Abstract
Recurrent mutations within EGR2 were recently reported in advanced-stage chronic lymphocytic leukemia (CLL) patients and associated with a worse outcome. To study their prognostic impact, 2403 CLL patients were examined for mutations in the EGR2 hotspot region including a screening (n = 1283) and two validation cohorts (UK CLL4 trial patients, n = 366; CLL Research Consortium (CRC) patients, n = 490). Targeted deep-sequencing of 27 known/postulated CLL driver genes was also performed in 38 EGR2-mutated patients to assess concurrent mutations. EGR2 mutations were detected in 91/2403 (3.8%) investigated cases, and associated with younger age at diagnosis, advanced clinical stage, high CD38 expression and unmutated IGHV genes. EGR2-mutated patients frequently carried ATM lesions (42%), TP53 aberrations (18%) and NOTCH1/FBXW7 mutations (16%). EGR2 mutations independently predicted shorter time-to-first-treatment (TTFT) and overall survival (OS) in the screening cohort; they were confirmed associated with reduced TTFT and OS in the CRC cohort and independently predicted short OS from randomization in the UK CLL4 cohort. A particularly dismal outcome was observed among EGR2-mutated patients who also carried TP53 aberrations. In summary, EGR2 mutations were independently associated with an unfavorable prognosis, comparable to CLL patients carrying TP53 aberrations, suggesting that EGR2-mutated patients represent a new patient subgroup with very poor outcome.
INTRODUCTION
Chronic lymphocytic leukemia (CLL), the most common adult leukemia in the Western world,1 is a malignancy of mature B lymphocytes that accumulate in the blood, bone marrow and other lymphoid tissues.2,3 Although treatment has undergone profound improvements in recent years,4–6 CLL shows a remarkable clinical variability, which is likely to be reflective of a large biological heterogeneity.7,8 While numerous prognostic markers have been identified, the mutational status of the immunoglobulin heavy variable (IGHV) genes and certain, high-risk cytogenetic/genetic aberrations (that is, 11q, 17p deletions and TP53 mutations) have remained the strongest markers and are today applied in routine diagnostics.9–12
More recently, whole-genome sequencing and whole-exome sequencing studies have begun to unravel the molecular landscape of CLL, revealing a limited number of frequently mutated genes (for example, ATM, NOTCH1, SF3B1, TP53)13,14 with a long tail of genes altered in < 5% of cases (for example, CHD2,15 MED12,16 NFKBIE,17 POT1,18 RPS15,19 SETD2,20 XPO121). Though integration of molecular information has been proposed to improve classical risk stratification models,22–25 a substantial proportion of patients with a dismal clinical course will not be captured by these algorithms, hence indicating a need to identify additional molecular markers of disease aggressiveness.
Recurrent missense mutations within the EGR2 (early growth response 2) gene, a versatile transcription factor involved in differentiation of hematopoietic cells,26–28 were recently reported in ~8% of advanced-stage CLL patients and appeared to be associated with a worse outcome.29 Notably, EGR2 mutations were predominantly observed within the three zinc-finger domains located in exon 2. EGR2 is activated through ERK phosphorylation upon B-cell receptor (BcR) stimulation,26 and we have previously shown that EGR2-mutated CLL patients display altered expression of EGR2 down-stream target genes compared with patients wild-type for EGR2, thus pointing to a pathogenic role for EGR2 mutations through dysregulated BcR signalling.29 In addition, global DNA methylation investigations linked abnormal EGR2 activity with aberrant hypomethylation of transcription factor binding sites in CLL.30
In this study, we investigated the frequency, clinical and biological associations, and prognostic impact of EGR2 mutations in a large well-characterized screening cohort (n = 1283), two validation cohorts comprising untreated patients from the LRF UK CLL4 trial (n = 366) and patient samples from the CLL Research Consortium (CRC, n = 490), a Chinese CLL cohort (n = 233) and Richter’s syndrome patients (n = 31). EGR2 mutations were associated with younger age, more advanced disease and other molecular high-risk markers, and remained as an independent factor predicting poor outcome both in the screening and validation cohort. These findings suggest that EGR2 mutations define a new, poor-prognostic subgroup of the disease.
METHODS
Patients
Peripheral blood samples from 1283 CLL patients with tumor content ⩾ 60% (median 94%) were collected from collaborating institutions in the Czech Republic, France, Germany, Greece, Italy, the Netherlands, Sweden, and the United States and comprized the screening cohort. All CLL cases were diagnosed according to the iwCLL guidelines and displayed a typical CLL phenotype.31 Over 76% of samples were collected before treatment and within a median of 7 months from time of diagnosis (EGR2-mutated cases, median 2 months). Clinical and biological characteristics of the screening cohort are summarized in Table 1; this cohort had a lower median age at diagnosis and a higher proportion of IGHV-unmutated cases compared with ‘general’ CLL cohorts, likely reflecting that several of the participating institutions are referral centers. In addition, 366 CLL patients, entered into the multicenter trial UK LRF CLL4 (a randomized first-line comparison of chlorambucil, fludarabine and fludarabine plus cyclophosphamide) served as the first validation cohort (Supplementary Table S1). Details of the CLL4 treatment protocol have been previously reported.32 Four hundred and ninety patients collected within the CRC served as the second validation cohort (Supplementary Table S2) with 81% samples obtained before treatment start. Finally, 233 cases from a Chinese CLL patient cohort (Supplementary Table S3), as well as 31 patients with Richter’s syndrome were also screened for EGR2 mutations. Written consent was obtained in accordance with the Declaration of Helsinki and with ethical approval obtained from the local ethics committees.
Table 1.
Comparison of clinical and biological characteristics between EGR2 mutated and wild-type CLL patients within the screening cohort (n = 1283)
| EGR2 wild-type n = 1233 | EGR2 mutated n=50 | P-value | |
|---|---|---|---|
| Age | |||
| Median (years) | 62.1 | 57.4 | 0.0042 |
| <55 years—no. (%) | 278 (25%) | 17 (35%) | |
| >71 years—no. (%) | 194 (17%) | 5 (10%) | |
| No information | 105 | 1 | |
| Sex | |||
| Female—no. (%) | 393 (34%) | 19 (40%) | 0.3490 |
| Male—no. (%) | 769 (66%) | 28 (60%) | |
| No information | 71 | 3 | |
| Binet stage | |||
| A—no. (%) | 674 (70%) | 17 (44%) | 0.0005 |
| B/C—no. (%) | 290 (30%) | 22 (56%) | |
| No information | 269 | 11 | |
| Need of treatment | |||
| Yes—no. (%) | 726 (68%) | 42 (91%) | 0.0010 |
| No—no. (%) | 334 (32%) | 4 (9%) | |
| No information | 173 | 4 | |
| CD38+ | |||
| High (>30%)—no. (%) | 161 (27%) | 20 (67%) | < 0.0001 |
| Low (≤30%)—no. (%) | 438 (73%) | 10 (33%) | |
| No information | 634 | 20 | |
| IGHV | |||
| Mutated—(<98% identity)—no. (%) | 460 (39%) | 9 (19%) | 0.0041 |
| Unmutated (≥98% identity)—no. (%) | 710 (61%) | 39 (81%) | |
| No information | 63 | 2 | |
| del(13)(q14) | |||
| Absent—no. (%) | 665 (64%) | 35 (81%) | 0.0194 |
| Present—no. (%) | 374 (36%) | 8 (19%) | |
| No information | 194 | 7 | |
| del(11)(q22) | |||
| Absent—no. (%) | 852 (82%) | 29 (67%) | 0.0161 |
| Present—no. (%) | 187 (18%) | 14 (33%) | |
| No information | 194 | 7 | |
| + 12 | |||
| Absent—no. (%) | 927 (89%) | 36 (84%) | 0.2587 |
| Present—no. (%) | 112 (11%) | 7 (16%) | |
| No information | 194 | 7 | |
| TP53abn | |||
| Absent—no. (%) | 993 (86%) | 41 (84%) | 0.6394 |
| Present—no. (%) | 161 (14%) | 8 (16%) | |
| No information | 79 | 1 | |
| NOTCH1 mutation | |||
| Absent—no. (%) | 812 (91%) | 42 (91%) | 0.8773 |
| Present—no. (%) | 84 (9%) | 4 (9%) | |
| No information | 337 | 4 | |
| SF3B1 mutation | |||
| Absent—no. (%) | 804 (90%) | 38 (84%) | 0.2703 |
| Present—no. (%) | 93 (10%) | 7 (16%) | |
| No information | 336 | 5 | |
| Sampled before treatment | |||
| Yes—no. (%) | 665 (76%) | 35 (80%) | 0.5480 |
| No—no. (%) | 215 (24%) | 9 (20%) | |
| No information | 353 | 6 |
Abbreviation: TP53abn, TP53 aberrations (that is, mutations and/or deletions). Recurrent genomic aberrations were classified according to the Döhner classification.9 A two-sided student’s t-test was used to assess differences in age at diagnosis, while a Chi-square test was applied to evaluate all other variables in EGR2 wild-type versus mutated cases.
Analysis of EGR2 mutations
The EGR2 mutational hotspot region, covering the three zinc-finger domains located in exon 2, was screened as follows: Sanger sequencing was performed to analyze 1048 CLL and 31 Richter’s syndrome patients; 622 cases were investigated using targeted next-generation sequencing (NGS), 171 patients were assessed by both techniques, and in 490 patients MassARRAY iPLEX assay was applied. In addition, for 41 patients in the screening cohort EGR2 mutation status was derived from whole-exome sequencing data.19 Bidirectional Sanger sequencing was performed according to standard protocols (primers available on request).33 For targeted NGS, a 500 bp amplicon was bead-purified and library preparation was performed using the Nextera XT (Illumina, CA, USA) kit. Libraries were sequenced on the MiSeq instrument using v2 sequencing chemistry (Illumina). Applying standard settings, sequences were mapped using the alignment tool BWA (v.0.7.12).34 Variant calling was carried out using VarScan 2 (v.2.3.7)35 with a minimum variant allele frequency (VAF) of 0.5% and variants were annotated using Annovar.36 Samples from 366 patients enrolled in the UK LRF CLL4 trial were investigated by targeted NGS using a custom design TruSeq gene panel (Illumina) that included the entire coding region of EGR2.20 Approximately 490 CRC samples were analyzed using the MassARRAY iPLEX assay (Agena, CA, USA) for recurrent mutations at EGR2 amino acid positions 356, 384 and 411/12; all mutated samples were validated by targeted NGS of a 210 bp amplicon using MiSeq (Illumina). Samples with >5% VAF were considered mutated.
Analysis of concurrent mutations by targeted deep-sequencing
Thirty-eight EGR2-mutated patients (including 5 patients with VAF <5%) were analyzed using Haloplex technology (Agilent Technologies, CA, USA) according to the manufacturer’s protocol. Probes targeting all coding exons or hotspot regions of 27 known CLL driver genes and/or genes previously reported in EGR2-mutated CLL19,29,37–44 were designed using Agilent’s SureDesign service (https://earray.chem.agilent.com/suredesign/home.htm; Supplementary Table S4). Cluster generation and 125 cycle paired-end sequencing of the pooled library over one lane of the HiSeq 2500 instrument using v4 sequencing chemistry was performed (Illumina). Illumina sequencing adapters were removed using TrimGalore (v.0.3.7) and trimmed reads were aligned to the hg19 human reference genome (February 2009 assembly) using BWA (v.0.7.12). Variants were detected using VarScan2 with a VAF cutoff of 5% and a minimum 30 reads covering the variant was required. Non-synonymous single nucleotide variants and insertions/deletions (indels) that were not present in the 1000 genomes database were included for downstream analyses.
Statistical analysis
Mutational frequencies were assessed using two-sided, descriptive statistics. Overall survival (OS) was calculated from date of diagnosis or time from randomization (UK LRF CLL4) until last follow-up or death, while time-to-first-treatment (TTFT) was calculated from date of diagnosis until initial treatment. Kaplan–Meier analysis was performed to construct survival curves and the Cox–Mantel log rank test was used to determine differences between groups. Cox regression analysis was applied to compare the prognostic significance of EGR2 mutations in relation to other prognostic markers. A significance level of p < 0.05 was applied and all statistical analyses were performed using Statistica Software 13.0 (Dell Inc., OK, USA).
RESULTS
EGR2 mutations and their association with patient characteristics The overall prevalence of EGR2 mutations in this study was 3.8% (91/2403 patients). In detail, EGR2 mutations were detected in 50/1283 CLL patients (3.9%) of the screening cohort, in 12/366 patients (3.3%) of the UK CLL4 trial cohort, and in 18/490 of the CRC cohort (3.7%; Figure 1a; Supplementary Table S5). Further screening revealed that 9/233 (3.9%) patients in the Chinese cohort, and 2/31 (6.5%) Richter’s syndrome patients carried an EGR2 mutation (Figure 1a;Supplementary Table S5).
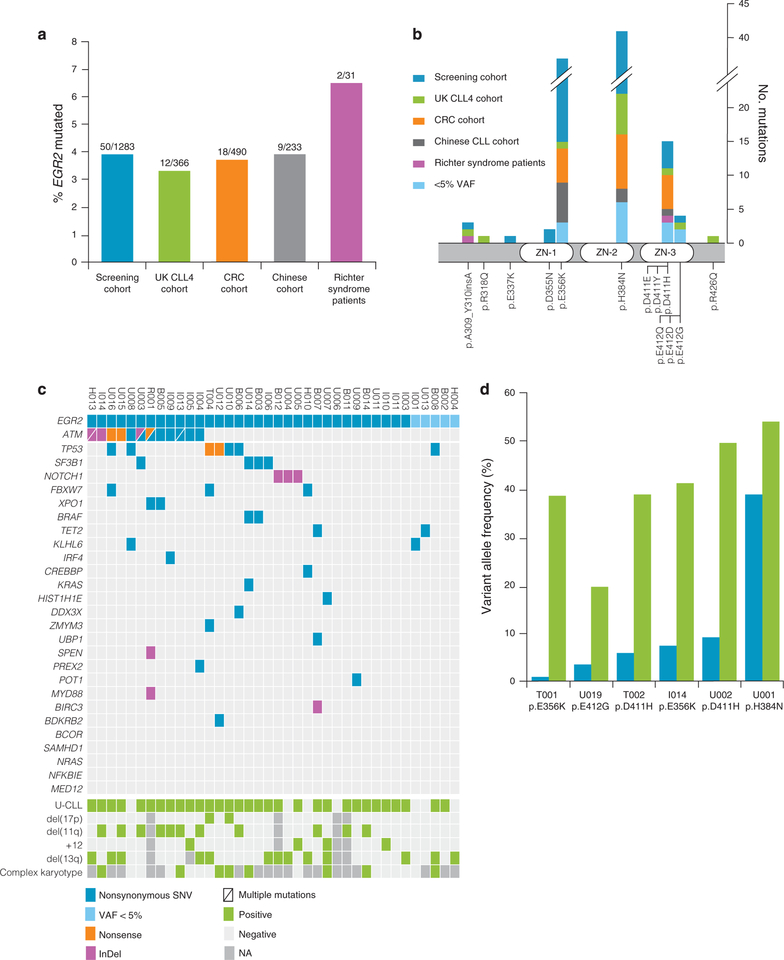
Figure 1.
Frequency, localization and dynamics of EGR2 mutations. (a) Frequency of EGR2 mutations in investigated cohorts. (b) Localization of mutations identified in EGR2. (c) Co-existing mutations in 38 EGR2-mutated CLL patients. (d) Clonal dynamics of six EGR2-mutated CLL patients. The first 5 patients received FCR therapy between the first (blue bars) and second (green bars) time point sample, while the last patient remained untreated at the second time point (7 years between the samples).
All mutations represented heterozygous missense mutations except for a recurrent in-frame 3-bp insertion identified in 3 cases. The majority of EGR2 mutations (85/91, 93%) were localized to the DNA-binding sites of the 3 zinc-finger domains, and predominantly affected codons E356, H384 and D411 (Figure 1b). These domains are highly conserved between orthologues in different species (Supplementary Figure S1). The somatic nature of EGR2 mutations affecting codons E356, H384, D411, and E412 has been previously confirmed.13,14,19,29 With the exception of R318Q, all identified amino acid substitutions were predicted damaging with a SIFT score <0.05.45
For 49 EGR2-mutated samples from the screening cohort and all patients from the two validation cohorts, information on allelic frequency was available from deep-sequencing and the median VAFs were 38.9%, 36.3% (UK CLL4 trial) and 39% (CRC cohort), respectively (range, 5.6–62%). In addition, 33 patients were found to exhibit low-frequency EGR2 variants at the aforementioned hotspot codons with a VAF ranging from 0.5 to 5%. However, only 14/33 (42%) of these variants could be verified in an independent experiment (Supplementary Table S6). These low-frequency EGR2 mutations were excluded from subsequent survival analyses.
We next evaluated the correlation between clinico-biological characteristics and EGR2 mutation status in all 1283 patients included in the screening cohort (Table 1). Compared with wild-type cases, EGR2-mutated patients were significantly younger (57 vs 62 years; P = 0.0042), more often presented with an advanced Binet stage at diagnosis (Binet B/C 56 vs 30%; P = 0.0005), carried unmutated IGHV genes (81 vs 61%; P = 0.0041) and del(11)(q22) (33 vs 18%; P = 0.0161), as well as expressed high levels of CD38 (67 vs 27%; P < 0.0001).
Co-existing mutations and clonal dynamics of EGR2-mutated CLL Through targeted enrichment, we investigated 27 known CLL driver genes and/or genes previously reported in EGR2-mutated patients19,29,37–44 in 38 EGR2-mutated CLL patients (including 5 cases with VAF < 5%). Overall, a mean coverage of 4100 reads per targeted region was achieved with at least 500 reads in 95% and 1000 reads in 90% per nucleotide base of the targeted regions (Supplementary Table S7), hence allowing reliable detection of co-existing mutations. A total of 92 nonsynonymous alterations were detected (Supplementary Table S8); 15 cases harbored one, 8 showed two, 5 patients had three and a single patient presented with four additional gene mutations (Figure 1c), while no additional mutations were detected in the remaining 9 EGR2-mutated cases. Mutations occurred most frequently in ATM (12/38, 31.6%), TP53 (7/38, 18.4%) and SF3B1 (4/38, 10.5%). Of note, 4/12 ATM-mutated cases had two ATM mutations and another 5 cases showed del(11q), resulting in a high frequency of multiple ATM aberrations in EGR2-mutated CLL (9/12, 75%). Alterations affecting the NOTCH signalling pathway were found in 7 patients (18.4%; NOTCH1 (n = 3), FBXW7 (n = 3) and SPEN (n = 1); Figure 1c). Similar findings were observed when comparing identified mutation frequencies with published whole-exome sequencing data from 964 EGR2-wildtype patients,13,14 showing a significant enrichment of ATM (31.6 vs 10.9%, P < 0.001, two-sided Fisher’s exact), TP53 (18.4 vs 5.4%, P = 0.005, two-sided Fisher’s exact) and FBXW7 (7.9 vs 1.8%, P = 0.036, two-sided Fisher’s exact) mutations.
To gain insights into the clonal dynamics of EGR2-mutated CLL patients, we studied the VAF derived from whole-exome sequencing data before fludarabine, cyclophosphamide, and rituximab treatment and at relapse in five patients with available samples from both time points. A sixth patient was sampled one year after diagnosis and again 7 years later; however remaining untreated at both time points. All samples at both times had been negatively selected for CD5+/CD19+ cells to ensure a high tumor content (>95%). In all six patients, the clone harboring an EGR2 mutation expanded during the clinical course and became the dominant clone at relapse or at follow-up (Figure 1d).
Clinical impact of EGR2 mutations
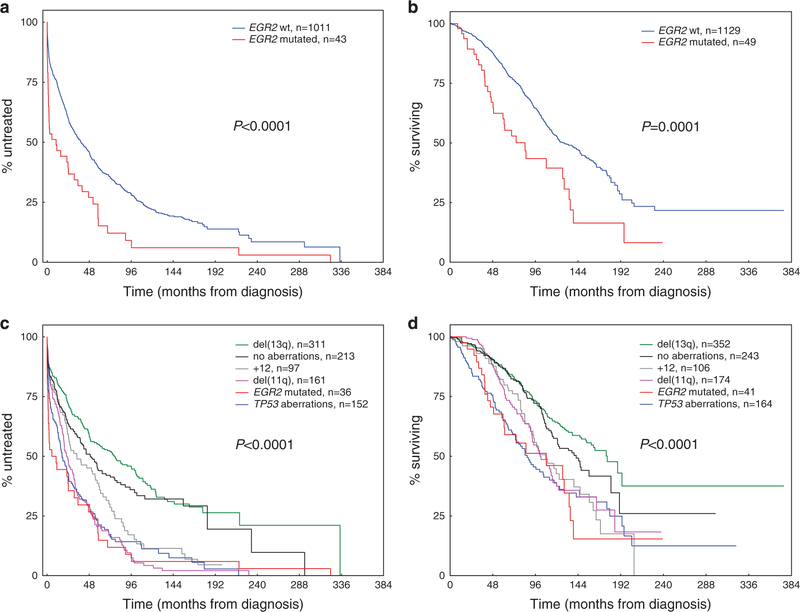
In the screening cohort (1178 cases with available clinical data), the median follow-up time for patients who remained alive was 87.5 months (interquartile range, 49.1 to 138.3 months). Patients with mutated EGR2 (VAF >5%) had a significantly worse TTFT (median, 7.8 vs 38.5 months; HR 1.86, 95% CI 1.35–2.57, P < 0.001; Figure 2a) and OS as compared with EGR2-wild-type patients (median, 74.7 vs 127.2 months; HR 2.03, 95% Ci 1.41–2.92, P<0.001; Figure 2b). No survival difference was observed neither between patients with TP53abn and EGR2 mutations (P = 0.900; Figures 2c and d), nor between EGR2-mutated patients with or without concomitant ATM lesions (P=0.665; Supplementary Figure S2). Notably, within the subgroup of 691 U-CLL patients, EGR2-mutated cases (n = 39) showed a significantly inferior OS than wild-type cases (P = 0.009;Supplementary Figure S3). Similarly, among the 164 patients with TP53abn, a high-risk group defined by a concomitant EGR2 mutation with a shorter OS was identified (P = 0.023; Supplementary Figure S4).
Figure 2.
Clinical impact of EGR2 mutations in 1178 CLL patients from the screening cohort. (a) Time-to-first-treatment and (b) overall survival in the screening cohort according to EGR2 mutation status. (c) Time-to-first-treatment and (d) overall survival in the screening cohort according to the established hierarchy for genomic aberrations9 and EGR2 mutation status. Patients with TP53abn and concomitant EGR2 mutation are grouped into the TP53abn group (TTFT, n = 7; OS, n = 8).
In multivariate analysis, EGR2 mutations remained an independent negative prognostic marker both for TTFT (HR 1.44, 95% CI 1.01–2.06;P = 0.047) and OS (HR 1.72, 95% CI 1.12–2.65, P = 0.014; Table 2), when including EGR2 mutation status, age, gender, Binet stage, IGHV mutational status, del(11q) and TP53abn in the model. This independent effect on shorter TTFT and OS remained significant also when including additional molecular markers to the model such as NOTCH1 and SF3B1 mutations (Supplementary Table S9).
Table 2.
Multivariate Cox proportional hazard analysis of time-to-first-treatment (TTFT, cases, n = 898; events, n = 624) and overall survival (OS, cases, n = 863; events, n = 371) in the screening cohort
| Variable | TTFT | OS | ||||
|---|---|---|---|---|---|---|
| Hazard ratio | 95% Confidence interval | P-value | Hazard ratio | 95% Confidence interval | P-value | |
| EGR2 mutation status | 1.44 | 1.01–2.06 | 0.047 | 1.72 | 1.12–2.65 | 0.014 |
| Age | 1.04 | 0.89–1.22 | 0.611 | 2.20 | 1.78–2.72 | <0.001 |
| Gender | 1.11 | 0.94–1.31 | 0.230 | 1.32 | 1.05–1.65 | 0.016 |
| Binet stage | NA | NA | NA | 2.26 | 1.82–2.81 | <0.001 |
| IGHV mutation status | 4.18 | 3.41–5.12 | <0.001 | 3.28 | 2.54–4.25 | <0.001 |
| del(11q)(q22) | 1.05 | 0.87–1.27 | 0.620 | 0.84 | 0.64–1.09 | 0.190 |
| TP53abn | 1.31 | 1.06–1.62 | 0.013 | 1.66 | 1.26–2.18 | <0.001 |
Abbreviations: NA, not applicable; TP53abn, TP53 aberrations (that is, mutations and or deletions).
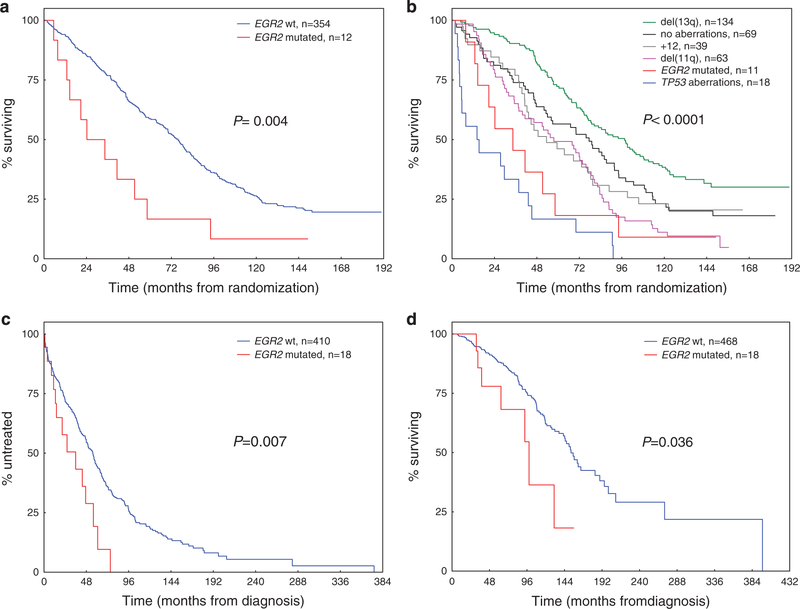
In the UK CLL4 trial cohort (n = 366), the median follow-up time for patients who remained alive was 145 months (interquartile range, 127.7–157 months). Among the 12 EGR2-mutated cases treated within the CLL4 trial, 7 patients were randomly assigned to the fludarabine plus cyclophosphamide arm, 5 patients received chlorambucil and none fludarabine treatment. In univariate analysis, EGR2 mutations were significantly associated with a reduced median OS from time of randomization of 24.6 versus 75.7 months for mutated vs. wild-type patients, respectively (HR 1.71, 95% CI 1.30–2.25, P = 0.004; Figures 3a and b). Multivariate analysis confirmed EGR2 mutation status as an independent risk factor for OS (HR 1.95, 95% CI 1.02–3.73, P = 0.043;Table 3).
Figure 3.
Clinical impact of EGR2 mutations in 366 patients from the UK LRF CLL4 trial and 486 patients from the CRC cohort. (a) Overall survival in the UK CLL4 validation cohort according to EGR2 mutation status. (b) Overall survival in the UK CLL4 validation cohort according to the established hierarchy for genomic aberrations9 and EGR2 mutation status. Patients with TP53abn and concomitant EGR2 mutation are grouped into the TP53abn group (n = 1). (c) Time-to-first-treatment and (d) overall survival in the CRC validation cohort according to EGR2 mutation status.
Table 3.
Multivariate Cox proportional hazard analysis of overall survival in the UK LRF CLL4 patients (cases, n = 297; events, n = 231)
| Variable | Hazard ratio | 95% Confidence interval | P-value |
|---|---|---|---|
| EGR2 mutation status | 1.95 | 1.02–3.73 | 0.043 |
| Age | 1.72 | 1.32–2.24 | <0.001 |
| Gender | 1.44 | 1.04–1.99 | 0.029 |
| Binet stage | 1.21 | 0.86–1.71 | 0.271 |
| IGHV mutation status | 2.22 | 1.64–2.99 | <0.001 |
| del(11q)(q22) | 1.46 | 1.07–1.99 | 0.018 |
| TP53abn | 4.85 | 2.90–8.11 | <0.001 |
Abbreviation: TP53abn, TP53 aberrations (that is, mutations and/or deletions).
In the CRC cohort (486 cases with available clinical data), the median follow-up time for patients who remained alive was 64.9 months (interquartile range, 35.6–103.7 months). In univariate analysis, EGR2 mutations were significantly associated with a shorter TTFT (median, 35.8 vs 55.7 months;HR 2.08, 95% CI 1.21–3.57, P = 0.007;Figure 3c) and reduced median OS of 98.3 versus 152.9 months for mutated vs wild-type patients, respectively (HR: 2.22, 95% CI 1.03–4.77, P = 0.036; Figure 3d). Lack of cytogenetic/molecular data in a substantial proportion of CRC patients precluded testing of the above multivariate model. Nevertheless, multivariate analysis including age, gender, IGHV and EGR2 mutation status, confirmed EGR2 as an independent risk factor for TTFT (HR 1.92, 95% CI 1.11–3.32, P = 0.020), while only a borderline significance was seen for OS (HR 1.90, 95% CI 0.88–4.12, P = 0.10; Supplementary Table S10), probably because of relatively few events and shorter median follow-up time in this cohort.
DISCUSSION
Missense mutations within the EGR2 gene were recently reported in progressive and/or relapsing CLL patients and hence indicated to be associated with a worse clinical outcome.29 Here, by investigating large well-characterized cohorts, we not only confirm and significantly extend this observation, but also reveal that EGR2-mutated CLL patients display distinctive clinicobiological features and a rapidly progressive disease course. Indeed, survival analysis in our screening cohort demonstrated a significant, negative prognostic impact of EGR2 mutations with markedly short TTFT and OS, similar to patients with TP53abn, that remained as an independent negative factor in multivariate analysis. While EGR2 mutations were confirmed as a high-risk marker of short TTFT and OS in the CRC cohort, they were also shown to independently predict short OS from time of randomization in the UK CLL4 trial cohort. Importantly, the negative prognostic impact of EGR2 mutations was also evident in aggressive subgroups, such as U-CLL and patients with TP53 aberrations, which displayed a particularly short OS. Taken together, our data support that EGR2 mutations define a new subgroup of patients with a particularly dismal outcome.
This comprehensive analysis identified EGR2 mutations in 3.8% of 2403 investigated patients with similar mutation frequencies in the different cohorts analyzed (range, 3.3–3.9%), although different techniques for mutation screening were used. This relatively low mutation rate reflects the known genetic heterogeneity in CLL, with only a handful of genes mutated in 10–20% of cases, but with a long list of gene mutations occurring in less than 5% of CLL cases.13,14 Notably, no difference was observed between European/American and Chinese CLL patients with respect to their EGR2 mutation frequencies as suggested for other known CLL drivers such as SF3B1.46 EGR2 mutations were less often found in subsets carrying stereotyped BcR (4/326 = 1.2%, P = 0.027) in contrast to SF3B1, NOTCH1, TP53 and NFKBIE mutations that recently were reported to be enriched in specific stereotyped subsets.17,47–49
EGR2 mutations were often detected with a high mutant allele burden (median VAF 38.9%; Supplementary Table S11), indicating that these aberrations occur early in CLL development.29 From our targeted NGS panel, we noted that patients with EGR2 mutations frequently displayed concurrent mutations in DNA damage response, including, ATM and TP53 and in NOTCH signalling pathway, that is, NOTCH1 and FBXW7, indicating that aberrations within these pathways are important contributors to the evolution of the aggressive phenotype in EGR2-mutated patients. In fact, the majority of EGR2-mutated patients (75%) showed multiple ATM aberrations, which is considerably higher than reported in EGR2 wild-type, ATM-mutated CLL patients (30–40%).50–52 Bi-allelic ATM inactivation is known to be associated with a shorter TTFT and OS.50 Collectively, our data indicate an accumulation of several distinct poor-prognostic markers in EGR2-mutated CLL.
Although most EGR2 mutations were deemed to be clonal, we observed a minor proportion of patients (n = 14) with a low EGR2 mutation burden (<5% VAF, confirmed by independent experiments). Survival analysis revealed a trend for shorter OS in these low-burden cases compared with wild-type patients (P = 0.11; Supplementary Figure S5). Furthermore, serial sampling in five treated CLL cases revealed an expansion of the EGR2-mutated subclone over the clinical course and at relapse (Figure 1d; Supplementary Figure S6). Interestingly, one additional case that remained untreated also showed an expansion of the EGR2-mutated clone. This preferential selection of molecularly defined subclones under pressure of chemotherapy is similar to other poor-prognostic markers such as TP53 in CLL53 and BCOR in myelodysplastic syndromes.54 Larger studies are now warranted to further analyze the potential clinical impact of low-frequency EGR2 mutations as was recently shown for TP53 and NOTCH1.53,55
Similar to the pivotal studies,29,30 EGR2 mutations were clustered in DNA binding domains pointing to a pathogenic role for the hotspots in codons E356, H384 and D411. The functional role of mutated EGR2 in leukemogenesis is however still poorly understood. We recently showed that mutations in the EGR2 DNA binding domain affect cell cycle behavior and lead to altered transcriptional activity and dysregulated BcR signalling.29 Oakes et al. also identified altered EGR2 activity as a mediator for hypomethylation of distinct transcription factor binding sites.30 Whether these effects are restricted to mutations localized in the three sites within the EGR2 DNA binding domain remains to be addressed. Nevertheless, considering the potential role of mutated EGR2 in altering BcR signalling, it will be particularly relevant to study the efficacy of BcR inhibitors in this patient subgroup.
In summary, our novel data highlight EGR2 mutations as an adverse prognostic biomarker for CLL. EGR2 appears to identify a subgroup of CLL patients with a particularly dismal outcome similar to patients with TP53abn. On confirmation of our current data in other cohorts, in particular in patients treated with novel agents (for example, ibrutinib and venetoclax), EGR2 mutation analysis should be considered for inclusion in the current work-up of CLL to identify high-risk patients.
Supplementary Material
ACKNOWLEDGEMENTS
This study was supported by grants #2015_A09 from the Else-Kröner-Fresenius-Stiftung, #DA1787/1-1 from the Deutsche Forschungsgemeinschaft, the Lady Tata Memorial Trust (all F.D.), the Swedish Cancer Society, the Swedish Research Council, Uppsala University, Uppsala University Hospital, the Lion’s Cancer Research Foundation, Uppsala, Marcus Borgström’s Foundation, Uppsala, and Selander’s Foundation, Uppsala, the research grants MSMT CR CEITEC2020 (LQ1601), and AZV MZCR 15-31834A/2015, H2020 ‘AEGLE, An analytics framework for integrated and personalized healthcare services in Europe’, and H2020 ‘MEDGENET, Medical Genomics and Epigenomics Network’ (No.692298), both funded by the European Commission. JCS was funded by Bloodwise (11052, 12036), the Kay Kendall Leukaemia Fund (873), Cancer Research UK (C34999/A18087, ECMC C24563/A15581), Wessex Medical Research and the Bournemouth Leukaemia Fund. This work was also supported by the Oxford Partnership Comprehensive Biomedical Research Centre with funding from the UK Department of Health’s National Institute of Health Research (NIHR) Biomedical Research Centre funding scheme. The views expressed in this publication are those of the authors and not necessarily those of the Department of Health. FD is participant in the BIH-Charité Clinical Scientist Program funded by the Charité University Medical Center Berlin and the Berlin Institute of Health.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
REFERENCES
- 1.Institute. NC. SEER stat fact sheets: chronic lymphocytic leukemia. Available at http://seer.cancer.gov/statfacts/html/clyl.html (accessed 15 May 2013).
- 2.Fabbri G, Dalla-Favera R. The molecular pathogenesis of chronic lymphocytic leukaemia. Nat Rev Cancer 2016; 16: 145–162. [DOI] [PubMed] [Google Scholar]
- 3.Zenz T, Mertens D, Kuppers R, Dohner H, Stilgenbauer S. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer 2010; 10: 37–50. [DOI] [PubMed] [Google Scholar]
- 4.Byrd JC, Brown JR, O’Brien S, Barrientos JC, Kay NE, Reddy NM et al. Ibrutinib versus ofatumumab in previously treated chronic lymphoid leukemia. N Engl J Med 2014; 371: 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furman RR, Sharman JP, Coutre SE, Cheson BD, Pagel JM, Hillmen P et al. Idelalisib and rituximab in relapsed chronic lymphocytic leukemia. N Engl J Med 2014; 370: 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts AW, Davids MS, Pagel JM, Kahl BS, Puvvada SD, Gerecitano JF et al. Targeting BCL2 with venetoclax in relapsed chronic lymphocytic leukemia. N Engl J Med 2016; 374: 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sutton LA, Rosenquist R. The complex interplay between cell-intrinsic and cell-extrinsic factors driving the evolution of chronic lymphocytic leukemia. Semin Cancer Biol 2015; 34: 22–35. [DOI] [PubMed] [Google Scholar]
- 8.Damm F, Nguyen-Khac F, Fontenay M, Bernard OA. Spliceosome and other novel mutations in chronic lymphocytic leukemia and myeloid malignancies. Leukemia 2012; 26: 2027–2031. [DOI] [PubMed] [Google Scholar]
- 9.Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med 2000; 343: 1910–1916. [DOI] [PubMed] [Google Scholar]
- 10.Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood 1999; 94: 1840–1847. [PubMed] [Google Scholar]
- 11.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood 1999; 94: 1848–1854. [PubMed] [Google Scholar]
- 12.Sutton LA, Rosenquist R. Deciphering the molecular landscape in chronic lymphocytic leukemia: time frame of disease evolution. Haematologica 2015; 100: 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puente XS, Bea S, Valdes-Mas R, Villamor N, Gutierrez-Abril J, Martin-Subero JI et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature 2015; 526: 519–524. [DOI] [PubMed] [Google Scholar]
- 14.Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J et al. Mutations driving CLL and their evolution in progression and relapse. Nature 2015; 526: 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez D, Bretones G, Quesada V, Villamor N, Arango JR, Lopez-Guillermo A et al. Mutations in CHD2 cause defective association with active chromatin in chronic lymphocytic leukemia. Blood 2015; 126: 195–202. [DOI] [PubMed] [Google Scholar]
- 16.Kampjarvi K, Jarvinen TM, Heikkinen T, Ruppert AS, Senter L, Hoag KW et al. Somatic MED12 mutations are associated with poor prognosis markers in chronic lymphocytic leukemia. Oncotarget 2015; 6: 1884–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mansouri L, Sutton LA, Ljungstrom V, Bondza S, Arngarden L, Bhoi S et al. Functional loss of IkappaBepsilon leads to NF-kappaB deregulation in aggressive chronic lymphocytic leukemia. J Exp Med 2015; 212: 833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramsay AJ, Quesada V, Foronda M, Conde L, Martinez-Trillos A, Villamor N et al. POT1 mutations cause telomere dysfunction in chronic lymphocytic leukemia. Nat Genet 2013; 45: 526–530. [DOI] [PubMed] [Google Scholar]
- 19.Ljungstrom V, Cortese D, Young E, Pandzic T, Mansouri L, Plevova K et al. Whole-exome sequencing in relapsing chronic lymphocytic leukemia: clinical impact of recurrent RPS15 mutations. Blood 2016; 127: 1007–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parker H, Rose-Zerilli MJ, Larrayoz M, Clifford R, Edelmann J, Blakemore S et al. Genomic disruption of the histone methyltransferase SETD2 in chronic lymphocytic leukaemia. Leukemia 2016; 30: 2179–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeromin S, Weissmann S, Haferlach C, Dicker F, Bayer K, Grossmann V et al. SF3B1 mutations correlated to cytogenetics and mutations in NOTCH1, FBXW7, MYD88, XPO1 and TP53 in 1160 untreated CLL patients. Leukemia 2014; 28:108–117. [DOI] [PubMed] [Google Scholar]
- 22.Oscier DG, Rose-Zerilli MJ, Winkelmann N, Gonzalez de Castro D, Gomez B, Forster J et al. The clinical significance of NOTCH1 and SF3B1 mutations in the UK LRF CLL4 trial. Blood 2013; 121: 468–475. [DOI] [PubMed] [Google Scholar]
- 23.Stilgenbauer S, Schnaiter A, Paschka P, Zenz T, Rossi M, Dohner K et al. Gene mutations and treatment outcome in chronic lymphocytic leukemia: results from the CLL8 trial. Blood 2014; 123: 3247–3254. [DOI] [PubMed] [Google Scholar]
- 24.Baliakas P, Hadzidimitriou A, Sutton LA, Rossi D, Minga E, Villamor N et al. Recurrent mutations refine prognosis in chronic lymphocytic leukemia. Leukemia 2015; 29: 329–336. [DOI] [PubMed] [Google Scholar]
- 25.Rossi D, Rasi S, Spina V, Bruscaggin A, Monti S, Ciardullo C et al. Integrated mutational and cytogenetic analysis identifies new prognostic subgroups in chronic lymphocytic leukemia. Blood 2013; 121: 1403–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yasuda T, Sanjo H, Pages G, Kawano Y, Karasuyama H, Pouyssegur J et al. Erk kinases link pre-B cell receptor signaling to transcriptional events required for early B cell expansion. Immunity 2008; 28: 499–508. [DOI] [PubMed] [Google Scholar]
- 27.Li S, Miao T, Sebastian M, Bhullar P, Ghaffari E, Liu M et al. The transcription factors Egr2 and Egr3 are essential for the control of inflammation and antigen-induced proliferation of B and T cells. Immunity 2012; 37: 685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herglotz J, Unrau L, Hauschildt F, Fischer M, Kriebitzsch N, Alawi M et al. Essential control of early B-cell development by Mef2 transcription factors. Blood 2016; 127: 572–581. [DOI] [PubMed] [Google Scholar]
- 29.Damm F, Mylonas E, Cosson A, Yoshida K, Della Valle V, Mouly E et al. Acquired initiating mutations in early hematopoietic cells of CLL patients. Cancer Discov 2014; 4: 1088–1101. [DOI] [PubMed] [Google Scholar]
- 30.Oakes CC, Seifert M, Assenov Y, Gu L, Przekopowitz M, Ruppert AS et al. DNA methylation dynamics during B cell maturation underlie a continuum of disease phenotypes in chronic lymphocytic leukemia. Nat Genet 2016; 48: 253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Dohner H et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood 2008; 111: 5446–5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Catovsky D, Richards S, Matutes E, Oscier D, Dyer MJ, Bezares RF et al. Assessment of fludarabine plus cyclophosphamide for patients with chronic lymphocytic leukaemia (the LRF CLL4 Trial): a randomised controlled trial. Lancet 2007; 370: 230–239. [DOI] [PubMed] [Google Scholar]
- 33.Damm F, Kosmider O, Gelsi-Boyer V, Renneville A, Carbuccia N, Hidalgo-Curtis C et al. Mutations affecting mRNA splicing define distinct clinical phenotypes and correlate with patient outcome in myelodysplastic syndromes. Blood 2012; 119: 3211–3218. [DOI] [PubMed] [Google Scholar]
- 34.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010; 26: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012; 22: 568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010; 38: qe164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med 2011; 365: 2497–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quesada V, Conde L, Villamor N, Ordonez GR, Jares P, Bassaganyas L et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet 2011; 44: 47–52. [DOI] [PubMed] [Google Scholar]
- 39.Fabbri G, Rasi S, Rossi D, Trifonov V, Khiabanian H, Ma J et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J Exp Med 2011; 208: 1389–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Messina M, Del Giudice I, Khiabanian H, Rossi D, Chiaretti S, Rasi S et al. Genetic lesions associated with chronic lymphocytic leukemia chemo-refractoriness. Blood 2014; 123: 2378–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 2013; 152: 714–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 2011; 475: 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuh A, Becq J, Humphray S, Alexa A, Burns A, Clifford R et al. Monitoring chronic lymphocytic leukemia progression by whole genome sequencing reveals heterogeneous clonal evolution patterns. Blood 2012; 120: 4191–4196. [DOI] [PubMed] [Google Scholar]
- 44.Ojha J, Secreto C, Rabe K, Ayres-Silva J, Tschumper R, Dyke DV et al. Monoclonal B-cell lymphocytosis is characterized by mutations in CLL putative driver genes and clonal heterogeneity many years before disease progression. Leukemia 2014; 28: 2395–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009; 4: 1073–1081. [DOI] [PubMed] [Google Scholar]
- 46.Xia Y, Fan L, Wang L, Gale RP, Wang M, Tian T et al. Frequencies of SF3B1, NOTCH1, MYD88, BIRC3 and IGHV mutations and TP53 disruptions in Chinese with chronic lymphocytic leukemia: disparities with Europeans. Oncotarget 2015; 6: 5426–5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sutton LA, Young E, Baliakas P, Hadzidimitriou A, Moysiadis T, Plevova K et al. Different spectra of recurrent gene mutations in subsets of chronic lymphocytic leukemia harboring stereotyped B-cell receptors. Haematologica 2016; 101: 959–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strefford JC, Sutton LA, Baliakas P, Agathangelidis A, Malcikova J, Plevova K et al. Distinct patterns of novel gene mutations in poor-prognostic stereotyped subsets of chronic lymphocytic leukemia: the case of SF3B1 and subset #2. Leukemia 2013; 27: 2196–2199. [DOI] [PubMed] [Google Scholar]
- 49.Malcikova J, Stalika E, Davis Z, Plevova K, Trbusek M, Mansouri L et al. The frequency of TP53 gene defects differs between chronic lymphocytic leukaemia subgroups harbouring distinct antigen receptors. Br J Haematol 2014; 166: 621–625. [DOI] [PubMed] [Google Scholar]
- 50.Skowronska A, Parker A, Ahmed G, Oldreive C, Davis Z, Richards S et al. Biallelic ATM inactivation significantly reduces survival in patients treated on the United Kingdom Leukemia Research Fund Chronic Lymphocytic Leukemia 4 trial. J Clin Oncol 2012; 30: 4524–532. [DOI] [PubMed] [Google Scholar]
- 51.Sutton LA, Ljungstrom V, Mansouri L, Young E, Cortese D, Navrkalova V et al. Targeted next-generation sequencing in chronic lymphocytic leukemia: a high-throughput yet tailored approach will facilitate implementation in a clinical setting. Haematologica 2015; 100: 370–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Navrkalova V, Sebejova L, Zemanova J, Kminkova J, Kubesova B, Malcikova J et al. ATM mutations uniformly lead to ATM dysfunction in chronic lymphocytic leukemia: application of functional test using doxorubicin. Haematologica 2013; 98: 1124–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malcikova J, Stano-Kozubik K, Tichy B, Kantorova B, Pavlova S, Tom N et al. Detailed analysis of therapy-driven clonal evolution of TP53 mutations in chronic lymphocytic leukemia. Leukemia 2015; 29: 877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Damm F, Chesnais V, Nagata Y, Yoshida K, Scourzic L, Okuno Y et al. BCOR and BCORL1 mutations in myelodysplastic syndromes and related disorders. Blood 2013; 122: 3169–3177. [DOI] [PubMed] [Google Scholar]
- 55.Nadeu F, Delgado J, Royo C, Baumann T, Stankovic T, Pinyol M et al. Clinical impact of clonal and subclonal TP53, SF3B1, BIRC3, NOTCH1, and ATM mutations in chronic lymphocytic leukemia. Blood 2016; 127: 2122–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.