Abstract
We describe a variation of droplet digital PCR (ddPCR) for DNA structural variant analysis, where expensive fluorescent probes are replaced by low-cost EvaGreen, a DNA binding dye, hence reducing cost without reducing sensitivity. ddPCR is a method that employs water-oil microfluidics and fluorescence technology to quantify target nucleic acids with extreme precision and sensitivity. Traditional ddPCR uses standard Taqman probe assays to determine target DNA concentrations, however these two-color fluorescent oligonucleotide probes are expensive and require extensive optimization. EvaGreen’s DNA binding properties allow target products to be effectively quantified at a significantly lower cost. This protocol employs the EvaGreen ddPCR assay to confirm structural variants on a ddPCR system and produces results comparable to the standard, far more expensive Taqman approach.
Keywords: ddPCR, rare variant detections, copy number variant, EvaGreen, structural variant
INTRODUCTION
Detection of copy-number variants in human DNA is a significant problem in human genetics, even after the advent of next-generation sequencing techniques. Although next-generation sequencing techniques have significantly enhanced our ability to detect copy-number variants, this detection method has a significant false-positive detection rate. Droplet digital polymerase chain reaction (ddPCR) is an exceptionally sensitive and powerful tool that enables absolute quantification of nucleic acids and is routinely used to confirm next-generation DNA sequencing identified variants. ddPCR protocols to identify copy-number variants commonly involve simultaneous PCR with two pairs of oligonucleotides. One primer of each pair has a unique fluorescent label. However, costs and optimization are significantly reduced by employing an alternative method of differentiating target and reference amplicons, through use of the nonspecific DNA binding properties of EvaGreen (EG) dye. This protocol provides an easier, lower cost method for performing ddPCR to detect copy-number variants.
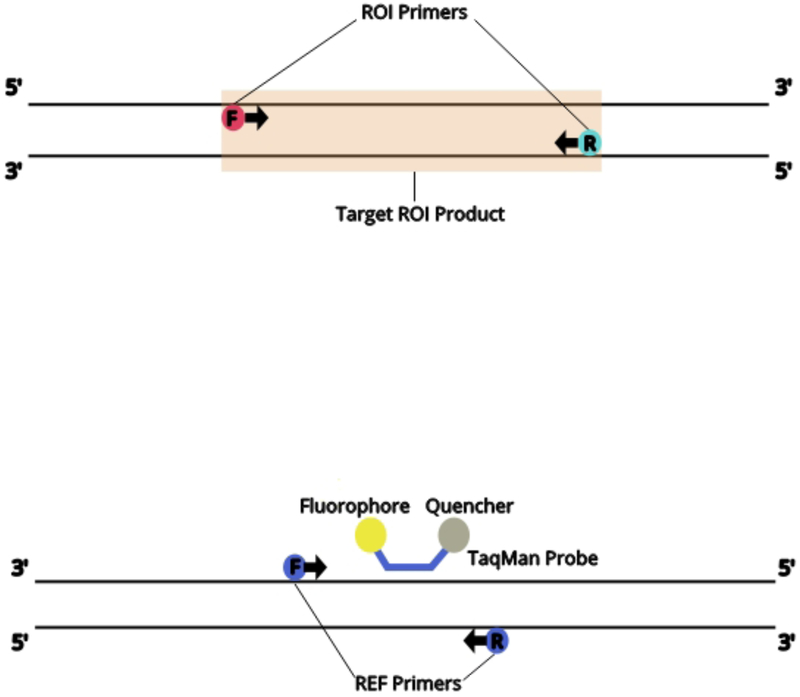
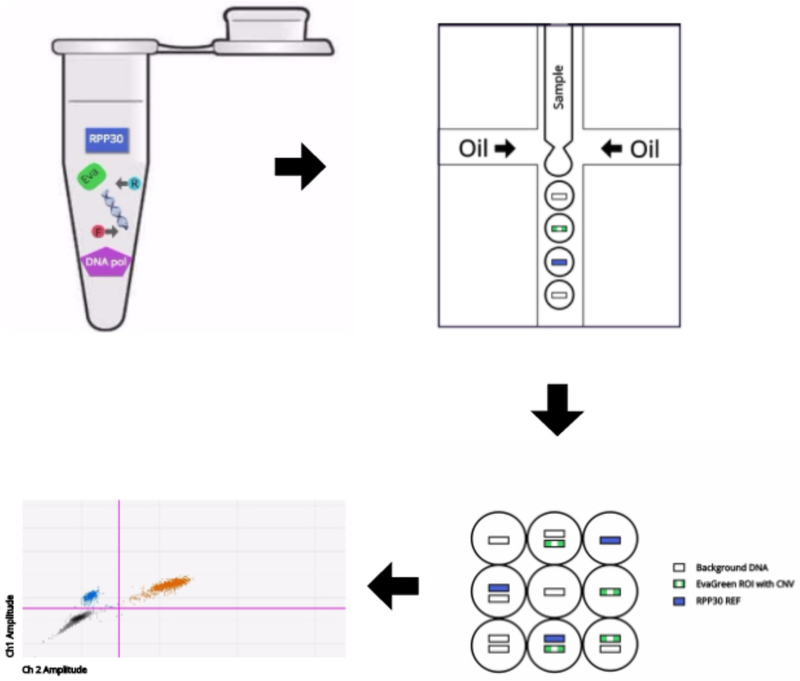
ddPCR with EvaGreen for SV analysis workflow begins with the same materials needed for a standard PCR assay (ref needed). Genomic DNA, dNTPs, primers, DNA polymerase and buffer are all combined. Additonally, ddPCR EvaGreen Supermix (Bio-Rad) is added to the PCR reaction. Although EvaGreen replaces the TaqMan probes targeting the specific region of interest (ROI’s) for each DNA sample, a common Taqman probe was used to target a standard reference gene across all samples. A DNA fragment from the human gene RPP30 is recommended as standard reference PCR product. The reaction is set up as a duplex PCR in which a primer pair is targeted for the region of interest (ROI) and the primer/probe mix is targeted for a standard reference (REF) gene (See Fig 1A). There are two fluorophores (EvaGreen and VIC) that correspond to ROI and REF targets, and fluorescent signal is generated when respective target products are present. Through microfluidics, the reaction mix is fractionated into 20,000 droplets in which each represents individual PCR reactions of ~1 nl each. There is a random distribution of ROI and/or REF targets within the droplets such that there will be droplets that contain 0, 1, or 2 target copies (See Fig 1B).
Figure 1.
Overview of the ddPCR with EvaGreen assay. (A) Schematic of the region of interest (ROI, upper) for copy number variant analysis and the reference (REF, lower). (B) Schematic of the ddPCR with EvaGreen workflow.
After amplification, the fluorescence of each droplet is read by a droplet reader. Droplets containing target ROI or REF will fluoresce in the corresponding channel (positive droplets), while those without target will not (negative droplets). The counts of positive and negative droplets for each target are related to the target’s concentration in the sample by the Poisson distribution:
where λ is the average number of copies per droplet and p is the ratio of positive droplets to the total number of droplets (Hindson et al., 2011). The copies per droplet λ, can be converted to the number of copies per ul by knowing the volume of reaction per droplet, which is about 1 nl (Mazaika et al 2014). Through this method, the concentration of the target nucleic acids (ROI or REF) can be calculated without the use of standard samples.
STRATEGIC PLANNING
Designing Assays
Primers are designed to amplify 90 to 110 bp within the target region (Fig 1). The PCR assay is designed in a similar manner as previously reported (Mazaika et al, 2014). However, only one fluoresceinated probe is required.
Preparing DNA
Use 10–50ng of DNA per reaction. The ddPCR assay works at a relatively broad input range compared to real time PCR. Reaction mixtures comprised of 50ng DNA is optimal to obtain PCR products in <40% of all 5,000–10,000 droplets. To optimize the results, a dilution series can be performed. DNA can be digested prior to generating droplets, due to the theoretical interference of the viscosity of undigested genomic DNA in properly partitioning droplets. However, we find that for most PCR reactions using most genomic DNA, there is no benefit to digesting the DNA with a restriction enzyme before use.
BASIC PROTOCOL 1
SV DETECTION USING BIO-RAD’s QX200 DIGITAL PCR PLATFORM
Although this protocol has been optimized on the Bio-Rad QX200 system, the same principles should apply to other ddPCR systems. The workflow consists of preparing DNA, setting up reactions, making droplets, thermal cycling, and analyzing on the droplet reader.
Materials
DNA
Nuclease free water
2x ddPCR EvaGreen Supermix (includes hot start DNA polymerase, dNTPs including dUTP; Bio-Rad)
20x ROI target primer (see Figure 1A)
20x REF target (RPP30) primer/Taqman probe mix (see Reagents and Solutions recipe)
2x buffer control for EvaGreen (Bio-Rad)
Droplet Generation Oil for Evagreen (Bio-Rad)
96-well plates
96-well plate centrifuge
DG8 droplet generator cartridges (single-use; Bio-Rad)
DG8 droplet generator cartridge holder (Bio-Rad)
DdPCR droplet generation (DG) oil (Bio-Rad)
DG8 gaskets (single-use; Bio-Rad)
QX200 droplet generator (Bio-Rad)
Eppendorf twin.tec semi-skirted 96 well plate
Heat sealer
Heat sealing PCR foil
Thermal cycler
Bio-Rad QX200 droplet reader
QuantaSoft software
Prepare the DNA
-
1
Quantify DNA
-
2
Dilute DNA to 10–50ng/μl
Assemble PCR reactions
-
3Assemble components of reaction in standard 96-well plate as described:
- 12.5 μL of 2x Bio-Rad ddPCR EvaGreen supermix (contains buffer, DNA polymerase, dideoxynucleotide triphosphates)
- 1.25 μL of 20x ROI target primers (forward and reverse)
- 1.25 μL of 20x RPP30 (REF target) primer mix
- 1–5μl DNA (~50ng) (from step 2)
- Fill to total volume of 25 μl with nuclease-free water
Reactions are assembled in a total volume of 25 μl, although only 20 μl is used in droplet formation. Excess volume ensures that no air bubbles are transferred to the droplet generator cartridge.
See Reagents and Solutions for oligo sequences of the REF target assay that we suggest for SV analysis (RPP30).
-
4
Centrifuge plate of reactions at 150 X g for 15 seconds at room temperature to ensure contents of reaction are at bottom of the well.
-
5
Mix reactions by pipetting 15 to 20 μl up and down ~10 times to ensure completely homogenous mixture
-
6
If necessary, centrifuge briefly to ensure contents at bottom of well
Droplet generation via QX200 droplet generator
-
7
Insert Bio-Rad microfluidics cartridge into cartridge holder and slide holder to snap closed.
-
8
Pipet 70 μl droplet generation oil for EvaGreen into each of the wells of cartridge designated “oil” by cartridge holder.
-
9
Pipet 20 μl of assembled reaction from step 6 into cartridge wells designated sample. Avoid pipetting any air bubbles into the well as this may prevent droplet generation.
When dispensing sample into cartridge, place pipet tip at bottom corner of well and release plunger slowly to prevent bubbles from forming at bottom of well. Do not depress plunger of pipet past the first stop when dispensing. A small amount of sample (<1ul) may remain in the pipet tip after dispensing to the first stop.
-
10
If the number of reaction is less than eight per cartridge, fill the remaining sample wells with 20 μl of control buffer (supplied at 2x) and nuclease-free water so that all of the sample wells are occupied during droplet formation.
-
11
Manually inspect each sample well for air bubbles and carefully remove bubbles with a pipet tip if necessary.
The droplet generator may fail to make droplets when air bubbles are present in the well, as the air may clog the microfluidic channel.
-
12
Attach the consumable gasket to the top of the holder/cartridge.
-
13
Insert the holder/cartridge into the QX100 or QX200 Droplet Generator and close the cover.
All eight positions must have oil and sample/buffer in the wells. QX100 or QX 200 Droplet Generator can be used with the EvaGreen unit, but when reading droplets, QX200 Droplet Reader must be used.
-
14
Wait 2 min for droplet generation to complete.
-
15
Open the door and remove holder/cartridge from the QX100 or QX200 Droplet Generator
-
16
Remove the gasket and manually inspect droplet wells to ensure droplet generation was successful.
When droplets are made successfully, the samples will appear slightly opaque.
-
17
Transfer entire volume of the droplet well (approximately 40 μl) from the cartridge holder into an Eppendorf twin.tec semi-skirted 96-well plate PCR plate.
We suggest using a multichannel pipet and pipetting slowly in order to prevent droplets from being disturbed or broken. When dispensing sample into plate, depress plunger slowly so that solution is transferred droplet by droplet. Pipet tip should never directly contact the bottom of the well, as this may cause droplets to break.
-
18
Seal the plate with an Easy Pierce thermal foil seal.
Do not centrifuge plate once droplets have been generated.
Thermal Cycling
-
19
Thermal cycle the plate in standard 96-well thermal cycler using the protocol as per Bio-Rad’s standard EvaGreen protocol (See Table 1).
| Cycling Step | Temperature,°C | Time | Ramp Rate | # of Cycles |
|---|---|---|---|---|
| Enzyme activation | 95 | 5 min | 2°C/sec | 1 |
| Denaturation | 95 | 30 sec | 40 | |
| Annealing/extension | 60 | 1 min | 40 | |
| Signal stabilization | 4 | 5 min | 1 | |
| 90 | 5 min | 1 | ||
| Hold | 4 | Infinite | 1 |
Droplet reader: set up and reading
-
20
Check that the droplet reader has sufficient droplet reader oil in supply bottle and that the waste container does not need to be emptied.
Indicator light will blink amber when oil needs to be refilled or waste emptied. Always add 50 ml of 10% bleach to the empty waste container.
-
21
Secure the PCR plate containing the droplets in the plate reader holder.
-
2223 Enter experiment information into the template and apply the following to each well:
- Experiment
- For CNV analysis choose number of copies of reference gene
- Sample name
- Define assay 1 (FAM) as EvaGreen
- Define assay 2 (VIC) as standard reference gene (RPP30)
- Select type of run (ddPCR supermix with probes)
EvaGreen is spectrally similar to the FAM fluorophore with an emission wavelength of ~500/530 nm when bound to DNA.
-
23
Save template
-
24
Start plate run.
-
25
Select the dye pair in use (FAM/VIC) and direction for the wells to be ready (by column or by row).
There is an option to select EvaGreen dye; however we use a Taqman probe for the reference gene, so we must select the option that will be able to detect its fluorescence.
Droplet reader: analysis
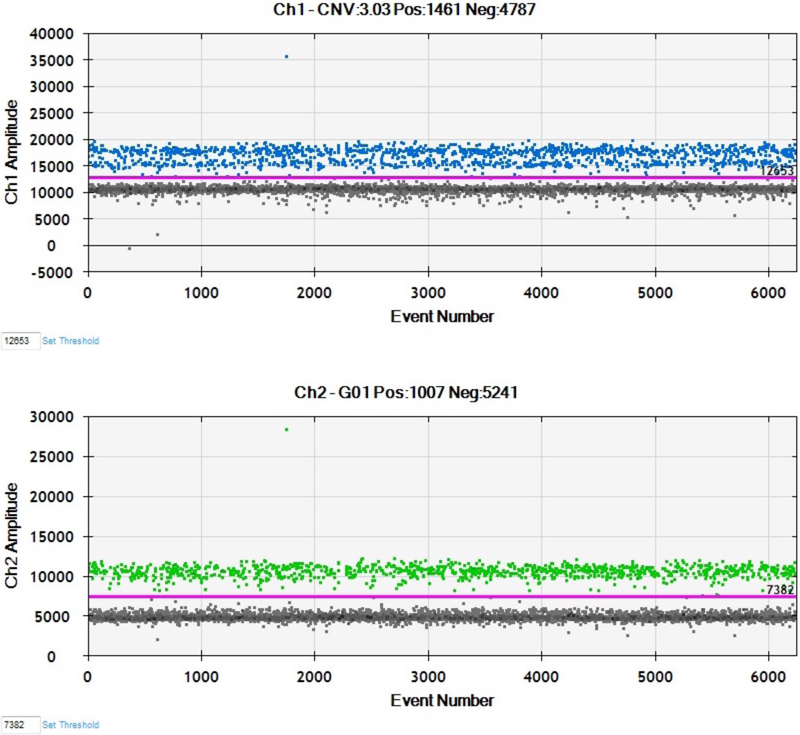
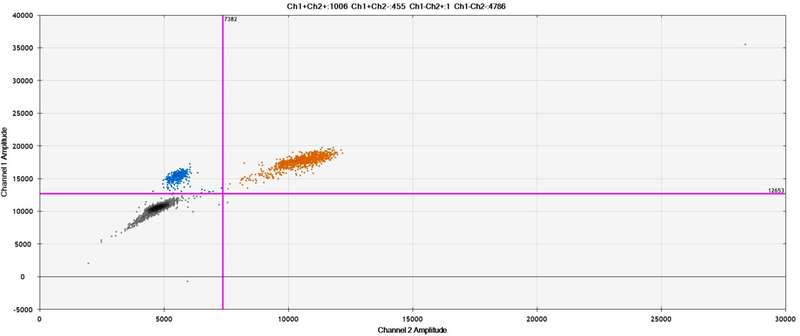
Figures 2A and 2B presented here are generated by the QuantaSoft software, which displays each droplet by a single dot in the 1-D and 2-D amplitude views. The QuantaSoft software will automatically call a fluorescence threshold for each well to distinguish between positive and negative droplets. The droplets with fluorescence above threshold will present in color as to show positive signal, and all droplets below threshold will be black as to show negative signal (See Fig 2A).
Figure 2.
Sample results from ddPCR with EvaGreen assay. (A) 1D plots for FAM (upper) and VIC (lower) channels. (B) 2D plot showing combined three clusters of droplets: blue, EvaGreen-positive (ROI); orange, EvaGreen- and VIC-positive(i.e., REF gene RRP30); and black, negative.
-
26
After the plate is run (approximately 2 min/well), manually inspect results of each well.
In the case that QuantaSoft software does not automatically set a fluorescence threshold and a “No call” status appears for a well, the threshold must be manually set.
Interpretation of results
In both 1-D and 2-D views, the droplets are graphically represented by a single dot. The 2-D view is most useful for interpreting results with EvaGreen because it can visually display the EvaGreen (blue), EvaGreen +RPP30 (orange), and negative (black) signal with three prominent clusters according to their fluorescence amplitude (See Fig 2B).
-
27
Open the “Analyze” section to find the counts for each quadrant.
-
28
Determine the average number of target ROI (EvaGreen)
-
29
Determine the average number of target REF (RPP30)
-
30
Calculate ratio of target ROI to target REF
We present results for a trio from a representative SV Experiment (See Table 2). EvaGreen and RPP30 counts must be calculated Individually using the Poisson function, and the ratio of EvaGreen to RPP30 must be determined to conclude presence and type of SV. In this example, the proband contained a de novo deletion.
Table 2.
Sample CNV Results using ddPCR with EvaGreen Assa
| Sample | Ch1+Ch2+ (Eva_RPP30) | Ch1+Ch2− (Eva) | Ch1−Ch2+ (RPP30) | Ch1−Ch2− (Negative) | Ratio |
|---|---|---|---|---|---|
| 1 | 1004 | 458 | 1 | 4786 | 0.43 |
| 1–01 | 1294 | 947 | 0 | 2563 | 0.70 |
| 1–02 | 1148 | 834 | 0 | 3548 | 0.70 |
REAGENTS AND SOLUTIONS
Nuclease-free water should be used in all recipe steps. For common stock solutions, see Appendix 2D. For suppliers, see Suppliers Appendix.
Primer/TaqMan probe mix, 20×
20x primer/TaqMan probe mix REF target is a combination of primers and fluorescent probes at the following concentrations:
Forward primer: 18 μM
Reverse primer: 18 μM
Labeled probe(s): 5 μM
RPP30 assay (Hindson et al., 2011): Primer and probe sequences
Forward primer: GATTTGGACCTGCGAGCG
Reverse primer: GCGGCTGTCTCCACAAGT
Probe: VIC-CTGACCTGAAGGCTCT-MGB-NFQ
TaqMan by Applied Biosystems designs its probes with a 5’ reporter (VIC) and 3’ MGB-NFQ, the minor groove binder/non-fluorescent quencher. MGB-NFQ provides lower background signal for increased precision in droplet quantification.
COMMENTARY
Background Information
This protocol describes an alternate procedure for validating copy-number variants using ddPCR that reduces cost and effort without reducing accuracy. The accuracy of this method, like all ddPCR methods, rests on the qualitative assessment of the fluorescence of each droplet. Because each droplet’s fluorescence is determined by the amount of PCR product in the droplet, the method is much less sensitive to PCR-efficiency differences between test and control DNA sequences than qPCR. In addition the method is essentially independent of DNA concentration measurements that can confound qPCR methods.
In ddPCR the amount of DNA in a droplet is usually assessed using Taqman probes, which only fluoresce when the probe sequence anneals to DNA. However, developing a unique Taqman probe that only fluoresces after PCR product has been generated in a droplet can be problematic. The method described here, uses the DNA-binding dye EvaGreen, which fluoresces only when bound to DNA. Importantly EvaGreen will fluoresce when bound to either the test sequence or the ‘control’ RPP30 sequence. Hence after thermal amplification, droplets will contain: a) no PCR product and will fluoresce in neither the VIC or FAM channels (channels 1 and 2); b) PCR product from the test region and will fluoresce due to EvaGreen (FAM or channel 2) but not due to the RPP30 Taqman probe (VIC) and channel 1 will be read as 0; or c) PCR product from the control RPP30 primers which will fluoresce due to the Taqman probe (VIC, channel 1) and due to EvaGreen (FAM, channel 2) so that these droplets have 2 color-fluorescence. In short, the ratio of EvaGreen (FAM) only droplets to Evagreen (FAM) and VIC (Taqman probe for RPP30) determines the ratio of test DNA to control DNA. When the ratio of test DNA containing droplets: control DNA containing droplets is 1, there is no evidence of a copy-number variant. If the test DNA contains a deletion, the ratio of test DNA containing droplets (FAM only): control DNA containing droplets (FAM and VIC) should be 0.5. If the test DNA contains a duplication of one allele, the ratio of test DNA containing droplets (FAM) : control DNA containing droplets (FAM and VIC) should be 1.5.
Critical Parameters
This procedure employs ddPCR. One critical parameter is that the two sets of PCR primers must amplify the test and control DNAs only at the desired target sites. That is, template DNA when placed in a PCR reaction with each of these primer pairs separately and the products fractionated by electrophoresis, should yield a single band. These studies can be confounded by PCR primers that yield multiple bands derived from multiple sites in the genomic DNA. Methods for optimizing PCR reactions are described elsewhere (ref CPMB, CPHG).
The goal of this method is to assess the ratio of test DNA to control DNA in a sample. This measurement is achieved by comparing the numbers of ddPCR droplets that have test DNA to control DNA. A fundamental principle of ddPCR is that the droplets are not ‘saturated’ that is that there is only one DNA molecule per droplet (either test DNA molecule or control DNA molecule). Hence a critical parameter is that there should be many empty droplets. Ideally, there should be <30% of droplets that produce a signal with EvaGreen.
Troubleshooting
There are several possible outcomes, which indicate a failed experiment:
-
a)
No fluorescing droplets. The PCR reactions have failed, either because there was essentially no (or very little template DNA), or because an essential PCR reaction component (primers, nucleotides, Taq polymerase etc) are missing. A good test to define the problem is to use the template DNA in PCR reactions with each of the two primer pairs.
-
b)
Too many fluorescing droplets (i.e. more than 50% of droplets are ‘positive’). Either the DNA concentration is higher than expected or the one of the primer pairs is giving multiple products (see Critical Parameters).
-
c)
The ratio of FAM droplets : VIC + FAM droplets is not in the expected range (i.e. 0.4–0.7, 0.9–1.2, 1.4–1.6). This does happen occasionally. Usually this reflects inadequate numbers of fluorescent positive droplets and can be corrected by repeating the experiment using more template DNA. We have speculated that digesting the DNA with a restriction enzyme may help resolve such issues, but we have not observed this improvement in practice.
Statistical Analyses
The ratio of test DNA containing droplets: control DNA containing droplets provides an accurate measure of the copy-number of the test DNA. The numbers of test DNA containing droplets and control DNA containing droplets should be corrected using the Poisson distribution to define the ‘concentration’ of each droplet type. The Poisson distribution corrects for the possibility that both the test DNA and the control DNA molecules are present in the same droplet. As the fraction of empty droplets increases, the utility of the Poisson distribution “correction” diminishes.
Understanding Results
The critical measurement is the ratio of FAM positive droplets: FAM + VIC positive droplets. If the ratio is 1.0 there is no copy-number variant in the test region; ratio = 1.5 the copy-number variant is a duplication and ratio = 0.5 the copy-number variant is a deletion. In practice the ratios generally are close to the theoretical values but may vary from one test site to another. In our experience the ratio ranges for deletions from 0.4–0.7, for no copy-number variant from 0.9–1.2 and for duplications from 1.4–1.6. As noted above (Troubleshooting), if the ratio is outside of these ranges the experiment should be repeated.
Time Considerations
The entire experiment should take no more than 2 days after the DNA is isolated from cells or tissue. PCR reactions can be set up in 2–4 hours depending upon the number of samples. Droplets are made in about 3 minutes per column of 8 samples. Thermal cycling requires about 2 hours. Droplets from a 96 well plate can be analyzed in ~2 hours.
Significance Statement.
This protocol provides a method for assessing copy-number (deletions or insertions) variants in DNA using digital droplet PCR (ddPCR). Whereas, traditional ddPCR for detecting structural variants this method employs two fluoresceinated oligonucleotide probes, one for the test sequence and one for a control, this procedure replaces one fluoresceinated probe with a DNA binding dye EvaGreen. This procedure reduces cost and simplifies optimization without reducing sensitivity.
LITERATURE CITED
- Bio-Rad. QX100 Droplet Digital PCR System Guide. Bio-Rad; Pleasanton, Calif: 2012. [Google Scholar]
- Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Bright IJ, Lucero MY, Hiddessen AL, Legler TC, Kitano TK, Hodel MR, Petersen JF, Wyatt PW, Steenblock ER, Shah PH, Bousse LJ, Troup CB, Mellen JC, Wittmann DK, Erndt NG, Cauley TH, Koehler RT, So AP, Dube S, Rose KA, Montesclaros L, Wang S, Stumbo DP, Hodges SP, Romine S, Milanovich FP, White HE, Regan JF, Karlin-Neumann GA, Hindson CM, Saxonov S, Colston BW. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604–8610. [DOI] [PMC free article] [PubMed] [Google Scholar]
KEY REFERENCE (optional)
- Mazaika E, Homsy J. Digital Droplet PCR: CNV Analysis and Other Applications. Curr Protoc Hum Genet. 2014; 82: 7.24.1–7.24.13. [DOI] [PMC free article] [PubMed] [Google Scholar]