Abstract
Despite extensive usage of gene therapy medicinal products (GTMPs) in clinical studies and recent approval of chimeric antigen receptor (CAR) T cell therapy, little information has been made available on the precise molecular characterization and possible variations in terms of insert integrity and vector copy numbers of different GTMPs during the complete production chain. Within this context, we characterize αβT cells engineered to express a defined γδT cell engineered to express a defined γδT receptor (TEG) currently used in a first-in-human clinical study (NTR6541). Utilizing targeted locus amplification in combination with next generation sequencing for the vector producer clone and TEG001 products, we report on five single-nucleotide variants and nine intact vector copies integrated in the producer clone. The vector copy number in TEG001 cells was on average a factor 0.72 (SD 0.11) below that of the producer cell clone. All nucleotide variants were transferred to TEG001 without having an effect on cellular proliferation during extensive in vitro culture. Based on an environmental risk assessment of the five nucleotide variants present in the non-coding viral region of the TEG001 insert, there was no altered environmental impact of TEG001 cells. We conclude that TEG001 cells do not have an increased risk for malignant transformation in vivo.
Keywords: engineered T cells, gene therapy medicinal product, GTMP, γδT cells, TEG001, targeted locus amplification, TLA, vector copy number
Straetemans and colleagues characterized αβT cells engineered to express a defined γδT cell receptor (TEG001) used in a first-in-human clinical study, with TLA in combination with NGS and qPCR. They provide a rapid strategy to evaluate the presence, integrity, and persistence of genetic information along the production chain of any GTMP.
Introduction
Adoptive cell transfer with T cells engineered to attack tumor cells, a class of so-called gene therapy medicinal products (GTMPs), involves the genetic reprogramming of T cells with defined immune receptors such as chimeric antigen receptors (CARs) or T cell receptors (TCRs). Recently, CAR T cells targeting CD19+ hematological malignancies have been approved by the US Food and Drug Administration (FDA) and the European Medicines Agency (EMA) in 2017 and 2018, respectively.1 This success has accelerated the already rapid improvements of gene transfer technologies and synthetic biology that offer a wide range of possibilities to design T cells with enhanced functions. One of the major limitations of currently explored and approved CAR T cell strategies is the lack of tumor-specific targets, limiting this therapy to date to mainly B cell-related malignancies. Despite great pre-clinical efforts to find new targets, only a few candidate CAR T targets, such as B cell maturation antigen (BCMA) and the interleukin-3 (IL-3) receptor (CD123), as well as a handful of tumor-specific αβTCRs targeting cancer-testis antigens, have reached the clinical stage of development.2,3 Alternative tumor-targeting strategies are needed; therefore, we introduced the concept of metabolic cancer targeting through an anti-tumor receptor derived from γδT cells and proposed to utilize αβT cells engineered to express a defined γδT cell receptor (TEGs).4,5 The TEG concept allows for the selection of the most potent anti-tumor γδTCR combined with the cytotoxic, proliferative, and memory formation capacities of αβT cells. Furthermore, TEG cells display identical tumor-reactivity as their parental γδT cell clone,5,6 suggesting that all properties from the γδTCR involved in anti-tumor reactivity are transferred to TEG cells. This concept has been recently expanded to Vδ2 negative γδTCRs expressed in αβT cells,7 allowing theoretically in the future potential dual reactivity of these types of immune receptor in TEG format as proposed by Melandri et al.8 Vγ9Vδ2 T cells sense via their non-major histocompatibility complex (MHC)-restricted TCR phosphoantigen (pAg) and small GTPase (RhoB)-dependent joint spatial and conformational changes in CD277 (CD277J) at the cell surface of cancer cells9, 10, 11 (for review, see Sebestyen et al.12). TEG001 cells are engineered to express a Vγ9Vδ2 T cell-derived receptor and, as a consequence, target a broad range of tumor types independent of genetic background including primary acute myeloid leukemia (AML) and multiple myeloma (MM) cells in vitro and in patient-derived xenograft (PDX) models but leave healthy cells unharmed.4,5,13, 14, 15
In order to further translate the TEG concept into first-in-human trials, we reported recently on the development of a Good Manufacturing Practice-compliant production protocol for the manufacturing of TEG001 cells using autologous T cells and retroviral-based gene transfer technology.14,16 European guidelines request for GTMPs such as TEG001 the precise characterization of all different production chains in terms of integrity of insert DNA, integration site, copy numbers, and potential tumorigenesis.17 This requirement was based on the observation that different vectors, such as retrovirus, lentivirus,18 or non-viral transposons,19 generate different integration site patterns and the observation that genetic modification of hematopoietic stem cells can induce leukemia.20,21 In contrast, genetic modification of mature mouse and human T cells using viral vectors did so far not lead to substantial toxicities.22, 23, 24, 25 For clinical studies utilizing CAR T cell products, such detailed molecular characterizations have mainly been published once side effects have been observed. For example, the disruption of the TET2 gene by CAR transgene integration was reported to induce greater CAR T cell proliferation capacity.26 Thus, we conclude that although different extensive studies have been published on individual products mainly after side effects have been observed, only little public information has been made available for many currently clinically tested or approved GTMPs (see Table S1 for a comprehensive literature overview). Another observation we made regarding the currently available dataset is that insert integrity published in reports was measured only indirectly by evaluation of the expression of the introduced transgene in the final cell product, for instance, by flow cytometric analysis of CAR or TCR expression.27, 28, 29 Thus, to our best knowledge, there is no comprehensive information available regarding the molecular characterization of integrity of CAR or TCR transgenes on the nucleotide level throughout the complete production chain as requested by authorities, and thus from vector producer cells until final medicinal product. Finally, there are a number of reports on the manufacturing and quality-control tests of engineered T cell products that include establishment of transgene copy numbers in the final product of pre-clinical production runs,30, 31, 32, 33, 34 but only a minority of studies share vector copy numbers of infused CAR T products27,35, 36, 37, 38 (Table S1). This lack of public knowledge makes it very difficult to put insert integrity and vector copy numbers of novel GTMPs within the context of existing GTMP products and clinical data linked to these products. Creating an extensive public database for GTMP production chains would allow assessing safety and potential risks of defined insertion sites, additional molecular alterations, or higher copy numbers. Therefore, we report now on the molecular integrity of the integrated DNA sequences throughout TEG001 manufacturing used for an ongoing clinical study. In addition, we propose that the applied targeted locus amplification (TLA) technology39 is an elegant and rapid technology for full molecular characterization of the transgene of interest from TEG001 vector plasmid until medicinal product.
Results
Mutational Variants in Non-coding Regions of the TEG001 Insert Integrated in the Master Cell Bank
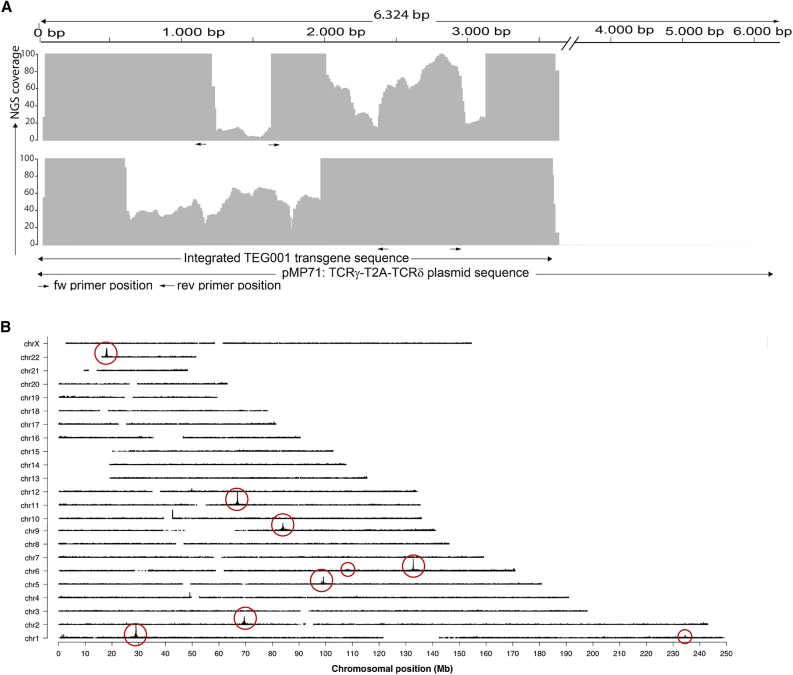
Accurate transfer of the transgene DNA sequence into the genome of target cells is of utmost importance in a GTMP. Nevertheless, variations can theoretically occur in retroviral-based gene transfer technology as a consequence of the nature of the retroviral life cycle that has been continuously evolved to escape host immune defense.40,41 In order to characterize potential major variations of the integrated TEG001 sequence (later referred as TEG001 insert; see Figure S1A) throughout the production process (Figure S1B), we utilized TLA technology in combination with next generation sequencing (NGS) (Figure S2). First, we set out to analyze the TEG001 insert in the virus producer cells, derived from the TEG001 master cell bank (MCB) clone 73 (Figure S1B). Two primer sets were designed targeting the TEG001 transgene to generate two independent datasets, followed by processing of the MCB cells and sequencing of PCR products. TLA technology further allowed alignment of captured sequences with the TEG001 reference plasmid sequence to determine the sequence identity of the TEG001 insert in the MCB clone. The coverage, i.e., the number of sequencing reads that align to each position, is plotted in Figure 1A. Complete coverage was obtained across the TEG001 insert from position 2 to 3592 in the original plasmid reference sequence, indicating that this sequence was integrated. Although the coverage for primer set 1 between the primer positions, namely, positions 1163–1614 at the reference sequence, was low, the coverage in that region for primer set 2 was high enough (above 30-fold) to then analyze the sequence integrity.39 No breakpoints or structural rearrangements that would have presented themselves as fusion sequences between two different parts of the insert were detected in the TEG001 insert above a 1% frequency threshold. Next, we investigated whether small sequence variants were present in the TEG001 insert above a 5% mutant allele frequency threshold and if they were found in the reads obtained by both primer sets. These small sequence variants are defined as single-nucleotide variants or small (<10-bp) insertions and deletions (InDels). There were no small sequence variants found in the protein encoding TCRγ5-T2A-δ5 DNA sequence; however, interestingly, four point mutations and one insertion were detected in the non-coding region of the TEG001 insert. The mutant allele frequencies (% mutation) ranged from 8% to 14%, suggesting that the single-nucleotide variants in this clonal cell population were present only in a fraction of the integrated transgene copies (Table 1).
Figure 1.
Transgene Integration in Master Cell Bank Clone 73
(A) TLA sequence coverage of the TEG001 insert in MCB across the pMP71:TCRγ5-T2A-δ5 plasmid reference sequence. The gray vertical bars represent the number of NGS reads that align to each plasmid position and indicate the integrated TEG001 transgene sequence. The coverage generated with primer set 1 is depicted in the top panel and with primer set 2 in the bottom panels. The positions of the primers are represented by black arrows below each coverage profile. The y axis is limited to 100×. (B) TLA sequence coverage across the human genome using the TEG001 insert specific primer set 2. The different chromosomes are indicated on the y axis, and the chromosomal position on the x axis. Encircled are the regions containing the TEG001 transgene integration sites. Locations in smaller circles show less obvious peaks, but fusion sequences to these positions do confirm the integration sites there.
Table 1.
Small Sequence Variants
| Position |
||||||
|---|---|---|---|---|---|---|
| 415 |
568 |
578 |
606 |
3552 |
||
| Reference Nucleotide | ||||||
| C |
T |
T |
A |
C |
||
| Mutation | ||||||
| G | C | C | +1C | G | ||
| Plasmid | coverage | 8,655 | 7,316 | 7,327 | 7,273 | 5,208 |
| % mutation | 0 | 0 | 0 | 0 | 0 | |
| MCB | coverage | 2,051 | 1,750 | 1,785 | 1,802 | 336 |
| % mutation | 10 | 9 | 14 | 10 | 8 | |
| TEG001-28 | coverage | 3,389 | 2,039 | 2,032 | 1,971 | 571 |
| % mutation | 11 | 15 | 19 | 12 | 10 | |
| TEG001-31 | coverage | 2,810 | 17,60 | 1,727 | 1,707 | 480 |
| % mutation | 13 | 17 | 20 | 10 | 11 | |
| TEG001-32 | coverage | 4,857 | 3,064 | 3,013 | 2,939 | 851 |
| % mutation | 12 | 16 | 17 | 9 | 14 | |
| TEG001-33 | coverage | 42,660 | 33,494 | 33,461 | 32,897 | 7,017 |
| % mutation | 12 | 19 | 17 | 8 | 11 | |
| TEG001-32_22w | coverage | 20,881 | 16,291 | 17,290 | 16,880 | 3,543 |
| % mutation | 8 | 23 | 14 | 5 | 8 | |
| TEG001-33_24w | coverage | 2,516 | 2,299 | 2,360 | 2,316 | 396 |
| % mutation | 8 | 3 | 2 | 3 | 7 | |
Position of the five nucleotide variants and their allele frequencies in the plasmid (as control), the MCB, the TEG001 drug product samples, and the cultured TEG001 samples are shown. Position: position in the reference sequence at which the variant is found; reference nucleotide: the nucleotide present in the reference sequence at this position; mutation: the variant nucleotide identified at the indicated position, +1C indicates the insertion of 1 C after the reference A. Coverage at this position is the average between the two primer sets. % mutation, mutant allele frequency; that is, the percentage of reads that contained the variant and the average of results of the two primer sets.
Multiple TEG001 Insert Integration Sites within the MCB
The MCB derived from producer cell clone 73 was selected on highest virus titer production.16 Therefore, and because the five small sequence variants were detected only with a mutant allele frequency below 15%, we assumed that the TEG001 insert was integrated multiple times within this clone. In order to assess the number of TEG001 inserts integrated, TLA data originally used for the characterization of the insert integrity, as described above, were now mapped across the human genome. Because in TLA data the highest coverage is obtained on the sequences in closest proximity of the location of the primer set, coverage peaks could be used to identify the genomic position of the TEG001 transgene. TLA sequence coverage revealed nine TEG001 transgene integration sites in the MCB, seven with high coverage peaks and two with lower coverage peaks, as depicted by circles in the coverage plot (Figure 1B). In each of the nine locations, which comprise the TEG001 insert, the exact integration sites were identified based on fused DNA sequences of host genome with TEG001 transgene sequences (Table S2). All host genome-TEG001 transgene fusions were found on two identical positions in the TEG001 transgene long terminal repeat (LTR) sequence, namely, positions 558 and 3108 in the original TEG001 plasmid sequence. No transgene-transgene fusions were found. For eight out of nine integration sites, 4 bp of the human genome were duplicated and flanking the proviral DNA, in line with the biology of Moloney murine leukemia virus (MoMLV)-based retroviral integration42 (illustrated in Figure S3). For one out of nine integrations sites, the integration at chromosome 1 (chr1), 624 bp of the human genome sequence were duplicated instead of 4 bp; however, the fusion sites on the transgenic site were similar to the other integrations, and therefore there was no indication that the transgenic sequence was altered in this integration site. Based on these results, it was estimated that nine copies of the TEG001 transgene were accurately integrated in the MCB derived from cell clone 73.
Phenotypical Description of TEG001 Drug Product
The differentiation and homing profile of an engineered T cell product is of importance for its in vivo function following adoptive cell transfer.43 Therefore, we extended our previous analysis16 on TEG001 cells by using a definition based on CD62L and CD45RO44 to describe the differentiation state. The GMP-grade manufacturing protocol yielded a TEG001 product with mostly effector memory phenotype (65%–75%) but also a substantial fraction of central memory cells (10%–15%) (Figure S4).
Transgene Copy Number in TEG001 Drug Product
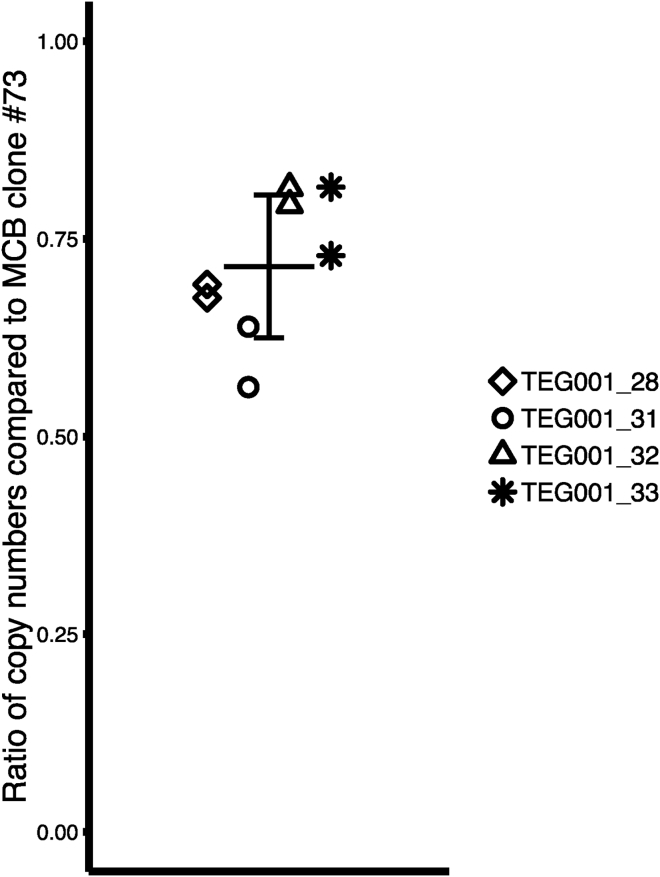
No clinical adverse events have been reported when using retroviral vectors to engineer mature T cells.24 However, theoretically, the genotoxicity risk will increase when the number of engineered T cells infused into a patient will increase, although it is unclear whether the number of integrations sites per cell adds to this risk.45 Although not formally assessed and stated, in discussions with the FDA five integration sites per cell are considered without increased safety risk when compared with other gene therapy products.46 Therefore, we developed a qPCR to determine the average TEG001 vector copy number compared with the MCB producer clone 73. The transgene-specific qPCR primer/probe set was designed on the T2A region of the TEG001 transgene (primer set 1). We determined the transgene copy numbers in four TEG001 cell products, which were on average a factor 0.72 (SD 0.09) below that of the MCB clone 73 (Figure 2). The results of primer set 1 were validated with a different transgene-specific qPCR with primers and probe (primer set 2) annealing to Gag-derived sequences in the 5′ LTR region of the TEG001 transgene. Detected transgene copy numbers compared with MCB clone 73 were comparable for primer sets 1 and 2 (Figure S5A). Finally, sensitivity of the qPCR was assessed by spiking TEG001_33 cells at different frequencies into a background of healthy donor PBMCs. A clear correlation (adjusted R2 = 0.999, p < 0.001) between the frequency of TEG001_33 cells in the tested sample and the detected copy numbers was found (Figure S5B). These data suggest that we did not only develop a sensitive and reproducible method to detect persisting TEG001 in humans during the ongoing clinical study, but that the clinical study will also have a safety impact on other clinical studies by testing whether higher copy numbers for GTMPs associate with additional long-term toxicity in humans.
Figure 2.
TEG001 Transgene Copy Number in Drug Product
Ratios of integrated copy numbers per unit/DNA of the TEG001 transgene in four independently produced TEG001 products are compared with copy number per unit/DNA of MCB clone 73 as determined by qPCR. The mean and standard deviation of the ratio of the TEG001 products compared with the MCB clone is indicated. Values represent replicates of two qPCR experiments.
Mutational Variants in the Non-coding Region of the TEG001 Inserts Are Transferred to the TEG001 Drug Product
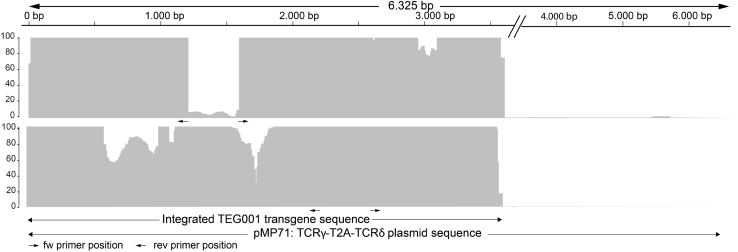
TEG001 production includes an in vitro expansion for 10 days after the retroviral transduction hit, before harvesting, purification, and formulation of engineered T cells into the TEG001 drug product (Figure S1B). As a consequence, also the final TEG001 drug product needs to be characterized for insert integrity, as well as frequency of mutational variants. In line with suggestions from authorities, we utilized for this analysis TEG001 drug product cells derived from four independent large-scale production runs and again performed TLA in order to first determine insert integrity. Again, the same two primer sets initially used for the analysis of the MCB were employed for the analyses, and coverage of the insert was determined by alignment with the plasmid reference sequence (Figure 3). Primer set 1 yielded 30-fold coverage or higher in all samples except for the region in between the two primers of the primer pair, positions 1163–1614. In line with insert integration in the MCB, the transgene was integrated in TEG001 drug product cells from position 2 to 3592. Again, no structural variants were detected within the insert, only the five single-nucleotide variants that were present in the MCB were transferred into TEG001 cells, and no new nucleotide variants were detected (Table 1). The mutant allele frequencies in the TEG001 samples were in the same range (from 8%–19%) as those in the MCB.
Figure 3.
Transgene Integration in TEG001 Drug Product
Representative picture of TLA sequence coverage of the TEG001 insert in a TEG001 drug product sample (TEG001-28). Coverage is depicted across the pMP71:TCRγ5-T2A-δ5 plasmid reference sequence; see legend to Figure 1A. Top panel: coverage generated with primer set 1; bottom panel: coverage generated with primer set 2.
Analysis of the Small Sequence Variants
Although virus titers and protein expression of TEG001 transgene were confirmed,16 the small sequence variants found in the non-coding viral region of the TEG001 insert may have biological consequences for the medicinal product. Therefore, the five variants were further assessed in detail as indicated in Table 2. In short, all five variants occurred outside of the promoter region, and no additional start or stop codon was introduced that may lead to an alternative open reading frame or as such have influence on protein transcription. Two mutations were found in the primer binding site at positions 568 and 578 of the reference sequence. The primer binding site is essential for viral replication because it is the site where the tRNA from the host cell binds as primer to initiate the reverse transcription and replication process. In the pMP71-based TEG001 vector, the primer binding site was derived from the murine embryonic stem cell virus-5′ untranslated region (MESV-5′ UTR)47 to provide a non-methylated 5′ UTR to improve stability of transgene expression in murine stem cells compared with the MoMLV primer binding site.48 The mutations found may abolish this function in murine stem cells. Effect of this specific primer binding site on protein stability in human differentiated T cells is not known. High virus titers measured by transgene expression on indicator cells imply that not only transcriptional activity was intact, but also protein stability and expression were not affected,16 even in long-term cultures of TEG001 (Figure S6). Based on these theoretical evaluations, it was concluded that the five small sequence variants did not result in an elevated safety risk of the TEG001 medicinal product for patients and the environment.
Table 2.
Environmental Impact of Five Nucleotide Variants Identified in the TEG001 Insert
| Mutation | Element | Impact |
|---|---|---|
| Position 415 C to G |
junction of U3 to R region in 5′ LTR | The mutation occurred outside the promoter region and therefore does not affect promoter activity. The mutation does not lead to the introduction of an additional start codon and will not lead to a new open reading frame (ORF). The mutation does not lead to the introduction of an additional stop codon and as such has no influence on protein transcription. It is unlikely that this mutation affects packaging, integration, and transcription, and is therefore considered neutral to the biology of the retroviral vector. In conclusion, there is no altered or additional environmental risk as a result of this mutation. |
| Position 568 T to C |
primer binding site | Because the mutations at positions 568 and 578 are both just after the 5′ LTR but in the primer binding site, their impact is considered together. The primer binding site is essential for replication because it is the site where the tRNA from the host cell binds as primer to initiate the inverse transcription and replication process. In the pMP71 vector, the primer binding site was derived from the murine embryonic stem cell virus-5′ untranslated region47 to provide a non-methylated 5′ UTR to improve stability of transgene expression in murine stem cells compared with the Moloney murine leukemia virus primer binding site.48 The mutations found may abolish this function in murine stem cells. The effect of this specific primer binding site on protein stability in human differentiated T cells is not known. High virus titers measured by transgene expression on indicator cells indicate that not only transcriptional activity was intact but also protein stability and expression were not affected. The mutations do not lead to the introduction of an additional start codon and as such will not lead to a new ORF. The mutations do not lead to the introduction of an additional stop codon and as such have no influence on protein transcription. It is unlikely that this mutation affects packaging, and integration resulted from their position. In conclusion, there is no altered or additional environmental risk as a result of these two mutations. |
| Position 578 T to C |
primer binding site | |
| Position 606 A +1C |
after primer binding site and before packaging sequence (Ψ) | The insertion occurred outside the primer binding site and packaging sequence, and therefore it is unlikely that it affects packaging, integration, and transcription. The insertion does not lead to the introduction of an additional start codon and will not lead to a new ORF. The insertion does not lead to the introduction of an additional stop codon and as such has no influence on protein transcription. In conclusion, there is no altered or additional environmental risk as a result of this insertion. |
| Position 3552 C to G |
junction of U3 to R region in 3′ LTR | This mutation is identical to the mutation at position 415; see above. In conclusion, there is no altered or additional environmental risk as a result of this mutation. |
Mutational Variants Do Not Skew the TEG001 Medicinal Product in Long-Term Cultures
As indicated in the theoretical analysis of mutational variants (Table 2), we would not expect an impact of mutational variants on the growth rate of individual T cells within the TEG001 medicinal product. In order to formally assess whether this assumption is correct and the variations in the non-coding region of the TEG001 insert would indeed not skew T cell proliferation over the original sequence, T cells from two individual TEG001 production runs (TEG001-32 and TEG001-33) were further expanded in vitro for up to 24 weeks with a well-established rapid expansion protocol.
TLA analysis was then repeated. In addition, compared with the reference TEG001 sequence, again the same five variants were identified in the cultured TEG001 cells (Table 1). When comparing the corresponding TEG001 drug product with the cultured samples, all frequencies decreased except only one variant in the cultured sample of TEG001-33, which increased slightly from 16% to 23%. Thus, with all limitations of an in vitro rapid expansion procedure, which can per definition not entirely predict outgrowth of individual clones in vivo, but rather reflects a forced expansion through external stimulation, sequence variants do not substantially change in frequency over time. Therefore, these five sequence variants most likely do not correlate with an elevated safety risk.
TEG001 Insert Integration in Medicinal Drug Product
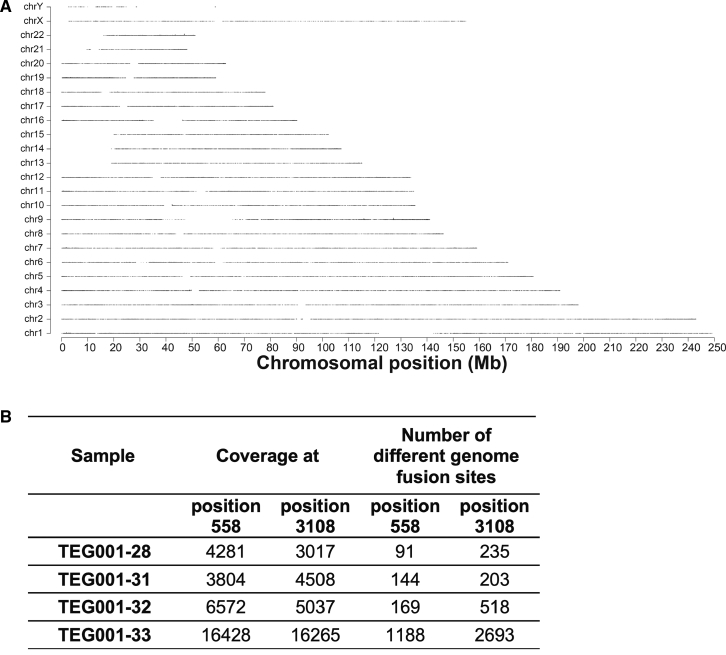
The four final TEG001 drug product samples analyzed with TLA were further used to perform an integration site analysis. Genomic coverage profiles showed, as expected, a heterogeneous integration pattern on a genome-wide scale because no large coverage peaks were detected in all samples (Figure 4A). In line with this observation, many different unique integration sites, identified in the data as reads that consist in part of transgene sequence and in part of genomic sequence (genome-transgene fusion reads), supported the notion of heterogeneity (Figure 4B).
Figure 4.
Heterogeneic Integration Pattern of TEG001 Insert in TEG001 Drug Product
(A) TLA sequence coverage across the human genome using the TEG001 specific primer set 1. No coverage peaks are visible. The different chromosomes are indicated on the y axis, and the chromosomal position on the x axis. One representative sample out of four is shown (TEG001-28). (B) Many integration sites are found in TEG001 drug product samples as an indication of a heterogeneic integration pattern. Coverage and number of different genome fusion sites are indicated per sample for the two positions in the LTR sequence that are fused to the human host genome. The data from primer sets 1 and 2 are combined.
Discussion
The viral vector particle, the vector backbone, and the transgene are the three components considered to be responsible for the biological effects of a GTMP;17 however, reports on a precise and complete molecular characterization of transgenes in GTMPs throughout the full production chain are scarce (Table S1). Therefore, we comprehensively characterized and report here the presence, integrity, and persistence of genetic information from plasmid vector into T cells in order to cover the whole GTMP production chain and assess early potential threats. Rapid and consistent preclinical evaluation of a complete production chain of a GTMP required also a new strategy in order to allow a sensitive but also reproducible early detection of small differences, which is applicable to any GTMP. By utilizing a combination of TLA and NGS, we report here that the protein encoding TEG001 transgene DNA sequence (TCRγ-T2A-TCRδ) consistently remained intact throughout the production process into TEG001 medicinal product. TLA technology allowed us to detect very small sequence variants in non-coding regions of the TEG001 insert in the MCB and TEG001 drug product, which appeared, however, not to lead to a growth advantage in long-term cultured TEG001 cells in vitro.
In contrast with previously reported technologies for integration site analysis, such as linear-amplification-mediated PCR (LAM-PCR) techniques,49,50 tagmantation PCR (tag-PCR),51 a complete pipeline named INSPIIRED,52,53 or others54,55 that amplify a small fragment of the transgene, TLA can most importantly combine integrity control of the complete transgene with integration site analysis. As such, this technology can be a valuable tool not only for quality control of a cellular engineered product and persistence in vivo but also for genotoxicity analysis in GTMP studies in case of an unexplained increase in clonal frequency of engineered cells in vivo. A qPCR technique has been applied in this study to relate the transgene copy number in TEG001 cells to that of the MCB clone. Interestingly, the number of integrated TEG001 inserts in the producer cell clone was estimated at nine copies per cell, and although the transgene copies per cell in TEG001 cells was calculated as being a factor 0.72 below, it still exceeded the five copy numbers per cell in TEG001. No more than five transgene copies per cell has been for many years considered as the ultimate threshold for acceptance of copy numbers for GTMPs, based on established quality-control tests used to release a lentiviral-based engineered T cell product targeting HIV-infected cells.46 Consequently, CAR T products do not exceed this threshold.27,30,32,33,36 However, most recently, higher copy numbers became acceptable if defined reasoning has been provided. In contrast with CAR T cells, the introduced γδTCR needs to outcompete the endogenous TCR for CD3 molecules to be expressed at the cell surface,4 implying that higher copy numbers may be required for TEG001 than for CAR T products. In addition, our production process includes an enrichment step, which allows selection for engineered immune cells with only highest expression of the transgene.14,16 Therefore, the ongoing TEG001 trial will not only be pivotal for testing a new type of metabolic cancer targeting with engineered immune cells but also addresses important safety aspects when transgene copy numbers exceed so far considered thresholds.
Inevitably, viral gene engineering techniques imply a risk for the occurrence of mutational and structural variants in the transgene DNA sequence integrated in the host genome. Selection of a producer clone based on vector titer and transgene expression can select against a negative effect of sequence variants on packaging of the vector and protein expression and stability; however, a more in-depth molecular analysis of the transgene is required for clinical application of a GTMP. We observed that two out of the five small-nucleotide variants, located in the non-coding regions of integrated TEG001 transgene, were in the primer binding site. This retroviral region is likely to be highly sensitive for mutations because of its function as primer binding site for host tRNA.40 In case tRNA molecules with an incomplete complementarity to the primer binding site bind and initiate reverse transcription, mutations are introduced in this region. A third variant, a single insertion, was found in the region in between the primer binding site and the packaging signal (Ψ). The other two mutations are at the junction of the R and U regions that is homolog for both the 5′ and 3′ LTRs, and are therefore likely the result of a single mutational event during reverse transcription. These five small-nucleotide variants did not in our perspective imply an altered or additional environmental safety risk of the TEG001 drug product over the reference viral sequences and as such were accepted by the competent authorities.
Our integration site analysis performed on TEG001 medicinal product revealed a heterogeneous integration pattern at the genome level, and heterogeneity decreased following a significant in vitro culture period. We conclude that none of the five small-nucleotide variants present in TEG001 cells lead to a growth advantage, indicative of the absence of an induction of an oncogenic transformation. These data support the impressive number of 1,000 patients treated with TCR and CAR-engineered T cells without the report of an oncogenic transformation of infused engineered T cells.2 Nevertheless, close monitoring of any adverse events remains indispensable and is also mandatory as requested by authorities.45 Thus, although very detailed molecular and genetic analyses were performed following clinical events post CAR T infusion,26,56 our suggested robust and cost-efficient genetic quality-control assay will reduce the risk for unintended side effect of GTMPs. TLA can be used to combine integration site analysis, complete transgene integrity analysis, and analysis of the neighboring DNA up to tens to hundreds of kilobases throughout the complete production chain;39 however, the lead time of these sequencing-based techniques may currently be a limiting factor for implementation of these into release test programs for GTMPs.
In conclusion, we have reported an extensive molecular characterization of TEG001 transgene integrity that resulted in the approval of a phase I clinical study that did not only allow to investigate the safety and tolerability of TEG001 in patients with relapsed/refractory AML, high-risk myelodysplastic syndrome, and relapsed/refractory MM, but also will provide a valuable framework for future GTMPs.
Materials and Methods
TEG001 manufacturing process
We reported recently on the development of a GMP-compliant TEG001 product.16 The production process included the generation of a 293VecRD114 packaging cell clone stably integrated with the TCRγ5-T2A-δ5 transgene (TEG001 transgene). The TEG001 transgene is composed of codon-optimized human TCR sequences from a Vγ9Vδ2T cell clone (clone 5) obtained from a healthy donor5 flanked by 5′ and 3′ LTR sequences and is 3,634 bp in size. In the transgene cassette, the individual γTCR and δTCR chains have been connected with a self-cleaving T2A ribosomal skipping sequence14 (Figure S1A). Following selection of clone 73 with the highest virus titer production and generation of the MCB, the GMP-grade retroviral vector stock was produced (see Figure S1B). The TEG001 manufacturing process includes the collection of T cells via leukapheresis, followed by T cell activation and transduction with the retroviral vector. The viral vector is equipped with viral machinery to reverse transcribe its RNA into DNA encoding the γδTCR to be stably integrated into the patients’ conventional αβT cells. After large-scale expansion, the cell product is harvested, concentrated, and purified to deplete untransduced cells. In-process monitoring and quality-control release tests are implemented before TEG001 is ready for infusion (Figure S1B).
Sample Collection
Plasmid DNA from the TEG001 vector was used as starting material for the production of the MCB clone 73, and a sample of this DNA was used as reference sample in TLA. The vector is named pMP71:TCRδ-T2A-TCRγ and contains: (1) the insert DNA sequence, i.e., the proviral DNA that will be inserted into the genome of the host cell; (2) the transgene DNA sequence as part of the insert, i.e., the TCRγ-T2A-TCRδ transgene cassette; and (3) the vector backbone DNA sequence not integrated into the host genome.
A total of 5 × 106 frozen cells from MCB clone 73 were used, as well as 5 × 106 TEG001 cells derived from four independent full-scale TEG001 production runs (run 28, run 31, run 32, and run 33) for TLA. Cells were taken from the TEG001 drug product and as a consequence, these cells were expanded for 10 days following retroviral transduction. TEG001 cells derived from two independent TEG001 production runs (run 32 and run 33) were expanded for 22 weeks (TEG001-32 22w) and 24 weeks (TEG001-33 24w), respectively, according to a rapid expansion protocol.57
Flow Cytometry
Antibodies used for flow cytometry were pan-γδTCR-phycoerythrin (PE) (clone IMMU510; Beckman Coulter), pan-αβTCR-allophycocyanin (APC) (clone IP26; eBioscience), CD8α-PerCP-Cy5.5 (RPA-T8; BioLegend), CD4-V450 (clone RPA-T4; BD Biosciences), CD62L-FITC (fluorescein isothiocyanate) (Dreg-56; Life Technologies), and CD45RO-PE-Cy7 (UCHL-1; BD Biosciences). Samples were analyzed on BD FACSCanto II or BD LSRFortessa flow cytometer using FACSDiva software (BD Biosciences). Lymphocytes were gated based on forward scatter (FSC) and side scatter (SSC). TEGs were defined as lymphocytes being positive for γδTCR.
TLA and NGS
TLA was performed as described previously39 and depicted in Figure S2. Shortly, cells were crosslinked using formaldehyde, and DNA was digested with NlaIII. Next, samples were ligated, crosslinks reversed, and DNA purified. To obtain circular chimeric DNA molecules for PCR amplification, the DNA molecules were trimmed with NspI and ligated at a DNA concentration of 5 ng/μL to promote intramolecular ligation. NspI was used for its RCATGY recognition sequence that encompasses the CATG recognition sequence of NlaIII. Consequently, only a subset of NlaIII (CATG) sites was (re-) digested, generating DNA fragments of approximately 2 kb and allowing the amplification of entire restriction fragments. Following ligation, DNA was purified, and eight 25-μL PCRs, each containing 100 ng template, were pooled for sequencing. The following inverse primer sets were used in the PCRs: set 1, 5′-ATCGCCGAGACCAAGCTG-3′ and 5′-CGCCATACACGCACAGGG-3′; set 2, 5′-GCCATCGTGCACACCGAG-3′ and 5′-GCCGTAGATGTCCTTCTCCC-3′. The primer sets were used in individual TLA amplifications. PCR products were purified and library prepped using the Illumnia NexteraXT protocol and sequenced on an Illumnia Miseq sequencer.
Mapping and Sequence Alignment
Reads were mapped using BWA-SW, which is a Smith-Waterman alignment tool. This allows partial mapping, which is optimally suited for identifying break-spanning reads. The generated data were mapped against the human genome version hg19 and the TEG001 vector pMP71:TCRδ-T2A-TCRγ sequence. The resulting mapped BAM files were analyzed using IGV software.58 For the identification of the integration sites in the MCB sample, both coverage peaks and the identified breakpoint reads were used. For the breakpoint reads in the MCB sample, the threshold was set to 1%.
qPCR
After thawing of frozen samples of the TEG001 MCB clone 73, TEG001 products and healthy donor PBMC genomic DNA (gDNA) were isolated using QIAamp DNA Blood Mini Kit (QIAGEN) according to the manufacturer’s instructions. Two gene-specific primer sets for TaqMan qPCR were used: primer set 1 for the T2A linker region of the TEG001 transgene (forward primer: 5′-CTGCAACGGCGAGAAGAG-3′, reverse primer: 5′-GCTGATCCGCTCCATGTTAAT-3′, probe: 5′-FAM_TTTCTTCCACATCGCCGCAGGTC_ TAMRA-3′), and primer set 2 for the Gag sequences in the 5′ LTR region of the transgene (forward primer: 5′-CTGTATCAGTTAACCTACCCGAG-3′, reverse primer: 5′-GGCGTAAAACCGAAAGCAAA-3′, probe: 5′-FAM_ CCCAACAAAGCCACGTACCCCT_TAMRA-3′). To correct for the loss of DNA during handling of the samples in preparation for the qPCR assay, we used TaqMan Copy Number Reference Assay RNase P (Applied Biosystems). The RNase P gene has two copies per diploid cell and is used as a reference in GTMP vector copy studies.34 All qPCR analyses were performed with TaqMan Universal PCR Master Mix (Applied Biosystems) on the ViiA7 Real-Time PCR system (Applied Biosystems). Each data point was evaluated in triplicates with mean values used for analysis. A seven-point standard curve to detect copy numbers per unit/DNA was generated consisting of 106 to 101 copies of the vector spiked into a background of 100 ng healthy donor PBMC gDNA. A five-point standard curve of 200 to 0.02 ng healthy donor PBMC gDNA for the reference assay was generated. Standard curves were included in every experiment and met predefined criteria (qPCR efficiency between 90% and 110%, adjusted R2 ≥ 0.99). A total of 100 ng of gDNA of the MCB clone 73 and the TEG products was used as input in a parallel amplification reaction for both qPCR of the transgene and RNaseP reference assay. Ratios of copy number per unit/DNA of TEG001 products compared with MCB clone 73 were calculated with the method of Pfaffl, which takes the efficiencies of the qPCRs into account.59
Author Contributions
Conceptualization, T.S. and J.K.; Methodology, T.S. and J.K.; Analysis, A.J., J.B., and M.S.; Investigation, A.J., T.A., R.D., K.J., J.B., and M.S.; Writing, T.S., A.J., and J.K.; Critical Review, Z.S., J.B., M.S., and M.d.W.; Project Administration, A.D.D.v.M.; Supervision, T.S. and J.K.; Funding Acquisition, J.K. and Z.S. All named authors have made a sufficient contribution to this paper and all agreed to submit the paper in the present form.
Conflicts of Interest
A.J., J.K., and Z.S. are inventors on different patents with γδT cell receptor sequences, recognition mechanisms, and isolation strategies. J.K. receives research funding from and is SA and shareholder of Gadeta (https://www.gadeta.nl/). J.B. and M.S. are employees of Cergentis. All other authors declare no competing interests.
Acknowledgments
Funding for this study was provided by ZonMw grant 43400003 and VIDI-ZonMW grant 917.11.337; KWF grants UU 2010-4669, UU 2013-6426, UU 2014-6790, UU 2015-7601, and UU 2018-11393; and Gadeta to J.K.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.11.030.
Contributor Information
Trudy Straetemans, Email: g.c.m.straetemans@umcutrecht.nl.
Jurgen Kuball, Email: j.h.e.kuball@umcutrecht.nl.
Supplemental Information
References
- 1.Chabannon C., Kuball J., Bondanza A., Dazzi F., Pedrazzoli P., Toubert A., Ruggeri A., Fleischhauer K., Bonini C. Hematopoietic stem cell transplantation in its 60s: A platform for cellular therapies. Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aap9630. eaap9630. [DOI] [PubMed] [Google Scholar]
- 2.June C.H., Sadelain M. Chimeric Antigen Receptor Therapy. N. Engl. J. Med. 2018;379:64–73. doi: 10.1056/NEJMra1706169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ping Y., Liu C., Zhang Y. T-cell receptor-engineered T cells for cancer treatment: current status and future directions. Protein Cell. 2018;9:254–266. doi: 10.1007/s13238-016-0367-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marcu-Malina V., Heijhuurs S., van Buuren M., Hartkamp L., Strand S., Sebestyen Z., Scholten K., Martens A., Kuball J. Redirecting αβ T cells against cancer cells by transfer of a broadly tumor-reactive γδT-cell receptor. Blood. 2011;118:50–59. doi: 10.1182/blood-2010-12-325993. [DOI] [PubMed] [Google Scholar]
- 5.Gründer C., van Dorp S., Hol S., Drent E., Straetemans T., Heijhuurs S., Scholten K., Scheper W., Sebestyen Z., Martens A. γ9 and δ2CDR3 domains regulate functional avidity of T cells harboring γ9δ2TCRs. Blood. 2012;120:5153–5162. doi: 10.1182/blood-2012-05-432427. [DOI] [PubMed] [Google Scholar]
- 6.Scheper W., van Dorp S., Kersting S., Pietersma F., Lindemans C., Hol S., Heijhuurs S., Sebestyen Z., Gründer C., Marcu-Malina V. γδT cells elicited by CMV reactivation after allo-SCT cross-recognize CMV and leukemia. Leukemia. 2013;27:1328–1338. doi: 10.1038/leu.2012.374. [DOI] [PubMed] [Google Scholar]
- 7.Kierkels G.J.J., Scheper W., Meringa A.D., Johanna I., Beringer D.X., Janssen A., Schiffler M., Aarts-Riemens T., Kramer L., Straetemans T. Identification of a tumor-specific allo-HLA-restricted γδTCR. Blood Adv. 2019;3:2870–2882. doi: 10.1182/bloodadvances.2019032409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melandri D., Zlatareva I., Chaleil R.A.G., Dart R.J., Chancellor A., Nussbaumer O., Polyakova O., Roberts N.A., Wesch D., Kabelitz D. The γδTCR combines innate immunity with adaptive immunity by utilizing spatially distinct regions for agonist selection and antigen responsiveness. Nat. Immunol. 2018;19:1352–1365. doi: 10.1038/s41590-018-0253-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sebestyen Z., Scheper W., Vyborova A., Gu S., Rychnavska Z., Schiffler M., Cleven A., Chéneau C., van Noorden M., Peigné C.M. RhoB Mediates Phosphoantigen Recognition by Vγ9Vδ2 T Cell Receptor. Cell Rep. 2016;15:1973–1985. doi: 10.1016/j.celrep.2016.04.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gu S., Sachleben J.R., Boughter C.T., Nawrocka W.I., Borowska M.T., Tarrasch J.T., Skiniotis G., Roux B., Adams E.J. Phosphoantigen-induced conformational change of butyrophilin 3A1 (BTN3A1) and its implication on Vγ9Vδ2 T cell activation. Proc. Natl. Acad. Sci. USA. 2017;114:E7311–E7320. doi: 10.1073/pnas.1707547114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harly C., Guillaume Y., Nedellec S., Peigné C.M., Mönkkönen H., Mönkkönen J., Li J., Kuball J., Adams E.J., Netzer S. Key implication of CD277/butyrophilin-3 (BTN3A) in cellular stress sensing by a major human γδ T-cell subset. Blood. 2012;120:2269–2279. doi: 10.1182/blood-2012-05-430470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sebestyen Z., Prinz I., Déchanet-Merville J., Silva-Santos B., Kuball J. Translating gammadelta (γδ) T cells and their receptors into cancer cell therapies. Nat. Rev. Drug Discov. 2019 doi: 10.1038/s41573-019-0038-z. Published online September 6, 2019. [DOI] [PubMed] [Google Scholar]
- 13.Braham M.V.J., Minnema M.C., Aarts T., Sebestyen Z., Straetemans T., Vyborova A., Kuball J., Öner F.C., Robin C., Alblas J. Cellular immunotherapy on primary multiple myeloma expanded in a 3D bone marrow niche model. OncoImmunology. 2018;7:e1434465. doi: 10.1080/2162402X.2018.1434465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Straetemans T., Gründer C., Heijhuurs S., Hol S., Slaper-Cortenbach I., Bönig H., Sebestyen Z., Kuball J. Untouched GMP-Ready Purified Engineered Immune Cells to Treat Cancer. Clin. Cancer Res. 2015;21:3957–3968. doi: 10.1158/1078-0432.CCR-14-2860. [DOI] [PubMed] [Google Scholar]
- 15.Johanna I., Straetemans T., Heijhuurs S., Aarts-Riemens T., Norell H., Bongiovanni L., de Bruin A., Sebestyen Z., Kuball J. Evaluating in vivo efficacy - toxicity profile of TEG001 in humanized mice xenografts against primary human AML disease and healthy hematopoietic cells. J. Immunother. Cancer. 2019;7:69. doi: 10.1186/s40425-019-0558-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Straetemans T., Kierkels G.J.J., Doorn R., Jansen K., Heijhuurs S., Dos Santos J.M., van Muyden A.D.D., Vie H., Clemenceau B., Raymakers R. GMP-Grade Manufacturing of T Cells Engineered to Express a Defined γδTCR. Front. Immunol. 2018;9:1062. doi: 10.3389/fimmu.2018.01062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biasco L., Rothe M., Büning H., Schambach A. Analyzing the Genotoxicity of Retroviral Vectors in Hematopoietic Cell Gene Therapy. Mol. Ther. Methods Clin. Dev. 2017;8:21–30. doi: 10.1016/j.omtm.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serrao E., Engelman A.N. Sites of retroviral DNA integration: From basic research to clinical applications. Crit. Rev. Biochem. Mol. Biol. 2016;51:26–42. doi: 10.3109/10409238.2015.1102859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vigdal T.J., Kaufman C.D., Izsvák Z., Voytas D.F., Ivics Z. Common physical properties of DNA affecting target site selection of sleeping beauty and other Tc1/mariner transposable elements. J. Mol. Biol. 2002;323:441–452. doi: 10.1016/s0022-2836(02)00991-9. [DOI] [PubMed] [Google Scholar]
- 20.Cavazzana-Calvo M., Payen E., Negre O., Wang G., Hehir K., Fusil F., Down J., Denaro M., Brady T., Westerman K. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hacein-Bey-Abina S., Garrigue A., Wang G.P., Soulier J., Lim A., Morillon E., Clappier E., Caccavelli L., Delabesse E., Beldjord K. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Recchia A., Bonini C., Magnani Z., Urbinati F., Sartori D., Muraro S., Tagliafico E., Bondanza A., Stanghellini M.T., Bernardi M. Retroviral vector integration deregulates gene expression but has no consequence on the biology and function of transplanted T cells. Proc. Natl. Acad. Sci. USA. 2006;103:1457–1462. doi: 10.1073/pnas.0507496103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newrzela S., Cornils K., Li Z., Baum C., Brugman M.H., Hartmann M., Meyer J., Hartmann S., Hansmann M.L., Fehse B., von Laer D. Resistance of mature T cells to oncogene transformation. Blood. 2008;112:2278–2286. doi: 10.1182/blood-2007-12-128751. [DOI] [PubMed] [Google Scholar]
- 24.Scholler J., Brady T.L., Binder-Scholl G., Hwang W.T., Plesa G., Hege K.M., Vogel A.N., Kalos M., Riley J.L., Deeks S.G. Decade-long safety and function of retroviral-modified chimeric antigen receptor T cells. Sci. Transl. Med. 2012;4:132ra53. doi: 10.1126/scitranslmed.3003761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biasco L., Ambrosi A., Pellin D., Bartholomae C., Brigida I., Roncarolo M.G., Di Serio C., von Kalle C., Schmidt M., Aiuti A. Integration profile of retroviral vector in gene therapy treated patients is cell-specific according to gene expression and chromatin conformation of target cell. EMBO Mol. Med. 2011;3:89–101. doi: 10.1002/emmm.201000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fraietta J.A., Nobles C.L., Sammons M.A., Lundh S., Carty S.A., Reich T.J., Cogdill A.P., Morrissette J.J.D., DeNizio J.E., Reddy S. Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells. Nature. 2018;558:307–312. doi: 10.1038/s41586-018-0178-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalos M., Levine B.L., Porter D.L., Katz S., Grupp S.A., Bagg A., June C.H. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci. Transl. Med. 2011;3:95ra73. doi: 10.1126/scitranslmed.3002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chapuis A.G., Egan D.N., Bar M., Schmitt T.M., McAfee M.S., Paulson K.G., Voillet V., Gottardo R., Ragnarsson G.B., Bleakley M. T cell receptor gene therapy targeting WT1 prevents acute myeloid leukemia relapse post-transplant. Nat. Med. 2019;25:1064–1072. doi: 10.1038/s41591-019-0472-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Loenen M.M., de Boer R., van Liempt E., Meij P., Jedema I., Falkenburg J.H., Heemskerk M.H. A Good Manufacturing Practice procedure to engineer donor virus-specific T cells into potent anti-leukemic effector cells. Haematologica. 2014;99:759–768. doi: 10.3324/haematol.2013.093690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lock D., Mockel-Tenbrinck N., Drechsel K., Barth C., Mauer D., Schaser T., Kolbe C., Al Rawashdeh W., Brauner J., Hardt O. Automated Manufacturing of Potent CD20-Directed Chimeric Antigen Receptor T Cells for Clinical Use. Hum. Gene Ther. 2017;28:914–925. doi: 10.1089/hum.2017.111. [DOI] [PubMed] [Google Scholar]
- 31.Hollyman D., Stefanski J., Przybylowski M., Bartido S., Borquez-Ojeda O., Taylor C., Yeh R., Capacio V., Olszewska M., Hosey J. Manufacturing validation of biologically functional T cells targeted to CD19 antigen for autologous adoptive cell therapy. J. Immunother. 2009;32:169–180. doi: 10.1097/CJI.0b013e318194a6e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blaeschke F., Stenger D., Kaeuferle T., Willier S., Lotfi R., Kaiser A.D., Assenmacher M., Döring M., Feucht J., Feuchtinger T. Induction of a central memory and stem cell memory phenotype in functionally active CD4+ and CD8+ CAR T cells produced in an automated good manufacturing practice system for the treatment of CD19+ acute lymphoblastic leukemia. Cancer Immunol. Immunother. 2018;67:1053–1066. doi: 10.1007/s00262-018-2155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X., Naranjo A., Brown C.E., Bautista C., Wong C.W., Chang W.C., Aguilar B., Ostberg J.R., Riddell S.R., Forman S.J., Jensen M.C. Phenotypic and functional attributes of lentivirus-modified CD19-specific human CD8+ central memory T cells manufactured at clinical scale. J. Immunother. 2012;35:689–701. doi: 10.1097/CJI.0b013e318270dec7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedman K.M., Garrett T.E., Evans J.W., Horton H.M., Latimer H.J., Seidel S.L., Horvath C.J., Morgan R.A. Effective Targeting of Multiple B-Cell Maturation Antigen-Expressing Hematological Malignances by Anti-B-Cell Maturation Antigen Chimeric Antigen Receptor T Cells. Hum. Gene Ther. 2018;29:585–601. doi: 10.1089/hum.2018.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davila M.L., Riviere I., Wang X., Bartido S., Park J., Curran K., Chung S.S., Stefanski J., Borquez-Ojeda O., Olszewska M. Efficacy and toxicity management of 19-28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci. Transl. Med. 2014;6:224ra25. doi: 10.1126/scitranslmed.3008226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brentjens R.J., Davila M.L., Riviere I., Park J., Wang X., Cowell L.G., Bartido S., Stefanski J., Taylor C., Olszewska M. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci. Transl. Med. 2013;5:177ra38. doi: 10.1126/scitranslmed.3005930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cameron B.J., Gerry A.B., Dukes J., Harper J.V., Kannan V., Bianchi F.C., Grand F., Brewer J.E., Gupta M., Plesa G. Identification of a Titin-derived HLA-A1-presented peptide as a cross-reactive target for engineered MAGE A3-directed T cells. Sci. Transl. Med. 2013;5:197ra103. doi: 10.1126/scitranslmed.3006034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Porter D.L., Levine B.L., Kalos M., Bagg A., June C.H. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N. Engl. J. Med. 2011;365:725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Vree P.J., de Wit E., Yilmaz M., van de Heijning M., Klous P., Verstegen M.J., Wan Y., Teunissen H., Krijger P.H., Geeven G. Targeted sequencing by proximity ligation for comprehensive variant detection and local haplotyping. Nat. Biotechnol. 2014;32:1019–1025. doi: 10.1038/nbt.2959. [DOI] [PubMed] [Google Scholar]
- 40.Hu W.S., Hughes S.H. HIV-1 reverse transcription. Cold Spring Harb. Perspect. Med. 2012;2 doi: 10.1101/cshperspect.a006882. a006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parthasarathi S., Varela-Echavarría A., Ron Y., Preston B.D., Dougherty J.P. Genetic rearrangements occurring during a single cycle of murine leukemia virus vector replication: characterization and implications. J. Virol. 1995;69:7991–8000. doi: 10.1128/jvi.69.12.7991-8000.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Serrao E., Ballandras-Colas A., Cherepanov P., Maertens G.N., Engelman A.N. Key determinants of target DNA recognition by retroviral intasomes. Retrovirology. 2015;12:39. doi: 10.1186/s12977-015-0167-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gattinoni L., Klebanoff C.A., Palmer D.C., Wrzesinski C., Kerstann K., Yu Z., Finkelstein S.E., Theoret M.R., Rosenberg S.A., Restifo N.P. Acquisition of full effector function in vitro paradoxically impairs the in vivo antitumor efficacy of adoptively transferred CD8+ T cells. J. Clin. Invest. 2005;115:1616–1626. doi: 10.1172/JCI24480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Restifo N.P., Gattinoni L. Lineage relationship of effector and memory T cells. Curr. Opin. Immunol. 2013;25:556–563. doi: 10.1016/j.coi.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aiuti A., Cossu G., de Felipe P., Galli M.C., Narayanan G., Renner M., Stahlbom A., Schneider C.K., Voltz-Girolt C. The committee for advanced therapies’ of the European Medicines Agency reflection paper on management of clinical risks deriving from insertional mutagenesis. Hum. Gene Ther. Clin. Dev. 2013;24:47–54. doi: 10.1089/humc.2013.119. [DOI] [PubMed] [Google Scholar]
- 46.Dropulic B., Schonely K., Slepushkin V., Lu X., Andre K., Boehmer J., Bengston K., Doub M., Cohen R., Berlinger D. QC Release Testing of an HIV-1 based Lentiviral Vector Lot and Transduced Cellular Product. Bioprocess. J. 2003;2:39–47. [Google Scholar]
- 47.Hildinger M., Abel K.L., Ostertag W., Baum C. Design of 5′ untranslated sequences in retroviral vectors developed for medical use. J. Virol. 1999;73:4083–4089. doi: 10.1128/jvi.73.5.4083-4089.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grez M., Akgün E., Hilberg F., Ostertag W. Embryonic stem cell virus, a recombinant murine retrovirus with expression in embryonic stem cells. Proc. Natl. Acad. Sci. USA. 1990;87:9202–9206. doi: 10.1073/pnas.87.23.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paruzynski A., Arens A., Gabriel R., Bartholomae C.C., Scholz S., Wang W., Wolf S., Glimm H., Schmidt M., von Kalle C. Genome-wide high-throughput integrome analyses by nrLAM-PCR and next-generation sequencing. Nat. Protoc. 2010;5:1379–1395. doi: 10.1038/nprot.2010.87. [DOI] [PubMed] [Google Scholar]
- 50.Schmidt M., Schwarzwaelder K., Bartholomae C., Zaoui K., Ball C., Pilz I., Braun S., Glimm H., von Kalle C. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR) Nat. Methods. 2007;4:1051–1057. doi: 10.1038/nmeth1103. [DOI] [PubMed] [Google Scholar]
- 51.Hamada M., Nishio N., Okuno Y., Suzuki S., Kawashima N., Muramatsu H., Tsubota S., Wilson M.H., Morita D., Kataoka S. Integration Mapping of piggyBac-Mediated CD19 Chimeric Antigen Receptor T Cells Analyzed by Novel Tagmentation-Assisted PCR. EBioMedicine. 2018;34:18–26. doi: 10.1016/j.ebiom.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sherman E., Nobles C., Berry C.C., Six E., Wu Y., Dryga A., Malani N., Male F., Reddy S., Bailey A. INSPIIRED: A Pipeline for Quantitative Analysis of Sites of New DNA Integration in Cellular Genomes. Mol. Ther. Methods Clin. Dev. 2016;4:39–49. doi: 10.1016/j.omtm.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berry C.C., Nobles C., Six E., Wu Y., Malani N., Sherman E., Dryga A., Everett J.K., Male F., Bailey A. INSPIIRED: Quantification and Visualization Tools for Analyzing Integration Site Distributions. Mol. Ther. Methods Clin. Dev. 2016;4:17–26. doi: 10.1016/j.omtm.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang G.P., Berry C.C., Malani N., Leboulch P., Fischer A., Hacein-Bey-Abina S., Cavazzana-Calvo M., Bushman F.D. Dynamics of gene-modified progenitor cells analyzed by tracking retroviral integration sites in a human SCID-X1 gene therapy trial. Blood. 2010;115:4356–4366. doi: 10.1182/blood-2009-12-257352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brady T., Roth S.L., Malani N., Wang G.P., Berry C.C., Leboulch P., Hacein-Bey-Abina S., Cavazzana-Calvo M., Papapetrou E.P., Sadelain M. A method to sequence and quantify DNA integration for monitoring outcome in gene therapy. Nucleic Acids Res. 2011;39:e72. doi: 10.1093/nar/gkr140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruella M., Xu J., Barrett D.M., Fraietta J.A., Reich T.J., Ambrose D.E., Klichinsky M., Shestova O., Patel P.R., Kulikovskaya I. Induction of resistance to chimeric antigen receptor T cell therapy by transduction of a single leukemic B cell. Nat. Med. 2018;24:1499–1503. doi: 10.1038/s41591-018-0201-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Voss R.H., Kuball J., Theobald M. Designing TCR for cancer immunotherapy. Methods Mol. Med. 2005;109:229–256. doi: 10.1385/1-59259-862-5:229. [DOI] [PubMed] [Google Scholar]
- 58.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pfaffl A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Research. 2001 doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.