SUMMARY
Cirmtuzumab is a humanized monoclonal antibody (mAb) that targets ROR1, an oncoembryonic orphan receptor for Wnt5a found on cancer stem cells (CSCs). Aberrant expression of ROR1 is seen in many malignancies and has been linked to Rho-GTPase activation and cancer stem cell self-renewal. For patients with chronic lymphocytic leukemia (CLL), self-renewing, neoplastic B cells express ROR1 in 95% of cases. High-level leukemia cell expression of ROR1 is associated with an unfavorable prognosis. We conducted a phase 1 study involving 26 patients with progressive, relapsed, or refractory CLL. Patients received four biweekly infusions, with doses ranging from 0.015 to 20 mg/kg. Cirmtuzumab had a long plasma half-life and did not have dose-limiting toxicity. Inhibition of ROR1 signaling was observed, including decreased activation of RhoA and HS1. Transcriptome analyses showed that therapy inhibited CLL stemness gene expression signatures in vivo. Cirmtuzumab is safe and effective at inhibiting tumor cell ROR1 signaling in patients with CLL.
Graphical Abstract
In Brief
Choi et al. find that cirmtuzumab, a humanized mAb specific for the cancer stem cell antigen ROR1, was well tolerated and stable in clinical testing in patients with relapsed chronic lymphocytic leukemia. Treatment inhibited activation of Rho-GTPase and HS1 in vivo and reversed the stemness gene expression signatures in leukemia cells.
INTRODUCTION
Receptor tyrosine kinase-like orphan receptor 1 (ROR1) is a type I transmembrane protein that is physiologically expressed in early embryogenesis and plays a critical role in organogenesis (Masiakowski and Carroll, 1992; Yoda et al., 2003). Expression of ROR1 attenuates rapidly after embryonic development, becoming virtually undetectable on post-partem tissues, with the exception of a few B cell precursors called hematogones (Broome et al., 2011). In contrast, ROR1 is expressed on a variety of cancers, particularly those that are less differentiated, and is associated with early relapse after therapy or metastasis (Zhang et al., 2012a, 2012b). Expression of ROR1 in ovarian cancer appears highest on a subpopulation of tumor cells that also have markers of cancer stem cells (CSCs) (Zhang et al., 2014) and are positively correlated with a metastatic trajectory (Gonzalez et al., 2018). Neoplastic cells from the majority of patients with chronic lymphocytic leukemia (CLL) express ROR1 (Cui et al., 2016; Fukuda et al., 2008).
While CLL is not thought of as a prototypical CSC-mediated malignancy, patients with CLL have aberrant hematopoietic stem cells that can give rise to monoclonal B cells that resemble CLL in mice (Damm et al., 2014; Kikushige et al., 2011). Moreover, CLL B cells manifest stemness properties, including niche dependency and the ability to self-renew or differentiate in response to microenvironmental signals (Kipps et al., 2017). CLL cells also may assimilate features of a recently described “stemness” index, which quantifies the degree to which cancers acquire progenitor-like stemness features associated with oncogenic de-differentiation (Crews et al., 2016; Malta et al., 2018; Milanovic et al., 2018; Zipeto et al., 2016). Indeed, expression of ROR1 in mouse models of CLL is associated with activation of gene expression/signaling networks implicated in embryonic and tumor cell proliferation and CSC self-renewal (Widhopf et al., 2014). Similarly, the expression of ROR1 in human CLL is associated with activation of comparable networks, and high-level leukemia cell expression of ROR1 correlates with relatively short treatment-free and overall survival (Cui et al., 2016).
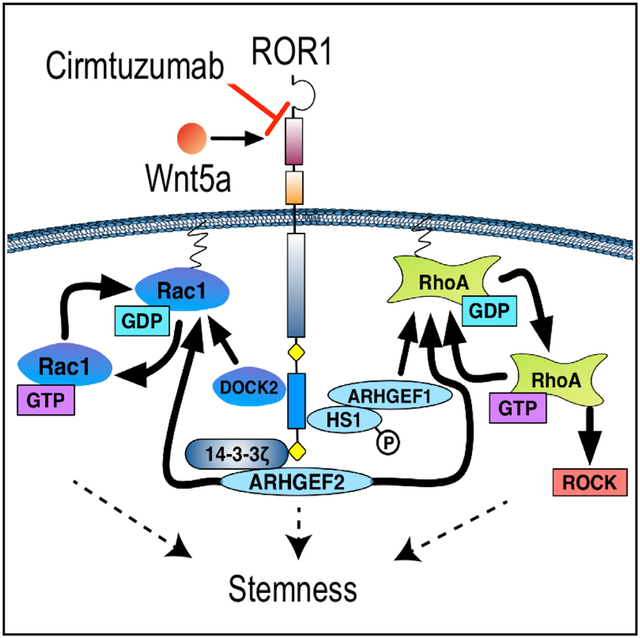
We found that ROR1 could serve as a receptor for Wnt5a (Fukuda et al., 2008), which can stimulate ROR1-dependent leukemia cell activation of Rho-GTPases (Yu et al., 2016). Wnt5a induces ROR1 to recruit 14-3-3ζ (Yu et al., 2017a) and Dedicator of cytokinesis 2 (DOCK2) (Hasan et al., 2018), which facilitate activation of Rac1/2 to enhance leukemia cell proliferation. Wnt5a also induces hematopoietic-lineage-specific protein 1 (HS1) to undergo tyrosine phosphorylation and recruitment to the proline-rich domain of ROR1, allowing the phosphorylation of HS1 in CLL to be used as a surrogate biomarker of ROR1 activation (Hasan et al., 2017). Prior studies found that HS1 played a central role in the trafficking and homing of leukemia B cells and that phosphorylation of HS1 was associated with an adverse prognosis in patients with CLL (Scielzo et al., 2005, 2010; ten Hacken et al., 2013).
Inhibition of ROR1 signaling may have therapeutic activity in patients with cancer. Inhibition or silencing of ROR1 in breast cancer suppressed epithelial mesenchymal transition (EMT) and repressed cancer migration and metastases (Cui et al., 2013). Inhibition of ROR1 inhibited the maintenance of CSC in ovarian cancer patient-derived xenografts (PDXs) and impaired CSC self-renewal as evidenced by the decreased capacity of PDXs to re-engraft immune-deficient mice (Zhang et al., 2014). Moreover, knockdown of ROR1 in glioblastoma suppressed expression of EMT-related genes, inhibited metastasis, and induced differentiation/senescence of CSCs (Jung et al., 2016).
Analysis of sera of patients immunized with autologous leukemia cells transduced to express CD154 revealed that some patients with favorable clinical outcomes produced anti-ROR1 autoantibodies that could neutralize the pro-survival effects of Wnt5a on leukemia cells in vitro (Fukuda et al., 2008). This provided a rationale for developing monoclonal antibodies (mAbs) targeting ROR1 for potential therapy of patients with ROR1-expressing cancers. Accordingly, we developed cirmtuzumab from an antibody originally selected from a library of anti-ROR1 monoclonal antibodies based on its capacity to inhibit ROR1 signaling. Cirmtuzumab has a high affinity and specificity for a distinctive epitope in the extracellular domain of human ROR1. Studies have demonstrated that such an antibody could impair engraftment of ROR1+ leukemia (Widhopf et al., 2014), block Wnt5a-induced ROR1 signaling, inhibit phosphorylation of HS1 and activation of Rho-GTPases (Hasan et al., 2017, 2018; Yu et al., 2016, 2017a, 2017b), repress genes associated EMT (Cui et al., 2013), and impair the capacity of primary ovarian cancer cells to form spheroids or establish tumor xenografts in immune-deficient mice (Zhang et al., 2014). Investigational New Drug (IND)-enabling studies demonstrated no cross-reactivity with normal human tissues and no toxicity when administered to rodents or non-human primates (Choi et al., 2015). The phase 1 study reported herein was designed to determine the safety and tolerability of cirmtuzumab, to assess whether it was suitable for phase 2 clinical evaluation, and to explore its capacity to inhibit ROR1 signaling and stemness associated with malignant reprogramming of neoplastic B cells into CSC.
RESULTS
Patient Characteristics
Twenty-six patients enrolled in the study between September 2014 and September 2017. Treatment consisted of four biweekly infusions of cirmtuzumab at doses ranging from 15 μg/kg to 20 mg/kg (Figure 1A). All patients had relapsed or refractory disease and objective signs of disease progression requiring therapy, as per international working group guidelines (Hallek et al., 2018). All patients had received prior anti-CD20 monoclonal antibody therapy. Most patients had at least one adverse prognostic feature, including CLL cell expression of unmutated immunoglobulin heavy chain variable region genes, complex karyotype, and/or deletions in the short arm of chromosome 17 (del(17p)). Each patient had elevated plasma levels of Wnt5a, which was significantly higher than that found in healthy age-matched adults (Figure 1B). The pre-treatment CLL cells of patients included in this study had between 2 × 103 and 1.1 × 104 ROR1 molecules of equivalent fluorescence (MESF) per cell (Figure 1C), which is comparable to the levels of CD20 on CLL cells (Huh et al., 2001). Fifteen patients had CLL cells that had a MESF with more than 5.8 × 103 molecules/cell, which correlated with the fluorescence threshold used to define the high-level expression of ROR1 associated with adverse prognosis in prior studies (Cui et al., 2016). Additional demographic information, patient characteristics, and dose assignments per cohort are provided in Tables S1 and S2.
Figure 1. Study Design, Pharmacokinetics, and ROR1 Pharmacodynamics.
(A) Eligible patients had relapsed or refractory chronic lymphocytic leukemia and progressive disease with an indication for therapy. The short duration of cirmtuzumab administration (four biweekly infusions) was sufficient to determine the primary aim of the study, which was to determine the maximum tolerated dose or biologically active dose of cirmtuzumab.
(B) Average number of ROR1 molecules per CLL cell (as indicated on the y axis) for individual patients treated in cohorts 1 through 7 and the average number of ROR1 molecules per CLL cell for all patients (±SD) as indicated on the x axis. A dotted line indicates the threshold for defining high-level ROR1, which was found to be associated with adverse prognosis (Cui et al., 2016).
(C) Concentration of cirmtuzumab in plasma of representative patients. Concentration (μg/mL) is indicated on the y axis, and time (days) is indicated on the x axis. Arrows indicate days of infusion of cirmtuzumab. Time points collected after the last infusion of cirmtuzumab indicate that significant levels are present for approximately 100–150 days, with a half-life of 32.4 days. Values indicated were determined by interpolation using a four-parameter logistic nonlinear regression model compared to a standard curve generated by serial dilutions of a known concentration of cirmtuzumab mAb.
(D) Wnt5a present in the plasma of patients with CLL who enrolled in the clinical trial (n = 26) is significantly higher than age-matched controls (n = 4) (unpaired Student’s t test, p = 0.0007).
(E) The relative levels of ROR1 on CLL cells of treated patients were normalized with respect to expression levels of ROR1 found on paired pre-treatment samples over time in days after the initial infusion, as indicated on the x axis.
Pharmacokinetics
To assess pharmacokinetics, we used an ELISA to determine the plasma concentrations of human IgG that was able to bind immobilized ROR1. Cirmtuzumab was detected within 60 min after each infusion. Peak concentrations increased with each infusion, including those at the same dose. Plasma levels of cirmtuzumab increased proportionally with dose, with peak concentrations exceeding 400 μg/mL for patients who received doses of 20 mg/kg (Figure 1D). Plasma collected approximately 3 months after the last infusion still had detectable cirmtuzumab at levels of ≥1 μg/mL following doses of 2 mg/kg or higher. We calculated the plasma half-life of cirmtuzumab to be 32.4 days based on the pharmacokinetic data of patients in cohort 6 (Figure 1D).
Safety and Tolerability
Cirmtuzumab infusions were safe and well tolerated. Adverse events (AEs) and laboratory abnormalities are listed in Table 1 regardless of attribution. There was no dose-limiting toxicity (DLT) and no serious AEs. The main recurrent laboratory abnormalities included anemia, thrombocytopenia, and neutropenia, which were primarily attributed to the underlying CLL. One instance of grade 3 alanine aminotransferase (ALT) elevation coincided with a patient having biliary colic associated with cholelithiasis, so it was not attributed to cirmtuzumab; the associated symptoms and ALT elevation did not recur when this patient was given subsequent infusions of cirmtuzumab. We observed three instances of self-limiting grade 3 lipase elevations (greater than two times the upper limit of normal); none were accompanied by abdominal pain or symptoms of pancreatitis. All lipase elevations resolved spontaneously without intervention within 24 hr, including one case that resolved within 2 hr and did not recur with subsequent infusions of cirmtuzumab. These events were not considered clinically significant or directly related to the study drug, particularly given the prolonged half-life of cirmtuzumab of over 32 days. If these AEs were cirmtuzumab related, the amylase and lipase elevations would have persisted for a longer period. Also, when these same patients received subsequent infusions of cirmtuzumab, they did not experience any recurrent elevations in serum lipase or amylase or other signs or symptoms of pancreatitis.
Table 1.
Adverse Events
| CTCAE (Ver 4.03) Grade | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | Total n = 26 | ||||||
| Adverse Event (Regardless of Attribution) | n | (%) | n | (%) | n | (%) | n | (%) | n | (%) |
| Anemia | 15 | (58) | 5 | (19) | 2 | (8) | - | - | 22 | (85) |
| Thrombocytopenia | 10 | (38) | 6 | (23) | 1 | (4) | - | - | 17 | (65) |
| Neutropenia | 6 | (23) | 4 | (15) | 1 | (4) | - | - | 11 | (42) |
| Fatigue | 6 | (23) | 1 | (4) | - | - | - | - | 7 | (27) |
| Upper respiratory infection | - | - | 7 | (27) | - | - | - | - | 7 | (27) |
| Diarrhea | 7 | (27) | - | - | - | - | - | - | 7 | (27) |
| Cough | 6 | (23) | - | - | - | - | - | - | 6 | (23) |
| Headache | 6 | (23) | - | - | - | - | - | - | 6 | (23) |
| Dyspnea | 5 | (19) | - | - | - | - | - | - | 5 | (19) |
| Serum lipase increased | 1 | (4) | 1 | (4) | 3 | (8) | - | - | 5 | (19) |
| Nausea | 4 | (16) | - | - | - | - | - | - | 4 | (16) |
| Constipation | 4 | (16) | - | - | - | - | - | - | 4 | (16) |
| Urinary tract infection | - | - | 3 | (12) | - | - | - | - | 3 | (12) |
| Akathisia | 3 | (12) | - | - | - | - | - | - | 3 | (12) |
| Dizziness | 3 | (12) | - | - | - | - | - | - | 3 | (12) |
| Bloating | 3 | (12) | - | - | - | - | - | - | 3 | (12) |
| Insomnia | 3 | (12) | - | - | - | - | - | - | 3 | (12) |
| Urinary Frequency | 3 | (12) | - | - | - | - | - | - | 3 | (12) |
| Creatinine increased | 3 | (12) | - | - | - | - | - | - | 3 | (12) |
| Hyperuricemia | 3 | (12) | - | - | - | - | - | - | 3 | (12) |
| Skin infection | - | - | 1 | (4) | 1 | (4) | - | - | 2 | (8) |
| Amylase increase | - | - | 1 | (4) | 1 | (4) | - | - | 2 | (8) |
| Serum ALT increased | 1 | (4) | - | - | 1 | (4) | - | - | 2 | (8) |
| Conjunctivitis | 1 | (4) | 1 | (4) | - | - | - | - | 2 | (8) |
| Anorexia | 1 | (4) | 1 | (4) | - | - | - | - | 2 | (8) |
| Maculopapular rash | 1 | (4) | 1 | (4) | - | - | - | - | 2 | (8) |
| Hypokalemia | 2 | (8) | - | - | - | - | - | - | 2 | (8) |
| Vomiting | 2 | (8) | - | - | - | - | - | - | 2 | (8) |
| Flushing | 2 | (8) | - | - | - | - | - | - | 2 | (8) |
| Hyperkalemia | 2 | (8) | - | - | - | - | - | - | 2 | (8) |
| Urinary Urgency | 2 | (8) | - | - | - | - | - | - | 2 | (8) |
| Hyperhidrosis | 2 | (8) | - | - | - | - | - | - | 2 | (8) |
| Gastroesophageal reflux | 2 | (8) | - | - | - | - | - | - | 2 | (8) |
Abbreviations: ALT, alanine aminotransferase; CTCAE, Common Terminology Criteria for Adverse Events.
Overall, no patient discontinued treatment due to an AE. Five patients discontinued cirmtuzumab prior to completing the four planned infusions. One patient in the first cohort discontinued treatment due to progressive disease following one dose of cirmtuzumab at 15 μg/kg. The four other patients elected to stop treatment when oral targeted therapies became available.
Assessment of ROR1/Wnt5a Signaling
Leukemia cells were assessed for surface expression of ROR1 following treatment using a fluorochrome-conjugated mAb that recognized a non-cross-reactive epitope of ROR1 distinct from that bound by cirmtuzumab. We noted a dose-dependent reduction in surface ROR1 on CLL cells of treated patients (Figures 1E and S1); patients in lower-dose-treatment cohorts had no or only modest reductions in surface ROR1 after 1 week (mean 95% of baseline, range 74%–114%), which returned to that of pre-treatment levels. However, the CLL cells of patients receiving cirmtuzumab at doses ≥2 mg/kg had, on average, a 33% reduction in detectable surface ROR1 (range 48%–84% of baseline). This difference in mean ROR1 reduction between low dose and ≥2 mg/kg was statistically significant (p = 0.001).
As phosphorylation of HS1 is an indicator of active ROR1 signaling, leukemia cells from patients in cohort 6 (16 mg/kg) were assessed for levels of phosphorylated HS1 (pHS1). We noted decreased levels of leukemia cell pHS1 at 24 hr after the first infusion of cirmtuzumab (Figure 2A). Consistent with a protracted pharmacokinetics, we observed inhibition of leukemic cell phosphorylation of pHS1 and reductions in pHS1/HS1 ratios, lasting 4 to 6 months after completion of therapy. The ratio of pHS1/HS1 approached pre-treatment levels when the cirmtuzumab plasma concentrations decreased to ≤10 μg/mL (concentrations of ≥10 μg/mL were not sustained with cirmtuzumab doses <2 mg/kg). Some patients elected to enroll in an extension study and received additional infusions of cirmtuzumab several months after completing this phase I protocol. In each case, pHS1/HS1 levels again decreased within 24 hr after re-treatment. We also looked for activated RhoA in the same samples via pull-down assay and immunoblot analyses (Figure 2B). Compared to pre-treatment samples, leukemia cells collected on day 2 (24 hr after the initial dose) and day 43 (24 hr after the fourth dose) each had substantial reductions in activated Rho-GTPase. Collectively, these results indicate that cirmtuzumab inhibited leukemia cell activation of Rho-GTPases and phosphorylation of HS1.
Figure 2. Inhibition of Rho-GTPase Activation.
(A) Levels of phosphorylated HS1 and total HS1 were assessed via immunoblot analyses on the CLL cells of patients in cohort 6 (16 mg/kg). There were marked reductions in the ratios of pHS1/HS1 within 24 hr of dosing. The pHS1/HS1 ratios rose after several months when the levels of cirmtuzumab in the plasma became undetectable. Reduction in pHS1/HS1 again were noted within 24 hr after re-treatment months later.
(B) Activated RhoA from patients in cohort 6 (16 mg/kg) was assessed by western blot after pull-down of GTP-Rho complexed with rhotekin-Rho binding domain beads. There was a reduction in Rho-GTPase activation noted 24 hr after dosing.
(C) Gene set enrichment analysis (GSEA) plots of genes activated by Rac1 and/or RhoA on pre-treatment versus paired post-cirmtuzumab CLL cells at D28 of each patient in cohort 6 (n = 3). Gene set size (SIZE), enrichment score, normalized ES (NES), and FDR q value (FDR q) are indicated.
(D) GSEA on the transcriptomes of paired pre- and post-treatment CLL cells at D28 evaluating for pre- and post-treatment differences in the expression of the 11,699 set of genes associated with stemness (Malta et al., 2018).
To assess for changes in leukemia cell gene expression following treatment, we isolated CLL cells to >95% purity from each patient in cohort 6 and extracted RNA for next-generation sequencing. This allowed us to compare the transcriptomes of matched sets of CLL cells obtained before therapy or on day 28 after the second dose of cirmtuzumab. Gene set enrichment analysis (GSEA) showed that post-treatment samples of each patient had reduced expression of genes associated with activation of Rac1 or RhoA compared to that of matched pre-treatment CLL samples (Figure 2C). Moreover, our analysis showed significant repression of genes associated with activation of Rac1, and a trend toward inhibition of genes associated with activation of RhoA, after adjustment for gene set size and multiple hypothesis testing. Compared to matched pre-treatment samples, the D28 post-treatment samples had significant repression of 7 of 8 gene signatures associated embryonic stem (ES) cell identity (Ben-Porath et al., 2008), Myc target genes (Fernandez et al., 2003; Li et al., 2003), or ES-cell-associated genes with promoters bound and activated in ES cells by Nanog, Oct4, Sox2, or NOS target regulators of ES cell identity (Boyer et al., 2005) (Figure S2). Only one of the two sets of genes overexpressed in ES cells compared to other cells (Assou et al., 2007) was significantly reduced in post-treatment samples. Most notably, GSEA showed that the post-treatment leukemia cells had a highly significant reversal of a recently described gene expression signature associated with stemness and oncogenic dedifferentiation, which we observed in pre-treatment CLL cells (FDR q < 0.001) (Figure 2D) (Malta et al., 2018).
Response and Time to CLL Progression
CLL disease burden was assessed based on aggregate lymph node size, spleen size, absolute lymphocyte count (ALC) in the blood, and hemoglobin and platelet counts per international working group (iwCLL) guidelines (Hallek et al., 2018). Overall, 22 patients were evaluable for response assessment; four patients who discontinued cirmtuzumab early without meeting criteria for progressive disease were not considered evaluable. No patients met criteria for complete or partial remission. Seventeen of 22 evaluable patients had stable disease. Five patients had progressive disease. Of these 5 patients, three received cirmtuzumab doses of ≤240 μg/kg, one received 8 mg/kg, and one received 20 mg/kg.
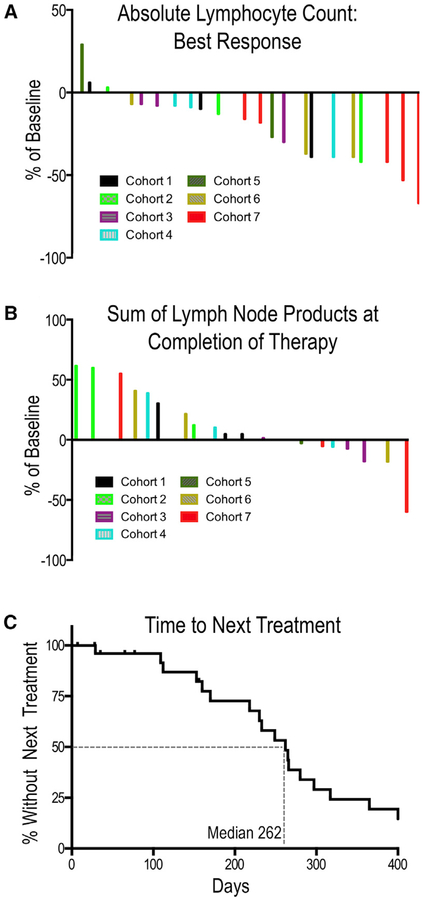
Most patients experienced reductions in the ALC with a trend toward greater proportionate reductions in patients who received higher doses of cirmtuzumab (Figure 3A). Lymph node sizes were assessed radiographically 2 months after the final cirmtuzumab dose, with calculation of the sum of the bi-dimensional area of target lesions (Sum of Lymph Node Products, see Figure 3B). Three patients (primarily those treated with doses of ≤240 μg/kg) had a greater than 50% increase in the total lymph node size, meeting working group criteria for progressive disease (Hallek et al., 2018). One patient in cohort 7 who received cirmtuzumab at 20 mg/kg had a greater than 50% reduction in lymphadenopathy.
Figure 3. Clinical Effects.
(A) Waterfall plot of best absolute lymphocyte count (ALC) on trial relative to pre-treatment baseline.
(B) Waterfall plot of the sum of lymph node products assessed by radiography 2 months after the final infusion of cirmtuzumab. Patients who discontinued therapy prior to the completion of four doses or who initiated other forms of treatment prior to the 2-month response assessment were not required to have imaging and were not included in the final response evaluation.
(C) Time to next treatment after CLL progression. Despite entering the study with progressive disease requiring therapy, patients did not require subsequent therapy for an extended duration after receiving four doses of cirmtuzumab. The median time to next treatment after CLL progression was 262 days. Four patients were censored because they started subsequent therapy prior to CLL progression and prior to completing the scheduled doses of cirmtuzumab. One patient switched therapy upon approval of venetoclax by the FDA; one patient had an isolated increase in the ALC (consistent with a redistributive lymphocytosis) but had concerns about progression and requested to come off trial to pursue other treatment options; one patient who noted eligibility for new clinical trial requested to switch treatment; and one patient had stable but bulky lymphadenopathy and chose to come off trial to seek other treatment. Three patients have not started subsequent therapy at the time of this analysis.
There were other signs of clinical benefit. We had pre- and post-treatment marrow biopsies from one patient; this revealed a significant reduction CLL cell infiltration of the marrow after therapy (Figure S3). Another patient had reduction in a CLL related pleural effusion with therapy. Furthermore, patients had stable disease upon completion of treatment with four doses of cirmtuzumab. The median time after therapy until when patients required additional therapy was 262 days (Figure 3C), corresponding to the time when plasma levels of cirmtuzumab became undetectable.
DISCUSSION
Development of small molecule inhibitors, cellular therapies, or mAbs that selectively and safely target therapy-resistant CSCs would satisfy a significant unmet medical need. Although there have been significant improvements in progression-free survival (PFS) with the advent of targeted therapies (such as ibrutinib), malignancies like CLL have a high propensity for relapse (Kipps et al., 2017). Toxicity limits the long-term tolerability of some of these agents, and resistance due to acquired mutations is a concern (Woyach et al., 2014). Moreover, patients typically relapse rapidly after discontinuation of therapy and have a relatively poor prognosis (Jain et al., 2015, 2017). For CLL and other cancers, novel therapeutic strategies are needed that target CSCs or “stemness” (Malta et al., 2018), which is associated with oncogenic dedifferentiation, metastases, and relapse after therapy.
ROR1 is an attractive target because it is expressed on neoplastic cells of intractable cancers but not on normal post-partem tissues (Zhang et al., 2012b). Although a pre-clinical report with other anti-ROR1 mAbs suggested that ROR1 was expressed on adipose, pancreatic, parathyroid, gastric, and other normal tissues (Balakrishnan et al., 2017), the epitopes recognized by these mAbs appear different from the one recognized by cirmtuzumab. Moreover, the observed pharmacokinetics and lack of toxicity in this clinical trial is consistent with cirmtuzumab’s lack of reactivity with normal human post-partem tissues in preclinical, IND-enabling studies (Choi et al., 2015). Moreover, treated patients did not experience weight loss, pancreatitis, hyperglycemia, or other drug-dependent toxicities, consistent with the notion that cirmtuzumab lacks off-target reactivity with normal tissues.
Cirmtuzumab was developed from a library of anti-ROR1 mAbs based on its distinctive capacity to inhibit ROR1 signaling. Therefore, its pharmacodynamic properties may be comparable to those associated with various small-molecule inhibitors of intracellular kinases but with a substantially longer half-life. Dosing each month may sustain levels of cirmtuzumab that can repress activation of Rho-GTPases and other downstream signaling events, such as phosphorylation of HS1. Consistent with cirmtuzumab having a long plasma half-life, the levels of pHS1 were depressed for several months following the final infusion of cirmtuzumab. These data and the observed pharmacokinetics indicate that treated patients did not develop neutralizing antibodies that shorten the half-life of cirmtuzumab or interfere with its capacity to bind to ROR1 or inhibit leukemia cell ROR1 signaling in vivo.
This phase 1 clinical trial was designed primarily to assess safety, pharmacokinetics, and biologic activity of cirmtuzumab after just four biweekly infusions. As such, it is impossible to compare the clinical activity of cirmtuzumab with that of targeted drugs like ibrutinib, which are administered daily until disease progression or intolerance. Nonetheless, it is encouraging that we observed a median time to next treatment (TTNT) (following CLL progression) of 259 days (8.6 months), considering that patients had progressive disease requiring treatment upon study enrollment. This duration of treatment-free survival is comparable to that of similar patients treated with single-agent anti-CD20 monoclonal antibodies. The median TTNT after four doses of single-agent rituximab was reported to be 12.5 weeks in a Nordic study (Itälä et al., 2002), 20 weeks in a study by Huhn and colleagues (Huhn et al., 2001), and 8 months after high doses of rituximab (O’Brien et al., 2001). Single-agent obinutuzumab had a similar post-treatment response duration in the phase 2 portion of the Gauguin study (Cartron et al., 2014). For patients with relapsed/refractory CLL, the median duration of remission was 8.9 months, and only 15% of patients met partial response (PR) criteria. Finally, comparable patients with relapsed/refractory CLL, who were treated with ofatumumab (2,000 mg for 12 doses over 6 months), had a median PFS of approximately 6 months (Wierda et al., 2011).
The results of this phase 1 clinical trial may have broader implications, as ROR1 is also a marker of CSCs and stemness in solid tumors. Prior studies have shown that treatment with cirmtuzumab can reduce the capacity of ovarian cancer PDXs to metastasize or re-engraft immune-deficient mice, indicating that targeting ROR1 affects CSC self-renewal (Zhang et al., 2014). The favorable safety profile of cirmtuzumab may allow for its use in combination with other anti-cancer agents or immunotherapies that fail to target CSCs.
Although the data collected in this study do not define a clear dose dependency with regard to clinical activity, we did observe dose dependency in the down modulation of ROR1 and reductions in the ratios of pHS1/HS1 when plasma levels of cirmtuzumab were ≥10 mg/mL, indicating that the drug has measurable in vivo activity. Phase 1b/2 trials that are powered to investigate a potential relationship between cirmtuzumab dose, biomarker modulation, and clinical outcome are now accruing patients at multiple centers.
In summary, this is the first clinical trial of any type targeting ROR1. The results support further development of cirmtuzumab as a specific and safe inhibitor of ROR1/Wnt5a signaling. The safety profile of cirmtuzumab makes it an ideal agent to combine with other therapies. We previously found that cirmtuzumab has complementary anti-tumor activity with ibrutinib because ibrutinib does not inhibit the capacity of Wnt5a to induce phosphorylation of HS1 or the migration of CLL cells (Hasan et al., 2017; Yu et al., 2017b). A phase 1b/2 combination clinical trial has been initiated based on these findings in patients with CLL or mantle cell lymphoma. In addition, clinical trials are planned for other intractable CSC-driven malignancies in which ROR1 is aberrantly expressed.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests of reagents can be directed to and fulfilled by the Lead Contact, Dr. Thomas Kipps (tkipps@ucsd.edu). A Material Transfer Agreement is required.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Ethics Committee Approval and Consent
The clinical trial protocol and informed consent forms were approved by the Institutional Review Board of the University of California, San Diego with respect to compliance with research and human subject regulations. Preclinical data and clinical trial design were also reviewed the United States Food and Drug Administration prior to Investigational New Drug (IND) application filing. The clinical protocol was registered with ClinicalTrials.Gov () prior to study initiation. Informed consent from all patients was obtained in accordance with the Declaration of Helsinki.
Subject Details, Eligibility, and Dose Allocation
26 human subjects enrolled in the clinical trial. Each patient had to have a diagnosis of CLL as per working-group criteria (Hallek et al., 2018). Patients had relapsed or refractory disease, and were without available approved therapies. Patients who were ≤70 years of age were required to have had prior purine-analog or alkylator-based therapy, or have CLL cells with deletion of chromosome 17p (Del 17p), or compromised renal function, marrow function, or performance status that precluded use of standard chemotherapy. All patients had progressive or symptomatic disease that required CLL therapy as per working-group guidelines (Hallek et al., 2018); this included at least one of the following: symptomatic or progressive splenomegaly; symptomatic or progressive lymphadenopathy; progressive anemia (hemoglobin ≤ 11 g/dL); progressive thrombocytopenia (platelets ≤ 100,000/mL); rapid lymphocyte doubling time; or constitutional “B” symptoms. Patients with recent or concurrent chemotherapy or investigational therapy were excluded, as were patients with active infection.
The median age was 72 (range 58–88). 14 (54% of patients) were male, 12 (46%) were female. There was not an association between sex or gender identity, and cirmtuzumab response, toleration, or time to next treatment. Further patient characteristics are detailed in Table S1.
The clinical trial was a dose-escalation study with 12 pre-defined dose levels (15 mcg/kg to 20 mg/kg) in 7 patient cohorts. Patients in the first four cohorts had intra-patient dose escalation, with the stipulation that a grade ≥ 2 adverse events (which occurred in cohort 4) would revert the dose escalation scheme to a standard 3 + 3 design without intra-patient dose escalation, starting at the dose at which the grade ≥ 2 toxicity occurred. The subsequent cohorts had a fixed dose level administered biweekly for a maximum of four doses. There were three evaluable patients at each of the first 11 dose levels. A total of six patients were treated at the highest dose level (20 mg/kg). Table S2 indicates patient disposition by cohort.
METHOD DETAILS
Cirmtuzumab Treatment
Patients were treated with cirmtuzumab via IV infusion on days 1, 15, 29, and 43 at the cohort-assigned dose (Table S2). Infusion duration was between 1.5 to 3 hr, with increasing rates of infusion similar to other agents of this class. The initial infusion included a 10-min test dose to monitor for allergic reaction.
Cirmtuzumab Pharmacokinetic Assessment
Concentration of cirmtuzumab in plasma of patients was assessed using an ELISA assay generated for human IgG binding to immobilized ROR1. Briefly, wells were coated overnight with 2.5 mg/ml of recombinant human ROR1-extracellular domain (ROR1-ECD), washed, and blocked with sample buffer (1 × BBS + 1% BSA) at 37 °C for 90 min. Serial dilutions of all samples were added to duplicate wells, incubated at ambient temperature for 60 min, washed, and then detected with mouse anti-human IgG1 Fc-HRP (Southern Biotech) and TMB microwell peroxidase substrate (SeraCare). Development was terminated by addition of 1M O-phosphoric acid and absorbance was read on a SpectraMax340 Microplate Reader (Molecular Devices). Values were determined by interpolation using a 4-parameter logistic nonlinear regression model, compared to a standard curve generated by serial dilutions of a known concentration of cirmtuzumab mAb.
ROR1 Immunophenotyping
The expression of ROR1 was detected by flow cytometry using cryopreserved samples from patients enrolled in the trial, and as previously described (Cui et al., 2016). Cells were stained with Alexa-647-conjugated anti-ROR1 mAb (4A5). Fluorochrome-conjugated, isotype control mAbs of irrelevant specificity were used to account for nonspecific staining. Data were acquired on a FACSCalibur (BD Biosciences, San Jose, CA, USA) or Attune NxT Flow Cytometer. The expression level of ROR1 was described by the delta mean fluorescence intensity (DMFI), which is the mean fluorescence intensity of CLL cells stained for ROR1 minus the mean fluorescence intensity of the same cells stained with isotype control. The molecules of ROR1 per cell was determined with the Quantum MESF (Molecules of Equivalent Soluble Fluorochrome (MESF) microspheres (Moskalensky et al., 2015). Alexa Fluor 647 conjugated beads were run on the same day as stained cell samples to establish a calibration curve relating channel values to MESF units or DMFI.
Plasma Wnt5a Measurement
Wnt5a was detected by Human WNT5A ELISA kit (OKEH00723, Aviva Systems Biology, CA, USA). Patient plasma was centrifuged for 15 min at 1,000 × g at 4 °C within 30 min of collection. Samples were stored at minus 20 °C until analysis. Diluted samples were incubated in WNT5A Microplates for 2 hr at 37 °C prior to addition of biotinylated WNT5A detector antibody, and subsequent addition of Avidin-HRP Conjugate and TMB Substrate per manufacturer instructions. O.D. absorbance at 450 nm was read, and relative OD versus serial standard curve was determined.
HS1 Assay
HS1 and pHS1 protein levels were assessed via immunoblot analysis with whole cell lysates (40–80 mg) prepared in lysis buffer [20 mM HEPES (pH 7.9), 25% (vol/vol) glycerol, 0.5 N NaCl, 1 mM EDTA, 1% Nonidet P-40, 0.5 mM DTT, and 0.1% deoxycholate], containing protease and phosphatase inhibitors (Roche). Immunoblots were probed using anti-HS1 (Cell Signaling Technology, Cat#4503S) or antiphosphorylated HS1 (OriGene, Cat#TA314001) and as described previously (Hasan et al., 2017).
RhoA Assay
GTP-bound active RhoA was pulled down from lysed CLL cells with Rhotekin-RBD beads (Cytoskeleton), and then subjected to immunoblot analysis, as described previously (Yu et al., 2016, 2017a). Immunoblots of whole-cell lysates were used to assess for total RhoA. The integrated optical density (IOD) of bands was evaluated by densitometry and analyzed using Gel-Pro Analyzer 4.0 software (Media Cybernetics, MD).
RNA Extraction and RNA-Seq
PBMC of 3 CLL samples were collected before therapy (Pre-Rx) and at day 28 of cirmtuzumab treatment (D28). Each D28 sample was collected after patients had received 2 doses of 16mg/kg cirmtuzumab. Negative isolation of CLL cells to R 95% purity was performed prior to RNA isolation. Total RNA was extracted using TRIzol reagent (Life Technologies). Data were analyzed by Rosalind (https://rosalind.onramp.bio/), with a HyperScale architecture developed by OnRamp BioInformatics (San Diego, CA). Reads were trimmed using cutadapt (Martin and Wang, 2011). Quality scores were assessed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc; Andrews, 2010). Reads were aligned to the Homo sapiens genome build hg19 using STAR (Dobin et al., 2013). Individual sample reads were quantified using Htseq (Anders et al., 2015) and normalized via Relative Log Expression (RLE) using DESeq2 R library (Love et al., 2014). DEseq2 was also used to calculate fold changes and p values.
QUANTIFICATION AND STATISTICAL ANALYSIS
The primary clinical trial end points were to determine the maximum tolerated dose (MTD), or recommended phase 2 dose (R2PD), and to assess safety. Adverse event data were reviewed by an independent Data and Safety Monitoring Board at the University of California, San Diego. Severity of non-hematologic AEs was assessed according to Common Terminology Criteria for Adverse Events (CTCAE) version 4.03. Severity of hematology AEs was according to CLL international working group criteria (Hallek et al., 2018). AEs were tabulated based on most severe occurrence of each AE for each patient, and percentage of patients experiencing each AE (n = 26).
Secondary end-points included treatment-emergent adverse events, clinical response rate, and PFS. Median PFS was estimated using the Kaplan-Meier method (GraphPad Prism version 6.0 was used for statistical analysis). Clinical activity was assessed using standard working-group criteria for CLL response assessment (Hallek et al., 2018).
PK studies used non-compartmental modeling. PD assessment of ROR1 expression, HS1 phosphorylation, and RhoA activation compared baseline, response assessment visit, and other time points prior to initiation of next treatment. pHS1 and RhoA assessment focused on patients who received 16 mg/kg (n = 3).
Gene Set Enrichment Analysis
Gene set enrichment analysis (GSEA) compared pre-treatment (Pre-Rx) and day 28 (D28) levels for the same patients in the 16 mg/kg cohort (n = 3). Using GSEA software (Subramanian et al., 2005) we conducted GSEA on the primary RNaseq data pre-Rx and day 28 of cirmtuzumab treatment. We focused GSEA on the stemness signature gene set recently identified by one-class logistic regression machine-learning algorithm (Malta et al., 2018) and 8 gene sets reflecting embryonic stem (ES) cell identity previously described by Ben-Porath and colleagues (Ben-Porath et al., 2008). These 8 gene sets, fall into three groups: (i) ES expressed genes: two sets of genes overexpressed in ES cells compared to other cells and tissues according to a multi-study compilation and meta-analysis (Assou et al., 2007). (ii) Nanog, Oct4, Sox2 and NOS targets: four sets of genes whose promoters are bound and activated in human ES cells by each of these regulators of ES cell identity, or co-activated by all (Boyer et al., 2005), (iii) Myc targets: two sets of genes bound and activated by c-Myc, identified in two independent studies (Fernandez et al., 2003; Li et al., 2003). GSEA also was performed focusing on Rac1 pathway in BIOCARTA database (Liberzon et al., 2011), and RhoA pathway in Ingenuity Pathway database (IPA, QIAGEN Redwood City, https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/). Each gene set was considered significant when the false discovery rate (FDR) was less than 25% (Subramanian et al., 2005). The FDR q value was adjusted for gene set size and multiple hypothesis testing.
DATA AND SOFTWARE AVAILABILITY
The accession number for the RNA-seq data reported in this paper is GEO: GSE114382 (https://www.ncbi.nlm.nih.gov/geo/).
Supplementary Material
KEY RESOURCES TABLE
| Reagent or Resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| Mouse Anti-human IgG1 Fc-HRP | Southern Biotech | Cat#9054–05; RRID: AB_2687484 |
| 4A5, Alexa 647 conjugated | In house | N/A |
| IgG2b, A647 conjugated isotype control mAb | BD Biosciences | Cat#557903; RRID: AB_396928 |
| Rabbit Anti-HS1 (human specific) | Cell Signaling Technology | Cat#4503S; RRID: AB_2096977 |
| Rabbit polyclonal Anti-phospho HS1 (Y378) | OriGene | Cat#TA314001 |
| Mouse monoclonal anti-RhoA | Cytoskeleton | Cat#ARH04 |
| Anti-mouse IgG, HRP-linked | Cell Signaling Technology | Cat#7076; RRID: AB_330924 |
| Anti-rabbit IgG, HRP-linked | Cell Signaling Technology | Cat#7074; RRID: AB_2099233 |
| Human IgG Control Ab | Sigma | Cat#I5154; RRID: AB_1163610 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| ROR1-extracellular domain | In house | N/A |
| Rhotekin-RBD beads | Cytoskeleton | Cat#RT02 |
| Quantum MESF Kit, Alexa Fluor 647 | Bangs Laboratories | Cat#555p |
| Deposited Data | ||
| Raw and analyzed RNA-seq data | NCBI GEO | GEO: GSE114382 |
| Software and Algorithms | ||
| Gel-Pro Analyzer 4.0 software | Media Cybernetics | http://www.mediacy.com |
| FlowJo (v.2.7.4) | FlowJo | https://www.flowjo.com |
| Gene set enrichment analysis | Broad Institute | http://software.broadinstitute.org/gsea/index.jsp |
| QuickCal v.2.3 analysis template | Bangs Laboratories | https://www.bangslabs.com/ |
| Graphpad Prism v.6.0 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Other | ||
| TMB microwell peroxidase substrate kit | SeraCare | Cat#5120–0053 |
| BSA | Fisher | Cat#BP1600 |
| ELISA high-protein binding plates | Costar | Cat#3690 |
| WNT5A ELISA kit | Aviva Systems Biology | Cat#OKEH00723, RRID: SCR_013560 |
| Cirmtuzumab | Pacific GMP/Abzena | N/A |
Highlights.
Cirmtuzumab targets ROR1 signaling on CLL and reduces activation of RhoA and HS1
26 patients received cirmtuzumab (up to 20 mg/kg) without dose-limiting toxicity
Cirmtuzumab inhibits expression of stemness gene expression signature in CLL
4 biweekly infusions of cirmtuzumab prolonged time to next treatment (TTNT)
ACKNOWLEDGMENTS
The clinical trial was supported by a grant from the California Institute of Regenerative Medicine (CIRM) (DR3-06924), the CIRM Alpha Stem Cell clinic (CIRM AC1-07764), and the Moores Cancer Center Specialized Cancer Center Support Grant (NIH NCI 2P30CA023100-28). This work was also supported by the UC San Diego Foundation Blood Cancer Research Fund (BCRF; F-3133), the Koman Family Foundation, a SCOR grant (7005-14) from the Leukemia and Lymphoma Society, and a PO1 grant (5P01CA081534-14) from the NIH for the CLL Research Consortium. The RNA sequencing was conducted at the IGM Genomics Center, University of California, San Diego, La Jolla, CA (Grant No. P30CA023100).
We also acknowledge appreciation for Dr. Gabriel Pineda’s expert assistance with isoelectric focusing-based confirmation of antibody purity. We would also like to acknowledge the contributions of Dr. Langdon Miller, Dr. James Breitmeyer, Mary Breitmeyer, David Johnson, and Dr. Brian Lannutti of Oncternal Therapeutics; Charlene Gutierrez, Tuan Tran, Susette Gorak, Kimi Denoble, Betty Cabrera, Dr. Tiffany Juarez, Sam Zhang, and Rebecca McAlpin of UC San Diego; our infusion unit and clinic nurses, pharmacists, and social workers; the Sanford Stem Cell Clinical Center and Alpha Clinic staff; and most of all, the patients who participated in this clinical trial.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes three figures and two tables and can be found with this article online at https://doi.org/10.1016/j.stem.2018.05.018.
DECLARATION OF INTERESTS
Cirmtuzumab was developed by Dr. Kipps and his laboratory and licensed by the University of California to Oncternal Therapeutics, Inc. M.Y.C., J.E.C., and T.J.K. received honoraria and research funding from Pharmacyclics. C.J. received research funding from Johnson & Johnson and Celgene.
REFERENCES
- Anders S, Pyl PT, and Huber W (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S (2010). FastQC: a quality control tool for high throughput sequence data (Babraham Bioinformatics).
- Assou S, Le Carrour T, Tondeur S, Ström S, Gabelle A, Marty S, Nadal L, Pantesco V, Réme T, Hugnot JP, et al. (2007). A meta-analysis of human embryonic stem cells transcriptome integrated into a web-based expression atlas. Stem Cells 25, 961–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan A, Goodpaster T, Randolph-Habecker J, Hoffstrom BG, Jalikis FG, Koch LK, Berger C, Kosasih PL, Rajan A, Sommermeyer D, et al. (2017). Analysis of ROR1 protein expression in human cancer and normal tissues. Clin. Cancer Res. 23, 3061–3071., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, and Weinberg RA (2008). An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet 40, 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. (2005). Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broome HE, Rassenti LZ, Wang HY, Meyer LM, and Kipps TJ (2011). ROR1 is expressed on hematogones (non-neoplastic human B-lymphocyte precursors) and a minority of precursor-B acute lymphoblastic leukemia. Leuk. Res 35, 1390–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartron G, de Guibert S, Dilhuydy MS, Morschhauser F, Leblond V, Dupuis J, Mahe B, Bouabdallah R, Lei G, Wenger M, et al. (2014). Obinutuzumab (GA101) in relapsed/refractory chronic lymphocytic leukemia: final data from the phase 1/2 GAUGUIN study. Blood 124, 2196–2202. [DOI] [PubMed] [Google Scholar]
- Choi MY, Widhopf GF 2nd, Wu CC, Cui B, Lao F, Sadarangani A, Cavagnaro J, Prussak C, Carson DA, Jamieson C, and Kipps TJ (2015). Pre-clinical specificity and safety of UC-961, a first-in-class monoclonal antibody targeting ROR1. Clin. Lymphoma Myeloma Leuk. 15 (Suppl), S167–S169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews LA, Balaian L, Delos Santos NP, Leu HS, Court AC, Lazzari E, Sadarangani A, Zipeto MA, La Clair JJ, Villa R, et al. (2016). RNA splicing modulation selectively impairs leukemia stem cell maintenance in secondary human AML. Cell Stem Cell 19, 599–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui B, Zhang S, Chen L, Yu J, Widhopf GF 2nd, Fecteau JF, Rassenti LZ, and Kipps TJ (2013). Targeting ROR1 inhibits epithelial-mesenchymal transition and metastasis. Cancer Res. 73, 3649–3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui B, Ghia EM, Chen L, Rassenti LZ, DeBoever C, Widhopf GF 2nd, Yu J, Neuberg DS, Wierda WG, Rai KR, et al. (2016). High-level ROR1 associates with accelerated disease progression in chronic lymphocytic leukemia. Blood 128, 2931–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm F, Mylonas E, Cosson A, Yoshida K, Della Valle V, Mouly E, Diop M, Scourzic L, Shiraishi Y, Chiba K, et al. (2014). Acquired initiating mutations in early hematopoietic cells of CLL patients. Cancer Discov. 4, 1088–1101. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, and Amati B (2003). Genomic targets of the human c-Myc protein. Genes Dev. 17, 1115–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Chen L, Endo T, Tang L, Lu D, Castro JE, Widhopf GF 2nd, Rassenti LZ, Cantwell MJ, Prussak CE, et al. (2008). Antisera induced by infusions of autologous Ad-CD154-leukemia B cells identify ROR1 as an oncofetal antigen and receptor for Wnt5a. Proc. Natl. Acad. Sci. USA 105, 3047–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez VD, Samusik N, Chen TJ, Savig ES, Aghaeepour N, Quigley DA, Huang YW, Giangarrà V, Borowsky AD, Hubbard NE, et al. (2018). Commonly occurring cell subsets in high-grade serous ovarian tumors identified by single-cell mass cytometry. Cell Rep. 22, 1875–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Döhner H, Hillmen P, Keating M, Montserrat E, Chiorazzi N, et al. (2018). Guidelines for diagnosis, indications for treatment, response assessment and supportive management of chronic lymphocytic leukemia. Blood. Published online March 14, 2018 10.1182/blood-2017-09-806398. [DOI] [PubMed] [Google Scholar]
- Hasan MK, Yu J, Chen L, Cui B, Widhopf Ii GF, Rassenti L, Shen Z, Briggs SP, and Kipps TJ (2017). Wnt5a induces ROR1 to complex with HS1 to enhance migration of chronic lymphocytic leukemia cells. Leukemia 31, 2615–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan MK, Yu J, Widhopf GF 2nd, Rassenti LZ, Chen L, Shen Z, Briggs SP, Neuberg DS, and Kipps TJ (2018). Wnt5a induces ROR1 to recruit DOCK2 to activate Rac1/2 in chronic lymphocytic leukemia. Blood. Published online April 20, 2018 10.1182/blood-2017-12-819383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh YO, Keating MJ, Saffer HL, Jilani I, Lerner S, and Albitar M (2001). Higher levels of surface CD20 expression on circulating lymphocytes compared with bone marrow and lymph nodes in B-cell chronic lymphocytic leukemia. Am. J. Clin. Pathol 116, 437–443. [DOI] [PubMed] [Google Scholar]
- Huhn D, von Schilling C, Wilhelm M, Ho AD, Hallek M, Kuse R, Knauf W, Riedel U, Hinke A, Srock S, et al. ; German Chronic Lymphocytic Leukemia Study Group (2001). Rituximab therapy of patients with B-chronic cell lymphocytic leukemia. Blood 98, 1326–1331. [DOI] [PubMed] [Google Scholar]
- Itälä M, Geisler CH, Kimby E, Juvonen E, Tjonnfjord G, Karlsson K, and Remes K (2002). Standard-dose anti-CD20 antibody rituximab has efficacy in chronic lymphocytic leukaemia: results from a Nordic multicentre study. Eur. J. Haematol 69, 129–134. [DOI] [PubMed] [Google Scholar]
- Jain P, Keating M, Wierda W, Estrov Z, Ferrajoli A, Jain N, George B, James D, Kantarjian H, Burger J, and O’Brien S (2015). Outcomes of patients with chronic lymphocytic leukemia after discontinuing ibrutinib. Blood 125, 2062–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain P, Thompson PA, Keating M, Estrov Z, Ferrajoli A, Jain N, Kantarjian H, Burger JA, O’Brien S, and Wierda WG (2017). Long-term outcomes for patients with chronic lymphocytic leukemia who discontinue ibrutinib. Cancer 123, 2268–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung EH, Lee HN, Han GY, Kim MJ, and Kim CW (2016). Targeting ROR1 inhibits the self-renewal and invasive ability of glioblastoma stem cells. Cell Biochem. Funct 34, 149–157. [DOI] [PubMed] [Google Scholar]
- Kikushige Y, Ishikawa F, Miyamoto T, Shima T, Urata S, Yoshimoto G, Mori Y, Iino T, Yamauchi T, Eto T, et al. (2011). Self-renewing hematopoietic stem cell is the primary target in pathogenesis of human chronic lymphocytic leukemia. Cancer Cell 20, 246–259. [DOI] [PubMed] [Google Scholar]
- Kipps TJ, Stevenson FK, Wu CJ, Croce CM, Packham G, Wierda WG, O’Brien S, Gribben J, and Rai K (2017). Chronic lymphocytic leukaemia. Nat Rev Dis Primers 3, 17008. [DOI] [PubMed] [Google Scholar]
- Li Z, Van Calcar S, Qu C, Cavenee WK, Zhang MQ, and Ren B (2003). A global transcriptional regulatory role for c-Myc in Burkitt’s lymphoma cells. Proc. Natl. Acad. Sci. USA 100, 8164–8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, and Mesirov JP (2011). Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malta TM, Sokolov A, Gentles AJ, Burzykowski T, Poisson L, Weinstein JN, Kaminska B, Huelsken J, Omberg L, Gevaert O, et al. ;Cancer Genome Atlas Research Network (2018). Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell 173, 338–354.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JA, and Wang Z (2011). Next-generation transcriptome assembly. Nat. Rev. Genet 12, 671–682. [DOI] [PubMed] [Google Scholar]
- Masiakowski P, and Carroll RD (1992). A novel family of cell surface receptors with tyrosine kinase-like domain. J. Biol. Chem 267, 26181–26190. [PubMed] [Google Scholar]
- Milanovic M, Fan DNY, Belenki D, D ä britz JHM, Zhao Z, Yu Y, Dörr JR, Dimitrova L, Lenze D, Monteiro Barbosa IA, et al. (2018). Senescence-associated reprogramming promotes cancer stemness. Nature 553, 96–100. [DOI] [PubMed] [Google Scholar]
- Moskalensky AE, Chernyshev AV, Yurkin MA, Nekrasov VM, Polshchitsin AA, Parks DR, Moore WA, Filatenkov A, Maltsev VP, and Orlova DY (2015). Dynamic quantification of antigen molecules with flow cytometry. J. Immunol. Methods 418, 66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien SM, Kantarjian H, Thomas DA, Giles FJ, Freireich EJ, Cortes J, Lerner S, and Keating MJ (2001). Rituximab dose-escalation trial in chronic lymphocytic leukemia. J. Clin. Oncol 19, 2165–2170. [DOI] [PubMed] [Google Scholar]
- Scielzo C, Ghia P, Conti A, Bachi A, Guida G, Geuna M, Alessio M, and Caligaris-Cappio F (2005). HS1 protein is differentially expressed in chronic lymphocytic leukemia patient subsets with good or poor prognoses. J. Clin. Invest 115, 1644–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scielzo C, Bertilaccio MT, Simonetti G, Dagklis A, ten Hacken E, Fazi C, Muzio M, Caiolfa V, Kitamura D, Restuccia U, et al. (2010). HS1 has a central role in the trafficking and homing of leukemic B cells. Blood 116, 3537–3546. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Hacken E, Scielzo C, Bertilaccio MT, Scarfò L, Apollonio B, Barbaglio F, Stamatopoulos K, Ponzoni M, Ghia P, and Caligaris-Cappio F (2013). Targeting the LYN/HS1 signaling axis in chronic lymphocytic leukemia. Blood 121, 2264–2273. [DOI] [PubMed] [Google Scholar]
- Widhopf GF 2nd, Cui B, Ghia EM, Chen L, Messer K, Shen Z, Briggs SP, Croce CM, and Kipps TJ (2014). ROR1 can interact with TCL1 and enhance leukemogenesis in Em-TCL1 transgenic mice. Proc. Natl. Acad. Sci. USA 111, 793–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierda WG, Padmanabhan S, Chan GW, Gupta IV, Lisby S, and Osterborg A; Hx-CD20–406 Study Investigators (2011). Ofatumumab is active in patients with fludarabine-refractory CLL irrespective of prior rituximab: results from the phase 2 international study. Blood 118, 5126–5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woyach JA, Furman RR, Liu TM, Ozer HG, Zapatka M, Ruppert AS, Xue L, Li DH, Steggerda SM, Versele M, et al. (2014). Resistance mechanisms for the Bruton’s tyrosine kinase inhibitor ibrutinib. N. Engl. J. Med 370, 2286–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda A, Oishi I, and Minami Y (2003). Expression and function of the Ror-family receptor tyrosine kinases during development: lessons from genetic analyses of nematodes, mice, and humans. J. Recept. Signal Transduct. Res 23, 1–15. [DOI] [PubMed] [Google Scholar]
- Yu J, Chen L, Cui B, Widhopf GF 2nd, Shen Z, Wu R, Zhang L, Zhang S, Briggs SP, and Kipps TJ (2016). Wnt5a induces ROR1/ROR2 heterooligomerization to enhance leukemia chemotaxis and proliferation. J. Clin. Invest 126, 585–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Chen L, Chen Y, Hasan MK, Ghia EM, Zhang L, Wu R, Rassenti LZ, Widhopf GF, Shen Z, et al. (2017a). Wnt5a induces ROR1 to associate with 14-3-3z for enhanced chemotaxis and proliferation of chronic lymphocytic leukemia cells. Leukemia 31, 2608–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Chen L, Cui B, Wu C, Choi MY, Chen Y, Zhang L, Rassenti LZ, Widhopf Ii GF, and Kipps TJ (2017b). Cirmtuzumab inhibits Wnt5a-induced Rac1 activation in chronic lymphocytic leukemia treated with ibrutinib. Leukemia 31, 1333–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Chen L, Cui B, Chuang HY, Yu J, Wang-Rodriguez J, Tang L, Chen G, Basak GW, and Kipps TJ (2012a). ROR1 is expressed in human breast cancer and associated with enhanced tumor-cell growth. PLoS ONE 7, e31127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Chen L, Wang-Rodriguez J, Zhang L, Cui B, Frankel W, Wu R, and Kipps TJ (2012b). The onco-embryonic antigen ROR1 is expressed by a variety of human cancers. Am. J. Pathol 181, 1903–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Cui B, Lai H, Liu G, Ghia EM, Widhopf GF 2nd, Zhang Z, Wu CC, Chen L, Wu R, et al. (2014). Ovarian cancer stem cells express ROR1, which can be targeted for anti-cancer-stem-cell therapy. Proc. Natl. Acad. Sci. USA 111, 17266–17271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipeto MA, Court AC, Sadarangani A, Delos Santos NP, Balaian L, Chun HJ, Pineda G, Morris SR, Mason CN, Geron I, et al. (2016). ADAR1 activation drives leukemia stem cell self-renewal by impairing Let-7 biogenesis. Cell Stem Cell 19, 177–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the RNA-seq data reported in this paper is GEO: GSE114382 (https://www.ncbi.nlm.nih.gov/geo/).