Abstract
Nucleic acid detection is vital for agricultural applications including trait detection during breeding, pest surveillance, and pathogen identification. Here, we use a modified version of the CRISPR-based nucleic acid detection platform SHERLOCK to quantify levels of a glyphosate resistance gene in a mixture of soybeans and to detect multiple plant genes in a single reaction. SHERLOCK is rapid (∼15 min), quantitative, and portable, and can process crude soybean extracts as input material for minimal nucleic acid sample preparation. This field-ready SHERLOCK platform with color-based lateral flow readout can be applied for detection and quantitation of genes in a range of agricultural applications.
Introduction
The sensitive, specific, and rapid detection of nucleic acids is important for the diagnosis and monitoring of disease and for tracking animal and plant traits in agricultural applications.1 Plant pathogens pose a dire threat to food security and are estimated to be responsible for 12.5% of losses,2 and early detection methods are necessary to restrict the extent of loss and prevent spread to neighboring fields.3,4 Moreover, as additional approaches are used for the selective breeding of plants for desirable traits, such as yield, robustness to harsh conditions, and disease resistance, methods are needed for the rapid detection of such traits.
Current detection methods, including polymerase chain reaction (PCR) and isothermal methods such as recombinase polymerase amplification (RPA)5 and loop-mediated isothermal amplification,6 suffer from a variety of limitations such as inhibition by crude plant extracts, requiring complex instrumentation7 and low specificity.3,6,8–10 Technologies that combine single-molecule sensitivity and single-nucleotide specificity with high multiplexing, portability, ease of use, and low cost are needed to scale diagnostic capacity to meet the demands of worldwide pathogen and trait detection.
We recently developed a nucleic acid detection platform called SHERLOCK11 that provides portable, programmable, and rapid nucleic acid detection by combining isothermal amplification via RPA with the CRISPR and CRISPR-associated (CRISPR-Cas) RNA-guided endoribonuclease, Cas13,12–15 which has been used for a variety of RNA-targeting applications biochemically11,16 and in cells.17,18 SHERLOCK takes advantage of the conditional promiscuous RNase activity of Cas13, referred to as collateral effect,12 where Cas13 enzymes cleave non-CRISPR RNA (crRNA) targeted RNA species in solution upon target RNA recognition.
By combining Cas13 with a quenched fluorescent RNA reporter12,13 or RNA lateral flow reporter,16 SHERLOCK can generate a fluorescent or colorimetric lateral flow readout upon Cas13 recognition of target nucleic acid species with single molecule sensitivity (2 aM input concentration in 1 μL of sample) and specificity for single-nucleotide discrimination. We recently developed the SHERLOCKv2 platform, which combines same-sample multiplexing, lateral flow visual readouts, quantitation, and Csm6 amplification of signal detection.16 In this report, we describe the development of the SHERLOCK method for agricultural applications, focusing on soybean genotyping and trait quantification.
Materials and Methods
Protein expression and purification of Cas13 and Csm6 orthologs
LwaCas13a expression and purification was carried out as described.11 PsmCas13b and Csm6 orthologs were expressed and purified with a modified protocol. In brief, bacterial expression vectors were transformed into Rosetta™ 2(DE3)pLysS Singles Competent Cells (Millipore). A 12.5 mL starter culture was grown overnight in Terrific Broth 4 growth media (TB; Sigma–Aldrich), which was used to inoculate 4 L of TB for growth, shaking at 37°C and 300 rpm until an OD600 of 0.5. At this time, protein expression was induced by supplementation with IPTG (Sigma–Aldrich) to a final concentration of 500 μM, and cells were cooled to 18°C for 16 h for protein expression. Cells were then centrifuged at 5,000 g for 15 min at 4°C. The cell pellet was harvested and stored at −80°C for later purification.
All subsequent steps of the protein purification were performed at 4°C. The cell pellet was crushed and re-suspended in lysis buffer (20 mM Tris-HCl, 500 mM NaCl, 1 mM DTT, pH 8.0) supplemented with protease inhibitors (Complete Ultra EDTA-free tablets), lysozyme (500 μg/1 mL), and benzonase followed by high-pressure cell disruption using the LM20 Microfluidizer system at 27,000 psi. Lysate was cleared by centrifugation at 10,000 g for 1 h at 4°C. The supernatant was applied to 5 mL of StrepTactin Sepharose (GE Healthcare) and incubated with rotation for 1 h followed by washing of the protein-bound StrepTactin resin three times in lysis buffer. The resin was re-suspended in SUMO digest buffer (30 mM Tris-HCl, 500 mM NaCl, 1 mM DTT, 0.15% Igepal [NP-40], pH 8.0), along with 250 IU of SUMO protease (250 mg/mL), and incubated overnight at 4°C with rotation. The suspension was applied to a column for elution and separation from resin by gravity flow. The resin was washed twice with one column volume of lysis buffer to maximize protein elution. The elute was diluted in cation exchange buffer (20 mM HEPES, 1 mM DTT, 5% glycerol, pH 7.0; pH 7.5 for EiCsm6 and LsCsm6) to lower the salt concentration in preparation for cation exchange chromatography to 250 mM.
For cation exchange and gel filtration purification, protein was loaded onto a 5 mL HiTrap SP HP cation exchange column (GE Healthcare) via fast protein liquid chromatography (FPLC; AKTA PURE, GE Healthcare) and eluted over a salt gradient from 250 mM to 2 M NaCl in elution buffer (20 mM HEPES, 1 mM DTT, 5% glycerol, pH 7.0). The resulting fractions were tested for the presence of recombinant protein by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and fractions containing the protein were pooled and concentrated via a centrifugal filter unit (50MWCO; Millipore) to 1 mL in S200 buffer (10 mM HEPES, 1 M NaCl, 5 mM MgCl2, 2 mM DTT, pH 7.0). The concentrated protein was loaded onto a gel filtration column (Superdex® 200 Increase 10/300 GL; GE Healthcare) via FPLC. The resulting fractions from gel filtration were analyzed by SDS-PAGE, and fractions containing protein were pooled and buffer exchanged into storage buffer (600 mM NaCl, 50 mM Tris-HCl, pH 7.5, 5% glycerol, 2 mM DTT) and frozen at −80°C for storage.
Accession numbers and plasmid maps for all proteins purified in this study are available in Supplementary Table S1.
Crude nucleic acid extraction from soybeans
Rapid nucleic acid extraction was performed, as previously described.19 Briefly, 20 mg of crushed soybeans was added to 200 μL of extraction buffer (500 mM NaOH and 10 mM EDTA), vortexed for 5 s, and incubated for 1 min at room temperature. After a 1:10 dilution of the supernatant, 0.4 μL of extracted genomic DNA was added to a 20 μL RPA reaction and used for SHERLOCK.
crRNA preparation
For preparation of crRNAs, constructs were ordered as ultramer DNA (Integrated DNA Technologies) with an appended T7 promoter sequence. crRNA DNA was annealed to a short T7 primer (final concentrations 10 μM) and incubated with T7 polymerase overnight at 37°C using the HiScribe T7 Quick High Yield RNA Synthesis Kit (New England Biolabs). crRNAs were purified using RNAXP clean beads (Beckman Coulter) at 2× ratio of beads to reaction volume, with an additional 1.8× supplementation of isopropanol (Sigma–Aldrich).
All crRNA sequences used in this study are available in Supplementary Table S2.
RPA
Primers for RPA were designed using NCBI Primer-BLAST20 using default parameters, with the exception of amplicon size (between 100 and 140 nt), primer melting temperatures (between 54°C and 67°C), and primer size (between 30 and 35 nt). Primers were then ordered as DNA (Integrated DNA Technologies).
RPA reactions run were as instructed with TwistAmp® Basic (TwistDx), with the exception that 280 mM of MgAc was added prior to the input template. Reactions were run with 1 μL of input for 10 min at 37°C, unless otherwise described.
For SHERLOCK quantification of nucleic acid, RPA primer concentration tested at a lower 240 nM concentration.
When multiple targets were amplified with RPA, primer concentration was adjusted to a final concentration of 480 nM. That is, 120 nM of each primer for two primer pairs were added for duplex detection.
All RPA primers used in this study are available in Supplementary Table S3.
Fluorescent cleavage assay
Detection assays were performed with 45 nM of purified Cas13, 22.5 nM of crRNA, quenched fluorescent RNA reporter (either 125 nM RNAse Alert v2; Thermo Scientific, 125 nM poly U reporter; Integrated DNA Technologies; 250 nM poly A repoter; TriLink BioTechnologies), 0.5 μL of murine RNase inhibitor (New England Biolabs), 25 ng of background total human RNA (purified from HEK293FT culture), and varying amounts of input nucleic acid target, unless otherwise indicated, in nuclease assay buffer (20 mM HEPES, 60 mM NaCl, 6 mM MgCl2, pH 6.8). Reactions were allowed to proceed for between 30 min and 3 h at 37°C (unless otherwise indicated) on a fluorescent plate reader (BioTek), with fluorescent kinetics measured every 5 min.
All cleavage reporters used in this study are available in Supplementary Table S4.
SHERLOCK nucleic acid detection
Detection assays were performed with 45 nM of purified Cas13, 22.5 nM of crRNA, quenched fluorescent RNA reporter (either 125 nM RNAse Alert v2; Thermo Scientific, 125 nM poly U reporter; Integrated DNA Technologies; 250 nM poly A repoter; TriLink BioTechnologies), 0.5 μL of murine RNase inhibitor (New England Biolabs), 25 ng of background total human RNA (purified from HEK293FT culture), and 1 μL of RPA reaction in nuclease assay buffer (20 mM HEPES, 60 mM NaCl, 6 mM MgCl2, pH 6.8), rNTP mix (1 mM final; New England Biolabs), 0.6 μL T7 polymerase (Lucigen), and 3 mM MgCl2. Reactions were allowed to proceed for between 30 min and 3 h at 37°C (unless otherwise indicated) on a fluorescent plate reader (BioTek), with fluorescent kinetics measured every 5 min.
Cas13-Csm6 fluorescent cleavage assay
Cas13-Csm6 combined fluorescent cleavage assays were performed, as described for standard Cas13 fluorescent cleavage reactions with the following modifications. Csm6 protein was added to 10 nM of final concentration, 400 nM of Csm6 fluorescent reporter (poly A reporter; TriLink BioTechnologies), and 500 nM of Csm6 activator (Integrated DNA Technologies) unless otherwise indicated. Because of the interference of rNTPs with Csm6 activity, the in vitro transcription was performed in the RPA pre-amplification step and then 1 μL of this reaction was added as input to the Cas13-Csm6 cleavage assay.
All Csm6 activators used in this study are available in Supplementary Table S5.
Lateral flow readout of Cas13 activity using FAM-biotin reporters
For lateral flow detection, the RPA was run for 10 min, and the SHERLOCK-LwaCas13a reactions were run for 20 min, unless otherwise indicated, and the reaction was set up as indicated above, except with the fluorescent reporter replaced with 1 μM final concentration of FAM-RNA-biotin reporter (Integrated DNA Technologies). After incubation, the entire 20 μL of LwaCas13a reaction was added to 100 μL of HybriDetect 1 assay buffer (Milenia) and run on HybriDetect 1 lateral flow strips (Milenia).
Results
Detection and quantitation of gene traits in soybeans
Nucleic acid detection in soybean plants is important for trait detection during breeding, as well as protecting crops from significant yield losses due to pathogens, such as Soybean mosaic virus.21 Several detection methods have been developed for detection of soybean pathogens22 or common traits, such as the gene providing resistance to glyphosate.23–25 However, these methods suffer from a number of limitations, including requiring instrumentation, poor sensitivity above attomolar concentrations, and incubation times >30 min.
To develop a CRISPR-based diagnostic for soybean nucleic acid detection using SHERLOCK, we designed an assay for detecting the gene encoding 5-enolpyruvylshikimate 3-phosphate synthase (EPSPS) from Agrobacterium sp. strain CP4 (CP4 EPSPS), which confers resistance to glyphosate. Because glyphosate-resistant (GR) and wild-type (WT) soybeans are readily available, CP4 EPSPS detection is an excellent model for detecting the presence of a nucleic acid target in a specific population of soybeans versus WT beans.
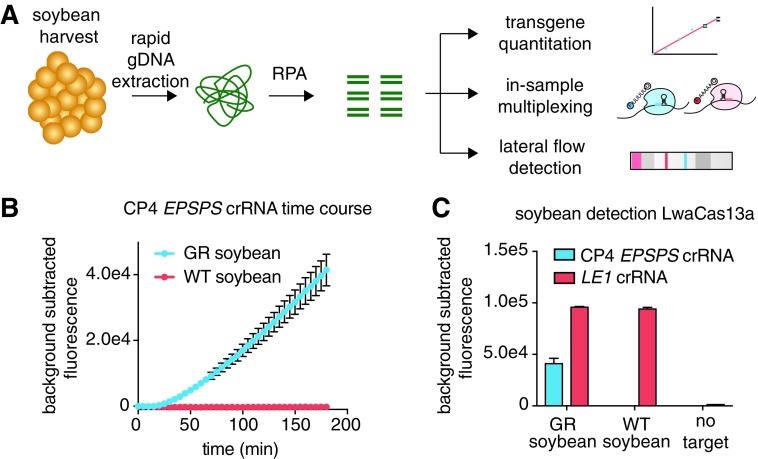
To enable a simple, portable test, we first established a rapid DNA extraction strategy for soybean (Glycine max) seeds that allows for direct SHERLOCK detection without prior DNA purification (Fig. 1A). By producing ground seed material with simple hand tools and rehydrating this material in extraction solution, we accomplished efficient extraction of genomic DNA and nucleic acid pre-amplification by RPA. We developed a Cas13 assay for detection by designing crRNAs against the CP4 EPSPS gene and the housekeeping gene lectin (LE1) as a control. We found that Cas13 detection via fluorescence on pre-amplified crude soybean extracts was able to identify the CP4 EPSPS gene in GR soybeans accurately (Fig. 1B and C and Supplementary Fig. S1A).
FIG. 1.
Rapid soybean gene detection with SHERLOCK. (A) A schematic of SHERLOCK in combination with a rapid genomic DNA extraction method allowing for detection of soybean genes in a quantitative, multiplexed, and portable manner via lateral flow strips. (B) Detection of the CP4 EPSPS gene using SHERLOCK and LwaCas13a in glyphosate-resistant (GR) soybeans and wild type (WT) soybeans over time. (C) SHERLOCK detection of the CP4 EPSPS gene and a positive control gene LE1 using LwaCas13a and a fluorescent reporter.
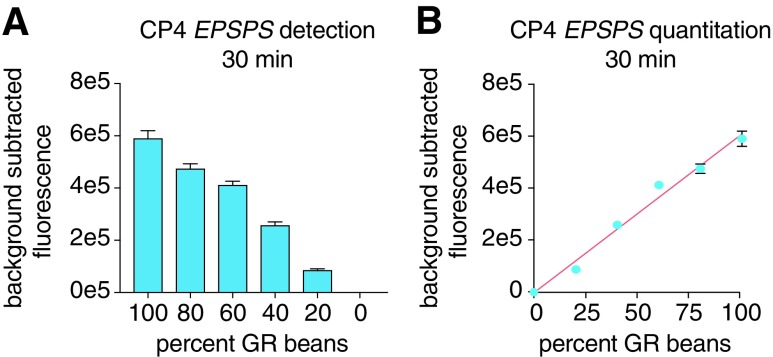
To evaluate the ability of SHERLOCK to quantify a specific target in heterogeneous soybean mixtures, we optimized SHERLOCK to quantify CP4 EPSPS from combinations of WT and GR soybeans. Using isolated genomic DNA from seed mixtures, we were able to distinguish 20% differences in CP4 EPSPS gene amount and establish a standard curve for GM content estimation in 30 min (Fig. 2A and B and Supplementary Fig. S2A and B).
FIG. 2.
Quantitative nucleic acid detection from soybean extract with SHERLOCK. (A) SHERLOCK signal detection of the CP4 EPSPS gene from soybean mixtures containing varying amounts of GR soybeans at the 30 min time point. (B) Quantitative SHERLOCK detection of the percent of the CP4 EPSPS gene in a complex mixture of soybeans (R2 = 0.98).
Multiplexed detection of genes in soybeans
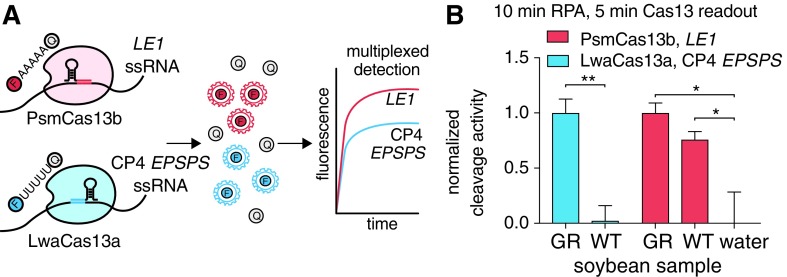
Concurrent detection of LE1 or other housekeeping genes is important as a positive control and for loading normalization, but it is inconvenient to run a reaction for each individual crRNA, particularly in cases where sample amount is limiting or DNA content varies between aliquots. By characterizing the base cleavage preferences of Cas13 orthologs, we found orthologs with mutually exclusive base preferences, allowing for collateral cleavage to be measured by orthogonal reporters in different spectral channels16 (Fig. 3A). We therefore developed an assay around the Cas13a from Leptotrichia wadei (LwaCas13a) using a poly-uridine RNA reporter and the Cas13b from Prevotella sp. MA2016 (PsmCas13b) using a poly-adenine reporter. Using a LwaCas13a crRNA complementary to the CP4 EPSPS gene and a PsmCas13b crRNA against the LE1 gene, we were able to detect both genes in the same reaction and correctly classified the GR soybeans as having the CP4 EPSPS gene (Fig. 3B). The in-sample detection of LE1 allowed us to ascertain that soybean material was present, even though the CP4 EPSPS gene was not detected.
FIG. 3.
Multiplexed detection of two genes from soybean extracts with SHERLOCK. (A) Schematic of in-sample multiplexed detection of CP4 EPSPS and LE1 genes using two-color SHERLOCK with LwaCas13a and PsmCas13b. (B) In-sample multiplexed detection of the CP4 EPSPS and LE1 genes using two-color SHERLOCK with LwaCas13a and PsmCas13b. LE1 detection of soybeans is compared to a no input water control sample.
Portable detection of soybean traits using lateral flow SHERLOCK
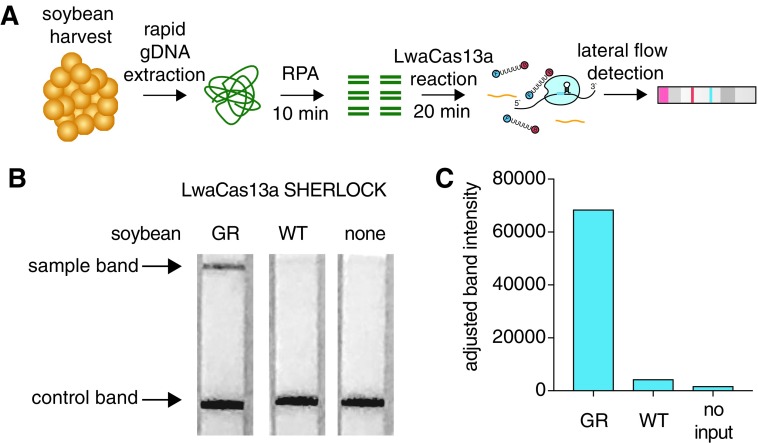
In many field applications, instrumentation may not be available for readout of a fluorescent signal. For easier visual detection, we created a reporter in SHERLOCKv216 to be compatible with lateral flow strip-based readouts by replacing the quenched fluorescent RNA reporter with an RNA functionalized with biotin and FAM on opposite ends (Fig. 4A). In the absence of reporter RNA cleavage, the RNA reporter is adsorbed at a streptavidin line and captures anti-FAM antibodies labeled with gold nanoparticles. If the RNA reporter is destroyed by the collateral effect, then antibody will flow through to a second capture line. To demonstrate this concept with rapid GR soybean detection, we pre-amplified the CP4 EPSPS nucleic acid target with RPA from crude soybean extract in 10 min and then performed a LwaCas13a detection reaction with the lateral flow RNA reporter in 20 min, resulting in lateral flow signal only in DNA from resistant seeds (Fig. 4B and C).
FIG. 4.
Portable gene detection from soybean extracts with SHERLOCK. (A) Schematic of rapid soybean nucleic acid detection using SHERLOCK lateral flow strips. (B) Rapid detection of the CP4 EPSPS gene within 30 min on lateral flow strips using SHERLOCK and LwaCas13a. (C) Quantitation of the sample band intensities from the lateral flow strips in g.
We also found that signal detection of the CP4 EPSPS gene can be enhanced by approximately three times by combining the type III CRISPR-associated endoribonuclease Csm626,27 into the SHERLOCK reaction16 (Supplementary Fig. S3). By using LwaCas13a collateral activity to generate a hexadenylate substrate with a 2′,3′-cyclic phosphate to stimulate Csm6 cleavage activity, we can activate both the Csm6 from Enterococcus italicus (EiCsm6) and the Csm6 from Lactobacillus salivarius (LsCsm6) to cause signal amplification and thus greater signal detection in the SHERLOCK assay (Supplementary Fig. S3).
Discussion
We applied the SHERLOCK technology to rapid identification of traits in soybean crops, demonstrating that nucleic acid targets can be readily amplified and detected from crude soybean extracts. SHERLOCK is easier to deploy than related PCR and immunoassay tests, as SHERLOCK works with rapidly extracted samples, is faster, and does not require complex instrumentation. In addition, we developed the test for simultaneously assaying both the CP4 EPSPS gene as well as the control LE1 gene, enabling results that are more reliable. Isothermal RPA tests have been developed for detecting soybean traits and pathogens,28 but they saturate in signal quickly and are not quantitative as a result. SHERLOCK, however, enables accurate quantification of a specific DNA target in a population of soybeans, providing results that are more relevant to the user in heterogeneous mixtures of soybeans. As quantifying trait generation during breeding can be difficult, SHERLOCK will enable trait heterozygosity to be determined, as well as facilitating the detection of trait stacking.29 While we used a thermal heating block for these assays, a simple heating device combined with a simple lateral flow reader may enable on-site and portable monitoring of traits or pathogens in future applications.
Technologies for the rapid detection of nucleic acids have a multitude of applications in agriculture, and while we have demonstrated the use of SHERLOCK for rapid and portable trait detection in plants during breeding, the same principles could be applied to numerous other contexts in agriculture. For example, early detection of plant pathogens or pests could enable rapid responses by farmers to reduce the use of pesticides or herbicides and control outbreaks around the world. Beyond plants, monitoring of livestock populations for alterations to the microbiome30 or for the emergence of pathogens or antibiotic resistance31 could inform feeding regimens or provide surveillance of emerging pathogens in herds. SHERLOCK could also be applied later in the production chain to monitor food spoilage or contamination.
In summary, the SHERLOCK technology provides a useful platform for many biotechnological and agricultural applications.
Supplementary Material
Acknowledgments
We thank J. Strecker and I.M. Slaymaker for protein purification assistance; D.B.T. Cox for assistance with Cas13b gene synthesis; B. Franklin, V. Verdine, and A.H. Le for additional experimental assistance; J.J. Collins for useful scientific discussion; and R. Macrae, R. Belliveau, E. Blackwell, and the entire Zhang lab for discussions and support. O.O.A. is supported by a Paul and Daisy Soros Fellowship and a NIH F30 NRSA 1F30-CA210382. F.Z. is a New York Stem Cell Foundation–Robertson Investigator. F.Z. is supported by NIH grants (1R01-HG009761, 1R01-MH110049, and 1DP1-HL141201); the Howard Hughes Medical Institute; the New York Stem Cell and Mathers Foundations; the Poitras Center for Affective Disorders Research at MIT; the Hock E. Tan and K. Lisa Yang Center for Autism Research at MIT; and J. and P. Poitras, R. Metcalfe, and D. Cheng. J.S.G., O.O.A, M.K., and F.Z. are co-inventors on patent applications filed by the Broad Institute relating to work in this manuscript. The authors plan to make the reagents widely available to the academic community through Addgene and to provide software tools via the Zhang lab website (www.genome-engineering.org) and GitHub (github.com/fengzhanglab).
Author Disclosure Statement
J.S.G., O.O.A, M.J.K., and F.Z. are co-inventors on patent applications filed by the Broad Institute relating to work in this study. J.S.G., O.O.A., and F.Z. are co-founders of Sherlock Biosciences. F.Z. is a co-founder and advisor of Beam Therapeutics, Editas Medicine, Pairwise Plants, and Arbor Biotechnologies. O.O.A. and J.S.G. are advisors for Beam Therapeutics. J.S.G. is a campus advisor of Benchling, Inc.
Supplementary Material
References
- 1. Zhang DB, Guo JC. The development and standardization of testing methods for genetically modified organisms and their derived products. J Integr Plant Biol 2011;53:539–551. DOI: 10.1111/j.1744-7909.2011.01060.x [DOI] [PubMed] [Google Scholar]
- 2. Oerke EC. Crop losses to pests. J Agric Sci 2006;144:31–43. DOI: 10.1017/S0021859605005708 [DOI] [Google Scholar]
- 3. Lau HY, Wu H, Wee EJ, et al. Specific and sensitive isothermal electrochemical biosensor for plant pathogen DNA detection with colloidal gold nanoparticles as probes. Sci Rep 2017;7:38896 DOI: 10.1038/srep38896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Khater M, de la Escosura-Muniz A, Merkoci A. Biosensors for plant pathogen detection. Biosens Bioelectron2017;93:72–86. DOI: 10.1016/j.bios.2016.09.091 [DOI] [PubMed] [Google Scholar]
- 5. Piepenburg O, Williams CH, Stemple DL, et al. DNA detection using recombination proteins. PLoS Biol 2006;4:e204 DOI: 10.1371/journal.pbio.0040204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ward LI, Harper SJ. Loop-mediated isothermal amplification for the detection of plant pathogens. Methods Mol Biol 2012;862:161–170. DOI: 10.1007/978-1-61779-609-8_13 [DOI] [PubMed] [Google Scholar]
- 7. Schrader C, Schielke A, Ellerbroek L, et al. PCR inhibitors - occurrence, properties and removal. J Appl Microbiol 2012;113:1014–1026. DOI: 10.1111/j.1365-2672.2012.05384.x [DOI] [PubMed] [Google Scholar]
- 8. Wang Y, Wang Y, Zhang L, et al. Multiplex, rapid, and sensitive isothermal detection of nucleic-acid sequence by endonuclease restriction-mediated real-time multiple cross displacement amplification. Front Microbiol 2016;7:753 DOI: 10.3389/fmicb.2016.00753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang Y, Wang Y, Ma AJ, et al. Rapid and sensitive isothermal detection of nucleic-acid sequence by multiple cross displacement amplification. Sci Rep 2015;5:11902 DOI: 10.1038/srep11902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kiddle G, Hardinge P, Buttigieg N, et al. GMO detection using a bioluminescent real time reporter (BART) of loop mediated isothermal amplification (LAMP) suitable for field use. BMC Biotechnol 2012;12:15 DOI: 10.1186/1472-6750-12-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gootenberg JS, Abudayyeh OO, Lee JW, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017;356:438–442. DOI: 10.1126/science.aam9321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abudayyeh OO, Gootenberg JS, Konermann S, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016;353:aaf5573 DOI: 10.1126/science.aaf5573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. East-Seletsky A, O'Connell MR, Knight SC, et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 2016;538:270–273. DOI: 10.1038/nature19802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shmakov S, Abudayyeh OO, Makarova KS, et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell 2015;60:385–397. DOI: 10.1016/j.molcel.2015.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smargon AA, Cox DB, Pyzocha NK, et al. Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol Cell 2017;65:618–630.e7. DOI: 10.1016/j.molcel.2016.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gootenberg JS, Abudayyeh O, Kellner M, et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cpf1, and Csm6. Science 2018;360:439–444. DOI: 10.1126/science.aaq0179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Abudayyeh OO, Gootenberg JS, Essletzbichler P, et al. RNA targeting with CRISPR-Cas13. Nature 2017;550:280–284. DOI: 10.1038/nature24049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cox DBT, Gootenberg JS, Abudayyeh OO, et al. RNA editing with CRISPR-Cas13. Science 2017;358:1019–1027. DOI: 10.1126/science.aaq0180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang R, Zhang F, Wang L, et al. Instant, visual, and instrument-free method for on-site screening of GTS 40-3-2 soybean based on body-heat triggered recombinase polymerase amplification. Anal Chem 2017;89:4413–4418. DOI: 10.1021/acs.analchem.7b00964 [DOI] [PubMed] [Google Scholar]
- 20. Ye J, Coulouris G, Zaretskaya I, et al. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 2012;13:134 DOI: 10.1186/1471-2105-13-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Whitham SA, Qi M, Innes RW, et al. Molecular soybean–pathogen interactions. Annu Rev Phytopathol 2016;54:443–468. DOI: 10.1146/annurev-phyto-080615-100156 [DOI] [PubMed] [Google Scholar]
- 22. Botelho SR, Martins TP, Duarte MF, et al. Development of methodologies for virus detection in soybean and wheat seeds. MethodsX 2016;3:62–68. DOI: 10.1016/j.mex.2016.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang XM, Teng D, Guan QF, et al. Detection of Roundup Ready soybean by loop-mediated isothermal amplification combined with a lateral-flow dipstick. Food Control 2013;29:213–220. DOI: 10.1016/j.foodcont.2012.06.007 [DOI] [Google Scholar]
- 24. Wu HH, Zhang Y, Zhu CQ, et al. Presence of CP4-EPSPS component in Roundup Ready soybean-derived food products. Int J Mol Sci 2012;13:1919–1932. DOI: 10.3390/ijms13021919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guan XY, Guo JC, Shen P, et al. Visual and rapid detection of two genetically modified soybean events using loop-mediated isothermal amplification method. Food Anal Method 2010;3:313–320. DOI: 10.1007/s12161-010-9132-x [DOI] [Google Scholar]
- 26. Kazlauskiene M, Kostiuk G, Venclovas C, et al. A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science 2017;357:605–609. DOI: 10.1126/science.aao0100 [DOI] [PubMed] [Google Scholar]
- 27. Niewoehner O, Garcia-Doval C, Rostol JT, et al. Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature 2017;548:543–548. DOI: 10.1038/nature23467 [DOI] [PubMed] [Google Scholar]
- 28. Chandu D, Paul S, Parker M, et al. Development of a rapid point-of-use DNA test for the screening of Genuity® Roundup Ready 2 Yield® soybean in seed samples. Biomed Res Int 2016;2016:3145921 DOI: 10.1155/2016/3145921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang X, Jiang D, Yang D. Fast-tracking determination of homozygous transgenic lines and transgene stacking using a reliable quantitative real-time PCR assay. Appl Biochem Biotechnol 2015;175:996–1006. DOI: 10.1007/s12010-014-1322-3 [DOI] [PubMed] [Google Scholar]
- 30. Clemmons BA, Voy BH, Myer PR. Altering the gut microbiome of cattle: considerations of host–microbiome interactions for persistent microbiome manipulation. Microb Ecol 2019;77:523–536. DOI: 10.1007/s00248-018-1234-9 [DOI] [PubMed] [Google Scholar]
- 31. Mathew AG, Cissell R, Liamthong S. Antibiotic resistance in bacteria associated with food animals: a United States perspective of livestock production. Foodborne Pathog Dis 2007;4:115–133. DOI: 10.1089/fpd.2006.0066 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.