Although Epstein-Barr virus (EBV) BFRF1 and BFLF2 are homologs of conserved viral nuclear egress complex (NEC) in all human herpesviruses, unique amino acid sequences and functions were identified in both proteins. In this study, the nuclear targeting and BFRF1-interacting domains were found within the N terminus of BFLF2. We showed that amino acids (aa) 82 to 106 are the major region required for BFLF2 to interact with BFRF1. However, the coimmunoprecipitation (Co-IP) data and glutathione transferase (GST) pulldown experiments revealed that multiple regions of both proteins contribute to reciprocal interactions. Different from the canonical nuclear localization signal (NLS) in other herpes viral homologs, BFLF2 contains a novel importin-dependent nuclear localization signal, including R47, K50, and R52 and several neighboring discontinuous arginine and histidine residues. Using a bacmid complementation system, we show that both the nuclear targeting and the novel nuclear localization signal within aa 82 to 106 of BFLF2 are required for virion secretion.
KEYWORDS: BFLF2, BFRF1, EBV, Epstein-Barr virus, importin beta, NLS, nuclear egress, nuclear egress complex, nuclear localization signal, herpesviruses
ABSTRACT
Epstein-Barr virus (EBV) genomic DNA is replicated and packaged into procapsids in the nucleus to form nucleocapsids, which are then transported into the cytoplasm for tegumentation and final maturation. The process is facilitated by the coordination of the viral nuclear egress complex (NEC), which consists of BFLF2 and BFRF1. By expression alone, BFLF2 is distributed mainly in the nucleus. However, it colocalizes with BFRF1 at the nuclear rim and in cytoplasmic nuclear envelope-derived vesicles in coexpressing cells, suggesting temporal control of the interaction between BFLF2 and BFRF1 is critical for their proper function. The N-terminal sequence of BFLF2 is less conserved than that of alpha- and betaherpesvirus homologs. Here, we found that BFLF2 amino acids (aa) 2 to 102 are required for both nuclear targeting and its interaction with BFRF1. Coimmunoprecipitation and confocal analysis indicated that aa 82 to 106 of BFLF2 are important for its interaction with BFRF1. Three crucial amino acids (R47, K50, and R52) and several noncontinuous arginine and histidine residues within aa 59 to 80 function together as a noncanonical nuclear localization signal (NLS), which can be transferred onto yellow fluorescent protein (YFP)-LacZ for nuclear targeting in an importin β-dependent manner. Virion secretion is defective in 293 cells harboring a BFLF2 knockout EBV bacmid upon lytic induction and is restored by trans-complementation of wild-type BFLF2, but not NLS or BFRF1-interacting defective mutants. In addition, multiple domains of BFRF1 were found to bind BFLF2, suggesting multiple contact regions within BFRF1 and BFLF2 are required for proper nuclear egress of EBV nucleocapsids.
IMPORTANCE Although Epstein-Barr virus (EBV) BFRF1 and BFLF2 are homologs of conserved viral nuclear egress complex (NEC) in all human herpesviruses, unique amino acid sequences and functions were identified in both proteins. In this study, the nuclear targeting and BFRF1-interacting domains were found within the N terminus of BFLF2. We showed that amino acids (aa) 82 to 106 are the major region required for BFLF2 to interact with BFRF1. However, the coimmunoprecipitation (Co-IP) data and glutathione transferase (GST) pulldown experiments revealed that multiple regions of both proteins contribute to reciprocal interactions. Different from the canonical nuclear localization signal (NLS) in other herpes viral homologs, BFLF2 contains a novel importin-dependent nuclear localization signal, including R47, K50, and R52 and several neighboring discontinuous arginine and histidine residues. Using a bacmid complementation system, we show that both the nuclear targeting and the novel nuclear localization signal within aa 82 to 106 of BFLF2 are required for virion secretion.
INTRODUCTION
Epstein-Barr virus (EBV) is a ubiquitous gammaherpesvirus that infects most of the human population worldwide. EBV is also associated with several neoplastic diseases, including Burkitt’s lymphoma, Hodgkin’s lymphoma, nasopharyngeal carcinoma, and a subset of gastric cancers (for a review, see reference 1). After mild or asymptomatic primary infection, EBV becomes latent in the host and can be reactivated in response to various stimuli. The immediate early transactivators, Zta and Rta, can turn on the early genes for viral DNA replication in the nucleus and, later, the structural proteins for viral nucleocapsid assembly and virus maturation. In the late replication stage, the intact nuclear envelope structure is a major barrier that the virus must overcome for translocation of viral nucleocapsids from the nucleus into the cytoplasm for tegumentation, final envelopment, and release (2).
The nuclear envelope is composed of two lipid bilayers, a perinuclear space between the inner and outer nuclear membranes (INM and ONM, respectively) with a width around 100 to 500 Å, and the internal protein mesh network (nuclear lamina). These structures are held together by the nuclear pore complexes (NPCs), which also function as gates for material transport between the cytoplasm and the nucleus (3). Each NPC comprises a vast protein complex, with an approximate molecular weight of 125 MDa, and is assembled from a class of proteins named “nucleoporins.” The nucleoporins contain phenylalanine-glycine (FG) repeats and can function as receptors for transferring macromolecules into the nucleus via an energy-dependent mechanism (4). The classical nuclear localization signal (NLS) is composed of either one or two short sequences of basic-rich amino acids (5, 6), such as “PKKKRRV” in SV40 large T antigen and “KRPAATKKAGQAKKKK” in the protein nucleoplasmin (6, 7). In the canonical nuclear import cycle, importin α recognizes the NLS-containing protein cargos in the cytoplasm and interacts with importin β, which regulates the interaction with FG-containing nucleoporins to pass through the NPC (8).
After viral DNA replication and packaging into procapsids, the nuclear envelope becomes an obstacle for nucleocapsids to egress from the nucleus into the cytoplasm for the subsequent maturation process. The average size of the nuclear pores is approximately 40 nm, whereas the herpesvirus nucleocapsids are more than 100 nm. Herpesviruses, therefore, have evolved with novel mechanisms to reorganize the nuclear envelope structure for the transport of nucleocapsids. The nuclear egress of alphaherpesvirus, facilitated by the nuclear egress complex (NEC; UL34/UL31), is the most well studied. With a single transmembrane domain, UL34 is believed to translocate from the rough endoplasmic reticulum (ER) after protein synthesis, move through the NPC, and become distributed at the inner nuclear membrane (9, 10). In the absence of UL34, UL31 is a phosphorylated protein that is expressed predominantly in the nucleus (11). In addition to interacting with UL34, UL31 plays multiple roles in the herpes simplex virus 1 (HSV-1) life cycle, including viral DNA replication, packaging, cleavage, and nuclear egress of nucleocapsids (12, 13). The NEC component pair UL34/UL31 is conserved in all human herpesviruses, including UL50/UL53 in human cytomegalovirus (HCMV) and BFRF1/BFLF2 in EBV (14, 15).
Coupled with NEC formation, viral or cellular kinases are involved in the phosphorylation of lamin A/C, leading to the partial disassembly of the nuclear lamina during herpesvirus nuclear egress. Our previous studies indicated that EBV BGLF4 kinase phosphorylates lamin A/C in a CDK1-mimicking manner to cause partial disassembly of the nuclear lamina (16). We found that the ESCRT system is recruited by the EBV nuclear egress protein BFRF1 and Alix complex to form intracytoplasmic vesicles, facilitating the transport of nucleocapsids across the nuclear envelope (17). Based on imaging studies, we postulate that some EBV nucleocapsids egress from the nucleus through double-membrane structures that may contain nuclear lamin A/C and NPC. Ubiquitination of BFRF1 and Itch E3 ligase regulates the ability of BFRF1 to form vesicles (18). On the other hand, BFRF1 is a pleiotropic protein that with multiple functions; its knockout bacmid carries out normal viral DNA replication but is defective for DNA cleavage and packaging into capsids (14). However, the precise functional domains within BFLF2 have not been characterized.
Compared to the well-studied nuclear egress of alphaherpesviruses, which includes three distinct steps, primary envelopment, de-envelopment, and secondary envelopment, the nuclear egress of EBV remains enigmatic. By sequence alignment and structural prediction analysis, unique sequences, distinct from other herpesviruses, have been identified in BFRF1 and BFLF2. Here, we sought to identify the nuclear targeting and BFRF1-interacting domains of BFLF2 and characterize their contribution to the virus maturation process.
RESULTS
The nuclear localization signal and BFRF1-interacting domain of BFLF2 are located within the region aa 2 to 102.
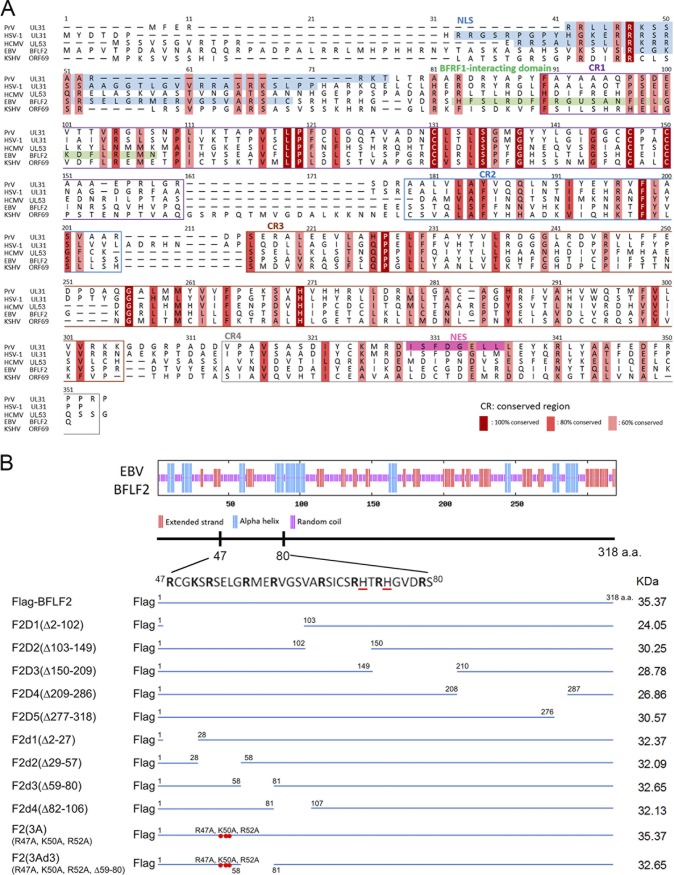
To identify the nuclear targeting and BFRF1-interacting domains, the secondary structures of BFLF2 were predicted by aligning its amino acid sequence with those of other HSV-1 UL31 homologs, including UL31 of pseudorabies virus (PrV), UL53 of HCMV, and ORF69 of another gammaherpesvirus, Kaposi's sarcoma-associated herpesvirus (KSHV) (Fig. 1A). The predicted nuclear localization signals are located in the N termini of several UL31 homologs. For example, two regions with positively charged amino acids, 21RRRSR25 and 35RRASRK40, were identified as a classical bipartite NLS in HSV-1 UL31 (19). Although UL31 homologs share some similar features, the N-terminal region is much less conserved within EBV BFLF2 (Fig. 1A). Several positively charged amino acids, such as lysines and arginines, are located within amino acids (aa) 2 to 102 (Fig. 1A and B, top), suggesting an NLS may be present in the N terminus of BFLF2.
FIG 1.
Schematic presentation of the predicted secondary structures of BFLF2 and Flag-BFLF2 wild-type and mutant constructs. (A) Protein sequence alignment of BFLF2 with herpesviral homologs. Sequence alignment of BFLF2 homologs; amino acid sequences of pseudorabies virus (PrV) (GenBank/EMBL/DDBJ accession number Q911V7), HSV-1 (P10215) UL31, HCMV pUL53 residues 1 to 292 (P16794), EBV BFLF2 (P03183), and KSHV ORF69 (YP_001129427) were compared using the ClustalW alignment tool of Geneious 8.0.5 (Biomatters, Ltd., Auckland, New Zealand). Homology between the different UL31 homologs is indicated by the following color scheme: dark red shading with white letter, consensus derived from completely conserved residues with a threshold of three; tangerine shading, consensus about 80% conservation; light red shading, consensus about 60% conservation. In this schematic representation of UL31 homologs alignment, the NLSs are highlighted by an azure background. The putative BFRF1-interacting domain is marked with a green box. In addition, conserved regions CR1, CR2, CR3, and CR4 are indicated in purple, blue, brown, and gray boxes, respectively. The identified nuclear export signal (NES) of PrV UL31 is marked by pink shading. (B) The secondary structure of BFLF2 was analyzed by the GOR prediction program (http://www.expasy.ch/tools/). Random coils, alpha helices, and extended strands are indicated as purple, blue, and red lines, respectively. Amino acid sequences between aa 47 and 80 of the BFLF2 gene are shown at the bottom. Discontinuous basic amino acids, arginine (R) or lysine (K), are indicated and histidine (H) is highlighted by red underlines. Serial deletions of Flag-BFLF2 mutants, including F2D1 (Δ2-102), F2D2 (Δ103-149), F2D3 (Δ150-209), F2D4 (Δ209-286), and F2D5 (Δ277-318), and F2d1 (Δ2-27), F2d2 (Δ29-57), F2d3 (Δ59-80), and F2d4 (Δ82-106), were generated by single primer mutagenesis. The 3A mutant had arginine 47, lysine 50, and arginine 52 changed into alanines and was used as the template to generate the 3Ad3 mutant with a deletion of d3 (Δ59-80). Predicted molecular weight of each construct is indicated at the right.
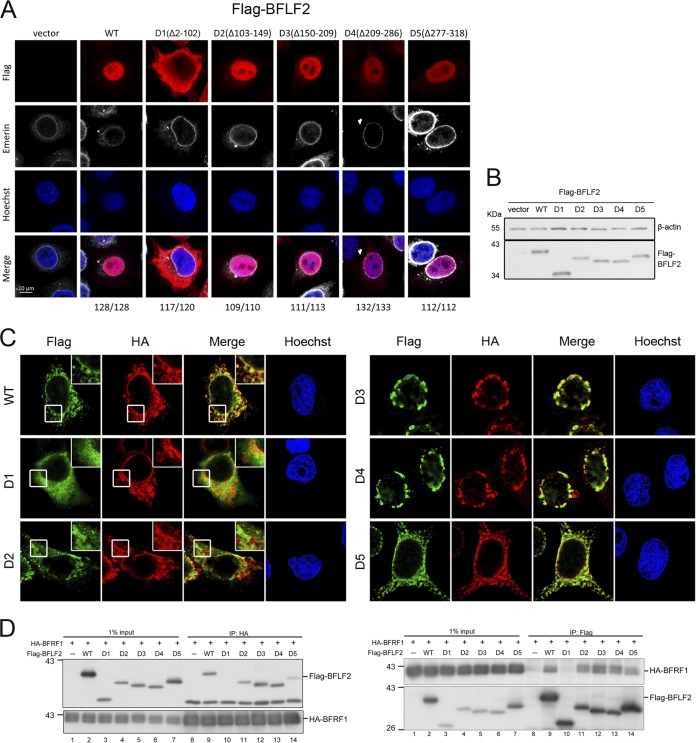
To map the regions contributing to the BFLF2 nuclear localization and BFRF1 interaction, we constructed systemic truncated mutants of BFLF2, based on the conserved region (CR) of alignment data and predicted secondary structure (Fig. 1A and B). First, Flag-tagged F2D1 (Δ2-102 aa), F2D2 (Δ103-149 aa), F2D3 (Δ150-209 aa), F2D4 (Δ209-286 aa), or F2D5 (Δ277-318 aa) was transfected into HeLa cells. Using emerin staining to indicate the nuclear envelope, we observed from confocal images that Flag-F2D1, but not other mutants, was defective in nuclear localization in transiently transfected cells (Fig. 2A and B). Coexpression of hemagglutinin (HA)-BFRF1 and Flag-BFLF2 induced the formation of nuclear envelope-derived cytoplasmic vesicles as we previously described (Fig. 2C and reference 17). When coexpressed with HA-BFRF1, Flag-F2D1 showed the cytoplasmic diffuse pattern, whereas HA-BFRF1 was detected at the nuclear rim and in the cytoplasmic vesicles, similarly to its expression alone. It suggests that Flag-F2D1 was defective in BFRF1 interaction and nuclear targeting. When coexpressed with HA-BFRF1, Flag-F2D2 showed irregular clustering patterns and partially colocalized with the cytoplasmic BFRF1 signals, suggesting that aa 103 to 149 of BFLF2 are involved in proper vesicle formation. On the other hand, some BFRF1/BFLF2 complexes seemed to be trapped on the nuclear envelope when cell coexpressed HA-BFRF1 and Flag-F2D3 or Flag-F2D4, suggesting these regions between aa 150 to 209 and aa 209 to 286 contribute to the function of NEC to generate nuclear envelope-derived cytoplasmic vesicles (Fig. 2C). Flag-F2D5 displayed a colocalization pattern with BFRF1 in cytoplasmic vesicles, similar to that of wild-type BFLF2. The same setting of coexpression lysates was used for reciprocal coimmunoprecipitations, and the data indicate that Flag-F2D1 lost the ability to interact with HA-BFRF1 in both directions of immunoprecipitation (Fig. 2D, lane 10 in both panels). Interestingly, Flag-F2D5 showed a weaker interaction with Flag-BFRF1, suggesting aa 277 to 318 may have an accessory function for the interaction (Fig. 2D, lane 14 in both panels). Taken together, aa 2 to 102 of BFLF2 are required for both nuclear targeting and interaction with BFRF1, whereas amino acids between 150 and 209 or aa 209 and 286 are required for proper formation of nuclear enveloped-derived cytoplasmic vesicles containing EBV nuclear egress complexes. Amino acids 277 to 318 of BFLF2 may play an accessory function for BFRF1 interaction.
FIG 2.
The region aa 2 to 102 of BFLF2 is required for nuclear targeting and interaction with BFRF1. (A) Plasmids expressing Flag-BFLF2 wild type, F2D1, F2D2, F2D3, F2D4, or F2D5 were transfected into slide-cultured HeLa cells. At 24 h posttransfection (hpt), cells were fixed with 4% paraformaldehyde, immunostained for Flag (red) and emerin (white), stained with Hoechst 33258 to indicate cellular DNA (blue), and analyzed by confocal microscopy. The cell numbers showing the representative pattern are indicated at the bottom. (B) At the same time, a portion of cell lysates was analyzed by immunoblotting against Flag and β-actin. The experiments shown in panels A and B were performed 3 times. Representative data from three independent experiments are shown. (C) Plasmids expressing HA-BFRF1 together with Flag-BFLF2 wild type, mutant F2D1 (Δ2-102), F2D2 (Δ103-149), F2D3 (Δ150-209), F2D4 (Δ209-286), or F2D5 (Δ277-318) were transfected into slide-cultured HeLa cells. At 24 hpt, the cells were fixed, immunostained for Flag (green) and HA (red), stained with Hoechst 33258 to indicate cellular DNA (blue), and analyzed by confocal microscopy. Cells showing representative staining patterns are displayed. The insets of Flag-WT, Flag-F2D1, and Flag-F2D2 are the enlarged images to show different cytoplasmic staining patterns. (D) The HA-BFRF1 plasmid was transfected with Flag-BFLF2 wild type, F2D1, F2D2, F2D3, F2D4, or F2D5 into HeLa cells. The lysates were immunoprecipitated with antibody against Flag and HA. The immune complexes were then resolved by SDS-PAGE and immunoblotted with antibodies against Flag or HA. Representative data from two independent experiments are shown.
Amino acids 28 to 58 are most critical for the nuclear localization of BFLF2, and the region aa 81 to 107 in BFLF2 is required for interaction with Flag-BFRF1.
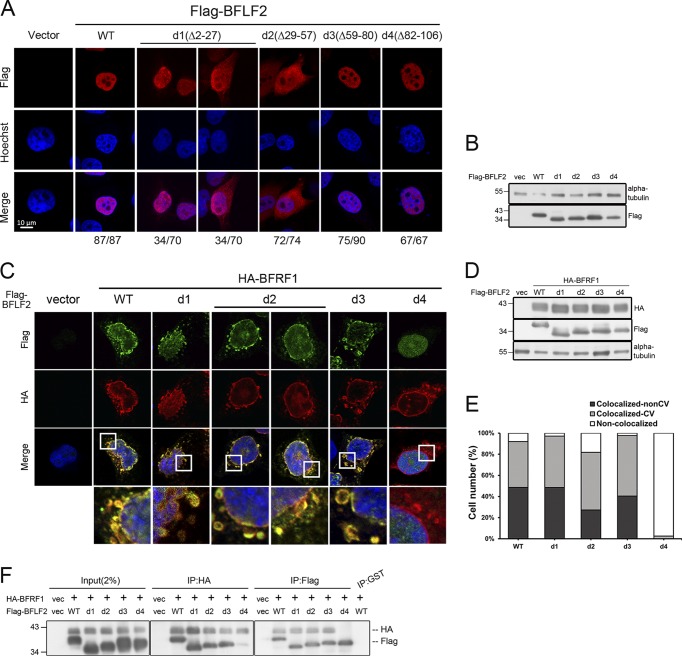
We noticed that the N terminus of BFLF2 is less conserved in the sequence alignment with other homologs for its extended N-terminal residues (Fig. 1A). The HSV-1 bipartite NLS 21RRRSR25 and 35RRASRK40 were then aligned to the region of BFLF2, 47 RCGKSRSELGRMERVGSVARSICSRHTRHGVDRS80, which contains discontinuous basic amino acids and two histidines (in boldface font). To dissect the BFLF2 nuclear localization signal and BFRF1-interacting domain within aa 2 to 102, a series of four deletion mutants were generated, including F2d1 (Δ2-27 aa), F2d2 (Δ29-57 aa), F2d3 (Δ59-80 aa), and F2d4 (Δ82-106 aa). We wondered either a monopartite or a bipartite NLS may be present within aa 29 to 57 or aa 58 to 81 of BFLF2, since these regions are lysine rich and arginine rich, and the region of aa 82 to 106 may contribute to interaction with BFRF1 because of its predicted α helical structure (Fig. 1B, top). To this end, HeLa cells were transfected with plasmids expressing Flag-BFLF2 wild type, F2d1, F2d2, F2d3, or F2d4 and analyzed by confocal microscopy to reveal the subcellular distribution pattern (Fig. 3A and B). At 24 h posttransfection, F2d3 and F2d4 were detected mainly in the nucleus, similarly to Flag-BFLF2 wild type; whereas Flag-F2d2 showed enhanced cytoplasmic distribution and weak nuclear staining in 72 of 74 cells, suggesting the absence of aa 29 to 57 significantly affected the nuclear targeting of BFLF2 (Fig. 3A and B). Interestingly, we found the deletion of aa 2 to 27 slightly affected BFLF2 nuclear localization (34/70 transfected cells retained the nuclear distribution, while 34/70 cells showed major nuclear distribution with weak cytoplasm signal), suggesting aa 2 to 27 may play an accessory role in the nuclear targeting of BFLF2.
FIG 3.
Amino acids 29 to 57 of BFLF2 are required for nuclear targeting and aa 81 to 107 of BFLF2 are required for BFRF1 interaction. (A) Plasmid pcDNA3.0 or plasmid expressing Flag-BFLF2 wild type, F2d1, F2d2, F2d3, and F2d4 were transfected individually into slide-cultured HeLa cells. At 24 hpt, cells were fixed and stained for Flag (red) and cellular DNA (blue). The images were analyzed by confocal microscopy. The cells with intranuclear or cytoplasmic distribution of BFLF2 were counted. The representative patterns are displayed. (B) Lysates from cells shown in panel A were harvested and analyzed by immunoblotting with antibodies against Flag and α-tubulin, which served as a loading control. (C) Plasmid expressing HA-BFRF1 was transfected with Flag-BFLF2 wild type or serial small-deletion mutants into slide-cultured HeLa cells. At 24 hpt, slides were fixed, stained with antibody against Flag (green) and HA (red), stained with Hoechst indicating cellular DNA (blue), and analyzed by confocal microscopy. (D) Lysates from cells shown in panel C were analyzed by immunoblotting against Flag, HA, and α-tubulin. Both experiments shown in panels C and D were performed two times, and representative data are shown. (E) Bar graph shows the percentages of cells with subcellular colocalization of BFRF1 and BFLF2 at the nuclear envelope or within cytoplasmic vesicles (CV; n = 22 to 42) from the experiment shown in panel C. (F) Plasmid expressing HA-BFRF1 was transfected with Flag-BFLF2 wild type or serial deletion mutants into HeLa cells. At 24 hpt, the lysates were harvested and immunoprecipitated with antibody against HA (top) or Flag (bottom). The immune complexes were then resolved by 10% SDS-PAGE and immunoblotted with antibodies against HA or Flag. The experiment was performed three times, and representative data from three independent experiments are shown.
The region aa 82 to 106 in BFLF2 is required for interaction with Flag-BFRF1.
To map the region required for interaction with BFRF1, Flag-BFLF2 wild type, F2d1, F2d2, F2d3, or F2d4 was transfected together with HA-BFRF1 into slide-cultured HeLa cells. At 24 h posttransfection (hpt), the cells were fixed, stained, and analyzed by confocal microscopy. We observed that Flag-F2d1 and F2d3 colocalized with Flag-BFRF1 at the nuclear rim or in the cytoplasmic vesicles of HeLa cells, similarly to BFLF2 wild type. Of note, two staining patterns of F2d2 with Flag-BFRF1 were observed. Approximately one-half of the cells contained smooth cytoplasmic vesicles, whereas the other half of the cells contained some intracytoplasmic aggregates. Thus, F2d4 distributed in the nucleus and did not colocalize with Flag-BFRF1 (Fig. 3C and D). The percentage of cells showing the colocalization of BFRF1 and Flag-F2d4 was significantly decreased (Fig. 3E), indicating that aa 82 to 106 are required for the colocalization of Flag-BFLF2 with HA-BFRF1. In addition, coimmunoprecipitation was performed using cotransfected cell lysates to examine the interaction between Flag-BFLF2 and HA-BFRF1. Reciprocal coimmunoprecipitations and immunoblotting further indicated that F2d4 interacts with HA-BFRF1 weakly compared to that with wild-type Flag-BFLF2 and other deletion mutants, suggesting sequences between aa 82 to 106 of BFLF2 are required for proper interaction with BFRF1 (Fig. 3F).
Amino acids 47R, 50K, and 52R act coordinately with aa 57 to 80 of BFLF2 for the nuclear targeting of BFLF2.
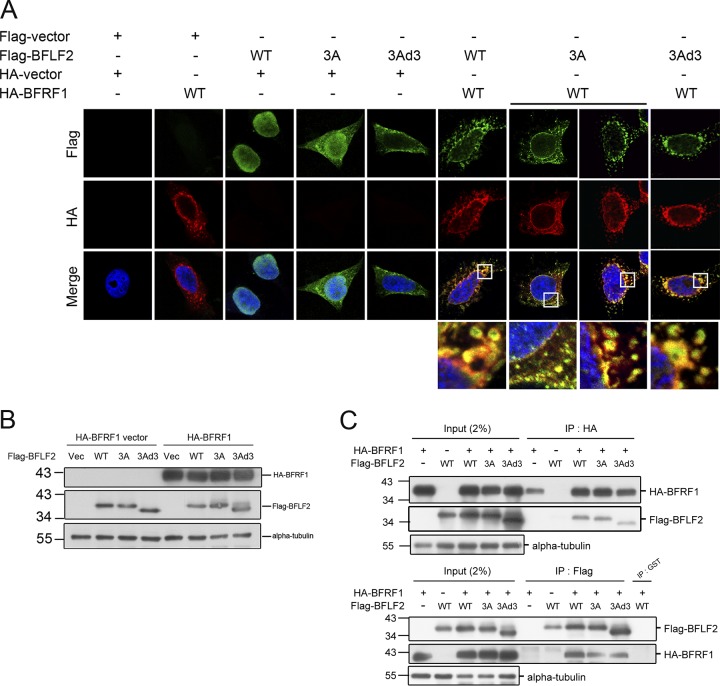
To further characterize the NLS of BFLF2, we substituted three positively charged amino acids within the d2 region, lysine 47, arginine 50, and lysine 52, with alanine residues, which was designated F2(3A) (R47, 52A, K50A). With similar expression levels in immunoblotting, F2(3A) was expressed in both the nucleus and cytoplasm in confocal analysis (Fig. 4A and B), similarly to F2d2. With the defect of nuclear localization, F2(3A) colocalized with HA-BFRF1 at the nuclear rim and in some cytoplasmic signals surrounding the nucleus. Notably, some green puncta of Flag-F2(3A) did not colocalize with red HA-BFRF1, suggesting these three amino acids (arginine47, lysine50, and arginine52) are not only crucial for BFLF2 nuclear localization but also important for its proper incorporation into BFRF1-containing vesicles (Fig. 4A and B, see enlarged insets in A). In addition, some of the cytoplasmic colocalized signals in F2(3A)-expressing cells were much smaller than those of BFLF2(wild type [WT])-expressing cells, and these cytoplasmic yellow signals in F2(3A)-expressing cells did not form proper vesicles, suggesting some interactions between Flag-BFLF2(3A) and HA-BFRF1 may occur in the cytoplasm.
FIG 4.
Amino acids R47, K50, and R52 and the region aa 59 to 80 are required for the nuclear localization of Flag-BFLF2, but not for BFRF1 interaction. (A) The plasmid expressing Flag-BFLF2 wild type, F2(3A) (R47A, K50A, and R52A mutant), or F2(3Ad3) (3A mutant and aa 59 to 80 deletion) was transfected with a plasmid expressing HA-BFRF1 into slide-cultured HeLa cells. At 24 hpt, the cells were fixed and stained for Flag (green), HA (red), and cellular DNA (with Hoechst 33258; blue). The slides were analyzed by confocal microscopy. Cells showing representative staining patterns are displayed. (B) The lysates from cells with the same setting as that for panel A were harvested and analyzed by immunoblotting against Flag, HA, and alpha-tubulin. (C) A plasmid expressing HA-BFRF1 was transfected with a plasmid expressing Flag-BFLF2 wild type, F2(3A), or F2(3Ad3) into HeLa cells. The lysates were immunoprecipitated with antibody against HA or Flag. The immune complexes were then resolved and immunoblotted with antibodies against Flag or HA. The experiment was performed two times, and representative data from two independent experiments are shown.
Considering that F2(3A) distributed partially in the nucleus, we thought that additional residues may coordinately regulate the nuclear localization of BFLF2. To this end, another BFLF2 mutant with mutations of arginine47, lysine50, and arginine52 and a deletion of aa 57 to 80 was generated as F2(3Ad3). In the confocal analysis, Flag-F2(3Ad3) was totally defective for nuclear localization when expressed alone, suggesting the nuclear localization of BFLF2 is regulated by a long stretch of sequences (Fig. 4A and B). However, when coexpressed with HA-BFRF1, F2(3Ad3) colocalized with HA-BFRF1 in a portion of irregular cytoplasm vesicles and at the nuclear rim, suggesting the interaction may occur outside the BFRF1-containing vesicles in the cytoplasm or at the cytoplasmic facet of the nuclear envelope (Fig. 4A). Furthermore, reciprocal immunoprecipitations showed that Flag-F2(3A) and Flag-F2(3Ad3) interacted with HA-BFRF1 (Fig. 4C). The results here suggest that these three amino acids (arginine47, lysine50, and arginine52), together with aa 57 to 80, may form a noncanonical NLS for BFLF2, and nuclear localization of BFLF2 was not essential for its interaction with BFRF1. Different from other herpes NEC homologs for which both proteins need to interact at the internal nuclear membrane before the production of cytoplasmic vesicles, it is very unique that BFRF1 expression alone can form cytoplasmic vesicles containing inner nuclear membrane marker emerin and nuclear pore complexes (NPCs) (17).Therefore, it is possible that the cytoplasmic retaining Flag-F2(3Ad3) is recruited to a portion of BFRF1-containing vesicles in the cytoplasm.
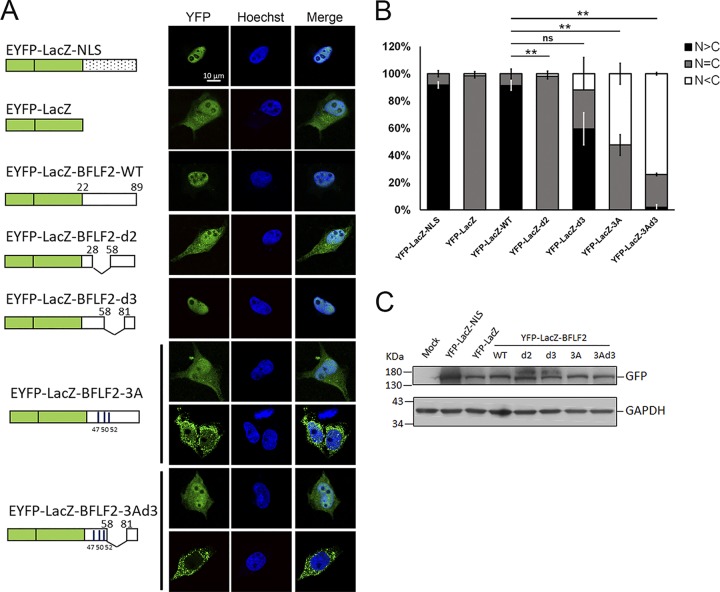
NLS of BFLF2 can be transferred onto the EYFP-LacZ reporter for nuclear targeting.
To test whether the noncanonical NLS of BFLF2 can be transferred onto another protein to promote nuclear targeting, the DNA fragments encoding aa 22 to 89 of wild-type BFLF2 or various nuclear targeting mutants were fused to enhanced yellow fluorescent protein (EYFP)-LacZ plasmid and transfected into HeLa cells to detect their subcellular distribution (Fig. 5). With the EYFP-LacZ fused to SV40 NLS as a positive control, wild-type EYFP-LacZ-BFLF2 showed a predominantly nuclear distribution compared to the diffuse pattern of EYFP-LacZ (Fig. 5A). With a similar protein expression level (Fig. 5B), EYFP-LacZ-BFLF2-d2 was distributed in both the cytoplasm and the nucleus, whereas the majority of EYFP-LacZ-BFLF2-d3 showed more nuclear distribution (72%) (Fig. 5B). For EYFP-LacZ-BFLF2-3A, two different populations were observed: approximately 55% of the cells showed a diffuse staining pattern, whereas approximately 45% of cells had a predominantly cytoplasmic distribution of EYFP signals. For EYFP-LacZ-BFLF2-3Ad3, the majority of cells (70%) contained exclusively cytoplasmic distribution of YFP signals (Fig. 5A). The proportion with cytosol-only localization was increased in EYFP-LacZ-BFLF2-3A- and EYFP-LacZ-BFLF2-3Ad3-expressing cells (Fig. 5B). Thus, we confirmed that the noncanonical NLS of BFLF2 can be transferred onto the reporter protein to function for nuclear targeting. The 3 amino acids 47R, 50K, and 52R and the region of aa 57 to 80 of BFLF2 function additively for nuclear translocation.
FIG 5.
Amino acids R47, K50, and R52 and the region aa 57 to 80 coordinately regulate the nuclear localization of EYFP-LacZ. (A) Plasmids expressing EYFP-LacZ-BFLF222–89 (wild type), BFLF222–89d2 (Δ29-57), BFLF222–89d3 (Δ59-80), BFLF222–893A (R47A,K50A,R52A), or BFLF222–893Ad3 (R47A,K50A,R52A, Δ59-80) were transfected into slide-cultured HeLa cells. EYFP-Lac-NLS containing the SV40 NLS and EYFP served as positive and negative controls for showing the subcellular distribution of YFP signals. At 24 hpt, the cells were fixed with 4% paraformaldehyde and stained with Hoechst 33258 to indicate cellular DNA (blue), and the YFP signals (green) were analyzed by confocal microscopy. Representative staining patterns are displayed. The experiment was performed two times, and representative data are shown. (B) The bar graph shows the percentages of subcellular distribution of YFP-LacZ in cells. Error bars were calculated with data from two independent experiments. The statistically significant differences were calculated by the paired Student's t test and are indicated at the top. **, P < 0.01; ns, no significant differences. (C) Lysates from cells with the same settings as that for panel A were harvested and analyzed by immunoblotting against GFP and GAPDH to indicate similar expression levels of different constructs.
Nuclear translocation of BFLF2 depends on importin β, and the NLS-directed nuclear accumulation of BFLF2 is enhanced by low-pH buffer treatment.
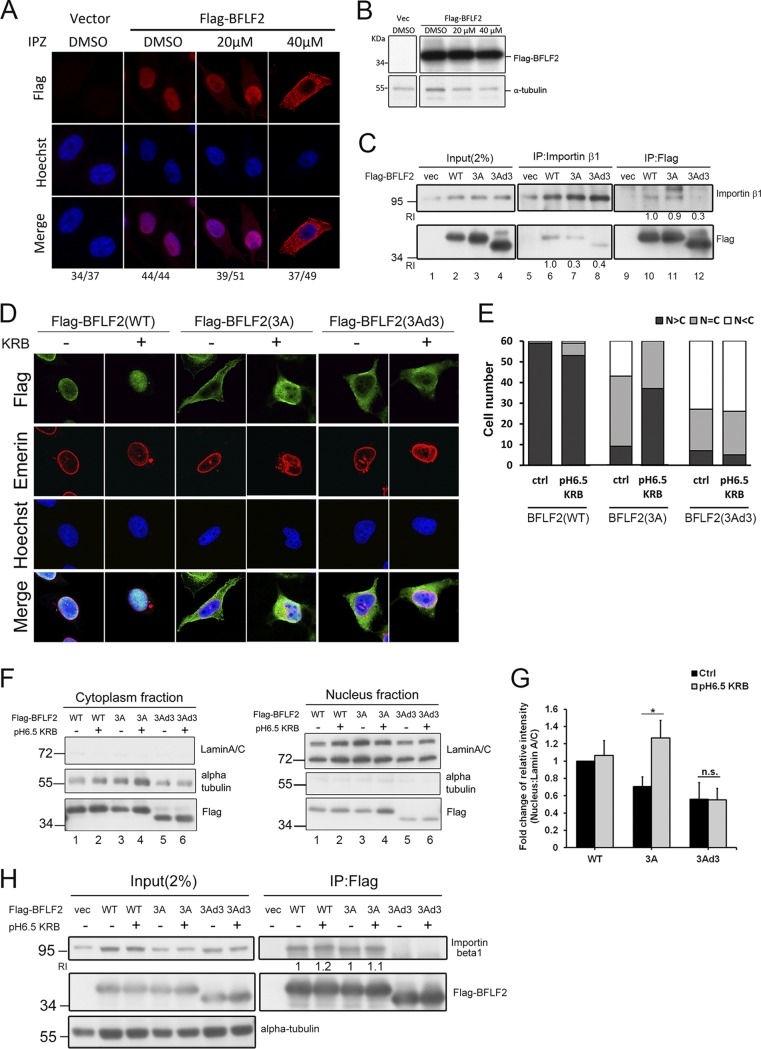
We were then curious to know whether this nonclassical NLS in BFLF2 functions in the importin β-dependent nuclear transport mechanism. A recently developed importin β inhibitor, importazole (IPZ) (20), which specifically blocks importin β-mediated nuclear import, was used to examine the nuclear targeting of BFLF2. Under 20 μM IPZ treatment, approximately one-half of Flag-BFLF2-expressing cells showed a nucleus staining pattern compared to dimethyl sulfoxide (DMSO) control-treated cells showing 100% nuclear distribution of Flag-BFLF2 in confocal image analysis. In the presence of 40 μM IPZ, most of the cells showed cytoplasmic distribution of Flag-BFLF2 (76% [37/49]) (Fig. 6A and B). It indicates that BFLF2 may contain a noncanonical NLS which is composed of discontinuous positively charged amino acids and has a different composition from the classical NLS of other herpesviruses. In addition, reciprocal coimmunoprecipitations were performed to confirm the interaction between Flag-BFLF2 and importin β. The importin β coimmunoprecipitated signals of Flag-F2(3A) or Flag-F2(3Ad3) were weaker, and less importin β immunoprecipitated with Flag-F2(3Ad3) (Fig. 6C, lanes 6, 7, 8, 10, 11, and 12). It indicates that the interaction is attenuated for Flag-F2(3A) and is much weaker for Flag-F2(3Ad3) compared to that of wild-type Flag-BFLF2.
FIG 6.
The nuclear localization of BFLF2 is importin β1-dependent, and low-pH KRB buffer solution enhances the nuclear accumulation of BFLF2. (A) Slide-cultured HeLa cells were transfected with vector or Flag-BFLF2. At 6 h posttransfection, IPZ (20 μM, 40 μM) or DMSO was added to the medium. After incubation for another 24 h, the cells were fixed and stained for Flag (red), and cellular DNA was stained with Hoechst 33258 (blue). The cells were analyzed by confocal microscopy. The experiment was performed two times, and representative data are shown. Cells showing representative staining patterns are displayed. (B) Lysates were harvested from cells used for panel A and analyzed by immunoblotting against Flag and α-tubulin. (C) HeLa cells were transfected with Flag-BFLF2, Flag-F2(3A), or Flag-F2(3Ad3), and the lysates were harvested for reciprocal coimmunoprecipitation with Flag or importin β antibodies to indicate the interaction between BFLF2 protein and importin β. The immunocomplexes were then resolved by SDS-PAGE and immunoblotted with antibodies against Flag and importin β1. The experiment was performed two times, and representative data are shown. Relative intensities (RI) of immunoprecipitated Flag-F2(3A) or Flag-F2(3Ad3) adjusted by the protein levels of immunoprecipitated importin β (lanes 7 and 8) were compared to that of Flag-BFLF2(WT) (lane 6). Similar quantitations were performed for coimmunoprecipitation with Flag antibody (lanes 10 to 12). (D) Slide-cultured HeLa cells were transfected with Flag-BFLF2, F2(3A), or F2(3Ad3). At 24 hpt, pH 6.5 KRB buffer or fresh medium was added to the dish. After incubation for 1 h, the cells were fixed and stained for Flag (green), emerin (red), and cellular DNA (with Hoechst 33258; blue). The cells were analyzed by confocal microscopy. Cells showing representative staining patterns are displayed. (E) Bar graph shows the percentages of cells displaying indicated subcellular distribution of BFLF2 (n = 60). (F) Immunoblotting of the lysates from cells used for data shown in panel D was performed using anti-Flag, anti-lamin A/C, or anti-α-tubulin antibodies after subcellular fractionation for cytoplasmic (left) and nuclear (right) fractions. The experiment was performed three times, and representative data are shown. (G) Bar graph shows the fold changes in relative intensities of Flag-BFLF2 compared to that of lamin A/C in the nuclear fraction with non-KRB treatment (Ctrl). Error bars were calculated with data from 3 independent experiments. The statistically significant differences were calculated by the paired Student's t test and are indicated at the top. *, P < 0.05: n.s., no significant differences. (H) Lysates from cells with the same setting as those in panel D were harvested and immunoprecipitated with antibody against importin β1. The immunocomplexes were then resolved by SDS-PAGE and immunoblotted with antibodies against Flag and importin β1. RI indicates the relative intensities of immunoprecipitated importin β adjusted by the protein levels of immunoprecipitated Flag-F2(3A) and compared to that of Flag-BFLF2(WT), which was not treated with pH 6.5 KRB buffer. Similar quantitations were performed for coimmunoprecipitation with Flag antibody.
On the other hand, previous studies indicated that some proteins are imported into the nucleus by interacting with importin β directly through ionic interactions (21). For example, the NLS of c-Met receptor tyrosine kinase relies on histidines, for which the positive charge changes depending on pH, rather than the lysines or arginines. Because the pKa of histidine is 6.0 to 6.2, it can be protonated under the lower pH condition inside the cell (22 – 24). The amount of nuclear c-Met protein and the interaction between the NLS fragment of c-Met and importin β both increased significantly when the cytosolic pH was low. We noticed that there are some histidines in the region aa 47 to 80 of BFLF2. Thus, we speculated that decreased cytosolic pH may increase the average positive charge of the histidine residue(s) and enhance the interaction between importin and the histidine-containing NLS of BFLF2, thereby affecting its nuclear translocation. To figure out whether the protonation of the basic amino acids and histidines in the region aa 47 to 80 of BFLF2 may enhance the interaction with importin β, HeLa cells transfected with Flag-BFLF2, Flag-F2(3A), or Flag-F2(3Ad3) were treated with pH 6.5 Krebs-Ringer bicarbonate (KRB) buffer to decrease the actual cytosolic pH in cells to 6.5 after incubation. After a 1-h treatment, Flag-F2(3A) was imported into the nucleus after the cytosolic pH decreased, whereas Flag-F2(3Ad3) remained in cytoplasm in confocal analysis (Fig. 6D). Cell counting from the confocal image showed that most of Flag-F2(WT) was retained in the nucleus and the number of cells containing nuclear Flag-F2(3A) increased after pH 6.5 KRB buffer treatment, suggesting amino acids within aa 47 to 80 also contribute to nuclear transport of BFLF2 in the absence of 47R, 50K, and 52R. However, the nuclear distribution of Flag-F2(3Ad3) was not further enhanced after the treatment, suggesting the effects of pH 6.5 KRB rely on the presence of aa 47 to 80 (Fig. 6D and E).
Subcellular fractionation was also performed to show the increased nuclear fraction of Flag-F2(3A) after pH 6.5 KRB buffer treatment, whereas the levels of Flag-F2(3Ad3) remained low in the nuclear fraction (Fig. 6F, right, lanes 3 to 6). The proportions of the intranuclear fractions of Flag-BFLF2 WT and Flag-F2(3A), but not Flag-F2(3Ad3), were increased after pH 6.5 KRB treatment (Fig. 6G). Furthermore, the level of importin β1 coimmunoprecipitated with Flag-BFLF2 was slightly increased in both Flag-F2(WT) and Flag-F2(3A) groups, but not in Flag-F2(3Ad3) after acidic buffer treatment (Fig. 6H). Even though the pH 7.4 lysis buffer may reverse the low-pH effect and attenuate the enhanced interaction, the quantitated data showed that the signal of the 3A mutant increased slightly compared to that of the untreated control. The data presented here indicate that the importin β-dependent pathway is required for the nuclear targeting of BFLF2. Treatment with pH 6.5 KRB enhanced the nuclear accumulation of BFLF2, suggesting histidines at aa 71 and 74 may contribute additively to the nuclear targeting of BFLF2.
Multiple regions of BFRF1 interact with BFLF2 and are required to bring BFLF2 out of the nucleus to cytoplasmic vesicles.
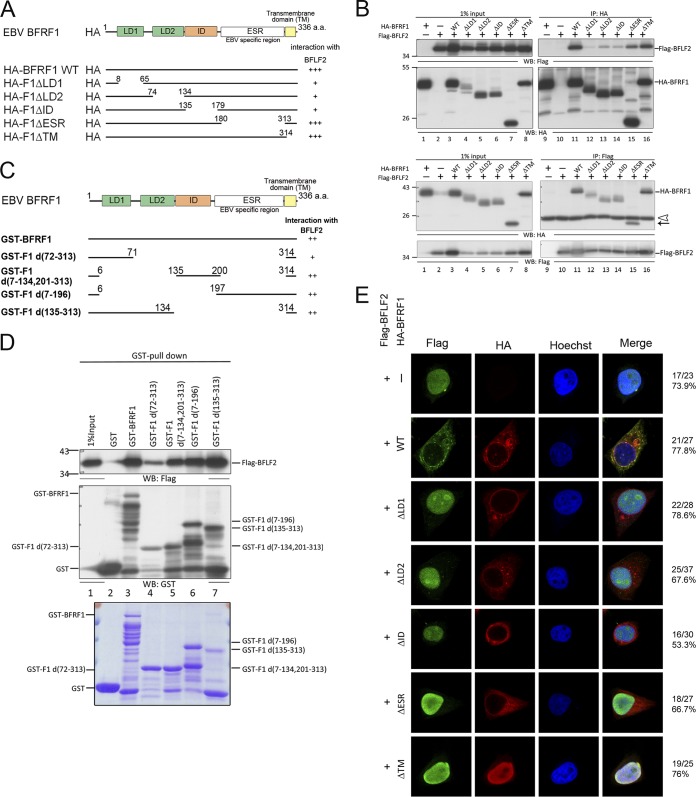
We noticed that in addition to Flag-F2D1, F2D5 also displayed weaker signals in coimmunoprecipitation assays (Fig. 2D). Because the interaction of NEC proteins during the nuclear egress process may involve multiple conformational changes, it is possible that multiple domains of NEC proteins may interact with each other at different stages. Through the protein alignment of BFRF1 with other herpesviral homologs, we had predicted a possible BFLF2-interacting domain (ID) located at the region of aa 135 to 179 (17). To confirm the prediction, serial deletion mutants of HA-BFRF1 (Fig. 7A) and Flag-BFLF2 were transfected into HeLa cells. The functional domains of BFRF1 were designated as in a previous study (17): LD1 and LD2 were two predicted domains that are involved in ESCRT recruitment, whereas the EBV-specific region (ESR) is a region not found in other herpesviruses. At 24 h posttransfection, the lysates were harvested for coimmunoprecipitation experiments. The results from both directions of coimmunoprecipitation showed that every deletion mutant of HA-BFRF1 had an interacting signal with BFLF2 (Fig. 7B). However, deletion of the N-terminal region of BFRF1, including LD1 and LD2 domains and ID, showed much weaker signals for coimmunoprecipitation (co-IP) (Fig. 7B, lanes 12, 13, and 14). To further demonstrate the contribution of individual regions, glutathione transferase (GST) pulldown assays using individual BFRF1 domains were used to confirm the interaction. The expression of different GST fusion proteins was checked by SDS-PAGE and Coomassie brilliant blue staining (Fig. 7C to E). Here, all GST fusion BFRF1 proteins containing different domains displayed interacting signals to pull down BFLF2 with various intensities (Fig. 7D, top). The GST-F1d(7-196) construct which contains the unique EBV-specific region (ESR) alone also pulled down Flag-BFLF2 efficiently (Fig. 7D, top, lane 5). The above-described data thus suggested that multiple regions of BFRF1 interact with BFLF2.
FIG 7.
Multiple domains of BFRF1 interact with BFLF2. (A) Schematic representation of predicted functional domains of BFRF1 and serial deletion mutants of HA-BFRF1. The abilities to interact with BFLF2 as detected by coimmunoprecipitation are summarized on the right. (B) Flag-BFLF2-expressing plasmid was transfected with plasmid expressing wild-type or serial deletion HA-BFRF1 mutants into HeLa cells. At 24 hpt, the lysates were harvested and immunoprecipitated with antibody against HA (top) or Flag (bottom). The immunocomplexes were then resolved by 10% SDS-PAGE and immunoblotted with antibodies against HA or Flag. Two independent experiments were performed, and representative data are shown. The position of HA-F1ΔESR is indicated by an arrow. The IgG L chain is indicated by an open triangle. (C) Schematic representation of full-length and truncated GST-BFRF1 constructs. (D) Plasmid expressing Flag-BFLF2 was transfected into HeLa cells. At 24 hpt, the lysates were harvested and incubated with GST-tagged BFRF1 fusion proteins to pull down BFLF2. The interacting complexes were then resolved by 10% SDS-PAGE and immunoblotted with antibodies against Flag or GST (top). The input GST-BFRF1 full-length and truncated mutants were harvested from isopropyl-β-d-thiogalactopyranoside (IPTG)-induced BL21 were displayed by SDS-PAGE and stained with Coomassie brilliant blue (bottom). The experiment was performed two times, and representative data are shown. (E) The same plasmids as for panel B were transfected into slide-cultured HeLa cells. At 24 hpt, the slides were fixed and stained with antibodies against Flag (green) and HA (red) and then stained with Hoechst to indicate cellular DNA (blue). The slides were analyzed by confocal microscopy. The experiment was performed twice, and the numbers of cells showing the representative pattern of each group of cells are indicated from one independent experiment.
To address whether these BFRF1 mutants are able to bring BFLF2 out of the nucleus to the nuclear rim or cytoplasmic vesicles, serial deletion mutants of HA-BFRF1 and Flag-BFLF2 were cotransfected into slide-cultured HeLa cells. Confocal image analysis at 24 hpt showed that Flag-BFLF2 was expressed mainly in the nucleus. Similar to the pattern observed in Fig. 4A, Flag-BFLF2 was distributed at the nuclear rim and colocalized with BFRF1 in the cytoplasmic vesicles in coexpressing cells (Fig. 7E), whereas all BFRF1 deletion mutants failed to bring BFLF2 out the nucleus to the nuclear rim or colocalize with BFLF2 in the cytoplasm (Fig. 7E). Furthermore, HA-F1ΔID showed a major distribution at the nuclear rim, whereas the HA-F1ΔTM, which contains a deletion of the transmembrane domain, was recruited by BFLF2 into nucleus. The results suggest the contributions of multiple regions are required for BFRF1 to function together with BFLF2 in forming proper nuclear egress vesicles.
The nuclear targeting and BFRF1-interacting domains of BFLF2 are required for virion release.
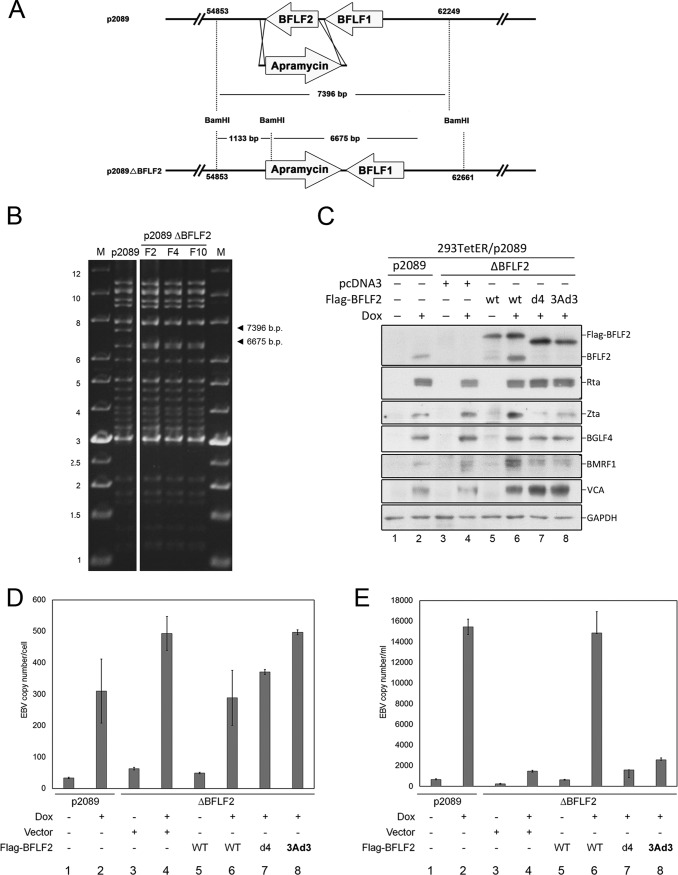
To determine the functional contribution of the nuclear targeting and BFRF1-interacting domains of BFLF2 to the virus replication process, BFLF2 was knocked out of the p2089 EBV bacmid through PCR targeting (Fig. 8A). In pulsed-field gel electrophoresis, the BamHI-digested wild-type p2089 showed a fragment of 7,396 bp containing BFLF2. After PCR targeting, BFLF2 was disrupted and exchanged for an apramycin resistance open reading frame. The 7,396-bp fragment was cleaved into two smaller fragments of 1,133 bp and 6,675 bp after BamHI digestion (Fig. 8B). The EBV bacmid p2089 or the p2089ΔBFLF2 was transfected into 293TetER cells, which carry a tet-on EBV Rta cassette that can be induced for Rta expression with doxycycline (25). After selection, green fluorescent protein (GFP)-positive cell colonies were combined into different pools to establish 293TetER/p2089ΔBFLF2 stable cells. After doxycycline (Dox) treatment for 60 h, the lytic proteins, including Zta, Rta, BMRF1, BGLF4, and VCA, were detected by Western blotting (Fig. 8C, lanes 1 and 2). Expression of Flag-BFLF2, Flag-F2(3A), or Flag-F2(3Ad3) was only detected in the 293TetER/p2089ΔBFLF2 stable cells after complementation (Fig. 8C, lanes 6, 7, and 8). It is known that BFLF2 promotes the egress of EBV nucleocapsids from the nucleus into the cytoplasm for the subsequent maturation process without affecting viral DNA replication (14). Indeed, the quantitative PCR (qPCR) result indicates that 293TetER/p2089ΔBFLF2 cells had slightly more viral DNA than 293TetER/p2089 cells (Fig. 8D, lanes 1 to 4), possibly due to a defect in the release of mature virions. Simultaneously, the level of extracellular virion DNA was lower in the culture supernatant of 293TetER/p2089ΔBFLF2 bacmid cells and was restored after Flag-BFLF2 complementation (Fig. 8E, lanes 5 and 6). However, when the BFRF1-interacting domain deletion mutant Flag-F2d4 or the NLS mutant Flag-F2(3Ad3) was transfected into 293TetER/p2089ΔBFLF2 cells, the level of intracellular DNA was slightly higher than that of wild-type Flag-BFLF2, suggesting that the replicated viral DNA was trapped in the cells in the absence of functional BFLF2 (Fig. 8D, lanes 6 to 8). Furthermore, levels of secreted virion were significantly decreased when Flag-BFLF2 mutants F2d4 or F2(3Ad3) were transfected into the 293TetER/p2089ΔBFLF2 cells (Fig. 8E, lanes 7 and 8). The data described here indicate that both the NLS and the most critical BFRF1-interacting domains of BFLF2 are required for EBV maturation and secretion.
FIG 8.
Complementation of 293TetER/p2089 ΔBFLF2 cells with F2(3Ad3) or F2d4 mutant was defective in restoring virion secretion. (A) Schematic summary of the BFLF2 mutant EBV bacmid cloning strategy, as described in Materials and Methods. To delete the BFLF2 open reading frame (ORF) of Maxi-EBV, the recombination primers (LMRC741 and LMRC742) were used to amplify the apramycin cassette, and the PCR products were transformed into DH10B/p2089/pKD46 by electroporation. Finally, the ORF of BFLF2 was disrupted by PCR targeting. (B) The wild-type and BFLF2 knockout bacmids were digested with BamHI and displayed by agarose gel electrophoresis to shown the identical fragments. In addition, cleavage of p2089 wild type produced a fragment of around 7.4 kb, which was cleaved into 1.1-kb and 6.6-kb fragments in p2089ΔBFLF2 bacmid. The digestions were displayed by pulsed-field gel electrophoresis. (C) 293TetER/p2089 wild-type and BFLF2 knockout cells were transfected with 1 μg of pcDNA3.0, Flag-tagged wild-type BFLF2, F2(3Ad3), or F2d4 plasmid and treated with doxycycline (50 ng/ml) at 18 h posttransfection. After doxycycline induction for 60 h, the cells were harvested for Western blotting of viral lytic proteins. (D) Simultaneously, similar sets of complementation cells were harvested for detecting the intracellular EBV DNA copy numbers by qPCR for the EBV BamHI W fragment and human β-globin (HBG). (E) Virion secretion of individual transfection was determined by qPCR as described in Materials and Methods. The experiments shown in panels C, D, and E were performed three times, and representative data from three independent experiments are shown.
DISCUSSION
The nuclear envelope is the highest-ordered, complicated, and dynamic organelle in the eukaryotic cell. Thus, the life cycle of large herpesviruses provides us a chance to reveal various aspects of nuclear envelope modification and nucleocytoplasmic transport regulation. The NEC of herpesviruses is required for nuclear egress of newly formed nucleocapsids. Despite the homology to alpha- and betaherpesviruses, gammaherpesviruses evolved with distinct mechanisms for the NEC complex to transport the nucleocapsids from the nucleus into the cytoplasm for the subsequent maturation process. Most of the studied herpesviral NECs form UL34/UL31-like heterodimers to promote the nuclear-to-cytoplasmic transport of nucleocapsids. However, we previously showed that EBV BFRF1 alone is able to recruit the ESCRT system through interaction with Alix protein to induce nuclear envelope-derived cytoplasmic vesicles (17). Ubiquitination and E3 ligase Itch are both required for this process (25). Thus, additional unique features of the EBV NECs are revealed in this study.
Here, the nuclear localization signal of BFLF2 was mapped to 47 RCGKSRSELGRMERVGSVARSICSRHTRHGVDRS80, which contains two histidines (in boldface font) in addition to discontinuous basic amino residues, namely, arginines and lysines. The region between aa 47 to 80 is a noncanonical NLS and functions in an importin β-dependent manner. Thus, we identified the most important BFRF1-interacting region of BFLF2 between aa 82 to 106, while the C-terminal aa 287 to 318 also contribute to the interaction. On the other hand, multiple domains of BFRF1 are required for bringing BFLF2 out of the nucleus and distributing it into cytoplasmic vesicles with BFRF1. Furthermore, both the NLS and the most critical BFRF1-interacting domains of BFLF2 are required for EBV maturation and secretion (Fig. 8).
We also fused the NLS region of BFLF2 onto the nuclear targeting reporter EYFP-LacZ. Three amino acids (Lys47, Lys50, and Lys52) and the segment between aa 58 and 81 of BFLF2 were demonstrated to function synergistically for nuclear targeting. Through a literature search and the acidic buffer treatment, we further confirmed that this nonclassical NLS is regulated by histidines in a pH-dependent manner (26). Such an NLS is present in the C-terminal juxtamembrane domain of the c-Met receptor tyrosine kinase and relies on histidines rather than lysines or arginines for the nuclear translocation (1068HVVIGPSSLIVH1079). Most interestingly, this NLS then serves as a pH sensor; Met binding to importin β and nuclear localization are enhanced when the cellular pH is lowered (21). It is thus interesting to know whether there is also a pH variation while EBV nucleocapsids are enveloped at the juxtanuclear membrane regions. Furthermore, the amino acid sequence alignment showed that there are some histidines and basic residues near the classical bipartite NLS of UL31 of HSV-1 (Fig. 1A); thus, it remains to be analyzed whether those basic residues also contribute to nuclear targeting in herpesviruses.
In addition, nuclear localization of UL31 homologs appeared to be required for the interaction with UL34 homologs to form UL34/UL31-like heterodimers at inner nuclear membranes to promote the nuclear egress of nucleocapsids in HSV and HCMV (UL50/UL53) (19, 27, 28). It is a unique feature that EBV BFRF1 alone is able to induce nuclear envelope-derived cytoplasmic vesicles (17). Thus, the nuclear localization-defective BFLF2 mutants, including Flag-F2(3A) and Flag-F2(3Ad3), still interacted with HA-BFRF1 in coimmunoprecipitation experiments and colocalized with BFRF1 in a subset of cytoplasmic vesicles (Fig. 4A). It suggests that once translocated into the cytoplasm, the BFRF1-containing vesicles may interact with the nuclear targeting-defective BFLF2 mutants in the cytoplasm. Notably, the colocalization signals of Flag-F2(3A) or Flag-F2(3Ad3) with BFRF1 are distinct from those of the wild-type Flag-BFLF2 (Fig. 4A, enlarged insets). In the presence of wild-type Flag-F2, the F2 green signals were wrapped by red BFRF1 signals in most of the cytoplasmic vesicles, whereas cytoplasmic Flag-F2(3A) and Flag-F2(3Ad3) staining patterns were not so homogenous and did not always associate with red HA-BFRF1 staining. It indicates that nuclear localization is not required for the BFLF2/BFRF1 interaction. However, the complementation experiment in 293TetER/p2089ΔBFLF2 cells further confirmed that neither Flag-F2(3Ad3), which failed to translocate into nucleus, nor Flag-F2d4, which cannot interact with BFRF1 efficiently, was able to restore virus release similarly to wild-type BFRF1 (Fig. 8).
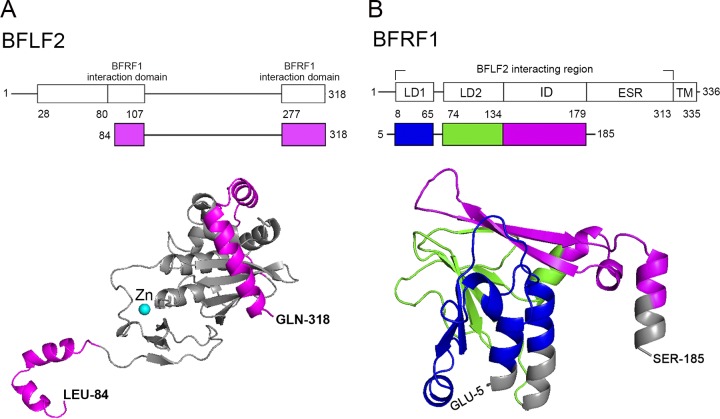
By characterizing various deletion mutants of BFRF1 and BFLF2, our data also showed that BFLF2 and BFRF1 possibly interact with each other through multiple regions (Fig. 3C and 7A to D). Indeed, both crystal structures of UL34/UL31 of HSV-1 and UL50/UL53 of HCMV revealed that there are two interfaces in the UL34/UL31 heterodimer (29, 30). UL53 homologs uses α-helix I and II to interact with UL50 homologs in interface I. Our coimmunoprecipitation and confocal image data confirmed that aa 82 to 106 of BFLF2 are required for proper interaction with BFRF1 (Fig. 3C, E, and F). The prediction of the three-dimensional structure of BFLF2 using HCMV UL53 as the template also showed that the BFRF1-interacting domain (aa 82 to 106) contains two α helices, which are highly conserved in the CR1 (conserved region 1) of herpesviruses (Fig. 9A). It was reported that residues surrounding the zinc finger of UL53 of HCMV contribute to the interaction with UL50 (30). The UL53 mutants containing substitutions of its zinc-coordinating cysteine result in partial defects in interaction with UL50. These zinc-coordinating residues are also conserved in CR1 of herpesviruses. In BFLF2, these residues are C129, C145, and C148, which are located within the aa 103 to 149 region.
FIG 9.
Summary of three-dimensional (3D) structural prediction of EBV nuclear egress complex according to herpesviral homologs. (A) The 3D structural prediction of BFLF2 was based on HCMV 5DOB chain A. The region of amino acids 1 to 75 is not shown due to the variation of N termini in UL31 homologs. The structure predictions were produced by (PS)2-v2:Protein Structure Prediction Server and were performed and presented by PyMOL, which is a molecular visualization system. Two regions of BFLF2 involved in BFRF1 interaction are indicated on the top and highlighted in pink in the predicated 3D structure. (B) The 3D structural prediction of BFRF1 was based on HCMV 5DOB chain B. The EBV-specific region (ESR), consisting of amino acids 180 to 313 could not be predicted. The LD1 domain is showed in green, the LD2 domain is shown in blue, and the ID domain is shown in pink. The regions involved in BFRF1 interaction are indicated on the top of BFRF1 functional domains and in the 3D modeling.
On the other hand, the crystal structure showed that UL50 homologs enable α-helix I, II, and IV to interact with UL53 homologs in interface I, and its β-sheet VI, VIII, and IX interact with UL53 homologs in interface II (29, 30). Our predicted three-dimensional structure of BFRF1 using CMV UL50 as a template also showed that the LD1 domain contains two α helices, while the LD2 domain and ID contain three β sheets (Fig. 9B). These structural analyses and our data suggested that LD1, LD2, and ID are required to interact with full-length BFLF2. Furthermore, we found that even though several domains of BFRF1 coimmunoprecipitated with BFLF2, all domains were still required to pull out full-length BFLF2 from the nucleus (Fig. 7D and E). It thus suggests that different domains may function sequentially for proper NEC function and the formation of cytoplasmic vesicles. Notably, the EBV-specific region (ESR; which is not found in other alpha- and betaherpesviral homologs and cannot be predicted for the structures) appeared to interact with BFLF2 efficiently in the GST pulldown experiment (Fig. 7D). A stretch of positive-charged amino acids was found in the ESR region, while the exact interaction mechanism remains to be revealed. It is further confirmed by the complementation experiments showing that the most important BFRF1-interacting domain within aa 80 to 106 and the nuclear localization signal within aa 29 to 80 are required for virus maturation (Fig. 8).
Overall, both conserved and unique features are demonstrated for EBV NEC proteins BFRF1 and BFLF2 in this study. Since EBV is a human oncogenic virus, it remains to be revealed whether repetitive reactivation of EBV in latent cells may cause accumulating effects on the integrity of the nuclear envelope and contribute to the oncogenesis or genome instabilities of tumor development.
MATERIALS AND METHODS
Plasmid construction.
All plasmids and primers used in this study are listed in Tables 1 and 2. In brief, Flag-tagged BFLF2 was generated by cloning BamHI-Flag-BFLF2-NotI into pcDNA3.0 (Clontech) and was kindly provided by Hsiu-Ming Shih (Academia Sinica, Taiwan). Flag-BFLF2 mutants, including pLSC12 [F2D1 (Δ2-102 aa)], pYZS2 [F2D2 (Δ103-149 aa)], pLSC15 [F2D3 (Δ150-209 aa)], pYZS3 [F2D4 (Δ209-286 aa)], and pYZS4 [F2D5 (Δ277-318 aa)], and small deletion mutants, including pYCD12 [F2d1 (Δ2-27 aa)], pYTL3 [F2d2 (Δ29-57 aa)], pYTL4 [F2d3 (Δ59-80 aa)], pYTL5 [F2d4 (Δ82-106 aa)], pYTL1 [F2(3A) (R47,52A,K50A)], and pYTL6 [F2(3Ad3) (R47,52A,K50A, Δ59-80 aa)] were generated using the single primer mutagenesis protocol (31), with pcDNA3-Flag-BFLF2 as the template and LMRC876, LMRC877, LMRC878, LMRC910, LMRC880, LMRC998, LMRC1006, LMRC1007, LMRC1008, and LMRC1009 as shown in Table 1 as the primers. For the assay of the nuclear translocation signal, we used pEYFP-LacZ-fused BFLF2 fragment aa 22 to 89, the pEYFP-C1-LacZ control plasmid, and the positive-control pEYFP-C1-LacZ-NLS (containing the SV40 NLS, kindly provided by Mitsuhiro Kawata, Kyoto Prefectural University of Medicine, Japan) (32). The pYCD5 (BFLF2, BFLF222–89) and its mutants, including pYCD6 [F222–89d2 (Δ29-57 aa)], pYCD7 [F222–89d3 (Δ59-80 aa)], pYCD8 [F222–89 3A (R47,52A,K50A)], and pYCD9 [F222–89 3Ad3 (R47,52A,K50A, Δ59-80 aa)], were generated by PCR amplification of the coding region between aa 22 to 89 with pcDNA3-Flag-BFLF2, pYTL3, pYTL4, pYTL1, and pYTL6 as the templates and LMRC1081 and LMRC1082 as primers; the PCR products were inserted into pEYFP-C1-LacZ at EcoRI and BamHI sites. For BFRF1-related constructs, HA-BFRF1 was generated by cloning XhoI-HA-BFRF1-NotI into pcDNA3.0 (Invitrogen) and was kindly provided by Hsiu-Ming Shih (Academia Sinica, Taiwan). All mutants generated by mutagenesis, including pLPT4, pLPT5, pLPT6, pLPT7, and pLPT8, were previously described (17). pGTL1, which expresses GST-BFRF1, was generated by PCR amplification with pcDNA3.0-HA-BFRF1 as the template and LMRC891 and LMRC892 as the forward and reverse primers, and then cloned into pGEX4T1 vector. Other GST-BFRF1 constructs, including, pGTL12 [GST-BFRF1d(72-313)], pGTL13 [GST-BFRF1d(135-313)], pYTJ1 [GST-BFRF1d(7-196)] and pYTJ3 [GST-BFRF1d(7-134,201-313)] were generated by mutagenesis with template of pGTL1 (pGEX4T1-BFRF1) and the LMRC933, LMRC935, LMRC1062, and LMRC1064 primers specified in Table 2.
TABLE 1.
Plasmids used in this study
| Name | Plasmid description | Template or source | Primer(s) | Enzymes or method |
|---|---|---|---|---|
| pYCD11 | pcDNA3.0-Flag-vector | pcDNA3.0 | LMRC1083, LMRC1084 | HindIII, KpnI |
| pcDNA3.0-Flag-BFLF2 | From Hsiu-Ming Shih (Academia Sinica, Taiwan) | |||
| pLSC12 | pcDNA3.0-Flag-BFLF2 D1 (Δ2-102) | pcDNA3.0-Flag-BFLF2 | LMRC876 | Mutagenesis |
| pYZS2 | pcDNA3.0-Flag-BFLF2 D2 (Δ103-149) | pcDNA3.0-Flag-BFLF2 | LMRC877 | Mutagenesis |
| pLSC15 | pcDNA3.0-Flag-BFLF2 D3 (Δ150-209) | pcDNA3.0-Flag-BFLF2 | LMRC878 | Mutagenesis |
| pYZS3 | pcDNA3.0-Flag-BFLF2 D4 (Δ209-276) | pcDNA3.0-Flag-BFLF2 | LMRC910 | Mutagenesis |
| pYZS4 | pcDNA3.0-Flag-BFLF2 D5 (Δ277-318) | pcDNA3.0-Flag-BFLF2 | LMRC880 | Mutagenesis |
| pYTL1 | pcDNA3.0-Flag-BFLF2 3A (R47,52A,K50A) | pcDNA3.0-Flag-BFLF2 | LMRC998 | Mutagenesis |
| pYCD12 | pcDNA3.0-Flag-BFLF2 dl (Δ2-27) | pcDNA3.0-Flag-BFLF2 | LMRC1006 | Mutagenesis |
| pYTL3 | pcDNA3.0-Flag-BFLF2 d2 (Δ29-57) | pcDNA3.0-Flag-BFLF2 | LMRC1007 | Mutagenesis |
| pYTL4 | pcDNA3.0-Flag-BFLF2 d3 (Δ59-80) | pcDNA3.0-Flag-BFLF2 | LMRC1008 | Mutagenesis |
| pYTL5 | pcDNA3.0-Flag-BFLF2 d4 (Δ82-106) | pcDNA3.0-Flag-BFLF2 | LMRC1009 | Mutagenesis |
| pYTL6 | pcDNA3.0-Flag-BFLF2 3Ad3 (R47,52A,K50A, Δ59-80) | pYTL4 | LMRC998 | Mutagenesis |
| pEYFP-C1-LacZ-NLS | From Mitsuhiro Kawata (Kyoto Prefectural University of Medicine, Japan) | |||
| pYCS2 | pEYFP-C1-LacZ | EcoRI, BamHI | ||
| pYCD5 | pEYFP-C1-LacZ-BFLF2 WT22–89 | pcDNA3.0-Flag-BFLF2 | LMRC1081, LMRC1082 | EcoRI, BamHI |
| pYCD6 | pEYFP-C1-LacZ-BFLF2 d222–89 (Δ29-57) | pcDNA3.0-Flag-BFLF2 d2 (Δ29-57) | LMRC1081, LMRC1082 | EcoRI, BamHI |
| pYCD7 | pEYFP-C1-LacZ-BFLF2 d322–89 (Δ59-80) | pcDNA3.0-Flag-BFLF2 d3 (Δ59-80) | LMRC1081, LMRC1082 | EcoRI, BamHI |
| pYCD8 | pEYFP-C1-LacZ-BFLF2 3A22–89 (R47,52A,K50A) | pcDNA3.0-Flag-BFLF2 3A (R47,52A,K50A) | LMRC1081, LMRC1082 | EcoRI, BamHI |
| pYCD9 | pEYFP-C1-LacZ-BFLF2 3Ad322–89 (R47,52A,K50A, Δ59-80) | pcDNA3.0-Flag-BFLF2 3Ad3 (R47,52A,K50A, Δ59-80) | LMRC1081, LMRC1082 | EcoRI, BamHI |
| pYCD10 | pcDNA3.0-HA-vector | pcDNA3.0 | LMRC1085, LMRC1086 | HindIII, KpnI |
| pcDNA3.0-HA-BFRF1 | From Hsiu-Ming, Shih (Academia Sinica, Taiwan) | |||
| pLPT4 | pcDNA3.0-HA-BFRF1 ΔLD1 | pcDNA3.0-HA-BFRF1 | LMRC833 | Mutagenesis |
| pLPT5 | pcDNA3.0-HA-BFRF1 ΔLD2 | pcDNA3.0-HA-BFRF1 | LMRC834 | Mutagenesis |
| pGTL1 | pGEX4T1-BFRF1 | pcDNA3.0-HA-BFRF1 | LMRC891, LMRC892 | Not I, BamHI |
| pGTL12 | pGEX4T1-BFRF1 d(72-313) | pGEX4T1-BFRF1 | LMRC932 | Mutagenesis |
| pGTL13 | pGEX4T1-BFRF1 d(135-313) | pGEX4T1-BFRF1 | LMRC933, LMRC935 | Mutagenesis |
| pYTJ1 | pGEX4T1-BFRF1 d(7-196) | pGEX4T1-BFRF1 | LMRC1062 | Mutagenesis |
| pYTJ2 | pGEX4T1-BFRF1 d(7-134) | pGEX4T1-BFRF1 | LMRC1063 | Mutagenesis |
| pYTJ3 | pGEX4T1-BFRF1 d(7-134, 201-313) | pGEX4T1-BFRF1 d(7-134) | LMRC1064 | Mutagenesis |
TABLE 2.
Primers used in this study
| Name | Sequence (5′→3′) | Purpose |
|---|---|---|
| LMRC741 | CCAATCCAACACGAGGCAAGTTTTAAGAGTTAAAAGCAAATTCCGGGGATCCGTCGACC | BFLF2 knockout, EBV bacmid |
| LMRC742 | TATCAAACTTCCCAGGTCTACGTGTGAAAAGTAAACCCGTGTAGGCTGGAGCTGCTTC | |
| LMRC833 | ATGGCGAGCCCGGAAGAGAGGGCCTTTAAGCTGAAGAACTGC | pLPT4 |
| LMRC834 | TAAGCTGAAGAACTGCAACAAAAGCCCCCTGGTCTTCC | pLPT5 |
| LMRC835 | GCCAGGATGACTTCATTAAGCTCGTCATGGATATGATTAGTGATAT | pLPT6 |
| LMRC836 | ACGCCCAGAAGGCCTCGCGGACACCTTATCTGGCACGGGT | pLPT7 |
| LMRC837 | TGGCGTTATTCTTGGCGCGCCTAGAGGGCCCTATTCTATAGT | pLPT8 |
| LMRC876 | TGATGACA AAGTCGACATGCGTGAGATGAACACCCCCA | pLSC12 |
| LMRC877 | GAGCTGGGCAAAGATTTTCTGAATAGGTCCCAGGTTCATGTG | pYZS2 |
| LMRC878 | CTCACTGCGAGATCTGCATCCACTTTTGTGGCACCATCTG | pLSC15 |
| LMRC880 | GGTCAGCCCCAGAGACACGTAGAGCTCGCTGATCAGCC | pYZS3 |
| LMRC891 | CGGGATCCATGGCGAGCCCGGAAGAG | pGTL1 |
| LMRC892 | TATCAAATAGCCGGCTCAGGTCCACCTCAGAAACA | pGTL1 |
| LMRC910 | CTACGCTTACCTGGTCACCGATGCCATCTACGAGGCCA | pYZS4 |
| LMRC932 | GGCCTTTAAGCTGAAGAACACACCTTATCTGGCACGGG | pGTL12 |
| LMRC933 | GGTTCCGCGTGGATCCATGTGCAACTACCCCTCCTCGC | pGTL13 |
| LMRC935 | ATCTGGTTCCGCGTGGATCCATGGTCATGGATATGATTAGTGATAT | pGTL13 |
| LMRC998 | TAGCGTGAAGTCGGTGTCCGCGTGTGGAGCATCTGCCTCAGAGCTGGGAAGAATG | pYTL1 |
| LMRC1006 | TGACAAAGTCGACATGCACCACAGAAACTACA | pYTL2 |
| LMRC1007 | CTAATGCATCCGCACATGGAAAGGGTTGGC | pYTL3 |
| LMRC1008 | GAGCTGGGAAGAATGCATTTTTCACTACGGG | pYTL4 |
| LMRC1009 | TGTAGACAGATCCCATACCCCCATACATGTC | pYTL5 |
| LMRC1062 | GCGAGCCCGGAAGAGGTCGAGGATACGGGCG | pYTJ1 |
| LMRC1063 | GCGAGCCCGGAAGAGAAAAGCCCCCTGGTCTTC | pYTJ2 |
| LMRC1064 | GTCAAGTCGAGGATACGACACTTATCTGGCACGG | pYTJ3 |
| LMRC1081 | CCGGAATTCCGCCGCCTAATGCATCCG | pYCD5, pYCD6, pYCD7, pYCD8, pYCD9 |
| LMRC1082 | CGCGGATCCTCAATTAATCAACCTGAAGAAGTCCCGTAGTG |
Cell culture and transfection.
HeLa cells were derived from human cervical epithelial cells (ATCC CCL-2). The 293TetER cells are Rta inducible and were generated from the 293Tet-on cell line (Invitrogen), carrying a Flag-Rta plasmid (33). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal calf serum and 2 mM l-glutamine and supplemented with penicillin (100 U/ml) and streptomycin (100 μg/ml) at 37°C with 5% CO2.
For transfection, HeLa cells were transfected with plasmids expressing Flag-BFLF2 or GFP-BFRF1 by using Lipofectamine 2000 (Invitrogen) in Opti-MEM medium (Gibco-BRL), according to the manufacturer’s instructions. 293TetER/p2089 bacmid cells were transfected with plasmids expressing wild-type or mutant Flag-BFLF2 by using the calcium phosphate–N,N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid (BES)-buffered solution (BBS) transfection protocol (34).
To select doxycycline-inducible EBV bacmid-positive cell lines, 293TetER cells (5 × 105 cells/well) were seeded in 6-well culture plates and transfected with 7 μg of EBV p2089 wild-type or mutant bacmid using the Lipofectamine 2000 protocol and selected with hygromycin B (100 μg/ml) as previously described (34, 35). The selected EBV wild-type or mutant clones were treated with doxycycline (50 ng/ml) to induce lytic replication; the lytic cycle progression was confirmed by Western blotting.
Immunoblotting.
Cells were lysed in RIPA lysis buffer (1% NP-40, 50 mM Tris [pH 7.5], 150 mM NaCl, 5 mM EDTA, 0.1% SDS, and 0.5% deoxycholate), the cell lysates were resolved by SDS-PAGE, and the proteins were transferred onto nitrocellulose membranes. The immunoblotting was performed as described previously (35). In-house antibodies, including mouse anti-Rta (467), anti-Zta (IB4), anti-BGLF4 (2616), anti-GST, anti-BMRF1 (88A9), and anti-VCA (L1), were used as primary antibodies, as previously described (16). The anti-BFLF2 antibody (CI) was kindly provided by Roberta Gonnella (Università La Sapienza, Italy). Other primary antibodies were anti-Flag antibody (M2; Sigma-Aldrich), anti-HA (HA.11; Covance), anti-GFP (GTX113617; GeneTex), anti-KPNB1 (3E9; Abcam), anti-lamin A/C (636; Santa Cruz Biotechnology), anti-α-tubulin (DM1A; Calbiochem), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Biodesign), and anti-β-actin (Sigma-Aldrich), together with species-specific horseradish peroxidase (HRP)-conjugated secondary antibody (Jackson ImmunoResearch and Chemicon).
Immunofluorescence assay.
HeLa cells (4 × 105) were seeded onto fluorescence-negative glass slides in 10-cm petri dishes and incubated for 24 h before transfection. At 24-h posttransfection (hpt), the cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS; 145 mM NaCl, 1.56 mM Na2HPO4, 1 mM KH2PO4, pH 7.2) at room temperature (RT) for 20 min. The cells were permeabilized with 0.1% Triton X-100 in PBS at RT for 5 min, stained with mouse anti-Flag (M2; Sigma-Aldrich), rabbit anti-Flag (AF1001; Viogene-Biotek), mouse anti-HA (HA.11, MMS-101R; Covance), or rabbit anti-emerin (SC-15378; Santa Cruz Biotechnology) at 37°C for 1.5 h, washed with PBS, and subsequently incubated with fluorochrome-conjugated secondary antibody (Jackson ImmunoResearch) for another 1.5 h. DNA was stained with Hoechst (33258) at RT for 5 min, and the cells were covered with H1000 mounting medium (Vector Labs). The cells were observed by confocal microscopy (Carl Zeiss LSM780 and LSM880) in the imaging core at the First Core Lab of National Taiwan University College of Medicine.
Coimmunoprecipitation assay.
For immunoprecipitation of HA-BFRF1 and Flag-BFLF2, HeLa cells (2 × 106) were seeded in a 10-cm dish and were cotransfected with plasmids expressing Flag-BFLF2 wild type or mutants and HA-BFRF1. At 24 hpt, cells were lysed with NP-40 buffer (1% NP-40, 150 mM NaCl, 2 mM EDTA, 50 mM Tris-HCl, pH 8.0) containing a protease inhibitor cocktail (Roche) with gentle shaking for 2 h at 4°C. The cell debris was clarified by centrifugation at 16,000 × g for 10 min at 4°C, and the supernatant was precleared by incubation with protein A-Sepharose beads (GE Healthcare) for 40 min at 4°C. The immunocomplexes (500 μg) were incubated with 1 μg of anti-Flag or anti-HA antibodies at 4°C overnight, following by an incubation with 20 μl of protein A-Sepharose beads for 2 h at 4°C. The immunocomplexes were collected and washed with NP-40 buffer and 1× PBS 2 times and then detected with anti-Flag or anti-HA antibodies by Western blot analysis. For immunoprecipitation (IP) of Flag-BFLF2 and endogenous importin β1, HeLa cells (2 × 106) were seeded in a 10-cm dish and were transfected with plasmid expressing Flag-BFLF2 wild type or mutants. At 24 hpt, cells were lysed with RIPA buffer (1% NP-40, 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris-HCl, pH 7.4) containing the protease inhibitor cocktail and passed through a 27-gauge syringe 10 times. The cell debris was clarified by centrifugation at 16,000 × g for 10 min at 4°C, and the supernatant was precleared by incubation with protein A-Sepharose beads (GE Healthcare) for 40 min at 4°C. The immunocomplexes (500 μg) were incubated with 1 μg of anti-Flag at 4°C overnight, followed by an incubation with 20 μl of protein A-Sepharose beads for 2 h at 4°C. The immunocomplexes were collected and washed with RIPA buffer and 1× PBS 2 times and then detected with anti-Flag or anti-importin β1 antibodies by Western blot analysis.
Importin β inhibitor importazole treatment.
A plasmid expressing Flag-BFLF2 wild type was transfected into 8 × 105 slide-cultured HeLa cells in a 10-cm petri dish. At 6 hpt, importin β inhibitor (20 μM, 40 μM, 80 μM importazole [IPZ], catalog number 401105-10MGC; Merck Millipore) or dimethyl sulfoxide (DMSO) was added to the medium. The cells were harvested after a further 24 h of incubation and subjected to immunostaining and confocal image analysis.
Decrease of cytosolic pH for detection of NLS function.
The decrease of cytosolic pH was performed as described previously (21). In brief, HeLa cells transfected with various Flag-BFLF2 constructs were treated either with pH 6.5 Krebs-Ringer bicarbonate (KRB) solution (140 mM NaCl, 3 mM KCl, 0.5 mM NaH2PO4, 0.5 mM MgSO4, 1.5 mM CaCl2, 10 mM HEPES, 2 mM Na2CO3, and 5.5 mM glucose) or culture medium as a control. Cells were then incubated at 37°C under a humidified atmosphere of 5% CO2 for 1 h. Cells were then harvested for further assays.
Cellular fractionation.
Cells were harvested and washed with PBS, resuspended in hypotonic buffer (5 mM Tris [pH 7.4], 60 mM KCl, 1.5 mM MgCl2, and 0.1 mM EGTA) containing freshly added protease inhibitor cocktail (Roche) and then incubated at 4°C for 5 min. Cells were sheared with a needle (27 gauge) and then centrifuged (500 × g, 5 min, 4°C). The supernatant was used as the cytoplasmic fraction. The pellet was resuspended in the hypotonic buffer, sheared again using a needle, and then centrifuged (500 × g, 5 min, 4°C). The pellet was washed three times with hypotonic buffer, resuspended in RIPA buffer containing freshly added protease inhibitors, and incubated on ice for 10 min. The supernatant was collected after centrifugation (16,000 × g, 10 min) and used as the nuclear fraction.
Construction of the BFLF2 knockout EBV bacmid.
The wild-type virus bacmid p2089 and BFRF1 knockout bacmids were kindly provided by Henri-Jacques Delecluse (36). The BFLF2 knockout bacmid was constructed by PCR targeting as previously described (37). In brief, the apramycin resistance cassette was amplified by PCR from the recombination primers LMRC741 and LMRC742. The apramycin gene products were transformed into E. coli DH10B, containing Maxi-EBV bacmid p2089 and Red recombinase plasmid pKD46, by electroporation. The recombination of the PCR products with the wild-type EBV genome resulted in the exchange between the BFLF2 gene region 54,853 to 62,249 of EBV B95.8 and the apramycin cassette. The p2089ΔBFLF2 bacmid was selected by apramycin and confirmed by BamHI digestion.
Extraction of intracellular EBV DNA and viral DNA in secreted virions.
Basically, the DNA extraction was conducted as described previously (18). In brief, 293TetER/p2089ΔBFLF2 cells (5 × 105) were transfected with vector pcDNA3, Flag-BFLF2 wild type, or mutants via 2× BBS transfection. The lysates from 293TetER/p2089 wild type or ΔBFLF2 complementation cells were lysed in 400 μl digestion buffer (0.1% SDS, 0.125 mg/ml proteinase K, 50 mM Tris-HCl [pH 8.0], 10 mM EDTA [pH 8.0]) and incubated at 55°C for 3 h. The sample was treated with RNase A and extracted with phenol-chloroform-isoamyl alcohol (25:24:1), followed by ethanol precipitation, and the pellet was dissolved in double-distilled water (ddH2O). For extraction of virion DNA, 293TetER/p2089ΔBFLF2 cells (5 × 105) were transfected with vector pcDNA3, Flag-BFLF2 wild type, or mutants via 2× BBS transfection. At 60 h postdoxycycline induction, the culture supernatants were collected and subjected to centrifugation at 15,000 × g for 30 min at 4°C to remove cell debris, followed by DNase I treatment to eliminate contamination of cellular DNA, as described previously (38). The viral genome was concentrated using a QIAmp MinElute Virus Spin kit (Qiagen) according to the manufacturer’s instructions.
Quantitative real-time PCR analysis.
To measure the viral DNA of in the cells and in the culture supernatants, quantitative real-time PCR SensiFAST SYBR No-ROX kit (Bioline) and the Bio-Rad CFX Connect was used to detect the EBV BamHI W fragment and human beta-globin (HBG) gene in the human genome (35). The standard curve for qPCR was generated by a 10-fold serial dilution of a mixture of 104 copies of genomic DNA of 293TetER cells and 5 × 106 copies of purified EBV bacmid DNA. A linear calibration curve was then generated by plotting threshold cycle (CT) values (y axis) against log10 BamHI W copy number (x axis) from which the number of EBV genomes in the individual samples was determined. All standards and samples, together with EBV-positive and EBV-negative controls, were analyzed in duplicates. The primers were forward 5′-CCCTGGTATAAAGTGGTCCT-3′ and reverse 5′-AAGTCCACTTACCTCTGG-3′ for BamHI W. The HBG primers used were forward 5′-GCTTCTGACACAACTGTGTTCACTAGC-3′ and reverse 5′-CACCAACTTCATCCACGTTCACC-3′.
ACKNOWLEDGMENTS
We thank H. J. Delecluse (German Cancer Research Centre [DKFZ]) for the EBV bacmid p2089, PCR template plasmid PIJ773, and Red recombination plasmid. We thank Roberta Gonnella (University of Rome La Sapienza, Department of Experimental Medicine) for the BFLF2 antibody. We also thank the cell imaging core of the First Core Labs in the National Taiwan University College of Medicine for confocal image analysis.
This study was supported by National Health Research Institutes (NHRI-EX107_10701BI, NHRI-EX108_10701BI) and partially supported by Ministry of Science and Technology, Taiwan (MOST 104-2320-B-002-054-MY3) and National Taiwan University College of Medicine (105C101-A2-523, 106C101F2, 105F025-03).
REFERENCES
- 1.Young LS, Yap LF, Murray PG. 2016. Epstein-Barr virus: more than 50 years old and still providing surprises. Nat Rev Cancer 16:789–802. doi: 10.1038/nrc.2016.92. [DOI] [PubMed] [Google Scholar]
- 2.Richard M, Longnecker EK, Cohen JI. 2013. Epstein-barr virus, p 1898–1954. In Knipe DM, Howley PM (ed), Fields virology, 6th ed Lippincott Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 3.Wente SR, Rout MP. 2010. The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol 2:a000562. doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D'Angelo MA, Hetzer MW. 2008. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol 18:456–466. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalderon D, Richardson WD, Markham AF, Smith AE. 1984. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- 6.Robbins J, Dilworth SM, Laskey RA, Dingwall C. 1991. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 7.Kosugi S, Hasebe M, Matsumura N, Takashima H, Miyamoto-Sato E, Tomita M, Yanagawa H. 2009. Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J Biol Chem 284:478–485. doi: 10.1074/jbc.M807017200. [DOI] [PubMed] [Google Scholar]
- 8.Gorlich D, Kostka S, Kraft R, Dingwall C, Laskey RA, Hartmann E, Prehn S. 1995. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr Biol 5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- 9.Hellberg T, Paßvogel L, Schulz KS, Klupp BG, Mettenleiter TC. 2016. Nuclear egress of herpesviruses: the prototypic vesicular nucleocytoplasmic transport. Adv Virus Res 94:81–140. doi: 10.1016/bs.aivir.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Paßvogel L, Trübe P, Schuster F, Klupp BG, Mettenleiter TC. 2013. Mapping of sequences in pseudorabies virus pUL34 that are required for formation and function of the nuclear egress complex. J Virol 87:4475–4485. doi: 10.1128/JVI.00021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang YE, Roizman B. 1993. The product of the UL31 gene of herpes simplex virus 1 is a nuclear phosphoprotein which partitions with the nuclear matrix. J Virol 67:6348–6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reynolds AE, Ryckman BJ, Baines JD, Zhou Y, Liang L, Roller RJ. 2001. U(L)31 and U(L)34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J Virol 75:8803–8817. doi: 10.1128/jvi.75.18.8803-8817.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reynolds AE, Wills EG, Roller RJ, Ryckman BJ, Baines JD. 2002. Ultrastructural localization of the herpes simplex virus type 1 U(L)31, U(L)34, and U(S)3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J Virol 76:8939–8952. doi: 10.1128/jvi.76.17.8939-8952.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Granato M, Feederle R, Farina A, Gonnella R, Santarelli R, Hub B, Faggioni A, Delecluse HJ. 2008. Deletion of Epstein-Barr virus BFLF2 leads to impaired viral DNA packaging and primary egress as well as to the production of defective viral particles. J Virol 82:4042–4051. doi: 10.1128/JVI.02436-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milbradt J, Auerochs S, Sevvana M, Muller YA, Sticht H, Marschall M. 2012. Specific residues of a conserved domain in the N terminus of the human cytomegalovirus pUL50 protein determine its intranuclear interaction with pUL53. J Biol Chem 287:24004–24016. doi: 10.1074/jbc.M111.331207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee C-P, Huang Y-H, Lin S-F, Chang Y, Chang Y-H, Takada K, Chen M-R. 2008. Epstein-Barr virus BGLF4 kinase induces disassembly of the nuclear lamina to facilitate virion production. J Virol 82:11913–11926. doi: 10.1128/JVI.01100-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee C-P, Liu P-T, Kung H-N, Su M-T, Chua H-H, Chang Y-H, Chang C-W, Tsai C-H, Liu F-T, Chen M-R. 2012. The ESCRT machinery is recruited by the viral BFRF1 protein to the nucleus-associated membrane for the maturation of Epstein-Barr virus. PLoS Pathog 8:e1002904. doi: 10.1371/journal.ppat.1002904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee C-P, Liu G-T, Kung H-N, Liu P-T, Liao Y-T, Chow L-P, Chang L-S, Chang Y-H, Chang C-W, Shu W-C, Angers A, Farina A, Lin S-F, Tsai C-H, Bouamr F, Chen M-R. 2016. The ubiquitin ligase Itch and ubiquitination regulate BFRF1-mediated nuclear envelope modification for Epstein-Barr virus maturation. J Virol 90:8994–9007. doi: 10.1128/JVI.01235-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Funk C, Ott M, Raschbichler V, Nagel CH, Binz A, Sodeik B, Bauerfeind R, Bailer SM. 2015. The herpes simplex virus protein pUL31 escorts nucleocapsids to sites of nuclear egress, a process coordinated by its N-terminal domain. PLoS Pathog 11:e1004957. doi: 10.1371/journal.ppat.1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soderholm JF, Bird SL, Kalab P, Sampathkumar Y, Hasegawa K, Uehara-Bingen M, Weis K, Heald R. 2011. Importazole, a small molecule inhibitor of the transport receptor importin-beta. ACS Chem Biol 6:700–708. doi: 10.1021/cb2000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaudhary SC, Cho M-G, Nguyen TT, Park K-S, Kwon M-H, Lee J-H. 2014. A putative pH-dependent nuclear localization signal in the juxtamembrane region of c-Met. Exp Mol Med 46:e119. doi: 10.1038/emm.2014.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kampmann T, Mueller DS, Mark AE, Young PR, Kobe B. 2006. The role of histidine residues in low-pH-mediated viral membrane fusion. Structure 14:1481–1487. doi: 10.1016/j.str.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 23.Li S, Hong M. 2011. Protonation, tautomerization, and rotameric structure of histidine: a comprehensive study by magic-angle-spinning solid-state NMR. J Am Chem Soc 133:1534–1544. doi: 10.1021/ja108943n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rötzschke O, Lau JM, Hofstätter M, Falk K, Strominger JL. 2002. A pH-sensitive histidine residue as control element for ligand release from HLA-DR molecules. Proc Natl Acad Sci U S A 99:16946–16950. doi: 10.1073/pnas.212643999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen YL, Chen YJ, Tsai WH, Ko YC, Chen JY, Lin SF. 2009. The Epstein-Barr virus replication and transcription activator, Rta/BRLF1, induces cellular senescence in epithelial cells. Cell Cycle 8:58–65. doi: 10.4161/cc.8.1.7411. [DOI] [PubMed] [Google Scholar]
- 26.Gomes DA, Rodrigues MA, Leite MF, Gomez MV, Varnai P, Balla T, Bennett AM, Nathanson MH. 2008. c-Met must translocate to the nucleus to initiate calcium signals. J Biol Chem 283:4344–4351. doi: 10.1074/jbc.M706550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hagen C, Dent KC, Zeev-Ben-Mordehai T, Grange M, Bosse JB, Whittle C, Klupp BG, Siebert CA, Vasishtan D, Bäuerlein FJB, Cheleski J, Werner S, Guttmann P, Rehbein S, Henzler K, Demmerle J, Adler B, Koszinowski U, Schermelleh L, Schneider G, Enquist LW, Plitzko JM, Mettenleiter TC, Grünewald K. 2015. Structural basis of vesicle formation at the inner nuclear membrane. Cell 163:1692–1701. doi: 10.1016/j.cell.2015.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leigh KE, Sharma M, Mansueto MS, Boeszoermenyi A, Filman DJ, Hogle JM, Wagner G, Coen DM, Arthanari H. 2015. Structure of a herpesvirus nuclear egress complex subunit reveals an interaction groove that is essential for viral replication. Proc Natl Acad Sci U S A 112:9010–9015. doi: 10.1073/pnas.1511140112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bigalke JM, Heldwein EE. 2015. Structural basis of membrane budding by the nuclear egress complex of herpesviruses. EMBO J 34:2921–2936. doi: 10.15252/embj.201592359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lye MF, Sharma M, El Omari K, Filman DJ, Schuermann JP, Hogle JM, Coen DM. 2015. Unexpected features and mechanism of heterodimer formation of a herpesvirus nuclear egress complex. EMBO J 34:2937–2952. doi: 10.15252/embj.201592651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makarova O, Kamberov E, Margolis B. 2000. Generation of deletion and point mutations with one primer in a single cloning. Biotechniques 29:970–972. doi: 10.2144/00295bm08. [DOI] [PubMed] [Google Scholar]
- 32.Kaku N, Matsuda K, Tsujimura A, Kawata M. 2008. Characterization of nuclear import of the domain-specific androgen receptor in association with the importin alpha/beta and Ran-guanosine 5’-triphosphate systems. Endocrinology 149:3960–3969. doi: 10.1210/en.2008-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su MT, Liu IH, Wu CW, Chang SM, Tsai CH, Yang PW, Chuang YC, Lee CP, Chen MR. 2014. Uracil DNA glycosylase BKRF3 contributes to Epstein-Barr virus DNA replication through physical interactions with proteins in viral DNA replication complex. J Virol 88:8883–8899. doi: 10.1128/JVI.00950-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen C, Okayama H. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol 7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su M-T, Wang Y-T, Chen Y-J, Lin S-F, Tsai C-H, Chen M-R. 2017. The SWI/SNF chromatin regulator BRG1 modulates the transcriptional regulatory activity of the Epstein-Barr virus DNA polymerase processivity factor BMRF1. J Virol 91:e02114-16. doi: 10.1128/JVI.02114-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delecluse HJ, Pich D, Hilsendegen T, Baum C, Hammerschmidt W. 1999. A first-generation packaging cell line for Epstein-Barr virus-derived vectors. Proc Natl Acad Sci U S A 96:5188–5193. doi: 10.1073/pnas.96.9.5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chua HH, Lee HH, Chang SS, Lu CC, Yeh TH, Hsu TY, Cheng TH, Cheng JT, Chen MR, Tsai CH. 2007. Role of the TSG101 gene in Epstein-Barr virus late gene transcription. J Virol 81:2459–2471. doi: 10.1128/JVI.02289-06. [DOI] [PMC free article] [PubMed] [Google Scholar]