Abstract
Metaphase spindles exert pole-directed forces on still-connected sister kinetochores. The spindle must counter these forces with extensive forces to prevent spindle collapse. In small spindles, kinetochore microtubules (KMTs) connect directly with the poles, and countering forces are supplied either by interdigitating MTs that form interpolar bundles or by astral MTs connected to the cell cortex. In bigger spindles, particularly those without structured poles, the origin of extensive forces is less obvious. We have used electron tomography of well-preserved metaphase cells to obtain structural evidence about interactions among different classes of MTs in metaphase spindles from Chlamydomonas rheinhardti and two strains of cultured mammalian cells. In all these spindles, KMTs approach close to and cross-bridge with the minus ends of non-KMTs, which form a framework that interdigitates near the spindle equator. Although this structure is not pole-connected, its organization suggests that it can support kinetochore tension. Analogous arrangements of MTs have been seen in even bigger spindles, such as metaphase spindles in Haemanthus endosperm and frog egg extracts. We present and discuss a hypothesis that rationalizes changes in spindle design with spindle size based on the negative exponential distribution of MT lengths in dynamically unstable populations of tubulin polymers.
INTRODUCTION
At metaphase, sister chromatids are pulled toward sister poles, so each centromere is under tension. These forces stabilize the connections between kinetochores and microtubules (MTs) (Ault and Nicklas, 1989; Yoo et al., 2018), and in later mitosis, the same forces contribute to chromosome segregation. Although the net force on a metaphase chromosome averages to zero, tension at the centromeres must be balanced by extensive forces acting wherever the kinetochore-attached MTs (KMTs) pull on the spindle. Here, we use images from electron tomography (ET) of high pressure-frozen, freeze-substitution fixed cells to examine the trajectories of MTs in metaphase spindles from the green algae Chlamydomonas reinhardtii and two strains of cultured mammalian cells (RPE1 and PtK2), seeking information about interactions among the MT classes that might contribute to a spindle’s mechanical stability.
Our observations have been made in the context of a large literature on the structure of spindles in diverse organisms. For practical reasons, the spindles most extensively studied by electron microscopy (EM) are small (reviewed in McIntosh and Hays, 2016). All these spindles have metaphase pole-to-pole lengths ≤2 µm, much shorter than spindles in bigger cells that range up to ∼50 µm, a size that implies a large difference in the amount of tubulin polymer. MT organization in small spindles is strikingly consistent and carries strong implications for spindle mechanics. Kinetochores associate directly with the plus end of one or more KMTs, and these run directly from kinetochore to pole. The poles in these cells are plate-like structures that bind the minus ends of both KMTs and non-KMTs by coupling a γ-tubulin ring complex at each MT’s minus end to a web of interconnected proteins in the spindle pole itself (Kilmartin and Goh, 1996; Muller et al., 2005; reviewed in Winey and Bloom, 2012). Many MTs that are not kinetochore-associated cluster in one or more bundles that run from pole to pole. These are the “interpolar” MTs, which emanate from each spindle pole and interdigitate near the spindle midplane. (For budding yeast, see Winey et al., 1995, and O’Toole et al., 1999; for fission yeast, see Ding et al., 1993; Grishchuk et al., 2007; and Ward et al., 2014; for a cellular slime mold, see McIntosh et al., 1985; and for diatoms, see McDonald et al., 1977, and Tippit et al., 1978; summarized in Figure 1.) Physiological studies of diatoms (Leslie and Pickett-Heaps, 1983) and fission yeasts (Khodjakov et al., 2004; Courtheoux et al., 2009) have used microbeams of light to ablate parts of the interpolar spindle, leading the poles to collapse toward one another. These experiments show that the interpolar spindle provides the outward forces necessary for spindle stability by balancing kinetochore tension with interpolar extension. Moreover, interpolar bundles isolated from both diatoms (McDonald et al., 1986) and fission yeast (Masuda et al., 1990) will elongate in vitro on the addition of Mg2-ATP, showing that this structure can also contribute to the spindle elongation characteristic of anaphase B.
FIGURE 1:
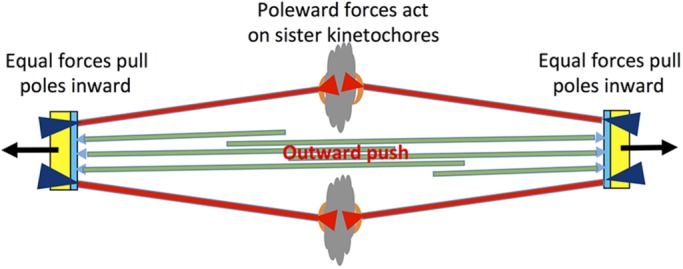
Diagram illustrating some of the forces present in small (≤2 µm) metaphase spindles. Each spindle pole body (yellow and blue) nucleates MTs, some of which bind kinetochores (red), while others overlap with MTs from opposite pole to form an interpolar spindle (green). Pole-directed forces act on sister kinetochores (red arrowheads in chromatin) and equal forces pull the poles inward (purple arrowheads at poles), so the net force on each KMT is zero. The overlapping ipMTs (green) provide an outward push on the poles, counterbalancing the inward pull on the poles by KMTs. In some cell types, an additional outward force is provided by astral MTs (black arrows at poles).
The fungi Fusarium and Ustilago achieve metaphase stability with a different strategy. At least part of the supporting action that maintains and increases pole separation in these organisms is provided by astral MTs that connect the cytoplasmic face of the spindle pole with actin microfilaments in the cell cortex, thanks to dynein–dynactin and a few coupling proteins (Aist and Berns, 1981; Aist et al., 1991; Fink et al., 2006).
In larger mitotic spindles and in some meiotic spindles, direct connections between kinetochores and poles are less evident and in some cases do not exist. In the micronuclei of Tetrahymena, there is no structured pole. The KMTs end micrometers away from the spindle’s ends, and even during anaphase these MTs do not approach the poles (LaFountain and Davidson, 1980). Higher plants too have no structured pole with which to make connections (Hepler, 1980; Jensen, 1982), rendering dubious the idea of a mechanical connection between chromosomes and “poles.” In the spindles of some cells that do possess poles, such as grasshopper spermatocytes, a bundle of KMTs can be severed a few micrometers from its kinetochore, yet during anaphase, that kinetochore will continue to move poleward without an apparent pole connection (Nicklas, 1989). The zygotic spindle of sand dollar eggs contains well-developed asters growing from classic spindle poles. However, both asters and poles of a metaphase spindle can be removed with a micropipet, yet the remaining spindle is stable and chromosome segregation proceeds (Hiramoto and Nakano, 1988).
The situation in cultured mammalian cells is more complex. Mammalian cells contain a structured pole from which many MTs arise. Although this structure can be damaged by laser irradiation and the spindle will still form (Khodjakov et al., 2000), the presence of a polar structure affects spindle geometry and aids in spindle formation. The kinetochores on mammalian chromosomes bind multiple MTs that extend poleward, forming so-called K-fibers. In metaphase PtK2 cells some of these KMTs extend far enough to reach the polar material, but about half of them end before going that far (Rieder, 1981; McDonald et al., 1992). Indeed, studies using microirradiation indicate that a direct connection between a mammalian kinetochore and its pole is not necessary for chromosome-to-pole motion (Elting et al., 2014; Sikirzhytski et al., 2014). Other experiments with similar technology reveal the ability of dynein and NuMA to gather the pole-proximal ends of both KMTs and non-KMTs into the vicinity of the pole, demonstrating a mechanically significant connection between poles and K-fibers in PtK cells (Elting et al., 2017). The structural basis for this connection seems to involve linkages between MT minus ends through dynein and NuMA to pole-associated material.
Other microbeam studies in mammalian spindles have revealed additional aspects of spindle mechanics. In PtK cells, ablation of the MTs between the separating chromosomes of anaphase B induces the chromosomes to separate faster (Aist et al., 1993). Likewise, depletion of the MT cross-linking protein, PRC1, from RPE1 cells leads to an increase in the speed of anaphase chromosome motion (Pamula et al., 2019). Relocation of PRC1 from the spindle midzone to the plasma membrane alters spindle mitotic dynamics (Milas et al., 2018), suggesting that these mammalian cells, like Fusarium and Ustilago, include a pulling action at the spindle poles. However, recent ablation studies during metaphase provide clear evidence that bundles of interdigitating MT that span the spindle’s midplane are mechanically linked to the kinetochore-proximal region of K-fibers in opposite half-spindles (Elting et al., 2014). Moreover, sliding between the antiparallel MTs in these bundles can push sister kinetochores apart in early anaphase (Polak et al., 2017; Vukušic´ et al., 2017; Tolic´, 2018), an action reminiscent of the interpolar spindles of diatoms and fission yeasts. Work on PtK spindles by serial section EM has provided some structural evidence for interactions between KMTs and non-kinetochore MTs (McDonald et al., 1992; Mastronarde et al., 1993), but just how extensive such connections might be has not been determined. Here, we present results from partial reconstructions by ET of spindles from Chlamydomonas and two strains of mammalian cells in culture; these data identify likely sources of the mechanical coupling between KMTs and the rest of the spindle.
RESULTS
Metaphase spindles in Chlamydomonas
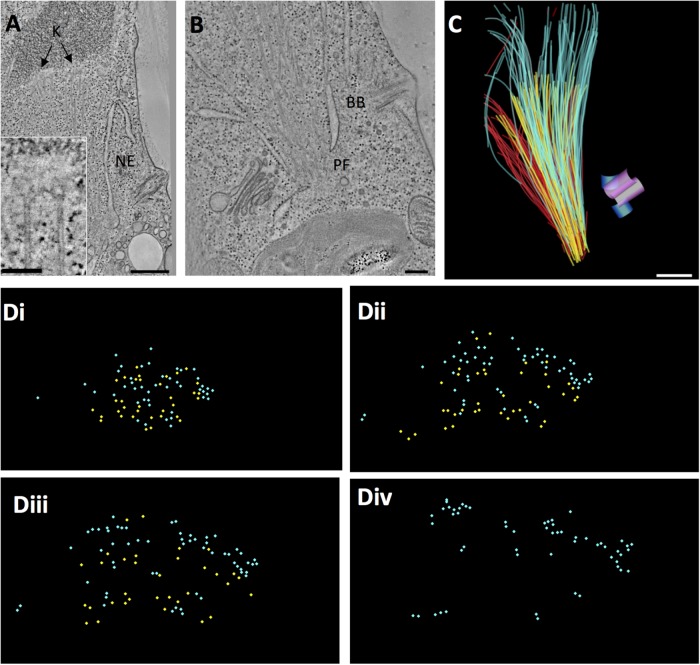
The spindle of this algae resides within an almost complete nuclear envelope (Johnson and Porter, 1968; O’Toole and Dutcher, 2014) (Figure 2, A and B). There is, however, a fenestra at each spindle pole (Figure 2B, PF), which allows the minus ends of the longest MTs to pass into the cytoplasm where they cluster and end in a region with no discernible structure (Figure 2B and Supplemental Movie S1). This lack of a structured pole is much like the situation in higher plants. Chlamydomonas chromosomes have visible kinetochores (Figure 2, A and insert), each of which interacts with 1–4 KMTs (2.66 ± 0.84, N = 48 kinetochores from three cells). Basal bodies in this organism duplicate during mitosis, but they remain anchored under the plasma membrane at some distance from the spindle (Figure 2, B and C). This cell has therefore allowed us to explore structural relationships among KMTs and non-KMTs in a spindle with no structured pole.
FIGURE 2:
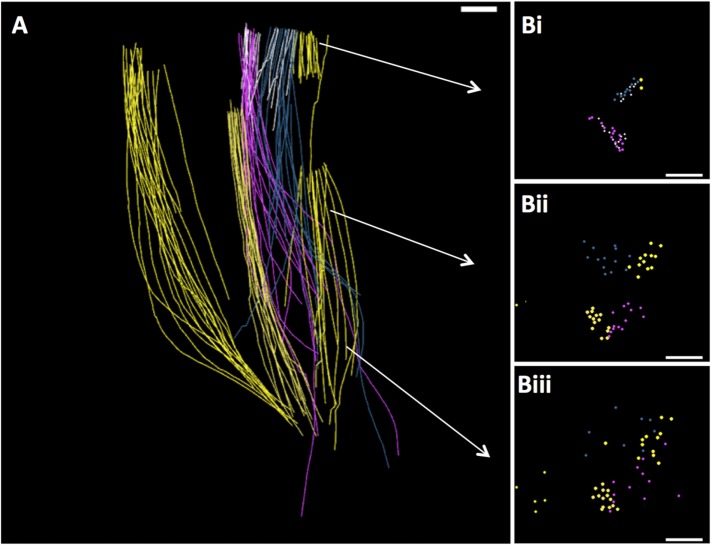
Chlamydomonas metaphase spindle. (A) Tomographic slice showing features of a metaphase half spindle. The nuclear envelope (NE) stays mostly intact during mitosis. Chromosomes with kinetochores (K) bind several MTs. Bar = 500 nm; 100 nm for insert. (B) The nucleus develops a fenestra at each pole (PF), through which spindle MTs project into the cytoplasm, ending in a region with no apparent structure. Meanwhile, the cell’s basal bodies (BB) remain at the plasma membrane. Bar = 200 nm. (C) Projection of a 3D model of a partial metaphase spindle reconstructed from four serial, 250-nm-thick sections. KMTs (yellow; n = 38), mcMTs (light blue; n = 68), and non-KMTs that either ended before reaching the chromosomes or went out of the volume of the reconstruction (red; n = 114). The mother basal bodies (pink cylinders) and forming daughters (blue cylinders) are some distance from the spindle pole. Bar = 500 nm. This reconstruction and its model are shown in Supplemental Movie S1. (D) The 3D model shown in C was resampled to display MT locations along the spindle axis from near the pole (i) to midspindle (ii and iii) to just beyond the metaphase plate (iv). mcMTs (light blue) commingle with KMTs (yellow) along their lengths. Supplemental Movie S2 displays this resampled model.
Movie S1.
Serial tomographic slices through a volume containing part of a Chlamydomonas metaphase spindle. The tomographic volume was built from four, 250 nm sections. The nuclear envelope, chromosomes, basal bodies and numerous spindle MTs can be seen. A 3D projection of the model shows KMTs (yellow) and mcMTs (light blue). Red indicates MTs that either end before reaching the chromosomes or end outside the reconstructed volume. The duplicated basal bodies are at the plasma membrane, not at the spindle pole. Scale bar = 500nm.
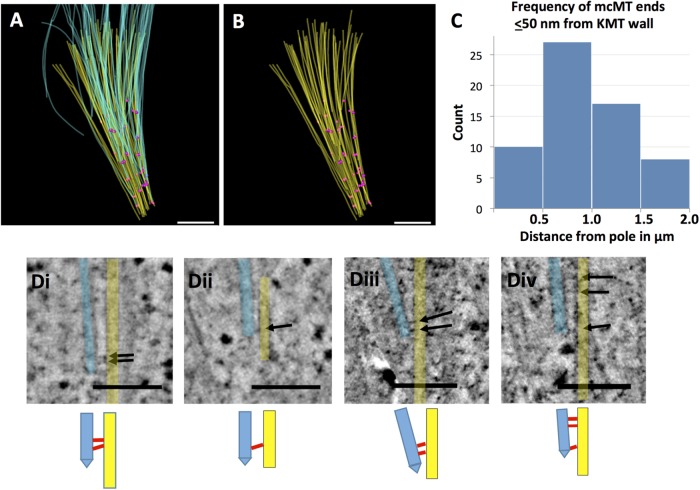
Previous studies have shown that pole-proximal MT ends are distributed throughout the Chlamydomonas spindle (O’Toole and Dutcher, 2014). To determine the three-dimensional (3D) relationships among different classes of MTs, we modeled these fibers in three metaphase cells (see Table 1 for details). We identified the ones that ended at kinetochores (KMTs, yellow in Figure 2, C and D); those that crossed the metaphase plate, which we call midplane-crossing MTs (mcMTs, blue in Figure 2, C and D); as well as those that did not encounter a kinetochore and were incomplete in the volume reconstructed (Figure 2C, red). (The latter class of MTs is not treated further here.) These relationships can be visualized both in longitudinal orientation (Figure 2C and Supplemental Movie S1) and in transverse view (Figure 2D and Supplemental Movie S2, which show only KMTs and mcMTs). The mcMTs commingle with KMT bundles along the length of the spindle, and almost all of their pole-proximal ends lie immediately next to a KMT. We have used the MTK program from the IMOD software suite to generate maps of the positions where mcMT ends lie close to a KMT. Following a study of the frequencies of distances between MTs at points of close approach, we selected ≤50 nm, center-to-center, as an appropriate value for a study of potential MT–MT-end interactions (Figure 3, A and B, pink circles). The models in Figure 3, A and B, contain 51 pole proximal mcMT ends and 62 places where a mcMT end lies within 50 nm of a KMT (several mcMT ends lie within 50 nm of more than one KMT). A histogram that plots the frequency of these regions of close approach relative to distance from the nearer spindle pole shows that the mcMT ends in KMT bundles are distributed over ∼2 µm along the spindle axis and are not limited to the polar region (Figure 3C). Using these maps to identify such regions in the 3D image data, we have found that bridges between mcMTs and KMTs are common as the MTs approach one another; often these bridges are near the capped end of the mcMT (Figure 3D, arrows and cartoons). These structural features were also found in two other Chlamydomonas metaphase spindles, as shown in Figure 4 and Supplemental Figure S1. The bridges are 18 ± 5 nm long and about 2 nm thick (N = 43, images from three cells) with two outliers ∼35 nm long. This range of size is big enough to suggest that there may be more than one kind of bridge in view.
TABLE 1:
Structure data obtained.
| Species and number of cells examined | Number of 250–300-nm sections reconstructed | Dimensions of volume imaged | Approximate fraction of spindle reconstructed |
|---|---|---|---|
| Chlamydomonas | |||
| Three metaphases | Four sections ( Figures 2 and 3) | 2.5 × 4.3 × 1.0 µm3 | One-quarter |
| Four sections ( Figure 4) | 2.4 × 4.7 × 1.0 µm3 | One-quarter | |
| Six sections (Supplemental Figure S1) | 2.6 × 5.2 × 1.5 µm3 | One-quarter | |
| RPE1 | |||
| Two metaphases | Five sections ( Figures 5 and 6) | 6.6 × 6.6 × 1.5 µm3 | One-fifth |
| Four sections (Supplemental Figure S2) | 5.5 × 11.0 × 1.0 µm3 | One-fifth | |
| PtK | |||
| One metaphase | Six sections ( Figures 7 and 8) | 13.4 × 13.1 × 1.8 µm3 | One-quarter |
Section thickness was estimated at the time of microtomy. This value multiplied times the number of sections gave the thickness of the reconstruction. Areas imaged were determined from the numbers and sizes of pixels.
FIGURE 3:
Chlamydomonas metaphase spindles contain numerous mcMTs whose pole-proximal ends lie among the bundles of KMTs. (A) Model showing KMTs (yellow) and mcMTs (light blue). Pink circles mark the positions of mcMT pole-proximal ends that lie within 50 nm of a KMT (n = 62). (B) Model showing KMTs (yellow) and positions of close approach of a mcMT end (pink circles). Bar in A and B = 500 nm. (C) Histogram showing the frequency of places where mcMTs end ≤50 nm from a KMT as a function of distance from the spindle pole. (D) Examples of capped mcMTs (light blue) whose pole-proximal end lies near KMTs (yellow). Arrows indicate connections between these MTs. Bar = 150 nm. Below each panel is a cartoon showing our interpretation of the interMT connections.
FIGURE 4:
A Chlamydomonas metaphase spindle in longitudinal view. (A) Projection of a 3D model of a metaphase spindle superimposed on a tomographic slice. KMTs (yellow; n = 70), mcMTs from either pole (white; n = 34, light blue; n = 43), and MTs that either ended before reaching the chromosomes or went out of the volume of the reconstruction (red; n = 106). Bar = 500 nm. Serial tomographic slices through the volume and the 3D model are shown in Supplemental Movie S3. (B) Projections of 3D model showing KMTs (yellow), mcMTs (white, light blue), and their pole proximal ends (pink circles; n = 26). (C) The model shown in B was resampled to display model points along the spindle axis. Clear bundles of mcMTs from either pole (white, light blue) commingle with KMTs (yellow). Supplemental Movie S4 displays this resampled model. (D) Examples of pole-proximal ends of mcMTs (light blue) adjacent to KMTs (yellow). Arrows indicate connections between these MTs. Bar = 75 nm. (E) Diagram of MT-dependent forces in a metaphase spindle that lacks structure poles. Now tension on kinetochores (red arrowheads) is balanced by an outward push from the mcMTs (blue arrowheads) that are connected directly to the walls of KMTs (black cross-bridges).
Movie S2.
The model shown in Figure 2C and Movie 1 was resampled along the spindle axis to display the model points in cross section. The movie begins at the spindle pole, goes to the metaphase plate then back to the pole. Yellow circles indicate positions of KMTs and blue circles indicate mcMTs. A comingling of these two MT classes can be seen throughout the spindle.
Movie S3.
Serial tomographic slices through a volume containing the midzone of a Chlamydomonas metaphase spindle. The tomographic volume as built from four, serial 250 thick sections. The model shows KMTs (yellow), and red indicates MTs that either end before reaching the chromosomes or end outside the reconstructed volume. mcMTs from either pole are colored in white and light blue, respectively. Bar = 500nm.
The midzones of Chlamydomonas metaphase spindles contain bundles of MTs that associate with either pole and span the metaphase plate (mcMTs; light blue, white), as shown in Figure 4A and Supplemental Movie S3, which also display KMTs (yellow) and non-KMTs (red). When this spindle model is sampled along its axis to view MTs in cross-section (Figure 4C and Supplemental Movie S4), the mcMTs (light blue, white) are seen clustering with one another and intermingling with the KMT bundles (yellow). Although the spindle poles were not complete in this reconstructed volume, the ends of many mcMTs are distributed throughout the spindle, and some lie close to KMTs (Figure 4B, pink circles). Images of these ends and apparent connections between mcMTs and KMT are shown in Figure 4D. In sum, these results identify interdigitating mcMTs as structural analogs of ipMTs in small spindles. However, the mcMTs in Chlamydomonas do not extend far enough to reach the spindle poles; they end along the spindle, commonly making connections with KMTs. This design is reminiscent of smaller spindles, except that physical connections between KMTs and other spindle MTs are made in the spindle, not at the poles (Figure 4E).
Movie S4.
The model in Figure 3A and Movie 3 was resampled along the spindle axis to display the model points in cross section. Yellow circles indicate positions of KMTs, blue and white circles indicate mcMTs from either pole, respectively. Bundles of mcMTs are well organized in the spindle midzone and these MTs comingle with KMTs.
We have examined our reconstructions for indications of helicity in the arrangement of MTs in Chlamydomonas spindles. No such twist is apparent (Figures 2 – 4; Supplemental Figure S1; Supplemental Movies S1–S4), but in partial reconstructions, such a long-range feature may be difficult to detect.
Metaphase spindles in RPE1 cells
RPE1 is a near-diploid cell line, established from human retinal pigment epithelium. We have prepared these cells for ET by high-pressure freezing and freeze-substitution fixation; preliminary models from these reconstructions have been described (O’Toole et al., 2018). The architecture of the spindle is well maintained by our preservation techniques, and even tomograms that include only a part of the spindle allow modeling of different classes of MTs: KMTs in yellow and non-KMTs in red as shown in Supplemental Movie S5 (see Table 1 for specimen details). One strength of our study is the evidence it provides for the density of spindle MTs, a feature that cannot be discerned with fluorescence microscopy. A limitation of our partial reconstructions is that many of the MTs seen are situated with one or both ends outside the volume considered, so their lengths and identity cannot be established. For example, Supplemental Movie S5 contains 76 KMTs, only 60 of which are complete in the volume studied; the latter are shown in Figure 5A. Likewise, the movie shows 1248 non-KMTs, only 383 of which are complete.
FIGURE 5:
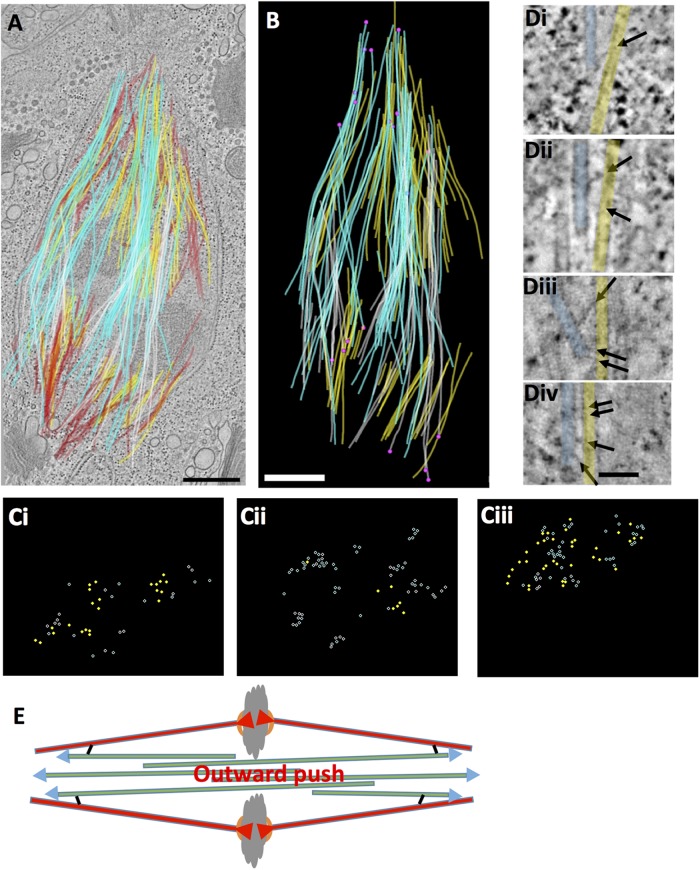
mcMT interactions with KMTs in an RPE1 spindle. (A) A model built from five serial, 300-nm-thick sections, representing ∼1/5th of a metaphase spindle. KMTs (yellow; n = 86), mcMTs from either pole (white, n = 109, light blue, n = 134) and the centriole pair (blue cylinders). (B) Pink circles show the position of pole-proximal MT ends that lie ≤50 nm from a KMT (n = 32). Some mcMT ends make multiple close approaches. Bars in A and B = 500 nm. (C) Histogram showing the distribution of places where mcMTs end ≤50 nm from a kinetochore MT as a function of distance from the spindle pole. (D) Gallery of pole-proximal ends of mcMT (blue) within 50 nm of a KMT (yellow). Arrows mark structures we identify as bridges. Bar = 50 nm.
Movie S5.
Serial, tomographic slices and projected 3D model of a spindle from the human cell line, RPE1. The tomographic volume was built from five, serial 300nm thick sections and comprises ~1/5 the total spindle volume. KMTs are yellow (n=76), nonKMTs are red (n=1252). Pole distal and pole proximal ends are marked by blue and green circles, respectively. A centriole pair (blue cylinders) marks the centrosome of the spindle pole. Bar = 1um.
Bundles of KMTs (yellow) are clear in both Supplemental Movie S5 and Figure 5, as well as in another RPE1 reconstruction shown in Supplemental Figure S2. The number of KMTs at each kinetochore ranges from 10 to 15 (12.6 ± 1.7, N = 93), results from 13 kinetochores in two cells. (Note that whereas 16 K-fibers can be seen in these two cells, three of the corresponding kinetochores were not complete in the volume reconstructed, so they were not scored for KMT number.) Several bundles of mcMTs are evident in Figure 5 (light blue and white); a subset of these equator-passing MTs have pole proximal ends in a K-fiber (Figure 5B). We used the MTK program to identify positions of close approach between the walls of KMTs and the ends of these mcMTs (pink circles in Figure 5B; n = 32). (Since our reconstructions are incomplete, this identification of mcMTs is conservative.) A majority of these sites are within ∼4 µm of the pole, but a few are within 2 µm of a kinetochore (Figure 5C and Supplemental Movie S6). Figure 5D shows a gallery of possible bridging structures at these sites of close approach. Again, these bridges are pleomorphic, ranging between 15 and 40 nm in length. The distribution of these sites suggests that this region of the spindle is mechanically significant for supporting the compressive forces generated by the tension applied to sister kinetochores. Supplemental Figure S3 casts further light on the issue of MT end positions and the proximity of mcMTs ends to KMTs. Supplemental Figure S3A shows the distribution of any pole-proximal ends (purple spheres; n = 1061), Supplemental Figure S3B shows any MT end that was close to a KMT (red spheres; n = 73) and Supplemental Figure S3C shows the ends of only mcMT that are close to KMTs (pink spheres; n = 32). Supplemental Figure S3D graphs the frequency of all minus MT ends as a function of their distance from the spindle pole.
Each mcMT bundle in these reconstructions displays an initial association with a particular kinetochore fiber (Figure 6, A and B; Supplemental Movie S6). Moreover, as these MTs pass the metaphase plate, they lie immediately next to the chromatin and appear to interact with it. However, as the bundles progress toward the pole, they fan out, so their component mcMTs comingle with several nearby bundles of KMTs (Figure 6). Thus, the association of non-KMT bundles solely with one pair of sister K-fibers, which has been described in the light microscope (Polak et al., 2017), is not confirmed in ET reconstructions that display individual MTs over greater distances. This statement is also supported by the partial reconstruction of a different RPE1 cell in metaphase (Supplemental Figure S2, A and B). Again, some mcMTs cluster and associate with sister bundles of KMTs on either side of the metaphase plate, but as these MT extend toward the poles, they fan out to interact with multiple K-fibers. Note that this structural feature should enhance the ability of mcMT bundles to support the tension generated at kinetochores.
FIGURE 6:
RPE1 mcMT bundles commingle with kinetochore fibers. (A) Two bundles of mcMTs (pink, blue) associating with KMTs (yellow) extracted from the model in Figure 5. Bar = 500 nm. (B) The model was resampled to display MT positions along the spindle axis. Cross-sections are shown at positions indicated by the arrows. Tight bundles of mcMTs are initially associated with a particular K-fiber, but as they continue toward the spindle pole, they diverge and commingle with other K-fibers. Bars = 500 nm. Supplemental Movie S6 shows the resampled model, displaying the comingling of these MT bundles.
Movie S6.
The model in Figure 6A was resampled along the spindle axis to display the model points in cross section. The movie begins at the pole and ends at the metaphase plate. Yellow circles indicate positions of KMTs, blue and pink circles indicate two bundles of mcMTs associated with the same pole. White circles represent mcMTs from the opposite pole. Bundles of mcMTs are well organized in the spindle midzone and these MTs initially associate laterally with specific KMT bundles. The mcMTs comingle with KMTs, but nearer the pole, they diverge to mingle with other KMT bundles.
Examination of Figures 5 and 6, Supplemental Figure S2, and Supplemental Movies S5 and S6 reveals that the spindles of RPE1 cells, like those in Chlamydomonas show no convincing evidence of helicity in the arrangement of their MTs. There is some twist in the bundle of mcMTs colored blue in Supplemental Movie S6, but none of the other bundles repeats this feature.
A metaphase spindle from a PtK 2 cell
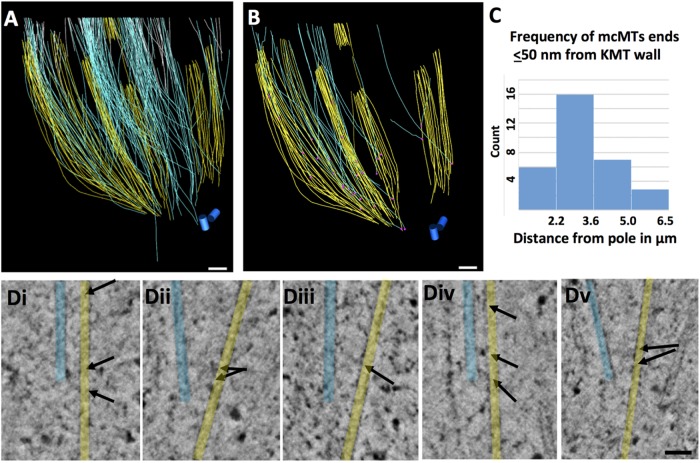
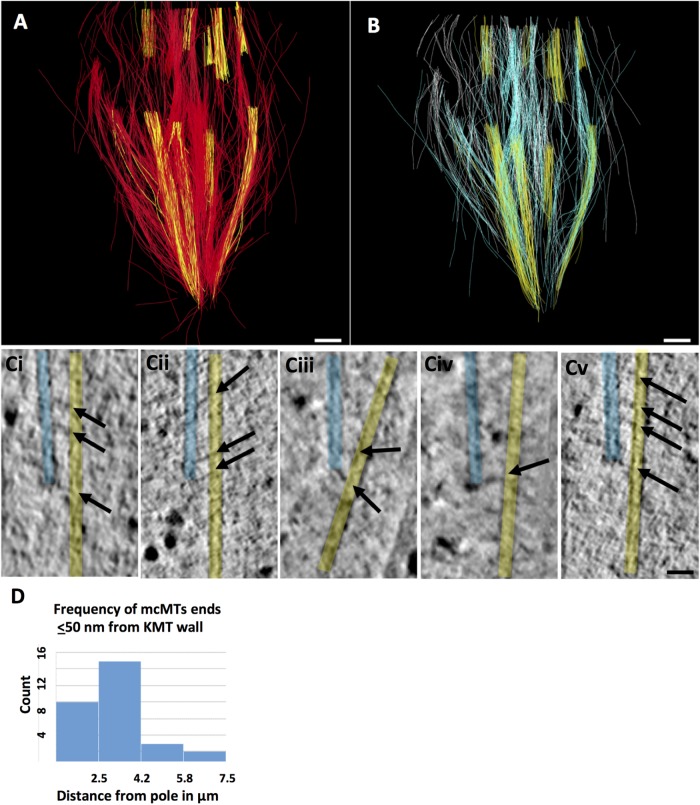
Partial tomographic reconstruction of a metaphase PtK2 spindle has yielded analogous information for this cell type. Figure 7A shows more than 1500 MTs that have been traced in this set of six serial, supermontaged tomograms (Supplemental Movie S7; see Table 1 for specimen details). The seven kinetochores in this reconstruction associate with 18–28 KMTs (23.4 ± 3.3, N = 164 KMTs), numbers similar to those previously reported (McDonald et al., 1992; McEwen et al., 1997). Figure 7B again shows KMTs in yellow and mcMTs in blue and white to reflect the different half-spindles with which their pole-proximal (likely minus) ends are associated. There are 657 pole-proximal ends of non-KMTs in this reconstructed volume (purple dots in Supplemental Figure S4A). Each bundle of KMTs includes numerous non-KMTs, 181 of which end within 50 nm of a KMT wall (red dots in Supplemental Figure S3B). Ninety-six of these non-KMTs could be identified as mcMTs and had minus ends in the volume of the reconstruction. Of these, 41 minus ends were within 50 nm of a KMT (Supplemental Figure S4C). A gallery of pole-proximal ends adjacent to KMTs shows bridging structures similar to those observed in Chlamydomonas and RPE1 spindles (Figure 7C). The distribution of these mcMT minus ends along the spindle axis is shown in Figure 7D. As in RPE1 cells, the majority of them are in the polar region, but a few are within 2 µm of kinetochores (Supplemental Figure S4B, green arrow). This localization is consistent with observations from several labs for this cell type (R.B. Nicklas, Duke University, Durham, NC; S. Dumont, University of California, San Francisco; I. Tolic, Rud¯er Boškovic´ Institute, Zagreb, Croatia); they saw that ∼2 µm was the minimum length of K-fiber that could sustain chromosome-to-pole motion (Elting et al., 2017; Vukušic´ et al., 2017). Our structural observations are also consistent with our previous observations on PtK cells at lower 3D resolution, made by serial section EM (McDonald et al., 1992; Mastronarde et al. 1993).
FIGURE 7:
PtK2 metaphase spindle. (A) Model built from six serial 300 nm sections resulting in a total reconstructed volume of 13.4 × 13.1 × 1.8 µm3, representing about one-quarter of the metaphase spindle. Eight K-fibers were identified (yellow), and non-KMTs are in red. Bar = 0.5 µm. Supplemental Movie S7 shows serial, tomographic slices through this volume and the 3D model. (B) KMTs (yellow) and two sets of mcMTs (blue and white) with all other non-KMTs removed. (C) Gallery of pole-proximal MT ends (light blue) within 50 nm of a KMT (yellow). Arrows point to bridges between these MTs. Bar = 50 nm. (D) Histogram showing the frequency of mcMT ends lying ≤50 nm from a kinetochore MT as a function of position along the spindle axis relative to one pole.
Movie S7.
Serial, tomographic slices and projected 3D model of a spindle from the mammalian tissue culture cell line, PtK2. The tomographic volume was built from six, serial 300nm thick sections. KMTs are shown in yellow (n=255), nonKMTs are in red (n=1384).
This spindle shows a slight tendency for the MTs to twist around the pole-to-pole axis, although the tendency is subtle (Supplemental Movie S7). This structural feature may correspond to the helicity described in U2OS cells by light microscopy (Tolic´ et al., 2019). We have also examined this reconstruction to ask whether the tracking of individual spindle MTs bears out the idea that bridging fibers run specifically between sister-sets of KMTs. Figure 8, A and B, shows one example of a pair of sister kinetochores, their bundles of KMTs (yellow), and the mcMTs that lie in their vicinity (blue and white). As in RPE1 cells, the images from ET differ from comparable light micrographs in that the bundles of mcMTs fan out, so they do not bridge only sister KMTs. In sum, the structural work using ET extends what has been seen by serial section EM and inferred from light microscopy on living cells; it provides a structural basis for the support framework that can provide extensive forces in mammalian spindles.
FIGURE 8:
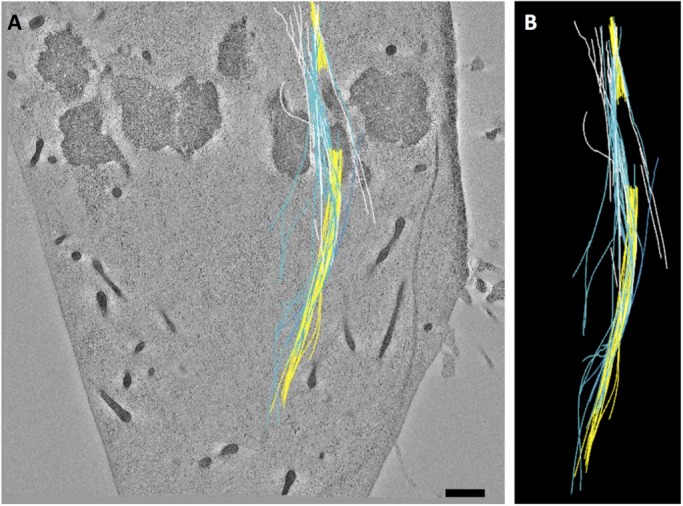
PtK2 mcMT bundles associate with kinetochore fibers. (A) A bundle of mcMTs (blue, white) in association with sister kinetochore bundles (yellow). (B) The modeled MTs extracted from image data.
DISCUSSION
We have identified structural connections between metaphase KMTs and regions at or near the pole-proximal ends of non-KMTs in spindles from two species, suggesting that mechanical interactions within the spindle’s body may play an important part in the mechanical stability of these spindles. In small spindles with well-structured poles, like those in yeasts, different classes of MTs connect directly with the pole, allowing the polar plate to link the constrictive and extensive actions of different MTs, helping the spindle to a stable mechanical equilibrium. When poles are absent, as in higher plants, algae like Chlamydomonas, and some meiotic spindles (Redemann et al., 2018), mechanical links between MT classes must be made elsewhere; bridges between KMTs and mcMTs can do this job. Even in the complex spindles found in mammalian spindles, where some MTs are linked mechanically with the poles, the observed connections between KMTs, and mcMTs may play an important role in metaphase spindle stability.
Mechanical solutions employed by even larger spindles
EM of serial sections cut from spindles in Haemanthus endosperm has revealed well-developed K-fibers that can include >100 MTs/kinetochore (Jensen, 1982). These spindles also contain large bundles of non-KMTs that run approximately parallel to the spindle axis, passing by the chromosomes during all stages of mitosis. During metaphase and anaphase there is a clear commingling of KMTs with non-KMTs in the region just poleward from each kinetochore, opening the possibility of mechanically significant interactions between them, though neither structural nor morphometric evidence for connections between these MT classes is yet available. Given the situation we describe in Chlamydomonas, it seems likely that analogous interactions provide the mechanical support necessary to withstand pole-directed tension at kinetochores in higher plants as well as an algae (Figure 4E).
Spindles in blastomeres from Caenorhabditis elegans provide a complement to the cases described above. Their metaphase spindles have been reconstructed in their entirety by ET (Redemann et al., 2017). These 3D studies demonstrate that most KMTs are not long enough to reach the spindle poles; they terminate in a network of non-KMTs that emanates from the spindle poles, suggesting that force-bearing linkages to the pole are indirect. Moreover, almost all non-KMTs in this metaphase spindle are too short to reach the spindle equator, so there is little evidence for mcMTs at metaphase. These spindles appear to lack a framework of mcMTs to support tension at the centromeres, although such a structure is visible by fluorescence microscopy in early anaphase (Saunders et al., 2007). Here, as in PtK cells, the MT bundles found between the separating chromosome slow, rather than drive, anaphase spindle elongation. Anaphase B appears to be effected by pulling forces that act through astral MTs, which connect each spindle pole with cortex-associated dynein (Grill et al., 2001). Cortical forces are also probably acting with sufficient strength during metaphase to support the tension that acts on kinetochores and would otherwise pull the poles toward the spindle equator.
The spindles formed in extracts from Xenopus eggs are an even more extreme example of supporting centromeric tension by indirect linkages. These spindles are 40–50 µm long and contain tens of thousands of MTs that are short relative to the spindle’s length (Brugués et al., 2012). During prometaphase, MTs form in the neighborhood of the chromosomes, then reorient and reposition to form the spindle (Karsenti and Vernos, 2001). Although kinetochores have not yet been identified by EM, these spindles contain many bundles of MTs, some of which are probably kinetochore associated (Tranfield et al., 2014; Weber et al., 2014). Clever use of laser-mediated cutting of spindle MTs that have been labeled with small amounts of fluorescent mammalian tubulin, followed by detailed study of the space and time dependence of fluorescence redistributions, has shown that these spindles are formed by assemblies of MTs that point in opposite directions. Average MT lengths vary from ∼3 µm near the poles to ∼13 µm far from the pole (Brugués et al., 2012). Time-dependent visualization of speckles induced in these spindles by sparse labeling of the MTs with fluorescent tubulin shows that the polymers are in slow but continuous flux toward one spindle pole or the other (Mitchison et al., 2004), making a two-way conveyer belt. There are no well-defined structures that could serve as spindle poles, so the support for kinetochore tension comes from relatively short non-KMTs that are linked together to form the necessary framework.
Why are direct connections between kinetochores and poles few or absent in big spindles?
The design of small spindles seems efficient and effective for forming a mechanically stable metaphase. Why is this design not used in the bigger spindles described here? A direct polar connection is of course impossible in cells with no structured pole, so linkages between MTs become necessary. Mammalian spindles possess structured poles, but many spindle MTs, both KMTs and non-KMTs, terminate before reaching the poles. In spindles that are bigger still, MTs long enough to extend from pole to kinetochore, or beyond to the spindle midplane, are very rare, defining a need for bridge-mediated connections to make a stable metaphase structure.
MTs in nonspindle systems can be almost arbitrarily long, probably as a result of the right MT-associated proteins. For example, flagellar MTs in sperm of Drosophila bifurca extend ≥5 mm (Pitnick et al., 1995). Why, then, do not big spindles retain the efficient design of small spindles but instead resort to coupling short MTs to make a framework that can support kinetochore tension? One factor may be that spindle MTs are necessarily dynamic. They form for the occasion of division but disappear before the following interphase. Moreover, dynamic instability is important for the likelihood that MTs will encounter an appropriate load, for example, a kinetochore (Kirschner and Mitchison, 1986; Magidson et al., 2011) or an mcMT from the opposite pole. Labile MTs displaying dynamic instability show a distribution of lengths that is well described by a negative exponential function (Verde et al., 1992; Redemann et al., 2017), so the most numerous MTs are short. This situation may be exacerbated by MT-severing enzymes that cut spindle components in some systems, making the average MT length even shorter (Srayko et al., 2006). To get a significant number of long, dynamic MTs, one must therefore make a very large number of short ones. When the distance from one pole to the far side of the spindle midplane is large, pole-initiated MTs long enough to cross the spindle midplane are expensive, given the tubulin needed to form the many short MTs characteristic of an exponential distribution of lengths. Thus, even in mammalian spindles, whose half-spindles are commonly ∼5 µm, only a very few pole-initiated MTs extend far enough to cross the midplane. To build a robust interpolar structure in a big spindle, the augmin complex can function to initiate MTs along the way from the pole to the midplane (Kamasaki et al., 2013). Many of the resulting MTs can then interdigitate with their counterparts from the opposite side of the spindle, yet none of them needs be too long. Indeed, when augmin levels are reduced by RNAi, the structure and function of a mammalian spindle are seriously compromised (Kamasaki et al., 2013).
Augmin-initiated MTs have the additional advantage in that they commonly occur along the walls of existing MTs (Kamasaki et al., 2013). This behavior may endow them with the ability to form functionally significant mcMT bundles, like the ones described here and by the Tolic´ lab. Indeed, some of the links between KMTs and the ends of non-KMTs described here may be the augmin complex bound to a KMT wall. The same logic could apply in any big spindle, although current evidence from genome sequences has not identified augmin-like molecules in nematodes. Other molecules, like Tangled1 from plants, are known to promote MT-MT binding, particularly the association of an MT end with an MT wall (Martinez et al., 2019), similar to the connections seen here. There are probably additional molecular players with similar properties that are yet to be identified. Whatever the molecular mechanisms, it seems that when cells need a large and labile spindle, they abandon the strategy that works in small spindles and make mechanically equivalent structures from shorter MTs, connected to make a framework that can support kinetochore tension.
MATERIALS AND METHODS
The cells used in this study were cultured and prepared for EM by high-pressure freezing, followed by standard methods of freeze-substitution fixation, embedding, sectioning, and staining. Briefly, Chlamydomonas reinhardtii (strain 137c mt+) was grown in liquid culture at 25°C on a 14:10 h light/dark cycle to enrich for mitotic cells (Umen and Goodenough, 2001). The cells were collected by centrifugation after shifting to the dark, then high-pressure frozen (HPM-010, Bal-Tec AG, Liechtenstein). The frozen cells were then freeze-substituted in 1% OsO4 plus 0.1% uranyl acetate in acetone and embedded in Epon/Araldite as described (O’Toole et al., 2003). Serial, 250–300-nm-thick sections were collected onto Formvar-coated copper slot grids and poststained in 2% uranyl acetate and Reynolds lead citrate.
Mammalian cells (RPE1 obtained from the American Type Culture Collection through M. Winey, University of Colorado, Boulder, and PtK2 from S. Dumont, University of California, San Diego, authenticated by transcriptome sequencing; Udy et al., 2015) were grown by standard methods and plated on sapphire disks (Morphew and McIntosh, 2003). When ∼80% confluent, cells were cryo-immobilized in a HPM-010 or a Wohlwend Compact 02 (Technotrade International) high-pressure freezer and then fixed by freeze-substitution at –90°C in acetone containing 1% OsO4 and 0.1% uranyl acetate; embedding was in Epon-Araldite. Mitotic cells were identified in the light microscope, remounted, serially sectioned, and stained, as above. Cellular regions were selected and imaged with an F30 intermediate voltage electron microscope (Thermo Fisher Scientific, Waltham MA) using Gatan (Pleasanton, CA) CCD cameras to record images with 1–2-nm pixels of serial tilts from ±60o in increments of 1o–1.5o. Single- or dual-axis tilt series were acquired from single frames or up to 3 × 3 montages using the SerialEM acquisition program (Mastronarde, 2005).
Tomograms were computed using the IMOD software package (Kremer et al., 1996; Mastronarde, 1997). For areas >10 µm, a supermontage was obtained using tools within the Navigator control and the user interface of SerialEM. For large area acquisition, the Navigator allows one to set up supermontage maps with appropriate image overlap, from which montage tilt series are automatically acquired. The individual 3 × 3 montage tilt series are then aligned, tomograms are computed using standard methods, and the resulting volumes are flattened. The lateral edges of the individual montages are then stitched together to create the supermontage. The result is a seamless, lateral stitching of the individual montage panels to create the supermontage volume (O’Toole et al., 2018). Tilt series were collected from serial sections (four to six sections) and the computed tomograms were aligned and joined to increase the volume of the reconstructed spindle. In total, three Chlamydomonas, two RPE1, and one PTK partial metaphase spindles were reconstructed (see Table 1).
The thickness of each reconstruction was determined by the thickness of the sections cut and the number of serial tomograms was made. The areas were assessed by the number of image pixels of a given size. The thickness of each section after imaging was about two-thirds the value estimated by microtomy because plastic sections collapse in the electron beam before the start of data collection for tomography (McEwen and Marko, 1999). All models, based on features drawn on tomographic slices, were therefore expanded in thickness by three-halves to compensate for this thinning.
MTs were manually traced in these reconstructions using the 3dmod program in the IMOD software package (Kremer et al., 1996), and graphic models of spindle features were added as separate objects. The resulting 3D models were projected as a series of 2D serial slices to show the complex arrangement of MT in the spindles. In some cases, the "resamplemod" program was used to sample the model points along the spindle axis and view the model in cross-section. Images and movies of the models and of slices cut from the tomograms were again prepared in IMOD. The places of close approach between MTs were identified with the MTK program in the IMOD suite. This program uses models of MT trajectories to identify points of close apposition and outputs a model object at each point of close approach. Its interactive structure allows one to specify features of particular interest, such as the minus ends of mcMTs (reference objects), and thereby seek the 3D coordinates of places where they are close to KMTs (neighboring objects). For the output model, we used a maximum close approach of 50 nm. The output model displays a connecting line between the mcMT end and the KMT with a central midpoint to identify places of close approach. The 3D distance between MT ends and a model point marking the spindle pole was determined using the imod-dist program in the IMOD suite. A point at the minus end of each mcMT end was modeled. The reference point for the pole of the Chlamydomonas spindle was estimated by marking a single point at the convergence of spindle MT ends. The reference point for a pole in a mammalian spindle was marked at the centrioles. The imod-dist program was then used to compute the 3D distances between the pole and the locations of mcMT ends.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Grants GM033787 to J.R.M. and P41GM103431 to Andreas Hoenger, University of Colorado, Boulder. ET was performed in the Electron Microscopy Core Facility at the University of Colorado, Boulder.
Abbreviations used
- EM
electron microscopy
- ET
electron tomography
- KMT
kinetochore microtubule
- mcMT
midplane-crossing microtubule
- MT
microtubule
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-07-0405) on December 11, 2019.
REFERENCES
- Aist JR, Bayles CJ, Tao W, Berns MW. (1991). Direct experimental evidence for the existence, structural basis and function of astral forces during anaphase B in vivo. J Cell Sci (Pt 2), 279–288. [DOI] [PubMed] [Google Scholar]
- Aist JR, Berns MW. (1981). Mechanics of chromosome separation during mitosis in Fusarium (Fungi imperfecti): new evidence from ultrastructural and laser microbeam experiments. J Cell Biol , 446–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aist JR, Liang H, Berns MW. (1993). Astral and spindle forces in PtK2 cells during anaphase B: a laser microbeam study. J Cell Sci (Pt 4), 1207–1216. [DOI] [PubMed] [Google Scholar]
- Ault JG, Nicklas RB. (1989). Tension, microtubule rearrangements, and the proper distribution of chromosomes in mitosis. Chromosoma , 33–39. [DOI] [PubMed] [Google Scholar]
- Brugués J, Nuzzo V, Mazur E, Needleman DJ. (2012). Nucleation and transport organize microtubules in metaphase spindles. Cell , 554–564. [DOI] [PubMed] [Google Scholar]
- Courtheoux T, Gay G, Gachet Y, Tournier S. (2009). Ase1/Prc1-dependent spindle elongation corrects merotely during anaphase in fission yeast. J Cell Biol , 399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R, McDonald KL, McIntosh JR. (1993). Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J Cell Biol , 141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elting MW, Hueschen CL, Udy DB, Dumont S. (2014). Force on spindle microtubule minus ends moves chromosomes. J Cell Biol , 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elting MW, Prakash M, Udy DB, Dumont S. (2017). Mapping load-bearing in the mammalian spindle reveals local kinetochore fiber anchorage that provides mechanical isolation and redundancy. Curr Biol , 2112–2122.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink G, Schuchardt I, Colombelli J, Stelzer E, Steinberg G. (2006). Dynein-mediated pulling forces drive rapid mitotic spindle elongation in Ustilago maydis. EMBO J , 4897–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill SW, Gonczy P, Stelzer EH, Hyman AA. (2001). Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature , 630–633. [DOI] [PubMed] [Google Scholar]
- Grishchuk EL, Spiridonov IS, McIntosh JR. (2007). Mitotic chromosome biorientation in fission yeast is enhanced by dynein and a minus-end-directed, kinesin-like protein. Mol Biol Cell , 2216–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler PK. (1980). Membranes in the mitotic apparatus of barley cells. J Cell Biol , 490–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramoto Y, Nakano Y. (1988). Micromanipulation studies of the mitotic apparatus in sand dollar eggs. Cell Motil Cytoskeleton , 172–184. [DOI] [PubMed] [Google Scholar]
- Jensen CG. (1982). Dynamics of spindle microtubule organization: kinetochore fiber microtubules of plant endosperm. J Cell Biol , 540–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson UG, Porter KR. (1968). Fine structure of cell division in Chlamydomonas reinhardi. Basal bodies and microtubules. J Cell Biol , 403–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamasaki T, O’Toole E, Kita S, Osumi M, Usukura J, McIntosh JR, Goshima G. (2013). Augmin-dependent microtubule nucleation at microtubule walls in the spindle. J Cell Biol , 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenti E, Vernos I. (2001). The mitotic spindle: a self-made machine. Science , 543–547. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Cole RW, Oakley BR, Rieder CL. (2000). Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol , 59–67. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, La Terra S, Chang F. (2004). Laser microsurgery in fission yeast; role of the mitotic spindle midzone in anaphase B. Curr Biol , 1330–1340. [DOI] [PubMed] [Google Scholar]
- Kilmartin JV, Goh PY. (1996). Spc110p: assembly properties and role in the connection of nuclear microtubules to the yeast spindle pole body. EMBO J , 4592–4602. [PMC free article] [PubMed] [Google Scholar]
- Kirschner M, Mitchison T. (1986). Beyond self-assembly: from microtubules to morphogenesis. Cell , 329–342. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. (1996). Computer visualization of three-dimensional image data using IMOD. J Struct Biol , 71–76. [DOI] [PubMed] [Google Scholar]
- LaFountain JR, Davidson LA. (1980). An analysis of spindle ultrastructure during anaphase of micronuclear division in Tetrahymena. Cell Motil , 41–61. [DOI] [PubMed] [Google Scholar]
- Leslie RJ, Pickett-Heaps JD. (1983). Ultraviolet microbeam irradiations of mitotic diatoms: investigation of spindle elongation. J Cell Biol , 548–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson V, O’Connell CB, Loncarek J, Paul R, Mogilner A, Khodjakov A. (2011). The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell , 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez P, Dixit R, Balkunde RS, OLeary SE, Brakke KA, Rasmussen CG. (2019). TANGLED1 mediates interactions between microtubules that may promote spindle organization and phragmoplast guidance to the division site in maize. BioRxiv 711796. [Google Scholar]
- Mastronarde DN. (1997). Dual-axis tomography: an approach with alignment methods that preserve resolution. J Struct Biol , 343–352. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. (2005). Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol , 36–51. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN, McDonald KL, Ding R, McIntosh JR. (1993). Interpolar spindle microtubules in PTK cells. J Cell Biol , 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda H, Hirano T, Yanagida M, Cande WZ. (1990). In vitro reactivation of spindle elongation in fission yeast nuc2 mutant cells. J Cell Biol , 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald KL, O’Toole ET, Mastronarde DN, McIntosh JR. (1992). Kinetochore microtubules in PTK cells. J Cell Biol , 369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald KL, Pfister K, Masuda H, Wordeman L, Staiger C, Cande WZ. (1986). Comparison of spindle elongation in vivo and in vitro in Stephanopyxis turris. J Cell Sci (Suppl 5), 205–227. [DOI] [PubMed] [Google Scholar]
- McDonald K, Pickett-Heaps JD, McIntosh JR, Tippit DH. (1977). On the mechanism of anaphase spindle elongation in Diatoma vulgare. J Cell Biol , 377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BF, Heagle AB, Cassels GO, Buttle KF, Rieder CL. (1997). Kinetochore fiber maturation in PtK1 cells and its implications for the mechanisms of chromosome congression and anaphase onset. J Cell Biol , 1567–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BF, Marko M. (1999). Three-dimensional transmission electron microscopy and its application to mitosis research. Methods Cell Biol , 81–111. [DOI] [PubMed] [Google Scholar]
- McIntosh JR, Hays T. (2016). A brief history of research on mitotic mechanisms. Biology (Basel) , 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR, Roos UP, Neighbors B, McDonald KL. (1985). Architecture of the microtubule component of mitotic spindles from Dictyostelium discoideum. J Cell Sci , 93–129. [DOI] [PubMed] [Google Scholar]
- Milas A, Jagric´ M, Martincˇic´ J, Tolic´ IM. (2018). Optogenetic reversible knocksideways, laser ablation, and photoactivation on the mitotic spindle in human cells. Methods Cell Biol , 191–215. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Maddox P, Groen A, Cameron L, Perlman Z, Ohi R, Desai A, Salmon ED, Kapoor TM. (2004). Bipolarization and poleward flux correlate during Xenopus extract spindle assembly. Mol Biol Cell , 5603–5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morphew MK, McIntosh JR. (2003). The use of filter membranes for high-pressure freezing of cell monolayers. J Microsc , 21–25. [DOI] [PubMed] [Google Scholar]
- Muller EG, Snydsman BE, Novik I, Hailey DW, Gestaut DR, Niemann CA, O’Toole ET, Giddings TH, Sundin BA, Davis TN. (2005). The organization of the core proteins of the yeast spindle pole body. Mol Biol Cell , 3341–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas RB. (1989). The motor for poleward chromosome movement in anaphase is in or near the kinetochore. J Cell Biol , 2245–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole ET, Dutcher SK. (2014). Site-specific basal body duplication in Chlamydomonas. Cytoskeleton (Hoboken) , 108–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole ET, Giddings TH, McIntosh JR, Dutcher SK. (2003). Three-dimensional organization of basal bodies from wild-type and delta-tubulin deletion strains of Chlamydomonas reinhardtii. Mol Biol Cell , 2999–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole E, van der Heide P, McIntosh JR, Mastronarde D. (2018). Large-scale electron tomography of cells using SerialEM and IMOD. In: Cellular Imaging: Electron Tomography and Related Techniques, Cham, Switzerland: Springer, 95–116. [Google Scholar]
- O’Toole ET, Winey M, McIntosh JR. (1999). High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol Biol Cell , 2017–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamula MC, Carlini L, Forth S, Verma P, Suresh S, Legant WR, Khodjakov A, Betzig E, Kapoor TM. (2019). High-resolution imaging reveals how the spindle midzone impacts chromosome movement. J Cell Biol , 2529–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitnick S, Spicer GS, Markow TA. (1995). How long is a giant sperm? Nature , 109. [DOI] [PubMed] [Google Scholar]
- Polak B, Risteski P, Lesjak S, Tolic´ IM. (2017). PRC1-labeled microtubule bundles and kinetochore pairs show one-to-one association in metaphase. EMBO Rep , 217–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redemann S, Baumgart J, Lindow N, Shelley M, Nazockdast E, Kratz A, Prohaska S, Brugués J, Fürthauer S, Müller-Reichert T. (2017). C. elegans chromosomes connect to centrosomes by anchoring into the spindle network. Nat Commun , 15288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redemann S, Lantzsch I, Lindow N, Prohaska S, Srayko M, Müller-Reichert T. (2018). A switch in microtubule orientation during C. elegans meiosis. Curr Biol , 2991–2997.e2. [DOI] [PubMed] [Google Scholar]
- Rieder CL. (1981). The structure of the cold-stable kinetochore fiber in metaphase PtK1 cells. Chromosoma , 145–158. [DOI] [PubMed] [Google Scholar]
- Saunders AM, Powers J, Strome S, Saxton WM. (2007). Kinesin-5 acts as a brake in anaphase spindle elongation. Curr Biol , R453–R454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikirzhytski V. et al (2014). Direct kinetochore-spindle pole connections are not required for chromosome segregation. J Cell Biol , 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srayko M, O’toole ET, Hyman AA, Müller-Reichert T. (2006). Katanin disrupts the microtubule lattice and increases polymer number in C. elegans meiosis. Curr Biol , 1944–1949. [DOI] [PubMed] [Google Scholar]
- Tippit DH, Schulz D, Pickett-Heaps JD. (1978). Analysis of the distribution of spindle microtubules in the diatom Fragilaria. J Cell Biol , 737–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolic´ IM. (2018). Mitotic spindle: kinetochore fibers hold on tight to interpolar bundles. Eur Biophys J , 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolic´ IM, Novak M, Pavin N. (2019) Helical twist and rotational forces in the mitotic spindle. Biomolecules , E132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranfield EM, Heiligenstein X, Peristere I, Antony C. (2014). Correlative light and electron microscopy for a free-floating spindle in Xenopus laevis egg extracts. Methods Cell Biol , 111–128. [DOI] [PubMed] [Google Scholar]
- Udy DB, Voorhies M, Chan PP, Lowe TM, Dumont S. (2015). Draft de novo transcriptome of the rat kangaroo Potorous tridactylus as a tool for cell biology. PLoS One , e0134738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen JG, Goodenough UW. (2001). Chloroplast DNA methylation and inheritance in Chlamydomonas. Genes Dev , 2585–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F, Dogterom M, Stelzer E, Karsenti E, Leibler S. (1992). Control of microtubule dynamics and length by cyclin A- and cyclin B-dependent kinases in Xenopus egg extracts. J Cell Biol , 1097–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukušic´ K, Bud¯a R, Bosilj A, Milas A, Pavin N, Tolic´ IM. (2017). Microtubule sliding within the bridging fiber pushes kinetochore fibers apart to segregate chromosomes. Dev Cell , 11–23.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JJ, Roque H, Antony C, Nédélec F. (2014). Mechanical design principles of a mitotic spindle. Elife , e03398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber B, Tranfield EM, Höög JL, Baum D, Antony C, Hyman T, Verbavatz J-M, Prohaska S. (2014). Automated stitching of microtubule centerlines across serial electron tomograms. PLoS One , e113222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Bloom K. (2012). Mitotic spindle form and function. Genetics , 1197–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O’Toole ET, Mastronarde DN, Giddings TH, McDonald KL, McIntosh JR. (1995). Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol , 1601–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo TY, Choi J-M, Conway W, Yu C-H, Pappu RV, Needleman DJ. (2018). Measuring NDC80 binding reveals the molecular basis of tension-dependent kinetochore-microtubule attachments. Elife , e36392. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.