Summary
Regulatory T cells (Tregs) subdue immune responses. Central to Treg activation are changes in lipid metabolism that support their survival and function. Fatty acid binding proteins (FABPs) are a family of lipid chaperones required to facilitate uptake and intracellular lipid trafficking. One family member, FABP5, is expressed in T cells, but its function remains unclear. We show that in Tregs, genetic or pharmacologic inhibition of FABP5 function causes mitochondrial changes underscored by decreased OXPHOS, impaired lipid metabolism, and loss of cristae structure. FABP5 inhibition in Tregs triggers mtDNA release and consequent cGAS-STING-dependent type I IFN signaling, which induces heightened production of the regulatory cytokine IL-10 and promotes Treg suppressive activity. We find evidence of this pathway, along with correlative mitochondrial changes in tumor infiltrating Tregs, which may underlie enhanced immunosuppression in the tumor microenvironment. Together, our data reveal that FABP5 is a gatekeeper of mitochondrial integrity that modulates Treg function.
Keywords: Treg, FABP5, lipids, tumor, suppression, mtDNA, cGAS-STING, type I IFN, IL-10, immunometabolism
Graphical Abstract

Highlights
-
•
FABP5 inhibition in Tregs alters mitochondria and enhances suppression
-
•
Disrupting FABP5 in Tregs results in mtDNA release and type I IFN signaling
-
•
cGAS/-STING-dependent type I IFN signals promote Treg IL-10 production
-
•
Tumor Tregs exhibit mitochondrial alterations and a type I IFN gene signature
Field et al. show that fatty acid binding protein 5 (FABP5) maintains mitochondrial integrity in regulatory T cells (Tregs). FABP5 inhibition results in mtDNA release, which triggers expression of IL-10 and promotes Treg suppressive capacity. These findings may have implications for therapeutically targeting Tregs in autoimmunity and cancer.
Context and Significance
The inability of immune cells to access nutrients or engage particular metabolic pathways can lead to altered functional output. Since inappropriate immune function can cause disease, there is a lot of interest in understanding how immune cell metabolic programming controls cell function. Max Planck researchers investigated the metabolism of regulatory T cells (Tregs), cells that prevent autoimmunity but also suppress anti-cancer immunity. They found that changes to the mitochondria as a consequence of altered lipid metabolism led to greater suppressive capacity, a phenotype also evident in tumor Tregs. These findings highlight a link between mitochondria and lipid metabolism in controlling Treg function and suggest that understanding this pathway may hold promise for the future treatment of autoimmune diseases or cancer.
Introduction
Tregs are responsible for modulating the immune system, maintaining tolerance to self-antigens, and preventing autoimmune disease (Sakaguchi, 2004). Tregs are defined by the lineage-specifying transcription factor Foxp3, and its expression is essential for their differentiation and suppressor function (Fontenot et al., 2003). Through a variety of mechanisms, Tregs can inhibit effector T cell proliferation and cytokine production and thereby play a vital role in limiting immune-mediated inflammation (Sakaguchi, 2004). While the mechanisms of suppression remain to be fully defined, they include the production of soluble mediators, involving cytokines such as IL-10 and TGF-β, that dampen effector T cell proliferation (Chaudhry et al., 2011, Li et al., 2007, Rubtsov et al., 2008). While Tregs protect the host by limiting exacerbated immune responses, they can also have a negative impact on host survival in settings such as cancer, where their immunosuppressive function limits effector T cell responses against tumor cells and allows tumor growth (Chaudhary and Elkord, 2016, Wang et al., 2017).
The link between cellular metabolism and functional output is well established in the immune system, where aberrant engagement of metabolic pathways in a variety of immune cell subsets can lead to abnormal cell function and disease progression (O’Neill et al., 2016, Pearce and Pearce, 2013). Tregs have been shown to rely on lipid metabolism for survival and function (Gerriets et al., 2015, Michalek et al., 2011) and, more specifically, to acquire and utilize extracellular free fatty acids to meet their metabolic demands (Berod et al., 2014). Mechanistically, raptor-mTORC1 signals in Tregs augment lipid and cholesterol metabolism to allow cell proliferation and surface expression of important molecules mediating immune suppression such as CTLA-4 and ICOS (Zeng et al., 2013). Tregs in tumors have been shown to rely on a combination of glycolysis and fatty acid synthesis and oxidation, allowing their survival and proliferation in the hostile tumor environment (Pacella et al., 2018). Together, these data highlight the important role of lipid metabolism in Tregs; however, a complete understanding of how lipids regulate Treg development and function is lacking (Newton et al., 2016).
Within cells, lipids are bound to lipid chaperones, such as fatty acid binding proteins (FABPs) (Furuhashi and Hotamisligil, 2008, Storch and McDermott, 2009). FABPs are a family of 14-15 kDa proteins that coordinate lipid trafficking and responses within cells by reversibly binding hydrophobic ligands, such as saturated and unsaturated fatty acids (Furuhashi and Hotamisligil, 2008). FABPs play important roles in fatty acid uptake from the microenvironment, trafficking lipids to specific cellular compartments, including the nucleus, peroxisomes, endoplasmic reticulum (ER), and mitochondria (Furuhashi and Hotamisligil, 2008, Lee et al., 2018, Storch and Corsico, 2008).
FABP5 is one of the most highly expressed FABPs in T cells (Rolph et al., 2006). FABP5 inhibition has been shown to minimize IL-17 cytokine production and skew T cells toward a Treg phenotype in vitro, and consistent with this, FABP5 inhibition in vivo has been reported to attenuate EAE (Rao et al., 2015). FABP5 has also been shown to be important for tissue-resident memory T cells (Pan et al., 2017) and macrophages (Moore et al., 2015, Zhang et al., 2014), but mechanistically FABP function is not clearly understood. Given the reported importance of increased lipid metabolism, including increased FAO in Treg cell function (Michalek et al., 2011), we set out to examine whether FABP5 plays a pivotal role in these processes.
Results
FABP5 Blockade Inhibits In Vitro Treg Proliferation and Mitochondrial Metabolism
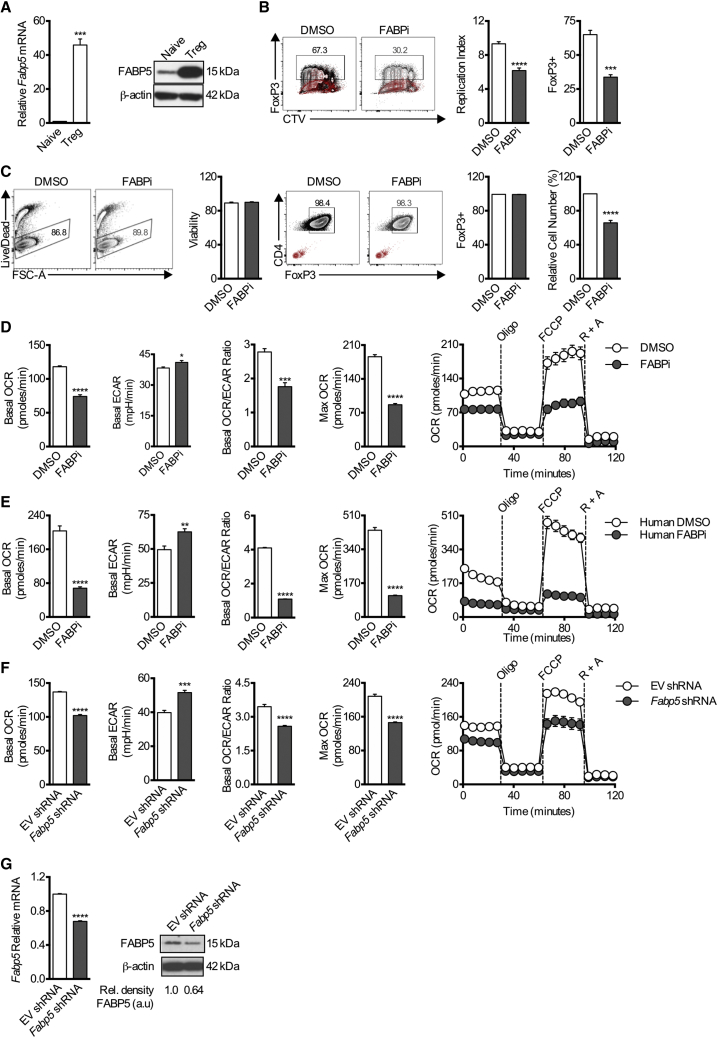
We examined FABP5 expression in Tregs generated from naive CD4+ T cells in vitro. FABP5 was more highly expressed in Tregs compared to naive CD4+ T cells (Figure 1A). The expression of Fabp3, Fabp4, and Fabp5 was also assessed in in vitro-differentiated Th1, Th2, Th17, and Tregs (Figure S1A). Fabp3 expression was comparable across all Th cell subsets, Fabp4 expression was highest in in vitro-induced Th17 cells, and Fabp5 was highest in in vitro-induced Tregs. Fabp3 and Fabp5 were more highly expressed in Th1 and Th17 cells compared to naive CD4+ T cells, and Fabp5 was most highly expressed in Th2 and Tregs compared to naive CD4+ T cells (Figure S1B). We next labeled naive CD4+ T cells with cell trace violet and cultured them under Treg polarizing conditions in the presence or absence of the FABP inhibitor BMS309403, which targets the fatty acid binding pockets of FABP3, FABP4, and FABP5 (Furuhashi et al., 2007, Sulsky et al., 2007). Both cellular proliferation and Foxp3 expression were inhibited by BMS309403, suggesting a role for FABP5 in Treg differentiation (Figure 1B). As a control, we also replicated this experiment using Th2 cells, as they also expressed Fabp5 at higher levels. No difference was evident in the induction of Gata3 in Th2 cells cultured in the presence of BMS309403 versus vehicle control; however, as in Tregs, cellular proliferation was inhibited (Figure S1C). Further, no increase in LDH in the media supernatant was observed following FABP5 inhibition, suggesting that the decreased cellularity was a consequence of impeded proliferation as opposed to cytotoxicity (Figure S1C). Because chronic administration of BMS309403 retarded Foxp3 expression and limited cellular proliferation in this in vitro-induced Treg system, we sought to assess the effects of acutely inhibiting FABP5 in fully differentiated Tregs. Using this approach, we were able to circumvent the confounding effects of chronic BMS309403 exposure on Treg differentiation and proliferation and to explore the role of FABP5 in Treg function. Here, we differentiated Tregs in vitro for 3 days before incubating the cells with BMS309403 overnight. In this setting, there was a reduction in cell number, but cell viability and Foxp3 expression were preserved (Figure 1C). We next assessed cellular bioenergetics and found that after BMS309403 treatment, Tregs exhibited decreased basal oxygen consumption rates (OCR), OCR/ECAR (extracellular acidification rate) ratio, and maximal respiratory capacity (evident after exposure to the uncoupler FCCP) (Figure 1D), indicating decreased mitochondrial activity. Accordingly, basal ECAR was increased when cells were treated with BMS309403, indicating a switch from oxidative phosphorylation to glycolysis after exposure to this inhibitor (Figure 1D). To extend these findings beyond mouse Tregs, we differentiated human Tregs in vitro before acute treatment with BMS309403. Consistent with the mouse Tregs, we also observed decreased OCR and enhanced ECAR (Figure 1E). Finally, we also tested whether the metabolic effects evident after FABP5 inhibition were reversible. When cells that had been cultured overnight with BMS309403 were washed and allowed to recover for a further 24 h in the absence of the inhibitor, the OCR and ECAR of the cells reverted to the levels measured in Tregs that had not been treated with the inhibitor. Conversely, maintaining cells in the presence of BMS309403 limited cellular bioenergetics (Figure S2A).
Figure 1.
Tregs Express FABP5 during In Vitro Differentiation, and Blockade Affects Differentiation and Metabolism
Naive CD4+ T cells were cultured for 4 days under Treg cell-differentiation conditions.
(A) Mean relative expression (±SEM) of Fabp5 mRNA in in vitro-differentiated Tregs compared to naive CD4+ T cells (n = 3). FABP5 protein expression was assessed on D4. Results represent two independent experiments.
(B) Representative flow plots and quantification of Foxp3 expression and CTV dilution (±SEM) in Tregs cultured in the presence or absence of the FABP5 inhibitor BMS309403 (n = 4). Results represent three independent experiments. Gating controls are depicted in red.
(C) Naive CD4+ T cells were cultured for 3 days under Treg differentiation conditions before overnight BMS309403 treatment, and mean viability, relative cell number, and Foxp3 expression (±SEM) were measured (n = 4). Results represent >6 independent experiments. Gating controls are depicted in red.
(D–F) Mean (±SEM) basal OCR, basal ECAR, OCR/ECAR ratio and maximal respiration (after FCCP) of in vitro-differentiated mouse Tregs (n = 5). Results represent >6 independent experiments. (D) In vitro-differentiated human Tregs (n = 6). Results represent three independent experiments (E) or in vitro-differentiated mouse Tregs expressing Fabp5 shRNA (n = 5). Results represent two independent experiments. (F) cultured in the presence or absence of BMS309403 overnight at baseline, and in response to oligomycin (Oligo), FCCP, and rotenone and antimycin A (R + A).
(G) qPCR and protein expression of FABP5 following shRNA knockdown. Results represent two independent experiments. ∗p < 0.05, ∗∗∗p < 0.005, ∗∗∗∗p < 0.001. P values were calculated using a two-tailed, unpaired t test.
In contrast to what we observed after acute pharmacologic inhibition with BMS309403, in vitro-generated Tregs from mice chronically deficient for FABP4 and FABP5 (Fabp4/5 dKO mice) had increased basal OCR, decreased basal ECAR, and enhanced maximal respiratory capacity (Figure S2B). These results suggested that either BMS309403 had off-target effects or that compensatory mechanisms allowed Tregs to develop normally in Fabp4/5 dKO mice by utilizing alternative pathways after Fabp4/5 germline deletion. Supporting the latter hypothesis, in vitro-generated Tregs from Fabp4/5 dKO mice had increased gene expression of Fabp3 compared to cells from WT mice (Figure S2C), suggesting the possibility that this protein might be upregulated to compensate for germline deletion of Fabp4/5. In contrast, WT Treg cells failed to increase Fabp3 expression in response to acute FABP inhibition (Figure S2D). Nevertheless, to circumvent possible off-target effects of BMS309403, we took a different genetic approach for acute loss of Fabp5 function in WT T cells by expressing a short hairpin RNA (shRNA) against Fabp5 to knock down FABP5 expression. Consistent with the results observed after acute pharmacologic inhibition, we observed decreased basal OCR, OCR/ECAR ratio, and maximal respiration, along with increased ECAR, in Tregs expressing Fabp5 shRNA (Figure 1F). ShRNA-mediated knockdown of Fabp5 mRNA and protein was confirmed in these cells (Figure 1G). Importantly, no significantly increased expression of Fabp3 or Fabp4 was observed following shRNA-mediated knockdown of Fabp5, (Figure S2D). Thus, acute genetic depletion of FABP5 matched the results observed after treating cells with BMS309403 overnight, suggesting that the acute loss of FABP function in both settings led to the respiratory defects observed in Tregs.
Memory CD8+ T cells (Tmem) have been shown to use oxidative metabolism fueled by glucose-derived lipids (O’Sullivan et al., 2014), and a recent report has shown the requirement for FABP4/5 in the maintenance of skin resident CD8+ Tmem cells, but not in central CD8+ Tmem cells (O’Sullivan et al., 2014, Pan et al., 2017). To explore a role for FABP5 in in vitro-generated Tmem cells, we activated CD4+ or CD8+ T cells in vitro in the presence of IL-2 for 3 days before further differentiation in IL-15 for 3 days (Carrio et al., 2004). When cells were switched to IL-15, they were also cultured in the presence of BMS309403 or vehicle control for 3 days. In contrast to in vitro-generated Tregs, in vitro-generated CD4+ and CD8+ Tmem cells exposed to BMS309403 showed no defect in mitochondrial respiration (Figures S3A and S3B), and CD8+ Tmem cells exhibited comparable cytokine production (Figure S3C), although there was a small reduction in the expression of CD44 and CD62L (Figure S3D). These results are in agreement with published data suggesting that FABP5 may not be critical for non-tissue-resident Tmem cells like central CD8+ Tmem cells (Pan et al., 2017), as these cells predominantly generate lipids from glucose rather than acquire extracellular lipids for energy generation (O’Sullivan et al., 2014). Furthermore, these data increasingly support that BMS309403 is not generally toxic to lymphocytes and has specific effects on Tregs.
Mitochondrial Disruption after FABP5 Inhibition in Tregs Is a Result of Defective Lipid Metabolism
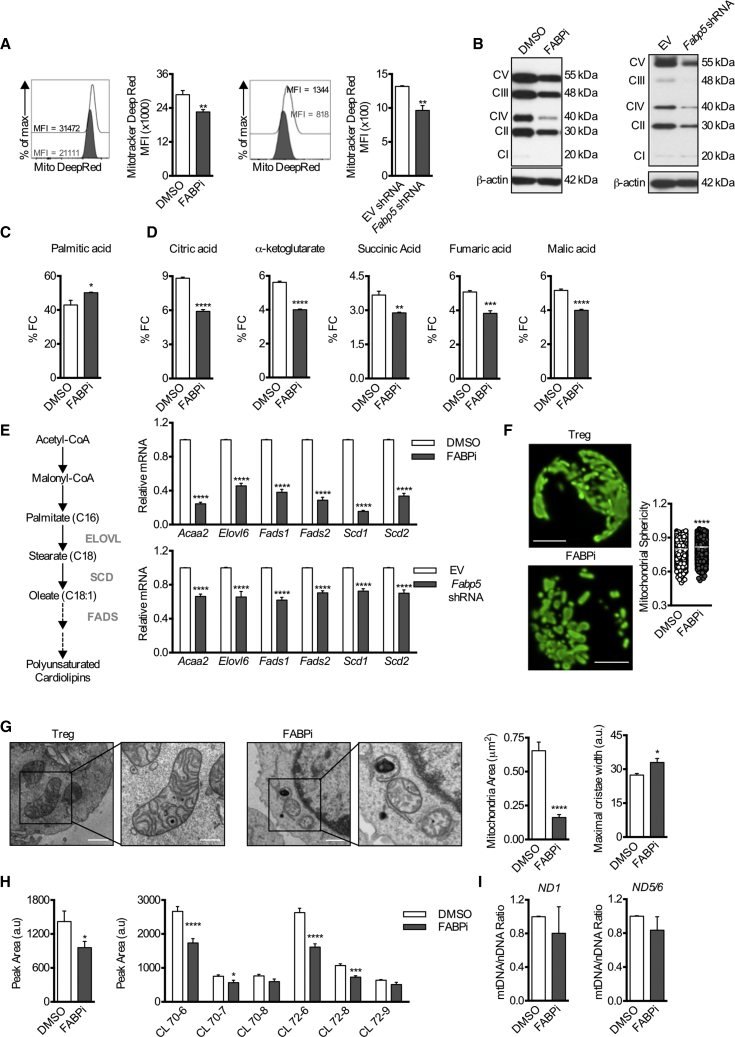
We next sought to determine the nature of the respiratory defects in Tregs after FABP5 inhibition. Both pharmacologic inhibition of FABP5 and acute shRNA knockdown of Fabp5 resulted in decreased MitoTracker deep red staining, which can indicate reduced mitochondrial mass and/or respiration (Mot et al., 2016) (Figure 2A). As mitochondria are essential organelles for respiration, further analysis also revealed decreased protein expression of the electron transport chain (ETC) complexes (Figure 2B), which was consistent with the observed reduction in OXPHOS in Tregs after BMS309403 treatment or acute Fabp5 shRNA knockdown (Figures 1D–1F).
Figure 2.
Impaired OXPHOS, Lipid Metabolism, and Loss of Cristae Structure Underlie Mitochondrial Alterations in Tregs Following FABP5 Inhibition
Naive CD4+ T cells were cultured for 3 days under Treg cell differentiation conditions before overnight BMS309403 treatment or were transduced with lentivirus expressing Fabp5 shRNA.
(A) Representative histogram and mean (±SEM) quantification of MitoTracker deep red dye (n = 4).
(B) Protein expression of the electron transport chain (ETC) complexes. Results represent three independent experiments.
(C) Fractional contribution of 13C-Palmitate-derived carbons to the intracellular palmitate pool in Tregs following acute FABP5 inhibition (n = 4).
(D) Fractional contribution of 13C-Palmitate-derived carbons to intermediates of the TCA cycle in Tregs following acute FABP5 inhibition (n = 4). Results represent two independent experiments.
(E) Schematic of the generation of monounsaturated cardiolipins from acetyl-CoA by the lipid elongation and saturation pathway. Mean (±SEM) expression of genes involved in the lipid metabolism, elongation, and desaturation pathway was assessed by qPCR (n = 4). Results represent two independent experiments.
(F) Tregs were differentiated in vitro from naive CD4+ T cells isolated from PhAM mice for 3 days before overnight BMS309403 treatment. Scale bar, 2 μm. Representative images and quantification of mitochondrial sphericity (n = 4) are shown. Results represent two independent experiments.
(G) Electron micrographs of mitochondria and quantification of mitochondrial area and mitochondrial cristae width from Tregs following acute FABP5 blockade (n = 4). Scale bar, 2 μm in the left panels and 500 nm in the right panels. Results represent two independent experiments.
(H) Left: mean (±SEM) quantification of total cardiolipin content. Right: cardiolipin species in Tregs following overnight treatment with DMSO or BMS309403 (n = 4).
(I) PCR analysis of mitochondrial DNA content (mtDNA/nDNA) in Tregs following overnight treatment with DMSO or BMS309403. Mitochondrial ND1 or ND5/6 were normalized to nuclear gene expression of 18 s. Results represent two independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005, ∗∗∗∗p < 0.001. P values were calculated using a two-tailed, unpaired t test (A, C, D, F, left panel, and I) or a two-way ANOVA with Bonferroni correction (E and H, right panel).
To assess lipid utilization and subsequent lipid oxidation in mitochondria, we cultured Tregs in the presence or absence of BMS309403 with 13C-palmitate and traced carbons into TCA cycle intermediates. Tracing with 13C-palmitate showed accumulation of palmitate-derived carbons in intracellular palmitate stores in the presence of BMS309403; however, there was a reduction in the presence of 13C-palmitate-derived carbons in TCA cycle intermediates (Figures 2C and 2D), indicating reduced β-oxidation and alterations in lipid utilization. Cardiolipin is a unique phospholipid found only in the inner mitochondrial membrane that is important for mitochondrial function, bioenergetics, and respiratory supercomplex formation (Paradies et al., 2014). Substrates for cardiolipin biosynthesis are generated from intracellular lipid elongation and desaturation (Peck et al., 2016) (Figure 2E). Given the observed mitochondrial defects, we assessed the fatty acid elongation and desaturation pathway after FABP5 inhibition. Genes encoding elongation and desaturation of fatty acids were decreased following BMS309403 treatment or Fabp5 shRNA knockdown (Figure 2E). These results indicated that lipid metabolism was markedly perturbed in Tregs after FABP5 inhibition, and they showed that defects in FABP5 function also affected expression of downstream genes important for lipid processing, suggesting a negative feedback mechanism between lipid metabolism and transcription.
Morphological changes in mitochondria and cristae width correlate with cellular metabolic changes (Buck et al., 2016). Live-cell confocal microscopy of in vitro-generated Tregs from photoactivatable mitochondria (PhAM) mice exhibited a diffuse mitochondrial network (Figure 2F). In contrast, acute BMS309403 treatment resulted in mitochondria with a punctate morphology, measured by a significant increase in mitochondrial sphericity (Figure 2F), a phenotype found in T cells with decreased respiration and increased aerobic glycolysis (Buck et al., 2016). Electron microscopy of in vitro-generated Tregs showed large, elongated mitochondria with tight cristae (Figure 2G). However, following BMS309403 treatment, mitochondria were smaller with increased cristae width (Figure 2G), a phenotype consistent with mitochondrial defects (Cogliati et al., 2013). In keeping with the decreased expression of genes controlling the lipid elongation and desaturation pathway (Figure 2E), and with the changes in mitochondrial structure (Figures 2F and 2G) and function (Figures 1D–1F), we found decreased levels of cardiolipin species in Tregs after BMS309403 treatment (Figure 2H) but no difference in the ratio of mitochondrial to nuclear DNA (mtDNA/nDNA) (Figure 2I). Together, these data illustrate that FABP5 has a function in maintaining lipid metabolism for mitochondrial function and integrity in Tregs. In contrast to the effects observed in Tregs, in vitro-generated Tmem cells showed no defect in cardiolipin biosynthesis after BMS309403 treatment (Figures S3E and S3F), consistent with our data showing no loss of oxidative metabolism in these cells with BMS309403 (Figures S3A and S3B) and supporting the notion that they might not require FABP5 to function.
FABP5 Inhibition Increases Treg Suppression
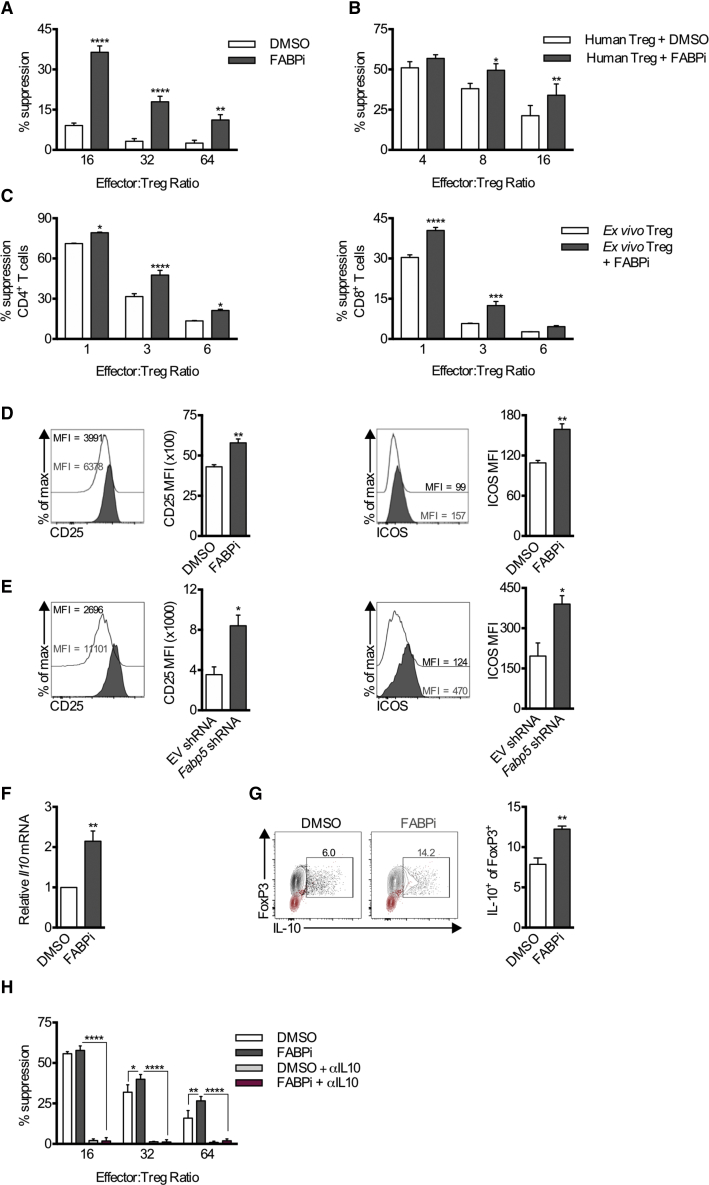
The requirement for oxidative phosphorylation for the suppressive function of Tregs has been previously reported (Beier et al., 2015, Gerriets et al., 2016, Weinberg et al., 2019). Therefore, we expected that the decreased mitochondrial function evident in Tregs after FABP5 inhibition would limit their suppressive capacity. However, we were surprised to find that in vitro-differentiated Tregs treated with BMS309403 exhibited an increased ability to suppress proliferation of responder CD4+ T cells compared to vehicle-treated Tregs (Figure 3A). Increased suppression was also observed from isolated human peripheral Tregs following in vitro expansion and acute treatment with BMS309403 (Figure 3B).
Figure 3.
FABP5 Inhibition of Fully Differentiated Tregs Increases Suppression and Expression of the Regulatory Cytokine IL-10
Treg suppression assays were performed following acute FABP5 blockade.
(A) Quantification of the mean suppression (±SEM) of responder effector cells from in vitro-differentiated Tregs following overnight BMS309403 treatment (n = 4). Results represent five independent experiments.
(B) Quantification of mean suppression (±SEM) from human Tregs following BMS309403 treatment (n = 4). Results represent the combined results from 4 independent donors.
(C) Quantification of mean suppression (±SEM) from ex vivo Tregs following 30 min BMS309403 treatment (n = 4). Results represent two independent experiments.
(D) Representative histograms and mean (±SEM) quantification of CD25 and ICOS expression on Tregs following acute FABP5 blockade (n = 4). Results represent six independent experiments.
(E) Representative histograms and mean (±SEM) quantification of CD25 and ICOS expression on Tregs following lentiviral knockdown of Fabp5. Results represent two independent experiments.
(F) Mean (±SEM) expression of Il10 gene expression measured by qPCR (n = 4).
(G) Mean (±SEM) protein expression of IL-10 in Tregs after overnight BMS309403 treatment (n = 5). Results represent two independent experiments. Gating controls are depicted in red.
(H) Mean quantification (±SEM) of suppression from in vitro-generated Tregs following overnight BMS309403 treatment in the presence or absence of anti-IL-10 antibody (αIL10) (n = 4). Results represent four independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005, ∗∗∗∗p < 0.001. P values were calculated using a two-way ANOVA with Bonferroni correction (A, B, C, and H) or a two-tailed, unpaired t test (D, E, F, and G).
The metabolic requirements of both murine and human natural Tregs (nTregs) in vivo have been shown to differ in some aspects from in vitro-differentiated Tregs due to differences in tonic signals and environmental cues (Procaccini et al., 2016). Therefore, we isolated nTregs directly from the periphery of naive mice and acutely treated the cells with BMS309403 prior to assessing their suppressive capacity compared to vehicle-treated nTregs. Again, BMS309403-treated ex vivo nTregs had an increased ability to suppress proliferation of responder CD4+ and CD8+ T cells compared to vehicle-treated nTregs (Figure 3C). In vitro-generated Tregs treated with BMS309403, or expressing Fabp5 shRNA, showed increased expression of CD25 and ICOS, two proteins expressed on highly suppressive Tregs (Figures 3D and 3E) (Redpath et al., 2013, Vocanson et al., 2010, Zeng et al., 2013). We next measured expression of IL-10, an inhibitory cytokine expressed by Tregs, and found that it was increased after BMS309403 treatment (Figures 3F and 3G). Finally, we sought to assess whether IL-10 led to the increased suppression observed after BMS309403 treatment (Figures 3A–3C). Treg-mediated suppression was abolished by the addition of anti-IL-10 antibody (αIL-10) to the suppression assay, indicating that IL-10 was responsible for mediating suppression of responder cells in this assay, with or without FABP5 inhibition (Figure 3H).
Induction of Type I Interferon Signaling in Tregs after FABP5 Inhibition
To ascertain why FABP5 inhibition led to greater suppressive capacity, we performed RNA-seq analysis on in vitro-generated Tregs after acute BMS309403 treatment. Among the top 40 most differentially regulated genes, 20 genes were related to type I interferon (IFN) signaling (Figure 4A). Furthermore, ingenuity pathway analysis (IPA) revealed pattern recognition of bacteria and viruses, and activation of IRF by cytosolic pattern recognition receptors, as the top differentially regulated pathways (Figure 4B). Type I IFNs have been implicated in the stability and function of Tregs under stress conditions, and they have been shown to induce IL-10 gene expression to enhance Treg suppression within the tumor microenvironment (Trinchieri, 2007, Zhang et al., 2011b). Confirming the increased expression of genes related to the type I IFN signaling pathway (Mx1, Ifi44, Rsad2, Ifit3b, Isg15, Cxcl11, Oas2, Mx2, Oasl1, Oas1a, Usp18, Atf3, Ifna4, Ifi204, Ifit3, Cxcl10, Lmna, Il23a, Ddx60, Oasl2), we observed increased phosphorylation of STAT1Tyr701 (pSTAT1Tyr701), indicating enhanced type I IFN signaling after BMS309403 treatment or shRNA knockdown of Fabp5 (Figure 4C).
Figure 4.
Disrupting FABP5 in Tregs Triggers mtDNA Release and Consequent cGAS-STING-Dependent Type I IFN Signaling, Which Induces Heightened IL-10 Expression
Naive CD4+ T cells were cultured for 3 days under Treg cell-differentiation conditions before overnight BMS309403 treatment.
(A) Heatmap of the top 40 differentially regulated genes identified by RNA-seq.
(B) Ingenuity pathway analysis of the top regulated pathways in Tregs after acute BMS309403 treatment.
(C) Protein expression of pSTAT1Tyr701 in Tregs after FABP5 inhibition or Fabp5 shRNA. Results represent two independent experiments.
(D) Representative confocal photomicrograph of mitochondrial staining and DNA in Tregs after overnight BMS309403 treatment. Scale bar, 2 μm.
(E) Mean quantification (±SEM) of extra-nuclear and extra-mitochondrial nucleoid structures (n = 4). Results represent two independent experiments.
(F) Quantification of cytosolic mtDNA content of Tregs after BMS309403 treatment (n = 3). Results represent two independent experiments.
(G) Mean (±SEM) expression of type I IFN-related genes in Tregs after BMS309403 treatment with siRNA-mediated silencing of cGAS or STING measured by qPCR (n = 4). Results represent two independent experiments.
(H) Mean (±SEM) expression of Il10 gene expression measured by qPCR (n = 4) in Tregs following overnight IFNα exposure (n = 4). Results represent two independent experiments.
(I) Mean (±SEM) expression of Il10 gene expression measured by qPCR (n = 4) in IFNAR KO Tregs after overnight BMS309403 treatment (n = 4). Results represent two independent experiments.
(J) Quantification of mean suppression (±SEM) from in vitro-differentiated IFNAR KO Tregs following overnight BMS309403 treatment (n = 4). Results represent three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005, ∗∗∗∗p < 0.001. P values were calculated using a two-tailed, unpaired t test (E and H) or two-way ANOVA with Bonferroni correction (F, G, I, and J). Statistics shown in (G) are relative to the FABPi group.
Perturbations in mitochondrial integrity have been shown to drive type I IFN signaling via release of mtDNA into the cytosol (West et al., 2015). In accordance with the decreased mitochondrial respiration and loss of cristae integrity observed (Figures 1D–1F, 2A, and 2G), we sought to determine whether mtDNA signaled to promote a type I IFN response after FABP5 inhibition in Tregs. We performed confocal microscopy on in vitro-generated Tregs after BMS309403 treatment. Confirming the data from Tregs expressing PhAM (Figure 2F), Tregs stained with MitoTracker deep red showed a more punctate mitochondrial morphology after BMS309403 treatment as compared to vehicle-treated cells (Figure 4D). In addition, we observed extra-nuclear and extra-mitochondrial DNA nucleoids throughout the cytoplasm of Tregs after BMS309403 treatment (Figures 4D and 4E). We performed cytosolic fractionation and recovered DNA from the cytosol of Tregs after BMS309403 treatment and assessed mtDNA by qPCR. We found increased mtCytB, mtND1, and mtND4 in BMS309403-treated Treg cytosol (Figure 4F) compared to vehicle-treated Tregs, suggesting that mtDNA was released into the cytosol after FABP5 inhibition and that this led to the induction of type I IFNs and subsequent type I IFN-regulated gene expression.
The cGAS-STING pathway acts as an important sensor of cytosolic DNA in response to viruses and has also been implicated in the sensing of mtDNA in fibroblasts (West et al., 2015). To test whether cytosolic sensing of mtDNA induced the type I IFN response after BMS309403 treatment, we expressed cGAS or STING siRNA in in vitro-generated Tregs. In keeping with our results in Figure 4A, we found that BMS309403 treatment of Tregs resulted in increased expression of Ifna4 as well as downstream genes associated with type I IFN signaling Isg15, Ifi44, and Rsad2; however, the expression of these genes was reduced to basal levels following siRNA knockdown of cGAS or STING (Figure 4G). These results indicated that cytosolic DNA sensing by cGAS and STING is critical for the type I IFN signaling evident in Tregs after FABP5 inhibition.
We hypothesized that the changes in mitochondrial structure upon FABP5 inhibition could contribute to mtDNA release and the type I IFN response in these cells. Opa1 is an important protein for regulating mitochondrial morphology and cristae structure in T cells, and cells isolated from mice with tissue specific deletions in Opa1 exhibit disorganized cristae and reduced OXPHOS (Buck et al., 2016, Cogliati et al., 2013), much like Tregs after FABP5 inhibition. We isolated CD4+ T cells from mice with a T-cell-specific deletion of Opa1 (Opa1 KO) and differentiated these cells into Tregs in vitro. Although Opa1 KO Tregs exhibited decreased basal OCR, a decreased OCR/ECAR ratio, and decreased maximal respiration as compared to WT Tregs (Figure S4A), a bioenergetic profile similar to Tregs after FABP5 inhibition (Figure 2), there was no increased pSTAT1Tyr701 in Opa1 KO Tregs (Figure S4B), nor was there increased cytosolic mtDNA (Figure S4C). These data suggested that although there were profound mitochondrial alterations in these cells, mtDNA did not trigger a type I IFN response in Opa1 KO Tregs in this setting. These observations suggest that even overt changes in cristae morphology in conjunction with respiratory defects per se do not necessarily lead to mtDNA release.
Additionally, we considered that BMS309403 might directly affect the type I IFN receptor (IFNAR) to induce type I IFN signaling or that type I IFN was required to limit Treg metabolism. However, direct exposure of in vitro-polarized Tregs to IFNα had no negative effect on metabolism (Figure S5A). In line, similar decreases in oxidative metabolism were observed in both WT and IFNAR KO Tregs after BMS309403 treatment (Figure S5B), supporting the idea that the mitochondrial dysfunction following FABP5 inhibition is the upstream signal required to promote type I IFN signaling and not that type I interferons limit cellular metabolism in this setting.
Type I IFN signaling has been shown to promote the induction of Il10 gene expression in Tregs in vivo (Stewart et al., 2013); thus, we sought to determine if exogenous IFNα would recapitulate this phenotype in vitro. We found that overnight exposure of in vitro-generated Tregs to IFNα induced Il10 gene expression (Figure 4H). In contrast to the increased Il10 gene expression measured in WT Tregs following acute FABP5 inhibition or exogenous IFNα exposure (Figures 3F and 4H), no increased Il10 gene expression was measured in IFNAR KO Tregs after BMS309403 treatment (Figure 4I). Accordingly, IFNAR KO in vitro-generated Tregs did not have increased suppressive capacity following BMS309403 treatment (Figure 4J). Finally, to further support the relationship between FABP5, type I IFN signaling, and IL-10 expression, we isolated nTregs directly from the periphery of naive mice and transduced these cells with shRNA against Fabp5. Similar to our results using in vitro-generated Tregs, we found that in nTregs, decreased expression of Fabp5 led to increased expression of Isg15, Rsad2, and Il10 after FABP5 knockdown (Figure S5C).
To further support the STING-mediated promotion of type I IFN signals and IL-10 expression independent of FABP5 in this setting, we differentiated Tregs in vitro for 3 days before treating cells with the STING agonist DMXAA (Prantner et al., 2012) or vehicle control overnight. In agreement with our other data, induction of Ifna4, Ifi44, Isg15, Rsad2, and Il10 gene expression was significantly increased; however, Fabp5 gene expression remained stable (Figure S5D). We also observed increased protein expression of IL-10 in vitro in response to DMXAA administration (Figure S5E).
Our earlier results showed reversion of inhibited oxidative metabolism in Tregs following BMS309403 washout (Figure S2A); thus, we sought to determine if type I IFN signaling was also decreased after withdrawal of this FABP inhibitor. Following BMS309403 washout, type I IFN genes were significantly decreased to near-basal levels, illustrating the link between defective mitochondrial metabolism mediated by FABP5 inhibition and the induction of type I IFN signaling (Figure S5F). Taken together, these results suggest that mitochondrial abnormalities can result in release of mtDNA to drive cGAS-STING signaling that induces autocrine type I IFN signaling to promote IL-10 expression and enhance Treg suppression.
The Tumor Microenvironment Enhances Type I IFN Signaling in Tregs
The tumor microenvironment (TME) can be nutrient sparse, and glucose limitation within this environment has been shown to have negative consequences on T cell effector function (Chang et al., 2015, Ho et al., 2015). Given that lipid uptake and metabolism are important in Tregs, we considered that perhaps a scarcity of nutrients in the TME could also impact lipid metabolism in these cells. We sought to assess whether tumor Tregs had evidence of mitochondrial dysfunction and subsequent type I IFN signaling.
A previously published RNA-seq analysis of the Treg phenotype in primary human breast carcinoma showed that type I IFN signaling was one of the most significantly dysregulated pathways in tumor Tregs relative to peripheral blood Tregs or Tregs in normal breast tissue (Plitas et al., 2016). We used this published dataset to directly explore the expression of FABP5, IL10, ISG15, IFI44, and RSAD2, and we found increased expression of these genes in human tumor Tregs compared to Tregs from the peripheral blood. (Figure 5A, Plitas et al., 2016). To explore these findings in a murine model, we injected mice with E.G7-OVA tumor cells and 14 days later isolated Tregs from both the spleen and solid tumor and analyzed gene expression. In line with the published human study (Plitas et al., 2016), we measured increased gene expression of Fabp5 in tumor infiltrating Tregs as compared to splenic Tregs as well as Il10 and type I IFN-stimulated genes Isg15, Ifi44, and Rsad2 (Figure 5B). We also observed decreased BODIPY-labeled C16 uptake in vivo (Figure 5C) and found Tregs within the TME to have less mitochondrial mass and/or respiration than splenic Tregs (Figure 5D), observations that would correlate with altered mitochondrial function and lipid metabolism. Furthermore, we measured increased CD25 and ICOS expression (marks of highly suppressive Treg cells) (Figure 5E), and IL-10 production (Figure 5F), by tumor Tregs as compared to splenic Tregs, which mirrored our results from in vitro-generated Tregs following acute FABP5 inhibition. Finally, confocal microscopy of tumor Tregs revealed extra-mitochondrial DNA nucleoids throughout the cytoplasm and an increase in mitochondrial sphericity as compared to splenic Tregs, similar to the results observed in in vitro-differentiated Tregs following BMS309403 treatment (Figure 5G).
Figure 5.
Tregs in the Tumor Microenvironment Exhibit a Suppressive Phenotype with Mitochondrial Alterations and Type I IFN Signaling
(A) Gene expression levels in Tregs isolated from human breast cancer relative to Tregs from healthy donor PBMC generated from previously published data (Plitas et al., 2016).
(B) Mice were injected with E.G7-OVA, and Tregs were isolated from the tumor or spleen on D14. Mean (±SEM) gene expression of type I IFN-related genes from intratumoral Tregs, normalized to Tregs isolated from the spleen (n = 5), is shown. Results represent two independent experiments.
(C) Representative histograms and mean (±SEM) quantification of Treg lipid uptake in vivo following labeled C16 administration. Results are representative of 5 mice from one experiment.
(D) Representative histograms and mean (±SEM) quantification of Mitotracker deep red staining. Results represent two independent experiments.
(E) Representative histograms and mean (±SEM) quantification of CD25 and ICOS from Tregs isolated from the spleen or tumor. Results represent two independent experiments.
(F) Representative flow plots and mean (±SEM) quantification of IL-10 expression from Tregs isolated from the spleen or tumor. Results represent two independent experiments. Gating controls are depicted in red.
(G) Representative confocal photomicrographs of mitochondria and DNA of Tregs isolated from the tumor or spleen and quantification of mitochondrial sphericity. Scale bar, 2 μm. Results represent one experiment.
(H) Mice were injected with E.G7-OVA, and serum, spleens, and tumors were excised on D14. The total lipid content of serum or interstitial fluid of the spleen or tumor was quantified by MS. Results are representative of 8 mice from one experiment.
(I) Mean (±SEM) quantification of Fabp5 gene expression and protein expression in Tregs following overnight culture in lipid or glucose depleted media (n = 4).
(J) Mean (±SEM) quantification of labeled C16 uptake in vitro in Tregs following overnight lipid starvation (n = 4) or from nTregs ex vivo from the spleen or tumor of mice (n = 5) as per (F). Results represent two independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005, ∗∗∗∗p < 0.001. P values were calculated using a two-way ANOVA with Bonferroni correction (A and B) or two-tailed, unpaired t test (C, D, E, G, H, I, and J).
The results we observed with in vitro Tregs following FABP5 inhibition were similar to our observations in tumor Tregs. Curiously, however, both human and mouse tumor Tregs displayed increased Fabp5 gene expression. Given our results showing that acute loss of function of FABP5 led to mitochondrial defects, we initially expected that these cells would have decreased expression of FABP5. We therefore sought to reconcile these seemingly disparate findings and understand how tumor Tregs could have enhanced Fabp5 gene expression, but disrupted mitochondria and type I IFN signaling, that would be more indicative of a loss of FABP5 function. We hypothesized that in vitro, there is an abundance of lipids in the media; however, these lipids are unable to be utilized efficiently by the cell when FABP5 is inhibited, either pharmacologically or genetically. Within the tumor microenvironment, we hypothesized that although Fabp5 is even more highly expressed, there may not be sufficient lipids available for the cell to acquire. Both of these scenarios would hypothetically lead to decreased lipid uptake by Tregs. Following this logic, we speculated that lipid availability and acquisition might serve a regulatory role to modulate the expression of genes involved in lipid trafficking, such as Fabp5. Therefore, in a tumor, decreased lipid content of the extracellular milieu might act to augment Fabp5 expression, but a lack of available lipid substrate would still mimic a loss of FABP5 function. To begin to test these ideas, we injected mice with E.G7-OVA and allowed tumors to develop for 14 days before harvesting the serum, spleen, and tumors to quantify the available extracellular lipid content. Spleens and tumors were digested, and lipids were isolated from equal volumes of interstitial fluid. Lipid content was quantified in the serum of all mice and in the interstitial fluid of the spleen and tumor. Comparatively, we found that there was a distinct paucity of lipids within the interstitial fluid of the tumor (Figure 5H).
To test whether the availability of lipids in the extracellular environment could influence Fabp5 gene expression, we cultured Tregs in vitro for 3 days before harvesting cells and re-culturing the cells overnight in nutrient-replete media, glucose-deficient media, or lipid-depleted media. In the absence of sufficient lipids, Fabp5 gene and protein expression increased; however, the glucose-deficient media did not elevate FABP5 protein expression (Figure 5I). To determine if the increased Fabp5 gene expression had a functional consequence, we re-plated Tregs from nutrient-replete or lipid-depleted media, or ex vivo nTregs isolated directly from spleens or tumors, back into complete media and quantified the uptake of BODIPY-labeled C16 after 30 min. BODIPY-labeled C16 uptake was greatest in cells that had been exposed to a low-lipid environment, either in vitro or within the tumor, as compared to control cells (Figure 5J). Together, these results support the notion that within the tumor microenvironment, a lack of available lipids from the extracellular milieu results in increased Fabp5 expression in tumor Tregs. Therefore, the lipid-restrictive environment of the tumor leads to mitochondrial perturbations and subsequent type I IFN signaling in Tregs that lead to greater IL-10 production and enhanced suppressive capacity.
Discussion
As hydrophobic molecules, lipids require chaperones to maintain solubility and travel throughout both the body and the cell. We found the lipid chaperone FABP5 to be highly expressed in Tregs. Acute pharmacological inhibition or shRNA knockdown of Fabp5 resulted in dysregulation of the mitochondrial network, loss of ETC complexes, and decreased mitochondrial OXPHOS, with a consequent switch to glycolysis. The mitochondrial defects after FABP5 inhibition accompanied impaired expression of genes involved in the lipid elongation and desaturation pathway, correlating with decreased synthesis of cardiolipin, a lipid required for maintenance of mitochondrial integrity (Zhang et al., 2011a) and ETC supercomplex formation (Ikon and Ryan, 2017). This mitochondrial dysfunction led to increased Treg suppressive capacity via the induction of type I IFN signaling and enhanced subsequent IL-10 production, which resulted in heightened proliferative suppression of responder cells in vitro. While we did not explore the role of FABP5 in promoting nuclear translocation of transcription factors, we propose that the reduction in the expression of lipid elongation and desaturation genes could be a consequence of decreased PPARγ expression, which, although not highlighted in our results, was downregulated in our RNA-seq data following FABP5 inhibition. Of note, Fabp5 is a direct PPARγ target; thus, a break in this cycle is likely to have far-reaching consequences for the biology of these cells.
Our in vitro data showed that inhibiting FABP5 in Tregs augmented their suppressive capacity. At first glance, however, the increased gene expression of Fabp5 in vivo in both human and mouse Tregs in the tumor microenvironment seemed inconsistent with these findings. However, our data revealed a paucity of lipids in the tumor compared to splenic microenvironment, suggesting that tumor Tregs may augment FABP5 in response to low-lipid environments. The relative scarcity of nutrients within not only the TME but also other tissue sites, such as the skin or adipose tissue, may also result in the increased expression of nutrient transporters, as cells struggle to meet their metabolic demands. Along these lines, under hypoxic conditions, and within the tumor microenvironment, CD8+ T cells have been shown to increase expression of the glucose transporter Glut1 (Cretenet et al., 2016).
Lipid trafficking to mitochondrial membranes is likely to play an important part in the regulation and maintenance of mitochondrial structure and function (Scharwey et al., 2013). Damaged mitochondria have been shown to release mtDNA into the cytosol, which activates the cytosolic DNA sensor cGAS and its adaptor STING, resulting in the induction of type I IFNs and subsequent expression of type I IFN-stimulated genes (Szczesny et al., 2018, West et al., 2015). Our data show that this process can be precipitated by loss of function of FABP5, either through inhibition, reduced expression, or by a lack of necessary lipid substrates. In these settings, utilizing PhAM reporter mice, or following intracellular staining protocols and confocal microscopy, we observed distinct punctate mitochondria associated with loss of FABP5 function, corroborating our previously published results showing that this mitochondrial shape in T cells correlates with a decrease in OXPHOS and a concomitant induction of aerobic glycolysis (Buck et al., 2016). Type I IFNs are known to have significant effects on Tregs (Piconese et al., 2015). Type I IFN signaling has been shown to promote Treg function under stress conditions (Metidji et al., 2015), and the accumulation of IL-10 producing Tregs in tumors requires type I IFN signaling (Kawano et al., 2018, Stewart et al., 2013). Further, our results showing increased IL-10 expression following DMXAA administration in Tregs in vitro suggest that Treg function should perhaps be further investigated in settings where DMXAA is used as an anti-tumor therapy in vivo (Baguley, 2003, Zhou et al., 2002). Our findings demonstrate that Treg-intrinsic mitochondrial damage, by inducing type I IFN production, can play a significant role in increased IL-10 production and thereby shape Treg suppressive capacity.
All FABPs bind to and have relatively high affinity for long-chain fatty acids but possess differences in ligand specificity, binding affinity, and binding mechanism (Chmurzyńska, 2006). FABP3 has a high affinity for omega-6 polyunsaturated fatty acids (PUFAs) (Balendiran et al., 2000, Liu et al., 2010, Veerkamp et al., 1999), FABP4 has a high affinity for oleate and has been shown to interact directly with hormone-sensitive lipase (HSL) to promote lipolysis of lipid droplets (Smith et al., 2007). Intriguingly, although FABP5 has a high affinity for saturated fatty acids, such as stearate, and monounsaturated fatty acids, such as oleate (Hohoff et al., 1999), FABP5 also has a similar binding affinity for retinoic acid (RA). Retinoic acid inhibits cell growth by binding to the nuclear RA receptor (RAR), but it also promotes cell growth upon binding to PPARβ/γ (Schug et al., 2007). The ratio of FABP5 to cellular RA binding protein II (CRABPII) determines the intracellular localization of retinoic acid, with a high FABP5-CRABPII ratio leading to RA activation of PPARβ/γ. Conversely, a low FABP5-CRABPII ratio leads to RAR activation (Schug et al., 2008). Expression of the stimulated by retinoic acid 6 (Stra6) gene was downregulated in our RNA-seq analysis from Tregs after BMS309403 treatment, suggesting that the limited flux of lipids into the cell may account for reduced cellular proliferation as opposed to activation of the RAR. It is interesting to note that RA promotes Treg induction from both naive CD4+ and CD4+CD44+ T cells (Hill et al., 2008, Mucida et al., 2009). Thus, the metabolic requirements of Tregs both in vitro and in vivo are likely to be dynamic and change throughout the course of differentiation and may also be dependent on the site of activation and nutrient microenvironment.
Confocal microscopy revealed punctate mitochondria with perturbed cristae after FABP5 inhibition, results that hinted at a possible role for mitochondrial remodeling in permitting the release of mtDNA into the cytosol. Opa1 is known to be critical for the fusion of the inner mitochondrial membrane, and T cells deficient for Opa1 have been shown to have defective mitochondrial metabolism (Buck et al., 2016, Lee et al., 2017). Following in vitro differentiation from naive CD4+ T cells, Opa1 KO Tregs showed decreased basal and maximal respiration as expected; however, no increased pSTAT1Tyr701 was detected, indicating no induction of type I IFN signaling in this setting. Furthermore, no increased cytosolic mtDNA was detected. These results showed that mtDNA release was not associated with the profound mitochondrial defects observed in the Opa1 KO Tregs, indicating that the effects of FABP5 loss of function that allow mtDNA release might be quite subtle. It will be interesting in future experiments to determine if acute inhibition of Opa1 in Tregs, either genetically or pharmacologically, could lead to type I IFN signaling via mtDNA release. Likewise, it may be useful to determine the suppressive capacity of Opa1 KO Tregs as compared to WT Tregs, which could decouple the metabolic phenotype of the Opa1 KO from the functional outcome observed in WT Tregs following FABP5 inhibition. Opa1 overexpression has been shown to ameliorate the phenotype of mice bearing mutations in Ndufs4 and Cox15, two genes critical for the formation of ETC complex I and IV, respectively (Civiletto et al., 2015). It remains to be investigated whether overexpression of Opa1, or of the mitochondrial outer membrane fusion machinery MFN1 and MFN2, protects Tregs from mitochondrial dysfunction induced following FABP5 inhibition.
Tregs differentiated in vitro in the presence of TGF-β demonstrate high oxidative metabolism, and low glycolytic metabolism, with a preference for fatty acid oxidation (Michalek et al., 2011). Conversely, in vivo thymic-derived Tregs engage glycolysis and glutaminolysis at comparable levels to effector T cells, with high mTORC1 activity, despite high Foxp3 expression (Newton et al., 2016). Exposure of thymic-derived Tregs to TGF-β in vitro was shown to switch metabolism from glycolysis and glutaminolysis to OXPHOS (Priyadharshini et al., 2018). In addition to its effects on Tregs, TGF-β can promote OXPHOS in podocytes and hepatocellular carcinoma, highlighting a key role for this cytokine in the regulation of metabolism (Abe et al., 2013, Soukupova et al., 2017). Thus, it would be interesting to examine the effects of FABP5 blockade on Tregs generated in vitro in the absence of TGF-β. Differentiation of human Tregs in vitro can be achieved with suboptimal TCR stimulation, and these cells maintain suppressive capacity. While these cells utilize lipids and FAO to meet their metabolic demands in vitro, they are also dependent on the induction of glycolysis to mediate the splicing of Foxp3 isoforms (De Rosa et al., 2015). Inhibition of mTORC1 with rapamycin can also promote the differentiation of human and murine Tregs in vitro in the absence of TGF-β (Battaglia et al., 2012, Ogino et al., 2012). Given the different culture methods used to achieve Treg differentiation in vitro, it would be informative to assess the expression of FABP5 in these cells across different culture conditions, and the consequence of FABP5 inhibition or knockdown, to further interrogate the role of cytokine and metabolic influences in determining Treg metabolism and function.
Many studies have shown that Tregs require OXPHOS for suppressive capacity (Beier et al., 2015, Gerriets et al., 2016, Weinberg et al., 2019). In our studies, OXPHOS was decreased in Tregs as a result of FABP5 loss of function, but nevertheless, suppressive activity was improved. We found that Tregs had a compensatory increase in glycolysis after FABP5 inhibition, and these cells also had enhanced suppression. In line with these observations, it has been shown that ex vivo-isolated human Tregs are highly glycolytic (Procaccini et al., 2016). Despite the loss of mitochondrial structure and ETC complexes, Tregs were still equally viable for 36 h following FABP5 inhibition, as compared to vehicle-treated cells. This is important, as the initiation of apoptosis has been shown to block mtDNA-induced type I IFN signaling (Rongvaux et al., 2014, White et al., 2014). This is a critical checkpoint to ensure that cellular apoptosis remains immunologically silent and does not trigger innate immune activation. Although our experiments showing that Tregs cultured in BMS309403 recovered metabolic homeostasis when the drug was washed out, it remains to be seen if Tregs undergo apoptosis following longer-term FABP5 inhibition and whether the induction of type I IFN signaling and subsequent IL-10 production wane over time. We speculate that long-term mitochondrial alterations mediated by this pathway could lead to increased Treg cell death. In this case, our data would align with recent findings that tumor Tregs have greater suppressive capacity linked to inadvertent death due to oxidative stress (Maj et al., 2017). In that study, it was shown that apoptotic Tregs mediated immunosuppression via adenosine rather than IL-10 (Maj et al., 2017). Together, these findings support the view that Tregs possess multiple suppressive mechanisms and, in certain settings, may not always have to be what is considered “optimally” fit to perform suppressor functions (Schmidt et al., 2012). Our study illuminates a novel link between mitochondrial health and lipid metabolism and how this serves as a Treg-intrinsic checkpoint for the expression of type I IFN/IL-10-mediated suppressive mechanisms.
Limitations of Study
Future additional work will be aimed at further exploring the metabolism of tumor infiltrating Tregs. However, based on the mitochondrial network shown by confocal microscopy and decreased MitoTracker red staining, we propose that tumor Tregs would have limited capacity for OXPHOS compared to splenic Tregs. Furthermore, this study has not shown the direct expression of type I IFN by Tregs in tumors, but rather, it has shown that they have mitochondrial dysfunction and a type I IFN-stimulated gene signature. More work will have to be done to determine the relevance of Treg cell-derived type I IFN expression during cancer.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD4-PerCpCy5.5 Clone GK1.5 (1:800) | Biolegend | #100434 |
| CD4-ApcCy7 Clone GK1.5 (1:500) | Biolegend | #100413 |
| CD4-BV711 Clone RM4-5 (1:1000) | Biolegend | #100594 |
| CD8-PerCpCy5.5 Clone 53-6.7 (1:400) | Biolegend | #100734 |
| CD25-AF700 Clone PC61 (1:400) | Biolegend | #102024 |
| CD25-BV421 Clone PC61 (1:1000) | Biolegend | #102034 |
| CD25-BV711 Clone PC61 (1:800) | Biolegend | #740714 |
| CD44-PeCy7 Clone IM7 (1:400) | Biolegend | #103030 |
| CD45.1-PE Clone A20 (1:200) | Biolegend | #110708 |
| CD45.2-FITC Clone 104 (1:200) | Biolegend | #109806 |
| CD62L-Pacific Blue Clone MEL-14 (1:200) | Biolegend | #104424 |
| Foxp3-eF450 Clone FJK-16S (1:100) | eBioscience | #48-5773-82 |
| ICOS-BV785 Clone C398.4A (1:400) | Biolegend | #313534 |
| IFNγ-FITC Clone XMG1.2 (1:200) | Biolegend | #505806 |
| IL-10 Purified Clone JES5-16E3 (1:100) | Biolegend | #505001 |
| TNFα-PE Clone MP6-XT22 (1:200) | Biolegend | #506306 |
| β-actin (1:10,000) | CST | 4970S |
| FABP5 (D1A7T) Rabbit mAb (1:2000) | CST | #3326 |
| Total OXPHOS Rodent WB Antibody Cocktail (1:2000) | Abcam | Ab110413 |
| Stat1 (D1K9Y) Rabbit mAb (1:2000) | CST | #14994 |
| Phospho-Stat1 (Tyr701) (58D6) Rabbit mAb (1:5000) | CST | #9167 |
| InVivoMab anti-mouse CD3 (5 μg/mL) | BioXCell | BP0001-1 |
| InVivioMab anti-mouse CD28 (0.5 μg/mL) | BioXCell | BE0015-1 |
| InVivoMab anti-mouse IL-12p40 (10 μg/mL) | BioXCell | BE0051 |
| InVivoMab anti-mouse IL-4 (10 μg/mL) | BioXCell | BE0199 |
| InVivoMab anti-mouse IFN-γ (10 μg/mL) | BioXCell | BE0055 |
| Biological Samples | ||
| Healthy Control Blood | UniKlinik Freiburg | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| BMS309403 | Cayman Chemical | #10010206 |
| C16-BODIPY | Thermo Fisher | D3821 |
| Cell Trace Violet | Thermo Fisher | C34557 |
| DMXAA | Sigma | D5817 |
| Mitotracker Deep Red | Thermo Fisher | M22426 |
| Live Dead Fixable Blue | Thermo Fisher | L23105 |
| IFNα | PBL Assay Science | #12100-1 |
| Recombinant murine IL-1β | Peprotech | 211-11B-50 |
| Recombinant human IL-2 | Peprotech | 200-02-1000 |
| Recombinant mouse IL-4 | Peprotech | 214-14-100 |
| Recombinant mouse IL-6 | Peprotech | 216-16-50 |
| Recombinant mouse IL-7 | Peprotech | 217-17-50 |
| Recombinant mouse IL-12 | Peprotech | 210-12-50 |
| Recombinant mouse IL-15 | Peprotech | 210-15-15 |
| Recombinant human TGFβ | Peprotech | 100-21-50 |
| Oligomycin | Sigma | #1404-19-9 |
| FCCP | Sigma | #370-86-5 |
| Rotenone | Sigma | #83-79-4 |
| Antimycin A | Sigma | #1397-94-0 |
| Critical Commercial Assays | ||
| Seahorse Extracellular Flux Analyzer XFe 96 | Agilent | www.agilent.com |
| Deposited Data | ||
| GEO Submission (GSE126245) | https://www.ncbi.nlm.nih.gov/geo/ | |
| Experimental Models: Cell Lines | ||
| E.G7-OVA | ATCC | ATCC Cat# CRL-2113, RRID: CVCL_3505 |
| Oligonucleotides | ||
| siRNA for murine cGAS | GE Dharmacon | #E-055608-00-0005 |
| siRNA for murine STING | GE Dharmacon | #E-055528-00-0005 |
| mtCytb FW (TTCATGTCGGACGAGGCTTA) | Sigma | N/A |
| mCytb RV (GTTTATTGGGGATTGAGCGTAG) | Sigma | N/A |
| mtND1 FW (CTAGCAGAAACAAACCGGGC) | Sigma | N/A |
| mtND1 RV (GTATGGTGGTACTCCCGCTG) | Sigma | N/A |
| mtND4 FW (ACAACACACACCTTAGACGCT) | Sigma | N/A |
| mtND4 RV (TGTGGATCCGTTCGTAGTTGG) | Sigma | N/A |
| Taqman 18 s Ribosomal RNA | Thermo Fisher | Mm03928990 |
| Taqman Fabp3 | Thermo Fisher | Mm02342495 |
| Taqman Fabp4 | Thermo Fisher | Mm00445878 |
| Taqman Fabp5 | Thermo Fisher | Mm00783731 |
| Taqman Il10 | Thermo Fisher | Mm01288386 |
| Taqman Isg15 | Thermo Fisher | Mm01705338 |
| Taqman Ifi44 | Thermo Fisher | Mm00505670 |
| Taqman Rsad2 | Thermo Fisher | Mm00491265 |
| Taqman Ifna4 | Thermo Fisher | Mm00833969 |
| Taqman Acaa2 | Thermo Fisher | Mm00624282 |
| Taqman Elovl6 | Thermo Fisher | Mm00851223 |
| Taqman Fads1 | Thermo Fisher | Mm00507605 |
| Taqman Fads2 | Thermo Fisher | Mm00517221 |
| Taqman Scd1 | Thermo Fisher | Mm00772290 |
| Taqman Scd2 | Thermo Fisher | Mm01208542 |
| Software and Algorithms | ||
| Wave Software Version 2.4 | Agilent | www.aglient.com |
| FlowJo v10 | FlowJo | www.flowjo.com |
| GraphPad Prism 6 | GraphPad Software | www.graphpad.com |
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be made available upon reasonable request by the Lead Contact, Erika L. Pearce (pearce@ie-freiburg.mpg.de). This study did not generate new unique reagents.
Experimental Model and Subject Details
Mouse Models
C57BL/6J (RRID: IMSR_JAX:000664), PhAM (RRID: IMSR_JAX:018397), IFNAR KO (MMRRC_32045) and TIGER/FIR mice (RRID: IMSR_JAX:008379) and CD45.1 congenic (RRID: IMSR_JAX:002014) mouse strains were purchased from The Jackson Laboratory. Opa1 conditional floxed mice (obtained from Dr. Hiromi Sesaki at Johns Hopkins University School of Medicine, Baltimore, MD) were crossed to CD4-Cre mice (obtained from Dr. Yongwon Choi at the University of Pennsylvania) to generate Opa1flox/flox CD4-Cre mice (maintained on a C57BL/6J background). All mice were maintained at the Max Planck Institute for Immunobiology and Epigenetics or the University of Minnesota and cared for according to the Institutional Animal Use and Care Guidelines.
Primary Cell culture
Total CD4+ T cells were positively selected by MACS isolation from spleen and peripheral LNs of C57BL/6 mice. Cells were cultured on anti-CD3 coated plates (5 μg/mL) at a density of 1 × 106/mL with 0.5 μg/mL anti-CD28, 100 U/mL IL-2, 5 ng/mL TGF-β and 10 ug/mL anti-IL-4, anti-IL-12 and anti-IFN-γ in 1640 RPMI media supplemented with 10% Hyclone serum, 1% Penstrep and 1% L-Glut and 55 μM beta-mercaptoethanol. After 72 h, cells were split 1:2 either in the presence or absence of BMS309403 (50 μM, solubilized in DMSO) or DMXAA (200 μM, solubilized in DMSO), or vehicle control. In some experiments, cells were cultured in the presence of BMS309403 or DMSO from the start of culture. For washout experiments, cells were harvested on day 4, washed to remove any residual BMS309403 or DMSO and cultured for a further 24 h. Th1 cells were differentiated on anti-CD3 coated plates (5 μg/mL) at a density of 1 × 106/mL with 0.5 μg/mL anti-CD28, 100 U/mL IL-2, 10 ng/mL IL-12 and 10 ug/mL anti-IL-4. Th2 cells were differentiated on anti-CD3 coated plates (5 μg/mL) at a density of 1 × 106/mL with 0.5 μg/mL anti-CD28, 100 U/mL IL-2, 10 ng/mL IL-4 and 10 ug/mL anti-IL-12 and anti-IFN-γ. Th17 cells were differentiated on anti-CD3 coated plates (5 μg/mL) at a density of 1 × 106/mL with 0.5 μg/mL anti-CD28, 10 μg/mL IL-1β, 10 ng/mL IL-6, 5 ng/mL TGF-β and 10 ug/mL anti-IL-4 and anti-IFN-γ.
For human studies, T cells were isolated from buffy coats that were kindly provided by the Institute for Transfusion Medicine and Gene Therapy, Medical Center – University of Freiburg (donor consent, anonymized) or at the University of Minnesota. Healthy donor blood was diluted 1:1 with PBS and layered over lymphoprep in SepMate PBMC Isolation tubes. Samples were centrifuged at 1200 x g for 15 min and PBMCs were poured off into a new tube. Human CD4+ T cells were positively isolated from the PBMC fraction by MACS separation. CD4+ T cells were cultured in vitro with human CD3/CD28 T cell activator beads (StemCell). 100 U/mL rhIL-2 and 5 ng/mL rhTGF-β for 7 days in 1640 RPMI media supplemented with 10% Hyclone serum, 1% Penstrep and 1% L-Glut and 55 μM beta-mercaptoethanol. After 7 days, human cells were split 1:2 either in the presence or absence of BMS309403 (250 μM, solubilized in DMSO) or vehicle control over night before analysis. Human cells were found to have a higher tolerance for FABP5 inhibition (Murine cells were found to be sensitive to BMS309403 at 50 μM where as human cells required 250 μM for efficacy). For CD4+ and CD8+ Tmem cell cultures, isolated cells were plated on anti-CD3 coated plates with 0.5 μg/mL anti-CD28 and 100 U/mL rhIL-2 for 3 days before being harvested and re-plated in the presence of rmIL-15 for a further 3 days. All cell culture was performed under 5% CO2 atmospheric oxygen, at 37°C in a humidified incubator.
Cell Lines
The murine EL4 lymphoblast cell line expressing OVA (E.G7-OVA) was purchased from ATCC (ATCC Cat# CRL-223). Cells were cultured for two passages before being implanted in the left flank of naive female mice. Further authentication of the cell line was not performed. The sex of the cell line is female. Cells were maintained in 1640 RPMI media supplemented with 10% GIBCO serum, 1% Penstrep and 1% L-Glut and 55 μM beta-mercaptoethanol, under 5% CO2 atmospheric oxygen, at 37°C in a humidified incubator.
Method Details
In Vivo Tumor Experiments
Mice were injected subcutaneously into the left flank with 1 × 106 E.G7-OVA in sterile PBS. After 14 days, mice were sacrificed and spleens and tumors harvested for further phenotyping. For lipid uptake, labeled C16 was administered to mice at 50 mg/kg 30 minutes prior to sacrifice.
qPCR
RNA was isolated from cell pellets using the QIAGEN easy kit as per the manufacturers instructions. RNA was reverse transcribed to cDNA by AB cDNA synthesis kit as per the manufacturers instructions. qPCR was performed using the iTaq Universal Probes Supermix (Biorad) and Taqman primers (Thermo Fisher Scientific). Fold change was assessed using the 2ˆ-(ddCT) method using 18S as a reference gene.
RNA Seq
Total RNA was extracted from cell pellets using the QIAGEN easy kit as per the manufacturer’s instructions and quantified using Qubit 2.0 (Thermo Fisher Scientific). Libraries were prepared using the TruSeq stranded mRNA kit (Illumina) and sequenced in a HISeq 3000 (Illumina) by the Deep-Sequencing Facility at the Max Planck Institute for Immunobiology and Epigenetics. Sequenced libraries were processed with the Galaxy platform and deepTools (Afgan et al., 2016, Ramírez et al., 2016), using STAR (Dobin et al., 2013) for trimming and mapping and featureCounts (Liao et al., 2014) to quantify mapped reads. Raw mapped reads were processed in R (Lucent Techonologies) with DESeq2 (Love et al., 2014) to determine differentially expressed genes and generate normalized read counts to visualize as heatmaps using Morpheus (Broad Institute). Pathway enrichment analysis was performed using Ingenuity Pathway Analysis (QIAGEN) which makes predictions using pairwise comparisons between conditions, incorporating statistically significant changes in the gene expression within each of the comparisons.
Total Lipid and Cardiolipin Quantification
Mouse blood was collected post-euthanasia and allowed to clot at room temp for 1 hour Samples were spun at 10,000 x g for 10 min and serum was isolated. Spleens and tumors were excised and mechanically dissociated in 2.5 mg/mL collagenase I and collagenase II with 5 ug/mL DNase before incubation at 37°C for 1 h. Samples were centrifuged at 500 x g for 4 min to remove solid debris and equal volumes of fluid were taken for lipid isolation and analysis. Lipids were quantified using the MTBE extraction protocol. Briefly, frozen cell pellets were resuspended in 100 μL ice cold PBS and transferred to 8 mL glass tubes before the addition of methanol. Methyl tert-butyl ether was added and tubes were shaken for 1 h at 4°C. Water was added to separate the phases before centrifugation at 1,000 x g for 10 min. The upper organic phase was collected and dried in a Genevac EZ2 speed vac. Samples were resuspended in 2:1:1 isopropanol:acetonitrile:water prior to analysis. LC-MS was carried out using an Agilent Zorbax Eclipse Plus C18 column using an Agilent 1290 Infinity II UHPLC inline with an Agilent 6495 Triple Quad QQQ-MS. Lipids were identified by fragmentation and retention time, and were quantified using Agilent Mass Hunter software. Cardiolipins were quantile normalized using the preprocesscore R package. Serum lipids were normalized to volume while tumor and spleen lipid quantity were normalized to organ weight.
Metabolic Phenotyping
Oxygen consumption rate (OCR) and extraellular acidification rate (ECAR) were measured using the Seahorse XFe bioanalyser. 1.5x105 T cells per well (> 3 wells per sample) were spun onto poly-D-lysine coated seahorse 96 well plates and preincubated in Seahorse XF media (non-buffered RMPI + 10 μM L-glutamine + 10 μM sodium pyruvate + 25 mM glucose) at 37°C for a minimum of 30 min in the absence of CO2. OCR and ECAR were measured under basal conditions, and after the addition of the following drugs: 1 μM oligomycin, 1.5 μM flurorcarbonyl cyanide phenylhydrazone (FCCP) and 100 nM rotenone + 1μM antimycin A as indicated. Measurements were taken using a 96 well Extracellular Flux Analyzer (Seahorse bioscience).
Flow Cytometry
Fluorochrome-conjugated monoclonal antibodies were purchased form eBioscience or BDBioscience. Staining was performed in PBS containing 2% FBS (GIBCO) + 2mM EDTA (Thermo Fisher), on ice. Dead cells were excluded with the Life/Dead Fixable Blue Cell Stain Kit (Thermo Fisher). For analysis of mitochondrial mass, MitoTracker Deep Red (Thermo Fisher) staining was performed in complete media at 37°C + 5% CO2 for 30 min. Lipid uptake was assessed by incubation with 1 μM C16-BODIPY (Thermo Fisher) in complete RPMI for 30 min at 37°C + 5% CO2. Intracellular staining was performed using the eBioscience Foxp3 staining kit as per manufacturers instructions. Samples were stored in 1% PFA, diluted in flow buffer before acquisition on LSR Fortessa flow cytometers with FACSDiva Software. After exclusion of doublets, and dead cells, Tregs were identified as CD4+Foxp3+ cells. Appropriate gating and negative controls were used in each experiment to identify cell populations.
Confocal and Electron Microscopy Imaging
The analysis of mitochondrial morphology in live cells was performed as described (Buck et al., 2016). Briefly, in vitro differentiated Treg cells from PhAM mice were treated as described and imaged live on glass bottom dishes (MAtTek) coated with poly-D-lysine in complete medium using a Zeiss spinning disk confocal with an Evolve (EMCCD) camera. Cells were kept in a humidified incubation chamber at 37°C with 5% CO2 during image collection. Images were deconvolved using Huygens imaging software and analyzed using the surface features of Imaris imaging software.
For the analysis of mtDNA nucleoids, in vitro differentiated or ex vivo Tregs FACS sorted from tumors or spleens were stained with Mitotracker Deep Red (Invitrogen) for 20 min at 37°C and adhered to coverslips coated with poly-D-lysine (Sigma) in complete medium. After washing in PBS, cells were fixed with 4% formaldehyde for 15 min, permeabilized with 0.05% Triton X-100 in PBS for 5 min and blocked with PBS containing 3% FBS for 30 min. Then, cells were stained with anti-DNA antibody (Progen, clone AC-30-10) for 90 min, stained with appropriate secondary antibody for 60 min (Invitrogen) and stained with Hoechst 42 (Molecular Probes) for 10 min. Cells were washed twice with PBS containing 1.5% FBS between each step. Coverslips were mounted with Prolong Gold anti-fade reagent (Molecular Probes). Approximately 10 fields of view from distinct biological replicates were captured at random with an LSM880 - Airyscan microscope with a Plan-Apochromat 63x 1.4 oil objective. Images were deconvolved using Huygens imaging software and analyzed using the surface and particle features of the Imaris imaging software.
For electron microscopy, 6x106 cells were fixed in 2.5% glutaraldehyde in 100 mM sodium cocodylate, washed in cocodylate buffer. After dehydration samples were embedded in Eponate 12 resin and sections were cut. Images were acquired using a JOEL 1200 EX transmission electron microscope with an ATMP digital camera or a FEI Tecnai 12 Transmission electron microscope equipped with a TIETZ digital camera. Mitochondria size and cristae width was measured using ImageJ software and averaged over 30 independent images.
Western Blotting
For western blot analysis, cells were washed with ice cold PBS and lysed in 1 x Cell Signaling lysis buffer (20 mM Tris-HCL [pH 7.5], 150 mM NaCl, 1 mM Na2EDTA, 1mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1mM Na3VO4, 1 μg/mL leupeptin), supplemented with 1mM PMSF. Samples were freeze thawed three times followed by centrifugation at 20,000 x g for 10 min at 4°C. Cleared protein lysate was denatured with LDS loading buffer for 10 min at 70°C and loaded onto precast 4% to 12% bis-tris protein gels. Proteins were transferred onto nitrocellulose membranes using the iBLOT2 system following the manufacturers protocols. Membranes were blocked with 5% w/v milk and 0.1% Tween-20 in TBS and incubated with the appropriate antibodies in 5% w/v BSA in TBS with 0.1% Tween-20 overnight at 4°C. All primary antibody incubations were followed by incubation with secondary HRP-conjugated antibody (Pierce) in 5% milk and 0.1% Tween-20 in TBS and visualized using SuperSignal West Pico or Femto Chemiluminescent Substrate (Pierce) on Biomax MR Film (Kodak). Optical density of the signals on the film was quantified using grayscale measurements in ImageJ software (NIH) and converted to fold change, normalized to the loading control.
shRNA Delivery
Isolated CD4+ T cells were transduced with empty vector (EV) or Fabp5 shRNA expressing lentivirus by centrifugation for 90 min (1,000 x g) in the presence of polybrene (8 μg/mL) and HEPES (10 mM). Following centrifugation, supernatant was replaced with fresh viral supernatant containing polybrene (8 μg/mL), HEPES (10 mM) and IL-7 10 U/mL and cells were incubated overnight at 37°C + 5% CO2.
siRNA Delivery
For siRNA transfection, we used the smartpool accell siRNA for murine cGAS (GE Dharmacon Cat# E-055528-00-0005) or STING (GE Dharmacon Cat# E-055608-00-0005) according to the manufacturer’s recommendations with minor modifications. Briefly isolated CD4+ T cells were cultured overnight at 37°C + 5% CO2 in Accell delivery medium (Cat# B-005000) supplemented with 1% fetal calf serum (Hyclone) and IL-7 at 10 U/mL.
Mouse Treg Suppression Assays
For assessment of the suppressive function of in vitro generated Tregs, cells were treated overnight with 50 μM BMS309403 or left untreated. Cells were harvested and washed to remove any residual BMS309403, before a serial dilution of cells was made. CD4+ T cells from CD45.1 congenic mice were labeled with Cell Trace Violet and added to Tregs. BMDCs were added on top of Treg and congenic responders with 0.5 μg/mL anti-CD3. In some experiments, anti-IL-10 was added to a final concentration of 10 μg/mL. For assessment of Tregs ex vivo, cells were purified from the spleen and peripheral lymph nodes of C57BL/6 mice using the EasySep CD4+ negative selection followed by CD25 positive selection to yield 99.9% CD4+CD25+ T cells). Following purification, cells were treated with 50 μM BMS309403 for 90 min at 37°C + 5% CO2. Conventional T cells and APCs were purified from C57BL/6 congenic mice and labeled with Cell Trace Violet. A serial dilution of Tregs was made before conventional T cells and APCs were added for 72 h. Suppression was calculated as:
Human Treg Suppression Assay
Naive thymic derived Tregs were isolated from human PBMCs by CD25 bead enrichment followed by FACS for CD4+CD252+CD127-CD45RA+ naive Tregs. Cells were initially expanded on KT64/86 cells for two weeks before freezing until required (Hippen et al., 2011). For the assays shown, tTreg were thawed, re-stimulated with anti-CD3/28 beads, and expanded for 10 days. Prior to the suppression assays, tTregs were harvested and incubated for 30 min with 250 μM BMS309403 at 37°C + 5% CO2. Cells were washed before the assay to remove any residual BMS309403. To assess suppression, PBMCs were purified, labeled with CFSE (carboxyfluorescein succinimidly ester) (Invitrogen, Carlsbad, CA), and stimulated with anti-CD3 mAb coated beads (Dynal, Invitrogen, Calrsbad, CA) ± tTreg (1:4 to 1:16 tTregs/PBMCs). On day 4, cells were stained with antibodies to CD4 and CD8 and suppression was determined from the Division Index (FlowJo, Treestar)
Quantification and Statistical Analysis
Flow cytometry data was analyzed using FlowJo 10 (BD Biosciences). Statistical Analysis was performed using Prism 6 software (GraphPad) and results are represented as Mean ± SEM, unless otherwise indicated. Comparisons for two groups were calculated using unpaired two-tailed Student’s t tests. Comparisons of more than two groups were calculated using one-way ANOVA with Bonferroni’s multiple comparison tests. Comparisons of grouped data were calculated using two-way ANOVA with Bonferroni’s multiple comparisons tests. We observed normal distribution and no difference in variance between groups in individual comparisons. Selection of sample size was based on extensive experience with metabolic assays. The sample size for in vivo tumor experiments was based on previous experience with the tumor cell line. The number of independent experiments performed, and the p values for each experiment are reported in the corresponding figure legends. For both in vitro and in vivo experiments, no initial exclusion criteria were used and no animals or replicates were excluded from the study.
Data and Code Availability
The RNA sequencing data generated in this study have been deposited in the Gene Expression Omnibus (GEO) repository (accession number: GEO: GSE126245).
Acknowledgments
We thank G. Hotamisligil for sharing the Fabp4/5 dKO mice and Johan Friden for assistance with the graphical abstract. This work was supported by grants from the National Institutes of Health R01 HL11879 and R37 AI34495 (B.R.B.), P01 CA065493 (B.R.B.; K.L.H.), R01 AI110481 (E.J.P.), R01 CA181125 (E.L.P.), and the Max Planck Society. F.B. and M.C. are supported by a Humboldt Fellowship from the Humboldt Foundation. D.J.P. is supported by a Sir Henry Wellcome Fellowship from the Wellcome Trust.
Author Contributions
C.S.F., F.B., R.L.K., D.E.S., K.L.H., M.L., E.J.P., B.R.B., and E.L.P. designed the research and analyzed data. C.S.F., F.B., R.L.K., A.M.C., G.T., D.E.S., K.L.H., M.L., D.J.P., M.C., J.E.-H., and K.M.G. performed experiments and analyzed data. C.S.F. and E.L.P. wrote the manuscript.
Declaration of Interests
E.L.P. is a SAB member of Immunomet, and E.L.P. and E.J.P. are founders of Rheos Medicines. B.R.B. receives remuneration as an advisor to Kamon Pharmaceuticals, Inc., Five Prime Therapeutics Inc., Regeneron Pharmaceuticals, Magenta Therapeutics, and BlueRock Therapeutics; research support from Fate Therapeutics, RXi Pharmaceuticals, Alpine Immune Sciences, Inc., Abbvie Inc., BlueRock Therapeutics Leukemia and Lymphoma Society, Childrens’ Cancer Research Fund, and KidsFirst Fund and is a co-founder of Tmunity.
Published: December 26, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cmet.2019.11.021.
Supplemental Information
References
- Abe Y., Sakairi T., Beeson C., Kopp J.B. TGF-β1 stimulates mitochondrial oxidative phosphorylation and generation of reactive oxygen species in cultured mouse podocytes, mediated in part by the mTOR pathway. Am. J. Physiol. Renal Physiol. 2013;305:F1477–F1490. doi: 10.1152/ajprenal.00182.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afgan E., Baker D., van den Beek M., Blankenberg D., Bouvier D., Čech M., Chilton J., Clements D., Coraor N., Eberhard C. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44(W1):W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baguley B.C. Antivascular therapy of cancer: DMXAA. Lancet Oncol. 2003;4:141–148. doi: 10.1016/s1470-2045(03)01018-0. [DOI] [PubMed] [Google Scholar]
- Balendiran G.K., Schnutgen F., Scapin G., Borchers T., Xhong N., Lim K., Godbout R., Spener F., Sacchettini J.C. Crystal structure and thermodynamic analysis of human brain fatty acid-binding protein. J. Biol. Chem. 2000;275:27045–27054. doi: 10.1074/jbc.M003001200. [DOI] [PubMed] [Google Scholar]
- Battaglia M., Stabilini A., Tresoldi E. Expanding human T regulatory cells with the mTOR-inhibitor rapamycin. Methods Mol. Biol. 2012;821:279–293. doi: 10.1007/978-1-61779-430-8_17. [DOI] [PubMed] [Google Scholar]
- Beier U.H., Angelin A., Akimova T., Wang L., Liu Y., Xiao H., Koike M.A., Hancock S.A., Bhatti T.R., Han R. Essential role of mitochondrial energy metabolism in Foxp3+ T-regulatory cell function and allograft survival. FASEB J. 2015;29:2315–2326. doi: 10.1096/fj.14-268409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berod L., Friedrich C., Nandan A., Freitag J., Hagemann S., Harmrolfs K., Sandouk A., Hesse C., Castro C.N., Bähre H. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat. Med. 2014;20:1327–1333. doi: 10.1038/nm.3704. [DOI] [PubMed] [Google Scholar]
- Buck M.D., O’Sullivan D., Klein Geltink R.I., Curtis J.D., Chang C.H., Sanin D.E., Qiu J., Kretz O., Braas D., van der Windt G.J. Mitochondrial dynamics controls t cell fate through metabolic programming. Cell. 2016;166:63–76. doi: 10.1016/j.cell.2016.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrio R., Bathe O.F., Malek T.R. Initial antigen encounter programs CD8+ T cells competent to develop into memory cells that are activated in an antigen-free, IL-7- and IL-15-rich environment. J. Immunol. 2004;172:7315–7323. doi: 10.4049/jimmunol.172.12.7315. [DOI] [PubMed] [Google Scholar]
- Chang C.H., Qiu J., O’Sullivan D., Buck M.D., Noguchi T., Curtis J.D., Chen Q., Gindin M., Gubin M.M., van der Windt G.J. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell. 2015;162:1229–1241. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary B., Elkord E. Regulatory t cells in the tumor microenvironment and cancer progression: role and therapeutic targeting. Vaccines (Basel) 2016;4:E28. doi: 10.3390/vaccines4030028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry A., Samstein R.M., Treuting P., Liang Y., Pils M.C., Heinrich J.M., Jack R.S., Wunderlich F.T., Brüning J.C., Müller W., Rudensky A.Y. Interleukin-10 signaling in regulatory T cells is required for suppression of Th17 cell-mediated inflammation. Immunity. 2011;34:566–578. doi: 10.1016/j.immuni.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmurzyńska A. The multigene family of fatty acid-binding proteins (FABPs): function, structure and polymorphism. J. Appl. Genet. 2006;47:39–48. doi: 10.1007/BF03194597. [DOI] [PubMed] [Google Scholar]
- Civiletto G., Varanita T., Cerutti R., Gorletta T., Barbaro S., Marchet S., Lamperti C., Viscomi C., Scorrano L., Zeviani M. Opa1 overexpression ameliorates the phenotype of two mitochondrial disease mouse models. Cell Metab. 2015;21:845–854. doi: 10.1016/j.cmet.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogliati S., Frezza C., Soriano M.E., Varanita T., Quintana-Cabrera R., Corrado M., Cipolat S., Costa V., Casarin A., Gomes L.C. Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell. 2013;155:160–171. doi: 10.1016/j.cell.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cretenet G., Clerc I., Matias M., Loisel S., Craveiro M., Oburoglu L., Kinet S., Mongellaz C., Dardalhon V., Taylor N. Cell surface Glut1 levels distinguish human CD4 and CD8 T lymphocyte subsets with distinct effector functions. Sci. Rep. 2016;6:24129. doi: 10.1038/srep24129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rosa V., Galgani M., Porcellini A., Colamatteo A., Santopaolo M., Zuchegna C., Romano A., De Simone S., Procaccini C., La Rocca C. Glycolysis controls the induction of human regulatory T cells by modulating the expression of FOXP3 exon 2 splicing variants. Nat. Immunol. 2015;16:1174–1184. doi: 10.1038/ni.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot J.D., Gavin M.A., Rudensky A.Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- Furuhashi M., Hotamisligil G.S. Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat. Rev. Drug Discov. 2008;7:489–503. doi: 10.1038/nrd2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuhashi M., Tuncman G., Görgün C.Z., Makowski L., Atsumi G., Vaillancourt E., Kono K., Babaev V.R., Fazio S., Linton M.F. Treatment of diabetes and atherosclerosis by inhibiting fatty-acid-binding protein aP2. Nature. 2007;447:959–965. doi: 10.1038/nature05844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerriets V.A., Kishton R.J., Nichols A.G., Macintyre A.N., Inoue M., Ilkayeva O., Winter P.S., Liu X., Priyadharshini B., Slawinska M.E. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J. Clin. Invest. 2015;125:194–207. doi: 10.1172/JCI76012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerriets V.A., Kishton R.J., Johnson M.O., Cohen S., Siska P.J., Nichols A.G., Warmoes M.O., de Cubas A.A., MacIver N.J., Locasale J.W. Foxp3 and Toll-like receptor signaling balance Treg cell anabolic metabolism for suppression. Nat. Immunol. 2016;17:1459–1466. doi: 10.1038/ni.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J.A., Hall J.A., Sun C.M., Cai Q., Ghyselinck N., Chambon P., Belkaid Y., Mathis D., Benoist C. Retinoic acid enhances Foxp3 induction indirectly by relieving inhibition from CD4+CD44hi Cells. Immunity. 2008;29:758–770. doi: 10.1016/j.immuni.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippen K.L., Merkel S.C., Schirm D.K., Sieben C.M., Sumstad D., Kadidlo D.M., McKenna D.H., Bromberg J.S., Levine B.L., Riley J.L. Massive ex vivo expansion of human natural regulatory T cells (T(regs)) with minimal loss of in vivo functional activity. Sci. Transl. Med. 2011;3:83ra41. doi: 10.1126/scitranslmed.3001809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho P.C., Bihuniak J.D., Macintyre A.N., Staron M., Liu X., Amezquita R., Tsui Y.C., Cui G., Micevic G., Perales J.C. Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor t cell responses. Cell. 2015;162:1217–1228. doi: 10.1016/j.cell.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohoff C., Börchers T., Rüstow B., Spener F., van Tilbeurgh H. Expression, purification, and crystal structure determination of recombinant human epidermal-type fatty acid binding protein. Biochemistry. 1999;38:12229–12239. doi: 10.1021/bi990305u. [DOI] [PubMed] [Google Scholar]
- Ikon N., Ryan R.O. Cardiolipin and mitochondrial cristae organization. Biochim. Biophys. Acta Biomembr. 2017;1859:1156–1163. doi: 10.1016/j.bbamem.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano Y., Zavidij O., Park J., Moschetta M., Kokubun K., Mouhieddine T.H., Manier S., Mishima Y., Murakami N., Bustoros M. Blocking IFNAR1 inhibits multiple myeloma-driven Treg expansion and immunosuppression. J. Clin. Invest. 2018;128:2487–2499. doi: 10.1172/JCI88169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Smith S.B., Yoon Y. The short variant of the mitochondrial dynamin OPA1 maintains mitochondrial energetics and cristae structure. J. Biol. Chem. 2017;292:7115–7130. doi: 10.1074/jbc.M116.762567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G.S., Pan Y., Scanlon M.J., Porter C.J.H., Nicolazzo J.A. Fatty acid-binding protein 5 mediates the uptake of fatty acids, but not drugs, into human brain endothelial cells. J. Pharm. Sci. 2018;107:1185–1193. doi: 10.1016/j.xphs.2017.11.024. [DOI] [PubMed] [Google Scholar]
- Li M.O., Wan Y.Y., Flavell R.A. T cell-produced transforming growth factor-beta1 controls T cell tolerance and regulates Th1- and Th17-cell differentiation. Immunity. 2007;26:579–591. doi: 10.1016/j.immuni.2007.03.014. [DOI] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liu R.Z., Mita R., Beaulieu M., Gao Z., Godbout R. Fatty acid binding proteins in brain development and disease. Int. J. Dev. Biol. 2010;54:1229–1239. doi: 10.1387/ijdb.092976rl. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maj T., Wang W., Crespo J., Zhang H., Wang W., Wei S., Zhao L., Vatan L., Shao I., Szeliga W. Oxidative stress controls regulatory T cell apoptosis and suppressor activity and PD-L1-blockade resistance in tumor. Nat. Immunol. 2017;18:1332–1341. doi: 10.1038/ni.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metidji A., Rieder S.A., Glass D.D., Cremer I., Punkosdy G.A., Shevach E.M. IFN-α/β receptor signaling promotes regulatory T cell development and function under stress conditions. J. Immunol. 2015;194:4265–4276. doi: 10.4049/jimmunol.1500036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalek R.D., Gerriets V.A., Jacobs S.R., Macintyre A.N., MacIver N.J., Mason E.F., Sullivan S.A., Nichols A.G., Rathmell J.C. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J. Immunol. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore S.M., Holt V.V., Malpass L.R., Hines I.N., Wheeler M.D. Fatty acid-binding protein 5 limits the anti-inflammatory response in murine macrophages. Mol. Immunol. 2015;67(2 Pt B):265–275. doi: 10.1016/j.molimm.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mot A.I., Liddell J.R., White A.R., Crouch P.J. Circumventing the Crabtree Effect: a method to induce lactate consumption and increase oxidative phosphorylation in cell culture. Int. J. Biochem. Cell Biol. 2016;79:128–138. doi: 10.1016/j.biocel.2016.08.029. [DOI] [PubMed] [Google Scholar]
- Mucida D., Pino-Lagos K., Kim G., Nowak E., Benson M.J., Kronenberg M., Noelle R.J., Cheroutre H. Retinoic acid can directly promote TGF-beta-mediated Foxp3(+) Treg cell conversion of naive T cells. Immunity. 2009;30:471–472. doi: 10.1016/j.immuni.2009.03.008. author reply 472–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton R., Priyadharshini B., Turka L.A. Immunometabolism of regulatory T cells. Nat. Immunol. 2016;17:618–625. doi: 10.1038/ni.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill L.A., Kishton R.J., Rathmell J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 2016;16:553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan D., van der Windt G.J., Huang S.C., Curtis J.D., Chang C.H., Buck M.D., Qiu J., Smith A.M., Lam W.Y., DiPlato L.M. Memory CD8(+) T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity. 2014;41:75–88. doi: 10.1016/j.immuni.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino H., Nakamura K., Iwasa T., Ihara E., Akiho H., Motomura Y., Akahoshi K., Igarashi H., Kato M., Kotoh K. Regulatory T cells expanded by rapamycin in vitro suppress colitis in an experimental mouse model. J. Gastroenterol. 2012;47:366–376. doi: 10.1007/s00535-011-0502-y. [DOI] [PubMed] [Google Scholar]
- Pacella I., Procaccini C., Focaccetti C., Miacci S., Timperi E., Faicchia D., Severa M., Rizzo F., Coccia E.M., Bonacina F. Fatty acid metabolism complements glycolysis in the selective regulatory T cell expansion during tumor growth. Proc. Natl. Acad. Sci. USA. 2018;115:E6546–E6555. doi: 10.1073/pnas.1720113115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Tian T., Park C.O., Lofftus S.Y., Mei S., Liu X., Luo C., O’Malley J.T., Gehad A., Teague J.E. Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature. 2017;543:252–256. doi: 10.1038/nature21379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradies G., Paradies V., Ruggiero F.M., Petrosillo G. Oxidative stress, cardiolipin and mitochondrial dysfunction in nonalcoholic fatty liver disease. World J. Gastroenterol. 2014;20:14205–14218. doi: 10.3748/wjg.v20.i39.14205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce E.L., Pearce E.J. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013;38:633–643. doi: 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck B., Schug Z.T., Zhang Q., Dankworth B., Jones D.T., Smethurst E., Patel R., Mason S., Jiang M., Saunders R. Inhibition of fatty acid desaturation is detrimental to cancer cell survival in metabolically compromised environments. Cancer Metab. 2016;4:6. doi: 10.1186/s40170-016-0146-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piconese S., Pacella I., Timperi E., Barnaba V. Divergent effects of type-I interferons on regulatory T cells. Cytokine Growth Factor Rev. 2015;26:133–141. doi: 10.1016/j.cytogfr.2014.10.012. [DOI] [PubMed] [Google Scholar]
- Plitas G., Konopacki C., Wu K., Bos P.D., Morrow M., Putintseva E.V., Chudakov D.M., Rudensky A.Y. Regulatory T Cells Exhibit Distinct Features in Human Breast Cancer. Immunity. 2016;45:1122–1134. doi: 10.1016/j.immuni.2016.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prantner D., Perkins D.J., Lai W., Williams M.S., Sharma S., Fitzgerald K.A., Vogel S.N. 5,6-Dimethylxanthenone-4-acetic acid (DMXAA) activates stimulator of interferon gene (STING)-dependent innate immune pathways and is regulated by mitochondrial membrane potential. J. Biol. Chem. 2012;287:39776–39788. doi: 10.1074/jbc.M112.382986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyadharshini B., Loschi M., Newton R.H., Zhang J.W., Finn K.K., Gerriets V.A., Huynh A., Rathmell J.C., Blazar B.R., Turka L.A. Cutting edge: TGF-β and phosphatidylinositol 3-kinase signals modulate distinct metabolism of regulatory T cell subsets. J. Immunol. 2018;201:2215–2219. doi: 10.4049/jimmunol.1800311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procaccini C., Carbone F., Di Silvestre D., Brambilla F., De Rosa V., Galgani M., Faicchia D., Marone G., Tramontano D., Corona M. The proteomic landscape of human ex vivo regulatory and conventional T cells reveals specific metabolic requirements. Immunity. 2016;44:712. doi: 10.1016/j.immuni.2016.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44(W1) doi: 10.1093/nar/gkw257. W160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao E., Singh P., Li Y., Zhang Y., Chi Y.I., Suttles J., Li B. Targeting epidermal fatty acid binding protein for treatment of experimental autoimmune encephalomyelitis. BMC Immunol. 2015;16:28. doi: 10.1186/s12865-015-0091-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redpath S.A., van der Werf N., Cervera A.M., MacDonald A.S., Gray D., Maizels R.M., Taylor M.D. ICOS controls Foxp3(+) regulatory T-cell expansion, maintenance and IL-10 production during helminth infection. Eur. J. Immunol. 2013;43:705–715. doi: 10.1002/eji.201242794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolph M.S., Young T.R., Shum B.O., Gorgun C.Z., Schmitz-Peiffer C., Ramshaw I.A., Hotamisligil G.S., Mackay C.R. Regulation of dendritic cell function and T cell priming by the fatty acid-binding protein AP2. J. Immunol. 2006;177:7794–7801. doi: 10.4049/jimmunol.177.11.7794. [DOI] [PubMed] [Google Scholar]
- Rongvaux A., Jackson R., Harman C.C., Li T., West A.P., de Zoete M.R., Wu Y., Yordy B., Lakhani S.A., Kuan C.Y. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell. 2014;159:1563–1577. doi: 10.1016/j.cell.2014.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov Y.P., Rasmussen J.P., Chi E.Y., Fontenot J., Castelli L., Ye X., Treuting P., Siewe L., Roers A., Henderson W.R., Jr. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- Sakaguchi S. Naturally arising CD4+ regulatory t cells for immunologic self-tolerance and negative control of immune responses. Annu. Rev. Immunol. 2004;22:531–562. doi: 10.1146/annurev.immunol.21.120601.141122. [DOI] [PubMed] [Google Scholar]
- Scharwey M., Tatsuta T., Langer T. Mitochondrial lipid transport at a glance. J. Cell Sci. 2013;126:5317–5323. doi: 10.1242/jcs.134130. [DOI] [PubMed] [Google Scholar]
- Schmidt A., Oberle N., Krammer P.H. Molecular mechanisms of treg-mediated T cell suppression. Front. Immunol. 2012;3:51. doi: 10.3389/fimmu.2012.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug T.T., Berry D.C., Shaw N.S., Travis S.N., Noy N. Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell. 2007;129:723–733. doi: 10.1016/j.cell.2007.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug T.T., Berry D.C., Toshkov I.A., Cheng L., Nikitin A.Y., Noy N. Overcoming retinoic acid-resistance of mammary carcinomas by diverting retinoic acid from PPARbeta/delta to RAR. Proc. Natl. Acad. Sci. USA. 2008;105:7546–7551. doi: 10.1073/pnas.0709981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.J., Thompson B.R., Sanders M.A., Bernlohr D.A. Interaction of the adipocyte fatty acid-binding protein with the hormone-sensitive lipase: regulation by fatty acids and phosphorylation. J. Biol. Chem. 2007;282:32424–32432. doi: 10.1074/jbc.M703730200. [DOI] [PubMed] [Google Scholar]
- Soukupova J., Malfettone A., Hyroššová P., Hernández-Alvarez M.I., Peñuelas-Haro I., Bertran E., Junza A., Capellades J., Giannelli G., Yanes O. Role of the transforming growth factor-β in regulating hepatocellular carcinoma oxidative metabolism. Sci. Rep. 2017;7:12486. doi: 10.1038/s41598-017-12837-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C.A., Metheny H., Iida N., Smith L., Hanson M., Steinhagen F., Leighty R.M., Roers A., Karp C.L., Müller W., Trinchieri G. Interferon-dependent IL-10 production by Tregs limits tumor Th17 inflammation. J. Clin. Invest. 2013;123:4859–4874. doi: 10.1172/JCI65180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storch J., Corsico B. The emerging functions and mechanisms of mammalian fatty acid-binding proteins. Annu. Rev. Nutr. 2008;28:73–95. doi: 10.1146/annurev.nutr.27.061406.093710. [DOI] [PubMed] [Google Scholar]
- Storch J., McDermott L. Structural and functional analysis of fatty acid-binding proteins. J. Lipid Res. 2009;50(Suppl):S126–S131. doi: 10.1194/jlr.R800084-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulsky R., Magnin D.R., Huang Y., Simpkins L., Taunk P., Patel M., Zhu Y., Stouch T.R., Bassolino-Klimas D., Parker R. Potent and selective biphenyl azole inhibitors of adipocyte fatty acid binding protein (aFABP) Bioorg. Med. Chem. Lett. 2007;17:3511–3515. doi: 10.1016/j.bmcl.2006.12.044. [DOI] [PubMed] [Google Scholar]
- Szczesny B., Marcatti M., Ahmad A., Montalbano M., Brunyánszki A., Bibli S.I., Papapetropoulos A., Szabo C. Mitochondrial DNA damage and subsequent activation of Z-DNA binding protein 1 links oxidative stress to inflammation in epithelial cells. Sci. Rep. 2018;8:914. doi: 10.1038/s41598-018-19216-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinchieri G. Interleukin-10 production by effector T cells: Th1 cells show self control. J. Exp. Med. 2007;204:239–243. doi: 10.1084/jem.20070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerkamp J.H., van Moerkerk H.T., Prinsen C.F., van Kuppevelt T.H. Structural and functional studies on different human FABP types. Mol. Cell. Biochem. 1999;192:137–142. [PubMed] [Google Scholar]
- Vocanson M., Rozieres A., Hennino A., Poyet G., Gaillard V., Renaudineau S., Achachi A., Benetiere J., Kaiserlian D., Dubois B. Inducible costimulator (ICOS) is a marker for highly suppressive antigen-specific T cells sharing features of TH17/TH1 and regulatory T cells. J Allergy Clin. Immunol. 2010;126:280–289. doi: 10.1016/j.jaci.2010.05.022. 289.e1-7. [DOI] [PubMed] [Google Scholar]
- Wang H., Franco F., Ho P.C. Metabolic regulation of Tregs in cancer: opportunities for immunotherapy. Trends Cancer. 2017;3:583–592. doi: 10.1016/j.trecan.2017.06.005. [DOI] [PubMed] [Google Scholar]
- Weinberg S.E., Singer B.D., Steinert E.M., Martinez C.A., Mehta M.M., Martínez-Reyes I., Gao P., Helmin K.A., Abdala-Valencia H., Sena L.A. Mitochondrial complex III is essential for suppressive function of regulatory T cells. Nature. 2019;565:495–499. doi: 10.1038/s41586-018-0846-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A.P., Khoury-Hanold W., Staron M., Tal M.C., Pineda C.M., Lang S.M., Bestwick M., Duguay B.A., Raimundo N., MacDuff D.A. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520:553–557. doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M.J., McArthur K., Metcalf D., Lane R.M., Cambier J.C., Herold M.J., van Delft M.F., Bedoui S., Lessene G., Ritchie M.E. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell. 2014;159:1549–1562. doi: 10.1016/j.cell.2014.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H., Yang K., Cloer C., Neale G., Vogel P., Chi H. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature. 2013;499:485–490. doi: 10.1038/nature12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Guan Z., Murphy A.N., Wiley S.E., Perkins G.A., Worby C.A., Engel J.L., Heacock P., Nguyen O.K., Wang J.H. Mitochondrial phosphatase PTPMT1 is essential for cardiolipin biosynthesis. Cell Metab. 2011;13:690–700. doi: 10.1016/j.cmet.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Yuan S., Cheng G., Guo B. Type I IFN promotes IL-10 production from T cells to suppress Th17 cells and Th17-associated autoimmune inflammation. PLoS ONE. 2011;6:e28432. doi: 10.1371/journal.pone.0028432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Sun Y., Rao E., Yan F., Li Q., Zhang Y., Silverstein K.A., Liu S., Sauter E., Cleary M.P., Li B. Fatty acid-binding protein E-FABP restricts tumor growth by promoting IFN-β responses in tumor-associated macrophages. Cancer Res. 2014;74:2986–2998. doi: 10.1158/0008-5472.CAN-13-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Kestell P., Baguley B.C., Paxton J.W. 5,6-dimethylxanthenone-4-acetic acid (DMXAA): a new biological response modifier for cancer therapy. Invest. New Drugs. 2002;20:281–295. doi: 10.1023/a:1016215015530. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing data generated in this study have been deposited in the Gene Expression Omnibus (GEO) repository (accession number: GEO: GSE126245).