Abstract
Background
Synaptic degeneration and accumulation of amyloid β-peptides (Aβ) are hallmarks of the Alzheimer diseased brain. Aβ is synaptotoxic and produced by sequential cleavage of the amyloid precursor protein (APP) by the β-secretase BACE1 and by γ-secretase. If APP is instead cleaved by the α-secretase ADAM10, Aβ will not be generated. Although BACE1 is considered to be a presynaptic protein and ADAM10 has been reported to mainly localize to the postsynaptic density, we have previously shown that both ADAM10 and BACE1 are highly enriched in synaptic vesicles of rat brain and mouse primary hippocampal neurons.
Results
Here, using brightfield proximity ligation assay, we expanded our previous result in primary neurons and investigated the in situ synaptic localization of ADAM10 and BACE1 in rat and human adult brain using both pre- and postsynaptic markers. We found that ADAM10 and BACE1 were in close proximity with both the presynaptic marker synaptophysin and the postsynaptic marker PSD-95. The substrate APP was also detected both pre- and postsynaptically. Subcellular fractionation confirmed that ADAM10 and BACE1 are enriched to a similar degree in synaptic vesicles and as well as in the postsynaptic density.
Conclusions
We show that the α-secretase ADAM10 and the β-secretase BACE1 are located in both the pre- and postsynaptic compartments in intact brain sections. These findings increase our understanding of the regulation of APP processing, thereby facilitating development of more specific treatment strategies.
Keywords: Alzheimer disease, Amyloid precursor protein, Secretases, Synapse, Nerve terminal
Background
Alzheimer disease (AD) is a slowly progressive neurodegenerative disorder leading to impairment in memory and other cognitive functions [1]. In the Alzheimer diseased brain, abnormal aggregation of the amyloid β-peptide (Aβ) leads to the formation of senile plaques. Together with neurofibrillary tangles of hyperphosphorylated tau, these plaques form the characteristic pathological hallmarks of AD [2]. Aβ has also been shown to cause synaptic dysfunction and degeneration [3–5], which are among the earliest pathological events during the course of the disease and correlate well with cognitive decline [1, 6]. The synaptotoxicity is particularly prominent for oligomeric forms of Aβ and might affect neuronal function long before the appearance of plaques [2, 7].
Aβ is produced from the amyloid precursor protein (APP) by sequential cleavage performed by β- and γ-secretase. Non-amyloidogenic cleavage of APP by α-secretase instead of β-secretase precludes Aβ formation since the α-secretase cleavage site of APP is located within the Aβ sequence [2]. The β-site APP cleaving enzyme 1 (BACE1) is the main β-secretase of neurons [8–12] while α-cleavage mainly is performed by a disintegrin and metalloproteinase 10 (ADAM10) [13, 14]. BACE1 levels and activity are increased in AD brain compared to control [15] as well as in the brains of transgenic mice models of AD, where it accumulates around amyloid plaques and in dystrophic neurites [16, 17]. In addition, a decrease in ADAM10 activity has been shown in AD patients when compared to control subjects [18].
An important issue for our understanding of the physiological and pathological role of Aβ is to elucidate how its production is regulated. A potential regulatory mechanism for Aβ production is the subcellular localization of APP and its secretases. Knowledge about the subcellular localization can open up for therapeutic opportunities by targeting inhibitors to specific subcellular sites or by manipulating the trafficking of the proteins. The importance of protein and vesicle trafficking in AD is emphasized by the fact that risk genes for AD, identified in genome wide association studies, are involved in these processes [19].
BACE1 is active in acidic environments, mainly in endosomes, and is generally considered to be located presynaptically in neurons [17, 20–23]. ADAM10, on the other hand, is mainly regarded to be localized to Golgi and the plasma membrane and to be part of the postsynaptic density (PSD) [24–26]. Based on these earlier studies, the amyloidogenic processing thus appears to take place presynaptically whereas the non-amyloidogenic processing would be considered to occur postsynaptically. However, using subcellular fractionation of rat brain and proximity ligation assay (PLA) in primary neurons, we have previously shown that both ADAM10 and BACE1 are highly enriched in synaptic vesicles (SVs). Aβ can also be produced and stored in SVs, although we could not detect any enrichment of Aβ production in pure synaptic vesicles [27–29]. However, since primary neurons do not recapitulate the situation in adult brain and due to the fact that homogenization and subcellular fractionation can introduce artefacts, it would also be important to study the localization of ADAM10 and BACE1 in sections from intact adult brain. In addition, not only the presynaptic but also the postsynaptic localization of these enzymes should be investigated. Furthermore, no studies have, to our knowledge, investigated the localization of ADAM10 and BACE1 in human brain. Normal immunohistochemistry can never reach a spatial resolution less than 200 nm and can therefore not provide detailed information about subcellular localization. In addition, novel methods such as super-resolution microscopy are not yet fully developed to be used in brain tissue and are hampered by the extensive auto-fluorescence in postmortem aged human brain. Therefore, we used brightfield proximity ligation (PLA) as an alternative approach to investigate the proximity of ADAM10 and BACE1, as well as their substrate APP, to the presynaptic marker synaptophysin and the postsynaptic marker PSD-95. In PLA, secondary antibodies are conjugated to oligonucleotides that, if the proteins of interest are within 40 nm distance from each other, can ligate to each other and be amplified and visualized [30]. The close proximity required thus provides much more detailed information than conventional immunohistochemistry. Using this method, as well as subcellular fractionation, we found that ADAM10 and BACE1 are located both pre- and postsynaptically in the adult rat brain as well as in human brain and that the distribution of the enzymes appears to be similar. Furthermore, we detected close proximity of APP with ADAM10, BACE1, synaptophysin and PSD95, suggesting that APP can be cleaved by ADAM10 and BACE1 both pre- and postsynaptically.
Results
In this study, we took advantage of the highly sensitive method PLA to visualize the in situ localization of ADAM10 and BACE1 in intact adult rat and human brain. With brightfield PLA, two proteins in close proximity (< 40 nm) can be visualized in situ. Thus, this method provides much more detailed information than normal immunohistochemistry and also circumvents the problem of auto-fluorescence, which is particularly prominent in aged human brain. We performed all PLA experiments in both hippocampal and cortical sections of rat and human brain, but since the results were similar, we have chosen only to present the data from the hippocampal sections.
Detecting pre- and postsynaptic ADAM10 and BACE1 in adult rat brain
To follow up on our previous study demonstrating close proximity of ADAM10 and BACE1 to the synaptic vesicle marker synaptophysin in mouse primary hippocampal neurons [27], we here investigated the synaptic localization of ADAM10 and BACE1 in situ in thin sections of intact adult rat hippocampus. In addition to investigating the proximity of these enzymes to the presynaptic marker synaptophysin, we also investigated the proximity to the postsynaptic marker PSD-95 and whether the pre- and postsynaptic distribution differ between ADAM10 and BACE1. Using the brain from one rat, we performed PLA for the combinations ADAM10 + synaptophysin (Fig. 1a), ADAM10 + PSD-95 (Fig. 1b), BACE1 + synaptophysin (Fig. 1c) and BACE1 + PSD-95 (Fig. 1d). All these combinations gave rise to more signals compared to the negative controls where only one of the antibodies were used (Fig. 1e–g), revealing the presence of ADAM10 and BACE1 at both the pre- and postsynaptic side of the synapse in adult rat brain hippocampus.
Fig. 1.
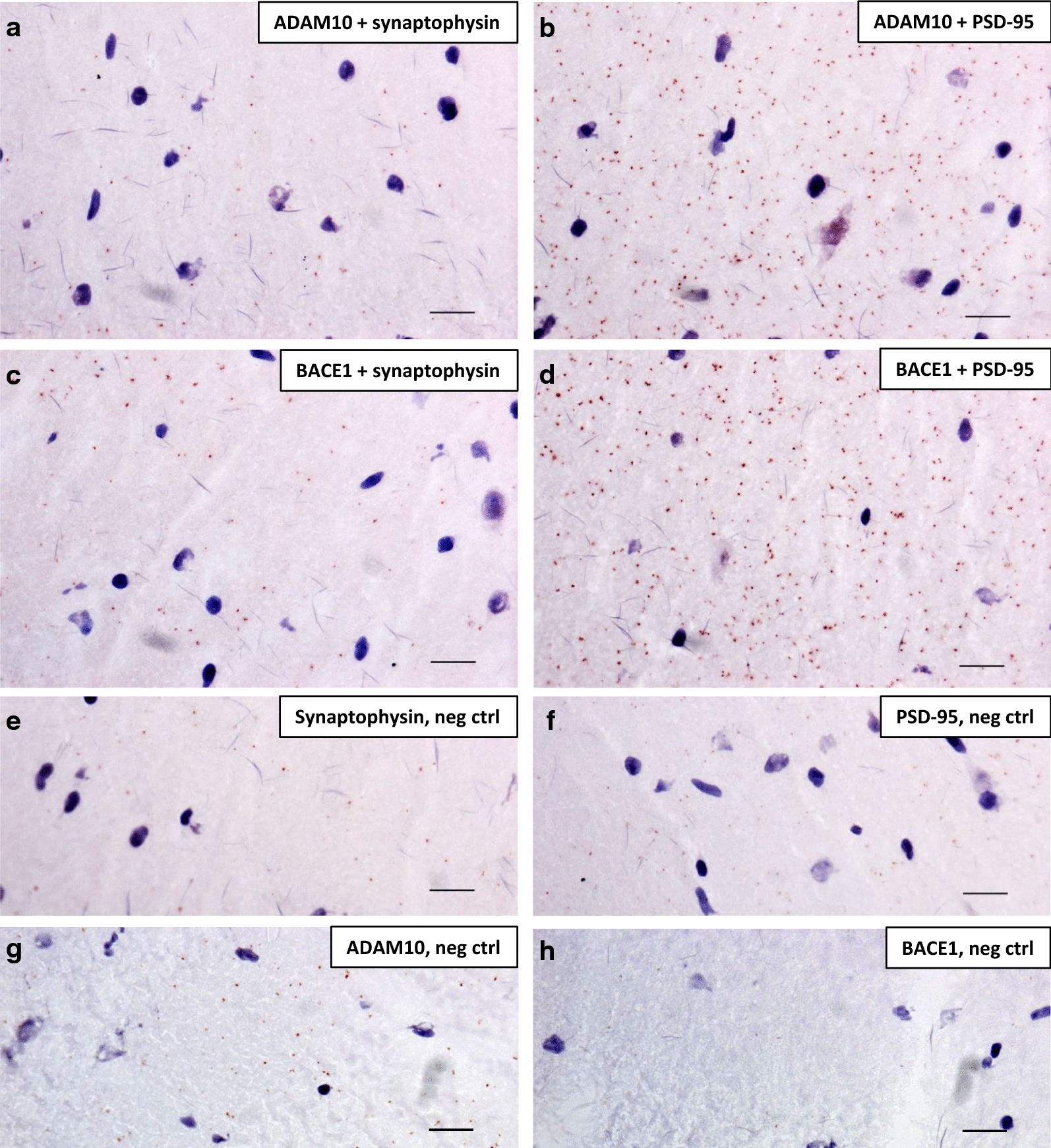
Proximity ligation assay showing pre- and postsynaptic localization of ADAM10 and BACE1 in adult rat brain. Adult rat brain hippocampal sections were labelled with primary antibodies (anti-ADAM10 or anti-BACE1 made in rabbit and anti-synaptophysin or anti-PSD-95 made in mouse) followed by secondary oligonucleotide-linked probes. Each PLA signal (brown dot) denotes two proteins within 40 nm distance from each other; a ADAM10 and synaptophysin, b ADAM10 and PSD-95, c BACE1 and synaptophysin and d BACE1 and PSD-95. In the negative control samples only one primary antibody (anti-synaptophysin (e), anti-PSD-95 (f), anti-ADAM10 (g) or anti-BACE1 (h)) was used. Brain tissue and cell nuclei were visualized by a nuclear stain solution containing Mayer´s haematoxylin. Each experiment was performed five times (technical replicates) and representative images are shown. Scale bar 20 µm
The number of PLA signals generated with PSD-95 was generally higher than with synaptophysin for both of the secretases (Fig. 1a–g). This could indicate that both ADAM10 and BACE1 are present in higher amounts at the postsynaptic side. However, we cannot rule out that the higher number of signals is due to higher concentrations of PSD-95 than synaptophysin and/or higher affinity of the PSD-95 antibody.
From this descriptive experiment we did not observe any differences between ADAM10 and BACE1 in the number of signals detected together with synaptophysin and PSD-95. Hence, the pre- and postsynaptic distribution of ADAM10 and BACE1 in adult rat brain appears to be similar.
Pre- and postsynaptic distribution of APP in adult rat brain
Apart from the localization of the secretases, we also elucidated the synaptic localization of the substrate APP using brightfield PLA. APP + synaptophysin (Fig. 2a) and APP + PSD-95 (Fig. 2b) resulted in higher number of signals than the negative control in which the primary antibodies were omitted (Fig. 2c). Thus, APP is in close proximity to both synaptophysin and PSD-95 in adult rat brain hippocampus (Fig. 2). Again, a higher number of signals was observed for PSD-95 than for synaptophysin but it is evident that APP is located both pre- and postsynaptically in rat brain. In addition, we performed brightfield PLA of ADAM10 and BACE1 with their substrate APP and, as expected, concluded that both secretases are in close proximity to APP in adult rat brain hippocampus (Additional file 1: Figure S1).
Fig. 2.
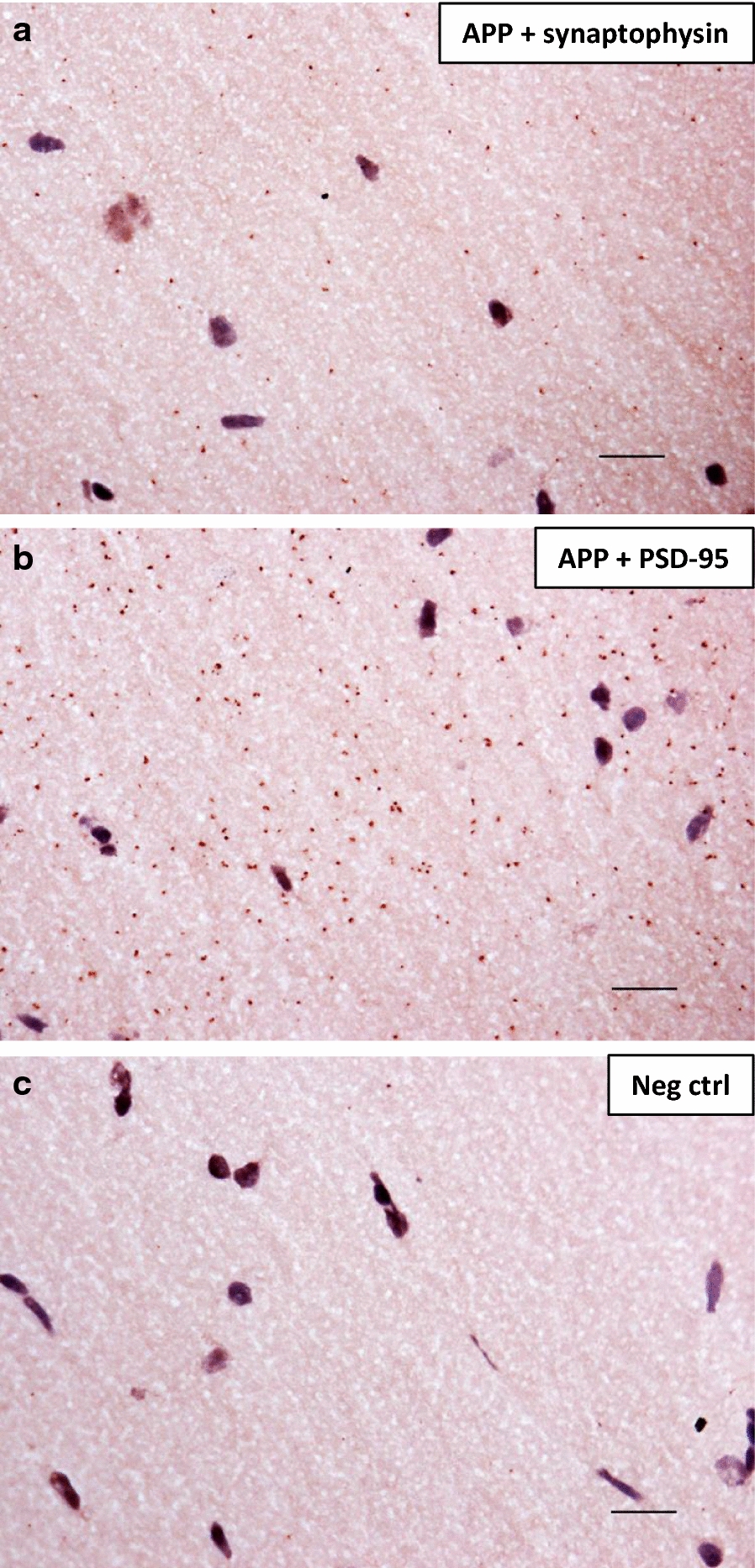
Proximity ligation assay showing pre- and postsynaptic localization of APP in adult rat brain. Adult rat brain hippocampal sections were subjected to PLA and each signal (brown dot) generated denotes two proteins within 40 nm distance from each other. a APP and synaptophysin, b APP and PSD-95. c Negative control with no primary antibodies. Brain tissue and cell nuclei were visualized by a nuclear stain solution containing Mayer´s haematoxylin. Each experiment was performed three times (technical replicates) and representative images are shown. Scale bar 20 µm
Pre- and postsynaptic localization of ADAM10 and BACE1 in fractionated rat brain
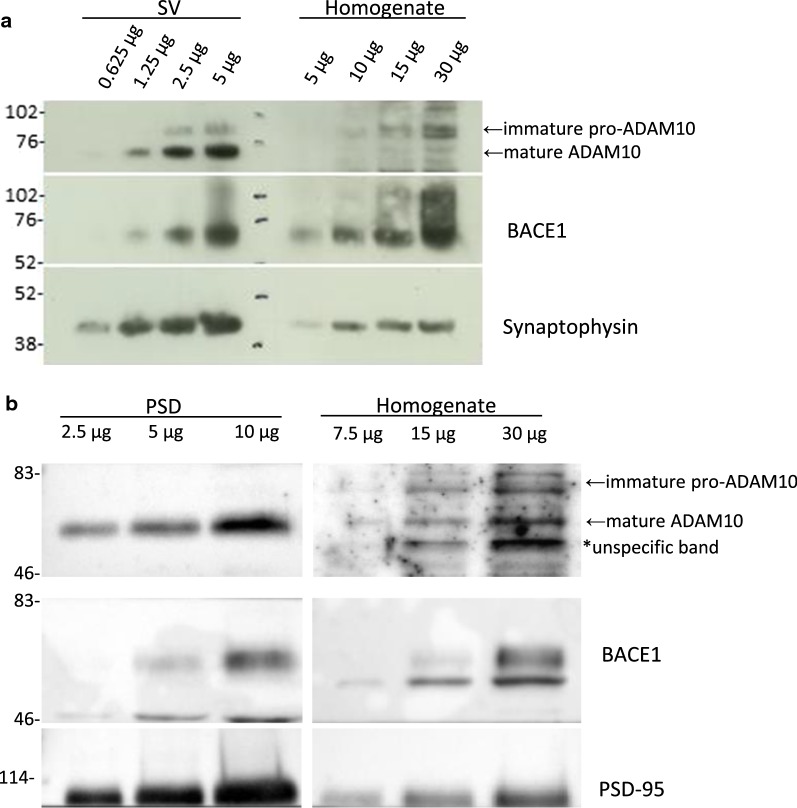
To corroborate our PLA results, we isolated pure SV and PSD fractions from adult rat brain and used western blotting to assess the presence and potential enrichment of ADAM10 and BACE1 in these fractions. The purity of these fractions (e.g. absence of postsynaptic elements in the SV fraction and vice versa) has been shown previously [26, 27, 31]. To quantify the amounts of ADAM10 and BACE1, we loaded different concentrations of the fractions on an SDS-PAGE gel to make standard curves from which to calculate the relative amounts of the secretases. In agreement with our previously published data [27], we observed enrichment of both ADAM10 and BACE1 in the SV fractions compared to homogenate (Fig. 3a). The enrichment of mature ADAM10 (predicted size 62–64 kDa [32]) in the SV fraction compared to homogenate was on average 95-fold (n = 4 different SV preparations, individual data: 200-fold, 110-fold, 42-fold and 27-fold) and the enrichment of BACE1 was on average 24-fold (n = 4 different SV preparations, individual data: 43-fold, 27-fold, 16-fold and ninefold) but varied considerably between preparations. In accordance with our above mentioned publication [27], we also observed a similar enrichment of APP C-terminal fragments (CTFs) in the SV fractions, while full length APP was less enriched (data not shown). Interestingly, only the mature form of ADAM10 was detected in the PSD fraction.
Fig. 3.
Detection of ADAM10 and BACE1 in synaptic vesicle and postsynaptic density fractions of adult rat brain. Indicated amounts of total brain homogenate (H), synaptic vesicle (SV) (a) and postsynaptic density (PSD) (b) fractions were loaded on a gel and subjected to western blotting using ADAM10, BACE1, and synaptophysin (a) or PSD-95 (b) antibodies. The SV fractionation was performed four times (from different pools of 20 rat brains) and a representative image is shown. The PSD fractionation was only performed once (from a pool of 30 rat brains)
A qualitative analysis of the enrichment of ADAM10 and BACE1 in the PSD fractions was carried out where different concentrations of homogenate and PSD fraction were loaded on a SDS-PAGE. The results show that both enzymes are present in the postsynaptic fraction and that they are enriched to a similar degree (Fig. 3b).
Pre- and postsynaptic distribution of ADAM10 and BACE1 in human brain
Since humans develop AD spontaneously whereas rats and mice do not, and since substantial translational problems between mouse models and human patients have been observed during the last years, it is important to also study human brain. We therefore performed the same sets of PLA experiments in human postmortem brain as we did in rat brain. Many PLA signals were generated when ADAM10 and BACE1 were probed against both synaptophysin (Fig. 4a–d) and PSD-95 (Fig. 4e–h) in AD or control brain hippocampus, whereas only limited number of signals were detected in the negative control samples where the primary antibodies had been omitted (Fig. 4i–j). We repeated the experiment in cortical sections from 3 AD and 3 control brains and in all cases we detected both pre- and postsynaptic localization of both ADAM10 and BACE1 (data not shown). Due to large inter-experimental variations, we were unfortunately not able to quantify potential differences in the pre- and postsynaptic distribution of the secretases between AD and control brain. However, as for rat brain, no evident differences in the pre- and postsynaptic distribution between ADAM10 and BACE1 could be observed.
Fig. 4.
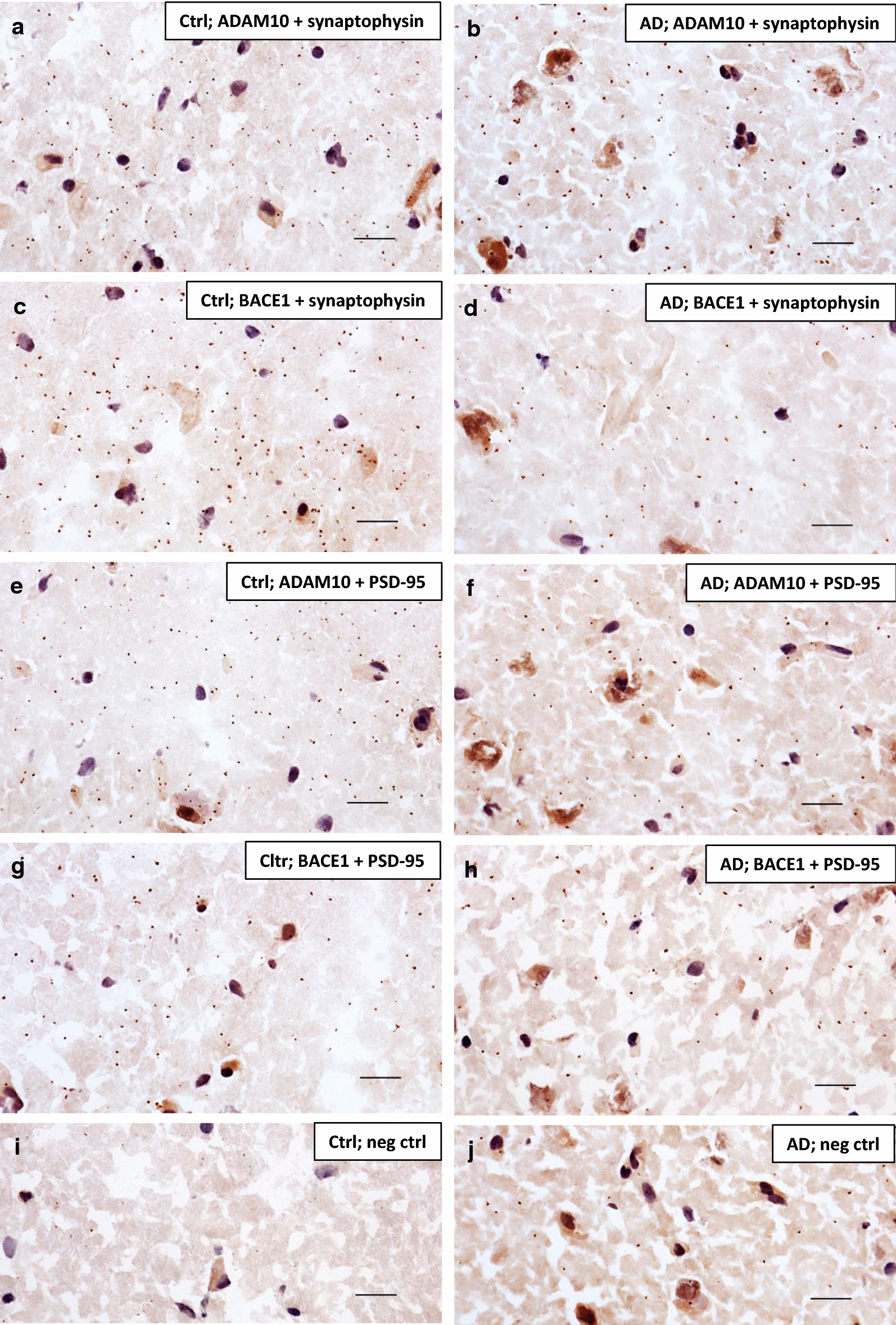
Proximity ligation assay showing pre- and postsynaptic localization of ADAM10 and BACE1 in human postmortem AD and control brain hippocampus. Human control (ctrl) (a, c, e, g, i) and AD (b, d, f, h, j) hippocampal sections were subjected to PLA and each signal (brown dot) generated denotes two proteins within 40 nm distance from each other, here ADAM10 and synaptophysin (a, b), BACE1 and synaptophysin (c, d), ADAM10 and PSD-95 (e, f) or BACE1 and PSD-95 (g, h). Primary antibodies were excluded from the negative control sections (i, j). Brain tissue and cell nuclei were visualized by a nuclear stain solution containing Mayer´s haematoxylin. Each experiment was performed at least three times (technical replicates) and representative images are shown. Scale bar 20 µm
Visualising pre- and postsynaptic APP in human brain
As in rat brain (Fig. 2), we detected PLA signals when investigating the proximity of APP and synaptophysin (Fig. 5a, b) as well as APP and PSD-95 (Fig. 5c, d) also in human AD and control hippocampus. On the contrary, very few signals were detected in the control experiments where the primary antibodies were omitted (Fig. 5e, f). Again, we could unfortunately not quantify the results to compare AD to control. APP also co-localized with both ADAM10 and BACE1 in human AD and control brain hippocampus (Additional file 2: Figure S2).
Fig. 5.
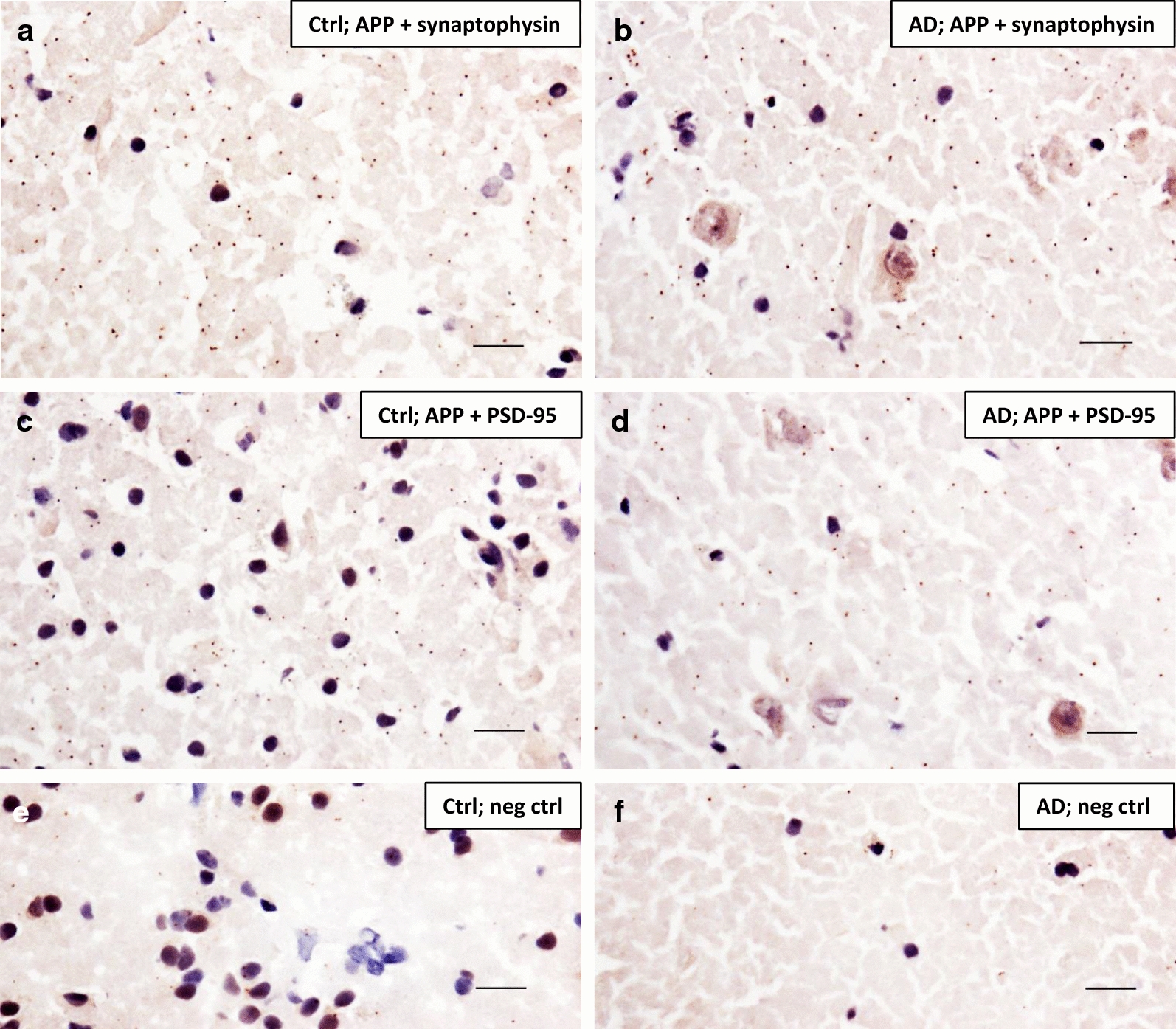
Proximity ligation assay showing pre- and postsynaptic localization of APP in human postmortem AD and control brain hippocampus. Human control (ctrl) (a, c) and AD (b, d) hippocampal sections were subjected to PLA and each signal (brown dot) generated denotes two proteins within 40 nm distance from each other. a, b APP and synaptophysin, c, d APP and PSD-95. Primary antibodies were excluded from the negative control (e, f) sections. Brain tissue and cell nuclei were visualized by a nuclear stain solution containing Mayer´s haematoxylin. Each experiment was performed three times (technical replicates) and representative images are shown. Scale bar 20 µm
Discussion
Due to the central role of Aβ in the pathology of AD and its toxic effects on synaptic function, it is essential to understand how its production is regulated. A key regulatory step for Aβ production is the subcellular location of APP and the secretases responsible for its initial processing. Thus, elucidating the location of these secretases is of major importance. Here we show that the α-secretase ADAM10, the β-secretase BACE1 and their substrate APP are located both pre- and postsynaptically with a similar distribution pattern.
The expression of both ADAM10 and BACE1 mRNA is widespread in the human brain [33, 34] and the protein level as well as the activity of BACE1 are increased in AD compared to control brain [15]. At the synapse, BACE1 is generally thought of as a pre-synaptic enzyme [17, 20–23] while ADAM10 is known to be present in the postsynaptic membrane [26, 35]. However, we have recently demonstrated that not only BACE1, but also ADAM10, is greatly enriched in the presynaptic compartment, more specifically in synaptic vesicles [27]. We have also previously shown that both ADAM10 and BACE1 are present in postsynaptic fractions of mouse brain and that their enrichment correlates with the purity of the postsynaptic fraction [26].
Using a different fractionation protocol than ours, Pliássova et al. [36, 37] found most ADAM10 to be postsynaptic and most BACE1 to be localized outside of the presynaptic active zone in rodent and human brain. They also report that APP mainly is presynaptic but also postsynaptic [36, 37] and that the synaptic levels of APP were decreased during aging while ADAM10 and BACE1 levels were not affected. Since the fractionation protocols and the homogenization process can affect the results, we also used intact brain and assessed the in situ localization of the secretases and APP in the present study.
Because of the importance of the hippocampus for learning and memory and the fact that it is extensively affected by neuronal death as well as plaques and tangles in the AD brain [2, 38], we focused mainly on the hippocampus in this study although all experiments were replicated in cortical sections as well. In both of these brain regions in rat and human adult brain we observed close proximity of both ADAM10 and BACE1 to the presynaptic marker synaptophysin as well as to the postsynaptic marker PSD-95, indicating the presence of these secretases both pre- and postsynaptically. Although our study did not provide enough statistical power to allow for quantitative analysis, the proportion of pre- and postsynaptic localization appeared similar for ADAM10 and BACE1. In line with this observation, Wang et al. [39] reported that endogenous ADAM10 and BACE1 can physically interact with each other. Unfortunately, due to large inter-experimental variations, we were unable to quantify potential differences in the pre- and postsynaptic distribution of the secretases between AD and control brain which was our initial intention. Reasons for this could be long postmortem intervals and difficulties in defining exactly the same region in the different sections.
The postsynaptic localization of BACE1 is particularly interesting since some BACE1 substrates, e.g. Seizure 6 (Sez6) [40, 41] have been found postsynaptically. The importance of Sez6 for postsynaptic functions of BACE1 was demonstrated by Zhu et al. who used two-photon microscopy to show that dendritic spine deficits introduced by BACE1 inhibition do not arise in Sez6−/− dendrites [41]. Their study also suggests that the synaptic deficits caused by BACE1 inhibitors would be of postsynaptic origin. On the other hand, ADAM10 also cleaves substrates which are located both pre- and postsynaptically as well as transsynaptically [14]. Therefore, the pre- and postsynaptic localization of both ADAM10 and BACE1 which we present evidence for in this study and in our previous studies [26, 27] is of physiological relevance.
Interestingly, in this study we show that the synaptic distribution pattern of ADAM10, BACE1 and APP seem to be similar both in rat and human brain. Our results also indicate that the proteins required for the initial step of Aβ generation are located both pre- and postsynaptically and that the balance between amyloidogenic and non-amyloidogenic processing is similar in pre- and postsynaptic compartments. Yet, for Aβ to be generated, APP is cleaved not only by BACE1 but also by γ-secretase. Previous work in our laboratory using super-resolution microscopy have demonstrated both pre- and postsynaptic localization of active γ-secretase [42]. However, the exact location of γ-secretase might be different from that of ADAM10 and BACE1 since the components of the γ-secretase complex are not enriched in SVs compared to total brain homogenate [27]. Moreover, only few PLA signals were generated when we probed for the γ-secretase complex and synaptophysin in primary neurons [27]. Nevertheless, a recent publication from our group showed, by using super-resolution microscopy, that a significant proportion of Aβ42 itself overlapped with the synaptic vesicle marker synaptophysin although other pre-synaptic structures might also contain Aβ [29].
The AD field is in urgent need of new and better therapies [43]. Reducing Aβ production by selectively inhibiting BACE1 cleavage of APP, and not of other substrates, is an attractive strategy. This can, for example, be achieved by targeting inhibitors to subcellular locations with enriched levels of both BACE1 and APP. Likewise, increasing ADAM10 cleavage of APP, which consequently would reduce Aβ production, could also be an interesting approach. This and other studies investigating the subcellular and synaptic distribution of the APP cleaving secretases are therefore vital for the development of such approaches and consequently for the advancement of AD therapy research.
Conclusions
We conclude that both ADAM10 and BACE1 as well as APP, can be found on both the pre- and postsynaptic side of the synapse and that the distribution between these locations are similar for ADAM10 and BACE1. Although descriptive and not quantitative, the findings presented in this study are of great relevance for the understanding of synaptic APP processing and hence for the development of treatment strategies for AD.
Methods
Postmortem material
The use of human brain material in this study was conformed to the Declaration of Helsinki and approved by the regional ethical review board of Stockholm (2015/1803-31/2). Fresh frozen brain tissue from hippocampus of one AD and one control subject and cortex of three AD and three control subjects was obtained from the Harvard Brain Tissue Resource Center at the NIH Brain Bank, US. All subjects were female, 71 to 81 years of age, and the tissues were collected with postmortem delays of 8 to 27 h. The AD subjects fulfilled Braak stage V and VI while the control subjects were Braak stage I and II.
Animals
All animals used in this study were handled according to the Swedish, German or Italian national guidelines and the guidelines of Karolinska Institutet, the University of Göttingen and the University of Milano as well as the current European Law (Directive 2010/63/EU). Animals were maintained on a 12-h light/dark cycle in a temperature-controlled room (20–22 °C) in spacious cages with free access to food and water. Tubes were provided as hideouts and paper tissues were available for the rats’ occupation and nest building. The male Wistar rats (Charles River) used for PLA were directly sacrificed by increasing levels of carbon dioxide and were instantly decapitated. The Sprague–Dawley rats (Charles River) used for the SV-fractionation were sacrificed by cervical dislocation followed by decapitation while the Sprague–Dawley rats (Charles River) used for the PSD-fractionation were anaesthetized with isoflurane (3–4%, inhalation for 2–3 min) before they were sacrificed by carbon dioxide and decapitated. The use of rat brain in this study was approved by the Animal research ethical committee of southern Stockholm (S21-14) and the Italian Ministry of Health (#326/2015). No experiments were performed on live animals. For PLA, the brains were dissected out and immediately snap frozen in liquid nitrogen or isopentane precooled to − 80 °C. A Leica CM 3050 cryostat was used to cut 10 µm thin brain sections which were mounted on Superfrost™ Plus Gold slides (Thermo Fisher Scientific) and stored at − 20 °C until use.
Antibodies and reagents
Primary antibodies for PLA and western blotting are described in Table 1. Secondary antibodies (horseradish peroxidase (HRP)-Goat anti-mouse and HRP-Goat anti-rabbit) were purchased from Bio-Rad and GE Healthcare. Secondary antibodies conjugated to PLA probes were purchased from Sigma-Aldrich (Merck).
Table 1.
List of antibodies used for western blotting (WB) and proximity ligation assay (PLA)
| Antibody name | Company | Dilution (WB) | Dilution (PLA) |
|---|---|---|---|
| Anti-ADAM10 | Abcam (ab39153) | 1:4000 | 1:4000 |
| Anti-Amyloid beta precursor protein C-terminus [Y188] | Abcam (ab32136) | 1:5000 | 1:5000 |
| Anti-APP C-terminus [C1/6.1] | BioLegend (802801) | – | 1:1000 |
| Anti-BACE1 | Cell Signaling Technology (D10E5) | 1:1000 | 1:200 |
| Anti-PSD-95 | Abcam (ab2723) | – | 1:20 000 |
| Anti-PSD-95 | Neuromab, k28/43 (75-028) | 1:1000 | – |
| Anti-Synaptophysin | Enzo (VAM-SV011F) | 1:10 000 | 1:3000 |
Proximity ligation assay
Brightfield PLA was essentially performed according to the protocol of the manufacturer (Sigma-Aldrich). Brain sections from Wistar rats or postmortem human tissue were fixed for 10 min at room temperature in 4% paraformaldehyde, washed with phosphate buffered saline (PBS) and permeabilized with 0.4% CHAPSO for another 10 min at room temperature. Endogenous peroxidase activity was blocked with hydrogen peroxide solution by 10 min incubation at room temperature and the slices were then washed in washing solution A. Unspecific binding sites for antibodies were blocked for 30 min with blocking solution at 37 °C and the slices were subsequently incubated with primary antibodies overnight at 4 °C. The following day, secondary probes attached to oligonucleotides were added and, after washing, the oligonucleotides of the bound probes where ligated, amplified and visualized by addition of the detection reagent and substrate solution. After addition of the nuclear stain solution, containing Mayer´s haematoxylin, the brain sections where dehydrated in ascending concentrations of ethanol and finally xylene and subsequently mounted. The PLA signals were detected using a light microscope (Nikon Eclipse E800M) and manually counted using the Image J software. As negative controls for the PLA experiments in Fig. 1, we omitted one of the primary antibodies. However, we found that similar amounts of signals appear in the negative controls when performing PLA without any primary antibodies. Hence, as negative control for the rest of the PLA experiments, we decided to totally exclude primary antibodies. All experiments in one group for the hippocampal experiments were performed on brain sections from the same rat, AD or control brain, but the experiments were also performed in cortical sections from several brains (three AD and three control brains).
Preparation of pure synaptic vesicles
Pure synaptic vesicles (SVs) were prepared from Sprague–Dawley rat forebrain as described before [31, 44]. Briefly, 20 rat brains were homogenized in homogenization buffer containing 320 mM sucrose and 4 mM HEPES–KOH (pH 7.4) in a Teflon-glass homogenizer (10 strokes at 900 rpm). The resulting homogenate (H) was centrifuged for 10 min at 1000g to remove cell debris and pellet nuclei (P1). Supernatants (S1) were pooled and centrifuged for 15 min at 10,000 g to obtain a crude synaptosomal fraction (P2), and crude brain cytosol (S2).
Synaptosomes were osmotically lysed by adding 9 vol. of ice-cold ddH2O containing protease-inhibitors (Roche) and homogenized in a Teflon-glass homogenizer (3 strokes at 2000 rpm). The lysed synaptosomes were centrifuged for 20 min at 25,000g at 4 °C to spin down lysed synaptosomal membranes (LP1). The SV containing supernatant (LS1), containing SVs and synaptosomal cytosol, was further ultracentrifuged for 2 h at 200,000g to separate SVs (LP2) from the synaptic cytosolic fraction (LS2). The LP2 fraction was resuspended in 40 mM sucrose and subjected to a continuous sucrose gradient centrifugation (from 0.05 M sucrose to 0.8 M sucrose) for 4 h at 82,500g and 4 °C. SVs from the gradient interface were further purified using size-exclusion chromatography on a CPG-column (using controlled pore glass beads), which separates large membrane structures (Peak 1) from SVs.
Purification of postsynaptic density
To isolate PSDs from Sprague Dawley rat hippocampi, a modification of the method of Carlin et al. [45] was used as described by Gardoni et al. [46, 47]. Homogenization of 30 rat hippocampi was carried out by 10 strokes in a Teflon-glass homogenizer in 4 volumes of 0.32 M sucrose containing 1 mM HEPES, 1 mM MgCl2, 1 mM NaHCO3, 1 mM NaF and 0.1 mM phenylmethylsulfonylfluoride (PMSF) (pH 7.4) in the presence of protease inhibitors (Complete; Roche/ Phosphatase Inhibitor Cocktail 2; Sigma-Aldrich), and centrifuged at 1000×g for 5 min at 4 °C. The supernatant was centrifuged at 13,800×g for 15 min at 4 °C to obtain a fraction containing mitochondria and synaptosomes. The pellet was resuspended in 2.4 volumes of 0.32 M sucrose containing 1 mM HEPES and Complete, overlaid on a sucrose gradient (0.85–1.0–1.2 M), and centrifuged at 82,500×g for 2 h at 4 °C. The fraction between 1.0 and 1.2 M sucrose was removed, diluted with an equal volume of 0.5% TritonX-100 in 0.16 M sucrose containing 1 mM HEPES and stirred at 4 °C for 15 min. This solution was centrifuged at 82,500×g for 45 min. The resulting pellet was resuspended in 0.32 M sucrose containing 1 mM HEPES and Complete buffer, overlaid on a sucrose gradient (1.0–1.5–2.1 M), and centrifuged at 100,000×g for 2 h at 4 °C. The fraction between 1.5 and 2.1 M sucrose was removed and diluted with an equal volume of 1% TritonX-100, 150 mM KCl and Complete. PSDs were collected by centrifugation at 100,000×g at 4 °C for 45 min and stored at − 80 °C.
SDS-PAGE and western blotting
Different concentrations of SV or PSD preparations and matching total brain homogenate were separated by SDS-PAGE on 4–12% polyacrylamide Bis–Tris gels (SV) (Invitrogen) or 7% SDS-PAGE/Tris–Glycine gels (PSD) and transferred to PVDF (SV) or nitrocellulose membranes (PSD) (Bio-Rad). The membranes were incubated with primary antibodies (Table 1) followed by secondary antibodies coupled to HRP (GE-Healthcare or Bio-Rad). Signals were visualized with SuperSignal West Pico enhanced chemiluminescent reagent (Pierce), Immobilon Western Chemiluminescent HRP Substrate (Millipore) or Clarity™ Western ECL substrate (Bio-Rad) and exposure to film (GE-Healthcare), LAS-3000 CCD camera (Fujifilm) or computer-assisted imaging (ChemiDoc system and Image lab 4.0 software; Bio-Rad).
Supplementary information
Additional file 1. Proximity ligation assay showing co-localization of ADAM10 and BACE1 with their substrate APP in adult rat brain. Adult rat brain hippocampal sections were subjected to PLA and each signal (brown dot) generated denotes two proteins within 40 nm distance from each other. (a) APP and ADAM10, (b) APP and BACE1. (c) Negative control with no primary antibodies. Brain tissue and cell nuclei were visualized by a nuclear stain solution containing Mayer´s haematoxylin. Each experiment was performed three times and representative images are shown. Scale bar 20 µm.
Additional file 2. Proximity ligation assay showing co-localization of ADAM0 and BACE1 with their substrate APP in human post mortem AD and control brain hippocampus. Human control (ctrl) (a, c and e) and AD (b, d and f) hippocampal sections were subjected to PLA and each signal (brown dot) generated denotes two proteins within 40 nm distance from each other. (a–b) APP and ADAM10, (c–d) APP and BACE1. Primary antibodies were excluded from the negative control (e–f) sections. Brain tissue and cell nuclei were visualized by a nuclear stain solution containing Mayer´s haematoxylin. Each experiment was performed three times and representative images are shown. Scale bar 20 µm.
Acknowledgements
Postmortem brain tissue was provided by the Harvard Brain Tissue Resource Center at the NIH NeuroBioBank—https://neurobiobank.nih.gov/.
Abbreviations
- Aβ
Amyloid β-peptide
- AD
Alzheimer disease
- ADAM10
A disintegrin and metalloproteinase 10
- APP
Amyloid precursor protein
- BACE1
β-Site APP cleaving enzyme 1
- CTF
c-Terminal fragment
- H
Homogenate
- HRP
Horseradish peroxidase
- PLA
Proximity ligation assay
- PMSF
Phenylmethylsulfonylfluoride
- PSD
Post-synaptic density
- PVDF
Polyvinylidene difluoride
- Sez6
Seizure 6
- SV
Synaptic vesicle
Authors’ contributions
JLL, BW, LOT, MDL, EM and SF planned the study. JLL, LV, ASM and SA performed the experiments. JLL and SF analyzed the data and wrote the manuscript. All authors proof-read the manuscript. All authors read and approved the final manuscript.
Funding
JLL was supported by Gun och Bertil Stohnes stiftelse’s Research Fellowship, LM was supported by the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No 676144 (Synaptic Dysfunction in Alzheimer Disease, SyDAD) and LOT was supported by Alzheimerfonden. This work was further supported by Swedish Research Council, No 2018-02843, Margaretha af Ugglas Foundation, Hjärnfonden, Demensfonden, Stiftelsen för Gamla Tjänarinnor, O.E. och Edla Johanssons vetenskapliga stiftelse and Progetto di Ricerca di Interesse Nazionale (PRIN2015N4FKJ4).
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author or from the archive at Karolinska Institutet on reasonable request.
Ethics approval and consent to participate
The use of human brain material in this study was conformed to the Declaration of Helsinki and approved by the by the regional ethical review board of Stockholm (2015/1803–31/2). Fresh frozen brain tissue from hippocampus of one AD and one control subject and cortex of three AD and three control subjects was obtained from the Harvard Brain Tissue Resource Center at the NIH Brain Bank, US who states that “All brain tissue is procured, stored, and distributed according to applicable state and federal guidelines and regulations involving consent, protection of human subjects and donor anonymity”.
All animals used in this study were handled according to the Swedish, German or Italian national guidelines and the guidelines of Karolinska Institutet, the University of Göttingen and the University of Milano as well as the current European Law (Directive 2010/63/EU). Male Wistar/Sprague–Dawley rats (Charles River) were sacrificed either directly by cervical dislocation or carbon dioxide, followed by decapitation or by first anesthetizing the rats with isoflurane (3–4%, inhalation for 2–3 min) followed by euthanization with carbon dioxide. The use of rat brain in this study was approved by the Animal research ethical committee of southern Stockholm (S21-14) and the Italian Ministry of Health (#326/2015).
Consent for publication
We do not have any information from the NIH Brain Bank whether a consent to publication have been provided but as stated above, donor anonymity is guaranteed and it therefore not possible to trace back to the patients. The patients are also deceased.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12868-020-0554-0.
References
- 1.Selkoe DJ. Alzheimer's disease is a synaptic failure. Science. 2002;298(5594):789–791. doi: 10.1126/science.1074069. [DOI] [PubMed] [Google Scholar]
- 2.Selkoe DJ. Alzheimer's disease. Cold Spring Harb Perspect Biol. 2011;3(7):a004457. doi: 10.1101/cshperspect.a004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambert MP, Barlow AK, Chromy BA, Edwards C, Freed R, Liosatos M, et al. Diffusible, nonfibrillar ligands derived from Abeta1-42 are potent central nervous system neurotoxins. Proc Natl Acad Sci USA. 1998;95(11):6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS, et al. Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416(6880):535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 5.Shankar GM, Li S, Mehta TH, Garcia-Munoz A, Shepardson NE, Smith I, et al. Amyloid-beta protein dimers isolated directly from Alzheimer's brains impair synaptic plasticity and memory. Nat Med. 2008;14(8):837–842. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Terry RD, Masliah E, Salmon DP, Butters N, DeTeresa R, Hill R, et al. Physical basis of cognitive alterations in Alzheimer's disease: synapse loss is the major correlate of cognitive impairment. Ann Neurol. 1991;30(4):572–580. doi: 10.1002/ana.410300410. [DOI] [PubMed] [Google Scholar]
- 7.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297(5580):353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 8.Hussain I, Powell D, Howlett DR, Tew DG, Meek TD, Chapman C, et al. Identification of a novel aspartic protease (Asp 2) as beta-secretase. Mol Cell Neurosci. 1999;14(6):419–427. doi: 10.1006/mcne.1999.0811. [DOI] [PubMed] [Google Scholar]
- 9.Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, et al. Beta-secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286(5440):735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- 10.Sinha S, Anderson JP, Barbour R, Basi GS, Caccavello R, Davis D, et al. Purification and cloning of amyloid precursor protein beta-secretase from human brain. Nature. 1999;402(6761):537–540. doi: 10.1038/990114. [DOI] [PubMed] [Google Scholar]
- 11.Lin X, Koelsch G, Wu S, Downs D, Dashti A, Tang J. Human aspartic protease memapsin 2 cleaves the beta-secretase site of beta-amyloid precursor protein. Proc Natl Acad Sci USA. 2000;97(4):1456–1460. doi: 10.1073/pnas.97.4.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan R, Bienkowski MJ, Shuck ME, Miao H, Tory MC, Pauley AM, et al. Membrane-anchored aspartyl protease with Alzheimer's disease beta-secretase activity. Nature. 1999;402(6761):533–537. doi: 10.1038/990107. [DOI] [PubMed] [Google Scholar]
- 13.Postina R, Schroeder A, Dewachter I, Bohl J, Schmitt U, Kojro E, et al. A disintegrin-metalloproteinase prevents amyloid plaque formation and hippocampal defects in an Alzheimer disease mouse model. J Clin Invest. 2004;113(10):1456–1464. doi: 10.1172/JCI20864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhn PH, Wang H, Dislich B, Colombo A, Zeitschel U, Ellwart JW, et al. ADAM10 is the physiologically relevant, constitutive alpha-secretase of the amyloid precursor protein in primary neurons. EMBO J. 2010;29(17):3020–3032. doi: 10.1038/emboj.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukumoto H, Cheung BS, Hyman BT, Irizarry MC. Beta-secretase protein and activity are increased in the neocortex in Alzheimer disease. Arch Neurol. 2002;59(9):1381–1389. doi: 10.1001/archneur.59.9.1381. [DOI] [PubMed] [Google Scholar]
- 16.Zhao J, Fu Y, Yasvoina M, Shao P, Hitt B, O'Connor T, et al. Beta-site amyloid precursor protein cleaving enzyme 1 levels become elevated in neurons around amyloid plaques: implications for Alzheimer's disease pathogenesis. J Neurosci. 2007;27(14):3639–3649. doi: 10.1523/JNEUROSCI.4396-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kandalepas PC, Sadleir KR, Eimer WA, Zhao J, Nicholson DA, Vassar R. The Alzheimer's beta-secretase BACE1 localizes to normal presynaptic terminals and to dystrophic presynaptic terminals surrounding amyloid plaques. Acta Neuropathol. 2013;126(3):329–352. doi: 10.1007/s00401-013-1152-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colciaghi F, Borroni B, Pastorino L, Marcello E, Zimmermann M, Cattabeni F, et al. [alpha]-Secretase ADAM10 as well as [alpha]APPs is reduced in platelets and CSF of Alzheimer disease patients. Mol Med. 2002;8(2):67–74. doi: 10.1007/BF03402076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karch CM, Goate AM. Alzheimer's disease risk genes and mechanisms of disease pathogenesis. Biol Psychiatry. 2015;77(1):43–51. doi: 10.1016/j.biopsych.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikin AF, Annaert WG, Takei K, De Camilli P, Jahn R, Greengard P, et al. Alzheimer amyloid protein precursor is localized in nerve terminal preparations to Rab5-containing vesicular organelles distinct from those implicated in the synaptic vesicle pathway. J Biol Chem. 1996;271(50):31783–31786. doi: 10.1074/jbc.271.50.31783. [DOI] [PubMed] [Google Scholar]
- 21.Kinoshita A, Fukumoto H, Shah T, Whelan CM, Irizarry MC, Hyman BT. Demonstration by FRET of BACE interaction with the amyloid precursor protein at the cell surface and in early endosomes. J Cell Sci. 2003;116(Pt 16):3339–3346. doi: 10.1242/jcs.00643. [DOI] [PubMed] [Google Scholar]
- 22.Rajendran L, Schneider A, Schlechtingen G, Weidlich S, Ries J, Braxmeier T, et al. Efficient inhibition of the Alzheimer's disease beta-secretase by membrane targeting. Science. 2008;320(5875):520–523. doi: 10.1126/science.1156609. [DOI] [PubMed] [Google Scholar]
- 23.Del Prete D, Lombino F, Liu X, D'Adamio L. APP is cleaved by Bace1 in pre-synaptic vesicles and establishes a pre-synaptic interactome, via its intracellular domain, with molecular complexes that regulate pre-synaptic vesicles functions. PLoS ONE. 2014;9(9):e108576. doi: 10.1371/journal.pone.0108576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lammich S, Kojro E, Postina R, Gilbert S, Pfeiffer R, Jasionowski M, et al. Constitutive and regulated alpha-secretase cleavage of Alzheimer's amyloid precursor protein by a disintegrin metalloprotease. Proc Natl Acad Sci USA. 1999;96(7):3922–3927. doi: 10.1073/pnas.96.7.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gutwein P, Mechtersheimer S, Riedle S, Stoeck A, Gast D, Joumaa S, et al. ADAM10-mediated cleavage of L1 adhesion molecule at the cell surface and in released membrane vesicles. FASEB J. 2003;17(2):292–294. doi: 10.1096/fj.02-0430fje. [DOI] [PubMed] [Google Scholar]
- 26.Marcello E, Gardoni F, Mauceri D, Romorini S, Jeromin A, Epis R, et al. Synapse-associated protein-97 mediates alpha-secretase ADAM10 trafficking and promotes its activity. J Neurosci. 2007;27(7):1682–1691. doi: 10.1523/JNEUROSCI.3439-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lundgren JL, Ahmed S, Schedin-Weiss S, Gouras GK, Winblad B, Tjernberg LO, et al. ADAM10 and BACE1 are localized to synaptic vesicles. J Neurochem. 2015;135(3):606–615. doi: 10.1111/jnc.13287. [DOI] [PubMed] [Google Scholar]
- 28.Lundgren JL, Ahmed S, Winblad B, Gouras GK, Tjernberg LO, Frykman S. Activity-independent release of the amyloid beta-peptide from rat brain nerve terminals. Neurosci Lett. 2014;566C:125–130. doi: 10.1016/j.neulet.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 29.Yu Y, Jans DC, Winblad B, Tjernberg LO, Schedin-Weiss S. Neuronal Abeta42 is enriched in small vesicles at the presynaptic side of synapses. Life Sci Alliance. 2018;1(3):e201800028. doi: 10.26508/lsa.201800028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soderberg O, Leuchowius KJ, Gullberg M, Jarvius M, Weibrecht I, Larsson LG, et al. Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay. Methods. 2008;45(3):227–232. doi: 10.1016/j.ymeth.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 31.Takamori S, Holt M, Stenius K, Lemke EA, Gronborg M, Riedel D, et al. Molecular anatomy of a trafficking organelle. Cell. 2006;127(4):831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 32.Anders A, Gilbert S, Garten W, Postina R, Fahrenholz F. Regulation of the alpha-secretase ADAM10 by its prodomain and proprotein convertases. FASEB J. 2001;15(10):1837–1839. doi: 10.1096/fj.01-0007fje. [DOI] [PubMed] [Google Scholar]
- 33.Kandalepas PC, Vassar R. Identification and biology of beta-secretase. J Neurochem. 2012;120(Suppl 1):55–61. doi: 10.1111/j.1471-4159.2011.07512.x. [DOI] [PubMed] [Google Scholar]
- 34.Saftig P, Lichtenthaler SF. The alpha secretase ADAM10: A metalloprotease with multiple functions in the brain. Prog Neurobiol. 2015;135:1–20. doi: 10.1016/j.pneurobio.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 35.Marcello E, Saraceno C, Musardo S, Vara H, de la Fuente AG, Pelucchi S, et al. Endocytosis of synaptic ADAM10 in neuronal plasticity and Alzheimer's disease. J Clin Invest. 2013;123(6):2523–2538. doi: 10.1172/JCI65401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pliassova A, Lopes JP, Lemos C, Oliveira CR, Cunha RA, Agostinho P. The association of amyloid-beta protein precursor with alpha- and beta-secretases in mouse cerebral cortex synapses is altered in early Alzheimer's disease. Mol Neurobiol. 2016;53(8):5710–5721. doi: 10.1007/s12035-015-9491-9. [DOI] [PubMed] [Google Scholar]
- 37.Pliassova A, Canas PM, Xavier AC, da Silva BS, Cunha RA, Agostinho P. Age-related changes in the synaptic density of amyloid-beta protein precursor and secretases in the human cerebral cortex. J Alzheimers Dis. 2016;52(4):1209–1214. doi: 10.3233/JAD-160213. [DOI] [PubMed] [Google Scholar]
- 38.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82(4):239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 39.Wang X, Wang C, Pei G. alpha-secretase ADAM10 physically interacts with beta-secretase BACE1 in neurons and regulates CHL1 proteolysis. J Mol Cell Biol. 2018;10(5):411–422. doi: 10.1093/jmcb/mjy001. [DOI] [PubMed] [Google Scholar]
- 40.Munro KM, Nash A, Pigoni M, Lichtenthaler SF, Gunnersen JM. Functions of the Alzheimer's disease protease BACE1 at the synapse in the central nervous system. J Mol Neurosci. 2016;60(3):305–315. doi: 10.1007/s12031-016-0800-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu K, Xiang X, Filser S, Marinkovic P, Dorostkar MM, Crux S, et al. Beta-site amyloid precursor protein cleaving enzyme 1 inhibition impairs synaptic plasticity via seizure protein 6. Biol Psychiatry. 2018;83(5):428–437. doi: 10.1016/j.biopsych.2016.12.023. [DOI] [PubMed] [Google Scholar]
- 42.Schedin-Weiss S, Caesar I, Winblad B, Blom H, Tjernberg LO. Super-resolution microscopy reveals gamma-secretase at both sides of the neuronal synapse. Acta Neuropathol Commun. 2016;4:29. doi: 10.1186/s40478-016-0296-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winblad B, Amouyel P, Andrieu S, Ballard C, Brayne C, Brodaty H, et al. Defeating Alzheimer's disease and other dementias: a priority for European science and society. Lancet Neurol. 2016;15(5):455–532. doi: 10.1016/S1474-4422(16)00062-4. [DOI] [PubMed] [Google Scholar]
- 44.Huttner WB, Schiebler W, Greengard P, De Camilli P. Synapsin I (protein I), a nerve terminal-specific phosphoprotein. III. Its association with synaptic vesicles studied in a highly purified synaptic vesicle preparation. J Cell Biol. 1983;96(5):1374–1388. doi: 10.1083/jcb.96.5.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carlin RK, Grab DJ, Cohen RS, Siekevitz P. Isolation and characterization of postsynaptic densities from various brain regions: enrichment of different types of postsynaptic densities. J Cell Biol. 1980;86(3):831–845. doi: 10.1083/jcb.86.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gardoni F, Caputi A, Cimino M, Pastorino L, Cattabeni F, Di Luca M. Calcium/calmodulin-dependent protein kinase II is associated with NR2A/B subunits of NMDA receptor in postsynaptic densities. J Neurochem. 1998;71(4):1733–1741. doi: 10.1046/j.1471-4159.1998.71041733.x. [DOI] [PubMed] [Google Scholar]
- 47.Gardoni F, Schrama LH, van Dalen JJ, Gispen WH, Cattabeni F, Di Luca M. AlphaCaMKII binding to the C-terminal tail of NMDA receptor subunit NR2A and its modulation by autophosphorylation. FEBS Lett. 1999;456(3):394–398. doi: 10.1016/S0014-5793(99)00985-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Proximity ligation assay showing co-localization of ADAM10 and BACE1 with their substrate APP in adult rat brain. Adult rat brain hippocampal sections were subjected to PLA and each signal (brown dot) generated denotes two proteins within 40 nm distance from each other. (a) APP and ADAM10, (b) APP and BACE1. (c) Negative control with no primary antibodies. Brain tissue and cell nuclei were visualized by a nuclear stain solution containing Mayer´s haematoxylin. Each experiment was performed three times and representative images are shown. Scale bar 20 µm.
Additional file 2. Proximity ligation assay showing co-localization of ADAM0 and BACE1 with their substrate APP in human post mortem AD and control brain hippocampus. Human control (ctrl) (a, c and e) and AD (b, d and f) hippocampal sections were subjected to PLA and each signal (brown dot) generated denotes two proteins within 40 nm distance from each other. (a–b) APP and ADAM10, (c–d) APP and BACE1. Primary antibodies were excluded from the negative control (e–f) sections. Brain tissue and cell nuclei were visualized by a nuclear stain solution containing Mayer´s haematoxylin. Each experiment was performed three times and representative images are shown. Scale bar 20 µm.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author or from the archive at Karolinska Institutet on reasonable request.