Coronaviruses are a family of positive-sense RNA viruses with human and agricultural significance. Characterizing the mechanisms by which coronavirus infection dictates pathogenesis or counters the host immune response would provide targets for the development of therapeutics. Here, we show that the aryl hydrocarbon receptor (AhR) is activated in cells infected with a prototypic coronavirus, mouse hepatitis virus (MHV), resulting in the expression of several effector genes. AhR is important for modulation of the host immune response to MHV and plays a role in the expression of TiPARP, which we show is required for maximal viral replication. Taken together, our findings highlight a previously unidentified role for AhR in regulating coronavirus replication and the immune response to the virus.
KEYWORDS: AhR, IDO1, TiPARP, coronavirus, mouse hepatitis virus
ABSTRACT
The aryl hydrocarbon receptor (AhR) is a cytoplasmic receptor/transcription factor that modulates several cellular and immunological processes following activation by pathogen-associated stimuli, though its role during virus infection is largely unknown. Here, we show that AhR is activated in cells infected with mouse hepatitis virus (MHV), a coronavirus (CoV), and contributes to the upregulation of downstream effector TCDD-inducible poly(ADP-ribose) polymerase (TiPARP) during infection. Knockdown of TiPARP reduced viral replication and increased interferon expression, suggesting that TiPARP functions in a proviral manner during MHV infection. We also show that MHV replication induced the expression of other genes known to be downstream of AhR in macrophages and dendritic cells and in livers of infected mice. Further, we found that chemically inhibiting or activating AhR reciprocally modulated the expression levels of cytokines induced by infection, specifically, interleukin 1β (IL-1β), IL-10, and tumor necrosis factor alpha (TNF-α), consistent with a role for AhR activation in the host response to MHV infection. Furthermore, while indoleamine 2,3-dioxygenase (IDO1) drives AhR activation in other settings, MHV infection induced equal expression of downstream genes in wild-type (WT) and IDO1−/− macrophages, suggesting an alternative pathway of AhR activation. In summary, we show that coronaviruses elicit AhR activation by an IDO1-independent pathway, contributing to upregulation of downstream effectors, including the proviral factor TiPARP, and to modulation of cytokine gene expression, and we identify a previously unappreciated role for AhR signaling in CoV pathogenesis.
IMPORTANCE Coronaviruses are a family of positive-sense RNA viruses with human and agricultural significance. Characterizing the mechanisms by which coronavirus infection dictates pathogenesis or counters the host immune response would provide targets for the development of therapeutics. Here, we show that the aryl hydrocarbon receptor (AhR) is activated in cells infected with a prototypic coronavirus, mouse hepatitis virus (MHV), resulting in the expression of several effector genes. AhR is important for modulation of the host immune response to MHV and plays a role in the expression of TiPARP, which we show is required for maximal viral replication. Taken together, our findings highlight a previously unidentified role for AhR in regulating coronavirus replication and the immune response to the virus.
INTRODUCTION
Coronaviruses (CoVs) are a group of positive-sense, single-stranded RNA viruses responsible for agricultural and human disease with a high economic burden and outbreak potential. Understanding the pathways driving CoV replication and pathogenesis is crucial to combat CoVs with high mortality, such as severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) CoVs. Mouse hepatitis virus (MHV), a prototypic CoV, causes hepatitis and/or encephalitis, depending on the strain. Previous studies have detailed many of the cellular pathways critical for or elicited by MHV infection (1, 2). Several antiviral mechanisms are induced by MHV infection, but MHV encodes proteins that counter these host processes. For instance, interferons (IFNs), especially type I IFNs (IFN-I), are vital to limiting MHV infection in mice (3, 4), but IFN-I production and signaling are inhibited during MHV infection in multiple cell types (5 – 7). MHV also inhibits the functions of downstream IFN-stimulated genes (ISGs). For example, the MHV macrodomain, a virus-encoded ADP-ribosylhydrolase, reverses cellular ADP ribosylation by IFN-I-induced poly(ADP-ribose) polymerases (PARPs) that limit viral replication (8, 9).
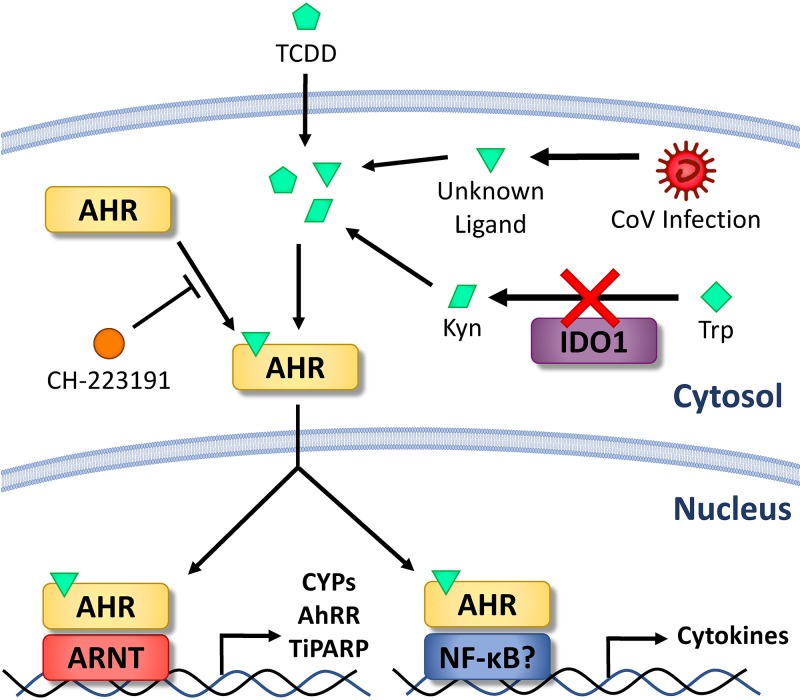
The aryl hydrocarbon receptor (AhR) is a receptor/transcription factor that has been shown to direct multiple host responses. While initially believed to operate only in the context of the cellular response to toxins, AhR is now recognized as a significant regulator of the host immune response, as well. AhR in the cytosol is activated by binding ligands that are exogenous, such as the toxin 2,3,7,8-tetrachlorodibenzodioxin (TCDD), or endogenous, such as cellular metabolites (Fig. 1). This triggers AhR translocation to the nucleus, where AhR complexes with AhR nuclear translocator (ARNT) or other binding partners to induce expression of several different proteins (downstream effectors) responsible for degrading the xenobiotic agent and for limiting potential cellular damage. Upregulated genes include those encoding cytochrome P450 enzymes (CYPs) that catabolize exotoxin, negatively regulating AhR activation by depleting AhR ligands (10). Another inhibitory protein, the AhR repressor (AhRR), is also upregulated and competes with ARNT for AhR dimerization in the nucleus (11). PARP7, also known as TCDD-inducible PARP (TiPARP), is highly induced by AhR activation, as well, indicating a relationship to cellular ADP ribosylation. TiPARP is responsible for mitigating pathology after TCDD administration to mice, at least in part due to feedback inhibition of AhR (12).
FIG 1.
Schematic diagram of AhR activation in MHV-infected cells. Ligands that can bind to AhR can be exogenous (pentagons), such as toxins like TCDD, or endogenous metabolic products (parallelograms), such as kynurenine derived by IDO1-catalyzed tryptophan degradation. CoV infection produces an unknown AhR-activating ligand independent of IDO1 (triangles). AhR in the cytosol is activated upon ligand binding and translocates to the nucleus to bind to genomic DNA. The specific genes targeted and induced by AhR are influenced by AhR binding partners, including ARNT and NF-κB. Modulated genes include those encoding AhR downstream effectors, such as CYPs, AhRR, and TiPARP, or immune proteins, such as cytokines. AhR activation/ligand binding can be inhibited chemically by treatment with CH-223191.
In the context of the immune response, endogenous metabolites are likely the primary ligands that drive AhR activation. The prototypical endogenous ligand is kynurenine, a tryptophan catabolite produced by indoleamine 2,3-dioxygenase 1 (IDO1) or IDO2 in immune cells or tryptophan 2,3-dioxygenase (TDO) (encoded by the TDO2 gene) in the liver (13, 14). IDO1, the best characterized of these enzymes, is induced by inflammatory factors, such as IFN-I or IFN-II, transforming growth factor beta (TGF-β), and interleukin 6 (IL-6) (15). AhR also activates IDO1 expression and enhances its activity through multiple pathways (16), and the resulting IDO1-AhR-IDO1 positive-feedback loop prolongs the effects of AhR activation (17, 18). Though kynurenine is believed to be the predominant derivative of tryptophan that drives AhR activation, multiple other degradation products of tryptophan or other biomolecules are synthesized independently of IDO1/2 or TDO or can be sourced from the gut microflora, diet, or even UV-mediated photo-oxidation (19). Prostaglandins, cyclic AMP (cAMP), and oxidative species may also activate AhR, though with unknown physiological or pathological significance (20 – 22).
Many studies have demonstrated that AhR activation during immunostimulation and inflammation generally exerts an immunosuppressive effect via multiple mechanisms (23). AhR activation by chemical agonists has been shown to influence the differentiation (24 – 28) and cytokine/chemokine production (25, 26, 29 – 32) of T cells, dendritic cells, and macrophages. AhR has also been shown to bind to and modulate the transcription specificity of NF-κB in multiple experimental settings, which could contribute to cytokine modulation (30, 33 – 35). Other studies have shown that AhR activation in immune cells is driven by IDO1, and the resulting IDO1-AhR-IDO1 positive-feedback loop helps to establish immunotolerance (18, 36, 37). In contrast, less is known about the role of AhR during virus infection. While previous work has explored pathways affected by AhR activation during influenza A virus (IAV), herpes simplex virus (HSV), hepatitis C virus (HCV), and Epstein-Barr virus (EBV) infection (38 – 41), the virological impact of AhR is still largely uncharacterized. Here, we show that CoV replication in macrophages results in AhR activation in an IDO1-independent manner, leading to increased expression of several downstream effectors and to modulation of the cytokine response. We also show that TiPARP, induced by AhR, is a proviral factor in CoV-infected cells.
RESULTS
MHV-A59 infection induces TiPARP expression through IFN-I-dependent and -independent mechanisms.
Infection of bone-marrow-derived macrophages (BMDMs) with the neurotropic JHM strain of MHV (MHV-JHM) results in increased expression of several IFN-I-inducible PARPs (PARP7, -9 through -12, and -14) (8). However, PARP7 (TiPARP) was also induced during MHV-JHM infection of IFNAR−/− BMDMs, suggesting that other factors besides IFN-I mediate TiPARP upregulation during infection (8). To expand on these results, we infected delayed brain tumor (DBT) astrocytoma cells (Fig. 2A) or 17Cl-1 fibroblast-like cells (Fig. 2B), both of which minimally produce IFN during MHV infection (6, 7), with the A59 strain of MHV (MHV-A59). We used MHV-A59 because it replicates to higher titers than MHV-JHM in vitro (42). Upregulation of most PARPs, including PARP9, -11, -12, and -14, was lost or attenuated in these cell lines during infection, contrasting with the robust PARP upregulation profile seen in MHV-A59-infected wild-type (WT) BMDMs (Fig. 2C). Despite the diminished expression of many IFN-dependent PARPs, TiPARP was highly upregulated following MHV-A59 infection in all three cell types, suggesting a conserved mechanism of induction. Furthermore, induction of TiPARP in IFNAR−/− BMDMs was conserved after infection with MHV-A59, consistent with previous findings with MHV-JHM (Fig. 2D). Although not further studied here, PARP13 was also upregulated in infected IFNAR−/− cells. Overall, these results indicate that TiPARP is upregulated by another pathway during infection in the absence of IFN-I signaling.
FIG 2.
MHV-A59 infection upregulates TiPARP in cell lines and primary cells in the absence of IFN-I signaling. (A and B) DBT (A) or 17Cl-1 (B) cell lines were infected with MHV-A59 at an MOI of 0.1 PFU/cell. Cells were collected at 18 hpi, and RNA was quantified by qRT-PCR with primers for the indicated PARPs. (C and D) WT (C) or WT and IFNAR−/− (D) BMDMs were mock infected or infected with MHV-A59 at an MOI of 5 (C) or 0.1 (D) PFU/cell. Cells were collected at 18 hpi, and mRNA was analyzed for quantification by qRT-PCR with primers for the indicated PARPs. The data show the results of one experiment representative of three independent experiments; n = 3. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ns, not significant; nd, not detectable. The data are expressed as means and SEM.
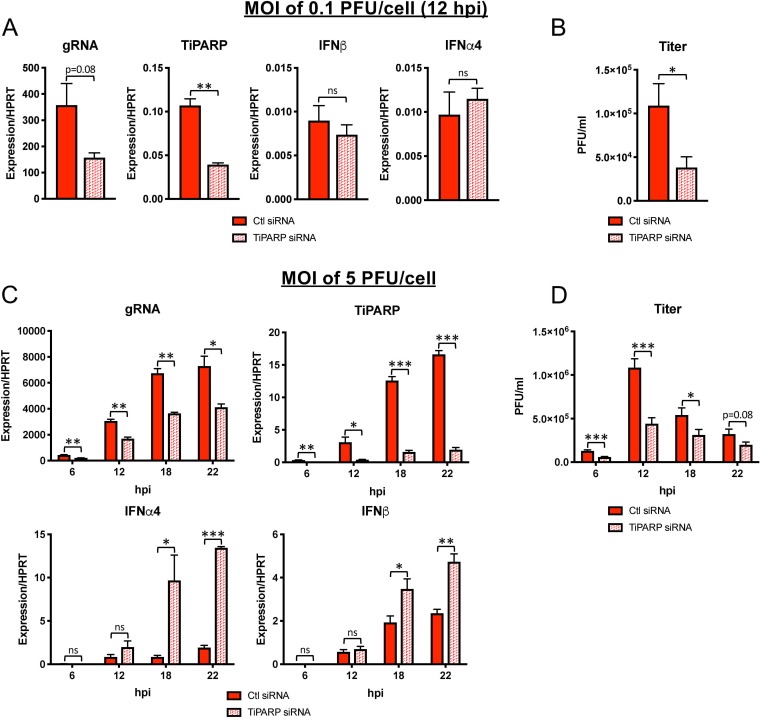
TiPARP knockdown restricts MHV-A59 replication.
We previously noted that viral genomic RNA (gRNA) levels during MHV-JHM infection were reduced in BMDMs treated with small interfering RNA (siRNA) directed toward TiPARP (8). To confirm this phenotype, we treated BMDMs with TiPARP-specific siRNA prior to infection with MHV-A59 at low (0.1 PFU/cell) and high (5 PFU/cell) multiplicities of infection (MOI) and measured replication by quantification of viral genomic content and infectious virus (Fig. 3). At low MOI at 12 h postinfection (hpi), TiPARP knockdown decreased viral gRNA levels (Fig. 3A) and titers (Fig. 3B), though the former only trended toward statistical significance. MHV infection at an MOI of 5 PFU/cell showed significantly decreased gRNA levels and virus titers in TiPARP knockdown cells (Fig. 3C and D), indicating a role for TiPARP in facilitating MHV-A59 infection. These differences in gRNA and virus titers persisted even at later time points p.i., when cell viability was decreased, resulting in decreased infectious-virus titers. Furthermore, while IFN-α4 and IFN-β mRNA levels were unaffected by TiPARP deficiency at low or high MOI at 6 or 12 hpi, they were increased at 18 and 22 hpi (Fig. 3A and C). Together, our results suggest that TiPARP augments MHV replication throughout infection and negatively regulates IFN-I expression during later stages of infection.
FIG 3.
MHV replication is diminished following TiPARP knockdown in BMDMs. WT BMDMs were transfected with siRNA targeting TiPARP and were then infected at 30 h posttransfection with MHV-A59 at an MOI of 0.1 (A and B) or 5 (C and D) PFU/cell. RNA was quantified by qRT-PCR with primers specific for viral gRNA or for the indicated transcripts (A and C), or virus titers were determined by freezing-thawing cells, followed by plaque assay on HeLa cells expressing the MHV receptor (B and D). RNA and cell collection occurred at 12 hpi for an MOI of 0.1 PFU/cell (A and B) or at 6, 12, 18, and 22 hpi for an MOI of 5 PFU/cell (C and D). (A and C) The data show the results of one experiment representative of at least two independent experiments; n = 3. (B and D) The data show combined results of 2 experiments; n = 6. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ns, not significant. The data are expressed as means and SEM.
MHV-A59 replication in vitro and in vivo induces expression of effector genes downstream of AhR activation.
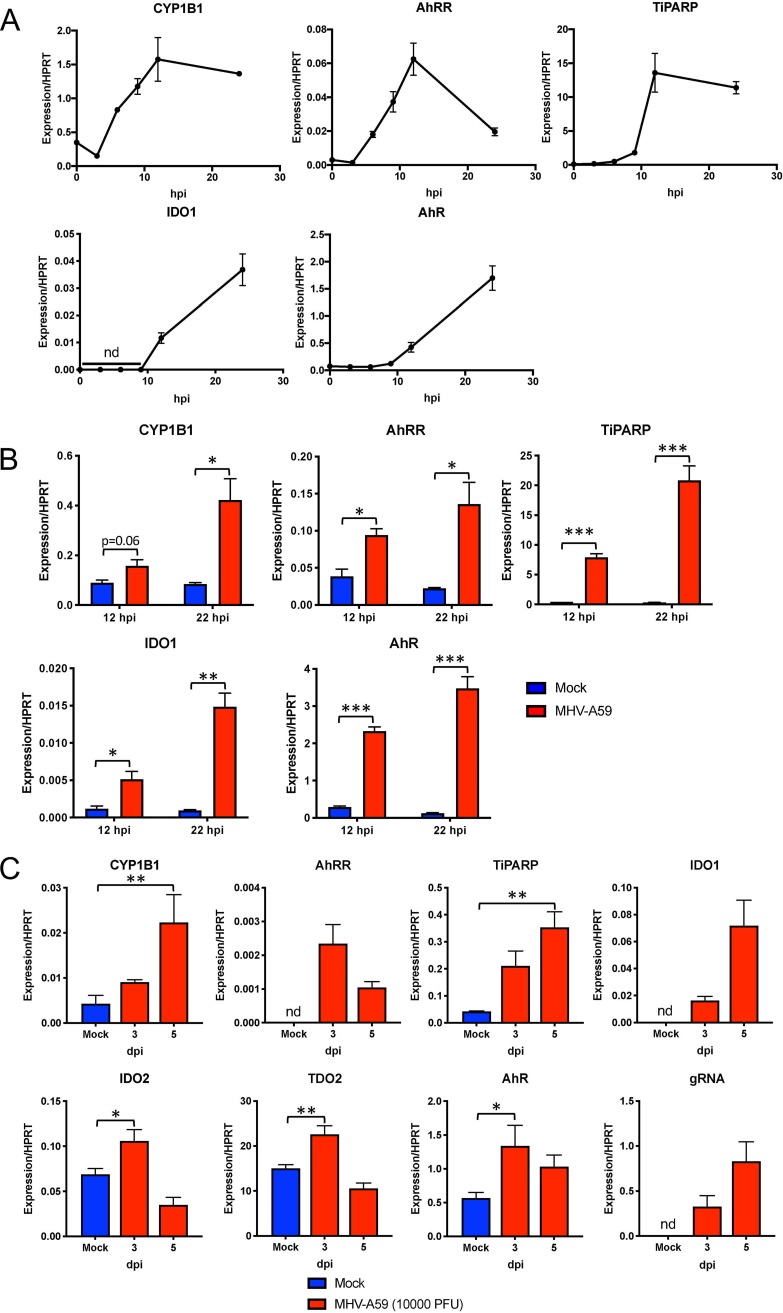
Because expression of TiPARP is well established to be induced by ligand-activated AhR (43), we hypothesized that MHV-A59 infection resulted in AhR activation. To assess AhR activation during MHV-A59 infection in BMDMs at multiple time points, we quantified the mRNA expression of known effectors downstream of AhR, including CYP1A1, CYP1A2, CYP1B1, TiPARP, and AhRR (Fig. 1). We also analyzed gene expression of AhR itself and of the AhR ligand-producing enzymes IDO1, IDO2, and TDO, as IDO1 gene expression can also be induced by AhR activation. We found that, while CYP1A1, CYP1A2, IDO2, and TDO2 mRNAs were undetectable at all time points, CYP1B1, AhRR, TiPARP, IDO1, and AhR mRNAs were all upregulated over the course of infection (Fig. 4A). To determine if this response is conserved in other immune cell types, we quantified the transcription of these downstream effectors in bone marrow-derived dendritic cells (BMDCs) (Fig. 4B). At both 12 and 22 hpi, CYP1B1, AhRR, TiPARP, IDO1, and AhR were all upregulated in BMDCs, suggesting that AhR activation occurs following MHV-A59 infection in multiple cell types.
FIG 4.
MHV infection results in upregulation of AhR downstream effector genes in BMDMs and BMDCs and in vivo. (A) BMDMs were infected with MHV-A59 at an MOI of 5 PFU/cell and collected at the indicated time points. mRNA levels of downstream effector AhR or IDO1 genes were then quantified by qRT-PCR. (B) BMDCs were infected with MHV-A59 at an MOI of 5 PFU/cell and collected at 12 or 22 hpi. mRNA levels of the indicated genes were quantified by qRT-PCR. (A and B) The data show the results of one experiment representative of two independent experiments; n = 3. (C) C57BL/6 mice were intraperitoneally infected with 104 PFU of MHV-A59, and perfused livers were harvested at 3 and 5 dpi. RNA was isolated, and mRNA levels of downstream effectors, AhR, or kynurenine-producing enzymes were quantified by qRT-PCR. The data show the results of one experiment representative of three independent experiments; n = 4. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; nd, not detectable. The data are expressed as means and SEM.
To determine if our in vitro results could be recapitulated in vivo, we infected C57BL/6 mice with MHV-A59 via intraperitoneal injection (Fig. 4C). At 3 and 5 days postinfection (dpi), levels of CYP1B1, TiPARP, AhRR, and IDO1 mRNA increased in the livers of infected mice, suggesting that MHV elicited AhR activation. Furthermore, upregulation of these downstream genes paralleled viral replication at 3 and 5 dpi, as assessed by measuring viral gRNA levels. In contrast to our results in BMDMs, IDO2 and TDO2 mRNAs were detectable in liver samples and modestly increased at 3 dpi.
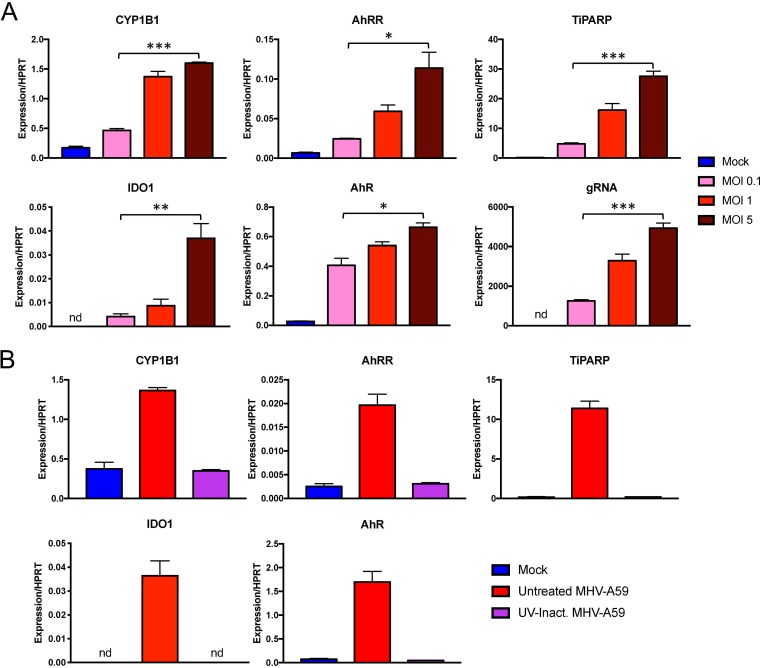
Consistent with the results obtained using infected livers, the level of AhR activation was correlated with virus replication in BMDMs, as downstream effector mRNA levels were less upregulated when cells were infected at lower MOI (Fig. 5A). In addition, UV-inactivated virus infection was unable to induce expression of AhR downstream effectors, indicating that AhR activation during MHV infection is completely dependent on viral replication (Fig. 5B).
FIG 5.
Virus replication is correlated with and required for expression of AhR downstream effectors during infection. (A) BMDMs were infected with MHV-A59 at the indicated MOIs. Cells were then collected at 12 hpi, and mRNA levels of the downstream effectors AhR and IDO1 were quantified by qRT-PCR. (B) BMDMs were infected with untreated or UV-inactivated virus at an MOI of 5 PFU/cell. Expression of the indicated genes was quantified by qRT-PCR. The data show the results of one experiment representative of two independent experiments; n = 3. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; nd, not detectable. The data are expressed as means and SEM.
AhR antagonist and agonist treatment modulates expression of downstream effector genes during infection.
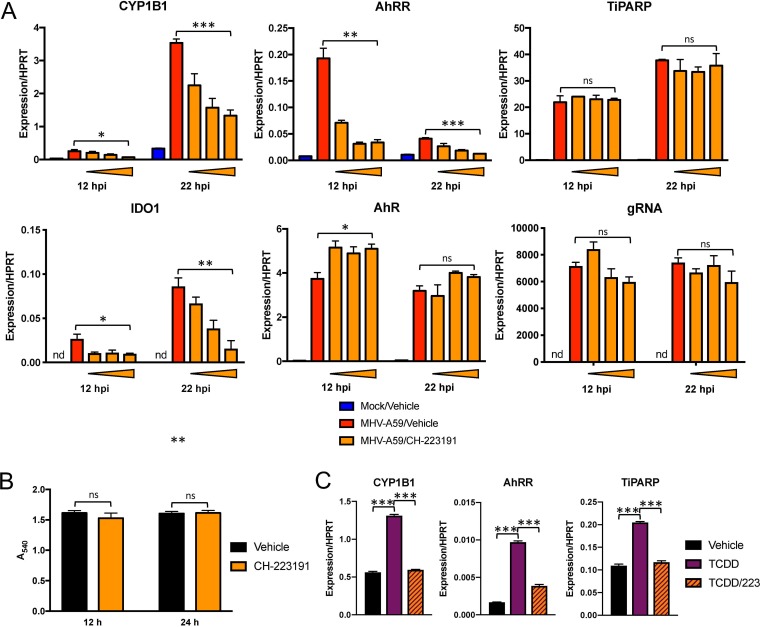
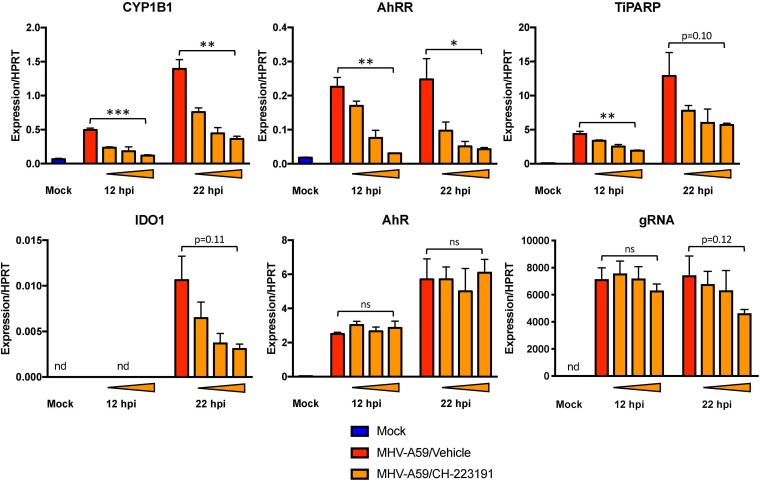
To examine whether AhR activation and not an alternative factor facilitated expression of these downstream effectors, we infected cells following chemical inhibition of AhR (Fig. 6A). We opted for treatment with CH-223191, a well-described chemical inhibitor that prevents ligand binding to AhR (Fig. 1) but does not inhibit other receptors, such as the estrogen receptor (44, 45). We first confirmed that CH-223191 inhibited chemical AhR activation by agonist TCDD in BMDMs without altering cell viability (Fig. 6B and C). During MHV-A59 infection, AhR inhibitor treatment resulted in dose-dependent attenuation of CYP1B1, AhRR, and IDO1. Surprisingly, TiPARP induction was not diminished at any concentration of inhibitor. Furthermore, CH-223191 treatment actually increased AhR mRNA levels slightly at 12 hpi but had no effect on gRNA levels, indicating that the CH-223191-mediated reduction in downstream effector expression was due to inhibition of AhR activation itself rather than decreased AhR expression or increased virus replication.
FIG 6.
Chemical inhibition of AhR during MHV infection results in attenuated expression of downstream genes. (A) WT BMDMs were pretreated with vehicle (0.01% DMSO) or 0.2, 1, or 5 μM CH-223191 and infected with MHV-A59 at an MOI of 5 PFU/cell. RNA was collected at 12 or 22 hpi and quantified for mRNA levels of the downstream effectors AhR and IDO1 and gRNA by qRT-PCR. The data show the results of one experiment representative of three independent experiments; n = 3. (B) Viability of BMDMs treated with vehicle or 5 μM CH-223191 was quantified by MTT assay at 12 and 24 h posttreatment. The data show the results of one experiment representative of two independent experiments; n = 4. (C) CH-223191 inhibitor efficacy was determined by preincubating uninfected BMDMs with vehicle (0.01% DMSO) or 5 μM CH-223191, followed by addition of vehicle or 10 nM TCDD. At 12 h, RNA was harvested, and mRNA levels of downstream effectors were determined by qRT-PCR. The data show the results of one experiment representative of two independent experiments; n = 3. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ns, not significant; nd, not detectable. The data are expressed as means and SEM.
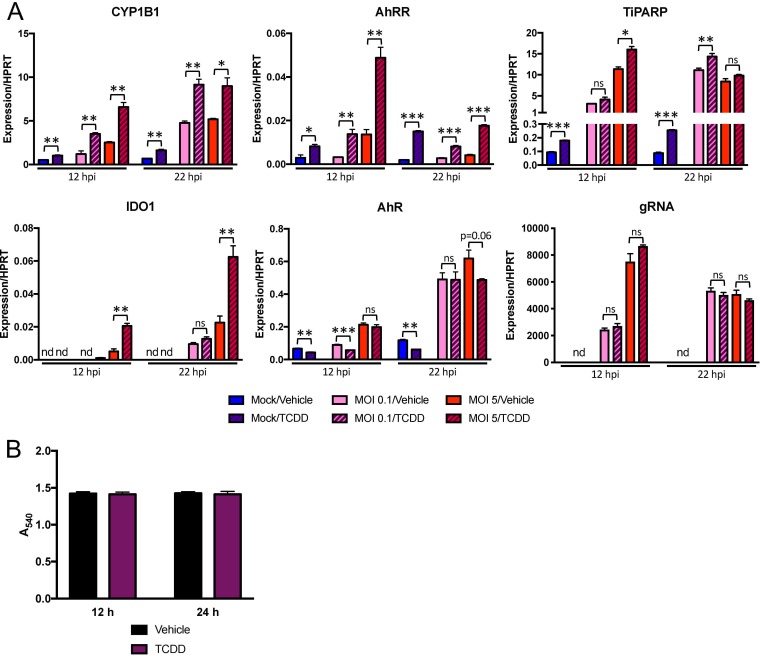
To complement these results and to determine if concurrent chemical activation during infection could further activate AhR, we treated BMDMs with TCDD and quantified induction of the same downstream effector genes that were attenuated by AhR inhibition (Fig. 7A). After confirming that TCDD did not affect cell viability (Fig. 7B), we found that agonist treatment increased expression of CYP1B1 and AhRR compared to vehicle treatment in both mock- and MHV-infected BMDMs. TiPARP mRNA increased following TCDD treatment of uninfected cells and only marginally, if at all, after infection. IDO1 required infection for any expression and was potentiated by TCDD at 22 hpi. Finally, TCDD treatment with or without infection resulted in small decreases in AhR expression in mock- and MHV-infected cells at 12 hpi but did not affect gRNA levels, again suggesting that agonist-induced changes in downstream effector expression was due primarily to AhR activation.
FIG 7.
Chemical activation of AhR in the absence and presence of infection enhances expression of downstream genes. (A) WT BMDMs were treated with vehicle (0.01% DMSO) or 10 nM TCDD and concurrently mock infected or infected with MHV-A59 at an MOI of 0.1 or 5 PFU/cell. RNA was collected at 12 or 22 hpi and quantified for mRNA levels of the downstream effectors AhR and IDO1 and gRNA by qRT-PCR. The data show the results of one experiment representative of two independent experiments; n = 3. (B) Viability of BMDMs treated with vehicle or 10 nM TCDD was quantified by MTT assay at 12 and 24 h posttreatment. The data show the results of one experiment representative of two independent experiments; n = 4. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ns, not significant; nd, not detectable. The data are expressed as means and SEM.
TiPARP is regulated by both IFN and the AhR during MHV infection.
TiPARP expression was induced by AhR agonist treatment (Fig. 7A) but did not change following inhibitor treatment during infection (Fig. 6A), suggesting MHV could also induce TiPARP expression by an AhR-independent mechanism. Because IFN-I treatment in BMDMs can induce TiPARP expression (8), we next examined whether IFN-I upregulated TiPARP expression after inhibition of AhR activation. Using MHV-infected IFNAR−/− BMDMs treated with CH-223191, we observed a dose-dependent decrease in the expression of CYP1B1, AhRR, and IDO1 (Fig. 8), confirming that AhR activation did not require IFN signaling. In addition, AhR inhibitor treatment reduced TiPARP induction in IFNAR−/− BMDMs, indicating that AhR and IFN-I were compensatory in inducing TiPARP during infection. However, CH-223191-mediated reduction of TiPARP mRNA levels in IFNAR−/− BMDMs was less than that of CYP1B1, AhRR, or IDO1, suggesting that an additional, as yet unknown factor modulated its expression. Finally, gRNA levels in IFNAR−/− BMDMs trended toward a modest reduction following AhR inhibition (P = 0.12), possibly reflecting decreases in TiPARP expression. Taken together, our data show that MHV-induced AhR activation is responsible for upregulation of CYP1B1, AhRR, and IDO1 in IFN-replete and -deficient cells and of TiPARP in the absence of IFN-I signaling.
FIG 8.
Infection upregulates TiPARP through redundant mechanisms requiring AhR activation or IFN-I signaling. BMDMs from IFNAR−/− mice were pretreated with vehicle (0.01% DMSO) or 0.2, 1, or 5 μM CH-223191 and infected with MHV-A59 at an MOI of 5 PFU/cell. RNA was collected at 12 or 22 hpi and quantified for mRNA levels of the downstream effectors AhR and IDO1 and gRNA by qRT-PCR. The data show the results of one experiment representative of two independent experiments; n = 3. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ns, not significant; nd, not detectable. The data are expressed as means and SEM.
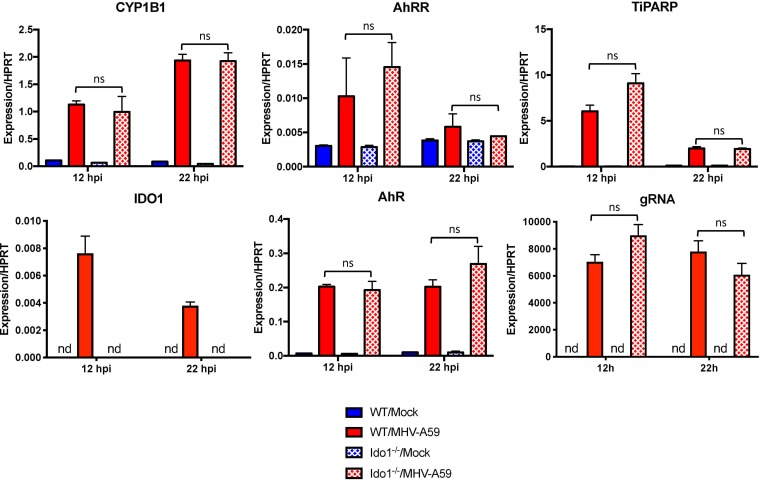
MHV infection activates AhR in an IDO1-independent manner.
While our results indicated that AhR activation during MHV-A59 infection modulates downstream IDO1 expression (Fig. 6 to 8), IDO1 can also act as an upstream regulator of AhR by catabolizing tryptophan to the AhR ligand kynurenine (Fig. 1) (13, 17). To determine if IDO1 has a role in AhR activation during MHV-A59 infection, we infected IDO1−/− BMDMs with MHV-A59 and quantified effector mRNA levels (Fig. 9B). Interestingly, levels of CYP1B1, AhRR, TiPARP, AhR, and gRNA were equivalent following infection in WT or IDO1−/− BMDMs. As expected, IDO1 mRNA was not detectable in deficient cells. To rule out compensatory effects of other enzymes known to produce kynurenine, we also assessed cells for IDO2 and TDO2 mRNAs but could detect neither in WT or IDO1−/− BMDMs. Together, our results suggest that MHV-A59 infection of BMDMs elicits AhR activation through a pathway independent of IDO1.
FIG 9.
IDO1 expression is dispensable for MHV-mediated AhR activation. BMDMs from WT or IDO1−/− mice were infected with MHV-A59 at an MOI of 5 PFU/cell. RNA was collected at 12 or 22 hpi, and expression of the indicated genes was quantified by qRT-PCR. The data show the results of one experiment representative of two independent experiments; n = 3. ns, not significant; nd, not detectable. The data are expressed as means and SEM.
MHV-induced AhR activation modulates cytokine production.
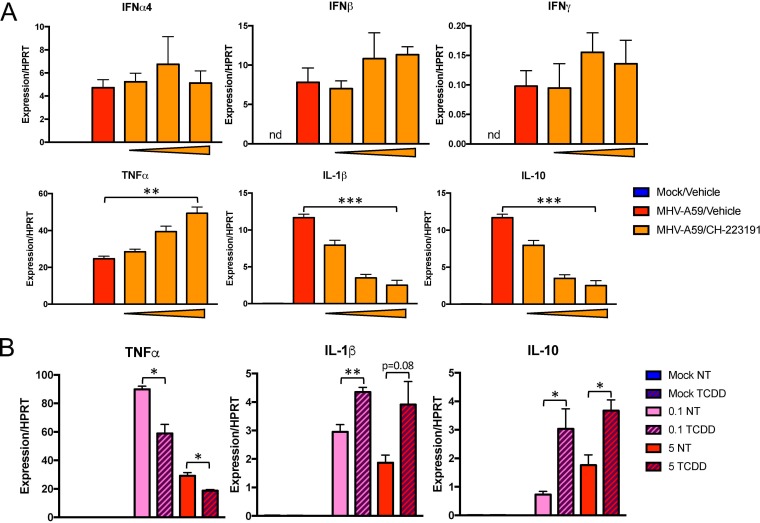
AhR activation can induce or modulate cytokine/chemokine production during the innate immune response (23). To determine whether AhR activation during CoV infection modulates cytokine expression, we infected CH-223191-treated BMDMs with MHV-A59 and quantified levels of IFN-α4, IFN-β, and IFN-γ mRNAs and of cytokines previously shown to be regulated by lipopolysaccharide (LPS)-induced AhR activation in macrophages (tumor necrosis factor alpha [TNF-α], IL-1β, and IL-10) (29, 30) (Fig. 10A). Inhibition of AhR had no impact on expression of IFN-α4/IFN-β/IFN-γ mRNAs. On the other hand, CH-223191 treatment resulted in a dose-dependent increase in the levels of TNF-α mRNA and a concurrent decrease in the levels of IL-1β and IL-10 mRNAs. Because AhR could be further activated by chemical means during infection (Fig. 7), we substantiated these findings by treating BMDMs with TCDD in the presence or absence of infection at low and high MOI (Fig. 10B). Consistent with our AhR inhibitor data, TNF-α mRNA was decreased and IL-1β and IL-10 mRNAs were increased by concurrent chemical activation of AhR. In summary, these results indicate that AhR activation modulates multiple cytokine expression levels during MHV-A59 infection.
FIG 10.
Inhibition or enhancement of AhR activation during MHV infection modulates cytokine expression. (A) WT BMDMs were pretreated with vehicle (0.01% DMSO) or 0.2, 1, or 5 μM CH-223191 and then infected at an MOI of 5 PFU/cell. RNA was collected and quantified at 22 hpi for mRNA levels of the indicated cytokines by qRT-PCR. The data show the results of one experiment representative of three independent experiments; n = 3. (B) WT BMDMs were treated with vehicle (0.01% DMSO) or 10 nM TCDD and concurrently infected with MHV-A59 at an MOI of 0.1 or 5 PFU/cell. RNA was collected and quantified at 22 hpi for mRNA levels of the indicated cytokines by qRT-PCR. The data show the results of one experiment representative of two independent experiments; n = 3. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ns, not significant; nd, not detectable. The data are expressed as means and SEM.
DISCUSSION
Here, we showed that MHV infection activates AhR, resulting in upregulation of multiple downstream effector genes, including CYP1B1, AhRR, and IDO1 genes, in infected BMDMs, BMDCs, and mouse livers (Fig. 4). We also demonstrated that another AhR downstream effector, TiPARP, has a proviral role in MHV replication, as genomic RNA levels and virus titers were decreased following TiPARP knockdown (Fig. 3). TiPARP induction is multifactorial, since IFN-I treatment induced TiPARP expression in BMDMs (8) but TiPARP expression was still observed in cell lines deficient in IFN expression and in IFNAR−/− BMDMs (Fig. 2). Our results showed that AhR activation directly induces TiPARP expression, as TCDD treatment enhanced TiPARP expression in uninfected cells and to a lesser extent in infected cells (Fig. 6). Further, TiPARP induction during infection was sensitive to chemical AhR inhibition in the absence, but not the presence, of IFN-I signaling in WT BMDMs (Fig. 6 and 8), consistent with redundant roles for AhR and IFN-I signaling in TiPARP expression during infection. Despite attenuation with inhibitor treatment, TiPARP expression in infected IFNAR−/− cells was still induced, indicating that other factors known to modulate TiPARP expression, such as estrogen receptor, glucocorticoid receptor, or TGF-β signaling pathways, could also impact its expression during CoV infection (46 – 48).
In addition to decreased replication, TiPARP knockdown also resulted in increased IFN-α4 and IFN-β mRNA expression (Fig. 3C). However, viral genomic RNA levels were reduced in TiPARP knockdown cells as early as 6 hpi, before IFN-I mRNA levels increased. Further study will be required to establish a causal relationship between the effects of TiPARP on IFN-I expression and replication. Our results align with those of a previous study showing that during IAV infection, TiPARP ADP ribosylates TBK1, resulting in increased replication and reduced IFN-I levels (40). However, TiPARP has also been shown to bind to Sindbis virus RNA to trigger an antiviral host response. Therefore, it will be important to determine whether TiPARP facilitation of MHV replication is dependent on its ADP-ribosylating and/or its RNA-binding activity.
In contrast with TiPARP, upregulation of the downstream effectors CYP1B1, AhRR, and IDO1 during infection is driven primarily by AhR activation. This is evidenced by the fact that expression of these effectors was enhanced by an AhR agonist and attenuated in a dose-dependent manner by an AhR-specific inhibitor (Fig. 6 and 7). While AhR activation has been studied in immune cells following agonist or immunostimulant treatment, its role in the context of virus infection is relatively understudied, and only a few studies have details of host pathways affected by AhR activation. EBV encodes the viral protein EBNA-3, which binds to ARNT and enhances transactivation of downstream effectors by AhR (41). AhR facilitates HCV infection by inducing expression of CYPs that aid in the formation of lipid droplets required for virus production (38). As mentioned above, AhR functions in a proviral manner in IAV infection by inducing TiPARP (40). In contrast, AhR activation in HSV- or HIV-infected macrophages restricts replication by inhibiting expression of cyclins and cyclin-dependent kinases (39). Thus, AhR activation during viral infection has differing effects and modulates multiple cellular pathways, many of which remain uncharacterized.
Surprisingly, while AhR is thought to be activated primarily by kynurenine synthesized via IDO1 in inflammatory states and can drive positive-feedback cycles in immune cells (17, 18), our results demonstrate that MHV infection activates AhR independently of IDO1 (Fig. 1 and 9). We detected gene expression of IDO2 and TDO2 in infected murine liver samples (Fig. 4C), consistent with the notion that TDO is largely confined to the liver (14). While TDO or IDO2 could be driving AhR activation by producing kynurenine during in vivo infection, IDO2 and TDO mRNAs were undetectable in IDO1−/− BMDMs. This suggests that kynurenine is produced by a novel IDOI/IDO2/TDO-independent pathway in BMDMs or that the cells do not utilize kynurenine as the primary AhR ligand during CoV infection. AhR can be activated by several biochemical species, including metabolites of tryptophan or bilirubin or other metabolites of uncharacterized biosynthetic pathways (49 – 52). Other potential initiating pathways include prostaglandin synthesis, cAMP production, and general oxidative stress (20 – 22). Alternatively, during infection, a host or viral protein could directly bind to AhR, enhancing its transactivation activity in a manner similar to that of EBV-encoded EBNA-3, though this may still require concomitant ligand binding (41). Nonetheless, the levels of the ligand or binding partner activating AhR in BMDMs would likely scale with replication, as downstream effector transcription was correlated with the virus load (Fig. 5). Additional investigation will be necessary to detail the exact mechanism of this IDO-independent AhR activation pathway. Furthermore, while IDO1 does not regulate AhR activation during MHV infection in BMDMs, it could still be modulating other cellular pathways as part of the host response. For instance, IDO1 has been shown to regulate immune cell function by mediating tryptophan starvation, resulting in activation of mTOR, a mediator of several metabolic and immune pathways (53, 54).
Finally, chemical activation or inhibition of AhR during infection resulted in changes in the mRNA levels of multiple cytokines (Fig. 10), suggesting that AhR functions to modulate the cytokine response to MHV infection. Specifically, AhR negatively regulated TNF-α and positively regulated IL-1β and IL-10, cytokines that are also upregulated in the brains of MHV-infected mice (2). These changes were not due to alterations in viral replication, as the viral load was unchanged by AhR activation or inhibition. The mechanism driving cytokine changes during MHV infection is likely complex but probably involves interaction of activated AhR with NF-κB (30, 33 – 35, 55) (Fig. 1). The effects of these cytokine expression changes will need to be investigated in vivo to determine their roles in pathogenesis, because their functions are protean: some of these cytokines are additionally posttranscriptionally regulated, and activating ligands may differ in cultured BMDMs versus infected mice. Thus, it is difficult to conclude at present that AhR drives a strictly pro- or anti-inflammatory phenotype. Rather, AhR activation may serve to fine tune the innate immune response to MHV infection.
MATERIALS AND METHODS
Mice.
Pathogen-free C57BL/6 WT and IFNAR−/− mice were purchased from Jackson Laboratories, and IDO1−/− mice were obtained as a generous gift from Mark Santillan (University of Iowa, Iowa City, IA). All mice were bred and maintained in the animal care facility at the University of Iowa as approved by the University of Iowa Institutional Animal Care and Use Committee (IACUC) following guidelines set forth in the Guide for the Care and Use of Laboratory Animals.
Cell cultures.
DBT cells, 17Cl-1 cells, and HeLa cells expressing the MHV receptor carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) (HeLa-MHVR cells) were grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin. Bone marrow cells obtained from WT, IFNAR−/−, and IDO1−/− C57BL/6 mice were differentiated into macrophages (BMDMs) by incubating the cells with 10% L929 cell supernatant and 10% FBS in RPMI medium for 7 or 8 days. The BMDMs were washed and replaced with fresh medium every day after the 4th day. Bone marrow cells obtained from WT C57BL/6 mice were differentiated into dendritic cells (BMDCs) by incubating them with 50 μg/ml granulocyte-macrophage colony-stimulating factor (GM-CSF) and 20 μg/ml IL-4. Extra medium was added at day 3 to refresh the cells, the medium was completely changed on day 6, and the cells were used for infections on day 7.
Virus infection.
MHV-A59 (56) was propagated on 17Cl-1 cells in the same manner described previously (8). BMDMs were infected with virus at the indicated MOI with a 45-min adsorption phase. At the indicated time points, the cells were lysed with TRIzol for RNA isolation or the cells were frozen with supernatants for determination of titers on HeLa cells. To generate replication-deficient virus, MHV-A59 stocks were UV inactivated using a biosafety cabinet UV lamp for 30 min at room temperature. Inactivation was confirmed by plaque assay. For AhR agonist studies, medium with 10 nM TCDD was added after mock adsorption or adsorption with MHV-A59. For chemical inhibitor studies, BMDMs were pretreated with 0.2, 1, or 5 μM CH-223191 (Sigma-Aldrich) vehicle (0.01% dimethyl sulfoxide [DMSO]) for 2 to 6 h prior to infection or to treatment with 10 nM TCDD. After TCDD treatment or infection, medium containing inhibitors was added back to the cells. For mouse infections, 5- to 8-week-old mice were anesthetized with ketamine/xylazine and inoculated intraperitoneally with 104 PFU of MHV-A59 in 300 μl DMEM or mock infected (DMEM only). Mice were sacrificed at 3 and 5 dpi, and livers were harvested and stored in TRIzol (Thermo Fisher Scientific).
siRNA transfection.
BMDMs were transfected with 50 pmol/ml of siRNA with Viromer Blue (Lipocalyx) following the manufacturer’s protocol. Media were replaced 4 h after transfection, and cells were infected 28 h posttransfection. The sequences of negative (nonspecific) control Dicer-substrate short interfering RNAs (DsiRNA) were as follows: sense, CGUUAAUCGCGUAUAAUACGCGUAT, and antisense, AUACGCGUAUUAUACGCGAUUAACGAC. The sequences of DsiRNA oligonucleotides directed toward TiPARP (Integrated DNA Technologies [IDT]) were as follows: sense, GAAGAUAAAAGUUAUCGAAUCAUTT, and antisense, AAAUGAUUCGAUAACUUUUAUCUUCUG.
RT-qPCR analysis.
RNA was purified using TRIzol (Thermo Fisher Scientific) on Direct-Zol columns (Zymo Research) or by phase separation as instructed by the manufacturer. cDNA was synthesized using Moloney murine leukemia virus (MMLV) reverse transcriptase (Thermo Fisher Scientific) and quantified with a real-time thermocycler using PowerUp SYBR green master mix (Thermo Fisher Scientific). The real-time quantitative PCR (RT-qPCR) primers are listed in Table 1. The primers spanned exon-exon junctions when possible to avoid quantification of any residual genomic DNA. A control without reverse transcriptase was also analyzed to confirm the absence of any contaminating DNA. Target genes were normalized to housekeeping gene hypoxanthine-guanine phosphoribosyltransferase (HPRT) by the following equation: ΔCT = CT (gene of interest) − CT (HPRT), where CT is the threshold cycle. All the results are shown as ratios to HPRT calculated as −2ΔCT.
TABLE 1.
RT-qPCR primers
| Gene | Primer (5′→3′) |
|
|---|---|---|
| Forward | Reverse | |
| HPRT | GCGTCGTGATTAGCGATGATG | CTCGAGCAAGTCTTTCAGTCC |
| PARP1 | CAGGAGAGTCAGCGATCTTGG | ACCCATTCCTTTCGGCTAGG |
| PARP2 | TGGAAGGCGAGTGCTAAATG | GGGCTTTGCCCTTTAACAGC |
| PARP3 | TGCGGCATGTTTGGAAAGTG | GTGCATGGTGGTAACATAGCC |
| PARP4 | AGTGCTACAGCCCGTTTCC | CACAGCTTTCAGTTGTGGGC |
| PARP5a | CCCTGAGGCCTTACCTACCT | TCAAGACCCGCAACTTCTCC |
| PARP5b | TGATGGCAGAAAGTCAACTCCA | GCCACAGGTCCATTGCATTC |
| PARP6 | GTACCTTGATGGACCAGAGCC | GCCAGCTCGGAACTTCTTGA |
| PARP7 | ATTTACAGACACTTGGTGGGG | GGCACTTGGATGAAGTCCTGA |
| PARP8 | CACTTCCGAAACCACTTCGC | TAGGATACACTTTTGGGGCCG |
| PARP9 | GCATTTGCTAAAGAGCACAAGGA | AAAGCACCACTATTACCGCTGA |
| PARP10 | CGAAACGGCACACTCTACGG | GAGACCCTCAAAGGAGGTGC |
| PARP11 | GGCTGTCTTTGGAAAAGGAACC | GCACTCGAGCAAGAAACATGG |
| PARP12 | AGACCGGGAAGAACTGTAGGA | TTTGGAAGGAGCAAGAGCCG |
| PARP13 | AGTAGTCCCACTGGTTTTGGC | TGCAACTCTGTGGCTTGTGG |
| PARP14 | TGCTGAAGCTGTCAAGACTACA | ACAATGGCATGGGTCGTAGC |
| PARP16 | CTTTGACCCGGCCAACTCC | AAACAGAGAAGTCTTGTTCAGGTG |
| gRNA | AGGGAGTTTGACCTTGTTCAG | ATAATGCACCTGTCATCCTCG |
| AhR | CCACTGACGGATGAAGAAGGA | ATCTCGTACAACACAGCCTCTC |
| CYP1A1 | CAGGACATTTGAGAAGGGCCAC | GCTTCCTGTCCTGACAATGC |
| CYP1A2 | TGGAGCTGGCTTTGACACAGT | GCCATGTCACAAGTAGCAAAATG |
| CYP1B1 | GGCTTCATTAACAAGGCGCT | CACTGATGAGCGAGGATGGA |
| AhRR | GTTGGATCCTGTAGGGAGCA | AGTCCAGAGGCTCACGCTTA |
| IDO1 | AGGATGCGTGACTTTGTGGA | TCCCAGACCCCCTCATACAG |
| IDO2 | CTCAGACTTCCTCACTTAATCG | GCTGCTCACGGTAACTCT |
| TDO2 | GTGAACGACGACTGTCATACCG | GCTGGAAAGGGACCTGGAAT |
| IFNα4 | TCCATCAGCAGCTCAATGAC | AGGAAGAGAGGGCTCTCCAG |
| IFN-β | TCAGAATGAGTGGTGGTTGC | GACCTTTCAAATGCAGTAGATTCA |
| IFN-γ | CGGCACAGTCATTGAAAGCCTA | GTTGCTGATGGCCTGATTGTC |
| TNF-α | TCAGCCGATTTGCTATCTCA | CGGACTCCGCAAAGTCTAAG |
| IL-1β | ACTGTTTCTAATGCCTTCCC | ATGGTTTCTTGTGACCCTGA |
| IL-10 | ATTTGAATTCCCTGGGTGAGAAG | CACAGGGGAGAAATCGATGACA |
Cell viability assay.
The viability/metabolism of BMDMs treated with 10 nM TCDD or 5 μM CH-223191 for 12 or 24 h was assessed using a Vybrant 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) cell proliferation assay (Thermo Fisher Scientific) according to the manufacturer’s instructions. Cell viability was measured by absorbance at 540 nm (A540).
Statistics.
An unpaired two-tailed Student t test was used to determine statistically significant differences in means between groups. All the graphs show means and standard errors of the mean (SEM). The n value represents the number of biological replicates for each figure. Multiple trials were combined into a single figure when expression values or titers were comparable. Significant P values are annotated. If the P value was less than 0.15 but greater than 0.05, the numerical value is listed above the graph bars and was considered to be trending toward significance.
ACKNOWLEDGMENTS
We thank members of the Perlman laboratory and Wendy Maury and Aloysius Klingelhutz for valuable discussions, Mark Santillan for mice, Alan Sariol for assistance with mouse experiments, and Aloysius Klingelhutz for critical readings of the manuscript.
This study was supported in part by grants from the NIH (ROI NS36592 [S.P.], CoBRE P20 GM113117-02 [A.R.F.], and K22 AI134993-01 [A.R.F.]) and the University of Kansas (A.R.F.).
REFERENCES
- 1.Fehr AR, Perlman S. 2015. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol 1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergmann CC, Lane TE, Stohlman SA. 2006. Coronavirus infection of the central nervous system: host-virus stand-off. Nat Rev Microbiol 4:121–132. doi: 10.1038/nrmicro1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roth-Cross JK, Bender SJ, Weiss SR. 2008. Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J Virol 82:9829–9838. doi: 10.1128/JVI.01199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ireland DD, Stohlman SA, Hinton DR, Atkinson R, Bergmann CC. 2008. Type I interferons are essential in controlling neurotropic coronavirus infection irrespective of functional CD8 T cells. J Virol 82:300–310. doi: 10.1128/JVI.01794-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou H, Perlman S. 2006. Preferential infection of mature dendritic cells by mouse hepatitis virus strain JHM. J Virol 80:2506–2514. doi: 10.1128/JVI.80.5.2506-2514.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou H, Perlman S. 2007. Mouse hepatitis virus does not induce Beta interferon synthesis and does not inhibit its induction by double-stranded RNA. J Virol 81:568–574. doi: 10.1128/JVI.01512-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Versteeg GA, Bredenbeek PJ, van den Worm SH, Spaan WJ. 2007. Group 2 coronaviruses prevent immediate early interferon induction by protection of viral RNA from host cell recognition. Virology 361:18–26. doi: 10.1016/j.virol.2007.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grunewald ME, Chen Y, Kuny C, Maejima T, Lease R, Ferraris D, Aikawa M, Sullivan CS, Perlman S, Fehr AR. 2019. The coronavirus macrodomain is required to prevent PARP-mediated inhibition of virus replication and enhancement of IFN expression. PLoS Pathog 15:e1007756. doi: 10.1371/journal.ppat.1007756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fehr AR, Channappanavar R, Jankevicius G, Fett C, Zhao J, Athmer J, Meyerholz DK, Ahel I, Perlman S. 2016. The conserved coronavirus macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome coronavirus infection. mBio 7:e01721-16. doi: 10.1128/mBio.01721-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gutiérrez-Vázquez C, Quintana FJ. 2018. Regulation of the immune response by the aryl hydrocarbon receptor. Immunity 48:19–33. doi: 10.1016/j.immuni.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mimura J, Ema M, Sogawa K, Fujii-Kuriyama Y. 1999. Identification of a novel mechanism of regulation of Ah (dioxin) receptor function. Genes Dev 13:20–25. doi: 10.1101/gad.13.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmed S, Bott D, Gomez A, Tamblyn L, Rasheed A, Cho T, MacPherson L, Sugamori KS, Yang Y, Grant DM, Cummins CL, Matthews J. 2015. Loss of the mono-ADP-ribosyltransferase, Tiparp, increases sensitivity to dioxin-induced steatohepatitis and lethality. J Biol Chem 290:16824–16840. doi: 10.1074/jbc.M115.660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Opitz CA, Litzenburger UM, Sahm F, Ott M, Tritschler I, Trump S, Schumacher T, Jestaedt L, Schrenk D, Weller M, Jugold M, Guillemin GJ, Miller CL, Lutz C, Radlwimmer B, Lehmann I, von Deimling A, Wick W, Platten M. 2011. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 478:197–203. doi: 10.1038/nature10491. [DOI] [PubMed] [Google Scholar]
- 14.Mellor AL, Munn DH. 2004. IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nat Rev Immunol 4:762–774. doi: 10.1038/nri1457. [DOI] [PubMed] [Google Scholar]
- 15.Murakami Y, Hoshi M, Imamura Y, Arioka Y, Yamamoto Y, Saito K. 2013. Remarkable role of indoleamine 2,3-dioxygenase and tryptophan metabolites in infectious diseases: potential role in macrophage-mediated inflammatory diseases. Mediators Inflamm 2013:391984. doi: 10.1155/2013/391984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pallotta MT, Fallarino F, Matino D, Macchiarulo A, Orabona C. 2014. AhR-mediated, non-genomic modulation of IDO1 function. Front Immunol 5:497. doi: 10.3389/fimmu.2014.00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Litzenburger UM, Opitz CA, Sahm F, Rauschenbach KJ, Trump S, Winter M, Ott M, Ochs K, Lutz C, Liu X, Anastasov N, Lehmann I, Hofer T, von Deimling A, Wick W, Platten M. 2014. Constitutive IDO expression in human cancer is sustained by an autocrine signaling loop involving IL-6, STAT3 and the AHR. Oncotarget 5:1038–1051. doi: 10.18632/oncotarget.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Q, Harden JL, Anderson CD, Egilmez NK. 2016. Tolerogenic phenotype of IFN-gamma-induced IDO+ dendritic cells is maintained via an autocrine IDO-kynurenine/AhR-IDO loop. J Immunol 197:962–970. doi: 10.4049/jimmunol.1502615. [DOI] [PubMed] [Google Scholar]
- 19.Wincent E, Amini N, Luecke S, Glatt H, Bergman J, Crescenzi C, Rannug A, Rannug U. 2009. The suggested physiologic aryl hydrocarbon receptor activator and cytochrome P4501 substrate 6-formylindolo[3,2-b]carbazole is present in humans. J Biol Chem 284:2690–2696. doi: 10.1074/jbc.M808321200. [DOI] [PubMed] [Google Scholar]
- 20.Oesch-Bartlomowicz B, Huelster A, Wiss O, Antoniou-Lipfert P, Dietrich C, Arand M, Weiss C, Bockamp E, Oesch F. 2005. Aryl hydrocarbon receptor activation by cAMP vs. dioxin: divergent signaling pathways. Proc Natl Acad Sci U S A 102:9218–9223. doi: 10.1073/pnas.0503488102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seidel SD, Winters GM, Rogers WJ, Ziccardi MH, Li V, Keser B, Denison MS. 2001. Activation of the Ah receptor signaling pathway by prostaglandins. J Biochem Mol Toxicol 15:187–196. doi: 10.1002/jbt.16. [DOI] [PubMed] [Google Scholar]
- 22.Wincent E, Bengtsson J, Mohammadi Bardbori A, Alsberg T, Luecke S, Rannug U, Rannug A. 2012. Inhibition of cytochrome P4501-dependent clearance of the endogenous agonist FICZ as a mechanism for activation of the aryl hydrocarbon receptor. Proc Natl Acad Sci U S A 109:4479–4484. doi: 10.1073/pnas.1118467109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rothhammer V, Quintana FJ. 2019. The aryl hydrocarbon receptor: an environmental sensor integrating immune responses in health and disease. Nat Rev Immunol 19:184–197. doi: 10.1038/s41577-019-0125-8. [DOI] [PubMed] [Google Scholar]
- 24.Gandhi R, Kumar D, Burns EJ, Nadeau M, Dake B, Laroni A, Kozoriz D, Weiner HL, Quintana FJ. 2010. Activation of the aryl hydrocarbon receptor induces human type 1 regulatory T cell-like and Foxp3(+) regulatory T cells. Nat Immunol 11:846–853. doi: 10.1038/ni.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quintana FJ, Basso AS, Iglesias AH, Korn T, Farez MF, Bettelli E, Caccamo M, Oukka M, Weiner HL. 2008. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature 453:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- 26.Apetoh L, Quintana FJ, Pot C, Joller N, Xiao S, Kumar D, Burns EJ, Sherr DH, Weiner HL, Kuchroo VK. 2010. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat Immunol 11:854–861. doi: 10.1038/ni.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quintana FJ, Murugaiyan G, Farez MF, Mitsdoerffer M, Tukpah AM, Burns EJ, Weiner HL. 2010. An endogenous aryl hydrocarbon receptor ligand acts on dendritic cells and T cells to suppress experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A 107:20768–20773. doi: 10.1073/pnas.1009201107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bankoti J, Rase B, Simones T, Shepherd DM. 2010. Functional and phenotypic effects of AhR activation in inflammatory dendritic cells. Toxicol Appl Pharmacol 246:18–28. doi: 10.1016/j.taap.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sekine H, Mimura J, Oshima M, Okawa H, Kanno J, Igarashi K, Gonzalez FJ, Ikuta T, Kawajiri K, Fujii-Kuriyama Y. 2009. Hypersensitivity of aryl hydrocarbon receptor-deficient mice to lipopolysaccharide-induced septic shock. Mol Cell Biol 29:6391–6400. doi: 10.1128/MCB.00337-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimura A, Naka T, Nakahama T, Chinen I, Masuda K, Nohara K, Fujii-Kuriyama Y, Kishimoto T. 2009. Aryl hydrocarbon receptor in combination with Stat1 regulates LPS-induced inflammatory responses. J Exp Med 206:2027–2035. doi: 10.1084/jem.20090560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masuda K, Kimura A, Hanieh H, Nguyen NT, Nakahama T, Chinen I, Otoyo Y, Murotani T, Yamatodani A, Kishimoto T. 2011. Aryl hydrocarbon receptor negatively regulates LPS-induced IL-6 production through suppression of histamine production in macrophages. Int Immunol 23:637–645. doi: 10.1093/intimm/dxr072. [DOI] [PubMed] [Google Scholar]
- 32.Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld JC, Stockinger B. 2008. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature 453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 33.Vogel CF, Matsumura F. 2009. A new cross-talk between the aryl hydrocarbon receptor and RelB, a member of the NF-kappaB family. Biochem Pharmacol 77:734–745. doi: 10.1016/j.bcp.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogel CF, Khan EM, Leung PS, Gershwin ME, Chang WL, Wu D, Haarmann-Stemmann T, Hoffmann A, Denison MS. 2014. Cross-talk between aryl hydrocarbon receptor and the inflammatory response: a role for nuclear factor-kappaB. J Biol Chem 289:1866–1875. doi: 10.1074/jbc.M113.505578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salisbury RL, Sulentic CE. 2015. The AhR and NF-kappaB/Rel proteins mediate the inhibitory effect of 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin on the 3’ immunoglobulin heavy chain regulatory region. Toxicol Sci 148:443–459. doi: 10.1093/toxsci/kfv193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salazar F, Awuah D, Negm OH, Shakib F, Ghaemmaghami AM. 2017. The role of indoleamine 2,3-dioxygenase-aryl hydrocarbon receptor pathway in the TLR4-induced tolerogenic phenotype in human DCs. Sci Rep 7:43337. doi: 10.1038/srep43337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bessede A, Gargaro M, Pallotta MT, Matino D, Servillo G, Brunacci C, Bicciato S, Mazza EM, Macchiarulo A, Vacca C, Iannitti R, Tissi L, Volpi C, Belladonna ML, Orabona C, Bianchi R, Lanz TV, Platten M, Della Fazia MA, Piobbico D, Zelante T, Funakoshi H, Nakamura T, Gilot D, Denison MS, Guillemin GJ, DuHadaway JB, Prendergast GC, Metz R, Geffard M, Boon L, Pirro M, Iorio A, Veyret B, Romani L, Grohmann U, Fallarino F, Puccetti P. 2014. Aryl hydrocarbon receptor control of a disease tolerance defence pathway. Nature 511:184–190. doi: 10.1038/nature13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohashi H, Nishioka K, Nakajima S, Kim S, Suzuki R, Aizaki H, Fukasawa M, Kamisuki S, Sugawara F, Ohtani N, Muramatsu M, Wakita T, Watashi K. 2018. The aryl hydrocarbon receptor-cytochrome P450 1A1 pathway controls lipid accumulation and enhances the permissiveness for hepatitis C virus assembly. J Biol Chem 293:19559–19571. doi: 10.1074/jbc.RA118.005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kueck T, Cassella E, Holler J, Kim B, Bieniasz PD. 2018. The aryl hydrocarbon receptor and interferon gamma generate antiviral states via transcriptional repression. Elife 7:e38867. doi: 10.7554/eLife.38867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamada T, Horimoto H, Kameyama T, Hayakawa S, Yamato H, Dazai M, Takada A, Kida H, Bott D, Zhou AC, Hutin D, Watts TH, Asaka M, Matthews J, Takaoka A. 2016. Constitutive aryl hydrocarbon receptor signaling constrains type I interferon-mediated antiviral innate defense. Nat Immunol 17:687–694. doi: 10.1038/ni.3422. [DOI] [PubMed] [Google Scholar]
- 41.Kashuba EV, Gradin K, Isaguliants M, Szekely L, Poellinger L, Klein G, Kazlauskas A. 2006. Regulation of transactivation function of the aryl hydrocarbon receptor by the Epstein-Barr virus-encoded EBNA-3 protein. J Biol Chem 281:1215–1223. doi: 10.1074/jbc.M509036200. [DOI] [PubMed] [Google Scholar]
- 42.Navas S, Weiss SR. 2003. Murine coronavirus-induced hepatitis: JHM genetic background eliminates A59 spike-determined hepatotropism. J Virol 77:4972–4978. doi: 10.1128/jvi.77.8.4972-4978.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma Q, Baldwin KT, Renzelli AJ, McDaniel A, Dong L. 2001. TCDD-inducible poly(ADP-ribose) polymerase: a novel response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Biochem Biophys Res Commun 289:499–506. doi: 10.1006/bbrc.2001.5987. [DOI] [PubMed] [Google Scholar]
- 44.Kim SH, Henry EC, Kim DK, Kim YH, Shin KJ, Han MS, Lee TG, Kang JK, Gasiewicz TA, Ryu SH, Suh PG. 2006. Novel compound 2-methyl-2H-pyrazole-3-carboxylic acid (2-methyl-4-o-tolylazo-phenyl)-amide (CH-223191) prevents 2,3,7,8-TCDD-induced toxicity by antagonizing the aryl hydrocarbon receptor. Mol Pharmacol 69:1871–1878. doi: 10.1124/mol.105.021832. [DOI] [PubMed] [Google Scholar]
- 45.Zhao B, Degroot DE, Hayashi A, He G, Denison MS. 2010. CH223191 is a ligand-selective antagonist of the Ah (dioxin) receptor. Toxicol Sci 117:393–403. doi: 10.1093/toxsci/kfq217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reddy TE, Pauli F, Sprouse RO, Neff NF, Newberry KM, Garabedian MJ, Myers RM. 2009. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res 19:2163–2171. doi: 10.1101/gr.097022.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen WV, Delrow J, Corrin PD, Frazier JP, Soriano P. 2004. Identification and validation of PDGF transcriptional targets by microarray-coupled gene-trap mutagenesis. Nat Genet 36:304–312. doi: 10.1038/ng1306. [DOI] [PubMed] [Google Scholar]
- 48.Kininis M, Chen BS, Diehl AG, Isaacs GD, Zhang T, Siepel AC, Clark AG, Kraus WL. 2007. Genomic analyses of transcription factor binding, histone acetylation, and gene expression reveal mechanistically distinct classes of estrogen-regulated promoters. Mol Cell Biol 27:5090–5104. doi: 10.1128/MCB.00083-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heath-Pagliuso S, Rogers WJ, Tullis K, Seidel SD, Cenijn PH, Brouwer A, Denison MS. 1998. Activation of the Ah receptor by tryptophan and tryptophan metabolites. Biochemistry 37:11508–11515. doi: 10.1021/bi980087p. [DOI] [PubMed] [Google Scholar]
- 50.Bittinger MA, Nguyen LP, Bradfield CA. 2003. Aspartate aminotransferase generates proagonists of the aryl hydrocarbon receptor. Mol Pharmacol 64:550–556. doi: 10.1124/mol.64.3.550. [DOI] [PubMed] [Google Scholar]
- 51.Song J, Clagett-Dame M, Peterson RE, Hahn ME, Westler WM, Sicinski RR, DeLuca HF. 2002. A ligand for the aryl hydrocarbon receptor isolated from lung. Proc Natl Acad Sci U S A 99:14694–14699. doi: 10.1073/pnas.232562899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Phelan D, Winter GM, Rogers WJ, Lam JC, Denison MS. 1998. Activation of the Ah receptor signal transduction pathway by bilirubin and biliverdin. Arch Biochem Biophys 357:155–163. doi: 10.1006/abbi.1998.0814. [DOI] [PubMed] [Google Scholar]
- 53.Metz R, Rust S, Duhadaway JB, Mautino MR, Munn DH, Vahanian NN, Link CJ, Prendergast GC. 2012. IDO inhibits a tryptophan sufficiency signal that stimulates mTOR: a novel IDO effector pathway targeted by D-1-methyl-tryptophan. Oncoimmunology 1:1460–1468. doi: 10.4161/onci.21716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Orabona C, Grohmann U. 2011. Indoleamine 2,3-dioxygenase and regulatory function: tryptophan starvation and beyond. Methods Mol Biol 677:269–280. doi: 10.1007/978-1-60761-869-0_19. [DOI] [PubMed] [Google Scholar]
- 55.Vogel CF, Sciullo E, Matsumura F. 2007. Involvement of RelB in aryl hydrocarbon receptor-mediated induction of chemokines. Biochem Biophys Res Commun 363:722–726. doi: 10.1016/j.bbrc.2007.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yount B, Denison MR, Weiss SR, Baric RS. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J Virol 76:11065–11078. doi: 10.1128/jvi.76.21.11065-11078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]