Abstract
The ability of LuxR-type proteins to regulate transcription is controlled by bacterial pheromones, N-acylhomoserine lactones (AHLs). Most LuxR-family proteins require their cognate AHLs for activity, and some of them require AHLs for folding and stability, and for protease-resistance. However, a few members of this family are able to fold, dimerize, bind DNA, and regulate transcription in the absence of AHLs; moreover, these proteins are antagonized by their cognate AHLs. One such protein is YenR of Yersinia enterocolitica, which is antagonized by N-3-oxohexanoyl-l-homoserine lactone (OHHL). This pheromone is produced by the OHHL synthase, a product of the adjacent yenI gene. Another example is CepR2 of Burkholderia cenocepacia, which is antagonized by N-octanoyl-L-homoserine lactone (OHL), whose synthesis is directed by the cepI gene of the same bacterium. Here, we describe the high-resolution crystal structures of the AHL binding domains of YenR and CepR2. YenR was crystallized in the presence and absence of OHHL. While this ligand does not cause large scale changes in the YenR structure, it does alter the orientation of several highly conserved YenR residues within and near the pheromone-binding pocket, which in turn caused a significant movement of a surface-exposed loop.
Keywords: Burkholderia cenocepacia, CepR2, pheromones, Quorum sensing, transcriptional regulation, Yersinia enterocolitica, YenR
1. INTRODUCTION
Many species of bacteria are able to synchronize gene expression with sibling cells using mechanisms that involve diffusible chemical signals, or pheromones. Bacteria use these molecules to regulate diverse behaviors, including bioluminescence, horizontal DNA transfer, biofilm formation, pathogenesis, and the production of virulence factors, antimicrobials and other secondary metabolites.1–4 These signals enable bacteria to estimate their population density, a process often referred to as quorum sensing. In the past few years, entirely new classes of pheromones have been described, and more are likely to be discovered in the near future. The fact that diffusible signals allow large numbers of bacteria to act in synchrony may blur the distinction between single cells and multicellular organisms.
The most intensively studied proteobacterial pheromones are N-acylhomoserine lactones (AHLs). These signal molecules have invariant homoserine lactone head groups linked to diverse hydrophobic acyl groups that differ in length, oxidation, and the level of saturation.3,4 While most of these side chains are linear acyl groups ranging from 4 to 18 carbons in length, a few have branched chains or aromatic moieties.5–7 The first described member of this family is N-3-oxohexanoyl-L-homoserine lactone (OHHL) of the marine bioluminescent bacterium Aliivibrio fischeri. OHHL is synthesized by LuxI and sensed by the OHHL-dependent transcription factor LuxR.8,9 LuxR and its homologues have two domains, an N-terminal domain that binds the pheromone, and a C-terminal domain that binds specific DNA sequences that lie near target promoters.10,11 As the population density of A. fischeri cells increases, the concentration of external OHHL increases. When the concentration of this signal reaches the nanomolar threshold, its passive efflux from the cells becomes balanced by an influx, so that it can interact with LuxR. LuxR-OHHL complexes bind to a sequence within the promoter of the luxICDABEG operon and activate its transcription, resulting in bioluminescence. One of the induced genes is luxI, which encodes the OHHL synthase itself, leading to positive feedback, and potentially a bistable switch.
Proteins homologous to LuxI and LuxR are widely distributed among the Proteobacteria. Perhaps the best studied examples are the TraI/TraR system of Agrobacterium tumefaciens, which uses N-3-oxooctanoyl-L-homoserine lactone (OOHL) as a signal and regulates the conjugation and replication genes of the Ti plasmid,12 and the LasI/LasR system of Pseudomonas aeruginosa, which uses the signal N-3-oxododecyl-L-homoserine lactone (OdDHL) to regulate a large number of genes including several that encode pathogenesis-associated proteins.13 P. aeruginosa also encodes a second quorum-sensing system composed of RhlR and RhlI, and an orphan AHL receptor called QscR that detects OdDHL.13
Most LuxR-type proteins cannot function in the absence of AHLs and at least some of these require AHLs for folding into a soluble, stable and protease-resistant form.14–17 However, some LuxR-type receptors function preferentially in the absence of their cognate pheromone and their DNA-binding activities are inhibited by these cognate signals.18 We refer to LuxR-type proteins having this property as “quorum-hindered.” Some LuxR-type proteins of this type form a well-defined monophyletic clade within the LuxR family, including EsaR of Pantoea stewartii and YenR of Yersinia enterocolitica, the subject of the current study. The CepR2 protein of Burkholderia cenocepacia is also quorum-hindered,19 but is only distantly related to EsaR and YenR.
Y. enterocolitica is an enteric Gammaproteobacterium that can colonize the gastro-intestinal tract by ingestion of contaminated food or water or by handling of infected animals.20 It is closely related to Y. pestis, the causative agent of Bubonic Plague, and a select agent of concern in biowarfare or bioterrorism.21 Y. enterocolitica colonizes the small intestine of humans and other mammals, causing a disease referred to as yersiniosis, whose effects are variable, depending partly on the age of the victim, and can include watery or bloody diarrhea, acute pain (sometimes misdiagnosed as appendicitis), and fever.20,22 Symptoms usually appear 4 to 7 days after exposure and may last 1 to 3 weeks or longer. After recovery, many infected individuals become asymptomatic carriers. These bacteria can also invade the Peyers patches of the small intestine and spread to lymph nodes. They can form abscesses in the spleen and liver of immunocompromised patients. Infection of the intestine is usually self-limiting and does not require treatment, while septicemia can be treated with antibiotics.
B. cenocepacia is a member of the so-called Burkholderia cepacia complex (BCC), which are opportunistic pathogens of humans and, in particular, are a serious threat to cystic fibrosis (CF) patients.23–25 Colonization of the CF lung by B. cenocepacia tends to occur in patients already infected with another opportunistic pathogen, P. aeruginosa. It is thought that an infection caused by both organisms can have a synergistic effect and result in serious clinical complications. Furthermore, B. cenocepacia is inherently resistant to many antibiotics, making it virtually impossible to eradicate from the lungs of CF patients.23 Infections with B. cenocepacia may have variable clinical outcomes ranging from asymptomatic carriage to a sudden fatal deterioration in lung function.23
The yenR gene of Y. enterocolitica is adjacent to its yenI gene, which encodes the OHHL synthase. The two genes are transcribed convergently and overlap at their 3′ ends by two codons. The 3′ untranslated region of each transcript is necessarily complementary to the coding region of the other, suggesting that expression of the yenR and yenI genes may be antagonistic, either via RNA polymerase collisions or by hybridization of the two countertranscribed mRNAs. The possibility that expression of these genes is antagonistic is potentially significant given that the protein product of one gene also antagonizes the other.
The 5′ end yenR gene is adjacent to a divergent gene designated yenS. The yenS gene encodes a small RNA whose expression is stimulated by apo-YenR. YenS inhibits the expression of YenI and also plays a positive role in swarming motility.26 YenR activates transcription preferentially in the absence of OHHL, though it retains some activity in the presence of saturating levels of this pheromone.26 Purified apo-YenR binds non-cooperatively to two 20-bp long sites that lie upstream of yenS. DNA binding occurs in the absence of OHHL, and YenR is largely released from the DNA by addition of the OHHL pheromone.26
The cepR2 gene lies within a genomic island that is present only in some strains of B. cenocepacia.27 This island includes an AraC-type transcription regulator homolog encoded by cepS, four genes predicted to direct the synthesis of a secondary metabolite, and four genes that are predicted to direct the efflux of this metabolite. All B. cenocepacia strains encode cepI, which linked to cepR. CepR is an OHL-dependent transcriptional activator.16,28 It is striking therefore that CepR and CepR2 detect the same pheromone but respond in opposite ways, in that CepR is a ligand-dependent transcriptional activator, while CepR2 is a ligand-hindered transcriptional repressor. In both cases, the ligand stimulates expression of target genes. Although these proteins detect the same ligand, they are not closely related, as their ligand binding domains are only 30% identical and 47% similar. While CepR2 is found only in a few isolates of B. cenocepacia, CepR and CepI orthologs are widely conserved throughout the genus.
To date, the structures of five LuxR-type proteins have been determined. The first was full length TraR of Agrobacterium tumefaciens, crystallized with the pheromone 3-oxooctanoyl-homoserine lactone (OOHL) and target DNA.29,30 CviR of Chromobacter violaceum was crystallized as a full length dimer complexed with the antagonist 4-(chlorophenoxy)-butanoyl-HSL, as a full length monomer with the ligand hexanoyl-homoserine lactone, and as dimers of the N-terminal domain complexes with a variety of AHLs.31 The N-terminal domain of LasR of P. aeruginosa was crystallized with 3-oxo-dodecanoyl-HSL (OdDHL).32,33 Full length QscR of the same organism was crystallized with OdDHL.34 Full length SdiA was crystallized with OOHL, and with OHHL, and with an unexpected compound octanoyl-rac-glycerol.35 This compound was apparently co-purified with SdiA from extracts of E. coli, although there are few if any reports that E. coli can synthesize such a compound. In a separate study, full length SdiA was crystallized without ligand, though the binding pocket contained two molecules of tetraethylene glycol, a component of the crystallization buffer.36
Quorum-hindered members of the LuxR family provide an attractive opportunity to study protein-pheromone interactions in the presence and absence of ligand. Those LuxR-type proteins that require AHLs for activity generally also require them for folding into a soluble and stable, protease-resistant state.14,15,17 Therefore, it has not been possible to purify those proteins in the absence of their pheromones, and thus impossible to compare such proteins in their liganded and unliganded forms. In contrast, YenR is fully soluble in both states, allowing us to compare apo-YenR with YenR-OHHL complexes at near atomic resolution. The structure of apo-CepR2 provides a second example of a LuxR-type protein that folds and functions in the absence of an AHL.
2. MATERIALS AND METHODS
2.1. Cloning and purification of YenR-LBD and CepR2-LBD
The N-terminal domain of YenR was fused at its C-terminus to a hexahistidyl tag by PCR amplification of the gene using primers 5′-GATATACATATGATAATTGAC-3′ and 5′-GGGAGATCTTCAGTGATGGTGATGGTGATGGTCTCGCTCTATTTTCGGGT-3′. The resulting PCR fragment had an NdeI site at the start codon of yenR and a BglII site at the opposite end. This fragment included codons 1–179 of yenR followed by six His-encoding codons, followed by TGA, which encodes a stop codon, followed by the BglII site. This fragment was digested with NdeI and BglII, and introduced into plasmid pRSETA after digestion with NdeI and BglII. The resulting plasmid, pGF102, was introduced into E. coli strain BL21 (DE3) carrying plasmid pMAGIC, which encodes the rare E. coli tRNA (Arg [AGG/AGA]).
The CepR2-LBD (1–170) gene was amplified from B. cenocepacia J2315 genomic DNA with KOD DNA polymerase using the forward primer 5′-TACTTCCAATCCAATGCCATGGACCTGACAATACTGCACG ACT–3′ and the reverse primer 5′-TTATCCACTTCCAATGTTACGCG ATCGGATCGGCGAG-3′ with conditions and reagents provided by the vendor (Novagen, Madison, WI). The gene was cloned into plasmid pMCSG68 using a modified ligation-independent cloning protocol.37,38 The pMCSG68 vector encodes a TEV protease cleavage site and creates a construct with a cleavable His6-tag fused into the N-terminus of the target protein and adds three artificial residues (Ser-Asn-Ala) on that end. The resulting plasmid was introduced into E. coli strain BL21 (DE3) carrying plasmid pMAGIC.
YenR-LBD and CepR2-LBD were purified from the respective strains by immobilized nickel-affinity chromatography (IMAC). The cells were cultured using selenomethionine (SeMet) containing enriched M9 medium and under conditions known to inhibit methionine biosynthesis.39 The cells were cultured at 37°C to an OD600 of 1 and protein expression was induced with 0.5 mM IPTG in the presence of SeMet (90 mg per L of culture) at 18°C. The cells were grown overnight with shaking at 18°C. The harvested cells were re-suspended in 4 volumes of lysis buffer (50 mM HEPES pH 8.0, 500 mM NaCl, 20 mM imidazole, 10 mM β-mercaptoethanol, and 5% v/v glycerol) and stored at −80°C. The thawed cells were lysed by sonication after the addition of inhibitors of proteases (Sigma, P8849). The lysate was clarified by centrifugation at 30,000g (RC5C-Plus centrifuge, Sorvall) for 90 min, followed by filtration through a 0.45 μm filter (Gelman). The filtrate was purified by IMAC using a 5-mL HisTrap HP column charged with Ni+2 ions followed by buffer-exchange chromatography on a HiPrep 26/10 desalting column (both GE Healthcare Life Sciences), carried out using an ÄKTAxpress system (GE Healthcare Life Sciences). The YenR-LBD with an un-cleavable His-tag in the C-terminus was then further purified by size-exclusion chromatography using a HiLoad superdex 200 26/600 PG column on an ÄKTAxpress system. For the CepR2-LBD, the N-terminal His6-tag was cleaved using recombinant TEV protease expressed from the vector pRK508. The protease was added to the target protein at a mass ratio of 1:30 and the mixture was incubated at 4°C for 48 h. The CepR2-LBD protein was then purified by applying a second IMAC step using the same Ni+2 ion charged column. The flowthrough and the wash fractions contain cleaved CepR2-LBD while the His6-tag, uncleaved proteins, His-tagged TEV protease were retained by the column. Both proteins were dialyzed against a solution containing 20 mM HEPES (pH 8.0), 250 mM NaCl, 2 mM DTT and concentrated using Amicon Ultra-15 centrifugal filter units (Millipore) to 48 mg/ml for YenR-LBD and to 14 mg/ml for CepR2-LBD.
2.2. Protein crystallization and data collection
Initial crystallization screens were set up using a Mosquito robot (TTP Labtech) and sitting drop vapor diffusion technique in a 96-well CrystalQuick plate (Greiner). The crystallization conditions for the proteins were screened using commercially available 96-well format screening kits including MCSG1–4 (Microlytic, Woburn, MA), PEG/Ion, and Index (Hampton) by vapor diffusion in hanging drops by mixing 0.4 μl of the protein solution with 0.4 μl of reservoir solution of crystallization conditions. The YenR-LBD protein was crystallized from MCSG-2 suite condition H9 containing 0.1 M sodium acetate pH 4.6 (pH was adjusted using HCl) and ammonium sulfate and equilibrated at 289 K over 140 μl of the same reservoir solution. Crystals were washed for a few seconds in reservoir solution plus 15–25% glycerol, ethylene glycol or glycerol-ethylene glycol mixture as a cryo-protectant, then flash-cooled in liquid nitrogen prior to data collection. The crystals of the YenR-LBD-OHHL complex were prepared by soaking the YenR-LBD crystals in the same reservoir condition plus 50 mM OHHL for 15 min. These soaked crystals were washed with the same cryo-protectant solution for a few seconds and flash-cooled in liquid nitrogen prior to data collection. The CepR2-LBD protein was crystallized in PEG/Ion suite condition F9 (0.2 M Sodium formate, 20% w/v polyethylene glycol 3350).
All diffraction data were collected at 100 K on the ADSC quantum Q315r charged coupled device detector in the 19-ID beamline of the Structural Biology Center at the Advanced Photon Source, Argonne National Laboratory.40 Single wavelength anomalous dispersion (SAD) data near the selenium absorption peak (0.9792 Å) were collected from SeMet-substituted proteins. All diffraction data were processed by using the HKL3000 suite of programs.41 For the YenR-LBD, 21587 unique reflections up to 2.20 Å resolution with the space group C2221 were obtained. For the YenR-LBD-OHHL crystal, 29198 reflections were collected, with the same (as the apo-form) space group C2221 up to 2.00 Å resolution. For CepR2-LBD, the 2.75 Å, P21 data with 48,922 reflections were collected. Data collection statistics are summarized in Table 1.
TABLE 1.
Data collection and refinement statistics
| YenR-LBD | YenR-LBD-OHHL | CepR2 | |
|---|---|---|---|
| Data collection | |||
| Space group | C2221 | C2221 | P21 |
| Cell dimensions | |||
| a, b, c (Å) | 64.37, 123.40, 106.91 | 63.88, 122.78, 107.60 | 120.28, 74.59, 118.03 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 117.3, 90 |
| Protein molecules/ASU | 2 | 2 | 6 |
| Temperature (K) | 100 | 100 | 100 |
| Radiation source | APS, 19-ID | APS, 19-ID | APS, 19-ID |
| Wavelength (Å) | 0.9792 | 0.9792 | 0.9792 |
| Resolution (Å)a | 2.20 (2.23–2.20) | 2.00 (2.03–2.00) | 2.75 (2.80–2.75) |
| Unique reflections | 21587 (1061) | 29198 (1422) | 48922 (2403) |
| Rmergeb | 0.098 (0.703) | 0.082 (0.616) | 0.095 (0.798) |
| 〈I〉/〈σI〉 | 17.1/3.1 | 23.9/5.3 | 12.6/1.9 |
| Completeness (%) | 97.3 (95.8) | 99.9 (100) | 99.1 (99.4) |
| Redundancy | 5.5 (4.9) | 6.5 (6.6) | 3.1 (2.9) |
| Refinement | |||
| Resolution (Å) | 2.20 | 2.00 | 2.75 |
| Reflections: work/test set | 19505/1054 | 26360/2774 | 40872/2158 |
| Rcryst/Rfreec | 0.175/0.214 | 0.171/0.203 | 0.184/0.218 |
| No. of atoms: protein/ligandsd/water | 2690/64/75 | 2718/93/136 | 7911/61/100 |
| Average B factor (Å2): protein/ligands/water | 45.9/77.0/43.1 | 34.7/49.2 (24.9)e/38.8 | 51.9/72.4/37.9 |
| Bond lengths (Å) | 0.008 | 0.009 | 0.004 |
| Bond angles (°) | 0.892 | 0.997 | 0.661 |
| Ramachandran plot | |||
| Most favored | 96.9 | 98.8 | 97.3 |
| Outliers | 0.0 | 0.0 | 0.41 |
| PDB code | 5L07 | 5L09 | 5L10 |
| Crystallization conditions | 0.1 M sodium acetate pH 4.5, 2.5 M ammonium sulfate, 16°C | 0.1 M sodium acetate pH 4.5, 2.5 M ammonium sulfate, 16°C | 0.2 M Sodium formate, 20% w/v Polyethylene glycol 3,350 |
ASU, Asymmetric Unit.
Values in parentheses correspond to the highest-resolution shell.
Rmerge = ΣhklΣi|Ii(hkl)- 〈I(hkl)〉|/ΣhklΣi|〈Ii(hkl)〉|, where Ii(hkl) is the intensity for the ith measurement of an equivalent reflection with indices h, k, and l.
Rcryst = Σhkl||Fobs| - |Fcalc||/Σhkl |Fobs|, where Fobs and Fcalc are observed and calculated structure factors, respectively. Rfree is calculated analogously for the test reflections, which were randomly selected and excluded from the refinement.
Ligands include all atoms excluding protein and water atoms.
Average B factor for OHHL.
2.3. Structure determination, refinement, and analysis
YenR-LBD and CepR2-LBD structures were solved by the SAD phasing method using selenium near absorption peak data. The YenR-LBD-OHHL structure was determined by molecular replacement using the YenR-LBD structure as a search model. All procedures for SAD phasing, phase improvement by density modification, and initial protein model building as well as molecular replacement phasing were done by SHELXC/D/E,42,43 MLPHARE,44 dm,45 HKL Builder (or BUCCANEER),46 and MOLREP47 as a part of the structure module of the HKL3000 software package.41 The mean figure of merit and the phasing power of the phase set for YenR-LBD were 0.282 and 1.22 for 50–2.40 Å data and for CepR2-LBD, they were 0.315 and 1.27 for 50–3.86 Å data. After the density modification (DM) and the phase extension to observed data resolution limits the structures were built using HKL builder. For the YenR-LBD structure, the HKL Builder built 309 out of 358 residues (2 protein molecules of 179 residues in the asymmetric unit). A total of 979 out of 1020 residues (6 protein molecules of 170 residues in the asymmetric unit) were built by HKL Builder for CepR2-LBD. For the YenR-LBD-OHHL complex structure, a monomer of the refined YenR-LBD model (5–169) was used as a search model for the molecular replacement using MOLREP.47 For the refinement, iterative alternating rounds of phenix.refine48 and COOT49 were used for computation and manual adjustment, respectively, until the models converged with the corresponding data. The YenR-LBD was refined with all data to Rcrystal/Rfree of 17.5/21.4% to 2.20 Å, and 17.1/20.3% to 2.00 Å, and 18.4/21.8% to 2.75 Å for YenR-LBD-OHHL and CepR2-LBD, respectively. The final model of the YenR-LBD structure contains two protein chains (chains A and B) with residues A:5–169 (92–94 missing) and B:5–169 (93–94 missing) and 4 sulfate molecules, 5 acetate molecules, 6 ethylene glycol molecules and 75 well-ordered water molecules. The final YenR-LBD-OHHL structure has two protein chains (chains A and B) of A:2–164 (92–94 missing) and B:5–169, with each protein molecule holding a well-defined OHHL in its binding site as well as 8 ethylene molecules, 4 acetate molecules, 3 sulfate molecules and 136 well-ordered water molecules. The final model of the CepR2-LBD structure consists of three protein dimers (chains A-F, A:−1–170, B:−1–168, C:0–167, D:0–167, E:−1–167, and F:0–167). In addition, there are 7 glycerol molecules, 5 formate molecules, one ethylene molecule, and 100 ordered water molecules. Both glycerol and ethylene molecules may be from the cryo-solvent used which contains both components. The final detailed refinement parameters are shown in Table 1. The stereochemistry of the structures were checked with a Ramachandran plot and Molprobity50 and the final structures and the data for YenR-LBD, YenR-LBD-OHHL, and CepR2-LBD were deposited in the PDB with PDBIDs of 5L07, 5L09, 5L10, respectively.
3. RESULTS
3.1. YenR tertiary structure
We overproduced and purified full length YenR and the YenR N-terminal ligand binding domain (YenR-LBD, residues 1–179), which was predicted to contain the full pheromone-binding domain and the interdomain linker. Full length apo-YenR tended to precipitate at high protein concentrations and was not characterized further. In contrast, apo-YenR-LBD remained highly soluble and was purified by immobilized nickel-affinity chromatography. Apo-YenR-LBD was crystallized by vapor diffusion in sitting drops as described above. These crystals were then incubated in a buffer containing OHHL to obtain crystals of protein-pheromone complexes. The crystal structure of apo-YenR-LBD was determined at 2.20 Å resolution while YenR-LBD-OHHL complexes were determined at 2.00 Å.
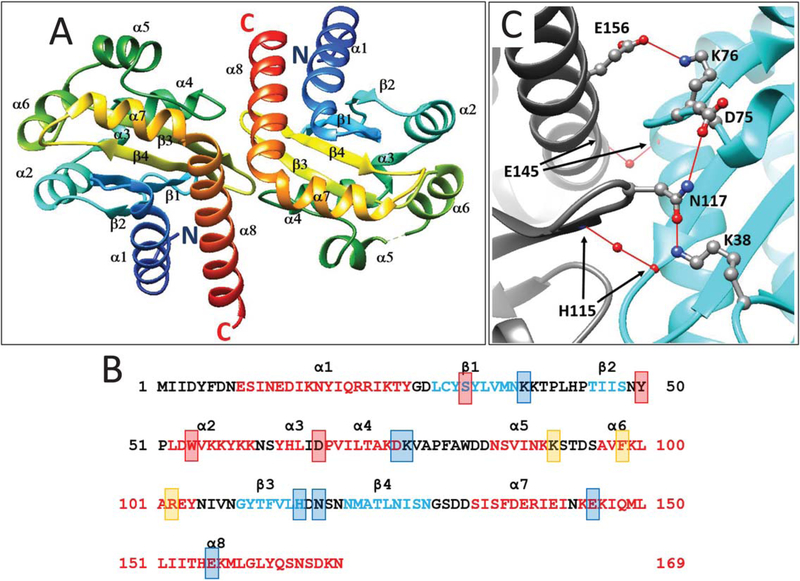
YenR-LBD forms dimers both in the presence and absence of pheromone and shows two-fold symmetry (Figure 1A). Each subunit of the dimer folds into an α/β/α sandwich. The core of each subunit consists of four antiparallel β strands (β1, residues 29–37, β2, residues 45–48, β3, residues 109–115, and β4, residues 120–128) that form a slightly twisted sheet. These are flanked on one side by three α helices (α1, residues 9–26, α7, residues 133–142, and α8, residues 144–169) and on the other side by five short helices (α2, residues 52–60, α3, residues 63–66, α4, residues 68–75, α5, residues 85–90, and α6, residues 96–104, see Figure 1B).
FIGURE 1.
A. Ribbon diagram of a dimer of the YenR-LBD in the absence of pheromone. Colors indicate position from the N terminus (blue) to C terminus (red). α helices and β strands are indicated and numbered from N terminus to C terminus. B. Amino acid sequence of YenR-LBD. Helices α1-α8 are depicted using red letters while strands β1-β4 are depicted using blue letters. Amino acid residues that make hydrogen bonds with OHHL are depicted using red shaded boxes, while those important for subunit interactions are shown using blue shaded boxes, and those at the opening of the OHHL binding cavity are shown using orange boxes. C. Subunit interface of the YenR-LBD dimer. A salt bridge is formed between a side-chain oxygen of Glu156 of each subunit and the side-chain amine of Lys76 of the opposite subunit. For clarity, only one such bond is shown. A hydrogen bond is formed between the side-chain oxygen of Asn117 of one subunit and the side-chain amine of Lys38 of the opposite subunit, while another hydrogen bond is formed between the side-chain amine of Asn117 and a side-chain oxygen of Asp75. A water-mediated hydrogen bond is formed between the backbone nitrogen of His115 of each subunit and the backbone oxygen of His115 of the opposite subunit. A second water-mediated hydrogen bond is formed between the backbone oxygen of Glu145 of each subunit [Color figure can be viewed at wileyonlinelibrary.com]
3.2. The dimerization interface
The two YenR-LBD subunits contact one another primarily via α8 helix and by three loops of each subunit (Figure 1C). One loop lies between β1 and β2 (residues 38–44), another between α4 and α5 (residues 76–84), and the third between β3 and β4 (residues 116–119). The total solvent excluded interaction surface is 848 Å2 for the apo-form and 896 Å2 for the ligand-bound form, as calculated by PISA server. The two α8 helices of each subunit contact one another at their N-terminal ends and are arranged at approximately 90° angles so that the C-termini of the two helices lie >50 Å apart.
Glu156 of each subunit forms a salt bridge with Lys76 of the opposite subunit (Figure 1C; for clarity, only one of the two symmetrical bonds is shown, see also Figure 1B), while Asn117 of each subunit forms hydrogen bonds with Lys38, Lys74 and Asp75 of the opposite subunit. A water-mediated hydrogen bond is formed between Glu145 of each subunit (Figure 1C), while a second water-mediated hydrogen bond is formed between His115 of each subunit. Numerous hydrophobic contacts are also observed on the subunit interface (not shown).
3.3. The OHHL binding site
In the apoprotein structure, one subunit contains one water molecule in the presumed ligand binding pocket, while the other contains two water molecules and one molecule of acetate (not shown). The acetate likely originated from the crystallization conditions. These and presumably other small molecules must prevent the inward collapse of the binding cavity in the apo-form, as there is little net expansion of this binding site in the pheromone-bound form (see below).
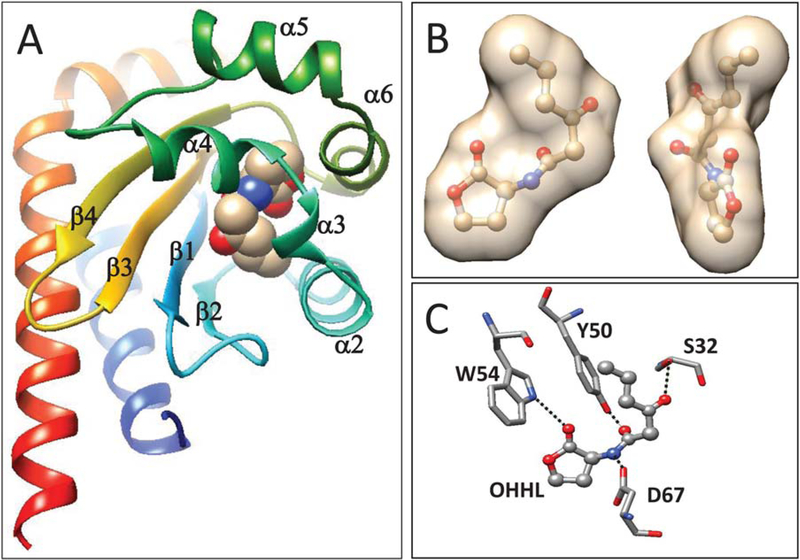
When crystals of the apoprotein were incubated in a buffer containing OHHL, each subunit bound one molecule of the pheromone. Well-defined electron density was found within the protein precisely at the predicted OHHL binding site, strongly suggesting that it corresponds to the bound pheromone molecule. As seen with several other LuxR-type proteins, the protein completely enveloped the pheromone such that there is virtually no contact between pheromone and bulk solvent (Figure 2A,B). The ring of OHHL makes hydrophobic contacts with Val69, Trp82, Phe98, and Ile106, while the acyl chain of OHHL makes hydrophobic contacts with Tyr58, Tyr50, and Leu34 (not shown). YenR also makes four hydrogen bonds with the ligand (Figures 1B and 2C). Trp54 forms a hydrogen bond (3.03 Å) between its ring nitrogen and the ring keto group (O4) of OHHL. Asp67 forms a hydrogen bond (2.75 Å) between its carboxyl group (OD2) and the OHHL imino group (N). Tyr50 forms a hydrogen bond (2.82 Å) between its hydroxyl group (OH) and the 1-oxo group (O2) of OHHL. Finally, Ser32 forms a hydrogen bond (2.80 Å) between its hydroxyl group (OH) and the 3-oxo group of OHHL. The first three of these contacts are completely conserved in all other LuxR-type proteins for which structures are available (see below). The bond between Ser32 and OHHL is somewhat unusual among YenR homologs, which tend to make water-mediated contacts to the 3-oxo group (see Discussion). The structures of apo-YenR-LBD and YenR-LBD-OHHL complexes are largely superimposable, though a loop between β4 and α7 shifts considerably (see below).
FIGURE 2.
Binding of OHHL to YenR. A: OHHL lies within a cavity with a four-stranded β sheet on one side and five short α helices on the other. B. YenR completely engulfs OHHL so that there is no contact between the pheromone and bulk solvent. The figure shows the inner surface of the protein in the presence of OHHL. C. YenR makes four direct hydrogen bonds to OHHL. Ser32 binds the 3-oxo group of the acyl group, Try50 binds the 1-oxo group of the acyl chain, Trp54 binds the ring oxo group, and Asp67 binds the imino group [Color figure can be viewed at wileyonlinelibrary.com]
3.4. OHHL-induced changes in the YenR structure
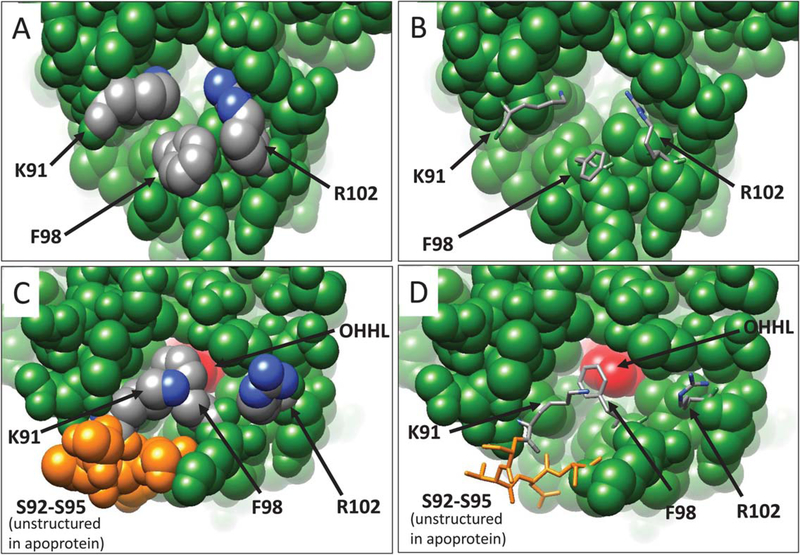
YenR is one of only two LuxR-type proteins to be crystallized in the presence and absence of its pheromone (the other being SdiA, see Discussion). As described above, YenR-LBD fully envelops OHHL and allows no significant contact between the pheromone and bulk solvent. However, apo-YenR detects and binds OHHL in vivo and in vitro,26 so its binding site must be accessible to this pheromone. Comparing the structures of apoprotein and the ligand-bound form provides evidence for a channel to the binding site in the apoprotein that closes upon pheromone binding (Figure 3). Lys91, Phe98, and Arg102 lie at the opening of this putative channel. In the apoprotein, Phe98 protrudes outward toward the solvent (Figure 3A,B). This is a rather unusual conformation for a hydrophobic side-chain; perhaps Phe98 acts as a lure to attract the hydrophobic pheromone. In contrast, the side chains of Lys91 and Arg102 are directed inward, toward the binding site. The positive charges of these residues helps maintain the electrostatic potential and hydrophilicity of the channel, allowing solvent and small molecules to gain access to the binding site. In addition, four residues connecting helix 5 (α5) with helix 6 (α6) (Ser92, Thr93, Asp94, Ser95) are disordered in the crystal structure, indicating that these residues are highly flexible. These four residues are invisible in Figure 3A,B. The flexibility of these residues and the outward orientation of Phe98 seems to keep the channel open. Moreover, the side chains of Lys91 and Arg102 may be quite flexible, and if so, their movement out of the channel would further widen it.
FIGURE 3.
YenR apoprotein has an aqueous channel that maintains access to the OHHL binding site. The side chains of Lys91 and Arg102 are oriented inward and may be flexible in solution (Figure 3A,B). In contrast, Phe98 is oriented outward to solvent in the apoprotein. Ser92-Ser95 are disordered and not visible in the apoprotein. In the presence of OHHL, Phe98 swings inward and makes hydrophobic contacts with the pheromone, while Lys91 and Arg102 swing outward to the solvent (Figure 3C,D). Ser92-Ser95 are structured in the presence of OHHL (Figure 3C,D, residues in orange). These changes effectively seal the pheromone within the protein. In parts A and C, all residues are depicted as space-filling spheres, while in parts B and D, critical residues are depicted as stick figures. OHHL is shown in red
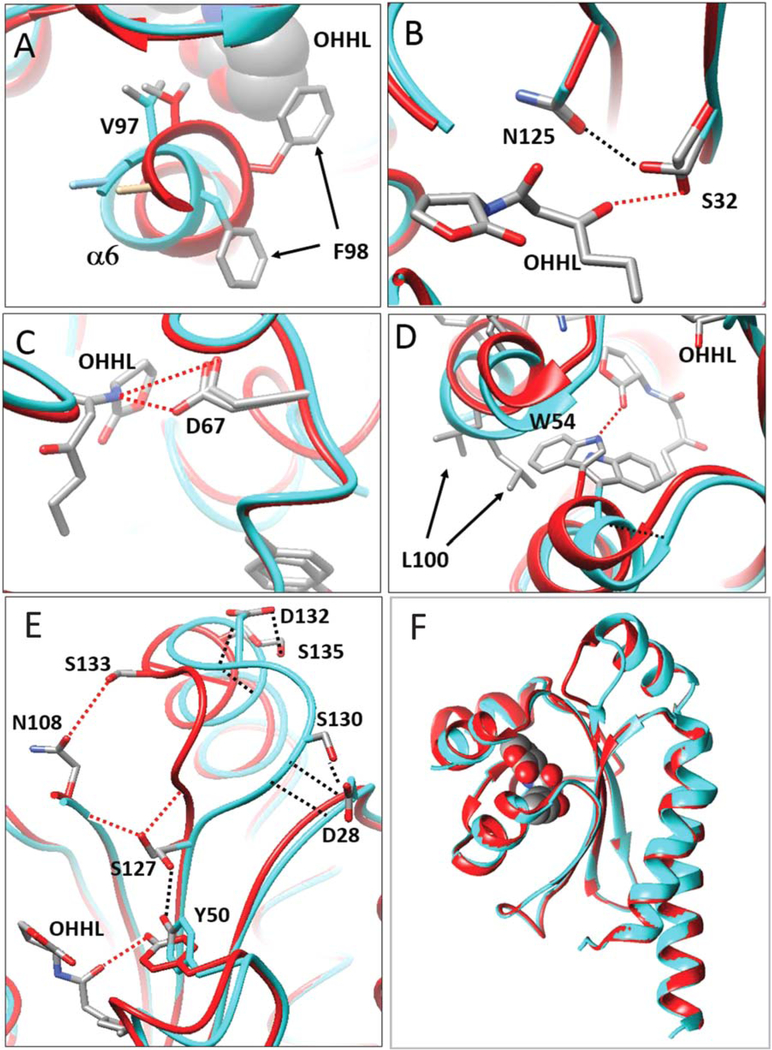
Binding of OHHL causes at least four changes in this channel. First, Phe98 swivels dramatically inward, away from the solvent, and toward the pheromone (Figures 3C,D and 4A). In this conformation, the aromatic ring of Phe98 makes hydrophobic contacts with lactone ring carbons of OHHL. Second, the movement of Phe98 toward OHHL causes a shift of the N-terminal region of helix α6, pulling it toward the pheromone, and most likely stabilizing the ligand-bound form (Figure 4A). In this conformation, Val97 contributes to the hydrophobic environment of the binding site. Third, the side chains of Lys91 and Arg102 move away from the binding site and toward the solvent (Figure 3C,D), thereby decreasing the hydrophilicity of the environment. Fourth, the four disordered residues in apo-form (Ser92-Ser95) become more ordered. These residues are fully visible in subunit B (Figures 3C and 4D, depicted in orange) while in the subunit A, two residues (Ser92, Thr93) remain disordered (not shown). The inward movement of Phe98 and Val97, the outward movement of Lys91 and Arg102, and the increased order of Ser92-Ser95 all contribute to enclose OHHL within the binding site. All of these changes were found in both subunits, although the movement of Phe98 and Val97 is somewhat more pronounced in the B subunit than in the A subunit.
FIGURE 4.
OHHL changes the conformation of residues at the binding site. Apoprotein is depicted in cyan, while the ligand-bound form is depicted in red. (A) OHHL causes Phe98 to swing inward, causing a distortion of helix a6. In this conformation, Val97 is drawn close to OHHL, contributing to the hydrophobic environment. (B) Ser32 makes a hydrogen bond with OHHL, while in the apoprotein, Ser32 swivels 90° to bond with Asn125. (C) Asp67 forms a hydrogen bond with OHHL, and makes little or no movement in response to OHHL. (D) Trp54 extends well into the binding pocket of the apoprotein, while OHHL causes Trp54 to swivel approximately 120°, avoiding a steric clash, and allowing a hydrogen bond between it and OHHL. This outward movement necessitates a movement of the nearby residue Leu100, which protrudes into solvent. (E) Binding of OHHL causes a movement of a loop between β4 and α7. In the apoprotein, Tyr50 bonds with Ser127, and the loop (cyan) is pulled to the right by bonds between Asp28 and Gly129-Ser130, and by bonds between Ser135 and Asp131-Asp132 (black lines). OHHL bonds with Tyr50, releasing the bond between Try50 and Ser127. Ser127 then can swivel and bond to the backbone of Asn108 and Gly129 (red lines). This pulls the loop (red) toward Asn108. This conformation is further stabilized by bonds between Ser133 and Asn108, and between Asn128 and Asp131 (not shown). (F) The loop described in part E is the only extensive conformational change in the protein induced by OHHL. All other tertiary structures can be closely superimposed [Color figure can be viewed at wileyonlinelibrary.com]
As described above, OHHL makes hydrogen bonds to four YenR residues, Ser32, Tyr50, Trp54, and Asp67. We have compared the environments of these four residues in the presence and absence of pheromone. In the ligand-bound form, Ser32 makes a hydrogen bond with the 3-oxo group of OHHL (Figures 2C and 4B, red dashed line). In the apoprotein, Ser32 swivels approximately 90° and forms a hydrogen bond with Asn125 (Figure 4B, black dashed line). In one subunit of the apoprotein, Ser32 makes an additional bond with water (not shown). In the ligand-bound protein, Asp67 makes a hydrogen bond with the imino nitrogen of OHHL (Figures 2C and 4C, red dashed line). In one subunit of the apoprotein, the carboxyl group of Asp67 forms a bond with water, while in the other subunit, it bonds with acetate (not shown).
More dramatic shifts occur at Tyr50 and Trp54. In the apoprotein, Trp54 extends well into the OHHL binding site to partly fill the volume of the cavity. Binding of OHHL causes Trp54 to swivel approximately 120°, avoiding a steric clash with the pheromone and allowing for a hydrogen bond to the ring keto group (Figure 4D, red dashed line). This movement distorts the helix α2, disrupting a hydrogen bond between the backbone nitrogen of Trp54 and the backbone carbonyl group of Pro51 (Figure 4D). This outward movement of Trp54 also necessitates the movement of Leu100 toward the solvent so as to avoid a steric clash (Figure 4D).
Another dramatic shift occurs at Tyr50, with conformational consequences quite distant from the binding site. In the ligand-bound form, Tyr50 forms a hydrogen bond with the 1-oxo group of the pheromone (Figure 4E, red line), while in the apoprotein, Tyr50 moves away from the binding site and bonds to Ser127 (blue line). This requires the movement and rotation of both amino acids. In the pheromone-bound form, Ser127 bonds to the backbone of Asn108 and Gly129 (red line). This causes the movement of a loop that lies between β4 and α7 (Gly129 to Ser132) (Figure 4E). In the apoprotein, this loop is stabilized by hydrogen bonding between Asp132 and Ser135, between Ser135 and Asp131, between Asp28 and Gly129, and between Asp28 and Ser130 (Figure 4E, black lines). In the ligand-bound form, these bonds are broken as the loop shifts toward Asn108. In this new conformation, the loop is stabilized by hydrogen bonding between Asn108 and Ser133, between Ser127 and Asn108, (Figure 4E, red lines) and by two hydrogen bonds between Asn128 and Asp131 (not shown). This displacement occurs in both subunits of the dimer, thus maintaining overall symmetry. The movement of this loop therefore appears to be a direct consequence of hydrogen bonding between Tyr50 and OHHL. This movement is the only global change in YenR conformation caused by pheromone binding (Figure 4F).
We have previously hypothesized that OHHL might decrease the affinity of the YenR subunits for each other, weakening the dimer interface.26 Such a model is not supported by the current structural data. In fact, the YenR-OHHL complex has a slightly larger interface than that of the apo-form (896 Å2 vs. 848 Å2). There is a conspicuous absence of movement of the backbone in the dimerization interface and very little movement of side chains in this region (not shown).
3.5. Structure of apo-CepR2-LBD
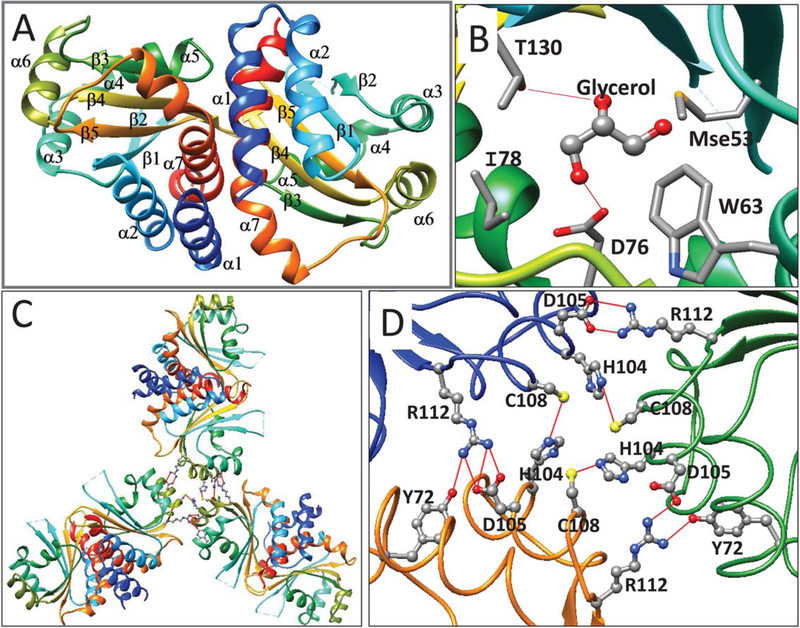
Efforts of crystalize full length CepR2 have so far been unsuccessful. However, CepR2-LBD was highly soluble in solution and was crystallized as an apoprotein. Efforts to infuse these crystals with the cognate ligand, octanoyl homoserine lactone (OHL) have been unsuccessful. The asymmetric unit consists of a trimer of dimers (see below). Each monomer resembles other LuxR-type proteins in that it consists of an α/β/α sandwich, with five antiparallel strands forming a twisted β sheet that is flanked by three α helices (α1, α2, and α7) on one side and four (α3, α4, α5, and α6) on the other (Figure 5A, see also Supporting Information Figure S2). Pairs of subunits dimerize with two-fold symmetry.
FIGURE 5.
(A) Ribbon diagram of apo-CepR2-LBD. Coloring and labelling are as described in Figure 1. (B) CepR2-LBD is complexed with one molecule of glycerol in the presumptive OHL binding site. Glycerol forms hydrogen bonds to Thr130 and Asp76 and makes van der Waals contacts to Ile78, Trp63 and Mse53. Trp63, Asp76, and Thr130 correspond to residues of homologous proteins that make hydrogen bonds with AHLs (see Supporting Information Figure S2). (C) The CepR-LBD asymmetric unit contains three dimers and shows three-fold rotational symmetry. D. Trimers of dimers of CepR2-LBD are stabilized by interprotein hydrogen bonds between Cys108 and His104 and between Arg112 and Asp105. In two subunits, Tyr72 also bonds to Arg112 [Color figure can be viewed at wileyonlinelibrary.com]
The dimerization interface consists of two perpendicular helices (α1, residues 4–15 and α7, residues 145–163) and two loops (residues 84–88 and 121–125) of each of the two subunits (not shown). The total solvent excluded interaction surface is 1421 Å2 as calculated by PISA server. Several interdimer hydrogen bonds are formed between His7, Asp11, and Gln14 of each subunit (helix α1), which make direct or water-mediated hydrogen bonds with Ser144 or Arg147 (helix α7) or Arg87 of the opposite subunit (not shown). An additional hydrogen bond is formed between Ser152 of each subunit and the backbone oxygen of Ala148 of the opposite subunit. Finally, Arg122 of each subunit forms hydrogen bonds with Ser84 and Tyr120 of the opposite subunit (not shown).
Crystals of apo-CepR2-LBD contained a molecule of glycerol in each subunit at the sites that are presumed to bind OHL (Figure 5B). Binding was stabilized by hydrogen bonds between glycerol and Thr130 and Asp76. Thr130 corresponds to Thr129 of TraR, which makes a water-mediated hydrogen bond with OOHL,29 while Asp76 corresponds to highly conversed aspartate residues that bond with the imine group of AHLs (see below). Binding of glycerol is stabilized by van der Waals interactions with Met53 (SeMet53), Trp63, and Ile78 (Figure 5B). Glycerol was used as a part of the cryo-protectant when crystals were cryocooled in liquid nitrogen prior to collecting X-ray data. It is tempting to speculate that LuxR-type proteins such as CepR2 and YenR, which must fold and function in the absence of AHL-type pheromones, may be stabilized by binding of small molecules such as glycerol. A similar idea has been proposed for SdiA, which was purified with bound octanoyl-rac-glycerol.35
As described above, the asymmetric unit contains three dimers. These dimers are arranged with a three-fold rotational symmetry (Figure 5C) and are stabilized by intermolecular hydrogen bonding between Tyr72, His104, Asp105, Cys108, and Arg112, most of which lie within or near helix α6 (Figure 5D). It is not known whether the full length CepR2 can form hexamers in vivo or in solution. The majority of LuxR proteins that have been characterized thus far form dimers in vivo and in vitro.18
4. DISCUSSION
4.1. Structural diversity among LuxR-type proteins
The tertiary structures of YenR-LBD and CepR2-LBD generally resemble those of other LuxR-type proteins, in that all consist of α/β/α sandwiches with the pheromone binding pocket (or presumed binding site in case of CepR2) located between the concave surface of a curved β sheet and a group of α helices. The hydrogen bonds formed between these proteins and their pheromones are also quite conserved. Perhaps surprisingly however, the quaternary structures of these proteins are quite diverse. The LuxR-type proteins that have been crystallized are generally dimeric, but the dimerization interfaces vary considerably (Supporting Information Figure S1). In every protein, there is a long helix at the C-terminus of the domain that contributes to the dimerization interface (shown in red in Supporting Information Figure S1). These helices in YenR and CepR2 intersect at 90° angles and make minimal contact. In contrast, the corresponding two helices of TraR are approximately parallel and make extensive contacts along their entire lengths. YenR and CepR2 subunits make additional contacts via loops at residues 74–79 and 115–118. In contrast, the loop of TraR that corresponds to residues 74–79 of YenR faces toward the solvent. The loop of TraR that corresponds to residues 115–118 of YenR contacts the opposite subunit, but in a completely different region, along the N-terminal helix. YenR and CepR2 resemble LasR, QscR, and SdiA in that all have the C-terminal helices arranged at right angles, while TraR resembles CviR in that dimerization occurs primarily along two parallel helices (Supporting Information Figure S1). This diversity in dimer interfaces could be a reflection of their somewhat low level of sequence conservation. For example, YenR-LBD and TraR-LBD show only 11.7% identity, and many pairs of these proteins show only 17–22% sequence identity.
These proteins also differ at their extreme N-termini. Most of these proteins have two helices (shown in blue) that lie between the N-terminus and the first β strand (β1). The only exception is YenR, which is shorter at its N-terminus, and has just one helix (Supporting Information Figures S1 and S2). The orientations of the N-termini with respect to the rest of the domain are also quite diverse. In the cases of CepR2, SdiA, TraR, and CviR, the N-terminus of each subunit interacts with the opposing subunit and contributes to the dimerization interface. In the case of YenR, LasR, and QscR, the N-terminal helices are on opposite faces of the dimer and make no contact (Supporting Information Figure S1).
Interactions between these proteins and the homoserine lactone ring of AHLs are conserved in some ways and dissimilar in others thus explaining pheromone specificity. As described above, all AHL-type pheromones have identical homoserine lactone rings and diverse acyl groups (or in a few cases aromatic groups). Interactions with the HSL moieties are strongly conserved, while interactions with the acyl group are less so. The ring oxo groups of AHLs invariably form a hydrogen bond with a Trp residue that corresponds to Trp54 of YenR. Similarly, the imino group of HSLs invariably form a hydrogen bond with an Asp residue that corresponds to Asp67 of YenR. The 1-oxo group of HSLs forms a hydrogen bond with a Tyr residue that corresponds to Tyr50 of YenR. This tyrosine is conserved in six of the seven proteins with available structures. The exception is CepR2, which has a histidine at this position that could in principle make a similar hydrogen bond.
In contrast, interactions between these receptors and the acyl chains of their ligands show much greater diversity. For example, many of these pheromones include a 3-oxo moiety as a part of the acyl chain, and the interactions between these groups and their receptors vary considerably. As described above, YenR residue Ser32 makes a direct hydrogen bond with the 3-oxo group of OHHL. In contrast, TraR residue Thr129 makes a water-mediated hydrogen bond with the 3-oxo group of OOHL. SdiA residue Ser134 makes a water-mediated hydrogen bond to the 3-oxo group of OOHL. LasR residues Tyr56 and Tyr64 make a water-mediated hydrogen bond to OdDHL, while QscR residues Tyr58 and Tyr66 make a water-mediated hydrogen bond to OdDHL. The overall trajectory of the acyl chains within their receptors also varies. In Supporting Information Figure S3, the conformation of the HSL rings were fixed in order to more clearly visualize differences in the positions of acyl groups. Most of these bound ligands have similar conformations, with the exception of OdDHL bound to LasR and QscR. These pheromones have considerably longer acyl chains that curve within the protein pocket. The 3-oxo groups of OdDHL bound to LasR and QscR are oriented quite differently from the 3-oxo groups of AHLs bound to YenR, TraR, and SdiA. In that regard, it is interesting to note that TraR, when overexpressed, can detect extremely long-chain AHLs,51,52 and it would be interesting to determine how these long chains are accommodated, and specifically whether these AHLs are fully buried within the protein, or whether part of the acyl group protrudes out of the protein and makes contact with solvent.
Here, we provide the first structural analysis of any quorum-hindered LuxR family member, that is, proteins that are fully functional in transcription regulation only as apo-proteins. YenR also is only the second member of the LuxR family to be crystallized both as a pheromone complex and as an apo-protein. SdiA has been crystallized in complex with two different AHL pheromones and an AHL-free protein that is complexed with a molecule of octanoyl-rac-glycerol at the AHL binding site, which may mimic an AHL.35 In a different study, SdiA was crystallized with either glycerol at the AHL binding site, or with two molecules of tetraethylene glycol, another possible AHL mimic.36 All other LuxR-type structures contain either AHLs or similar analogs. In contrast, apo-YenR described here has only water in the ligand binding site of one subunit and water and acetate in the other subunit. Most likely, these binding sites may contain additional water molecules or other small molecules that were not sufficiently ordered to be resolved. Comparing the structures of YenR in the presence and absence of ligand allowed us to get a detailed picture of how the ligand impacts the YenR structure.
Like all other LuxR-type proteins that have been crystallized, YenR completely engulfs its pheromone, such that the pheromone makes no significant contact with bulk solvent. In previous studies, we have shown that another member of this family, TraR, must bind its pheromone during or immediately after its synthesis, and that failure to bind pheromone leads to rapid proteolysis.14 Furthermore, overproduction of the molecular chaperone GroESL leads to increased accumulation of TraR in A. tumefaciens53 while a GroESL mutation decreases TraR activity in Sinorhizobium meliloti.54 Both studies indicate a role for GroESL in the TraR folding and pheromone binding. When LuxR, CepR, and LasR are overexpressed in E. coli, their folding requires their cognate AHLs and each of these proteins forms insoluble inclusion bodies in the absence of pheromone,14–17 suggesting that they too need AHLs to fold correctly. In contrast, those LuxR proteins that are active as apoproteins must be able to fold and function without the ligand. At the same time, the binding site must be sufficiently accessible to bind OHHL in vivo. This is supported by our finding that the binding site is accessible in YenR crystals. OHHL binding closes this opening and makes the cavity inaccessible to solvent. We noted above that in the apoprotein, residues Lys91, Phe98 and Arg102 can provide access to the binding site. In addition, the residues between Trp83 and Val108 contain two helices that contribute residues to the OHHL binding site. Trp83 and Val108 are close to one another; if these regions are sufficiently flexible, they might create a hinge that would allow the region between them to open slightly, allowing pheromone to enter the binding site. If so, bound OHHL could help to stabilize these helices in a closed position. OHHL makes hydrophobic contacts with Trp82, Phe98, Ala101, and Ile106, all of which lie within this region. Binding of this portion of YenR to OHHL would therefore sequester these nonpolar molecules from the solvent and might therefore enhance binding of OHHL. It remains a mystery that YenR and CepR2 can fold as apoproteins, while most LuxR homologs cannot.
4.2. Implications for DNA binding
Binding of OHHL to YenR-LBD caused somewhat modest global conformational changes in the protein dimer structure. These changes do not reveal the mechanism by which the pheromone converts YenR from an active form to a less active one. We have previously speculated that OHHL might act by decreasing the affinity of YenR subunits for each other, enhancing dissociation into monomers that are unable to bind DNA.18 If so, this would be difficult to detect in a protein crystal, and is contrary to the observed increase in surface of the dimer interface caused by OHHL. However, one might expect to see at least small changes in the binding interface, and we saw virtually none. Alternatively, OHHL could support a conformational change that is propagated to the C-terminal DNA binding domain. The helix at the C-terminus of the YenR-LBD is considerably longer than its counterparts of other proteins (Supporting Information Figure S2), and the C-terminal residues of the dimer are exceptionally far apart from each other. These C-terminal residues were less structured in the holoprotein than in the apoprotein, though this could be due to a crystallography artefact. Another possible way that OHHL could decrease YenR activity would be to decrease its abundance and sensitivity to proteolysis. This would be analogous (though opposite) to the effect of OOHL on TraR activity, as this pheromone dramatically increases protein abundance and protease resistance.
Bacterial proteins that regulate transcription can function in diverse ways. Some function as repressors while others function as activators, and a few can do both.55–57 Most transcriptional regulators are interconverted between functional and nonfunctional forms by either low molecular weight ligands or by protein modifications such as phosphorylation. Among transcriptional activators, there are many examples of such proteins that function only when ligand-bound, and these would include most LuxR homologs. In contrast, there are very few activators known to function preferentially as apoproteins.58–60 YenR and its close relatives, which activate transcription preferentially as apoproteins, seem to belong to this rare category of transcription activators.
5. CONCLUSION
While most LuxR-type transcription factors require there cognate pheromones for activity and even for folding and protease-resistance, a few members function preferentially as apoproteins. We describe the structures of the ligand binding domains of two such proteins, YenR and CepR2. These are the first “quorum-hindered” LuxR-type proteins ever crystallized, and virtually the first LuxR-type proteins crystallized as apoproteins. YenR was solved in the presence and absence of its ligand and we were therefore able to visualize a channel in the apoprotein between bulk solvent and the ligand binding site, and show how this channel is sealed upon ligand binding. We also showed how ligand binding can cause conformational changes far from the binding site that could help to explain how these pheromones convert this protein to an inactive form.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge helpful discussions with all the members of our two laboratories. The authors wish to thank members of the SBC at Argonne National Laboratory for their help with data collection at the 19-ID beamline. This work was supported by National Institutes of Health Grants GM094585 and GM115586 and by the U.S. Department of Energy, Office of Biological and Environmental Research, under contract DE-AC02–06CH11357.
Funding information
National Institutes of Health, Grant/Award Number: GM094585 and GM115586; U.S. Department of Energy, Office of Biological and Environmental Research, Grant/Award Number: DE-AC02-06CH11357
Footnotes
SUPPORTING INFORMATION
Additional Supporting Information may be found online in the supporting information tab for this article.
REFERENCES
- [1].Winans SC. A new family of quorum sensing pheromones synthesized using S-adenosylmethionine and Acyl-CoAs. Mol Microbiol. 2011;79(6):1403–1406. [DOI] [PubMed] [Google Scholar]
- [2].Pappas KM, Weingart CL, Winans SC. Chemical communication in proteobacteria: biochemical and structural studies of signal synthases and receptors required for intercellular signalling. Mol Microbiol. 2004;53(3):755–769. [DOI] [PubMed] [Google Scholar]
- [3].Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Ann Rev Cell Develop Biol. 2005;21(1):319–346. [DOI] [PubMed] [Google Scholar]
- [4].Ng W-L, Bassler BL. Bacterial quorum-sensing network architectures. Ann Rev Genet. 2009;43:(1):197–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Schaefer AL, Greenberg EP, Oliver CM, et al. A new class of homoserine lactone quorum-sensing signals. Nature. 2008;454(7204): 595–599. [DOI] [PubMed] [Google Scholar]
- [6].Lindemann A, Pessi G, Schaefer AL, et al. Isovaleryl-homoserine lactone, an unusual branched-chain quorum-sensing signal from the soybean symbiont Bradyrhizobium japonicum. Proc Natl Acad Sci USA. 2011;108(40):16765–16770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ahlgren NA, Harwood CS, Schaefer AL, Giraud E, Greenberg EP. Aryl-homoserine lactone quorum sensing in stem-nodulating photosynthetic bradyrhizobia. Proc Natl Acad Sci USA. 2011;108(17): 7183–7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Eberhard A, Burlingame AL, Eberhard C, Kenyon GL, Nealson KH, Oppenheimer NJ. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry. 1981;20(9):2444–2449. [DOI] [PubMed] [Google Scholar]
- [9].Engebrecht J, Nealson K, Silverman M. Bacterial bioluminescence: Isolation and genetic analysis of functions from Vibrio fischeri. Cell. 1983;32(3):773–781. [DOI] [PubMed] [Google Scholar]
- [10].Choi SH, Greenberg EP. The C-terminal region of the Vibrio fischeri LuxR protein contains an inducer-independent lux gene activating domain. Proc Natl Acad Sci USA. 1991;88(24):11115–11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hanzelka BL, Greenberg EP. Evidence that the N-terminal region of the Vibrio fischeri LuxR protein constitutes an autoinducer-binding domain. J Bacteriol. 1995;177(3):815–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].White CE, Winans SC. Cell–cell communication in the plant pathogen Agrobacterium tumefaciens. Philos Trans R Soc Lond B: Biol Sci. 2007;362(1483):1135–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Schuster M, Joseph Sexton D, Diggle SP, Peter Greenberg E. Acylhomoserine lactone quorum sensing: from evolution to application. Ann Rev Microbiol. 2013;67(1):43–63. [DOI] [PubMed] [Google Scholar]
- [14].Zhu J, Winans SC. The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. Proc Natl Acad Sci USA. 2001;98 (4):1507–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Urbanowski ML, Lostroh CP, Greenberg EP. Reversible acylhomoserine lactone binding to purified Vibrio fischeri LuxR protein. J Bacteriol. 2004;186(3):631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Weingart CL, White CE, Liu S, et al. Direct binding of the quorum sensing regulator CepR of Burkholderia cenocepacia to two target promoters in vitro. Mol Microbiol. 2005;57(2):452–467.15978077 [Google Scholar]
- [17].Schuster M, Urbanowski ML, Greenberg EP. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc Natl Acad Sci USA. 2004;101(45):15833–15839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Tsai C-S, Winans SC. LuxR-type quorum sensing regulators that are detached from common scents. Mol Microbiol. 2010;77(5):1072–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ryan GT, Wei Y, Winans SC. A LuxR-type repressor of Burkholderia cenocepacia inhibits transcription via antiactivation and is inactivated by its cognate acylhomoserine lactone. Mol Microbiol. 2013; 87(1):94–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].CDC. http://www.cdc.gov/nczved/divisions/dfbmd/diseases/yersinia/.2009.
- [21].CDC. http://www.aphis.usda.gov/programs/ag_selectagent/ag_bioterr_toxinslist.html, 2014.
- [22].Smego AR, Frean J, Koornhof JH. Yersiniosis I: microbiological and clinicoepidemiological aspects of plague and non-plague yersinia infections. Eur J Clin Microbiol Infect Dis. 1999;18(1):1–15. [DOI] [PubMed] [Google Scholar]
- [23].Isles A, Maclusky I, Corey M, et al. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J Pediatr. 1984;104(2):206–210. [DOI] [PubMed] [Google Scholar]
- [24].Vandamme P, Holmes B, Vancanneyt M, et al. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int J Syst Bacteriol. 1997;47(4):1188–1200. [DOI] [PubMed] [Google Scholar]
- [25].Mahenthiralingam E, Bischof J, Byrne SK, et al. DNA-Based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J Clin Microbiol. 2000;38(9):3165–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Tsai C-S, Winans SC. The quorum-hindered transcription factor YenR of Yersinia enterocolitica inhibits pheromone production and promotes motility via a small non-coding RNA. Mol Microbiol. 2011; 80(2):556–571. [DOI] [PubMed] [Google Scholar]
- [27].Malott RJ, O’grady EP, Toller J, Inhülsen S, Eberl L, Sokol PA. A Burkholderia cenocepacia Orphan LuxR Homolog Is Involved in Quorum-Sensing Regulation. J Bacteriol. 2009;191(8):2447–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wei Y, Ryan GT, Flores-Mireles AL, Costa ED, Schneider DJ, Winans SC. Saturation mutagenesis of a CepR binding site as a means to identify new quorum-regulated promoters in Burkholderia cenocepacia. Mol Microbiol. 2011;79(3):616–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhang R-G, Pappas T, Brace JL, et al. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature. 2002;417(6892):971–974. [DOI] [PubMed] [Google Scholar]
- [30].Vannini A, Volpari C, Gargioli C, et al. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 2002;21(17):4393–4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chen G, Swem LR, Swem DL, et al. A strategy for antagonizing quorum sensing. Mol Cell. 2011;42(2):199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bottomley MJ, Muraglia E, Bazzo R, Carfì A. Molecular insights into quorum sensing in the human pathogen Pseudomonas aeruginosa from the structure of the virulence regulator LasR bound to its autoinducer. J Biol Chem. 2007;282(18):13592–13600. [DOI] [PubMed] [Google Scholar]
- [33].Zou Y, Nair SK. Molecular basis for the recognition of structurally distinct autoinducer mimics by the Pseudomonas aeruginosa LasR quorum-sensing signaling receptor. Chem Biol. 2009;16(9): 961–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lintz MJ, Oinuma K-I, Wysoczynski CL, Greenberg EP, Churchill MEA. Crystal structure of QscR, a Pseudomonas aeruginosa quorum sensing signal receptor. Proc Natl Acad Sci USA. 2011;108(38):15763–15768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Nguyen Y, Nguyen NX, Rogers JL, et al. Structural and mechanistic roles of novel chemical ligands on the SdiA quorum-sensing transcription regulator. mBio. 2015;6(2). doi: 10.1128/mBio.02429-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kim T, Duong T, Wu C-A, et al. Structural insights into the molecular mechanism of Escherichia coli SdiA, a quorum-sensing receptor. Acta Crystallogr D Biol Crystallogr. 2014;70(3):694–707. [DOI] [PubMed] [Google Scholar]
- [37].Stols L, Gu M, Dieckman L, Raffen R, Collart FR, Donnelly MI. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr Purif. 2002;25(1):8–15. [DOI] [PubMed] [Google Scholar]
- [38].Kim Y, Babnigg G, Jedrzejczak R, et al. High-throughput protein purification and quality assessment for crystallization. Methods. 2011;55(1):12–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ferla MP, Patrick WM. Bacterial methionine biosynthesis. Microbiology. 2014;160(Pt 8):1571–1584. [DOI] [PubMed] [Google Scholar]
- [40].Rosenbaum G, Alkire RW, Evans G, et al. The Structural Biology Center 19ID undulator beamline: facility specifications and protein crystallographic results. J Synchrotron Radiat. 2006;13(Pt 1):30–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 8):859–866. [DOI] [PubMed] [Google Scholar]
- [42].Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 10 Pt 2):1772–1779. [DOI] [PubMed] [Google Scholar]
- [43].Sheldrick G Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr. 2010;66(4):479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Otwinowski Z Daresbury Study Weekend Proceedings. Warrington, UK: SERC Daresbury Laboratory; 1991. p 80–85. [Google Scholar]
- [45].Cowtan KD, Main P. Improvement of macromolecular electron-density maps by the simultaneous application of real and reciprocal space constraints. Acta Crystallogr D Biol Crystallogr. 1993;49(Pt 1):148–157. [DOI] [PubMed] [Google Scholar]
- [46].Cowtan K Fitting molecular fragments into electron density. Acta Crystallogr D Biol Crystallogr. 2008;64(Pt 1):83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Crystallogr. 1997;30(6):1022–1025. [Google Scholar]
- [48].Adams PD, Afonine PV, Bunkóczi G, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt2):213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1): 2126–2132. [DOI] [PubMed] [Google Scholar]
- [50].Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32(Web Server issue):W615–W619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zhu J, Beaber JW, Moré MI, Fuqua C, Eberhard A, Winans SC. Analogs of the autoinducer 3-oxooctanoyl-homoserine lactone strongly inhibit activity of the TraR protein of Agrobacterium tumefaciens. J Bacteriol. 1998;180(20):5398–5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Zhu J, Chai Y, Zhong Z, Li S, Winans SC. Agrobacterium bioassay strain for ultrasensitive detection of n-acylhomoserine lactone-type quorum-sensing molecules: detection of autoinducers in Mesorhizobium huakuii. Appl Environ Microbiol. 2003;69(11):6949–6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Chai Y, Winans SC. The chaperone groESL enhances the accumulation of soluble, active TraR protein, a quorum-sensing transcription factor from Agrobacterium tumefaciens. J Bacteriol. 2009;191(11): 3706–3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Marketon MM, González JE. Identification of Two Quorum-Sensing Systems in Sinorhizobium meliloti. J Bacteriol. 2002;184(13):3466–3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Seshasayee A, Sivaraman K, Luscombe N. An overview of prokaryotic transcription factors. Subcell Biochem. 2011;52:7–23. [DOI] [PubMed] [Google Scholar]
- [56].Schleif R AraC protein, regulation of the l-arabinose operon in Escherichia coli, and the light switch mechanism of AraC action. FEMS Microbiol Rev. 2010;34(5):779–796. [DOI] [PubMed] [Google Scholar]
- [57].O’Halloran TV. Transition metals in control of gene expression. Science. 1993;261(5122):715–725. [DOI] [PubMed] [Google Scholar]
- [58].My L, Ghandour Achkar N, Viala JP, Bouveret E. Reassessment of the genetic regulation of fatty acid synthesis in Escherichia coli: global positive control by the dual functional regulator FadR. J Bacteriol. 2015;197(11):1862–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Zhang Y-M, Rock CO. Transcriptional regulation in bacterial membrane lipid synthesis. J Lipid Res. 2009;50(Suppl):S115–S119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Chang C, Tesar C, Li X, Kim Y, Rodionov DA, Joachimiak A. A novel transcriptional regulator of L-arabinose utilization in human gut bacteria. Nucleic Acids Res. 2015;43(21):10546–10559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.