Abstract
Studies have found that RNA-binding proteins (RBPs) and long non-coding RNAs (lncRNAs) are dysregulated and play an important regulatory role in the development of tumors. Based on The Cancer Genome Atlas (TCGA) database, our findings from experiments, and the evidence of previous studies, we screened DiGeorge syndrome critical region gene 8 (DGCR8), ZFAT antisense RNA 1 (ZFAT-AS1), and caudal type homeobox 2 (CDX2) as research candidates. In the present study, DGCR8 and CDX2 were highly expressed and ZFAT-AS1 was markedly downregulated in glioma tissues and cells. DGCR8 or CDX2 knockdown or ZFAT-AS1 overexpression suppressed glioma cell proliferation, migration, and invasion and facilitated apoptosis. DGCR8 might decrease ZFAT-AS1 expression by attenuating its stability in a manner of inducing its cleavage. Importantly, ZFAT-AS1 could inhibit CDX2 transcription by mediating the methylation of histone H3 on lysine 27 (H3K27me3) modification induced by PRC2 in the CDX2 promoter region. In addition, CDX2 transcriptionally activated DGCR8 expression by binding to its promoter regions, forming a positive feedback loop of DGCR8/ZFAT-AS1/CDX2. In conclusion, DGCR8/ZFAT-AS1 promotes CDX2 transcription in a PRC2 complex-dependent manner to facilitate the malignant biological behavior of glioma cells.
Keywords: glioma, DGCR8, ZFAT-AS1, H3K27me3, CDX2
Graphical Abstract
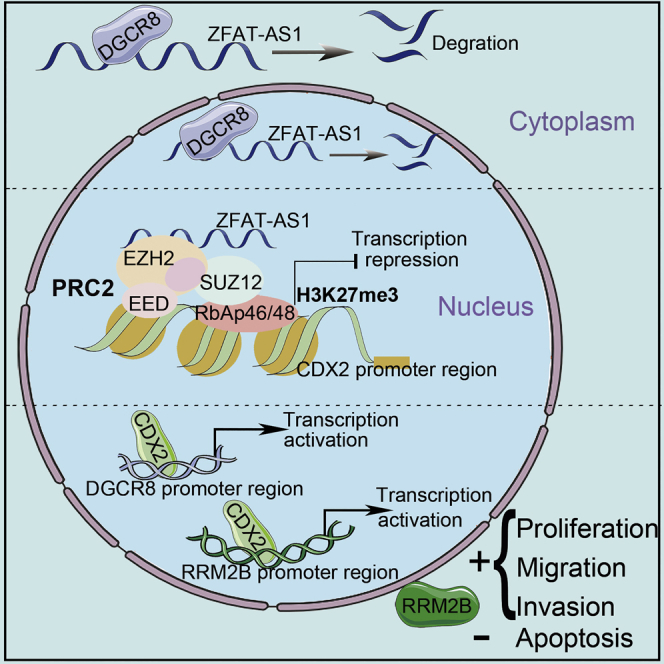
Yixue Xue et al. demonstrate that DGCR8 can decrease ZFAT-AS1 expression by attenuating its stability in a manner of inducing its cleavage. Importantly, ZFAT-AS1 can inhibit CDX2 transcription by mediating the H3K27me3 modification induced by PRC2 in the CDX2 promoter region, which can affect the malignant biological behavior of glioma cells.
Introduction
Glioma is the most common primary malignant tumor of the human CNS.1 Although the applications of surgery, chemotherapy, and radiotherapy have made significant progress in the treatment of glioma in recent years, the prognosis of patients with glioma remains poor, with the lowest 5-year survival rate among all cancers;2,3 in addition, the median survival time of patients with gliomas of high pathological grade is approximately 15 months.4,5 Therefore, molecular targeting has become a focus of current studies being conducted on glioma therapeutics.
RNA-binding proteins (RBPs) bind to double-stranded or single-stranded RNA in cells, and their targets are structure-specific components important to the fate of RNA in terms of processing, transport, localization, stability, or function; furthermore, these targets are involved in the formation of ribonucleoprotein complexes (RNPs).6,7 DiGeorge syndrome critical region gene 8 (DGCR8) is a double-stranded RNA-binding protein that acts as a non-catalytic subunit of the microprocessor complex. With the catalytic subunit Drosha, DGCR8 forms a Drosha-DGCR8 complex, which splices primary microRNAs (pri-miRNAs) to regulate the synthesis of miRNAs.8 Sand et al.9 have found that DGCR8 is highly expressed in human gastrointestinal cancer cell lines (AGS, KYSE30, and HepG2). DGCR8 mRNA has been found to be upregulated in invasive ductal breast carcinoma, and its expression is higher in the high pathological and metastatic groups than in the low pathological and non-metastatic groups, suggesting that upregulation of DGCR8 may be involved in the development and invasiveness of invasive ductal breast carcinoma.10 Additionally, Drosha-DGCR8 is able to regulate the stability of long non-coding RNAs (lncRNAs), including MALAT1, affecting their expression.8,11
lncRNAs are involved in epigenetic regulation, gene transcription, and post-transcriptional gene regulation,12,13 and they play an important regulatory role in the development of tumors, including glioma.14,15 For example, MEG3 shows markedly low expression in glioma tissues and exerts tumor-suppressive effects.16 ZEB1-AS1 is highly expressed in glioma tissues, promoting tumorigenesis, and its elevated expression is correlated with a poor prognosis.17 TSLC1-AS1 plays the role of tumor suppressor genes in gliomas.18 lncRNA ZFAT antisense RNA 1 (ZFAT-AS1) is an imprinted gene that is located on the long arm of chromosome 8 of the human genome and is an antisense transcript of the ZFAT gene. ZFAT is a zinc finger and AT-hook domain-containing coding gene, which encodes a protein that likely binds to DNA and functions as a transcriptional regulator involved in apoptosis and cell survival.19 It has been reported that RBP SRSF1 regulates cell migration, VEGFA mRNA splicing, and angiogenesis in glioblastoma multiforme by binding to circSMARCA5.20,21 However, the regulatory role and potential mechanisms of DGCR8 and ZFAT-AS1 affecting the biological behavior of glioma have not yet been elucidated.
The polycomb repressive complex 2 (PRC2) is a highly conserved protein complex composed of multiple subunits. PRC2 can inhibit gene transcriptional activity by catalyzing methylation of histone H3 on lysine 27 (H3K27me3) modification.22 A previous study found that lncRNA-HOTAIR promoted the transition of histone H3 lysine 27 acetylation to methylation by recruiting the PRC2 complex and promoted epithelial-mesenchymal transition of gastric cancer.23
CDX2, a member of the caudal-related homeobox transcription factor gene family, encodes an intestine-specific nuclear transcription factor that regulates the balance of proliferation and differentiation of intestinal epithelial cells.24 The aberrant expression of this gene is associated with intestinal inflammation and tumorigenesis. Studies have found that overexpression of CDX2 inhibits the proliferation and tumor formation of colon cancer cells.25 CDX2 expression decreased progressively with the advancing stage of gastric cancers and showed a negative correlation with the depth of tumor invasion.26
RRM2B encodes one of two forms of the ribonucleotide reductase R2 subunit and is necessary for normal DNA repair and mtDNA synthesis in non-proliferating cells. Studies have found that RRM2B is highly expressed in non-small cell lung cancer and esophageal squamous cell carcinoma, and it is a biomarker for tumor invasion and poor prognosis.27,28 The mRNA and protein levels of RRM2B in cervical cancer tissues and cells are significantly upregulated, and this upregulation influences the progression, metastasis, and poor prognosis of tumors. In addition, it also plays an oncogenic role.29,30
In the present study we attempted to prove that cell proliferation, migration, and invasion facilitated by DGCR8/ZFAT-AS1 in vitro might be ascribed to PRC2-induced H3K27me3 modification in the CDX2 promoter region. Additionally, we aimed to elucidate whether DGCR8 and ZFAT-AS1 played an important role in the progression of glioma.
Results
DGCR8 Promoted Malignant Biological Behavior of Glioma Cells, but ZFAT-AS1 Suppressed Malignant Biological Behavior of Glioma Cells
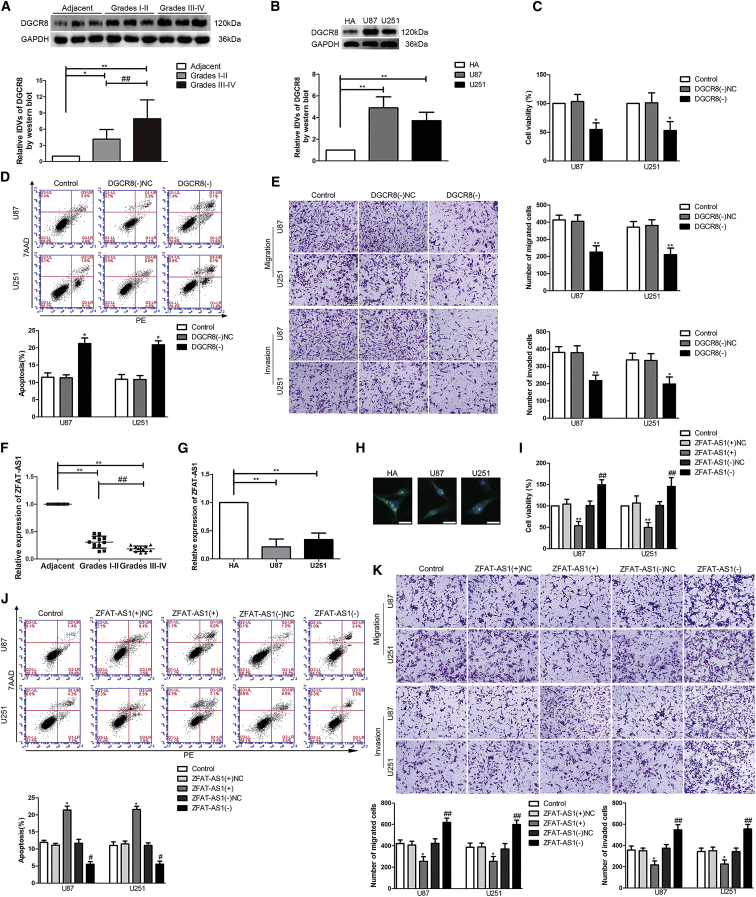
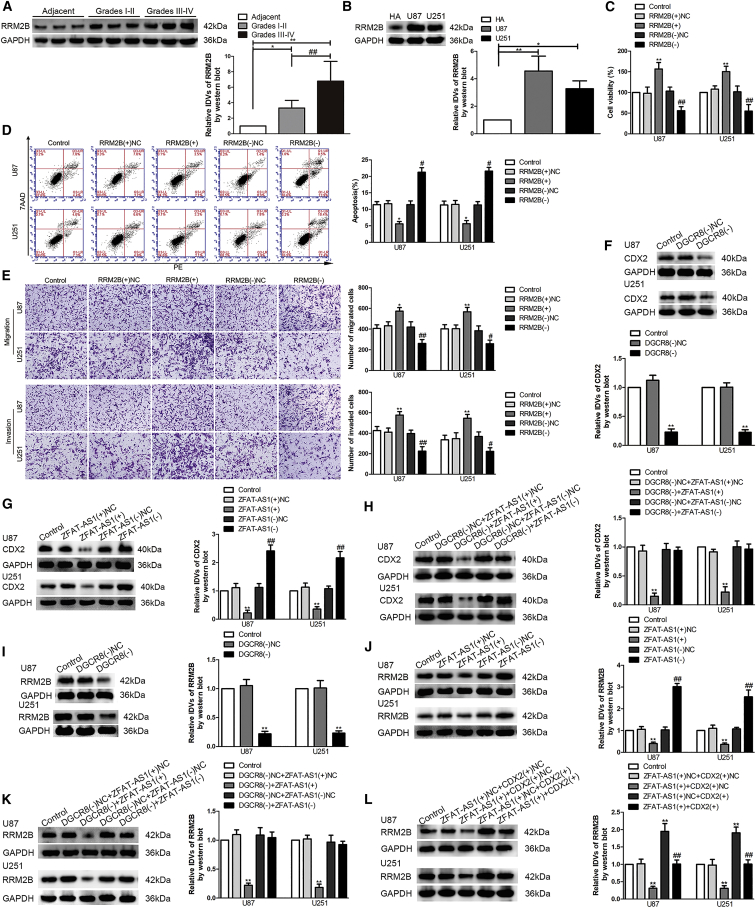
According to the data obtained from The Cancer Genome Atlas (TCGA), the effect of DGCR8 expression on the survival of patients with low-grade glioma was significant (p = 0.018), and DCGR8 mRNA was highly upregulated in U87 and U251 cells among the four glioma cell lines of U87, U373, U251, and A172 (Figures S1A–S1C). DGCR8 protein levels were detected in normal brain parenchyma deriving from regions adjacent to the glioma (Adjacent), different grades of glioma tissues, and human astrocyte (HA) cells and U87 and U251 glioma cells (Figures 1A and 1B). As shown in Figure 1A, compared with Adjacent, DGCR8 protein levels were significantly elevated in glioma tissues, and its protein levels increased as the pathological grade rose. In addition, compared with HA cells, significant upregulation of DGCR8 protein levels was also found in both U87 and U251 glioma cells (Figure 1B).
Figure 1.
Expression and Effect of DGCR8 and ZFAT-AS1 in Glioma
(A) Normal brain parenchyma deriving from regions adjacent to the glioma (Adjacent) and glioma tissues of different grades were analyzed for DGCR8 protein levels by western blot. The integrated density values (IDVs) of the blot bands were statistically analyzed. Data are presented as the mean ± SD (n = 12, each group). *p < 0.05, **p < 0.01, ##p < 0.01 by Student’s t test. (B) DGCR8 protein levels were detected by western blot in normal human astrocytes (HAs) and glioma cell lines (U87 and U251). Data are presented as the mean ± SD (n = 3, each group). **p < 0.01 by Student’s t test. (C) A Cell Counting Kit-8 (CCK-8) assay was used to detect the effects of DGCR8 knockdown on the proliferation of U87 and U251 glioma cells. DGCR8(−), group treated with plasmids with the short hairpin RNA against DGCR8; DGCR8(−)NC, group treated with negative control empty vector plasmids with non-targeting sequence. (D) Flow cytometry analysis was used to detect the apoptotic percentages of U87 and U251 cells after DGCR8 knockdown. (E) A cell migration and invasion assay (transwell assay) was used to detect the effect of DGCR8 on cell migration and invasion of U87 and U251 cells. Scale bars, 50 μm. Data are presented as the mean ± SD (n = 3, each group). *p < 0.05 versus DGCR8(−)NC group; **p < 0.01 versus DGCR8(−)NC group by one-way ANOVA. (F) ZFAT-AS1 expression was analyzed by quantitative real-time PCR in Adjacent and glioma tissues of different grades. Data are presented as the mean ± SD (n = 12, each group). **p < 0.01, ##p < 0.01 by Student’s t test. (G) ZFAT-AS1 expression was detected by quantitative real-time PCR in normal HA, U87, and U251 cells. Data are presented as the mean ± SD (n = 3, each group). **p < 0.01 by Student’s t test. (H) Fluorescence in situ hybridization (FISH) analysis of the location of ZFAT-AS1 (green) in the cytoplasm and nuclear fractions (blue) of HA, U87, and U251 cells. Nuclei were labeled with DAPI. Scale bars, 20 μm. (I) The effect of ZFAT-AS1 on U87 and U251 proliferation was evaluated by the CCK-8 assay. ZFAT-AS1(+), group treated with plasmids with ZFAT-AS1 full-length sequence; ZFAT-AS1(+)NC, group treated with negative control empty vector plasmids with a non-targeting sequence. (J) Flow cytometry analysis of the apoptosis of U87 and U251 cells with altered expression of ZFAT-AS1. (K) The effect of ZFAT-AS1 on cell migration and invasion of U87 and U251 cells was evaluated by transwell assays. Scale bars, 50 μm. Data are presented as the mean ± SD (n = 3, each group). *p < 0.05 versus ZFAT-AS1(+)NC group; **p < 0.01 versus ZFAT-AS1(+)NC group; #p < 0.05 versus ZFAT-AS1(−)NC group; ##p < 0.01 versus ZFAT-AS1(−)NC group by one-way ANOVA.
By using western blot results, we assessed the influence of DGCR8 on proliferation, apoptosis, migration, and invasion of glioma cells. First, stable DGCR8 knockdown U87 and U251 cells were constructed. The results of a Cell Counting Kit-8 (CCK-8) assay showed that cell viability of U87 and U251 cells was prominently reduced when DGCR8 was knocked down (Figure 1C). However, flow cytometry analysis showed that knockdown of DGCR8 elevated the apoptosis rate of U87 and U251 cells (Figure 1D). The migration and invasion abilities were decreased after DGCR8 knockdown (Figure 1E).
The expression profile from lncRNA analysis of U87 and U251 cells upon DGCR8 knockdown revealed the upregulation of several lncRNAs. The lncRNAs with an approximately 1- to 2-fold change were validated using quantitative real-time PCR. Interestingly, ZFAT-AS1 showed an ∼10-fold change (Figures S1D–S1F). ZFAT-AS1 expression levels in different glioma tissues, HA cells, and U87 and U251 glioma cells were measured by quantitative real-time PCR. Compared with Adjacent, ZFAT-AS1 expression was downregulated in glioma tissues of different grades, and its expression levels decreased as the pathological grade increased (Figure 1F). In addition, ZFAT-AS1 expression levels were reduced in U87 and U251 cells compared with HA cells (Figure 1G). The fluorescence in situ hybridization (FISH) showed that ZFAT-AS1 was localized both in cytoplasm and nucleus (Figure 1H).
To analyze the potential role of ZFAT-AS1 in biological behavior of glioma cells, we next overexpressed and knocked down ZFAT-AS1 in U87 and U251 glioma cells. A significant reduction of cell viability, migration, and invasion of both U87 and U251 cells was led by ZFAT-AS1 overexpression, but ZFAT-AS1 overexpression promoted cell apoptosis (Figure 1I–1K). Whereas ZFAT-AS1 knockdown promoted cell viability, migration, and invasion of U87 and U251 cells, it suppressed cell apoptosis (Figure 1I–1K).
DGCR8 Downregulated ZFAT-AS1 Expression via Inducing Its Cleavage, and ZFAT-AS1 Participated in DGCR8-Knockdown-Mediated Inhibition on Malignant Biological Behavior of Glioma Cells
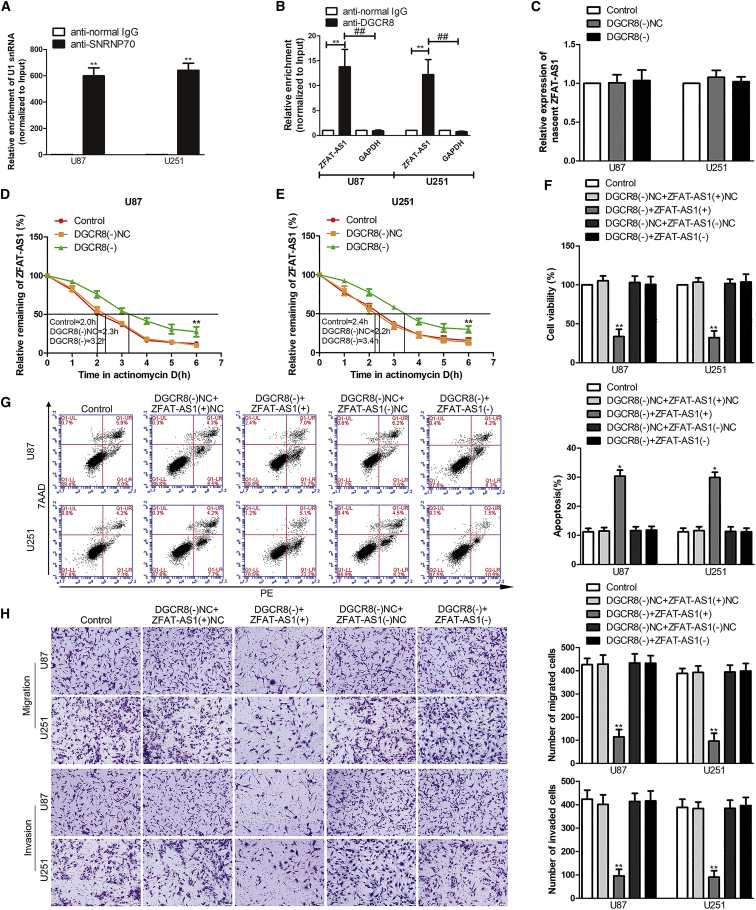
To clarify the relationship between DGCR8 and ZFAT-AS1, using the bioinformatics database starBase v2.0, it was predicted that ZFAT-AS1 harbored a binding sequence of DGCR8 and FUS, but the influence of FUS on ZFAT-AS1 was not significant (Figures S1G and S1H). To validate this hypothesis, we performed an RNA immunoprecipitation (RIP) assay. Results of the RIP assay showed that the enrichment of ZFAT-AS1 was significantly increased in the anti-DGCR8 group compared with the negative control anti-normal immunoglobulin (Ig)G group and the negative RNA control GAPDH (Figures 2A and 2B). To further elucidate the mechanism of DGCR8 that could downregulate ZFAT-AS1 expression, we next analyzed nascent ZFAT-AS1 and its half-life in stable DGCR8 knockdown U87 and U251 cells by quantitative real-time PCR. Figures 2C–2E show that transcription of nascent ZFAT-AS1 has no significant change after DGCR8 knockdown. However, the half-life of ZFAT-AS1 was significantly prolonged when DGCR8 was knocked down.
Figure 2.
ZFAT-AS1 Participates in DGCR8-Knockdown-Mediated Inhibition on Malignant Biological Behavior of Glioma Cells
(A and B) An RNA immunoprecipitation (RIP) assay confirmed that DCGR8 bound to ZFAT-AS1. Relative enrichment was measured using quantitative real-time PCR. (A) Relative enrichment of U1 small nuclear RNA (snRNA) in SNRNP70 relative to normal IgG immunoprecipitates. (B) Relative enrichment of ZFAT-AS1 in anti-DHCR8 relative to normal IgG immunoprecipitates. Normal mouse IgG was used as a negative control, SNRNP70 was used as a positive control, and GAPDH was used as a negative RNA control. Data are presented as the mean ± SD (n = 3, each group). **p < 0.01 versus anti-normal IgG group; ##p < 0.01 versus GAPDH group by Student’s t test. (C) The nascent ZFAT-AS1 was measured by quantitative real-time PCR. DGCR8(−), group treated with plasmids with the short hairpin RNA against DGCR8; DGCR8(−)NC, group treated with the negative control empty vector plasmids with a non-targeting sequence. (D and E) Graphs show ZFAT-AS1 levels at different times treated by actinomycin D in U87 (D) and U251 (E) cells. Data are presented as mean ± SD (n = 3, each group). **p < 0.01 versus DGCR8(−)NC group by one-way ANOVA. (F) Effect of DGCR8 and ZFAT-AS1 on the proliferation of U87 and U251 cells. (G) Flow cytometry analysis to evaluate cell apoptosis of U87 and U251 cells. (H) Effect of DGCR8 and ZFAT-AS1 on the migration and invasion of U87 and U251 cells. Scale bars, 50 μm. Data are presented as the mean ± SD (n = 3, each group). *p < 0.05 versus DGCR8(−)NC + ZFAT-AS1(+)NC group; **p < 0.01 versus DGCR8(−)NC + ZFAT-AS1(+)NC group by one-way ANOVA.
Having found that DGCR8 could regulate ZFAT-AS1 expression, we speculated whether ZFAT-AS1 was involved in DGCR8-knockdown-mediated inhibition on malignant biological behavior of glioma cells. The stable DGCR8 knockdown cells were transfected with ZFAT-AS1(+), ZFAT-AS1(−), or their corresponding negative control plasmids. ZFAT-AS1 overexpression enhanced the suppressive effect on cell viability of U87 and U251 cells induced by DGCR8 knockdown, while ZFAT-AS1 knockdown reversed the suppressive effect (Figure 2F). Similar effects were observed on cell migration and invasion (Figure 2H). However, ZFAT-AS1 overexpression enhanced the promotion of U87 and U251 cell apoptosis induced by DGCR8 knockdown. ZFAT-AS1 knockdown rescued the influence of DGCR8 knockdown on cell apoptosis (Figure 2G). The results described above supported that an involvement of ZFAT-AS1 was in DGCR8-knockdown-mediated inhibition on malignant biological behavior of glioma cells.
Knockdown of CDX2 Suppressed Glioma Cell Proliferation, Migration, and Invasion and Accelerated Apoptosis
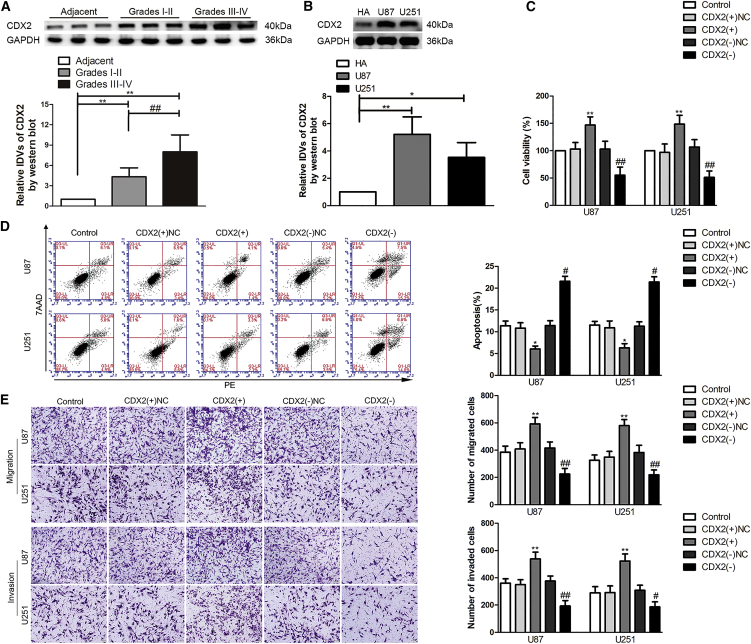
Compared with Adjacent, CDX2 protein levels were significantly highly expressed in different grades of glioma tissues and were elevated more in grades III–IV glioma tissues than in grades I–II glioma tissues (Figure 3A). Consistently, CDX2 protein expression was upregulated in U87 and U251 glioma cells (Figure 3B). To assess CDX2 function on the malignant biological behavior of glioma cells, cells were treated with CDX2(+) or CDX2(−) plasmids. CDX2 overexpression induced a significant increase of cell viability, migration, and invasion of U87 and U251 glioma cells, but a reduction of cell apoptosis. Whereas CDX2 knockdown suppressed U87 and U251 cell viability, migration, and invasion, it promoted cell apoptosis (Figures 3C–3E).
Figure 3.
CDX2 Endogenous Expression and Its Effect on the Proliferation, Apoptosis, Migration, and Invasion in Glioma Cells
(A) CDX2 protein expression levels in normal brain parenchyma deriving from regions adjacent to the glioma (Adjacent) and glioma tissues of different grades. The integrated density values (IDVs) of the blot bands were statistically analyzed. Data are presented as the mean ± SD (n = 12, each group). **p < 0.01, ##p < 0.01 by Student’s t test. (B) Protein expression levels of CDX2 in HA, U87, and U251 cells. Data are presented as the mean ± SD (n = 3, each group). *p < 0.05, **p < 0.01 by Student’s t test. (C) Effect of CDX2 on the proliferation of U87 and U251 cells. CDX2(−), group treated with plasmids with the short hairpin RNA against CDX2; CDX2(+), group treated with plasmids with CDX2 full-length sequence; CDX2(−)NC and CDX2(+)NC), groups treated with negative control empty vector plasmids with non-targeting sequence, respectively. (D) Apoptotic percentages of U87 and U251 cells were detected after CDX2 overexpression or knockdown. (E) Effect of CDX2 on cell migration and invasion of U87 and U251 cells. Scale bars, 50 μm. Data are presented as the mean ± SD (n = 3, each group). *p < 0.05 versus CDX2(+)NC group; **p < 0.01 versus CDX2(+)NC group; #p < 0.05 versus CDX2(−)NC group; ##p < 0.01 versus CDX2(−)NC group by one-way ANOVA.
ZFAT-AS1 Inhibited the Transcription of CDX2 by Binding and Recruiting the PRC2 Complex to Induce H3K27me3 Modification in the CDX2 Promoter Region
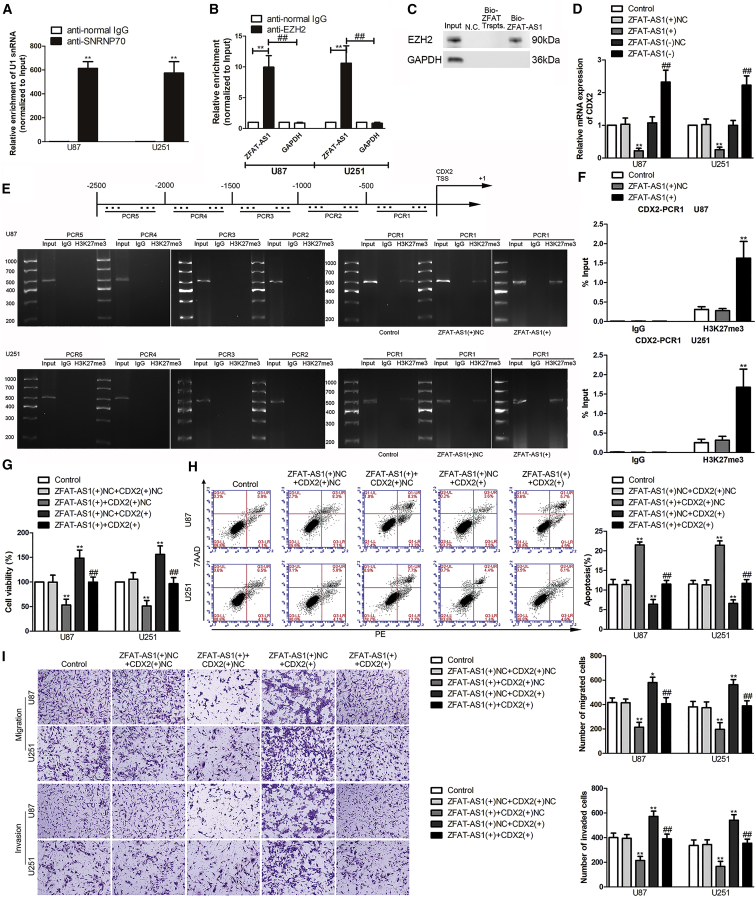
PRC2 inhibits gene transcriptional activity by catalyzing H3K27me2/3. Emerging evidence suggests that lncRNA is becoming an important participant in the function of PRC2. The results of a RIP assay showed that the enrichment of ZFAT-AS1 was significantly increased in the anti-EZH2 group compared with the negative control anti-normal IgG group and the negative RNA control GAPDH (Figures 4A and 4B); consistently, EZH2 bound to ZFAT-AS1 in an RNA pull-down assay (Figure 4C). The results above confirmed that ZFAT-AS1 bound to the important subunit EZH2 of the PRC2 complex. Figure 4D displayed that the mRNA of CDX2 was decreased following ZFAT-AS1 overexpression. Therefore, to further clarify whether PRC2 mediated ZFAT-AS1 regulation of CDX2 transcription, we performed a chromatin immunoprecipitation (ChIP) assay. 2,500 bp upstream of the CDX2 transcription start site (TSS) were on average divided into five fragments and then were designed as primers. The primers are provided in Table S2, and PCR was performed with the precipitated DNA. The results showed that H3K27me3 was enriched in the 500-bp region (PCR1) upstream of the TSS, but it was not present in negative control. Further experiments showed that the H3K27me3 reaction bands were significantly brighter in the ZFAT-AS1(+) group than in the ZFAT-AS1(+)NC group (Figure 4E). At the same time, quantitative real-time PCR was utilized to measure the percentage of PCR1 products amplified with the DNA precipitated by H3K27me3 relative to its corresponding input. The primers are provided in Table S3. Figure 4F shows that the PCR1 products were significantly increased after ZFAT-AS1 overexpression. Next, chromatin isolation by RNA purification (ChIRP) was conducted. ZFAT-AS1 obtained 35%–87% retrieval using tiling probes from the RNA fraction recovered, the promoter region 0–500 bp of CDX2 obtained ∼80% retrieval, and the promoter region 500–1000 bp obtained ∼4% retrieval from the DNA fraction recovered (Figures S3A–S3D). To further clarify the regulation between ZFAT-AS1 and CDX2, the stable ZFAT-AS1-overexpressed cells were transfected with CDX2(+) or its corresponding negative control plasmids. As shown in Figures 4G–4I, CDX2(+) could rescue the inhibitory effects on cell proliferation, migration, and invasion and the promotion effects on cell apoptosis caused by ZFAT-AS1(+) alone.
Figure 4.
The Molecular Mechanism of ZFAT-AS1 Inhibits CDX2 Transcription and Expression by Binding PRC2 to Induce H3K27me3 Modification in the CDX2 Promoter Region
(A and B) An RNA immunoprecipitation (RIP) assay confirmed that EZH2 bound to ZFAT-AS1. Relative enrichment was measured using quantitative real-time PCR. (A) Relative enrichment of U1 snRNA in SNRNP70 relative to normal IgG immunoprecipitates. (B) Relative enrichment of ZFAT-AS1 in anti-EZH2 relative to normal IgG immunoprecipitates. Normal mouse IgG was used as a negative control, SNRNP70 was used as a positive control, and GAPDH was used as a negative RNA control. Data are presented as the mean ± SD (n = 3, each group). **p < 0.01 versus anti-normal IgG group; ##p < 0.01 versus GAPDH group by Student’s t test. (C) RNA pull-down confirmed that ZFAT-AS1 bound to EZH2. EZH2 was detected using western blot. NC was representative of biotin-labeled negative RNA control (poly(A)25 RNA). Bio-ZFAT Trspts and Bio-ZFAT-AS1 are representative of 3′ end desthiobiotinylated ZFAT transcripts and ZFAT-AS1, respectively. (D) CDX2 mRNA in ZFAT-AS1 overexpression or knockdown cells. NC, group treated with negative control empty vector plasmids with non-targeting sequence. Data are presented as the mean ± SD (n = 3, each group). **p < 0.01 versus ZFAT-AS1(+)NC group; ##p < 0.01 versus ZFAT-AS1(−)NC group by one-way ANOVA. (E) H3K27me3 reaction stripe was detected in the CDX2 promoter region 500 bp (PCR1) upstream of the transcription start site (TSS) in U87 and U251 cells of control, ZFAT-AS1(+)NC, and ZFAT-AS1(+) groups. Schematic representation of the human CDX2 promoter region 2,500 bp upstream of TSS, which was designated as +1, is shown. PCR was conducted with the resulting precipitated DNA. (F) Percentage of PCR1 relative to input was analyzed by quantitative real-time PCR. Data are presented as the mean ± SD (n = 3, each group). **p < 0.01 versus ZFAT-AS1(+)NC group by one-way ANOVA. (G) Effect of ZFAT-AS1 and CDX2 on the proliferation of U87 and U251 cells. (H) Cell apoptosis of U87 and U251 cells. (I) Effect of ZFAT-AS1 and CDX2 on the migration and invasion of U87 and U251 cells. Scale bars, 50 μm. Data are presented as the mean ± SD (n = 3, each group). *p < 0.05 versus ZFAT-AS1(+)NC + CDX2(+)NC group; **p < 0.01 versus ZFAT-AS1(+)NC + CDX2(+)NC group; ##p < 0.01 versus ZFAT-AS1(+) + CDX2(+)NC group by one-way ANOVA.
RRM2B, as a Tumor Initiation Factor, Facilitated Glioma Cell Proliferation, Migration, and Invasion and Repressed Apoptosis
To reveal the precise mechanism involved in CDX2-regulated malignant biological behavior of glioma cells, we selected RRM2B as a potential tumor-related initiation factor. We measured the endogenous expression and functional role of RRM2B in glioma cells. The protein levels of RRM2B were significantly elevated in glioma tissues and were more upregulated in grades III–IV glioma tissues than in grades I–II glioma tissues (Figure 5A). The protein levels of RRM2B were higher in U87 and U251 cells than in HA cells (Figure 5B). We subsequently detected the influence on glioma cell proliferation, apoptosis, migration, and invasion following RRM2B overexpression or knockdown. Compared with the RRM2B(+)NC group, the cell viability ratio and migrated and invaded cells numbers of U87 and U251 cells were significantly advanced in the RRM2B(+) group, but the cell apoptosis rate was reduced in the RRM2B(+) group. Conversely, RRM2B knockdown weakened the abilities of cell viability, migration, and invasion in U87 and U251 cells, but it accelerated cell apoptosis (Figures 5C–5E). The above data supported that RRM2B functioned as an oncogene factor to facilitate the malignant biological behavior of glioma cells.
Figure 5.
RRM2B Endogenous Expression and Its Effect on the Proliferation, Migration, Invasion, and Apoptosis in Glioma Cells
(A) Protein expression levels of RRM2B in normal brain parenchyma deriving from regions adjacent to the glioma (Adjacent) and glioma tissues of different grades. The integrated density values (IDVs) of the blot bands were statistically analyzed. Data are presented as the mean ± SD (n = 12, each group). *p < 0.05, **p < 0.01, ##p < 0.01 by Student’s t test. (B) Protein expression levels of CDX2 in HAs and U87 and U251 glioma cells. Data are presented as the mean ± SD (n = 3, each group). *p < 0.05; **p < 0.01 by Student’s t test. (C) Effect of RRM2B on the proliferation of U87 and U251 cells by the CCK-8 assay. NC, group treated with negative control empty vector plasmids with non-targeting sequence. (D) Apoptotic percentages of U87 and U251 were detected after RRM2B overexpression or knockdown. (E) Effect of RRM2B on cell migration and invasion of U87 and U251 cells by transwell assays. Scale bars, 50 μm. Data are presented as the mean ± SD (n = 3, each group). *p < 0.05 versus RRM2B(+)NC group; **p < 0.01 versus RRM2B(+)NC group; #p < 0.05 versus RRM2B(−)NC group; ##p < 0.01 versus RRM2B(−)NC group by one-way ANOVA. (F) CDX2 protein expression levels after DGCR8 knockdown. Data are presented as the mean ± SD (n = 3, each group). **p < 0.01 versus DGCR8(−)NC group by one-way ANOVA. (G) CDX2 protein expression levels after ZFAT-AS1 overexpression or knockdown. Data are presented as the mean ± SD (n = 3, each group). **p < 0.01 versus ZFAT-AS1(+)NC group; ## p < 0.01 versus ZFAT-AS1(−)NC group by one-way ANOVA. (H) CDX2 protein expression levels after regulation by DGCR8 and ZFAT-AS1. Data are presented as the mean ± SD (n = 3, each group). **p < 0.01 versus DGCR8(−)NC + ZFAT-AS1(+)NC group by one-way ANOVA. (I) RRM2B protein expression levels after DGCR8 knockdown. Data are presented as the mean ± SD (n = 3, each group). **p < 0.01 versus DGCR8(−)NC group by one-way ANOVA. (J) RRM2B protein expression levels after ZFAT-AS1 overexpression or knockdown. Data are presented as the mean ± SD (n = 3, each group). **p < 0.01 versus ZFAT-AS1(+)NC group; ##p < 0.01 versus ZFAT-AS1(−)NC group by one-way ANOVA. (K) RRM2B protein expression levels after regulation by DGCR8 and ZFAT-AS1. Data are presented as the mean ± SD (n = 3, each group). **p < 0.01 versus DGCR8(−)NC + ZFAT-AS1(+)NC group by one-way ANOVA. (L) The RRM2B protein expression levels after regulation by ZFAT-AS1 and CDX2. Data are presented as the mean ± SD (n = 3, each group). **p < 0.01 versus ZFAT-AS1(+)NC + CDX2(+)NC group; ##p < 0.01 versus ZFAT-AS1(+) + CDX2(+)NC group by one-way ANOVA.
Western blotting results showed that CDX2 protein levels were markedly decreased following DGCR8 knockdown (Figure 5F). CDX2 protein levels were also markedly decreased following ZFAT-AS1 overexpression but increased with ZTAT-AS1 knockdown (Figure 5G). We next detected CDX2 protein levels in the stable co-transfected DGCR8(−) and ZFAT-AS1(+) or ZFAT-AS1(−) cells. The stable co-transfected DGCR8(−) and ZFAT-AS1(+) cells had a more significant inhibitory effect on the protein expression level of CDX2, whereas ZFAT-AS1 knockdown restored the inhibition of CDX2 protein expression induced by DGCR8 knockdown alone (Figure 5H). Collectively, the findings described above supported that DGCR8 knockdown lowered CDX2 protein levels by upregulating ZFAT-AS1.
Interestingly, from the results shown in Figures 5I–5L, we also found that RRM2B protein levels were decreased with DGCR8 knockdown or ZFAT-AS1 overexpression, and RRM2B protein levels were elevated after ZFAT-AS1 knockdown; nevertheless, ZFAT-AS1 knockdown reversed the reduction of RRM2B protein induced by DGCR8 knockdown alone, and CDX2 overexpression restored the inhibition of RRM2B protein expression induced by ZFAT-AS1 overexpression. Ultimately, we concluded that DGCR8 might regulate RRM2B expression through CDX2.
CDX2 Directly Bound to the Promoter Region of RRM2B and DGCR8 and Transcriptionally Activated Their Expression
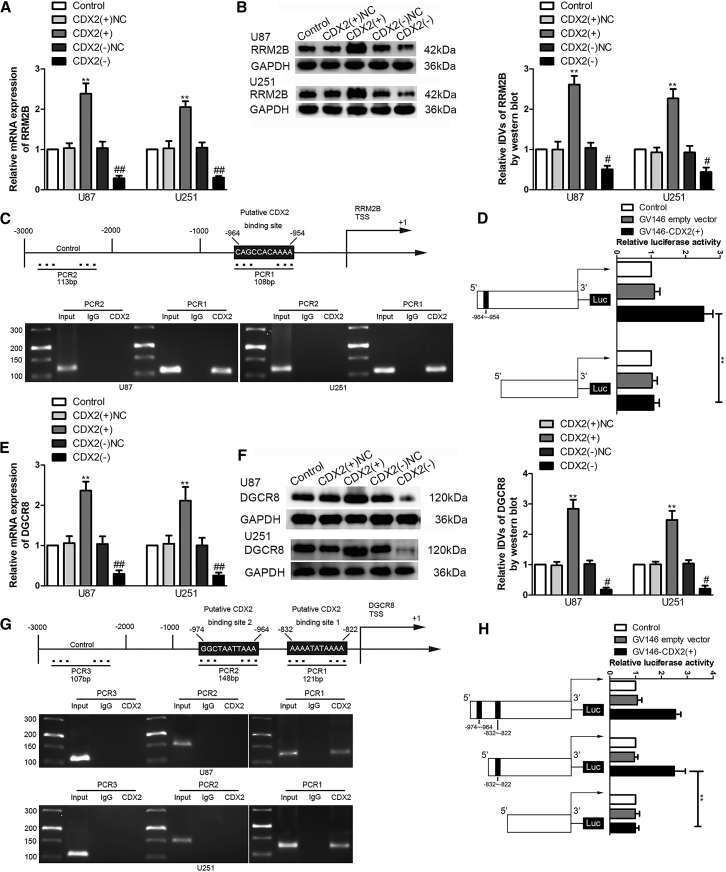
Importantly, the mRNA and protein expression levels of RRM2B were elevated by CDX2 overexpression, but a significant reduction was caused by CDX2 knockdown (Figures 6A and 6B). A putative binding site for CDX2 was predicted in the promoter region −1,000 to 0 bp of RRM2B using the bioinformatics database JASPAR. The ChIP assay and luciferase reporter assay were performed to validate the interaction between the CDX2 and RRM2B promoter region. As shown in Figures 6C and 6D, the results of the two assays above supported that CDX2 could bind to the RRM2B promoter region and activate its transcription, and RRM2B was a downstream target of CDX2.
Figure 6.
CDX2 Directly Bound with the Promoter Region of RRM2B and DGCR8 and Transcriptionally Activated Their Expression
The RRM2B mRNA (A) and protein expression (B) were detected after CDX2 overexpression or knockdown. NC, group treated with negative control empty vector plasmids with non-targeting sequence. The integrated density values (IDVs) of the blot bands were statistically analyzed. Data are presented as the mean ± SD (n = 3, each group). **p < 0.01 versus CDX2(+)NC group; #p < 0.05 versus CDX2(−)NC group; ##p < 0.01 versus CDX2(−)NC group by one-way ANOVA. (C) CDX2 bound to the promoter of RRM2B in U87 and U251 cells. A schematic representation of the human RRM2B promoter region 1,000 bp upstream of TSS, which was designated as +1, is shown. The putative CDX2 binding site is indicated. PCR was conducted with the resulting precipitated DNA. (D) Schematic depiction of the different reporter plasmids and relative luciferase activity. The relative luciferase activity was conducted after cells were co-transfected with the RRM2B promoter (−1,000 to 0 bp) (or RRM2B promoter-deleted putative CDX2 binding site) with GV146 empty vector or GV146-CDX2(+). Data are presented as the mean ± SD (n = 3, each group). **p < 0.01 by one-way ANOVA. DGCR8 mRNA (E) and protein expression (F) were detected after CDX2 overexpression or knockdown. Data are presented as the mean ± SD (n = 3, each group). **p < 0.01 versus CDX2(+)NC group; #p < 0.05 versus CDX2(−)NC group; ##p < 0.01 versus CDX2(−)NC group by one-way ANOVA. (G) CDX2 bound to the promoter of DGCR8 in U87 and U251 cells. A schematic representation of the human DGCR8 promoter region 1,000 bp upstream of TSS, which was designated as +1, is shown. Putative CDX2 binding sites were indicated. PCR was conducted with the resulting precipitated DNA. (H) Schematic depiction of the different reporter plasmids and relative luciferase activity. The relative luciferase activity was conducted after cells were co-transfected with DGCR8 promoter (−1,000 to 0 bp) (or DGCR8 promoter-deleted putative CDX2 binding sites) with GV146-CDX2(+) or GV146 empty vector. Data are presented as the mean ± SD (n = 3, each group). **p < 0.01 by one-way ANOVA.
Interestingly, we also found that the mRNA and protein expression levels of DGCR8 were also elevated with CDX2 overexpression but decreased with CDX2 knockdown (Figures 6E and 6F). Similarly, from the results shown in Figures 6G and 6H, CDX2 could bind to the DGCR8 promoter region binding site 1 and activate its transcription. Together, DGCR8 regulated the expression of CDX2 through ZFAT-AS1, and CDX2 could transcriptionally activate the expression of DGCR8, forming a positive feedback loop of DGCR8/ZFAT-AS1/CDX2.
DGCR8 Knockdown Combined with ZFAT-AS1 Overexpression Produced the Optimum Tumor-Suppressive Effect and the Longest Survival Time In Vivo
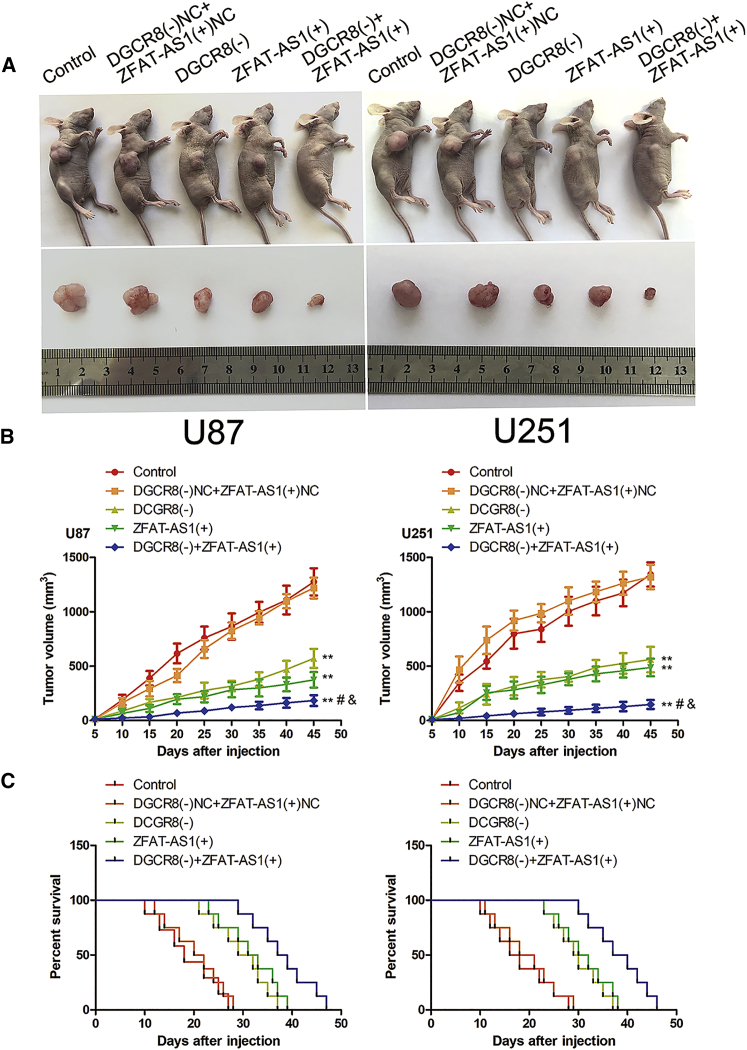
To determine the functions of DGCR8 and ZFAT-AS1 on tumor growth ability in vivo, we subcutaneous injected glioma cells of DGCR8 knockdown, ZFAT-AS1 overexpression, or a combination of the two to establish mouse xenograft models. As shown in Figures 7A and 7B, compared with the DGCR8(−)NC + ZFAT-AS1(+)NC group, the volumes of xenograft gliomas were prominently smaller in the DGCR8(−), ZFAT-AS1(+), and DGCR8(−) + ZFAT-AS1(+) groups. Additionally, combination with the two resulted in a minimum xenograft glioma volume.
Figure 7.
Tumor Xenograft Studies
(A) The nude mice carrying tumors from respective groups are shown. The sample tumors from respective groups are shown. (B) Tumor growth curves are shown. Tumor volume was calculated every 5 days after injection, and the tumor was taken after 45 days. DGCR8(−), group treated with plasmids with the short hairpin RNA against DGCR8; DGCR8(−)NC, group treated with negative control empty vector plasmids with non-targeting sequence. Data are presented as the mean ± SD (n = 8, each group). **p < 0.01 versus DGCR8(−)NC + ZFAT-AS1(+)NC group; #p < 0.05 versus DGCR8(−) group; &p < 0.05 versus ZFAT-AS1(+) group by one-way ANOVA. (C) Survival curves from representative nude mice injected into the right striatum are shown (n = 8, each group).
For survival analysis, we stereotactically implanted the above glioma cells into the right striatum of nude mice. As results in Figure 7C show, mice in the DGCR8(−), ZFAT-AS1(+), and DGCR8(−) + ZFAT-AS1(+) groups possessed longer survival time than did the DGCR8(−)NC + ZFAT-AS1(+)NC group. Furthermore, mice in the DGCR8(−) + ZFAT-AS1(+) group had the longest survival time.
Schematic Cartoon of the Mechanism of DGCR8/ZFAT-AS1 Regulation of CDX2 and RRM2B in Glioma
Collectively, these results demonstrate that DGCR8 upregulation plays an important role in glioma development and progression through promotion of glioma cell proliferation, migration, and invasion, which partly depends on ZFAT-AS1 regulation of transcription factor CDX2 expression via binding and recruiting PRC2. Additionally, upregulation of DGCR8 and RRM2B in glioma cells is partly due to CDX2 binding to their promoter region and promoting transcription.
Discussion
Recent studies have found that RBPs are significantly dysregulated and play an important role in tumor progression.31, 32, 33 RBP SRSF1 bound to circSMARCA5 regulates VEGFA mRNA splicing and angiogenesis in glioblastoma multiforme.20 It has also been found that circSMARCA5 may inhibit the migration of glioblastoma multiforme cells by tethering the RBP SRSF1.21 In the present study, DGCR8 protein levels were markedly elevated in glioma tissues and in U87 and U251 cells. DGCR8 knockdown significantly suppressed proliferation, migration, and invasion and accelerated the apoptosis of glioma cells. These findings support that DGCR8 facilitates the malignant progression of glioma cells. Similarly, DGCR8 is highly expressed in ovarian cancer tissues and SKOV3 cells, and DGCR8 knockdown suppresses the proliferation, migration, and invasion of ovarian cancer cells;34 DGCR8 expression is elevated in colorectal carcinomas tissues, and this regulates the occurrence of colorectal carcinomas;35 and DGCR8 is upregulated in epithelial skin cancer36 and pleomorphic adenomas of the salivary gland.37
Increasing evidence demonstrates that lncRNAs play an important regulatory role in the development and progression of tumors.38 We found that ZFAT-AS1 was markedly expressed at low levels in glioma tissues and in U87 and U251 cells. ZFAT-AS1 overexpression in glioma cells significantly suppressed malignant biological behavior such as proliferation, migration, and invasion, and it accelerated apoptosis, whereas the converse was observed upon ZFAT-AS1 knockdown. These observations suggested that ZFAT-AS1 might function as a tumor suppressor in gliomas. Previous reports have shown that ZFAS1 is highly expressed in glioma tissues and cells, and ZFAS1 downregulation significantly inhibits glioma cells proliferation, migration, and invasion, and it accelerates apoptosis.39,40 The expression of ZFAS1 is downregulated in breast cancer and acts as a tumor-suppressive gene;41 in contrast, ZFAS1 is upregulated in colorectal, gastric, hepatocellular, bladder, and ovarian cancers, acting as an oncogene; and ZFAS1 is dysregulated in various malignant tumors and is involved in tumor proliferation, invasion, metastasis, apoptosis, cell cycle, and drug resistance.42
Many proteins interact with the transcribed RNAs to form RNPs. RBPs, in the form of RNPs, modulate the fate of binding RNAs by regulating transcription, editing, splicing, polyadenylation, translocation, and turnover.43 A previous study found that in addition to miRNAs that are the targets of DGCR8, DGCR8 also targeted other RNAs, including mRNA, small nucleolar RNA (snoRNA), and lncRNAs.8 In the current study, it was predicted that ZFAT-AS1 harbored a binding site for DGCR8 using a bioinformatics database (starBase v2.0). Results of the RIP assay validated that DGCR8 could bind to ZFAT-AS1. Accordingly, it was reported that DGCR8 could bind to MALAT1 and several other lncRNAs, including ZFAT-AS1. Moreover, the microprocessor (Drosha-DGCR8) could cleave MALAT1 in vivo. DGCR8 binds to and controls the stability of MALAT1, and the binding of DGCR8 could indeed induce RNA cleavage.8 More importantly, the present study showed that after DGCR8 knockdown, expression of ZFAT-AS1 was increased, whereas the nascent ZFAT-AS1 was not transcribed but the half-life of ZFAT-AS1 was prolonged. Therefore, DGCR8 may affect the expression level of ZFAT-AS1 by inducing its cleavage, rather than by affecting the transcription of its nascent RNA.
Studies have found that CDX2 inhibits colon cancer cell proliferation and tumor formation.25 During the development of mice, the spontaneous loss of CDX2 heterozygosity causes the transformation of squamous epithelial cells, leading to colon tumors.44,45 In the current study, we discovered that CDX2 expression was increased in glioma tissues, U87 cells, and U251 cells, and CDX2 knockdown significantly inhibited glioma cell proliferation, migration, and invasion, and it promoted apoptosis.
Polycomb group (PcG) proteins form PRCs, which are involved in the epigenetic modification of chromatin structure, promote gene silencing; PRC1 and PRC2 are defined as the two major PRCs that play primary roles in PcG silencing. The four core subunits of PRC2 include EZH1/2, SUZ12, EED, and RbAp46/48, of which EZH2 (histone-lysine N-methyltransferase) is a catalytic subunit that catalyzes H3K27me2/3.22,46, 47, 48 H3K27me3 is an epigenetic marker of chromatin silencing.49 Studies have shown that epigenetic processes play a vital role in cancer, including DNA methylation, histone modification, genetic imprinting, chromatin remodeling, and non-coding RNA regulation.50,51 Parodi et al.52 reported that DNA methylation could interfere with gene expression from a distance through the methylation or demethylation of the regulatory regions of miRNAs; additionally, the multiplicity of miRNA targets may result in the simultaneous alteration of many biological pathways such as cell proliferation, apoptosis, migration, and differentiation. Recent studies have shown that lncRNAs can recruit PcG proteins to regulate gene expression.53,54 Notably, lncRNAs are frequently involved in regulatory processes of epigenetic modifications such as histone methylation and acetylation.55
In the present study, the RIP assay and RNA pull-down demonstrated the binding of ZFAT-AS1 and EZH2. It has been reported that CDX2 is one of the developmental transcription factor families bound by SUZ12, and SUZ12 is bound to the CDX2 promoter region. SUZ12 binding represents PRC2 binding at almost all target genes,47 and this suggests that CDX2 is a target gene of PRC2. ChIP assays revealed that H3K27me3 enrichment was only present in −500 to 0 bp of the CDX2 promoter region and was increased following ZFAT-AS1 overexpression, while CDX2 mRNA and proteins levels were significantly reduced. Reportedly, 50% of the SUZ12 binding sites in human embryonic stem cells correspond to CpG islands, and the H3K27me3 enrichment region corresponds to CpG islands in 79% of cases.47,56,57 The CpG island of CDX2 promoter region 2,500 to 0 bp was located in −319 to −213 bp based on the bioinformatics software MethPrimer 2.0, which was consistent with the results of H3K27me3 enrichment in −500 to 0 bp of the CDX2 promoter region. In addition, the ChIRP assay revealed that ZFAT-AS1 was present at the promoter region of CDX2. Collectively, these results supported that ZFAT-AS1 mediated H3K27me3 by binding and recruiting PRC2 to the CDX2 promoter region, and it inhibited CDX2 transcription, thereby inhibiting the proliferation, migration, and invasion of glioma cells and promoting apoptosis. However, the specific interaction patterns between ZFAT-AS1 and EZH2 and how ZFAT-AS1 recruits the PRC2 complex to the promoter region of CDX2 remain to be elucidated. Similar to our findings, a previous study has shown that MRCCAT1 mediates the NPR3 promoter region H3K27me3 by binding to EZH2, and in turn inhibits NPR3 transcription.58 By associating with PRC2, MALAT1 detaches EZH2 bound to the HIV-1 LTR promoter, leading to the removal of H3K27me3 and reversal of epigenetic silencing of HIV-1 transcription.59 lncRNAs from the antisense strand of p21 gene inhibit the transcription of p21 by inducing H3K27 methylation in its promoter region.60 It has also been reported that altered expression of miRNAs and methylation of their promoters were correlated in neuroblastoma.61
Typically, the copy number of RRM2B is increased in a variety of tumors.62,63 Researchers have speculated that RRM2B is a tumor initiation factor.64 In the current study, we found that RRM2B was upregulated in glioma tissues, U87 cells, and U251 cells. RRM2B significantly enhanced glioma cell proliferation, migration, and invasion, and it prevented the apoptosis of glioma cells, supporting that RRM2B played an oncogenic role in glioma development. In addition, our findings supported that DGCR8 knockdown lowered CDX2 protein levels by upregulating ZFAT-AS1, and we also concluded that DGCR8 regulated RRM2B protein expression through CDX2. Collectively, we concluded that DGCR8 affected glioma malignant biological behavior by regulating RRM2B expression.
Previous studies have reported that CDX2 transcriptionally activates the expression of glycogen synthase kinases GSK-3β and Axin2 by directly binding to the promoter of GSK-3β and an enhancer upstream of Axin2.25 Comparison of the adult mouse and human intestinal cells revealed that CDX2 binds to and activates different target genes during development.65 To elucidate the mechanism through which CDX2 regulates RRM2B expression, we used the bioinformatics software JASPAR and identified that the RRM2B promoter region possessed a potential binding site for CDX2. Results of the ChIP assay and luciferase reporter system experiments validated that CDX2 could directly bind to the RRM2B promoter region and activate its transcription. Therefore, according to these findings, we concluded that CDX2 affected the biological behavior of glioma cells by activating the transcription and expression of the target gene RRM2B.
Interestingly, based on the results of the JASPAR and ChIP analyses, the luciferase reporter system assay validated that CDX2 transcriptionally activated the expression of DGCR8 by binding to its promoter region. These findings suggested that CDX2 could regulate the expression of upstream DGCR8, forming a positive feedback loop to regulate the biological behavior of glioma cells. Similar to our findings, Teng et al.66 reported that RUNX1 elevates the promoter activity of HCP5, and Su et al.67 reported that SOX3 could transcriptionally activate SOX2OT expression by binding to its promoter region, forming a positive feedback loop to affect the malignant biological behavior of glioma cells.
Finally, our in vivo study showed that DGCR8 knockdown and ZFAT-AS1 overexpression, as well a combination of the two, significantly reduced the xenograft glioma volumes and were associated with longer survival. However, compared to the DGCR8 knockdown and ZFAT-AS1 overexpression groups, a combination of the two resulted in minimal xenograft glioma volume and had the longest survival time. These findings implied that DGCR8 knockdown combined with ZFAT-AS1 overexpression possessed a potential clinical value.
In conclusion, in the present study, we found that DGCR8 and CDX2 were highly expressed and that ZFAT-AS1 was markedly downregulated in glioma tissues and cells. Highly expressed DGCR8 decreased ZFAT-AS1 stability to downregulate its expression, and then reduced the modification of H3K27me3 in the CDX2 promoter region mediated by ZFAT-AS1, and it relieved the transcriptional inhibition on CDX2, resulting in the increase of CDX2 expression. CDX2 transcriptionally activated the expression of RRM2B and DGCR8, forming a positive feedback regulatory loop to collectively regulate the malignant biological behavior of glioma cells. This study revealed a new molecular mechanism underlying the DGCR8/ZFAT-AS1/CDX2/RRM2B feedback loop regulating glioma cell malignant biological behavior, and it also provided new insights into the molecular exploration of glioma etiology.
Materials and Methods
Human Brain Tissues and Glioma Specimens
Normal brain parenchyma deriving from regions adjacent to the glioma (Adjacent) and glioma specimens were collected from Shengjing Hospital of China Medical University. All patients voluntarily signed informed consent, and this study received approval from the Ethics Committee of Shengjing Hospital of China Medical University. Adjacent (n = 12) specimens acquired from patients with glioma were used as negative controls. All samples being resected were rapidly frozen in liquid nitrogen. According to the 2007 World Health Organization (WHO) classification, the pathological grades of glioma tissue specimens were classified into four grades: WHO I and II (n = 12) and WHO III and IV (n = 12).
Cell Culture
The HEK293T and human glioma cell lines (U87 and U251) were purchased from the Shanghai Institutes for Biological Sciences Cell Resource Center. DMEM (HyClone, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, USA) was employed to maintain HEK293T, U87, and U251 cells. HA (ZY-1028) cells were purchased from Shanghai Zeye Biotechnology and cultured in complete growth medium: RPMI 1640 + 10% FBS + 1% penicillin-streptomycin solution. All cells were cultured in a humidified Calorstat at 37°C, 5% CO2.
Quantitative Real-Time PCR
Quantitative real-time PCR was carried as previously described.68 A One-Step SYBR PrimeScript RT-PCR kit (Takara Bio, Japan) was used to evaluate the expression of ZFAT-AS1 and mRNA of DGCR8, CDX2, and RRM2B with a 7500 Fast real-time PCR system (Applied Biosystems, USA). The primers are provided in Table S1. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous control. The relative quantification of above was calculated to the 2−ΔΔCt value.
FISH
In brief, HA, U87, and U251 cells were fixed on slides in 4% formaldehyde (Sigma) for 15 min and then washed with PBS (containing 1% diethyl pyrocarbonate [DEPC], Dingguo, China) three times. After digestion with PCR-grade proteinase-K (Roche Diagnostics, Mannheim, Germany), the hybridization mix was prepared with ZFAT-AS1 probe (green-labeled, Servicebio, Wuhan, China) (5′-CAACATCTTGCTGAAGTGTCCA-3′) in hybridization solution. Then, the slides were washed with washing buffer (containing 1% DEPC) and the sections were stained with anti-digoxin rhodamine conjugate (1:100, Exon Biotech, Guangzhou, China) at 37°C for 1 h away from light. The sections were stained with DAPI (Beyotime Institute of Biotechnology, Jiangsu, China) for nuclear staining subsequently. All fluorescence images were captured using an Olympus BX51 fluorescence microscope (Olympus, Tokyo, Japan).
Microarray Analysis of Human lncRNA Expression Profile
For the expression profile of lncRNA, analysis was performed by Kangchen Biotech (Shanghai, P.R. China) using an Agilent chip platform. Briefly, 60-mer oligonucleotide probes and chip (ID no. G4112F-014850) designed by Agilent Technologies were used. Total RNA was extracted from U87 and U251 cells of stable transfected DGCR8(−)NC and DGCR8(−), which was reverse transcribed to cDNA. It was Cy3-labeled after cRNA being transcribed from obtained cDNA in vitro, and the labeled cRNA was purified. Each slide was hybridized with Cy3-labeled cRNA in a hybridization oven. After hybridization, the slides were washed in staining dishes. The slides were scanned using an Agilent microarray scanner. The data were extracted using Feature Extraction software 10.7 (Agilent Technologies). Lastly, the differently expressed genes were obtained from samples in three groups after data analysis.
Western Blot
Western blot was carried as previously described.68 Lastly, integrated density values (IDVs) were analyzed with Gel-Pro professional image analysis software (Media Cybernetics, USA). The primary antibodies are as follows: DGCR8 (Cat# 60084-1-Ig, RRID: AB_2090985, Proteintech Group), CDX2 (Cat# sc-166830, RRID: AB_2260278, Santa Cruz Biotechnology), RRM2B (Cat# 18005-1-AP, Proteintech Group), EZH2 (ab191250, Abcam, UK), and GAPDH (Cat# 60004-1-Ig, RRID: AB_2107436, Proteintech Group).
Cell Transfections
The short hairpin RNAs against DGCR8 (DGCR8(−); sequence: site #1, 5′-GGAGTATGCAGTGCTCGATGA-3′; site #2, 5′-GGTCCAGGCCATACTTCTTGG-3′; site #3, 5′-GCATCCTGCACGAGTACATGC-3′; site #4, 5′-GCGGGTGGTGTAAGAACAAGA-3′), CDX2 (CDX2(−); sequence: site #1, 5′-GGACAAGGACGTGAGCATGTA-3′; site #2, 5′-GACGAAAGACAAATATCGAGT-3′; site #3, 5′-GAAGAAGTTGCAGCAGCAACA-3′), or RRM2B (RRM2B(−); site #1, 5′-GGTTTGTCATCTTTCCAATCC-3′; site #2, 5′-GCAGATGAGAAGTACTTCATC-3′; site #3, 5′-GCTGCTATATTCTGGCTAAAG-3′; site #4, 5′-GCTTGCCTGATGTTCCAATAC-3′) were introduced in pGPU6-GFP-Neo vectors by GenePharma (Shanghai, China) and their respective non-targeting sequence (negative control [NC], DGCR8(−)NC, CDX2(−)NC, or RRM2B(−)NC) empty vectors plasmids were also constructed. ZFAT-AS1-targeted short hairpin RNA (ZFAT-AS1(−)), and its respective non-targeting sequence (ZFAT-AS1(−)NC) empty vectors plasmids were constructed by GeneChem (Shanghai, China) with U6-MCS-IRES-puromycin vectors. The plamids with CDX2 full-length sequence (CDX2(+)), RRM2B full-length sequence (RRM2B(+)), and their respective empty vector plasmids without sequence (CDX2(+)NC or RRM2B(+)NC) were constructed by GeneChem with CMV-MCS-IRES-EGFP-SV40-neomycin vectors and CMV-MCS-EGFP-SV40-neomycin vectors. The plasmids with ZFAT-AS1 full-length sequence (ZFAT-AS1(+)) and its corresponding empty vector plasmids without sequence (ZFAT-AS1(+)NC) were constructed by GenScript (Piscataway, NJ, USA) with pcDNA3.1 vectors. U87 and U251 cells were seeded into 24-well plates (Corning, NJ, USA) and transfected with the above plasmids when cell confluence reached 70%–80% using Lipofectamine 3000 reagent (Life Technologies, Carlsbad, CA, USA) following the manufacturer’s protocols. The applicable stable transfected cells were screened by neomycin or puromycin. (The stable transfection was carried out after the site with the highest knockdown efficiency selected by quantitative real-time PCR after 48 h of transient transfection was carried out with the above plasmids using Lipofectamine 3000 reagent following the manufacturer’s protocols). The efficiency of transient or stable transfection was detected by quantitative real-time PCR or western blotting (Figure S2). Also, the stable DGCR8 knockdown cells were transfected with ZFAT-AS1(+), ZFAT-AS1(−), or their corresponding negative control plasmids, respectively, and the stable ZFAT-AS1 overexpressed cells were transfected with CDX2(+) or its corresponding negative control plasmids, respectively.
Cell Viability Assay
Glioma cell viability was evaluated by the Cell Counting Kit-8 (CCK-8) assay (Beyotime Institute of Biotechnology, Jiangsu, China). 100 μL of cell suspensions (2 × 104 cells/mL) was seeded in 96-well plates and incubated for 48 h at 37°C. Then, 10 μL of CCK-8 solution was added into each well and the cells were incubated for 2 h at 37°C. Next, the absorbance was gauged at 450 nm through the SpectraMax M5 microplate reader (Molecular Devices, USA).
Evaluation of Apoptosis by Flow Cytometry
Cell apoptosis was assessed by an ApoScreen annexin V apoptosis kit-phycoerythrin (PE) (SouthernBiotech, USA). Cells were washed with PBS, centrifuged twice, and then resuspended with cold 1× annexin V binding buffer and harvested cells (1 × 106 cells/mL). Based on the manufacturer’s instructions, cells were added to 10 μL of conjugated annexin V-PE and gently eddied and protected from light and incubated at 2°C–8°C for 15 min. Subsequently, 380 μL of cold 1× annexin V binding buffer was added, followed by the addition 10 μL of 7-aminoactinomycin (7-AAD). Lastly, the cells were analyzed with FACScan flow cytometry (BD Biosciences, USA) to determine the apoptotic fractions.
Cell Migration and Invasion Assay (Transwell Assay)
A cell migration and invasion assay was performed as previously described.67 Cell numbers of three randomly chosen fields were counted and imaged under a microscope.
Nascent RNA Capture
Following the manufacture’s protocols, nascent RNAs was detected by Click-iT nascent RNA capture kit (Thermo Fisher Scientific, USA). Then, 5-ethymyl uridine (EU) was clicked into nascent RNAs, after which EU-nascent RNAs were captured by streptavidin magnetic beads for the succeeding quantitative real-time PCR.
RNA Stability Evaluation
De novo synthesis of RNA was inhibited by adding actinomycin D (ActD; NobleRyder, China) to cell culture medium. Total RNA was isolated at different time points, respectively, and tested by quantitative real-time PCR. Compared with zero time, the half-life of RNA is determined by its level decreasing to 50% at a certain time point.
RIP Assay
Based on the manufacturer’s protocols, a RIP assay was carried using an EZ-Magna RNA-binding protein immunoprecipitation kit (Millipore, USA) in this study. Lysates of U87 cells were incubated with compounds of RIP buffer, magnetic beads, and human anti-DGCR8 (Proteintech Group, Cat# 60084-1-Ig, RRID: AB_2090985) (or anti-EZH2 [Abcam, UK]), negative control normal mouse IgG (Millipore, USA), and positive control anti-SNRNP70 (Millipore, USA). Next, the above compounds were incubated with proteinase K. Then, immunoprecipitated RNA was purified, and the RNA concentration and quality were assessed by a NanoDrop spectrophotometer (Thermo Scientific). Lastly. the purified RNA was subjected to quantitative real-time PCR analysis using specific primers to illustrate the presence of the binding targets. GAPDH was used as the negative RNA control.
RNA Pull-Down
A Pierce magnetic RNA-protein pull-down kit (Thermo Fisher Scientific, USA) was used to detect the interaction between ZFAT-AS1 and EZH2 protein according to the manufacturer’s instructions. ZFAT transcripts, ZFAT-AS1, and negative RNA control (poly(A)25 RNA) were labeled using desthiobiotinylated cytidine bisphosphate and T4 RNA ligase according to the Pierce RNA 3′-End Desthiobiotinylation Kit procedure. The biotin-labeled negative RNA control (poly(A)25 RNA), ZFAT transcripts, and ZFAT-AS1 were captured using streptavidin magnetic beads in RNA capture buffer for 30 min at room temperature. Beads were washed twice in 20 mM Tris (pH 7.5), once in protein-RNA binding buffer, and U87 extract was added. Samples were incubated for 45 min at 4°C, washed three times with wash buffer, and eluted after 15 min of incubation at 37°C with biotin elution buffer. RNA pull-down specificity was assessed by western blotting with samples normalized by volume, and the input was U87 lysates.
Reporter Vector Constructs and Luciferase Reporter Assay
The reporter vector construction was carried out as previously described.68 HEK293T cells were co-transfected with constructed reporter vectors and GV146-CDX2(+) (or GV146 empty vector) using Lipofectamine 3000. The plasmid of GV146-CDX2(+) (GeneChem, Shanghai, China) contained human CDX2 full-length sequence. The relative luciferase activities were detected after co-transfection 48 h using a single luciferase reporter gene assay kit (YPH, Beijing, China) following the manufacturer’s protocols.
ChIP Assay
ChIP assays were conducted as previously described67 using a SimpleChIP enzymatic ChIP kit (Cell Signaling Technology, USA). The antibodies used in assays were as follows: anti-CDX2 antibody (Cat# sc-166830, RRID: AB_2260278, Santa Cruz Biotechnology), anti-H3K27me3 antibody (Cat# ab6002, RRID: AB_305237, Abcam), and normal rabbit IgG. Immunoprecipitated DNA from anti-H3K27me3 (or anti-CDX2) and anti-IgG was amplified by PCR with primers. The primers for each PCR set, annealing temperatures, and the sizes of PCR products are provided in Tables S2 and S4. For each PCR, the corresponding input was taken in parallel for PCR validation. Then, immunoprecipitated DNA from anti-H3K27me3 was quantitated by quantitative real-time PCR with SYBR Premix Ex TaqII (Tli RNase H Plus) (Takara Bio), and the primers are provided in Table S3. The results are displayed as the percentage of immunoprecipitated over input values (% input).
ChIRP
ChIRP (n = 3) was performed as described,69 using antisense DNA oligonucleotide probes specific for full-length ZFAT-AS1 (designed on https://www.singlemoleculefish.com and listed in Table S5). A set of probes against LacZ RNA was also generated as the mock control.69 All probes were biotinylated at the 3′ end (RiboBio, Guangzhou, China). Eluted RNA was subjected to quantitative reverse-transcription PCR for the detection of enriched transcripts. DNA fragments were recovered and analyzed by qPCR. Data were normalized to the corresponding DNA input control and are represented as percentage of input. For PCR analysis, cDNA tiling oligonucleotide probes against ZFAT-AS1 or the CDX2 promoter region (Table S5) were used.
Xenograft Mouse Model In Vivo
Athymic nude mice (BALB/c) were 4 weeks old and purchased from the Cancer Institute of the China Academy of Medical Science for the in vivo study. All experiments were performed strictly following the Animal Welfare Act and approved by the Ethics Committee of China Medical University. The stable transfected and expressing cells (DGCR8(−)NC + ZFAT-AS1(+)NC, DGCR8(−), ZFAT-AS1(+), and DGCR8(−) + ZFAT-AS1(+)) were selected. For subcutaneous xenografts, 3 × 105 cells were subcutaneously injected into the right flank area of each nude mouse. The volumes of tumor were gauged every 5 days until 45 days and calculated according to the estimate formula: volume (mm3) = length × width2/2. At 45 days after injection, the mice were sacrificed and then tumors were dissected and separated. For survival analysis, nude mice were stereotactically implanted at a 3 × 105 glioma cells into the right striatum. The number of surviving nude mice was recorded, and Kaplan-Meier survival curve was applied for the survival analysis.
Statistical Analysis
All experimental data were manifested as mean ± SD. GraphPad Prism v5.01 (GraphPad, CA, USA) was applied for statistical analysis. Group comparisons were made with a Student’s t test or one-way ANOVA. Differences were considered statistically significant when p < 0.05.
Author Contributions
Y.X. and Y.L. designed the study. F.Z., X.R., J.M., X.L., J.Z., L.L., and S.S. performed the experiments and acquired the data. F.Z., L.S., D.W., C.Y., H.C, Z.L., and Z.F. analyzed the data. Y.X. and F.Z. contributed to discussion and writing of the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work is supported by grants from the Natural Science Foundation of China (81872503 and 81872073), Liaoning Science and Technology Plan Project (2017225020 and 2015225007), Project of Key Laboratory of Neuro-oncology in Liaoning Province (112-2400017005), and by a special developmental project guided by the Central Government of Liaoning Province (2017011553-301). We would like to thank Editage for English language editing.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.11.015.
Supplemental Information
References
- 1.Furnari F.B., Fenton T., Bachoo R.M., Mukasa A., Stommel J.M., Stegh A., Hahn W.C., Ligon K.L., Louis D.N., Brennan C. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 2.Davis F.G., Freels S., Grutsch J., Barlas S., Brem S. Survival rates in patients with primary malignant brain tumors stratified by patient age and tumor histological type: an analysis based on Surveillance, Epidemiology, and End Results (SEER) data, 1973–1991. J. Neurosurg. 1998;88:1–10. doi: 10.3171/jns.1998.88.1.0001. [DOI] [PubMed] [Google Scholar]
- 3.Penas-Prado M., Gilbert M.R. Molecularly targeted therapies for malignant gliomas: advances and challenges. Expert Rev. Anticancer Ther. 2007;7:641–661. doi: 10.1586/14737140.7.5.641. [DOI] [PubMed] [Google Scholar]
- 4.Kalpathy-Cramer J., Gerstner E.R., Emblem K.E., Andronesi O., Rosen B. Advanced magnetic resonance imaging of the physical processes in human glioblastoma. Cancer Res. 2014;74:4622–4637. doi: 10.1158/0008-5472.CAN-14-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van den Bent M., Chinot O.L., Cairncross J.G. Recent developments in the molecular characterization and treatment of oligodendroglial tumors. Neuro-oncol. 2003;5:128–138. doi: 10.1215/S1522-8517-02-00028-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerstberger S., Hafner M., Tuschl T. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014;15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh G., Pratt G., Yeo G.W., Moore M.J. The clothes make the mRNA: past and present trends in mRNP fashion. Annu. Rev. Biochem. 2015;84:325–354. doi: 10.1146/annurev-biochem-080111-092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macias S., Plass M., Stajuda A., Michlewski G., Eyras E., Cáceres J.F. DGCR8 HITS-CLIP reveals novel functions for the microprocessor. Nat. Struct. Mol. Biol. 2012;19:760–766. doi: 10.1038/nsmb.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sand M., Skrygan M., Georgas D., Arenz C., Gambichler T., Sand D., Altmeyer P., Bechara F.G. Expression levels of the microRNA maturing microprocessor complex component DGCR8 and the RNA-induced silencing complex (RISC) components argonaute-1, argonaute-2, PACT, TARBP1, and TARBP2 in epithelial skin cancer. Mol. Carcinog. 2012;51:916–922. doi: 10.1002/mc.20861. [DOI] [PubMed] [Google Scholar]
- 10.Fardmanesh H., Shekari M., Movafagh A., Alizadeh Shargh S., Poursadegh Zonouzi A.A., Shakerizadeh S., Poursadegh Zonouzi A., Hosseinzadeh A. Upregulation of the double-stranded RNA binding protein DGCR8 in invasive ductal breast carcinoma. Gene. 2016;581:146–151. doi: 10.1016/j.gene.2016.01.033. [DOI] [PubMed] [Google Scholar]
- 11.Meseure D., Vacher S., Lallemand F., Alsibai K.D., Hatem R., Chemlali W., Nicolas A., De Koning L., Pasmant E., Callens C. Prognostic value of a newly identified MALAT1 alternatively spliced transcript in breast cancer. Br. J. Cancer. 2016;114:1395–1404. doi: 10.1038/bjc.2016.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai M.C., Spitale R.C., Chang H.Y. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang G., Lu X., Yuan L. lncRNA: a link between RNA and cancer. Biochim. Biophys. Acta. 2014;1839:1097–1109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 16.Wang P., Ren Z., Sun P. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J. Cell. Biochem. 2012;113:1868–1874. doi: 10.1002/jcb.24055. [DOI] [PubMed] [Google Scholar]
- 17.Lv Q.L., Hu L., Chen S.H., Sun B., Fu M.L., Qin C.Z., Qu Q., Wang G.H., He C.J., Zhou H.H. A long noncoding RNA ZEB1-AS1 promotes tumorigenesis and predicts poor prognosis in glioma. Int. J. Mol. Sci. 2016;17:E1431. doi: 10.3390/ijms17091431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin X., Yao J., Geng P., Fu X., Xue J., Zhang Z. lncRNA TSLC1-AS1 is a novel tumor suppressor in glioma. Int. J. Clin. Exp. Pathol. 2014;7:3065–3072. [PMC free article] [PubMed] [Google Scholar]
- 19.Metsalu T., Viltrop T., Tiirats A., Rajashekar B., Reimann E., Kõks S., Rull K., Milani L., Acharya G., Basnet P. Using RNA sequencing for identifying gene imprinting and random monoallelic expression in human placenta. Epigenetics. 2014;9:1397–1409. doi: 10.4161/15592294.2014.970052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barbagallo D., Caponnetto A., Brex D., Mirabella F., Barbagallo C., Lauretta G., Morrone A., Certo F., Broggi G., Caltabiano R. circSMARCA5 regulates VEGFA mRNA splicing and angiogenesis in glioblastoma multiforme through the binding of SRSF1. Cancers (Basel) 2019;11:E194. doi: 10.3390/cancers11020194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barbagallo D., Caponnetto A., Cirnigliaro M., Brex D., Barbagallo C., D’Angeli F., Morrone A., Caltabiano R., Barbagallo G.M., Ragusa M. circSMARCA5 inhibits migration of glioblastoma multiforme cells by regulating a molecular axis involving splicing factors SRSF1/SRSF3/PTB. Int. J. Mol. Sci. 2018;19:E480. doi: 10.3390/ijms19020480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simon J.A., Kingston R.E. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 23.Song Y., Wang R., Li L.W., Liu X., Wang Y.F., Wang Q.X., Zhang Q. Long non-coding RNA HOTAIR mediates the switching of histone H3 lysine 27 acetylation to methylation to promote epithelial-to-mesenchymal transition in gastric cancer. Int. J. Oncol. 2019;54:77–86. doi: 10.3892/ijo.2018.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beck F., Stringer E.J. The role of Cdx genes in the gut and in axial development. Biochem. Soc. Trans. 2010;38:353–357. doi: 10.1042/BST0380353. [DOI] [PubMed] [Google Scholar]
- 25.Yu J., Liu D., Sun X., Yang K., Yao J., Cheng C., Wang C., Zheng J. CDX2 inhibits the proliferation and tumor formation of colon cancer cells by suppressing Wnt/β-catenin signaling via transactivation of GSK-3β and Axin2 expression. Cell Death Dis. 2019;10:26. doi: 10.1038/s41419-018-1263-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park D.Y., Srivastava A., Kim G.H., Mino-Kenudson M., Deshpande V., Zukerberg L.R., Song G.A., Lauwers G.Y. CDX2 expression in the intestinal-type gastric epithelial neoplasia: frequency and significance. Mod. Pathol. 2010;23:54–61. doi: 10.1038/modpathol.2009.135. [DOI] [PubMed] [Google Scholar]
- 27.Uramoto H., Sugio K., Oyama T., Hanagiri T., Yasumoto K. P53R2, p53 inducible ribonucleotide reductase gene, correlated with tumor progression of non-small cell lung cancer. Anticancer Res. 2006;26(2A):983–988. [PubMed] [Google Scholar]
- 28.Okumura H., Natsugoe S., Yokomakura N., Kita Y., Matsumoto M., Uchikado Y., Setoyama T., Owaki T., Ishigami S., Aikou T. Expression of p53R2 is related to prognosis in patients with esophageal squamous cell carcinoma. Clin. Cancer. Res. 2006;12:3740–3745. doi: 10.1158/1078-0432.CCR-05-2416. [DOI] [PubMed] [Google Scholar]
- 29.Bo H., Weiyang C., Zhang Y. p53R2 is oncogenic in human cervical cancer. Cell Cycle. 2017;16:1323–1324. doi: 10.1080/15384101.2017.1337978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang C., Xu R., Li X.X., Wang Y.Y., Liang W.Q., Zeng J.D., Zhang S.S., Xu X.Y., Yang Y., Zhang M.Y. p53R2 overexpression in cervical cancer promotes AKT signaling and EMT, and is correlated with tumor progression, metastasis and poor prognosis. Cell Cycle. 2017;16:1673–1682. doi: 10.1080/15384101.2017.1320629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z.L., Li B., Luo Y.X., Lin Q., Liu S.R., Zhang X.Q., Zhou H., Yang J.H., Qu L.H. Comprehensive genomic characterization of RNA-binding proteins across human cancers. Cell Rep. 2018;22:286–298. doi: 10.1016/j.celrep.2017.12.035. [DOI] [PubMed] [Google Scholar]
- 32.Lukong K.E., Chang K.W., Khandjian E.W., Richard S. RNA-binding proteins in human genetic disease. Trends Genet. 2008;24:416–425. doi: 10.1016/j.tig.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 33.Castello A., Fischer B., Hentze M.W., Preiss T. RNA-binding proteins in Mendelian disease. Trends Genet. 2013;29:318–327. doi: 10.1016/j.tig.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 34.Guo Y., Tian P., Yang C., Liang Z., Li M., Sims M., Lu L., Zhang Z., Li H., Pfeffer L.M., Yue J. Silencing the double-stranded RNA binding protein DGCR8 inhibits ovarian cancer cell proliferation, migration, and invasion. Pharm. Res. 2015;32:769–778. doi: 10.1007/s11095-013-1219-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim B., Lee J.H., Park J.W., Kwon T.K., Baek S.K., Hwang I., Kim S. An essential microRNA maturing microprocessor complex component DGCR8 is up-regulated in colorectal carcinomas. Clin. Exp. Med. 2014;14:331–336. doi: 10.1007/s10238-013-0243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jafari N., Dogaheh H.P., Bohlooli S., Oyong G.G., Shirzad Z., Alibeiki F., Asl S.H., Zargar S.J. Expression levels of microRNA machinery components Drosha, Dicer and DGCR8 in human (AGS, HepG2, and KEYSE-30) cancer cell lines. Int. J. Clin. Exp. Med. 2013;6:269–274. [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X., Cairns M., Rose B., O’Brien C., Shannon K., Clark J., Gamble J., Tran N. Alterations in miRNA processing and expression in pleomorphic adenomas of the salivary gland. Int. J. Cancer. 2009;124:2855–2863. doi: 10.1002/ijc.24298. [DOI] [PubMed] [Google Scholar]
- 38.Li J., Li Z., Leng K., Xu Y., Ji D., Huang L., Cui Y., Jiang X. ZEB1-AS1: a crucial cancer-related long non-coding RNA. Cell Prolif. 2018;51:e12423. doi: 10.1111/cpr.12423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lv Q.L., Chen S.H., Zhang X., Sun B., Hu L., Qu Q., Huang Y.T., Wang G.H., Liu Y.L., Zhang Y.Y., Zhou H.H. Upregulation of long noncoding RNA zinc finger antisense 1 enhances epithelial-mesenchymal transition in vitro and predicts poor prognosis in glioma. Tumour Biol. 2017;39 doi: 10.1177/1010428317695022. 1010428317695022. [DOI] [PubMed] [Google Scholar]
- 40.Gao K., Ji Z., She K., Yang Q., Shao L. Long non-coding RNA ZFAS1 is an unfavourable prognostic factor and promotes glioma cell progression by activation of the Notch signaling pathway. Biomed. Pharmacother. 2017;87:555–560. doi: 10.1016/j.biopha.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 41.Askarian-Amiri M.E., Crawford J., French J.D., Smart C.E., Smith M.A., Clark M.B., Ru K., Mercer T.R., Thompson E.R., Lakhani S.R. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA. 2011;17:878–891. doi: 10.1261/rna.2528811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang X., Yang Z., Li Z. Zinc finger antisense 1: a long noncoding RNA with complex roles in human cancers. Gene. 2019;688:26–33. doi: 10.1016/j.gene.2018.11.075. [DOI] [PubMed] [Google Scholar]
- 43.Müller-McNicoll M., Neugebauer K.M. How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat. Rev. Genet. 2013;14:275–287. doi: 10.1038/nrg3434. [DOI] [PubMed] [Google Scholar]
- 44.Chawengsaksophak K., James R., Hammond V.E., Köntgen F., Beck F. Homeosis and intestinal tumours in Cdx2 mutant mice. Nature. 1997;386:84–87. doi: 10.1038/386084a0. [DOI] [PubMed] [Google Scholar]
- 45.Tamai Y., Nakajima R., Ishikawa T., Takaku K., Seldin M.F., Taketo M.M. Colonic hamartoma development by anomalous duplication in Cdx2 knockout mice. Cancer Res. 1999;59:2965–2970. [PubMed] [Google Scholar]
- 46.Schuettengruber B., Cavalli G. Recruitment of Polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136:3531–3542. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- 47.Lee T.I., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 49.Hawkins P.G., Morris K.V. RNA and transcriptional modulation of gene expression. Cell Cycle. 2008;7:602–607. doi: 10.4161/cc.7.5.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taby R., Issa J.P. Cancer epigenetics. CA Cancer J. Clin. 2010;60:376–392. doi: 10.3322/caac.20085. [DOI] [PubMed] [Google Scholar]
- 51.Sharma S., Kelly T.K., Jones P.A. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parodi F., Carosio R., Ragusa M., Di Pietro C., Maugeri M., Barbagallo D., Sallustio F., Allemanni G., Pistillo M.P., Casciano I. Epigenetic dysregulation in neuroblastoma: a tale of miRNAs and DNA methylation. Biochim. Biophys. Acta. 2016;1859:1502–1514. doi: 10.1016/j.bbagrm.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 53.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.C., Hung T., Argani P., Rinn J.L. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Su W., Xu M., Chen X., Chen N., Gong J., Nie L., Li L., Li X., Zhang M., Zhou Q. Long noncoding RNA ZEB1-AS1 epigenetically regulates the expressions of ZEB1 and downstream molecules in prostate cancer. Mol. Cancer. 2017;16:142. doi: 10.1186/s12943-017-0711-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mohn F., Weber M., Rebhan M., Roloff T.C., Richter J., Stadler M.B., Bibel M., Schübeler D. Lineage-specific Polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 58.Li J.K., Chen C., Liu J.Y., Shi J.Z., Liu S.P., Liu B., Wu D.S., Fang Z.Y., Bao Y., Jiang M.M. Long noncoding RNA MRCCAT1 promotes metastasis of clear cell renal cell carcinoma via inhibiting NPR3 and activating p38-MAPK signaling. Mol. Cancer. 2017;16:111. doi: 10.1186/s12943-017-0681-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qu D., Sun W.W., Li L., Ma L., Sun L., Jin X., Li T., Hou W., Wang J.H. Long noncoding RNA MALAT1 releases epigenetic silencing of HIV-1 replication by displacing the polycomb repressive complex 2 from binding to the LTR promoter. Nucleic Acids Res. 2019;47:3013–3027. doi: 10.1093/nar/gkz117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morris K.V., Santoso S., Turner A.M., Pastori C., Hawkins P.G. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maugeri M., Barbagallo D., Barbagallo C., Banelli B., Di Mauro S., Purrello F., Magro G., Ragusa M., Di Pietro C., Romani M., Purrello M. Altered expression of miRNAs and methylation of their promoters are correlated in neuroblastoma. Oncotarget. 2016;7:83330–83341. doi: 10.18632/oncotarget.13090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang X., Zhenchuk A., Wiman K.G., Albertioni F. Regulation of p53R2 and its role as potential target for cancer therapy. Cancer Lett. 2009;276:1–7. doi: 10.1016/j.canlet.2008.07.019. [DOI] [PubMed] [Google Scholar]
- 63.Pontarin G., Ferraro P., Bee L., Reichard P., Bianchi V. Mammalian ribonucleotide reductase subunit p53R2 is required for mitochondrial DNA replication and DNA repair in quiescent cells. Proc. Natl. Acad. Sci. USA. 2012;109:13302–13307. doi: 10.1073/pnas.1211289109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aye Y., Li M., Long M.J., Weiss R.S. Ribonucleotide reductase and cancer: biological mechanisms and targeted therapies. Oncogene. 2015;34:2011–2021. doi: 10.1038/onc.2014.155. [DOI] [PubMed] [Google Scholar]
- 65.Kumar N., Tsai Y.H., Chen L., Zhou A., Banerjee K.K., Saxena M., Huang S., Toke N.H., Xing J., Shivdasani R.A. The lineage-specific transcription factor CDX2 navigates dynamic chromatin to control distinct stages of intestine development. Development. 2019;146 doi: 10.1242/dev.172189. dev172189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Teng H., Wang. P., Xue Y., Liu X., Ma J., Cai H., Xi Z., Li Z., Liu Y. Role of HCP5-miR-139-RUNX1 feedback loop in regulating malignant behavior of glioma cells. Mol. Ther. 2016;24:1806–1822. doi: 10.1038/mt.2016.103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Su R., Cao S., Ma J., Liu Y., Liu X., Zheng J., Chen J., Liu L., Cai H., Li Z. Knockdown of SOX2OT inhibits the malignant biological behaviors of glioblastoma stem cells via up-regulating the expression of miR-194-5p and miR-122. Mol. Cancer. 2017;16:171. doi: 10.1186/s12943-017-0737-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shen S., Yu H., Liu X., Liu Y., Zheng J., Wang P., Gong W., Chen J., Zhao L., Xue Y. PIWIL1/piRNA-DQ593109 regulates the permeability of the blood-tumor barrier via the MEG3/miR-330-5p/RUNX3 axis. Mol. Ther. Nucleic Acids. 2018;10:412–425. doi: 10.1016/j.omtn.2017.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chu C., Qu K., Zhong F.L., Artandi S.E., Chang H.Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.