Abstract
Background
The analysis of circulating free tumour DNA (ctDNA) in blood, commonly referred as liquid biopsy, is being used to characterise patients with solid cancers. Tumour-specific genetic variants can also be present in DNA isolated from other body fluids, such as urine. Unlike blood, urine sampling is non-invasive, can be self-performed, and allows recurrent longitudinal monitoring. The features of tumour DNA that clears from the glomerular filtration barrier, named trans-renal tumour DNA (trtDNA), are largely unexplored.
Patients and methods
Specimens were collected from 24 patients with KRAS or BRAF mutant metastatic colorectal cancer (mCRC). Driver mutations were assessed by droplet digital PCR (ddPCR) in ctDNA from plasma and trtDNA from urine. Whole exome sequencing (WES) was performed in DNA isolated from tissue, plasma and urine.
Results
Out of the 24 CRC cases, only four had sufficient DNA to allow WES analyses in urine and plasma. We found that tumour alterations primarily reside in low molecular weight fragments (less than 112 bp). In patients whose trtDNA was more than 2.69% of the urine derived DNA, cancer-specific molecular alterations, mutational signatures and copy number profiles identified in urine DNA are comparable with those detected in plasma ctDNA.
Conclusions
With current technologies, WES analysis of trtDNA is feasible in a small fraction of mCRC patients. Tumour-related genetic information is mainly present in low molecular weight DNA fragments. Although the limited amounts of trtDNA poses analytical challenges, enrichment of low molecular weight DNAs and optimised computational tools can improve the detection of tumour-specific genetic information in urine.
Keywords: liquid biopsy, urine trans-renal DNA, non-invasive, colorectal cancer, plasma ctDNA
Key questions.
What is already known about this subject?
Analysis of tumour-derived DNA isolated from urine has been mainly applied to cancers of the urogenital tract. Few studies found KRAS alterations in urine trans-renal tumour DNA (trtDNA) of colorectal cancer patients using a quantitative mutation-enrichment next generation sequencing (NGS) method. These data were compared with archived tumour tissue and plasma circulating free tumour DNA (ctDNA) samples from patients with advanced metastatic colorectal cancer (mCRC), and it was observed a correlation between the molecular profile of trtDNAs and clinical outcomes.
What does this study add?
We defined an optimised protocol to isolate trtDNA from urine and performed whole exome sequencing analysis of urine trtDNA as well as matched plasma ctDNA with tumour tissue of four mCRCs. We calculated genetic mutational concordance, estimated tumour content and defined cancer-related mutational signatures. Most notably, we found that tumour-related genetic information is primarily present in low molecular weight trtDNA fragments.
How might this impact on clinical practice?
Our method could improve the sensitivity and feasibility of NGS technology. Urine trtDNA is a completely non-invasive test, which can complement blood-based ctDNA studies in screening out the general population to identify high risk individuals. As compared with blood, urine collection can be self-performed and, in principle, endlessly repeated, allowing more effective longitudinal monitoring of treatment response and minimal residual disease detection after surgery. Our study paves the way for using trtDNA in clinic; however, currently, trtDNA profiling is informative only in a small subset of colorectal cancer patients.
Introduction
Blood is the main source for the analysis of tumour biomarkers in patients with solid cancers. Serum proteins are routinely employed to track tumour burden in specific settings, but their clinical utility has inherent limitations including lack of specificity and sensitivity.1 On the other hand, plasma contains circulating tumour-derived nucleic acids and recent studies have indicated that they can be used for early detection,2 minimal residual disease quantification, tumour genotyping and molecular assessment of drug resistance.3 4 Tumour-derived DNA (circulating free tumour DNA (ctDNA)) has also been identified in other body fluids, such as pleural effusions or cerebrospinal fluid of patients affected by thoracic or central nervous system tumours, respectively.5–8 However, the collection of these biospecimens, including blood, cannot be self-performed and requires dedicated equipment as well as trained personnel. Urine has been proposed as an alternate non-invasive and cost-effective source of cancer biomarkers, since urine collection can be self-performed and endlessly repeated (to monitor cancer progression and drug response) at any location and with a minimal effort. Fragments of urinary DNA originate either from urogenital tract cells, shedding during urine transit, or from circulating free DNA (cfDNA) passing through the glomerular barrier, which is also known as trans-renal DNA (trDNA). High molecular-weight (HMW) DNA fragments are usually over 1 kbp and are predominantly released into the urine by cells present in the genito-urinary tract, such as necrotic cells or lymphocytes.9–11 On the contrary, low-molecular-weight (LMW) DNA fragments, which are in the range of 10–200 bp, are though to derive from the blood-filtered ctDNA.9–11
The physiopathological mechanisms involved in the glomerular filtration of cfDNA and, hence, in the formation of LMW DNA (a.k.a trans-renal tumour DNA or trtDNA), have not yet been fully elucidated; although, it is known that the quantity of LMW DNA depends on the permeability of the basal membrane and on the slit membranes between podocytes pedicles during glomerular filtration.
So far, urine has been mainly exploited for the analysis of biomarkers in patients affected by urological malignancies.11 Studies of urinary cfDNA from cancer patients with non-urological tumours have been more limited and have mainly focused on the assessment of selected genetic alterations. Indeed, proof-of-concept studies revealed the possibility to use urine trtDNA to inform about the pharmacodynamics of tyrosine kinase inhibitors in epidermal growth factor receptor (EGFR) mutant lung cancer patients.12 13 Similarly, in a case report study we found that tumour burden could be tracked in urine trtDNA by detecting a CAD-ALK gene rearrangement in a metastatic colorectal cancer (mCRC) patient treated with an ALK inhibitor.14RAS mutations, which are routinely assessed in mCRC patients to guide treatment selection, could be also detected in urine trtDNA using a quantitative, mutation-enrichment NGS method, and showed a good concordance with tumour tissue DNA or matched plasma ctDNA samples.15
All of the above studies employed urine cfDNA for assessing specific alterations, and it remains to be established whether urinary cfDNA is sufficient to have a comprehensive characterisation of solid tumour genotyping. To our knowledge, the entire coding sequence of tumours from urine-derived DNA has not been previously reported. Here we describe how we improved the current protocols to determine the molecular landscape via whole exome sequencing (WES) analysis in urine trtDNA of four mCRC tumours, assessing their mutational concordance between tissue, plasma and urine, calculating their tumour loads, and defining their cancer-related mutational signatures.
Methods
Patients
Patients were selected according to the following criteria: (1) patients had to have histologically confirmed diagnosis of mCRCs; (2) with a tumour burden defined as the sum of the longest diameters of tumour in at least three measurable lesions, higher than 2 cm in total; (3) availability of matched tissue in formalin-fixed paraffin-embedded (FFPE) or fresh tissue, blood, peripheral blood mononuclear cells (PBMC) and urine samples; (4) KRAS and BRAF positive mutational status in tissue was also required. Importantly, the patients should not have had any comorbidity in the urinary tract.
Using droplet digital PCR (ddPCR) analysis the concordance for KRAS or BRAF mutations was 100% (24/24) between tissue and plasma.
Tumour specimens (FFPE), plasma and urine samples of 24 patients were collected from histologically confirmed mCRC patients, treated at Grande Ospedale Metropolitano Niguarda (NCC) or Istituto Nazionale Tumori (INT), Milan, Italy. Availability of tumour sample qualitatively and quantitatively suitable for molecular analyses was a requirement for being considered in the present study.
DNA isolation from FFPE, plasma and urine
Genomic DNA and plasma derived cfDNA were isolated from fresh tissue, FFPE and blood-isolated PBMC, as previously described.16
For trtDNA extraction, at least 100 mL of urine was concentrated to 4 mL using Vivacell 100 concentrators (Sartorius Corp) and incubated with 700 ul of Q-sepharose Fast Flow quaternary ammonium resin (GE Healthcare). We modified the previously used protocol for trtDNA isolation from urine and performed a double-step ultrafiltration to separate the low molecular weight from the high molecular weight fragments, and treated them as individual samples until the end of the procedure.14 trtDNA fragment size distribution was assessed using the 2100 Bioanalyzer High-Sensitivity DNA assay kit (Agilent Technologies) according to the manufacturer’s instructions.
ddPCR analysis
Isolated cfDNA was amplified using ddPCR Supermix for Probes (Bio-Rad) with KRAS and BRAF assays (PrimePCR ddPCR Mutation Assay, Bio-Rad). ddPCR was then carried out according to the manufacturer’s protocol and the results were reported as percentage or fractional abundance of mutant DNA alleles to total (mutant plus wild type DNA alleles).
NGS library preparation
Libraries from PBMC and the fresh tissue available for CRC-UD09 patient were prepared starting from 100 up to 120 ng of extracted DNA by means of Nextera Rapid Capture Exome kit, according to manufacturer’s protocol and as previously described.16
For NGS on liquid biopsies from CRC patients, library preparation was performed using up to 150 ng of cfDNA from plasma or urine samples. cfDNA has been treated with NEBNext Ultra DNA Library Prep Kit for Illumina. For subsequent steps of library prep workflow, Nextera Rapid Capture Exome kit reagents have been used as previously reported.16
All libraries were sequenced on the Illumina NextSeq500 sequencer (Illumina) and 150 bp paired end reads were generated.
WES bioinformatic analysis
Genetic discovery analysis was performed using a previously described NGS pipeline designed for WES analyses of paired cancer genomes which identifies somatic variations, insertions and deletions (indels) and copy number alterations.7 16–18 Details on bioinformatics analysis are provided in online supplementary file 2.
esmoopen-2019-000572supp002.pdf (58.6KB, pdf)
The final median depth obtained was 210X (on 18 WES), with more than 97.03% (see online supplementary file) of the targeted region covered (after filtering). Furthermore, only mutations with 5% significance level obtained with a Fisher test, supported with a minimum depth of 5X and at least 1% allelic frequency were considered.
esmoopen-2019-000572supp001.pdf (54.9KB, pdf)
Using the information of somatic single nucleotide variants, a series of mutational profiles were extracted and genetic signatures were calculated using MuSiCa tool.19
Data are available at the link: https://www.ebi.ac.uk/ena/data/view/PRJEB33785
Accession number: PRJEB33785.
Unique name: ena-STUDY-CANDIOLO CANCER INSTITUTE-31-07-2019-11:20:38:154–19.
Results
Urine DNA differential isolation
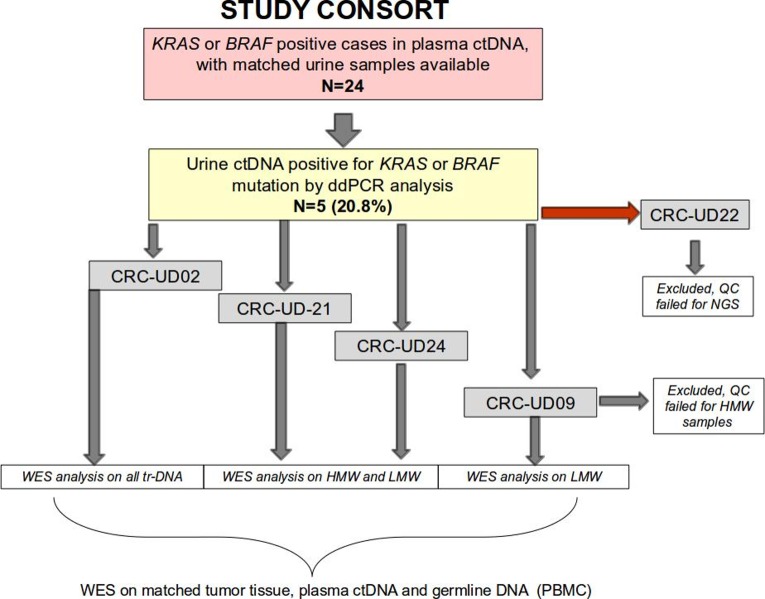
To proceed with trtDNA analysis, we further improved a protocol we previously employed for urine trtDNA isolation to enrich for tumour-derived DNA content. Specifically, we performed a double-step ultrafiltration to separate LMW fragments from the HMW ones by treating the two fractions as individual sample until the end of the DNA isolation procedure (figure 1, online supplementary figures S1 and S2). The new ultrafiltration step separated the majority of the LMW fragments from those longer than 400 base pairs. We found that this method avoids the massive DNA loss that is encountered with other techniques. All urine trDNA were isolated and tested with ddPCR (online supplementary table S1). In five cases out of 24 (20.8%) (online supplementary table S2), KRAS or BRAF mutations were identified in both urine and matched plasma (details are reported in online supplementary file 1, online supplementary tables S1 and S2). Therefore, only those five cases where considered for further analysis.
Figure 1.
Stratification of patient cohort. Diagram showing study cohort of 24 patients from whom we obtained matched urine and blood samples gathered from two Italian institutions. Four out of five patients were analysed by WES analysis, two were positive for KRAS and two for BRAF mutations, one patient was filtered out due to failure to pass NGS quality check. CRC, colorectal cancer; ctDNA, circulating free tumour DNA; ddPCR, droplet digital PCR; HMW, high molecular weight; LMW, low molecular weight; WES, whole exome sequencing.
esmoopen-2019-000572supp005.pdf (64.7KB, pdf)
esmoopen-2019-000572supp006.pdf (46.7KB, pdf)
Prior to NGS, we performed a quality control to assess suitability of DNA for WES analysis. The case CRC-UD22 failed our quality control and was excluded. For the other four cases (clinical characteristics reported in online supplementary table S2), WES was performed on DNA coming from urine, tissue, plasma, and matched PBMC samples. The germ-line WES was used as a reference for bioinformatics analysis (details are reported in online supplementary file 2). When the amount of DNA was conductive (cases CRC-UD24 and CRC-UD21), WES analysis was performed on both HMW and LMW enriched-fragments (cut-off: 400 bp) trtDNA fractions. In the case CRC-UD09 we only analysed the LMW-enriched compartment, since there was not sufficient amount of DNA in the HMW one to perform WES, while for the case CRC-UD02, WES analysis was performed on the entire trtDNA recovered from the urine fluid because the recovered amount of HMW fragments was too low (figure 1 and online supplementary figure 2).
esmoopen-2019-000572supp004.pdf (33.4KB, pdf)
SNPs, mutational concordance and tumour content analysis of matched plasma/urine samples in CRC patients
After verifying the coverage in all the sequencing runs (online supplementary table 3), we confirmed that the samples analysed belonged to the same cancer patient by comparing their single nucleotide polymorphisms (SNPs, dbSNP V.147). In all cases the allelic matching was confirmed (online supplementary table 4).
esmoopen-2019-000572supp007.pdf (42.7KB, pdf)
esmoopen-2019-000572supp008.pdf (44.2KB, pdf)
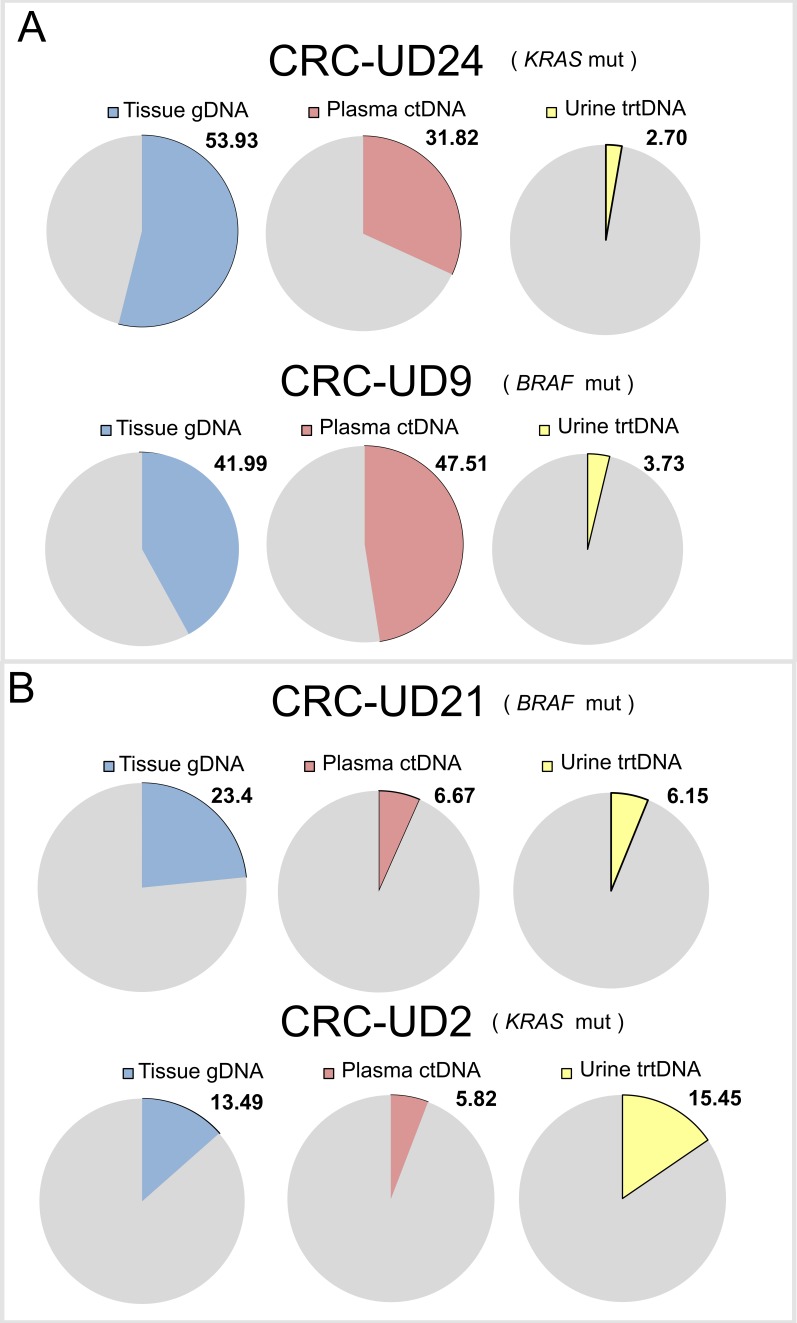
We next identified trunk alterations (i.e., KRAS or BRAF mutations) in tissue genomic DNA (gDNA) in all four mCRC cases and used them to estimate the tumour DNA content in each sample (figure 2). We then established the level of concordance between the alterations detected in urine and in matched plasma samples. For this analysis we examined all the molecular alterations detected in matched tumour tissue and we obtained a value of genetic concordance between plasma and urine (defined as mutation positivity for the alterations confirmed in tumour tissue), ranging from 27.9% to 57.4% (figure 3A, B), which we found to be influenced by the tumour DNA content in each liquid specimen. In detail, CRC-UD24 and CRC-UD09 had high tumour DNA content in plasma, but low in the matched urine (figure 2A), while the cases CRC-UD21 and CRC-UD02 had a comparable tumour DNA content in both urine and matched plasma samples (figure 2B).
Figure 2.
Tumour DNA content in matched tissue, plasma and urine samples from four CRC patients. Pie charts showing tumour content measured using fractional abundance of driver alterations: BRAF p.V600E mutation for cases CRC-UD09 and CRC-UD21, KRAS p.G13D and p.G12D, respectively for CRC-UD24 and CRC-UD02. Number indicates mutational frequency of the gene molecular alteration used to calculate tumour content. CRC, colorectal cancer; ctDNA, circulating free tumortumour DNA; gDNA: genomic DNA; trtDNA, trans-renal tumour DNA.
Figure 3.
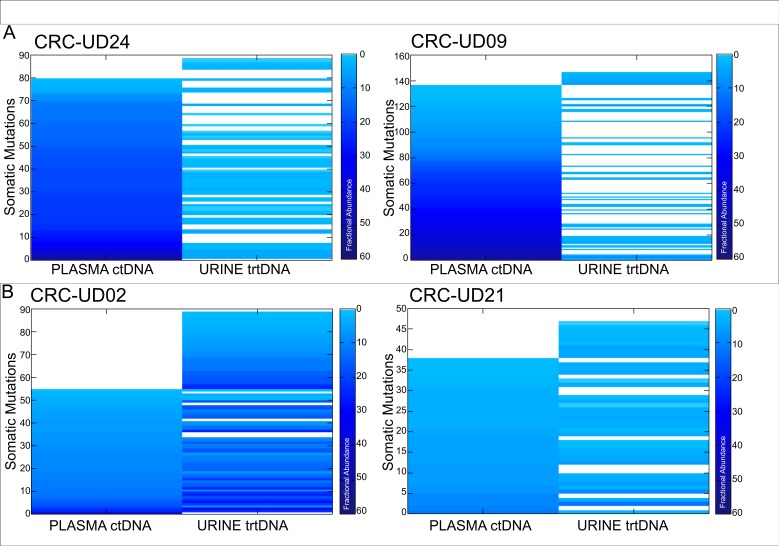
Concordance analysis of matched plasma/urine samples in CRC patients. Mutations found in the tumour tissue and in both matched plasma and urine ctDNA are shown. ‘Concordance’ indicates the number of somatic mutations shared between plasma and urine samples. For cases CRC-UD21 and CRC-UD24, the average frequency of each mutation identified in two urine samples is reported. Coloured-scale is used to show fractional abundance for each mutation. (A) Cancer patients with low tumour content identified in urine ctDNA. (B) Cancer patients with high tumour content identified in urine ctDNA. CRC, colorectal cancer; ctDNA, circulating free tumour DNA; trtDNA, trans-renal tumour DNA.
Indeed, when tumour DNA content was low and closer to the WES limit of detection (nearly 1%), the amount of variations identified varied noticeably. In fact, in the first two cases (CRC-UD24 and CRC-UD09), the number of detected variations in the urine trtDNAs were few, 40/89 mutations (44.9%) and 41/147 mutations (27.9%), as well as total tumour DNA content which was 2.7% and 3.7%, respectively (figure 2A and figure 3A). In the second group (CRC-UD21 and CRC-UD02) the total number of mutations identified in urine, 6.15% and 15.4%, was comparable to the one found in the plasma, 6.7% and 5.8% (figure 2B). This led to an increased number of shared somatic mutations between the two specimens: 27/47 mutations (57.4%) in CRC-UD21, and 49/89 mutations (55.4%) in CRC-UD02 (figure 3B).
Copy number alterations, mutational profiles and signature analysis in matched plasma and urine samples
Overall, the copy number profiles in plasma and urine samples were comparable. However, we noticed that a case (CRC-UD09) displayed an increased copy number of the chromosome seven in the plasma ctDNA sample, but not in the matched urine (online supplementary figure 3). This discrepancy might be due to the different tumour content, which was much lower (3.73%) in the urine compared with the matched plasma (47.51%) (figure 2A).
esmoopen-2019-000572supp009.pdf (76KB, pdf)
We defined mutational signatures as combinations of types of mutations resulting from mutagenesis processes such as variations in DNA replication, DNA enzymatic editing, exposures to DNA damaging agents and tissue culture conditions. Thus, we characterised the mutational signatures of the four mCRC patients in the plasma ctDNA and matched urine trtDNA samples, identifying in the plasma as well as in the urine samples a combination of signatures 1, 6 and 10 (online supplementary figure 4). Overall there was high concordance between signatures present in urine and plasma (online supplementary figure 5).
esmoopen-2019-000572supp010.pdf (39.1KB, pdf)
esmoopen-2019-000572supp011.pdf (74.3KB, pdf)
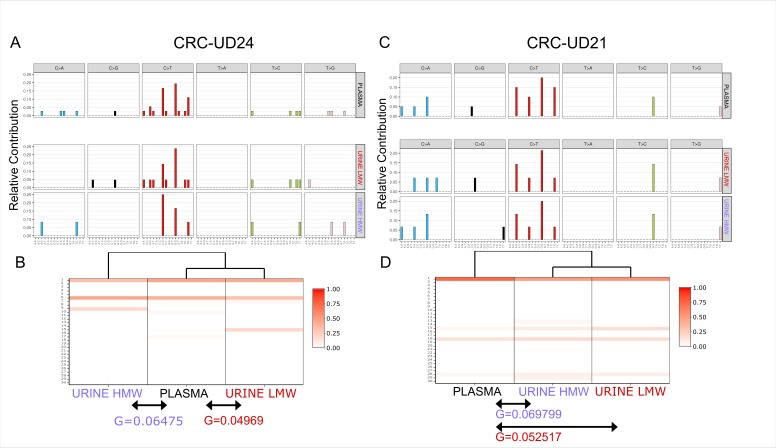
Next, we compared the mutational profile obtained from high and low-enriched molecular weight DNA fragments in the cases CRC-UD24 and CRC-UD21 and observed a higher similarity in the genetic profiles between the plasma and urine LMW-enriched DNA fragments (figure 4A,C). This confirmed our previous observations.
Figure 4.
Signature and mutational profiles of samples enriched for high and low molecular weight fragments. (A) and (C) Mutational profiles of enriched HMW and LMW urine samples and matched plasma ctDNA for cases CRC-UD24 (left) and CRC-UD21 (right). (B) and (D) Signature enrichment of HMW-enriched and LMW-enriched urine samples and matched plasma ctDNA is shown. Dendrogram is shown on the top, with a percentage indicating the similarity of each of the 30 signatures to the samples shown in the centre, while the G parameter (below the arrows) indicates the sum of the squares of differences between each signature observed in plasma ctDNA and the matched signatures in enriched HMW and LMW urine trtDNA samples. CRC, colorectal cancer; ctDNA, circulating free tumour DNA; HMW, high molecular weight; LMW, low molecular weight; trtDNA, trans-renal tumour DNA.
Finally, we calculated the G parameter, defined as the sum of the squares of the differences of each signature between plasma and HMW-enriched (G=0.06475 and G=0.069799, respectively) as well as between plasma and LMW-enriched (G=0.04969 and G=0.052517, respectively). In both CRC-UD24 and CRC-UD21 the genetic similarity between the LMW-enriched urine and the matched plasma was higher than that identified in HMW-enriched DNA fragments (figure 4B,D).
Tumour-specific alterations occur more frequently in shorter reads
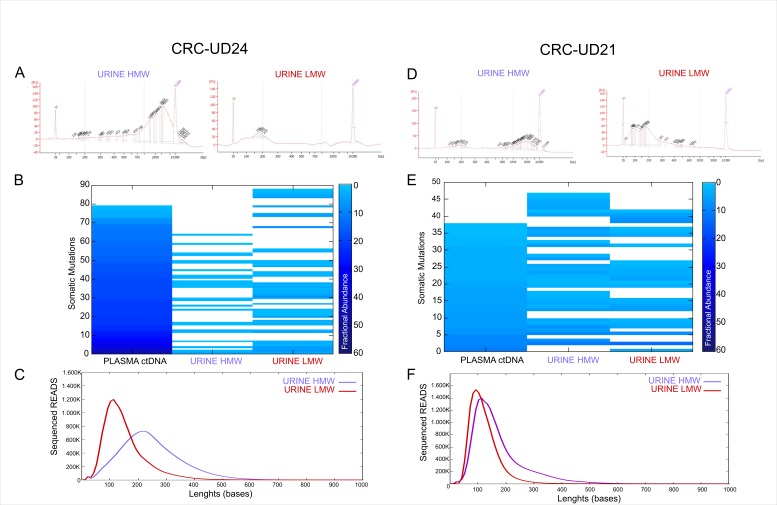
In the cases CRC-UD24 and CRC-UD21, in which we were able to analyse the low and high-enriched molecular weight fragments, separately, we sought to differentiate the molecular variations identified in the HMW-enriched and LMW-enriched DNA fragments in urine sample (figure 5A,D) as well as in the matched tumour tissue and plasma. A concordance of 40/89 mutations (45%) and 25/89 mutations (28%) was obtained in LMW-enriched and HMW-enriched DNA fragments, respectively (figure 5B). This confirms an enrichment of DNA from tumour origin in the low molecular weight DNA fragments, as also shown by the average read lengths analysis (figure 5C).
Figure 5.
Low molecular weight (LMW) DNA fragments in urine samples are enriched in tumour content. (A) and (D) Distribution of DNA fragment lengths in urine sample processed to enrich high molecular weight fragments (HMW in purple, on the left) from low molecular weight fragments (LMW in red, on the right) before sequencing. (B) and (E) Mutations found in plasma ctDNA and matched urine when enriched for high and low molecular weight DNA fragments. Only mutations also found in the matched tumour tissue are shown. The concordance frequency (%) was calculated on the number of shared variations identified in both plasma and urine ctDNA on the total number of variations identified. Blue-coloured scale indicates different fractional abundances reported for each mutation. (C) and (F) DNA fragments’ length distribution in urine trtDNA sample enriched for HMW and LMW. Data were obtained by measuring all reads lengths sequenced and aligned to the human genome reference V.19. CRC, colorectal cancer; ctDNA, circulating free tumour DNA.
In CRC-UD21 we obtained a concordance level of 22/47 (46%) in LMW-enriched and of 27/47 (57%) in HMW-enriched DNA (figure 5E) due to a minor enrichment of short fragments for DNA of tumour origin (figure 5F).
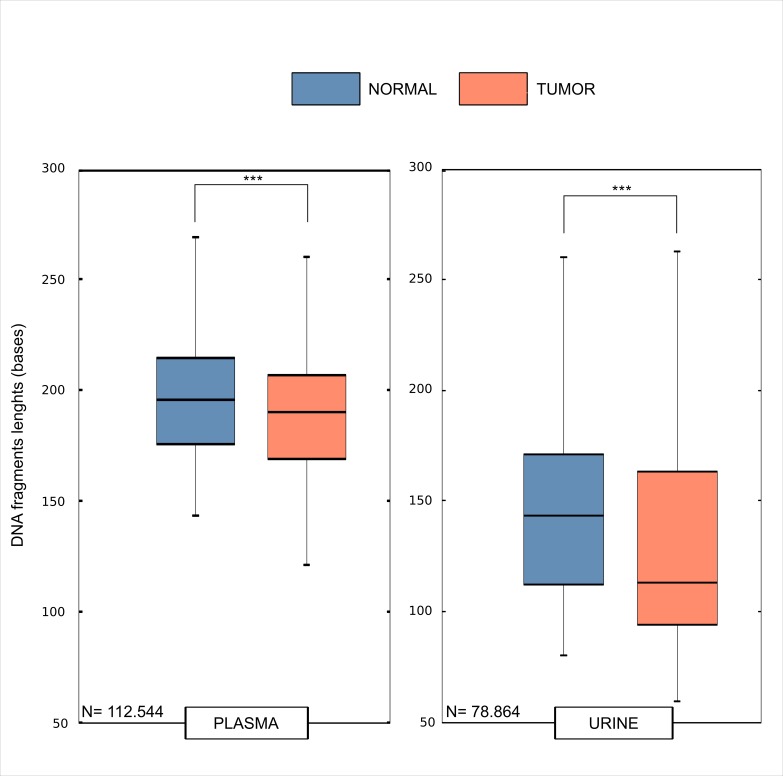
Intrigued by these results, we tested whether there were any differences in DNA fragments length shed by tumour cells (therefore carrying mutated reads) and those originated from normal cells (wild type reads). To formally test this, we used NGS data and compared the read length of the urine fragments with or without the mutations previously identified in the primary tumours. We observed a significant difference (Wilcoxon rank test, p value <0.00001) between the tumour DNA fragments lengths in plasma (figure 6) of six bases (190 bp vs 196 bp, respectively).
Figure 6.
Tumour-specific alterations occur more frequently in shorter reads. Distributions of mutated reads (tumour, red) and wild type reads (normal, blue) are shown in boxplots. For each somatic mutation of four patients, the reads encompassing somatic mutations were used to calculate the distributions of lengths. N depicts the total number of reads used to calculate the distributions for both plasma and urine trtDNA samples. trtDNA, trans-renal tumour DNA.
We then performed the same analysis on the DNA fragments derived from the urine and observed again a difference (Wilcoxon rank test, p value <0.00001) of 31 bases (112 bp vs 143 bp) between the tumour read lengths in urine as compared with their WT counterpart, confirming our initial observation that tumour-specific alterations occur more frequently in shorter reads (figure 6).
Discussion
ctDNA isolated from various body fluids has been exploited as a novel biomarker in the clinical management of cancer patients.3 In this context, urine has great advantages: it is completely non-invasive and easily accessible, it requires no specialised facility or equipment for collection, which makes urine highly convenient and a cost-effective biomarker source. Blood collection, on the contrary, can be limited in term of accessibility, frequencies, and volume drawn for ethical or clinical reasons, limiting real time patient monitoring.
We and others previously evaluated the concordance between molecular tumour alterations in urine-derived trtDNA, tumour tissue and plasma.15 20 These studies included KRAS mutations and gene fusions assessments in CRC, BRAF mutations in histiocytic disorders, and EGFR mutations in non-small cell lung cancer.21
The paucity of urine-based liquid biopsy studies might be explained by the limitations of this sample ctDNA source being more diluted by non-tumoural cfDNA. Consequently, more sensitive assays are mandatory for trtDNA detection.
In the present study, to obtain a molecular portrait of the patients’ DNA with maximum sensitivity to biologically profile DNA characteristics, we modified the previously used protocol for trtDNA isolation from urine14 with a double-step ultrafiltration, allowing an enrichment of low molecular weight fragments. Such concentration of the LMW-enriched DNA fragments was shown to increase the sensitivity towards DNA from tumour origin.20 In fact, Su and colleagues used a size-selection based on magnetics beads which in our hands (data not shown) was associated with loss of material during the washes. Therefore, we recommend a size-selection via dual steps centrifugation which, despite being less efficient in selecting fragments, was able to maintain a very high recovery rate.
Interestingly, two patients (CRC-UD24 and CRC-UD09) had high tumour DNA content in plasma, but low in the matched urine, while other two cases (CRC-UD21 and CRC-UD02) had a comparable tumour DNA content in both urine and matched plasma samples.
We hypothesize that the higher tumour burden, detected in plasma ctDNA in the cases CRC-UD24 and CRC-UD09, could be due to the plasma ctDNA from blood, which was not entirely filtered by the kidney barrier (i.e., long DNA fragments and DNA from necrotic cells did not go through the glomerular capillaries). This resulted in an overall reduction of tumour-derived DNA quantity in the urine, as effectively detected. Furthermore, the differential amount of DNA released from the cells of the urinary tract (which conceivably could be individual-specific) could also have affected the dilution effect on tumour-derived DNA fragments. This could have influenced (positively or negatively) the detection of tumour-specific molecular alterations. It is reasonable to believe that these two scenarios could vary among different individuals or different collection timepoints in the same subject. This could explain why the cases CRC-UD21 and CRC-UD02 had comparable tumour contents (tumour DNA fragments) in the plasma and matched urine fluid.
We speculate that although the urine trtDNA is a result of the glomerular filtration of the plasma ctDNA, this phenomenon could be due to our urine isolation protocol which selects and enriches for the low-molecular weight fragments.
We also wanted to highlight that we collected first-morning void urine for DNA extraction and analysis, following the gold standard protocol for urine analysis. However, it would be reasonable to argue that the first-morning void urine (which has been in contact with the bladder overnight) might contain higher amount of normal DNA from urothelium cells, contributing to the dilution of the tumour-derived fragments. However, in another case, UD-CRC21, the (double-step DNA) isolation procedure did not result in an efficient DNA fragment separation, influencing our mutational calling capabilities.
Most importantly, we found that tumour mutations are mainly supported by shorter reads. This evidence emerged due to the improved NGS-computational pipeline we developed which allowed us to observe that tumour-derived DNA is mainly present in low molecular weight fragments. This finding has implications for future studies that aim to improve the sensitivity of trtDNA analysis and opens unprecedented opportunities for early detection, therapy monitoring and measurement of minimal residual disease after surgery.
In conclusion, this study is a proof of concept that clearly showed the feasibility of the use of urine samples for genetic profiling of non-urogenital tract tumours, mCRC patients. Moreover, we found that DNA fragments of low molecular weight are preferentially derived from tumour cells, highlighting the relevance of developing protocols that improve their isolation to increase the recovery rate of these fragments. While urine-based liquid biopsy is promising and has potential advantages over blood-based tests, additional technical improvements are required before entering as a routine text in clinic to monitor the genotype of tumours originating not in the urogenital tract.
esmoopen-2019-000572supp003.pdf (62.5KB, pdf)
Acknowledgments
We thank the patients and their families. We thank Nicole M Reilly for critical reading of the manuscript.
Footnotes
Contributors: GS, GC and AB designed and supervised the study. BM, AB, MM and LB performed the experiments. GS, GC and FDN analysed data. GC conducted bioinformatics data analyses. AS-B, AC, SS, SM, FP, AM and FM treated patients and provided clinical samples. GS, GC and AB wrote the manuscript. All authors read and approved the final submitted manuscript.
Funding: The research leading to these results has received funding from Fondazione AIRC under 5 per Mille 2018 ID. 21091 program—P.I. Bardelli Alberto (AB and SS); AIRC IG 2015 n. 16788 (AB); AIRC IG 2018—ID. 21923 project—PI Bardelli Alberto (AB). This work was also supported by European Community’s Seventh Framework Programme under grant agreement no. 602901 MErCuRIC (AB); H2020 grant agreement no. 635342-2 MoTriColor (AB and SS); IMI contract n. 115749 CANCER-ID (AB); AIRC-CRUK-FC AECC Accelerator Award contract 22795 (AB); Progetto NET-2011-02352137 Ministero della Salute (AB); Fondazione Piemontese per la Ricerca sul Cancro-ONLUS 5 per mille 2014 e 2015 Ministero della Salute (AB and SS). GS was funded by Roche per la Ricerca grant 2017 and AIRC three years fellowship 2017. AIRC Investigator Grant [Project ID 20685] to SS. Fondazione Oncologia Niguarda Onlus, grant Terapia Molecolare dei Tumori (AS-B, SS, AC), grant studies to Develop Therapies Against Colorectal Cancer in Young Adults (AS-B, SS, AC); grant Fondazione Regionale Ricerca Biomedica – Regione Lombardia – (FRRB) IANG CRC (AS-B, SS, AC).
Competing interests: AB is a consultant for Biocartis and Guardant Health. AS-B is an advisory board member for Amgen, Bayer and Sanofi.
Patient consent for publication: The study was conducted according to the provisions of the Declaration of Helsinki, and all patients signed and provided their informed consent before sample collection.
Ethics approval: Samples were obtained through protocols approved by local Ethical Committees as follows: Comitato Etico Grande Ospedale Metropolitano Niguarda Ca’ Granda, Milano, Italy and IRB Fondazione IRCCS Istituto Nazionale Tumori (INT), Milano, Italy, study protocol 117/15.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data are available in a public, open access repository.
References
- 1.Duffy MJ, van Dalen A, Haglund C, et al. . Clinical utility of biochemical markers in colorectal cancer: European group on tumour markers (EGTM) guidelines. Eur J Cancer 2003;39:718–27. 10.1016/s0959-8049(02)00811-0 [DOI] [PubMed] [Google Scholar]
- 2.Chan KCA, Woo JKS, King A, et al. . Analysis of plasma Epstein–Barr virus DNA to screen for nasopharyngeal cancer. N Engl J Med 2017;377:513–22. 10.1056/NEJMoa1701717 [DOI] [PubMed] [Google Scholar]
- 3.Siravegna G, Marsoni S, Siena S, et al. . Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 2017;14:531–48. 10.1038/nrclinonc.2017.14 [DOI] [PubMed] [Google Scholar]
- 4.Bettegowda C, Sausen M, Leary RJ, et al. . Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24 10.1126/scitranslmed.3007094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimura H, Fujiwara Y, Sone T, et al. . Egfr mutation status in tumour-derived DNA from pleural effusion fluid is a practical basis for predicting the response to gefitinib. Br J Cancer 2006;95:1390–5. 10.1038/sj.bjc.6603428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seoane J, De Mattos-Arruda L, Le Rhun E, et al. . Cerebrospinal fluid cell-free tumour DNA as a liquid biopsy for primary brain tumours and central nervous system metastases. Ann Oncol 2019;30:211–8. 10.1093/annonc/mdy544 [DOI] [PubMed] [Google Scholar]
- 7.Siravegna G, Geuna E, Mussolin B, et al. . Genotyping tumour DNA in cerebrospinal fluid and plasma of a HER2-positive breast cancer patient with brain metastases. ESMO Open 2017;2:e000253 10.1136/esmoopen-2017-000253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siravegna G, Mussolin B, Buscarino M, et al. . Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med 2015;21:795–801. 10.1038/nm.3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melkonyan HS, Feaver WJ, Meyer E, et al. . Transrenal nucleic acids: from proof of principle to clinical tests. Ann N Y Acad Sci 2008;1137:73–81. 10.1196/annals.1448.015 [DOI] [PubMed] [Google Scholar]
- 10.Su Y-H, Wang M, Brenner DE, et al. . Human urine contains small, 150 to 250 nucleotide-sized, soluble DNA derived from the circulation and may be useful in the detection of colorectal cancer. J Mol Diagn 2004;6:101–7. 10.1016/S1525-1578(10)60497-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Umansky SR, Tomei LD. Transrenal DNA testing: progress and perspectives. Expert Rev Mol Diagn 2006;6:153–63. 10.1586/14737159.6.2.153 [DOI] [PubMed] [Google Scholar]
- 12.Chen S, Zhao J, Cui L, et al. . Urinary circulating DNA detection for dynamic tracking of EGFR mutations for NSCLC patients treated with EGFR-TKIs. Clin Transl Oncol 2017;19:332–40. 10.1007/s12094-016-1534-9 [DOI] [PubMed] [Google Scholar]
- 13.Husain H, Melnikova VO, Kosco K, et al. . Monitoring daily dynamics of early tumor response to targeted therapy by detecting circulating tumor DNA in urine. Clin Cancer Res 2017;23:4716–23. 10.1158/1078-0432.CCR-17-0454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siravegna G, Sartore-Bianchi A, Mussolin B, et al. . Tracking a CAD-ALK gene rearrangement in urine and blood of a colorectal cancer patient treated with an ALK inhibitor. Ann Oncol 2017;28:1302–8. 10.1093/annonc/mdx095 [DOI] [PubMed] [Google Scholar]
- 15.Fujii T, Barzi A, Sartore-Bianchi A, et al. . Mutation-Enrichment Next-Generation Sequencing for Quantitative Detection of KRAS Mutations in Urine Cell-Free DNA from Patients with Advanced Cancers. Clin Cancer Res 2017;23:3657–66. 10.1158/1078-0432.CCR-16-2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corti G, Bartolini A, Crisafulli G, et al. . A genomic analysis workflow for colorectal cancer precision oncology. Clin Colorectal Cancer 2019;18:91–101. 10.1016/j.clcc.2019.02.008 [DOI] [PubMed] [Google Scholar]
- 17.Siravegna G, Lazzari L, Crisafulli G, et al. . Radiologic and genomic evolution of individual metastases during HER2 blockade in colorectal cancer. Cancer Cell 2018;34:148–62. 10.1016/j.ccell.2018.06.004 [DOI] [PubMed] [Google Scholar]
- 18.Orzan F, De Bacco F, Crisafulli G, et al. . Genetic evolution of glioblastoma stem-like cells from primary to recurrent tumor. Stem Cells 2017;35:2218–28. 10.1002/stem.2703 [DOI] [PubMed] [Google Scholar]
- 19.Díaz-Gay M, Vila-Casadesús M, Franch-Expósito S, et al. . Mutational signatures in cancer (MuSiCa): a web application to implement mutational signatures analysis in cancer samples. BMC Bioinformatics 2018;19:224 10.1186/s12859-018-2234-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su Y-H, Song J, Wang Z, et al. . Removal of high-molecular-weight DNA by carboxylated magnetic beads enhances the detection of mutated K-ras DNA in urine. Ann N Y Acad Sci 2008;1137:82–91. 10.1196/annals.1448.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hyman DM, Diamond EL, Vibat CRT, et al. . Prospective blinded study of BRAFV600E mutation detection in cell-free DNA of patients with systemic histiocytic disorders. Cancer Discov 2015;5:64–71. 10.1158/2159-8290.CD-14-0742 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
esmoopen-2019-000572supp002.pdf (58.6KB, pdf)
esmoopen-2019-000572supp001.pdf (54.9KB, pdf)
esmoopen-2019-000572supp005.pdf (64.7KB, pdf)
esmoopen-2019-000572supp006.pdf (46.7KB, pdf)
esmoopen-2019-000572supp004.pdf (33.4KB, pdf)
esmoopen-2019-000572supp007.pdf (42.7KB, pdf)
esmoopen-2019-000572supp008.pdf (44.2KB, pdf)
esmoopen-2019-000572supp009.pdf (76KB, pdf)
esmoopen-2019-000572supp010.pdf (39.1KB, pdf)
esmoopen-2019-000572supp011.pdf (74.3KB, pdf)
esmoopen-2019-000572supp003.pdf (62.5KB, pdf)