-nt siRNA (B), or 0.5 μM ssRNA (C) at 25°C for 30 min. The complexes were separated on 1.5% native-TBE agarose gel, transferred to membranes, and then incubated with anti-DIG antibody conjugated with alkaline phosphatase. MBP-FB2 and MBP alone were used as positive and negative controls, respectively.
Together, these data show that the K124A/K128A and K139A/K142A residues are critical for the dsRNA/siRNA-binding and VSR activities of SFV capsid.
Construction and recovery of VSR-deficient SFV.
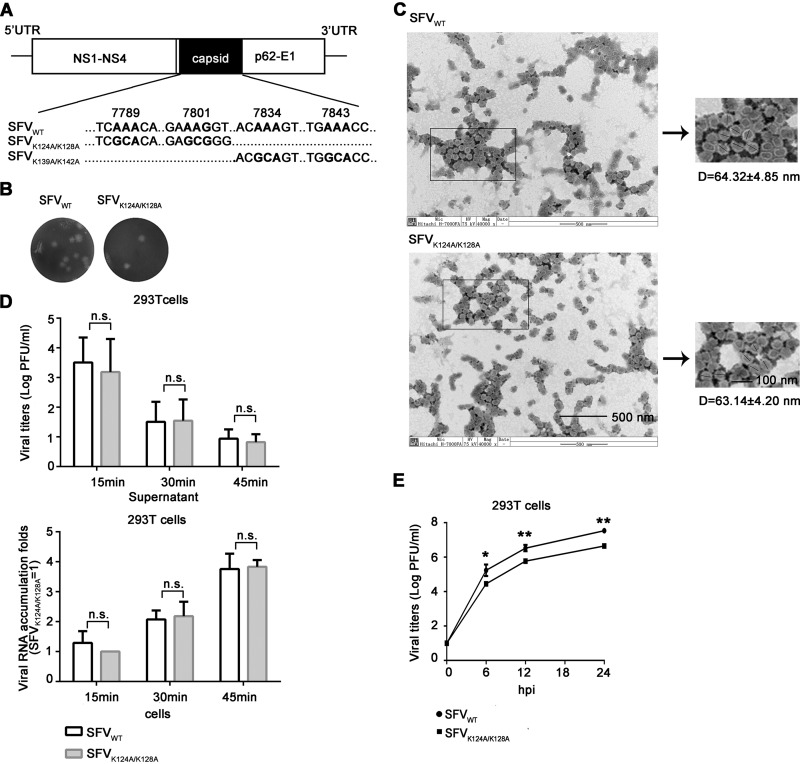
To find out whether SFV capsid protein indeed suppresses antiviral RNAi during virus infection, we introduced the K124A/K128A and K139A/K142A mutation into the capsid coding region of the infectious clone of SFV (Fig. 6A). The wild-type (SFVWT) and K124A/K128A mutant (SFVK124A/K128A) viruses were successfully recovered and the plaque morphology of these two viruses was similar (Fig. 6B), whereas K139A/K142A mutant virus displayed a lethal phenotype.
FIG 6.
Construction and recovery of VSR-deficient SFV. (A) SFV genome and the mutation sites of K124A/K128A and K139A/K142A. (B) Plaque morphology of SFVWT and SFVK124A/K128A. (C) The virions of SFVWT and SFVK124A/K128A were examined by TEM at magnifications of ×40,000. (D) 293T cells were infected with SFVWT and SFVK124A/K128A at an MOI of 5. At 15, 30, and 45 min postinfection, the viruses remaining in the supernatant and the viral RNAs entered into cells were examined by plaque assays and qRT-PCR, respectively. The RNA levels of SFVK124A/K128A in 293T cells at 15 min postinfection are defined as 1. All data represent the means and standard deviations of three independent experiments. n.s., not significant. (E) 293T cells were infected with SFVWT or SFVK124A/K128A (MOI = 0.1), respectively. Viral titers were measured at the indicated times using a standard plaque assay in Vero cells. All data represent the means and standard deviations of three independent experiments. *, P < 0.05; **, P < 0.01 (as measured by two-way analysis of variance [ANOVA]; GraphPad Prism).
Because alphaviral capsid protein is critical for the formation of nucleocapsid and virion maturation, we sought to exclude the possibility that the K124A/K128A mutation may affect other important functions of SFV capsid, such as the process of virion assembly and the virus entry into cells. Our previous data showed that SFV capsidK124A/K128A still kept the ssRNA-binding activity (Fig. 5C), implying that the interaction between SFV capsidK124A/K128A and viral genomic RNAs, which is important for nucleocapsid assembly, is not affected. Moreover, we purified the virions of SFVWT and SFVK124A/K128A and then examined the morphology of both WT and mutant virions via transmission electron microscopy (TEM). Our results showed that the K124A/K128A mutation did not affect the morphology and diameter of viral particles (Fig. 6C).
Subsequently, we sought to determine whether the K124A/K128A mutation affected the entry efficiency of SFV into cells. To this end, 293T cells were infected with SFVWT or SFVK124A/K128A at a multiplicity of infection (MOI) of 5; at 15, 30, and 45 min postinfection, the viruses remaining in the supernatant and the viral RNAs within cells were examined by plaque assays and qRT-PCR, respectively. Our data show that the levels of the remaining virions in the supernatant and the viral RNAs within cells of SFVK124A/K128A were comparable to those of SFVWT at each time point (Fig. 6D), indicating that the K124A/K128A mutation did not affect the entry of SFV into cells.
After determining that the K124A/K128A mutation did not affect the virion assembly and viral entry of SFV, we determined the one-step growth curve of WT and mutant viruses in human 293T cells, showing that SFVK124A/K128A exhibited weaker growth patterns than did SFVWT in 293T cells (Fig. 6E). This result indicates that the inactivation of VSR function led to the restricted replication of SFV.
The replication defect of VSR-deficient SFV can be rescued by the deficiency of RNAi in 293T cells.
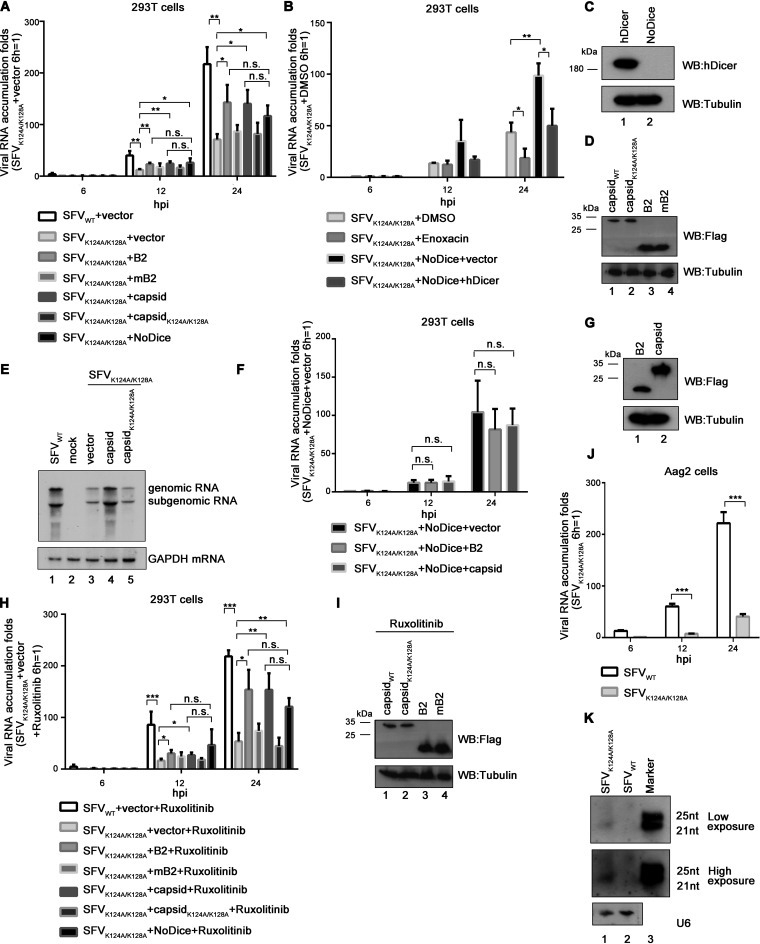
To further explore the VSR function of SFV capsid during viral infection, 293T cells were infected with WT or VSR-deficient SFV, and viral RNA accumulation were determined at 6, 12, and 24 h postinfection (hpi), respectively. As expected, the viral RNA accumulation of SFVK124A/K128A was lower than that of SFVWT in 293T cells (Fig. 7A). The genetic ablation of the RNAi pathway by Dicer knockout in 293T cells (NoDice) rescued the replication of SFVK124A/K128A (Fig. 7A). Interestingly, the RNA accumulation of SFVK124A/K128A was significantly reduced in 293T cells treated with enoxacin compared to that in the controlled 293T cells (Fig. 7B). Of note, enoxacin is a well-known RNAi enhancer and functions at steps after siRNA production by Dicer in the RNAi pathway (9, 42). Expectedly, ectopic expression of human Dicer (hDicer) in 293T-NoDice cells resulted in the reduced RNA accumulation of SFVK124A/K128A (Fig. 7B and C).
FIG 7.
The deficiency of RNAi rescued the defective replication of VSR-deficient SFV. (A) 293T or 293T-NoDice cells were transfected with either empty plasmid or a plasmid encoding NoV B2, mB2 (B2R59Q), SFV capsid, or capsidK124A/K128A as indicated. At 24 hpt, the cells were infected with SFVWT or SFVK124A/K128A at an MOI of 1. At 6, 12, and 24 hpi, the levels of SFV genomic RNAs in cells were determined by qRT-PCR, and the level of SFVK124A/K128A RNA in 293T cells at 6 hpi was defined as 1. All data represent the means and standard deviations of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (as measured by two-way ANOVA; GraphPad Prism). (B) 293T or NoDice 293T cells were transfected with either empty plasmid or a plasmid encoding human Dicer (hDicer) as indicated. At 24 hpt, the cells were treated with enoxacin (100 μM) for 1 h and then infected with SFVWT at an MOI of 1. At 6, 12, and 24 hpi, the levels of SFV genomic RNAs in cells were determined by qRT-PCR, and the level of SFVK124A/K128A RNA in 293T cells treated with DMSO at 6 hpi was defined as 1. All data represent the means and standard deviations of three independent experiments. (C and D) The expression of capsid, capsidK124A/K128A, NoV B2, mB2, or hDicer proteins in 293T or 293T-NoDice cells was detected by Western blotting. (E) 293T cells were transfected with SFV capsid or capsidK124A/K128A as indicated and then infected with SFVWT or SFVK124A/K128A at an MOI of 1. At 24 hpi, the total RNAs were extracted, the levels of SFV genomic and subgenomic RNAs were examined via Northern blotting with DIG-labeled RNA probe targeting nt 723 to 1314 of the SFV E2 coding region. GAPDH mRNA was used the loading control. (F) 293T-NoDice cells were transfected with either empty plasmid or a plasmid encoding NoV B2, SFV capsid as indicated. At 24 hpt, the cells were infected with SFVK124A/K128A at an MOI of 1. At 6, 12, and 24 hpi, the levels of SFV genomic RNAs in cells were determined by qRT-PCR, and the level of SFVK124A/K128A RNA in 293T cells transfected with either empty plasmid at 6 hpi was defined as 1. (G) The expression of SFV capsid, and NoV B2 in 293T-NoDice cells was detected by Western blotting. (H) 293T or 293T-NoDice cells were transfected with either empty plasmid or a plasmid encoding NoV B2, mB2 (B2R59Q), SFV capsid, or capsidK124A/K128A as indicated. At 24 hpt, the cells were treated with ruxolitinib (10 μM) for 1 h and then infected with SFVWT or SFVK124A/K128A at an MOI of 1. At 6, 12, and 24 hpi, the levels of SFV genomic RNAs in cells were determined by qRT-PCR, and the level of SFVK124A/K128A RNA in 293T cells at 6 hpi was defined as 1. (I) The expression of capsid, capsidK124A/K128A, NoV B2, or mB2 in ruxolitinib-treated 293T cells was detected by Western blotting. (J) Aag2 cells were infected with SFVWT or SFVK124A/K128A at an MOI of 1. At 6, 12, and 24 hpi, the levels of SFV genomic RNAs in cells were determined by qRT-PCR, and the level of SFVK124A/K128A RNA in MLF cells at 6 hpi was defined as 1. (K) Aag2 cells were infected with SFVWT or SFVK124A/K128A at an MOI of 10. At 24 hpi, the total RNAs were extracted, and the levels of vsiRNAs were examined via Northern blotting with a DIG-labeled RNA probe targeting nt 1 to 50 of antigenomic SFV RNA. U6 was used a loading control.
Moreover, the viral RNA replication of SFVK124A/K128A in 293T cells was increased by the ectopic expression of SFV capsid or NoV B2, but not the VSR-deficient mutant of capsid (capsidK124A/K128A) or NoV B2 (B2R59Q, named mB2) (Fig. 7A and D). Moreover, the rescuing effect of ectopically expressed capsid on the replication of VSR-deficient SFV was confirmed by using Northern blotting with a DIG-labeled RNA probe that recognized both genomic and subgenomic RNAs of SFV (Fig. 7E).
Furthermore, we found that ectopic expression of NoV B2 or SFV capsid could not rescue the RNA accumulation of SFVK124A/K128A in 293T-NoDice cells (Fig. 7F and G). These results indicate that the rescuing effect of ectopically expressed foreign VSRs on the replication of SFVK124A/K128A is indeed RNAi dependent.
To exclude the potential impact of IFN-I response, we treated 293T or 293T-NoDice cells infected by SFVK124A/K128A with ruxolitinib, a JAK1 and JAK3 inhibitor, to block IFN-I. We found that the ectopic expression of foreign VSRs or a deficiency of Dicer could still enhance the RNA accumulation of SFVK124A/K128A in ruxolitinib-treated cells (Fig. 7H and I), confirming that the rescuing effect is irrespective of IFN-I.
In addition, we examined the production of vsiRNAs in SFVK124A/K128A-infected Aedes mosquito Aag2 cells. The deficiency in VSR also resulted in substantial attenuation of the viral RNA accumulation of SFVK124A/K128A in Aag2 cells (Fig. 7J), a result consistent with the findings in 293T cells. SFV vsiRNAs in Aag2 cells were detected via Northern blotting with RNA probe complementary to the 3′-end 1 to 50 nt of the negative-stranded antigenomic RNA. Our findings showed that the production of SFV vsiRNAs was dramatically enhanced in Aag2 cells infected with SFVK124A/K128A but not SFVWT (Fig. 7K).
DISCUSSION
Alphaviruses infect and replicate in both invertebrate vectors and mammalian hosts. Efficient transmission of these viruses depends on their activities to counteract the antiviral immune response in both mosquito vectors and human hosts (31). RNAi is an antiviral immune response conserved in both invertebrates and mammals, whereas many viruses encode VSRs as the countermeasure (2, 5). However, it was still unclear whether alphavirus encodes a VSR to antagonize antiviral RNAi in the context of viral infection in mammalian cells. To date, the identification of a bona fide VSR encoded by a mammalian virus requires to answer the following questions: (i) whether disabling VSR by reverse genetic can cause viral replication defect during authentic viral infection and (ii) whether the genetic ablation of the RNAi pathway can rescue the replication defect of the VSR-disabled mutant virus.
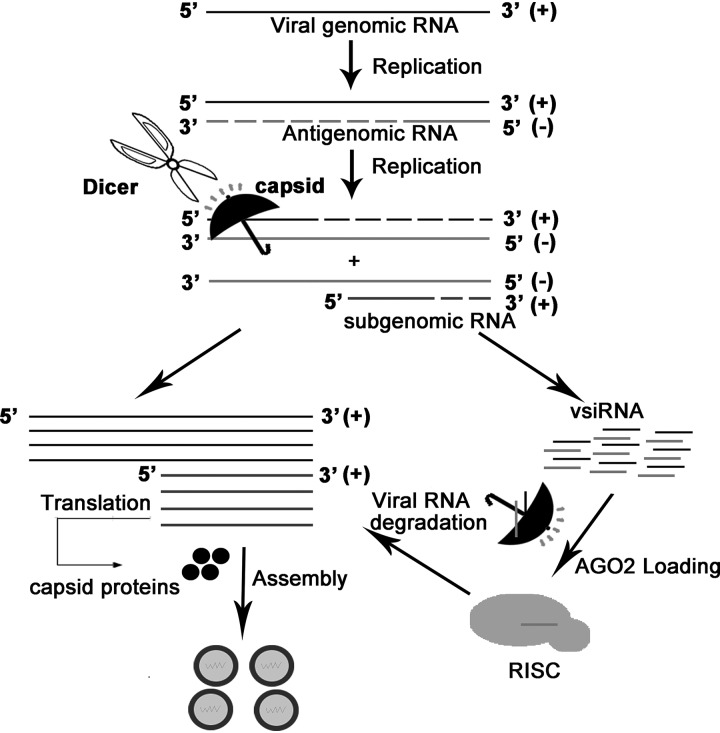
In the present study, we screened the viral proteins of SFV, a prototypic alphavirus, for VSR activity in insect and mammalian cells. We found that SFV capsid protein possesses VSR activity. The in vitro EMSA analysis shows that SFV capsid has both dsRNA- and siRNA-binding activities that are indispensable for its VSR function. More importantly, the VSR deficiency of SFV capsid resulted in substantial restriction of viral replication in mammalian cells. This defective replication of VSR-deficient mutant SFV can be rescued by the deficiency of RNAi by either ectopically expressing foreign VSRs or blocking the RNAi pathway, which is irrespective of the IFN-I system. In addition, when the capsid-mediated RNAi suppression was genetically disabled in SFV, the production of vsiRNAs was enhanced in infected mosquito cells. Thus, we propose a hypothesis that SFV capsid may protect viral RNA from antiviral RNAi at two stages: (i) blocking vRI-dsRNA from Dicer cleavage through dsRNA-binding activity and (ii) binding to vsiRNA to suppress the incorporation of vsiRNA to RISC (Fig. 8).
FIG 8.
Model for the suppression of RNAi in mammalian and mosquito cells by SFV capsid protein. When SFV entry the host cells, the single-stranded genomic (+)RNAs are used as templates to produce the negative genomic (–)RNAs. The (–)RNAs can be used as templates to produce more progeny (+)RNAs and subgenomic RNAs. The vRI-dsRNAs formed by the 5′-terminal nascent (+)RNAs or/subgenomic RNAs and the (–)RNA template could be recognized and cleaved by Dicer, thus triggering antiviral RNAi. SFV capsid protein can suppress antiviral RNAi by sequestrating dsRNA and siRNA as indicated.
In addition to SFV capsid, a remaining question is whether alphaviruses encode another VSR(s). Previous study has identified CHIKV nsP2 and nsP3 suppressed shRNA-induced RNAi in insect and mammalian cells (43). However, the potential VSR activity of nsP2 and nsP3 was not tested in the context of an authentic viral infection. Overexpression of any protein with an effective dsRNA- or siRNA-binding activity may suppress RNAi in some cases, which does not mean that it acts as a VSR during viral infection. In addition, viruses may encode more than one VSR, as seems to be the case for some plant viruses, such as citrus tristeza virus (p20, p23, and coat protein) (44) and potyviruses (P1 and HcPro) (45).
Alphaviral RNA packaging requires active viral RNA synthesis, implying that the replication and encapsidation are tightly connected (46). This may explain why only the genomic RNA can be specifically packaged into the virion and suggests that alphaviral structural proteins could be directly involved in viral RNA replication. Indeed, SFV capsid is a versatile structure protein that can bind to the large ribosomal subunit in the cytoplasm and associate with the viral RNAs to form the viral nucleocapsid (38, 39). In this present study, we provide evidence that SFV capsid is a potent VSR, which plays an important role in the viral RNA replication in insect and mammalian cells. Our findings are consistent with the previous findings that VSR activities were observed in viral structural coat proteins encoded by numerous viruses, including Turnip crinkle virus coat protein, YFV capsid protein, and coronavirus nucleocapsid protein, and their VSR activities have been found to be associated with their dsRNA-binding capacities (22, 24, 47).
SFV capsid is made up of two domains, the RNA binding N-terminal segment (aa 1 to 118) and the C-terminal globular protease domain (aa 119 to 267), which is critical for the dimerization of capsid (40). Indeed, our mutational analyses showed that mutations of K124A/K128A or K139A/K142A abolished the dsRNA-/siRNA-binding of SFV capsid but not ssRNA-binding activities. These data indicate that these residues are not directly involved in the association of SFV capsid with viral genomic RNAs but are possibly involved in the process of protein dimerization. Our results are consistent with the previous findings that dimerization is required for RNAi suppression activities of many VSRs, such as NoV B2 and EV-A71 3A (8, 48). The future work will demonstrate the relationship between the VSR activity and the dimerization of SFV capsid.
In summary, our study demonstrates that SFV capsid functions as a VSR in facilitating viral replication by blocking either dsRNA from Dicer cleavage or vsiRNA loading into RISC in mammalian cells. Moreover, it shows for the first time that an alphavirus can antagonize antiviral RNAi in the context of viral infection, extending our knowledge about the interaction between alphavirus and antiviral RNAi immunity.
MATERIALS AND METHODS
Plasmids and RNAs.
For the expression of SFV proteins, their ORFs were cloned into insect expression vector pAc5.1/V5-HisB, respectively. To express proteins in 293T cells, the ORF of SFV capsid was constructed into the pRK-Flag. The full-length cDNA of the FHV RNA1 and FHV RNA1 ΔB2 were described previously (24). For the purification of the MBP fusion capsid protein, its ORF was inserted into the pMAL-c2X vector. The EGFP-siRNA (siEGFP) was chemically synthesized by Rui Bo, Guangzhou, China. The full-length SFV cDNA clone (strain SFV4) was constructed in pCMV-Myc vector driven by cytomegalovirus promoter (kindly provided by Tero Ahola, University of Helsinki).
Cell culture.
HEK293T cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS; Gibco), 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C in an incubator with 5% CO2. The 293T-NoDice cell line was kindly provided by Bryan R. Cullen (Durham, NC). Drosophila S2 or Aag2 cells were cultured in Schneider insect medium with 10% FBS at 27°C. Before transfection with FuGene HD reagent (Roche, Basel, Switzerland), the medium was changed to DMEM or Schneider insect medium containing 2% FBS without any antibiotic. Cells were harvested at 48 hpt.
Construction and recovery of SFV mutant virus.
To construct the SFV mutant clones, we obtained nt 6636 to 8748 of the full-length SFV cDNA clone via PCR and cloned it into the pMd-18T vector. The K124A/K128A and K139A/K142A mutations were then introduced into this plasmid via overlap PCR. The resulting mutant fragments were inserted into the full-length cDNA clone by double enzyme digestion (SbfI and XbalI). These infectious cDNA clones were then transfected into the 293T cells using the FuGene HD reagent (Roche), and the rescued viruses were harvested 48 hpt. The virus titers were measured by plaque assays.
Western blotting.
Cells were harvested in lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 0.25% deoxycholate, and a protease inhibitor cocktail [Rhochel]). The lysates were then subjected to 12% SDS-PAGE and Western blotting according to our standard procedures (49). The antibodies used in this study are included anti-tubulin (Protein Tech Group, 1:3,000), anti-His (Protein Tech Group, 1:10,000), anti-Flag (Protein Tech Group, 1:5,000), and anti-Dicer (Protein Tech Group, 1:2,000).
Northern blotting and qRT-PCR.
Total RNAs were extracted using TRIzol reagent (Thermo) according to the manufacturer’s instructions. For the detection of EGFP mRNA, 5 μg of total RNAs was subjected to denatured 1.5% agarose gels with 2.2 M formaldehyde. The separated RNAs were transferred onto the Hybond-A nylon membrane (GE Healthcare) and fixed by 120°C for 15 min. The membranes were then hybridized with DIG-labeled probes in Hybridization Ovens at 65°C overnight. The membranes were next incubated with anti-DIG antibody conjugated with alkaline phosphatase and exposed to a luminescent image analyzer LAS4000 (Fuji Film). Probes for the detection of EGFP, Rp49, and GAPDH mRNA were complementary to nt 520 to 700, nt 273 to 490, and nt 760 to 1060 of their ORF regions, respectively. The probe for the detection FHV RNA1 and subgenomic RNA3 was complementary to nt 2738 to 3058 of the B2 coding region, and the probe for the detection of SFV genome and subgenomic RNA was complementary to nt 723 to 1314 of the E2 protein coding region. These probes were labeled with DIG-UTP (Roche) by in vitro transcription. For the detection of small RNAs, 20 μg of total RNAs was subjected to 7 M urea–15% PAGE and transferred to a Hybond-A nylon membrane (GE Healthcare). The membrane was chemically cross-linked in 1-ethly-3-(3-dimethylaminopropyl)-carbodiimide (EDC) at 60°C. For detection of vsiRNA, Aag2 cells were infected with SFVWT or SFVK124A/K128A at an MOI of 10 for 24 hpt, and then the total RNAs were subjected to 7 M urea–15% PAGE for Northern blotting. The probes targeting EGFP siRNA, U6, and vsiRNA were synthesized by TaKaRa or by in vitro transcription, and their sequences are listed in Table 1 . qRT-PCR using SYBR mix (TaKaRa) was carried out to detect the expression of EGFP mRNA, β-actin mRNA, and SFV NS1 mRNA. The primers and oligonucleotides used in this study are shown in Table 1 .
TABLE 1.
Primers and oligonucleotides used in this study
| Primer or oligonucleotide | Sequence (5′–3′)a |
|---|---|
| Construction of SFV proteins | |
| pAc-v5/His-NS1-sense | GAATTCTATGGCCGCCAAAGTGCATGTTGATAT (EcoRI) |
| pAc-v5/His-NS1-anti | CTCGAGCGTGCACCTGCGTGATACTCTAGTTC (XhoI) |
| pAc-v5/His-NS2-sense | GCGGCCGCATGGGGGTCGTGGAAACACCTCGCA (NotI) |
| pAc-v5/His-NS2-anti | TCTAGACTACACCCGGCCGTGTGCATGGCTTCTC (XbalI) |
| pAc-v5/His-NS3-sense | GAATTCTATGGCACCATCCTACAGAGTTAAGAGAGC (EcoRI) |
| pAc-v5/His-NS3-anti | TCTAGACTTGCACCCGCGCGGCCTAGTCGCAGGAC (XbalI) |
| pAc-v5/His-NS4-sense | GAATTCTATGTATATTTTCTCCTCGGACACACTGGCA (EcoRI) |
| pAc-v5/His-NS4-anti | CTCGAGCGACGCACCAATCTAGGACCGCCGTAGAG (XhoI) |
| pAc-v5/His-capsid-sense | GCGGCCGCATGAATTACATCCCTACGCAAACGTTTT (NotI) |
| pAc-v5/His-capsid-anti | CTCGAGCGCCACTCTTCGGACCCCTCGGGGGTCAC (XhoI) |
| pAc-v5/His-E3-sense | GAATTCTATGTCCGCCCCGCTGATTACTGCCATGT (EcoRI) |
| pAc-v5/His-E3-anti | TCTAGAGCGCCGGTGTCTTGTTCCGTTTCGGCACG (XbalI) |
| pAc-v5/His-E2-sense | GCGGCCGCATGAGCGTGTCGCAACACTTCAACGTG (NotI) |
| pAc-v5/His-E2-anti | CTCGAGCGTGCGTGCGCCCGCGGGGCGCAGCAGAG (XhoI) |
| pAc-v5/His-6k-sense | GCGGCCGCCCATGGCTAGTGTGGCAGAGACTATGGC (NotI) |
| pAc-v5/His-6k-anti | CTCGAGCGAGCTCTGGCGGTTGCCCCGAGGCTCAGT (XhoI) |
| pAc-v5/His-E1-sense | GCGGCCGCATGTACGAACATTCGACAGTAATGCCGA (NotI) |
| pAc-v5/His-E1-anti | CTCGAGCGTCTGCGGAGCCCAATGCAAGTGACCACA (XhoI) |
| pRK-capsid-sense | GTCGACGATGAATTACATCCCTACGCAAACGTTTTAC (SalI) |
| pRK-capsid-anti | GCGGCCGCTTACCACTCTTCGGACCCCTCGGGGGTCA (NotI) |
| pMal-c2x-capsid-sense | GAATTCATGAATTACATCCCTACGCAAACGTTTTAC (EcoRI) |
| pMal-c2x-capsid-anti | GTCGACCCACTCTTCGGACCCCTCGGGGGTCACTC (SalI) |
| Construction of capsid mutants and SFV mutant | |
| K124A/K128A-sense | TCTTCGAAGTCGCACACGAAGGAGCGGTCACTGG (–) |
| K124A/K128A-anti | CCAGTGACCGCTCCTTCGTGTGCGACTTCGAAGA (–) |
| K139A/K142A-sense | GCCTGGTGGGCGACGCAGTCATGGCACCTGCCCA (–) |
| K139A/K142A-anti | TGGGCAGGTGCCATGACTGCGTCGCCCACCAGGC (–) |
| pMD18-T-K124A/K128A-sense | TCTAGAGACGGACATTGCATCATTC (XbalI) |
| pMD18-T-K124A/K128A-anti | CCTGCAGGAATTCACCCGGTGGGCA (SbfI) |
| qRT-PCR | |
| EGFP-sense | GACAAGCAGAAGAACGGCATC |
| EGFP-anti | CGGACTGGGTGCTCAGGTA |
| SFV-sense | CCGGAGGACGCACAGAAGTTG |
| SFV-anti | TGCGACGGCCACAATCGGAAG |
| Human actin-sense | AGAGCTACGAGCTGCCTGAC |
| Human actin-anti | AGCACTGTGTTGGCGTACAG |
| Aedes aegypti Rp49-sense | GCTATGACAAGCTTGCCCCCA |
| Aedes aegypti Rp49-anti | TCATCAGCACCTCCAGCT |
| Amplification of templates for in vitro transcription dsRNAs and ssRNAs | |
| EGFP-ds400-sense | TAATACGACTCACTATAGATGGTGAGCTAGGGCGAGGA |
| EGFP-ds400-anti | TAATACGACTCACTATAGGCTTGTGCCCCAGGATGTTGC |
| EGFP-ds200-sense | TAATACGACTCACTATAGATGGTGAGCTAGGGCGAGGA |
| EGFP-ds200-anti | TAATACGACTCACTATAGGCGGCTGAAGCACTGCACGC |
| dsAgo2–sense | TAATACGACTCACTATAGATGGGAAAAAAAGATAAGAACAAGC |
| dsAgo2–anti | TAATACGACTCACTATAGCACCTTGTTGACCTTGTTTAGTCCAC |
| EGFP-ss200-sense | TAATACGACTCACTATAGATGGTGAGCTAGGGCGAGGA |
| EGFP-ss200-anti | CTATAGTGAGTCGTATTAGCGGCTGAAGCACTGCACGC |
| Amplification of templates for in vitro transcription RNA probes | |
| EGFP-probe-sense | GATCCGCCACAACATCGAGGACGGC |
| EGFP-probe-anti | TAATACGACTCACTATAGTTACTTGTACAGCTCGTCCATGCC |
| S2-Rp49-sense | CCGTGAATACTGTGGTGAAATTGCC |
| S2-Rp49-anti | TAATACGACTCACTATAGGTTTTTTTTTCACTTTTAACGTTTCA |
| GAPDH-probe-sense | GGCGTGATGGCCGCGGGGCTCTCC |
| GAPDH-probe-anti | TAATACGACTCACTATAGGCAAAGGTGGAGGAGTGGGTGTCGC |
| SFV-probe-sense | GCTGCCAAATCAAAACGAACCCTGTCAG |
| SFV-probe-anti | TAATACGACTCACTATAGGTCTGCGGAGCCCAATGCAAGTGACC |
| FHV-probe-sense | ATGCCAAGCAAACTCGCGCTAATCCAGGAACTTCCCG |
| FHV-probe-anti | TAATACGACTCACTATAGGCCTCTAGGTATGCCACCACGCTGGGTTTCTC |
| siEGFP-sense | GCUGACCCUGAAGUUCAUCUU |
| siEGFP-anti | GAUGAACUUCAGGGUCAGCUU |
| SFV-vsiRNA-probe-sense | TAATACGACTCACTATAGATGGCGGATGTGTGACATACACGACGCCAAAAGATTTTGTTCCAGCTCCT |
| SFV-vsiRNA-probe-anti | AGGAGCTGGAACAAAATCTTTTGGCGTCGTGTATGTCACACATCCGCCATCTATAGTGAGTCGTATTA |
| EGFP-siRNA Probe | DIG-GCAAGCUGACCCUGAAGUUCAUC-DIG |
| U6-Probe | DIG-GCAGGGGCCAUGCUAAUCUUCU-DIG |
Restriction enzymes are indicated in parentheses where applicable. –, No restriction enzyme. DIG, digoxigenin labeled.
Expression and purification of capsid protein.
The coding regions of SFV capsid protein and FHV B2 were cloned into pMAL-c2X. Escherichia coli BL21 (Invitrogen) transformed with the expression plasmids was grown to log phase at 37°C and induced with 0.8 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 22°C for 6 h. The cells were harvested by centrifugation and resuspended in lysis buffer (1 M Tris-HCl [pH 7.5], 0.2 M NaCl, 0.5 M EDTA [pH 8.0], 10 mM β-mercaptoethanol, 5% absolute ethyl alcohol, 10% glycerinum), followed by sonication and centrifugation to remove the debris. Finally, the proteins in the supernatant were purified by using amylose resin (New England BioLabs) according to the manufacturer’s instructions.
Electrophoretic mobility shift assay and RNase III cleavage assay.
The MBP fusion capsid was reacted with DIG-labeled RNAs (0.5 μM 200-nt dsRNA, 0.5 μM 200-nt ssRNA, or 1 μM 22-nt siRNA) in a reaction buffer containing 40 mM MgCl2, 50 mM NaCl, 25 mM HEPES (pH 7.5), 3 mM dithiothreitol, and 1 U of RNase inhibitor; the total volume was 10 μl. After incubation for 30 min at 25°C, the reaction mixtures were subjected to 1.5% native-TBE agarose gel and then transferred to a Hybond-A nylon membrane (GE Healthcare). The membranes were washed with maleic acid buffer for 10 min and then incubated with anti-DIG antibody conjugated to alkaline phosphatase (Roche) for 30 min.
For the RNase III cleavage assay, 1 μM 200-nt dsRNA was incubated with 1 U of RNase III (Invitrogen) and MBP fusion proteins in reaction buffer according to the manufacturer’s instructions at 37°C for 30 min. The mature siRNAs processed by RNase III were extracted from the reaction complex by using TRIzol reagent (Thermo), subjected to 7 M urea–15% PAGE, and then transferred to a Hybond-A nylon membrane.
Virus infection and plaque assays.
At the day of infection, the medium was changed with 2% FBS–DMEM, and then the viruses were added to 293T or Aag2 cells at an MOI of 1. Total RNAs were extracted at 6, 12, and 24 hpi and subjected to Northern blotting and qRT-PCR analysis. For the rescue experiments, 293T cells were first transfected with the plasmid encoding the indicated proteins for 24 hpt and treated with ruxolitinib (10 μM; Selleck) or Enoxacin (100 μM; Selleck) for 1 h and then infected with viruses. To examine the entry efficiency of SFV into cells, 293T cells were infected with WT or mutant SFV at an MOI of 5; at 15, 30 and 45 min postinfection, the viruses remaining in the supernatant and the viral RNAs that had invaded the cells were examined by plaque assay and qRT-PCR, respectively.
For the plaque assays, Vero cells in 12-well plates were infected with a 10-fold serial dilution of viruses. The cells were cultured at 37°C for 2 h to allow the adsorption of all the viruses. The supernatant was then replaced with 1× MEM containing 2% FBS and 1% penicillin-streptomycin with isopycnic 1% low-melting-point agarose (Sigma-Aldrich). After incubation at 37°C for 72 h, the cells were fixed with 10% formaldehyde and stained with 0.5% crystal violet at 4°C for 2 h.
Sucrose density gradient ultracentrifugation and transmission electron microscopy.
The cellular supernatant was filtrated by using the 0.45-μm filtering membrane and subjected to ultracentrifugation of 150,000 × g for 3 h at 4°C in an SW28 rotor. After centrifugation, the precipitate was resuspended in NTE buffer (10 mM Tris-HCl [pH 7.5], 120 mM NaCl, 1 mM EDTA) and then put into a 10 to 60% continuous sucrose gradient for ultracentrifugation at 150,000 × g for 3 h at 4°C in an SW41 rotor. The 30 and 40% sucrose solution containing virions was collected and further subjected to ultracentrifugation at 150,000 × g for 2 h at 4°C in an SW41 rotor. In the end, the precipitate was resuspended in 50 μl of NTE buffer.
For the TEM, virions were adsorbed to glow-discharged electron microscope grids and negatively stained with purified terephthalic acid according to our standard procedures (50). Samples were imaged by using a 100-kV Hitachi H-7000FA transmission electron microscope.
ACKNOWLEDGMENTS
We are grateful to Tero Ahola (Helsinki, Finland) and Bryan R. Cullen (Durham, NC) for reagents and to Pei Zhang and An-na Du from the Core Facility and Technical Support, Wuhan Institute of Virology, for their help with producing TEM micrographs.
This study was supported by the National Science and Technology Major Project of China (2018ZX10101004 to X.Z.), the Strategic Priority Research Program of Chinese Academy of Sciences (XDB29010300 to X.Z.), the National Natural Science Foundation of China (81873964 to Y.Q., 31800140 to J.M., and 31670161 to X.Z.), the Science and Technology Bureau of Wuhan (2018060401011309 to X.Z.), the Advanced Customer Cultivation Project of Wuhan National Biosafety Laboratory (2018ACCP-MS11 to Y.Q.), and the Yunde Hou Academician Fund from National Institute For Viral Disease Control and Prevention (2019HYDQNJJ10 to J.M.). X.Z. is supported by the Newton Advanced Fellowship from the Academy of Medical Sciences, UK (NAF005\1002).
REFERENCES
- 1.Ding SW. 2010. RNA-based antiviral immunity. Nat Rev Immunol 10:632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 2.Guo Z, Li Y, Ding SW. 2019. Small RNA-based antimicrobial immunity. Nat Rev Immunol 19:31–44. doi: 10.1038/s41577-018-0071-x. [DOI] [PubMed] [Google Scholar]
- 3.Blair CD. 2011. Mosquito RNAi is the major innate immune pathway controlling arbovirus infection and transmission. Future Microbiol 6:265–277. doi: 10.2217/fmb.11.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aliyari R, Wu Q, Li HW, Wang XH, Li F, Green LD, Han CS, Li WX, Ding SW. 2008. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe 4:387–397. doi: 10.1016/j.chom.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maillard PV, Veen AG, Poirier EZ, Reis e Sousa C. 2019. Slicing and dicing viruses: antiviral RNA interference in mammals. EMBO J 38:e100941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Lu J, Han Y, Fan X, Ding SW. 2013. RNA interference functions as an antiviral immunity mechanism in mammals. Science 342:231–234. doi: 10.1126/science.1241911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maillard PV, Ciaudo C, Marchais A, Li Y, Jay F, Ding SW, Voinnet O. 2013. Antiviral RNA interference in mammalian cells. Science 342:235–238. doi: 10.1126/science.1241930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qiu Y, Xu Y, Zhang Y, Zhou H, Deng YQ, Li XF, Miao M, Zhang Q, Zhong B, Hu Y, Zhang FC, Wu L, Qin CF, Zhou X. 2017. Human virus-derived small RNAs can confer antiviral immunity in mammals. Immunity 46:992–1004.e1005. doi: 10.1016/j.immuni.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 9.Xu YP, Qiu Y, Zhang B, Chen G, Chen Q, Wang M, Mo F, Xu J, Wu J, Zhang RR, Cheng ML, Zhang NN, Lyu B, Zhu WL, Wu MH, Ye Q, Zhang D, Man JH, Li XF, Cui J, Xu Z, Hu B, Zhou X, Qin CF. 2019. Zika virus infection induces RNAi-mediated antiviral immunity in human neural progenitors and brain organoids. Cell Res 29:265–273. doi: 10.1038/s41422-019-0152-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Basavappa M, Lu J, Dong S, Cronkite DA, Prior JT, Reinecker HC, Hertzog P, Han Y, Li WX, Cheloufi S, Karginov FV, Ding SW, Jeffrey KL. 2016. Induction and suppression of antiviral RNA interference by influenza A virus in mammalian cells. Nat Microbiol 2:16250. doi: 10.1038/nmicrobiol.2016.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Q, Wang X, Ding SW. 2010. Viral suppressors of RNA-based viral immunity: host targets. Cell Host Microbe 8:12–15. doi: 10.1016/j.chom.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chao JA, Lee JH, Chapados BR, Debler EW, Schneemann A, Williamson JR. 2005. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol 12:952–957. doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- 13.Li H, Li WX, Ding SW. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- 14.Lu R, Maduro M, Li F, Li HW, Broitman-Maduro G, Li WX, Ding SW. 2005. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature 436:1040–1043. doi: 10.1038/nature03870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi N, Zhang L, Qiu Y, Wang Z, Si J, Liu Y, Xiang X, Xie J, Qin CF, Zhou X, Hu Y. 2012. Targeting of dicer-2 and RNA by a viral RNA silencing suppressor in Drosophila cells. J Virol 86:5763–5773. doi: 10.1128/JVI.07229-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qi N, Cai D, Qiu Y, Xie J, Wang Z, Si J, Zhang J, Zhou X, Hu Y. 2011. RNA binding by a novel helical fold of b2 protein from Wuhan nodavirus mediates the suppression of RNA interference and promotes b2 dimerization. J Virol 85:9543–9554. doi: 10.1128/JVI.00785-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nayak A, Kim DY, Trnka MJ, Kerr CH, Lidsky PV, Stanley DJ, Rivera BM, Li KH, Burlingame AL, Jan E, Frydman J, Gross JD, Andino R. 2018. A viral protein restricts Drosophila RNAi immunity by regulating Argonaute activity and stability. Cell Host Microbe 24:542–557.e549. doi: 10.1016/j.chom.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haasnoot J, de Vries W, Geutjes EJ, Prins M, de Haan P, Berkhout B. 2007. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog 3:e86. doi: 10.1371/journal.ppat.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bennasser Y, Le SY, Benkirane M, Jeang KT. 2005. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity 22:607–619. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 20.Chen W, Zhang Z, Chen J, Zhang J, Zhang J, Wu Y, Huang Y, Cai X, Huang A. 2008. HCV core protein interacts with Dicer to antagonize RNA silencing. Virus Res 133:250–258. doi: 10.1016/j.virusres.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 21.Kakumani PK, Ponia SS, S RK, Sood V, Chinnappan M, Banerjea AC, Medigeshi GR, Malhotra P, Mukherjee SK, Bhatnagar RK. 2013. Role of RNA interference (RNAi) in dengue virus replication and identification of NS4B as an RNAi suppressor. J Virol 87:8870–8883. doi: 10.1128/JVI.02774-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samuel GH, Wiley MR, Badawi A, Adelman ZN, Myles KM. 2016. Yellow fever virus capsid protein is a potent suppressor of RNA silencing that binds double-stranded RNA. Proc Natl Acad Sci U S A 113:13863–13868. doi: 10.1073/pnas.1600544113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karjee S, Minhas A, Sood V, Ponia SS, Banerjea AC, Chow VT, Mukherjee SK, Lal SK. 2010. The 7a accessory protein of severe acute respiratory syndrome coronavirus acts as an RNA silencing suppressor. J Virol 84:10395–10401. doi: 10.1128/JVI.00748-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui L, Wang H, Ji Y, Yang J, Xu S, Huang X, Wang Z, Qin L, Tien P, Zhou X, Guo D, Chen Y. 2015. The nucleocapsid protein of coronaviruses acts as a viral suppressor of RNA silencing in mammalian cells. J Virol 89:9029–9043. doi: 10.1128/JVI.01331-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anonymous. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev 58:806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCarthy MK, Morrison TE. 2016. Chronic chikungunya virus musculoskeletal disease: what are the underlying mechanisms? Future Microbiol 11:331–334. doi: 10.2217/fmb.15.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X, Tharmarajah K, Taylor A. 2017. Ross River virus disease clinical presentation, pathogenesis and current therapeutic strategies. Microbes Infect 19:496–504. doi: 10.1016/j.micinf.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Laine M, Luukkainen R, Toivanen A. 2004. Sindbis viruses and other alphaviruses as cause of human arthritic disease. J Intern Med 256:457–471. doi: 10.1111/j.1365-2796.2004.01413.x. [DOI] [PubMed] [Google Scholar]
- 29.Yuan Z. 2018. Investigation of viral pathogen profiles in some natural hosts and vectors in China. Virol Sin 33:1–4. doi: 10.1007/s12250-018-0021-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quetglas JI, Ruiz-Guillen M, Aranda A, Casales E, Bezunartea J, Smerdou C. 2010. Alphavirus vectors for cancer therapy. Virus Res 153:179–196. doi: 10.1016/j.virusres.2010.07.027. [DOI] [PubMed] [Google Scholar]
- 31.Samuel GH, Adelman ZN, Myles KM. 2018. Antiviral immunity and virus-mediated antagonism in disease vector mosquitoes. Trends Microbiol 26:447–461. doi: 10.1016/j.tim.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia H, Wang Y, Atoni E, Zhang B, Yuan Z. 2018. Mosquito-associated viruses in China. Virol Sin 33:5–20. doi: 10.1007/s12250-018-0002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fros JJ, Pijlman GP. 2016. Alphavirus infection: host cell shut-off and inhibition of antiviral responses. Viruses 8:166. doi: 10.3390/v8060166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atkins GJ, Sheahan BJ, Dimmock NJ. 1985. Semliki Forest virus infection of mice: a model for genetic and molecular analysis of viral pathogenicity. J Gen Virol 66:395–408. doi: 10.1099/0022-1317-66-3-395. [DOI] [PubMed] [Google Scholar]
- 35.Mazzon M, Castro C, Thaa B, Liu L, Mutso M, Liu X, Mahalingam S, Griffin JL, Marsh M, McInerney GM. 2018. Alphavirus-induced hyperactivation of PI3K/AKT directs pro-viral metabolic changes. PLoS Pathog 14:e1006835. doi: 10.1371/journal.ppat.1006835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim KH, Rumenapf T, Strauss EG, Strauss JH. 2004. Regulation of Semliki Forest virus RNA replication: a model for the control of alphavirus pathogenesis in invertebrate hosts. Virology 323:153–163. doi: 10.1016/j.virol.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 37.Rupp JC, Sokoloski KJ, Gebhart NN, Hardy RW. 2015. Alphavirus RNA synthesis and nonstructural protein functions. J Gen Virol 96:2483–2500. doi: 10.1099/jgv.0.000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wengler G. 2009. The regulation of disassembly of alphavirus cores. Arch Virol 154:381–390. doi: 10.1007/s00705-009-0333-9. [DOI] [PubMed] [Google Scholar]
- 39.Zheng Y, Kielian M. 2015. An alphavirus temperature-sensitive capsid mutant reveals stages of nucleocapsid assembly. Virology 484:412–420. doi: 10.1016/j.virol.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi HK, Lu G, Lee S, Wengler G, Rossmann MG. 1997. Structure of Semliki Forest virus core protein. Proteins 27:345–359. doi:. [DOI] [PubMed] [Google Scholar]
- 41.Yang D, Buchholz F, Huang Z, Goga A, Chen CY, Brodsky FM, Bishop JM. 2002. Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc Natl Acad Sci U S A 99:9942–9947. doi: 10.1073/pnas.152327299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shan G, Li Y, Zhang J, Li W, Szulwach KE, Duan R, Faghihi MA, Khalil AM, Lu L, Paroo Z, Chan AW, Shi Z, Liu Q, Wahlestedt C, He C, Jin P. 2008. A small molecule enhances RNA interference and promotes microRNA processing. Nat Biotechnol 26:933–940. doi: 10.1038/nbt.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mathur K, Anand A, Dubey SK, Sanan-Mishra N, Bhatnagar RK, Sunil S. 2016. Analysis of chikungunya virus proteins reveals that nonstructural proteins nsP2 and nsP3 exhibit RNA interference (RNAi) suppressor activity. Sci Rep 6:38065. doi: 10.1038/srep38065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu R, Folimonov A, Shintaku M, Li WX, Falk BW, Dawson WO, Ding SW. 2004. Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc Natl Acad Sci U S A 101:15742–15747. doi: 10.1073/pnas.0404940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anandalakshmi R, Pruss GJ, Ge X, Marathe R, Mallory AC, Smith TH, Vance VB. 1998. A viral suppressor of gene silencing in plants. Proc Natl Acad Sci U S A 95:13079–13084. doi: 10.1073/pnas.95.22.13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jose J, Snyder JE, Kuhn RJ. 2009. A structural and functional perspective of alphavirus replication and assembly. Future Microbiol 4:837–856. doi: 10.2217/fmb.09.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manfre AJ, Simon AE. 2008. Importance of coat protein and RNA silencing in satellite RNA/virus interactions. Virology 379:161–167. doi: 10.1016/j.virol.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 48.Korber S, Shaik Syed Ali P, Chen JC. 2009. Structure of the RNA-binding domain of Nodamura virus protein B2, a suppressor of RNA interference. Biochemistry 48:2307–2309. doi: 10.1021/bi900126s. [DOI] [PubMed] [Google Scholar]
- 49.Miao M, Yu F, Wang D, Tong Y, Yang L, Xu J, Qiu Y, Zhou X. 2019. Proteomics profiling of host cell response via protein expression and phosphorylation upon dengue virus infection. Virol Sin 34:549–562. doi: 10.1007/s12250-019-00131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dai S, Zhang T, Zhang Y, Wang H, Deng F. 2018. Zika virus baculovirus-expressed virus-like particles induce neutralizing antibodies in mice. Virol Sin 33:213–226. doi: 10.1007/s12250-018-0030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]