Abstract
Piwi and piRNA form the piRNA‐induced silencing complex (piRISC) to repress transposons. In the current model, Armitage (Armi) brings the Piwi–piRISC precursor (pre‐piRISC) to mitochondria, where Zucchini‐dependent piRISC maturation occurs. Here, we show that Armi is necessary for Piwi–pre‐piRISC formation at Yb bodies and that Armi triggers the exit of Piwi–pre‐piRISC from Yb bodies and the translocation to mitochondria. Piwi–pre‐piRISC resist leaving Yb bodies until Armi binds Piwi–pre‐piRISC through the piRNA precursors. The lack of the Armi N‐terminus also blocks the Piwi–pre‐piRISC exit from Yb bodies. Thus, Armi determines Piwi–piRISC processing, in a multilayered manner, from precursor formation and quality control to inter‐organelle translocation for maturation.
Keywords: Armitage, Gasz, mitochondrion, piRNA, Yb body
Subject Categories: RNA Biology
Armitage determines Piwi–piRISC processing, in a multilayered manner, from precursor formation and quality control to inter‐organelle translocation for Zucchini‐dependent piRISC maturation on mitochondria.
Introduction
PIWI‐interacting RNAs (piRNAs) repress transposons to protect the germline genome from their harmful invasion 1, 2, 3. To this end, piRNAs stoichiometrically associate with members of the PIWI protein family to form piRNA‐induced silencing complexes (piRISCs) and target them to their RNA substrates via Watson–Crick (RNA–RNA) base pairing 1, 2, 3. Drosophila expresses three PIWI members, Piwi, Aubergine (Aub), and Argonaute3 (Ago3); the lack of any of these members causes transposon derepression, leading to severe defects in gonadal development and infertility, showing their functional non‐redundancy 4, 5, 6.
In Drosophila ovarian somatic cells (OSCs), transposon‐repressing piRNAs originate from a soma‐specific piRNA cluster flamenco (flam) on the X chromosome 7, 8, 9. The flam locus is rich in transposon remnants, whose directions of insertion at the locus are mostly opposite to the direction of flam transcription 7, 10. Therefore, by their nature, flam‐derived piRNAs are created to act as antisense oligonucleotides to silence their parental transposons. OSCs express Piwi but no other PIWI proteins 7, 11. The Piwi–piRISC assembly takes place in the cytoplasm. The complex is then imported into the nucleus and represses transposons at the transcriptional level 12, 13.
Nine proteins have so far been designated as piRNA biogenesis factors in OSCs: female sterile (1) Yb (Yb), Armitage (Armi), Sister of Yb (SoYb), Vreteno (Vret), Shutdown (Shu), Zucchini (Zuc), Minotaur (Mino), Daedalus (Daed), and Gasz (germ cell protein with ankyrin repeats, sterile alpha motif, and leucine zipper) 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27. These factors can be classified into two groups by their apparent cellular localization in OSCs. Yb, Armi, SoYb, Vret, and Shu are detected at the perinuclear non‐membranous organelles called Yb bodies 16, 22, 23, 24, 25, whose assembly is required for transposon‐repressing piRNA production 28. In contrast, Zuc, Mino, Daed, and Gasz are localized at the outer membrane of mitochondria via their own transmembrane targeting signals (TMs) 17, 18, 20, 21, 26. However, the rest of the protein bodies project toward the cytosol, allowing them to act in piRNA biogenesis together with other cytosolic factors.
Yb is a Tudor domain‐containing protein 25, which self‐associates and binds to the flam RNA transcripts (i.e., transposon‐repressing piRNA precursors) through the cis‐elements within the substrates 28, 29, 30. This multivalent association between Yb and flam RNAs induces liquid–liquid phase separation, leading to the assembly of Yb bodies 28. Armi, a superfamily 1 (SF1) RNA helicase exhibiting ATP‐dependent, 5′‐to‐3′ directional RNA‐unwinding activity 31, 32, subsequently joins Yb bodies by associating with Yb, allowing Armi to bind specifically to flam RNAs 31, 33. In the absence of Yb, Yb bodies disappeared and Armi remains cytosolic 22, 24. Under such circumstance, Armi binds cellular RNAs promiscuously and produces piRNAs from the bound RNAs 31. This means that piRNA production in OSCs does not always require Yb (and Yb‐body formation). However, the expression levels of piRNAs become low and those piRNAs are mostly non‐transposon‐targeting; thus, transposons are not sufficiently silenced.
Zuc is an endonuclease responsible for excising mature piRNAs successively from the 5′ end of the precursors, giving rise to phased piRNAs 15, 19, 29. According to the current model, the precursors in this context are pre‐bound with Piwi through the 5′ end (i.e., Piwi–pre‐piRISC), while Armi binds downstream regions to relax the RNA substrates for Zuc cleavage, facilitating the piRNA production 31, 33. A recent study showed that Gasz and Daed collaboratively recruit Armi to the mitochondrial surface to bring Piwi–pre‐piRISC proximal to Zuc 20. The report also provided a model showing that Piwi is bound with piRNA precursors at Yb bodies. However, the regulatory mechanism(s) underlying the formation of Piwi–pre‐piRISC at Yb bodies and the translocation from Yb bodies to mitochondria, the site for piRISC maturation, remains elusive. In this study, to shed light on the Piwi–piRISC processing pathway, we conducted a number of biochemical analyses using cultured OSCs 11, 34.
Results
Armi aberrantly accumulates on mitochondria in the absence of Zuc but is relocated to Yb bodies upon Gasz loss
We first set out to confirm the importance of Gasz in piRNA biogenesis in OSCs. Gasz was downregulated by RNAi (Fig EV1A) for which northern blotting was performed. The probe used was for idefix‐piRNA arising from the flam locus. In parallel with this, Zuc was depleted as a positive control 21, 35. Without Gasz, mature idefix‐piRNA almost disappeared but the piRNA precursors strongly accumulated, as was the case upon Zuc depletion (Fig 1A). These findings corroborated the notion that Gasz acts in conjunction with Daed as the platform for Zuc‐dependent piRNA maturation from piRNA precursors 20.
Figure EV1. Loss of Zuc and Gasz impacts the cellular localization of Armi and Piwi in OSCs.
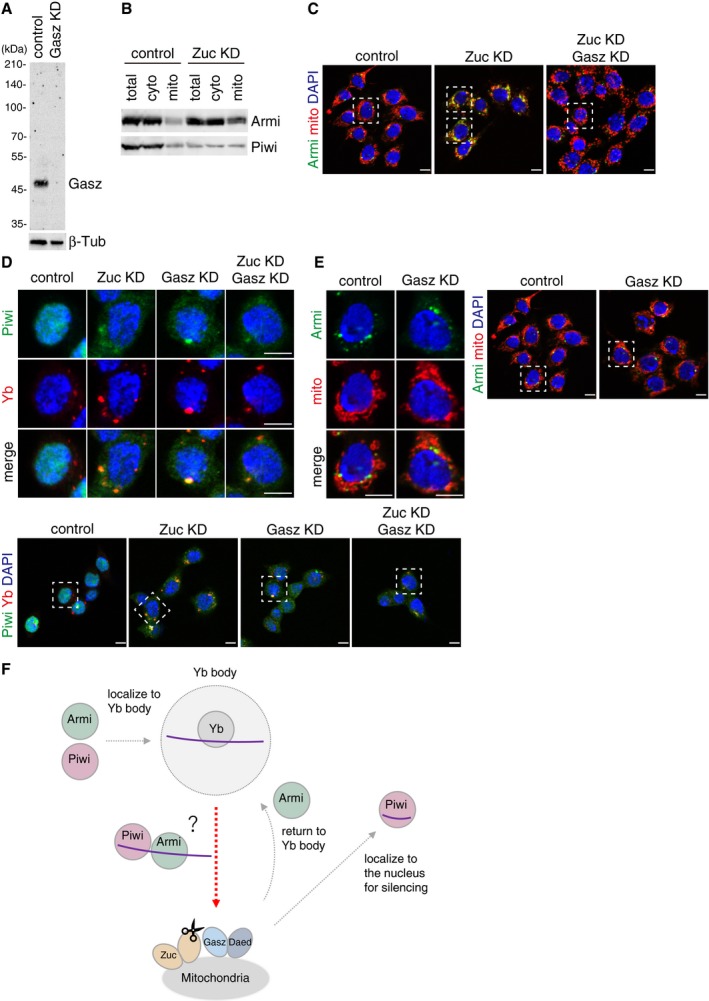
- Western blotting using anti‐Gasz monoclonal antibody shows that the Gasz signal disappeared upon Gasz RNAi treatment in OSCs. Anti‐Gasz monoclonal antibody was produced in this study. β‐Tubulin (β‐Tub) was detected as a loading control.
- Western blotting shows that the levels of Armi and Piwi in the cytoplasmic fraction after mitochondrial isolation (cyto) were little impacted by Zuc depletion, while the level of Armi in the mitochondrial fraction (mito) increased.
- Larger immunostaining views of the cell images shown in Fig 1C and D. The cells appeared in the main figures are boxed with white dotted line. In “Zuc KD” panel, the upper cell appeared in Fig 1C, while the lower one appeared in Fig 1D. Armi behaved similarly in the cells other than the one shown in Fig 1C and D. The scale bar represents 5 μm.
- Upper: Localization of Piwi (green) in OSCs upon Zuc and Gasz depletion. Yb bodies are shown in red. Lower: Larger immunostaining views of the cell images shown in upper panels. The cells appeared in the upper panels are boxed with white dotted line. Piwi was mislocalized in the cytosol upon the loss of Zuc and/or Gasz. Some fraction of Piwi was detected at Yb bodies as has previously been reported 24. The scale bar represents 5 μm. DAPI (blue) shows the nuclei.
- Left: Localization of Armi (green) in OSCs upon Gasz depletion. Mitochondria are shown in red. Right: Larger immunostaining views of the cell images shown on the left. The scale bar represents 5 μm. DAPI (blue) shows the nuclei.
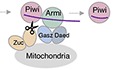
- Piwi localizes to Yb bodies after Armi has localized there. Upon Piwi–piRISC formation by Zuc on the outer membrane of mitochondria, Armi goes back to Yb bodies, while Piwi–piRISC is translocated to the nucleus for silencing. The details and requirements of inter‐organelle translocation of Armi and Piwi–pre‐piRISC remain unknown (red dotted line with an arrowhead).
Figure 1. Loss of Zuc and Gasz impacts the cellular localization of Armi and Piwi in OSCs.
- Northern blotting shows that Gasz depletion (Gasz KD) led to the accumulation of flam‐piRNA precursors but reduced the level of mature idefix‐piRNA arising from the precursors, as in the case of Zuc depletion (Zuc KD). U6 snRNA was detected as a loading control.
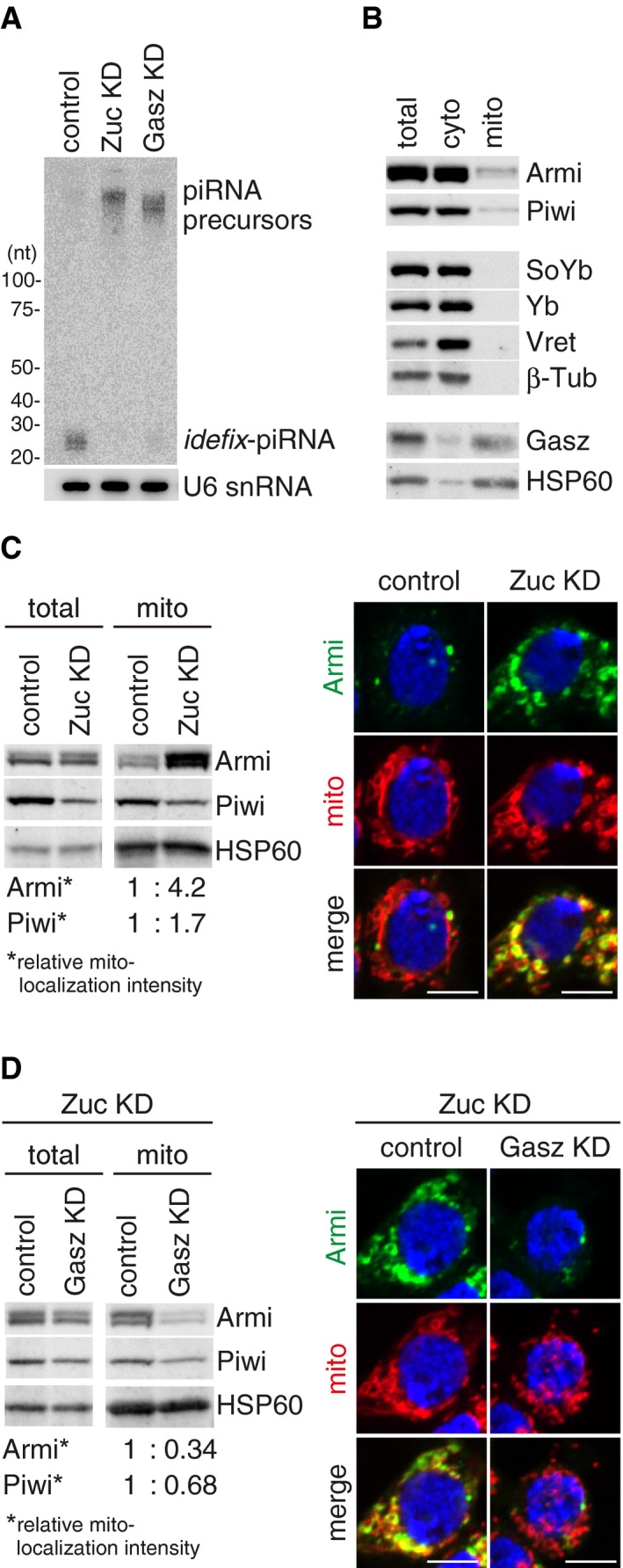
- Western blotting showing the protein levels of endogenous Armi, Piwi, SoYb, Yb, Vret, and Gasz in total OSC lysate (total), the mitochondrial fraction (mito), and the cytoplasmic fraction after mitochondrial isolation (cyto). β‐Tubulin (β‐Tub) and HSP60 were detected as markers for the “cyto” and “mito” fractions, respectively.
- Left: Western blotting comparing the abundances of Armi and Piwi in total OSC lysate (total) and the mitochondrial fraction (mito) before (control) and after (Zuc KD) Zuc knockdown in OSCs. Relative mito‐localization intensity (*) shows the amount of proteins in “mito” normalized by the amount of proteins in “total” and HSP60. Right: Immunofluorescence shows that the Armi signal (green) is strongly overlapped with the mitochondrial signal (red) in Zuc‐depleted OSCs (Zuc KD). Armi is localized to Yb bodies in control OSCs (control). The scale bar represents 5 μm. DAPI (blue) shows the nuclei.
- Left: Western blotting comparing the abundances of Armi and Piwi in total OSC lysate (total) and the mitochondrial fraction (mito) before (control) and after (Gasz KD) Gasz depletion in Zuc‐depleted OSCs (Zuc KD). Relative mito‐localization intensity (*) shows the amount of proteins in “mito” normalized by the amount of proteins in “total” and HSP60. Right: Immunofluorescence shows that the Armi mitochondrial signal (green) found in Zuc‐depleted cells (Zuc KD/control) mostly disappeared after additional Gasz depletion in the cells (Zuc KD/Gasz KD), but was found at Yb bodies. Mitochondrial signal is shown in red. The scale bar represents 5 μm. DAPI (blue) shows the nuclei.
We next isolated mitochondria from OSC cytoplasmic lysate and examined the levels of endogenous Gasz, Piwi, and four Yb‐body components (i.e., Armi, SoYb, Yb, and Vret) in the fraction and also in the cytoplasmic lysate after mitochondrial isolation. Gasz was detected in the mitochondrial fraction, as was the mitochondrial marker protein HSP60 (Fig 1B). Piwi and Armi were also present in the mitochondrial fraction, although their levels were much lower than those in the mitochondrion‐depleted cytoplasmic fraction. SoYb, Yb, and Vret were nearly exclusively cytoplasmic. These results support the earlier notion of Armi's involvement in piRISC (piRNA) maturation on the mitochondrial surface in OSCs 20, 31, 33.
We then compared the levels of mitochondrial Piwi and Armi in the presence and absence of Zuc. The level of mitochondrial Piwi was increased upon Zuc depletion (~1.7‐fold) (Figs 1C and EV1B). Strikingly, the accumulation of Armi in the mitochondrial fraction was higher upon Zuc depletion (~4.2‐fold). The data from immunofluorescence analyses agreed with this: The Armi signal was strongly overlapped with the mitochondrial signal upon Zuc depletion (Figs 1C and EV1C), while Armi in normal OSCs was detected at Yb bodies, as we originally reported 24. Yb bodies were present even in Zuc‐lacking OSCs (Fig EV1D). Piwi was mostly mislocalized to the cytoplasm, due to the loss of piRNA loading (Fig EV1D) 24. The mitochondrial localization of Piwi was unclear (Fig EV1D). This might be attributable to the reduced level of Piwi due to the loss of piRNA loading (Figs 1C and EV1B). Zuc knockdown caused slightly mitochondrial clustering in OSCs (Figs 1C and EV1C), as has been reported previously 23, although at present the effect of this phenomenon on piRNA biogenesis remains unclear.
Curiously, additional knockdown of Gasz in Zuc‐lacking OSCs restored the Yb‐body localization of Armi (Figs 1D and EV1C). The findings from Western blotting analysis agreed with this observation (Fig 1D). Changes in Piwi abundance and localization were not so drastic between before and after Gasz knockdown in Zuc‐lacking OSCs (Figs 1D and EV1D). Single depletion of Gasz had little impact on the Yb‐body localization of Armi (Fig EV1E), although Piwi was largely cytosolic in Gasz‐depleted OSCs (Fig EV1D). These results suggest that, without Gasz, regardless of the presence or absence of Zuc, Armi no longer remains on the mitochondrial surface and so returns to Yb bodies while leaving Piwi in the cytosol. Some fraction of Piwi was detected at Yb bodies, but to a lesser extent than that in Zuc‐depleted cells (Fig EV1D). Previous studies showed that Gasz loss perturbs mitochondrial morphology in OSCs 17, 20, which was also observed but to a lesser extent in this study (Fig EV1E).
By integrating all circumstantial evidence that has so far been reported in the field, we now claim that the intracellular movement of nascent Armi in OSCs is cytoplasm→Yb body→mitochondria→Yb body, while that of Piwi is cytoplasm→Yb body→mitochondria→nucleus (Fig EV1F). Upon piRISC formation by Zuc on the mitochondrial surface, Armi returns to Yb bodies, while Piwi–piRISC is imported into the nucleus, suggesting that they should be separated from each other upon Zuc cleavage. However, the relationship between Armi and Piwi and the requirements for their translocation from Yb bodies to mitochondria remain unclear (Fig EV1F). We therefore aim to address these important issues using OSCs in which Armi, Zuc, and also Piwi are depleted singularly or in various combinations from OSCs.
Gasz binds Armi–Piwi–pre‐piRISC on the outer membrane of mitochondria
To examine whether Armi and Piwi in the mitochondrial fraction are in a bound form with Gasz, we prepared the mitochondrial fraction, immunopurified the Gasz complex from mitochondrial lysate, and conducted Western blotting. Both Armi and Piwi were detected in the immunopurified material (Fig 2A), indicating that Armi, Piwi, and Gasz form a trimeric complex on the outer membrane of mitochondria. The abundance of Armi and Piwi in the Gasz complex was increased upon Zuc depletion (Figs 2B and EV2A) in accordance with the results shown in Fig 1C and D. We then performed northern blotting using idefix‐piRNA probe as in Fig 1A. flam‐piRNA precursors, albeit at very low levels, were detected in the mitochondrial Gasz complex in normal OSCs (Figs 2C and EV2B). The precursor signal was greatly increased upon Zuc depletion as were Armi and Piwi signals (Fig 2C). These results provide evidence that Piwi and Armi are bound with mitochondrial Gasz in a complex that includes flam‐piRNA precursors.
Figure 2. Mitochondrial Gasz interacts with Armi and Piwi bound with piRNA precursor.
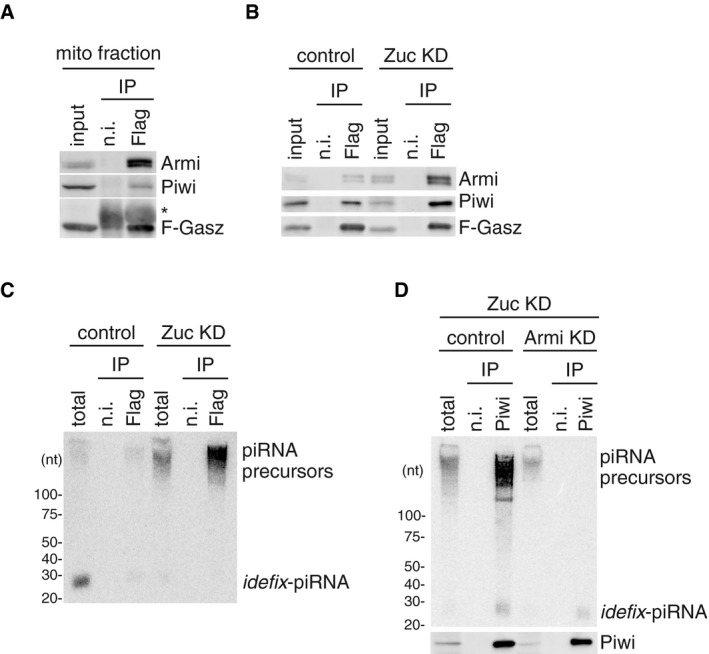
- Armi and Piwi co‐immunoprecipitated with Flag‐Gasz (F‐Gasz) from the mitochondrial fraction (mito fraction) of OSCs (input). F‐Gasz was ectopically expressed in the cells by transfection prior to immunoprecipitation. *: Heavy chains of antibodies used for immunoprecipitation. n.i.: Non‐immune IgG used as a negative control.
- Co‐immunoprecipitation from whole‐cell lysate of OSCs before (control) and after (Zuc KD) Zuc depletion and subsequent Western blotting show that the levels of Piwi and Armi in the Flag‐Gasz (F‐Gasz) complex increased upon Zuc depletion (Zuc KD). Armi signal with a longer exposure is shown in Fig EV2A.
- Northern blotting shows that the flam‐piRNA precursors in the Flag‐Gasz complex (Flag) accumulated upon Zuc depletion (Zuc KD). A gel image of the control lanes (control) with a long exposure is shown in Fig EV2B.
- Upper: The flam‐piRNA precursors associated with Piwi disappeared upon Armi depletion (Armi KD) in Zuc‐lacking OSCs (Zuc KD). Lower: The abundance of immunopurified Piwi is shown by Western blotting.
Figure EV2. Gasz binds Armi–Piwi–pre‐piRNA on the outer membrane of mitochondria.
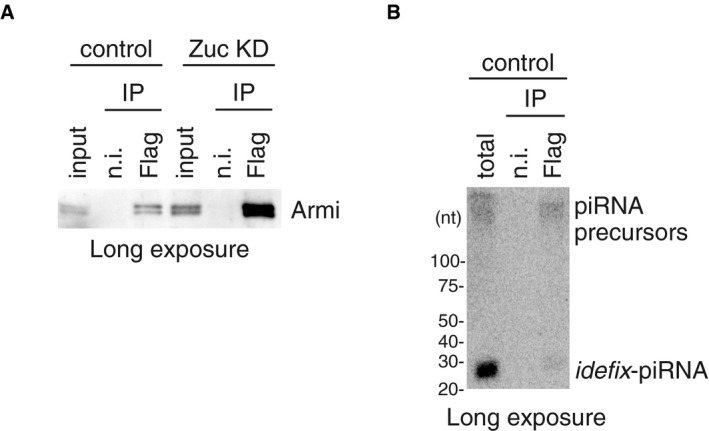
We previously showed that Piwi was barely localized to Yb bodies without Armi regardless of the presence of Yb bodies in OSCs 24. In this study, we observed that Piwi was no longer associated with piRNA precursors upon the loss of Armi (Fig 2D). Thus, Piwi requires Armi to be loaded with the precursors at Yb bodies. These findings revealed that Piwi–pre‐piRISC is formed at Yb bodies in an Armi‐dependent manner.
Departure of Armi from Yb bodies depends on Piwi
We depleted Piwi in Zuc‐depleted OSCs and performed immunofluorescence. Strikingly, the aberrant mitochondrial accumulation of Armi induced by Zuc loss was completely released upon additional Piwi depletion and Armi localization to Yb bodies was fully restored (Figs 3A and B, and EV3A). Western blotting confirmed that this was not due to unexpected reduction in the level of Armi (Fig EV3B). The immunofluorescence data also showed that Yb‐body localization of Armi occurs in a Piwi‐independent manner (Figs 3B and EV3A). Our previous study demonstrated that Piwi's Yb‐body localization depends on Armi 24. The hierarchy of Piwi and Armi in Yb‐body localization was clarified.
Figure 3. Departure of Armi from Yb bodies depends on Piwi.
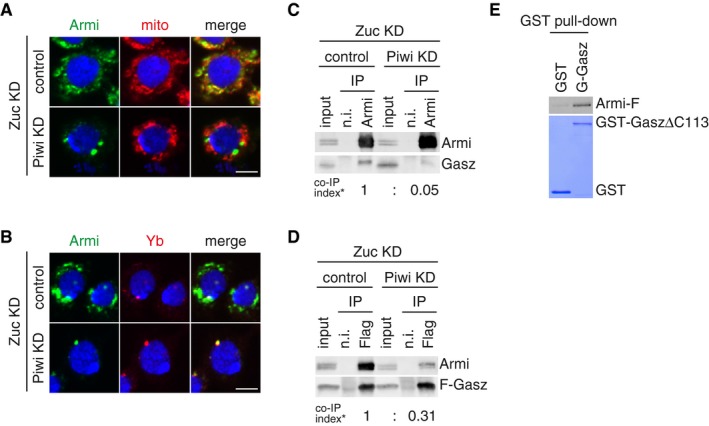
-
A, BDepletion of endogenous Piwi (Piwi KD) in Zuc‐lacking OSCs (Zuc KD) caused Armi (green) to relocate to Yb bodies. Localization of Armi at Yb bodies is shown in (B). Mitochondrial (upper panel) and Yb (lower panel) signals are shown in red. The scale bar represents 5 μm. DAPI (blue) shows the nuclei.
-
BThe Armi complex was immunopurified from Zuc‐lacking OSCs (Zuc KD) before (control) and after (Piwi KD) Piwi depletion and probed for detecting Armi and Gasz. Armi no longer stably associated with endogenous Gasz upon Piwi depletion in Zuc‐lacking OSCs. Co‐IP index (*) shows that the ratio of Gasz amount in the Armi complex isolated from Zuc‐depleted cells (Zuc KD/control) and Zuc‐ and Piwi‐depleted cells (Zuc KD/Piwi KD) is 1:0.05.
-
CThe Flag‐Gasz (F‐Gasz) complex was immunopurified from Zuc‐lacking OSCs (Zuc KD) before (control) and after (Piwi KD) Piwi depletion and probed for detecting Armi and F‐Gasz. F‐Gasz was ectopically expressed prior to immunoprecipitation. Co‐IP index (*) shows that the ratio of Armi amount in the F‐Gasz complex isolated from Zuc‐depleted cells (Zuc KD/control) and Zuc‐ and Piwi‐depleted cells (Zuc KD/Piwi KD) is 1:0.31.
-
DIn vitro pull‐down assays show that Armi‐Flag (Armi‐F) directly binds with GST‐GaszΔC113 but hardly with GST. Armi‐F was immunopurified from Schneider 2 (S2) cells under harsh conditions. GST and GST‐GaszΔC113 were visualized by CBB staining, while Armi‐F was detected by Western blotting using anti‐Flag antibodies.
Figure EV3. Departure of Armi from Yb bodies depends on Piwi.
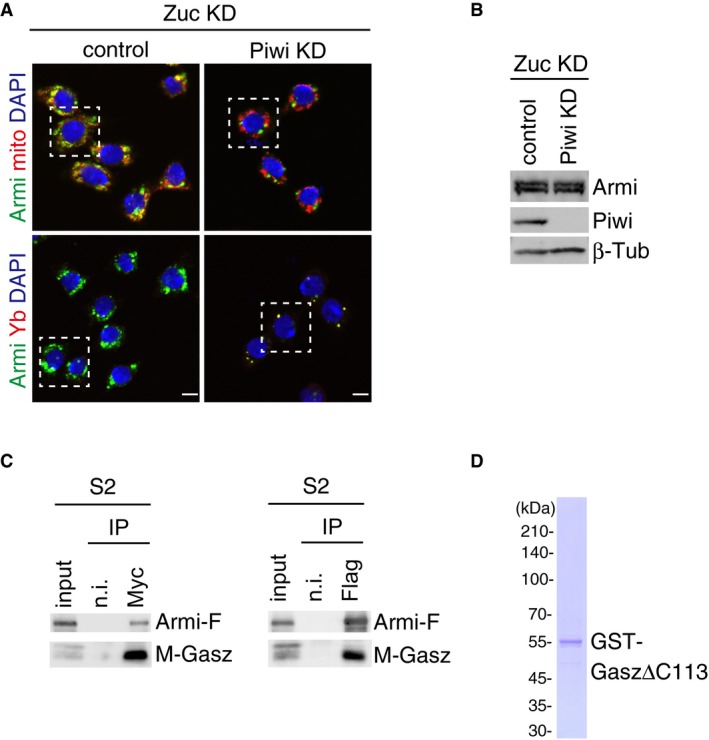
- Piwi depletion hardly affected the level of Armi in Zuc‐depleted OSCs. β‐Tubulin (β‐Tub) was detected as a loading control.
- Myc‐Gasz (M‐Gasz) and Armi‐Flag (Armi‐F) interacted with each other in Schneider 2 (S2) cells.
- Purified recombinant GST‐Gasz (ΔC113 lacking Leu349‐Ser461 containing the TM region at the C‐terminus) used in the experiment shown in Fig 3E. The protein was stained with CBB.
We also found that under these conditions, Armi no longer stably associated with Gasz (Fig 3C and D). We postulated two scenarios to explain this: Without Piwi, (i) Armi resists leaving Yb bodies and thus is stuck there, or (ii) Armi leaves Yb bodies and moves to mitochondria but cannot remain there, possibly because of the loss of the ability to attach to Gasz, and repeatedly travels back and forth between the two organelles.
A recent study demonstrated that Gasz and Armi interact with each other in Schneider 2 (S2) cells where Yb and Piwi are not expressed 20. The Gasz–Armi association in S2 cells was recapitulated in this study (Fig EV3C). We also demonstrate that recombinant Armi (purified from S2 cells under harsh conditions, preventing co‐purification of other proteins 31) bound with recombinant Gasz lacking the C‐terminal TM region (ΔC113), which was bacterially purified, without any other factors (Figs 3E and EV3D); this supports their ability to directly interact. These findings more strongly support the first scenario that Armi resists leaving Yb bodies unless Piwi localizes to Yb bodies to become pre‐piRISC there.
Departure of Armi from Yb bodies requires piRNA precursor loading onto Piwi
We next explored how the loss of the RNA‐binding activity of Piwi influences the cellular dynamics of Armi in OSCs. To make Piwi defective in RNA binding, we mutagenized the protein based on our structural study of Siwi, a PIWI member in silkworm 36. Tyr551 in Piwi was mutated to alanine to produce Piwi MID mutant lacking the 5′‐end piRNA binding. We previously produced Piwi PAZ mutant lacking the 3′‐end piRNA binding by mutating Tyr327 and Tyr328 to alanines 24. As we expected, the MID mutant was loaded with neither mature ideflx‐piRNA nor piRNA precursors, while the PAZ mutant was loaded with precursors, as was Piwi WT control (Fig 4A). We performed the experiments in Zuc‐lacking OSCs because under the circumstance the level of Piwi–pre‐piRISC was higher than that in normal OSCs (Fig 2C). Endogenous Piwi was also depleted simultaneously to minimize the effect on piRNA precursor loading onto Flag‐Piwi. The level of mature piRNA with Piwi PAZ mutant was much lower than that with Piwi WT. These results corroborated that the 5′‐binding pocket in the MID domain, but not the 3′‐binding pocket in the PAZ domain, is crucial for the Piwi–pre‐piRISC assembly in OSCs.
Figure 4. Departure of Armi from Yb bodies requires piRNA precursor loading onto Piwi.
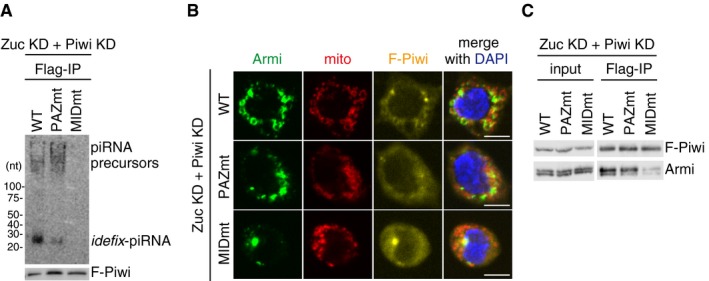
- Upper: Northern blotting shows that Flag‐Piwi WT (WT) and PAZ mutant (PAZmt) but not MID mutant (MIDmt) bound with flam‐piRNA precursors in Zuc‐depleted OSCs. Endogenous Piwi was depleted simultaneously with Zuc (Zuc KD + Piwi KD). Zuc was depleted by RNAi and so residual Zuc may have remained in cells. Therefore, a tiny amount of piRNAs was loaded onto Piwi. F‐Piwi WT, PAZmt, and MIDmt were made to be RNAi‐resistant by mutagenesis (see Materials and Methods). Lower: The abundance of immunopurified Flag‐Piwi (F‐Piwi) is shown by Western blotting.
- Armi localization to Yb bodies observed in Zuc‐ and Piwi‐depleted OSCs (Zuc KD + Piwi KD) (Fig 3A and B) was restored by ectopic expression of Flag‐Piwi WT and PAZmt but not of MIDmt (see also Fig EV4B). Armi, mitochondria, and F‐Piwi are shown in green, red, and yellow, respectively. The scale bar represents 5 μm. DAPI (blue) shows the nuclei.
- Western blotting shows that Flag‐Piwi (F‐Piwi) WT (WT) and PAZ mutant (PAZmt) but not MID mutant (MIDmt) bound with Armi in Zuc/Piwi‐depleted OSCs (Zuc KD + Piwi KD).
When the MID mutant was expressed in Zuc‐depleted OSCs instead of endogenous Piwi, Armi was localized to Yb bodies and failed to translocate to mitochondria (Figs 4B and EV4A and B). When either Piwi WT or the PAZ mutant was expressed instead of the MID mutant, no such phenomenon regarding Armi localization was observed (Figs 4B and EV4A and B). The PAZ mutant interacted with Armi as efficiently as Piwi WT did, but the MID mutant did not (Fig 4C). Both Piwi mutants were detected at Yb bodies (Fig EV4B). A similar outcome was obtained when Zuc was still expressed in OSCs as in normal cells (Fig EV4C). These findings suggest that, when Piwi is not bound with piRNA precursors, Armi fails to bind Piwi and resists leaving Yb bodies.
Figure EV4. Departure of Armi from Yb bodies requires piRNA precursor loading onto Piwi.
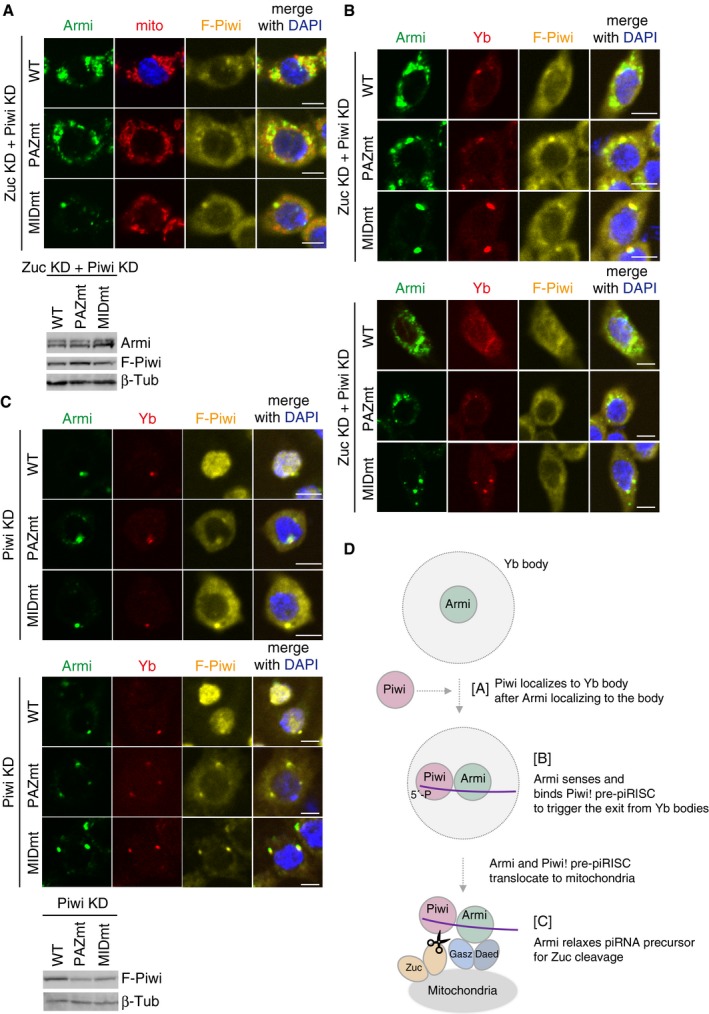
- Upper: Another set of cell images of the one shown in Fig 4B. The scale bar represents 5 μm. DAPI (blue) shows the nuclei. Lower: The expression levels of Flag‐Piwi (F‐Piwi) WT and PAZ/MID mutants (PAZmt/MIDmt) in Zuc/Piwi‐depleted OSCs (Zuc KD + Piwi KD). β‐Tubulin (β‐Tub) was detected as a loading control.
- Upper: Armi localization to Yb bodies observed in Zuc/Piwi‐depleted OSCs (Zuc KD + Piwi KD) (Fig 3A and B) was rescued by ectopic expression of Flag‐Piwi WT and PAZmt but not of MIDmt. “Rescued” means that Armi is now localized to mitochondria as in Zuc‐depleted OSCs (Fig 1C). The scale bar represents 5 μm. DAPI (blue) shows the nuclei. Lower: Another set of cell images of the one shown on the left. The scale bar represents 5 μm. DAPI (blue) shows the nuclei.
- Upper: The cellular localization of Flag‐Piwi (F‐Piwi) WT and PAZ/MID mutants (PAZmt/MIDmt) (yellow), Yb (red), and Armi (green) in Piwi‐depleted OSCs (Piwi KD). Middle: Another set of cell images of the one shown on the left. The scale bar represents 5 μm. DAPI (blue) shows the nuclei. Lower: The expression levels of Flag‐Piwi (F‐Piwi) WT and PAZ/MID mutants (PAZmt/MIDmt) in Piwi‐depleted OSCs (Piwi KD). β‐Tubulin (β‐Tub) was detected as a loading control.
- Armi has three functions. [A] Armi's Yb‐body localization is required for Piwi localization at Yb bodies. [B] Armi senses and binds Piwi–pre‐piRISC at Yb bodies. This action of Armi is required for inter‐organelle translocation of the complex from Yb bodies to mitochondria. [C] Armi relaxes piRNA precursors for Zuc cleavage on the mitochondria.
On the basis of these findings and previous ones, we propose that Armi plays three roles in Piwi–piRISC assembly in OSCs (Fig EV4D): controlling apo‐Piwi localization to Yb bodies 24, sensing Piwi–pre‐piRISC assembly at Yb bodies (this study), and translocating together with Piwi–pre‐piRISC to mitochondria, where it relaxes RNA substrates for Zuc‐dependent processing of piRISC maturation 31.
Translocation of Piwi–pre‐piRISC from Yb bodies requires the RNA‐binding activity of Armi
We then examined how the RNA‐binding activity of Armi impacts on the translocation of Piwi–pre‐piRISC. For this, we produced an RNA‐binding‐defective Armi mutant N756A (Fig EV5A), given the earlier observation that Asn462 in yeast Upf1 (corresponding to Asn756 in Armi) was predicted to play a crucial role in the Upf1–RNA interaction (on the basis of structural analysis) 37. Substitution of endogenous Armi with the N756A mutant in OSCs strongly impaired piRNA biogenesis (Fig EV5B).
Figure EV5. The behaviors of Armi N756A mutant and ΔN34 mutant.
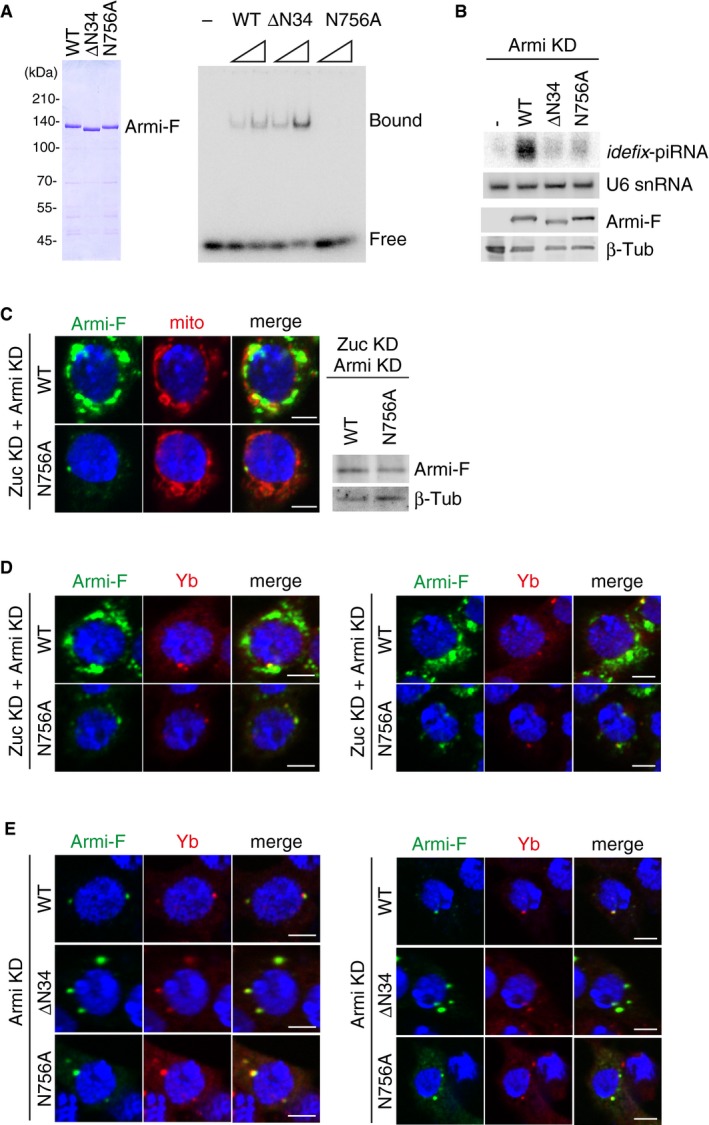
- Left: Purified recombinant Armi‐Flag (Armi‐F) WT and mutants (ΔN34 and N756A) used in the experiment. Right: RNA‐binding activity of Armi‐Flag WT, ΔN34, and N756A. Here, 16‐nt single‐stranded RNA was 5′‐end‐labeled and incubated with Armi‐Flag.
- Top: Northern blotting shows that siRNA‐resistant WT Armi‐Flag (Armi‐F), but not ΔN34 and N756A mutants, rescued the defects in idefix‐piRNA production caused by endogenous Armi depletion. U6 snRNA was used as a loading control. Bottom: The expression levels of Armi‐Flag (Armi‐F) WT, ΔN34, and N756A mutants. β‐Tubulin was used as a loading control.
- Left: Another set of cell images of the one shown in Fig 5B. The scale bar represents 5 μm. DAPI (blue) shows the nuclei. Right: The expression levels of Armi‐Flag (Armi‐F) WT and N756A mutant in Zuc/Armi‐depleted OSCs (Zuc KD + Armi KD). β‐Tubulin (β‐Tub) was detected as a loading control.
- Left: B. Armi‐Flag (Armi‐F) WT and the N756A mutant were expressed in OSCs where endogenous Armi and Zuc had been depleted by RNAi (Zuc KD + Armi KD). Armi‐F WT (green) localized onto mitochondria, whereas Armi‐F N756A mutant (green) localized to Yb bodies (red). The scale bar represents 5 μm. DAPI (blue) shows the nuclei. Right: Another set of cell images of the one shown on the left. The scale bar represents 5 μm. DAPI (blue) shows the nuclei.
- Left: Armi‐Flag (Armi‐F) WT and ΔN34/N756A mutants (green) localized at Yb bodies (red) in Armi‐depleted OSCs (Armi KD). Right: Another set of cell images of the one shown on the left. The scale bar represents 5 μm. DAPI (blue) shows the nuclei.
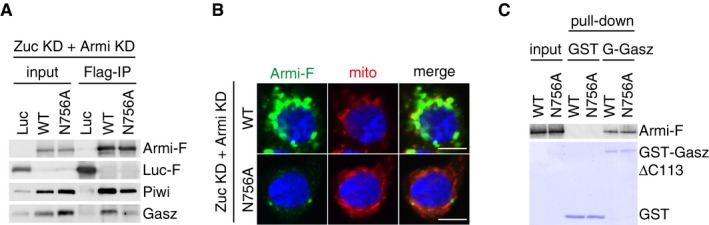
We then explored the behavioral aspects of this mutant in vivo. First, we tested whether the Armi mutant can assemble the trimeric complex with Gasz and Piwi on mitochondria in Zuc‐depleted OSCs. To this end, we expressed Armi‐Flag N756A mutant in OSCs where Zuc and Armi were depleted by RNAi and then performed immunoprecipitation. The Armi mutant associated with Piwi as efficiently as Armi WT did, but the ability to interact with Gasz was greatly reduced (Fig 5A). Immunofluorescence showed that this mutant does not accumulate to mitochondria and localizes to Yb bodies in Zuc‐ and Armi‐lacking OSCs (Figs 5B and EV5C and D). This situation was almost identical to that of Armi WT in Zuc‐ and Piwi‐lacking OSCs (Fig 3A and B). This localization of Armi mutant was not the consequence of Zuc depletion (Fig EV5E). Moreover, recombinant Armi N756A mutant maintained the ability to directly interact with Gasz in in vitro assays (Fig 5C). These results suggest that the localization of Armi itself to and retention at Yb bodies are independent of the RNA‐binding activity of Armi. These factors probably depend markedly on Yb–Armi protein–protein interaction 28.
Figure 5. Translocation of Piwi–pre‐piRISC from Yb bodies requires the RNA‐binding activity of Armi.
- Armi‐Flag (Armi‐F) WT and the N756A mutant were expressed in OSCs where endogenous Armi and Zuc had been depleted by RNAi (Zuc KD + Armi KD). Immunoprecipitation and subsequent Western blotting show that Armi‐F WT bound with Piwi and Gasz. The Armi N756A mutant bound with Piwi strongly but only weakly with Gasz. Luciferase‐Flag (Luc‐F) was used as a negative control.
- Armi‐Flag (Armi‐F) WT and the N756A mutant were expressed in OSCs where endogenous Armi and Zuc had been depleted by RNAi (Zuc KD + Armi KD). Armi‐F WT (green) localized onto mitochondria (red), whereas Armi‐F N756A mutant (green) localized to Yb bodies (see also Fig EV5D). The scale bar represents 5 μm. DAPI (blue) shows the nuclei.
- In vitro pull‐down assays show that both Armi‐Flag (Armi‐F) WT and N756A mutant directly bind with GST‐GaszΔC113 but not with GST. Armi‐F WT and mutant were immunopurified from Schneider 2 (S2) cells under harsh conditions. GST and GST‐GaszΔC113 were visualized by CBB staining, while Armi‐F WT and N756A mutant were detected by Western blotting using anti‐Flag antibodies.
The findings also support the interesting idea that the departure of Piwi–pre‐piRISC from Yb bodies, prior to its relocation to mitochondria, depends on the RNA‐binding activity of Armi but not on the Armi–Piwi interaction. This type of regulation relying on the RNA‐binding activity of Armi is biologically relevant because Armi without RNA‐binding activity would fail to unwind (relax) piRNA precursors for Zuc cleavage, blocking piRNA biogenesis. The regulation would provide major help in avoiding this. Another potential reason for relying on Armi's RNA‐binding activity might be related to Armi's promiscuous RNA binding 31, 33. When Armi is already bound with Piwi–pre‐piRISC via piRNA precursor, after leaving Yb bodies Armi would not be able to bind to other cellular RNAs at random. Thus, the restrictive system to ensure that the Piwi–pre‐piRISC departure from Yb bodies occurs only after Armi associates with Piwi–pre‐piRISC via both Piwi and piRNA precursor is very important for maintaining the integrity of piRNA biogenesis in OSCs.
The Armi mutant defective in RNA binding was able to interact with Piwi through protein–protein interaction (Fig 5A). However, Armi failed to associate with Piwi when the latter was devoid of RNAs (Fig 4C). These findings suggest that conformational change of Piwi occurs before and after its piRNA precursor loading and that Armi is sensitive to the conformational alteration of Piwi.
The present study revealed two requirements for the departure of the Armi–Piwi complex from Yb bodies prior to its relocation to mitochondria: (i) Piwi–piRNA precursor association through the 5′‐end pocket in the Piwi MID domain and (ii) Armi–Piwi–pre‐piRISC association through piRNA precursor. When the assembly is not complete, both Armi and Piwi resist leaving Yb bodies. Considering this along with our earlier notion that Armi is required for Piwi localization to Yb bodies, we claim that Armi determines piRISC processing, from the formation of Piwi–pre‐piRISC to its inter‐organelle translocation, in OSCs.
The N‐terminal end of Armi is essential for Armi–Piwi–pre‐piRISC to depart Yb bodies
Armi is detected as a doublet on Western blots (22; this study). The lower one of the two bands was previously predicted to be an Armi isoform lacking 34 N‐terminal amino acids, on the basis of 5′ RACE analysis 22. Two anti‐Armi monoclonal antibodies, 2F8A9 and 2D6E11, are now available (24; 2D6E11 was produced in the study but remains unreported). Of these, 2F8A9 recognized both Armi bands on Western blots, while 2D6E11 recognized solely the upper band (Fig EV6A). We expressed Armi WT and the ΔN34 mutant (lacking Met1‐Leu34) in endogenous Armi‐lacking OSCs individually and performed probing with the antibodies. 2F8A9 recognized both Armi WT and ΔN34 mutant, while 2D6E11 recognized only WT as expected (Fig EV6B).
Figure EV6. The N‐terminal end of Armi plays an important role in the departure of Piwi–pre‐piRISC from Yb bodies.
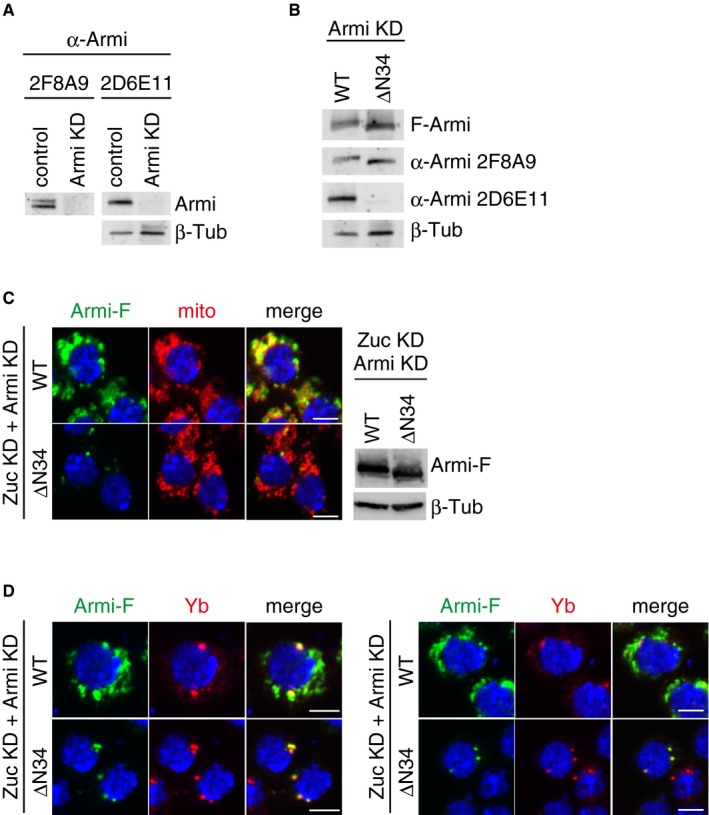
- Anti‐Armi antibody 2F8A9 recognized both Armi bands on Western blots, while 2D6E11 recognized solely the upper band. The signals of 2F8A9 were stripped away, and then, the same membrane was reprobed with 2D6E11. β‐Tubulin (β‐Tub) was detected as a loading control.
- Western blotting showed that 2F8A9 anti‐Armi antibody recognized both Armi‐Flag (Armi‐F) WT and ΔN34 mutant, while 2D6E11 recognized only WT. β‐Tubulin (β‐Tub) was detected as a loading control.
- Left: Another set of cell images of the one shown in Fig 6B. The scale bar represents 5 μm. Right: The expression levels of Armi‐Flag (Armi‐F) WT and ΔN34 mutants in Zuc/Armi‐depleted OSCs (Zuc KD + Armi KD). β‐Tubulin (β‐Tub) was detected as a loading control.
- Left: Armi‐Flag (Armi‐F) WT and the ΔN34 mutant were expressed in OSCs where endogenous Armi and Zuc had been depleted by RNAi (Zuc KD + Armi KD). Armi‐F WT (green) localized onto mitochondria, whereas Armi‐F ΔN34 mutant (green) localized to Yb bodies (red). The scale bar represents 5 μm. DAPI (blue) shows the nuclei. Right: Another set of cell images of the one shown on the left. The scale bar represents 5 μm. DAPI (blue) shows the nuclei.
The Armi ΔN34 mutant failed to restore piRNA production that had been abrogated by the loss of endogenous Armi, as did the Armi N756A mutant, although the ΔN34 mutant showed the ability to bind RNAs unlike the N756A mutant (Fig EV5A and B). Immunoprecipitation and Western blotting showed that the ΔN34 mutant associated with Piwi but not with Gasz (Fig 6A), as did the N756A mutant (Fig 5A). In addition, the ΔN34 mutant mostly localized to Yb bodies in Zuc‐ and Armi‐depleted OSCs (Figs 6B and EV6C and D). These results suggest that the N‐terminal end of Armi is essential for the ability of the Armi–Piwi–pre‐piRISC complex to depart Yb bodies. Another possibility is that Armi loses its ability to be bound with Gasz when the N‐terminus is lacking. However, in in vitro protein–protein interaction assays, recombinant Armi ΔN34 mutant bound with Gasz as efficiently as the WT control did (Fig 6C), ruling out this latter possibility. Interestingly, when we fused a Flag‐tag to the N‐terminus of Armi, instead of the Armi C‐terminus, the Armi–Gasz interaction was impaired (Fig 6D), supporting the importance of the N‐terminus of Armi in the piRISC processing pathway. Although at present the role of the N‐terminus remains unknown, if it were revealed, this would be very helpful to understand in more detail the molecular flow and regulation of the piRISC processing pathway. Such investigation is therefore currently underway in our laboratory.
Figure 6. The N‐terminal end of Armi plays an important role in the departure of Piwi–pre‐piRISC from Yb bodies.
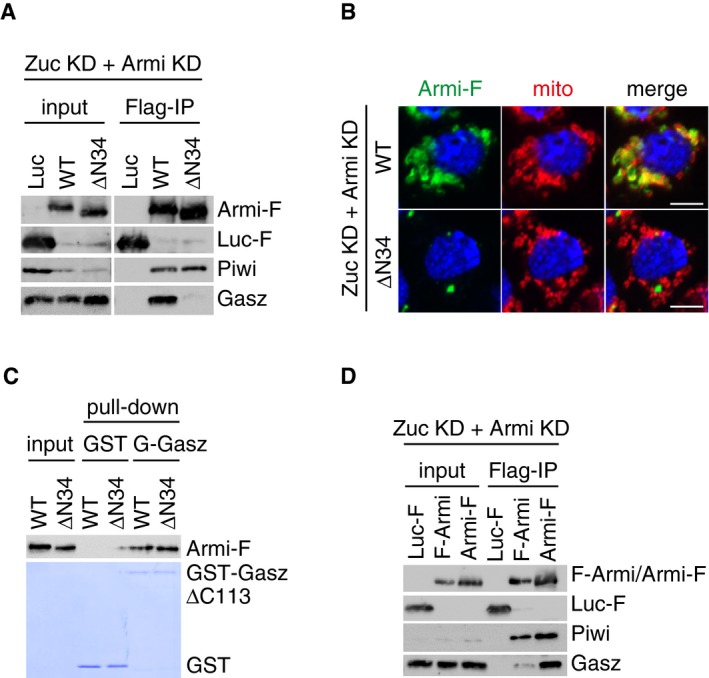
- Armi‐Flag (Armi‐F) WT and the ΔN34 mutant were expressed in OSCs where endogenous Armi and Zuc had been depleted by RNAi (Zuc KD + Armi KD). Immunoprecipitation and subsequent Western blotting show that Armi‐F WT but not Armi ΔN34 mutant bound with Gasz in the cells. Both Armi WT and ΔN34 mutant interacted with Piwi to similar extents. Luciferase‐Flag (Luc‐F) was used as a negative control.
- Armi‐Flag (Armi‐F) WT and the ΔN34 mutant were expressed in OSCs where endogenous Armi and Zuc had been depleted by RNAi (Zuc KD + Armi KD). Armi‐F WT (green) localized onto mitochondria (red), whereas Armi‐F ΔN34 mutant (green) localized to Yb bodies (see also Fig EV6D). The scale bar represents 5 μm. DAPI (blue) shows the nuclei.
- In vitro pull‐down assays show that both Armi‐Flag (Armi‐F) WT and ΔN34 mutant directly bind with GST‐GaszΔC113 but not with GST. Armi‐F WT and mutant were immunopurified from Schneider 2 (S2) cells under harsh conditions. GST and GST‐GaszΔC113 were visualized by CBB staining, while Armi‐F WT and mutant were detected by Western blotting using anti‐Flag antibodies.
- Immunoprecipitation and subsequent Western blotting show that Armi‐Flag (Armi‐F) but not Flag‐Armi (F‐Armi) strongly bound with Gasz in Zuc/Armi‐depleted OSCs (Zuc KD + Armi KD). Piwi bound with both Armi (F‐Armi and Armi‐F). Luciferase‐Flag (Luc‐F) was used as a negative control.
Discussion
In OSCs, transposon‐repressing piRISC formation starts with Yb‐body formation, which is initiated by multivalent cis‐trans association between Yb and flam RNAs 28. Other Yb‐body components are subsequently incorporated into the bodies in a hierarchical manner: Armi goes in first and then the heterodimer composed of SoYb and Vret joins the body structures. We also showed that without Armi, Piwi remains cytosolic and free from piRNAs 24. In the present study, we found that Armi localization to Yb bodies occurs in a Piwi‐independent manner (Fig 7). At present, the biological relevance of this hierarchical regulation is unclear. One possibility is that, without Armi, flam primary RNA transcripts would not be converted to shorter, secondary precursors, reducing the levels of Piwi–pre‐piRISC formed at Yb bodies. The nuclease(s) responsible for roughly digesting the primary RNA transcripts to secondary piRNA precursors remains to be identified. It would be of interest to test whether Armi localization to Yb bodies would indeed be required to produce such secondary flam‐piRNA precursors. If this were the case, the processor creating secondary piRNA precursors (i.e., creating 5′ ends of the precursors for Piwi binding) from primary RNA transcripts may be identified from Armi partners.
Figure 7. The model of dynamics of Armi and Piwi in OSCs.
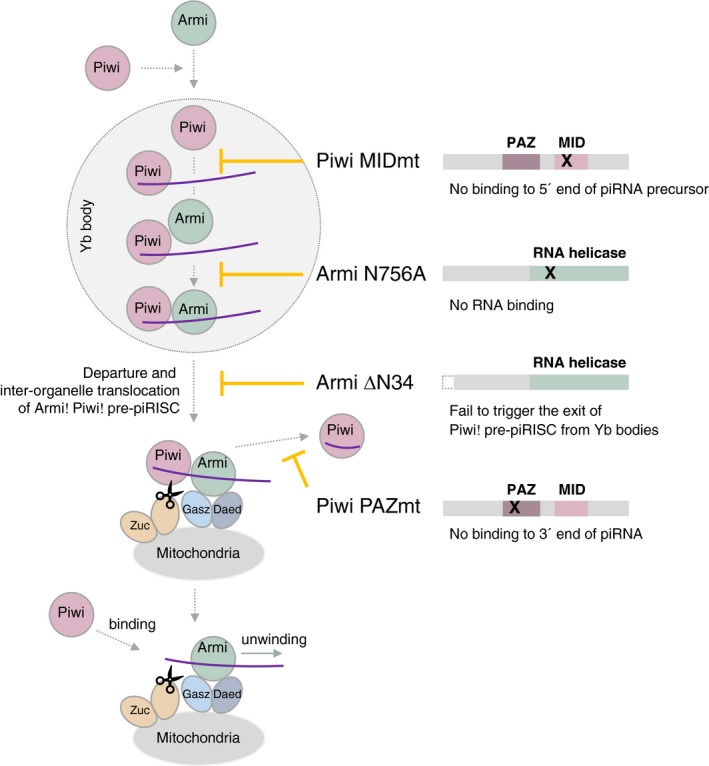
Armi localizes to Yb bodies Piwi independently, but Piwi localization to Yb bodies depends on Armi. At Yb bodies, Piwi is loaded with piRNA precursor (through their 5′ ends) to become pre‐piRISC. Armi joins the complex by binding the downstream region in the precursor and also Piwi. The Armi–Piwi–pre‐piRISC complex then leaves Yb bodies to head to mitochondria for Zuc cleavage. The Armi–Piwi–pre‐piRISC complex is docked onto mitochondria through the Gasz–Daed complex, under which Zuc cleaves the precursor and releases Piwi–piRISC to the cytosol, while Armi returns to Yb bodies for another round of reaction (not shown). Piwi–piRISC is translocated to the nucleus for transposon repression. Nascent Piwi may bind 5′ end of piRNA precursor produced by Zuc cleavage, while the RNA is still bound with Armi. Another Zuc cleavage gives rise to phased piRNAs. The Piwi mutant MIDmt does not bind piRNA precursor at Yb bodies, abrogating the whole processing. Another Piwi mutant PAZmt does bind piRNA precursor at Yb bodies, but after Zuc reaction, the 3′ end of piRNA is not bound by the PAZ domain because of the mutation, and so functional piRISC is not sufficiently produced in the cells. The Armi mutant N756A fails to bind piRNA precursor pre‐bound with Piwi at Yb bodies, leading to the failure of Piwi–pre‐piRISC departure from Yb bodies. Another Armi mutant ΔN34 is able to bind Piwi–pre‐piRISC at Yb bodies but fails to leave the bodies for an unknown reason. The whole processing of Piwi–piRISC formation is well regulated by the interdependence between Armi and Piwi and also the RNA‐binding activity of the two proteins. Functions of SoYb, Vret, Shu, and Mino (not shown) in the pathway remain to be determined.
The RNA‐binding activities of both Armi and Piwi were indispensable for proper Armi–Piwi–pre‐piRISC complex formation and also the exit of the complex from Yb bodies (Fig 7): Armi failed to associate with Piwi when Piwi was devoid of piRNA precursors and neither of these proteins exited the bodies. Even under conditions where Piwi–pre‐piRISC was properly formed at Yb bodies, when Armi lacked RNA‐binding activity, both proteins remained at the bodies. Furthermore, even after Armi successfully bound to Piwi–pre‐piRISC via piRNA precursor, the lack of Armi's 34 N‐terminal residues blocked the release of the complex from the bodies. Why is such multilayered, elaborate regulation required in piRISC processing (Fig 7)? One possible reason is that Yb, but not Armi, Zuc, or even Piwi, has a strong sequence preference in RNA binding 31. To control this situation where the selectivity of piRNA precursor is completely due to Yb, OSCs might employ Yb as the core factor of Yb‐body formation [because of the propensity to form LIPS‐driven multivalent concentrates (Yb bodies) when bound with huge flam RNA transcripts] and decide to use Yb bodies as the site for Armi to induce Piwi–pre‐piRISC formation and quality control before the complex heads to mitochondria where piRISC maturation finally occurs. In this way, the flam‐piRNA precursors are securely bound with Armi and Piwi, achieving the production of transposon‐repressing piRISC.
Materials and Methods
Cell culture
OSCs were cultured at 26°C in Shields and Sang M3 medium (USBiological) supplemented with 10% fetal bovine serum (Funakoshi), 0.6 mg/ml glutathione, 10 mU/ml insulin, and 10% fly extract. The fly extract was prepared as shown previously 28. S2 cells were cultured at 26°C in Schneider's Drosophila medium (Gibco) supplemented with 10% fetal bovine serum (Equitech‐Bio) and 1× penicillin−streptomycin−glutamine (Gibco) 31.
RNAi and plasmid transfection
For RNAi of OSCs, 5.0 × 106 cells were suspended in 20 μl of Solution SF of the Cell Line Nucleofector Kit SF (Lonza Bioscience) with 200–400 pmol siRNAs. Electroporation was performed by the DG‐150 program of a Nucleofector 96‐well Shuttle device (Lonza Bioscience). The sequences of the siRNAs used in this study are shown in Table EV1. For plasmid transfection or plasmid and siRNA co‐transfection, 1.5 × 107 cells were suspended in 100 μl of transfection buffer [180 mM sodium phosphate buffer for Church and Gilbert hybridization (pH 7.2), 5 mM KCl, 15 mM MgCl2, 50 mM D‐mannitol; modified from 38] with 2–10 μg of plasmids and 600–1,200 pmol siRNAs. Electroporation was performed by the N‐020 program of a Nucleofector 2b device (Lonza Bioscience). In the RNAi experiments, cells were transfected with siRNAs twice every 48 h and harvested 48 h later. In the rescue assays, endogenous gene was knocked down by RNAi 48 h before transfection of both plasmids and siRNA, and cells were harvested 48 h later.
Production of anti‐Gasz monoclonal antibody
Recombinant proteins of full‐length Gasz fused to glutathione S‐transferase (GST) (GST‐Gasz) were expressed in and purified from E. coli, and then injected into mice for immunization. The production and selection of hybridomas that produced anti‐Gasz monoclonal antibodies were performed as described previously 39.
Mitochondrial fractionation
Mitochondrial fractionation of OSC lysates was performed as previously reported 40 with some modifications. Briefly, 1.2 × 108 cells were suspended with 500 μl of buffer for first resuspension [30 mM Tris–HCl (pH 7.3), 225 mM D‐mannitol, 75 mM sucrose, 0.05 mM EGTA] and ruptured by passing 30 times through a 30‐gauge needle attached to a 1‐ml syringe. The lysates were centrifuged at 600 g for 5 min, and the pellets of nucleus and cell debris were discarded. This step was repeated twice. The lysates were centrifuged at 7,000 g for 10 min, and the supernatants were collected as the cytoplasmic fraction. The pellets were collected and resuspended with 500 μl of buffer for the second resuspension [30 mM Tris–HCl (pH 7.3), 225 mM D‐mannitol, 75 mM sucrose]. This step was repeated three times at 7,000 g for 10 min and two times at 10,000 g for 10 min. The final pellets were collected as the mitochondrial fraction. Proteins in each fraction were detected by Western blotting. For “mito” blot, we used mitochondria isolated from 5 volumes of total lysates used in the “total” blot.
Plasmid construction
To construct Myc‐Gasz‐, Flag‐Gasz‐, and GST‐Gasz‐expressing plasmids, a full‐length Gasz cDNA was amplified from OSC mRNAs by RT–PCR and cloned into pAcM, pAcF, and pGEX vectors, respectively. Expression vector of Armi‐Flag WT was produced by inverse PCR and infusion using the RNAi‐resistant Flag‐Armi WT vector 31 as a template. Expression vectors of Armi‐Flag mutants (N756A and ΔN34) were generated by inverse PCR using Armi‐Flag WT vector. Armi‐Flag means that a Flag peptide was added to the C‐terminal end of Armi. Flag‐Armi means that a Flag peptide was added to the N‐terminal end of Armi (Figs 6D and EV6B). Expression vectors of Flag‐tagged Piwi mutants (PAZmt and MIDmt) were generated by inverse PCR and infusion, using the RNAi‐resistant Myc‐Piwi PAZmt vector 24 and the RNAi‐resistant Flag‐Piwi WT vector 13 as templates, respectively. To construct Luc‐Flag‐expressing vector, a full‐length luciferase cDNA was amplified from firefly mRNAs by RT–PCR and cloned into pAcF vector. The primers used are summarized in Table EV1.
Western blotting
Western blotting was performed in principle as described previously 41. The primary antibodies used in this study were anti‐Armi monoclonal antibody (2F8A9, 1:1,000 dilution, 24, 2D6E11, supernatant of hybridoma cells), anti‐Piwi monoclonal antibody (3G11, 1:1,000 dilution, 42), anti‐Yb monoclonal antibody (8H12D12, 1:1,000 dilution, 35), anti‐Gasz monoclonal antibody (1:1,000 dilution, this study), anti‐Vret monoclonal antibody (1:1,000 dilution, 28), anti‐SoYb monoclonal antibody (1:1,000 dilution, 28), anti‐β‐tubulin monoclonal antibody (1:1,000 dilution, E7, Developmental Studies Hybridoma Bank), anti‐HSP60 monoclonal antibody (1:2,000 dilution, LK1, StressMarq Biosciences), anti‐Myc monoclonal antibody (9E10, 1:1,000 dilution, Fujifilm Wako Pure Chemical), and anti‐Flag monoclonal antibody (FLA‐1, 1:10,000 dilution, Medical & Biological Laboratories). Peroxidase‐conjugated anti‐mouse IgG antibodies (1:5,000 dilution, Cappel) were used as secondary antibodies. The signal intensity of bands was quantified by ImageJ (National Institutes of Health). All the experiments were performed at least three times to draw each conclusion.
Immunofluorescence analysis
Immunofluorescence analysis was essentially performed as described previously 24. Primary antibodies used in this study were anti‐Yb monoclonal antibody (8H12D12, 1:200 dilution, 1G2H3, 1:100 dilution, 35), anti‐Armi monoclonal antibody (2F8A9, 1:200 dilution), anti‐Piwi monoclonal antibody (4D2, 1:200 dilution, 42), and anti‐Flag monoclonal antibody (FLA‐1, 1:5,000 dilution). The secondary antibodies used in this study were Alexa 488‐conjugated anti‐mouse IgG1, Alexa 488‐conjugated anti‐mouse IgG2a, Alexa 488‐conjugated anti‐mouse IgG2b, Alexa 555‐conjugated anti‐mouse IgG1, Alexa 555‐conjugated anti‐mouse IgG2a, Alexa 555‐conjugated anti‐mouse IgG2b, and Alexa 633‐conjugated anti‐mouse IgG2a (all from Thermo Fisher Scientific). All secondary antibodies were used at 1:1,000 dilution. Before fixation, the cells were incubated with MitoTracker Orange CMXRos (1:2,000 dilution, Invitrogen) for 15 min at 26°C. The cells were mounted with VECTASHIELD Mounting Medium with DAPI (Vector Laboratories), followed by observation using an LSM 710 laser scanning confocal microscope (Carl Zeiss). More than 15 cells were analyzed, and representative images are shown.
Immunoprecipitation
Immunoprecipitation was primarily performed as described previously 24. Cells were suspended with binding buffer [30 mM HEPES‐KOH (pH 7.3), 150 mM KOAc, 5 mM Mg(OAc)2, 5 mM DTT, 1.0% Triton X‐100, 2 μg/ml leupeptin, 2 μg/ml pepstatin A, 0.5% aprotinin] and ruptured by passing 10 times through a 30‐gauge needle attached to a 1‐ml syringe. Cell lysate was incubated with antibodies bound to Dynabeads Protein G (Thermo Fisher Scientific) at 4°C for 1 h. The beads were washed five times with binding buffer. Eluted proteins were separated on SDS–polyacrylamide gels and detected by Western blotting (see above).
Northern blotting
Northern blotting was carried out as described previously 43. Total RNAs were isolated from OSCs using ISOGEN II (Fujifilm Wako Pure Chemical). RNAs in immunoprecipitated complex were eluted from beads with phenol–chloroform and precipitated with ethanol. The oligodeoxynucleotides used as probes are shown in Table EV1. All the experiments were performed at least three times to draw each conclusion.
Recombinant proteins
Purification of GST‐fused proteins was mainly performed as described previously 39. Briefly, GST‐fused proteins were expressed in E. coli and were resuspended with lysis buffer [20 mM Tris–HCl (pH 7.5), 500 mM NaCl, 1.0% Triton X‐100, 1× cOmplete™ ULTRA Tablets, EDTA‐free, glass vials Protease Inhibitor Cocktail (Sigma), 1 mM DTT] and lysed by sonication. The supernatants after centrifugation were bound to Glutathione Sepharose 4B (GE Healthcare) and washed five times with lysis buffer. The proteins were eluted with elution buffer [20 mM Tris–HCl (pH 9.0), 150 mM NaCl, 1.0% Triton X‐100, 1 mM DTT, 10 mM glutathione, 10% glycerol]. The eluted proteins were dialyzed in dialysis buffer [20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM DTT, 10% glycerol]. Recombinant Armi protein was purified as previously described 31.
Gel shift assay
Gel shift assays were carried out as previously described 31.
GST pull‐down assay
GST pull‐down assay was performed as previously reported 44 with some modifications. Briefly, GST‐fused proteins were bound to Glutathione Sepharose 4B in binding buffer [30 mM HEPES‐KOH (pH 7.3), 150 mM KOAc, 5 mM Mg(OAc)2, 5 mM DTT, 1.0% Triton X‐100, 2 μg/ml leupeptin, 2 μg/ml pepstatin A, 0.5% aprotinin] by rotation at 4°C for 1 h. Beads were washed with high‐salt wash buffer [30 mM HEPES‐KOH (pH 7.3), 500 mM NaCl, 150 mM KOAc, 5 mM Mg(OAc)2, 5 mM DTT, 1.0% Triton X‐100, 2 μg/ml leupeptin, 2 μg/ml pepstatin A, 0.5% aprotinin] twice and with binding buffer twice. The beads and Armi‐Flag proteins were rotated at 4°C for 1 h. Beads were washed with wash buffer four times. Proteins were eluted with SDS sample buffer, separated on SDS–polyacrylamide gels, and detected by Western blotting or CBB staining.
Author contributions
HY performed all experiments with help from MN and TK. HO produced anti‐Gasz antibodies. HY, TK, HI, and MCS designed the experiments, and analyzed and interpreted the data. HY and MCS wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Review Process File
Acknowledgements
We thank S. Hirakata and K. Saito for comments on this manuscript and technical support, respectively. We also thank all other members of the Siomi laboratory at the University of Tokyo. This study was supported by research grants from MEXT for M.C.S. (19H05466) and H.I. H.Y. is supported by the Japan Society for the Promotion of Science. T.K. is supported by the Graduate Program for Leaders in Life Innovation.
EMBO Reports (2020) 21: e48769
References
- 1. Czech B, Munafò M, Ciabrelli F, Eastwood EL, Fabry MH, Kneuss E, Hannon GJ (2018) piRNA‐guided genome defense: from biogenesis to silencing. Annu Rev Genet 52: 131–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ozata DM, Gainetdinov I, Zoch A, O'Carroll D, Zamore PD (2019) PIWI‐interacting RNAs: small RNAs with big functions. Nat Rev Genet 20: 89–108 [DOI] [PubMed] [Google Scholar]
- 3. Yamashiro H, Siomi MC (2018) PIWI‐interacting RNA in Drosophila: biogenesis, transposon regulation, and beyond. Chem Rev 118: 4404–4421 [DOI] [PubMed] [Google Scholar]
- 4. Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA (2001) Double‐stranded RNA‐mediated silencing of genomic tandem repeats and transposable elements in the melanogaster melanogaster germline. Curr Biol 11: 1017–1027 [DOI] [PubMed] [Google Scholar]
- 5. Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H (1998) A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self‐renewal. Genes Dev 12: 3715–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM et al (2009) Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137: 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ (2007) Discrete small RNA‐generating loci as master regulators of transposon activity in Drosophila . Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- 8. Zanni V, Eymery A, Coiffet M, Zytnicki M, Luyten I, Quesneville H, Vaury C, Jensen S (2013) Distribution, evolution, and diversity of retrotransposons at the flamenco locus reflect the regulatory properties of piRNA clusters. Proc Natl Acad Sci USA 110: 19842–19847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goriaux C, Desset S, Renaud Y, Vaury C, Brasset E (2014) Transcriptional properties and splicing of the flamenco piRNA cluster. EMBO Rep 15: 411–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Prud'homme N, Gans M, Masson M, Terzian C, Bucheton A (1995) Flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster . Genetics 139: 697–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC (2009) A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila . Nature 461: 1296–1299 [DOI] [PubMed] [Google Scholar]
- 12. Sato K, Siomi MC (2018) Two distinct transcriptional controls triggered by nuclear Piwi‐piRISCs in the Drosophila piRNA pathway. Curr Opin Struct Biol 53: 69–76 [DOI] [PubMed] [Google Scholar]
- 13. Yashiro R, Murota Y, Nishida KM, Yamashiro H, Fujii K, Ogai A, Yamanaka S, Negishi L, Siomi H, Siomi MC (2018) Piwi nuclear localization and its regulatory mechanism in Drosophila ovarian somatic cells. Cell Rep 23: 3647–3657 [DOI] [PubMed] [Google Scholar]
- 14. Czech B, Preall JB, McGinn J, Hannon GJ (2013) A Transcriptome‐wide RNAi screen in the Drosophila ovary reveals factors of the germline piRNA pathway. Mol Cell 50: 749–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Han BW, Wang W, Li C, Weng Z, Zamore PD (2015) piRNA‐guided transposon cleavage initiates Zucchini‐dependent, phased piRNA production. Science 348: 817–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Handler D, Olivieri D, Novatchkova M, Gruber FS, Meixner K, Mechtler K, Stark A, Sachidanandam R, Brennecke J (2011) A systematic analysis of Drosophila TUDOR domain‐containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. EMBO J 30: 3977–3993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Handler D, Meixner K, Pizka M, Lauss K, Schmied C, Gruber F, Brennecke J (2013) The genetic makeup of the Drosophila piRNA pathway. Mol Cell 50: 762–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ipsaro JJ, Haase AD, Knott SR, Joshua‐tor L, Hannon GJ (2012) The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature 491: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mohn F, Handler D, Brennecke J (2015) piRNA‐guided slicing specifies transcripts for Zucchini‐dependent, phased piRNA biogenesis. Science 348: 812–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Munafò M, Manelli V, Falconio FA, Sawle A, Kneuss E, Eastwood EL, Seah JWE, Czech B, Hannon GJ (2019) Daedalus and Gasz recruit Armitage to mitochondria, bringing piRNA precursors to the biogenesis machinery. Genes Dev 33: 844–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani MK, Bonnefond L, Matsumoto N, Nishizawa T, Nakanaga K, Aoki J et al (2012) Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature 491: 284–287 [DOI] [PubMed] [Google Scholar]
- 22. Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J (2010) An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila . EMBO J 29: 3301–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Olivieri D, Senti K‐A, Subramanian S, Sachidanandam R, Brennecke J (2012) The cochaperone Shutdown defines a group of biogenesis factors essential for all piRNA populations in Drosophila . Mol Cell 47: 954–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, Siomi H, Siomi MC (2010) Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila . Genes Dev 24: 2493–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Szakmary A, Reedy M, Qi H, Lin H (2009) The Yb protein defines a novel organelle and regulates male germline stem cell self‐renewal in Drosophila melanogaster . J Cell Biol 185: 613–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vagin VV, Yu Y, Jankowska A, Luo Y, Wasik KA, Malone CD, Harrison E, Rosebrock A, Wakimoto BT, Fagegaltier D et al (2013) Minotaur is critical for primary piRNA biogenesis. RNA 19: 1064–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zamparini AL, Davis MY, Malone CD, Vieira E, Zavadil J, Sachidanandam R, Hannon GJ, Lehmann R (2011) Vreteno, a gonad‐specific protein, is essential for germline development and primary piRNA biogenesis in Drosophila . Development 138: 4039–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hirakata S, Ishizu H, Fujita A, Tomoe Y, Siomi MC (2019) Requirements for multivalent Yb body assembly in transposon silencing in Drosophila . EMBO Rep 20: e47708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Homolka D, Pandey RR, Goriaux C, Brasset E, Vaury C, Sachidanandam R, Fauvarque M, Pillai RS (2015) PIWI slicing and RNA elements in precursors instruct directional primary piRNA biogenesis. Cell Rep 12: 418–428 [DOI] [PubMed] [Google Scholar]
- 30. Ishizu H, Iwasaki YW, Hirakata S, Ozaki H, Iwasaki W, Siomi H, Siomi MC (2015) Somatic primary piRNA biogenesis driven by cis‐acting RNA elements and trans‐acting Yb. Cell Rep 12: 429–440 [DOI] [PubMed] [Google Scholar]
- 31. Ishizu H, Kinoshita T, Hirakata S, Komatsuzaki C, Siomi MC (2019) Distinct and collaborative functions of Yb and Armitage in transposon‐targeting piRNA Biogenesis. Cell Rep 27: 1822–1835 [DOI] [PubMed] [Google Scholar]
- 32. Pandey RR, Homolka D, Chen KM, Sachidanandam R, Fauvarque MO, Pillai RS (2017) Recruitment of Armitage and Yb to a transcript triggers its phased processing into primary piRNAs in Drosophila ovaries. PLoS Genet 13: e1006956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ge DT, Wang W, Tipping C, Gainetdinov I, Weng Z, Zamore PD (2019) The RNA‐binding ATPase, Armitage, couples piRNA amplification in nuage to phased piRNA production on mitochondria. Mol Cell 74: 982–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Niki Y, Yamaguchi T, Mahowald AP (2006) Establishment of stable cell lines of Drosophila germ‐line stem cells. Proc Natl Acad Sci USA 103: 16325–16330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Murota Y, Ishizu H, Nakagawa S, Iwasaki YW, Shibata S, Kamatani MK, Saito K, Okano H, Siomi H, Siomi MC (2014) Yb Integrates piRNA intermediates and processing factors into perinuclear bodies to enhance piRISC assembly. Cell Rep 8: 103–113 [DOI] [PubMed] [Google Scholar]
- 36. Matsumoto N, Nishimasu H, Sakakibara K, Nishida KM, Hirano T, Ishitani R, Siomi H, Siomi MC, Nureki O (2016) Crystal structure of Silkworm PIWI‐clade Argonaute Siwi bound to piRNA. Cell 167: 484–497 [DOI] [PubMed] [Google Scholar]
- 37. Chakrabarti S, Jayachandran U, Bonneau F, Fiorini F, Basquin C, Domcke S, Le Hir H, Conti E (2011) Molecular mechanisms for the RNA‐dependent ATPase activity of Upf1 and its regulation by Upf2. Mol Cell 41: 693–703 [DOI] [PubMed] [Google Scholar]
- 38. Nye J, Buster DW, Rogers Gregory C (2014) The use of cultured Drosophila cells for studying the microtubule cytoskeleton. Methods Mol Biol 1136: 81–101 [DOI] [PubMed] [Google Scholar]
- 39. Nishida KM, Iwasaki YW, Murota Y, Nagao A, Mannen T, Kato Y, Siomi H, Siomi MC (2015) Respective functions of two distinct Siwi complexes assembled during PIWI‐interacting RNA biogenesis in Bombyx germ cells. Cell Rep 10: 193–203 [DOI] [PubMed] [Google Scholar]
- 40. Wieckowski MRMR, Giorgi C, Lebiedzinska M, Duszynski J, Pinton P (2009) Isolation of mitochondria‐associated membranes and mitochondria from animal tissues and cells. Nat Protoc 4: 1582–1590 [DOI] [PubMed] [Google Scholar]
- 41. Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC (2005) Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev 19: 2837–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC (2006) Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev 20: 2214–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Saito K, Ishizuka A, Siomi H, Siomi MC (2005) Processing of pre‐microRNAs by the Dicer‐1‐Loquacious complex in Drosophila cells. PLoS Biol 3: e235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nguyen TN, Goodrich JA (2006) Protein‐protein interaction assays: eliminating false positive interactions. Nat Methods 3: 135–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Table EV1
Review Process File


