Epstein-Barr virus infects most people across the world and causes several kinds of cancer. Zta is an important viral protein that makes the virus replicate by binding to its DNA and turning on the expression of some genes. We used a sensitive, unbiased approach to isolate and identify viral and cellular proteins that physically interact with Zta. This revealed 39 viral and cellular proteins. We found that one protein, termed NFATc2, was already known to be important for a very early step in viral replication. We identify that once this step has occurred, Zta reduces the effectiveness of NFATc2, and we suggest that this is important to prevent cells from dying before viral replication is complete and the mature virus is released from the cells.
KEYWORDS: Epstein-Barr virus, proteomics, transcriptional regulation
ABSTRACT
The human gammaherpesvirus Epstein-Barr virus (EBV) (human herpesvirus 4 [HHV4]) infects most adults and is an important contributor to the development of many types of lymphoid and epithelial cancers. Essential contributions of viral genes to viral replication are known, but the potential contributions of cell genes are less well delineated. A key player is the viral protein Zta (BZLF1, ZEBRA, or Z). This sequence-specific DNA-binding protein can disrupt EBV latency by driving the transcription of target genes and by interacting with the EBV lytic origin of replication. Here, we used an unbiased proteomics approach to identify the Zta-interactome in cells derived from Burkitt’s lymphoma. Isolating Zta and associated proteins from Burkitt’s lymphoma cells undergoing EBV replication, followed by tandem mass tag (TMT) mass spectrometry, resulted in the identification of 39 viral and cellular proteins within the Zta interactome. An association of Zta with the cellular protein NFATc2 was validated in independent experiments. Furthermore, the ability of Zta to attenuate the activity of an NFAT-dependent promoter was shown, which suggests a functional consequence for the association. The expression of Zta is itself regulated through NFAT activity, suggesting that Zta may contribute to a feedback loop that would limit its own expression, thus aiding viral replication by preventing the known toxic effects of Zta overexpression.
IMPORTANCE Epstein-Barr virus infects most people across the world and causes several kinds of cancer. Zta is an important viral protein that makes the virus replicate by binding to its DNA and turning on the expression of some genes. We used a sensitive, unbiased approach to isolate and identify viral and cellular proteins that physically interact with Zta. This revealed 39 viral and cellular proteins. We found that one protein, termed NFATc2, was already known to be important for a very early step in viral replication. We identify that once this step has occurred, Zta reduces the effectiveness of NFATc2, and we suggest that this is important to prevent cells from dying before viral replication is complete and the mature virus is released from the cells.
INTRODUCTION
Epstein-Barr virus (EBV) establishes lifelong latency in memory B cells following primary infection (1, 2). The virus is reactivated from latency to undergo lytic replication when the infected B cell is activated following the presentation of its cognate antigen (3 – 5). A cascade of viral gene expression ensues, resulting in viral genome replication, packaging, and egress from cells. Zta (BZLF1, EB1, ZEBRA, or Z), a viral transcription factor that often works in concert with a second viral transcription factor, Rta (BRLF1), activates the expression of many of these lytic cycle genes (6). Genome-wide analyses of Zta- and Rta-binding sites reveal many viral genes that are direct targets for transactivation (7 – 9). In addition to this important role, Zta binds to the lytic origin of viral genome replication (OriLyt) (7, 8, 10 – 12). Lytic genome replication is achieved by the action of six additional viral genes (13), with potential additional contributions from cellular proteins. In addition to binding to the viral genome at OriLyt, Zta interacts with some viral protein components of the EBV replisome (13), potentially facilitating the formation of the functional replisome at OriLyt.
Viral replication is inextricably linked to the cellular environment, but the precise contributions of cellular proteins to EBV replication are unclear. Unbiased proteomics screens have identified many cellular proteins that interact with Zta. For mtSSB (14), 53BP1 (15), TORC2 (16), and INO80 (17), evidence from gene knockdown approaches shows that each gene contributes to the ability of EBV to replicate within a cell.
Here, we sought to identify cellular proteins that interact with the full-length Zta protein during authentic viral lytic replication, following the application of a physiologically relevant stimulus to latently infected B cells. We anticipated that this may identify cellular proteins that contribute to any stage of EBV lytic replication, including but not exclusive to known transcription and DNA replication functions of Zta. This unbiased screen identified 39 candidate proteins: 7 viral and 32 cellular. The validity of the data set is shown by the inclusion of previously identified targets of Zta and our demonstration of the relevance of a novel component (NFATc2) for efficient EBV lytic replication.
RESULTS
Isolation of the Zta interactome in Akata BL cells.
An EBV-positive Burkitt’s lymphoma (BL) cell line (Akata) that harbors a latent EBV genome was stimulated to enter lytic replication by cross-linking the surface B cell receptor (BCR) using anti-human IgG to mimic the physiologically relevant event of antigen recognition (3). Entry into the lytic cycle was demonstrated by evaluating the expression of Zta protein (Fig. 1A) and changes in the abundance of the EBV genome (Fig. 1B). Analysis of the total population of cells entering the early and late lytic cycles was also monitored by fluorescence-activated cell sorting for Zta and gp110 (BALF4 or VCA), respectively (data not shown).
FIG 1.
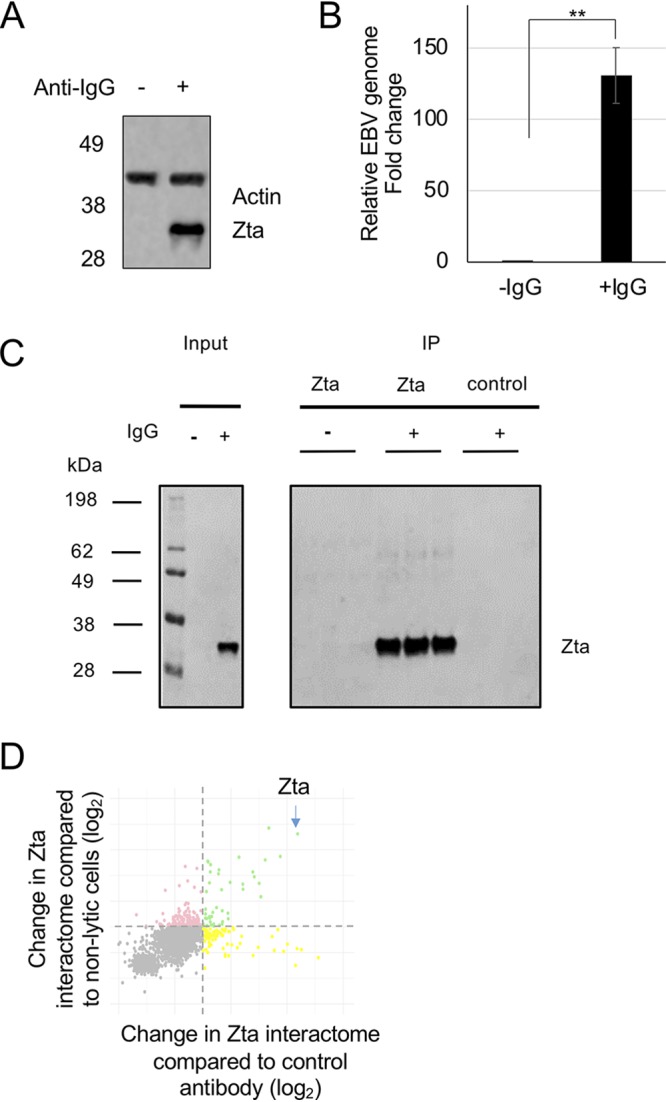
Identification of the Zta interactome in Akata cells undergoing EBV lytic replication. Akata cells were induced to initiate EBV replication by exposure to IgG for 24 h. (A) Zta protein expression was validated by Western blot analysis with the antibodies for the indicated proteins. The migration of molecular weight markers (in kilodaltons) is shown on the left. (B) Induction of EBV lytic replication was assessed using quantitative PCR to determine the change in the EBV genome load in the cells. **, P ≤ 0.01. (C) The immunoprecipitates for each of the triplicates in each arm of the study were analyzed by Western blotting for Zta protein. (D) A DSP-cross-linked protein extract was isolated following treatment with DNase. Following immunoprecipitation with control or Zta-specific antibodies, proteins were subjected to TMT labeling and mass spectrometry. The abundance changes are shown on a scatterplot relative to each control (log2). Those shown in green were enriched ≥2-fold with respect to either control.
The major challenge in isolating the Zta interactome lies with its strong DNA association. Native cell lysis results in low yields of Zta, yet harsh conditions are likely to dissociate the sought-for interactions. Zta-bound DNA has been previously extracted from cells by combining the use of an in-cell cross-linking agent, formaldehyde, followed by a harsh ionic detergent (8, 18, 19). Here, we sought a compromise approach. We used dithiobis-succinimidyl propionate (DSP), a bifunctional amine-reactive cross-linker with a spacer arm of 12 Å, to undertake in-cell cross-linking of proteins. This promoted the formation of high-molecular-weight Zta complexes, which can be released by subsequent reduction (data not shown). Following cross-linking, proteins were extracted into a nondenaturing buffer from the insoluble chromatin by digesting the chromatin with DNase.
This approach was used to isolate the Zta interactome from Akata cells undergoing lytic replication. Two controls were designed to enable reliable identification of the Zta interactome: extracts from latent Akata cells, precipitated with Zta antibody, and extracts from Akata cells induced to enter lytic replication, precipitated with a control antibody. Repeat experiments were undertaken in triplicate (Fig. 1C). Each of the nine samples was labeled with a different tandem mass tag (TMT) reagent and processed for mass spectrometry. Proteins that were enriched >2-fold compared to each control (P ≤ 0.05) were classified as specifically interacting with Zta in the lytic cycle (Fig. 1D).
Analysis of the proteins in the Zta interactome.
As anticipated, Zta was the highest-scoring protein identified during the proteomic analysis (Fig. 1D). In addition, seven viral proteins displayed reproducibly (P ≤ 0.05) high ratios in the Zta immunoprecipitation (IP) compared to either control (Table 1). Of these, both BGLF4 and BALF5 have been shown to interact with Zta previously (10, 20, 21). The identification of these proteins provides a validation of the experimental approach.
TABLE 1.
Viral proteins in the Zta interactome in Akata cells undergoing EBV lytic replicationa
| Gene name | Description | No. of unique peptides | No. of PSMs | Fold change in abundance compared to nonlytic cells | Fold change in abundance compared to control antibody |
|---|---|---|---|---|---|
| BBRF1 | Portal protein | 1 | 1 | 28.82 | 10.21 |
| BVRF2 | Capsid scaffolding protein | 12 | 15 | 13.12 | 4.88 |
| BGLF4 | Serine/threonine protein kinase | 5 | 5 | 11.54 | 2.28 |
| BALF2 | Major DNA-binding protein | 17 | 18 | 11.77 | 2.26 |
| BcLF1 | Major capsid protein | 17 | 18 | 10.72 | 2.42 |
| BALF5 | DNA polymerase | 5 | 5 | 8.81 | 2.40 |
| BMLF1 | mRNA export factor ICP27 homolog | 6 | 6 | 6.57 | 2.35 |
EBV proteins identified as part of the Zta interactome in Akata cells are shown, together with the fold changes in abundance relative to each control (P ≤ 0.05), the total number of identified peptide spectra matched (PSMs) for the protein, and a brief description of gene function.
Thirty-two cellular proteins were also reproducibly (P ≤ 0.05) enriched more than 2-fold in the Zta precipitation compared to both controls (Table 2). Analysis of common gene ontology (GO) (biological processes) revealed an enrichment for proteins involved in transcription, the nucleus, and the nucleoplasm (Fig. 2; see also Table S1 in the supplemental material). Of these functionally related proteins, one was chosen for further analysis, NFATc2.
TABLE 2.
Cell proteins in the Zta interactome in Akata cells undergoing EBV lytic replicationa
| Gene name | Description | No. of unique peptides | No. of PSMs | Fold change in abundance compared to nonlytic cells | Fold change in abundance compared to control antibody |
|---|---|---|---|---|---|
| FAM96B | Mitotic spindle-associated MMXD complex subunit MIP18 | 2 | 2 | 13.35 | 13.55 |
| TMED9 | Transmembrane emp24 protein transport domain-containing 9 | 1 | 2 | 8.8 | 6.88 |
| TIPRL | TIP41-like protein | 2 | 2 | 7.74 | 7.14 |
| NCOA5 | Nuclear receptor coactivator 5 | 20 | 34 | 6.65 | 3.92 |
| MMS19 | MMS19 nucleotide excision repair protein homolog | 7 | 7 | 6.47 | 8.05 |
| TMED10 | Full-length cDNA 5′ end of clone CS0DF013YM24 of fetal brain of Homo sapiens (human) variant (fragment) | 2 | 2 | 6.33 | 5.37 |
| FGFR2 | Adenosylhomocysteinase | 2 | 2 | 5.52 | 5.39 |
| CIAO1 | Probable cytosolic iron-sulfur protein assembly protein CIAO1 | 2 | 2 | 4.51 | 8.39 |
| NFATC2 | Nuclear factor of activated T cells, cytoplasmic 2 | 6 | 6 | 3.37 | 2.8 |
| HSPA8 | Heat shock cognate 71-kDa protein (fragment) | 15 | 70 | 3.23 | 2.13 |
| ARID1A | AT-rich interactive domain-containing protein 1A | 4 | 4 | 3.09 | 3.23 |
| RUNX3 | Runt-related transcription factor | 1 | 1 | 2.96 | 2.55 |
| ADAMTSL1 | ADAMTS-like protein 1 | 1 | 1 | 2.89 | 3.71 |
| HSPA9 | Stress 70 protein, mitochondrial | 9 | 24 | 2.83 | 2.13 |
| TLE3 | Transducin-like enhancer protein 3 | 4 | 4 | 2.65 | 2.48 |
| NFATC1 | Nuclear factor of activated T cells, cytoplasmic 1 | 5 | 6 | 2.63 | 2.35 |
| TMED2 | Transmembrane emp24 domain-containing protein 2 | 2 | 2 | 2.62 | 2.43 |
| TAF6 | Transcription initiation factor TFIID subunit 6 | 1 | 1 | 2.53 | 2.37 |
| SRSF9 | Serine/arginine-rich splicing factor 9 | 6 | 8 | 2.53 | 2.13 |
| HMG20A | High-mobility-group protein 20A | 2 | 2 | 2.51 | 2.20 |
| PABPC1 | Polyadenylate-binding protein 1 | 18 | 28 | 2.49 | 3.74 |
| MEF2B | Myocyte-specific enhancer factor 2B | 7 | 7 | 2.3 | 3.25 |
| RBMXL1 | RNA-binding motif protein, X-linked-like 1 | 4 | 26 | 2.29 | 2.4 |
| GATAD2B | cDNA FLJ37346 fis, clone BRAMY2021310, highly similar to transcriptional repressor p66 beta | 9 | 11 | 2.17 | 2.02 |
| PABPC4 | Polyadenylate-binding protein | 12 | 19 | 2.16 | 2.61 |
| YLPM1 | YLP motif-containing protein 1 | 21 | 64 | 2.16 | 3.22 |
| CPSF3L | Integrator complex subunit 11 | 4 | 4 | 2.11 | 2.45 |
| KHDRBS1 | KH domain-containing, RNA-binding, signal transduction-associated protein 1 | 14 | 47 | 2.09 | 3.00 |
| RBMX | RNA-binding motif protein, X chromosome | 9 | 36 | 2.09 | 2.02 |
| TCF20 | Transcription factor 20 | 23 | 24 | 2.07 | 2.21 |
| SMARCD2 | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 2 | 5 | 5 | 2.05 | 2.60 |
| PHF14 | PHD finger protein 14 | 14 | 15 | 2.05 | 2.21 |
Cell proteins identified as part of the Zta interactome in Akata cells are shown, together with the fold changes in abundance relative to each control (P ≤ 0.05), the total number of identified peptide spectra matched (PSMs) for the protein, and a brief description of gene function.
FIG 2.
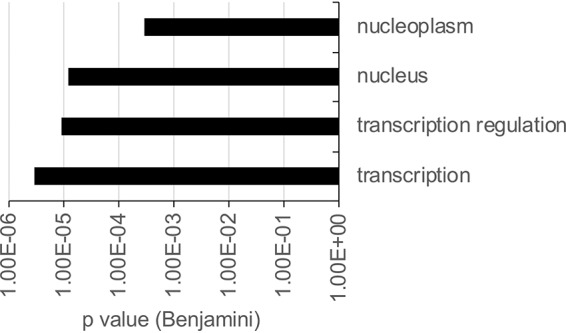
Zta interactome gene ontology analysis. Data from gene ontology analysis of the 37 human proteins found to interact with Zta by mass spectrometry are shown. The components of the most enriched group are shown in the chart (with a P value of ≤0.001).
Contribution of NFATc2 to EBV replication.
NFATc2 encodes a transcription cofactor that is activated through calcium-mediated signal transduction following dephosphorylation by calcineurin. It acts together with the AP1 transcription factor to activate gene expression via a composite DNA element, antigen response element 2 (ARRE2) (22 – 24). Using immunoprecipitation with non-cross-linked protein extracts from Akata cells induced to initiate the EBV lytic cycle, we demonstrate the coprecipitation of NFATc2 with Zta antibodies (Fig. 3A). To probe the specificity of the interaction further, we undertook additional immunoprecipitation experiments with two other nuclear DNA-binding proteins expressed in B cells, EBF1 and LEF1. Neither of these proteins coprecipitated NFATc2 protein (Fig. 3B and C).
FIG 3.
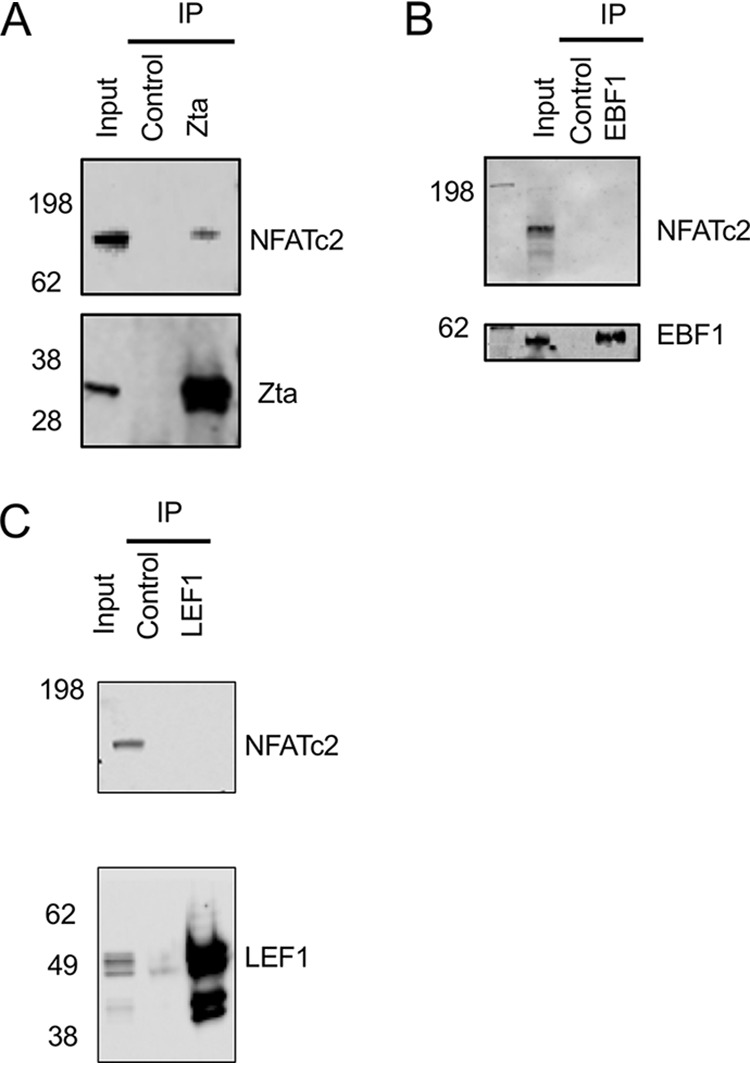
Association of NFATc2 with Zta in cells. Akata cells were induced to enter the lytic cycle for 24 h following exposure to anti-IgG, and proteins were extracted and analyzed as the input. Extracts were subjected to immunoprecipitation with the indicated antibodies and isotype controls and then analyzed by Western blotting for the proteins shown. (A) Immunoprecipitation with Zta antibody. (B) Immunoprecipitation antibody for EBF1. (C) Immunoprecipitation antibody with LEF1 antibody. The migration of molecular weight markers (in kilodaltons) is shown on the left.
To explore the contribution of NFATc2 to histidine-tagged Zta (hisZta)-mediated transcriptional regulation, we used a Zta-responsive viral reporter, BHFL1p-luciferase (Fig. 4). This promoter is transactivated by >200-fold when introduced into cells with a hisZta expression vector. When phorbol myristate acetate (PMA) and ionomycin are added to stimulate the activation of NFATc2/AP1, there is little impact on either basal transcription or Zta-mediated activation (Fig. 4A to C). This suggests that the NFATc2 interaction with Zta does not result in an alteration of the transactivation potential of Zta. To explore this further, the endogenous abundance of NFATc2 was decreased using a smart small interfering RNA (siRNA) pool against NFAT genes, and the impact on Zta-mediated activation of BHLF1p was determined. Although the smart pool reduced the NFATc2 protein abundance by 66%, there was no consequent decrease in the ability of Zta to activate gene expression (Fig. 4D to F).
FIG 4.
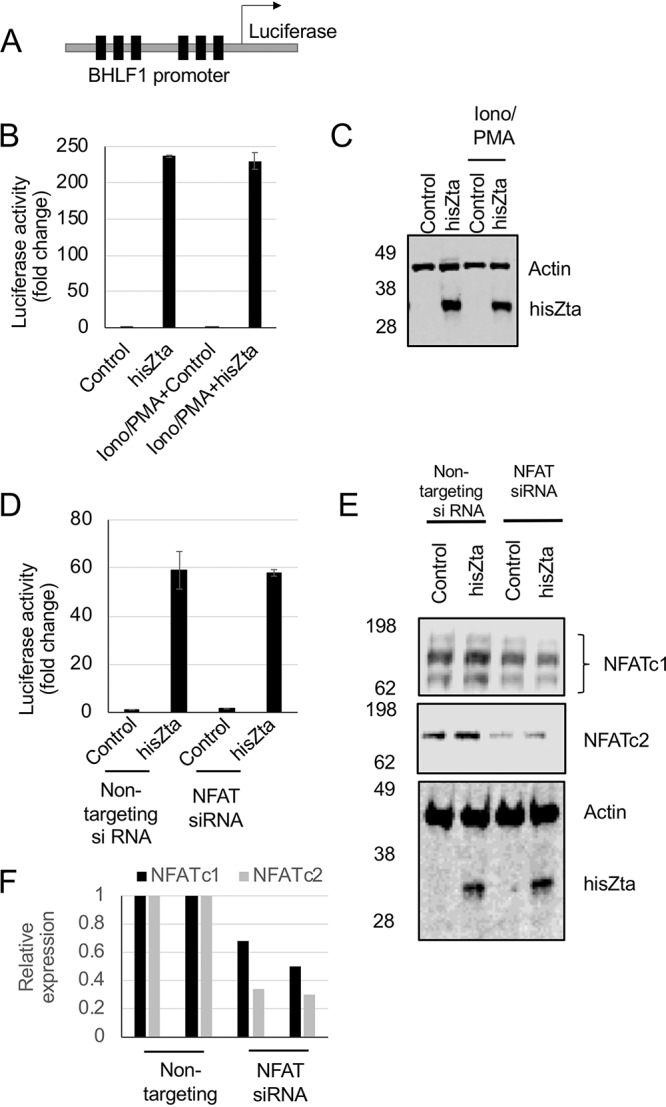
No impact of the calcium signaling pathway on transcriptional activation by Zta in B cells. DG75 B cells were transfected with a Zta reporter plasmid (BHLF1-luciferase) with and without cotransfection of expression vectors for hisZta. (A) The ZRE-driven reporter BHLF1-luciferase, with the locations of six Zta response elements (ZREs) indicated (as filled boxes). (B) Calcium signaling was stimulated or not through the addition of ionomycin-PMA. Forty-eight hours later, cells were harvested for luciferase assays and Western blot analysis. Luciferase reporter assays were undertaken, and activation by hisZta or the control vector was normalized to the total protein concentration. Error bars represent the means of triplicate readings ± standard deviations (SD). (C) Western blot analysis of Zta in panel B. (D) BHLF1-luciferase was cotransfected with Zta or a control vector and the NFATc1 and NFATc2 siRNA smart pool or nontargeting control siRNA by Neon electroporation in DG75 cells. Luciferase reporter assays were undertaken, and the activation of hisZta and the control vector, normalized to the total protein concentration, is shown. (E) Western blot analysis of the indicated proteins in panel D. (F) Following quantitation, the expression levels of NFATc1 and NFATc2 were determined. Error bars represent the means of triplicate readings ± SD.
An NFAT reporter assay was used to ask the reciprocal question, whether an NFAT-dependent promoter was impacted by the expression of hisZta (Fig. 5A). The NFAT reporter pGL3 NFAT-luciferase was introduced into DG75 B cells with control siRNA or the smart siRNA pool against NFAT genes. As anticipated, the combination of both ionomycin and PMA led to a substantial increase in the transcriptional activation of the NFAT reporter. The inclusion of the NFAT siRNA smart pool resulted in an ∼50% reduction in the expression of both NFATc1 and NFATc2. A similar decrease in ionomycin- and PMA-driven promoter activity was observed when the NFAT smart siRNA pool was coexpressed (Fig. 5B and C). This confirms that the ionomycin and PMA stimulation of the pGL3 NFAT-luciferase promoter is regulated by NFAT in these cells.
FIG 5.
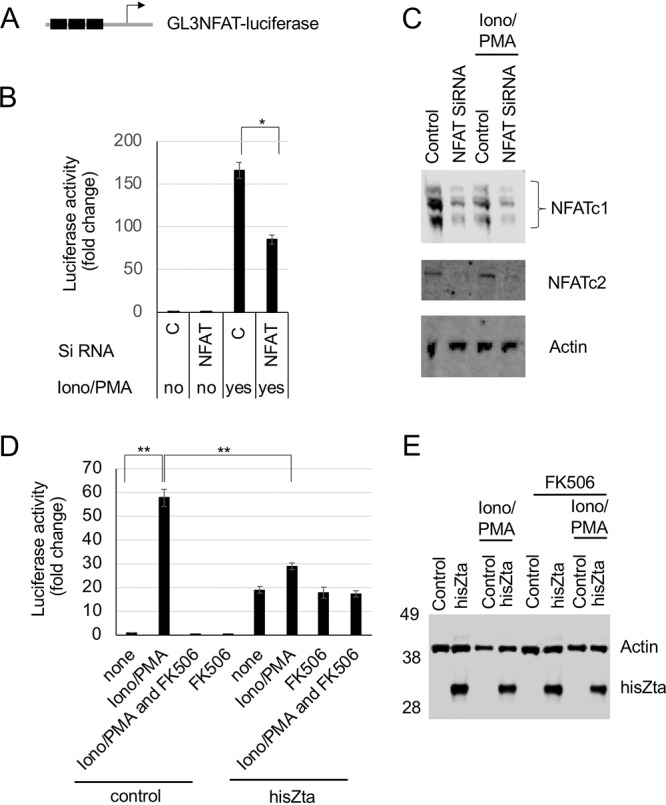
Impact of Zta on NFAT transcription activity. The pGL3-IL2 NFAT-luciferase reporter construct was cotransfected with hisZta, a control vector, or siRNA pools into DG75 cells. Ca2+ signaling was activated by ionomycin-PMA immediately after transfection, with the addition of the inhibitor FK506, as indicated. After 48 h, cells were harvested for luciferase reporter and Western blot analyses. (A) The luciferase reporter promoter. The NFAT ARRE2 elements are indicated as filled boxes. (B) Luciferase assay showing the fold changes in activity after ionomycin-PMA stimulation and the impact of NFATc1 and NFATc2 siRNAs. (C) Western blot showing the impact on NFATc1 and NFATc2 expression levels. (D) Luciferase assay showing the fold changes in activity after ionomycin-PMA stimulation and FK506 inhibition and the impact of Zta expression. (E) Western blot showing Zta expression. Error bars represent the means of triplicate readings ± standard deviations. *, P < 0.05; **, P < 0.01.
The inhibitor FK506 blocks the activation of calcineurin and Jun N-terminal protein kinase (JNK) (25), thereby inhibiting the activation of NFATc2/AP1; this was used here to identify calcineurin- and JNK-dependent regulatory events. The increased NFAT reporter promoter activity stimulated by ionomycin and PMA was negated by the addition of FK506, as expected (Fig. 5D). Cotransfection of hisZta with the NFAT reporter resulted in a rise in basal promoter activity. As AP1 sites have some sequence similarity with Zta response elements (ZREs), we questioned whether Zta may act directly through ARRE2. We analyzed the composite NFAT/AP1 site in this reporter for known Zta-binding sites (26), and we evaluated the potential for Zta to bind to the element using in vitro DNA-binding assays (Fig. 6). Neither line of investigation provided support for a direct Zta DNA interaction with this promoter. Zta also interacts with components of the basal transcriptional machinery, including the TATA-binding protein TFIID (27), and we suggest that the increase in basal expression may result from this. When the impact of hisZta expression on promoter activation observed following ionomycin and PMA stimulation was assessed, hisZta was shown to significantly and substantially reduce this activation by 25-fold (Fig. 5C and D). The impact of hisZta expression is also reflected when considering the ionomycin- and PMA-stimulated cells and comparing the difference in NFAT-dependent promoter activities in the presence and absence of hisZta expression, where a significant reduction in activity is also observed.
FIG 6.
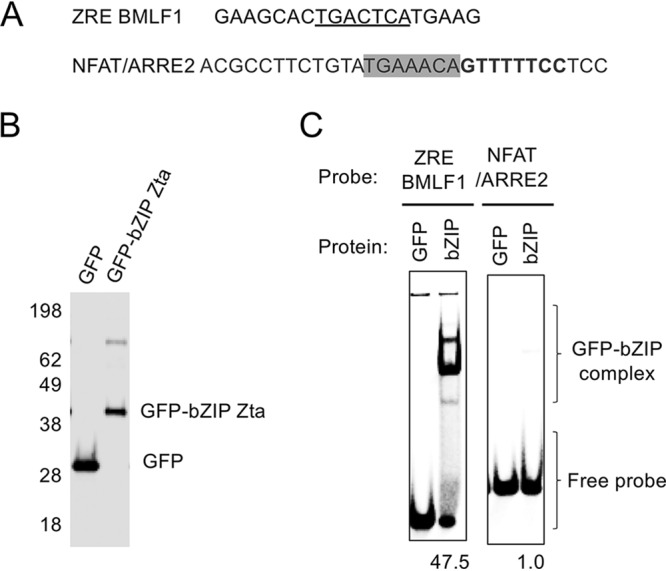
Zta bZIP domain interaction with a ZRE and the NFAT/ARRE2 element. The ability of the GFP-tagged Zta bZIP domain to interact with the ARRE2 element was compared to that of a ZRE element by an electrophoretic mobility shift assay (EMSA). (A) DNA sequence of each probe, with the known ZRE (underscored), the NFAT element (bold), and the AP1 site (boxed) shown. (B) GFP and GFP-bZIP Zta proteins were produced, and 1 μg was analyzed by staining. (C) Interaction of each GFP and GFP-bZIP Zta protein with the indicated ZREs. The relative level of the complex is shown below each GFP-bZIP Zta lane.
DISCUSSION
The identification of the Zta interactome opens a new avenue of understanding of Zta function during the EBV lytic replication cycle. The identification of two previously known targets of Zta (BGLF4 and BALF5) provides robust confirmation of the relevance of these targets and the additional proteins.
Of the five novel viral proteins, three are found in the viral capsid. The portal protein BBRF1 is a homolog of herpes simplex virus 1 (HSV-1) UL6, which is required for the encapsulation and cleavage of the viral genome. Specifically, the deletion of BBRF1 from the EBV genome has been shown to result in “empty” capsids containing no viral genome (28), and transmission electron microscopy of BBRF1 revealed the formation of a self-assembling structure consistent with other herpesvirus portals (29). Two further capsid proteins, BVRF2, a protease, and BcLF1, the major capsid protein, are both required for EBV capsid assembly (30). The interaction of Zta with these three capsid proteins suggested that, in addition to its well-characterized role in events leading to EBV genome replication, Zta might have additional roles downstream of genome replication, relating to the assembly or function of the capsid.
The importance of calcium signaling to the disruption of latency by the reactivation of the EBV lytic cycle following immunoglobulin stimulation is well established; inhibitors of critical aspects of Ca2+-mediated signal transduction pathways, cyclosporine (CsA) and tacrolimus (FK506), which converge on the inhibition of the Ca2+-dependent phosphatase calcineurin, both block EBV lytic reactivation (31). Calcium signaling acts on the BZLF1 promoter through many routes, including the actions of TORC2 and MEF2 (32). Recently, enhanced EBV lytic cycle reactivation was identified in cancer-associated EBV strains, and a sequence variation within the BZLF1 promoter that forms a functional response element for the NFAT transcription factor was found to contribute an additional route for activation through calcium signal transduction (33).
We were therefore intrigued to identify NFATc1 and NFATc2 as members of the Zta interactome. NFAT has four family members, which are activated by dephosphorylation by the Ca2+-dependent phosphatase calcineurin, resulting in nuclear translocation, followed by direct interaction with DNA response elements. The obvious approach to questioning the role of the Zta-NFAT interaction in the EBV lytic cycle is to inhibit the expression of NFAT. The calcineurin inhibitor FK506 suggests a route to question the contribution of NFATs to Zta function. However, the first step in EBV reactivation, the activation of Zta expression, is controlled through Ca2+-dependent signal transduction, and so inhibiting calcineurin-dependent signal transduction with FK506 will inhibit both Zta activation and all subsequent Zta-dependent events (31); we confirmed that this was so for Akata cells (data not shown). We therefore used simple promoter-reporters to dissect the potential for cross talk between Zta and the NFAT transcription factors. After validating that PMA and ionomycin stimulation of the NFAT-dependent promoter is reliant on NFAT expression in these cells, we found that Zta attenuates NFAT-mediated activation. In contrast, NFAT does not impact Zta-mediated activation through ZREs. This approach revealed that the impact of NFAT-Zta cross talk is unidirectional. We propose a model whereby the attenuation of NFAT-mediated activation by Zta occurs through the protein-protein interaction between Zta and NFAT that we identified using TMT mass spectrometry (Fig. 7).
FIG 7.
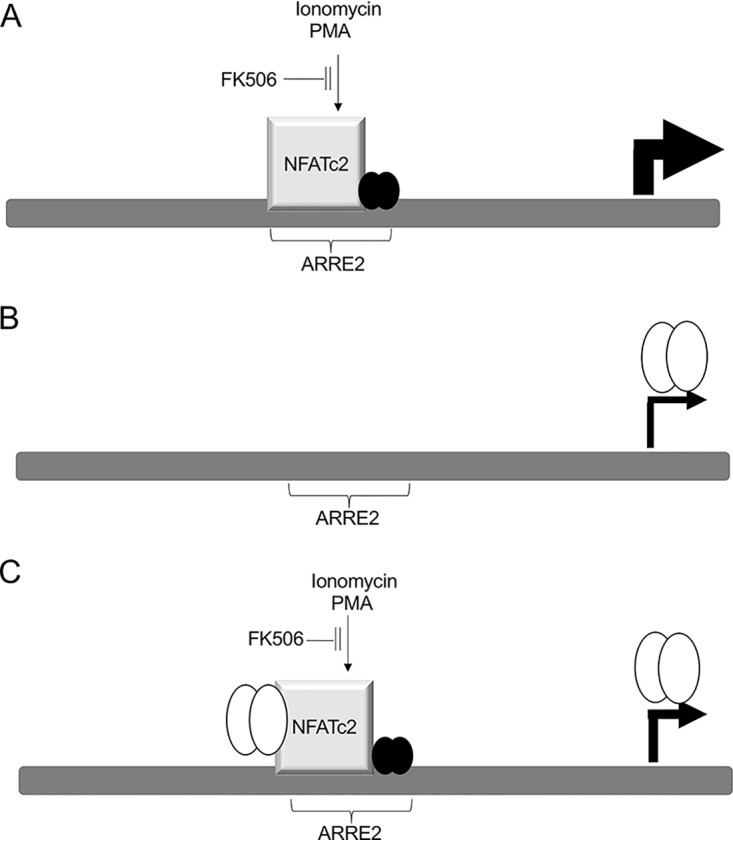
Impact of ionomycin, PMA, and Zta on the interleukin-2 (IL-2) promoter. The IL-2 promoter and one ARRE2 element are shown (dark gray boxes), along with locations of transcription factors (gray cubes, NFATc2; filled circles, AP1; open circles, Zta). (A) Following stimulation with ionomycin and PMA, transcription is activated through the interaction of NFATc2 and AP1 at the ARRE2 element. This is blocked by FK506. (B) Following the expression of Zta, transcription is activated to a lesser extent through an indirect means that is not inhibited by FK506. (C) By combining stimulation with ionomycin and PMA and the expression of Zta, transcription is activated to an intermediate extent. FK506 reduces expression to the level seen when Zta is activated alone. The interaction of Zta with NFATc2 may account for the reduction of the impact of ionomycin and PMA stimulation.
The attenuation of NFAT-mediated gene activation by Zta poses a conundrum when considering the contribution of NFAT to EBV lytic reactivation. We know that inhibition by the calcineurin inhibitor FK506 prevents EBV lytic cycle reactivation, implying a positive role for calcineurin-dependent targets such as NFAT during EBV lytic reactivation. However, the attenuation of NFAT-mediated gene activation by Zta suggests that NFAT activity may be obstructive to the EBV lytic cycle. A solution lies with careful consideration of the order of the events during EBV reactivation and the lytic cycle. It is known that Zta expression can have negative impacts on cell viability and that during the EBV lytic cycle, the expression of Zta is coupled to an ordered coexpression of the antiapoptotic viral protein v-BCL2 (BHRF1) (34). Indeed, there is evidence that Zta (BZLF1) mRNA expression is limited during the EBV lytic cycle; the abundance of the transcript is known to first peak and then fall following immunoglobulin cross-linking (35). We propose that we have identified a negative-feedback loop that acts to fine-tune Zta expression during the EBV lytic cycle and that this is mediated by the ability of Zta to attenuate NFAT activity. The Ca2+-dependent signal transduction that initiates EBV lytic reactivation stimulates NFAT transcription factors to activate the BZLF1 promoter through several DNA elements, including the NFAT response element. This promotes the expression of Zta protein, which, by acting through interactions with its DNA response elements, directly reprograms EBV and cellular gene expression. As Zta levels rise, we suggest that the interaction between Zta and NFATc2 results in attenuation of NFAT-dependent transcription, leading to reduced Zta expression. Together, this maintains the Zta abundance in cells within the required yet tolerated zone. The direct interactions between Zta and NFATc2 suggest a novel molecular mechanism to fine-tune the expression of Zta.
MATERIALS AND METHODS
Cells and transfections.
Akata Burkitt’s lymphoma cells (4) and DG75 cells (36) were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine (Fisher Scientific) at 37°C with 5% CO2. To induce the EBV lytic cycle in Akata cells, cells were resuspended at 2 × 106 cells/ml in medium, anti-human IgG (Dako) was added to 0.13% (vol/vol) of the final volume, and the cells were then incubated for 24 h.
For promoter-reporter experiments, electroporation (Bio-Rad) was used to introduce 5 μg of BHLF1-luciferase or GL3NFAT-luciferase together with 5 μg of the pcDNA3 or pcDNA3-hisZta plasmid into 1 × 107 DG75 cells. Forty-eight hours later, 90 μl of passive cell lysis buffer (Promega) was added to the cell pellets and incubated for 30 min on ice and then for 30 min at room temperature. The cell lysates were then centrifuged, and the supernatant was transferred to clean tubes. Ten microliters of triplicate samples was pipetted into a white 96-well plate. A Glomax (Promega) multidetection plate reader with an autosampler dispensed 50 μl of luciferase activation reagent (Promega) per well, and output light was read for 10 s with a delay for 2 s after the addition of the reagent. The protein concentration in the lysates was determined using a bicinchoninic acid (BCA) assay (Pierce). The concentration of each lysate was used to normalize the luciferase activity.
Either a smart pool of siRNA directed against NFATc1 and -c2 or nontargeting siRNA pool 1 (Dharmacon) was introduced into DG75 cells. Nontargeting smart pool 1 consists of the sequences UAGCGACUAAACACAUCAA, UAAGGCUAUGAAGAGAUAC, and AUGUAUUGGCCUGUAUUAG; the NFATc1-targeting smart pool consists of the sequences UCAGAAACUCCGACAUUGA, GGACAGCUAUCCGGUCGUG, GUUGAGAUCCCGCCAUUUC, and AGGAAGAACACACGGGUAC; and the NFATc2-targeting smart pool consists of the sequences CCAAUAAUGUCACCUCGAA, GCAGAAUCGUCUCUUUACA, GCGGGGAUCUUGAAGCUUA, and UCAUGUACUGCGAGAAUUU. The concentration of nontargeting siRNA pool 1 was 200 μM, that of human NFATc1 siRNA was 100 μM, and that of human NFATc2 siRNA was 100 μM. A total of 250 ng of the pcDNA3 or pcDNA3-hisZta expression vector and 250 ng of BHLF1-luciferase were delivered at the same time. The Neon transfection system (100-μl Neon kit) was used in this experiment. A total of 5 × 106 cells were resuspended in 104 μl buffer T. Six microliters of a DNA-and-RNA mix under each condition was prepared before addition to the cells. Electroporation was performed at 1,300 V for 30 ms with 1 pulse in a 100-μl Neon tip. Whole medium without antibiotics was used for culturing the cells. At 24 h posttransfection, ionomycin (final concentration, 1 μM) and PMA (final concentration, 20 ng/ml) were added, as indicated, and after another 48 h, cells were harvested and washed with Dulbecco's phosphate-buffered saline (D-PBS). Next, 1/10 of the cells were used for Western blotting, and the rest of the cells were used for the luciferase assay.
Cell viability was determined using alamarBlue (Invitrogen). Following overnight incubation, fluorescence was measured using a Glomax plate reader (Promega) with an excitation wavelength of 520 nm and an emission wavelength of 580 to 640 nm.
The EBV viral load was determined using an established assay (37).
An inhibitor of calcineurin, FK506 (Sigma), was solubilized in water.
Proteins.
Immunoprecipitations were carried out using lysates derived from 5 × 107 Akata cells without cross-linking of cells. Lysis in cell lytic reagent (Sigma) and DNase digestion were undertaken, as described below for proteomics preparation. However, the anti-Zta goat antibody SCZ (Santa Cruz) was added overnight, followed by the addition of protein G-Dynabeads without prior cross-linking of the antibody to the beads. For LEF1 immunoprecipitations, rabbit LEF1 antibody (catalog number 2286s) from CST was used, and for EBF1, mouse EBF antibody (C-8) (catalog number sc-137065; Santa Cruz) was used. Following washing, precipitated proteins were analyzed by Western blotting with monoclonal antibodies specific for NFATc2 (Santa Cruz) and the precipitated proteins.
For Western blot analysis, protein samples were separated in a 10% Bis-Tris NuPAGE gel in morpholinepropanesulfonic acid (MOPS) buffer. Following blocking for nonspecific binding, the membrane was incubated with primary antibodies at 4°C overnight, including BZ1 antibody (a kind gift from Martin Rowe) (mouse, 1:200), actin antibody (Sigma) (rabbit, 1:1,000), NFATc2 antibody (Santa Cruz) (mouse, 1:200), and green fluorescent protein (GFP) antibody (Invitrogen) (rabbit, 1:1,000). Following washing in phosphate-buffered saline (PBS)-Tween (0.1%), incubation continued with either fluorescent or enzyme-linked secondary antibodies. Donkey anti-rabbit (catalog number 800CW; Li-Cor) (1:5,000 to 1:10,000), goat anti-mouse (catalog number 680RD; Li-Cor) (1:5,000 to 1:10,000), and anti-multispecies horseradish peroxidase (HRP)-conjugated (VeriBlot; Abcam) (1:1,000) antibodies were incubated for 1 h at room temperature. The fluorescence signal was detected in either the 700-nm or 800-nm channel by using the Odyssey Fc system. The HRP-linked signal was detected with the Westernsure ECL substrate (Li-Cor) and the Odyssey Fc imager (Li-Cor).
Proteomics and immunoprecipitations.
For each replicate, 1 × 108 Akata cells were concentrated to 1 × 107 cells/ml and cross-linked following the addition of 0.2 mM DSP (Thermo Scientific) for 30 min at room temperature. The reaction was stopped by the addition of a Tris solution to a final concentration of 50 mM, the mixture was incubated for 15 min, and cells were then washed with D-PBS (Gibco). The cells were resuspended in CellLytic MT cell lysis reagent (Sigma) for 10 min at 4°C. After centrifugation, the nuclear pellets were then resuspended in lysis reagent (Sigma) with the addition of 250 U/ml of Benzonase (Sigma). The nuclear extracts were then diluted in IP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris [pH 8.0], 167 mM NaCl). Ten micrograms of SCZ Zta antibody or control goat IgG was added to 25 μl of protein G-Dynabeads and cross-linked following the addition of bis(sulfosuccinimidyl)suberate (BS3) (Thermo Scientific). These were then incubated with the nuclear extracts overnight at 4°C on a rotating wheel. Beads were washed with low-salt, then high-salt, and then lithium buffer as described previously for chromatin precipitation (38) and finally washed in a solution containing 10 mM Tris-Cl (pH 7.5), 150 mM NaCl, and 0.5 mM EDTA. The final wash buffer was removed, and the beads were resuspended in 50 mM triethyl ammonium bicarbonate prior to reduction [10 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) at 55°C for 1 h], alkylation (18.75 mM iodoacetamide, ambient, for 30 min), and digestion with trypsin (2.5 μg trypsin per sample; 37°C overnight). Proteins were eluted in 2× Laemmli sample buffer for 10 min at 95°C. Samples on the beads were reduced, alkylated, and digested with trypsin (2.5 μg trypsin per sample; 37°C overnight). The resulting peptides were labeled with tandem mass tag (TMT) 10-plex reagents according to the manufacturer’s protocol (Thermo Fisher Scientific), and the labeled samples were pooled. The pooled sample was evaporated to dryness and resuspended in buffer A (20 mM ammonium hydroxide, pH 10) prior to fractionation by high-pH reversed-phase (RP) chromatography using an Ultimate 3000 liquid chromatography system (Thermo Fisher Scientific). In brief, the sample was loaded onto an XBridge ethylene-bridged hybrid (BEH) C18 column (130 Å, 3.5 μm, 2.1 mm by 150 mm; Waters, UK) in buffer A, and peptides were eluted with an increasing gradient of buffer B (20 mM ammonium hydroxide in acetonitrile, pH 10) from 0 to 95% over 60 min. The resulting fractions (four in total) were evaporated to dryness and resuspended in 1% formic acid prior to analysis by nano-liquid chromatography-tandem mass spectrometry (LC-MS/MS) using an Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific).
High-pH RP fractions were further separated using an Ultimate 3000 nano-high-performance liquid chromatography (HPLC) system in line with an Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific). In brief, peptides in 1% (vol/vol) formic acid were injected onto an Acclaim PepMap C18 nano-trap column (Thermo Scientific). After washing with 0.5% (vol/vol) acetonitrile–0.1% (vol/vol) formic acid, peptides were resolved on a 250-mm by 75-μm Acclaim PepMap C18 reverse-phase analytical column (Thermo Scientific) over a 150-min organic gradient, using 7 gradient segments (1 to 6% solvent B over 1 min, 6 to 15% solvent B over 58 min, 15 to 32% solvent B over 58 min, 32 to 40% solvent B over 5 min, and 40 to 90% solvent B over 1 min, with a hold at 90% solvent B for 6 min and then reduction to 1% solvent B over 1 min), with a flow rate of 300 nl min−1. Solvent A was 0.1% formic acid, and solvent B was aqueous 80% acetonitrile in 0.1% formic acid. Peptides were ionized by nano-electrospray ionization at 2.0 kV using a stainless steel emitter with an internal diameter of 30 μm (Thermo Scientific) and a capillary temperature of 275°C.
All spectra were acquired using an Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific) controlled by Xcalibur 2.0 software (Thermo Scientific) and operated in data-dependent acquisition mode using an SPS-MS3 workflow. Fourier transform mass spectrometry (FTMS1) spectra were collected at a resolution of 120,000, with an automatic gain control (AGC) target of 200,000 and a maximum injection time of 50 ms. Precursors were selected with an intensity threshold of 5,000, according to the charge state (to include charge states 2 to 7), and with monoisotopic precursor selection. Previously interrogated precursors were excluded using a dynamic window (60 s ± 10 ppm). The MS2 precursors were isolated with a quadrupole mass filter set to a width of m/z 1.2. ion trap mass spectrometry (ITMS2) spectra were collected with an AGC target of 10,000, a maximum injection time of 70 ms, and a collision-induced dissociation (CID) collision energy of 35%. For FTMS3 analysis, the Orbitrap was operated at a resolution of 50,000 with an AGC target of 50,000 and a maximum injection time of 105 ms. Precursors were fragmented by high-energy collision dissociation (HCD) at a normalized collision energy of 60% to ensure maximal TMT reporter ion yield. Synchronous precursor selection (SPS) was enabled to include up to 5 MS2 fragment ions in the FTMS3 scan.
The raw data files were processed and quantified using Proteome Discoverer software v2.1 (Thermo Scientific) and searched against the UniProt human database (140,000 entries) plus Uniprot reference viral database EBV_proteome_B95_8 (UP000153037) and EBV_Proteome_GD1 (UP000103223) sequences using the SEQUEST algorithm. The peptide precursor mass tolerance was set at 10 ppm, and the MS/MS tolerance was set at 0.6 Da. Search criteria included oxidation of methionine (+15.9949) as a variable modification and carbamidomethylation of cysteine (+57.0214) and the addition of the TMT mass tag (+229.163) to peptide N termini and lysine as fixed modifications. Searches were performed with full tryptic digestion, and a maximum of 2 missed cleavages were allowed. The reverse database search option was enabled, and all data were filtered to satisfy a false discovery rate (FDR) of 5%.
Gene ontology (GO) analysis was undertaken through the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/) (39).
Data availability.
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE [1] partner repository with the data set identifier PXD013727. (ftp://ftp.pride.ebi.ac.uk/pride/data/archive/2019/06/PXD013727)
Supplementary Material
ACKNOWLEDGMENT
This study was funded with support from the China Sussex Scholarship Fund.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Epstein A. 2015. Why and how Epstein-Barr virus was discovered 50 years ago. Curr Top Microbiol Immunol 390:3–15. doi: 10.1007/978-3-319-22822-8_1. [DOI] [PubMed] [Google Scholar]
- 2.Shannon-Lowe C, Rickinson AB, Bell AI. 2017. Epstein-Barr virus-associated lymphomas. Philos Trans R Soc Lond B Biol Sci 372:20160271. doi: 10.1098/rstb.2016.0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takada K. 1984. Cross-linking of surface immunoglobulins induces Epstein-Barr virus in Burkitt’s lymphoma cell lines. Int J Cancer 33:27–32. doi: 10.1002/ijc.2910330106. [DOI] [PubMed] [Google Scholar]
- 4.Takada K, Ono Y. 1989. Synchronous and sequential activation of latently infected Epstein-Barr virus genomes. J Virol 63:445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takada K, Horinouchi K, Ono Y, Aya T, Osato T, Takahashi M, Hayasaka S. 1991. An Epstein-Barr virus-producer line Akata: establishment of the cell line and analysis of viral DNA. Virus Genes 5:147–156. doi: 10.1007/bf00571929. [DOI] [PubMed] [Google Scholar]
- 6.Kenney SC, Mertz JE. 2014. Regulation of the latent-lytic switch in Epstein-Barr virus. Semin Cancer Biol 26:60–68. doi: 10.1016/j.semcancer.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergbauer M, Kalla M, Schmeinck A, Gobel C, Rothbauer U, Eck S, Benet-Pages A, Strom TM, Hammerschmidt W. 2010. CpG-methylation regulates a class of Epstein-Barr virus promoters. PLoS Pathog 6:e1001114. doi: 10.1371/journal.ppat.1001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramasubramanyan S, Kanhere A, Osborn K, Flower K, Jenner RG, Sinclair AJ. 2012. Genome-wide analyses of Zta binding to the Epstein-Barr virus genome reveals interactions in both early and late lytic cycles and an epigenetic switch leading to an altered binding profile. J Virol 86:12494–12502. doi: 10.1128/JVI.01705-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heilmann AMF, Calderwood MA, Portal D, Lu Y, Johannsen E. 2012. Genome-wide analysis of Epstein-Barr virus Rta DNA binding. J Virol 86:5151–5164. doi: 10.1128/JVI.06760-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baumann M, Feederle R, Kremmer E, Hammerschmidt W. 1999. Cellular transcription factors recruit viral replication proteins to activate the Epstein-Barr virus origin of lytic DNA replication, oriLyt. EMBO J 18:6095–6105. doi: 10.1093/emboj/18.21.6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schepers A, Pich D, Hammerschmidt W. 1993. A transcription factor with homology to the AP-1 family links RNA transcription and DNA replication in the lytic cycle of Epstein-Barr virus. EMBO J 12:3921–3929. doi: 10.1002/j.1460-2075.1993.tb06070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schepers A, Pich D, Hammerschmidt W. 1996. Activation of oriLyt, the lytic origin of DNA replication of Epstein-Barr virus, by BZLF1. Virology 220:367–376. doi: 10.1006/viro.1996.0325. [DOI] [PubMed] [Google Scholar]
- 13.Fixman ED, Hayward GS, Hayward SD. 1995. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J Virol 69:2998–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiedmer A, Wang P, Zhou J, Rennekamp AJ, Tiranti V, Zeviani M, Lieberman PM. 2008. Epstein-Barr virus immediate-early protein Zta co-opts mitochondrial single-stranded DNA binding protein to promote viral and inhibit mitochondrial DNA replication. J Virol 82:4647–4655. doi: 10.1128/JVI.02198-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bailey SG, Verrall E, Schelcher C, Rhie A, Doherty AJ, Sinclair AJ. 2009. Functional interaction between Epstein-Barr virus replication protein Zta and host DNA damage response protein 53BP1. J Virol 83:11116–11122. doi: 10.1128/JVI.00512-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murata T, Sato Y, Nakayama S, Kudoh A, Iwahori S, Isomura H, Tajima M, Hishiki T, Ohshima T, Hijikata M, Shimotohno K, Tsurumi T. 2009. TORC2, a coactivator of cAMP-response element-binding protein, promotes Epstein-Barr virus reactivation from latency through interaction with viral BZLF1 protein. J Biol Chem 284:8033–8041. doi: 10.1074/jbc.M808466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaeffner M, Mrozek-Gorska P, Buschle A, Woellmer A, Tagawa T, Cernilogar FM, Schotta G, Krietenstein N, Lieleg C, Korber P, Hammerschmidt W. 2019. BZLF1 interacts with chromatin remodelers promoting escape from latent infections with EBV. Life Sci Alliance 2:e201800108. doi: 10.26508/lsa.201800108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramasubramanyan S, Osborn K, Flower K, Sinclair AJ. 2012. Dynamic chromatin environment of key lytic cycle regulatory regions of the Epstein-Barr virus genome. J Virol 86:1809–1819. doi: 10.1128/JVI.06334-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramasubramanyan S, Osborn K, Al-Mohammad R, Naranjo Perez-Fernandez IB, Zuo J, Balan N, Godfrey A, Patel H, Peters G, Rowe M, Jenner RG, Sinclair AJ. 2015. Epstein-Barr virus transcription factor Zta acts through distal regulatory elements to directly control cellular gene expression. Nucleic Acids Res 43:3563–3577. doi: 10.1093/nar/gkv212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Q, Hong Y, Dorsky D, Holley-Guthrie E, Zalani S, Elshiekh NA, Kiehl A, Le T, Kenney S. 1996. Functional and physical interactions between the Epstein-Barr virus (EBV) proteins BZLF1 and BMRF1: effects on EBV transcription and lytic replication. J Virol 70:5131–5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakayama S, Murata T, Murayama K, Yasui Y, Sato Y, Kudoh A, Iwahori S, Isomura H, Kanda T, Tsurumi T. 2009. Epstein-Barr virus polymerase processivity factor enhances BALF2 promoter transcription as a coactivator for the BZLF1 immediate-early protein. J Biol Chem 284:21557–21568. doi: 10.1074/jbc.M109.015685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Durand DB, Shaw JP, Bush MR, Replogle RE, Belagaje R, Crabtree GR. 1988. Characterization of antigen receptor response elements within the interleukin-2 enhancer. Mol Cell Biol 8:1715–1724. doi: 10.1128/mcb.8.4.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao A. 1994. NF-ATp: a transcription factor required for the co-ordinate induction of several cytokine genes. Immunol Today 15:274–281. doi: 10.1016/0167-5699(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 24.Jain J, McCaffrey PG, Valge-Archer VE, Rao A. 1992. Nuclear factor of activated T-cells contains Fos and Jun. Nature 356:801–804. doi: 10.1038/356801a0. [DOI] [PubMed] [Google Scholar]
- 25.Matsuda G, Nakajima K, Kawaguchi Y, Yamanashi Y, Hirai K. 2003. Epstein-Barr virus (EBV) nuclear antigen leader protein (EBNA-LP) forms complexes with a cellular anti-apoptosis protein Bcl-2 or its EBV counterpart BHRF1 through HS1-associated protein X-1. Microbiol Immunol 47:91–99. doi: 10.1111/j.1348-0421.2003.tb02790.x. [DOI] [PubMed] [Google Scholar]
- 26.Flower K, Hellen E, Newport MJ, Jones S, Sinclair AJ. 2010. Evaluation of a prediction protocol to identify potential targets of epigenetic reprogramming by the cancer associated Epstein Barr virus. PLoS One 5:e9443. doi: 10.1371/journal.pone.0009443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lieberman PM, Berk AJ. 1991. The Zta trans-activator protein stabilizes TFIID association with promoter DNA by direct protein-protein interaction. Genes Dev 5:2441–2454. doi: 10.1101/gad.5.12b.2441. [DOI] [PubMed] [Google Scholar]
- 28.Pavlova S, Feederle R, Gartner K, Fuchs W, Granzow H, Delecluse HJ. 2013. An Epstein-Barr virus mutant produces immunogenic defective particles devoid of viral DNA. J Virol 87:2011–2022. doi: 10.1128/JVI.02533-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Visalli RJ, Schwartz AM, Patel S, Visalli MA. 2019. Identification of the Epstein Barr virus portal. Virology 529:152–159. doi: 10.1016/j.virol.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 30.Henson BW, Perkins EM, Cothran JE, Desai P. 2009. Self-assembly of Epstein-Barr virus capsids. J Virol 83:3877–3890. doi: 10.1128/JVI.01733-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldfeld AE, Liu PF, Liu SF, Flemington EK, Strominger JL, Speck SH. 1995. Cyclosporin A and FK506 block induction of the Epstein-Barr virus lytic cycle by anti-immunoglobulin. Virology 209:225–229. doi: 10.1006/viro.1995.1247. [DOI] [PubMed] [Google Scholar]
- 32.Murata T. 2014. Regulation of Epstein-Barr virus reactivation from latency. Microbiol Immunol 58:307–317. doi: 10.1111/1348-0421.12155. [DOI] [PubMed] [Google Scholar]
- 33.Bristol JA, Djavadian R, Albright ER, Coleman CB, Ohashi M, Hayes M, Romero-Masters JC, Barlow EA, Farrell PJ, Rochford R, Kalejta RF, Johannsen EC, Kenney SC. 2018. A cancer-associated Epstein-Barr virus BZLF1 promoter variant enhances lytic infection. PLoS Pathog 14:e1007179. doi: 10.1371/journal.ppat.1007179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zuo J, Thomas WA, Haigh TA, Fitzsimmons L, Long HM, Hislop AD, Taylor GS, Rowe M. 2011. Epstein-Barr virus evades CD4+ T cell responses in lytic cycle through BZLF1-mediated downregulation of CD74 and the cooperation of vBcl-2. PLoS Pathog 7:e1002455. doi: 10.1371/journal.ppat.1002455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tierney RJ, Shannon-Lowe CD, Fitzsimmons L, Bell AI, Rowe M. 2015. Unexpected patterns of Epstein-Barr virus transcription revealed by a high throughput PCR array for absolute quantification of viral mRNA. Virology 474:117–130. doi: 10.1016/j.virol.2014.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ben-Bassat H, Goldblum N, Mitrani S, Klein G, Johansson B. 1976. Concanavalin A receptors on the surface membrane of lymphocytes from patients with African Burkitt’s lymphoma and lymphoma cell lines. Int J Cancer 17:448–454. doi: 10.1002/ijc.2910170406. [DOI] [PubMed] [Google Scholar]
- 37.Gallagher A, Armstrong AA, MacKenzie J, Shield L, Khan G, Lake A, Proctor S, Taylor P, Clements GB, Jarrett RF. 1999. Detection of Epstein-Barr virus (EBV) genomes in the serum of patients with EBV-associated Hodgkin’s disease. Int J Cancer 84:442–448. doi:. [DOI] [PubMed] [Google Scholar]
- 38.Godfrey A, Ramasubramanyan S, Sinclair AJ. 2017. The use of chromatin precipitation coupled to DNA sequencing (ChIP-Seq) for the analysis of Zta binding to the human and EBV genome. Methods Mol Biol 1532:191–206. doi: 10.1007/978-1-4939-6655-4_14. [DOI] [PubMed] [Google Scholar]
- 39.Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. 2003. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE [1] partner repository with the data set identifier PXD013727. (ftp://ftp.pride.ebi.ac.uk/pride/data/archive/2019/06/PXD013727)


