Abstract
Bacillus licheniformis strain SSA 61, originally isolated from Sambhar salt lake, was observed to grow even in the presence of 25 % salt stress. Osmoadaptive mechanisms of this halotolerant B. licheniformis strain SSA 61, for long-term survival and growth under salt stress, were determined. Proline was the preferentially accumulated compatible osmolyte. There was also increased accumulation of antioxidants ascorbic acid and glutathione. Among the different antioxidative enzymes assayed, superoxide dismutase played the most crucial role in defense against salt-induced stress in the organism. Adaptation to stress by the organism involved modulation of cellular physiology at various levels. There was enhanced expression of known proteins playing essential roles in stress adaptation, such as chaperones DnaK and GroEL, and general stress protein YfkM and polynucleotide phosphorylase/polyadenylase. Proteins involved in amino acid biosynthetic pathway, ribosome structure, and peptide elongation were also overexpressed. Salt stress-induced modulation of expression of enzymes involved in carbon metabolism was observed. There was up-regulation of a number of enzymes involved in generation of NADH and NADPH, indicating increased cellular demand for both energy and reducing power.
Introduction
Sambhar lake, India’s largest inland salt lake is situated west of Jaipur, Rajasthan, and forms a vast saline wetland. It is one of the largest inland saline depressions in western desert of India and has been declared a Ramsar site (wet-land of international importance) in 1990 due to its biological and biotic importance [19]. The total salt content of Sambhar lake brines was reported to range between 7 % and more than 30 % [31]. The main microflora observed have been Dunaliella sp. (green alga), certain cyanobacteria, photosynthetic purple bacteria, and halophilic archaebacteria [18, 31]; forms of life higher than protozoa have not been reported.
Saline habitats like salt lakes, coastal lagoons, or man-made salterns are predominantly inhabited by prokaryotic microorganisms, especially bacteria, which have adapted to these ecosystems. Most bacteria are unable to grow in the extreme environments of high salinity as found in Sambhar lake. However, some bacteria are able to survive under these extreme environments. Gram-positive bacteria are well represented in saline habitats, and members of the genera Bacillus and Micrococcus are dominant among other Gram-positive bacteria in saline soils [20].
Bacteria in these extreme environments are subject to fluctuations in salinity concentration due to alterations in soil moisture levels, and the microbes adapt themselves to changes in their environment by modulating their osmolyte concentration. These organisms also undergo physiological adaptations for better survival in the stressed environment. Osmoadaptive mechanisms in the halophilic bacteria have been extensively studied [28]. These prokaryotes possess physiologies adapted to a high saline environment and the intracellular machinery functions in presence of high salt. In contrast, the halotolerant bacteria grow well in the absence of salt; however, these are able to tolerate high salt concentrations. A number of physiological changes occur in a bacterium on exposure to salt stress such as increased uptake of potassium, accumulation of compatible osmolytes, and antioxidants, which enables it to adapt to growth under high salt concentration and to withstand adverse environmental conditions [16]. From a physiological point of view, the cellular adaptation and osmotic adjustment to stress are crucial factors that determine growth and survival of bacteria in the stressed environment. Stress responses are also characterized by induction/differential expression of general proteins involved in various metabolic pathways such as glycolysis, TCA cycle, and specific proteins involved in stress adaptation like chaperones DnaK and members of SigB regulon [25]. Antioxidative enzymes also play a crucial role in the survival of bacteria in the stressed environment [5].
Comprehensive knowledge about the uptake and intracellular synthesis of compatible osmolytes and enzymes involved therein, and physiological proteomics in response to salt stress is available for Bacillus subtilis [16]; however, limited information is available on the modulation of cellular physiologies for long-term adaptation to salt stress for Bacillus licheniformis. Shroeter et al. [27] have carried out in-depth proteomic and transcriptomic analysis on short-term response, for survival, of exponentially growing moderately halotolerant B. licheniformis on sudden exposure to salt stress over a time interval of 2 h. In the present investigation, we have allowed an extremely halotolerant B. licheniformis strain SSA 61 to grow in presence of salt stress, and made an effort to gain insight into the osmoadaptive mechanisms and long-term cellular adjustments occurring for survival and growth in presence of salt stress. Accumulation of compatible osmolytes and antioxidants, and up-regulation of cellular proteins under exposure to salt stress were investigated. An attempt was also made to identify the cellular proteins up-regulated under exposure to salt stress by B. licheniformis strain SSA 61. Oxidative stress is secondary stress response caused by the salt stress, hence, induction of antioxidative enzymes was also studied.
Materials and Methods
Effect of Salt on Growth
Salt-tolerant B. licheniformis strain SSA 61 (GenBank accession No. KF318306) isolated from Sambhar lake sediments was screened for tolerance to higher salt concentration in nutrient broth supplemented with 1 % glucose and NaCl concentration ranging from 5 to 25 %. Appropriate controls and three replications per treatment were maintained. The effect of salt on bacterial growth was determined by protein estimation by Lowry’s method [22].
Biochemical Characterization
Bacillus licheniformis strain SSA 61 was characterized for cell morphology, gram staining, spore formation, endospore production, and motility. It was biochemically characterized for oxidase, catalase, and urease activities; lipid, starch, gelatin and casein hydrolysis; carbohydrate fermentation test; ammonia production from peptone; indole and H2S production; nitrate reduction and citrate utilization as per standard methods [7].
Accumulation of Compatible Osmolytes and Antioxidants
Bacillus licheniformis strain SSA 61 was grown in nutrient broth supplemented with 1 % glucose and 10 % NaCl on an incubator shaker at 30 °C for 3 days. After incubation, the cells were pelleted and then lysed using 20 % TCA [15]. The supernatant obtained was analyzed for compatible osmolytes and antioxidants.
Intracellular amino acids were estimated by the method of Chen et al. [7] using ninhydrin reagent for color development. Proline was estimated by the method of Bates et al. [3]. Glycine betaine was measured by the method of Grieve and Grattan [12]. Reduction of dinitrophenylhydrazine by ascorbic acid to phenyl hydrazone in acidic medium was monitored at 530 nm using Perkin Elmer spectrophotometer, model Lambda E2201 for estimation of ascorbic acid [23]. Total glutathione in the supernatant was estimated by the method of Griffith [13].
Antioxidative Enzymes
Bacillus licheniformis strain SSA 61 was grown in nutrient broth supplemented with 1 % glucose and 10 % NaCl on an incubator shaker at 30 °C for 3 days. The cells were pelleted, resuspended in 5 ml of appropriate buffer, kept in an ice bath, and sonicated by giving total 10 strokes, each pulse of 30 s with gap of 45 s at output 80 using ultrasonicator model B. Braun Labsonic®U.
For superoxide dismutase (SOD), catalase, glutathione reductase, and peroxidase, 0.1 M phosphate buffer (pH 7.5) containing 0.5 mM EDTA was used. For ascorbate peroxidase, the above mentioned buffer was supplemented with 1 mM ascorbic acid.
SOD activity was assayed by the method of Dhindsa et al. [9] by monitoring the decrease in optical density at 560 nm due to inhibition of photochemical reduction of nitro-blue tetrazolium (NBT); 50 % decrease in absorbance as compared to a blank was expressed as one unit (U) of enzyme activity. Catalase activity was assayed by monitoring the decrease in optical density due to reduction of H2O2 [2]. Enzyme activity was expressed in terms of amount of H2O2 reduced/min/mg protein. Glutathione reductase activity was assayed by monitoring the increase in optical density at 412 nm [28]. The activity was expressed as Δ increase in absorbance/min/mg protein. Peroxidase activity was assayed using orthodianisidine as a substrate and monitoring the increase in optical density [10]. U of enzyme activity was defined as the amount of enzyme, which caused a change in the absorbance of 0.1/ min (OD/min). Ascorbate peroxidase activity was assayed by monitoring the decrease in absorbance of ascorbic acid at 290 nm due to oxidation of ascorbic acid to monodehydroascorbic acid and dehydroascorbic acid [24]. Enzyme activity was expressed in terms of the amount of ascorbic acid oxidized/min/mg protein. Appropriate controls were maintained in all the enzyme assays.
SDS-PAGE
Bacillus licheniformis strain SSA 61 was grown in nutrient broth supplemented with 1 % glucose, and 10 % and 15 % NaCl on an incubator shaker at 30 °C. The cells were lysed by sonication as described earlier. The whole-cell protein profiles were examined by SDS-PAGE (12 % polyacrylamide). Hundred ng of proteins of each sample along with unstained protein molecular marker (Fermentas, USA) was loaded on the gel and run on a midi gel electrophoresis at 100 V for 4 h and stained with Coomassie Brilliant Blue G 250 [21].
LC-Tandem MS Analysis
The differentially expressed protein bands were excised and subjected to LC–MS/MS analysis. The coomassie-stained gels were destained with a 1:1 mixture of 100 mM NH3HCO3:Acetonitrile. The proteins were then reduced ingel with tris-(2-carboxyethyl)-phosphine, alkylated with iodocetamide, and digested with trypsin (Sequence grade modified trypsin, 12.5 ng/lg; Promega, Madison, WI, USA). The resulting peptides were eluted from the gel pieces by extracting with equal parts 25 mM ammonium bicarbonate and acetonitrile, and then equal parts 5 % v/v formic acid and acetonitrile. The pooled extracts were concentrated on a centrifugal evaporator to an approximate volume of 2–5 μL. To the concentrated tryptic peptides, 15 μL of 5 % v/v formic acid was added, and the peptide samples were sequenced by data-dependent liquid chromatography (LC)-tandem mass spectrometry (MS/MS) with an LC Packings Nano HPLC system and an ABI Qstar XL Q-TOF mass spectrometer. The samples were loaded onto a reverse phase trap column (Acclaim PepMap 100 C18, 300 lm i.d. × 15 mm length) using a programmed autosampler. The trap column was washed with 0.1 % formic acid in 95 % water/5 % acetonitrile for desalting at a flow rate of 30 μL/min. After 3 min, the 10-port switching valve changed position such that the nanoflow pump back-flushed the trap column, and the flow was directed to a capillary C18 column (Acclaim PepMap 100 C18, 75 μm i.d. × 15 cm length) at a flow rate of 250 nL/ min. For the LC separation, the mobile phases consisted of A (0.1 % v/v formic acid in 95 % water/5 % acetonitrile) and B (0.1 % formic acid in 90 % acetonitrile/10 % water). After switching the valve, the gradient program held the mobile phase at 0 % B for 3 min, followed first by a linear gradient to 20 % B over 2 min, and then second by a linear gradient to 75 % B over 25 min. A clean-up at 100 % B for 5 min and re-equilibration of the column at 0 % B for 35 min concluded each run. The capillary column was connected to a nanoflow electrospray ion source and a mass spectrometer which collected tandem mass spectra for the entire 70 min run.
For the MS/MS data analysis, each run was set to acquire a full scan between 100 and 2,000 m/z followed by three MS/MS scans between 75 and 2,000 m/z of the top three most intense ions from the preceding MS scan. To ensure that the same high-abundance ions were not continually analyzed, ions were set to be excluded from MS/ MS scans for 0.5 min after they were analyzed. LC–MS/ MS data were processed with Analyst QS software to generate a Mascot Generic File for Mascot search. The database searches were carried out allowing a 2.0 Da (monoisotopic) peptide mass tolerance and a 0.8 Da (monoisotopic) fragment ion mass tolerance. The peptide charge was set to 2. Variable modification for oxidation of methionine residues was included. An allowance was made for up to two missed cleavages by trypsin.
Results and Discussion
Sambhar is inland salt lake and its waters have been used for centuries to make salt. Such saline environments are inhabited mainly by halophilic eubacteria and archaebacteria [34]. The bacterial communities may also include halotolerant bacteria which have become adapted to these extreme environments. In the present investigation, the B. licheniformis strain SSA 61 was observed to be a halotolerant bacterium which had become adapted to the saline environment.
Biochemical and Growth Characterization
Bacillus licheniformis strain SSA 61 was characterized based on different biochemical tests. The organism was Gram-positive, rod shaped, spore former, motile, catalase, and oxidase positive (Table 1). It was able to utilize dextrose and lactose with gas formation; however, no gas production was observed during utilization of sucrose. It was also able to utilize citrate. It was able to hydrolyze starch, casein, and lipids but was negative for other biochemical tests such as nitrate reduction, H2S, ammonia and indole production, urease activity, and gelatin hydrolysis.
Table 1.
Morphological and biochemical characterization of Bacillus licheniformis strain SSA 61
| Tests | Characters | Tests | Characters |
|---|---|---|---|
| Cell shape | Rod | Starch hydrolysis | Positive |
| Gram reaction | Gram-positive | Gelatin hydrolysis | Negative |
| Spore formation | Positive | Casein hydrolysis | Positive |
| Motility | Positive | Lipid hydrolysis | Positive |
| Catalase | Positive | H2S production | Negative |
| Oxidase | Positive | Ammonia from peptone | Negative |
| Dextrose utilization | Positive, gas formation | Nitrate reduction | Negative |
| Sucrose utilization | Positive, no gas formation | Indole production | Negative |
| Lactose utilization | Positive, gas formation | Citrate utilization | Positive |
| Urease | Negative |
Effect of Salt on Growth
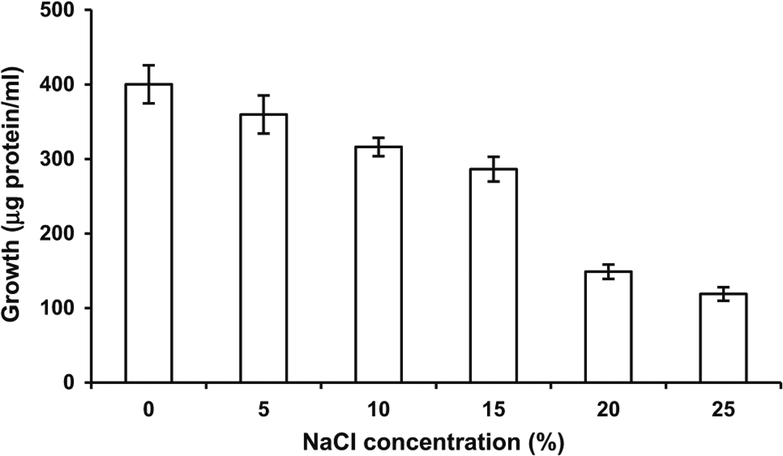
The culture was able to grow at salt concentrations ranging from 5 to 25 %, however, it showed higher growth in the absence of salt (Fig. 1). A decrease in bacterial growth was observed with an increase in salt concentration.
Fig. 1.
Effect of salt concentration on growth of Bacillus licheniformis strain SSA 61
Accumulation of Compatible Osmolytes and Antioxidants
There was not much change in total amino acids accumulated under control conditions and when the culture was exposed to salt stress during growth (Table 2), however, higher accumulation of proline and glycine betaine was observed under salt stress conditions. Although increased accumulation of glycine betaine (nearly 1.5 times) was observed under salt stress conditions, there was tremendous (nearly 5 times) increase in proline concentration within the bacterial cell, indicating it to be the preferential compatible osmolyte. Most of the halotolerant bacteria have evolved a number of adaptive processes, which help in osmotic adjustment for these to be able to survive such harsh conditions. To counterbalance osmotic difference, the salt-tolerant bacteria accumulate compatible solutes such as amino acids, proline, trehalose, and glycine betaine, which do not interfere with cell metabolism [28]. In B. licheniformis strain DSM 13T and B. subtilis, accumulation of these organic solutes under salt stress was due to both de novo synthesis and uptake from the environment [16, 27]. In the present investigation, since a rich medium was used to grow B. licheniformis strain SSA 61, it could not be ascertained whether increased accumulation was due to increased uptake, de novo synthesis or both.
Table 2.
Effect of higher concentration of salt on accumulation of compatible osmolytes and antioxidants by Bacillus licheniformis strain SSA 61
| Treatments | Amino acid (μg/mg protein) | Proline (μmoles/mg protein) | Glycine betaine (μg/mg protein) | Ascorbic acid (μmoles/mg protein) | Glutathione (μmoles/mg protein) |
|---|---|---|---|---|---|
| Control | 18.03 ± 1.08 | 24.59 ± 1.85 | 8.69 ± 0.92 | 258.0 ± 6.4 | 6.66 ± 0.46 |
| 10 % NaCl | 17.87 ± 0.03 | 121.28 ± 11.35 | 13.91 ± 0.85 | 970.6 ± 14.23 | 29.17 ± 2.45 |
Nearly fourfold increase in antioxidants ascorbic acid and glutathione was also observed under salt stress. The non-enzymatic antioxidants ascorbic acid and glutathione are known to play an important role in overcoming environmental stresses [6].
Induction of Antioxidative Enzymes
An increased induction of ascorbate peroxidase, glutathione reductase, and SOD activities was observed (Table 3), when the culture was exposed to salt stress during growth, with the highest induction observed for SOD. However, SOD activity was not detected in cells grown under control conditions. Up-regulation of oxidative stress-related proteins during exposure to abiotic stresses such as salt stress has been previously reported for B. subtilis and B. cereus [8, 16]. Due to environmental stresses, there is a build-up of reactive oxygen intermediates, and under these conditions B. licheniformis has been observed to up-regulate SOD protein production [4, 32]. SOD enzyme plays a crucial role in defense against the toxic effects of reactive oxygen intermediates produced, by scavenging them. There was a decrease in catalase and peroxidase enzyme activities under stress conditions.
Table 3.
Effect of salt stress on antioxidative enzyme activity of Bacillus licheniformis strain SSA 61
| Treatments | Antioxidative enzyme activity | ||||
|---|---|---|---|---|---|
| Superoxide dismutase (U/mg protein) | Catalase (μmoles/min/mg protein) | Peroxidase (mmoles/min/mg protein) | Ascorbate peroxidase (μmoles/min/mg protein) | Glutathione reductase (μmoles/min/mg protein) | |
| Control | ND | 1.28 ± 0.14 | 0.096 ± 0.006 | 0.563 ± 0.016 | 0.083 ± 0.002 |
| 10 % NaCl | 8.35 ± 1.17 | 0.7 ± 0.06 | ND | 0.68 ± 0.001 | 0.13 ± 0.02 |
Differential Protein Expression
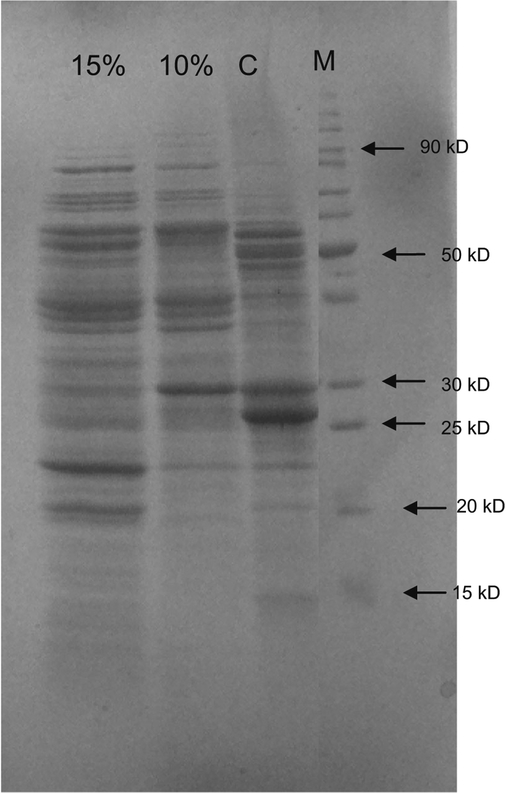
In order to determine the changes in protein expression by the B. licheniformis strain SSA 61, we examined the protein profiles obtained under control conditions and after exposure to two levels of salt stress (10 and 15 %) during growth. The protein profile of the bacterium grown under 10 % salt stress conditions showed up-regulation of sixteen and down-regulation of four proteins (Fig. 2; Table 4) as compared to control conditions. When the bacterium was grown under still higher salt stress of 15 %, there was up-regulation of three more proteins (19 proteins in total) as compared to 10 % salt stress conditions. One protein present under control and 10 % salt stress conditions was absent when B. licheniformis strain SSA 61 was exposed to 15 % salt stress during growth. One protein was observed to be newly induced under 15 % salt stress. Protein Identification and Sequencing by LC-Tandem MS
Fig. 2.
SDS-PAGE showing the effect of NaCl concentration on soluble cellular protein profile of Bacillus licheniformis strain SSA 61 exposed to salt stress
Table 4.
Profile of soluble salt stress proteins in Bacillus licheniformis SSA61
| s. no. | Character of salt stress protein | Salt concentration (%) | Molecular weight (kDa) |
|---|---|---|---|
| 1 | Newly induced | 15 | 35 |
| 2 | Repressed | 10 | 24, 46, 47, 48 |
| 15 | 29 | ||
| 3 | Overexpressed | 10 | 15, 16, 19, 25, 33, 34, 37, 39, 41, 43, 56, 66, 70, 79, 84, 87 |
| 15 | 20, 21, 22 |
The different protein bands showing considerable up-regulation when B. licheniformis strain SSA 61 was exposed to salt stress, during growth, were excised from 1-D SDS-PAGE and identified by LC–MS/MS analysis. All the overexpressed proteins shared highest identity with known and identified proteins from B. licheniformis strain ATCC 14580 in protein database. The identified proteins were classified into five metabolic groups. Among the four proteins involved in prokaryotic stress response, two proteins with chaperonic functions (molecular chaperone DnaK and chaperonin GroEL), one general stress protein (YfkM), and one protein involved in defense against oxidative stress (iron/manganese-containing SOD) were identified (Table 5). Earlier observations on up-regulation of DnaK and GroEL proteins in B. licheniformis under salt stress conditions [27] also support our findings. During stress, there is an accumulation of abnormal or misfolded proteins in the cell due to denaturation and errors in bio-synthesis. In response, the cell increases synthesis of molecular chaperones, which help in proper folding or refolding of proteins, and of proteases, which degrade the proteins that cannot be refolded [30]. These heat shock proteins, with chaperone like functions, are usually induced by multiple stresses and play an important role in withstanding and surviving stressful conditions by stabilizing proteins and thereby their functioning, during abiotic stresses such as salt, heat, cold, and osmotic stress [11]. YfkM is a SigB-regulated protein with probably endopeptidase activity, and may play an important role in degradation of misfolded and thereby non-functional proteins [1]. Along with observed increase in SOD activity, concomitant up-regulation of iron/manganese-containing SOD (SodA) protein, supported our observations. Even in B. licheniformis strain DSM 13T, within 10 min of exposure to salt stress, there was up-regulation of SOD protein and up-regulation of this protein was also observed 2 h after exposure [26].
Table 5.
Proteins bands up-regulated after exposure to salt stress in Bacillus licheniformis as identified by LC-Tandem MS analysis
| Homologous protein | Mascot* Score | Mw/PI** | Sequence*** coverage (%) | Acc. no. | Peptides matched |
|---|---|---|---|---|---|
| 50S ribosomal protein L5 [Bacillus licheniformis ATCC 14580 | 395 | 20,248/9.67 | 57 | YP_077412 | 4 |
| General stress protein YfkM [Bacillus licheniformis ATCC 14580] | 214 | 18,660/4.90 | 30 | YP_078040 | 3 |
| Superoxide dismutase [Bacillus licheniformis ATCC 14580] | 488 | 22,530/5.33 | 62 | YP_079829 | 3 |
| Aminotransferase, class IV YjlD [Bacillus licheniformis ATCC 14580] | 445 | 41,878/6.21 | 25 | YP_079296 | 4 |
| Ketol-acid reductoisomerase [Bacillus licheniformis ATCC 14580] | 427 | 37,445/5.50 | 20 | YP_080103 | 4 |
| Glycerol dehydrogenase [Bacillus licheniformis ATCC 14580] | 381 | 39,452/4.79 | 21 | YP_078053 | 5 |
| Iron-containing alcohol dehydrogenase YugJ [Bacillus licheniformis ATCC 14580] | 345 | 42,696/5.09 | 23 | YP_080422 | 4 |
| Succinyl-CoA synthetase subunit beta [Bacillus licheniformis ATCC 14580] | 332 | 41,544/5.09 | 16 | YP_079002 | 6 |
| Methylcitrate synthase [Bacillus licheniformis ATCC 14580] | 253 | 41,595/5.71 | 24 | YP_080206 | 5 |
| Glyceraldehyde-3-phosphate dehydrogenase [Bacillus licheniformis ATCC 14580] | 520 | 35,863/5.11 | 39 | YP_080753 | 6 |
| Isocitrate dehydrogenase [Bacillus licheniformis ATCC 14580] | 324 | 48,027/5.05 | 23 | YP_092619 | 6 |
| Phosphopyruvate hydratase [Bacillus licheniformis ATCC 14580] | 1025 | 46,610/4.67 | 55 | YP_080749 | 10 |
| Chaperonin GroEL [Bacillus licheniformis ATCC 14580] | 1450 | 57,534/4.71 | 53 | YP_077851 | 10 |
| Molecular chaperone DnaK [Bacillus licheniformis ATCC 14580] | 1232 | 65,852/4.78 | 48 | YP_092303 | 10 |
| Elongation factor G [Bacillus licheniformis ATCC 14580] | 859 | 76,276/4.83 | 31 | YP_077397 | 12 |
| Polynucleotide phosphorylase/polyadenylase [Bacillus licheniformis ATCC 14580] | 498 | 77,484/5.15 | 14 | YP_079067 | 7 |
| Transketolase [Bacillus licheniformis ATCC 14580] | 490 | 72,591/5.08 | 30 | YP_079202 | 10 |
| Alpha-ketoglutarate decarboxylase [Bacillus licheniformis ATCC 14580] | 651 | 10,6605/5.96 | 18 | YP_079421 | 11 |
| Aconitate hydratase [Bacillus licheniformis ATCC 14580] | 592 | 99,283/5.04 | 15 | YP_091630 | 8 |
Mascot score, used in peptide mass fingerprinting, is a similarity score, which is a probability-based algorithm to estimate the significance of a match. Protein scores greater than 100 are considered significant
MW is molecular weight and PI is isoelectric pH of protein
Sequence coverage is the percentage of the database protein sequence covered by matching peptides. The larger the sequence coverage, the more certain is the identification of protein. Sequence coverage is calculated by dividing the number of amino acids observed by the protein amino acid length
Polynucleotide phosphorylase/polyadenylase involved in mRNA degradation in bacteria was also up-regulated. This enzyme degrades RNA from 3′ to 5′ and is a major component of Escherichia coli degradosomes [26]. It has been demonstrated to play an important role in cold and oxidative stress adaptation of bacteria [14] and could also be involved in salt stress adaptation. In E. coli, it has been reported to play an essential role in stress adaptation by selectively degrading mRNAs for stress response proteins to prevent overproduction of these proteins, which is deleterious to cells [33].
Proteins implicated in amino acid biosynthetic pathways (aminotransferase class IV YjlD, ketol-acid reductoisomerase), pathway for production of biomolecules involved in nucleosides and amino acids biosynthesis (transketolase), formation of ribosomes (50S ribosomal protein L5), and peptide elongation (elongation factor G) were also up-regulated. All these proteins are involved in various steps of protein synthesis. De novo synthesis and up-regulation of expression of certain stress responsive proteins are essential for successful adaptation to environmental stresses [17].
Up-regulation of enzymes involved in carbohydrate and lipid metabolism during growth under salt stress was also observed. Many proteins identified were enzymes involved in either glycolysis (glyceraldehyde-3-phosphate dehydrogenase, phosphopyruvate hydratase) or TCA (aconitate hydratase, isocitrate dehydrogenase, succinyl-CoA synthetase, and alpha-ketoglutarate) pathways. Many among these and other enzymes up-regulated were involved in reducing power (NADH/NADPH) generation (glyceraldehyde-3-phosphate dehydrogenase, isocitrate dehydrogenase, succinyl-CoA synthetase, ketol-acid reductoisomerase, glycerol dehydrogenase, and iron-containing alcohol dehydrogenase). While NADH is a source of ATP through oxidative phosphorylation, NADPH is an important source of reducing power for antioxidative enzymes [18]. This indicated that during growth there was increased cellular requirement of ATP and reducing power for successful adaptation of the microbe to environmental stress. This was also evident from the fact that under control conditions there was not much difference in growth obtained in presence/absence of glucose in the medium. However, when exposed to salt stress, the bacterium showed better growth in presence of glucose and with an increase in salt concentration, this gap further widened. (Supplementary Fig. 1).
Conclusions
Bacillus licheniformis strain SSA 61 was grown in presence of salt (10 and 15 %) and long-term adaptation to salt stress by this microbe, isolated from Sambhar lake sediment, involved modulation of cellular physiology at various levels. There was increased accumulation of compatible osmolytes, antioxidants, and induction of antioxidative enzymes. Up-regulation of chaperones DnaK and GroEL, general stress protein YfkM and polynucleotide phosphorylase/polyadenylase indicated their involvement in cellular adaptation to salt stress. Enzymes involved in biosynthesis of amino acids, ribosome assembly, and peptide elongation were highly expressed. Enzymes of both glycolysis and TCA pathways were highly up-regulated along with certain other enzymes involved in generation of NADH and NADPH, indicating increased cellular demand for both energy and reducing power during growth in presence of salt stress.
Supplementary Material
Acknowledgments
The authors are thankful to Dr Shyamala Rajan, Argonne National Laboratory, Argonne, USA. Without her help, the collaboration between authors at Indian Agricultural Research Institute, New Delhi, India and authors at Argonne National Laboratory, Argonne, USA would not have been possible.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00284-014-0761-y) contains supplementary material, which is available to authorized users.
Contributor Information
Jyoti Kumar Thakur, Division of Soil Biology, Indian Institute of Soil Science, Bhopal 462 038, Madhya Pradesh, India.
G. S. Bandeppa, Division of Microbiology, Indian Agricultural Research Institute, New Delhi 10012, India
Md. Aslam Khan, Department of Biology, Faculty of Science, Jazan University, Jazan, Kingdom of Saudi Arabia.
References
- 1.Abdallah J, Caldas T, Kthiri F, Kern R, Richarme G (2007) YhbO protects cells against multiple stresses. J Bacteriol 189:9140–9144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aebi H (1984) Catalase. In: Packer L (ed) Methods in enzymology, vol 105 Academic Press, Orlando, pp 121–126 [DOI] [PubMed] [Google Scholar]
- 3.Bates L, Waldren RP, Teare ID (1973) Rapid determination of free proline for water-stress studies. Plant Soil 39:205–207 [Google Scholar]
- 4.Boyadzhieva I, Emanuilova E (2010) Induction of superoxide dismutase production in Bacillus licheniformis M20 by heat shock and oxidative stress. Compt Rend Acad Bulg Sci 63:1571–1576 [Google Scholar]
- 5.Cabiscol E, Tamarit J, Ros J (2000) Oxidative stress in bacteria and protein damage by reactive oxygen species. Int Microbiol 3:3–8 [PubMed] [Google Scholar]
- 6.Cappucino GJ, Sherman N (1992) Microbiology: a laboratory manual. The Benjamin Cummings Publishing Company INC, Redwood City [Google Scholar]
- 7.Chen Z, Cuin TA, Zhou M, Twomey A, Naidu BP, Shabala S (2006) Compatible solute accumulation and stress-mitigating effects of barley genotypes contrasting in their salt tolerance. J Exp Bot 58:4245–4255 [DOI] [PubMed] [Google Scholar]
- 8.den Besten HMW, Mols M, Moezelaar R, Zwietering MH, Abee T (2009) Phenotypic and transcriptomic analyses of mildly and severely salt-stressed Bacillus cereus ATCC 14579 cells. Appl Environ Microbiol 75:4111–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dhindsa RH, Plumb-Dhindsa P, Thorpe TA (1981) Leaf senescence correlated with increased level of membrane permeability, lipid peroxidation and decreased level of SOD and CAT. J Exp Bot 32:93–101 [Google Scholar]
- 10.Fry SC (1990) The growing plant cell wall: chemical and metabolic analysis. The Blackburn Press, New Jersey [Google Scholar]
- 11.Garnier M, Matamoros S, Chevret D, Pilet M-F, Leroi F, Tresse O (2010) Adaptation to cold and proteomic responses of the psychrotrophic biopreservative Lactococcus piscium strain CNCM I-4031. Appl Environ Microbiol 76:8011–8018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grieve CM, Grattan SR (1983) Rapid assay for determination of water soluble quaternary ammonium compounds. Plant Soil 70:303–307 [Google Scholar]
- 13.Griffith OW (1980) Determination of glutathione and glutathione disulfide using glutathione reductase and 2-vinylpyridine. Anal Biochem 106:207–212 [DOI] [PubMed] [Google Scholar]
- 14.Haddad N, Tresse O, Rivoal K, Chevret D, Nonglaton Q, Burns CM, Prévost H, Cappelier JM (2012) Polynucleotide phosphorylase has an impact on cell biology of Campylobacter jejuni. Front Cell Inf Microbiol 2:30. doi: 10.3389/fcimb.2012.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halverson LJ, Jones TM, Firestone MK (2000) Release of intracellular solutes by four soil bacteria exposed to dilution stress. Soil Sci Soc Am J 64:1630–1637 [Google Scholar]
- 16.Höper D, Bernhardt J, Hecker M (2006) Salt stress adaptation of Bacillus subtilis: a physiological proteomics approach. Proteomics 6:1550–1562 [DOI] [PubMed] [Google Scholar]
- 17.Jan G, Leverrier P, Pichereau V, Boyaval P (2001) Changes in protein synthesis and morphology during acid adaptation of Propionibacterium freudenreichii. Appl Environ Microbiol 67:2029–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kostadinova N, Vassilev S, Spasova B, Angelova M (2011) Cold stress in antarctic fungi targets enzymes of the glycolytic pathway and tricarboxylic acid cycle. Biotech Biotechnol Equip 25:50–57 [Google Scholar]
- 19.Kumar S (2008) Conservation of Sambhar lake—an important waterfowl habitat and a Ramsar site in India. In: Sengupta M, Dalwani R (eds) Proceedings of Taal 2007: The 12th world lake conference, pp 1509–1517 [Google Scholar]
- 20.Kunte HJ, Trüper HG, Stan-Lotter H (2009) Halophilic microorganisms. In: Horneck G, Baumstark-Khan C (eds) Astrobiology: the quest for the conditions of life; Springer, Berlin, pp 185–199 [Google Scholar]
- 21.Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 22.Lowry OH, Rosebrough NJ, Farr AL, Randal RJ (1951) Protein measurement with the Folin-Phenol reagents. J Biol Chem 193:265–275 [PubMed] [Google Scholar]
- 23.Mukherjee SP, Choudhuri MA (1983) Implications of water stress-induced changes in the leaves of indigenous ascorbic acid and hydrogen peroxide in Vigna seedlings. Physiol Plant 58:166–170 [Google Scholar]
- 24.Nakano Y, Asada K (1991) Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol 22:867–880 [Google Scholar]
- 25.Paul D (2013) Osmotic stress adaptations in rhizobacteria. J Basic Microbiol 53:101–110 [DOI] [PubMed] [Google Scholar]
- 26.Regonesi ME, Briani F, Ghetta A, Zangrossi S, Ghisotti D, Tortora P, Dehò G (2004) A mutation in polynucleotide phosphorylase from Escherichia coli impairing RNA binding and degradosome stability. Nucleic Acids Res 32:1006–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schroeter R, Hoffmann T, Voigt B, Meyers H, Bleisteiner M, Muntel J, Jürgen B, Albrecht D, Becher D, Lalks M, Evers S, Bongaerts J, Maurer K-H, Putzer H, Hecker M, Schweder T, Bremer E (2013) Stress response of the industrial workhorse Bacillus licheniformis to osmotic challenges. PLoS One 8:e80956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shivanand P, Mugeraya G (2011) Halophilic bacteria and their compatible solutes—osmoregulation and potential applications. Curr Sci 100:1516–1521 [Google Scholar]
- 29.Smith IK, Vierheller TL, Thorne CA (1988) RAssay of glutathione reductase in crude tissue homogenates using 5,5′-dithiobis(2-nitrobenzoic acid). Anal Biochem 175:408–413 [DOI] [PubMed] [Google Scholar]
- 30.Thomsen LE, Olsen JE, Foster JW, Ingmer H (2002) ClpP is involved in the stress response and degradation of misfolded proteins in Salmonella enterica serovar Typhimurium. Microbiol 148:2727–2733 [DOI] [PubMed] [Google Scholar]
- 31.Upasani V, Desai S (1990) Sambhar Salt Lake—Chemical composition of the brines and studies on haloalkaliphilic archaebacteria. Arch Microbiol 154:589–593 [Google Scholar]
- 32.Wiegand S, Voigt B, Albrecht D, Bongaerts J, Evers S, Hecker M, Daniel R (2013) Liesegang H (2013) Fermentation stage-dependent adaptations of Bacillus licheniformis during enzyme production. Microb Cell Fact 12:120. doi: 10.1186/1475-2859-12-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamanaka K, Inouye M (2001) Selective mRNA degradation by polynucleotide phosphorylase in cold shock adaptation in Escherichia coli. J Bacteriol 183:2808–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zahran HH (1997) Diversity, adaptation and activity of the bacterial flora in saline environments. Biol Fertil Soils 25:211–223 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.