Abstract
Short hairpin RNAs that are delivered by recombinant adeno-associated virus (rAAV) have the potential to elicit long-term RNAi therapy for human disease. However, the discovery that short hairpin sequences can cause truncation of the rAAV genome calls into question the efficiency and gene-silencing specificity of this strategy in humans. Here, we report that embedding the guide strand of a small silencing RNA into an artificial microRNA (miRNA) scaffold derived from mouse miRNA-33 ensures rAAV genomic integrity and reduces off-targeting by 10-fold, while maintaining effective in vivo target gene repression in mice.
Graphical Abstract
Efficient and specific gene silencing is a challenge for rAAV-mediated RNAi. Short hairpin-induced vector genomic truncations raised concerns for clinical transition. Xie and colleagues report that an artificial miRNA scaffold derived from mouse miR-33 ensures rAAV genomic integrity and reduces off-targeting 10-fold, while maintaining effective target gene repression in vivo in mice.
Introduction
The RNA interference (RNAi) pathway uses small RNA guides to direct Argonaute (AGO) proteins to slice an mRNA target in two, thereby silencing its expression.1 Exogenous small double-stranded RNAs can co-opt this process by mimicking endogenous triggers, enabling gene knockdown for research or therapy.2 Unlike chemically modified small interfering RNAs, short hairpin RNAs (shRNAs) can be delivered by recombinant adeno-associated viruses (rAAVs), viral vectors that can efficiently transfer foreign genes into many tissues and provide long-term transgene expression with no known pathogenic consequences.3 However, high levels of AAV-delivered shRNAs can perturb the RNAi machinery, leading to cellular toxicity.4,5 Reducing the amount of shRNA by lowering vector doses, selecting less efficient AAV serotypes, or using weaker RNA polymerase (Pol) II promoters, instead of a strong and constitutive H1 or U6 Pol III promoters, can prevent toxicity.5,6 Alternatively, embedding the antisense RNA in endogenous microRNA (miRNA) scaffolds may improve small RNA processing, express multiple silencing RNAs against one or multiple target genes, and alleviate RNAi toxicity.5,7, 8, 9, 10 However, each of these strategies has thus far compromised RNAi potency.5,11
Off-target activity poses a second hurdle for RNAi applications. Off-target gene silencing can complicate data interpretation in functional studies and raises safety concerns for RNAi-based therapies.12 Undesirable incorporation of the passenger strand into AGO and fortuitous complementarity of the guide strand seed sequence (positions g2–g8) with non-target mRNAs can trigger miRNA-like repression of off-target genes. Because the seed sequence is determined by the position of the 5′ end of the small RNA, heterogeneity at the 5′ end increases the potential for miRNA-like off-target activity and is a common problem with transcribed shRNAs.13, 14, 15 Identifying designs that promote accurate processing of shRNAs remains a crucial step for rAAV-delivered RNAi.
Beyond these shRNA-specific challenges, our previous work has shown that shRNA-encoded sequences redirect rAAV viral genome replication, generating truncated rAAV genomes lacking an intact shRNA cassette.16 This discovery highlights the importance of developing AAV-compatible gene-silencing vectors that retain high efficacy, confer minimal off-targeting, and yield a high proportion of intact viral genomes during vector production.
Results
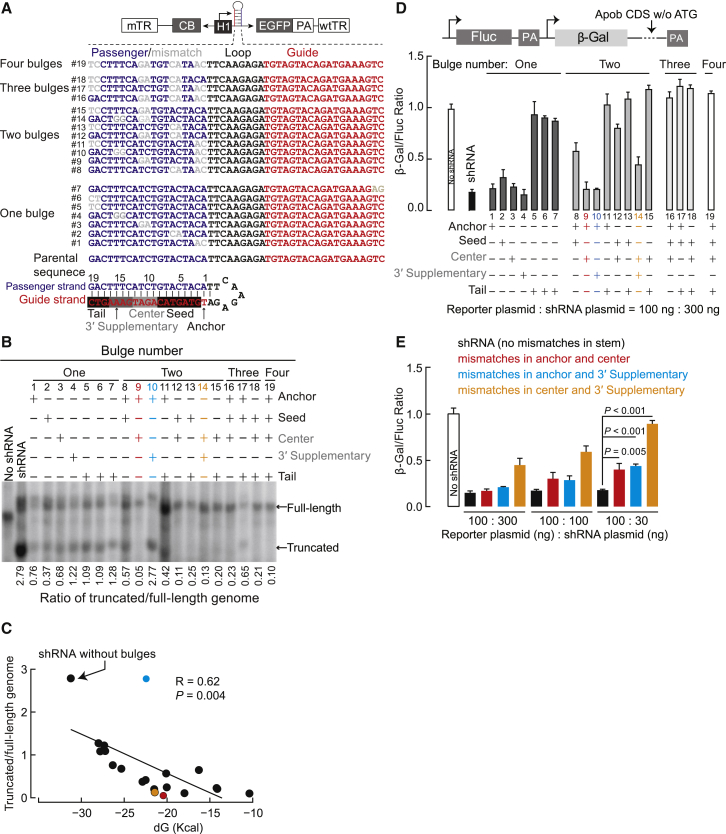
Thermodynamic Stability of shRNA-Encoding Sequences Influences AAV Genome Integrity
We previously reported that shRNA cassettes in rAAV genomes can hinder and redirect rAAV genome replication, leading to heterogeneous populations comprising a mixture of full-length genomes and truncated vector species that lack the capacity to silence mRNAs. Stable base pairing between sequences that encode the passenger and guide strands can cause truncation events.16 We hypothesized that introducing mismatches into the shRNA stem may improve rAAV genome integrity. To test this idea, we introduced up to four dinucleotide bulges at different positions in the passenger strand, while keeping the guide strand of an shRNA against mouse apolipoprotein B transcripts (shApob) unchanged (Figure 1A). The shApob cassettes that carry bulges were incorporated into the self-complementary (sc)AAV-CB-Egfp construct, within the intron residing between the Egfp coding sequence and the cytomegalovirus (CMV) enhancer/chicken β-actin (CB) promoter. We co-transfected these scAAV-shApob plasmids with pAdeno-helper and pRep2/Cap9 plasmids into HEK293 cells and analyzed low-molecular-weight DNAs. Southern blot analysis revealed that overall, shApob cassettes with bulges generated fewer truncated genomes than cassettes without bulges (Figure 1B). shApob cassettes with lower thermodynamic stabilities yielded fewer truncated genomes (Figure 1C). To quantify the gene-silencing efficiencies of shApob cassettes that carry bulges, we co-transfected shApob constructs with a pmiCHECK-Apob sensor plasmid containing an Apob-coding sequence placed in the 3′ UTR of the β-Gal reporter gene (Figures 1D and 1E). Lowering the thermodynamic stability of the shRNA stem improved rAAV genome integrity (Figures 1B and 1C), but at the cost of reduced RNAi efficacy (Figures 1D and 1E). The large abundance of unprocessed shRNA precursors and less antisense small RNA likely explain the low RNAi efficacies observed from bulged-shApob cassettes (Figure S1).
Figure 1.
Thermodynamic Stability of the shRNA Stem Influences rAAV Genome Homogeneity
(A) Diagram of the self-complementary (sc)AAV construct carrying shApob sequences within the intronic region between the CMV enhancer/chicken β-actin (CB) promoter and the Egfp transgene. Nucleotide mismatches are in gray. (B) Southern blot analysis of low-molecular-weight DNA extracted from HEK293 cells transfected with the scAAV-shApob plasmid, pRep2/Cap9 plasmid, and pAdeno-helper plasmid using 32P-labeled EGFP probe. (C) Correlation between the thermodynamic stabilities of short hairpin DNA and their ratios of truncated AAV genome to full-length genomes. The ΔG values were calculated with RNAfold. (D) Ratios of β-galactosidase (β-Gal) and Firefly luciferase (Fluc) in HEK293 cells co-transfected with shApob constructs and a sensor plasmid carrying the Apob coding sequence in the 3′ UTR of β-Gal. After 48 h, Fluc and β-Gal levels were measured, and the ratio between β-Gal and Fluc was calculated to reflect the gene-silencing efficacies. (E) Relative β-galactosidase activity in HEK293 transfected with sensor plasmid and RNAi plasmids at different ratios. Colored scheme in (C) (dots) correlates with the construct types indicated in the legend (E). The amount of sensor plasmid and shApob plasmid for transfection is indicated. mTR, mutant TR; TR, terminal repeat. Values are mean ± SD.
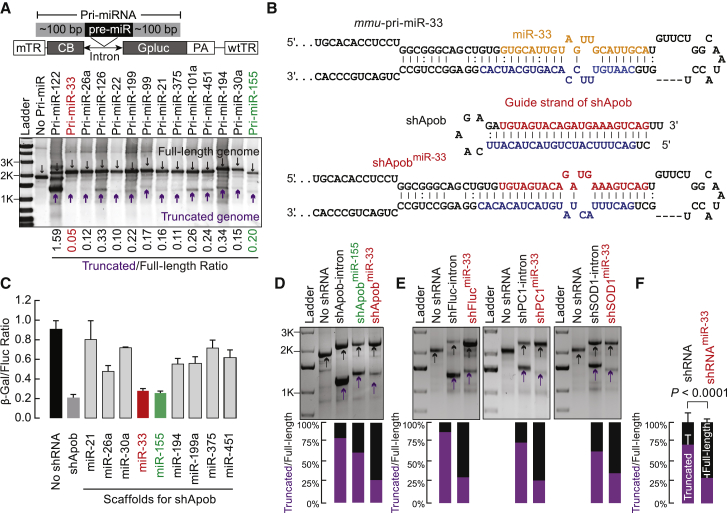
The miR-33 Scaffold Improves rAAV Genome Integrity
We tested whether natural pre-miRNA scaffolds could improve rAAV genome integrity without compromising efficacy. Unlike artificial shRNAs, endogenous miRNAs precursors (pre-miRNAs) typically bear mismatches within their stems. First, we designed and packaged capsids with rAAV constructs harboring DNA sequences derived from 15 different mouse primary miRNAs (pri-miRNAs), transcripts that undergo sequential processing into pre-miRNAs and then into miRNAs. Agarose gel analysis of viral DNA showed that the frequency of genome truncation varied among the tested pri-miRNAs (Figure 2A). We next used pri-miRNAs as scaffolds to house the Apob antisense sequence. For each artificial miRNA scaffold (shRNAmiRNA), the original miRNA guide was replaced with the shApob guide strand; bulges were incorporated into the passenger strands to mimic the native structure of the endogenous pre-miRNA. Each construct incorporated both the pre-miRNA loop and ∼100 nt of flanking sequence derived from the corresponding mouse pri-miRNAs. Figure 2B illustrates the shApob guide that is embedded within the mmu-pri-miR-33 scaffold. We measured RNAi potency for conventional shApob- and shApobmiRNA-expressing plasmids in HEK293 cells. Transcription of shRNAmiR-33 and shRNAmiR-155 was driven by the RNA Pol II CB promoter. These activities triggered reporter silencing comparable with that produced by conventional shRNA constructs transcribed by RNA Pol III from the H1 promoter (Figure 2C). We packaged each of the shRNAmiRNA-expressing constructs into AAV and examined genome integrity of the purified vectors. The shApob vector encapsidated 20% full-length genomes, whereas miR-33 (shApobmiR-33) and miR-155 (shApobmiR-155) scaffolds packaged 75% and 40% intact genomes, respectively (Figure 2D). AAV vectors harboring shRNAs against the firefly luciferase reporter gene (Fluc), mouse ectonucleotide pyrophosphatase/phosphodiesterase 1 (PC-1), or human cytosolic Cu/Zn superoxide dismutase 1 (SOD-1) exhibited 12%, 25%, and 36% full-length genomes, respectively. In contrast, their counterparts using the miR-33 scaffold increased the percentage of full-length genomes to 68%, 72%, and 63%, respectively (Figure 2E). By introducing bulges into the short hairpin DNA, the miR-33 scaffold improved rAAV genome integrity from 28% ± 10% to 69% ± 4%. Faint bands that were attributed to small truncated genomes were still observed (Figures 2D and 2E) and are likely caused by the palindromic sequences residing in other regions of the vector as previously reported.16
Figure 2.
The miR-33 Scaffold Reduces the Prevalence of Vector Genome Truncations Caused by Short DNA Hairpins
(A) Top: viral genome schematic of scAAV8 vectors carrying pri-miRNA fragments that contain pre-miRNA and ~100-bp flanking sequences at each end. Bottom: a representative 1% agarose gel of viral genomes isolated from packaged vectors. (B) Design of artificial miRNA constructs using the miR-33 scaffold. An Excel macro was developed to embed the shRNA into the miR-33 scaffold (Data S1). (C) Comparisons of gene-silencing efficiencies between miRNA scaffolds and conventional shApob in HEK293 cells. Plasmids expressing RNA inhibitors were co-transfected with a β-Gal sensor plasmid into HEK293 cells at 200 and 60 ng/well, respectively. Relative reduction in β-Gal activity reflects knockdown efficacy. (D) Agarose gel analysis of genomic DNA from purified vectors that carry no shRNA, shApob, artificial miR-155, or miR-33 against Apob (shApobmiR-155 or shApobmiR-33). (E) Agarose gel analyses of viral DNA extracted from purified vectors that harbor shRNA or shRNAmiR-33 against Fluc, PC-1, or SOD-1. (F) Summary of the ratios between truncated and full-length genomes from vectors harboring shRNA or shApobmiR-33 against Apob1, Fluc, PC-1, or SOD-1. Fisher’s exact test was used. (D–F) The ratios of truncated and full-length genomes were calculated using band intensities and normalized to their molecular sizes. Purple arrows indicate truncated genomes. Black arrows indicate full-length genomes. Values are mean ± SD.
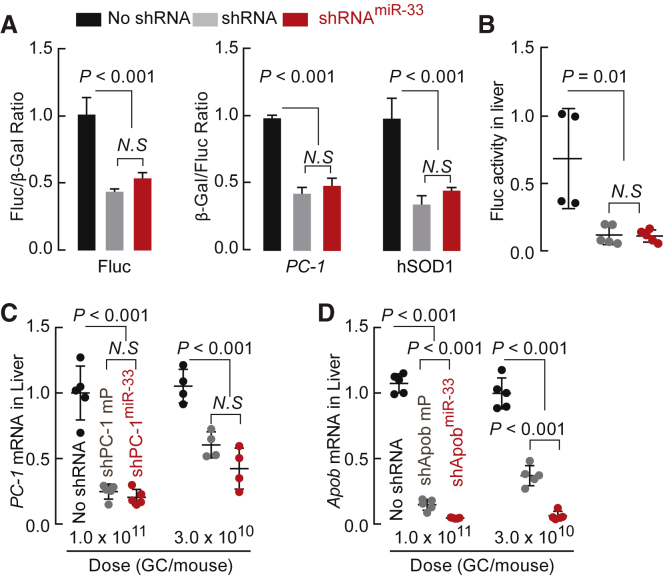
miR-33 Scaffolds Achieve Effective Gene Silencing in Cultured Cells and Mice
Next, we compared the gene-silencing efficacies of shRNAs and their counterpart miR-33 scaffolds in HEK293 cells and mice. Knockdown efficacy of the shRNAmiR-33 constructs targeting Apob (Figure 2C), Fluc, PC-1, and SOD-1 in HEK293 cells was as potent as the counterpart shRNAs driven by the strong U6 or H1 Pol III promoter (Figure 3A).
Figure 3.
miR-33-Scaffolded shRNAs Promote Efficient Gene Silencing in Cultured Cells and Mice
(A) Relative reporter activity in HEK293 cells transfected with Fluc, PC-1, and SOD1 gene-silencing constructs and their sensor plasmids. (B) Firefly luciferase activities detected in treated mouse liver extracts. Mice were co-administrated with rAAV9-Fluc reporter vector at 1.0 × 1011 GCs/mouse and the gene-silencing vectors rAAV9-shFluc (n = 5), shFlucmiR-33 (n = 5), or no shRNA (n = 4) at 5.0 × 1011 GCs/mouse. After 3 weeks, mice were sacrificed to assess firefly luciferase activity. (C) pc-1 mRNA levels in the livers of mice treated with rAAV9-shPC-1 or rAAV9-shPC-1miR-33. Five mice in the 1.0 × 1011 GCs/mouse group and four mice in the 3.0 × 1010 GCs/mouse group. (D) apob mRNA levels in the livers of mice (n = 5 in each group) treated with rAAV9-shApob or rAAV9-shApobmiR-33. rAAVs at the indicated doses were injected into adult C57BL/6 mice by tail vein. After 3 weeks, mice were sacrificed for PC-1 and Apob gene expression analysis using qRT-PCR. shFluc is driven by the U6 promoter. shApob, shPC-1, and shSOD-1 are driven by the H1 promoter. Values are mean ± SD. GC, genome copy.
Features that enable efficient processing of pri-miRNAs have been described previously.17,18 Incorporating some of these motifs into artificial miR-30 and miR-155 scaffolds has been shown to confer increased RNAi potency.9,19 The mmu-pri-miR-33 transcript contains nearly all features of an optimal miRNA,17,18 including a 35-bp stem, a UG motif at the 5′ end of the pre-miRNA, a mismatched GHG motif in the stem, a UGUG motif in the loop, and a CNNC motif downstream of the pre-miRNA 3′ end (Figure S2A). We converted the sub-optimal nucleotides in mmu-pri-miR-33 into the exact motifs of de novo designed miRNA18 to generate two additional scaffolds, miR-33-m and miR-33-p (Figure S2A, bottom). miR-33-m possesses a bulged-stem similar to endogenous miR-33, whereas miR-33-p carries a perfectly complementary stem. Dose-response analyses showed that the respective vector activities of the modified miR-33 scaffolds were more efficient than the non-optimized shApobmiR-33 vector (Figure S2B, top), comparable with the non-optimized shPC-1miR-33 vector (Figure S2B, middle), and less efficient than the non-optimized shSOD1miR33 vector (Figure S2B, bottom). Overall, the introduction of optimal miRNA motifs failed to consistently enhance the potency of the miR-33 scaffold.
We packaged constructs harboring shRNAmiR-33 within the intron or shRNA next to the mutant terminal repeat or within the intron into AAV capsids (Figure S3A). We positioned the shRNA next to the mutant terminal repeat to decrease the proportion of truncated genomes16 (Figures S3B–S3D, shRNA-mp in lane 3). The vectors were intravenously injected into adult mice. After 3 weeks, we used live bioluminescence imaging (Figure S4) and quantified luciferase activities in liver extracts (Figure 3B) to measure Fluc expression in mice previously co-administered a rAAV9-Fluc reporter vector and a gene-silencing vector. For mice receiving gene-silencing vectors against PC-1 or Apob-1, liver mRNA levels were assessed by quantitative reverse transcription-PCR (qRT-PCR; Figures 3C and 3D). Compared with AAV-shRNA vectors, shRNAmiR-33 vectors achieved comparable gene silencing of fluc and pc-1 mRNA, and a greater reduction of apob mRNA in mice. Thus, in cultured cells and mice, the miR-33 scaffold effectively mediates gene silencing that is comparable with conventional Pol III promoter-driven shRNAs.
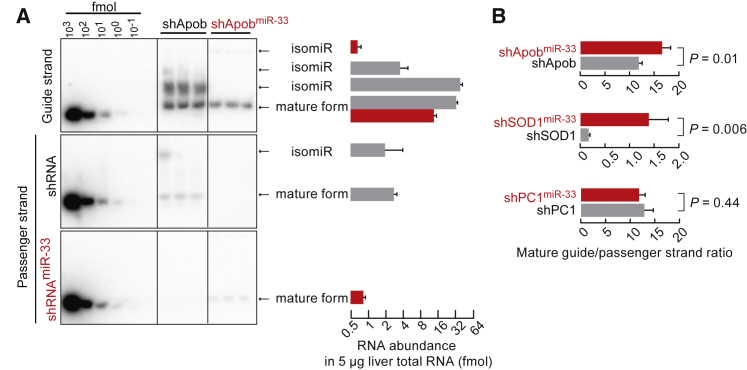
Small RNAs Transcribed from miR-33 Scaffolds Are Efficiently Processed in Cultured Cells
We next evaluated RNA processing in HEK293 cells transfected with shRNA or shRNAmiR-33 plasmids by northern blot analysis. For the shApob construct, we detected two isoforms of guide strand and one passenger strand isoform. In contrast, we detected one guide strand isoform and an absence of a passenger strand isoform for the shApobmiR-33 construct (Figure 4A). For the shSOD1 construct, we detected one isoform of guide strand and two isoforms of passenger strand, whereas for the shSOD1miR-33 construct, only the mature guide and passenger strand were detected (Figure S5A). For the shPC-1 construct, there were two isoforms for both strands, whereas the shPC-1miR-33 construct produced one guide strand isoform and one passenger strand isoform (Figure S5B). We concluded that shRNAmiR-33 plasmids generate less unprocessed guide and passenger strand isoforms compared with shRNA plasmids. Furthermore, the miR-33 scaffold increased the ratio of mature guide to passenger strand in two of three scaffolds tested (Figure 4B). The miR-33 scaffold was especially useful for improving poorly processed shRNAs, as in the case of the low guide/passenger strand ratio for shSOD1 (Figure 4B; Figure S5A). In summary, transcribed RNAs from miR-33 scaffolds were accurately and efficiently processed, yielding little unprocessed precursor, high guide-to-passenger strand ratios, and levels of mature guide strand comparable with conventional Pol III promoter-driven shRNAs.
Figure 4.
Small RNA Processing in HEK293 Cells
(A) Northern blot analysis of small RNAs in HEK293 cells transfected with shApob or shApobmiR-33 plasmids. Total RNA was extracted from the HEK293 cells 48 h after transfection. Five micrograms of total RNA was used for northern blot analysis. Synthetic oligonucleotides at indicated amounts were used as reference for quantification. shRNA and shRNAmiR-33 plasmids transcribe the same guide strand (top), but different passenger strands (middle and bottom). The transcribed RNAs from shApob and shApobmiR-33 are in gray and red, respectively. (B) Ratios of mature small RNA guide strand and passenger strand detected from shApob, shSOD-1, shPC-1, and their counterpart miR-33 scaffolds. Values are mean ± SD.
The miR-33 Scaffold Generates More Accurate Gene Silencing in Mice
To assess RNAi accuracy conferred by the miR-33-scaffolded cassettes, we sequenced exogenously expressed shRNAs, endogenous miRNAs, and mRNAs of mouse livers treated with AAV-shPC-1 or AAV-shPC-1miR-33 vectors (Figures 5A–5C).
Figure 5.
Small RNA and Transcriptome Analysis in Mouse Liver
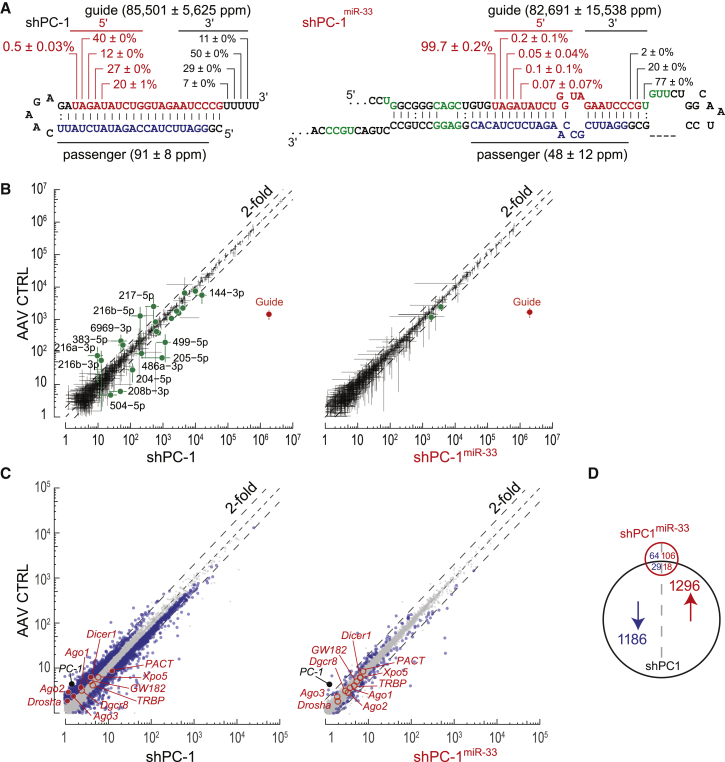
Adult C57BL/6 mice were treated with 1.0 × 1011 GCs rAAV9-shPC-1, rAAV9-shPC-1miR-33, or rAAV vector without shRNA by tail vein injection. After 3 weeks, mice were sacrificed, and liver RNA was extracted and used for the following small silencing RNA (A), endogenous miRNA (B), and global gene expression analyses (C). (A) Profiling of 5′ and 3′ distribution of guide (red) and passenger (blue) strands of silencing RNAs detected in mice by small RNA-seq (mean ± SD, n = 3). (B) Scatterplots comparing the abundance of endogenous miRNAs and small RNAs following treatment with AAV control (y axis) and AAV-shPC-1 (x axis, left plot) or AAV-shPC-1miR-33 (x axis, right plot). Each point represents the normalized miRNA reads in parts per million (ppm) averaged from three biological replicates. Error bar represents SD (n = 3). Differentially expressed miRNAs (adjusted p < 0.05) are in green solid dots. The guide strand is indicated in red. (C) Comparison of mRNA abundance in whole transcriptome analysis. Each point represents the mean fragments per kilobase of exon model per million reads mapped (FPKM) value of a single gene (n = 3). Significantly dysregulated mRNAs are represented in blue dots. Genes associated with the RNAi pathway are annotated with open red circles (no change) or red dots (significantly changed). PC-1 is annotated with a solid black dot. (D) A Venn diagram summarizing differentially expressed genes in shPC-1miR-33- and shPC-1-treated mice. GC, genome copy.
Both the conventional shRNA and the shRNAmiR-33 scaffold promoted efficient loading of the guide strand into AGO: passenger strands were <0.2% of small RNAs generated from mice receiving shPC-1 (guide strand, [85 ± 6] × 103 ppm; passenger strand, 91 ± 8 ppm; guide/passenger strand ratio, [953 ± 34]-fold) or shPC-1miR-33 (guide strand, [83 ± 16] × 103 ppm; passenger strand, 48 ± 12 ppm; guide/passenger strand ratio, [1,878 ± 402]-fold). However, the 5′ ends of the shPC-1-derived guide strands were highly heterogeneous, and the majority of 5′ ends were 3′-shifted by one to four nucleotides for AAV-shPC-1, producing four different seeds. Notably, 99.50% ± 0.03% of the AAV-shPC-1 guides differed from the desired sequence. In contrast, shPC-1miR-33 generated just 0.3% ± 0.2% incorrect 5′ ends among >2.2 ± 0.4 million guide strand reads (Figure 5A). Thus, shPC-1miR-33 effectively produces only a single guide sequence.
The conventionally designed shPC-1, but not the shPC-1miR-33 vector, also perturbed endogenous miRNA levels: small-RNA profiling of mouse livers treated with AAV-shPC-1 revealed 13 significantly dysregulated miRNAs (>2-fold change and false discovery rate [FDR] < 0.05), whereas no significant changes in miRNA expression were observed in mice receiving AAV-shPC-1miR-33 (Figure 5B).
Similarly, the conventional shPC-1, but not the shPC-1miR-33 vector, perturbed mRNA expression. Compared with a control AAV vector lacking a shRNA cassette, the AAV-shPC-1 vector altered expression of 2,541 genes (FDR < 0.05; 1,215 decreased and 1,326 increased). In contrast, injection of AAV-shPC-1miR-33 decreased expression of just 93 genes and increased expression of 124 genes. Not including pc-1, the shPC-1 sequence contains seed matches to many mRNAs whose steady-state abundance decreased: 535 contained a guide+1 seed match, 657 contained a guide+2 seed match, 770 contained a guide+3 seed match, and 980 contained a guide+4 seed match (Figure S6; Table S2). In contrast, 41 decreased mRNAs harbored a match to the shPC-1miR-33 seed sequence (Figure S6; Table S3). Among these dysregulated genes, 9 decreased and 12 increased genes were also found in patients with a PC-1 loss-of-function mutation (Table S4).20 Finally, shPC-1, but not shPC-1miR-33, changed the abundance of mRNAs encoding RNAi pathway proteins (AGO1, AGO2, AGO3, DICER1, DROSHA, and PACT), suggesting that AAV-shRNA vectors not only perturb global liver gene expression, but also disrupt the miRNA pathway itself (Figures 5C and 5D).
Discussion
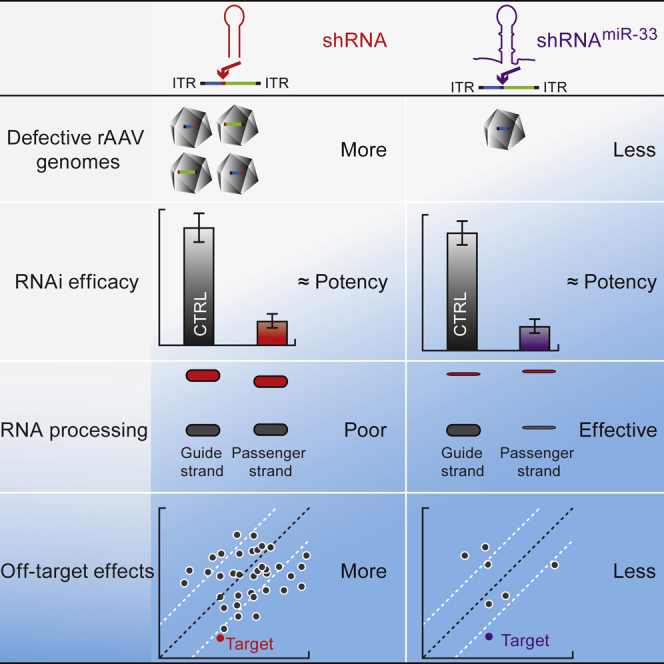
Efficient and specific gene silencing are critical challenges for RNAi therapeutics.21 shRNA-encoded sequences can redirect rAAV viral genome replication, generating truncated rAAV genomes lacking an intact shRNA cassette to produce silencing RNA.16 Here, we showed that Pol II CB promoter-driven miR-33-scaffolded shRNAs can confer gene knockdown in cultured cells and mice at levels that are comparable with conventional Pol III promoter-driven shRNA constructs, while achieving the accuracy that is lacking with conventional shRNAs. Importantly, the miR-33 scaffold exhibits reduced frequency of truncated viral vector genomes.
Consistent with the previous observations on Pol III promoter-driven shRNA,13, 14, 15 the 5′ ends of guide strands are highly heterogenous due to imprecise cutting by Dicer. The accuracy of Dicer processing can be greatly improved by optimizing the loop position of shRNAs.15 By contrast, the miR-33 scaffold generates a unique seed sequence. Other miRNA scaffolds also showed accurate processing of the guide strands.7,13,14 Similar to endogenous miRNAs, which have unique seed sequences, the artificial miRNAs are processed by Drosha and Dicer by mimicking the endogenous miRNA, ensuring accurate processing at the 5′ ends of mature siRNAs.
Artificial miRNAs have been used for RNAi, but their efficacies have generally been less than RNA Pol III-transcribed shRNAs.5,11 U6 and H1 Pol III promoter-driven SOD-1miR-155 cassettes were shown to be more efficient at gene knockdown than cassettes with SOD-1miR-155 that are driven by a Pol II CB promoter. However, they also led to elevated liver transaminases in macaques.22 The miR-33 scaffold efficiently and accurately reduces target expression, making it a promising platform for AAV-mediated RNAi gene therapy for fatal neurodegenerative diseases such as Huntington’s disease and amyotrophic lateral sclerosis.
Previous studies have proposed optimizing the shRNA design to alleviate off-target effects caused by incorporation of passenger strands into AGOs.23,24 Although several miRNA scaffolds have been reported to yield effective and safe gene silencing (e.g., miR-30,15 miR-155,7,15 and miR-22313), the miR-33 scaffold accurately processes the guide strand and yields the highest ratio of guide to passenger strand, eliminating the need for more complex strategies such as co-expressing RNA decoys.25
shRNAmiR-33, with its less thermostable stem, produced fewer truncated genomes compared with other miRNA scaffolds (Figures 2A and 2D). Moreover, the reduction in thermodynamic stability enhances the rate-limiting step of unwinding the siRNA duplex for noncleaving mammalian AGO1, AGO3, and AGO4 proteins.26,27 Screening or optimizing gene-silencing scaffolds with low thermostabilities marks a new direction for the development of high-efficiency, rAAV-compatible gene-silencing platforms. Although the miR-33 scaffold’s stem mismatches significantly reduce the proportion of truncated genomes, its stem-loop structure still causes some truncation events (Figures 2D and 2E). More comprehensive screening of endogenous pri-miRNA scaffolds along with optimized guide sequences may further reduce genome heterogeneity and increase RNAi potency. Optimization of current rAAV vector production systems to include modification of factors that can facilitate error-free DNA replication through palindromic sequences may improve rAAV homogeneity for clinical applications. Because of their small sizes, most gene-silencing expression cassettes are engineered into scAAV vectors, which can bypass the rate-limiting step of second-strand synthesis for robust transduction. In the context of single-stranded AAV genome, the high thermodynamic stability of shRNA-encoding sequences also leads to truncated vector genomes,16 which may further compromise the in vivo genome stability and functionality. The incorporation of guide sequences into the miR-33 scaffold can be easily accomplished using a simple Excel Macro we have developed (Data S1). The miR-33 scaffold also allows for transcription by RNA Pol II, creating safe inducible or tissue- or cell-type-specific knockdown.28,29 The miR-33 scaffold provides a valuable tool for achieving on-target, potent silencing that is deliverable via rAAVs or other viral vectors for research and clinical applications.
Materials and Methods
Vector Design, Construction, and Production
To generate the rAAV constructs harboring sequences encoding endogenous miRNAs, pri-miRNA fragments were PCR amplified from mouse genomic DNA and cloned into the PpuMI site in the intronic region of pAAVsc-CB-PI-Gluc plasmid.30 The identity of each pri-miRNA was verified by Sanger sequencing. shRNA fragments were generated by oligonucleotide annealing and cloned after the U6 or H1 promoter in pAAVsc-CB-Egfp.16 The shRNAmiR fragments were incorporated into the intronic region of pAAVsc-CB-PI-Gluc or pAAVsc-CB-Egfp. An Excel macro was developed and can be used to design shRNAmiR-33 cassettes (Data S1). The RNAi sensor plasmids were made by incorporating a partial target coding sequence (CDS) after the β-galactosidase reporter gene in the pmiCHECK plasmid.31 Table S1 shows the sequence of oligonucleotides for amplifying mmu-pri-miRNAs, generating shRNA fragments for gene silencing and partial CDS for constructing sensor plasmids. The shRNAmiR-33 was synthesized as gBlocks (IDT). AAV vectors used in this study were generated, purified, and titered as described previously.32
Cell Culture and Reporter Assay
HEK293 cells were cultured in DMEM supplemented with 10% FBS and 100 mg/L of penicillin-streptomycin (HyClone). Cells were maintained in a humidified incubator at 37°C and 5% CO2. Plasmids were transfected into cells in 24-well plates using Lipofectamine 2000 (Invitrogen). After 48 h, cells were lysed, and 10 μL lysates was used for the β-galactosidase assay using the Galacto-Star System (Applied Biosystems), according to the manufacturer’s instructions. Firefly luciferase activity was assessed using the Luciferase Assay System (Promega) with the same amount of lysate for β-galactosidase assay in accordance with the manufacturer’s instructions.
Southern Blot Analysis for Hirt DNA and Vector DNA Analysis
Low-molecular-weight Hirt DNA was extracted from triple plasmid-transfected HEK293 cells33 and digested with DpnI before hybridization. The Egfp fragment was labeled by 32P as probe using a random primer labeling kit (Takara). The blots were visualized using a FLA-7000 Imager (Fujifilm), and densitometry analyses were performed using MultiGauge software (Fujifilm). Viral DNAs were extracted from purified vectors following procedures for extraction of recombinant adenovirus genomic DNA.16,32 Vector DNAs equivalent to 1 × 1010 to 1 × 1011 genomes were loaded onto agarose gels, stained with ethidium bromide, and analyzed with Image Lab software (Bio-Rad).
Small RNA Northern Blot Analysis
HEK293 cells in 10-cm cell culture dishes were transfected with shRNA or shRNAmiR-33 plasmids using Lipofectamine 2000. After 48 h, total RNA was extracted using TRIzol (Invitrogen). Five micrograms of total RNA was used for small RNA northern blot analysis as described previously.30 Synthetic DNA oligonucleotides were used as reference for quantification analyses or labeled with γ-32P ATP using T4 polynucleotide kinase (NEB) as probes (Table S1).
Mouse Studies
Male C57BL/6 mice at 6–8 weeks old (Harlan, IN, USA) were sacrificed 3 weeks after intravenous injection of rAAV vectors at indicated doses. Bioluminescence levels were measured by IVIS-100 system, and liver tissues were harvested for downstream analysis. Firefly luciferase activities in mouse livers were assessed using the Luciferase Assay System (Promega) with 10 μg of liver protein. Total liver RNA was extracted using TRIzol (Invitrogen). qRT-PCR was performed as reported before.30 Primers used in the study are shown in Table S1. All animal procedures were performed according to the guidelines of the Institutional Animal Care and Use Committee of University of Massachusetts Medical School.
Small RNA Library Construction and Analysis
Total liver RNA (40 μg) was purified by 15% urea polyacrylamide gel electrophoresis (PAGE), selecting for 18- to 30-nt-long RNAs. Ligation of the 3′ adaptor (5′-rApp TGG AAT TCT CGG GTG CCA AGG /ddC/-3′) was done using truncated, K227Q mutant T4 RNA Ligase 2 at 25°C for ≥16 h. 5′ Adaptor (5′-GUU CAG AGU UCU ACA GUC CGA CGA UC-3′) was added using T4 RNA ligase (Ambion) at 25°C for ≥2 h, followed by reverse transcription using avian myeloblastosis virus (AMV) reverse transcriptase and PCR using Q5 polymerase (NEB). An Illumina NextSeq550 was used for high-throughput, single-end 50-nt sequencing. The 3′ adaptors of small RNA libraries were removed using Flexbar 3.0.3.34 After trimming, only reads with Phred scores at each position higher than 20 were kept for downstream analysis. Such reads were first aligned with Bowtie v.1.2.1.135 to miRNA hairpins36 supplemented with corresponding hairpin backbones of miR-33 and shRNA. Tailed reads were discovered with Tailor v.1.1.37 Differential analysis was done with DESeq v.3.5.38
RNA Sequencing Analysis
Strand-specific RNA sequencing (RNA-seq) libraries were prepared as described previously.39 Paired-end RNA-seq reads were first aligned to ribosomal RNA (BK000964.1) with Bowtie2.40 Non-rRNA reads were subsequently used for gene quantification with Salmon v.0.8.2.41 Differential analysis was performed using DESeq v.3.5.38 Data processing and figures were prepared with R v.3.4.1.42
Statistical Analysis
All results are shown as mean ± SD and compared between groups using two-tailed Student’s t test, except for Figure 1F data, which used Fisher’s exact test.
Accession Numbers
The raw sequencing data reported here have been deposited to the NCBI Sequence Read Archive (SRA) under the accession number SRA: SRP124896.
Author Contributions
J.X., P.D.Z., and G.G. designed the experiments. J.X., S.G., S.Z., Y.W., C.L., C.C., Q.S., R.H., J.L., H.Y., and J.K. performed the experiments. C.L. from Fornax Biotech LLC led the analysis of high-throughput sequencing data. P.W.L.T. and R.H. assisted in high-throughput sequencing data analysis. A.B. wrote the Excel macro for the design of shRNAmiR-33. P.W.L.T. and A.B. helped with manuscript editing. J.X. and C.L. prepared the figures. J.X., P.D.Z., and G.G. wrote the manuscript.
Conflicts of Interest
G.G. is a scientific co-founder of Aspa Therapeutics, Inc., holds equity in the company, and is an inventor on patents with potential royalties licensed to Aspa Therapeutics, Inc. G.G. and P.D.Z. are founders of Voyager Therapeutics, hold equity in the company, and are inventors on patents with potential royalties licensed to Voyager Therapeutics and other biopharmaceutical companies.
Acknowledgments
G.G. is supported by grants from the University of Massachusetts Medical School (an internal grant) and the NIH (R01NS076991-01, 1P01AI100263-01, 4P01HL131471-02, and R01HL097088).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.11.018.
Contributor Information
Jun Xie, Email: jun.xie@umassmed.edu.
Phillip D. Zamore, Email: phillip.zamore@umassmed.edu.
Guangping Gao, Email: guangping.gao@umassmed.edu.
Supplemental Information
References
- 1.Guo H., Ingolia N.T., Weissman J.S., Bartel D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 3.Gao G., Vandenberghe L.H., Wilson J.M. New recombinant serotypes of AAV vectors. Curr. Gene Ther. 2005;5:285–297. doi: 10.2174/1566523054065057. [DOI] [PubMed] [Google Scholar]
- 4.Grimm D., Streetz K.L., Jopling C.L., Storm T.A., Pandey K., Davis C.R., Marion P., Salazar F., Kay M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 5.McBride J.L., Boudreau R.L., Harper S.Q., Staber P.D., Monteys A.M., Martins I., Gilmore B.L., Burstein H., Peluso R.W., Polisky B. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc. Natl. Acad. Sci. USA. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giering J.C., Grimm D., Storm T.A., Kay M.A. Expression of shRNA from a tissue-specific pol II promoter is an effective and safe RNAi therapeutic. Mol. Ther. 2008;16:1630–1636. doi: 10.1038/mt.2008.144. [DOI] [PubMed] [Google Scholar]
- 7.Pfister E.L., Chase K.O., Sun H., Kennington L.A., Conroy F., Johnson E., Miller R., Borel F., Aronin N., Mueller C. Safe and Efficient Silencing with a Pol II, but Not a Pol lII, Promoter Expressing an Artificial miRNA Targeting Human Huntingtin. Mol. Ther. Nucleic Acids. 2017;7:324–334. doi: 10.1016/j.omtn.2017.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boudreau R.L., Martins I., Davidson B.L. Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo. Mol. Ther. 2009;17:169–175. doi: 10.1038/mt.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fowler D.K., Williams C., Gerritsen A.T., Washbourne P. Improved knockdown from artificial microRNAs in an enhanced miR-155 backbone: a designer’s guide to potent multi-target RNAi. Nucleic Acids Res. 2016;44:e48. doi: 10.1093/nar/gkv1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maepa M.B., Ely A., Grayson W., Arbuthnot P. Sustained Inhibition of HBV Replication In Vivo after Systemic Injection of AAVs Encoding Artificial Antiviral Primary MicroRNAs. Mol. Ther. Nucleic Acids. 2017;7:190–199. doi: 10.1016/j.omtn.2017.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boudreau R.L., Monteys A.M., Davidson B.L. Minimizing variables among hairpin-based RNAi vectors reveals the potency of shRNAs. RNA. 2008;14:1834–1844. doi: 10.1261/rna.1062908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson A.L., Linsley P.S. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat. Rev. Drug Discov. 2010;9:57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 13.Guda S., Brendel C., Renella R., Du P., Bauer D.E., Canver M.C., Grenier J.K., Grimson A.W., Kamran S.C., Thornton J. miRNA-embedded shRNAs for Lineage-specific BCL11A Knockdown and Hemoglobin F Induction. Mol. Ther. 2015;23:1465–1474. doi: 10.1038/mt.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maczuga P., Lubelski J., van Logtenstein R., Borel F., Blits B., Fakkert E., Costessi A., Butler D., van Deventer S., Petry H. Embedding siRNA sequences targeting apolipoprotein B100 in shRNA and miRNA scaffolds results in differential processing and in vivo efficacy. Mol. Ther. 2013;21:217–227. doi: 10.1038/mt.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu S., Jin L., Zhang Y., Huang Y., Zhang F., Valdmanis P.N., Kay M.A. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell. 2012;151:900–911. doi: 10.1016/j.cell.2012.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie J., Mao Q., Tai P.W.L., He R., Ai J., Su Q., Zhu Y., Ma H., Li J., Gong S. Short DNA Hairpins Compromise Recombinant Adeno-Associated Virus Genome Homogeneity. Mol. Ther. 2017;25:1363–1374. doi: 10.1016/j.ymthe.2017.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Auyeung V.C., Ulitsky I., McGeary S.E., Bartel D.P. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell. 2013;152:844–858. doi: 10.1016/j.cell.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang W., Bartel D.P. The Menu of Features that Define Primary MicroRNAs and Enable De Novo Design of MicroRNA Genes. Mol. Cell. 2015;60:131–145. doi: 10.1016/j.molcel.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fellmann C., Hoffmann T., Sridhar V., Hopfgartner B., Muhar M., Roth M., Lai D.Y., Barbosa I.A., Kwon J.S., Guan Y. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 2013;5:1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 20.Chourabi M., Liew M.S., Lim S., H’mida-Ben Brahim D., Boussofara L., Dai L., Wong P.M., Foo J.N., Sriha B., Robinson K.S. ENPP1 Mutation Causes Recessive Cole Disease by Altering Melanogenesis. J. Invest. Dermatol. 2018;138:291–300. doi: 10.1016/j.jid.2017.08.045. [DOI] [PubMed] [Google Scholar]
- 21.Davidson B.L., McCray P.B., Jr. Current prospects for RNA interference-based therapies. Nat. Rev. Genet. 2011;12:329–340. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borel F., Gernoux G., Sun H., Stock R., Blackwood M., Brown R.H., Jr., Mueller C. Safe and effective superoxide dismutase 1 silencing using artificial microRNA in macaques. Sci. Transl. Med. 2018;10:eaau6414. doi: 10.1126/scitranslmed.aau6414. [DOI] [PubMed] [Google Scholar]
- 23.Boudreau R.L., Spengler R.M., Davidson B.L. Rational design of therapeutic siRNAs: minimizing off-targeting potential to improve the safety of RNAi therapy for Huntington’s disease. Mol. Ther. 2011;19:2169–2177. doi: 10.1038/mt.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding H., Liao G., Wang H., Zhou Y. Asymmetrically designed siRNAs and shRNAs enhance the strand specificity and efficacy in RNAi. J. RNAi Gene Silencing. 2007;4:269–280. [PMC free article] [PubMed] [Google Scholar]
- 25.Mockenhaupt S., Grosse S., Rupp D., Bartenschlager R., Grimm D. Alleviation of off-target effects from vector-encoded shRNAs via codelivered RNA decoys. Proc. Natl. Acad. Sci. USA. 2015;112:E4007–E4016. doi: 10.1073/pnas.1510476112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu S., Jin L., Zhang F., Huang Y., Grimm D., Rossi J.J., Kay M.A. Thermodynamic stability of small hairpin RNAs highly influences the loading process of different mammalian Argonautes. Proc. Natl. Acad. Sci. USA. 2011;108:9208–9213. doi: 10.1073/pnas.1018023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park J.H., Shin C. Slicer-independent mechanism drives small-RNA strand separation during human RISC assembly. Nucleic Acids Res. 2015;43:9418–9433. doi: 10.1093/nar/gkv937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Y.S., Xie J., Wang D., Kim J.M., Tai P.W.L., Gravallese E., Gao G., Shim J.H. Bone-targeting AAV-mediated silencing of Schnurri-3 prevents bone loss in osteoporosis. Nat. Commun. 2019;10:2958. doi: 10.1038/s41467-019-10809-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sweeney C.G., Kearney P.J., Fagan R.R., Smith L.A., Bolden N.C., Zhao-Shea R., Rivera I.V., Kolpakova J., Xie J., Gao G. Conditional, inducible gene silencing in dopamine neurons reveals a sex-specific role for Rit2 GTPase in acute cocaine response and striatal function. Neuropsychopharmacology. 2019 doi: 10.1038/s41386-019-0457-x. Published online July 5, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie J., Ameres S.L., Friedline R., Hung J.H., Zhang Y., Xie Q., Zhong L., Su Q., He R., Li M. Long-term, efficient inhibition of microRNA function in mice using rAAV vectors. Nat. Methods. 2012;9:403–409. doi: 10.1038/nmeth.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He X., Xie J., Zhang D., Su Q., Sai X., Bai R., Chen C., Luo X., Gao G., Pan W. Recombinant adeno-associated virus-mediated inhibition of microRNA-21 protects mice against the lethal schistosome infection by repressing both IL-13 and transforming growth factor beta 1 pathways. Hepatology. 2015;61:2008–2017. doi: 10.1002/hep.27671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao G., Sena-Esteves M. Introducing genes into mammalian cells: viral vectors. In: Green M.R., Sambrook J., editors. Volume 2. Cold Spring Harbor Laboratory Press; 2012. pp. 1209–1313. (Molecular Cloning: A Laboratory Manual). [Google Scholar]
- 33.Wang X.S., Ponnazhagan S., Srivastava A. Rescue and replication signals of the adeno-associated virus 2 genome. J. Mol. Biol. 1995;250:573–580. doi: 10.1006/jmbi.1995.0398. [DOI] [PubMed] [Google Scholar]
- 34.Roehr J.T., Dieterich C., Reinert K. Flexbar 3.0—SIMD and multicore parallelization. Bioinformatics. 2017;33:2941–2942. doi: 10.1093/bioinformatics/btx330. [DOI] [PubMed] [Google Scholar]
- 35.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozomara A., Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chou M.T., Han B.W., Hsiao C.P., Zamore P.D., Weng Z., Hung J.H. Tailor: a computational framework for detecting non-templated tailing of small silencing RNAs. Nucleic Acids Res. 2015;43:e109. doi: 10.1093/nar/gkv537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Z., Theurkauf W.E., Weng Z., Zamore P.D. Strand-specific libraries for high throughput RNA sequencing (RNA-Seq) prepared without poly(A) selection. Silence. 2012;3:9. doi: 10.1186/1758-907X-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.R Development Core Team . R Foundation for Statistical Computing; 2017. R: A language and environment for statistical computing. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.