Abstract
CRISPR activation (CRISPRa) is a burgeoning technology for programmable gene activation, but its potential for tissue regeneration has yet to be fully explored. Bone marrow-derived mesenchymal stem cells (BMSCs) can differentiate into osteogenic or adipogenic pathways, which are governed by the Wnt (Wingless-related integration site) signaling cascade. To promote BMSC differentiation toward osteogenesis and improve calvarial bone healing by BMSCs, we harnessed a highly efficient hybrid baculovirus vector for gene delivery and exploited a synergistic activation mediator (SAM)-based CRISPRa system to activate Wnt10b (that triggers the canonical Wnt pathway) and forkhead c2 (Foxc2) (that elicits the noncanonical Wnt pathway) in BMSCs. We constructed a Bac-CRISPRa vector to deliver the SAM-based CRISPRa system into rat BMSCs. We showed that Bac-CRISPRa enabled CRISPRa delivery and potently activated endogenous Wnt10b and Foxc2 expression in BMSCs for >14 days. Activation of Wnt10b or Foxc2 alone was sufficient to promote osteogenesis and repress adipogenesis in vitro. Furthermore, the robust and prolonged coactivation of both Wnt10b and Foxc2 additively enhanced osteogenic differentiation while inhibiting adipogenic differentiation of BMSCs. The CRISPRa-engineered BMSCs with activated Wnt10b and Foxc2 remarkably improved the calvarial bone healing after implantation into the critical-sized calvarial defects in rats. These data implicate the potentials of CRISPRa technology for bone tissue regeneration.
Keywords: baculovirus, bone regeneration, CRISPRa, gene activation, Wnt10b, Foxc2
The potential of CRISPR activation (CRISPRa) for tissue regeneration remains to be explored. Hsu et al. developed a CRISPRa system delivered by a hybrid baculovirus, which enabled robust and prolonged coactivation of Wnt10b and Foxc2 in stem cells, thereby promoting osteogenesis in vitro and improving bone healing in vivo.
Introduction
Calvarial bone formation occurs in young infants, but adults lose the ability to generate calvarial bones, hence making the repair of large calvarial defects a challenging task.1 Bone repair requires complex interplay among osteoprogenitor cells, osteoinductive growth factors, osteoconductive matrix, and angiogenesis.2 To stimulate bone healing, osteoinductive genes can be delivered into osteoprogenitor cells, such as bone marrow-derived mesenchymal stem cells (BMSCs), to trigger osteogenic differentiation, followed by seeding into the osteoconductive scaffold and implantation into the defects. However, these approaches often fail to produce satisfactory calvarial bone healing.3,4
BMSCs are able to differentiate into competing lineages, including osteogenic and adipogenic pathways, and hold great promise in bone regeneration. One key to control the reciprocal relationship between osteogenic and adipogenic lineages is Wnt signaling, which stimulates osteogenesis and represses adipogenesis.5 Therefore, activation of the Wnt signaling cascade increases bone mass6 and improves bone healing.7 In this regard, WNT10B is one of the 19 secreted Wnt ligands and can bind to its cognate surface receptor to trigger the canonical Wnt signaling cascade. Conversely, Forkhead c2 (FOXC2) is a transcription factor that triggers the noncanonical Wnt pathway8 and also promotes osteogenesis and inhibits adipogenesis.9
CRISPR is a customizable, RNA-guided system that uses Cas9 nuclease and single-guide RNA (sgRNA) for targeted genome editing and has been explored for various applications.10, 11, 12, 13 Cas9 can be mutated to become a catalytically deactivated Cas9 (dCas9) protein and fused with a transcription activator (e.g., VP64) for CRISPR activation (CRISPRa) of the target gene.14 To potentiate the magnitude of stimulation, a synergistic activation mediator (SAM)-based CRISPRa system was developed,15 which consists of dCas9-VP64, a scaffold sgRNA, and an MPH (MS2 coat protein [MCP], p65, and activating domain of heat shock factor 1 [HSF1]) fusion protein effector. The scaffold sgRNA comprises a dCas9 binding domain, a spacer sequence that can be custom designed to bind the specific gene of interest, and two extra MS2 aptamers that recognize MCP. By expressing these components in the same cell, the scaffold sgRNA orchestrates with dCas9-VP64 to locate the genomic loci and recruits the MPH complex by the binding between MS2 aptamer and MCP to activate the gene expression. Such SAM-based CRISPRa has been exploited for basic research purposes,16, 17, 18 but its potential in tissue regeneration has yet to be fully explored. Moreover, delivery of the complex SAM-based CRISPRa system into same cells is difficult for the commonly used adeno-associated virus (AAV) and lentivirus, due to limited packaging capacity.
In contrast, baculovirus is a nonpathogenic insect virus with a packaging capacity exceeding 38 kb, due to its large genome (≈134 kb).19 Baculovirus can carry (transduce) transgenes into stem cells at efficiencies exceeding 95%,20, 21, 22 which is significantly more efficient than nonviral vectors. Therefore, baculovirus has been harnessed to modify stem cells genetically for the regeneration of cartilage,23,24 bone,25,26 nerve,27 and heart.28 Baculovirus neither replicates nor integrates its genome into the chromosomes of transduced cells, which minimizes possible genotoxicity but restricts the duration of transgene expression.19 To prolong the transgene expression, we previously developed a hybrid system comprising two baculovirus vectors: one expressing Cre recombinase (Bac-Cre) and the other substrate baculovirus harboring the transgene cassette flanked by loxP (locus of X-over P1) sites.29 After cotransduction of BMSCs with the two baculovirus vectors, the expressed Cre recognizes the loxP sequences and excises the loxP-flanking transgene cassette off the substrate baculovirus genome. The excised transgene cassette re-circularizes and forms an episomal DNA minicircle encompassing the transgene within the cells.29,30 This Cre/loxP-based hybrid baculovirus system was exploited to express a microRNA sponge or growth factor to augment stem cell differentiation and bone healing in vivo.7,31
Given the potentials of Wnt10b and Foxc2 in stimulating osteogenic differentiation, here, we explored the SAM-based CRISPRa to upregulate Wnt10b and Foxc2 selectively in BMSCs, alone or in combination, in attempts to stimulate calvarial bone regeneration. Given the large cloning capacity of the baculovirus, we constructed the Cre/loxP-based hybrid baculovirus to deliver the SAM-based CRISPRa system into rat BMSCs. We showed that the hybrid baculovirus robustly activated endogenous Wnt10b and Foxc2 for a prolonged period of time. Coactivation of Wnt10b and Foxc2 effectively stimulated osteogenesis and repressed adipogenesis in vitro. Implantation of the CRISPRa-engineered BMSCs into the critical-sized calvarial defects significantly improved the bone healing.
Results
Design of Baculovirus System (Bac-CRISPRa) for CRISPRa Delivery
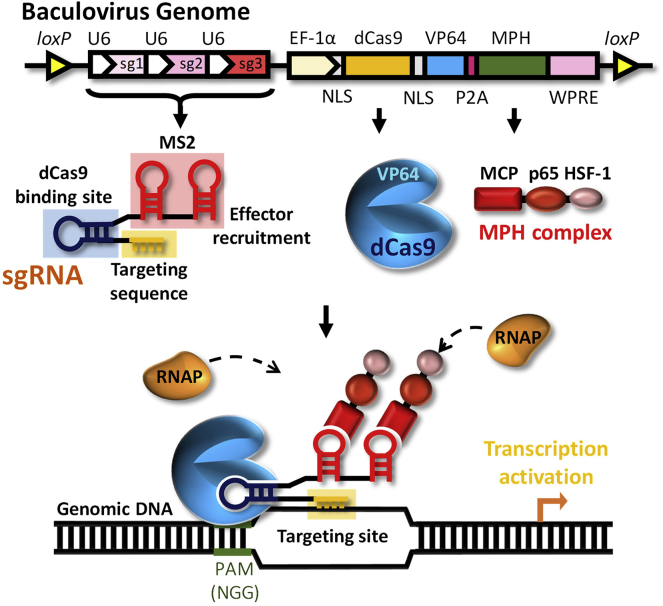
We first designed a baculovirus platform (Bac-CRISPRa) to deliver the SAM-based CRISPRa system for targeted activation of distinct endogenous genes (Figure 1). The baculovirus platform was designed to coexpress dCas9-VP64 and the MPH complex under the control of the rat elongation factor 1α (EF-1α) promoter. The dCas9-VP64 fusion protein contained two flanking nuclear localization signals (NLSs) for nuclear transport. The MPH complex consisted of MCP, p65, and HSF1 for gene activation and was separated from dCas9-VP64 by the porcine teschovirus-1 2A (P2A) peptide. Additionally, the baculovirus expressed three sgRNA under the human U6 (hU6) promoter. Each of the sgRNA comprised distinct targeting (spacer) sequences but identical dCas9 binding domain and two MS2 binding aptamers for MPH recruitment. We chose to express an array of three sgRNA to target the same gene because multiple sgRNA together with dCas9-VP64 potentiates the gene activation effects.32,33 We envisioned that baculovirus transduction would confer coexpression of dCas9-VP64, MPH complex, and three sgRNA in the same cells, after which, dCas9-VP64 associates with each sgRNA, binds to the specific genomic DNA sequence, and recruits the MPH complex and RNA polymerase (RNAP) to activate the gene expression (Figure 1). To prolong the effect of gene activation, we adopted the hybrid Cre/loxP-based baculovirus system and added two loxP sequences to flank the entire CRISPRa cassette.
Figure 1.
Bac-CRISPRa System for Endogenous Gene Activation
The CRISPRa system consists of dCas9-VP64, the MPH (MCP-p65-HSF1) activation effector, and the sgRNA array. The entire CRISPRa system was flanked by loxP sequences and encoded in the baculovirus genome. The dCas9 (with 5′ and 3′ NLS sequences), fused with the VP64 activation domain (dCas9-VP64), and MPH were separated by the P2A sequence and were driven by the rat CMV enhancer/rat EF-1α promoter. The three sgRNA expression cassettes consisted of the hU6 promoter, sgRNA backbone with two MS2 coat protein (MCP) binding motifs, and the spacers targeting different sequences of interest. Baculovirus transduction would confer coexpression of dCas9-VP64, the MPH complex, and three sgRNAs in the same cells, after which, dCas9-VP64 associates with each sgRNA, binds to the specific genomic DNA sequence, and recruits the MPH complex and RNA polymerase (RNAP) to activate the gene expression. PAM, protospacer adjacent motif; WPRE, woodchuck hepatitis post-transcriptional response element (for enhancing mRNA stability).
Construction and Validation of Baculoviruses for Wnt10b and Foxc2 Activation
We constructed two baculoviruses (Figure 2A) expressing identical dCas9-VP64 and MPH but distinct arrays of three sgRNAs targeting different regions of endogenous Wnt10b (Bac-W10b) and Foxc2 (Bac-Foxc2). Spacer sequences on the sgRNA were designed using an online tool with windows from ≈−400 to ≈−50 bp, relative to the transcription start site of target genes (Figure 2A; Table S1). Cotransduction of cells with the hybrid Bac-W10b (or Bac-Foxc2) and Bac-Cre29 that expressed the Cre recombinase (Figure 2A) would lead to the Cre-mediated excision of the CRISPRa system off the hybrid baculovirus, recircularization, and formation of the DNA minicircle (≈10 kb) encompassing the CRISPRa system (Figure S1).
Figure 2.
Construction and Validation of Baculoviruses for Wnt10b and Foxc2 Activation
(A) Schematic of Bac-W10b, Bac-Foxc2, and Bac-Cre vectors. Bac-W10b and Bac-Foxc2 harbored the expression cassettes for dCas9-VP64, MPH, and three sgRNAs to target the upstream of transcription start sites of Wnt10b (−74, −95, and −394) and Foxc2 (−97, −162, and −219). Bac-Cre expressed Cre recombinase under the rat EF-1α promoter. (B) Wnt10b expression. (C) Foxc2 expression. Rat BMSCs were singly transduced with Bac-W10b (MOI 200) or Bac-Foxc2 (MOI 200) or cotransduced with Bac-W10b/Bac-Cre (MOI 200/100) or Bac-Foxc2/Bac-Cre (MOI 200/100). The cells were analyzed for Wnt10b and Foxc2 transcription by quantitative real-time RT-PCR at 2, 5, 10, and 14 days post-transduction. The expression levels in the transduced cells were normalized to those in Mock-transduced cells to yield the relative gene expression. The data represent mean ± SD of three independent culture experiments. Student’s t test was used to analyze statistical significance.
After vector construction, we mock transduced or transduced rat BMSCs with different baculovirus combinations and analyzed the gene expression by quantitative real-time reverse transcriptase PCR (quantitative real-time RT-PCR) (Figures 2B and 2C). Compared with the Mock transduction control, single transduction with Bac-W10b (Wnt group) and Bac-Foxc2 (Foxc2 group) significantly activated the expression of Wnt10b (≈214.7-fold) and Foxc2 (≈3.4-fold) at 2 days post-transduction (dpt). Yet the magnitude of activation decreased with time due to the rapid degradation of the large baculovirus genome.21,29 In contrast, cotransduction with Bac-W10b/Bac-Cre (Wnt/Cre group) and Bac-Foxc2/Bac-Cre (Foxc2/Cre group) further substantiated the Wnt10b (≈274.6-fold) and Foxc2 (≈10.7-fold) activation at 2 dpt. At 14 dpt, the Wnt10b (7.8-fold) and Foxc2 (3.8-fold) in the Wnt/Cre and Foxc2/Cre groups still significantly (p < 0.05) exceeded those in the Wnt and Foxc2 groups. These data confirmed that the CRISPRa system delivered by the hybrid baculovirus effectively activated endogenous genes in BMSCs for a prolonged period of time.
Wnt10b and Foxc2 Activation by Bac-CRISPRa Promoted Osteogenesis and Inhibited Adipogenesis
To assess whether the activation of Wnt10b and Foxc2, alone or in combination, could improve osteogenic differentiation and suppress adipogenic differentiation, we cotransduced BMSCs using Bac-Cre with Bac-W10b (Wnt/Cre group), Bac-Foxc2 (Foxc2/Cre group), or Bac-W10b/Bac-Foxc2 (Wnt/Foxc2/Cre group). Cells were cultured for 14 days in osteoinduction medium to induce osteogenesis or in adipoinduction medium to trigger adipogenesis. Mock-transduced cells were cultured similarly and served as a control.
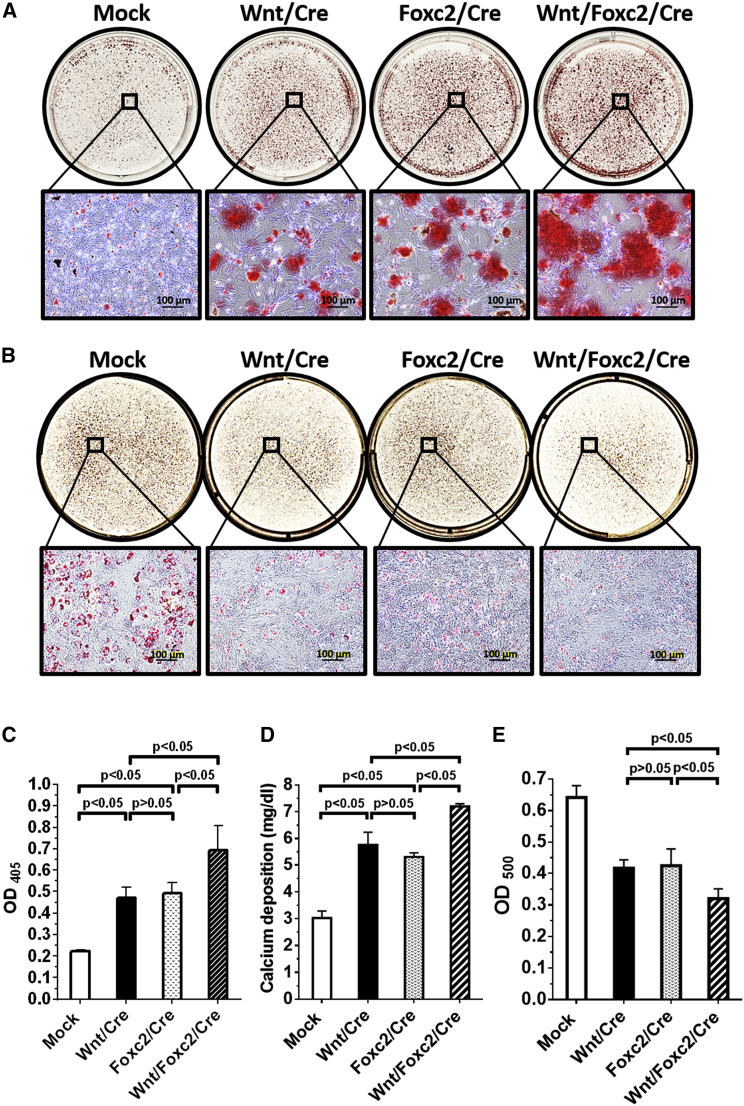
After 14 days of osteoinduction, Alizarin red staining illustrated evidently more matrix mineralization in the Wnt/Cre and Foxc2/Cre groups than in the Mock group (Figure 3A). After 14 days of adipoinduction, conversely, Oil Red O staining showed less oil-droplet accumulation in the Wnt/Cre and Foxc2/Cre groups than in the Mock group (Figure 3B). Quantification of matrix stained by Alizarin red (Figure 3C) and another calcium deposition assay (Figure 3D) further confirmed that the Wnt/Cre and Foxc2/Cre groups improved the mineralization. Likewise, quantification of oil droplet stained by Oil Red O attested that Wnt/Cre and Foxc2/Cre groups attenuated the oil-droplet formation (Figure 3E). Importantly, the qualitative (Figures 3A and 3B) and quantitative (Figures 3C–3E) analyses demonstrated that the Wnt/Foxc2/Cre group exhibited more potent matrix mineralization and less oil accumulation than the Wnt/Cre and Foxc2/Cre groups. These data altogether showed that the activation of endogenous Wnt10b or Foxc2 alone was sufficient to enhance osteogenesis and suppress adipogenesis in BMSCs, whereas coactivating Wnt10b and Foxc2 additively augmented the effects. Therefore, in subsequent experiments, we only compared the Wnt/Foxc2/Cre and Mock groups.
Figure 3.
Wnt10b and Foxc2 Activation Promoted Matrix Mineralization and Inhibited Oil Accumulation
(A) Alizarin red staining. (B) Oil Red O staining. (C) Quantification of Alizarin red staining. (D) Extracellular calcium deposition. (E) Quantification of Oil Red O staining. BMSCs were cotransduced with Bac-W10b/Bac-Cre (Wnt/Cre group) or Bac-Foxc2/ Bac-Cre (Foxc2/Cre group) at MOI 200/100. BMSCs were also transduced with Bac-W10b/Bac-Foxc2/Bac-Cre (Wnt/Foxc2/Cre group) at MOI 200/200/100. Cells were cultured for 14 days in osteoinduction medium to induce osteogenesis or in adipoinduction medium to trigger adipogenesis. Mock-transduced cells were cultured similarly and served as a control. At 14 dpt, the osteoinduced cells were stained with Alizarin red while the adipoinduced cells were stained with Oil Red O, followed by quantitative analysis. Alternatively, the osteoinduced cells were subject to a calcium deposition assay. The data represent mean ± SD of three independent culture experiments. Student’s t test was used to analyze statistical significance.
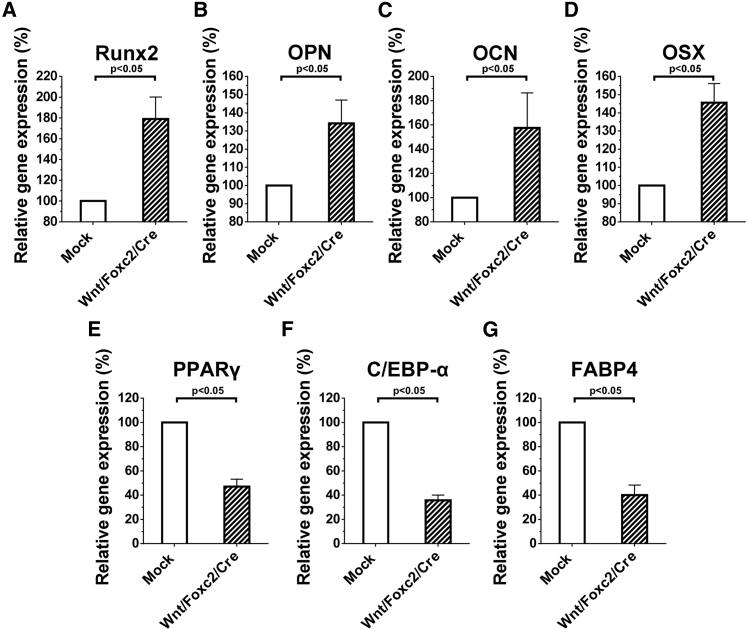
To gain more insights into the osteogenic differentiation, we repeated the osteoinduction experiments as in Figure 3A and analyzed the expression of various osteogenic markers by quantitative real-time RT-PCR: Runx2 (Runx2 runt-related transcription factor 2), Opn (osteopontin), Ocn (osteocalcin), and Osx (osterix). Figures 4A–4D depict that the Wnt/Foxc2/Cre group significantly (p < 0.05) upregulated the expression of all of these osteogenic marker genes. Likewise, we repeated the adipoinduction experiments as in Figure 3B and analyzed the expression of three adipogenic marker genes: Pparγ (peroxisome proliferator-activated receptor γ), C/ebp-α (CCAAT/enhancer binding protein α), and Fabp4 (fatty acid-binding protein 4) by quantitative real-time RT-PCR. Figures 4E–4G show that the Wnt/Foxc2/Cre group significantly (p < 0.05) attenuated the expression of all of these adipogenic genes.
Figure 4.
Upregulation of Osteogenic Genes and Downregulation of Adipogenic Genes by Wnt10b and Foxc2 Activation
(A) Runx2. (B) Opn. (C) Ocn. (D) Osx. (E) Pparγ. (F) C/ebp-α. (G) Fabp4. The rat BMSCs were transduced and cultured as in Figure 3. The gene-expression levels were analyzed by quantitative real-time RT-PCR at days 5 (Pparγ, C/ebp-α, Fabp4, and Runx2), 10 (Opn), or 14 (Ocn and Osx) and normalized to those of the mock-transduced BMSCs at the same day. The data represent mean ± SD of three independent culture experiments. Student’s t test was used to analyze statistical significance.
CRISPRa Coactivation of Wnt10b and Foxc2 Improved Calvarial Bone Healing
Figures 3 and 4 collectively demonstrate that CRISPRa-mediated coactivation of Wnt10b and Foxc2 in BMSCs augmented osteogenesis/mineralization and repressed adipogenesis in vitro. To demonstrate the in vivo healing, we transduced rat BMSCs with Bac-W10b/Bac-Foxc2/Bac-Cre (Wnt/Foxc2/Cre group), seeded the cells to gelatin scaffold, and implanted the cell/scaffold constructs to critical-size calvarial bone defects (6 mm in diameter) in rats (n = 7). As a control, mock-transduced BMSCs were seeded to scaffolds and implanted in the same manner (n = 5).
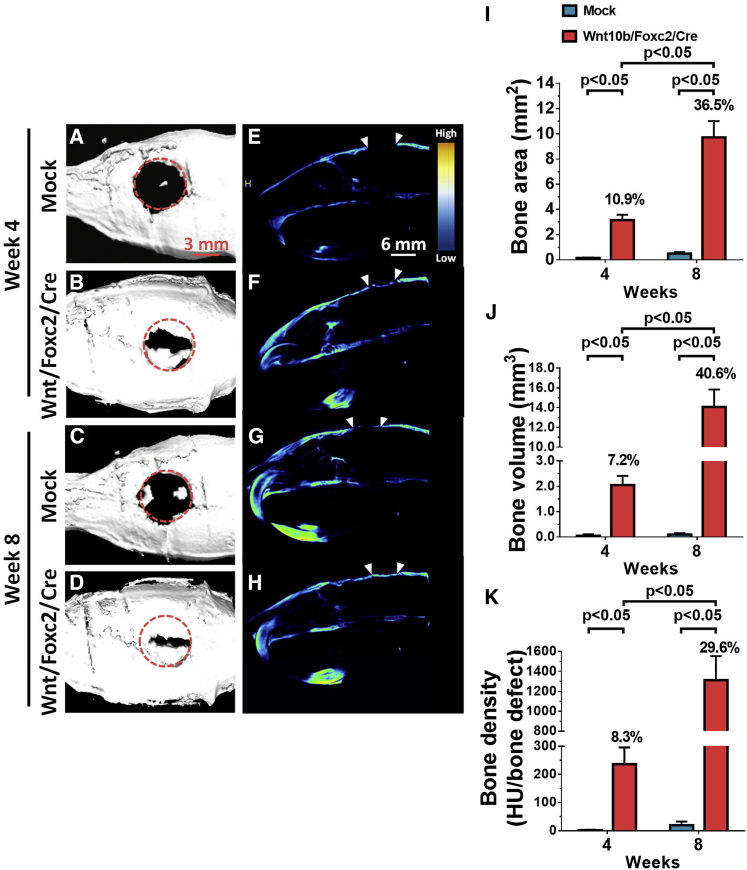
The top (Figures 5A–5D) and sagittal (Figures 5E–5H) views of micro-computed tomography (μCT) imaging illustrated that the Mock group barely triggered bone formation, even at week 8 (Figures 5C and 5G), indicating that BMSCs in gelatin scaffold were insufficient to heal such large calvarial bone defects. Conversely, the Wnt/Foxc2/Cre group elicited evident bone formation at the peripheral of the defect at week 4 (Figure 5B). The bone growth continued with enlarged bone area (Figure 5D) and apparent bone bridging (Figure 5H) at week 8. Quantitative analyses using the μCT images revealed only marginal increases in the bone area, bone volume, and bone density with time in the Mock group (Figures 5I–5K). In contrast, the bone area, volume, and density increased sharply with time in the Wnt/Foxc2/Cre group, filling 36.5% of the original defect area (Figure 5I) and ≈40.6% of the original defect volume (Figure 5J) at week 8, with the corresponding bone density (Figure 5K) reaching ≈29.6% that of the original defect.
Figure 5.
Calvarial Bone Healing Evaluated by μCT
(A–D) Top views of regenerated bones at weeks 4 and 8. (A) Mock, week 4. (B) Wnt/Foxc2/Cre, week 4. (C) Mock, week 8. (D) Wnt/Foxc2/Cre, week 8. (E–H) Sagittal views of regenerated bones at weeks 4 and 8. (E) Mock, week 4. (F) Wnt/Foxc2/Cre, week 4. (G) Mock, week 8. (H) Wnt/Foxc2/Cre, week 8. (I) Bone area. (J) Bone volume. (K) Bone density. BMSCs were mock transduced (Mock group, n = 5) or cotransduced with Bac-W10b/Bac-Foxc2/Bac-Cre (Wnt10b/Foxc2/Cre group, n = 7), seeded to gelatin scaffolds, and implanted into the calvarial bone defect (6 mm in diameter). The μCT images were captured at weeks 4 and 8 and analyzed for bone area, volume, and density. Two-way ANOVA was used to analyze statistical significance.
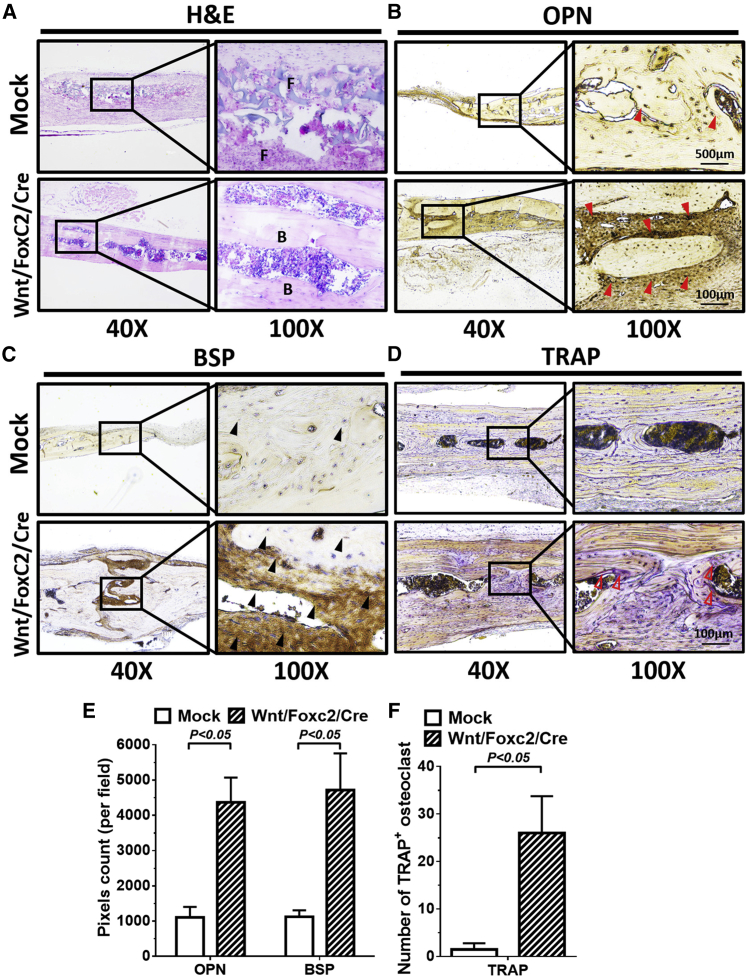
After μCT imaging at week 8, calvarial bone specimens were harvested for histochemical and immunohistochemical staining. As illustrated by H&E staining (Figure 6A), the Mock group was mostly filled with disordered fibrous tissues, whereas the Wnt/Foxc2/Cre group was filled with abundant ordered and organized bone matrix. OPN and bone sialoprotein (BSP) are markers of osteoblasts and osteocytes, respectively.34 Immunohistochemical staining demonstrated ample deposition of OPN (Figure 6B) and BSP (Figure 6C) in the Wnt/Foxc2/Cre groups but not in the Mock group.
Figure 6.
Histological and Immunohistochemical Staining Evaluated
(A) H&E staining. (B) Osteopontin (OPN) staining. (C) Bone sialoprotein (BSP) staining. (D) TRAP staining. (E) Statistical analysis of staining results (pixels count/field). The rats were sacrificed at 8 weeks postimplantation, and the calvarial bones were removed and sectioned for histological or immunohistochemical staining. (F) Statistical analysis of number of TRAP+ osteoclasts. Red arrowheads indicate OPN; black arrowheads indicate BSP. Open arrowheads indicate TRAP. F, fibrous tissue; B, bone tissue. For quantitative analysis, five fields from each section were analyzed using ImageJ software. Representative images of five animals are shown and the data represent the mean ± SD. Student’s t test was used to analyze statistical significance.
Bone remodeling is critical for successful bone repair in the long term and requires the osteoclast activity; thus, we also performed histochemical staining for tartrate-resistant acid phosphatase (TRAP), which is active in osteoclasts and is a marker of bone remodeling.35 Figure 6D illustrates much denser TRAP staining in the Wnt/Foxc2/Cre groups than in the Mock group. Quantitative analysis of the staining results confirmed that the Wnt/Foxc2/Cre group conferred more abundant accumulation of OPN and BSP (Figure 6E) and more TRAP+ osteoclasts (Figure 6F) than the Mock group. Figures 5 and 6 attest that coactivation of Wnt10b and Foxc2 in BMSCs improved the bone formation and remodeling processes.
Discussion
Since its advent, CRISPRa has been exploited for such applications as interrogation of gene regulatory networks,36,37 genetic screening,16,38, 39, 40 engineering of signaling pathways,41 and cell-fate manipulation.32,42 Despite the promise of CRISPRa in fundamental research in vitro, the potentials of CRISPRa for in vivo healing or regeneration of tissues remain poorly explored. One recent study harnessed CRISPRa to activate genes in vivo to promote the wound healing of corneal endothelial injury.43 Other studies employed CRISPRa to ameliorate the muscular dystrophy symptoms in mice44 or to stimulate the regeneration of injured sciatic nerves in rat models.45 More recently, we developed a CRISPRai system that enables simultaneous activation of Sox9 and repression of Pparγ in BMSCs, so as to promote bone healing after implantation of the engineered BMSCs.46
Here, we developed a Bac-CRISPRa vector that exploits the Cre/loxP-based hybrid baculovirus for the delivery of SAM-based CRISPRa into rat BMSCs to activate Wnt10b and Foxc2 (Figures 1 and 2A). Among the several CRISPRa systems, we chose the SAM-based CRISPRa system because it enables more robust gene activation than other CRISPRa systems.17,47 The hybrid Bac-CRISPRa system enabled efficient baculovirus-mediated gene delivery and formation of the minicircle that encompassed the CRISPRa system for gene activation (Figure S1). The resultant DNA minicircle (≈10 kb) was smaller than the baculoviral genome (≈134 kb) and was devoid of bacterial components,30 thus avoiding intracellular nuclease attack and allowing for prolonged existence and gene expression within the cells.7,27 With the same CRISPRa module (dCas9 and MPH) but different sgRNA combinations, the hybrid Bac-CRISPRa system robustly activated Wnt10b and Foxc2 for at least 14 days, albeit to varying degrees (274.6-fold versus 10.7-fold; Figures 2B and 2C). The sgRNA was designed following the same rules (targeting the template strand upstream of the transcriptional star site);38,48 thus, the differential activation magnitude probably stemmed from the discrepancy in the basal expression level and local chromatin structure in Wnt10b and Foxc2.
Despite the lower magnitude of activation, Foxc2 activation (Foxc2/Cre group; Figure 3) promoted matrix mineralization and inhibited oil-droplet accumulation in BMSCs as effectively as Wnt10b activation (Wnt/Cre group; Figure 3). WNT10B is known to trigger canonical Wnt/β-catenin signaling, which leads to stabilization and nuclear transport of the transcription factor β-catenin.6 The activated Wnt/β-catenin pathway promotes osteogenic differentiation and represses adipogenic differentiation by activating master osteogenic transcription factor RUNX2 and inhibiting the adipogenic master regulatory genes C/EBP-α and PPARγ.49 Conversely, FOXC2 provokes the noncanonical Wnt signaling pathway through a mechanism independent of β-catenin. FOXC2 activates Wnt4,50,51 which is a Wnt ligand that promotes osteogenic differentiation of BMSCs by activating the p38 mitogen-activated protein kinase (MAPK) pathway52 and triggers bone formation by inhibiting nuclear factor-κB.53 Moreover, FOXC2 activates the expression of osteoinductive growth factor bone morphogenetic protein (BMP)-450 and blocks adipogenic differentiation.54 FOXC2 also promotes the secretion of stromal cell-derived factor 1 (SDF-1) and activates the chemokine receptor 4 (CXCR4).55 SDF-1 can bind to CXCR4 and initiate extracellular signal-regulated kinase 1/2 (ERK1/2) pathways, thus enhancing the levels of Runx2 by preventing Runx2 degradation.31 These β-catenin-independent, noncanonical mechanisms induced by Foxc2 might collectively contribute to BMSC osteogenesis as effectively as the canonical Wnt pathway induced by Wnt10b (Figure S2).
When both Wnt10b and Foxc2 were coactivated (Wnt/Foxc2/Cre group), the canonical and noncanonical Wnt pathways converged to enhance further the osteogenic differentiation and inhibited adipogenic differentiation of BMSCs in vitro (Figures 3 and 4). Consequently, implantation of the BMSCs into the critical-sized calvarial bone defects significantly improved the bone formation and bone remodeling in vivo (Figures 5 and 6). In addition to promoting osteogenesis, Foxc2 also induces the vascular endothelial growth factor (VEGF) expression and stimulates new blood-vessel formation,56 which might contribute to the potent calvarial bone healing in the Wnt/Foxc2/Cre group (Figure S2). It should be noted, however, that Foxc2 plays pleiotropic roles in cell proliferation and may be involved in cancer progression.55 Therefore, caution should be used when choosing Foxc2 as the activation target. Fortunately, baculovirus genome and the episomal DNA minicircle were degraded with time;29,57 thus, the Bac-CRISPRa system only temporarily activated Wnt10b and Foxc2 (Figure 2), which reduced the potential risk.
Recent decades have witnessed the marriage of gene therapy and regenerative medicine, wherein exogenous genes encoding growth factors or transcription factors are delivered into stem cells and implanted into the damaged tissue to stimulate regeneration.4,58 Albeit effective and promising, intricate and regulatable gene expression is desirable, yet to date, the exogenous gene is often driven by a constitutive promoter, and the expression level is difficult to control. In contrast to the conventional cell-based gene-therapy approach, the Bac-CRISPRa system enables facile and tunable control of gene-activation level by altering the baculovirus dose and by changing the sgRNA design. The number of sgRNAs in the sgRNA array and the spacer sequences can be varied to target distinct sites in a given genomic locus to recruit activators differentially, so as to fine tune the expression level.59,60
Furthermore, the Bac-CRISPRa system is programmable and capable of activating multiple target genes if several sgRNAs are accommodated in the 134-kb baculovirus genome.45 Besides Wnt10b and Foxc2, other growth factors, such as BMP-2; VEGF; Wnt ligands, such as Wnt3a6 and Wnt661; as well as transcription factors, such as RUNX2 and msh homeobox homolog 2 (Msx2),62 are known to promote bone healing. FOXC2 is also shown to cooperate with BMP-28 or long noncoding RNA H1951 to promote synergistically bone matrix mineralization by stem cells. The Bac-CRISPRa system may be designed to coactivate a combinatory library of these factors, which could further optimize calvarial bone healing and enable identification of the combination of factors contributing to the optimal regeneration.
In summary, we developed the Bac-CRISPRa system to activate the two genes (Wnt10b and Foxc2) governing both canonical and noncanonical Wnt pathways, as well as complementary pathways. The robust and prolonged coactivation of Wnt10b and Foxc2 in BMSCs potentiated osteogenesis and repressed adipogenesis in vitro, thereby substantiating the calvarial bone healing in vivo.
Materials and Methods
Isolation and Culture of BMSCs
All procedures involving animal experiments were approved by the Institutional Animal Care and Use Committee of National Tsing Hua University and performed in compliance with the Guide for the Care and Use of Laboratory Animals (Ministry of Science and Technology, Taiwan). BMSCs were isolated from the tibial and femoral bone marrow of 4-week-old Sprague-Dawley rats (Lesco Biotech, Taiwan), as described earlier31 and in the Supplemental Methods. BMSCs of passages 3–5 were used for subsequent experiments.
Construction and Preparation of Baculovirus Vectors
The baculovirus vectors were constructed using pBac-LEW31 as the starting backbone, which contained two loxP sites flanking a cytomegalovirus (CMV) enhancer/rat EF-1α promoter, multiple cloning site (MCS), and woodchuck hepatitis post-transcriptional response element (WPRE). The Streptococcus pyogenes dCas9 gene was PCR amplified from pcDNA-dCas9-p300 Core (Addgene #6135763) with a 5′ and 3′ flanking NLS. The resultant NLS-dCas9-NLS fragment was subcloned into the MCS of pBac-LEW, downstream of the CMV enhancer/rat EF-1α promoter, to generate pBac-dCas9. The activation domain of transcription activator VP64 was PCR amplified from pHAGE EF-1α dCas9-VP64 (Addgene #5091833) and inserted downstream of the NLS-dCas9-NLS sequence in pBac-dCas9 to yield pBac-dCas9_VP64. The MCP-p65-HSF1 (MPH) fusion gene was PCR amplified from pMS2-P65-HSF1_GFP (Addgene #6142315) with a P2A sequence at the 5′ end. The P2A-MPH gene fragment was subcloned into pBac-dCas9_VP64, downstream of dCas9-VP64, to generate pBac-dCas9_VP64_MPH.
The sgRNA cassette, including the hU6 promoter, a spacer insertion linker, and the sgRNA scaffold with two MS2 coat protein (MCP) recognition aptamer sequences, was PCR amplified from the sgRNA (MS2) cloning backbone (Addgene #6142415). The spacer sequences targeting Wnt10b and Foxc2 were designed using a guide RNA design tool CRISPR-ERA (http://crispr-era.stanford.edu/). The 20-nt spacer sequences with the highest targeting specificity scores (from −400 to −50 relative to the transcription start site [Table S1]) were chosen, chemically synthesized, annealed, and inserted into BbsI-digested pTA-sgRNA. The resultant sgRNA sequences were subcloned into another MCS in pBac-dCas9_VP64_MPH to yield pBac-Wnt10b or pBac-Foxc2.
pBac-Wnt10b and pBac-Foxc2 were used to generate baculovirus vectors Bac-Wnt10b and Bac-Foxc2, respectively, using the Bac-To-Bac system (Thermo Fisher Scientific). The baculovirus Bac-Cre that expressed Cre recombinase under the rat EF-1α promoter was constructed previously.29 Baculovirus vectors were amplified by infecting insect cell Sf-9. The virus supernatant was harvested at 4 days postinfection by centrifugation, and virus titers were determined by the end-point dilution method.30
Baculovirus Transduction, Osteoinduction, and Adipoinduction
Baculovirus transduction of rat BMSCs was performed as described.30 Briefly, rat BMSCs were seeded to 6-well plates (2 × 105 cells/well) for in vitro experiments or 15-cm dish (2.5 × 106 cells/dish) for animal experiments. Cells were cultured overnight in α-minimal essential medium (α-MEM) containing 10% fetal bovine serum (FBS), 100 IU/ml penicillin, and 100 IU/ml streptomycin. In parallel, the baculovirus supernatant was diluted with fresh Grace’s medium (Sigma) for which the volume depended on the MOI and baculovirus titer. The diluted virus was further mixed with NaHCO3-free α-MEM at a volumetric ratio of 1:4.64 For mock transduction, virus-free Grace’s medium was mixed with NaHCO3-free α-MEM at a volumetric ratio of 1:4.
To initiate virus transduction, rat BMSCs were washed twice with PBS (pH 7.4), added with the corresponding virus solution (0.5 mL/well for 6-well plates and 7.5 m/dish for 15-cm dishes) at the desired MOI, and gently shaken on a rocking plate at room temperature for 6 h. After transduction, the virus solution was replaced with osteoinduction medium (α-MEM containing 10% FBS, 100 IU/ml penicillin, 100 IU/ml streptomycin, 0.1 μM dexamethasone, 10 mM β-glycerol phosphate, and 50 μM ascorbic acid 2-phosphate) containing 3 μM sodium butyrate,23 and cells continued to be cultured at 37°C.
After 24 h, the cells were harvested for animal experiments. Alternatively, transduced cells continued to be cultured by replacing the medium with fresh osteoinduction medium (for gene-activation analysis or osteoinduction). The medium was replenished every 2–3 days until analysis. Conversely, to induce adipogenic differentiation, the medium was replaced in the same manner but with adipoinduction medium prepared using adipocyte differentiation basal medium and adipogenesis supplement (StemPro Adipogenesis Differentiation Kit; Gibco).
Quantitative Real-Time RT-PCR
Total RNA was isolated from the cells using the Quick-RNA Miniprep kit (Zymo Research) and reverse transcribed to cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The cDNA was subjected to quantitative real-time PCR (StepOnePlus Real-Time PCR Systems; Applied Biosystems) using primers specific to Wnt10b, Foxc2, Runx2, Opn, Ocn, Osx, Pparγ, C/ebp-α, and Fabp4 (Table S2). The gene-expression levels in the transduced cells were normalized against Gapdh and referenced to that of the mock-transduced BMSCs.
Bone Matrix Mineralization, Calcium Deposition, and Oil-Droplet Accumulation
Bone matrix mineralization and calcium deposition are important signs of osteogenic differentiation.65 After culturing rat BMSCs in osteoinduction medium for 14 days, the matrix mineralization was stained by Alizarin Red S (A5533; Sigma-Aldrich), followed by quantitative analysis. The calcium deposition was measured using the Calcium Liquicolor Complete Test kit (Human). Alternatively, the cells were cultured in adipoinduction medium for 14 days, and oil-droplet formation was stained by Oil Red O, followed by quantitative analysis. All of these procedures are described in detail in Supplemental Methods.
Fabrication of rBMSCs/Scaffold Constructs and Surgical Procedures
To fabricate the rBMSCs/scaffold constructs for in vivo bone healing, the Spongostan gelatin sponge (porosity ≈97%, cat. #MS0003; Ethicon) was cut into disks (diameter ≈6 mm) and submerged in PBS for 30 min. The mock-transduced and transduced rBMSCs were trypsinized from 15-cm dishes at 1 dpt, resuspended in α-MEM, seeded onto the gelatin scaffold (5 × 106 cells/scaffold), and allowed to adhere for 4 h. The rBMSCs/scaffold constructs were cultured with osteoinduction medium containing 3 mM sodium butyrate for 24 h.
In parallel, 6-week-old female Sprague-Dawley rats were anesthetized by intramuscular injection of Zoletil 50 (25 mg/kg body weight; Virbac Animal Health) and 2% Rompun (0.15 mL/kg body weight; Bayer Health Care), followed by intramuscular injection of the antibiotic cefazolin (160 mg/kg body weight). A midline sagittal incision (2 cm) on the scalp was made to expose the parietal bone, and the pericranium was removed carefully by blunt scraping. A critical-size (6 mm in diameter) defect in the middle of the parietal bone was created using a disposable biopsy punch (Integra Miltex) without disturbing the underlying dura mater. Damage to the dura mater could lead to poor regeneration. To minimize damage to the skull and adjacent blood vessels, sterile saline solution was sprayed to the skull to reduce the temperature upon creating the defects by drilling. The constructs were implanted onto the defect and gently pressed, followed by suturing with a 4-0 absorbable stitch (Polysorb; Coviden). The animals received a second intramuscular injection of cefazolin and a topical administration of neomycin and bacitracin zinc at the surgery site.
μCT Imaging Analysis
The calvarial bone regeneration was evaluated using Nano SPECT/CT (Cold Spring Harbor) at the tube voltage of 100 kV and 16 μm resolution. The 3-dimensional images of calvarial bone regeneration were reconstructed using Amira software (Visualization Science Group). The regenerating bone area (square millimeter), bone volume (cubic millimeter), and bone density (average Hounsfield Unit [HU]) were evaluated using PMOD software (PMOD Technologies) within a chosen disk-shaped volume of interest (VOI; 6 mm in diameter and 1 mm in height) that matched the original defect. The data were normalized to the original defect area (28.3 mm2), volume (28.3 mm3), and density (≈4,600 HU) to yield the percentage of bone regeneration.
Histological and Immunohistochemical Staining
After μCT scanning, the calvarial bone specimens were removed and immersed in Osteosoft (Merck) for 15–20 days for complete decalcification and dehydrated in a series of graded concentration of ethanol from 70% to 100%. Details for subsequent H&E staining, histochemical staining of TRAP, immunohistochemical staining of OPN, and BSP are described in Supplemental Methods.
Statistical Analysis
All quantitative data were analyzed using two-way ANOVA or Student’s t test using a two-tailed distribution. The in vitro data represent the mean ± SD of at least three independent experiments. p < 0.05 was considered significant.
Author Contributions
M.-N.H. and K.-L.H. designed and performed experiments and wrote the manuscript. F.-J.Y., P.-L.L., A.V.T., M.-W.L., N.T.K.N., and C.-C.S. performed experiments. S.-M.H. provided cells and supervised experiments. Y.-H.C. and Y.-C.H. designed experiments, supervised project progress, and wrote the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
The data shown in this paper are available in the article and its supplemental data files or available from the authors upon request. The authors acknowledge financial support from the Ministry of Science and Technology (MOST; 105-2923-E-007-002-MY3, 107-2221-E-007-046-MY3, and 108-3017-F-007-003) and CGMH Intramural Project (CRRPG3E0173 and CMRPG310181). This work was also financially supported by the Frontier Research Center on Fundamental and Applied Sciences of Matters and from the Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE; 108QR001I5), Taiwan.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.11.029.
Contributor Information
Yu-Han Chang, Email: yhchang@cloud.cgmh.org.tw.
Yu-Chen Hu, Email: yuchen@che.nthu.edu.tw.
Supplemental Information
References
- 1.Szpalski C., Barr J., Wetterau M., Saadeh P.B., Warren S.M. Cranial bone defects: current and future strategies. Neurosurg. Focus. 2010;29:E8. doi: 10.3171/2010.9.FOCUS10201. [DOI] [PubMed] [Google Scholar]
- 2.Lopes D., Martins-Cruz C., Oliveira M.B., Mano J.F. Bone physiology as inspiration for tissue regenerative therapies. Biomaterials. 2018;185:240–275. doi: 10.1016/j.biomaterials.2018.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shapiro G., Lieber R., Gazit D., Pelled G. Recent advances and future of gene therapy for bone regeneration. Curr. Osteoporos. Rep. 2018;16:504–511. doi: 10.1007/s11914-018-0459-3. [DOI] [PubMed] [Google Scholar]
- 4.Lu C.-H., Chang Y.-H., Lin S.-Y., Li K.-C., Hu Y.-C. Recent progresses in gene delivery-based bone tissue engineering. Biotechnol. Adv. 2013;31:1695–1706. doi: 10.1016/j.biotechadv.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 5.Leucht P., Lee S., Yim N. Wnt signaling and bone regeneration: Can’t have one without the other. Biomaterials. 2019;196:46–50. doi: 10.1016/j.biomaterials.2018.03.029. [DOI] [PubMed] [Google Scholar]
- 6.Bennett C.N., Longo K.A., Wright W.S., Suva L.J., Lane T.F., Hankenson K.D., MacDougald O.A. Regulation of osteoblastogenesis and bone mass by Wnt10b. Proc. Natl. Acad. Sci. USA. 2005;102:3324–3329. doi: 10.1073/pnas.0408742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li K.-C., Chang Y.-H., Yeh C.-L., Hu Y.-C. Healing of osteoporotic bone defects by baculovirus-engineered bone marrow-derived MSCs expressing MicroRNA sponges. Biomaterials. 2016;74:155–166. doi: 10.1016/j.biomaterials.2015.09.046. [DOI] [PubMed] [Google Scholar]
- 8.Zhang W., Zhang X., Li J., Zheng J., Hu X., Xu M., Mao X., Ling J. Foxc2 and BMP2 induce osteogenic/odontogenic differentiation and mineralization of human stem cells from Apical Papilla. Stem Cells Int. 2018;2018:2363917. doi: 10.1155/2018/2363917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.You W., Fan L., Duan D., Tian L., Dang X., Wang C., Wang K. Foxc2 over-expression in bone marrow mesenchymal stem cells stimulates osteogenic differentiation and inhibits adipogenic differentiation. Mol. Cell. Biochem. 2014;386:125–134. doi: 10.1007/s11010-013-1851-z. [DOI] [PubMed] [Google Scholar]
- 10.Li L., Hu S., Chen X. Non-viral delivery systems for CRISPR/Cas9-based genome editing: Challenges and opportunities. Biomaterials. 2018;171:207–218. doi: 10.1016/j.biomaterials.2018.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y., Shen S., Zhao G., Xu C.-F., Zhang H.-B., Luo Y.-L., Cao Z.T., Shi J., Zhao Z.B., Lian Z.X., Wang J. In situ repurposing of dendritic cells with CRISPR/Cas9-based nanomedicine to induce transplant tolerance. Biomaterials. 2019;217:119302. doi: 10.1016/j.biomaterials.2019.119302. [DOI] [PubMed] [Google Scholar]
- 12.Li X.L., Li G.H., Fu J., Fu Y.W., Zhang L., Chen W., Arakaki C., Zhang J.P., Wen W., Zhao M. Highly efficient genome editing via CRISPR-Cas9 in human pluripotent stem cells is achieved by transient BCL-XL overexpression. Nucleic Acids Res. 2018;46:10195–10215. doi: 10.1093/nar/gky804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sung L.-Y., Wu M.-Y., Lin M.-W., Hsu M.-N., Truong V.A., Shen C.-C., Tu Y., Hwang K.Y., Tu A.P., Chang Y.H., Hu Y.C. Combining orthogonal CRISPR and CRISPRi systems for genome engineering and metabolic pathway modulation in Escherichia coli. Biotechnol. Bioeng. 2019;116:1066–1079. doi: 10.1002/bit.26915. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert L.A., Larson M.H., Morsut L., Liu Z., Brar G.A., Torres S.E., Stern-Ginossar N., Brandman O., Whitehead E.H., Doudna J.A. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C., Hsu P.D., Habib N., Gootenberg J.S., Nishimasu H. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joung J., Engreitz J.M., Konermann S., Abudayyeh O.O., Verdine V.K., Aguet F., Gootenberg J.S., Sanjana N.E., Wright J.B., Fulco C.P. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature. 2017;548:343–346. doi: 10.1038/nature23451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J., Rajan S.S., Friedrich M.J., Lan G., Zou X., Ponstingl H., Garyfallos D.A., Liu P., Bradley A., Metzakopian E. Genome-scale CRISPRa screen identifies novel factors for cellular reprogramming. Stem Cell Reports. 2019;12:757–771. doi: 10.1016/j.stemcr.2019.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shao J., Wang M., Yu G., Zhu S., Yu Y., Heng B.C., Wu J., Ye H. Synthetic far-red light-mediated CRISPR-dCas9 device for inducing functional neuronal differentiation. Proc. Natl. Acad. Sci. USA. 2018;115:E6722–E6730. doi: 10.1073/pnas.1802448115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Airenne K.J., Hu Y.-C., Kost T.A., Smith R.H., Kotin R.M., Ono C., Matsuura Y., Wang S., Ylä-Herttuala S. Baculovirus: an insect-derived vector for diverse gene transfer applications. Mol. Ther. 2013;21:739–749. doi: 10.1038/mt.2012.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chuang C.-K., Wong T.-H., Hwang S.-M., Chang Y.-H., Chen G.Y., Chiu Y.-C., Huang S.F., Hu Y.C. Baculovirus transduction of mesenchymal stem cells: in vitro responses and in vivo immune responses after cell transplantation. Mol. Ther. 2009;17:889–896. doi: 10.1038/mt.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lo W.-H., Hwang S.-M., Chuang C.-K., Chen C.-Y., Hu Y.-C. Development of a hybrid baculoviral vector for sustained transgene expression. Mol. Ther. 2009;17:658–666. doi: 10.1038/mt.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin C.-Y., Wang Y.-H., Li K.-C., Sung L.-Y., Yeh C.-L., Lin K.-J., Yen T.C., Chang Y.H., Hu Y.C. Healing of massive segmental femoral bone defects in minipigs by allogenic ASCs engineered with FLPo/Frt-based baculovirus vectors. Biomaterials. 2015;50:98–106. doi: 10.1016/j.biomaterials.2015.01.052. [DOI] [PubMed] [Google Scholar]
- 23.Chen H.-C., Sung L.-Y., Lo W.-H., Chuang C.-K., Wang Y.-H., Lin J.-L., Hu Y.C. Combination of baculovirus-mediated BMP-2 expression and rotating-shaft bioreactor culture synergistically enhances cartilage formation. Gene Ther. 2008;15:309–317. doi: 10.1038/sj.gt.3303087. [DOI] [PubMed] [Google Scholar]
- 24.Lu C.-H., Yeh T.-S., Yeh C.-L., Fang Y.-H.D., Sung L.-Y., Lin S.-Y., Yen T.C., Chang Y.H., Hu Y.C. Regenerating cartilages by engineered ASCs: prolonged TGF-β3/BMP-6 expression improved articular cartilage formation and restored zonal structure. Mol. Ther. 2014;22:186–195. doi: 10.1038/mt.2013.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin C.-Y., Chang Y.-H., Li K.-C., Lu C.-H., Sung L.-Y., Yeh C.-L., Lin K.J., Huang S.F., Yen T.C., Hu Y.C. The use of ASCs engineered to express BMP2 or TGF-β3 within scaffold constructs to promote calvarial bone repair. Biomaterials. 2013;34:9401–9412. doi: 10.1016/j.biomaterials.2013.08.051. [DOI] [PubMed] [Google Scholar]
- 26.Lin C.-Y., Lin K.-J., Kao C.-Y., Chen M.-C., Lo W.H., Yen T.C., Chang Y.H., Hu Y.C. The role of adipose-derived stem cells engineered with the persistently expressing hybrid baculovirus in the healing of massive bone defects. Biomaterials. 2011;32:6505–6514. doi: 10.1016/j.biomaterials.2011.05.059. [DOI] [PubMed] [Google Scholar]
- 27.Hsu M.-N., Liao H.-T., Li K.-C., Chen H.-H., Yen T.-C., Makarevich P., Parfyonova Y., Hu Y.C. Adipose-derived stem cell sheets functionalized by hybrid baculovirus for prolonged GDNF expression and improved nerve regeneration. Biomaterials. 2017;140:189–200. doi: 10.1016/j.biomaterials.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 28.Yeh T.-S., Fang Y.H., Lu C.-H., Chiu S.-C., Yeh C.-L., Yen T.-C., Parfyonova Y., Hu Y.C. Baculovirus-transduced, VEGF-expressing adipose-derived stem cell sheet for the treatment of myocardium infarction. Biomaterials. 2014;35:174–184. doi: 10.1016/j.biomaterials.2013.09.080. [DOI] [PubMed] [Google Scholar]
- 29.Sung L.-Y., Chen C.-L., Lin S.-Y., Hwang S.-M., Lu C.-H., Li K.-C., Lan A.S., Hu Y.C. Enhanced and prolonged baculovirus-mediated expression by incorporating recombinase system and in cis elements: a comparative study. Nucleic Acids Res. 2013;41:e139. doi: 10.1093/nar/gkt442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sung L.-Y., Chen C.-L., Lin S.-Y., Li K.-C., Yeh C.-L., Chen G.-Y., Lin C.Y., Hu Y.C. Efficient gene delivery into cell lines and stem cells using baculovirus. Nat. Protoc. 2014;9:1882–1899. doi: 10.1038/nprot.2014.130. [DOI] [PubMed] [Google Scholar]
- 31.Lo S.-C., Li K.-C., Chang Y.-H., Hsu M.-N., Sung L.-Y., Vu T.A., Hu Y.C. Enhanced critical-size calvarial bone healing by ASCs engineered with Cre/loxP-based hybrid baculovirus. Biomaterials. 2017;124:1–11. doi: 10.1016/j.biomaterials.2017.01.033. [DOI] [PubMed] [Google Scholar]
- 32.Black J.B., Adler A.F., Wang H.G., D’Ippolito A.M., Hutchinson H.A., Reddy T.E., Pitt G.S., Leong K.W., Gersbach C.A. Targeted epigenetic remodeling of endogenous loci by CRISPR/Cas9-based transcriptional activators directly converts fibroblasts to neuronal cells. Cell Stem Cell. 2016;19:406–414. doi: 10.1016/j.stem.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kearns N.A., Genga R.M.J., Enuameh M.S., Garber M., Wolfe S.A., Maehr R. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development. 2014;141:219–223. doi: 10.1242/dev.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Staines K.A., MacRae V.E., Farquharson C. The importance of the SIBLING family of proteins on skeletal mineralisation and bone remodelling. J. Endocrinol. 2012;214:241–255. doi: 10.1530/JOE-12-0143. [DOI] [PubMed] [Google Scholar]
- 35.Greenblatt M.B., Tsai J.N., Wein M.N. Bone turnover markers in the diagnosis and monitoring of metabolic bone disease. Clin. Chem. 2017;63:464–474. doi: 10.1373/clinchem.2016.259085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jost M., Chen Y., Gilbert L.A., Horlbeck M.A., Krenning L., Menchon G., Rai A., Cho M.Y., Stern J.J., Prota A.E. Combined CRISPRi/a-based chemical genetic screens reveal that Rigosertib is a microtubule-destabilizing agent. Mol. Cell. 2017;68:210–223.e6. doi: 10.1016/j.molcel.2017.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rubin A.J., Parker K.R., Satpathy A.T., Qi Y., Wu B., Ong A.J., Mumbach M.R., Ji A.L., Kim D.S., Cho S.W. Coupled single-cell CRISPR screening and epigenomic profiling reveals causal gene regulatory networks. Cell. 2019;176:361–376.e17. doi: 10.1016/j.cell.2018.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilbert L.A., Horlbeck M.A., Adamson B., Villalta J.E., Chen Y., Whitehead E.H., Guimaraes C., Panning B., Ploegh H.L., Bassik M.C. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bester A.C., Lee J.D., Chavez A., Lee Y.R., Nachmani D., Vora S., Victor J., Sauvageau M., Monteleone E., Rinn J.L. An integrated genome-wide CRISPRa approach to functionalize lncRNAs in drug resistance. Cell. 2018;173:649–664.e20. doi: 10.1016/j.cell.2018.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y., Yu C., Daley T.P., Wang F., Cao W.S., Bhate S., Lin X., Still C., 2nd, Liu H., Zhao D. CRISPR activation screens systematically identify factors that drive neuronal fate and reprogramming. Cell Stem Cell. 2018;23:758–771.e8. doi: 10.1016/j.stem.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y., Han J., Chen Z., Wu H., Dong H., Nie G. Engineering cell signaling using tunable CRISPR-Cpf1-based transcription factors. Nat. Commun. 2017;8:2095. doi: 10.1038/s41467-017-02265-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu P., Chen M., Liu Y., Qi L.S., Ding S. CRISPR-based chromatin remodeling of the endogenous Oct4 or Sox2 locus enables reprogramming to pluripotency. Cell Stem Cell. 2018;22:252–261.e4. doi: 10.1016/j.stem.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 43.Chang Y.K., Hwang J.S., Chung T.Y., Shin Y.J. SOX2 activation using CRISPR/dCas9 promotes wound healing in corneal endothelial cells. Stem Cells. 2018;36:1851–1862. doi: 10.1002/stem.2915. [DOI] [PubMed] [Google Scholar]
- 44.Liao H.-K., Hatanaka F., Araoka T., Reddy P., Wu M.-Z., Sui Y., Yamauchi T., Sakurai M., O’Keefe D.D., Núñez-Delicado E. In vivo target gene activation via CRISPR/Cas9-mediated trans-epigenetic modulation. Cell. 2017;171:1495–1507.e15. doi: 10.1016/j.cell.2017.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hsu M.-N., Liao H.-T., Truong V.A., Huang K.-L., Yu F.-J., Chen H.-H., Nguyen T.K.N., Makarevich P., Parfyonova Y., Hu Y.C. CRISPR-based activation of endogenous neurotrophic genes in adipose stem cell sheets to stimulate peripheral nerve regeneration. Theranostics. 2019;9:6099–6111. doi: 10.7150/thno.36790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Truong V.A., Hsu M.-N., Kieu Nguyen N.T., Lin M.-W., Shen C.-C., Lin C.-Y., Hu Y.C. CRISPRai for simultaneous gene activation and inhibition to promote stem cell chondrogenesis and calvarial bone regeneration. Nucleic Acids Res. 2019;47:e74. doi: 10.1093/nar/gkz267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chavez A., Tuttle M., Pruitt B.W., Ewen-Campen B., Chari R., Ter-Ovanesyan D., Haque S.J., Cecchi R.J., Kowal E.J.K., Buchthal J. Comparison of Cas9 activators in multiple species. Nat. Methods. 2016;13:563–567. doi: 10.1038/nmeth.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.La Russa M.F., Qi L.S. The new state of the art: Cas9 for gene activation and repression. Mol. Cell. Biol. 2015;35:3800–3809. doi: 10.1128/MCB.00512-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ross S.E., Hemati N., Longo K.A., Bennett C.N., Lucas P.C., Erickson R.L., MacDougald O.A. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289:950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- 50.Gozo M.C., Aspuria P.J., Cheon D.J., Walts A.E., Berel D., Miura N., Karlan B.Y., Orsulic S. Foxc2 induces Wnt4 and Bmp4 expression during muscle regeneration and osteogenesis. Cell Death Differ. 2013;20:1031–1042. doi: 10.1038/cdd.2013.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou P., Li Y., Di R., Yang Y., Meng S., Song F., Ma L. H19 and Foxc2 synergistically promotes osteogenic differentiation of BMSCs via Wnt-β-catenin pathway. J. Cell. Physiol. 2019;234:13799–13806. doi: 10.1002/jcp.28060. [DOI] [PubMed] [Google Scholar]
- 52.Chang J., Sonoyama W., Wang Z., Jin Q., Zhang C., Krebsbach P.H., Giannobile W., Shi S., Wang C.Y. Noncanonical Wnt-4 signaling enhances bone regeneration of mesenchymal stem cells in craniofacial defects through activation of p38 MAPK. J. Biol. Chem. 2007;282:30938–30948. doi: 10.1074/jbc.M702391200. [DOI] [PubMed] [Google Scholar]
- 53.Yu B., Chang J., Liu Y., Li J., Kevork K., Al-Hezaimi K., Graves D.T., Park N.H., Wang C.Y. Wnt4 signaling prevents skeletal aging and inflammation by inhibiting nuclear factor-κB. Nat. Med. 2014;20:1009–1017. doi: 10.1038/nm.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gerin I., Bommer G.T., Lidell M.E., Cederberg A., Enerback S., Macdougald O.A. On the role of FOX transcription factors in adipocyte differentiation and insulin-stimulated glucose uptake. J. Biol. Chem. 2009;284:10755–10763. doi: 10.1074/jbc.M809115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang J., Yue X. Role and importance of the expression of transcription factor FOXC2 in cervical cancer. Oncol. Lett. 2017;14:6627–6631. doi: 10.3892/ol.2017.7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.You W., Gao H., Fan L., Duan D., Wang C., Wang K. Foxc2 regulates osteogenesis and angiogenesis of bone marrow mesenchymal stem cells. BMC Musculoskelet. Disord. 2013;14:199. doi: 10.1186/1471-2474-14-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luo W.Y., Shih Y.S., Hung C.L., Lo K.W., Chiang C.S., Lo W.H., Huang S.F., Wang S.C., Yu C.F., Chien C.H., Hu Y.C. Development of the hybrid Sleeping Beauty: baculovirus vector for sustained gene expression and cancer therapy. Gene Ther. 2012;19:844–851. doi: 10.1038/gt.2011.129. [DOI] [PubMed] [Google Scholar]
- 58.Evans C.H. Gene delivery to bone. Adv. Drug Deliv. Rev. 2012;64:1331–1340. doi: 10.1016/j.addr.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Larouche J., Aguilar C.A. New technologies to enhance in vivo reprogramming for regenerative medicine. Trends Biotechnol. 2019;37:604–617. doi: 10.1016/j.tibtech.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 60.Komor A.C., Badran A.H., Liu D.R. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell. 2017;168:20–36. doi: 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cawthorn W.P., Bree A.J., Yao Y., Du B., Hemati N., Martinez-Santibañez G., MacDougald O.A. Wnt6, Wnt10a and Wnt10b inhibit adipogenesis and stimulate osteoblastogenesis through a β-catenin-dependent mechanism. Bone. 2012;50:477–489. doi: 10.1016/j.bone.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ichida F., Nishimura R., Hata K., Matsubara T., Ikeda F., Hisada K., Yatani H., Cao X., Komori T., Yamaguchi A., Yoneda T. Reciprocal roles of MSX2 in regulation of osteoblast and adipocyte differentiation. J. Biol. Chem. 2004;279:34015–34022. doi: 10.1074/jbc.M403621200. [DOI] [PubMed] [Google Scholar]
- 63.Hilton I.B., D’Ippolito A.M., Vockley C.M., Thakore P.I., Crawford G.E., Reddy T.E., Gersbach C.A. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shen H.-C., Lee H.-P., Lo W.-H., Yang D.-G., Hu Y.-C. Baculovirus-mediated gene transfer is attenuated by sodium bicarbonate. J. Gene Med. 2007;9:470–478. doi: 10.1002/jgm.1037. [DOI] [PubMed] [Google Scholar]
- 65.Niu H., Ma Y., Wu G., Duan B., Wang Y., Yuan Y., Liu C. Multicellularity-interweaved bone regeneration of BMP-2-loaded scaffold with orchestrated kinetics of resorption and osteogenesis. Biomaterials. 2019;216:119216. doi: 10.1016/j.biomaterials.2019.05.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.