Abstract
Introduction
SCLC accounts for approximately 250,000 deaths worldwide each year. Acquisition of adequate tumor biopsy samples is challenging, and liquid biopsies present an alternative option for patient stratification and response monitoring.
Methods
We applied whole genome next-generation sequencing to circulating free DNA (cfDNA) from 39 patients with limited-stage (LS) SCLC and 30 patients with extensive-stage SCLC to establish genome-wide copy number aberrations and also performed targeted mutation analysis of 110 SCLC associated genes. Quantitative metrics were calculated for copy number aberrations, including percent genome amplified (PGA [the percentage of genomic regions amplified]), Z-score (a measure of standard deviation), and Moran’s I (a measure of spatial autocorrelation). In addition CellSearch, an epitope-dependent enrichment platform, was used to enumerate circulating tumor cells (CTCs) from a parallel blood sample.
Results
Genome-wide and targeted cfDNA sequencing data identified tumor-related changes in 94% of patients with LS SCLC and 100% of patients with extensive-stage SCLC. Parallel analysis of CTCs based on at least 1 CTC/7.5 mL of blood increased tumor detection frequencies to 95% for LS SCLC. Both CTC counts and cfDNA readouts correlated with disease stage and overall survival.
Conclusions
We demonstrate that a simple cfDNA genome-wide copy number approach provides an effective means of monitoring patients through treatment and show that targeted cfDNA sequencing identifies potential therapeutic targets in more than 50% of patients. We are now incorporating this approach into additional studies and trials of targeted therapies.
Keywords: Small cell lung cancer, Circulating free DNA, Copy number, TP53, Patient monitoring
Introduction
SCLC is an aggressive neuroendocrine tumor accounting for 10% to 15% of the 1.8 million lung cancers diagnosed worldwide annually. SCLC is strongly associated with tobacco smoke carcinogen exposure1 and characterized by high growth rate and early metastatic spread. Most patients with SCLC present with extensive-stage (ES) SCLC with distant metastases and median overall survival (OS) of less than 1 year despite treatment with platinum-based chemotherapy.2 Patients with limited-stage (LS) SCLC, which is localized and encompassable within a single radiotherapy field, have a median survival of 18 months. Durable responses to treatment (>5 years) are seen in approximately 20% to 30% of patients with LS SCLC but only less than 2% of patients with ES SCLC.3
Genomic analysis of SCLC tumors have identified extensive copy number alterations (CNAs) and high mutation rates.4,5 There is almost universal inactivation and prevalent loss of the tumor suppressors tumor protein p53 gene (TP53) and retinoblastoma 1 gene (RB1) and frequent amplification of transcriptional regulators (SRY-box transcription factor 2 gene [SOX2], nuclear factor I B gene [NFIB], v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog gene [MYCL1], v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog gene [MYCN], and v-myc avian myelocytomatosis viral oncogene homolog gene [MYC]), with recurrent mutations of Notch and histone acetyltransferase genes (CREB binding protein gene [CREBBP] and E1A binding protein p300 gene [EP300]).4, 5, 6 Tumor biopsies, and particularly serial biopsies, in SCLC are a significant challenge,7,8 hampering our understanding of the rapidly evolving biology of SCLC and development of targeted therapies. Surgical resection of SCLC is rare, yet by necessity, most genomic studies have been performed on resected tumors4,9 and hence may be unrepresentative of most patients. Fine-needle aspirates or biopsy samples are often of poor quality and/or quantity or unavailable for research. Nevertheless, recent advances that have started to deliver new therapy candidates including exploitation of Notch ligand, delta-like ligand 3 to deliver a cytotoxic payload,10 synthetic lethal strategies targeting cell cycle regulation, oncogene-driven replication stress and DNA damage repair (DDR) responses are under evaluation in early clinical trials.11 Positive results are also emerging from trials of immune checkpoint targeting, including the phase III trial IMPower133, which combines immune checkpoint inhibition with cytotoxic chemotherapy which has shown significantly longer OS and progression-free survival in the treatment arm.12,13 Over the past decade, it has been established that a “liquid biopsy” sample from peripheral blood can facilitate analysis of tumor DNA present in either circulating tumor cells (CTCs)8 or circulating free DNA (cfDNA).7,14 Liquid biopsy data often correlates with the genomic variants in matched tumor tissue and can extend the genomic profile detected.8,15,16 Liquid biopsies overcome many limitations associated with tissue sampling and can be scheduled before, during, and after therapy, enabling assessment of tumor burden, treatment response, and genetic changes associated with drug resistance.17
A number of studies, including our own, have shown that CTC numbers are higher in SCLC than in other cancer types and constitute an independent prognostic biomarker for OS.18 We also demonstrated that a CNA signature, measured in single CTCs from pretreatment blood samples, predicts duration of response to chemotherapy.8 Despite widespread development of cfDNA molecular readouts for diagnostic, predictive, prognostic, and minimal residual disease detection across many cancer types,14,16,19 there have been few studies of ctDNA in SCLC.7,20, 21, 22, 23 For optimal use in SCLC clinical trials and eventual routine clinical implementation, a cfDNA-based assay should (1) quantify circulating tumor DNA (ctDNA) to monitor the presence and extent of disease and (2) identify ctDNA alterations that inform therapeutic decision making. Prior studies have demonstrated that SCLC ctDNA can be identified and profiled by detecting either a single gene of interest, TP53,7 or a panel of SCLC-associated genes,20,21 as well as for patient monitoring using a panel of 430 genes.22
Here we describe a flexible and sensitive workflow for ctDNA detection based on next-generation sequencing (NGS) of cfDNA that was developed for use in SCLC clinical trials (Fig. 1A). After the generation of a single whole genome NGS library from each SCLC cfDNA sample, an initial aliquot is used for the detection of CNA by means of shallow whole genome sequencing, and a second aliquot is used for target enrichment of a panel of 110 genes recurrently mutated in SCLC. We applied this simple workflow to a pilot study of 69 samples from patients with SCLC (39 with LS SCLC and 30 with ES SCLC) and detected tumor associated-changes in cfDNA, including both CNA and putative cancer driver mutations in 94% of patients with LS SCLC and 100% of patients with ES SCLC. Additionally, for six patients, we demonstrate that cfDNA CNA and targeted analysis provide an effective means of disease monitoring.
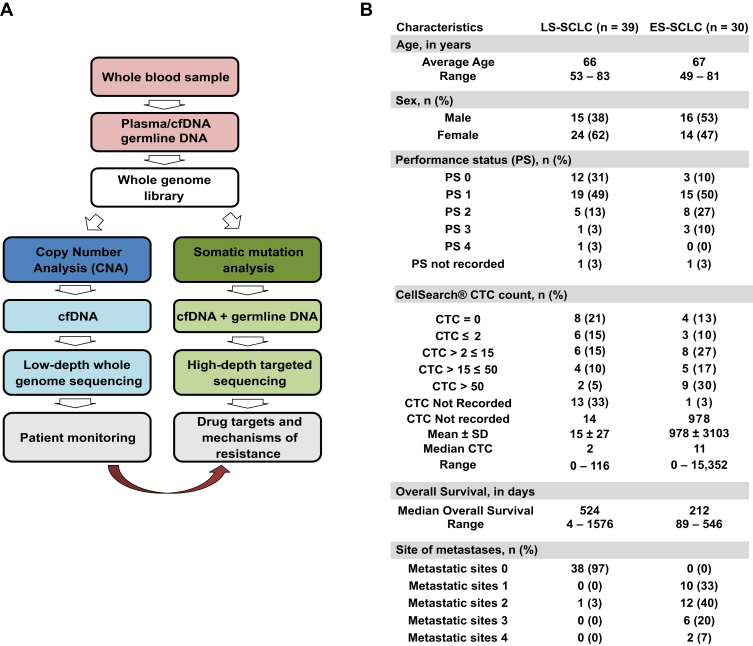
Figure 1.
(A) Workflow illustrating genomic analysis of SCLC whole blood. Blood samples for circulating free DNA (cfDNA) analysis was collected in Cell-Free DNA blood collection tubes (Streck), CellSave, Becton Dickinson (BD) Vacutainer K2EDTA, or BD Vacutainer Heparin and for germline DNA analysis in the BD Vacutainer K2EDTA vacutainer. (B) Demographics of 69 patients with SCLC included in this study. LS, limited-stage; ES, extensive-stage; CTC, circulating tumor cell.
Materials and Methods
Collection of Blood Samples from NCCs and Patients
Blood samples were collected in either Cell-Free DNA BCT tubes (Streck, Omaha, NE), CellSave, BD Vacutainer K2 ethylenediaminetetraacetic acid (K2EDTA), or BD Vacutainer heparin for cfDNA analysis along with a BD Vacutainer K2EDTA vacutainer (BD Biosciences, Heidelberg, Germany) for germline DNA analysis. All samples were collected either from volunteers without cancer, referred to as non-cancer controls (NNCs) (i.e., persons who do not currently have cancer or were not being treated for it, University of Manchester ethics committee approval no. 2017-2761-4606), or from patients with SCLC (ChemoRes and CONVERT Trials) after receipt of informed consent in compliance with the Declaration of Helsinki and Good Clinical Practice after approval from the internal review and the ethics boards of the Christie Hospital National Health Service Trust (research ethics committee [REC] approval no. 07/H1014/96). The ChemoRes trial was reviewed in the United Kingdom by the National Research Ethics Service Committee North West–Greater Manchester West, which granted ethics approval for the study on February 26, 2008 (REC reference no. 07/H1014/96). The CONVERT trial was reviewed in the UK by the National Research Ethics Service Committee North West–Greater Manchester Central, which granted ethics approval for the study on December 21, 2007 (REC reference no. 07/H1008/229). Samples were collected for this study from a total of 32 NCCs and 69 patients with SCLC (Fig. 1B). We have examined whether the inclusion of heparin plasma samples for cfDNA affects the overall readouts, and this is shown in Supplementary Fig. 1.
Preparation of CNA Control Nucleosomal DNA
For nucleosomal fragmentation we applied the EZ Nucleosomal DNA Prep Kit (Zymo Research, Orange, CA) to the SCLC cell line H446 and peripheral blood mononuclear cells (PBMCs). Serial dilutions were created to give H446 DNA as 100%, 20%, 10%, 5%, 1%, and 0% against a background of PBMC DNA (Supplementary Fig. 2A and B). Each diluted sample was subjected to whole genome NGS using inputs of 20 ng and 5 ng to approximate the range of input cfDNA seen in patient samples. All NGS samples were sequenced at low depth to evaluate CNAs.
Circulating Cell-Free DNA and Germline DNA Extraction
Blood sample processing, extraction of cfDNA and germline DNA, and subsequent DNA quantification were performed as previously described.16 In brief, cfDNA was isolated by using the QIAmp Circulating Nucleic Acid Kit (Qiagen) according to the manufacturer’s instructions and/or the QIAsymphony with the Circulating DNA Kit (Qiagen). Germline DNA was isolated from EDTA whole blood by using the QIAmp Blood Mini Kit (Qiagen) according to the manufacturer’s instructions and sheared to 200 to 300 base pairs on the Bioruptor Pico (Diagenode). Sheared germline DNA and cfDNA yields were quantified by using the TaqMan RNase P Detection Reagents Kit (Life Technologies).
NGS Library Preparation and Whole Genome Sequencing
Whole genome cfDNA and germline DNA libraries from the patients and from NCCs were carried out as previously described.16 In brief, whole genome libraries were generated from 2 ng to 25 ng of cfDNA or from 25 ng of sheared germline DNA by using Accel-NGS 2S DNA Library Kits for the Illumina platform (Swift Biosciences) according to the manufacturer’s instructions with the following modifications. Library amplification and indexing was carried out with KAPA HiFi HotStart Polymerase Chain Reaction (PCR) Kits (Kapa Biosystems) and NEBNext Index Primers for Illumina (New England Biolabs). Paired-end sequencing (300 cycles) was performed on the Illumina MiSeq benchtop sequencer (Illumina, San Diego, CA) by using the MiSeq Reagent Kit v2 (Illumina).
Targeted NGS Analysis
Targeted NGS of 110 SCLC associated genes (Supplementary Table 1) for whole genome libraries from cfDNA and corresponding germline DNA were carried out by using Agilent SureSelectXT essentially as described previously.16 In brief, 1 μg of each whole genome indexed library was pooled (up to 6 μg or six libraries) as input for custom capture (110-gene panel) on SureSelectXT Reagent Kits (Agilent) according to the manufacturer’s instructions. Captured libraries were amplified by using KAPA HiFi HotStart PCR Kits and quantified by using the KAPA Library Quantification qPCR kit (Roche). Libraries were paired-end sequenced on an Illumina NextSeq 500, 2 × 150 base pairs High Output V2 Kit (Illumina).
Somatic Mutation Detection from Targeted Resequencing Data
Somatic mutations and indels were called as was previously described.16 FASTQ files were generated from the sequencer’s output by using Illumina bcl2fastq2 software (version 2.17.1.14, Illumina) with the default chastity filter to select sequence reads for subsequent analysis. All sequencing reads were aligned to the human genome reference sequence (GRCh37) by using the Burrows-Wheeler aligner (version 0.7.12) maximal exact matches algorithm. Picard tools (version 2.1.0) were used to mark and/or remove PCR duplicates and to calculate sequencing metrics. Somatic point mutations were called by using both MuTect (version 1) and the commercial software, Biomedical Genomics Workbench (BGW) (version .5.0) (Qiagen), by comparing plasma cfDNA with germline control DNA. Somatic InDels were called by using both VarScan and BGW. Mutations called by two independent pipelines (MuTect + BGW or VarScan + BGW) were classed as high-confidence and kept. Only cfDNA variants with a variant allele fraction (VAF) at least three times that of the matching germline DNA VAF were called to filter out likely clonal haematopoiesis and germline variants from the cfDNA signal.24
The quality control measures were as follows. First, all samples were considered for analysis if they had a minimum coverage of 100× for cfDNA and the corresponding germline DNA. Second, if the error rate as measured by Qualimap was above 1.5% for cfDNA and germline DNA, these samples were rejected from further analysis (Supplementary Table 2).
CNA Metrics from Low-Coverage Whole Genome Sequencing Data
CNA analysis was performed by using HMMCopy after size selection of cfDNA fragments; down-sampling of uniquely mapped sequencing reads to 1 million and removal of noisy bins. Further CNA scores—PGA, z score, and Moran’s I (MI)—were calculated. The Supplementary Appendix provides detailed information of the bioinformatic analysis for CNA and score calculation.
Statistical Analyses
For the survival analysis, the continuous variables in the data set were not normally distributed, nor could they be normalized by using transformation. The data have been categorized by using the maximally selected rank statistic to select the optimal cutpoint. None of the models violated the proportionality of hazard assumption. Selection of the predictors for the multivariate model was done by using elastic net penalized regression with the significant univariate variables as input. Multivariate imputation by chained equations with predictive mean matching was used to handle the missing data. The hyperparameters of the elastic net were selected by using 10-fold cross validation. Selected predictors that are correlated have been excluded on the basis of their variance inflation factor.
Associations between disease stage and mutation profiles, CTC number, CNA metrics, and VAFs were tested by using Wilcoxon rank sum tests. Overall quantification of cfDNA (genome equivalents per milliliter of plasma), median fragment length of cfDNA, PGA, z score, MI, RB1 VAF, TP53 VAF, highest VAF all genes, and CTC count at baseline number were compared by using Spearman’s rho analysis. Associations of clinical variables (stage, sex, and performance status) and cfDNA readouts were compared by using Fisher’s exact test.
Cancer Genome Interpreter
All somatic nonsynonymous variants detected across the 62 cfDNA samples from patients with SCLC were fed into an online platform, Cancer Genome Interpreter (www.cancergenomeinterpreter.org),25 to interpret the potential of variants detected in our study. This platform also allows for identification of biomarkers of response to anticancer drugs and the evidence that support the same (Supplementary Tables 3–5).
Results
Evaluation of CNA Metrics and Sensitivity in Control Samples
CNA data generated by low-pass, whole genome NGS applied to a titration of PBMC/H446 nucleosomal DNA admixtures are shown in Supplementary Figure 2A and B and detailed in the Materials and Methods. In addition to visualizing copy number gains and losses across the genome (see Supplementary Fig. 2B), we generated and compared three CNA numerical outputs: 1) Percent genome amplified (PGA), 2) Z-score and 3) Moran's I, reflecting different aspects of genome-wide changes measurable in cfDNA: values reported were above the thresholds set by using NCCs (see Supplementary Fig. 2A and B). Percent genome amplified (PGA) sums the percentage of genome with a measurable copy number gain as determined by HMMCopy26, Z-score is a previously published cfDNA metric27,28, and MI is a measure of spatial autocorrelation often used to study stochastic processes such as in geographical sciences29 that we evaluated as a novel measure to gauge tumor content in cfDNA. A clear decrease in copy number visualization was observed as correlating with the proportion of H446 DNA present in the titration samples with a decrease in numerical values of each of the CNA metrics (see Supplementary Fig. 2A and B). PGA and Z-score measurements exceeded those of NCCs in mixtures containing 10% H446 DNA. However, the MI metric was more sensitive, identifying differences relative to NCCs in mixtures containing 5% H446 DNA (see Supplementary Fig. 2B).
CNA Analysis of Patient Samples
We applied our approach to samples from 30 patients with ES SCLC, 39 patients with LS SCLC, and 16 independent samples from NCCs. The blood samples from all 69 patients (see Fig. 1B) were drawn at diagnosis (before treatment). To increase the sensitivity of genome-wide CNA detection, we exploited recent observations that ctDNA in the blood of patients with cancer is slightly shorter than cfDNA from NCCs.30,31 To establish whether the reduced ctDNA fragment length seen with other solid cancers30,32 is also seen in SCLC, we examined the median fragment length of all ES SCLC, LS SCLC, and NCC cfDNA samples and showed that the fragment size distribution in each group was statistically distinct from that of the each of the others (Supplementary Fig. 2C). Further, because selection of NGS reads based on fragment size increased sensitivity for both PGA and Z-score outputs (Supplementary Table 6 and see also Supplementary Fig. 3), this size selection strategy was applied in all subsequent CNA analyses.
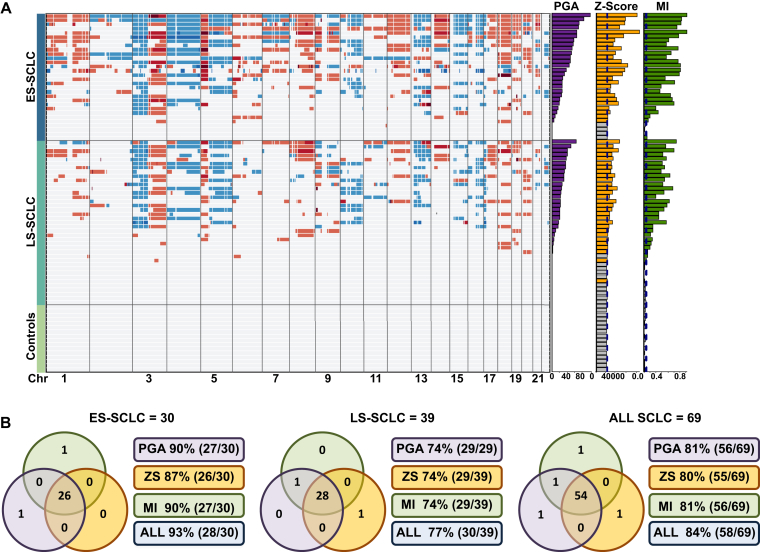
CNA readout (PGA, Z-score, and MI) thresholds were set by using 16 cfDNA NCC samples (C1–C16) and evaluated by applying them to an additional 16 test NCC samples (C17–C32) and all 69 clinical samples (Supplementary Table 7). The output from each of the three CNA metrics for all samples (Fig. 2A and B and Supplementary Table 8) shows clear CNA detectable in most of the SCLC cfDNA samples, whereas none of the 16 NCCs gave values above the predefined thresholds (see Fig. 2A and B). A comparison of the three CNA readout detection sensitivities revealed a high degree of overlap, with MI showing the highest overall sensitivity and z score showing the lowest (Supplementary Table 9 and see also Fig. 2A and B and Supplementary Table 8). Using CNA readouts, we detected tumor-related changes in 84% of all SCLC samples (58 of 69); 93% of the ES SCLC samples (28 of 30), and 77% of the LS SCLC samples (30 of 39) (see Fig. 2B).
Figure 2.
Copy number alterations (CNA) of patients with limited-stage (LS) SCLC, patients with extensive-stage (ES) SCLC, and controls. (A) Copy number heatmap of circulating free DNA from 69 patients with SCLC. The top grouping includes all patients with ES SCLC, the middle group includes all patients with LS SCLC, and the bottom group shows noncancer controls. Each row represents a circulating free DNA analysis of a single patient. For the copy number heatmap, the x axis represents each chromosome from chromosome 1 to chromosome 22 with regions in red indicating gains in copy number and regions in blue indicating loss of copy number. For each sample, the values of the whole genome CNA metrics percent genome amplified (PGA), Z-score (ZS), and Moran's I (MI) are plotted as bar graphs to the right of the main heatmap. For each group, the patient samples are arranged accordint to the descending value of PGA. (B) A comparison of the sensitivity of CNA metrics. The three sets of Venn diagrams show the number of samples identified as positive for tumor DNA by using PGA,ZS, and MI values for all patients, as well as for patients with ES SCLC and LS SCLC shown as separate groups. Chr, chromosome; ES, extensive-stage; LS, limited-stage; PGA, Percent genome amplified; ZS, Z-score; MI, Moran's I.
An examination of specific gains and losses identified in cfDNA revealed SCLC associated copy number losses (chromosomes 3p and 4) and gains (3q and 5p) in 90% of patients with ES SCLC (27 of 30) and 74% of patients with LS SCLC (29 of 39) (Supplementary Tables 10 and 11 and see also Fig. 2B). Previously, studies identified genes frequently lost or amplified in SCLC,4,6,12 and these were confirmed in our cfDNA CNA analysis. These SCLC-prevalent changes included copy number gains for SOX2 (36 of 69 samples), MYC (21 of 69 samples), NFIB (16 of 69 samples), and CD274 molecule gene (CD274) (14 of 69 samples) and copy number losses for contactin 3 gene (CNTN3) (41 of 69 samples), fragile histidine triad gene (FHIT) (40 of 69 samples), Ras association domain family member 1 gene (RASSF1) (38 of 69 samples), RB1 (24 of 69 samples), and kinesin heavy chain member 2A gene (KIF2A) (20 of 69 samples) (see Supplementary Tables 10 and 11). Statistically significant differences in CNA metrics were observed between ES SCLC and LS SCLC (Supplementary Fig. 4A).
Targeted Sequencing of 110 Genes
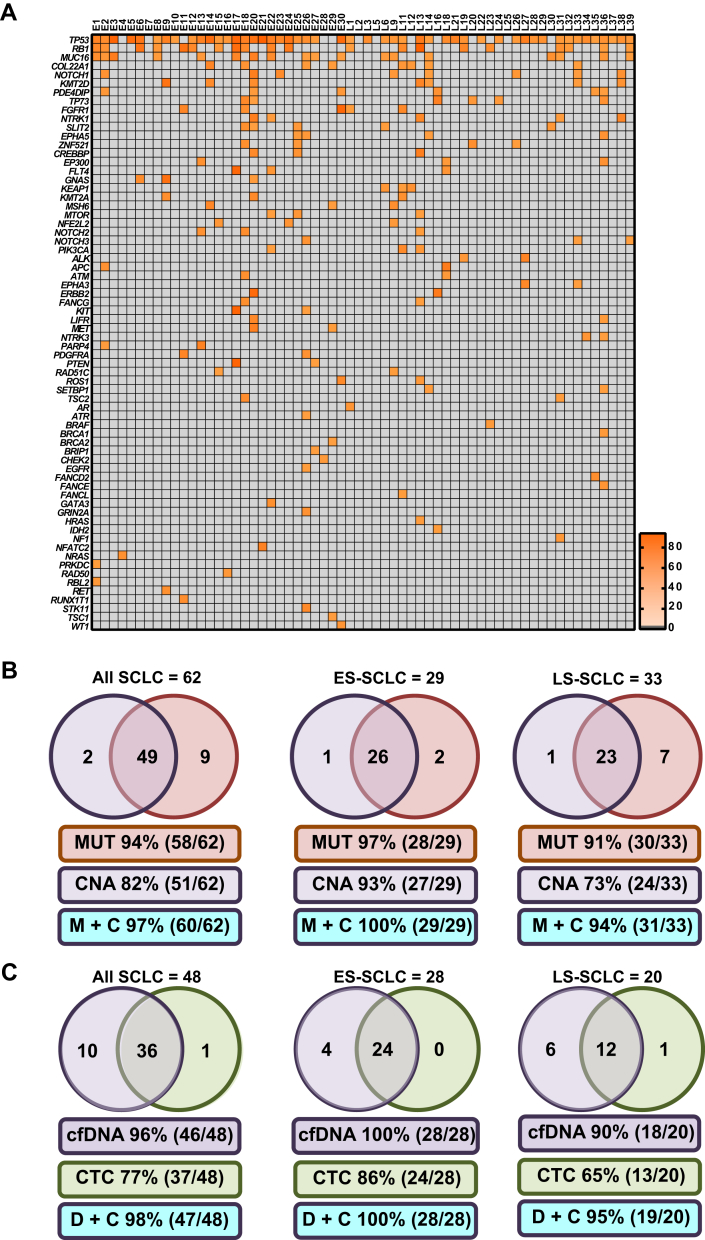
Targeted sequencing of 110 SCLC-associated genes (see Supplementary Table 1) was performed to detect somatic nonsynonymous mutations present in cfDNA. Of the 69 cfDNA samples analyzed for CNA, 62 (29 ES SCLC and 33 LS SCLC samples) passed quality control for targeted sequencing (see Supplementary Table 2). No somatic nonsynonymous mutations were detected in any of the 23 NCC samples tested. At least one nonsynonymous somatic mutation was detected in 94% of the 62-patient cohort (Fig. 3A and B and see also Supplementary Table 3): 97% of the 29 samples from patients with ES SCLC and 91% of the 33 samples from patients with LS SCLC. We detected TP53 mutations in 79% of the cohort (49 of 62): in 83% of the patients with ES SCLC (24 of 29) and 76% of the patients with LS SCLC (25 of 33) (Supplementary Tables 12 and 13 and see also Supplementary Tables 3, 8 and 9 and Fig. 3A). We also detected nonsynonymous somatic RB1 mutations in 34% of the patient samples (21 of 62), including those of 38% of patients with ES SCLC (11 of 29) and 30% of patients with LS SCLC (10 of 33) (see Supplementary Tables 3, 12, and 13 and Fig. 3A).
Figure 3.
Somatic mutations in SCLC circulating free DNA (cfDNA) and comparison of detection frequencies between cfDNA and circulating tumor cell (CTC) readouts. (A) The 62 patient cfDNA samples (29 patients with ES SCLC and 33 patients with LS SCLC) are arranged from left to right with each column representing mutation profile of an individual patient cfDNA sample. The genes are arranged from top to bottom according to the incidence of mutation of the gene in this cohort of patients and are displayed on the left y axis of the heatmap. The incidence of mutations occurring on a specific gene is annotated on the right y axis of the heatmap. (B) Detection frequencies were determined for the 62 patients whose samples passed the targeted next-generation sequencing quality control steps. Positive CNA samples were scored on the basis of any of the three copy number alteration (CNA) metric values rising above predetermined control thresholds, and for cfDNA targeted mutation analysis (MUT), the sample was scored positive if at least one mutation was detected. (C) Comparison of cfDNA analysis and CellSearch CTC enumeration. For 51 of the 69 cfDNA samples from the cohort of patients with SCLC, a parallel blood sample was collected at the same time point and CellSearch CTC enumeration was carried out. For cfDNA, samples were scored as positive samples if any of the CNA metric values were above predetermined control thresholds or if at least one mutation was present. Positive CTC samples were defined as any sample having at least one CellSearch CTC. Abbreviations: ES, extensive-stage; LS, limited-stage; MUT, Mutation; M+C, MUT and CNA; D+C, cfDNA and CTC; TP53, tumor protein p53 gene; RB1, retinoblastoma gene; MUC16, mucin 16, cell surface associated gene; COL22A1, collagen type XXII alpha 1 chain gene; NOTCH1, notch 1 gene; KMT2D, lysine methyltransferase 2D gene; PDE4DIP, phosphodiesterase 4D interacting protein gene; TP73, tumor protein p73 gene; FGFR1, fibroblast growth factor receptor 1 gene; NTRK1, neurotrophic receptor tyrosine kinase 1 gene; SLIT2, slit guidance ligand 2 gene; EPHA5, EPH receptor A5 gene; ZNF521, zinc finger protein 521 gene; CREBBP, CREB binding protein gene; EP300, E1A binding protein p300 gene; FLT4, fms related tyrosine kinase 4 gene; GNAS, GNAS complex locus gene; KEAP1, kelch like ECH associated protein 1 gene; KMT2A, lysine methyltransferase 2A gene; MSH6, mutS homolog 6 gene; MTOR, mechanistic target of rapamycin kinase gene; NFE2L2, nuclear factor, erythroid 2 like 2 gene; NOTCH2, notch gene; NOTCH3, notch gene; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha gene; ALK, ALK receptor tyrosine kinase gene; APC, APC, WNT signaling pathway regulator gene; ATM, ATM serine/threonine kinase gene; EPHA3, EPH receptor A3 gene; ERBB2, erb-b2 receptor tyrosine kinase 2 gene; FANCG, FA complementation group G gene; KIT, KIT proto-oncogene receptor tyrosine kinase gene; LIFR, LIF receptor subunit alpha gene; MET, MNNG HOS Transforming gene; NTRK3, neurotrophic receptor tyrosine kinase 3 gene; PARP4, poly(ADP-ribose) polymerase family member 4 gene; PDGFRA, platelet derived growth factor receptor alpha gene; PTEN, phosphatase and tensin homolog gene; RAD51C, RAD51 paralog C gene; SETBP1, SET binding protein 1 gene; TSC2, tuberous sclerosis 2 gene; AR, androgen receptor gene; ATR, ATR serine/threonine kinase gene; BRCA1, BRCA1, DNA repair associated gene; BRCA2, BRCA2, DNA repair associated gene; BRIP1, BRCA1 interacting protein C-terminal helicase 1 gene; CHEK2, checkpoint kinase 2 gene; FANCD2, Fanconi anemia complementation group D2 gene; FANCE, FA complementation group E gene; FANCL, FA complementation group L gene; GATA3, GATA binding protein 3 gene; GRIN2A, glutamate ionotropic receptor NMDA type subunit 2A gene; HRAS, Harvey rat sarcoma viral oncogene homolog gene; IDH2, isocitrate dehydrogenase (NADP(+)) 2, mitochondrial gene; NF1, neurofibromin 1 gene; NFATC2, nuclear factor of activated T cells 2 gene; PRKDC, protein kinase, DNA-activated, catalytic polypeptide gene; RAD50, RAD52 homolog, DNA repair protein gene; RBL2, retinoblastoma-like 2 gene; RET, ret proto-oncogene gene; RUNX1T1, RUNX1 translocation partner 1 gene; STK11, serine/threonine kinase 11 gene; TSC1, tuberous sclerosis 1 gene; WT1, Wilms tumor 1 gene.
Beyond TP53 and RB1, the most commonly mutated genes were gene (COL22A1), lysine methyltransferase 2D gene (KMT2D), notch 1 gene (NOTCH1), and mucin 16, cell surface associated gene (MUC16) (see Fig. 3A and Supplementary Tables 3, 12, and 13). An examination of genes implicated in DDR (see Supplementary Table 1 [marked in red]) showed that 85% of patients (90% of those with ES SCLC [26 of 29] and 82% of those with LS SCLC [27 of 33]) had a mutation in at least one of 18 DDR genes (see Fig. 3A). Also, when the 23 RAS/phosphoinositide 3-kinase pathway genes (see Supplementary Table 1 [marked in blue]) were examined, at least one somatic nonsynonymous mutation was found in 32% of patients (38% of those with ES SCLC [11 of 29] and 27% of those with LS SCLC [9 of 33]) (see Fig. 3A). In addition, 29% of patients with SCLC (38% of those with ES SCLC [11 of 29] and 21% of those with LS SCLC [seven of 33]) harbored at least one mutation in the 28 genes involved in transcriptional regulation (see Supplementary Table 1 [marked in green] and Fig. 3A).
When the mutation profiles of LS SCLC and ES SCLC were compared, there were no statistically significant differences in number of genes mutated or number of mutations between the two groups (Supplementary Fig. 3C). However, there were statistically significant quantitative differences in VAF, with higher levels in ES SCLC (p = 0.0188 [see Supplementary Fig. 3C]), which is consistent with the proportion of ctDNA present in total cfDNA being higher in ES SCLC.
Comparison of CNA, Targeted Mutation, and CTC Detection
Of the 62 patient samples with both CNA and targeted mutation data acquired, 58 (28 from patients with ES SCLC and 30 from patients with LS SCLC) had at least one somatic nonsynonymous mutation. For the single ES SCLC sample in which no detectable somatic variants were detected (E7), we detected amplification of a TP53-associated gene (tumor protein p53 regulating apoptosis inducing protein 1 gene [TP53AIP1]), whereas for the three LS SCLC samples with no detectable somatic variants (L3, L6, and L24), all CNA readouts were also negative (see Fig. 3A and B). The overall coverage when CNA and mutation detection were combined for all 62 eligible samples was 97% (60 of 62): 100% for the samples from patients with ES SCLC (29 of 29) and 94% of the samples from patients with for LS SCLC (31 of 33) (see Fig. 3B).
CTC number (by CellSearch, EpCAM positive, pan-cytokeratin positive, CD45 negative cells) is elevated in SCLC compared with in other cancers and is prognostic for OS.18 For the 62 patient samples subjected to both CNA and targeted sequencing, we also obtained a CTC count from a parallel blood sample taken at the same time point as for the cfDNA analysis in 48 patients (28 with ES SCLC and 20 with LS SCLC). For these 48 patients, the mean and median CTC numbers per 7.5 mL of blood were 523 and 5, respectively (range 0–15,352 [see Fig. 1B and Supplementary Fig. 3B), which is consistent with the data from previous studies.18 For 37 of these 48 patients (24 of the 28 with ES SCLC and 13 of the 20 with LS SCLC), CellSearch detected at least one CTC, whereas 46 of the same 48 patients (28 of the 28 with ES SCLC and 18 of the 20 with LS SCLC) had detectable ctDNA (see Fig. 3C and Supplementary Table 8).
Readouts and Potential Clinical Utility
Correlation of cfDNA Readouts with Clinical Outcomes
CNA metrics and VAFs differed significantly between patients with ES SCLC and those with LS SCLC (Supplementary Fig. 4A and C). Because CTC number (as determined by CellSearch) is a prognostic biomarker for OS in SCLC,18 we compared our cfDNA CNA readouts as well as TP53 VAF and highest VAF with CTC number, which showed a significant positive correlation (p ≤ 0.0001 in all cases [Supplementary Tables 14 and 15 and Supplementary Fig. 5A–C). In contrast, there was no statistical correlation found between either the number of genes mutated or the number of mutations present in cfDNA with CTC counts (see Supplementary Fig. 5 and Supplementary Tables 14 and 15). Similarly, there was no statistical correlation found between any of the cfDNA NGS–based readouts and cfDNA quantification per milliliter of plasma (Supplementary Fig. 6 and see also Supplementary Table 14).
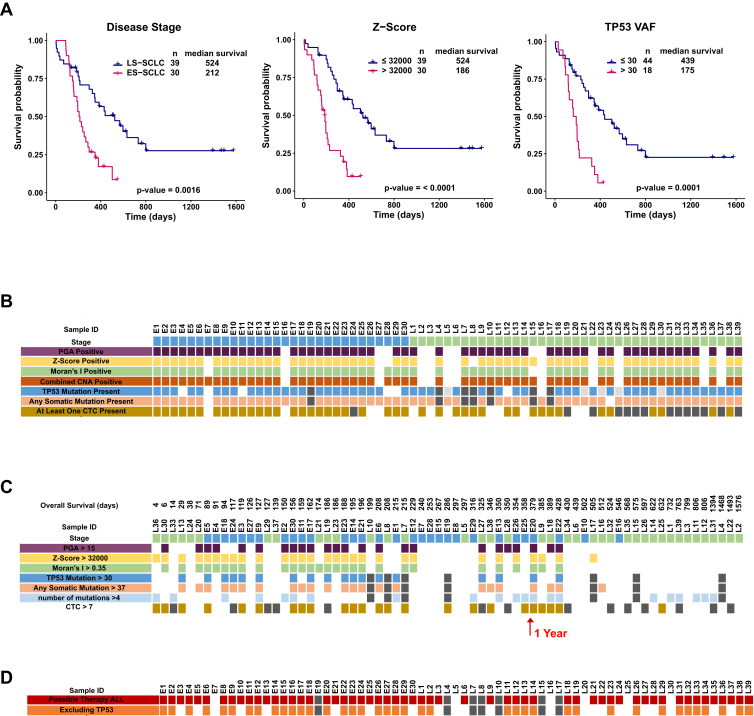
Significant statistical associations were observed between clinical stage and cfDNA readouts (see Supplementary Table 14). In univariate Cox regression analysis, the CNA readouts (p ≤ 0.001), TP53 VAF, and highest VAF along with number of CTCs and lactate dehydrogenase (LDH) were identified as significant predictors of shorter survival (Supplementary Tables 16 and 17). The results of the univariate and multivariate analyses of ctDNA-based scores in relation to each of the clinical covariates in the cohorts (Fig. 4 A–C, Supplementary Fig. 7A–D, and Supplementary Tables 16 and 17) show significant relationship between stage of disease, z score, and number of mutations per sample in multivariate analysis.
Figure 4.
Potential clinical utility of circulating free DNA (cfDNA) readouts. (A) Kaplan-Meier curves of all 69 patients for (1) disease stage, (2) z score, and (3) tumor protein p53 gene (TP53) variant allele fraction (VAF). (B) Copy number alteration (CNA) metrics (percent genome amplified [PGA], z score, and Moran's I [MI]), mutation analysis metrics (TP53 VAF and VAF of all genes), and circulating tumor cell (CTC) counts are marked positive on the heatmap if they are higher than the thresholds set based on 16 noncancer controls. (C) Overview of cfDNA readouts based on maximally selected rank statistic and ranked from left to right according to overall survival of the patient. CNA metrics (PGA, z score, and MI), mutation analysis metrics (TP53 VAF and VAF of all genes), and CTC counts are marked positive on the heatmap if they are higher than the thresholds set based on receiver operating characteristic curve analysis. (D) Overview of the number of cfDNA samples in which potential therapies were identified. Abbreviations: LS, limited-stage; ES, extensive-stage; ID, identifier.
Identification of Potential Actionable Mutations
To establish whether the mutations detected by our sequencing approach could have supported personalized therapy, we evaluated all mutations by using the Cancer Genome Interpreter (CGI25). Supplementary Table 3 presents findings for all 272 somatic nonsynonymous mutations detected and the frequency of mutation across all patients. As expected, many of the TP53 mutations present in 78% of the samples were identified as cancer driver mutations and some linked to potential therapies (see Supplementary Tables 3–5). Excluding TP53 mutations, CGI identified cancer drivers in 69% of samples, with 60% linked to potential therapies (see Fig. 4D and Supplementary Tables 3–5).
cfDNA Analysis for Disease Monitoring
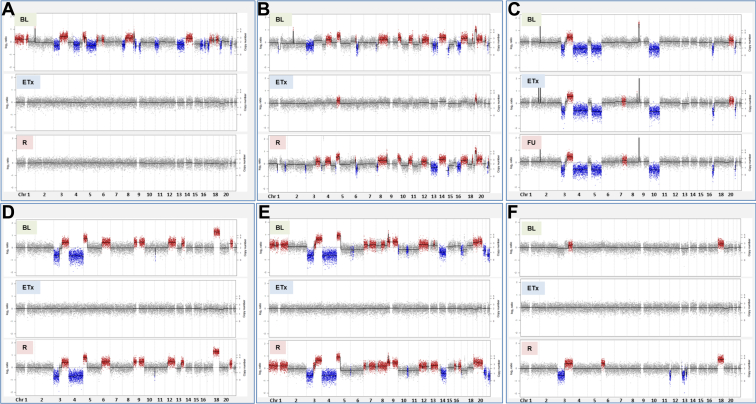
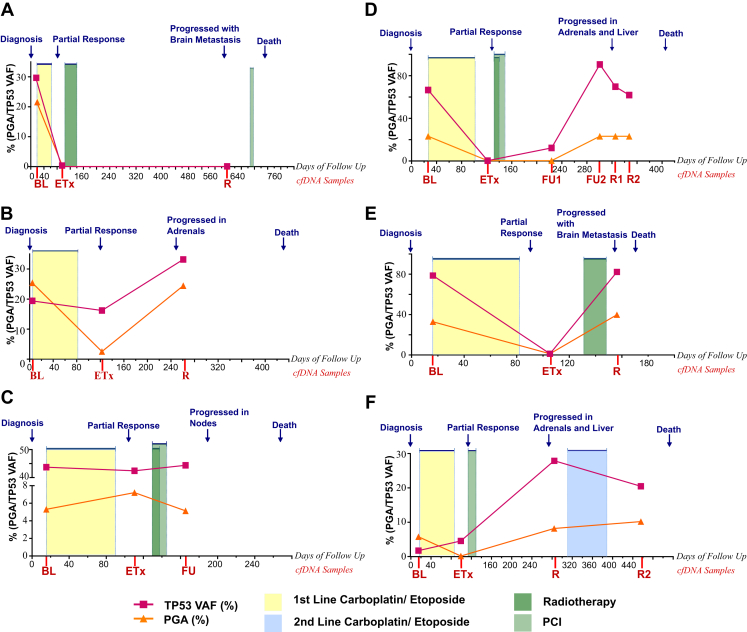
To establish whether the cfDNA approach we describe is suitable for patient monitoring, longitudinal samples were available from six patients (Supplementary Table 18 and Figs. 5 and 6 and see also Supplementary Table 5). In all six patients there was detectable tumor-specific CNA measurable at baseline, whereas at the end of treatment no CNA was detected for four patients, a 10-fold CNA reduction was seen for patient L19, and increased CNA was seen for patient L21 (Fig. 5 A–F, Fig. 6A–F, and Supplementary Table 18). CNA was detectable in four of the five available relapse samples and in the follow-up sample for patient L21 (see Fig. 5 B–F). For patient L18 there was no detectable CNA in the relapse sample, although there was disease progression with stable disease in lung and metastasis found in the brain (see Figs. 5A and 6A).
Figure 5.
Circulating free DNA copy number alteration (CNA) longitudinal analysis. Genome-wide CNA plots of three longitudinal samples from six patients with SCLC: L18 (A), L19 (B), L21 (C), E20 (D), E30 (E), and E29 (F). For each patient there are a baseline (BL) pretreatment sample and subsequent samples taken at end of treatment (eTx), samples taken at the follow-up time point (before clinical detection of relapse) (FU), and samples taken at relapse (R). For each CNA plot, copy number chromosomes (Chr) are indicated along the x axis, copy number and log-2 ratios are indicated along the y axis, gains are indicated as red, and copy number losses are shown as blue.
Figure 6.
Patient time courses. Summary of the clinical course and blood biomarker readouts of six patients with SCLC: L18 (A), L19 (B), L21 (C), E20 (D), E30 (E), and E29 (F). In each plot, the x axis indicates the days of follow-up with the time point corresponding to day 0 as diagnosis. The clinical events are indicated in blue along the top of each plot, the circulating free DNA (cfDNA) samples are indicated along the x axis as baseline (BL), end of treatment (eTx), follow-up 1 (prerelapse clinic appointment [FU1 or FU]), follow-up 2 (prerelapse clinic appointment [FU2]), and relapse (R). The treatment received and the duration of treatment by the patient are indicated by the colored boxes on each plot, with yellow representing first-line carboplatin/etoposide, blue for second-line carboplatin/etoposide, light green for prophylactic cranial irradiation (PCI), and dark green for radiotherapy. Abbreviations: PGA, percent genome amplified; TP53, tumor protein p53 gene; VAF, variant allele fraction.
Discussion
We examined the levels and composition of ctDNA in all stages of SCLC with a view to establishing a cfDNA analysis pipeline suitable for routine patient monitoring (see Fig. 1A). Whole genome CNA changes detectable in cfDNA were obtained by using an optimized bioinformatics workflow incorporating stringent elimination of unreliable genomic regions and selective analysis of shorter cfDNA fragments, thereby enriching for ctDNA33 (see Supplementary Fig. 3 and Supplementary Table 6). Global CNA was measured by using three separate readouts reflecting differing aspects of genomic change. Given that the pattern and extent of CNA differ from patient to patient, we reasoned that there may be differences in the CNA readouts reflecting the varying nature of each SCLC tumor. This prediction was borne out by the finding that 54 of 69 patient samples were ctDNA positive for all three readouts, one sample was detected by PGA and MI, one sample was detected by PGA alone, and one was detected by z score alone (see Fig. 2B). Examination of the two PGA-positive, Z-score–negative samples revealed small focal copy number gains restricted to 0.24% and 2.02% of the genome (see Fig. 2 and Supplementary Table 8), which were insufficient to register by using the genome-wide Z-score, demonstrating that combining the three readouts increases sensitivity by broadening the tumor-specific features measured. MI has the additional benefit that it is calculated on the basis of tumor-associated alterations in an individual cfDNA sample and it does not require the support of control or germline samples for its calculation. When all three CNA readouts (PGA, Z-score, and MI) were used, tumor-related CNA changes were seen in 84% of 69 patients with SCLC (93% of those with ES SCLC and 77% of those with LS SCLC [see Fig. 2A and B]). Similar CNA patterns were seen in the cfDNA samples in this study (see Fig. 2A and B and Supplementary Tables 10 and 11), as previously reported for SCLC tumors or SCLC CTCs.4,6,8,12,15
Recently, a panel of 430 genes has been applied to ctDNA of patients with SCLC and an association of pretreatment ctDNA VAFs was linked to OS.22 Here we show that each of the CNA readouts was also significantly associated with OS (see Supplementary Table 15), and Kaplan-Meier analysis suggested prognostic utility similar to that with CellSearch CTC enumeration18 (see Fig. 4A). Given the increased patient coverage of the cfDNA readouts for patients with LS and ES compared with CTC detection (see Figs. 3C and 4B), the simple CNA readouts described here provide a valuable means for prognosis and monitoring. It is important to point out that the approach taken by us to identify CNA patterns in pretreatment CTCs linked to treatment response8 cannot currently be applied to our cfDNA analysis because of varying dilution of ctDNA in cfDNA samples. However, targeted NGS of a single cfDNA sample has the advantage of providing a much simpler means of following specific mutations compared with the analysis of multiple CTCs.8 In summary, the analysis of CTCs and cfDNA provide differing yet overlapping information, and the aim of any given study will determine whether it is sufficient to analyze CTCs or cfDNA alone or whether both are required.
Our CNA results are in agreement with those of multiple studies demonstrating the effectiveness of CNA readouts applied to many types of cancer,17,27,34 but to our knowledge, this is the first CNA study analyzing a cohort of more than 60 patients with SCLC, and the results demonstrate that the approach developed by us will detect ctDNA at both early and late stages of SCLC. Although there are no comparable studies of SCLC cfDNA CNA, the higher sensitivities reached by using the combined CNA readouts (79%–93% depending on stage of disease) compare favorably with those reported with targeted NGS applied to SCLC cfDNA, which have been reported as 49% (n = 51),7 85% (n = 40),20 92.5% (n = 227),35 100% (n = 22),22 and 100% (n = 3).21 In addition, our CNA approach also led to the identification of a focal amplification of the TP53-regulated proapoptotic gene TP53AIP1 in SCLC (see Supplementary Table 10). Because increased expression of TP53AIP1 has been linked to poor prognosis in NSCLC36 but has not been previously reported in SCLC, our findings suggest that further investigation of TP53AIP1 in SCLC is warranted.
Targeted sequencing of cfDNA is increasingly being used to personalize treatment and monitor tumor evolution and therapy-emergent resistance. Here we show that after CNA analysis, the same whole genome libraries can be used to identify at least one somatic mutation in 93% of all SCLC samples (96% ES SCLC and 90% LS SCLC), which is equivalent to the detection rates previously reported for targeted sequencing of SCLC cfDNA.7,20, 21, 22,35 As expected, we detected frequent mutations in TP53 and RB1 and also saw a high frequency of the COL22A1, KMT2D, NOTCH1, and MUC16 genes. We also show that we could pick up at least one mutation in genes implicated in DDR in the cfDNA of 85% of patients with SCLC (90% of those with ES SCLC and 82% of those with LS SCLC [see Supplementary Table 3]), which may indicate selection for impaired DDR in SCLC, which is a hypothesis that we will evaluate in a larger study now under way. Furthermore, analysis of the cfDNA mutations in this study by using CGI identified cancer drivers in 69% of samples, with 60% linked to potential therapies, indicating that this targeted sequencing approach could support stratification of patients with SCLC in future clinical trials (see Supplementary Tables 3–5).
For a subgroup of six patients, we show that our cfDNA CNA approach also provides an effective means of disease monitoring in SCLC and that additional targeted sequencing of our 110-gene panel provides a potential means of patient stratification (see Figs. 5 and 6).11 For two patients (L21 and E20), detection of ctDNA using either CNA or targeted sequencing preceded radiological progression (Figs. 5C and D and 6C and D). For patient E20, mutational analysis identified the presence of the TP53 mutation found in the pretreatment sample 14.6 weeks before radiological progression (see Figs. 5C and D and 6 C and D). Taken together, these initial data suggest that our cfDNA CNA approach combined with highly targeted sequencing of TP53 will provide an effective means of monitoring of patients with SCLC and can guide selection of suitable samples for more extensive sequencing. Currently, efforts are ongoing to address the potential of disease monitoring by using cfDNA analyses in a larger cohort of samples.
In summary, as the molecular segmentation of SCLC disease emerges37 and rational drug development programs are now reaching early clinical testing,38 it is essential that a clinically implementable strategy for routine SCLC disease monitoring be established. The liquid biopsy approaches that we have reported here have been shown to provide effective baseline analysis and longitudinal monitoring of both LS and ES disease and are now being incorporated into extended SCLC studies and trials.
Acknowledgments
Funding for this study was from Cancer Research UK (CRUK) through the core CRUK Manchester Institute grant (C5759/A27412), the CRUK Manchester Centre (C5759/A25254), the CRUK Lung Cancer Center of Excellence (A25146), and the CRUK Manchester Experimental Cancer Medicines Centre (A20465). Dr. Mohan’s salary and consumable costs were funded through a translational research grant to Dr. Dive (10001080/AgrID486). Sample collection was undertaken via the CONVERT trial (Concurrent Once-Daily versus Twice-Daily RadioTherapy), and the ChemoRes trial (Molecular Mechanisms Underlying Chemotherapy Resistance, Therapeutic Escape, Efficacy, and Toxicity–Improving Knowledge of Treatment Resistance in Patients with Lung Cancer). This work was supported by the National Institute for Health Research (NIHR) Manchester Biomedical Research Centre (BRC), and the NIHR Christie Clinical Research Facility, and the Manchester Medical Research Council (MRC) Single Cell Research Centre (MR/M008908/1). The views expressed are those of the author(s) and not necessarily those of the National Health Service, the NIHR or the Department of Health. We sincerely thank the patients and their families for provision of blood samples for research. Drs. Brady, Dive, Blackhall, and Carr designed the study. Drs. Blackhall, Faivre-Finn, Mr. Carter, and Ms. Priest recruited and obtained consent from the patients, collected blood samples, and provided clinical data. Drs. Mohan, Rothwell, Brady, and Dive conceived and designed the experiments. Drs. Mohan, Ayub, and Mr. Smith performed the experiments. Drs. Mohan, Brady, Leong, Sahoo, Schofield, Mr. Kilerci, Drs. Miller, and Blackhall analyzed the data. Drs. Descamps and Zhou performed the statistical analysis. Drs. Mohan, Brady, Dive, and Blackhall interpreted the data. Drs. Mohan, Brady, Leong, Foy and Dive prepared the article. All data are available in the main text or the supplementary materials. Genome data has been deposited at the European Genome-Phenome Archive, which is hosted at the European Bioinformatics Institute (EBI) and the Centre for Genomic Regulation (CRG), under accession number EGAS00001003110.
Footnotes
Disclosure: Dr. Dive acts in a consultant or advisory role for Biocartis and AstraZeneca and receives research grants and/or support from AstraZeneca, Astex Pharmaceuticals, Bioven, Amgen, Angle PLC, Carrick Therapeutics, Merck AG, Taiho Oncology, GSK, Bayer, Boehringer Ingelheim, Roche, BMS, Novartis, Celgene, Epigene Therapeutics Inc., Menarini, and Clearbridge Biomedics, all outside the scope of work. Dr. Carr is an employee of AstraZeneca. The remaining authors declare no conflict of interest.
Note: To access the supplementary material accompanying this article, visit the online version of the Journal of Thoracic Oncology at www.jto.org and at https://doi.org/10.1016/j.jtho.2019.10.007.
Supplementary Data
References
- 1.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.van Meerbeeck J.P., Fennell D.A., De Ruysscher D.K.M. Small-cell lung cancer. Lancet. 2011;378:1741–1755. doi: 10.1016/S0140-6736(11)60165-7. [DOI] [PubMed] [Google Scholar]
- 3.Faivre-Finn C., Snee M., Ashcroft L. Concurrent once-daily versus twice-daily chemoradiotherapy in patients with limited-stage small-cell lung cancer (CONVERT): an open-label, phase 3, randomised, superiority trial. Lancet Oncol. 2017;18:1116–1125. doi: 10.1016/S1470-2045(17)30318-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.George J., Lim J.S., Jang S.J. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rudin C.M. Genomic and epigenomic targets in small cell lung cancer [abstract] Clin Cancer Res. 2014;20(suppl 2):IA03. [Google Scholar]
- 6.Rudin C.M., Durinck S., Stawiski E.W. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet. 2012;44:1111–1116. doi: 10.1038/ng.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernandez-Cuesta L., Perdomo S., Avogbe P.H. Identification of circulating tumor DNA for the early detection of small-cell lung cancer. EBioMedicine. 2016;10:117–123. doi: 10.1016/j.ebiom.2016.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter L., Rothwell D.G., Mesquita B. Molecular analysis of circulating tumor cells identifies distinct copy-number profiles in patients with chemosensitive and chemorefractory small-cell lung cancer. Nat Med. 2016;23:114–119. doi: 10.1038/nm.4239. [DOI] [PubMed] [Google Scholar]
- 9.Booton R., Blackhall F., Kerr K. Individualised treatment in non-small cell lung cancer: precise tissue diagnosis for all? Thorax. 2011;66:273–275. doi: 10.1136/thx.2010.138370. [DOI] [PubMed] [Google Scholar]
- 10.Rudin C.M., Pietanza M.C., Bauer T.M. Rovalpituzumab tesirine, a DLL3-targeted antibody-drug conjugate, in recurrent small-cell lung cancer: a first-in-human, first-in-class, open-label, phase 1 study. Lancet Oncol. 2017;18:42–51. doi: 10.1016/S1470-2045(16)30565-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sen T., Gay C.M., Byers L.A. Targeting DNA damage repair in small cell lung cancer and the biomarker landscape. Transl Lung Cancer Res. 2018;7:50–68. doi: 10.21037/tlcr.2018.02.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.George J., Saito M., Tsuta K. Genomic amplification of CD274 (PD-L1) in small-cell lung cancer. Clin Cancer Res. 2017;23:1220–1226. doi: 10.1158/1078-0432.CCR-16-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horn L. first-line atezolizumab plus chemotherapy in extensive-stage small-cell lung cancer. N Engl J Med. 2018;379:2220–2229. doi: 10.1056/NEJMoa1809064. [DOI] [PubMed] [Google Scholar]
- 14.Babayan A., Pantel K. Advances in liquid biopsy approaches for early detection and monitoring of cancer. Genome Med. 2018;10:21. doi: 10.1186/s13073-018-0533-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hodgkinson C.L., Morrow C.J., Li Y. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med. 2014;20:897–903. doi: 10.1038/nm.3600. [DOI] [PubMed] [Google Scholar]
- 16.Rothwell D.G., Avub M., Cook N. Utility of ctDNA to support patient selection for early phase clinical trials: the TARGET study. Nat Med. 2019;25 doi: 10.1038/s41591-019-0380-z. 7387–43. [DOI] [PubMed] [Google Scholar]
- 17.Mohan S., Heitzer E., Ulz P. Changes in colorectal carcinoma genomes under anti-EGFR therapy identified by whole-genome plasma DNA sequencing. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hou J.M., Krebs M.G., Lancashire L. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J Clin Oncol. 2012;30:525–532. doi: 10.1200/JCO.2010.33.3716. [DOI] [PubMed] [Google Scholar]
- 19.Belic J., Graf R., Bauernhofer T. Genomic alterations in plasma DNA from patients with metastasized prostate cancer receiving abiraterone or enzalutamide. Int J Cancer. 2018;143:1236–1248. doi: 10.1002/ijc.31397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Almodovar K., Iams W.T., Meador C.B. Longitudinal cell-free DNA analysis in patients with small cell lung cancer reveals dynamic insights into treatment efficacy and disease relapse. J Thorac Oncol. 2018;13:112–123. doi: 10.1016/j.jtho.2017.09.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaudhuri A.A., Lovejoy A.F., Chabon J.J. Circulating tumor DNA analysis during radiation therapy for localized lung cancer predicts treatment outcome. Int J Radiat Oncol Biol Phys. 2017;99:S1–S2. [Google Scholar]
- 22.Nong J., Gong Y., Guan Y. Circulating tumor DNA analysis depicts subclonal architecture and genomic evolution of small cell lung cancer. Nat Commun. 2018;9:3114. doi: 10.1038/s41467-018-05327-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Board R.E. Isolation and extraction of circulating tumor DNA from patients with small cell lung cancer. Ann N Y Acad Sci. 2008;1137:98–107. doi: 10.1196/annals.1448.020. [DOI] [PubMed] [Google Scholar]
- 24.Li B.T., Janku F., Jung B. Ultra-deep next-generation sequencing of plasma cell-free DNA in patients with advanced lung cancers: results from the Actionable Genome Consortium. Ann Oncol. 2019;30:597–603. doi: 10.1093/annonc/mdz046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamborero D., Rubio-Perez C., Deu-Pons J. Cancer Genome Interpreter annotates the biological and clinical relevance of tumor alterations. Genome Med. 2018;10:25. doi: 10.1186/s13073-018-0531-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan K.C., Jiang P., Zheng Y.W. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem. 2013;59:211–224. doi: 10.1373/clinchem.2012.196014. [DOI] [PubMed] [Google Scholar]
- 27.Heitzer E., Ulz P., Belic J. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med. 2013;5:30. doi: 10.1186/gm434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiu R.W., Chan K.C., Gao Y. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc Natl Acad Sci U S A. 2008;105:20458–20463. doi: 10.1073/pnas.0810641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moran P. Notes on continuous stochastic phenomena. Biometrika. 1950;37:17–23. [PubMed] [Google Scholar]
- 30.Jiang P., Chan C.W., Chan K.C. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci U S A. 2015;112:E1317–E1325. doi: 10.1073/pnas.1500076112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mouliere F., Chandrananda D., Piskorz A.M. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med. 2018;10:eaat4921. doi: 10.1126/scitranslmed.aat4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giacona M.B., Ruben G.C., Iczkowski K.A., Roos T.B., Porter D.M., Sorenson G.D. Cell-free DNA in human blood plasma: length measurements in patients with pancreatic cancer and healthy controls. Pancreas. 1998;17:89–97. doi: 10.1097/00006676-199807000-00012. [DOI] [PubMed] [Google Scholar]
- 33.Scheinin I., Sie D., Bengtsson H. DNA copy number analysis of fresh and formalin-fixed specimens by shallow whole-genome sequencing with identification and exclusion of problematic regions in the genome assembly. Genome Res. 2014;24:2022–2032. doi: 10.1101/gr.175141.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bardelli A., Corso S., Bertotti A. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov. 2013;3:658. doi: 10.1158/2159-8290.CD-12-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morgensztern D., Dewvarakonda S., Masood A. P1.07-035. Circulating cell-free tumor DNA (cfDNA) testing in small cell lung cancer [abstract] J Thorac Oncol. 2017;12:S717–S718. [Google Scholar]
- 36.Yamashita S.I., Masuda Y., Yoshida N. p53AIP1 expression can be a prognostic marker in non-small cell lung cancer. Clin Oncol. 2008;20:148–151. doi: 10.1016/j.clon.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 37.Gazdar A.F., Bunn P.A., Minna J.D. Small-cell lung cancer: what we know, what we need to know and the path forward. Nat Rev Cancer. 2017;17:725–737. doi: 10.1038/nrc.2017.87. [DOI] [PubMed] [Google Scholar]
- 38.Sabari J.K., Lok B.H., Laird J.H., Poirier J.T., Rudin C.M. Unravelling the biology of SCLC: implications for therapy. Nat Rev Clin Oncol. 2017;14:549. doi: 10.1038/nrclinonc.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.