Abstract
DUF89 family proteins occur widely in both prokaryotes and eukaryotes, but their functions are unknown. Here we define three DUF89 subfamilies (I, II, and III), with subfamily II being split into stand-alone proteins and proteins fused to pantothenate kinase (PanK). We demonstrated that DUF89 proteins have metal-dependent phosphatase activity against reactive phosphoesters or their damaged forms, notably sugar phosphates (subfamilies II and III), phosphopantetheine and its S-sulfonate or sulfonate (subfamily II-PanK fusions), and nucleotides (subfamily I). Genetic and comparative genomic data strongly associated DUF89 genes with phosphoester metabolism. The crystal structure of the yeast (Saccharomyces cerevisiae) subfamily III protein YMR027W revealed a novel phosphatase active site with fructose 6-phosphate and Mg2+ bound near conserved signature residues Asp254 and Asn255 that are critical for activity. These findings indicate that DUF89 proteins are previously unrecognized hydrolases whose characteristic in vivo function is to limit potentially harmful buildups of normal or damaged phosphometabolites.
Many protein families still have no known functions, and it is a foundational task of post-genomic biology to discover these functions, particularly for families whose members are widely distributed1. The DUF89 (Pfam01937) protein family is one such case. This family is large, diverse, and present in all domains of life, but no biochemical or biological activity has been robustly demonstrated for any of its members. The scant information available on the family may be summarized as follows.
DUF89 proteins occur in stand-alone form2 and as C-terminal fusions to pantothenate kinase (PanK) in plants and animals3,4. The crystal structure of the stand-alone DUF89 protein At2g17340 from Arabidopsis thaliana (PDB codes 1XFI and 2Q40) reveals a new protein fold and several conserved residues, including Asp220, Asn221, and Asp256 coordinating a metal ion (probably Mg2+), as well as other residues located near the metal-binding site2. The high sequence conservation around the metal-ion-coordinating residues points to the metal-binding site as central to DUF89 function2.
In S. cerevisiae, the DUF89 gene YMR027W is upregulated in response to treatment with the DNA-damaging agent methyl methanesulfonate5, and the knockout strain shows a phenotype indicative of increased DNA damage or decreased repair6. The human ortholog of YMR027W, C6orf211, is also implicated in the response to DNA damage7. C6orf211 is reportedly associated with a protein carboxyl methyltransferase activity7, but whether this protein indeed has such an activity remains to be determined.
The putative active site residues and bound divalent metal ion in the plant DUF89 structure2 are suggestive of a metallohydrolase8. Many such enzymes are known to rid cells of unwanted and potentially harmful metabolites or side products, in related metabolic damage-control processes termed ‘damage pre-emption’9 (or ‘housecleaning’10) and ‘directed overflow’11. Damage pre-emption processes remove side-products formed spontaneously or by enzyme errors9,10, whereas directed overflow eliminates excesses of normal metabolites11,12. The association of yeast YMR027W and human C6orf211 with response to DNA damage fits with a damage pre-emption function10.
Here we present bioinformatic, biochemical, genetic, and structural analyses of DUF89 proteins from all domains of life. These analyses identify three DUF89 subfamilies, show that members of each subfamily are metal-dependent phosphatases with probable in vivo damage-control functions, and define a new phosphatase active site for which a catalytic mechanism is proposed.
RESULTS
Sequence features define three DUF89 subfamilies
Forty representative DUF89-family sequences (including those with crystal structures) were aligned, compared, and analyzed phylogenetically. The family was cleanly separable into three subfamilies on the basis of conserved motifs (Fig. 1a) and phylogenetic trees (Fig. 1b). All subfamilies share the previously identified2 metal-binding aspartate and asparagine residues.
Figure 1 |. The three subfamilies of DUF89 proteins.
(a) The type and position of sequence motifs that characterize the whole family (DNxG) or individual subfamilies. (b) Neighbor-joining tree (1,000 bootstrap replicates) of representative DUF89 proteins; bootstrap values for the major nodes (*) were 100%. The proteins examined in this study are in red. SEED or GenBank identifiers for the proteins in the tree are given in Supplementary Table 7.
Subfamily I proteins (~260–320 amino acids) are distinguished by three motifs: CxxC near the N terminus, GNFE (or similar) ~40 residues from the C terminus, and KC ~25 residues from the C terminus. In the crystal structures of the subfamily I proteins PH1575 from Pyrococcus horikoshii (PDB code 2G8L) and AF1104 from Archaeoglobus fulgidus (PDB code 2FFJ), the side chains of the cysteines in the CxxC and KC motifs face each other across the rim of the putative substrate-binding cleft, and the GNFE motif lies deep in the cleft close to the metal-binding aspartate and asparagine. Subfamily I proteins occur only in anaerobic or microaerophilic archaea and bacteria.
Subfamily II proteins (~340–370 amino acids) have an EGMGR motif ~50 residues from the C terminus. This motif lies near the metal-binding residues in the putative substrate-binding cleft2. Subfamily II proteins occur only in eukaryotes, in two forms: as a stand-alone unit in plants, and as a C-terminal domain of pantothenate kinases in plants, animals, and chytrid fungi (Fig. 1a). The stand-alone and fused proteins form distinct clades within subfamily II (Fig. 1b).
Subfamily III proteins (~380–480 amino acids) have an RTxK motif ~40–50 residues from the C terminus; the threonine may be replaced by serine or cysteine. Subfamily III proteins occur in animals, fungi, green algae, apicomplexans, haptophytes, and certain bacteria.
Representatives of each subfamily were selected for biochemical and structural characterization: P. horikoshii PH1575 from subfamily I; Arabidopsis At2g17340 and the DUF89 domains of Arabidopsis pantothenate kinase 2 (PanK2) and human pantothenate kinase 4 (PanK4) from subfamily II; and yeast YMR027W from subfamily III. These proteins were expressed in Escherichia coli and purified (Supplementary Results, Supplementary Fig. 1).
DUF89 proteins have metal-dependent phosphatase activity
The above five proteins were screened for enzymatic activity against a panel of 96 typical phosphatase substrates (Supplementary Table 1) in the presence of 5 mM Mg2+ plus 0.5 mM Mn2+. Activity was detected with various substrates for all five proteins. A good substrate for each enzyme was then used to define metal-ion preference (Supplementary Fig. 2). In all cases, there was little phosphatase activity unless a metal was added, and even less when EDTA was present. Activity was strongly promoted by Co2+, Ni2+, Mg2+ and, in most cases, Mn2+, with the order of preference showing some variation. The activity of PH1575 was also strongly promoted by Zn2+ and Cu2+. Km values for metal ions were in the micromolar range: 1.9 μM (Zn2+) and 4.6 μM (Ni2+) for PH1575, and 7.5 μM (Co2+) and 8.8 μM (Zn2+) for YMR027W. We propose that Mn2+ and possibly Ni2+ represent biologically relevant metal ions for DUF89 phosphatases.
The enzymes were then rescreened against the panel of phosphorylated compounds using preferred metal cofactors. Each enzyme was active against several substrates, the best being fructose 1- and 6-phosphates for YMR027W, sugar phosphates and p-nitrophenyl phosphate (pNPP) for At2g17340, and pNPP for the other enzymes (Fig. 2). Michaelis–Menten kinetics were found for YMR027W, PH1575, and At2g17340, whereas At2g17340 showed sigmoidal kinetics with ribose 5-phosphate or 2-deoxyribose 5-phosphate (Supplementary Table 2 and Supplementary Fig. 3). The Km or S0.5 (the substrate concentration needed for one-half maximal velocity) values for the sugar phosphate substrates of YMR027W and At2g17340 were physiologically reasonable (~0.1–0.5 mM).
Figure 2 |. Substrate profiles of DUF89 proteins from each subfamily.
(a) Arabidopsis subfamily II protein At2g17340. (b) Arabidopsis subfamily II protein fused to PanK2. (c) Human subfamily II protein fused to Pank4. (d) P. horikoshii subfamily I protein PH1575. (e) Yeast subfamily III protein YMR027W. The panel of 96 substrates screened, and their abbreviations, are listed in Supplementary Table 1. Data are mean ± s.d. of four replicates for At2g17340, Arabidopsis PanK2, and human PanK4, and means of duplicates for PH1575 and YMR027W. Activities against substrates not shown were <1–5% of that of the best substrate of each enzyme.
These assays identify plausible physiological substrates for YMR027W (fructose phosphates) and At2g17340 (pentose and tetraose phosphates), but not for the PanK-fused proteins or for the subfamily I protein PH1575, all of which preferred the artificial substrate pNPP. We therefore applied comparative genomics to deduce likely natural substrates for these enzymes.
PanK DUF89 domains act on damaged phosphopantetheine
The domains of fusion proteins usually have associated functions13. The natural substrates of the plant and animal DUF89 domains fused to PanK are thus a priori likely to relate to coenzyme A (CoA) synthesis or salvage because PanK is central to CoA pathways (Fig. 3a). Plant and animal PanKs phosphorylate the CoA precursor pantothenate and the CoA and acyl carrier protein catabolite pantetheine14,15 (Fig. 3a). We therefore tested the phosphate esters produced by these reactions, and their known or probable chemical or enzymatic damage products16–19 (Fig. 3b), as substrates for DUF89 domains. The damage products were oxidized forms of 4′-phosphopantetheine (disulfide, S-sulfonate, and sulfonate) and the 2′,4′-cyclic phosphates of pantothenate and pantetheine. The Arabidopsis and human enzymes had activity against 4′-phosphopantetheine and its S-sulfonate or sulfonate that was comparable to—and in some cases above—that against pNPP (Fig. 3c), the best substrate from the activity screen (Fig. 2). The Arabidopsis enzyme was also active against 4′-phosphopantothenate. Neither enzyme had detectable activity (<1 nmol min−1 per mg protein) against the cyclic phosphates.
Figure 3 |. Phosphatase activities of pantothenate kinase DUF89 domains in relation to CoA synthesis and salvage.
(a) Pathways of CoA synthesis (solid arrows) and salvage (dashed arrows). Eukaryotic-type pantothenate kinases can mediate phosphorylation of pantothenate (synthesis pathway) or pantetheine (salvage pathway). (b) Structures of pantothenate, pantetheine, and their damage products. (c) Activities of the DUF89 domains of Arabidopsis and human pantothenate kinases against these substrates. Results are mean ± s.d. of three replicates.
Kinetic characterization of the Arabidopsis and human enzymes confirmed that they prefer CoA-related substrates to pNPP and markedly prefer one or other oxidized form of 4′-phosphopantetheine to 4′-phosphopantetheine itself (Supplementary Table 2). Thus, relative to 4′-phosphopantetheine, the specificity constant (kcat/Km) of the human enzyme is 5-fold higher for the sulfonate and that of the Arabidopsis enzyme is 25-fold higher for the S-sulfonate. In both cases, this is due mainly to a far lower Km for the oxidized form (in the low micromolar range). The Arabidopsis enzyme also had a high specificity constant and a low Km with 4′-phosphopantothenate. The Km value of the Arabidopsis enzyme for the S-sulfonate (2.9 μM) is consistent with the level of this compound (~0.13 μM on a plant-water basis) reported for plant tissue16.
We also tested the Arabidopsis PanK fusion domain for the ability to release 4′-phosphopantetheine or pantetheine from acyl carrier protein because the protein responsible for this activity is not known in eukaryotes14. No activity was found (Supplementary Table 3).
Yeast YMR027W and P. horikoshii PH1575 had little or no activity (<5 nmol min−1 per mg protein) against any CoA-related substrate. Arabidopsis At2g17340 had modest activity against phosphopantothenate (130 nmol min−1 per mg protein) but not against other CoA-related substrates.
Comparative genomics of subfamily I
Because the chromosomal context and occurrence profile of a gene can point to its function20,21, we analyzed DUF89 genes in >1,600 diverse prokaryotic genomes using the SEED database and its tools21. We found subfamily I genes in 145 genomes, all from anaerobes or microaerophiles. Context analysis strongly associated 36 (25%) of these genes with the purine synthesis pathway (Supplementary Fig. 4a). In genomes from two archaeal and three bacterial phyla, the subfamily I genes cluster with one or more of six purine synthesis genes (purF, purM, purE, purP, purO, and guaB) (Supplementary Fig. 4b). The substrates and products of the corresponding enzymes are all phosphate esters, and several are oxidatively unstable22,23. That subfamily I is linked to the O2-labile purine pathway in O2-sensitive organisms suggests that subfamily I proteins could hydrolyze oxidatively damaged intermediates or products of this pathway. Testing nucleotide derivatives as substrates for PH1575 supported this scenario inasmuch as oxidized (8-oxo) forms of dGTP and dATP were among the best substrates (Supplementary Fig. 5). As the kinetic constants for 8-oxo-dGTP were close to those for ADP (Supplementary Fig. 5), PH1575 presumably does not selectively attack low concentrations of this particular oxo-nucleotide in vivo; it might nonetheless selectively attack others.
In vivo hydrolysis of fructose phosphates
The preference of the subfamily III YMR027W protein for fructose 1-phosphate (Fig. 2) suggests that the protein has a role in damage control. Fructose 1-phosphate is not a classical yeast metabolite24, but it can derive from enzymatic damage processes, notably 1-phosphorylation of fructose via kinase side activities, or phosphatase attack on fructose 1,6-diphosphate25 (Fig. 4a). If formed, fructose 1-phosphate would require hydrolysis because high concentrations are toxic to yeast24. We therefore compared growth rates and sugar phosphate levels of wild type and a YMR027W deletion strain cultured on minimal medium with glucose or fructose as carbon source. The deletion strain had no obvious growth defect on either medium, but it showed significant accumulation of fructose 1-phosphate on glucose medium and even more on fructose (Fig. 4b and Supplementary Data Set 1). The accumulation was greater in cells harvested at an OD600 of 0.5 rather than 1, when the diauxic shift starts and hexose phosphate synthesis rates fall. The deletant showed no accumulation of fructose 6-phosphate (Fig. 4b) and little or no accumulation of other sugar phosphates (Supplementary Data Set 1), indicating that YMR027W is effectively selective for fructose 1-phosphate in vivo. These data validate a damage-control function for YMR027W in hexose phosphate metabolism.
Figure 4 |. Hexose metabolism in wild-type and ΔYMR027W yeast strains.
(a) Classical reactions of yeast hexose metabolism (black arrows), with potential damage reactions (red arrows) and potential DUF89-mediated damage-control reactions (blue arrows). (b) Targeted metabolomics analysis of fructose 1-phosphate and fructose 6-phosphate levels in ΔYMR027W and wild-type cells grown with fructose or glucose as carbon source to an OD600 of 0.5 or 1.0. Data are mean ± s.d. of six replicates. *, **, and *** denote differences between the ΔYMR027W strain and the wild type that are significant at P < 0.05, P < 0.01, and P < 0.001, respectively, according to Student’s t-test. Untargeted metabolomics data for other sugar phosphates are summarized in Supplementary Data Set 1.
Subfamily I proteins have an iron-containing chromophore
Whereas purified subfamily II and III proteins were colorless, the subfamily I protein PH1575 was red-brown in solution and had absorption peaks at 330 nm and 420 nm (Supplementary Fig. 6). Three other purified subfamily I proteins (A. fulgidus AF1104, Methanothermobacter thermautotrophicus MTH1744, and Desulfatibacillum alkenivorans Dalk1756; Supplementary Fig. 1) were found to have similar spectra (Supplementary Fig. 6). Such spectra are typical for proteins with a [2Fe-2S] cluster. Inductively coupled plasma mass spectrometry analysis showed that subfamily I protein PH1575 contained substantial Fe (0.25 g-atoms/mol), and that subfamily II and III proteins did not (Supplementary Table 4). The Fe level in PH1575 is lower than expected for a protein with an intact [2Fe-2S] cluster, but this is not unusual for recombinant iron–sulfur proteins26 and we observed that PH1575 lost chromophore during purification. PH1575 also contained substantial Zn (0.25 g-atoms/mol), as do other recombinant Fe-containing proteins27. Collectively, these data indicate that subfamily I proteins have an Fe-containing chromophore, possibly a [2Fe-2S] cluster. Subfamily II and III proteins contained low levels of Ni (Supplementary Table 4), which fits with their metal preferences (Supplementary Fig. 2). The available subfamily I structures (P. horikoshii PH1575 and A. fulgidus AF1104) lack a prosthetic group, but this is consistent with the loss of chromophore during purification noted above.
Crystal structure of DUF89 proteins
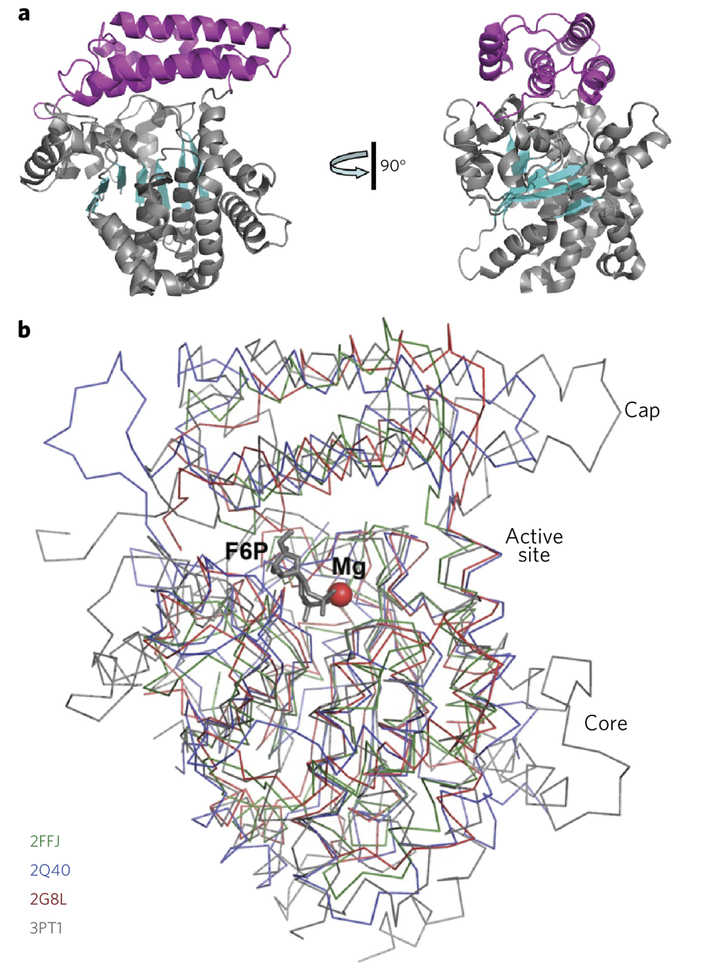
Yeast YMR027W (subfamily III) was crystallized using the sitting-drop vapor diffusion method, and its structure was solved to 1.80 Å resolution using the selenomethionine-substituted protein and single-wavelength anomalous dispersion (Supplementary Table 5). The structure of Arabidopsis At2g17340 (ref. 2) (subfamily II) and the structures of PH1575 (PDB code 2G8L) and AF1104 (PDB code 2FFJ) (subfamily I) were used for comparative analysis. The protomer structures of these proteins revealed a core domain with an α–β–α sandwich-like fold and an α-helix bundle cap domain (Fig. 5a and Supplementary Fig. 7). All four proteins have similar structures, the main differences being in the cap (Fig. 5b). The YMR027W structure is monomeric, whereas PH1575 and AF1104 are dimers. Analysis of the crystal contacts using the PDBePISA tool also indicated that YMR027W is a monomer and that PH1575 and AF1104 are dimers. Size-exclusion chromatography indicated that YMR027W is monomeric in solution (observed mass 52.8 kDa, predicted monomer mass 54.1 kDa) and that PH1575 and AF1104 are dimeric (observed mass 59.8 and 60.8 kDa, predicted monomer mass 32.6 and 32.0 kDa, respectively) (data not shown).
Figure 5 |. Overall structures of DUF89 protomers.
(a) The structure of YMR027W (PDB code 5BY0), shown in two orientations related by 90° rotation. The protein core domain is colored in gray (helices) and cyan (strands); the cap domain is colored in magenta. (b) Structural superimposition revealing a very similar fold of the protomers of four DUF89 structures. P. horikoshii PH1575 (PDB code 2G8L; in red), Arabidopsis At2g17340 (PDB code 2Q40; in blue), A. fulgidus AF1104 (PDB code 2FFJ; in green), and yeast YMR027W (PDB code 3PT1; in gray). A Mg2+ ion (red sphere) and a molecule of fructose 6-phosphate (F6P, gray) mark the active site cavity in the structure of YMR027W.
A Dali search for YMR027W structural homologs identified the other three DUF89 proteins as the best matches—Arabidopsis At2g17340, PH1575, and AF1104—although these proteins share only 13–19% sequence identity with YMR027W.
Active site of DUF89 phosphatases
In the YMR027W structure, the active site position is indicated by the bound metal ion, which we propose to be Mg2+ based on its presence in the crystallization solution (as for the metal ion in the At2g17340 structure2). Consistent with this proposal, the ion has octahedral coordination, being liganded by the side chains of the signature residues Asp254 (2.2 Å), Asn255 (2.2 Å), and Asp292 (2.1 Å) with the remaining three sites occupied by well-ordered (Supplementary Table 5) water molecules (~2.1 Å) (Fig. 6a). The side chain oxygens of the signature residues form the base of a trigonal pyramid (2.9–3.1 Å) with the Mg2+ ion located at the pyramid apex. Similar metal-ion coordination and distances are seen in the YMR027W–Mn2+ complex (Fig. 6b). The signature residues are also arranged similarly in the active sites of AF1104, PH1575, and At2g17340 (Fig. 6c and Supplementary Fig. 8). The AF1104 active site contains two sulfate ions and that of PH1575 includes an unknown ligand, possibly 2-methyl-2,4-pentanediol from the crystallization solution. These ligands probably mimic substrates or products.
Figure 6 |. Close-up views of the active site of DUF89 proteins.
(a) YMR027W with bound Mg2+. (b) YMR027W with bound Mn2+ (an anomalous difference Fourier maps at the Mn edge as shown in magenta at 7 σ) and two phosphate ions (P1, P2). (c) PH1575 with bound unknown ligand (UL). (d) YMR027W with bound fructose 6-phosphate and magnesium ion (in blue, Fo – Fc omit map of fructose 6-phosphate and Mg2+ at 3 σ). W, water molecules (in all panels).
Crystal soaking using YMR027W and the substrate fructose 6-phosphate produced a structure containing an additional electron density located in the active site near the Mg2+ ion (Fig. 6d). This structure (PDB code 3PT1) was solved to 1.77-Å resolution by single-wavelength anomalous diffraction (SAD) using the coordinates of the Se-substituted native protein (PDB code 5BY0). The additional density was modeled as the cyclic (furanose) form of fructose 6-phosphate, the main form in solution28. The phosphate group of fructose 6-phosphate (found in two conformations) is coordinated by the Mg2+ ion (2.3 Å) and the side chains of the signature residues Asp254 (2.5 Å and 2.9 Å) and Asn255 (3.3 Å), as well as Lys423 (2.7 Å) (Fig. 6d). On the other side of the molecule, the C1 hydroxyl interacts with the side chains of conserved Arg23 (2.9 Å) and Glu110 (2.4 Å), which also coordinates the C2 hydroxyl (3.2 Å), while the C3 hydroxyl group is located near Asp384 (2.6 Å). Because the furanose forms of fructose 6- and 1-phosphates are almost rotationally identical (the C6–C4 moiety of the 6-phosphate is highly similar to the C1–C3 moiety of the 1-phosphate), fructose 1-phosphate is likely to interact with YMR027W in much the same way as the 6-phosphate. The structure of the YMR027W-fructose 6-phosphate–Mg2+ complex (Fig. 6d) most probably represents the initial unproductive enzyme–fructose 6-phosphate complex that will be converted by an internal isomerization to the productive complex followed by fructose 6-phosphate dephosphorylation.
Interestingly, the structure of PH1575 showed that the three conserved cysteine residues (Cys7, Cys10, and Cys264) are near enough to each other (4.9–6.3 Å) (Fig. 6c) to coordinate the iron–sulfur cluster inferred from spectral and metal analysis data. Similar cysteine–cysteine distances are reported for MitoNEET, which contains a [2Fe-2S] cluster coordinated by three cysteines and a histidine29. The PH1575 structure has no equivalent histidine, but histidine could potentially be replaced by a glutamate residue (the semiconserved Glu6) as in Pyrococcus furiosus sulfide dehydrogenase30.
Site-directed mutagenesis
To identify residues needed for phosphatase activity, we mutated 13 conserved or semiconserved residues located in the catalytic cleft of YMR027W, 11 residues in or near the cleft of PH1575, and the DND signature residues in At2g17340. These residues were changed to alanine, and activities of the mutated proteins against pNPP were compared with wild type (Supplementary Figs. 9 and 10a). Alanine replacement of the signature residues of YMR027W (Asp254, Asn255, and Asp292), PH1575 (Asp156, Asn157, and Asp191), and At2g17340 (Asp220, Asn221, and Asp256) abolished catalytic activity, confirming that these residues form the DUF89 active site.
To test whether the three conserved cysteines in subfamily I are required for chromophore binding and enzyme activity, they were individually replaced by alanine in PH1575. None of these substitutions affected phosphatase activity (Supplementary Fig. 10a), but each of them abolished absorption at 300–500 nm (Supplementary Fig. 10b). Conversely, replacing the catalytic residues did not affect absorption (Supplementary Fig. 10c). These data indicate that the conserved trio of cysteines coordinates the Fe-containing chromophore, which is not directly involved in catalysis.
Of the other 15 residues mutated in YMR027W or PH1575, two are homologous to each other (Asn192/108 and Glu259/160, respectively); both were critical to activity, resulting in ≥90% activity loss when changed to alanine in either protein (Supplementary Figs. 9 and 10a). Ten of the 11 other residues mutated in one or other protein were also important for activity, with the change to alanine causing ≥80% loss of activity; the exception was Gln14 in PH1575, whose replacement by alanine had no effect (Supplementary Figs. 9 and 10a).
DISCUSSION
Our work establishes that DUF89 proteins are ubiquitous, metal-dependent phosphatases active against diverse metabolites. Like other metal-dependent phosphatases (such as the HAD31, HD32, and Nudix33 families), DUF89 phosphatases exist as stand-alone or fusion proteins and fall into distinct subfamilies that differ in substrate preference.
Although the DUF89 proteins studied all had wide substrate ranges in vitro, there is evidence that at least some of them—like other ‘housecleaning’ phosphatases31–33—have narrower ranges in vivo, and control metabolite damage by preventing harmful buildups of normal or damaged phosphometabolites9–11. The clearest case is the subfamily III protein, yeast YMR027W. In vitro, this enzyme preferred fructose 1-phosphate—a damage product rather than a canonical yeast metabolite (Fig. 4a)—and fructose 1-phosphate accumulated in YMR027W knockout cells (Fig. 4b). Fructose 1-phosphate is a strong glycating agent that causes DNA damage34; its buildup in the YMR027W knockout could thus account for this strain’s DNA damage phenotype6. The cytosolic and nuclear location of YMR027W35 would give it access to fructose 1-phosphate in vivo. YMR027W may, however, not be the only intracellular phosphatase that acts on fructose 1-phosphate in vivo because the fructose 1-phosphate accumulations in the knockout were modest (1.5-fold or less). The modest fructose 1-phosphate levels in the YMR027W knockout (≤65 ng/106 cells, equivalent to ~3 mM intracellular concentration) are only one-tenth of those that cause growth arrest in yeast cells overexpressing rat liver fructose 1-phosphotransferase24, which is consistent with the lack of an overt growth defect in the knockout. Moreover, the metabolic impacts of knocking out metabolite damage-control systems typically lead not to drastic growth phenotypes but rather to subtle reductions in fitness that may be inapparent under normal culture conditions9,12,36.
Like YMR027W, the stand-alone Arabidopsis DUF89 protein At2g17340 preferred sugar phosphate substrates in vitro, including ribose 5- and erythrose 4-phosphates. These aldose phosphates are extremely potent glycating agents37; an enzyme that selectively hydrolyzes excess levels of them could clearly help pre-empt glycation damage. The sigmoidal kinetics of At2g17340 acting on aldose phosphates (Supplementary Fig. 3) are consistent with such a role because sigmoidicity allows a steep rise in enzyme activity once substrate concentration exceeds a certain range.
The fusion of Arabidopsis and human subfamily II DUF89 domains to pantothenate kinase—which could confer in vivo specificity by proximity12,36—and their in vitro preference for normal or oxidatively damaged products of this enzyme (Fig. 3, Supplementary Table 2) provide strong indirect evidence that these DUF89 domains pre-empt damage in the CoA pathway. Eukaryotes regulate synthesis of 4′-phosphopantothenate (the first committed CoA synthesis intermediate) via feedback regulation of pantothenate kinase by CoA and its esters3,38, but they have less control over 4′-phosphopantetheine because it can come from salvage routes as well as synthesis (Fig. 3a). 4′-Phosphopantetheine and its oxidation products may therefore build up under certain conditions. Hydrolyzing excess 4′-phosphopantetheine could constitute a directed overflow mechanism11 to prevent its oxidation to the S-sulfonate, sulfonate, or other forms. Hydrolyzing 4′-phosphopantetheine sulfonate or S-sulfonate would forestall their conversion to inactive forms of CoA and acyl carrier protein—a risk because the CoA synthesis enzymes downstream of pantothenate kinase are promiscuous39.
Lastly, the genomic association of subfamily I with purine synthesis in O2-sensitive organisms suggests that subfamily I enzymes could hydrolyze oxidatively damaged purine nucleotides or their biosynthetic intermediates. The activity of PH1575 against 8-oxo nucleotides provided some support for the first possibility. The second possibility—dephosphorylation of damaged purine precursors—is attractive because of the high reactivity of these compounds22,23 but is ipso facto hard to test.
The O2 sensitivity of the organisms with subfamily I enzymes is noteworthy in light of the evidence that these enzymes have an iron-containing chromophore—most likely an iron–sulfur cluster, based on the optical spectra, metal data, and trio of cysteine ligands40, assuming a fourth non-cysteine ligand. These data do not fit with a molybdopterin chromophore, such as has been proposed for subfamily I protein PF1587 from P. furiosus (based on a preparation of uncertain purity whose Mo content was <<1 g-atom/mol)41. Because the chromophore was not needed for catalysis (Supplementary Fig. 10), a regulatory or structural role is implied. Other iron–sulfur clusters have such roles, including the [4Fe-4S] cluster in the purine synthesis enzyme PurF of Bacillus subtilis; this cluster has an O2-sensing role and regulates the turnover of the PurF protein40,42.
The structure of YMR027W in complex with fructose 6-phosphate and mutational studies identified a novel phosphatase active site, which includes the catalytic triad DND coordinating a divalent metal cation (Mg2+) that in turn binds the phosphate moiety of the substrate (Fig. 6a,d). The catalytic mechanism of known metal-dependent phosphohydrolases involves the activation of a catalytic water molecule by a general base in the active site, which can contain one, two, or three metal cofactors43. The DUF89 active site resembles that of the haloacid dehalogenase (HAD)-like phosphatases, and the 3D structures of DUF89 proteins have a structural fold, composed of core and cap domains, that resembles the overall structural organization of HAD phosphatases. HAD phosphatases use a two-step metal-dependent catalytic mechanism31 in which the aspartate nucleophile first attacks the substrate phosphoryl group, producing a phosphoaspartyl enzyme intermediate that is then hydrolyzed via a nucleophilic attack by a water molecule activated by another aspartate residue. The metal ion plays a key role through the correct positioning of the aspartate nucleophile and the substrate phosphate, as well as through charge neutralization of the trigonal bipyramidal transition state44. On the basis of the YMR027W structure in complex with fructose 6-phosphate (Fig. 6d), we propose that DUF89 phosphatases use a similar mechanism. In this model, the unprotonated side chain of Asp254 functions as a nucleophile directly attacking the substrate phosphoryl group and producing a phosphoaspartyl intermediate and fructose. In the second step, the catalytic water molecule activated by the Asp292 side chain initiates a nucleophilic attack on the phosphoaspartyl intermediate generating free phosphate and restoring the catalytic Asp254. Alternatively, the metal ion could function as a general base activating the catalytic water nucleophile (by reducing pKa and inducing deprotonation) as well as contributing to phosphate coordination and transition state stabilization. The activated hydroxyl initiates a nucleophilic attack on the substrate phosphorus atom, releasing free phosphate and the dephosphorylated product.
It should be noted that dephosphorylation of excess or damaged intermediates is not a complete damage-control solution because these compounds could potentially be rephosphorylated. However, dephosphorylation immediately forestalls damage by ejecting such compounds from pathways, and this process is analogous to the established roles of pyrophosphatases and phosphatases in ‘sanitizing’ the pools of other metabolites9,10,36. The fact that these damage-control hydrolases may need to be backed up by other enzymes that further metabolize their reaction products does not detract from their efficacy. Finally, that the DUF89 family is so large and widespread, yet until now had no known functions, highlights two key points about metabolite damage control9,10: it is crucial to life but often overlooked.
METHODS
Methods and any associated references are available in the online version of the paper.
ONLINE METHODS
Bioinformatics.
DNA and protein sequences were from GenBank or SEED21. Sequences were aligned with Multalin45 or Clustal W46. Phylogenetic trees were constructed from Clustal W alignments by the Neighbor-joining method using MEGA5 (ref. 47). A representative set of 1641 bacterial and archaeal genomes was analyzed using SEED tools21. Full results are encoded in the SEED subsystem ‘DUF89_Pfam01937’ (http://pubseed.theseed.org//SubsysEditor.cgi?page=ShowSubsystem&subsystem=DUF89_Pfam01937).
Chemicals.
Biochemicals and reagents, including those used for screening, were from Sigma-Aldrich or Fisher Scientific except for the following. Malachite Green (BIOMOL Green) reagent was from Enzo Life Science. Calcium D-pantetheine-S-sulfonate (70% w/v solution) was from Ikeda Corp. of America. Phosphopantothenate (PPa), phosphopantethine (PPaSS) and phosphopantetheine-S-sulfonate (PPaSSO3) were synthesized using Staphylococcus aureus PanK48 provided by H.W. Park (Tulane School of Medicine). The reaction products were filtered with Amicon 10K cutoff centrifugal filters and separated on a Microsorb C18 column (150 × 4.6 mm, 5 μm particle size, Agilent) kept at 35 °C. An isocratic system with mixing formic acid/water (0.25/99.75, v/v) with a flow rate of 0.6 ml/min or acetonitrile/formic acid/water (5/0.25/94.75) with a flow rate of 1.0 ml/min was used. The detection wavelength was 191 nm. Peak fractions were collected, pooled, dried in vacuo, and redissolved in water. Phosphopantetheine (PPaSH) was obtained by reducing PPaSS with sodium borohydride. Phosphopantetheine-sulfonate (PPaSO3) was obtained by oxidizing PPaSSO3 with performic acid49, then dried in vacuo and redissolved in water. The 2′,4′-cyclic phosphates of phosphopantothenate (cPPa) and phosphopantetheine (cPPaSH) were obtained by hydrolyzing CoA at 100 °C with 1 N NaOH for 10 min and 20 min, respectively. The reaction mixtures were neutralized, filtered, and separated by HPLC as above. All the synthesized substrates were shown to be chromatographically pure and (except for PPaSS and PPaSH) were further validated by liquid chromatography coupled with accurate mass high-resolution mass spectrometry in positive and negative ion modes. The concentrations of PPa, PPaSH, PPaSSO3, and PPaSO3 were estimated by determining the phosphate released after complete dephosphorylation, catalyzed by the Arabidopsis DUF89 protein. The concentrations of PPaSS, cPPa, and cPPaSH were estimated by HPLC (absorption at 191 nm) using unphosphorylated standards.
Escherichia coli expression constructs.
Primers are given in Supplementary Table 6. All constructs were sequence-verified. Full-length cDNAs for Arabidopsis PanK2 (At4g32180) and human PanK4 were obtained from the Riken BioResource Center (clone RAFL09–98-B15) and Thermo Scientific (clone 5244472), respectively. The Arabidopsis PanK2 and human PanK4 DUF89 domains were expressed by changing the Leu504 or Leu410 codon, respectively, to the start Met codon. Both sequences were amplified with Phusion High-Fidelity DNA polymerase (New England BioLabs). The amplified DNAs were purified, digested with BspI/HindIII (Arabidopsis) or NcoI/XhoI (human), and ligated into the matching sites of pET28b (Novagen), which added a C-terminal His6-tag. The D. alkenivorans DUF89 gene (Dalk1756) was recoded for E. coli expression (Supplementary Fig. 11), adding 5′ NcoI and 3′ XhoI sites. The recoded DNA was digested with NcoI/XhoI and ligated into the matching sites of pET28b, which added a C-terminal His6-tag. The genes encoding DUF89 proteins from yeast (YMR027W, Q04371), P. horikoshii (PH1575, O59272), A. fulgidus (AF1104, O29161), and M. thermautotrophicus (MTH1744, O27776) were cloned and mutated as described50. The At2g17340 expression construct was a maltose-binding protein (MBP) fusion in the pVP13-GW vector2.
Production and purification of proteins.
Plasmids encoding Arabidopsis PanK2, human PanK4, or D. alkenivorans DUF89 proteins were transformed into BL21-CodonPlus (DE3)-RIPL (Stratagene) cells. Cultures (0.5–1 l) were grown at 37 °C in LB medium containing 50 μg/ml kanamycin, 50 μg/ml spectinomycin, and 34 μg/ml chloramphenicol. When A600 reached 0.6–0.8, IPTG was added (final concentration 0.5 mM) and incubation was continued for 4 h at 22 °C (human) or overnight at 16 °C (Arabidopsis and D. alkenivorans). Cells were harvested by centrifugation (6,300g, 15 min), resuspended in 50 mM Tris-HCl, 300 mM NaCl, and 10 mM imidazole, pH 8.0, and sonicated. The lysate was centrifuged at 17,000g for 20 min and the supernatant was incubated with 1 ml of Ni2+-NTA 50% slurry (Qiagen) for 1 h at 4 °C. The slurry was poured into a column and allowed to drain. After washing with 50 ml of 50 mM Tris-HCl, 300 mM NaCl, and 20 mM imidazole, pH 8.0, proteins were eluted with 2.5 ml of this buffer containing 250 mM imidazole, desalted on PD-10 columns (GE Healthcare) equilibrated in 50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 10% (v/v) glycerol (human and Arabidopsis) or 50 mM Tris-HCl, pH 7.0, 10% glycerol (D. alkenivorans). The human and Arabidopsis proteins were further purified by anion exchange chromatography on a Mono Q 5/50 column (GE Healthcare), eluting with a 20-ml gradient from 100 to 300 mM NaCl in 20 mM Tris-HCl, pH 8.0; 1-ml fractions were collected and analyzed by SDS-PAGE and Coomassie Blue staining. DUF89-containing fractions were pooled, desalted into 50 mM Tris-HCl, pH 7.5, 10% (v/v) glycerol, and concentrated to ~1 mg/ml in Amicon Ultra-4 10K units (Millipore), then aliquoted, frozen in liquid N2, and stored at −80 °C. Yeast YMR027W, P. horikoshii PH1575, A. fulgidus AF1104, M. thermautotrophicus MTH1744, and the At2g17340-MBP fusion protein, were expressed in E. coli and purified as described2,50. Protein purity was checked by SDS-PAGE (Supplementary Fig. 1). Recombinant spinach holo-acyl carrier protein isoform I was prepared as described51. Purified E. coli acyl carrier protein hydrolase (AcpH) was from J.E. Cronan (University of Illinois).
Metal analyses.
The metal content of D. alkenivorans DUF89 was quantified by inductively coupled plasma-mass spectrometry (ICP-MS)52 at the University of Georgia Center for Applied Isotope Studies. The protein was purified and desalted into 20 mM Tris-HCl, pH 8.0 as above. Free divalent cations were then removed by adding Chelex 100 resin (Na+ form) (Bio-Rad) and incubating for 1 h at 4 °C. The slurry was poured into a clean column and nitric acid was added to the flowthrough to a final concentration of 2% (v/v). Precipitated protein was removed by centrifugation (11,000g, 10 min) and the supernatant was used for metal analysis. The blank was 20 mM Tris-HCl, pH 8.0, prepared the same way as the protein sample. The metal content of the other purified and dialyzed DUF89 proteins was determined using inductively coupled plasma-atomic emission spectrometry as described previously53.
Enzyme assays.
Purified DUF89 proteins were screened for the presence of several general enzymatic activities including phosphatase using pNPP as substrate and against 96 phosphorylated metabolites (Supplementary Table 1) using the Malachite Green reagent as previously described50. Assays were replicated two to four times. Screening assays (Fig. 2) contained 0.25 mM substrate; assay temperature was 30 °C, or 75 °C for the thermophilic P. horikoshii PH1575 enzyme. The metal type and concentration used for each enzyme were as follows: Arabidopsis At2g17340, Ni2+ 0.2 mM; Arabidopsis PanK2, Ni2+ 0.5 mM; human PanK4, Co2+ 0.5 mM; P. horikoshii PH1575, Ni2+ 0.5 mM; Yeast YMR027W, Co2+ 0.1 mM. The dependence of phosphatase activity on metal cations was determined using saturating concentrations of indicated substrates and cations (5 mM Mg2+ or 0.5 mM for other ions). Kinetic parameters were estimated by measuring rates of product formation at various substrate concentrations. The Km value for Co2+ for YMR027W was estimated similarly. Other metal Km values were determined by isothermal titration calorimetry; s.d. values were <10% of the mean. Kinetic parameters were calculated by fitting data to the Michaelis-Menten or Hill equation using GraphPad Prism Software (version 6.00 for Windows, GraphPad Software, San Diego, CA). In vivo fructose 1-phosphate concentrations in diploid yeast cells were calculated using cell volume data from the BioNumbers database (http://bionumbers.hms.harvard.edu/).
Assays with phosphopantothenate (PPa), phosphopantetheine (PPaSH) and their damaged products (PPaSS, PPaSSO3, PPaSO3, cPPa, and cPPaSH; Fig. 3c) were made in 50-μl reaction mixtures containing 50 mM Tris-HCl, pH 7.5, 0.5 mM metal (Ni2+ for Arabidopsis and P. horikoshii, Co2+ for human and yeast), 2 mM substrate, and 0.5 μg DUF89 protein. After incubation at 30 °C (55 °C for P. horikoshii PH1575) for 30 min, phosphate release was quantified as above. Assays were replicated three times. Activities against cyclic phosphates were assayed by HPLC. For kinetic analyses, the assay buffer was supplemented with 1 mg/ml bovine serum albumin alone (for Arabidopsis) or plus 10% (v/v) glycerol (for human).
Acyl carrier protein phosphodiesterase assays (20 μl) contained 50 mM Tris-HCl, pH 8.0, 1.0 mM MgCl2, 0.1 mM MnCl2, 1.0 mM DTT, and 50 μM spinach holo-acyl carrier protein. Reactions were started by adding 1 μg of DUF89 or 1 μg of E. coli AcpH54 (as a positive control). After incubation for 90 min at 30 °C, reactions were mixed with 480 μl of 50 mM Hepes-NaOH, pH 8.0, 5 mM EDTA, 20 μM DTT, 2 mM monobromobimane and incubated for 30 min at 30 °C. Methanesulfonic acid (2 M, 6.3 μl) was then added and excess monobromobimane was removed by partitioning against 1 ml of dichloromethane. After centrifugation (15,000g, 15 min) the aqueous phase was analyzed by fluorometric HPLC. Pantetheine and phosphopantetheine derivatives were analyzed (20-μl injections) using a Microsorb C18 column (150 × 4.6 mm, 5 μm particle size, Agilent) and a linear gradient of methanol in 0.25% (v/v) acetic acid titrated to pH 3.5 with 10 M NaOH. Gradient parameters were: 20% methanol for 3 min, an increase to 60% methanol over 15 min, then a return to 20% methanol over 2 min and re-equilibration in 20% methanol for 5 min. Flow rate was 1.2 ml/min. Detector excitation and emission wavelengths were 388 nm and 480 nm.
Yeast growth and metabolomics analysis.
Diploid yeast ΔYMR027W strain Y30602 and the corresponding wild type strain BY4743 (from Euroscarf) were cultured at 30 °C in SD medium containing appropriate supplements and 2% (w/v) glucose or fructose, monitoring growth at OD600nm. When OD600nm reached 0.5 or 1.0, cells were harvested, extracted, and processed for GC-MS analysis as described55; 1 OD600nm was taken as equivalent to 3 × 107 cells/ml (http://bionumbers.hms.harvard.edu/bionumber.aspx?&id=100986&ver=3). The statistical significance of differences in metabolite contents was evaluated using Student’s t-test.
Crystallization and structure determination of YMR027W.
Crystals of selenomethionine-substituted YMR027W–Mg2+ were grown at 22 °C using the sitting-drop vapor diffusion method by mixing 0.5 μl of protein solution (22 mg/ml, pretreated with chymotrypsin, 1/70, v/v) with 0.5 μl of reservoir solution containing 0.1 M Bis-tris (pH 5.5), 0.2 M MgCl2 and 25% PEG 3350. Crystals of YMR027W–Mn2+ were grown using 0.5 μl of reservoir solution containing 0.5 M ammonium dihydrogen phosphate, 50 mM MnCl2 and 25% PEG 3350. For crystal soaking, the crystals of native YMR027W–Mg2+ were grown as above and soaked in 10 mM fructose 6-phosphate (in 0.1 M Bis-tris, pH 5.5, 0.2 M MgCl2, 25% PEG 3350, 8% glycerol) for 10 min at room temperature. The crystals were cryoprotected by Paratone-N oil and flash-frozen in liquid N2. X-ray diffraction data for the YMR027W–Mg2+ (0.9794 Å) and YMR027W–Mn2+ (0.9794 and 1.897 Å) crystals were collected at the beam-lines 19-ID and 19-BM (Structural Biology Center, Advanced Photon Source, Argonne National Laboratory). Data were processed using the HKL-3000 suite of programs56, and the structure of YMR027W–Mg2+ was determined by the Se-methionine SAD phasing, density modification, and initial model building using the HKL-3000 software package57,58. X-ray diffraction data for the YMR027W–Mg2+–fructose 6-phosphate crystals were collected using the Rigaku FRE-Superbright rotating anode on a Rigaku R-Axis IV++ image plate (Rigaku Americas, TX). The data were processed using the programs MOSFLM59 and Scala60. The structures of the YMR027W–Mg2+–fructose 6-phosphate and YMR027W–Mn2+ complexes were determined by molecular replacement using the coordinates of the YMR027W–Mg2+ monomer as a search model and the program MOLREP of the CCP4 suite57. Cycles of manual model correction were carried out using COOT and refined with Phenix58. The final model was refined against all reflections except for 5% randomly selected reflections, which were used for monitoring Rfree. Data collection and refinement statistics are summarized in Supplementary Table 5.
Supplementary Material
Acknowledgments
This work was supported by US National Science Foundation grants MCB-1153413 and IOS-1025398 (to A.D.H.) and MCB-1153491 (to O.F.); by Genome Canada, the Ontario Genomics Institute (2009-OGI-ABC-1405), the Ontario Research Fund (ORF-GL2-01-004), and NSERC Strategic Network grant IBN (to A.F.Y.); by National Institutes of Health grant GM094585 and the US Department of Energy, Office of Biological and Environmental Research, contract DE-AC02-06CH11357 (to A.J.); and by the C.V. Griffin Sr. Foundation (to A.D.H.). J.S. was supported by US Department of Energy, Office of Basic Energy Sciences, grant DOE KC0304000. We thank J.E. Cronan and H.W. Park for enzymes, K. Kodama of Ikeda Corp. of America for pantetheine-S-sulfonate, S. Gerdes for bioinformatic help, E. van Schaftingen for advice on metabolism, and J.D. Rabinowitz for pilot metabolomics.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
Additional information
Any supplementary information, chemical compound information and source data are available in the online version of the paper.
References
- 1.Galperin MY & Koonin EV From complete genome sequence to ‘complete’ understanding? Trends Biotechnol. 28, 398–406 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bitto E, Bingman CA, Allard ST, Wesenberg GE & Phillips GN Jr. The structure at 1.7 A resolution of the protein product of the At2g17340 gene from Arabidopsis thaliana. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun 61, 630–635 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tilton GB, Wedemeyer WJ, Browse J & Ohlrogge J Plant coenzyme A biosynthesis: characterization of two pantothenate kinases from Arabidopsis. Plant Mol. Biol 61, 629–642 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Hörtnagel K, Prokisch H & Meitinger T An isoform of hPANK2, deficient in pantothenate kinase-associated neurodegeneration, localizes to mitochondria. Hum. Mol. Genet 12, 321–327 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Gasch AP et al. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol. Biol. Cell 12, 2987–3003 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alvaro D, Lisby M & Rothstein R Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet. 3, e228 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perry JJ et al. Human C6orf211 encodes Armt1, a protein carboxyl methyltransferase that targets PCNA and is linked to the DNA damage response. Cell Rep. 10, 1288–1296 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hernick M & Fierke CA Mechanisms of metal-dependent hydrolases in metabolism in Comprehensive Natural Products II: Chemistry and Biology (eds. Mander L & Liu HW) 47–581 (Elsevier, Amsterdam, 2010). [Google Scholar]
- 9.Linster CL, Van Schaftingen E & Hanson AD Metabolite damage and its repair or pre-emption. Nat. Chem. Biol 9, 72–80 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Galperin MY, Moroz OV, Wilson KS & Murzin AG House cleaning, a part of good housekeeping. Mol. Microbiol 59, 5–19 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Reaves ML, Young BD, Hosios AM, Xu YF & Rabinowitz JD Pyrimidine homeostasis is accomplished by directed overflow metabolism. Nature 500, 237–241 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frelin O et al. A directed-overflow and damage-control N-glycosidase in riboflavin biosynthesis. Biochem. J 466, 137–145 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suhre K Inference of gene function based on gene fusion events: the rosetta-stone method. Methods Mol. Biol 396, 31–41 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Gerdes S et al. Plant B vitamin pathways and their compartmentation: a guide for the perplexed. J. Exp. Bot 63, 5379–5395 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Abiko Y Investigations on pantothenic acid and its related compounds. IX. Biochemical studies.4. Separation and substrate specificity of pantothenate kinase and phosphopantothenoylcysteine synthetase. J. Biochem 61, 290–299 (1967). [DOI] [PubMed] [Google Scholar]
- 16.Yoshioka M & Tamura Z Bifidus factors in carrot. II. The structure of the factor in fraction IV Chem. Pharm. Bull (Tokyo: ) 19, 178–185 (1971). [Google Scholar]
- 17.Nakamura H & Tamura Z Studies on the metabolites in urine and feces of rat after oral administration of radioactive pantethine Chem. Pharm. Bull (Tokyo: ) 20, 2008–2016 (1972). [DOI] [PubMed] [Google Scholar]
- 18.Pierpoint WS, Hughes DE, Baddiley J & Mathias AP The phosphorylation of pantothenic acid by Lactobacillus arabinosus 17–5. Biochem. J 61, 368–374 (1955). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baddiley J & Thain EM Coenzyme-A. 6. The identification of pantothenic acid-4′ and −2′−4′ phosphates from a hydrolysate. J. Chem. Soc 1952, 3783–3789 (1952). [Google Scholar]
- 20.Hanson AD, Pribat A, Waller JC & de Crécy-Lagard V ‘Unknown’ proteins and ‘orphan’ enzymes: the missing half of the engineering parts list—and how to find it. Biochem. J 425, 1–11 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Overbeek R et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33, 5691–5702 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lukens L & Flaks J Intermediates in purine nucleotide synthesis. Methods Enzymol. 6, 671–702 (1963). [Google Scholar]
- 23.Chaudhuri B, Ingavale S & Bachhawat AK apd1+, a gene required for red pigment formation in ade6 mutants of Schizosaccharomyces pombe, encodes an enzyme required for glutathione biosynthesis: a role for glutathione and a glutathione-conjugate pump. Genetics 145, 75–83 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donaldson IA, Doyle TC & Matas N Expression of rat liver ketohexokinase in yeast results in fructose intolerance. Biochem. J 291, 179–186 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerin I et al. Identification of TP53-induced glycolysis and apoptosis regulator (TIGAR) as the phosphoglycolate-independent 2,3-bisphosphoglycerate phosphatase. Biochem. J 458, 439–448 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Weinberg MV, Jenney FE Jr., Cui X & Adams MW Rubrerythrin from the hyperthermophilic archaeon Pyrococcus furiosus is a rubredoxin-dependent, iron-containing peroxidase. J. Bacteriol 186, 7888–7895 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dauter Z, Wilson KS, Sieker LC, Moulis JM & Meyer J Zinc- and iron-rubredoxins from Clostridium pasteurianum at atomic resolution: a high-precision model of a ZnS4 coordination unit in a protein. Proc. Natl. Acad. Sci. USA 93, 8836–8840 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Midelfort CF, Gupta RK & Rose IA Fructose 1,6-bisphosphate: isomeric composition, kinetics, and substrate specificity for the aldolases. Biochemistry 15, 2178–2185 (1976). [DOI] [PubMed] [Google Scholar]
- 29.Hou X et al. Crystallographic studies of human MitoNEET. J. Biol. Chem 282, 33242–33246 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Hagen WR et al. Novel structure and redox chemistry of the prosthetic groups of the iron-sulfur flavoprotein sulfide dehydrogenase from Pyrococcus furiosus; evidence for a [2Fe-2S] cluster with Asp(Cys)3 ligands. J. Biol. Inorg. Chem 5, 527–534 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Allen KN & Dunaway-Mariano D Markers of fitness in a successful enzyme superfamily. Curr. Opin. Struct. Biol 19, 658–665 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aravind L & Koonin EV The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem. Sci 23, 469–472 (1998). [DOI] [PubMed] [Google Scholar]
- 33.Mildvan AS et al. Structures and mechanisms of Nudix hydrolases. Arch. Biochem. Biophys 433, 129–143 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Levi B & Werman MJ Fructose and related phosphate derivatives impose DNA damage and apoptosis in L5178Y mouse lymphoma cells. J. Nutr. Biochem 14, 49–60 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Huh WK et al. Global analysis of protein localization in budding yeast. Nature 425, 686–691 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Goyer A et al. A cross-kingdom Nudix enzyme that pre-empts damage in thiamin metabolism. Biochem. J 454, 533–542 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Fortpied J, Gemayel R, Stroobant V & van Schaftingen E Plant ribulosamine/erythrulosamine 3-kinase, a putative protein-repair enzyme. Biochem. J 388, 795–802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rock CO, Calder RB, Karim MA & Jackowski S Pantothenate kinase regulation of the intracellular concentration of coenzyme A. J. Biol. Chem 275, 1377–1383 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Rothmann M et al. Metabolic perturbation of an essential pathway: evaluation of a glycine precursor of coenzyme A. J. Am. Chem. Soc 135, 5962–5965 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson DC, Dean DR, Smith AD & Johnson MK Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem 74, 247–281 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Cvetkovic A et al. Microbial metalloproteomes are largely uncharacterized. Nature 466, 779–782 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Smith JL et al. Structure of the allosteric regulatory enzyme of purine biosynthesis. Science 264, 1427–1433 (1994). [DOI] [PubMed] [Google Scholar]
- 43.Dupureur CM Roles of metal ions in nucleases. Curr. Opin. Chem. Biol 12, 250–255 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Lu Z, Dunaway-Mariano D & Allen KN The catalytic scaffold of the haloalkanoic acid dehalogenase enzyme superfamily acts as a mold for the trigonal bipyramidal transition state. Proc. Natl. Acad. Sci. USA 105, 5687–5692 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Corpet F Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16, 10881–10890 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Larkin MA et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Tamura K et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leonardi R et al. A pantothenate kinase from Staphylococcus aureus refractory to feedback regulation by coenzyme A. J. Biol. Chem 280, 3314–3322 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Stanford M On the determination of cysteine as cysteic acid. J. Biol. Chem 238, 235–237 (1962). [Google Scholar]
- 50.Kuznetsova E et al. Genome-wide analysis of substrate specificities of the Escherichia coli haloacid dehalogenase-like phosphatase family. J. Biol. Chem 281, 36149–36161 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Broadwater JA & Fox BG Spinach holo-acyl carrier protein: overproduction and phosphopantetheinylation in Escherichia coli BL21(DE3), in vitro acylation, and enzymatic desaturation of histidine-tagged isoform I. Protein Expr. Purif 15, 314–326 (1999). [DOI] [PubMed] [Google Scholar]
- 52.Olivares JA Inductively coupled plasma-mass spectrometry. Methods Enzymol. 158, 205–222 (1988). [DOI] [PubMed] [Google Scholar]
- 53.Högbom M et al. A high throughput method for the detection of metalloproteins on a microgram scale. Mol. Cell. Proteomics 4, 827–834 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Thomas J & Cronan JE The enigmatic acyl carrier protein phosphodiesterase of Escherichia coli: genetic and enzymological characterization. J. Biol. Chem 280, 34675–34683 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Niehaus TD et al. Genomic and experimental evidence for multiple metabolic functions in the RidA/YjgF/YER057c/UK114 (Rid) protein family. BMC Genomics 16, 382 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Minor W, Cymborowski M, Otwinowski Z & Chruszcz M HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr 62, 859–866 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr 50, 760–763 (1994). [DOI] [PubMed] [Google Scholar]
- 58.Terwilliger TC & Berendzen J Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr 55, 849–861 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leslie AG The integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr 62, 48–57 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Evans P Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr 62, 72–82 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.